Abstract
Neuronal activity influences protein synthesis and neuronal growth. Availability of nutrients, especially leucine and arginine, regulates the mammalian target of rapamycin (mTOR) pathway that controls cell growth. We show that NMDA receptor activation markedly reduces arginine transport by decreasing surface expression of the cationic amino acid transporters (CAT) 1 and 3. Depletion of CAT1 and CAT3 by RNA interference blocks influences of NMDA receptor activation on the mTOR pathway and neuronal process formation. Thus, the CATs mediate influences of NMDA receptor activation on the mTOR pathway that regulates neuronal processes.
Keywords: cationic amino acid transporters, NMDA receptor, mTOR, neurite growth, neuronal activity, uptake
Introduction
Neuronal activity regulates process extension and synaptic plasticity via protein synthesis (Ruthazer et al., 2003; Colonnese et al., 2005; Sutton and Schuman, 2005; Colonnese and Constantine-Paton, 2006). The nutrient-sensitive mammalian target of rapamycin (mTOR) pathway that controls protein translation (Snyder et al., 1998; Sarbassov et al., 2006; Um et al., 2006; Wullschleger et al., 2006) mediates synaptic plasticity and elaboration of neuronal processes (Tang et al., 2002; Schratt et al., 2004; Takei et al., 2004; Jaworski et al., 2005; Kumar et al., 2005; Tavazoie et al., 2005; Gong et al., 2006; Raab-Graham et al., 2006). Neuronal activity regulates the mTOR pathway (Lenz and Avruch, 2005; Gong et al., 2006; Raab-Graham et al., 2006), but molecular mechanisms for these influences have not been readily evident. Because mTOR responds to altered nutrient status, reflected notably in the availability of arginine or leucine (Hara et al., 1998; Long et al., 2005), one possibility would be for neuronal activity to influence arginine transport.
Arginine can be accumulated by a variety of amino acid transport systems (Closs et al., 2004). Three relatively selective arginine transport proteins have been cloned and are designated cationic amino acid transporters (CATs) (Closs EI, 2002). CAT1 is widely distributed in many tissues with the exception of adult liver (Kim et al., 1991). CAT2 is expressed in two splicing variants, with the low-affinity CAT2A being constitutively expressed in liver and muscle (Closs et al., 1993; Finley et al., 1995) and the high-affinity CAT2B being induced by cytokines in macrophages, astrocytes, and myocytes in conjunction with the induction of inducible nitric oxide (NO) synthase (iNOS) (Simmons et al., 1996; Stevens et al., 1996; Nicholson et al., 2001). The relationship of CAT2B and iNOS may reflect the role of arginine as the precursor of NO. CAT3 is selectively expressed in brain and early embryonic tissues (Hosokawa et al., 1997; Ito and Groudine, 1997). A specific association of the CAT family to mTOR signaling has been identified in Drosophila (Colombani et al., 2003). Growth of Drosophila originates from the fat body (Colombani et al., 2003). Deletion of a growth modifier gene, Slimfast, from the fat body causes growth defects resembling those associated with nutrient deprivation or genetic deficiency in the mTOR pathway (Oldham et al., 2000; Zhang et al., 2000). CAT1 and CAT3 are mammalian homologs of Slimfast (Colombani et al., 2003).
In the present study, we show that the influence of NMDA receptor activation on mTOR activity and neuronal process extension is mediated by CATs. NMDA receptor activation elicits marked reductions in CAT1 and CAT3 surface expression in neuronal tissue associated with decrements in arginine transport. Suppression of CAT1 and CAT3 by RNA interference (RNAi) abolishes the influences of NMDA on mTOR function and elaboration of neuronal processes.
Materials and Methods
CAT1 and CAT3 expression constructs.
Full-length cDNAs of CAT1 and CAT3 were isolated from a rat cDNA library by PCR with two pairs of oligos (upstream, 5′GGTAGATCTATGGGC TGCAAAAACCTG and downstream, 5′ CGCAGAATTCGATTTGCACTGGTCCAA for CAT1; upstream, 5′ GGTAGATCTATGCTGTGGCAGGC and downstream, 5′ CGCAGAATT CGAAATACTATGAACACA for CAT3) using high-fidelity polymerase (Roche, Indianapolis, IN). CAT1 and CAT3 cDNAs were subcloned into enhanced green fluorescent protein (EGFP)–N1 vector (Clontech, Mountain View, CA) at BglII and EcoRI sites. Both cDNA sequences were confirmed by DNA sequencing.
Viral vectors and lentiviral production.
The lentiviral expression system used in the present study consists of three vectors (FUGW, pCMVΔ8.9, and pVSVG) and was originally described by Lois et al. (2002). In brief, RNAi expression cassettes containing H1 promoter (derived from pRNA-H1.1/Neo vector; Genscript, Piscataway, NJ) and RNAi sequences (CAT1, ttggcactctcctggcttact; CAT3, agctgactgtccaggctctat) were subcloned into FUGW vector at PacI site at upstream of EGFP expression cassette that is driven by a ubiquitin promoter. One Shot Stbl3 Chemically Competent Cells (Invitrogen, Carlsbad, CA) were used for subcloning and plasmid amplification. HEK293T cells (human embryonic kidney cells transfected with large T antigen of simian virus 40) were used for virus production. To make viral particles, three plasmids (FUGW, pCMVΔ8.9 plasmid, and envelope vector pVSVG) were transiently cotransfected into HEK293T cells by Lipofectamine 2000 (Invitrogen). At 24 h after transfection, cells were then treated with 10 mm sodium butyrate for 8 h. Culture medium was afterward replaced with fresh DMEM/10% FBS medium and incubated for an additional 48 h. Viruses released into the culture medium were concentrated by ultracentrifugation with SW 41 rotor (Beckman Instruments, Fullerton, CA) at 30,000 rpm for 1.5 h. After removal of the supernatant, pellets with viruses were dissolved in Opti-mem medium (Invitrogen) for overnight. Lentiviruses were titrated using HEK293 cells. For infection, 20–40 μl of lentiviruses (2 × 106 pfu/ml) were directly added into the culture medium in 6- or 12-well plates. The viral stocks were stored at −80°C.
Primary culture, transfection, and lentiviral infection.
Primary neurons were prepared from rat embryonic day 18–19 brains. Neurons were plated in 6- or 12-well plates and 35 mm Petri dishes with glass coverslips, which were coated with poly-d-lysine (40 μg/ml), at a density of ∼2–4 × 105 per well in Neurobasal media with B27 supplement. Neurons were infected with lentivirus at in vitro day 4 and used for experiments at approximately in vitro day 10. Neurons were transfected with plasmids by Lipofectamine 2000 (Invitrogen).
Arginine uptake assay.
Before uptake assay, neurons were rinsed with artificial CSF (ACSF) three times and then incubated for 5 min in ACSF buffer with 5 μm unlabeled l-arginine plus 0.25 μm [3H]l-arginine (5 μCi/ml; NEN, Boston, MA) and 1 mm leucine at 37°C. In case of NMDA and KCl treatment, Mg2+ was eliminated from ACSF. Extracellular arginine was removed by rinsing cells three times with ice-cold ACSF. The uptaked arginine was extracted with 70% methanol in ACSF on ice for 20 min. Approximately 10% of cell extracts was quantified by liquid scintillation using a Beckman Instruments LS6500. The time course of arginine uptake is linear during the first 20 min, and, thus, all uptake assays were performed in 5 min.
Reverse transcriptase-PCR for cationic amino acid transporters.
Total RNA was prepared from primary cortical culture at 10 d with Trizol (Invitrogen). Reverse transcription (RT)-PCR was performed with SuperScript One-Step (Invitrogen) with oligos as follows: CAT1 upstream primer, GCGCCGGAAAGTGGTGGACTGCAG; CAT1 downstream primer, CCCA CTGTCACATAGCTGTAGAGG; CAT2 upstream primer, GCCAAAGCCGATTCTGGCCC GAGT; CAT2 downstream primer, CAGTGTAATTCATTTTGAAGTACG; CAT3 upstream primer, GCAGGACCATCCATTGTGATCTGC; and downstream primer, CATGAGGCATGTTCA GTAAGATGG.
Surface biotinylation assay.
At 48 h after transfection with GFP-tagged CAT1 and CAT3 plasmids, neurons were rinsed twice with ACSF (in mm: 124 NaCl, 3 KCl, 1.25 Na2HPO4, 1.6 CaCl2, 1.8 MgSO4, 10 d-glucose, and 10 HEPES, pH 7.4) and then treated for 10 min with 50 μm dl(−)-2-amino-5-phosphonopentanoic acid (APV), 50 μm NMDA, 50 μm NMDA plus 20 μm MK801 [(+)-5-methyl-10,11-dihydro-5H-dibenzo [a,d] cyclohepten-5,10-imine maleate], or 40 mm KCl, respectively. Neurons were gradually cooled down to 4°C and labeled for 30 min at 4°C with EZ-link NHS-SS-biotin (300 μg/ml) (Pierce, Rockford, IL) in ACSF to biotinylate surface proteins. Neurons were washed three times in ice-cold ACSF, scraped in ACSF, and then pelleted at 16,000 × g for 5 min. Cell pellets were lysed in 200 μl of extraction buffer (25 mm HEPES, pH 7.4, 150 mm NaCl, 1 mm EDTA, proteinase inhibitors, and 1% SDS) and incubated on ice for 10 min. After centrifugation at 16,000 × g, supernatants were then diluted with 4× volumes of buffer [25 mm HEPES, pH 7.4, 150 mm NaCl, 1 mm EDTA, proteinase inhibitor set (Roche), and 1% Triton X-100]. The supernatants containing equal amounts of total proteins were then incubated with streptavidin beads. After washing in dilution buffer, streptavidin-bead pellets were boiled in sample buffer. Precipitated proteins were separated by a reducing SDS-PAGE. Twenty microliters of supernatant derived from post-streptavidin precipitation was loaded into a gel that is marked as lysate control. Biotinylated CAT1 and CAT3 proteins were detected by immunoblot with an anti-GFP antibody. Anti-raptor antibody was used to check the specificity of surface labeling. ECL blots were exposed to BioMax film (Eastman Kodak, Rochester, NY). Blots were quantified using ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
Image acquisition and processing.
Total internal reflection fluorescent microscopy (TIRFM) was performed using the Olympus Optical (Melville, NY) total internal reflection system, including an inverted epifluorescence microscope (IX-70; Olympus Optical) and a high-numerical-aperture (NA) objective (Apo 60×, NA 1.45; Olympus Optical) as described by Huang et al. (2005). Primary hippocampal neurons were plated on 35 mm dishes with a glass coverslip (refractive index n = 1.8 at 488 nm; Olympus Optical). The cells were transfected with GFP-tagged CAT1 and CAT3 plasmids and grown in Neurobasal medium with B27 supplement (Invitrogen) (Brewer et al., 1993). Before imaging, culture medium was rinsed twice and replaced with Neurobasal medium without Mg2+. GFP proteins were excited by light from an argon laser (488 nm; Melles Griot, Carlsbad, CA). Time-lapse sequences of images (1 frame/s) were acquired by a CCD camera (Hamamatsu Photonics, Bridgewater, NJ) controlled by MetaMorph (Universal Imaging, Downingtown, PA). Imaging data were processed by MetaMorph and Excel (Microsoft, Redmond, WA).
Immunoblots.
Primary cortical neurons at approximately in vitro day 9–11 were rinsed three times in Neurobasal medium that lacks arginine and lysine and then further incubated for 20 min in the same medium. Neurons were afterward treated for 15 min with reagents (50 μm NMDA, 20 μm MK801, 50 μm APV, or 200 nm rapamycin) in Neurobasal medium that lacks either Mg2+ or arginine and lysine. In case of KCl treatment, Neurobasal medium was made with 40 mm KCl in which NaCl concentration was proportionately reduced by 40 mm so the osmolarity is kept the same as in regular medium. Cells were lysed in 1× sample buffer (Invitrogen) with 5% β-mercaptoethanol, EDTA-free protease inhibitors (Roche), 1.5 mm Na3VO4, and 10 mm NaF. The primary antibodies against the phosphorylated S6 or raptor (Cell Signaling Technology, Beverly, MA) were diluted in a blocking buffer (3% bovine serum albumin and 0.05% Tween 20 in PBS). The blots were developed using Supersignal West Pico chemiluminescent substrate (Pierce).
Filamentous actin staining and quantification.
Low-density neurons at in vitro day 2–3 were infected with RNAi lentivirus and, 1–2 d later, were treated with APV, MK801, or cultured in Neurobasal medium lacking arginine and lysine. At approximately in vitro day 9–10, neurons were fixed in 4% paraformaldehyde in PBS at room temperature. Filamentous actin (F-actin) was stained with phalloidin conjugated with tetramethylrhodamine isothiocyanate (TRITC) (Sigma, St. Louis, MO). Images were acquired with a fluorescent microscope using 63× or 10× objectives. F-actin staining dots larger than twice the size of shaft were manually counted. Data for each group are averaged from 70–100 cells from 10 images acquired at low magnitude (10×).
Quantification of spines.
Low-density hippocampal neurons at in vitro day 10–11 were transfected with control or CAT1 and CAT3 RNAi vectors. Forty-eight hours after transfection, neurons were treated with 50 μm APV or cultured in neurobasal medium lacking arginine and lysine. Three days later, neurons were fixed in 4% paraformaldehyde in PBS at room temperature. Neuronal processes were visualized by expression of GFP. Individual images were acquired with the confocal microscope Meta 510 using a 63× objective. Each image was a Z-series of 16–30 images at 0.3 μm interval. Representative images were obtained by integrating whole stack using maximal projection. Neurites, spines, and protrusions (or filopodia) were automatically traced with Imaris software (Bitplane, Zurich, Switzerland) as demonstrated in supplemental Figure 5 (available at www.jneurosci.org as supplemental material). Neuronal processes <7 μm long are automatically selected. The number, length, and volume of spines and protrusions are automatically quantified by Imaris. The ratio of volume/length for spines with bulbous heads is bigger than the ratio for the skinny filopodia-like protrusions. Spines with bulbous heads that represent mature spines were quantified by cutoff at the ratio of 0.39 μm3/μm.
Results
NMDA receptor activation decreases arginine uptake and surface expression of CAT1 and CAT3
In cerebral cortical cultures, [3H]arginine uptake is linear for 20 min, after which levels plateau until at least 60 min (data not shown). Activation of NMDA receptors with 50 μm NMDA or depolarization with 40 mm KCl reduces [3H]arginine uptake by 40% (Fig. 1A). In contrast, the NMDA antagonist APV (50 μm) augments basal uptake by 45%. This action of APV indicates that basal levels of NMDA transmission regulate [3H]arginine uptake. The effects of NMDA or KCl on arginine uptake are fully reversible, because removal of NMDA or KCl restores arginine uptake (supplemental Fig. 1, available at www.jneurosci.org as supplemental material), suggesting that reduction of arginine uptake is not the result of NMDA-mediated excitotoxicity. The decrease in arginine uptake elicited by depolarization is selective in that KCl does not influence cysteine or methionine uptake; NMDA does not affect cysteine uptake and increases methionine uptake by 30% (Fig. 1B). KCl-induced neuronal depolarization affects multiple processes such as the release of neurotransmitters, Ca+ influx, and others. To further pinpoint factors responsible for the effect of KCl on arginine uptake, we used the NMDA receptor antagonist APV, the voltage-gated Na+ channel blocker TTX, and the voltage-gated Ca+ channel blocker nifedipine. None of them reverses the inhibitory effect of KCl on arginine uptake (supplemental Fig. 2A, available at www.jneurosci.org as supplemental material). AMPA receptors mediate the vast majority of synaptic transmission at excitatory synapses. We therefore investigated the effect of AMPA receptor activation on arginine uptake. Treatment with 20 μm AMPA elicits similar effects as NMDA treatment with AMPA actions blocked by the AMPA antagonist CNQX (supplemental Fig. 2B, available at www.jneurosci.org as supplemental material).
Figure 1.
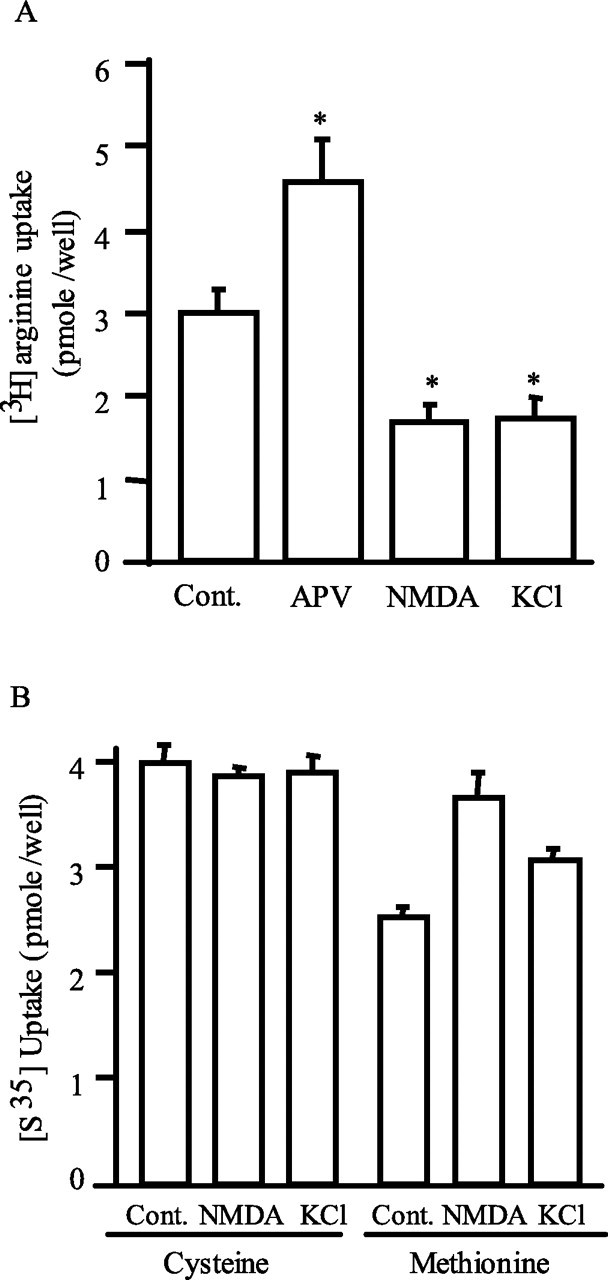
Neuronal activity-dependent uptake of l-arginine. A, Primary cortical neurons were treated for 5 min with 50 μm APV, 50 μm NMDA, or 40 mm KCl in uptake buffer (ACSF plus 0.25 μm [3H]arginine, 5 μm unlabeled arginine, and 1 mm leucine). Extracellular [3H]l-arginine was removed by rinsing neurons three times with the ice-cold ACSF. The amounts of accumulated [3H]l-arginine were as follows (in pmol/well): 2.96 ± 0.28, 4.53 ± 0.52, 1.64 ± 0.24, and 1.72 ± 0.24 for control, APV, NMDA, and KCl, respectively. Data are presented as mean ± SE; n = 6. *p < 0.01. B, Neuronal uptake of [35S]l-cysteine and [35S]l-methionine. The uptake assays were performed for 5 min in ACSF buffer containing 0.25 μm [35S]-labeled plus 5 μm unlabeled cysteine or methionine (in pmol/well: control, 4.00 ± 0.18; 50 μm NMDA, 3.88 ± 0.08; 40 mm KCl, 3.91 ± 0.16 for cysteine uptake vs control, 2.55 ± 0.09; 50 μm NMDA, 3.66 ± 0.25; 40 mm KCl, 3.09 ± 0.10 for methionine uptake). n = 3.
Arginine uptake is mediated via multiple amino acid transporters (Closs et al., 2004), including the Y+ system (CAT1, CAT2, and CAT3) that is primarily responsible for arginine uptake in primary cortical neurons (Stevens and Vo, 1998). RT-PCR reveals the presence of CAT1 and CAT3 but not CAT2 in cortical preparations, consistent with the known absence of CAT2 in nervous tissue (Fig. 2A,B). We examined surface levels of CAT1 and CAT3 in primary cortical neurons by biotinylation assays, which reveal that 30 min treatment of APV augments surface levels of both CAT1 and CAT3 by ∼40% (Fig. 2C–F). In contrast, NMDA depletes surface levels by ∼50%, an effect that is completely reversed by the NMDA antagonist MK801. Potassium depolarization decreases surface levels of CAT1 and CAT3 to approximately the same extent as NMDA. The closely similar effects of NMDA receptor activation on arginine uptake and the surface expression of CAT transporters suggest that the alterations in uptake are determined by the CATs. Raptor, a cytosolic protein, is not evident in the same blot, indicating that plasma membrane fraction is quite pure.
Figure 2.
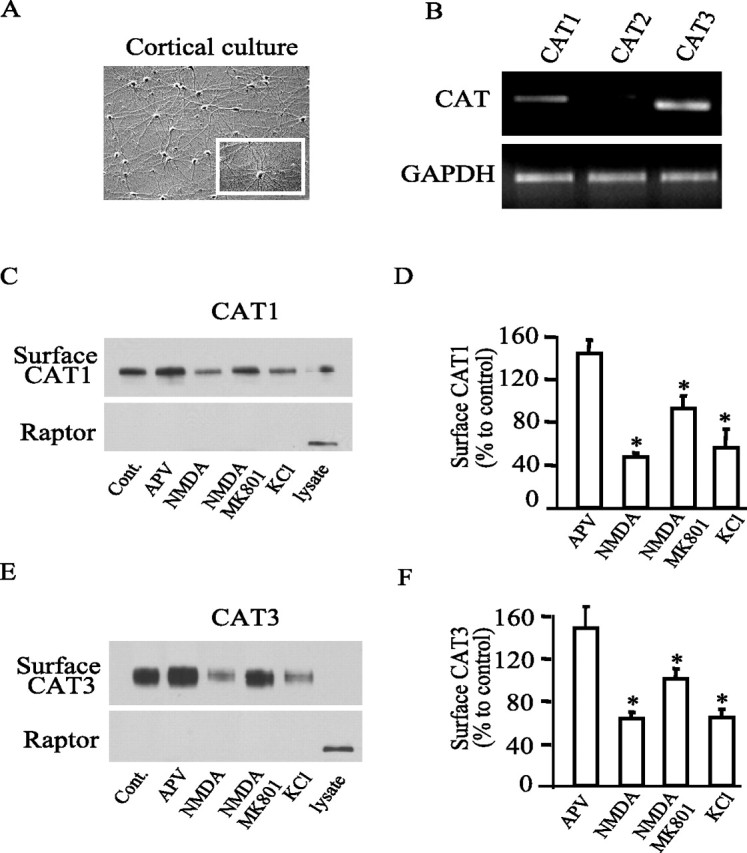
Neuronal activity-dependent internalization of CAT1 and CAT3. A, The image shows a typical primary cortical culture at day 12. B, RT-PCR indicates that cortical neurons express CAT1 and CAT3 but negligible CAT2. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a control. C, E, Surface biotinylation of CAT1 and CAT3. Neurons were transfected with GFP-tagged CAT1 and CAT3 and treated with 50 μm APV, 50 μm NMDA, 50 μm NMDA plus 20 μm MK801, or 40 mm KCl, respectively. Surface biotinylated CAT1 and CAT3 were detected by Western blot using anti-GFP antibody. Immunoblot with anti-raptor antibody indicates that biotin selectively labels plasma membrane surface proteins. D, F, Quantification of surface biotinylated CAT1 and CAT3. Data are normalized to control values and are means from three independent experiments (CAT1, 146.7 ± 12, 62.7 ± 3.7, 99.3 ± 5.8, 64 ± 4.6 vs CAT3, 144.7 ± 7, 47.3 ± 2.3, 92.7 ± 7, 56 ± 10.1 for 50 μm APV, 50 μm NMDA, 50 μm NMDA plus 20 μm MK801, or 40 mm KCl, respectively). *p < 0.01.
To explore the kinetics of changes in CAT1 and CAT3, we used TIRFM imaging, monitoring surface GFP-labeled CAT1 or CAT3 on NMDA activation of primary hippocampal cultures (Fig. 3). Within 10 s of NMDA application, we observe decreases of surface CAT1 and CAT3, effects that are maximal by 20–30 s. The extent of depletion is similar for CAT1 and CAT3. The very rapid decreases of surface GFP-labeled CAT1 and CAT3 indicate that NMDA receptor activation impacts on the trafficking of the transporters.
Figure 3.
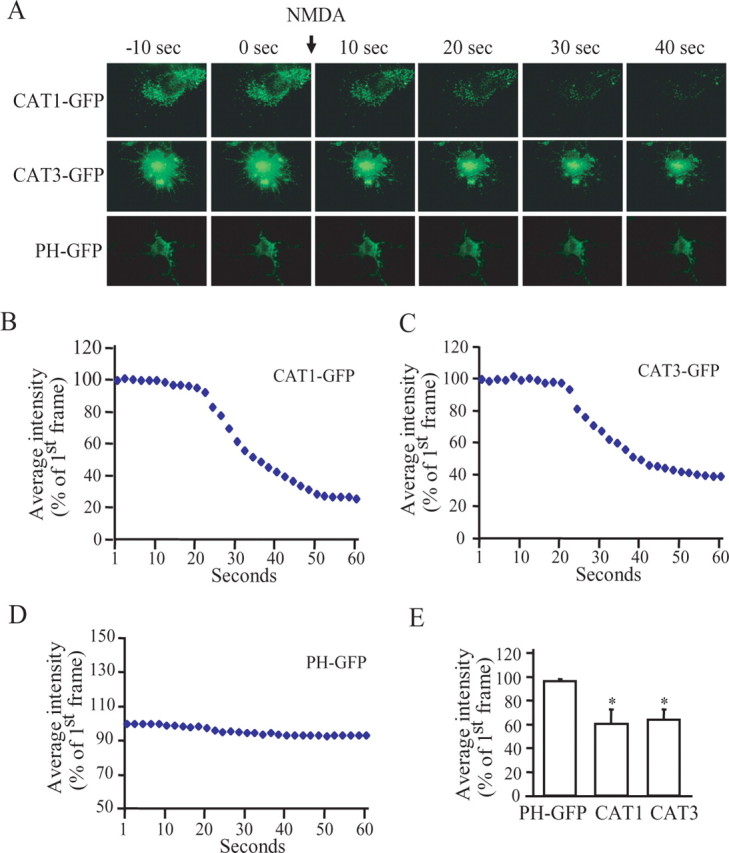
NMDA receptor activation-induced internalization of CAT1 and CAT3. A, Primary cultured neurons were transfected with GFP-tagged CAT1, CAT3, or plectin homology (PH) domain. CAT1, CAT3, and PH domain at membrane surface were visualized using TIRFM. Representative images are from time-lapse sequences recorded in 1 frame/s. NMDA treatment (50 μm) is marked with an arrow. B–D, Fluorescent intensity of each frame in a time-lapse sequence was normalized to the first frame. E, The change of fluorescence intensity induced by NMDA treatment. Data are the means of fluorescence intensity at frame 50 from four experiments (PH, 96 + 1.8; CAT1, 60.5 ± 11.9; and CAT3, 63.75 ± 8.6). *p < 0.01.
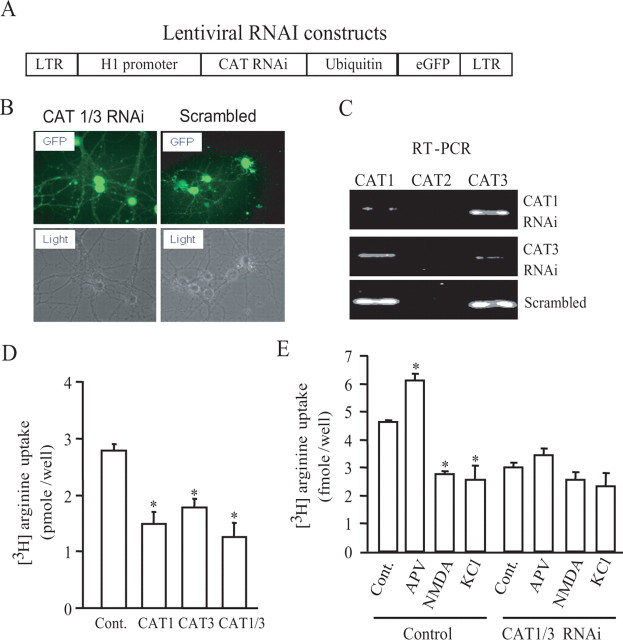
We used a lentivirus to deliver RNAi directed against CAT1 and CAT3 and monitored infection efficiency through GFP expression (Fig. 4A). We observe expression of the RNAi in virtually all neurons (Fig. 4B). RT-PCR reveals that specific RNAi treatment nearly abolishes expression of CAT1 and CAT3, whereas scrambled RNAi is ineffective (Fig. 4C).
Figure 4.
CAT1 and CAT3 mediate arginine uptake. A, Lentiviral constructs for RNAi. Expression of CAT RNAi is driven by H1 promoter. Neurons infected with lentiviral RNAi are marked with EGFP, which is driven by a ubiquitin promoter. LTR, Long terminal repeat. B, Approximately 85–98% of neurons are infected with lentivirus as indicated by expression of bright GFP at approximately day 4–6 after infection. C, RT-PCR measures CAT1, CAT2, and CAT3 transcripts in total RNA purified from neurons infected with RNAi lentivirus for 8 d. Lentivirus with scrambled RNAi was used as a control. D, Eight days after infection, neurons were used for arginine uptake assay. Knockdown of CAT1 and CAT3 reduces arginine uptake (control, 2.79 ± 0.11; CAT1, 1.48 ± 0.22; CAT3, 1.78 ± 0.16; CAT1/CAT3, 1.26 ± 0.26 in picomoles per well). n = 4; *p < 0.01. E, Neurons infected with both CAT1 and CAT3 or control virus were treated for 5 min with 50 μm APV, 50 μm NMDA, or 40 mm KCl. The uptakes of [3H]l-arginine by neurons infected with control lentivirus are as follows (in pmol/well): control, 4.63 ± 0.6; APV, 6.11 ± 0.26; NMDA, 2.79 ± 0.07; KCl, 2.57 ± 0.51 The uptakes by neurons infected with RNAi lentiviruses for CAT1 and CAT3 are as follows (in pmol/well): control, 2.99 ± 0.18; APV, 3.45 ± 0.23; NMDA, 2.58 ± 0.27; KCl, 2.35 ± 0.47. n = 4; *p < 0.01.
To examine the role of the two CATs in regulating arginine transport, we monitored [3H]arginine uptake in cultures using lentiviral RNAi for CAT1, CAT3, or both (Fig. 4D). RNAi depletions of CAT1 or CAT3 each produce an ∼45% decrease in basal [3H]arginine uptake, whereas combined treatment produces a greater effect. Thus, both CAT1 and CAT3 contribute substantially to basal arginine uptake. Combined RNAi for CAT1 and CAT3 abolishes the APV-induced increase in [3H]arginine uptake (Fig. 4E). To rule out the possibility that the effects of CAT1 and CAT3 RNAi on arginine uptake reflect cell toxicity after amino acid starvation, we monitored neuronal viabilities after RNAi treatment and did not find any noticeable changes (supplemental Fig. 3, available at www.jneurosci.org as supplemental material).
NMDA receptor activation regulates the mTOR pathway via the CAT1/3 arginine transport system
To determine whether arginine transport mediates influences of NMDA transmission on neuronal process disposition, we first examined the effects of NMDA on the phosphorylation status of S6, a downstream protein in the mTOR cascade of protein translation (Fig. 5A). NMDA markedly reduces levels of phospho-S6, effects that are blocked by APV and MK801. Rapamycin totally depletes phospho-S6, consistent with regulation by the mTOR pathway. Arginine removal from the medium also depletes phospho-S6, indicating a critical role for arginine transport in maintaining mTOR activity.
Figure 5.
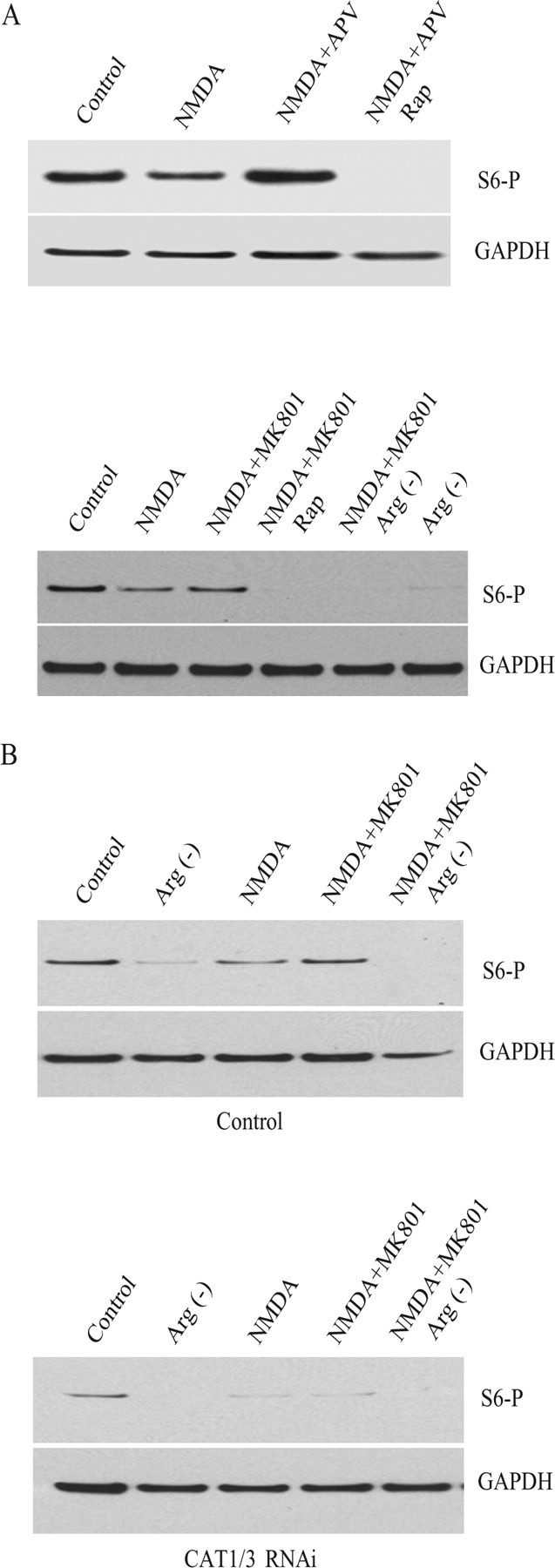
Arginine bioavailability regulates mTOR activity. A, Cortical neurons were treated for 20 min with 50 μm NMDA, 50 μm APV, 200 nm rapamycin (Rap), or 20 μm MK801, either alone or in combination in Neurobasal medium as indicated. Either Mg2+ or arginine and lysine were removed from Neurobasal medium for NMDA treatment or for the arginine-minus [Arg(−)] condition. Phosphorylated S6 proteins were visualized by Western blot with an antibody against phosphorylated S6. Anti-GAPDH was used as control. Experiments were repeated three times with a similar result. B, Knockdown of CAT1 and CAT3 by RNAi reduces S6 phosphorylation. Neurons infected with lentivirus for 6 d were treated with 50 μm NMDA in Neurobasal medium with or without arginine and lysine. Experiments were repeated twice with a similar result.
We wondered whether basal, physiologic NMDA receptor activation regulates the mTOR pathway. To explore this question, we treated cortical cultures with the NMDA antagonist APV and observed a pronounced increase in phospho-S6 as well as phospho-4E-binding protein (4E-BP), another downstream target of mTOR (supplemental Fig. 4, available at www.jneurosci.org as supplemental material). In the same experiment, we confirmed the NMDA-induced reduction of phosho-S6 levels and also observed a decrease in phospho-4E-BP.
To evaluate the importance of the CATs for the mTOR cascade, we monitored levels of phospho-S6 protein and phospho-4E-BP, whose phosphorylation is directly regulated by mTOR (Fig. 5B). Deletion of CAT1 and CAT3 markedly diminishes phospho-S6 and phospho-4E-BP levels under basal conditions and after treatment with APV (Fig. 5B) (supplemental Fig. 4, available at www.jneurosci.org as supplemental material).
Neuronal growth is regulated by NMDA receptor activation via arginine transporters
It is well established that the mTOR pathway regulates the cytoskeleton (Jacinto et al., 2004; Tavazoie et al., 2005), suggesting that it may influence neuronal process extension. To explore the influences of NMDA transmission on neuronal growth, we stained cortical cultures for F-actin (Fig. 6A). NMDA receptor antagonists APV and MK801 both elicit the formation of many punctate structures that, at high power, are identified as growth cones. Depletion of arginine from the medium reduces basal growth cone formation by ∼50% (Fig. 6A,D). APV-stimulated growth cone formation is markedly diminished by depletion of arginine (Fig. 6A,D). In these experiments, we were unable to examine the effects of NMDA itself, because the chronic exposure required for monitoring neuronal growth would be associated with NMDA-mediated excitotoxicity. The pronounced augmentation of growth cone formation caused by APV indicates that basal NMDA receptor activation in the cortical cultures suppresses growth cone formation.
Figure 6.
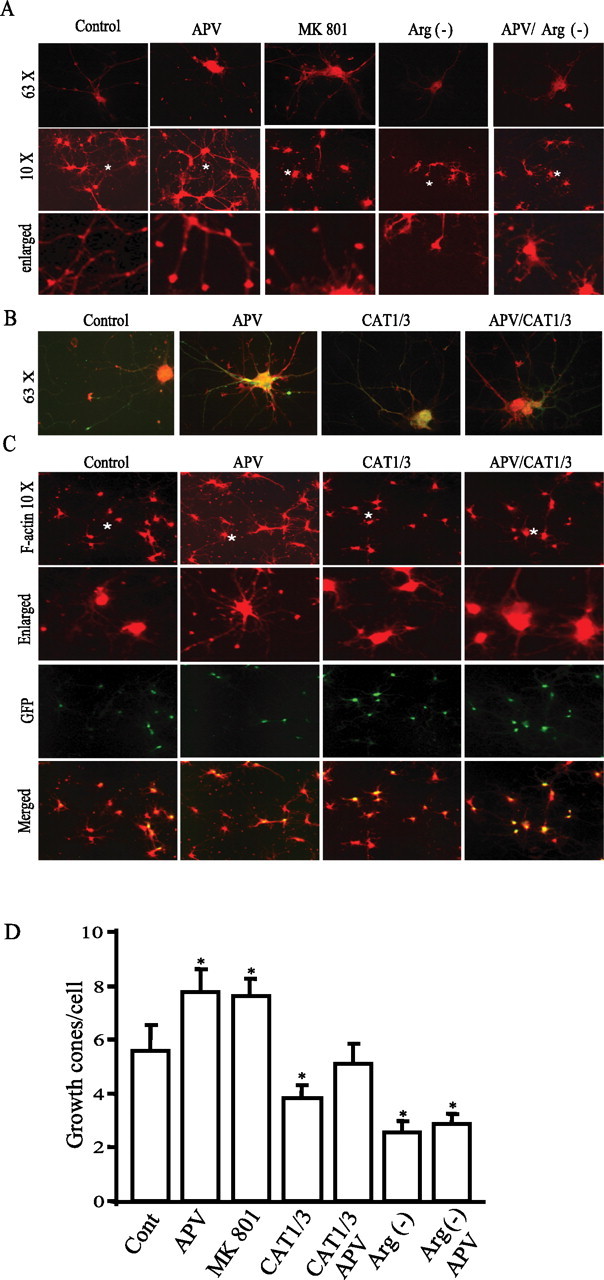
Arginine bioavailability regulates growth cone formation. A, F-actin staining and growth cone formation. Cortical neurons at in vitro day 4 were treated with 50 μm APV or 20 μm MK801 in Neurobasal medium with or without arginine/lysine. F-actin was stained with phalloidin conjugated with TRITC. Images in the top and middle rows were taken at high power (with 63× lens) and low power (with 10× lens), respectively. The F-actin puncta (fine dots) in images reflect growth cones. The images in the bottom row are derived from regions marked with a symbol * in the images in the middle row. B, C, Neurons at approximately in vitro day 3–4 were infected with RNAi lentivirus and then treated with 50 μm APV at 2 d after infection. Neurons at in vitro day 10 were stained with phalloidin conjugated with TRITC (63× in B and 10× in C). Colocalization reveals that almost all neurons stained with F-actin are GFP positive, indicating that they are infected with RNAi lentivirus. D, Quantification of growth cones. Data for each group is averaged from 70–100 cells from 10 images acquired at low magnitude (10×) (in growth cones/cell: control, 5.58 ± 0.98; APV, 7.78 ± 0.84; MK801, 7.59 ± 0.67; CAT1/3, 3.81 ± 0.49; CAT1/3 plus APV, 5.12 ± 0.75; arginine-minus [Arg(−)] condition, 2.56 ± 0.42; arginine-minus condition plus APV, 2.87 ± 0.36). *p < 0.05.
CAT1 and CAT3 are also critical for nerve growth. The pronounced increase of growth cone density elicited by APV is abolished by RNAi against CAT1 and CAT3 (Fig. 6B–D). Suppression of CAT1 and CAT3 also substantially reduces the basal number of growth cones, reinforcing the importance of arginine transport for basal neuronal growth.
Growth cones represent an early stage of axonal and dendritic process extension, whereas dendritic spines occur in more mature neurons. The mTOR pathway is well established to influence dendritic spine morphology (Jaworski et al., 2005; Kumar et al., 2005; Tavazoie et al., 2005). Accordingly, we evaluated dendritic spine morphology in hippocampal cultures at 15 d (Fig. 7A). We observe two types of spines as reported previously (Kumar et al., 2005; Tavazoie et al., 2005), one with a bulbous head that represents a mature spine that presumably harbors more synapses and another with skinny filopodia-like protrusions that are relatively unstable. Blockade of basal NMDA receptors increases the population of spines with bulbous heads (Fig. 7A,B). In contrast, arginine depletion or knockdown of CAT1 and CAT3 by RNAi reduces the number of spines with bulbous heads with a concurrent increase in the number of spines with thin filopodia-like protrusions (Fig. 7A–C).
Figure 7.
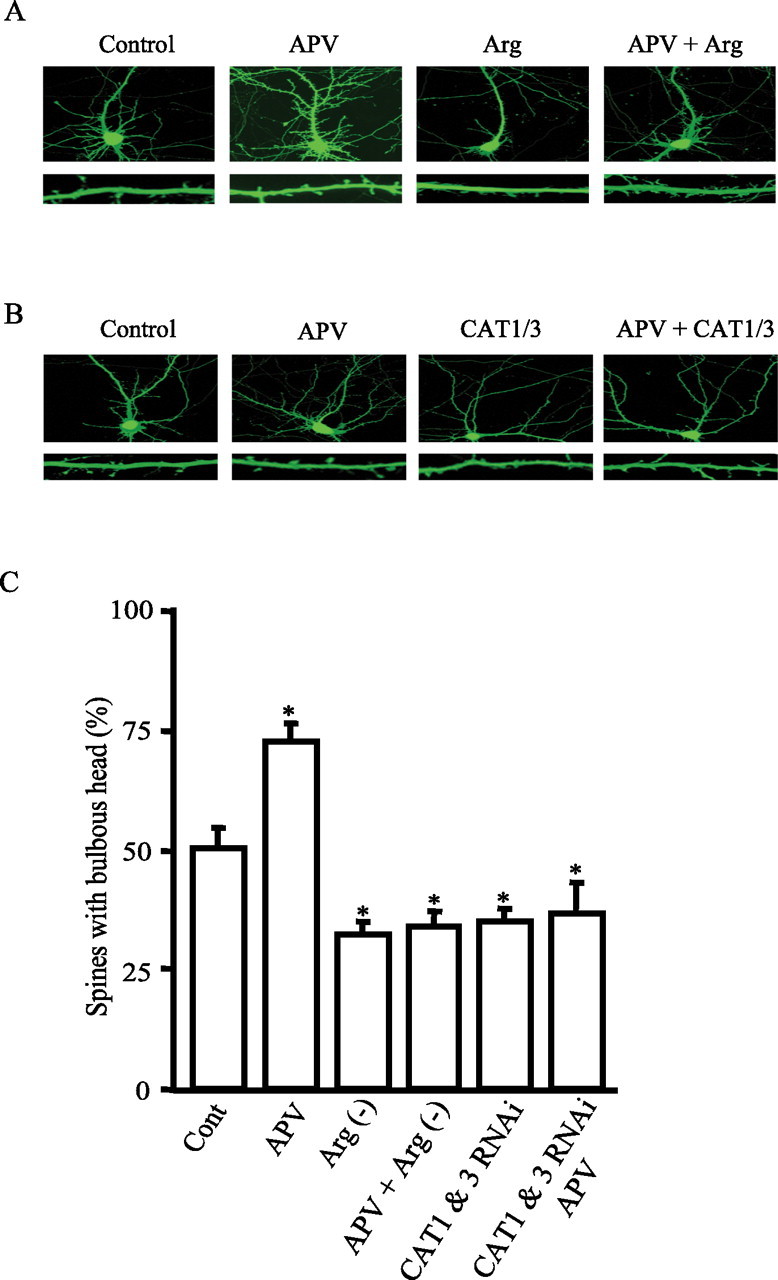
Arginine bioavailability regulates neuronal process disposition. A, Spine morphology. Representative images of hippocampal neurons (top) at in vitro day 15. Neurons were treated with 50 μm APV or cultured in media without arginine (Arg)/lysine for 3 d. Neuronal processes were visualized by expression of GFP. The images in the bottom row are enlarged spiny dendrites derived from the top row. B, Representative images of hippocampal neurons (at the top row) at in vitro day 15. Neurons were transfected with control or CAT1 and CAT3 RNAi vectors for 3 d with treatment of 50 μm APV for 3 d. Neuronal processes were visualized by expression of GFP. The images in the bottom row are enlarged spiny dendrites derived from the top row. C, Quantification of spines. Data for each group are averaged from five to six images acquired with 63× objective. Percentage of spines with bulbous heads is as follows: 50.5 ± 4.1 for control; 72.5 ± 4.0 for APV; 32.23 ± 3.0 for arginine-minus [Arg(−)] condition; 34 ± 2.9 for APV plus arginine-minus condition; 35 ± 2.86 for CAT1 and CAT3 RNAi; and 36.5 ± 6.6 for CAT1 and CAT3 RNAi plus APV (*p < 0.05).
Discussion
The major findings of our study are that NMDA neurotransmission markedly reduces arginine uptake by causing a rapid internalization of the arginine transporters CAT1 and CAT3. Basal levels of NMDA receptor activation impact this process, because NMDA receptor blockade substantially augments arginine transport. The alterations in arginine transport influence the mTOR pathway as well as neuronal processes. Suppression by RNAi of CAT1 and CAT3 blocks the influences of NMDA receptor activation on the mTOR pathway and growth cone/dendritic spine formation. Thus, neuronal activity-dependent arginine bioavailability physiologically regulates mTOR activity and neuronal processes.
Molecular mechanisms underlying the neuronal activity-dependent trafficking of arginine transporter remain unclear. PKC might play a role, because it regulates endocytosis of transporters in other mammalian cells (Rotmann et al., 2006). It remains unclear whether changes in surface levels of transporters after NMDA receptor activation reflect alterations in exocytotic or endocytotic pathways.
In our studies, APV alone stimulates arginine uptake and neurite growth, indicating that basal NMDA activation regulates these processes. It is unclear whether NMDA activation reflects synaptic or extrasynaptic receptors. Synaptic and extrasynaptic responses may involve different receptor subtypes. For instance, NMDA receptor NR1/2B is responsible for the most extrasynaptic receptor activities (Thomas et al., 2006).
Our findings are consistent with numerous studies demonstrating an influence of NMDA receptor activation on neuronal growth via mTOR pathway. For instance, Constantine-Paton and colleagues (Colonnese et al., 2005) observed pronounced suppression of neuronal processes and synapse formation in the sprouting of retinal axons in response to NMDA receptor activation, whereas NMDA receptor blockade had opposite effects (Colonnese and Constantine-Paton, 2006). Ruthazer et al. (2003) demonstrated that NMDA receptor activation mediates selective neuronal process elimination in the optic tectum. Chronic blockage of NMDA receptors in hippocampal slice cultures increases the synapse formation and more complex arborization of dendrites (Luthi et al., 2001). Sutton et al. (2004) observed regulation of dendritic protein synthesis by synaptic transmission. Late-phase long-term potentiation (LTP) expression is also regulated by mTOR (Tang et al., 2002; Cammalleri et al., 2003). Lenz et al. (2005) recently demonstrated that glutamatergic transmission regulates p70S6 kinase in cultured primary mouse neurons. Neuronal growth factors such as brain-derived neurotrophic factor regulate dendritic protein synthesis and neuron development by influences on the mTOR pathway (Schratt et al., 2004; Takei et al., 2004). Our data suggests that neuronal activity-dependent restriction of arginine uptake may play a role in NMDA receptor-dependent stabilization of synaptic connections and elimination of neuronal processes during early brain development (Rabacchi et al., 1992; Luthi et al., 2001; Ruthazer et al., 2003).
One of our major findings is that the CAT1 and CAT3 arginine transporters are rate-limiting regulators of the mTOR pathway and elaboration of neuronal processes. Protein translation mediated by the mTOR pathway influences synaptic plasticity in multiple ways. The early phase of LTP/long-term depression is regulated by rapid alterations in protein synthesis occurring locally in or near dendritic spines in the absence of morphological changes in dendrites, a process that might involve mTOR. Presumably long-lasting changes in synaptic plasticity are mediated by morphological changes in dendritic spines, which have been shown to involve mTOR signaling (Tavazoie et al., 2005). The signaling scheme we report here appears to use this latter process. Tang et al. (2002) recently reported that blockade by rapamycin of mTOR signaling prevented a late, but not an early, phase of LTP. mTOR is a major determinant of protein synthesis in response to nutrients, with arginine being one of the activators of the mTOR pathway (Weng et al., 1995; Hara et al., 1998; Kim et al., 2002; Long et al., 2005). Alterations in amino acid availability influence the binding of the translation repressor 4E-BP to the eukaryotic translation initiation factor 4E eIF-4E (Beretta et al., 1996; Burnett et al., 1998; Hara et al., 1998; Kumar et al., 2000; Kim et al., 2002) and the phosphorylation state of the ribosomal protein S6 kinase leading to changes in its enzymatic activity (Price et al., 1991; Burnett et al., 1998; Hara et al., 1998; Long et al., 2005). The molecular identity of the amino acid sensor that initiates the activation of mTOR remains elusive. Activation of mTOR signaling elicited by amino acids apparently involves class 3 phosphatidylinositol 3OH-kinase and the small GTPase Rheb (Nobukuni et al., 2005; Roccio et al., 2005). Conceivably, neuronal activity-dependent arginine bioavailability influences protein synthesis and synaptic plasticity in processes such as long-term potentiation.
CATs may be the major nodal point in regulating the mTOR pathway in multiple tissues, not just the nervous system. Colombani et al. (2003) identified a Drosophila nutrient signal in fat bodies wherein deletion of a gene designated Slimfast elicits the same phenotype as nutrient deprivation or deficiencies in the mTOR pathway (Montagne et al., 1999; Oldham et al., 2000; Zhang et al., 2000). Slimfast is an arginine transporter with 40% homology to mammalian CAT1 or CAT3. Genetic evidence implicates mammalian CATs in growth and development. CAT1-deficient mice display perinatal death or growth retardation with reduced body mass resembling deletions in the mTOR pathway (Perkins et al., 1997; Montagne et al., 1999; Colombani et al., 2003).
A selective role for arginine in the nutrient-sensing system may interface with other functions of arginine. Arginine is converted by NO synthase to NO and citrulline and by arginase to ornithine, the precursor of the polyamine system, which in turn influences protein synthesis (Morris, 2004). Conceivably, the bioavailability of arginine for the mTOR pathway may be determined by the relative activities of NO synthase and arginase. In this context, Sarbassov and Sabatini (2005) recently showed that mTOR activity is regulated by a redox mechanism.
Footnotes
This work was funded by United States Public Health Service Grant MH-18501 (S.H.S.) and Research Scientist Award DA-00074 (S.H.S.). We thank Drs. Richard Huganir and Kogo Takamiya for their generous gift of lentivirus constructs and Dr. Adam Resnick for invaluable comments.
References
- Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- Brewer GJ, Torricelli JR, Evege EK, Price PJ. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J Neurosci Res. 1993;35:567–576. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci USA. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammalleri M, Lutjens R, Berton F, King AR, Simpson C, Francesconi W, Sanna PP. Time-restricted role for dendritic activation of the mTOR-p70S6K pathway in the induction of late-phase long-term potentiation in the CA1. Proc Natl Acad Sci USA. 2003;100:14368–14373. doi: 10.1073/pnas.2336098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Closs EI. Expression, regulation and function of carrier proteins for cationic amino acids. Curr Opin Nephrol Hypertens. 2002;11:99–107. doi: 10.1097/00041552-200201000-00015. [DOI] [PubMed] [Google Scholar]
- Closs EI, Albritton LM, Kim JW, Cunningham JM. Identification of a low affinity, high capacity transporter of cationic amino acids in mouse liver. J Biol Chem. 1993;268:7538–7544. [PubMed] [Google Scholar]
- Closs EI, Simon A, Vekony N, Rotmann A. Plasma membrane transporters for arginine. J Nutr. 2004;134:2752S–2759S. doi: 10.1093/jn/134.10.2752S. [DOI] [PubMed] [Google Scholar]
- Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Leopold P. A nutrient sensor mechanism controls Drosophila growth. Cell. 2003;114:739–749. doi: 10.1016/s0092-8674(03)00713-x. [DOI] [PubMed] [Google Scholar]
- Colonnese MT, Constantine-Paton M. Developmental period for N-methyl-d-aspartate (NMDA) receptor-dependent synapse elimination correlated with visuotopic map refinement. J Comp Neurol. 2006;494:738–751. doi: 10.1002/cne.20841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonnese MT, Zhao JP, Constantine-Paton M. NMDA receptor currents suppress synapse formation on sprouting axons in vivo. J Neurosci. 2005;25:1291–1303. doi: 10.1523/JNEUROSCI.4063-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley KD, Kakuda DK, Barrieux A, Kleeman J, Huynh PD, MacLeod CL. A mammalian arginine/lysine transporter uses multiple promoters. Proc Natl Acad Sci USA. 1995;92:9378–9382. doi: 10.1073/pnas.92.20.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong R, Park CS, Abbassi NR, Tang SJ. Roles of glutamate receptors and the mammalian target of rapamycin (mTOR) signaling pathway in activity-dependent dendritic protein synthesis in hippocampal neurons. J Biol Chem. 2006;281:18802–18815. doi: 10.1074/jbc.M512524200. [DOI] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- Hosokawa H, Sawamura T, Kobayashi S, Ninomiya H, Miwa S, Masaki T. Cloning and characterization of a brain-specific cationic amino acid transporter. J Biol Chem. 1997;272:8717–8722. doi: 10.1074/jbc.272.13.8717. [DOI] [PubMed] [Google Scholar]
- Huang Y, Man HY, Sekine-Aizawa Y, Han Y, Juluri K, Luo H, Cheah J, Lowenstein C, Huganir RL, Snyder SH. S-nitrosylation of N-ethylmaleimide sensitive factor mediates surface expression of AMPA receptors. Neuron. 2005;46:533–540. doi: 10.1016/j.neuron.2005.03.028. [DOI] [PubMed] [Google Scholar]
- Ito K, Groudine M. A new member of the cationic amino acid transporter family is preferentially expressed in adult mouse brain. J Biol Chem. 1997;272:26780–26786. doi: 10.1074/jbc.272.42.26780. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- Jaworski J, Spangler S, Seeburg DP, Hoogenraad CC, Sheng M. Control of dendritic arborization by the phosphoinositide-3′-kinase-Akt-mammalian target of rapamycin pathway. J Neurosci. 2005;25:11300–11312. doi: 10.1523/JNEUROSCI.2270-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Kim JW, Closs EI, Albritton LM, Cunningham JM. Transport of cationic amino acids by the mouse ecotropic retrovirus receptor. Nature. 1991;352:725–728. doi: 10.1038/352725a0. [DOI] [PubMed] [Google Scholar]
- Kumar V, Pandey P, Sabatini D, Kumar M, Majumder PK, Bharti A, Carmichael G, Kufe D, Kharbanda S. Functional interaction between RAFT1/FRAP/mTOR and protein kinase C delta in the regulation of cap-dependent initiation of translation. EMBO J. 2000;19:1087–1097. doi: 10.1093/emboj/19.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Zhang MX, Swank MW, Kunz J, Wu GY. Regulation of dendritic morphogenesis by Ras–PI3K–Akt–mTOR and Ras–MAPK signaling pathways. J Neurosci. 2005;25:11288–11299. doi: 10.1523/JNEUROSCI.2284-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G, Avruch J. Glutamatergic regulation of the p70S6 kinase in cultured primary mouse neurons. J Biol Chem. 2005;280:38121–38124. doi: 10.1074/jbc.C500363200. [DOI] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Long X, Ortiz-Vega S, Lin Y, Avruch J. Rheb binding to mammalian target of rapamycin (mTOR) is regulated by amino acid sufficiency. J Biol Chem. 2005;280:23433–23436. doi: 10.1074/jbc.C500169200. [DOI] [PubMed] [Google Scholar]
- Luthi A, Schwyzer L, Mateos JM, Gahwiler BH, McKinney RA. NMDA receptor activation limits the number of synaptic connections during hippocampal development. Nat Neurosci. 2001;4:1102–1107. doi: 10.1038/nn744. [DOI] [PubMed] [Google Scholar]
- Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, Thomas G. Drosophila S6 kinase: a regulator of cell size. Science. 1999;285:2126–2129. doi: 10.1126/science.285.5436.2126. [DOI] [PubMed] [Google Scholar]
- Morris SM., Jr Enzymes of arginine metabolism. J Nutr. 2004;134:2743S–2747S. doi: 10.1093/jn/134.10.2743S. [DOI] [PubMed] [Google Scholar]
- Nicholson B, Manner CK, Kleeman J, MacLeod CL. Sustained nitric oxide production in macrophages requires the arginine transporter CAT2. J Biol Chem. 2001;276:15881–15885. doi: 10.1074/jbc.M010030200. [DOI] [PubMed] [Google Scholar]
- Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, Zwartkruis FJ, Thomas G. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci USA. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham S, Montagne J, Radimerski T, Thomas G, Hafen E. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev. 2000;14:2689–2694. doi: 10.1101/gad.845700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins CP, Mar V, Shutter JR, del Castillo J, Danilenko DM, Medlock ES, Ponting IL, Graham M, Stark KL, Zuo Y, Cunningham JM, Bosselman RA. Anemia and perinatal death result from loss of the murine ecotropic retrovirus receptor mCAT-1. Genes Dev. 1997;11:914–925. doi: 10.1101/gad.11.7.914. [DOI] [PubMed] [Google Scholar]
- Price DJ, Mukhopadhyay NK, Avruch J. Insulin-activated protein kinases phosphorylate a pseudosubstrate synthetic peptide inhibitor of the p70 S6 kinase. J Biol Chem. 1991;266:16281–16284. [PubMed] [Google Scholar]
- Raab-Graham KF, Haddick PC, Jan YN, Jan LY. Activity- and mTOR-dependent suppression of Kv1.1 channel mRNA translation in dendrites. Science. 2006;314:144–148. doi: 10.1126/science.1131693. [DOI] [PubMed] [Google Scholar]
- Rabacchi S, Bailly Y, Delhaye-Bouchaud N, Mariani J. Involvement of the N-methyl-d-aspartate (NMDA) receptor in synapse elimination during cerebellar development. Science. 1992;256:1823–1825. doi: 10.1126/science.1352066. [DOI] [PubMed] [Google Scholar]
- Roccio M, Bos JL, Zwartkruis FJ. Regulation of the small GTPase Rheb by amino acids. Oncogene. 2005;25:657–664. doi: 10.1038/sj.onc.1209106. [DOI] [PubMed] [Google Scholar]
- Rotmann A, Vekony N, Gassner D, Niegisch G, Strand D, Martine U, Closs EI. Activation of classical protein kinase C reduces the expression of human cationic amino acid transporter 3 (hCAT-3) in the plasma membrane. Biochem J. 2006;395:117–123. doi: 10.1042/BJ20051558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthazer ES, Akerman CJ, Cline HT. Control of axon branch dynamics by correlated activity in vivo. Science. 2003;301:66–70. doi: 10.1126/science.1082545. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Sabatini DM. Redox regulation of the nutrient-sensitive raptor-mTOR pathway and complex. J Biol Chem. 2005;280:39505–39509. doi: 10.1074/jbc.M506096200. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2006;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Nigh EA, Chen WG, Hu L, Greenberg ME. BDNF regulates the translation of a select group of mRNAs by a mammalian target of rapamycin-phosphatidylinositol 3-kinase-dependent pathway during neuronal development. J Neurosci. 2004;24:7366–7377. doi: 10.1523/JNEUROSCI.1739-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons WW, Closs EI, Cunningham JM, Smith TW, Kelly RA. Cytokines and insulin induce cationic amino acid transporter (CAT) expression in cardiac myocytes. Regulation of l-arginine transport and no production by CAT-1, CAT-2A, and CAT-2B. J Biol Chem. 1996;271:11694–11702. doi: 10.1074/jbc.271.20.11694. [DOI] [PubMed] [Google Scholar]
- Snyder SH, Lai MM, Burnett PE. Immunophilins in the nervous system. Neuron. 1998;21:283–294. doi: 10.1016/s0896-6273(00)80538-3. [DOI] [PubMed] [Google Scholar]
- Stevens BR, Vo CB. Membrane transport of neuronal nitric oxide synthase substrate L-arginine is constitutively expressed with CAT1 and 4F2hc, but not CAT2 or rBAT. J Neurochem. 1998;71:564–570. doi: 10.1046/j.1471-4159.1998.71020564.x. [DOI] [PubMed] [Google Scholar]
- Stevens BR, Kakuda DK, Yu K, Waters M, Vo CB, Raizada MK. Induced nitric oxide synthesis is dependent on induced alternatively spliced CAT-2 encoding L-arginine transport in brain astrocytes. J Biol Chem. 1996;271:24017–24022. doi: 10.1074/jbc.271.39.24017. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Local translational control in dendrites and its role in long-term synaptic plasticity. J Neurobiol. 2005;64:116–131. doi: 10.1002/neu.20152. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Wall NR, Aakalu GN, Schuman EM. Regulation of dendritic protein synthesis by miniature synaptic events. Science. 2004;304:1979–1983. doi: 10.1126/science.1096202. [DOI] [PubMed] [Google Scholar]
- Takei N, Inamura N, Kawamura M, Namba H, Hara K, Yonezawa K, Nawa H. Brain-derived neurotrophic factor induces mammalian target of rapamycin-dependent local activation of translation machinery and protein synthesis in neuronal dendrites. J Neurosci. 2004;24:9760–9769. doi: 10.1523/JNEUROSCI.1427-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang SJ, Reis G, Kang H, Gingras AC, Sonenberg N, Schuman EM. A rapamycin-sensitive signaling pathway contributes to long-term synaptic plasticity in the hippocampus. Proc Natl Acad Sci USA. 2002;99:467–472. doi: 10.1073/pnas.012605299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie SF, Alvarez VA, Ridenour DA, Kwiatkowski DJ, Sabatini BL. Regulation of neuronal morphology and function by the tumor suppressors Tsc1 and Tsc2. Nat Neurosci. 2005;8:1727–1734. doi: 10.1038/nn1566. [DOI] [PubMed] [Google Scholar]
- Thomas CG, Miller AJ, Westbrook GL. Synaptic and extrasynaptic NMDA receptor NR2 subunits in cultured hippocampal neurons. J Neurophysiol. 2006;95:1727–1734. doi: 10.1152/jn.00771.2005. [DOI] [PubMed] [Google Scholar]
- Um SH, D'Alessio D, Thomas G. Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell Metab. 2006;3:393–402. doi: 10.1016/j.cmet.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Weng QP, Andrabi K, Kozlowski MT, Grove JR, Avruch J. Multiple independent inputs are required for activation of the p70 S6 kinase. Mol Cell Biol. 1995;15:2333–2340. doi: 10.1128/mcb.15.5.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Zhang H, Stallock JP, Ng JC, Reinhard C, Neufeld TP. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 2000;14:2712–2724. doi: 10.1101/gad.835000. [DOI] [PMC free article] [PubMed] [Google Scholar]