Abstract
Inhibitory GABAergic interneurons of the mouse neocortex are a highly heterogeneous population of neurons that originate from the ventral telencephalon and migrate tangentially up into the developing cortical plate. The majority of cortical interneurons arise from a transient embryonic structure known as the medial ganglionic eminence (MGE), but how the remarkable diversity is specified in this region is not known. We have taken a genetic fate mapping strategy to elucidate the temporal origins of cortical interneuron subtypes within the MGE. We used an inducible form of Cre under the regulation of Olig2, a basic helix-loop-helix transcription factor highly expressed in neural progenitors of the MGE. We observe that the physiological subtypes of cortical interneurons are, to a large degree, unique to their time point of generation.
Keywords: cortex, electrophysiology, GABA, genetics, interneurons, transcription factor
Introduction
Mammalian neocortex comprises two major classes of neurons, excitatory glutamatergic pyramidal neurons and inhibitory GABAergic interneurons. Fate-mapping studies of the ventral telencephalon have revealed that the medial (MGE) and caudal (CGE) ganglionic eminences produce most of these GABAergic cortical interneurons (Wichterle et al., 2001; Nery et al., 2002; Xu et al., 2004; Butt et al., 2005; Wonders and Anderson, 2006). How the diverse subtypes of interneurons within each eminence are generated is unknown. This is particularly important for the MGE, which generates the large majority of all cortical interneurons, including parvalbumin (PV)- and somatostatin (SST)-expressing subtypes with highly divergent intrinsic electrophysiological profiles (Xu et al., 2004, 2005; Butt et al., 2005). Diversity in the excitatory pyramidal neurons is produced in a temporal sequence (Rakic, 1974). Similarly, at least with regard to laminar position, cortical interneurons are specified in a similar inside-out manner (Miller, 1985; Fairen et al., 1986; Valcanis and Tan, 2003; Xu et al., 2004; Yozu et al., 2004). However, whether their time of origin has a more profound role in determining their physiological subtype has not been investigated.
DNA analogs such as [3H]thymidine or 5-bromo-2′-deoxyuridine (BrdU) (Angevine and Sidman, 1961; Miller and Nowakowski, 1988) have been traditionally used to birthdate neurons. Indeed, it was through such an analysis that the correlation between the laminar position of cortical interneurons and their birth date was established (Miller, 1985; Fairen et al., 1986; Valcanis and Tan, 2003; Xu et al., 2004; Yozu et al., 2004). Extending these studies to examine whether time of birth also predicts the physiological subtypes of cortical interneurons is non-trivial. Focal injection methods suffer from the difficulty of reproducibly targeting specific embryonic structures, and BrdU labeling is incompatible with physiological methods, except by retrospective analysis. Recently, the advent of temporally inducible genetic fate mapping has provided an ideal means to reliably label progenitor populations at specific developmental times (Kimmel et al., 2000; Joyner and Zervas, 2006). Using this approach, we have assessed whether distinct cortical interneuron subtypes are generated at different developmental time points. Here, we study the distinct cohorts of cells arising from progenitors expressing high levels of the basic helix-loop-helix (bHLH) gene Olig2 (Lu et al., 2002; Zhou and Anderson, 2002; Lee et al., 2005). In general, the bHLH family of transcription factors are appealing targets for use in inducible genetic fate mapping because their expression is tightly regulated and often involved in the specification of neuronal subtypes (Bertrand et al., 2002; Ma et al., 2006).
In the forebrain, Olig2 is expressed in the progenitors of the ventral embryonic eminences, with the highest levels of expression occurring in the Nkx2.1-expressing MGE domain. We have taken advantage of this expression pattern to perform genetic fate mapping to elucidate the temporal origins of cortical interneurons. Our fate-mapping analysis reveals that different physiological subtypes of cortical interneurons arise at different embryonic time points. Thus, we conclude that cortical interneuron subtypes are determined by their time of origin.
Materials and Methods
Animals.
Olig2-CreER heterozygous (Takebayashi et al., 2002), Z/EG transgenic (Novak et al., 2000) males were crossed to 7- to 8-week-old wild-type Swiss Webster females (Taconic, Germantown, NY) for producing the litters. Four milligrams [embryonic day 9.5 (E9.5), E10.5, E12.5] or 5 mg (E15.5) of tamoxifen (Sigma, St. Louis, MO) prepared 20 mg/ml in corn oil (Sigma) were given to pregnant mothers at 1:00 P.M. ±30 min by the gavaging method with silicon-protected needles. When mothers did not deliver pups at noon on E19.5, a cesarean section was performed, and pups were fostered. PCR genotyping of Olig2-CreER allele was performed by using a set of three primers (forward, TCGAGAGCTTAGATCATCC; reverse, CACCGCCGCCCAGTTTGTCC; mutant reverse, GGACAGAAGCATTTTCCAGG) giving wild-type (247 bp) and mutant (172 bp) bands. Z/EG allele was genotyped by using the primers specific to the enhanced green fluorescent protein (EGFP) or by using LacZ staining solution to the animal's body piece.
Immunohistochemistory.
Molecular expression profiles of interneurons are characterized at postnatal day 21 (P21) in somatosensory barrel cortex. Three brains were analyzed for each time point of tamoxifen administration, and interneurons were characterized by immunohistochemistry (E9.5, n = 641; E10.5, n = 1123; E12.5, n = 815; E15.5, n = 268). Brains were fixed by transcardiac perfusion followed by 1 or 3 h of postfixation on ice with 4% formaldehyde/PBS solution. Brains were rinsed with PBS and cryoprotected by using 25% sucrose/PBS solution overnight at 4°C. Cryosections were prepared at 12 μm (for counting immunoprofiles) or 25 μm (for capturing images) thickness. Immunohistochemistry was performed by using a PBS solution containing 1.5% normal goat serum and 0.1% Triton X-100 for all procedures, except for the washing steps just with PBS. The basic strategy is as described previously (Butt et al., 2005). Nuclear counterstaining was performed with 100 ng/ml 4′,6-diamidino-2-phenylindole (DAPI) solution in PBS for 20 min. LacZ histochemical stainings on cryosections were performed as described previously (Nery et al., 2002). Antibodies were used in the following concentrations: rabbit anti-Olig2 (1:200; gift from Dr. C. Stiles, Dana-Farber Cancer Institute, Boston, MA), mouse anti-Nkx2.1 (TTF-1) (1:200; Progen, Heidelberg, Germany), mouse anti-Pax6 (1:200; Developmental Studies Hybridoma Bank, Iowa City, IA), mouse anti-Mash1 (1:1000; BD PharMingen, San Diego, CA), mouse anti-Ki67 (1:500; BD PharMingen), rat anti-platelet-derived growth factor receptor α (PDGFRα) (1:500; BD PharMingen), rabbit anti-pan-Dlx (1:1000; gift from Dr. J. Kohtz, Northwestern University, Chicago, IL), rabbit anti-GFP (1:1000; Invitrogen, Eugene, OR), rat anti-GFP (1:1000; Nacalai Tesque, Kyoto, Japan), mouse anti-GFAP (1:400; Chemicon, Temecula, CA), rabbit anti-NG2 (1:200; Chemicon), mouse anti-PV (1:1000; Sigma), rat anti-SST (1:250; Chemicon), rabbit anti-neuropepetide Y (1:500; Incstar, Stillwater, MN), rabbit anti-vasointestinal polypeptide (VIP) (1:500; Incstar), and mouse anti-calretinin (CR) (1:1500; Chemicon). Fluorescent images were captured using a cooled CCD camera (Princeton Scientific Instruments, Trenton, NJ) using MetaMorph software (Universal Imaging, Downingtown, PA).
Electrophysiology.
Electrophysiological analysis was performed on mice (P13–P22) as described previously (Butt et al., 2005). Briefly, animals were deeply anesthetized and decapitated, and the brain was quickly removed and transferred to ice-cold physiological Ringer's solution of the following composition (in mm): 125 NaCl, 2.5 KCl, 25 NaHCO3, 1.25 NaH2PO4, 1 MgCl2, 2 CaCl2, and 20 glucose. The brain was then fixed to a stage, and 250 μm slices were cut on a Vibratome (Vibratome 3000 EP; Vibratome, St. Louis, MO). Slices were then individually transferred to an incubation chamber containing oxygenated Ringer's solution at room temperature for a minimum period of 1 h before recording. During recording, slices were continually perfused with oxygenated Ringer's solution of the same composition.
Whole-cell tight-seal recordings were made from EGFP-positive neurons located in layers II–VI of the somatosensory and more medial cortices. Apart from their cortical area, we randomly selected EGFP-positive profiles typically 40–70 μm below the surface of the slice. Patch electrodes were made from borosilicate glass (resistance, 5–8 MΩ; Harvard Apparatus, Kent, UK) and filled with a solution containing (in mm) 128 K-gluconate, 4 NaCl, 0.3 GTP, 5 ATP, 0.0001 CaCl2, 10 HEPES, and 1 glucose. For postrecording immunohistochemistry and morphological reconstruction, 0.2% Lucifer yellow was also included.
All recordings were performed in current-clamp mode (Axoclamp 2B; Molecular Devices, Union City, CA) and analyzed off-line in Clampfit version 9.2. Standard electrophysiological protocols were followed throughout. Passive membrane properties were ascertained shortly after rupturing the patch and periodically during the course of the experiments to ensure that there was no significant deterioration in the health of the cell. Cells that showed significant rundown were discarded. Two types of basic protocol were used. (1) Short-duration (500 ms) depolarizing and hyperpolarizing current steps (0.1–0.2 Hz) were applied to the cells to characterize their endogenous electrophysiological profile, similar to Kawaguchi (1995) and colleagues. All parameters were measured on at least three occasions for each cell, and the average value was calculated. (2) Prolonged current injection (5 s) was applied at around threshold and suprathreshold values, and cells were classified according to the criteria established by Markram and colleagues (Gupta et al., 2000; Markram et al., 2004).
Note on interneuron nomenclature.
Recorded interneurons were characterized on the basis of 26 intrinsic electrophysiological parameters as well as morphology and laminar location. Analysis of all of these parameters revealed that the majority of interneurons could be allocated to 1 of 10 discrete populations. In terms of nomenclature, we have diverged from our previous system (Butt et al., 2005) in favor of that proposed in the conference at Petilla (2005) and, in particular, that outlined in the principles of in vitro classification. This classification recognizes six types of spike pattern: fast spiking (FS); non-adapting, non-fast spiking (NFS); adapting; accelerating; irregular; and intrinsic bursting (IB). Of the cells recorded, we found three broad types: FS, IB, and NFS. A key concern lies in that definitions and nomenclature based solely on intrinsic electrophysiological criteria do not adequately reflect the homogeneity in both physiology and morphology that we encountered. For example, although a number of late-spiking (LS) interneurons showed acceleration, others did not. However, they were sufficiently distinct in nearly every other electrophysiological criterion and most notably in their neurogliaform morphology that it was evident that they represented a distinct class of interneuron. Similarly with regard to non-FS interneurons, a large number of the populations showed variability in the degree of spike frequency adaptation but were otherwise extremely homogeneous in single spike dynamics, membrane properties, and morphology, so to split them along the lines of non-adapting and adapting appeared arbitrary. Although we have attempted to remain faithful to key tenants of the Petilla classification, we suggest that the developmental genetics provides a better context for defining discrete interneuron subtypes than criteria based on mature characteristics.
Results
Inducible genetic fate mapping of Olig2high precursors
To genetically identify the origins of cortical interneuron subtypes, we have focused on a bHLH gene, Olig2, that is expressed in the ventricular zone (VZ) of the ventral telencephalon (Lu et al., 2000; Takebayashi et al., 2000; Nery et al., 2001). We began by doing a detailed immunohistochemical analysis of Olig2 protein expression at multiple embryonic time points (see supplemental Fig. 1, available at www.jneurosci.org as supplemental material). We observed that, between E9.5 and E16.5, Olig2 expression levels in VZ progenitors is high within the MGE and low within the lateral ganglionic eminence (LGE) and the CGE (Fig. 1C; see also the illustration of embryonic forebrain in Fig. 1B). The high levels of Olig2 expression in the MGE coincide with the expression of the homeobox domain-containing gene Nkx2.1 (Fig. 1D). Nkx2.1 is not only expressed throughout the majority of the MGE but is known to be important for establishing this region, because MGE character is altered in the absence of this gene (Sussel et al., 1999). Furthermore, this region is known to give rise to a large percentage of cortical interneurons (Wichterle et al., 2001; Xu et al., 2004), which in turn represent a large degree of the mature physiological heterogeneity (Butt et al., 2005).
Figure 1.
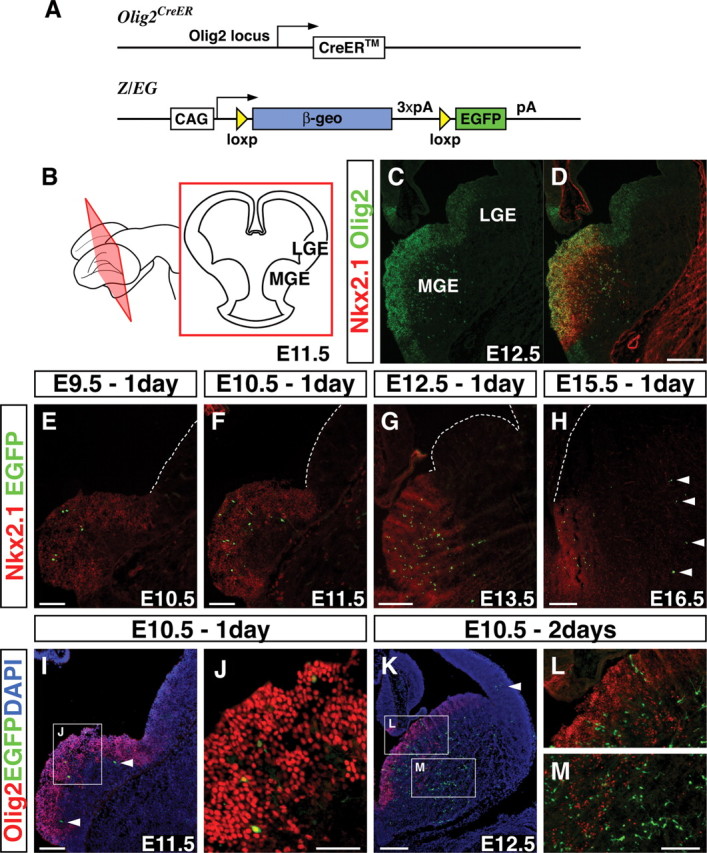
Temporally regulated genetic fate mapping of the Olig2high precursors. A, A schematic showing the genetic fate-mapping strategy used in the experiments by combining the Olig2-CreER driver and the Z/EG reporter lines. CAG, Hybrid promoter; pA, polyadenylation signal. B, Schematic of the E11.5 brain with a coronal section showing the location of the MGE and the LGE. C, D, Olig2 is expressed in the VZ progenitors at high levels within the Nkx2.1-expressing domain but only weakly in the Nkx2.1-negative domain of the ventral telencephalon. E–H, By administrating 4 mg (E9.5, E10.5, E12.5) or 5 mg (E15.5) of tamoxifen to timed pregnant mothers at E9.5 (E), E10.5 (F), E12.5 (G), and E15.5 (H) and analyzing 1 d later, labeled EGFP-expressing cells were found within the Nkx2.1-expressing domain. After E15.5 tamoxifen administration, some cells in the mantle were also labeled where Olig2 is expressed at this stage (H, arrowheads) (see also supplemental Fig. 1, available at www.jneurosci.org as supplemental material). I, J, At E11.5, 1 d after tamoxifen administration at E10.5, many labeled cells are Olig2 positive (I; high magnification in J). However, a few cells have already downregulated Olig2 by this time point (I, arrowheads). K, Two days after tamoxifen administration at E10.5, some labeled cells have already migrated into the cortex (arrowhead). Many EGFP-positive cells do not coexpress Olig2 2 d after tamoxifen administration (K), suggesting that these cells have already downregulated Olig2 (K–M). L, M, High magnifications of the regions in K. Scale bars: C, D, G, H, K, 200 μm; E, F, I, L, M, 100 μm; J, 50 μm.
Because we hypothesized that time of origin is critical to the generation of cortical interneuron diversity, we wanted to fate map the Olig2-expressing precursors at different embryonic time points. To accomplish this, we took advantage of two genetically modified alleles: (1) a driver line carrying a tamoxifen-inducible form of the site-specific Cre recombinase (CreER) under the control of the Olig2 locus (Takebayashi et al., 2002) (Fig. 1A); and (2) a Z/EG reporter line (Novak et al., 2000) (Fig. 1A) that expresses sufficient levels of EGFP for detection subsequent to the Cre-mediated removal of a stop cassette flanked by loxp sites (Fig. 1A). Using these alleles, we performed a mosaic fate-mapping analysis of Olig2-expressing precursors by transiently activating CreER through tamoxifen administration to pregnant mothers at four different embryonic time points: E9.5, E10.5, E12.5, and E15.5. The reporter initiates EGFP expression after 6 h, and maximal labeling is complete 12–24 h after tamoxifen administration (Hayashi and McMahon, 2002). To identify the initial population for our fate-mapping study, we have analyzed 24 h after tamoxifen administration and found that the majority of the EGFP-expressing cells are found within the domain of Nkx2.1 expression (Fig. 1E–H). This suggests that the labeled cells are confined to the spatially restricted domain where Olig2 shows high levels of expression (Fig. 1C,D). The only exceptions were a few cells in the mantle from the E15.5 tamoxifen administration (Fig. 1H, arrowheads) that most likely correspond to oligodendrocyte progenitors, because many of the fate-mapped cells coexpress the oligodendrocyte progenitor marker PDGFRα (data not shown). In fact, at E15.5, many Olig2-expressing cells are observed in the mantle coexpressing PDGFRα (see supplemental Fig. 1J, available at www.jneurosci.org as supplemental material).
Spatially restricted labeling within the Nkx2.1-expressing domain was achieved by adjusting the amount of tamoxifen to the pregnant mothers (4 mg: E9.5, E10.5, and E12.5; 5 mg: E15.5). Notably, administrating higher amounts of tamoxifen (8 mg) or using a different reporter line [e.g., R26R-stop-LacZ line (Soriano, 1999)] results in the labeling of cells with low levels of Olig2 expression in the LGE and CGE regions (see supplemental Fig. 2, available at www.jneurosci.org as supplemental material). Given that our objective was to confine our fate mapping to the cells within the MGE (i.e., the Olig2high region), only the dose of tamoxifen that resulted in near-exclusive labeling within the Nkx2.1-expression domain was used in our analysis.
At E11.5, 1 d after tamoxifen administration, many EGFP-expressing cells also coexpressed Olig2 (Fig. 1I,J). Those that did not coexpress Olig2 (Fig. 1I, arrowheads) were positioned just lateral to the region of high Olig2 expression, suggesting that they have recently downregulated Olig2 expression. At E12.5, 2 d after tamoxifen administration, many fate-mapped cells are found in the mantle of the ventral telencephalon (Fig. 1K,M), and some cells are already evident in the cortex (Fig. 1K, arrowhead). We found that most of the fate-mapped cells do not express Olig2 at this time point (Fig. 1K–M) and most of them have exited the progenitor domain. Because of this, when tamoxifen administration is performed at E12.5, a population distinct from that seen at E10.5 is labeled. Hence, by using Olig2-CreER driver and Z/EG reporter lines in combination with a specific dose of tamoxifen, we successfully label temporally distinct populations.
Temporally distinct layering of cortical interneurons reveals that our fate mapping targets precursors
Based on previous analysis (Miller, 1985; Fairen et al., 1986; Valcanis and Tan, 2003; Xu et al., 2004; Yozu et al., 2004), it is known that early-born interneurons occupy deep layers and late-born interneurons end up in more superficial layers of mature cortex similar to the inside-out pattern of pyramidal cell generation (Rakic, 1974). We therefore examined whether the time of tamoxifen administration correlates with the laminar distribution of the fate-mapped cortical interneurons (Fig. 2). To narrow our focus and to better compare our results to our previous fate-mapping analysis of cortical interneuron diversity (Butt et al., 2005), we restricted our analysis to the P21 somatosensory barrel field (Fig. 2A–D) [based on the atlas of the mouse brain (Paxinos and Franklin, 2001)]. We examined coronal sections using DAPI nuclear counterstaining to ascertain the laminar position of the fate-mapped cortical interneurons (Fig. 2A–D). EGFP-expressing cells with glial morphology were eliminated from our analysis (Fig. 2D, open arrowheads, and supplemental Fig. 3, available at www.jneurosci.org as supplemental material), and only cortical interneurons were analyzed (Fig. 2D, filled arrowhead and inset). Consistent with our short-term fate-mapping results, we observed that although those populations labeled at early developmental time points (E9.5, E10.5) preferentially populated deep cortical laminas (Fig. 2A,B,E), those cells labeled at the late developmental time point mainly populated superficial cortical layers (Fig. 2D,E). Our results are thus consistent with former birthdating studies examining GAD immunopositive cells within this same cortical area (Fairen et al., 1986). This suggests that our temporally regulated genetic fate-mapping method correlates highly with the “birth date” of the cortical interneurons and that our labeling is targeting precursors rather than progenitor populations.
Figure 2.
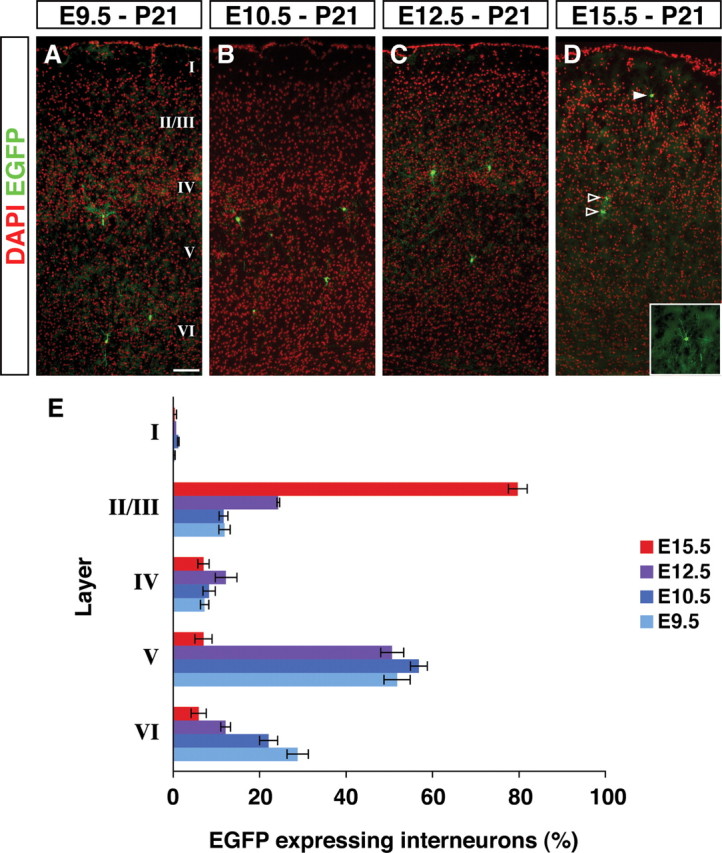
Early and late fate-mapped cortical interneurons follow an inside-out layering pattern. Layering of the fate-mapped cortical interneurons within the P21 somatosensory barrel field was analyzed. A–D, Representative examples of sections are shown from tamoxifen administration at E9.5 (A), E10.5 (B), E12.5 (C), and E15.5 (D). Analysis has been done based on the cortical layers shown by DAPI nuclear staining (pseudocolored in red) and EGFP immunostaining (green). Cells with interneuron morphologies (D, filled arrowhead and inset) were analyzed for their molecular expression and physiological character, but the glial cells (D, open arrowheads) were not considered further in our analysis. E, The layer distribution of cortical interneurons fate mapped from each embryonic time point were analyzed. Fate-mapped interneurons followed an early-deep and late-superficial trend. Error bars indicate SE. Scale bar, 100 μm.
Molecular profiles of the fate-mapped cortical interneurons
To examine cortical interneuron subtypes of the different temporal cohorts fate mapped from Olig2high precursors, we undertook an immunohistochemical analysis at P21. Interneuron markers have been partially explored in previous birthdating studies (Cavanagh and Parnavelas, 1988, 1989, 1990; Xu et al., 2004; Yozu et al., 2004). A recent study (Xu et al., 2006) highlighted species differences between mouse and rat in the molecular expression profiles of cortical interneurons. We therefore began by characterizing known interneuron markers in mouse somatosensory cortex (Fig. 3H). In accordance with previous work in the rat visual (Gonchar and Burkhalter, 1997) and frontal (Kubota et al., 1994; Kawaguchi and Kubota, 1997; Kawaguchi and Kondo, 2002) cortices, we confirmed that the PV, SST, and VIP interneurons constitute primarily non-overlapping populations (Fig. 3H). We did, however, note certain differences that may reflect species (rat vs mouse) or areal (visual, frontal vs somatosensory) variation. For instance, subpopulations of CR-expressing interneurons also coexpressed SST, in accordance with Xu et al. (2006). Similarly, not all of the VIP-expressing population was found to express CR (Fig. 3H). Both of these observations suggest that the dual labeling of cortical interneurons is required for more precise assignment of cells to specific subpopulations. For this reason, we have characterized cortical interneurons based on PV or SST/CR or VIP/CR profiles (Fig. 3H). This allowed us to classify cortical interneurons into seven groups based on their molecular expression profiles.
Figure 3.
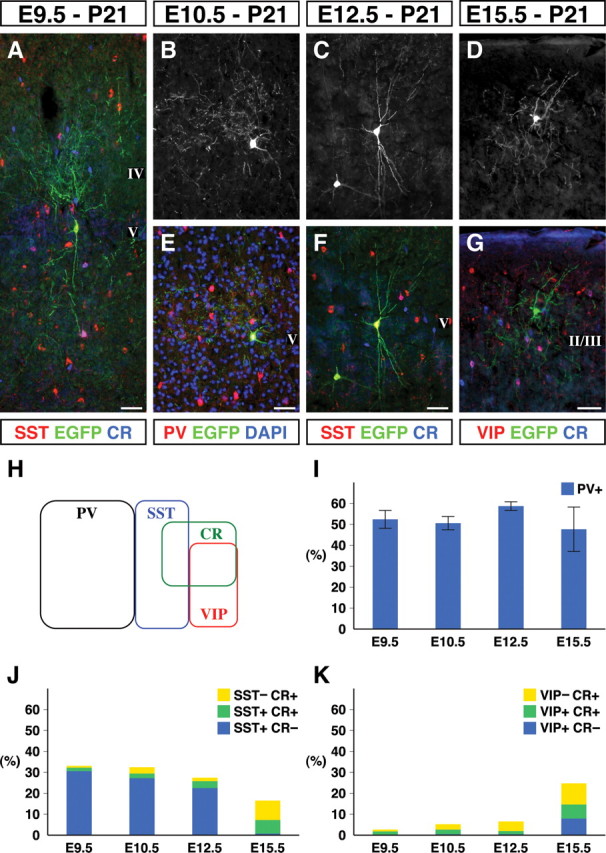
The morphologies and the molecular expression profiles of fate-mapped cortical interneurons. A–G, Images of mature fate-mapped cortical interneurons were visualized by their EGFP expression at P21 after tamoxifen administration at E9.5 (A), E10.5 (B, E), E12.5 (C, F), and E15.5 (D, G). These experiments resulted in the labeling of various interneuron subtypes including, SST+/CR− Martinotti cells (A, C, F), PV+ basket cells (B, E), and VIP−/CR− neurogliaform cells (D, G). Scale bars, 100 μm. H, The distribution of cortical interneuron markers at P21 in mouse somatosensory barrel cortex. PV, SST and VIP are expressed in mutually exclusive populations, whereas the CR+ population somewhat overlapped, except for the PV+ interneuron population. Based on these four markers, interneurons can be subdivided into seven groups. I, The PV+ FS interneurons comprised ∼50% of fate-mapped interneurons at all the time points examined. J, The SST+/CR− subtype comprised 30% of the interneurons fate mapped at early time points but are almost completely absent in the E15.5 fate mapping. J, K, In contrast, the VIP+ and CR+ subtypes were not fate mapped at early time points but became more prominent at the E15.5 time point of fate mapping. Each value with an SE represents data collected from three brains.
PV expression correlates strongly with the electrophysiological subclass of FS cortical interneurons. Indeed, in mouse cortex, previous work has shown that PV is widely found coexpressed with Kv3.1 (Chow et al., 1999), a voltage-gated potassium channel that is functionally required for interneurons to possess FS character (Erisir et al., 1999). Large numbers of PV-expressing interneurons (Fig. 3B,E) were labeled by tamoxifen administration at all the time points tested. In fact, ∼50% of the fate-mapped interneurons at each time point were PV positive (Fig. 3I). This suggests that the FS subtype of cortical interneurons is continuously generated throughout development.
In contrast, the numbers of SST-positive, CR-negative interneurons that include a number of non-FS interneuron subtypes comprised 30% of the populations fate mapped at early time points (Fig. 4A,J) but decreased in number at later time point (Fig. 3C,F,J) and were almost absent from the populations labeled with tamoxifen administration at E15.5 (Fig. 3J). Finally, CR-positive, SST-negative interneurons and VIP-positive interneurons that are often associated with bipolar, bitufted, or double-bouquet morphologies were mainly observed at the E15.5 tamoxifen administration and found enriched within the superficial layers of cortex (Fig. 3J,K). Based on observations in a previous analysis (Butt et al., 2005), in which the fate of cortical interneurons was examined through homotopic transplantation, it seems likely that these late-born populations arise from progenitors that do not express Nkx2.1.
Figure 4.
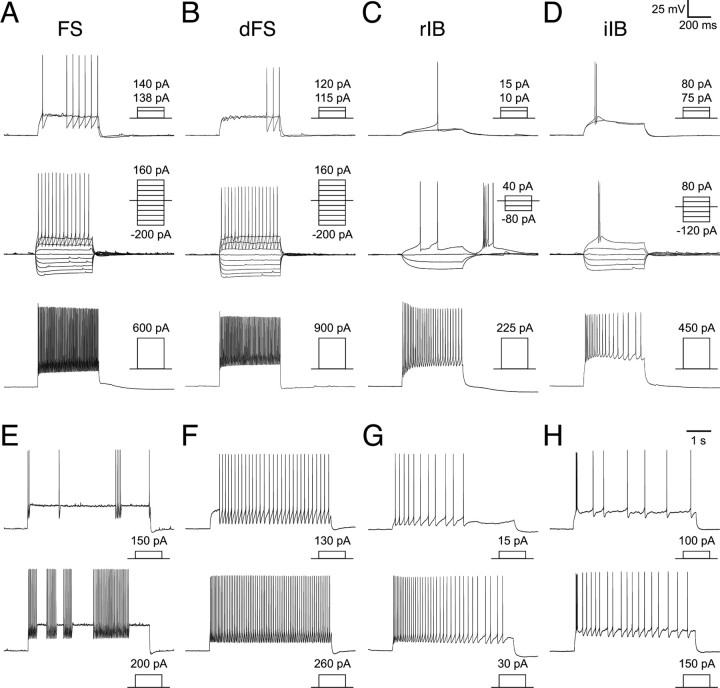
Intrinsic electrophysiological profiles of the fate-mapped cortical interneurons: FS and IB interneuron subtypes. A–H, The response of four subtypes of fate-mapped interneurons of FS and IB classes to short-duration (500 ms) current steps (A–D) or prolonged (5 s) near threshold current injection (E–H). In A–D, short-duration current injection protocols tested threshold current injection (top), response to hyperpolarizing current injection (middle), and suprathreshold current injection (bottom) that elicited discharge of action potentials at a near maximal firing frequency. Profiles are shown for a classical FS interneuron (A) with a stuttering firing pattern (E); a dFS interneuron (B) with a continuous firing pattern (F); an rIB interneuron (C) that exhibits pronounced adaptation in firing frequency (C, G); and an iIB interneuron (D) that exhibits a burst of two to three spikes and then sporadic spikes or pronounced adaptation in firing frequency (D, H).
Surprisingly, despite the known role for Olig2 in the generation of oligodendrocytes (Lu et al., 2002; Takebayashi et al., 2002; Yue et al., 2006), based on their morphologies, very few of early (E9.5: 2.6%, n = 658; E10.5: 1.7%, n = 1143; E12.5: 3.6%, n = 845) fate-mapped cells took on a glial fate. Because most of the cells fate mapped in our experiments arise from precursors within the Nkx2.1-expression domain (Fig. 1E–H), our results are consistent with recent findings showing that the cortical oligodendrocytes generated from Nkx2.1-expressing progenitors are essentially eliminated by P10 (Kessaris et al., 2006). However, our fate-mapping efforts after E15.5 tamoxifen administration resulted in many oligodendrocytes in the cortex (see supplemental Fig. 3A,B,F–I, available at www.jneurosci.org as supplemental material). This was not surprising because oligodendrocyte progenitors in the mantle region of both the ventral telencephalon and the cortex express Olig2 at high levels at E15.5 (see supplemental Fig. 1J–L, available at www.jneurosci.org as supplemental material), and some are labeled after short-term fate mapping (Fig. 1H, arrowheads). Interestingly, although cortical progenitors marked by Pax6 start expressing Olig2 at E16.5 (see supplemental Fig. 1K,L, available at www.jneurosci.org as supplemental material), we did not observe any pyramidal neurons fate mapped even when the highly recombinagenic R26R-stop-LacZ reporter line was used (data not shown). This suggests that the cortical progenitors that express Olig2 give rise entirely to glia.
Analysis of the intrinsic physiological properties of fate-mapped cortical interneurons
Even using immunohistochemical double labeling with the best-characterized markers of cortical interneurons, it is impossible to accurately assess the extent to which specific subtypes of cortical interneurons (Markram et al., 2004) arise from the MGE at different embryonic time points. This encouraged us to undertake a more rigorous physiological analysis. To this end, the intrinsic properties of a total of 203 EGFP-positive interneurons were recorded from P13–P21 animals that had previously undergone tamoxifen administration at E9.5 (n = 51), E10.5 (n = 50), E12.5 (n = 52), or E15.5 (n = 50). Consistent with our immunohistochemical analysis, a majority of EGFP-positive interneurons that we recorded were from the somatosensory cortex (see Materials and Methods for details). Of all the interneurons recorded, 97% belonged to one of 10 discrete interneuron populations (Table 1) defined on the basis of both their intrinsic electrophysiological profiles (Figs. 4, 5) and morphologies (Fig. 6). Tamoxifen administration at E15.5 exhibited EGFP-positive morphologies consistent with them being glia (see supplemental Fig. 3, available at www.jneurosci.org as supplemental material). Indeed, these cells did not fire action potentials in response to current injection (data not shown). Data from these cells were not included in our analysis.
Table 1.
Principle intrinsic characteristics of Olig2-CreER heterozygous, Z/EG transgenic interneurons (n = 203)
| FS (n = 71) | dFS (n = 25) | rIB (n = 17) | iIB (n = 9) | NFS1 (n = 16) | NFS2 (n = 17) | dNFS1 (n = 11) | dNFS2 (n = 6) | LS (n = 17) | iAD (n = 7) | |
|---|---|---|---|---|---|---|---|---|---|---|
| RMP | −68.4 | −67.1 | −58.5 | −63.8 | −63.6 | −60.5 | −62.3 | −62.5 | −65.9 | −66.0 |
| Rin | 246 | 221 | 514 | 548 | 273 | 572 | 562 | 514 | 669.1 | 542 |
| Tau | 13.9 | 13.4 | 42.6 | 47.9 | 21.9 | 38.5 | 45.6 | 36.1 | 32.9 | 17.9 |
| Spike threshold | −32.9 | −32.9 | −43.3 | −41.6 | −41.5 | −38.7 | −35.8 | −34.7 | −34.1 | −33.2 |
| Spike height | 66.0 | 61.2 | 77.0 | 73.4 | 81.2 | 71.8 | 68.7 | 76.2 | 65.3 | 67.8 |
| Spike width | 0.7 | 0.8 | 1.1 | 1.3 | 0.9 | 1.2 | 1.3 | 1.3 | 1.4 | 1.5 |
| AHP1 rt | 4.5 | 5.7 | 4.6 | 5.1 | 5.6 | 8.7 | 11.4 | 12.5 | 20.0 | 10.1 |
| AHP1 amp | −20.4 | −22.8 | −9.8 | −9.1 | −10.4 | −14.7 | −18.3 | −16.2 | −20.8 | −10.3 |
| AHP2 rt | 37.7 | 43.8 | 32.2 | 29.9 | 27.3 | 28.4 | 27.9 | |||
| AHP2 amp | −9.5 | −7.9 | −9.8 | −11.7 | −16.2 | −12.9 | −14.5 | |||
| Delay to spike | 68.3 | 376.5 | 146.8 | 191.0 | 104.4 | 104.4 | 327.1 | 256.9 | 399.5 | 54.5 |
| Maximum firing frequency | 160.0 | 156.1 | 77.8 | 68.4 | 99.9 | 85.6 | 85.5 | 73.3 | 74.1 | 72.9 |
| 2 × T frequency | 53.7 | 53.2 | 16.9 | 18.2 | 24.0 | 20.8 | 16.4 | 22.3 | 21.3 | 18.6 |
| Percentage of adaptation frequency | 17.6 | 15.5 | 44.4 | 41.7 | 36.2 | 32.1 | 29.5 | 33.1 | 33.4 | 45.1 |
| Voltage sag (Ih) | 2.9 | 2.6 | 3.8 | 2.5 | 5.0 | 4.3 | 5.0 | 3.5 | 3.1 | 0.9 |
| Rebound spike | N | N | Y (5.4) | N (2.4)a | N | N | N | N | N | N |
Cells not included are six CGE-like interneurons and one unspecified NFS. Rin, Input resistance (MΩ); RMP, resting membrane potential; rt, rise time; amp, amplitude; 2 × T, 2 × threshold current stop; N, no, Y, yes.
aBurst of two or more spikes from a hyperpolarized potential.
Figure 5.
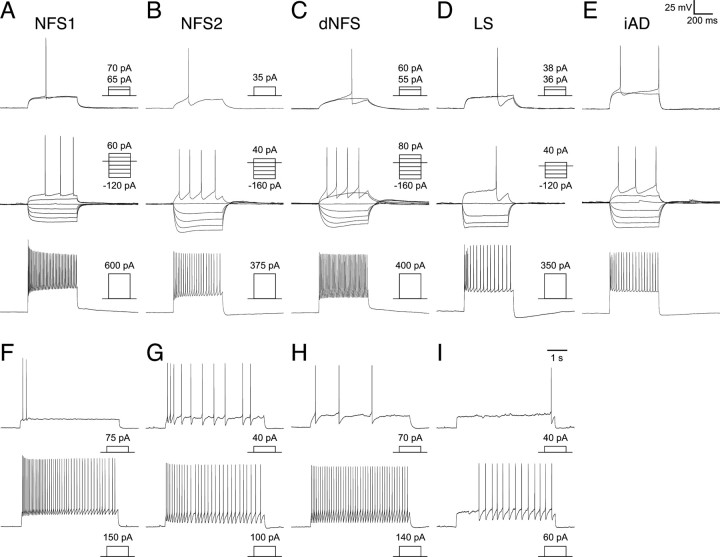
Intrinsic electrophysiological profiles of the fate-mapped cortical interneurons: NFS, LS, and iAD interneuron subtypes. As in Figure 4, the response of six subtypes of fate-mapped interneurons are described. Profiles are shown for an NFS1 interneuron (A, F) that showed adaptation in firing frequency; an NFS2 interneuron (B, G) showing biphasic AHPs and also showed adaptation in firing frequency; dNFS1 and dNFS2 interneurons (C, H) that varied in the degree of exhibiting adaptation in spike frequency; an LS interneuron that, in some case, showed pronounced initial adaptation in firing frequency and spike amplitude (D, I) but in others showed either a slight increase in firing frequency or no adaptation. E, iAD interneurons all analyzed have shown pronounced adaptation in both spike frequency and amplitude in the initial phase of the current injection.
Figure 6.
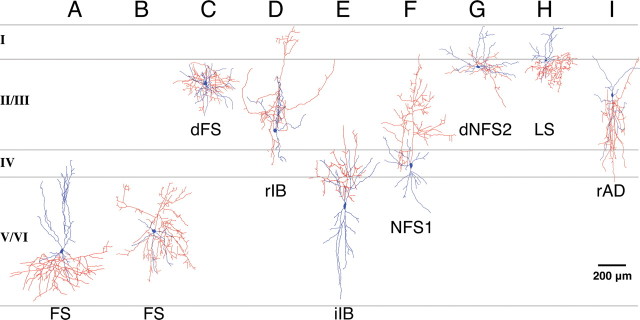
Examples of the morphological diversity of the fate-mapped cortical interneurons. EGFP-expressing interneurons were recorded in all layers, and their morphologies were recovered and reconstructed using camera lucida. Axons and dendrites are indicated in red and blue, respectively. Examples are not representative of quantitative laminar distribution. A, B, Classical FS interneurons were recovered from all of the layers and exhibited an array of dendritic and axonal arbors. A large basket cell (A) and nest basket cell (B) are shown. C, dFS interneurons were mainly found in superficial layers. D, rIB interneurons were preferentially recorded from early fate-mapping time points and were located in both deep and superficial layers. E, iIB interneurons were preferentially located in deep layers and often had axons emerging from pial-side dendrites. F, A number of NFS1 interneurons were analyzed with early fate-mapped populations (E10.5–E12.5) exhibiting bitufted morphologies with ascending axons as in the case of NFS1 interneurons. G, dNFS2 interneurons exhibited prominent lateral dendritic and axonal arbours. H, LS neurogliaform interneurons were mainly located in superficial layers. I, Six interneurons of the total analyzed [example of a rapidly adapting interneuron (rAD) is shown] exhibited bipolar or double-bouquet morphologies and were not readily assigned to one of the 10 subtypes we have identified. The lamina location for all of the interneurons shown is approximate.
The major population of interneurons observed in this study was FS subtype (Fig. 4A). They were characterized by fast, non-adapting spiking character and possessed action potentials of short half-width and large, fast, single-event afterhyperpolarizations (AHPs). In response to sustained current injection at or above threshold, FS interneurons either exhibited a stuttering spike discharge (Fig. 4E) or fired in a continuous manner (Fig. 4F). Two subpopulations of FS interneuron could be discriminated on the basis of the spiking at threshold: classical FS interneurons fired a spike at the onset of the current step (Fig. 4A), whereas delayed FS interneurons fired late in the threshold current step on a slow ramp depolarization (Fig. 4B). In addition, the latter were morphologically distinct in that they had numerous primary dendrites and dense, local axonal and dendritic arbors (Fig. 6C); classical FS interneurons represented a more diverse array of morphologies of small, large (Fig. 6A), and nest (Fig. 6B) basket cells.
Another distinctive interneuron subtype was classified on the basis that it fired a burst of two or more action potentials at threshold when stepped from a hyperpolarized potential (typically −80 mV). These interneurons previously termed burst-spiking nonpyramidal interneurons (Kawaguchi, 1995) were divided into two populations: rebound intrinsic bursting (rIB) (Fig. 4C) and initial intrinsic bursting (iIB) interneurons (Fig. 4D). The main difference between these two classes was that rIB interneurons exhibited significantly higher resting membrane potentials than iIB interneurons (Table 1) and when injected with hyperpolarizing current steps exhibited a prominent rebound-depolarizing hump and associated burst of action potentials (Fig. 4C). rIB interneurons do not, however, burst when injected with depolarizing current steps from resting membrane potential but fired single spikes at threshold (Fig. 4C). At suprathreshold current injections, rIB interneurons fired in a frequency adapting manner (Fig. 4C). The other population, iIB, fired a burst of two or more action potentials when depolarized from resting membrane potential (Fig. 4D) but exhibited no transient rebound depolarization or burst. iIB interneurons, however, showed pronounced adaptation in spike frequency at suprathreshold (Fig. 4H). Morphologically, both classes of IB interneurons had bitufted, sparsely spiny dendrites with an ascending axon that, in the cases in which it could be followed, projected to and arborized in layer I (Fig. 6D,E). The axon emerged from the pial side of the soma in all of the rIB interneurons but in six of nine iIB interneurons emerged from one of the pial-side primary dendrites (Fig. 6E).
Four of the remaining six subtypes were termed NFS, and these corresponded to previously described regular-spiking nonpyramidal (RSNP) interneurons (Kawaguchi, 1995; Butt et al., 2005). One subtype that has not previously been described in detail was the NFS type 1 (NFS1) interneuron population (Fig. 5A), which was highly distinctive in that they had large spike height, short half-width action potentials, and low input resistance. In these aspects, they shared somewhat similar characteristics to FS interneurons. However, different from FS subtype, they had small biphasic AHPs and showed adaptation in spike frequency (Fig. 5A). NFS2 interneurons are similar to the NR-RSNP subtype previously reported in the in vivo MGE transplantation study (Butt et al., 2005) and had biphasic AHPs often with a prominent notch between the fast and slow AHPs (Fig. 5B). Although NFS1 interneurons always exhibited considerable adaptation in spike frequency (Fig. 5A,F), NFS2 interneurons varied, with some showing hardly any adaptation at all; but unlike FS interneurons, they never fired at high frequency (>100 Hz) (Fig. 5B,G). Both classes of NFS interneurons had smooth bitufted dendrites, and in the majority of cases had ascending axons that branched around and above the soma as well as often projecting into superficial layers (Fig. 6F). Two subclasses of NFS interneurons showed a pronounced delay to spike at threshold (Fig. 5C), and we have termed them delayed NFS types 1 and 2 (dNFS1 and dNFS2). dNFS1 subtype was morphologically and electrophysiologically identical to the delayed-spiking interneurons recorded from a previous in vivo transplantation study (Butt et al., 2005). Morphologically they were bitufted with sparsely spiny dendrites and axonal arbors ramifying through the layers immediately above the soma (data not shown). dNFS2 (n = 6) differed significantly from dNFS1 in their morphological characteristics and had distinctive lateral dendrites running parallel to the layer II/I boundary, and in all cases, the axon emerged from one of the lateral dendrites (Fig. 6G). Similar to the dendritic arbor, the axon often extended horizontally and often into layer I, and in addition, in most cases, it also projected a single or maximally a couple of collaterals down into the underlying layers. Similar to dNFS1 interneurons, dNFS2 interneurons were immunopositive for SST (n = 3 of 4 tested; data not shown).
The final two interneuron subtypes possessed small cell bodies and were predominantly located in superficial layers. One subtype (n = 17) was the classical LS class that possesses a neurogliaform morphology (Fig. 6H) (Kawaguchi, 1995). This subtype showed a steady ramp depolarization (when stepped to near threshold) either from resting membrane potential (Fig. 5D,I) or from a steady-state hyperpolarized membrane potential (data not shown). In response to large depolarizing current injection, they showed pronounced adaptation (Fig. 5D) (see also Kawaguchi, 1995). Similar to the LS subtype, initial-adapting (iAD) interneurons also showed pronounced initial adaptation in both spike height and frequency (Fig. 5E). However, iAD interneurons did not possess the ramp depolarization at threshold and fired spikes early in the initial phase of the current step injection (Fig. 5E).
From our analysis of 203 cortical interneurons fate mapped, 6 of them were similar to the subtypes found in our former in vivo CGE transplantation study (Butt et al., 2005). They were rapidly-adapting interneurons and had bipolar or double-bouquet morphologies (Fig. 6I). This suggests that we very rarely fate map interneurons derived from the low Olig2-expression domain. Based on their intrinsic physiological properties, cortical interneurons fate mapped from the four embryonic time points of Olig2high precursors were classified into 10 subtypes.
The temporal basis for cortical interneuron subtype generation
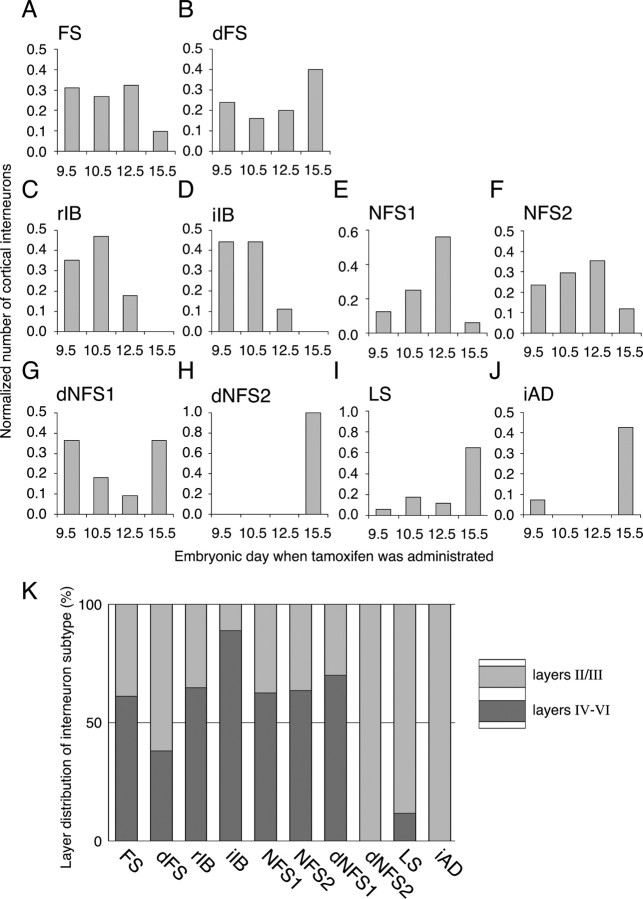
With this analysis, we are now in the position to ask whether there is a correlation between cortical interneuron subtype and the developmental stage from which they are fate mapped. Although the majority of classical FS interneurons were fate mapped from earlier time points (Fig. 7A), the delayed-subtype of fast-spiking interneurons (dFS) showed an increase in numbers at E15.5 (Fig. 7B). The distinct production peaks for FS and dFS interneurons in part could be explained by the layer distribution, in that whereas FS subtypes are fairly evenly distributed in both deep and superficial layers, the majority of dFS interneurons are located in layer II (Fig. 7K).
Figure 7.
Temporal and lamina distribution of the fate-mapped interneuron subtypes. A–J, Histograms showing the normalized distribution of each cell type over the four time points of tamoxifen administration (E9.5, E10.5, E12.5, E15.5). A, B, Classical FS (A) and dFS (B) interneurons together; FS interneurons were the major class found in this study, representing ∼50% of fate-mapped interneurons at all time points examined. dFS interneurons were found more from the E15.5 fate mapping (B). C, D, rIB (C) and iIB (D) subtypes of interneurons were fate mapped from only early time points. E, F, Within two of the NFS interneurons, NFS1 (E) peaked at the E12.5 time point. G–J, Three subtypes of dNFS2 (H), LS (I), and iAD (J) interneurons were mainly found in the E15.5 fate mapping. dNFS1 (G) interneurons were the only subtype to exhibit a bimodal distribution. K, The location of interneurons in deep versus superficial laminar distribution primarily reflected the time point at which interneurons were fate mapped. Interneuron subtypes predominantly fate mapped from the E15.5 time point (H–J) showed a preferential location in superficial layers (K).
The non-FS interneurons showed three broad, sequential temporal waves of generation. First, the majority of IB (rIB and iIB) interneurons with Martinotti morphologies were fate mapped from the E9.5 and E10.5 time points (Fig. 7C,D). Second, the NFS 1 and NFS2 interneurons peaked at the E12.5 time point (Fig. 7E,F). Third, the majority of dNFS2, LS, and iAD interneuron subtypes were fate mapped from the E15.5 time point (Fig. 7H–J), and those were preferentially located in the superficial layers (Fig. 7K). Only one interneuron subtype, the dNFS1 interneurons, exhibited a bimodal temporal distribution. As reported previously (Butt et al., 2005), these interneurons were preferentially located in the layers of Va and II.
The iIB interneurons were highly specific to layer V (n = 8 of 9) (Fig. 7K), and therefore it is not surprising that they were fate mapped from early time points given that laminar fate of the cortex is primarily determined by birth date (Miller, 1985). However, the rIB interneurons showed a more even distribution (6 of 17 were located in superficial layers) (Fig. 7K) despite the fact that they were labeled with the cohort of early fate-mapped NFS interneurons that occupy deep layers. On the other hand, the late fate-mapped interneurons (dNFS2, LS, and iAD) were almost entirely confined to the superficial layers (Fig. 7K). Hence, although early-born interneurons primarily occupy deep cortical layers, some interneuron subtypes that are generated at early time points can occupy more superficial layers. This suggests that the temporal origins of cortical interneurons are more closely linked to the physiological subtypes than to their laminar fate in the mature cortex.
Discussion
In this study, we have applied a genetic strategy to label and fate map precursors within the MGE at different time points. We show that the 10 previously identified subtypes of cortical interneurons are generated in a temporal sequence. Hence, we have demonstrated that their time of generation is a key feature underlying how different subtypes of cortical interneurons are specified. Moreover, our approach allows for the selective targeting of specific subpopulations of cortical interneurons before they can be identified by morphological, immunohistochemical, or physiological criteria. Thus, using this method, one can, with a high degree of fidelity, examine and genetically manipulate near identical neuronal subtypes that occupy similar spatial distributions from one animal to another, a proposition that until relatively recently was limited to invertebrate systems.
Genetic fate mapping of MGE precursors
A number of previous studies have used either an in vitro culture assay (Xu et al., 2004) or in vivo transplantation (Nery et al., 2002; Butt et al., 2005) to analyze the origins of cortical interneuron subtypes in the ventral eminences. These studies provided evidence that immunohistochemically (Nery et al., 2002; Xu et al., 2004; Butt et al., 2005) and electrophysiologically (Butt et al., 2005) distinct interneuron subtypes are generated from transient embryonic structures within the ventral telencephalon, termed the MGE and CGE. Moreover, this work suggested that, as occurs in the spinal cord, gene expression within these developmental regions may be causal in the generation of cortical interneuron diversity.
To begin to address the question of what role transcription factors play in determining different cortical interneuronal subtypes, we decided to extend our observations by labeling the precursors expressing a specific candidate gene, the bHLH transcription factor Olig2. We reasoned that the central role bHLH factors play in generating particular neuronal subtypes made this family of genes a promising one to study (Bertrand et al., 2002; Ma, 2006). As we have shown in this study, high levels of Olig2 expression coincides with the expression of homeodomain-containing gene Nkx2.1. The boundary between regions of Olig2 high versus low expression (as well as that of Nkx2.1) does not correspond to the morphological boundary dividing the MGE from the LGE (Fig. 1C,D). Similarly, the pallial and subpallial boundaries, as judged by morphology and gene expression, are also distinct (Yun et al., 2001). In fact, most morphologically defined embryonic structures are transient. Hence, although the MGE is no longer apparent by E15.5, based on Nkx2.1 expression it persists on through birth. Because the regional character of the developing forebrain is established and maintained by transcription factors, analyzing the fate of cortical interneuron populations is better done using genetic rather than morphological criteria. Indeed, the important role of Nkx2.1 in patterning is demonstrated by the near absence of MGE character in mice lacking this gene (Sussel et al., 1999).
MGE precursors undergo temporal shifts in fate
In principle, two broad developmental strategies could underlie the generation of different cortical interneuron subtypes from the MGE. Different interneuron classes might be generated within the MGE from discrete spatial subdomains. Alternatively, they might be generated through a shifting developmental fate of the MGE to sequentially give rise to discrete interneuron subtypes over time. The observation that the MGE is relatively homogenous in its expression of transcription factors makes it unlikely that subdomains within the progenitors akin to those observed in the spinal cord exist. Indeed, to our knowledge there exist only two molecular expression subdivisions within the MGE, the Nkx2.1/Olig2high domain and the dorsal domain that solely coexpresses Nkx6.2 (Stenman et al., 2003). Moreover, at present it is unclear whether the Nkx6.2-expression domain is fated to give rise to cortical interneurons. It is, however, possible that additional analysis will reveal additional molecular divisions within the MGE. Nonetheless, our present data argue that the diversity of neurons arising from the Olig2high precursors within the MGE is a result of temporal changes in the fate to give rise to interneurons of discrete subtypes.
The time span over which we observed interneurons to be derived from Olig2high precursors was from E9.5 until E15.5 tamoxifen administration. At early time points, interneurons were found predominantly in deep cortical layers, whereas at late time points, they were relatively scarce and confined to superficial layers, specifically the layer II/I boundary. Comparison with our previous in vivo transplantation data (Butt et al., 2005) reveals a number of similarities in the interneuron subtypes encountered at P14–P21. When the MGE (at E13.5) was transplanted in vivo, it gave rise to ∼50% of FS interneurons. This is remarkably similar to the numbers obtained using genetic fate mapping of Olig2high precursors at the corresponding time point (tamoxifen at E12.5) and at all of the other time points examined. The FS interneurons are continuously derived from Olig2high precursors throughout the period of interneuron generation. This is consistent with the broad distribution of PV-expressing FS interneurons throughout all of the cortical layers. In addition, in the present study, we reclassified a population of dFS. They were mainly found in the superficial layers and were biased to the later time points of fate mappings. Furthermore, the majority of NFS (previously termed RSNP) subtypes observed in our in vivo transplant study (Butt et al., 2005) were identified in this study. Indeed, only one population (NFS1 interneuron that peaked in number at E12.5) had not been previously detected in the in vivo transplants although a few subtypes (mainly those derived from late time points) were observed only in small numbers (LS, iAD) or found in the homotopic transplantation study of CGE. Given that our present genetic fate-mapping experiments sampled a much wider swath of developmental time points, this result is perhaps not that surprising. Nonetheless, the observation that VIP- and/or CR-expressing cortical interneurons arise from E15.5 Olig2-expressing precursors suggests that these populations may arise from an Nkx2.1-negative precursor population.
Are the changes in temporal fate of MGE precursors a result of intrinsic or extrinsic cues?
Our results demonstrate that MGE precursors give rise to distinct subtypes of cortical interneurons dependent on the developmental time point at which they are generated. A key question that must be addressed is the extent to which these populations are derived from single versus multiple progenitor populations. Two models could explain the present results. Perhaps the Olig2high precursors represent a single progenitor population fated to produce different interneuron subtypes over time. This would suggest that interneuron diversity in this population is generated by a mechanism similar to that used in Drosophila (Pearson and Doe, 2004) or cortex (Hanashima et al., 2004), in which the sequential expression of transcription factors results in a tightly controlled output of specific subtypes of neurons. Currently, there is no known evidence of a temporal shift in the expression of any transcription factors within the Olig2high precursors of MGE.
Alternatively, perhaps different interneuron subtypes are generated from distinct progenitor populations, each of which is controlled by a unique maturation program. This idea is consistent with the presence of a large subventricular zone (SVZ) within the MGE. Time-lapse movies of progenitors within the subpallium has shown that lineages within the ventral forebrain are derived by VZ progenitors seeding the SVZ with proliferative neuroblasts (S. Noctor and A. Kriegstein, personal communication). Perhaps the neuroblasts within the SVZ become restricted in the subclass(es) of cortical interneurons they will give rise to dependent on the timing of their exit from the VZ. An alternative possibility is that akin to what has been proposed in the cortex (Tarabykin et al., 2001; Zimmer et al., 2004), the VZ and SVZ could give rise to deep versus superficial interneuron populations. To distinguish between these possibilities, it will be important in the future to undertake lineage studies of the Olig2high progenitors (McCarthy et al., 2001). It has been suggested that the lineage of cortical interneuron progenitors are not restricted to certain subtypes, because cells derived from a common clone express different profiles of calcium-binding proteins, including PV, CR, and calbindin (Mione et al., 1994).
Our previous heterotopic transplantation of MGE and CGE progenitors demonstrated that the regional subtypes of cortical interneurons are already specified as progenitors (Nery et al., 2002, Butt et al., 2005). However, perhaps the temporal shifts in interneuron subtypes produced by the Olig2high precursors are controlled by extrinsic cues. Indeed, recent data from experiments involving the heterochronic injection of MGE cells into the telencephalic ventricles in utero (Valcanis and Tan, 2003) are consistent with this idea. Both younger (E12.5) and older (E15.5) MGE cells ectopically entering the cortical VZ were able to adapt to a novel environment by adopting a laminar position within the cortex appropriate to the age of the host animal (Valcanis and Tan, 2003). Although the physiological subtypes of these interneurons are not evaluated, these findings raise the possibility that the temporal shift in the production of cortical interneuron subtypes is controlled by environmental cues within the cortex itself. This model suggests that although MGE-derived progenitors are restricted in the broad class of interneurons they produce (Nery et al., 2002; Butt et al., 2005), their specific subtype is dependent on cues they receive on entry into the cortex (Valcanis and Tan, 2003). To explore this will require heterochronic transplantation studies combined with electrophysiological analysis.
To examine whether Olig2 has a role in cortical interneuron subtype specification, we have performed a conditional loss-of-function analysis by combining a floxed-Olig2 allele (Yue et al., 2006) with a pan-telencephalic Cre driver line, FoxG1-Cre (Hebert and McConnell, 2000). At P21, there was no obvious behavioral deficit and no significant change in any of the cortical interneuron markers examined, including PV, SST, VIP, and CR (G. Miyoshi and G. Fishell, unpublished observations). This either suggests that the requirement for Olig2 in interneurons was compensated for by other bHLH genes (most likely Olig1) or there is no role for Olig genes in cortical interneuron specification.
Using developmental genetics to track neuronal diversity
The diversity of mouse cortical interneurons has been apparent since the early 20th century anatomical work of Lorente de Nó (1992). However, it has only been recently appreciated that the majority, if not all, GABAergic interneurons in the mouse neocortex are derived from the embryonic Dlx-expressing ventral eminences (Anderson et al., 1997; Stuhmer et al., 2002). In this study, we have profiled interneurons generated from a restricted domain containing Olig2high precursors that primarily correspond to the Nkx2.1-expressing portion of the MGE. We have identified 10 subtypes of cortical interneurons that exhibit distinct temporal profiles with regard to their generation. Many of these interneurons are PV-expressing multipolar basket interneurons or SST-expressing interneurons with bitufted Martinotti morphologies: neuronal subtypes that predominantly innervate the soma or dendrites of other neurons, respectively (DeFelipe, 1997; Kawaguchi and Kubota, 1997; Somogyi et al., 1998). FS interneurons that we analyzed in this study possessed the diverse range of morphological and electrophysiological criteria previously reported by others (Kawaguchi, 1995; Cauli et al., 1997; Wang et al., 2002). If we cluster the variability observed in FS basket interneurons (here simply termed classical or dFS) excluding the variability of synaptic connectivity (Gupta et al., 2000), our observations closely match the previously described classification of FS subtype (Markram et al., 2004). The NFS interneuron subtypes we analyzed (aside from the NFS1 class) have also been described in studies characterizing the diversity in SST-expressing interneurons (Kawaguchi and Kubota, 1996; Kawaguchi and Kubota, 1998; Wang et al., 2004; Halabisky et al., 2006; Ma et al., 2006; Xu et al., 2006). We report here a novel subtype of NFS interneurons, the NFS1 (Fig. 5A). But in terms of electrophysiological profile, as well as laminar location, they are similar to the interneurons observed in the GAD67-EGFP transgenic line X94 (Ma, et al., 2006). Thus, we do not feel that the cortical interneurons in this study represent novel subtypes.
In conclusion, our fate-mapping study has demonstrated that we can reproducibly label discrete cortical interneuron populations and trace them through development into the mature cortex. This opens up new possibilities for both developmental neurobiologists and systems neuroscientists to study the specification and circuit integration of specific cortical interneuron populations. First, by using conditional null alleles for genes of interest instead of simply reporter lines, the contribution of specific genes to cortical interneuron development can be analyzed. Second, the introduction of functional components such as trans-synaptic tracers, as well as activity indicators or modulators, will allow us to characterize and even manipulate functional neuronal networks (Wulff and Wisden, 2005; Miyoshi and Fishell, 2006).
Footnotes
Research in the Fishell laboratory was supported by National Institutes of Health–National Institute of Mental Health–National Institute of Neurological Disorders and Stroke Grants R01 MH068469 and R01NS039007. G.M. was supported by a grant from the Japan Society for the Promotion of Science. S.J.B.B. was supported by a long-term fellowship awarded by the Human Frontier Science Program Organization. We thank all of the Fishell laboratory members for support and comments on this manuscript. We thank Staci Rakowiecki, Lihong Yin, Rebecca Wolsky, and Yuanyuan Huang for technical help with genotyping the mice. We thank Joshua Corbin, Jeremy Dasen, and Bernardo Rudy for critically reading this manuscript. We thank Dr. Kazuhiro Ikenaka for supplying us with the Olig2-CreER mice. We thank Drs. Charles Stiles and Jhumku Kohtz for kindly providing us Olig2 and pan-Dlx antibodies, respectively. We thank Dr. Richard Lu for generously providing us with the conditional Olig2 allele. Monoclonal antibodies from the Developmental Studies Hybridoma Bank were developed under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biological Science at The University of Iowa (Iowa City, IA).
References
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- Angevine JB, II, Sidman RL. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Butt SJ, Fuccillo M, Nery S, Noctor S, Kriegstein A, Corbin JG, Fishell G. The temporal and spatial origins of cortical interneurons predict their physiological subtype. Neuron. 2005;48:591–604. doi: 10.1016/j.neuron.2005.09.034. [DOI] [PubMed] [Google Scholar]
- Cauli B, Audinat E, Lambolez B, Angulo MC, Ropert N, Tsuzuki K, Hestrin S, Rossier J. Molecular and physiological diversity of cortical nonpyramidal cells. J Neurosci. 1997;17:3894–3906. doi: 10.1523/JNEUROSCI.17-10-03894.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh ME, Parnavelas JG. Development of somatostatin immunoreactive neurons in the rat occipital cortex: a combined immunocytochemical-autoradiographic study. J Comp Neurol. 1988;268:1–12. doi: 10.1002/cne.902680102. [DOI] [PubMed] [Google Scholar]
- Cavanagh ME, Parnavelas JG. Development of vasoactive-intestinal-polypeptide-immunoreactive neurons in the rat occipital cortex: a combined immunohistochemical-autoradiographic study. J Comp Neurol. 1989;284:637–645. doi: 10.1002/cne.902840410. [DOI] [PubMed] [Google Scholar]
- Cavanagh ME, Parnavelas JG. Development of neuropeptide Y (NPY) immunoreactive neurons in the rat occipital cortex: a combined immunohistochemical-autoradiographic study. J Comp Neurol. 1990;297:553–563. doi: 10.1002/cne.902970408. [DOI] [PubMed] [Google Scholar]
- Chow A, Erisir A, Farb C, Nadal MS, Ozaita A, Lau D, Welker E, Rudy B. K+ channel expression distinguishes subpopulations of parvalbumin- and somatostatin-containing neocortical interneurons. J Neurosci. 1999;19:9332–9345. doi: 10.1523/JNEUROSCI.19-21-09332.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFelipe J. Types of neurons, synaptic connections and chemical characteristics of cells immunoreactive for calbindin-D28K, parvalbumin and calretinin in the neocortex. J Chem Neuroanat. 1997;14:1–19. doi: 10.1016/s0891-0618(97)10013-8. [DOI] [PubMed] [Google Scholar]
- Erisir A, Lau D, Rudy B, Leonard CS. Function of specific K(+) channels in sustained high-frequency firing of fast-spiking neocortical interneurons. J Neurophysiol. 1999;82:2476–2489. doi: 10.1152/jn.1999.82.5.2476. [DOI] [PubMed] [Google Scholar]
- Fairen A, Cobas A, Fonseca M. Times of generation of glutamic acid decarboxylase immunoreactive neurons in mouse somatosensory cortex. J Comp Neurol. 1986;251:67–83. doi: 10.1002/cne.902510105. [DOI] [PubMed] [Google Scholar]
- Gonchar Y, Burkhalter A. Three distinct families of GABAergic neurons in rat visual cortex. Cereb Cortex. 1997;7:347–358. doi: 10.1093/cercor/7.4.347. [DOI] [PubMed] [Google Scholar]
- Gupta A, Wang Y, Markram H. Organizing principles for a diversity of GABAergic interneurons and synapses in the neocortex. Science. 2000;287:273–278. doi: 10.1126/science.287.5451.273. [DOI] [PubMed] [Google Scholar]
- Halabisky B, Shen F, Huguenard JR, Prince DA. Electrophysiological classification of somatostatin-positive interneurons in mouse sensorimotor cortex. J Neurophysiol. 2006;96:834–845. doi: 10.1152/jn.01079.2005. [DOI] [PubMed] [Google Scholar]
- Hanashima C, Li SC, Shen L, Lai E, Fishell G. Foxg1 suppresses early cortical cell fate. Science. 2004;303:56–59. doi: 10.1126/science.1090674. [DOI] [PubMed] [Google Scholar]
- Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- Hebert JM, McConnell SK. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev Biol. 2000;222:296–306. doi: 10.1006/dbio.2000.9732. [DOI] [PubMed] [Google Scholar]
- Joyner AL, Zervas M. Genetic inducible fate mapping in mouse: Establishing genetic lineages and defining genetic neuroanatomy in the nervous system. Dev Dyn. 2006;235:2376–2385. doi: 10.1002/dvdy.20884. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y. Physiological subgroups of nonpyramidal cells with specific morphological characteristics in layer II/III of rat frontal cortex. J Neurosci. 1995;15:2638–2655. doi: 10.1523/JNEUROSCI.15-04-02638.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Kondo S. Parvalbumin, somatostatin and cholecystokinin as chemical markers for specific GABAergic interneuron types in the rat frontal cortex. J Neurocytol. 2002;31:277–287. doi: 10.1023/a:1024126110356. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kubota Y. Physiological and morphological identification of somatostatin- or vasoactive intestinal polypeptide-containing cells among GABAergic cell subtypes in rat frontal cortex. J Neurosci. 1996;16:2701–2715. doi: 10.1523/JNEUROSCI.16-08-02701.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Kubota Y. GABAergic cell subtypes and their synaptic connections in rat frontal cortex. Cereb Cortex. 1997;7:476–486. doi: 10.1093/cercor/7.6.476. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kubota Y. Neurochemical features and synaptic connections of large physiologically-identified GABAergic cells in the rat frontal cortex. Neuroscience. 1998;85:677–701. doi: 10.1016/s0306-4522(97)00685-4. [DOI] [PubMed] [Google Scholar]
- Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, Richardson WD. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci. 2006;9:173–179. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel RA, Turnbull DH, Blanquet V, Wurst W, Loomis CA, Joyner AL. Two lineage boundaries coordinate vertebrate apical ectodermal ridge formation. Genes Dev. 2000;14:1377–1389. [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Hattori R, Yui Y. Three distinct subpopulations of GABAergic neurons in rat frontal agranular cortex. Brain Res. 1994;649:159–173. doi: 10.1016/0006-8993(94)91060-x. [DOI] [PubMed] [Google Scholar]
- Lee SK, Lee B, Ruiz EC, Pfaff, SL Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 2005;19:282–294. doi: 10.1101/gad.1257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorente de Nó R. The cerebral cortex of the mouse (a first contribution—the “acoustic” cortex) Somatosens Mot Res. 1992;9:3–36. doi: 10.3109/08990229209144760. [DOI] [PubMed] [Google Scholar]
- Lu QR, Yuk D, Alberta JA, Zhu Z, Pawlitzky I, Chan J, McMahon AP, Stiles CD, Rowitch DH. Sonic hedgehog–regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25:317–329. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Ma Q. Transcriptional regulation of neuronal phenotype in mammals. J Physiol (Lond) 2006;575:379–387. doi: 10.1113/jphysiol.2006.113449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Hu H, Berrebi AS, Mathers PH, Agmon A. Distinct subtypes of somatostatin-containing neocortical interneurons revealed in transgenic mice. J Neurosci. 2006;26:5069–5082. doi: 10.1523/JNEUROSCI.0661-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- McCarthy M, Turnbull DH, Walsh CA, Fishell G. Telencephalic neural progenitors appear to be restricted to regional and glial fates before the onset of neurogenesis. J Neurosci. 2001;21:6772–6781. doi: 10.1523/JNEUROSCI.21-17-06772.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MW. Cogeneration of retrogradely labeled corticocortical projection and GABA-immunoreactive local circuit neurons in cerebral cortex. Brain Res. 1985;355:187–192. doi: 10.1016/0165-3806(85)90040-9. [DOI] [PubMed] [Google Scholar]
- Miller MW, Nowakowski RS. Use of bromodeoxyuridine-immunohistochemistry to examine the proliferation, migration and time of origin of cells in the central nervous system. Brain Res. 1988;457:44–52. doi: 10.1016/0006-8993(88)90055-8. [DOI] [PubMed] [Google Scholar]
- Mione MC, Danevic C, Boardman P, Harris B, Parnavelas JG. Lineage analysis reveals neurotransmitter (GABA or glutamate) but not calcium-binding protein homogeneity in clonally related cortical neurons. J Neurosci. 1994;14:107–123. doi: 10.1523/JNEUROSCI.14-01-00107.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi G, Fishell G. Directing neuron-specific transgene expression in the mouse CNS. Curr Opin Neurobiol. 2006;16:577–584. doi: 10.1016/j.conb.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Nery S, Wichterle H, Fishell G. Sonic hedgehog contributes to oligodendrocyte specification in the mammalian forebrain. Development. 2001;128:527–540. doi: 10.1242/dev.128.4.527. [DOI] [PubMed] [Google Scholar]
- Nery S, Fishell G, Corbin JG. The caudal ganglionic eminence is a source of distinct cortical and subcortical cell populations. Nat Neurosci. 2002;5:1279–1287. doi: 10.1038/nn971. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. Ed 2. San Diego: Academic; 2001. The mouse brain in stereotaxic coordinates. [Google Scholar]
- Pearson BJ, Doe CQ. Specification of temporal identity in the developing nervous system. Annu Rev Cell Dev Biol. 2004;20:619–647. doi: 10.1146/annurev.cellbio.19.111301.115142. [DOI] [PubMed] [Google Scholar]
- Rakic P. Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science. 1974;183:425–427. doi: 10.1126/science.183.4123.425. [DOI] [PubMed] [Google Scholar]
- Somogyi P, Tamas G, Lujan R, Buhl EH. Salient features of synaptic organization in the cerebral cortex. Brain Res Brain Res Rev. 1998;26:113–135. doi: 10.1016/s0165-0173(97)00061-1. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Stenman JM, Wang B, Campbell K. Tlx controls proliferation and patterning of lateral telencephalic progenitor domains. J Neurosci. 2003;23:10568–10576. doi: 10.1523/JNEUROSCI.23-33-10568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuhmer T, Puelles L, Ekker M, Rubenstein JL. Expression from a Dlx gene enhancer marks adult mouse cortical GABAergic neurons. Cereb Cortex. 2002;12:75–85. doi: 10.1093/cercor/12.1.75. [DOI] [PubMed] [Google Scholar]
- Sussel L, Marin O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126:3359–3370. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- Takebayashi H, Yoshida S, Sugimori M, Kosako H, Kominami R, Nakafuku M, Nabeshima Y. Dynamic expression of basic helix-loop-helix Olig family members: implication of Olig2 in neuron and oligodendrocyte differentiation and identification of a new member, Olig3. Mech Dev. 2000;99:143–148. doi: 10.1016/s0925-4773(00)00466-4. [DOI] [PubMed] [Google Scholar]
- Takebayashi H, Nabeshima Y, Yoshida S, Chisaka O, Ikenaka K, Nabeshima Y. The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr Biol. 2002;12:1157–1163. doi: 10.1016/s0960-9822(02)00926-0. [DOI] [PubMed] [Google Scholar]
- Tarabykin V, Stoykova A, Usman N, Gruss P. Cortical upper layer neurons derive from the subventricular zone as indicated by Svet1 gene expression. Development. 2001;128:1983–1993. doi: 10.1242/dev.128.11.1983. [DOI] [PubMed] [Google Scholar]
- Valcanis H, Tan SS. Layer specification of transplanted interneurons in developing mouse neocortex. J Neurosci. 2003;23:5113–5122. doi: 10.1523/JNEUROSCI.23-12-05113.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gupta A, Toledo-Rodriguez M, Wu CZ, Markram H. Anatomical, physiological, molecular and circuit properties of nest basket cells in the developing somatosensory cortex. Cereb Cortex. 2002;12:395–410. doi: 10.1093/cercor/12.4.395. [DOI] [PubMed] [Google Scholar]
- Wang Y, Toledo-Rodriguez M, Gupta A, Wu C, Silberberg G, Luo J, Markram H. Anatomical, physiological and molecular properties of Martinotti cells in the somatosensory cortex of the juvenile rat. J Physiol (Lond) 2004;561:65–90. doi: 10.1113/jphysiol.2004.073353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H, Turnbull DH, Nery S, Fishell G, Alvarez-Buylla A. In utero fate mapping reveals distinct migratory pathways and fates of neurons born in the mammalian basal forebrain. Development. 2001;128:3759–3771. doi: 10.1242/dev.128.19.3759. [DOI] [PubMed] [Google Scholar]
- Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat Rev Neurosci. 2006;7:687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- Wulff P, Wisden W. Dissecting neural circuitry by combining genetics and pharmacology. Trends Neurosci. 2005;28:44–50. doi: 10.1016/j.tins.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Xu Q, Cobos I, De La Cruz E, Rubenstein JL, Anderson SA. Origins of cortical interneuron subtypes. J Neurosci. 2004;24:2612–2622. doi: 10.1523/JNEUROSCI.5667-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Wonders CP, Anderson SA. Sonic hedgehog maintains the identity of cortical interneuron progenitors in the ventral telencephalon. Development. 2005;132:4987–4998. doi: 10.1242/dev.02090. [DOI] [PubMed] [Google Scholar]
- Xu X, Roby KD, Callaway EM. Mouse cortical inhibitory neuron type that coexpresses somatostatin and calretinin. J Comp Neurol. 2006;499:144–160. doi: 10.1002/cne.21101. [DOI] [PubMed] [Google Scholar]
- Yozu M, Tabata H, Nakajima K. Birth-date dependent alignment of GABAergic neurons occurs in a different pattern from that of non-GABAergic neurons in the developing mouse visual cortex. Neurosci Res. 2004;49:395–403. doi: 10.1016/j.neures.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Yue T, Xian K, Hurlock E, Xin M, Kernie SG, Parada LF, Lu QR. A critical role for dorsal progenitors in cortical myelination. J Neurosci. 2006;26:1275–1280. doi: 10.1523/JNEUROSCI.4717-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun K, Potter S, Rubenstein JL. Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development. 2001;128:193–205. doi: 10.1242/dev.128.2.193. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- Zimmer C, Tiveron MC, Bodmer R, Cremer H. Dynamics of Cux2 expression suggests that an early pool of SVZ precursors is fated to become upper cortical layer neurons. Cereb Cortex. 2004;14:1408–1420. doi: 10.1093/cercor/bhh102. [DOI] [PubMed] [Google Scholar]