Abstract
Climbing fiber (CF) neurons in the inferior olivary nucleus (ION) extend their axons to Purkinje cells, playing a crucial role in regulating cerebellar function. However, little is known about their precise place of birth and developmental molecular machinery. Here, we describe the origin of the CF neuron lineage and the involvement of Ptf1a (pancreatic transcription factor 1a) in CF neuron development. Ptf1a protein was found to be expressed in a discrete dorsolateral region of the embryonic caudal hindbrain neuroepithelium. Because expression of Ptf1a is not overlapping other transcription factors such as Math1 (mouse atonal homolog 1) and Neurogenin1, which are suggested to define domains within caudal hindbrain neuroepithelium (Landsberg et al., 2005), we named the neuroepithelial region the Ptf1a domain. Analysis of mice that express β-galactosidase from the Ptf1a locus revealed that CF neurons are derived from the Ptf1a domain. In contrast, retrograde labeling of precerebellar neurons indicated that mossy fiber neurons are not derived from Ptf1a-expressing progenitors. We could observe a detailed migratory path of CF neurons from the Ptf1a domain to the ION during embryogenesis. In Ptf1a null mutants, putative immature CF neurons produced from this domain were unable to migrate or differentiate appropriately, resulting in a failure of ION formation. Apoptotic cells were observed in the mutant hindbrain. Furthermore, the fate of some cells in the Ptf1a lineage were changed to mossy fiber neurons in Ptf1a null mutants. These findings clarify the precise origin of CF neurons and suggest that Ptf1a controls their fate, survival, differentiation, and migration during development.
Keywords: bHLH, cell fate, glutamatergic neurons, hindbrain, transcription factor, inferior olive
Introduction
Climbing fiber (CF) neurons in the inferior olivary nucleus (ION) receive input from the cerebral cortex, the red nucleus, spinal cord, and other brainstem nuclei and project to the cerebellar Purkinje cells (Ruigrok and Cella, 1995). Through the regulation of Purkinje cell activity via climbing fibers, CF neurons are thought to modulate cerebellar function to control animal movement.
Previous studies suggested that precerebellar nuclei neurons, which consist of mossy fiber (MF) neurons and CF neurons, emerge from the dorsal part of the caudal hindbrain and migrate tangentially or circumferentially to their final loci (Bloch-Gallego et al., 1999; Yee et al., 1999; Kyriakopoulou et al., 2002). MF neurons send their axons to cerebellar granule cells conveying peripheral and cortical information to the cerebellum. Along the rostrocaudal axis, both MF and CF neurons are generated from the caudal hindbrain, around rhombomeres 6–8 (r6–r8), as suggested by avian grafting studies as well as mammalian fate map studies (Ambrosiani et al., 1996; Cambronero and Puelles, 2000; Farago et al., 2006; D. Kawauchi et al., 2006). Regarding the dorsoventral axis, the origin of MF neurons has been well studied. The four types of MF neurons, comprising neurons in the pontine nuclei (PN), pontine reticulotegmental nucleus (RTN), lateral reticular nucleus (LRN), and external cuneate nucleus (ECN), are derived from a restricted region of the hindbrain neuroepithelium, which expresses Math1 (mouse atonal homolog 1), a basic helix–loop–helix (bHLH)-type transcription gene (Landsberg et al., 2005; Wang et al., 2005).
Conversely, CF neurons were shown to emerge from a distinct progenitor pool in the hindbrain (Rodriguez and Dymecki, 2000; Nichols and Bruce, 2006). Although their birth place has been suggested to be located in the neuroepithelial region that expresses Wnt-1 (wingless-type MMTV integration site family 1) very weakly but not strongly (Landsberg et al., 2005), the precise location along the dorsoventral axis has not been defined. Moreover, transcription factor(s) responsible for CF neuron development have not been found.
Ptf1a (pancreatic transcription factor 1a), which encodes a bHLH-type transcription factor, was originally reported as a pancreatic determiner that drives undifferentiated cells in the foregut endoderm to differentiate into a pancreatic lineage (Krapp et al., 1998; Kawaguchi et al., 2002). Recently, pivotal roles of Ptf1a in nervous system development have been reported. This gene is involved in development of GABAergic neurons in the cerebellum and dorsal spinal cord (Glasgow et al., 2005; Hoshino et al., 2005) and amacrine and horizontal cells in retina (Fujitani et al., 2006; Nakhai et al., 2007).
In this study, we identified a neuroepithelial region in the embryonic caudal hindbrain that expresses Ptf1a. Through the creation of a fate map of these cells in the Ptf1a lineage, we discovered that CF neurons, but not MF neurons, originate from this region. Furthermore, loss of Ptf1a results in a disturbance of the CF neuron development, leading to absence of the ION formation, suggesting that Ptf1a is essential in this process. These findings provide clues toward understanding the development of CF neurons as well as the domain structure of the caudal hindbrain neuroepithelium along the dorsoventral axis.
Materials and Methods
Animals.
The Ptf1acre and Rosa26R (R26R) mouse lines were described previously (Soriano, 1999; Kawaguchi et al., 2002; Hoshino et al., 2005).
Antibodies and immunohistochemistry.
Primary antibodies used in this study were anti-Brn3a (brain-specific homeobox/POU domain protein 3A) (1:10, mouse monoclonal; Santa Cruz Biotechnology, Santa Cruz, CA), anti-Brn3a (1:10, rabbit polyclonal; Abcam, Cambridge, UK), anti-Brn3b (1:25, goat; Santa Cruz Biotechnology), anti-GABA (1:500, rabbit; Sigma, St. Louis, MO), anti-HuC/D (1:500, mouse monoclonal; Invitrogen, Carlsbad, CA), anti-β-galactosidase (β-gal) (1:400, goat; Biogenesis, Poole, UK), anti-β-galactosidase (1:1600, rabbit; Cappel, Aurora, OH), anti-glutaminase (1:600, rabbit) (Hoshino et al., 2005), anti-Ngn1 (Neurogenin1) (1:100, goat; Santa Cruz Biotechnology), anti-bromodeoxyuridine (BrdU) (1:75, mouse monoclonal; BD, Franklin Lakes, NJ), anti-Fluorogold (1:2000, rabbit; Chemicon, Temecula, CA), anti-cleaved Caspase-3 (1:100, rabbit; Cell Signaling Technology, Beverly, MA), and anti-Ptf1a (1:3000). The anti-Ptf1a antibody was raised against the N-terminal region of Ptf1a (amino acids 9–78) fused with glutathione S-transferase and affinity purified with the antigen. This antibody detects no signals in the Ptf1a-null (Ptf1acre/cre) mutants (data not shown). Immunohistochemistry was performed as described previously (Matsuo et al., 2002, 2003; Yoshizawa et al., 2005).
In situ hybridization in brain sections.
In situ hybridization was performed as described previously (Hoshino et al., 1999; Yoshizawa et al., 2002, 2003). The probes used in this report were Math1 [a kind gift from Dr. R. Kageyama, Kyoto University, Kyoto, Japan (Akazawa et al., 1995)] and Mbh2(mammalian BarH 2)/Barhl1 (BarH-like 1) [a kind gift from Dr. T. Saito, Chiba University, Chiba, Japan (Saito et al., 1998)].
Detection of β-galactosidase using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside.
Adult brains were perfused with 4% paraformaldehyde (PFA) in PBS and then fixed in the same fixative for 2 h. Embryonic brains were fixed with 4% PFA in PBS at 4°C for 3 h. After several washes with PBS, fixed brains were incubated in the 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) working solution [0.05% X-gal, 1 mm MgCl2, 3 mm K4Fe(CN)6, 3 mm K3Fe(CN)6, and 0.1% Triton X-100 in PBS] at 37°C overnight. Cryosectioned samples were reincubated with the X-gal working solution, when required.
X-gal staining followed by in situ hybridization.
After several washes with PBS, frozen sections were incubated in the RNase-free X-gal working solution at 37°C overnight. Subsequently, in situ hybridization was performed on these sections as described above.
BrdU incorporation experiment.
Pregnant mice [embryonic day 10.5 (E10.5) and E11.5] were given two 50 mg/kg intraperitoneal injections of BrdU with a 30 min interval. One hour after the first injection, the embryos were fixed, and frozen sections were subjected to immunostaining with an anti-BrdU antibody as described previously (Kawauchi et al., 2003, 2006). After the immunostaining, sections were further counterstained with 1:1000 diluted TOPRO-3 (Invitrogen) in 0.1% Triton X-100/PBS for 1 h to visualize individual nuclei. Signals of BrdU, β-gal, and TOPRO-3 were captured using a Leica (Nussloch, Germany) TCS SP laser scanning confocal microscope. Among β-gal-localizing nuclei within the ventricular zone, the numbers of BrdU-positive and -negative nuclei were counted. Five frozen sections per animal were subjected to this assay. Three animals were used for each genotype (Ptf1acre/+; R26R or Ptf1acre/cre; R26R). Statistical evaluation was performed with Student's t test.
Retrograde labeling of precerebellar neurons.
The animals were anesthetized by an intraperitoneal injection of 3.5% chloral hydrate (3.5 mg/10 g body weight) and immobilized in a stereotactic apparatus (Narishige, Tokyo, Japan). After incision of the skin overlying the occipital region, a small burr hole was made directly over the left hemisphere of the cerebellum of recipient mice using a dental drill, and four injections of 0.2 μl of 4% Fluorogold (Fluorochrome, Englewood, CO) into anterior and posterior sites of the left cerebellar hemisphere and vermis, respectively, were made by applying pressure through a micropipette attached to the barrel of a 1 μl Hamilton microliter syringe under an operating microscope. The micropipette was kept in place for 5 min before removal to minimize leakage. At 4 d after injection, the animals were reanesthetized and transcardially perfused with 4% PFA in phosphate buffer (PB). The brains were dissected, fixed in 4% PFA/20% sucrose in PB, embedded in 20% sucrose, and cut at a thickness of 15 μm with a cryostat. The sections were stained with the anti-Fluorogold antibody (1:2000, rabbit; Chemicon).
Detection of proteolytically activated form of Caspase-3.
Frozen sections (15 μm) of E13.5 hindbrains were subjected to immunostaining with the anti-cleaved Caspase-3 (Asp715) antibody (1:100, rabbit; Cell Signaling Technology). The cleaved Caspase-3-positive cells located in the ventral half of the hindbrain were counted in each section. Along the rostrocaudal axis, six sections of the caudal hindbrain were collected from each animal. Four heterozygous (Ptf1acre/+) and six homozygous (Ptf1acre/cre) brains were analyzed. The mean number of positive cells per one section were calculated and indicated in Figure 4Z. Student's t test was used to evaluate the statistical significance (p = 0.015).
Figure 4.
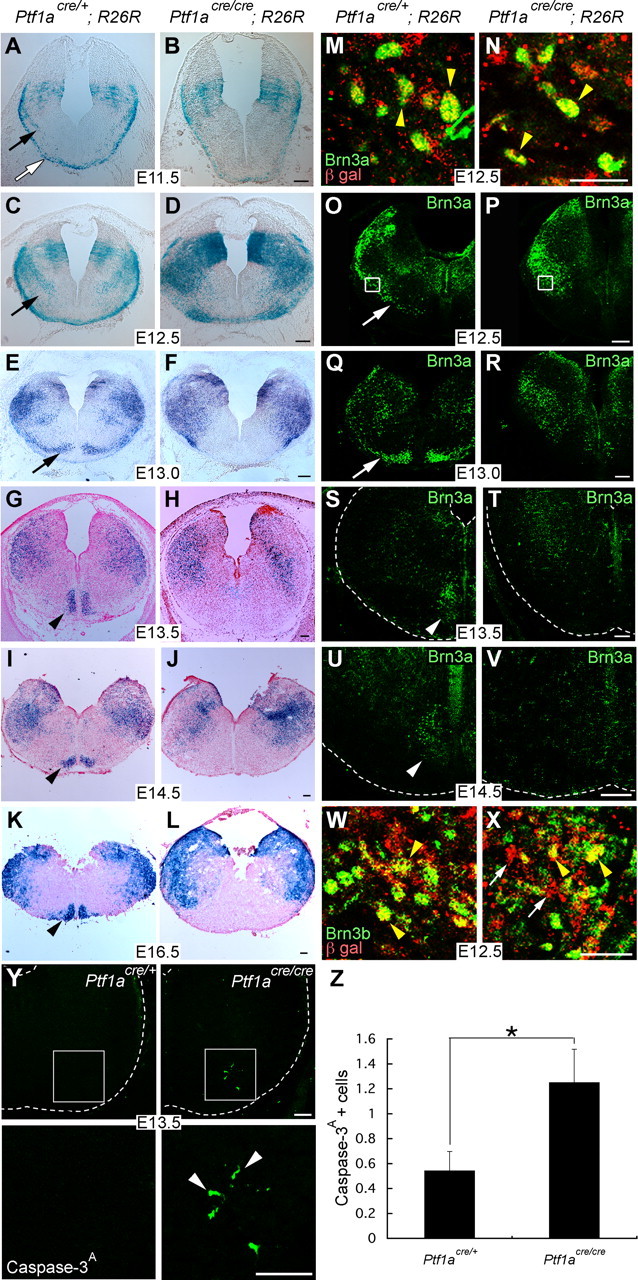
Dynamics of Ptf1a lineage cells in the caudal hindbrain of the heterozygous and homozygous animals for Ptf1a during embryogenesis. A–L, X-gal-stained transverse sections of the caudal hindbrain. M, N, Double staining with Brn3a and β-gal in the regions indicated by rectangles in O and P, respectively. Yellow arrowheads indicate double-positive cells. O–V, Transverse sections stained with Brn3a. White arrows indicate putative migrating neurons that give rise to CF neurons. White arrowheads indicate CF neurons in the ION. W, X, Double staining with Brn3b and β-gal around the regions indicated by rectangles in O and P, respectively. Yellow arrowheads indicate double-positive cells. White arrows indicate β-gal-positive cells that do not express Brn3b. Y, Top, A ventrolateral quarter of caudal hindbrain at E13.5 immunostained with an anti-activated Caspase-3. Bottom, High-magnification pictures of rectangular regions in the top. Arrowheads indicate apoptotic cells. Z, Quantification of the numbers of cleaved Caspase-3-positive cells located in the ventral half of a caudal hindbrain section. The numbers are 0.54 ± 0.16 and 1.25 ± 0.27 in heterozygotes and homozygotes, respectively (mean ± SEM). *p = 0.015, Student's t test. n = 24 in Ptf1acre/+, n = 36 in Ptf1acre/cre. Developmental stages and genotypes are indicated. Scale bars: A–L, O–V, Y, 100 μm; M, N, W, X, 20 μm.
Results
CF neurons are in the Ptf1a lineage
Ptf1a was initially found to be expressed in the relatively dorsal regions of the mouse embryonic neural tube (Obata et al., 2001), and more recently we have shown that it is expressed in the ventricular zone neuroepithelium of the cerebellar primordium (Hoshino et al., 2005). Because Ptf1a is expressed in neural precursor/progenitor cells and implicated for a crucial role in the development of several types of neurons (e.g., cerebellar and spinal cord GABAergic neurons, retinal amacrine and horizontal cells) (Glasgow et al., 2005; Hoshino et al., 2005; Fujitani et al., 2006; Nakhai et al., 2007), we undertook the making of an intensive fate map of cells derived from Ptf1a-expressing progenitors. Toward this aim, we used a recombination-based lineage tracing technique. The Ptf1acre allele was generated by the replacement of the Ptf1a protein-coding region with that of a Cre (enterobacteria phage P1, cyclization recombinase) recombinase targeted to the nucleus (Kawaguchi et al., 2002). We crossed Ptf1acre/+ with Gt(ROSA)26Sortm1sor (R26R) mice, which carry a modified lacZ gene driven by a cell-type-independent ROSA26 promoter (Soriano, 1999). In offspring obtained from this cross, Ptf1a-driven expression of Cre excises a stop cassette upstream of lacZ and activates β-gal expression, which results in the labeling of Ptf1a-expressing cells and their progeny (Kawaguchi et al., 2002; Hoshino et al., 2005).
In Ptf1acre/+; R26R adult mice, β-gal-positive cells were found in some brain regions, including the cerebellum, hindbrain, and hypothalamus (Fig. 1A–D). We examined serial transverse sections throughout the CNS. Some cells in the medial preoptic area (Fig. 1E,F), ventral medial hypothalamic nucleus (Fig. 1G,H), and the cerebellum (Fig. 1I) were β-gal positive, as has been observed previously (Glasgow et al., 2005; Hoshino et al., 2005). In transverse sections at the level of the pontine nuclei, no positive signals were detected (Fig. 1J). However, many β-gal-positive cells were observed in the dorsal cochlear nucleus, whereas significantly fewer positive cells were found in the ventral cochlear nucleus (Fig. 1K,L). In addition, many cells in the vestibular nuclei were also β-gal positive (Fig. 1K). At the level of the caudal hindbrain in which the ION is located, the spinal trigeminal nucleus, solitary nucleus, and ION were observed to contain β-gal-positive cells (Fig. 1M–O). Brain regions with Ptf1a lineage cells are summarized in supplemental Figure 1 (available at www.jneurosci.org as supplemental material) and compared with Math1 lineage cells, which was reported previously (Landsberg et al., 2005; Machold and Fishell, 2005; Wang et al., 2005).
Figure 1.
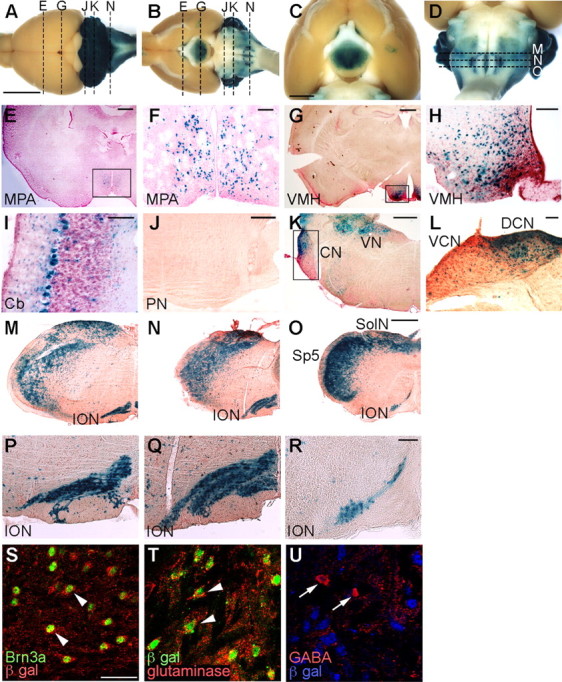
Cells in the Ptf1a lineage were visualized by X-gal staining of Ptf1acre/+; R26R adult brains. A–D, Dorsal (A) and ventral (B–D) views of whole-mount X-gal-stained brains. E–L, Transverse sections of X-gal-stained brains at the level of the medial preoptic area (E, F), ventral medial hypothalamus (G, H), cerebellum (I), pontine nuclei (J), and cochlear nuclei (K, L), which correspond to broken lines in A and B. F, H, L, Higher-magnification views of the rectangular regions indicated in E, G, K, respectively. M–O, X-gal-stained transverse sections of adult medulla oblongata of Ptf1acre/+; R26R at the levels of the rostral (M), middle (N), and caudal (O) ION, which correspond to the broken lines in D, respectively. P–R, Higher-magnification views of M–O, respectively. E–R, Transverse sections were counterstained with nuclear fast red or neutral red. S–U, Double immunolabeling visualized with indicated antibodies to the adult ION of Ptf1acre/+; R26R mice. Scale bars: A–D, 5 mm; E, G, J, K, M–O, 500 μm; F, H, I, L, P–R, 100 μm; S–U, 40 μm. MPA, Medial preoptic area; VMH, ventral medial hypothalamic nucleus; Cb, cerebellar cortex; PN, pontine nuclei, CN, cochlear nuclei; DCN, dorsal cochlear nucleus; VCN, ventral cochlear nucleus; VN, vestibular nuclei; SolN, solitary nucleus; Sp5, spinal trigeminal nucleus.
Because of our interest in the development of CF neurons, we examined all of the serial sections along the rostrocaudal axis in detail, with particular focus on β-gal-positive cells within the ION region (visualized by neutral red counterstaining). β-gal-positive cells were found throughout the entire length of the ION, from the rostral to caudal ends (Fig. 1M–R). Double immunolabeling with Brn3a (a marker for CF neurons, Fedtsova and Turner, 1995) and β-gal showed that most Brn3a-expressing cells were β-gal positive in the ION (Fig. 1S), suggesting that most CF neurons are derived from Ptf1a-expressing cells. Previously, it was reported that the ION contains not only glutamatergic CF neurons but also a small number of GABAergic neurons that probably contribute to the local circuitry (Fredette et al., 1992). Our double immunostaining experiments revealed that Brn3a-expressing cells in the ION are immunoreactive to glutaminase (glutamatergic neuron marker) but not to GABA (GABAergic neuron marker) (supplemental Fig. 2A–F, available at www.jneurosci.org as supplemental material). Accordingly, in the adult ION of Ptf1acre/+; R26R mice, most glutaminase-positive cells were marked by β-gal (Fig. 1T), whereas GABA-positive cells were not (Fig. 1U). These observations suggest that only glutamatergic CF neurons in the ION are from the Ptf1a lineage.
In the spinal trigeminal nucleus and solitary nucleus, some portion of β-gal-positive cells were immunoreactive to GABA but not glutaminase (data not shown), suggesting that the Ptf1a lineage cells in those nuclei include GABAergic neurons but not glutamatergic neurons.
MF neurons are not in the Ptf1a lineage
Whereas all climbing fibers originate in the ION, mossy fibers that project to cerebellar granule cells have multiple origins, such as the PN, RTN, LRN, and ECN. We investigated whether there exist some MF neurons in the Ptf1a lineage. To label both MF and CF neurons in a retrograde manner, a dye (Fluorogold) was injected into the cerebellar hemisphere and vermis of adult Ptf1acre/+; R26R mice, which were killed 4 d after injection (Fig. 2A). MF and CF neurons were labeled with an anti-Fluorogold antibody. Consistent with the data in Figure 1P–S, Fluorogold-labeled CF neurons in the ION were found to be expressing β-gal (Fig. 2B–D). Because CF neurons exist only in the ION, all Fluorogold-positive cells outside the ION should therefore be MF neurons. In the ECN and LRN, no Fluorogold-positive cells were labeled with β-gal (Fig. 2E–J), suggesting that MF neurons in the these nuclei are not in the Ptf1a lineage. Similar results were obtained in the PN and RTN (data not shown), consistent with the observation that no β-gal-positive cells are found in those nuclei (Fig. 1J). MF neurons in the spinal trigeminal nucleus (Okada et al., 2007), which were visualized with Fluorogold, were not β-gal positive (Fig. 2K–M). Furthermore, MF neurons in the dorsal nucleus (Clarke's column) in the spinal cord (Molander et al., 1989) were also not labeled with β-gal (Fig. 2N–P). These observations suggest that MF neurons, during the period of our study, are not derived from Ptf1a-expressing progenitor cells, whereas nearly all CF neurons are in the Ptf1a lineage.
Figure 2.
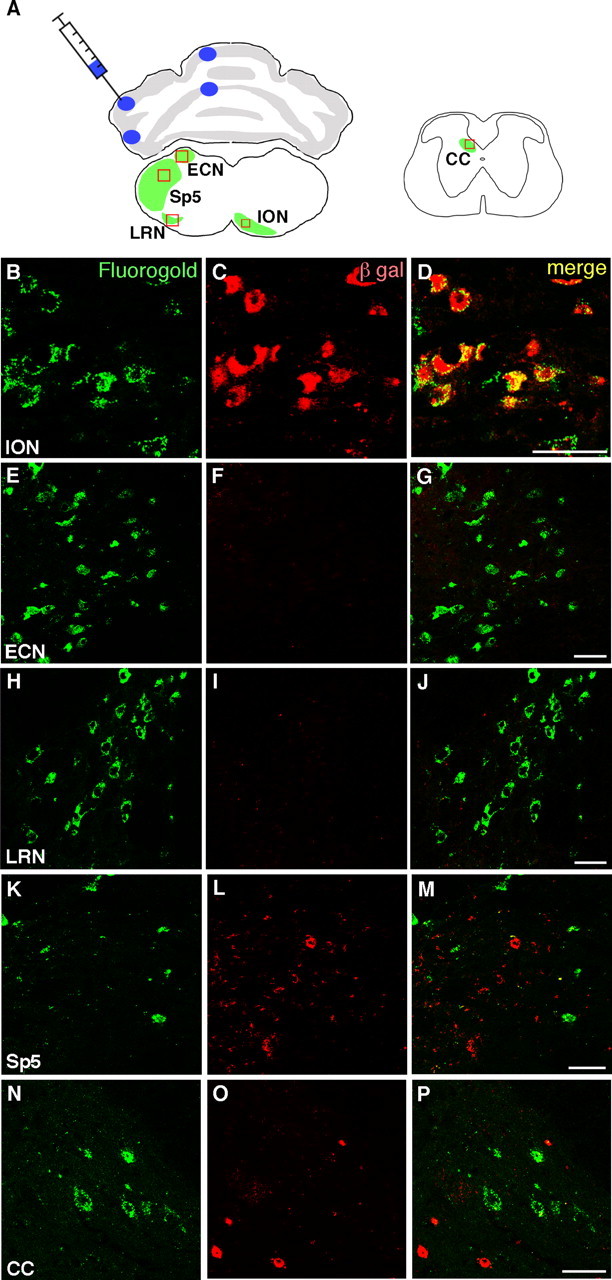
Cells in the Ptf1a lineage do not contain MF neurons. A, Schematic drawing that indicates the injection sites of Fluorogold and retrogradely labeled areas in the caudal hindbrain (left) and the spinal cord (right). Fluorogold was injected into four sites of the cerebellum: anterior and posterior sites of hemisphere and vermis. B–P, Double immunostaining with anti-Fluorogold (green) and β-gal (red) antibodies in the rectangular regions indicated in A. Merged images are also shown. Scale bars, 40 μm. Sp5, Spinal trigeminal nucleus; CC, Clarke's column (dorsal nucleus).
Ptf1a is expressed in a discrete neuroepithelial region of the caudal hindbrain
Previous studies suggested that CF neurons are derived from the neuroepithelium in the embryonic r6–r8 hindbrain region (Ambrosiani et al., 1996; Cambronero and Puelles, 2000; Farago et al., 2006; D. Kawauchi et al., 2006) and that they are produced at approximately E10.5–E11.5 (Pierce, 1973). Several transcriptional factors, such as Lmx1a (LIM homeobox transcription factor 1α), Math1, and Ngn1, are expressed in this region (Akazawa et al., 1995; Ben-Arie et al., 1996; Landsberg et al., 2005). Based on the nonoverlapping expression patterns of those transcriptional genes, Landsberg et al. (2005) defined domains of the caudal hindbrain neuroepithelium along the dorsoventral axis: the Lmx1a, Math1, and dorsal and ventral Ngn1 domains (Fig. 3C). We analyzed the expression of Ptf1a protein in the caudal hindbrain (at the r7 level) at E11.5 and compared it with the localization of the Math1 transcript and Ngn1 protein. As described previously, Math1 is expressed in the dorsal part of the hindbrain (Fig. 3A). Immunostaining of serial sections with anti-Ptf1a and Ngn1 antibodies revealed that Ptf1a was expressed in a region sandwiched by dorsal and ventral Ngn1 domains (Fig. 3B), although a small gap was still recognized between the dorsal Ngn1 domain and Ptf1a-expressing region. BrdU incorporation and immunolabeling studies revealed that these Ptf1a-expressing cells contained a mitotic population (Fig. 3D–F) and are not expressing HuC/D, a postmitotic neuronal marker (Fig. 3G–I), indicating that Ptf1a is not expressed in postmitotic neurons. The expression of Ptf1a in this region was also observed at E10.5, E12.5, and E13.5 but was absent after E14.5 (data not shown). These findings suggest that Ptf1a is expressed in the mitotic neuroepithelial cells located between the dorsal and ventral Ngn1 domains in the caudal hindbrain, and therefore we tentatively named this region the Ptf1a domain. Together with the finding that CF neurons are from the Ptf1a lineage, these observations indicate that CF neurons originate from the Ptf1a neuroepithelial domain of the caudal hindbrain.
Figure 3.
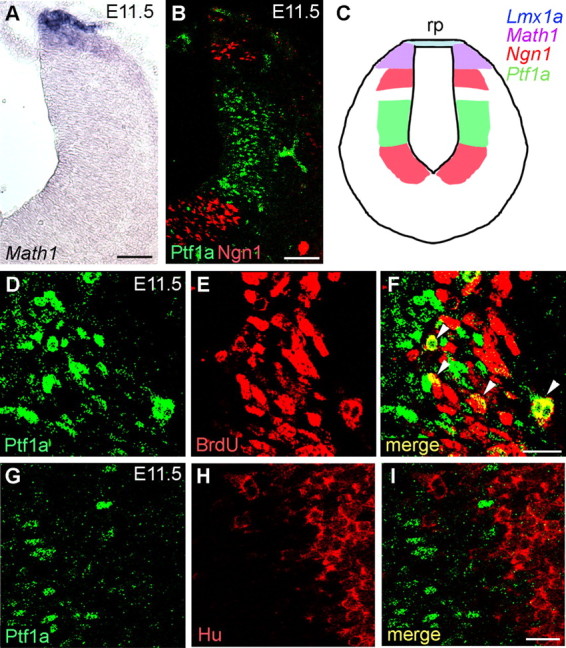
Expression of Ptf1a in the embryonic caudal hindbrain. A, B, Serial transverse frozen sections of r7 hindbrain at E11.5. Localization of Math1 transcripts (visualized by in situ hybridization) and Ptf1a and Ngn1 proteins (visualized by immunohistochemistry) are shown. C, Schematic diagram of expression of bHLH transcription factors/genes in the caudal hindbrain at E11.5. Lmx1a is reported to be expressed in the roof plate (rp) (Landsberg et al., 2005). D–F, Double immunostaining with anti-Ptf1a and BrdU antibodies within the Ptf1a domain of the E11.5 caudal hindbrain. Pregnant mice were given BrdU injections 1 h before embryo harvest and fixation. Many Ptf1a-positive cells incorporate BrdU (arrowheads in F), indicating that they are mitotic. G–I, Double immunostaining with anti-Ptf1a and HuC/D antibodies around the Ptf1a domain of the caudal hindbrain at E11.5. Ptf1a-positive cells do not express HuC/D, suggesting that they do not include postmitotic neurons. Scale bars: A, B, 100 μm; D–I, 20 μm.
Dynamic movement of CF neurons produced from the Ptf1a domain
We next used the β-gal-positive cells in the heterozygous brains (Ptf1acre/+; R26R) to investigate dynamic movement of Ptf1a lineage cells during embryonic stages. β-gal-positive cells could be detected as early as E10.5. At this stage, a small number of β-gal-positive cells were observed in the Ptf1a-expressing neuroepithelial region (data not shown). By E11.5, many cells just beneath the pial surface were expressing β-gal (Fig. 4A, white arrow). Although their location appears to coincide with the posterior precerebellar extramural stream, which has been suggested to be the path for MF neurons in LRN and ECN (Wang et al., 2005), those neurons are produced at later stages (approximately E12.5) and from a more dorsal region, the Math1 domain (Pierce, 1973; Landsberg et al., 2005; Wang et al., 2005). Furthermore, it was shown in Figure 2 that those MF neurons in LRN and ECN are not in the Ptf1a lineage. We suspect that some of the β-gal-positive cells just beneath the pial surface in Figure 4A may migrate contralaterally to give rise to neurons of spinal trigeminal nucleus, although we have no direct evidence of this at present. Actually, some neurons in the spinal trigeminal nucleus were shown to emerge from the contralateral neuroepithelium and migrate circumferentially just beneath the pial surface to reach their final position (Okada et al., 2007). In addition, it was reported that proliferation period for neurons of this nucleus is from E9.5 to E12.5 (Pierce, 1973).
In E12.5 and E13.0 heterozygous brains (Ptf1acre/+; R26R), we were able to identify β-gal-positive cells, which would give rise to CF neurons migrating along the intramural stream (Fig. 4C,E, arrows) (Altman and Bayer, 1987; Bourrat and Sotelo, 1988). This was confirmed by immunostaining of adjacent sections with Brn3a (Fig. 4O,Q) as well as double staining for β-gal and Brn3a (Fig. 4M). Most β-gal-positive CF neurons had reached the ventralmost region of the caudal hindbrain by E13.5 (Figs. 4G,I,K, 5G,I). This was confirmed by immunostaining of adjacent sections with Brn3a (Figs. 4S,U, 5K,M).
Figure 5.
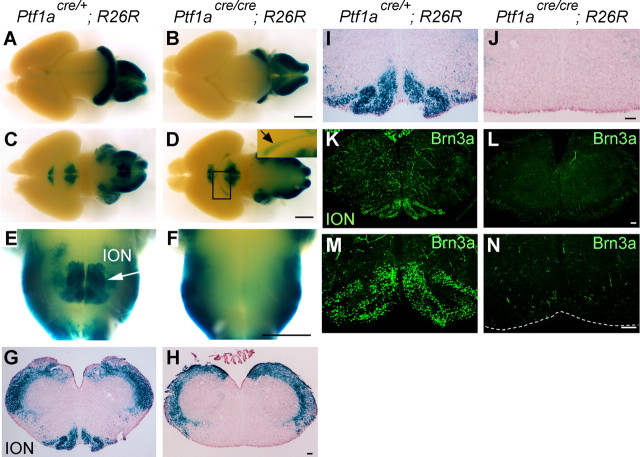
ION is lost in the Ptf1a-null mice. All samples are at E18.5. A–D, Whole-mount X-gal-stained brains of indicated genotypes. Dorsal (A, B) and ventral (C, D) views. The arrow in the inset in D indicates the optic nerve and optic tract stained with X-gal. E, F, High-magnification views of C and D around the ION, respectively. An arrow in E indicates the ION. G, H, X-gal-stained sections of caudal hindbrain of indicated genotypes. I, J, High magnifications of G and H, respectively. K, L, Immunostaining with Brn3a was performed on sections of the caudal hindbrain. M, N, High-magnification views of the ventralmost regions that were stained with Brn3a. Weak false-positive signals can be observed in small blood vessels, because the antibody used was mouse monoclonal. Scale bars: A–F, 1 mm; G–N, 100 μm.
Ptf1a is required for development of CF neurons
Because the Ptf1a homozygous null mice (Ptf1acre/cre) die just after birth (Kawaguchi et al., 2002), we examined heterozygous and homozygous embryos at E18.5, both of which contained R26R reporter alleles (Ptf1acre/+; R26R, Ptf1acre/cre; R26R, respectively). In whole-mount X-gal-stained brains, β-gal-positive areas were found in the medial preoptic area, hypothalamus, cerebellum, and parts of the hindbrain in both heterozygotes and homozygotes (Fig. 5A–D). Interestingly, the optic nerves and tracts were β-gal positive in the homozygotes but not in the heterozygotes (Fig. 5D, inset). This is likely attributable to the fate switching of retinal β-gal-positive cells from amacrine cells to ganglion cells in the Ptf1a mutants, as reported previously (Fujitani et al., 2006; Nakhai et al., 2007). In addition, β-gal-positive areas located at the ventromedial part of the caudal hindbrain could be observed in the heterozygotes (Fig. 5E) but barely in the homozygotes (Fig. 5F). In the heterozygous caudal hindbrain sections, many β-gal-positive areas, including the ION, were observed (Fig. 5G,I), whereas, in Ptf1a null brains (Ptf1acre/cre; R26R), β-gal-positive areas were not observed in the ventromedial region of the caudal hindbrain (Fig. 5H,J). Furthermore, Brn3a-positive cells were not observed in the caudal hindbrain in the mutants (Fig. 5K–N). The morphology of the heterozygous ION is indistinguishable from that of wild-type mice, in terms of location, size, lamellation structure, etc. (data not shown). These results suggest that Ptf1a is involved in development of CF neurons in the ION.
To investigate whether neuroepithelial proliferation in the Ptf1a domain is affected in the Ptf1a null mutants (Ptf1acre/cre; R26R), pregnant mice (E10.5 and E11.5) were given two intraperitoneal injections of BrdU with a 30 min interval. One hour after the first injection, the embryos were fixed, sectioned, and double stained with BrdU and β-gal. In addition, sections were also stained with TOPRO-3 to visualize nuclei (Fig. 6A–D) (supplemental Fig. 3A–D, available at www.jneurosci.org as supplemental material). BrdU incorporation rates (mean ± SEM) of β-gal-positive cells in the ventricular zone were 40.68 ± 3.98% at E10.5 and 46.44 ± 1.79% at E11.5 in Ptf1acre/+; R26R, and 48.99 ± 3.03% at E10.5 and 42.87 ± 1.80% at E11.5 in Ptf1acre/cre; R26R (supplemental Fig. 3E,F, available at www.jneurosci.org as supplemental material). Statistically, we could not find any significant difference in BrdU incorporation rates between two genotypes at either E10.5 or E11.5 (p > 0.1, t test). This indicates that proliferation of neuroepithelial cells in the Ptf1a domain, which are thought to include progenitor cells of CF neurons, is not significantly affected by loss of Ptf1a expression.
Figure 6.
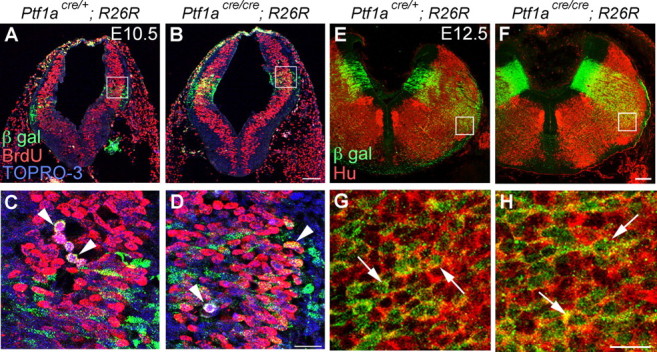
Neuroepithelial cells in the Ptf1a domain proliferate normally and produce neurons in the Ptf1a null mutants. A, B, Double immunolabeling with BrdU and β-gal of E10.5 caudal hindbrains that received BrdU injections 1 h before fixation. Sections were also counterstained with TOPRO-3 to visualize nuclei. C, D, High-magnification views of the rectangular regions in A and B, respectively. Arrowheads indicate β-gal-positive, BrdU-positive nuclei. E, F, Double immunostaining with β-gal and HuC/D in E12.5 caudal hindbrains. G, H, High-magnification views of rectangular regions in E and F, respectively. Arrows indicate representatives of Hu-positive, β-gal positive cells. Genotypes are indicated. Scale bars: A, B, E, F, 100 μm; C, D, G, H, 20 μm.
We performed double staining for β-gal and HuC/D in E12.5 Ptf1acre/+;R26R and Ptf1acre/cre; R26R embryos. Many cells double positive for β-gal and HuC/D were observed in the hindbrains of both heterozygous and homozygous embryos (Fig. 6E–H), suggesting that Ptf1a is not required for producing neurons from the neuroepithelium.
In homozygous (Ptf1acre/cre; R26R) embryos at E11.5 and E12.5, β-gal-positive cells were observed just beneath the pial surface (Fig. 4B,D) similar to the distribution pattern in Ptf1acre/+; R26R (Fig. 4A,C), although the distribution at E12.5 was slightly different from that of heterozygotes (Fig. 4D). At E13.0, we did not observe any ventrally migrating β-gal-positive cells for ION formation in the mutants (Fig. 4F). In contrast to Brn3a-positive cells, which could be observed migrating to form the ION in heterozygotes (Fig. 4O,Q, arrows), Brn3a-positive cells were found to be accumulated in the lateral region of the brainstem in E12.5 and E13.0 homozygotes (Fig. 4P,R). Double immunolabeling with Brn3a and β-gal showed that these Brn3a-positive cells were derived from the Ptf1a domain, which lost the Ptf1a expression (Fig. 4N, data not shown). We further confirmed that most β-gal-positive cells localized in the lateral region in E12.5 heterozygotes were immunoreactive to Brn3b (Fig. 4W), another marker for CF neurons. However, in E12.5 homozygotes, some β-gal-positive cells in that region are not expressing Brn3b (Fig. 4X, white arrows). The ratios of Brn3b-positive cells in β-gal-positive cells accumulated in the lateral region of the caudal hindbrain were 90.35 ± 7.90 and 61.86 ± 8.53% in heterozygotes and homozygotes, respectively, exhibiting a significant statistic difference (n = 4; p < 0.01). This suggests that a part of cells that should differentiate into CF neurons cannot express the CF neuron-specific marker protein Brn3b in homozygotes. Consistently, the number of Brn3a-positive cells decreased as development progressed, and, eventually, no β-gal- and Brn3a-positive ION neurons were found in the ventralmost regions of the caudal hindbrain in E13.5–E16.5 embryos (Fig. 4H,J,L,T,V). At E18.5, no Brn3a or Brn3b-expressing cells were observed anywhere in the mutant caudal hindbrains (Fig. 5L,N) (data not shown).
To detect apoptotic cells, we performed immunostaining with an antibody against the proteolytically activated form of Caspase-3, which can be used as an indicator of apoptosis (Fernandes-Alnemri et al., 1994; Ura et al., 2001). At E13.5, a significantly larger number of positive cells were found in the ventrolateral region of the caudal hindbrain of the mutants compared with heterozygotes (Fig. 4Y,Z).
These findings suggest that the Ptf1a neuroepithelial domain produces neurons that should differentiate into mature CF neurons, but, in Ptf1a mutants, those neurons could neither migrate ventrally nor maintain the expression of CF neuron-specific proteins, such as Brn3a and Brn3b. Moreover, these cells seemed to undergo apoptosis.
Fate of some cells in Ptf1a lineage was changed to MF neurons in the Ptf1a null mutants
In E18.5 heterozygous embryos (Ptf1acre/+; R26R), few β-gal-positive cells were observed in the PN regions (Fig. 7A,C), indicating that PN do not include cells in the Ptf1a lineage. This was also confirmed by our retrograde labeling of MF neurons in the adult PN by Fluorogold, as described above. In contrast, we observed significant β-gal-positive signals in PN regions of the whole-mount mutant embryos at E18.5 (Ptf1acre/cre; R26R) (Fig. 7B,D). In sections, many β-gal-expressing cells were localized in the lateral region of the mutant PN (Fig. 7F,H), whereas β-gal-positive cells were hardly observed in the heterozygous PN (Fig. 7E,G). Adjacent serial sections of samples in Figure 7, E and F, were stained with X-gal and subsequently subjected to in situ hybridization with Mbh2/Barhl1, a specific marker for MF neurons (Saito et al., 1998; Li et al., 2004; Wang et al., 2005). In the mutant PN, many X-gal-stained cells were Mbh2/Barhl1 positive (Fig. 7I,J), suggesting that the fate of these Ptf1a lineage cells was changed to MF neurons.
Figure 7.
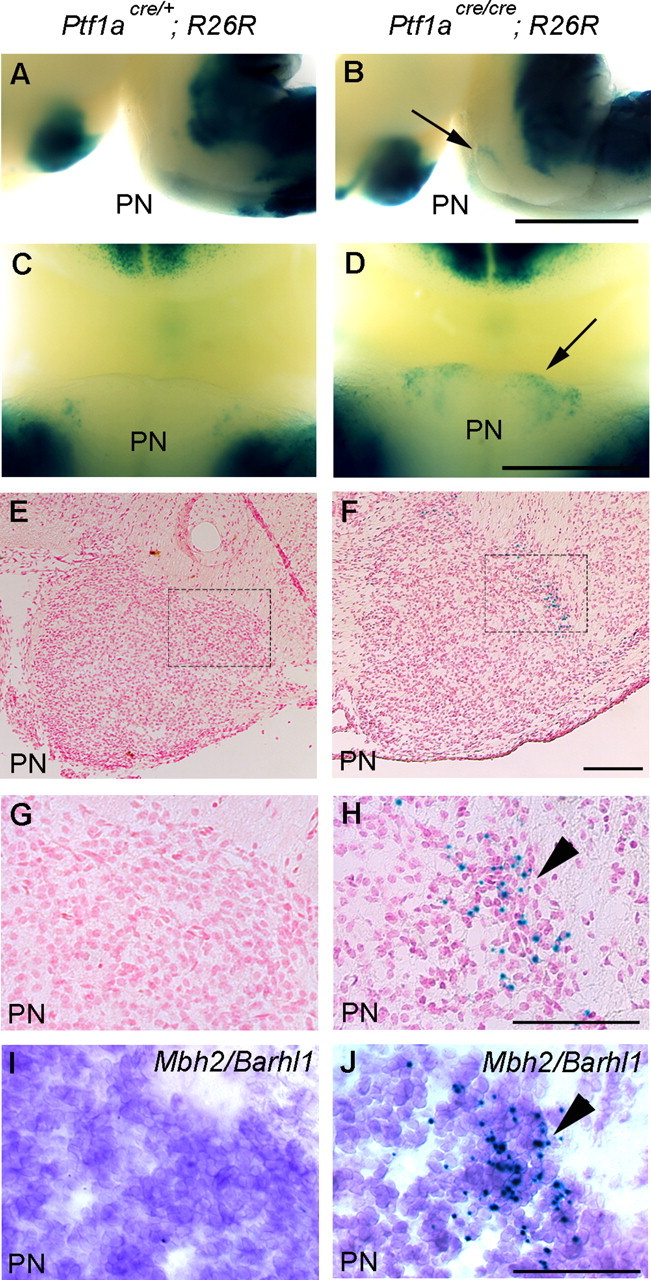
Fate change of Ptf1a lineage cells to MF neurons of the PN in Ptf1a null mutants at E18.5. Genotypes are indicated. A, B, Lateral views of whole-mount embryos stained with X-gal. C, D, Ventral views around the PN regions. Arrows in B and D indicate ectopically localized β-gal-positive cells in the mutant PN. E, F, Transverse sections of the PN regions stained with X-gal and nuclear fast red. G, H, High-magnification views of rectangular regions in E and F, respectively. I, J, Adjacent serial sections of G and H, respectively, which were stained with X-gal and subsequently subjected to in situ hybridization with Mbh2/Barhl1. Scale bars: A–D, 1 mm; E–J, 100 μm.
In homozygous embryos (Ptf1acre/cre; R26R) at E18.5, significant levels of β-gal-positive signals were observed in the dorsolateral region of the caudal hindbrain, which is thought to correspond to the ECN, whereas less intense signals were found in the same region of heterozygous embryos (Ptf1acre/+; R26R) (supplemental Fig. 4A,B, available at www.jneurosci.org as supplemental material). In situ hybridization with Mbh2/Barhl1 showed that MF neurons are present in the heterozygous and homozygous embryos in the ECN (supplemental Fig. 4C,D, available at www.jneurosci.org as supplemental material). Unfortunately, signals detected by in situ hybridization with Mbh2/Barhl1 after the incubation for the X-gal staining were very weak in the ECN region, and therefore we were unable to confirm that β-gal-positive cells in the mutant ECN are expressing Mbh2/Barhl1. However, X-gal staining, performed on the adjacent serial sections of supplemental Figure 4, C and D (available at www.jneurosci.org as supplemental material), revealed that many β-gal-positive cells were localized in the region in which many cells express Mbh2/Barhl1 in homozygotes, whereas much less β-gal-positive cells were found there in heterozygotes (supplemental Fig. 4E,F, available at www.jneurosci.org as supplemental material). This may suggest that cells in the Ptf1a lineage have changed their fate to MF neurons in the ECN.
Discussion
Previously, Landsberg et al. (2005) defined several neuroepithelial subregions in the caudal hindbrain along the dorsoventral axis according to the nonoverlapping expression of several transcriptional genes. From dorsal to ventral, the Lmx1a domain, Math1 domain, and dorsal and ventral Ngn1 domains were delineated. However, the region between the dorsal and ventral Ngn1 domains was not defined, because few transcription factors were known to be expressed there. In this report, we show that a bHLH-type transcription factor, Ptf1a, is expressed in this region.
Previously, several genetic lineage trace analyses revealed the fate of cells generated from discrete neuroepithelial domains in the caudal hindbrain. The Lmx1a domain was found to produce cells in the roof plate and choroid plexus, and the Math1 domain generates MF neurons of PN, RTN, LRN, and ECN (Ben-Arie et al., 2000; Rodriguez et al., 2000; Landsberg et al., 2005; Wang et al., 2005; Chizhikov et al., 2006). In this report, we showed that most CF neurons in the ION are generated from the Ptf1a domain. This is consistent with our previous finding that at least a portion of the ION neurons are derived from Ptf1a-expressing cells (Hoshino et al., 2005).
Landsberg et al. performed in vivo lineage trace analyses by using two variants of FLP (Flippase recombinase) with different recombinase activities that were expressed under the control of the Wnt-1 promoter, demonstrating that CF neurons are derived from the neuroepithelial region in which Wnt-1 is very weakly expressed. It has also been reported that CF neurons are not derived from the Math1 domain (Landsberg et al., 2005; Wang et al., 2005). By comparing the expression patterns of Wnt-1, Ngn1, and Math1, they deduced that CF neurons should originate from the dorsal Ngn1 domain of the caudal hindbrain, which seemed to express Wnt-1 weakly. However, it is very difficult to define the ventral edge of the Wnt-1-expressing area because Wnt-1 expression gradually decreases ventrally (McMahon et al., 1992; Landsberg et al., 2005). We believe that Wnt-1 is expressed extremely weakly in the Ptf1a domain (probably in the dorsal subregion of the Ptf1a domain), in which CF neurons are produced. Nichols and Bruce generated transgenic mice carrying a Wnt-1 enhancer/lacZ transgene and observed that the ION was not labeled by β-gal in those mice. This indicates that, in the transgenic animals, the activity of the Wnt-1 enhancer is below detectable levels in the region in which CF neurons arise, or the Ptf1a domain (Nichols and Bruce, 2006).
In this study, we show that Ptf1a is required for CF neuron development, because the ION is not formed in Ptf1a null mutants. Even in the absence of Ptf1a expression, the Ptf1a neuroepithelial domain can proliferate normally and produce neurons, as revealed by BrdU incorporation and HuC/D immunostaining studies. These facts suggest that Ptf1a is not required for neuroepithelial proliferation and neuron production. However, neurons produced from the mutant Ptf1a domain cannot differentiate into CF neurons appropriately. Those neurons failed to migrate to the ventralmost region of the hindbrain and were unable to maintain the expression of CF neuronal markers. Furthermore, significantly larger numbers of apoptotic cells were observed in the caudal hindbrain of the Ptf1a-null mutants. These findings suggest that Ptf1a may be involved in directing differentiation of neuronal progenitors into mature CF neurons as well as survival of those neurons.
Disorder in ION formation is also observed in several mutant mouse lines. In netrin-1-deficient mice, a portion of CF neurons cannot migrate to the ventromedial region of the caudal hindbrain, resulting in ectopic localization of CF neurons along the migratory path (Bloch-Gallego et al., 1999). In map1b−/− (microtubule-associated protein 1b knock-out mice) or map1b−/−; map2−/− mice, folding of the ION laminar structure is disorganized (Teng et al., 2001). Targeted disruption of cyclin dependent kinase 5 (Cdk5) results in failure to form the ION, probably attributable to a migration disorder in CF neurons (Ohshima et al., 2002). Similar phenotypes are also observed in double mutant mice for Cdk5 activators, p35 and p39 (Ko et al., 2001). All of these genes have been implicated in neuronal migration or guidance, rather than neuronal differentiation, whereas Ptf1a seems to participate in differentiation of CF neurons.
In Ptf1acre/cre; R26R mice, cells in the Ptf1a lineage (β-gal-positive cells) were ectopically localized in the PN, which expresses Mbh2/Barhl1, an MF neuron-specific marker. In the ECN of the mutants, the number of β-gal-positive cells was found to be dramatically increased. These observations suggest the possibility that, in the Ptf1a-null mutants, the fate of cells in the Ptf1a lineage may be changed to MF neurons in the PN and ECN. However, this does not directly indicate that cells that were committed to be CF neurons become MF neurons when Ptf1a expression is lost, because cells in the Ptf1a lineage include not only CF neurons but also other cell types, such as neurons in the spinal trigeminal nucleus, solitary nucleus, and dorsal cochlear nucleus. Along the rostrocaudal axis, it is known that MF neurons in the PN and ECN are generated from the caudal part of the hindbrain (Ambrosiani et al., 1996; Cambronero and Puelles, 2000; Farago et al., 2006; D. Kawauchi et al., 2006). CF neurons are also generated from the caudal part of the hindbrain, although their neuroepithelial origin along the dorsoventral axis is distinct from that of MF neurons. If the Math1 and Ptf1a neuroepithelial domains of the caudal hindbrain possess a common spatial identity relative to the rostrocaudal axis, it would be possible for cells that are committed to become CF neurons to change their fate to MF neurons in PN and ECN. However, we do not have direct evidence for this. In the Ptf1a mutant mice, changes of cell fates have been reported in the pancreas, retina, spinal cord, and cerebellum (Glasgow et al., 2005; Fujitani et al., 2006; Fukuda et al., 2006; Nakhai et al., 2007; Pascual et al., 2007).
In this study, the Ptf1a domain of the caudal hindbrain generates not only the CF neurons but also other cell types, such as neurons in the spinal trigeminal nucleus and solitary nucleus. Therefore, we believe that additional transcriptional factors are expressed within the Ptf1a domain and specific combinations of transcription factors that include Ptf1a may be involved in specifying precise neuronal subtypes. Alternatively, transcription factors expressed in immature neurons that have just exited the cell cycle may be involved in specifying neuronal subtypes.
In the cerebellum, granule cells are derived from the Math1-expressing rhombic lip (Ben-Arie et al., 1997), whereas Purkinje cells are produced from the ventricular zone in which Ptf1a is expressed (Hoshino et al., 2005). Development of both granule and Purkinje cells are dependent on Math1 and Ptf1a, respectively. Regarding the cerebellar afferent pathways, as pointed out previously (Bermingham et al., 2001), MF neurons and their targets (granule cells) are both derived from Math1-expressing neuroepithelial regions, although their birthplaces along the rostrocaudal axis are distinct. In this report, we show that both Purkinje cells and CF neurons, components of the other cerebellar afferent pathway, both arise from Ptf1a-expressing neuroepithelial regions, although their places of origin are distinct along the rostrocaudal axis. These correlations suggest a role for cell-fate-influencing transcription factors, such as Math1 and Ptf1a, in specifying multiple components of a neuronal pathway or in the formation of a specific neuronal network.
In cerebelless (cbll), the hypomorphic mutant of Ptf1a (Ptf1acbll/cbll), the ION, which contains CF neurons, seems to develop normally until postnatal day 2 (P2). However, during early postnatal stages (P3–P4), it suddenly disappears, accompanied by apoptosis (Hoshino et al., 2005). This contrasts with the Ptf1a null mutant (Ptf1acre/cre) in which CF neurons do not develop, resulting in a failure to form the ION during embryogenesis. Our quantitative reverse transcription-PCR analysis revealed that the levels of Ptf1a transcripts in Ptf1acbll/cbll embryos were dramatically reduced in the cerebellum (26.3-fold reduction), but the hindbrain was more mildly affected (4.5-fold reduction). The amount of Ptf1a expression remaining in the cbll hindbrain may be sufficient to allow development of CF neurons.
It was shown previously that Ptf1a is involved in differentiation of GABAergic neurons in the cerebellum and dorsal spinal cord (Glasgow et al., 2005; Hoshino et al., 2005). Furthermore, ectopic expression of Ptf1a in the dorsal telencephalon can convert the neuronal subtype from glutamatergic to GABAergic (Hoshino et al., 2005). These findings suggest that Ptf1a may affect the transcription of GABAergic neuron-related genes. However, in this report, we show that Ptf1a is also involved in the development of glutamatergic CF neurons. Moreover, Ptf1a was shown recently to be involved in development of retinal horizontal and amacrine cells that contain non-GABAergic cell types. These observations indicate that other transcriptional factors coexpressed with Ptf1a may modify the transcriptional targets of Ptf1a to specify neuronal subtypes. Additional studies are required for understanding the molecular machinery that specifies neuronal subtypes in the nervous system.
Footnotes
This work was supported by Grants-in-Aid for Scientific Research on Priority Areas “Elucidation of Glia–Neuron Network-Mediated Information Processing Systems” (Grant 18053014), “Molecular Brain Science 16067101” (Grant 18022019), “A Nuclear System to DECODE” (Grant 18055017), “Dynamics of Extracellular Environments” (Grant 18060019), and “G Protein Signal” (Grant 18057010); by grants from the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation; and by the Uehara Memorial Foundation. We thank T. Obata for technical assistance, C. V. E Wright for Ptf1acre mice, P. Soriano and R. Behringer for R26R mice, R. Kageyama and T. Saito for the Math1 and Mbh2/Barhl1 probes for in situ hybridization, respectively; M. Watanabe for an anti-glutaminase antibody; Y. Gotoh for technical advice on the apoptosis assay; and T. Kawauchi and R. Yu for helpful discussions.
References
- Akazawa C, Ishibashi M, Shimizu C, Nakanishi S, Kageyama R. A mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J Biol Chem. 1995;270:8730–8738. doi: 10.1074/jbc.270.15.8730. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Development of the precerebellar nuclei in the rat. I–IV. J Comp Neurol. 1987;257:477–552. doi: 10.1002/cne.902570402. [DOI] [PubMed] [Google Scholar]
- Ambrosiani J, Armengol JA, Martinez S, Puelles L. The avian inferior olive derives from the alar neuroepithelium of the rhombomeres 7 and 8: an analysis by using chick-quail chimeric embryos. NeuroReport. 1996;7:1285–1288. doi: 10.1097/00001756-199605170-00013. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, McCall AE, Berkman S, Eichele G, Bellen HJ, Zoghbi HY. Evolutionary conservation of sequence and expression of the bHLH protein Atonal suggests a conserved role in neurogenesis. Hum Mol Genet. 1996;5:1207–1216. doi: 10.1093/hmg/5.9.1207. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Hassan BA, Bermingham NA, Malicki DM, Armstrong D, Matzuk M, Bellen HJ, Zoghbi HY. Functional conservation of atonal and Math1 in the CNS and PNS. Development. 2000;127:1039–1048. doi: 10.1242/dev.127.5.1039. [DOI] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Wang VY, Fernandez M, Banfi S, Bellen HJ, Fritzsch B, Zoghbi HY. Proprioceptor pathway development is dependent on Math1. Neuron. 2001;30:411–422. doi: 10.1016/s0896-6273(01)00305-1. [DOI] [PubMed] [Google Scholar]
- Bloch-Gallego E, Ezan F, Tessier-Lavigne M, Sotelo C. Floor plate and netrin-1 are involved in the migration and survival of inferior olivary neurons. J Neurosci. 1999;19:4407–4420. doi: 10.1523/JNEUROSCI.19-11-04407.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourrat F, Sotelo C. Migratory pathways and neuritic differentiation of inferior olivary neurons in the rat embryo. Axonal tracing study using the in vitro slab technique. Brain Res. 1988;467:19–37. doi: 10.1016/0165-3806(88)90064-8. [DOI] [PubMed] [Google Scholar]
- Cambronero F, Puelles L. Rostrocaudal nuclear relationships in the avian medulla oblongata: a fate map with quail chick chimeras. J Comp Neurol. 2000;427:522–545. doi: 10.1002/1096-9861(20001127)427:4<522::aid-cne3>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Chizhikov VV, Lindgren AG, Currle DS, Rose MF, Monuki ES, Millen KJ. The roof plate regulates cerebellar cell-type specification and proliferation. Development. 2006;133:2793–2804. doi: 10.1242/dev.02441. [DOI] [PubMed] [Google Scholar]
- Farago AF, Awatramani RB, Dymecki SM. Assembly of the brainstem cochlear nuclear complex is revealed by intersectional and subtractive genetic fate maps. Neuron. 2006;50:205–218. doi: 10.1016/j.neuron.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Fedtsova NG, Turner EE. Brn-3.0 expression identifies early post-mitotic CNS neurons and sensory neural precursors. Mech Dev. 1995;53:291–304. doi: 10.1016/0925-4773(95)00435-1. [DOI] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Litwack G, Alnemri ES. CPP32, a novel human apoptotic protein with homology to Caenorhabditis elegans cell death protein Ced-3 and mammalian interleukin-1 beta-converting enzyme. J Biol Chem. 1994;269:30761–30764. [PubMed] [Google Scholar]
- Fredette BJ, Adams JC, Mugnaini E. GABAergic neurons in the mammalian inferior olive and ventral medulla detected by glutamate decarboxylase immunocytochemistry. J Comp Neurol. 1992;321:501–514. doi: 10.1002/cne.903210402. [DOI] [PubMed] [Google Scholar]
- Fujitani Y, Fujitani S, Luo H, Qiu F, Burlison J, Long Q, Kawaguchi Y, Edlund H, MacDonald RJ, Furukawa T, Fujikado T, Magnuson MA, Xiang M, Wright CV. Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development. 2006;133:4439–4450. doi: 10.1242/dev.02598. [DOI] [PubMed] [Google Scholar]
- Fukuda A, Kawaguchi Y, Furuyama K, Kodama S, Horiguchi M, Kuhara T, Koizumi M, Boyer DF, Fujimoto K, Doi R, Kageyama R, Wright CV, Chiba T. Ectopic pancreas formation in Hes1 -knockout mice reveals plasticity of endodermal progenitors of the gut, bile duct, and pancreas. J Clin Invest. 2006;116:1484–1493. doi: 10.1172/JCI27704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow SM, Henke RM, Macdonald RJ, Wright CV, Johnson JE. Ptf1a determines GABAergic over glutamatergic neuronal cell fate in the spinal cord dorsal horn. Development. 2005;132:5461–5469. doi: 10.1242/dev.02167. [DOI] [PubMed] [Google Scholar]
- Hoshino M, Sone M, Fukata M, Kuroda S, Kaibuchi K, Nabeshima Y, Hama C. Identification of the stef gene that encodes a novel guanine nucleotide exchange factor specific for Rac1. J Biol Chem. 1999;274:17837–17844. doi: 10.1074/jbc.274.25.17837. [DOI] [PubMed] [Google Scholar]
- Hoshino M, Nakamura S, Mori K, Kawauchi T, Terao M, Nishimura YV, Fukuda A, Fuse T, Matsuo N, Sone M, Watanabe M, Bito H, Terashima T, Wright CV, Kawaguchi Y, Nakao K, Nabeshima Y. Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron. 2005;47:201–213. doi: 10.1016/j.neuron.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- Kawauchi D, Taniguchi H, Watanabe H, Saito T, Murakami F. Direct visualization of nucleogenesis by precerebellar neurons: involvement of ventricle-directed, radial fibre-associated migration. Development. 2006;133:1113–1123. doi: 10.1242/dev.02283. [DOI] [PubMed] [Google Scholar]
- Kawauchi T, Chihama K, Nabeshima Y, Hoshino M. The in vivo roles of STEF/Tiam1, Rac1 and JNK in cortical neuronal migration. EMBO J. 2003;22:4190–4201. doi: 10.1093/emboj/cdg413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi T, Chihama K, Nabeshima Y, Hoshino M. Cdk5 phosphorylates and stabilizes p27kip1 contributing to actin organization and cortical neuronal migration. Nat Cell Biol. 2006;8:17–26. doi: 10.1038/ncb1338. [DOI] [PubMed] [Google Scholar]
- Ko J, Humbert S, Bronson RT, Takahashi S, Kulkarni AB, Li E, Tsai LH. p35 and p39 are essential for cyclin-dependent kinase 5 function during neurodevelopment. J Neurosci. 2001;21:6758–6771. doi: 10.1523/JNEUROSCI.21-17-06758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Knofler M, Ledermann B, Burki K, Berney C, Zoerkler N, Hagenbuchle O, Wellauer PK. The bHLH protein PTF1–p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 1998;12:3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakopoulou K, de Diego I, Wassef M, Karagogeos D. A combination of chain and neurophilic migration involving the adhesion molecule TAG-1 in the caudal medulla. Development. 2002;129:287–296. doi: 10.1242/dev.129.2.287. [DOI] [PubMed] [Google Scholar]
- Landsberg RL, Awatramani RB, Hunter NL, Farago AF, DiPietrantonio HJ, Rodriguez CI, Dymecki SM. Hindbrain rhombic lip is comprised of discrete progenitor cell populations allocated by Pax6. Neuron. 2005;48:933–947. doi: 10.1016/j.neuron.2005.11.031. [DOI] [PubMed] [Google Scholar]
- Li S, Qiu F, Xu A, Price SM, Xiang M. Barhl1 regulates migration and survival of cerebellar granule cells by controlling expression of the neurotrophin-3 gene. J Neurosci. 2004;24:3104–3114. doi: 10.1523/JNEUROSCI.4444-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24. doi: 10.1016/j.neuron.2005.08.028. [DOI] [PubMed] [Google Scholar]
- Matsuo N, Hoshino M, Yoshizawa M, Nabeshima Y. Characterization of STEF, a guanine nucleotide exchange factor for Rac1, required for neurite growth. J Biol Chem. 2002;277:2860–2868. doi: 10.1074/jbc.M106186200. [DOI] [PubMed] [Google Scholar]
- Matsuo N, Terao M, Nabeshima Y, Hoshino M. Roles of STEF/Tiam1, guanine nucleotide exchange factors for Rac1, in regulation of growth cone morphology. Mol Cell Neurosci. 2003;24:69–81. doi: 10.1016/s1044-7431(03)00122-2. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Joyner AL, Bradley A, McMahon JA. The midbrain-hindbrain phenotype of Wnt-1-/Wnt-1- mice results from stepwise deletion of engrailed-expressing cells by 9.5 days postcoitum. Cell. 1992;69:581–595. doi: 10.1016/0092-8674(92)90222-x. [DOI] [PubMed] [Google Scholar]
- Molander C, Xu Q, Rivero-Melian C, Grant G. Cytoarchitectonic organization of the spinal cord in the rat. II. The cervical and upper thoracic cord. J Comp Neurol. 1989;289:375–385. doi: 10.1002/cne.902890303. [DOI] [PubMed] [Google Scholar]
- Nakhai H, Sel S, Favor J, Mendoza-Torres L, Paulsen F, Duncker GI, Schmid RM. Ptf1a is essential for the differentiation of GABAergic and glycinergic amacrine cells and horizontal cells in the mouse retina. Development. 2007;134:1151–1160. doi: 10.1242/dev.02781. [DOI] [PubMed] [Google Scholar]
- Nichols DH, Bruce LL. Migratory routes and fates of cells transcribing the Wnt-1 gene in the murine hindbrain. Dev Dyn. 2006;235:285–300. doi: 10.1002/dvdy.20611. [DOI] [PubMed] [Google Scholar]
- Obata J, Yano M, Mimura H, Goto T, Nakayama R, Mibu Y, Oka C, Kawaichi M. p48 subunit of mouse PTF1 binds to RBP-Jkappa/CBF-1, the intracellular mediator of Notch signalling, and is expressed in the neural tube of early stage embryos. Genes Cells. 2001;6:345–360. doi: 10.1046/j.1365-2443.2001.00422.x. [DOI] [PubMed] [Google Scholar]
- Ohshima T, Ogawa M, Takeuchi K, Takahashi S, Kulkarni AB, Mikoshiba K. Cyclin-dependent kinase 5/p35 contributes synergistically with Reelin/Dab1 to the positioning of facial branchiomotor and inferior olive neurons in the developing mouse hindbrain. J Neurosci. 2002;22:4036–4044. doi: 10.1523/JNEUROSCI.22-10-04036.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Keino-Masu K, Masu M. Migration and nucleogenesis of mouse precerebellar neurons visualized by in utero electroporation of a green fluorescent protein gene. Neurosci Res. 2007;57:40–49. doi: 10.1016/j.neures.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Pascual M, Abasolo I, Mingorance-Le Meur A, Martinez A, Del Rio JA, Wright CV, Real FX, Soriano E. Cerebellar GABAergic progenitors adopt an external granule cell-like phenotype in the absence of Ptf1a transcription factor expression. Proc Natl Acad Sci USA. 2007;104:5193–5198. doi: 10.1073/pnas.0605699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce ET. Time of origin of neurons in the brain stem of the mouse. Prog Brain Res. 1973;40:53–65. doi: 10.1016/S0079-6123(08)60679-2. [DOI] [PubMed] [Google Scholar]
- Rodriguez CI, Dymecki SM. Origin of the precerebellar system. Neuron. 2000;27:475–486. doi: 10.1016/s0896-6273(00)00059-3. [DOI] [PubMed] [Google Scholar]
- Ruigrok TJH, Cella F. The rat nervous system. Ed 2. New York: Academic; 1995. Precerebellar nuclei and red nucleus; pp. 277–308. [Google Scholar]
- Saito T, Sawamoto K, Okano H, Anderson DJ, Mikoshiba K. Mammalian BarH homologue is a potential regulator of neural bHLH genes. Dev Biol. 1998;199:216–225. doi: 10.1006/dbio.1998.8889. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Teng J, Takei Y, Harada A, Nakata T, Chen J, Hirokawa N. Synergistic effects of MAP2 and MAP1B knockout in neuronal migration, dendritic outgrowth, and microtubule organization. J Cell Biol. 2001;155:65–76. doi: 10.1083/jcb.200106025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ura S, Masuyama N, Graves JD, Gotoh Y. MST1-JNK promotes apoptosis via caspase-dependent and independent pathways. Genes Cells. 2001;6:519–530. doi: 10.1046/j.1365-2443.2001.00439.x. [DOI] [PubMed] [Google Scholar]
- Wang VY, Rose MF, Zoghbi HY. Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron. 2005;48:31–43. doi: 10.1016/j.neuron.2005.08.024. [DOI] [PubMed] [Google Scholar]
- Yee KT, Simon HH, Tessier-Lavigne M, O'Leary DM. Extension of long leading processes and neuronal migration in the mammalian brain directed by the chemoattractant netrin-1. Neuron. 1999;24:607–622. doi: 10.1016/s0896-6273(00)81116-2. [DOI] [PubMed] [Google Scholar]
- Yoshizawa M, Hoshino M, Sone M, Nabeshima Y. Expression of stef, an activator of Rac1, correlates with the stages of neuronal morphological development in the mouse brain. Mech Dev. 2002;113:65–68. doi: 10.1016/s0925-4773(01)00650-5. [DOI] [PubMed] [Google Scholar]
- Yoshizawa M, Sone M, Matsuo N, Nagase T, Ohara O, Nabeshima Y, Hoshino M. Dynamic and coordinated expression profile of dbl-family guanine nucleotide exchange factors in the developing mouse brain. Gene Expr Patterns. 2003;3:375–381. doi: 10.1016/s1567-133x(03)00002-4. [DOI] [PubMed] [Google Scholar]
- Yoshizawa M, Kawauchi T, Sone M, Nishimura VY, Terao M, Chihama K, Nabeshima Y, Hoshino M. Involvement of a Rac activator, P-Rex1, in neurotrophin-derived signaling and neuronal migration. J Neurosci. 2005;25:4406–4419. doi: 10.1523/JNEUROSCI.4955-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]