Abstract
Cadherins function in the adhesion of presynaptic and postsynaptic membranes at excitatory synapses. Here we show that the cadherin-associated protein neural plakophilin-related arm protein (NPRAP; also called δ-catenin) binds via a postsynaptic density-95 (PSD-95)/discs large/zona occludens-1 (PDZ) interaction to AMPA receptor (AMPAR)-binding protein (ABP) and the related glutamate receptor (GluR)-interacting protein (GRIP), two multi-PDZ proteins that bind the GluR2 and GluR3 AMPAR subunits. The resulting cadherin–NPRAP–ABP/GRIP complexes serve as anchorages for AMPARs. Exogenous NPRAP that was bound to cadherins at adherens junctions of Madin-Darby canine kidney cells recruited ABP from the cytosol to form cadherin–NPRAP–ABP complexes, dependent on NPRAP interaction with the ABP PDZ domain 2. The cadherin–NPRAP–ABP complexes also bound GluR2. In cultured hippocampal neurons, dominant-negative mutants of NPRAP designed to disrupt tethering of ABP to NPRAP–cadherin complexes reduced surface levels of endogenous GluR2, indicating that interaction with cadherin–NPRAP–ABP complexes stabilized GluR2 at the neuronal plasma membrane. Cadherins, NPRAP, GRIP, and GluR2 copurified in the fractionation of synaptosomes and the postsynaptic density, two fractions enriched in synaptic proteins. Furthermore, synaptosomes contain NPRAP–GRIP complexes, and NPRAP localizes with the postsynaptic marker PSD-95 and with AMPARs and GRIP at spines of hippocampal neurons. Thus, tethering is likely to take place at synaptic or perisynaptic sites. NPRAP also binds PSD-95, which is a scaffold for NMDA receptors, for AMPARs in complexes with auxiliary subunits, the TARPs (transmembrane AMPA receptor regulator proteins), and for adhesion molecules. Thus, the interaction of scaffolding proteins with cadherin–NPRAP complexes may anchor diverse signaling and adhesion molecules at cadherins.
Keywords: GluR2, AMPA receptor, ABP, GRIP, cadherin, NPRAP
Introduction
Efficient glutamatergic transmission requires the alignment of presynaptic glutamate release machinery with postsynaptic glutamate receptors (GluRs). Cell adhesion molecules (CAMs) and synaptic scaffolding proteins contribute to this alignment (for review, see Washbourne et al., 2004). Interactions of the extracellular domains of CAMs hold presynaptic and postsynaptic membranes in close proximity. In the nerve terminal, scaffolding proteins bound to CAM intracellular domains tether the neurotransmitter release machinery, whereas postsynaptic scaffolding proteins, also bound to CAMs, anchor glutamate receptors. At the postsynaptic side, the scaffolding protein postsynaptic density-95 (PSD-95) binds neuroligin (for review, see Kim and Sheng, 2004), a postsynaptic CAM that interacts with presynaptic neurexins. At synapses, PSD-95 also binds NMDA receptors (NMDARs) and AMPA receptors (AMPARs) in complexes with the auxiliary AMPAR subunits the transmembrane AMPA receptor regulator proteins (TARPs) (Chen et al., 2000).
Cadherins are Ca2+-dependent homophilic synaptic CAMs that contribute to spine morphogenesis and synapse assembly and regulation (for review, see Takeichi and Abe, 2005). Cadherins complexed with β- and α-catenin link the synapse to the actin cytoskeleton and function in long-term potentiation (LTP) and the synaptic localization of a third glutamate receptor type, the kainate receptor (Coussen et al., 2002). However, a structural basis for cadherin association with AMPA receptors is not known.
AMPAR-binding protein (ABP) and glutamate receptor-interacting protein (GRIP) are PSD-95/discs large/zona occludens-1 (PDZ) domain-containing components of the PSD that bind the GluR2 and GluR3 AMPAR subunits (Dong et al., 1997; Srivastava et al., 1998; Barry and Ziff, 2002). We have identified neural plakophilin-related arm protein (NPRAP; also called δ-catenin) (Paffenholz and Franke, 1997; Kosik et al., 2005), as an ABP/GRIP interactor. NPRAP is an ARM repeat protein that binds to the juxtamembrane region of the cadherin intracellular domain. We show that NPRAP links ABP and GRIP to cadherins, creating an anchorage for GluR2. Because we find that NPRAP also binds to PSD-95, cadherin–NPRAP complexes may be linked to the PSD-95 scaffold, which is involved in tethering NMDAR and TARP–AMPAR complexes and which also interacts with neuroligins. Thus, NPRAP may be a key adaptor that enables diverse components of the postsynaptic signaling machinery to anchor at the sites of cadherins.
Materials and Methods
Yeast two-hybrid system and yeast mating assays.
The Matchmaker II kit (Clontech, Palo Alto, CA) was used for yeast two-hybrid screens according to the manufacturer's instructions, using the N-terminal 852 bp of ABP-S (Srivastava et al., 1998) corresponding to the first set of three PDZ domains, ABP-SetI, cloned in expression vector pAS2, as bait in yeast strain Y190. Approximately 500,000 of the total independent clones in the matchmaker cDNA library were screened. Yeast mating assays were performed as described above except using pGBKT7 (Clontech). Strain Y187 was transformed with these fusion plasmids, and transformed colonies were then mated with strain Y190 expressing the library clones that were recovered from the screens.
Glutathione S-transferase pull-down assays.
cDNA encoding the designated C-terminal amino acids of NPRAP was subcloned in the glutathione S-transferase (GST) expression vector, pGEX4T-1 (GE Healthcare, Piscataway, NJ). The GST–NPRAP fusion proteins were expressed in the BL-21 bacterial strain by induction with β-d-thiogalactopyranoside as described previously (Perez et al., 2001). In vitro translated protein was created using the TNT Quick Coupled Translation System (Promega, Madison, WI) according to the manufacturer's instructions. GST fusion proteins were immobilized on glutathione-Sepharose beads and then incubated with 35S-labeled ABP-S for 2–3 h at 4°C in buffer containing 20 mm HEPES, 300 mm NaCl, 1% Triton X-100, and 5% glycerol. Bound proteins were eluted by boiling in SDS gel sample buffer and were resolved on an SDS gel by electrophoresis and visualized by autoradiography.
Immunoprecipitations from transfected HEK 293T cells.
Full-length NPRAP cDNA and J19 anti-NPRAP antiserum (Paffenholz et al., 1999) were generously provided by Dr. Werner Franke (German Cancer Research Center, Heidelberg, Germany), and coding sequences for NPRAP were subcloned into pcDNA3 for heterologous cell expression.
Human embryonic kidney cells (HEK 293T) were grown in DMEM plus 10% fetal bovine serum. Cells were transfected by the calcium phosphate method. Approximately 40 h after transfection, cells were washed twice with 1× PBS and scraped in 1 ml of PBS. Cells were solubilized in lysis buffer (25 mm HEPES, pH 7.4, 150 mm NaCl, 1% Triton X-100) and protease inhibitor mixture from Complete Protease Inhibitor Cocktail Tablets (Roche Diagnostics, Mannheim, Germany) for 30 min on ice. Insoluble material was pelleted, and the supernatant was used for Western blots. Antibodies for immunoprecipitations [unless otherwise noted, 2 μg of antibody (Ab) was added to each reaction] were as follows: M2 anti-FLAG monoclonal antibody (Sigma-Aldrich, St. Louis, MO), MAB397 anti-GluR2 N-terminal antibody (Millipore, Billerica, MA), s.c.-7870 anti-E-cadherin antibody (used to immunoprecipitate endogenous protein; Santa Cruz Biotechnology, Santa Cruz, CA), and J19 anti-NPRAP antiserum at a 1:20 dilution. Antibodies were incubated with the lysate for 1.5 h at 4°C. Then, 20 μl of protein G or protein A beads (Santa Cruz Biotechnology), as determined by the IgG isotype of the antibody, was added to the reaction for an additional 1 h at 4°C, followed by one wash with lysis buffer and two washes with lysis buffer containing 500 mm NaCl. Bound proteins were eluted by boiling in SDS gel sample buffer, separated by 8% SDS-PAGE, and transferred onto nitrocellulose. The following antibodies were used in Western blots (unless otherwise noted, antibodies were used at a concentration of 1 μg/ml): anti-ABP Linker-2 antibody (Srivastava et al., 1998), J19 anti-NPRAP antiserum at a 1:100 dilution, s.c.-7870 anti-E-cadherin antibody, MAB397 anti-GluR2 N-terminal antibody, M2 anti-FLAG antibody (2.5 μg/ml; Sigma-Aldrich), and AB1504 anti-GluR1 C-terminal antibody (Millipore). Blots were visualized by enhanced chemiluminescence with the ECL kit (GE Healthcare).
Immunocytochemical and biochemical experiments with Madin-Darby canine kidney cells.
An N-terminal hemagglutinin (HA) epitope tag (10 codons) was added to full-length NPRAP in pcDNA3, to create an expression vector encoding HA–NPRAP. The NPRAP mutant that lacked all the Armadillo repeats (HA–NPRAPΔARM), HA–NPRAPΔ3, which lacks the PDZ binding motif consisting of the 3 C-terminal amino acids, and HA–ΔARMΔ3, which lacks this motif and the ARM repeats, were also cloned into pcDNA3. ABP-L–GFP, ABP-SΔSI–GFP, and GFP–ABP-SetI fusion constructs were described previously (DeSouza et al., 2002; Fu et al., 2003). The former two constructs contained green fluorescent protein (GFP) at the C terminus of ABP, whereas the third construct encoded GFP at the N terminus.
Madin-Darby canine kidney (MDCK) cells were obtained from the American Type Culture Collection (Manassas, VA) and were grown in DMEM plus 10% fetal bovine serum, and transfections were conducted with SuperFect Transfection Reagent (Qiagen, Valencia, CA) according to the manufacturer's instructions using 10 μg of DNA per reaction. Immunoprecipitations were described for HEK 293T cells. The following antibodies were used for the immunoprecipitations (2 μg per reaction): s.c.-7870 anti-E-cadherin antibody (Santa Cruz Biotechnology) and anti-δ-catenin antibody (BD Biosciences, Lexington, KY). These same antibodies were used for Western blots at a concentration of 1 μg/ml. In addition, anti-GFP antibody (1:4000 dilution) (Fu et al., 2003) was used on immunoblots to recognize GFP fusion proteins.
MDCK cells were also plated on glass coverslips in six-well dishes for immunocytochemistry. At ∼40% confluence, transfections were conducted with FuGene 6 Transfection Reagent (Roche Diagnostics) according to the manufacturer's instructions, maintaining a FuGene Reagent:DNA ratio of 3:1. Cells were grown until they were completely confluent on the coverslips (usually ∼24 h) and fixed by submerging coverslips in ice-cold methanol for 20 min at 4°C. Cells were washed three times with PBS and permeabilized with 0.2% Triton X-100 in PBS for 5 min, blocked with 10% BSA in PBS for 1 h at room temperature, and incubated with primary antibody diluted in 3% BSA/PBS for 1 h at room temperature. After three PBS washes for 5 min each, secondary antibody also diluted in 3% BSA/PBS was added for 1 h at room temperature. After final PBS washes, the coverslips were mounted on glass slides with mounting medium. Transfected NPRAP and related mutants were stained using anti-δ-catenin antibody at a 1:100 dilution. Results were confirmed with anti-HA antibody (Roche Diagnostics) at a concentration of 1 μg/ml. E-cadherin was stained with the same anti-E-cadherin antibody (5 μg/ml) as above. Secondary antibodies were used at a 1:300 dilution: Rhodamine Red X-conjugated donkey anti-rabbit IgG (red channel) and fluorescein isothiocyanate (FITC)-conjugated donkey anti-mouse IgG (green channel; Jackson ImmunoResearch, West Grove, PA). In experiments with GFP fusion proteins, Cy5-conjugated donkey anti-mouse antibody (blue channel; Jackson ImmunoResearch) was used at a 1:200 dilution to recognize the anti-δ-catenin antibody.
Preparation of primary dissociated neuronal cultures.
Rat hippocampal neurons in low-density culture were prepared as described previously (Osten et al., 1998). Hippocampal primary cultures were prepared from embryonic day 18 (E18) Sprague Dawley rat embryonic tissue by dissociation with trypsin and were plated on poly-l-lysine-coated glass coverslips at a density of 140,000 cells per coverslip and cultured with Neurobasal media with B27 (Invitrogen, Carlsbad, CA). At the first change of media, a one-time dose of the drug AraC (4 μm; Sigma-Aldrich) was added to inhibit growth of dividing cells. After 19–21 d, cells were processed for staining endogenous proteins or infected with recombinant Sindbis viruses. Cortical cultures were also prepared from E18 Sprague Dawley rat embryonic brain tissue. Cells were plated on 6 cm dishes at a density of 750,000 cells per dish. Cultures were harvested for immunoprecipitation of endogenous proteins after 2–3 weeks in vitro.
Production of recombinant Sindbis viruses, infection of hippocampal cultures, and immunocytochemical and biochemical assays.
Recombinant defective Sindbis viruses were constructed using the vector pSinRep5 (Invitrogen), as described previously (Osten et al., 2000). Coverslips of infected hippocampal neurons were processed for immunocytochemistry in a manner similar to that for MDCK cells. The following primary antibodies were used to visualize exogenous or endogenous proteins within infected cells: anti-HA antibody (1 μg/ml), AB1504 anti-GluR1 C-terminal antibody (1.5 μg/ml), and anti-synaptophysin antibody (1:300; Zymed, Carlsbad, CA). The following secondary antibodies were used: Rhodamine Red X-conjugated donkey anti-rabbit IgG (1:300; red channel), FITC-conjugated donkey anti-mouse IgG (1:300; green channel), and Cy 5-conjugated donkey anti-mouse IgG (1:200; blue channel; all three from Jackson ImmunoResearch).
Hippocampal neurons that were not infected by Sindbis viruses were processed for immunostaining using the following primary antibodies: mouse anti-δ-catenin (1:200; BD Biosciences), mouse anti-PSD-95 antibody (1:500; clone K28.43; NeuroMab, Davis, CA), rabbit anti-GluR2 antibody (1 μg/ml; Millipore), and anti-GRIP1 antibody (6 μg/ml; Millipore). Isotype-specific monoclonal secondary antibodies IgG1 (Alexa Fluor 568; 1:1000; Invitrogen), IgG2A (Alexa Fluor 488; 1:1000; Invitrogen), and polyclonal FITC-conjugated secondary antibody (1:250; Jackson ImmunoResearch) were added for 1 h at room temperature.
Coimmunoprecipitation.
Hippocampal neurons from 6 cm dishes were lysed in Triton X-100 lysis buffer, 150 mm NaCl, 20 mm HEPES, pH 7.4, 2 mm EDTA, 1% Triton X-100, 1× protease inhibitor mixture (Roche Diagnostics), and 100 μg/ml PMSF. Neuronal lysates were extracted for 1 h at 4°C and cleared at 16,000 × g for 10 min at 4°C and used for coimmunoprecipitation. Lysate (500 μg) was incubated with 1.5 μg of GRIP Ab (Millipore), 1.5 μg of PSD-95 Ab (Millipore), or 1.5 μg of synaptophysin Ab (Zymed) at 4°C overnight. Complexes were pulled down by adding protein A/G beads and were washed five times with lysis buffer. Samples were separated on 6% SDS-PAGE gels.
Analysis of whole-brain, synaptosomal, and PSD fractions.
PSD fractions and synaptosomal fractions were prepared as described previously (Jordan et al., 2004). Two micrograms of each fraction were loaded on 8% SDS-PAGE gel. Western blots were probed with different antibodies: GRIP (Millipore), δ-catenin (NPRAP; BD Biosciences), synaptophysin (Sigma-Aldrich), GluR2/3 (Millipore), and N-cadherin (BD Biosciences). The synaptosomal fraction was also solubilized for 1 h at 4°C with Triton X-100 lysis buffer and cleared at 16,000 × g for 10 min at 4°C. Supernatant containing 1 mg of protein was analyzed by immunoprecipitation with 2 μg of the noted antibodies and Western blotting.
Dominant-negative experiments.
Hippocampal neurons, grown in Neurobasal medium supplemented with B27, were infected at 15–20 d in vitro (DIV) with recombinant Sindbis viruses expressing dsRed NPRAP-CT or dsRed NPRAP-CTΔ3 in one set of experiments or expressing HA–NPRAP or HA–NPRAPΔ3 in a second series of experiments. Neurons were labeled live (37°C for 10 min) with monoclonal antibody to the GluR2 N-terminal domain (10 μg/ml; Millipore) to detect surface receptors. Neurons were then fixed, permeabilized, stained with polyclonal antibody to synaptophysin (Zymed) for dsRed construct experiments and with polyclonal antibody to HA (Santa Cruz Biotechnology) for the HA tagged vector experiments, and stained with secondary antibodies. Surface receptor levels were measured by quantitative confocal imaging of immunofluorescence of endogenous GluR2 in those neurons expressing the NPRAP constructs, as determined by dsRed fluorescence or HA immunofluorescence, as appropriate.
Analysis of surface expression of GluR2.
To detect surface endogenous GluR2, neurons were stained live with an anti-GluR2 N-terminal antibody (10 μg/ml; Millipore) for 15 min at 37°C and fixed with 4% paraformaldehyde/4% sucrose for 10 min at room temperature. Neurons were then stained with secondary antibody under unpermeabilized conditions where indicated. To analyze GluR2 surface fluorescence, 20 μm of dendrites from infected and uninfected neurons were carefully traced. The surface fluorescence from the chosen dendrite was analyzed with Simple32 Imaging software (C-Imaging Systems, Cranberry, NJ). Each experimental manipulation was performed at least three times. Error bars are SEs, and t test was performed to determine the significance.
Confocal microscopy and image acquisition.
For immunocytochemistry of heterologous cells and primary hippocampal neurons expressing heterologous proteins, acquisition and analysis of images was performed on a Nikon (Tokyo, Japan) PCM 2000 confocal microscope using Simple PCI software (C-Imaging Systems) for initial processing. Images of endogenous proteins in dissociated hippocampal neurons were captured with a Zeiss (Thornwood, NY) LSM510 Meta laser-scanning confocal microscope using a Plan-Apochromat 63× objective and LSM 510 Meta software.
Results
Interaction of NPRAP with ABP PDZ SetI
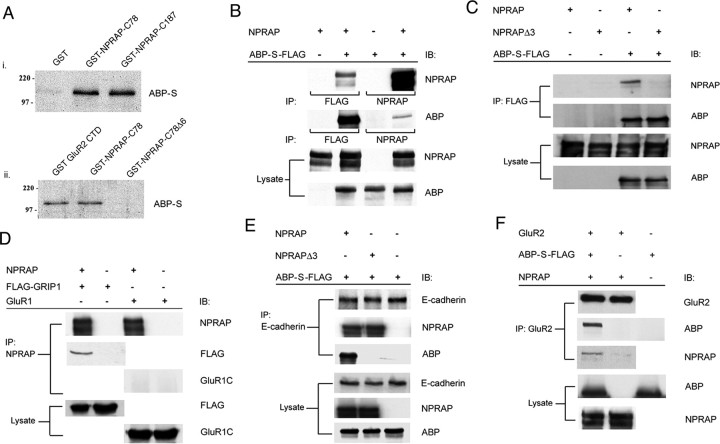
We screened an adult rat brain cDNA library for binding partners of ABP, using the yeast two-hybrid system, using the first three PDZ domains (SetI) of ABP as bait. Seven clones of the protein p0071 and two of the related NPRAP, both of which are members of the Armadillo family of proteins (Hatzfeld, 1999), were isolated in two successive screens. All clones included the proteins' extreme C terminus, which contains a class I PDZ binding motif, Ser-Trp-Val-COOH, that can potentially bind either of the first two PDZ domains of ABP (Srivastava et al., 1998). Sequencing established the presence of a stop codon after Ser-Trp-Val, confirming a discrepancy noted in the originally published p0071 sequence (Hatzfeld and Nachtsheim, 1996; Jaulin-Bastard et al., 2002). Of the several splice variant forms of ABP, two, ABP-L and pABP-L (which is palmitoylated), have seven PDZ domains, and a third form, ABP-S, has six PDZ domains (DeSouza et al., 2002). In a pull-down assay of the NPRAP–ABP interaction, fusions of GST to 187 or 78 aa of the NPRAP C terminus bound to in vitro translated 35S-labeled ABP-S, dependent on the NPRAP C-terminal 6 aa, which contain the PDZ binding motif (Fig. 1A). When ABP-S and the full-length NPRAP protein were coexpressed in HEK 293T cells, anti-NPRAP antiserum pulled out from cell lysates a complex containing both NPRAP and ABP-S (Fig. 1B). This interaction was dependent on the PDZ binding motif, which is deleted in mutant NPRAPΔ3 (Fig. 1C). NPRAP also formed a complex in HEK 293 cells with coexpressed GRIP, a protein highly homologous to ABP, but not with a coexpressed control protein, GluR1 (Fig. 1D). This demonstrates that NPRAP and ABP-S interact in vitro and in mammalian cells, dependent on the PDZ binding motif of NPRAP. The NPRAP C-terminal sequence is a class I PDZ domain interaction sequence (Songyang et al., 1997), and PDZ domains 1 and 2 of ABP-SetI are class I PDZs, whereas PDZ3 is class II (Srivastava et al., 1998). Yeast mating assays demonstrated that the NPRAP C terminus can associate with the isolated PDZ domain 2 and with constructs that include PDZ2 (PDZ1–PDZ3 and PDZ2–PDZ3) (Table 1). However, no binding was detected for either PDZ1 or PDZ3. Thus, PDZ2 mediates the interaction between ABP and the C terminus of NPRAP. It is noteworthy that although PDZ1 is also a class I PDZ domain, it failed to interact with NPRAP. In fact, PDZ1 reduced binding when the first two domains were mated with NPRAP. GRIP SetI also interacted with the C terminus of NPRAP in the mating assays. Interestingly, the set of three PDZ domains of PSD-95, which are all class I (Sheng and Sala, 2001), also gave a positive result (Table 1). The C terminus of p0071 bound PDZ domains with the same specificity as that of NPRAP (data not shown).
Figure 1.
Formation of GluR2–ABP/GRIP–NPRAP–cadherin complexes in vitro and in vivo. A, GST–NPRAP C-terminal domain (CTD) fusion proteins bind in vitro translated ABP-S via the NPRAP PDZ binding motif. i, Fusions of 78 or 187 NPRAP C-terminal amino acids to GST, but not GST alone, bind ABP-S in a GST pulldown assay. ii, The GluR2 C-terminal domain (a positive control) and the NPRAP 78 C-terminal amino acids, but not the latter with a 6 aa terminal deletion that removes the PDZ binding site, bind to ABP-S when fused to GST. B, NPRAP and ABP-S associate in HEK 293T cells. NPRAP and ABP-S with a FLAG epitope tag were expressed in HEK 293T cells by plasmid transfection, as indicated. Western blot analysis of immunoprecipitates of cell lysates using the indicated antibodies revealed the presence of NPRAP–ABP-S complexes. C, ABP-S interaction with NPRAP in HEK 293T cells depends on the NPRAP PDZ binding motif. Formation of ABP-S complexes with NPRAP was assayed as in B. NPRAP, but not NPRAPΔ3, which lacks the C-terminal PDZ binding motif, formed a complex with ABP-S. D, GRIP1 binds to NPRAP. Formation of NPRAP complexes with FLAG–GRIP1 in HEK 293T cells was assayed by coexpression and immunoprecipitation as in B, using the indicated antibodies. NPRAP associates with GRIP1 but not a control protein, GluR1. E, Cadherin, NPRAP, and ABP-S form a complex. NPRAP and ABP-S were expressed in HEK 293T cells as indicated, and cell lysates were prepared. Complexes containing endogenous E-cadherin were immunoprecipitated and analyzed by Western blotting for the presence of NPRAP and ABP-S. NPRAP associated with E-cadherin independently of the NPRAP PDZ binding motif, which is absent in mutant NPRAPΔ3. ABP-S was present in complexes containing E-cadherin, dependent on the expression of NPRAP and the integrity of the NPRAP PDZ binding motif. F, GluR2 forms a complex with NPRAP dependent on the presence of ABP-S. GluR2, ABP-S–FLAG, and NPRAP were coexpressed in HEK 293T cells as indicated, and complexes containing GluR2 were immunoprecipitated from cell lysates. Western blotting revealed the presence of NPRAP in these complexes, dependent on the presence of ABP-S. B–F, Blotting of lysates confirmed protein expression. IB, Immunoblot; IP, immunoprecipitation.
Table 1.
NPRAP and p0071 CTDs bind PDZ2 of ABP in yeast mating assays
| GAL4 binding domain fusion | Interaction phenotype with NPRAP |
|
|---|---|---|
| Growth on His− media | β-Galactosidase signal | |
| pGBKT7 alone | − | − |
| ABP PDZ1–PDZ3 | ++++ | + |
| ABP PDZ1 | − | − |
| ABP PDZ2 | +++ | + |
| ABP PDZ3 | − | − |
| ABP PDZ1–PDZ2 | + | − |
| ABP PDZ2–PDZ3 | ++++ | + |
| GRIP PDZ1–PDZ3 | ++++ | + |
| PSD-95 PDZ1–PDZ3 | ++++ | + |
Mating assays of PDZ domain interaction with the NPRAP C-terminal 309 aa or the p0071 C-terminal 188 aa were performed as described in Materials and Methods. In total, 18 matings were analyzed for ability to grow on media lacking histidine (His−), and viable colonies were then assayed for production of β-galactosidase. Plus (+) and minus (−) signs denote positive and negative results, respectively, from each set of matings. Relative amounts of growth are shown by a range of plus signs from one to four. The binding domain alone (listed as pGBKT7 alone) did not interact with either CTD, providing a negative control.
The NPRAP–ABP complex binds cadherin and GluR2
NPRAP binds the cytoplasmic tails of E- and N-cadherin, suggesting that NPRAP–ABP complexes may interact at adherens junctions with cadherins (Lu et al., 1999). To investigate such an interaction, we first asked whether NPRAP could bind both ABP and cadherin simultaneously. When NPRAP and ABP-S were expressed in 293T cells, immunoprecipitation of endogenous E-cadherin pulled down both NPRAP and ABP-S (Fig. 1E). ABP coprecipitation by anti-E-cadherin antibody was dependent on NPRAP and was eliminated when the NPRAP C-terminal 3 aa were deleted in the mutant NPRAPΔ3 (Fig. 1E). This demonstrated the formation of an E-cadherin–NPRAP–ABP complex, dependent on NPRAP C terminus–ABP interaction. We next asked whether ABP could bind NPRAP and GluR2 simultaneously. In Figure 1F, an antibody against GluR2 pulled down a complex containing GluR2, ABP, and NPRAP from lysates of 293T cells expressing these three proteins exogenously, but NPRAP was not pulled down when ABP-S was absent. This indicates that GluR2 can bind NPRAP–ABP complexes.
NPRAP draws ABP and GluR2 to adherens junctions
To investigate the role of adherens junctions in the localization of NPRAP–ABP complexes, we used MDCK cells, which express high levels of E-cadherin, and which at confluence form a uniform monolayer with prominent adherens junctions (Manniello et al., 1993; Lu et al., 1999). We asked whether the E-cadherin–NPRAP complex can form at these junctions and if so, whether it can associate with ABP. In MDCK cells, E-cadherin was found predominantly at the cell junctions (Fig. 2A), and ABP-L–GFP expressed on its own formed large cytoplasmic clusters, as was seen for ABP-S in HeLa cells (Fig. 2B) (DeSouza et al., 2002). Significantly, unlike ABP-L–GFP, exogenous NPRAP colocalized with E-cadherin at the plasma membrane (Fig. 2C) and with β-catenin (data not shown), as previously reported (Lu et al., 1999). This localization was not dependent on the NPRAP PDZ binding site (Fig. 2D). After coexpression with NPRAP, ABP-L–GFP shifted its location from the cytoplasmic clusters to the plasma membrane, at which ABP-L–GFP formed small puncta that extensively colocalized with NPRAP and E-cadherin (Fig. 2E). No shift was seen with NPRAPΔ3, indicating that recruitment of ABP to adherens junctions by NPRAP depended on the NPRAP–ABP PDZ interaction (Fig. 2F). These results show that a PDZ-dependent interaction between NPRAP and ABP can localize ABP at the membrane of adherens junctions and support the finding that NPRAP can bind both E-cadherin and ABP in a single complex. A similar result was obtained in HeLa cells, a transformed epithelial cell line that also forms adherens junctions (data not shown).
Figure 2.
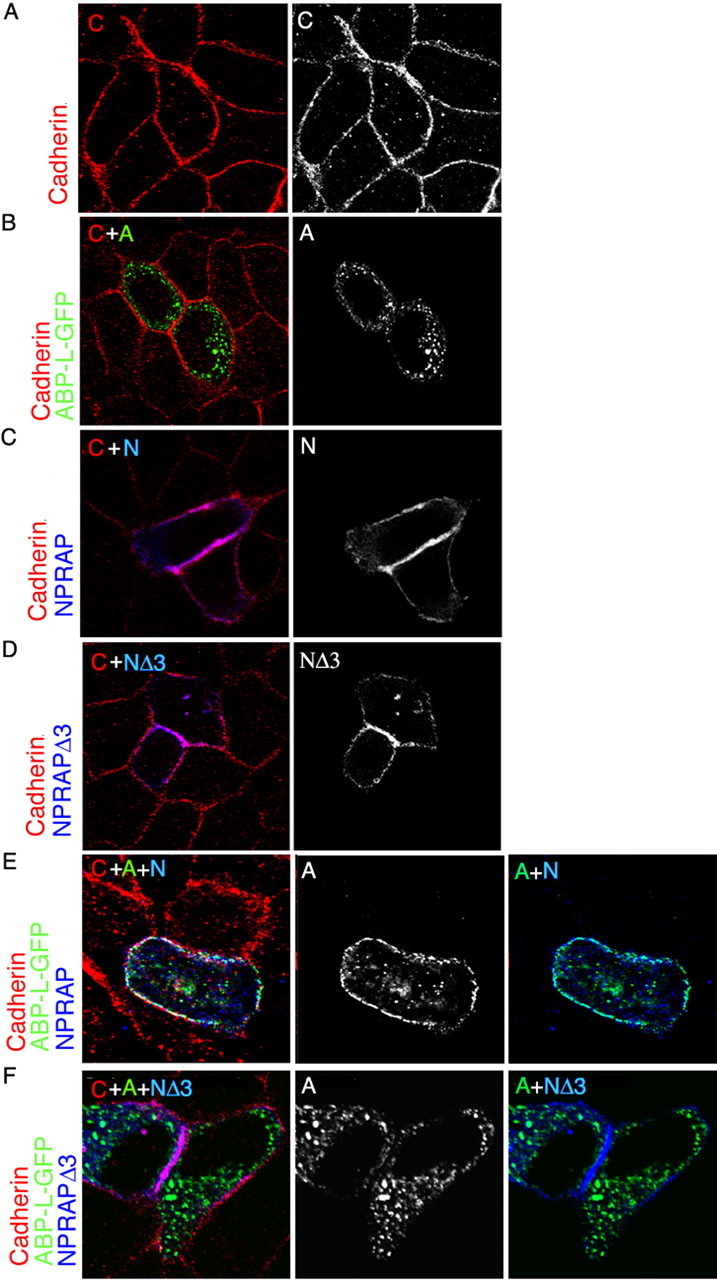
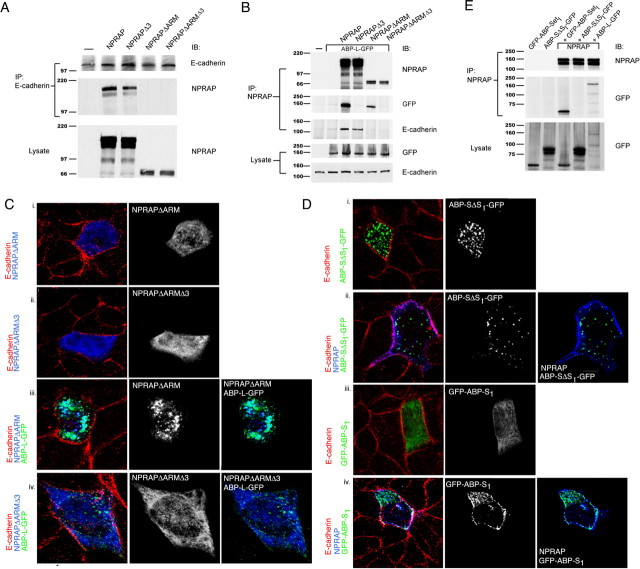
NPRAP localizes with E-cadherin at adherens junctions in MDCK cells and recruits ABP by a PDZ domain interaction. NPRAP and ABP-L were expressed in MDCK cells by transfection and were visualized together with endogenous E-cadherin by immunofluorescence as described in Materials and Methods. NPRAP colocalized with E-cadherin at adherens junctions. ABP-L–GFP colocalized with NPRAP at these junctions dependent on the NPRAP PDZ binding site. A, Endogenous E-cadherin localizes at adherens junctions in MDCK cells. B, ABP-L–GFP forms cytoplasmic clusters and does not colocalize with E-cadherin in MDCK cells in the absence of exogenous NPRAP. C, NPRAP colocalizes with E-cadherin in MDCK cells. D, The NPRAP mutant, NPRAPΔ3, which lacks the PDZ binding site, colocalizes with endogenous E-cadherin in MDCK cells. E, NPRAP and ABP-L–GFP were expressed by cotransfection in MDCK cells. NPRAP colocalized with endogenous E-cadherin and recruited ABP-L–GFP to these junctions. F, NPRAPΔ3 and ABP-L–GFP were expressed by cotransfection in MDCK cells. NPRAPΔ3 colocalizes with E-cadherin at adherens junctions but failed to recruit ABP-L–GFP, which remained in cytoplasmic clusters. A, ABP-L–GFP; C, cadherin; N, NPRAP.
ARM repeats mediate NPRAP interaction with E-cadherin and determine NPRAP distribution
NPRAP binds cadherins via its ARM repeats (Lu et al., 1999), and two NPRAP constructs containing the ARM repeats, NPRAP and NPRAPΔ3, when expressed by transfection coimmunoprecipitated with E-cadherin from MDCK cell extracts. However, mutants lacking the ARM repeats, NPRAPΔARM and NPRAPΔARMΔ3, did not bind E-cadherin (Fig. 3A). ABP interaction with NPRAP, which depended on the 3 C-terminal amino acids, did not require the ARM repeats because immunoprecipitation of NPRAPΔARM pulled down ABP-L (Fig. 3B). As expected, a mutant lacking both the ARM domains and the C-terminal amino acids, NPRAPΔARMΔ3, did not coimmunoprecipitate with ABP-L. Both NPRAPΔARM and NPRAPΔARMΔ3 were diffusely distributed throughout the cytoplasm, consistent with their inability to bind E-cadherin (Fig. 3Ci–ii). Most significantly, NPRAPΔARM did not shift ABP to the membrane, and instead, NPRAPΔARM redistributed into the ABP clusters (Fig. 3Ciii). NPRAPΔARMΔ3 and ABP-L, when coexpressed, retained the same patterns as when transfected separately (Fig. 3Civ), confirming that NPRAPΔARM distribution to ABP-L clusters depended on PDZ interaction. We conclude that NPRAP in association with cadherin can direct ABP to adherens junctions, but if the NPRAP–cadherin interaction is disrupted, ABP-L exerts the dominant role and directs NPRAP to cytoplasmic clusters.
Figure 3.
Role of ARM domains and PDZ binding site in NPRAP recruitment of ABP to adherens junctions in MDCK cells. NPRAP, ABP-L–GFP, and their mutants were expressed by transfection in MDCK cells as indicated. NPRAP formed a complex with and localized with E-cadherin at adherens junctions, dependent on the NPRAP ARM domain, which is absent in NPRAPΔARM. NPRAP recruited ABP to E-cadherin dependent on the NPRAP PDZ binding site, which is deleted in NPRAPΔ3. The mutant NPRAPΔARMΔ3 lacks both the ARM domains and the PDZ binding site. NPRAP formed a complex with ABP and recruited ABP to E-cadherin at adherens junctions dependent on ABP-SetI, which is present in GFP–ABP-SetI and absent in ABP-SΔSI–GFP. A, Endogenous E-cadherin was immunoprecipitated from extracts of transfected MDCK cells, and the presence of NPRAP or its mutants was assayed by Western blotting. E-cadherin coimmunoprecipitated NPRAP and NPRAPΔ3, but not NPRAPΔARM or NPRAPΔ3ΔARM, indicating that the ARM domain of NPRAP but not the PDZ binding site is necessary for NPRAP association with E-cadherin. B, NPRAP was immunoprecipitated from extracts of transfected MDCK cells, and the presence of E-cadherin and ABP-L–GFP was assayed by Western blotting. NPRAP associates with both ABP-L–GFP and E-cadherin. The NPRAP PDZ binding site and the ARM repeats are necessary for binding, respectively, to ABP-L–GFP and E-cadherin. C, ABP-L–GFP and NPRAP or its mutants were visualized by immunofluorescence in transfected MDCK cells. i, ii, NPRAP deletion mutants lacking the ARM repeats, NPRAPΔARM and NPRAPΔARMΔ3, do not colocalize with E-cadherin and are diffuse throughout the cytoplasm. iii, NPRAPΔARM colocalizes with ABP-L–GFP in the cytoplasm, but not with E-cadherin, which is localized to the adherens junction. iv, NPRAPΔARMΔ3 does not colocalize with ABP-L–GFP or E-cadherin. D, ABP-L–GFP and NPRAP or its mutants were expressed by transfection in MDCK cells. NPRAP, ABP-L–GFP, and endogenous E-cadherin were visualized by immunofluorescence. i, ABP lacking SetI, ABP-SΔSI–GFP, forms cytoplasmic clusters and does not colocalize with E-cadherin, which is at adherens junctions. ii, ABP-SΔSI–GFP does not colocalize with NPRAP or E-cadherin in MDCK cells. iii, GFP–ABP-SetI, when transfected on its own into MDCK cells, is diffusely distributed in the cytoplasm and does not colocalize with E-cadherin. iv, When GFP–ABP-SI and NPRAP are cotransfected into MDCK cells, the two colocalize with each other and with E-cadherin at adherens junctions. E, SetI of ABP is necessary and sufficient for ABP to associate with NPRAP in MDCK cells. After cotransfection with NPRAP, GFP–ABP-SetI and ABP-L–GFP coimmunoprecipitated with NPRAP from cell extracts, whereas ABP-SΔSI–GFP did not coimmunoprecipitate with NPRAP. IB, Immunoblot; IP, immunoprecipitation.
ABP-SetI is necessary and sufficient for NPRAP to target ABP to E-cadherin
We examined the role of ABP PDZ SetI in ABP localization at adherens junctions. The mutant ABP-SΔSI–GFP, which lacks SetI, localized in large cytoplasmic clusters, similar to full-length ABP (Fig. 3Di). However, it did not shift to the membrane after NPRAP coexpression (Fig. 3Dii). GFP–ABP-SetI on its own was diffusely distributed throughout the cytoplasm, but when expressed with NPRAP it concentrated at the plasma membrane and colocalized with NPRAP and E-cadherin (Fig. 3Diii–iv). Thus, ABP-SetI is both necessary and sufficient to interact with NPRAP and to redistribute to adherens junctions. Immunoprecipitation revealed that ABP-SetI, but not ABP-SΔSI–GFP, was pulled down with NPRAP in MDCK cells (Fig. 3E), confirming a physical interaction of ABP-SetI with NPRAP in these cells.
Endogenous NPRAP is synaptic and colocalizes with GRIP in hippocampal neurons
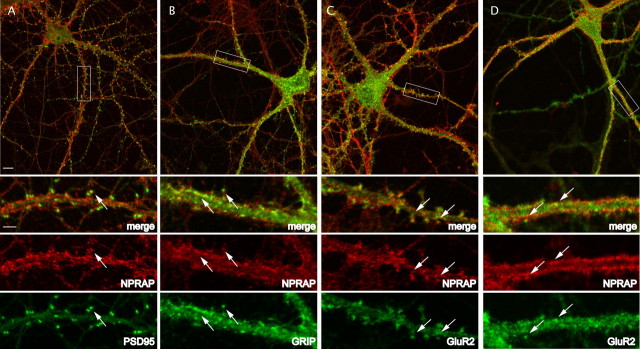
In cultured hippocampal neurons, endogenous NPRAP within wide dendrites was observed in puncta at both sides of the dendrite (Fig. 4). The great majority of spiny neurons expressed the postsynaptic synaptic marker PSD-95, and in a proportion of these neurons, NPRAP was also observed in spines, colocalizing with PSD-95 (Fig. 4A). The colocalization suggests that NPRAP is located at synapses along dendrites, in agreement with a dendritic localization for NPRAP in adult mouse brain (Ho et al., 2000), and implies a postsynaptic role for NPRAP. Attempts to assay endogenous ABP colocalization with NPRAP in cultured neurons using available ABP antibodies yielded low signals, so the localization of the highly homologous GRIP protein, which we showed also interacts with NPRAP, was examined instead. In spiny neurons, NPRAP overlapped with GRIP puncta in spines and along the dendritic membrane (Fig. 4B), consistent with NPRAP interaction with GRIP (Fig. 5B,D; see below). In spiny neurons, NPRAP also colocalized with GluR2 in spines (Fig. 4C), consistent with its interaction with GRIP–GluR2 complexes (Fig. 1F). We conclude that NPRAP colocalizes with a synaptic marker, PSD-95, and with AMPARs, and with a subset of GRIP that is synaptic. We also observed NPRAP in a large proportion of aspiny neurons, in which NPRAP was less punctate and was found along the boundary of the dendrite, at which it appeared to line the plasma membrane. In these neurons, NPRAP colocalized frequently with GluR2 (Fig. 4D). Thus, NPRAP appeared in two different patterns of localization, one in spines, in which it was punctate, and the second in aspiny neurons, in which it appeared along the perimeter of the dendrite in a more continuous distribution. In both instances, it colocalized with GluR2.
Figure 4.
Endogenous NPRAP is found at synapses of cultured hippocampal neurons and colocalizes with AMPA receptors. A–D, The designated proteins were localized in dissociated embryonic hippocampal neurons 19–21 d in culture by immunofluorescence as described in Materials and Methods. A, NPRAP colocalizes with the postsynaptic marker PSD-95. Scale bars: Top panels, 10 μm; bottom panels, 3 μm. B, NPRAP and GRIP colocalize at spines. C, NPRAP colocalizes in spines with GluR2, a marker for synaptic AMPARs. D, NPRAP and GluR2 colocalize along the dendritic membrane in aspiny neurons. Arrows identify sites of protein colocalization.
Figure 5.
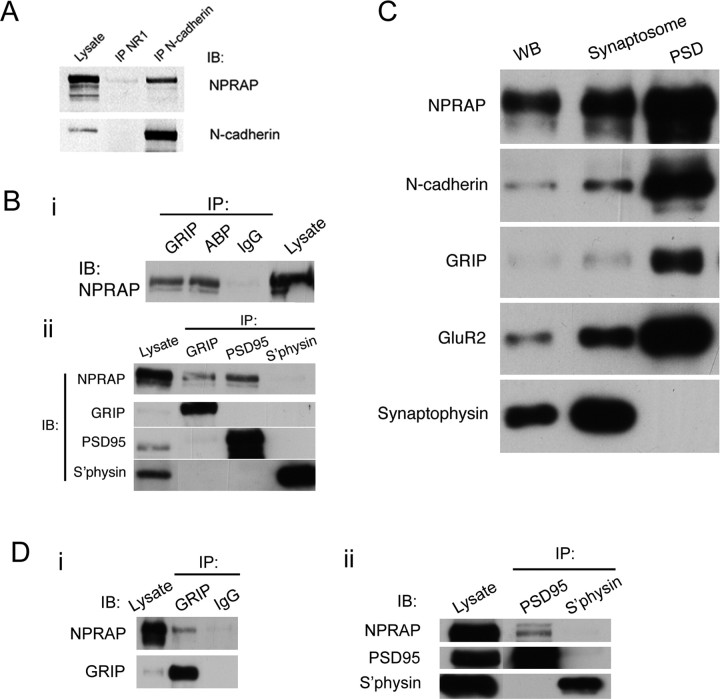
PSD enrichment and interaction of endogenous NPRAP with N-cadherin and the scaffolding proteins ABP, GRIP, and PSD-95 in brain and synaptosomal fractions. A, B, Triton X-100 lysates prepared from cultured cortical neurons, 28–35 DIV, were analyzed by immunoprecipitation. A, Endogenous N-cadherin, but not the NMDAR subunit NR1, coimmunoprecipitated endogenous NPRAP, as determined by Western blotting. This demonstrates that endogenous NPRAP interacts specifically with N-cadherin in neurons. B, The interaction of NPRAP with scaffolding proteins was assayed by immunoprecipitation of ABP, GRIP, and PSD-95 from neuron lysates and Western blotting for NPRAP. i, Immunoprecipitates of cultured cortical neuron lysates (400–500 μg) using antibodies to GRIP and ABP and with IgG (control) were analyzed with anti-NPRAP antibody. NPRAP coimmunoprecipitated with GRIP and ABP, demonstrating interaction in cultured neurons. The original lysate (20 μg) was analyzed in parallel. ii, Immunoprecipitates of cultured cortical neuron lysates (400–500 μg) with antibodies to GRIP, ABP, and synaptophysin (S'physin; a control) were analyzed in parallel with the original lysate (20 μg) by Western blotting. NPRAP coimmunoprecipitated with GRIP and PSD-95, but not with synaptophysin, demonstrating specific interactions. C, NPRAP, N-cadherin, GRIP, and GluR2 are enriched in the PSD. Whole-brain (WB) lysate and the synaptosome and PSD fractions were isolated from adult rat brain, and equal quantities of protein (2 μg) of each fraction were analyzed by Western blotting with the indicated antibodies. NPRAP, N-cadherin, GRIP, and GluR2 were enriched in the PSD, whereas synaptophysin, a control, was absent. D, Synaptosomes contain NPRAP in complexes with GRIP and PSD-95. Synaptosomes were isolated from adult rat brain, and Triton X-100 lysates were analyzed by Western blotting, either directly (2 μg) or after immunoprecipitation (1 mg). i, Immunoprecipitation with anti-GRIP antibody but not with a control IgG coimmunoprecipitated NPRAP. ii, Immunoprecipitation with anti-PSD-95 antibody but not with synaptophysin antibody (control) coimmunoprecipitated NPRAP. Blots were reprobed with antibodies to GRIP, PSD-95, and synaptophysin to verify protein presence in the lysates. IB, Immunoblot; IP, immunoprecipitation.
NPRAP forms complexes with scaffolding proteins
To assay for NPRAP complexes with cadherins or with ABP, GRIP, and PSD-95, which also bound NPRAP (Table 1), we coimmunoprecipitated endogenous proteins from brain lysates. NPRAP coimmunoprecipitated with N-cadherin (Fig. 5A) and with GRIP, ABP, and PSD-95, but not with synaptophysin or with a control IgG immunoprecipitation (Fig. 5Bi,ii). This confirmed that NPRAP interacts with cadherins, ABP, GRIP, and PSD-95 in primary neurons.
To assess the synaptic localization of these proteins, we assayed their presence in whole-brain Triton X-100 extracts and in synaptically enriched brain fractions (Fig. 5C). NPRAP, N-cadherin, GRIP, GluR2, and synaptophysin were all enriched in synaptosomes and, with the exception of the synaptic vesicle marker, synaptophysin, were greatly enriched in the PSD fraction (Fig. 5C). Furthermore, immunoprecipitation of GRIP and PSD-95, but not of IgG or synaptophysin, from lysed synaptosomes specifically coimmunoprecipitated NPRAP, indicating the presence of NPRAP–GRIP and NPRAP–PSD-95 protein complexes in this synaptically enriched fraction (Fig. 5Di,ii).
Exogenous NPRAP anchors ABP at spines of cultured neurons
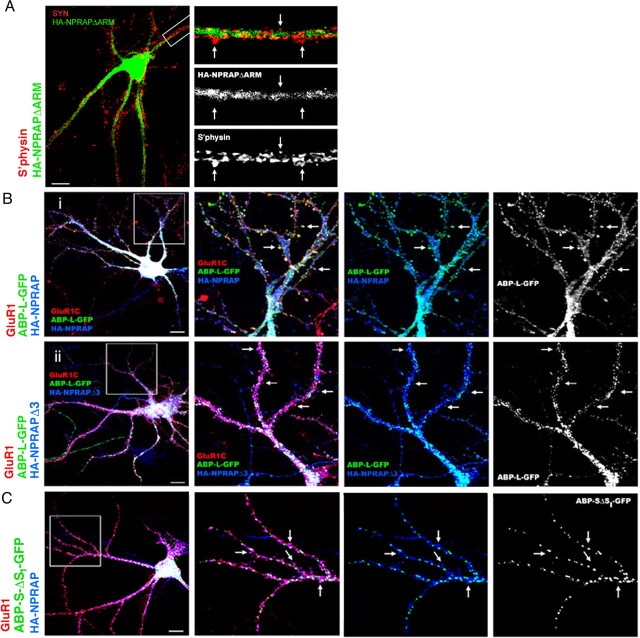
We next deduced the role of protein interactions in the localization of NPRAP in neurons. We first confirmed that NPRAP used its ARM domains, which bind to cadherins, to associate with spines. We previously observed that wild-type endogenous NPRAP was synaptic in such neurons (Fig. 4A). We then expressed HA-tagged NPRAPΔARM using Sindbis viral vectors and found that HA–NPRAPΔARM was diffuse and did not colocalize with the synaptic marker, synaptophysin (Fig. 6A). Thus the ARM domains tether NPRAP at spines, which is in agreement with ARM domain localization of NPRAP at the plasma membrane of MDCK cells.
Figure 6.
Exogenous NPRAP recruits ABP to spines of cultured neurons dependent on the PDZ binding site. Embryonic hippocampal neurons, 19–21 d in culture, were infected with Sindbis virus vectors expressing HA–NPRAP, HA–NPRAPΔ3, or HA–NPRAPΔARM and ABP-L–GFP or ABP-S-ΔS1–GFP, as indicated. Proteins were localized by immunofluorescence or in the case of ABP, by GFP fluorescence. A, HA–NPRAPΔARM expressed from a Sindbis virus vector fails to colocalize with synaptophysin (S'physin, SYN), a synaptic marker. B, HA–NPRAP, but not HA–NPRAPΔ3, recruits ABP-L–GFP to spines containing GluR1, an AMPAR marker, in cultured hippocampal neurons. i, In cultured neurons infected with viruses expressing ABP-L–GFP and HA–NPRAP, ABP-L–GFP and HA–NPRAP colocalize with GluR1 in spines. ii, In cultured neurons infected with viruses expressing ABP-L and HA–NPRAPΔ3, HA–NPRAPΔ3 but not ABP-L–GFP colocalizes with GluR1 in spines. C, After coexpression in cultured neurons with exogenous HA–NPRAP, ABP-S-ΔS1–GFP, which lacks the NPRAP binding site, SetI, does not colocalize with GluR1 in cultured neurons. Arrows identify sites of protein colocalization.
We next examined the role of NPRAP in anchoring ABP to spines. In Figure 6B, exogenous ABP-L–GFP (green) was coexpressed with exogenous HA–NPRAP or HA–NPRAPΔ3 (blue). Both NPRAP species distributed to spines, at which they colocalized with GluR1 (red) (Fig. 6Bi,ii). Exogenous ABP-L–GFP was also found in spines when it was coexpressed with wild-type NPRAP, as indicated by numerous white spine structures (Fig. 6Bi). However, ABP-L–GFP, when coexpressed with NPRAPΔ3, was absent from spines as indicated by the numerous magenta spine structures, which result from the overlap in spines of GluR1 with NPRAPΔ3 but not with ABP-L–GFP. ABP-L–GFP instead appeared in cytoplasmic clusters (Fig. 6Bii). A quantitative assessment showed that whereas only a small proportion of neurons expressing ABP-L–GFP alone or both ABP-L–GFP and NPRAPΔ3 showed a spiny distribution for ABP-L, a much larger proportion of neurons expressing ABP-L–GFP with wild-type NPRAP displayed a spiny distribution for ABP-L (Table 2). This supported a role for the NPRAP PDZ binding site in anchorage of ABP-L in spines.
Table 2.
In the presence of full-length NPRAP, ABP-L redistributes from intracellular compartments to spines within cultured neurons
| Expressed proteins | ABP-L | ABP-L plus NPRAP | ABP-L plus NPRAPΔ3 |
|---|---|---|---|
| Neurons with ABP-L in spines | 3 of 30 | 22 of 30 | 6 of 30 |
In order to quantify changes in the distribution of ABP-L–GFP when coexpressed with HA–NPRAP, the distribution of ABP-L in infected neurons was counted in three separate experiments for a total of 30 cells for each set of infections. The distribution of ABP-L was categorized ″blindly″ before the presence of NPRAP was known. Analysis was limited to infected neurons that displayed a spiny pattern, as determined by GluR1 staining. Columns showing cells results from cells infected with only ABP-L or ABP-L plus NPRAP were determined from the same coverslips.
When expressed alone, ABP-SΔSI–GFP, which lacks the ABP N-terminal leader and the first set of PDZ domains, formed punctate clusters rather than entering spines (data not shown), and it did not change its distribution when coexpressed with HA–NPRAP (Fig. 6C), suggesting that interaction of ABP-SetI with NPRAP is required for its localization in spines. These results indicate that targeting of ABP to spines requires SetI, which interacts with NPRAP.
NPRAP–ABP interaction anchors GluR2 at the plasma membrane
NPRAP that is bound through its ARM domains to cadherins may in turn anchor, via its C terminus, PDZ-containing scaffolding proteins including ABP and GRIP, which bind to AMPARs. PSD-95, which also binds to the NPRAP C terminus, can anchor AMPAR–TARP complexes (Chen et al., 2000). To examine the role of NPRAP interaction with PDZ scaffolding proteins in the surface localization of AMPA receptors, we performed dominant-negative NPRAP experiments. First, we expressed in neurons dsRed-tagged NPRAP-CT, which is the red-shifted GFP, dsRed, linked to the C-terminal 77 aa of NPRAP. We hypothesized that this construct might act as a dominant-negative mutant by binding to and sequestering ABP, GRIP, and PSD-95 (collectively, NPRAP-interacting scaffolds), hence disrupting anchorage of these scaffolds by NPRAP at synapses. If dsRed NPRAP-CT did block anchorage, AMPARs that were bound to PDZ5 of ABP/GRIP, as well as stargazin–AMPAR complexes that were bound to PSD-95, would be displaced from NPRAP, releasing them from their anchorage at the plasma membrane. The mutant, dsRed NPRAP-CTΔ3, which lacks the last 3 aa of NPRAP, provides a control that any such release is dependent on the PDZ binding site of dsRed NPRAP-CT. Uninfected control cells and dsRed NPRAP-CTΔ3-expressing cells showed similar levels of surface endogenous GluR2, which was measured by a live cell immunofluorescent staining protocol. However, cells expressing dsRed NPRAP-CT showed a significant, 30% decrease in surface GluR2 levels (Fig. 7A,B). This indicates that the surface localization of GluR2 is diminished when interactions of the NPRAP C-terminal PDZ binding site are disrupted.
Figure 7.
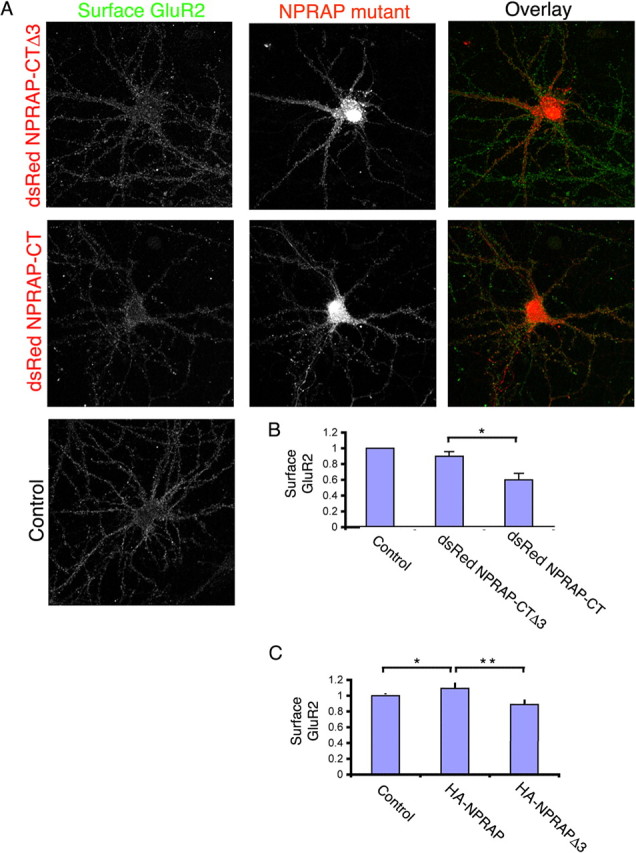
Dominant-negative NPRAP constructs decrease cell surface endogenous AMPAR. A, The NPRAP C-terminal constructs, dsRed NPRAP-CT or dsRed NPRAP-CTΔ3, which contain dsRed linked respectively to the N terminus of the C-terminal 77 aa of NPRAP or this C terminus lacking the final 3 aa, were expressed in cultured embryonic hippocampal neurons from Sindbis virus vectors. The levels of endogenous GluR2 at the cell surface were measured by live cell staining as described in Materials and Methods. B, Quantitation of surface, endogenous GluR2 in neurons expressing no virus (control), dsRed NPRAP-CT, or dsRed NPRAP-CTΔ3. The values are the averages of three experiments with 10–12 cells per experiment. *p = 0.022. C, The proteins HA–NPRAP and HA–NPRAPΔ3, which lack the NPRAP 3 C-terminal amino acids, including the PDZ binding site, were expressed in hippocampal neurons from Sindbis virus vectors. The surface levels of endogenous GluR2 in these cultures and in uninfected controls were measured by surface labeling of live cells (see Materials and Methods). Surface GluR2 levels decreased significantly in the HA–NPRAPΔ3-expressing cells relative to the HA–NPRAP cells. Cells expressing HA–NPRAP showed higher surface levels than the control. The results of two experiments (control, 36 cells; HA–NPRAP, 16 cells; HA–NPRAPΔ3, 17 cells) are shown. *p = 0.038; **p = 0.009.
Because dsRed NPRAP-CT could potentially displace receptors anchored to class I PDZ domains of scaffolding proteins that are not bound to cadherins, we performed an additional experiment. We expressed HA–NPRAPΔ3, which was expected to bind to cadherins and specifically block binding of endogenous NPRAP and any associated PDZ domain scaffolding proteins. In this manner, HA–NPRAPΔ3 would selectively disrupt anchorage of AMPA receptors tethered through NPRAP interactions. As a control, we expressed HA–NPRAP, which could enhance NPRAP-dependent receptor tethering. As shown in Figure 7C, the levels of surface GluR2 were decreased significantly in the HA–NPRAPΔ3-expressing cells relative to an uninfected control, whereas surface GluR2 levels in the HA–NPRAP expressing cells were elevated relative to this control. This supports the conclusion that cadherin-NPRAP interactions with GRIP/ABP or PSD-95 maintain GluR2 on the neuron surface.
Discussion
Synaptic cell adhesion proteins bind the PDZ scaffold
At excitatory synapses, CAMs, including the heterophilic neurexins and neuroligins (Cantallops and Cline, 2000) and the homophilic N-CAM, SynCAM, and cadherins (Bamji, 2005), interact across the synaptic cleft to provide adhesive interactions between the presynaptic and postsynaptic membranes (Yamagata et al., 2003). In this report, we show that the cadherin cell adhesion apparatus can anchor AMPARs in complexes with ABP/GRIP. Specifically, GluR2–ABP/GRIP complexes can bind through the ABP/GRIP second PDZ domain, PDZ2, to NPRAP, an ARM repeat protein that is a component of the postsynaptic density that binds to cadherins. Experiments using NPRAP dominant-negative constructs suggest that in our cell system, as much as one-third of AMPA receptors may be localized at the plasma membrane by such an anchorage. PSD-95 also binds to NPRAP, providing an additional scaffold that may be tethered by cadherins.
NPRAP complexes at synapses
Cadherins mediate homophilic, Ca2+-dependent cellular adhesion at adherens junctions. Experiments in MDCK cells, which provide a model for cadherins at adherens junctions, demonstrated the formation of cadherin–NPRAP–ABP complexes dependent on the expression of NPRAP and the integrity of the NPRAP–ABP interaction, and the formation of NPRAP–ABP-GluR2 complexes dependent on expression of ABP. This suggests that the proposed cadherin–NPRAP–ABP–GluR2 complexes can indeed form in cells. Two dominant-negative mutants of NPRAP were assayed for their effects on GluR2. One mutant does not interact with ABP, GRIP, or PSD-95, and the second binds these scaffolding proteins but does not bind to cadherins. Both mutants reduced GluR2 surface levels. This suggests that cadherin–NPRAP–ABP–GluR2 complexes contribute to the surface stabilization of GluR2. In neurons, adherens junction structures containing cadherins are found at synapses (Takeichi and Abe, 2005). This suggests that the AMPARs that are anchored to cadherins by NPRAP–GRIP/ABP complexes in neurons are synaptic. Significantly, we found that in whole brain, the components of the proposed cadherin–AMPAR scaffolding complex, N-cadherin, NPRAP, GRIP, and GluR2, were all enriched in the PSD, a postsynaptic structure. Complexes of endogenous NPRAP with cadherins and with the scaffolding proteins, ABP, GRIP, and PSD-95 could be detected in whole-brain lysates. Most significant, NPRAP–GRIP and NPRAP–PSD-95 complexes could be isolated from synaptosomes, which are highly enriched in synaptic components. Finally, NPRAP colocalized with PSD-95, a postsynaptic marker, with GRIP, a PSD component, and with GluR2, which is a marker for synapses containing AMPA receptors. Interestingly, two different distributions were observed. In spiny neurons, NPRAP was punctate, whereas in aspiny neurons it displayed a more continuous distribution along the surface of the dendrite, and in both cases it colocalized with GluR2. These experiments strongly suggest that the cadherin–NPRAP–ABP/GRIP anchorages for GluR2-containing AMPARs are found at synapses. However, we cannot exclude the possibility that the anchorage of AMPARs by this complex is perisynaptic or that a proportion of the anchored receptors are at other membrane sites.
We showed that in MDCK cells and cultured neurons, NPRAP interacts with cadherins via its ARM domains, in agreement with previous reports (Lu et al., 1999). NPRAP that is tethered in this manner at the cell surface to cadherins was able to recruit ABP from the cytosol to the plasma membrane of MDCK cells and to spines of cultured neurons. The forms of ABP studied here, ABP-L and ABP-S, are cytosolic in the absence of interaction with NPRAP–cadherin (Fu et al., 2003). Thus, recruitment of ABP to the membrane by NPRAP is likely to be a dynamic process. Indeed, when the ARM domains of NPRAP were deleted to abolish NPRAP binding to cadherins, ABP sequestered the NPRAP mutant in the cytosol.
A second ABP form, pABP-L, is palmitoylated in its N-terminal leader sequence, and the lipid modification targets pABP-L to the plasma membrane of heterologous cells and to spine heads of cultured neurons (DeSouza et al., 2002). A variant of GRIP is similarly palmitoylated (Yamazaki et al., 2001), and PSD-95 also associates with the synapse through N-terminal palmitate (Topinka and Bredt, 1998; El-Husseini Ael et al., 2002). Thus lipid modification provides an alternative to NPRAP interaction for localizing ABP, GRIP, and PSD-95 at spines.
Cadherin function at synaptic junctions
The present work suggests that cadherin intracellular domains bind NPRAP, which in turn anchors the ABP and GRIP scaffolds that bind AMPARs. GRIP and ABP bind numerous other signaling factors, including ephrins and Eph receptors (Bruckner et al., 1999; Contractor et al., 2002), a neuronal ras GEF (Ye et al., 2000), and liprin-α (Wyszynski et al., 2002). Thus, cadherins acting through NPRAP–GRIP/ABP complexes could serve as anchorage sites for diverse signaling components of the synapse. NPRAP also interacted with the excitatory synapse scaffolding protein PSD-95, as detected in yeast mating assays, and NPRAP–PSD-95 complexes were found in whole-brain and synaptosomal lysates. PSD-95 anchors NMDAR during the early stages of synaptogenesis (Perez-Otano and Ehlers, 2004), and although it does not bind AMPAR directly, it tethers AMPAR–TARP complexes at synapses (Chen et al., 2000) (for review, see Ziff, 2007).
PSD-95 also binds to cell adhesion molecules, including the neuroligins, whose interactions with neurexins have been shown to induce functional presynaptic structures (Scheiffele, 2003). Although not directly tested here, this raises the possibility that cadherin–NPRAP complexes could bind PSD-95–neuroligin complexes, linking these two adhesion structures. Members of the NPRAP/δ-catenin/p120 family bind to yet other PDZ proteins, including synaptic scaffolding molecule (S-SCAM) (Ide et al., 1999) and PAPIN (Deguchi et al., 2000) and to the LAP protein, densin180 (Izawa et al., 2002).
Notably, β-catenin, like NPRAP, targets S-SCAM to synapses (Nishimura et al., 2002). NPRAP is also found in complexes with NR2A and mGluR1-α (Jones et al., 2002) and is downregulated after AMPA receptor stimulation (Jones et al., 2002). NPRAP itself binds to the actin regulatory protein cortactin (Martinez et al., 2003) and induces cytoskeletal remodeling (Kim et al., 2002). The related p120 protein regulates the family of rho-GTPases, which are critical actin regulators (Anastasiadis, 2007; Fox and Peifer, 2007). Thus NPRAP itself, situated at the nexus of diverse scaffolds, CAMs, and signaling molecules, may play an important role in regulating synapse morphology or signaling.
Receptor tethering by catenins
ABP and GRIP form a scaffold that binds GluR2 and GluR3 (Dong et al., 1997; Srivastava et al., 1998). Dissociation of receptors from this scaffold by phosphorylation of GluR2 at S880 by PKC has been implicated in mechanisms of long-term depression (LTD) (Matsuda et al., 1999; Chung et al., 2000; Xia et al., 2000). Association of receptors with this scaffold may contribute to dedepression, a repotentiation of synapses after LTD (Daw et al., 2000). The cadherin–NPRAP–ABP/GRIP complex studied here may contribute specifically to these forms of synapse regulation, which involve GluR2 and GluR3. The cadherin intracellular domain also binds β-catenin, which binds through α-catenin to the actin cytoskeleton (for review, see Kobielak and Fuchs, 2004). A complex containing β-catenin, plus the NPRAP-related protein, p120, and Velis (mLin7), recruits the kainate receptor subunit, GluR6, to cadherin-induced adherens junctions (Coussen et al., 2002). In C. elegans, β-catenin binds the multi-PDZ complex, Lin2–Lin7–Lin10, which binds the glutamate receptor GLR1 (Rongo et al., 1998). In mammals, a related complex, mLin2/CASK, mLin7/Velis/Mals, mLin10/X11a/Mint, binds the kinesin KIF17 and may transport and tether NMDA receptors (Setou et al., 2000; Schnapp, 2003). Also, β-catenin and cadherin form a complex with AMPAR in neurons (Nuriya and Huganir, 2006). Significantly, GluR2 and cadherins also interact directly through their extracellular domains (Saglietti et al., 2007). Thus, in neurons, cadherins may have a general role in tethering glutamate receptors by binding catenin–PDZ scaffold peptide–receptor complexes or receptor subunits.
Control of cell adhesion
Formation of the NPRAP–ABP/GRIP complex could also influence cell adhesion. Although cadherins situated on opposite membranes interact with one another in trans, they also undergo cis-interactions that form cadherin dimers (Patel et al., 2003). NPRAP contains a coiled-coil region that may induce dimerization (Paffenholz and Franke, 1997), and ABP/GRIP form dimers through interactions involving Set2 (Srivastava et al., 1998; Fu et al., 2003). Interactions of NPRAP–ABP/GRIP multimers with the cadherin intracellular domains could contribute to the formation of cadherin arrays. Indeed, the p120 protein, which is related to NPRAP, has been shown to regulate cadherin stability (Anastasiadis, 2007; Fox and Peifer, 2007). Thus, NPRAP may be part of a larger, Ca2+-responsive adhesion complex involving cadherins and stabilized by both extracellular and intracellular interactions. Synaptic cadherins reorganize structurally after neuron depolarization (Tanaka et al., 2000), and peptides and antibodies that block cadherin intercellular homophilic interactions impair the induction of LTP (Tang et al., 1998). Significantly, overexpression of cadherins increased the surface expression of GluR1 (Nuriya and Huganir, 2006). This suggests that cadherins can themselves contribute to spine and synapse plasticity. Because knock-out of δ-catenin (NPRAP) leads to specific neural deficits resembling cri du chat syndrome, a form of mental retardation (Medina et al., 2000; Israely et al., 2004), disruption of the interactions described here may have a role in neurological disease.
Footnotes
This work was supported by National Institutes of Health (NIH) Grant AG13620 (E.B.Z.). S.R. was supported by a Bernard B. Levine postdoctoral fellowship, and J.S. was supported by the New York University Medical Scientist Training Program through Grant T32 GM07308. The Zeiss LSM510 Meta laser-scanning confocal microscope was funded by Shared Instrumentation Grant S10 RR017970 from the NIH. We thank B. States, G. Rameau, S. DeSouza, and S. Srivastava for plasmids; W. Franke for antibodies; T. Serra for aid in preparing this manuscript; and B. Fernholz and B. Jordan for critical readings of this manuscript.
References
- Anastasiadis, 2007.Anastasiadis PZ. p120-ctn: A nexus for contextual signaling via Rho GTPases. Biochim Biophys Acta. 2007;1773:34–46. doi: 10.1016/j.bbamcr.2006.08.040. [DOI] [PubMed] [Google Scholar]
- Bamji, 2005.Bamji SX. Cadherins: actin with the cytoskeleton to form synapses. Neuron. 2005;47:175–178. doi: 10.1016/j.neuron.2005.06.024. [DOI] [PubMed] [Google Scholar]
- Barry and Ziff, 2002.Barry MF, Ziff EB. Receptor trafficking and the plasticity of excitatory synapses. Curr Opin Neurobiol. 2002;12:279–286. doi: 10.1016/s0959-4388(02)00329-x. [DOI] [PubMed] [Google Scholar]
- Bruckner et al., 1999.Bruckner K, Pablo Labrador J, Scheiffele P, Herb A, Seeburg PH, Klein R. EphrinB ligands recruit GRIP family PDZ adaptor proteins into raft membrane microdomains. Neuron. 1999;22:511–524. doi: 10.1016/s0896-6273(00)80706-0. [DOI] [PubMed] [Google Scholar]
- Cantallops and Cline, 2000.Cantallops I, Cline HT. Synapse formation: if it looks like a duck and quacks like a duck. Curr Biol. 2000;10:R620–R623. doi: 10.1016/s0960-9822(00)00663-1. [DOI] [PubMed] [Google Scholar]
- Chen et al., 2000.Chen L, Chetkovich DM, Petralia RS, Sweeney NT, Kawasaki Y, Wenthold RJ, Bredt DS, Nicoll RA. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature. 2000;408:936–943. doi: 10.1038/35050030. [DOI] [PubMed] [Google Scholar]
- Chung et al., 2000.Chung HJ, Xia J, Scannevin RH, Zhang X, Huganir RL. Phosphorylation of the AMPA receptor subunit GluR2 differentially regulates its interaction with PDZ domain-containing proteins. J Neurosci. 2000;20:7258–7267. doi: 10.1523/JNEUROSCI.20-19-07258.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contractor et al., 2002.Contractor A, Rogers C, Maron C, Henkemeyer M, Swanson GT, Heinemann SF. Trans-synaptic Eph receptor-ephrin signaling in hippocampal mossy fiber LTP. Science. 2002;296:1864–1869. doi: 10.1126/science.1069081. [DOI] [PubMed] [Google Scholar]
- Coussen et al., 2002.Coussen F, Normand E, Marchal C, Costet P, Choquet D, Lambert M, Mege RM, Mulle C. Recruitment of the kainate receptor subunit glutamate receptor 6 by cadherin/catenin complexes. J Neurosci. 2002;22:6426–6436. doi: 10.1523/JNEUROSCI.22-15-06426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daw et al., 2000.Daw MI, Chittajallu R, Bortolotto ZA, Dev KK, Duprat F, Henley JM, Collingridge GL, Isaac JT. PDZ proteins interacting with C-terminal GluR2/3 are involved in a PKC-dependent regulation of AMPA receptors at hippocampal synapses. Neuron. 2000;28:873–886. doi: 10.1016/s0896-6273(00)00160-4. [DOI] [PubMed] [Google Scholar]
- Deguchi et al., 2000.Deguchi M, Iizuka T, Hata Y, Nishimura W, Hirao K, Yao I, Kawabe H, Takai Y. PAPIN. A novel multiple PSD-95/Dlg-A/ZO-1 protein interacting with neural plakophilin-related armadillo repeat protein/delta-catenin and p0071. J Biol Chem. 2000;275:29875–29880. doi: 10.1074/jbc.M005384200. [DOI] [PubMed] [Google Scholar]
- DeSouza et al., 2002.DeSouza S, Fu J, States BA, Ziff EB. Differential palmitoylation directs the AMPA receptor-binding protein ABP to spines or to intracellular clusters. J Neurosci. 2002;22:3493–3503. doi: 10.1523/JNEUROSCI.22-09-03493.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong et al., 1997.Dong H, O'Brien RJ, Fung ET, Lanahan AA, Worley PF, Huganir RL. GRIP: a synaptic PDZ domain-containing protein that interacts with AMPA receptors. Nature. 1997;386:279–284. doi: 10.1038/386279a0. [DOI] [PubMed] [Google Scholar]
- El-Husseini Ael et al., 2002.El-Husseini Ael D, Schnell E, Dakoji S, Sweeney N, Zhou Q, Prange O, Gauthier-Campbell C, Aguilera-Moreno A, Nicoll RA, Bredt DS. Synaptic strength regulated by palmitate cycling on PSD-95. Cell. 2002;108:849–863. doi: 10.1016/s0092-8674(02)00683-9. [DOI] [PubMed] [Google Scholar]
- Fox and Peifer, 2007.Fox DT, Peifer M. Cell adhesion: separation of p120's powers? Curr Biol. 2007;17:R24–R27. doi: 10.1016/j.cub.2006.11.040. [DOI] [PubMed] [Google Scholar]
- Fu et al., 2003.Fu J, deSouza S, Ziff EB. Intracellular membrane targeting and suppression of Ser880 phosphorylation of glutamate receptor 2 by the linker I-set II domain of AMPA receptor-binding protein. J Neurosci. 2003;23:7592–7601. doi: 10.1523/JNEUROSCI.23-20-07592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzfeld, 1999.Hatzfeld M. The armadillo family of structural proteins. Int Rev Cytol. 1999;186:179–224. doi: 10.1016/s0074-7696(08)61054-2. [DOI] [PubMed] [Google Scholar]
- Hatzfeld and Nachtsheim, 1996.Hatzfeld M, Nachtsheim C. Cloning and characterization of a new armadillo family member, p0071, associated with the junctional plaque: evidence for a subfamily of closely related proteins. J Cell Sci. 1996;109:2767–2778. doi: 10.1242/jcs.109.11.2767. [DOI] [PubMed] [Google Scholar]
- Ho et al., 2000.Ho C, Zhou J, Medina M, Goto T, Jacobson M, Bhide PG, Kosik KS. Delta-catenin is a nervous system-specific adherens junction protein which undergoes dynamic relocalization during development. J Comp Neurol. 2000;420:261–276. [PubMed] [Google Scholar]
- Ide et al., 1999.Ide N, Hata Y, Deguchi M, Hirao K, Yao I, Takai Y. Interaction of S-SCAM with neural plakophilin-related Armadillo-repeat protein/delta-catenin. Biochem Biophys Res Commun. 1999;256:456–461. doi: 10.1006/bbrc.1999.0364. [DOI] [PubMed] [Google Scholar]
- Israely et al., 2004.Israely I, Costa RM, Xie CW, Silva AJ, Kosik KS, Liu X. Deletion of the neuron-specific protein delta-catenin leads to severe cognitive and synaptic dysfunction. Curr Biol. 2004;14:1657–1663. doi: 10.1016/j.cub.2004.08.065. [DOI] [PubMed] [Google Scholar]
- Izawa et al., 2002.Izawa I, Nishizawa M, Ohtakara K, Inagaki M. Densin-180 interacts with delta-catenin/neural plakophilin-related armadillo repeat protein at synapses. J Biol Chem. 2002;277:5345–5350. doi: 10.1074/jbc.M110052200. [DOI] [PubMed] [Google Scholar]
- Jaulin-Bastard et al., 2002.Jaulin-Bastard F, Arsanto JP, Le Bivic A, Navarro C, Vely F, Saito H, Marchetto S, Hatzfeld M, Santoni MJ, Birnbaum D, Borg JP. Interaction between Erbin and a catenin-related protein in epithelial cells. J Biol Chem. 2002;277:2869–2875. doi: 10.1074/jbc.M109652200. [DOI] [PubMed] [Google Scholar]
- Jones et al., 2002.Jones SB, Lanford GW, Chen YH, Moribito M, Kim K, Lu Q. Glutamate-induced delta-catenin redistribution and dissociation from postsynaptic receptor complexes. Neuroscience. 2002;115:1009–1021. doi: 10.1016/s0306-4522(02)00532-8. [DOI] [PubMed] [Google Scholar]
- Jordan et al., 2004.Jordan BA, Fernholz BD, Boussac M, Xu C, Grigorean G, Ziff EB, Neubert TA. Identification and verification of novel rodent postsynaptic density proteins. Mol Cell Proteomics. 2004;3:857–871. doi: 10.1074/mcp.M400045-MCP200. [DOI] [PubMed] [Google Scholar]
- Kim and Sheng, 2004.Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- Kim et al., 2002.Kim K, Sirota A, Chen YH, Jones SB, Dudek R, Lanford GW, Thakore C, Lu Q. Dendrite-like process formation and cytoskeletal remodeling regulated by delta-catenin expression. Exp Cell Res. 2002;275:171–184. doi: 10.1006/excr.2002.5503. [DOI] [PubMed] [Google Scholar]
- Kobielak and Fuchs, 2004.Kobielak A, Fuchs E. Alpha-catenin: at the junction of intercellular adhesion and actin dynamics. Nat Rev Mol Cell Biol. 2004;5:614–625. doi: 10.1038/nrm1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik et al., 2005.Kosik KS, Donahue CP, Israely I, Liu X, Ochiishi T. Delta-catenin at the synaptic-adherens junction. Trends Cell Biol. 2005;15:172–178. doi: 10.1016/j.tcb.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Lu et al., 1999.Lu Q, Paredes M, Medina M, Zhou J, Cavallo R, Peifer M, Orecchio L, Kosik KS. Delta-catenin, an adhesive junction-associated protein which promotes cell scattering. J Cell Biol. 1999;144:519–532. doi: 10.1083/jcb.144.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manniello et al., 1993.Manniello A, Aresu O, Parodi B, Romano P, Iannotta B, Rondanina G, Viegi L, Ruzzon T. The cell line data base and the new catalogue: detailed information on 2650 human and animal cell lines. Cytotechnology. 1993;11(Suppl 1):S130–S133. [PubMed] [Google Scholar]
- Martinez et al., 2003.Martinez MC, Ochiishi T, Majewski M, Kosik KS. Dual regulation of neuronal morphogenesis by a delta-catenin-cortactin complex and Rho. J Cell Biol. 2003;162:99–111. doi: 10.1083/jcb.200211025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda et al., 1999.Matsuda S, Mikawa S, Hirai H. Phosphorylation of serine-880 in GluR2 by protein kinase C prevents its C terminus from binding with glutamate receptor-interacting protein. J Neurochem. 1999;73:1765–1768. doi: 10.1046/j.1471-4159.1999.731765.x. [DOI] [PubMed] [Google Scholar]
- Medina et al., 2000.Medina M, Marinescu RC, Overhauser J, Kosik KS. Hemizygosity of delta-catenin (CTNND2) is associated with severe mental retardation in cri-du-chat syndrome. Genomics. 2000;63:157–164. doi: 10.1006/geno.1999.6090. [DOI] [PubMed] [Google Scholar]
- Nishimura et al., 2002.Nishimura W, Yao I, Iida J, Tanaka N, Hata Y. Interaction of synaptic scaffolding molecule and β-catenin. J Neurosci. 2002;22:757–765. doi: 10.1523/JNEUROSCI.22-03-00757.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuriya and Huganir, 2006.Nuriya M, Huganir RL. Regulation of AMPA receptor trafficking by N-cadherin. J Neurochem. 2006;97:652–661. doi: 10.1111/j.1471-4159.2006.03740.x. [DOI] [PubMed] [Google Scholar]
- Osten et al., 1998.Osten P, Srivastava S, Inman GJ, Vilim FS, Khatri L, Lee LM, States BA, Einheber S, Milner TA, Hanson PI, Ziff EB. The AMPA receptor GluR2 C terminus can mediate a reversible, ATP-dependent interaction with NSF and alpha- and beta-SNAPs. Neuron. 1998;21:99–110. doi: 10.1016/s0896-6273(00)80518-8. [DOI] [PubMed] [Google Scholar]
- Osten et al., 2000.Osten P, Khatri L, Perez JL, Kohr G, Giese G, Daly C, Schulz TW, Wensky A, Lee LM, Ziff EB. Mutagenesis reveals a role for ABP/GRIP binding to GluR2 in synaptic surface accumulation of the AMPA receptor. Neuron. 2000;27:313–325. doi: 10.1016/s0896-6273(00)00039-8. [DOI] [PubMed] [Google Scholar]
- Paffenholz and Franke, 1997.Paffenholz R, Franke WW. Identification and localization of a neurally expressed member of the plakoglobin/armadillo multigene family. Differentiation. 1997;61:293–304. doi: 10.1046/j.1432-0436.1997.6150293.x. [DOI] [PubMed] [Google Scholar]
- Paffenholz et al., 1999.Paffenholz R, Kuhn C, Grund C, Stehr S, Franke WW. The arm-repeat protein NPRAP (neurojungin) is a constituent of the plaques of the outer limiting zone in the retina, defining a novel type of adhering junction. Exp Cell Res. 1999;250:452–464. doi: 10.1006/excr.1999.4534. [DOI] [PubMed] [Google Scholar]
- Patel et al., 2003.Patel SD, Chen CP, Bahna F, Honig B, Shapiro L. Cadherin-mediated cell-cell adhesion: sticking together as a family. Curr Opin Struct Biol. 2003;13:690–698. doi: 10.1016/j.sbi.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Perez et al., 2001.Perez JL, Khatri L, Chang C, Srivastava S, Osten P, Ziff EB. PICK1 targets activated protein kinase Cα to AMPA receptor clusters in spines of hippocampal neurons and reduces surface levels of the AMPA-type glutamate receptor subunit 2. J Neurosci. 2001;21:5417–5428. doi: 10.1523/JNEUROSCI.21-15-05417.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Otano and Ehlers, 2004.Perez-Otano I, Ehlers MD. Learning from NMDA receptor trafficking: clues to the development and maturation of glutamatergic synapses. Neurosignals. 2004;13:175–189. doi: 10.1159/000077524. [DOI] [PubMed] [Google Scholar]
- Rongo et al., 1998.Rongo C, Whitfield CW, Rodal A, Kim SK, Kaplan JM. LIN-10 is a shared component of the polarized protein localization pathways in neurons and epithelia. Cell. 1998;94:751–759. doi: 10.1016/s0092-8674(00)81734-1. [DOI] [PubMed] [Google Scholar]
- Saglietti et al., 2007.Saglietti L, Dequidt C, Kamieniarz K, Rousset MC, Valnegri P, Thoumine O, Beretta F, Fagni L, Choquet D, Sala C, Sheng M, Passafaro M. Extracellular interactions between GluR2 and N-cadherin in spine regulation. Neuron. 2007;54:461–477. doi: 10.1016/j.neuron.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Scheiffele, 2003.Scheiffele P. Cell-cell signaling during synapse formation in the CNS. Annu Rev Neurosci. 2003;26:485–508. doi: 10.1146/annurev.neuro.26.043002.094940. [DOI] [PubMed] [Google Scholar]
- Schnapp, 2003.Schnapp BJ. Trafficking of signaling modules by kinesin motors. J Cell Sci. 2003;116:2125–2135. doi: 10.1242/jcs.00488. [DOI] [PubMed] [Google Scholar]
- Setou et al., 2000.Setou M, Nakagawa T, Seog DH, Hirokawa N. Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science. 2000;288:1796–1802. doi: 10.1126/science.288.5472.1796. [DOI] [PubMed] [Google Scholar]
- Sheng and Sala, 2001.Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- Songyang et al., 1997.Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton A, Chan AC, Anderson JM, Cantley LC. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- Srivastava et al., 1998.Srivastava S, Osten P, Vilim FS, Khatri L, Inman G, States B, Daly C, DeSouza S, Abagyan R, Valtschanoff JG, Weinberg RJ, Ziff EB. Novel anchorage of GluR2/3 to the postsynaptic density by the AMPA receptor-binding protein ABP. Neuron. 1998;21:581–591. doi: 10.1016/s0896-6273(00)80568-1. [DOI] [PubMed] [Google Scholar]
- Takeichi and Abe, 2005.Takeichi M, Abe K. Synaptic contact dynamics controlled by cadherin and catenins. Trends Cell Biol. 2005;15:216–221. doi: 10.1016/j.tcb.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Tanaka et al., 2000.Tanaka H, Shan W, Phillips GR, Arndt K, Bozdagi O, Shapiro L, Huntley GW, Benson DL, Colman DR. Molecular modification of N-cadherin in response to synaptic activity. Neuron. 2000;25:93–107. doi: 10.1016/s0896-6273(00)80874-0. [DOI] [PubMed] [Google Scholar]
- Tang et al., 1998.Tang L, Hung CP, Schuman EM. A role for the cadherin family of cell adhesion molecules in hippocampal long-term potentiation. Neuron. 1998;20:1165–1175. doi: 10.1016/s0896-6273(00)80497-3. [DOI] [PubMed] [Google Scholar]
- Topinka and Bredt, 1998.Topinka JR, Bredt DS. N-terminal palmitoylation of PSD-95 regulates association with cell membranes and interaction with K+ channel Kv1.4. Neuron. 1998;20:125–134. doi: 10.1016/s0896-6273(00)80440-7. [DOI] [PubMed] [Google Scholar]
- Washbourne et al., 2004.Washbourne P, Dityatev A, Scheiffele P, Biederer T, Weiner JA, Christopherson KS, El-Husseini A. Cell adhesion molecules in synapse formation. J Neurosci. 2004;24:9244–9249. doi: 10.1523/JNEUROSCI.3339-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyszynski et al., 2002.Wyszynski M, Kim E, Dunah AW, Passafaro M, Valtschanoff JG, Serra-Pages C, Streuli M, Weinberg RJ, Sheng M. Interaction between GRIP and liprin-alpha/SYD2 is required for AMPA receptor targeting. Neuron. 2002;34:39–52. doi: 10.1016/s0896-6273(02)00640-2. [DOI] [PubMed] [Google Scholar]
- Xia et al., 2000.Xia J, Chung HJ, Wihler C, Huganir RL, Linden DJ. Cerebellar long-term depression requires PKC-regulated interactions between GluR2/3 and PDZ domain-containing proteins. Neuron. 2000;28:499–510. doi: 10.1016/s0896-6273(00)00128-8. [DOI] [PubMed] [Google Scholar]
- Yamagata et al., 2003.Yamagata M, Sanes JR, Weiner JA. Synaptic adhesion molecules. Curr Opin Cell Biol. 2003;15:621–632. doi: 10.1016/s0955-0674(03)00107-8. [DOI] [PubMed] [Google Scholar]
- Yamazaki et al., 2001.Yamazaki M, Fukaya M, Abe M, Ikeno K, Kakizaki T, Watanabe M, Sakimura K. Differential palmitoylation of two mouse glutamate receptor interacting protein 1 forms with different N-terminal sequences. Neurosci Lett. 2001;304:81–84. doi: 10.1016/s0304-3940(01)01766-9. [DOI] [PubMed] [Google Scholar]
- Ye et al., 2000.Ye B, Liao D, Zhang X, Zhang P, Dong H, Huganir RL. GRASP-1: a neuronal RasGEF associated with the AMPA receptor/GRIP complex. Neuron. 2000;26:603–617. doi: 10.1016/s0896-6273(00)81198-8. [DOI] [PubMed] [Google Scholar]
- Ziff, 2007.Ziff EB. TARPs and the AMPA receptor trafficking paradox. Neuron. 2007;53:627–633. doi: 10.1016/j.neuron.2007.02.006. [DOI] [PubMed] [Google Scholar]