Abstract
We previously identified Neuregulin1 (NRG1) as a gene contributing to the risk of developing schizophrenia. Furthermore, we showed that NRG1+/− mutant mice display behavioral abnormalities that are reversed by clozapine, an atypical antipsychotic drug used for the treatment of schizophrenia. We now present evidence that ErbB4 (v-erb-a erythroblastic leukemia viral oncogene homolog 4), the tyrosine kinase receptor for NRG1 in hippocampal neurons, interacts with two nonreceptor tyrosine kinases, Fyn and Pyk2 (proline-rich tyrosine kinase 2). NRG1 stimulation of cells expressing ErbB4 and Fyn leads to the association of Fyn with ErbB4 and consequent activation. Furthermore, we show that NRG1 signaling, through activation of Fyn and Pyk2 kinases, stimulates phosphorylation of Y1472 on the NR2B subunit of the NMDA receptor (NMDAR), a key regulatory site that modulates channel properties. NR2B Y1472 is hypophosphorylated in NRG1+/− mutant mice, and this defect can be reversed by clozapine at a dose that reverses their behavioral abnormalities. We also demonstrate that short-term synaptic plasticity is altered and theta-burst long-term potentiation is impaired in NRG1+/− mutant mice, and incubation of hippocampal slices from these mice with NRG1 reversed those effects. Attenuated NRG1 signaling through ErbB4 may contribute to the pathophysiology of schizophrenia through dysfunction of NMDAR modulation. Thus, our data support the glutamate hypothesis of schizophrenia.
Keywords: neuregulin, NMDAR, fyn, Src-family kinases, synaptic plasticity, schizophrenia
Introduction
Schizophrenia is a heritable, highly debilitating psychotic disorder that affects 0.5–1% of the general population (Task Force on DSM-IV, 2000). In a genome-wide scan performed in Iceland, we identified Neuregulin 1 (NRG1) as a gene conferring susceptibility to schizophrenia (Stefansson et al., 2002). This finding has now been replicated in seven of nine published studies conducted on populations outside of Iceland, although the exact markers and/or haplotypes showing the most significant association to schizophrenia vary (for review, see Harrison and Weinberger, 2005). Although no coding mutations have yet been found in this gene, postmortem transcript analysis has shown that several at-risk single nucleotide polymorphisms are associated with altered ratios of NRG1 mRNA isoforms in brains of schizophrenics (Hasimoto et al., 2004; Law et al., 2006). In addition, the mRNA for ErbB3 (v-erb-a erythroblastic leukemia viral oncogene homolog 3), a tyrosine kinase receptor for NRG1, is significantly underexpressed in prefrontal cortex of schizophrenics (Hakak et al., 2001; Aston et al., 2004), and a genetic interaction has been reported between NRG1 and ErbB4, another receptor for NRG1 (Norton et al., 2006). With the identification of NRG1 and a number of other susceptibility genes in the past several years, a molecular understanding of schizophrenia has begun to emerge (for review, see Harrison and Owen, 2003; Corfas et al., 2004). Interestingly, several of these candidate genes appear to play a role in glutamatergic neurotransmission through the NMDA receptor (NMDAR), and mutations in these genes may confer susceptibility to schizophrenia by impairing synaptic function (Konradi and Heckers, 2003).
The NRG1 gene encodes multiple protein isoforms that play a crucial role in the development of many organs, including the brain (Buonanno and Fischbach, 2001; Falls, 2003), and modulate neurotransmission in developing and adult synapses (Huang et al., 2000; Roysommuti et al., 2003; Gu et al., 2005; Kwon et al., 2005; Chen et al., 2006).
NRG1 signals through the ErbB family of receptor tyrosine kinases. ErbB4, the predominant receptor for NRG1 on neurons, has been shown to interact with PDZ [postsynaptic density-95 (PSD-95)/Discs large/zona occludens-1] domain-containing components of the postsynaptic density complex, such as PSD-95 (Garcia et al., 2000; Huang et al., 2000). PSD-95 also interacts with the NMDAR complex, and this interaction appears to be critical for the phosphorylation of the NMDAR by the Src family kinase (SFK) Fyn (Tezuka et al., 1999; Nakazawa et al., 2001). NMDARs are ligand and voltage-gated ion channels that are strongly affected by phosphorylation (Wang and Salter, 1994; Yu et al., 1997; Salter and Kalia, 2004). Clozapine, an antipsychotic drug, increases NMDA EPSCs in the nucleus accumbens and in pyramidal cells of the rat prefrontal cortex. This increase is dependent on protein kinase A, calcium/calmodulin-dependent kinase II, and SFKs. In the nucleus accumbens, clozapine only affects NR2B subunit-containing NMDARs (Ninan et al., 2003; Wittmann et al., 2005). Subtle misregulation of membrane potential, ligand binding, or tyrosine phosphorylation may have profound effects on NMDAR opening, thus influencing behaviors modulated by this channel (Moghaddam, 2003).
Although dopamine D2 receptor antagonists are widely used clinically to control the positive symptoms of schizophrenia (Freedman, 2003), dissociative anesthetics such as phencyclidine and ketamine that produce schizophrenia-like disorders target the NMDAR rather than the dopamine pathway. Whereas manipulation of dopamine, for example by chronic exposure to amphetamines, mimics the positive symptoms of schizophrenia, exposure to dissociative anesthetics acutely reproduces the more clinically challenging negative and cognitive symptoms of the disease (Coyle et al., 2003). Hence, a role for NMDAR hypofunction in schizophrenia has also been suggested on pharmacological grounds.
Here we demonstrate that NRG1 signaling stimulates NR2B Y1472 phosphorylation through the activation of the SFKs Fyn and Pyk2 (proline-rich tyrosine kinase 2). Furthermore, we present evidence that NR2B Y1472 is hypophosphorylated in NRG1+/− and ErbB4+/− mutant mice and that this can be corrected by treatment with clozapine at doses that reverse behavioral abnormalities. Last, NRG1+/− mice show altered hippocampal synaptic plasticity. We propose that attenuated NRG1 signaling may contribute to the pathophysiology of schizophrenia through dysfunction of NMDAR modulation.
Materials and Methods
Reagents.
All chemicals were purchased from Sigma (St. Louis, MO) unless otherwise noted. Tissue culture media and supplements were from Invitrogen (Carlsbad, CA). Recombinant human NRG1α2 epidermal growth factor (EGF) domain [amino acids 177–241; Reference Sequence (RefSeq) accession number NP_039258] (catalog #296-HR-050; R & D Systems, Minneapolis, MN) was used for all biochemical assays. NRG1 stimulation was performed in serum-free medium for 0–15 min. For all electrophysiology studies, slices were treated with the entire extracellular domain (ECD) of NRG1β1 (amino acids 1–241; RefSeq accession number NP_039250) (catalog #RP-318-PA; LabVision, Fremont, CA). Although both forms of NRG1 gave similar results in a reporter gene assay (described below), the β form of the protein predominates in brain. However, during the time these studies were being performed, the NRG1β1 protein was in short supply and was often backordered for months from the sole supplier. For this reason, we reserved it for the electrophysiological studies in which we felt the cellular environment warranted the use of the more relevant form of the protein (Ozaki et al., 1997).
Yeast two-hybrid screening.
A random-primed human hippocampus cDNA library was created using the ZAP-cDNA Gigapack III Gold Cloning kit (Stratagene, La Jolla, CA) and transformed into yeast. The ErbB4 bait was created by cloning the entire cytoplasmic domain into the vector pGBKT7 (Clontech, Palo Alto, CA) using homologous recombination in the yeast strain AH109 (Clontech) and selection on Leu− plates. Diploid yeast cells were created by mating 5 × 109 bait cells (AH109) with 1 × 108 pretransformed library cells (Y187) and plated on minimum media, without leucine, triptophan, and histidine (−LTH) agar. A total of 2.4 × 106 clones was screened. After incubation for 5–10 d at 30°C, colonies were picked and grown for 48 h at 30°C in −LT media before gridding onto −LTH agar supplemented with 5 mm 3-amino-triazole (3AT). β-Galactosidase activity was assayed using a Yeast β-Galactosidase Assay kit (Pierce, Rockford, IL) according to the instructions of the manufacturer. Prey plasmids were isolated using a 96-well MultiScreen system (Millipore, Billerica, MA) and insert sequences amplified by PCR (forward primer, 5′-TTGGAATCACTACAGGGATGTTTAATAC; reverse primer, 5′-CTCTGCAGTAATACGACTCACTATAGGG) before sequencing. For growth assays, yeast cells (Y190) were cotransformed with the appropriate plasmids using a Frozen EZ Yeast Transformation II kit (Zymo Research, Orange, CA), and grown for 6 d at 30°C on −LTH agar containing 25 mm 3AT. Controls consisted of cotransformation with p53 bait or with simian virus 40 prey plasmids (Clontech).
Plasmids and transfection.
Human ErbB4 in pcDNA3.1 (Invitrogen) was kindly provided by Prof. Kermit L. Carraway III (University of California Davis Cancer Center, Davis, CA). Plasmids containing the ErbB4 D843A and K751A mutations were prepared using a Quick Change XL Site-Directed Mutagenesis kit (Stratagene). A human Fyn plasmid was purchased from Invitrogen (clone identification number RG000030), and the C-terminal V5 epitope tag was added by subcloning. The serum response element (SRE)–Luc plasmid was obtained from Stratagene. Transfections were performed using Fugene6 (Roche, Indianapolis, IN).
Mammalian tissue culture.
All cells coexpressing ErbB4 and p59Fyn–V5 were generated by double transfection, using the ErbB4 and p59Fyn–V5 plasmids, and grown in a double selective media [using neomycin (G418) and Zeocin both from Invitrogen]. Protein expression in single-cell clones were confirmed using protein-specific antibodies and a Leica (Bannockburn, IL) deconvolution microscope.
COS-7 and HEK293 cells were grown in DMEM supplemented with 10% (v/v) fetal calf serum, 100 IU/ml penicillin, and 100 μm streptomycin, and CHO-K1 and BE(2)-M17 (European Collection of Cell Cultures, Wiltshire, UK) were grown in DMEM/Nutri-mix F-12 supplemented with 10% (v/v) fetal bovine serum (FBS) and 100 IU/ml penicillin and 100 μm streptomycin. All lines were maintained at 37°C, 5% CO2. Treatment of cells with the tyrosine kinase inhibitors PP2 [4-amino-5-(4-chlorophenyl)-7-(t-butyl)pyrazolo[3,4-d]pyrimidine] or PP3 (4-amino-7-phenylpyrazol[3,4-d]pyrimidine) (25 nm each; Calbiochem, San Diego, CA) were done overnight in serum-free media. BE(2)-M17 cells were differentiated by incubation with 10 μm retinoic acid (RA) for 3 d. The BE(2)-M17 cells were stimulated with 14.4 nm NRG1 for 10 min. These conditions were experimentally determined and gave the most robust activation of the ErbB4 signaling cascade in the cells and were therefore chosen to explore the effect of NRG1 stimulation on NR2B phosphorylation.
Colocalization assay.
Transiently transfected COS-7 cells were grown on glass slides to 70% confluency, treated with 7.2 nm NRG1α2 for 10 min, fixed in 4% paraformaldehyde, and permeabilized with 100% methanol for 1 min. Nonspecific binding sites were blocked with 10% FBS in PBS for 1 h. Fyn was tagged by a C-terminal V5 epitope (Fyn–V5) in these experiments. Primary antibodies [anti-ErbB4 (1:400; Santa Cruz Biotechnology, Santa Cruz, CA) and anti-V5 (1:500; Invitrogen)] were added overnight at 4°C. Secondary antibodies goat anti-rabbit-Alexa Fluor 488 and goat anti-mouse-Alexa Fluor 594 (Invitrogen), each at 1:500, were added for 4 h at 4°C. Cells were mounted in DL-2-amino-5-phosphonovaleric acid/1,4-diazabicyclo(2.2.2)octane (Millipore) and examined with a Leica deconvolution CTR MIC Microscope using Leica QFluoro Image Manager software.
Immunoprecipitation from mammalian tissue culture cell lysates.
CHO-K1 cells stably expressing ErbB4 and Fyn–V5 were treated with 7.2 nm NRG1α2 for 10 min and lysed using radioimmunoprecipitation assay radioimmunoprecipitation assay buffer [20 mm 3-(N-morpholino)-propanesulfonic acid, 0.5 mm EDTA, pH 7.0, 150 mm NaCl, 3.5 mm SDS, 1% NP-40, and 1% deoxycholate (DOC)] containing Complete Protease Inhibitor Cocktail (Roche) and Phosphatase Inhibitor Cocktail Set II (Calbiochem). Twenty-five units of Benzonase Nuclease (Novagen, Madison, WI) per 1 ml of lysate were added to each sample. Lysates were incubated with the appropriate antibodies (1 μg/ml lysate) for 2 h at 4°C [anti-glutamate (Glu) (deCODE Biostructures, Seattle, WA), anti-ErbB4, anti- hemagglutinin (HA), and anti-PSD-95 (all from Santa Cruz Biotechnology)]. Immunocomplexes were isolated using Protein G Sepharose 4 Fast Flow (GE Healthcare, Piscataway, NJ) and recovered by boiling for 7 min in 1× SDS-PAGE sample buffer.
Mouse hippocampus, immunoprecipitation, and Western blot.
Animals were killed by cervical dislocation because anesthetics were found to alter NR2B Y1472 phosphorylation. Hippocampus tissue was isolated by manual dissection and homogenized in DOC buffer (20 μm ZnCl2, 50 mm Tris, pH 9.0, and 1% DOC) (1 ml/50 mg of tissue) containing protease and phosphatase inhibitors as above. Twenty-five units of Benzonase Nuclease per 1 ml of lysate were added to each sample during a 60 min incubation at 4°C. For SDS-PAGE and Western blot, sample protein concentration was determined using a BCA Protein Assay Reagent kit (Pierce), adjusted to 2 μg/μl in 1× SDS-PAGE sample buffer, and then 20–25 μg of protein was loaded per gel lane. For immunoprecipitation, samples were adjusted to a final concentration of 2 μg/μl total protein in DOC buffer. Lysates were incubated with the appropriate antibodies (1 μg of antibody/250 μg of total protein) overnight at 4°C (anti-Glu, anti-ErbB4, anti-HA, and anti-PSD-95). Immunocomplexes were isolated using Protein G Sepharose 4 Fast Flow and recovered by boiling for 7 min in 1× SDS-PAGE sample buffer. Samples were subjected to one-dimensional SDS-PAGE followed by silver staining, in-gel digestion, and mass spectrometric analysis, or by Western blotting.
Western blotting.
Samples were separated by SDS-PAGE on 4–20% Tris-HCl Ready Gels (Bio-Rad, Hercules, CA), blotted onto nitrocellulose membranes (Schleicher and Schuell, Keene, NH) and blocked in TBST (10 mm Tris Base, pH 7.6, 150 mm NaCl, and 0.06% Tween 20) containing 3% (w/v) fat-free dried skimmed milk (Osta og Smjörsalan Sf, Reykjavík, Iceland). Blots were incubated with the following primary antibodies: anti-V5 (1:1000; Invitrogen); anti-Pyk2 (1:500; Santa Cruz Biotechnology); anti-ErbB4 (1:1000; Santa Cruz Biotechnology); anti-phosphotyrosine 4G10 (1:2000; Upstate Biotechnology, Lake Placid, NY); anti-phospho-p44/42 mitogen-activated protein kinase (MAPK) Thr202/Tyr204 (1:500; Cell Signaling Technology, Danvers, MA); anti-Fyn/Src PY420 (1:800; Invitrogen); anti-Fyn/Src PY531 (1:1000; Invitrogen); and anti-NMDA NR2B phospho-specific-Y1472 (1:250 or 1:500; Calbiochem) at 4°C overnight, followed by incubation with HRP-labeled secondary antibodies (1:2000; DakoCytomation, Carpinteria, CA; or 1:20000; Jackson Immunoresearch, West Grove, PA) for 1 h at room temperature. Blots were incubated with ECL Plus Western Blotting Detection System (GE Healthcare) or SuperSignal West Femto Maximum Sensitivity Substrate (Pierce) according to the instructions of the manufacturer, and signal was detected using Hyperfilm ECL (GE Healthcare). Multiple exposures of film were made to ensure that the dynamic range of the film had not been exceeded.
Mass spectrometry.
Bands selected from silver-stained SDS-PAGE gels were excised and destained using destainer solutions contained in the Silverquest Silver Staining kit (Invitrogen). The gel pieces were subjected to in-gel digestion according to a modified version of the method by Rosenfeld et al. (1992), as described by Edmondson et al. (2002). Liquid chromatography mass spectrometry/mass spectrometry (MS/MS) was used for the analysis of in-gel digests.
Nanoscale reversed-phase chromatography was performed using an Ultimate system with FAMOS autosampler (LC Packings, Amsterdam, The Netherlands). At a flow rate of 200 nl/min, proteolytic digests were loaded from a 10 μl sample loop onto a 75 μm inner diameter × 15 cm column packed with PepMap C18 material (LC Packings) connected directly to the spray needle of the microelectrospray spray ionization source (to which 3 kV was applied) of a Micromass Q-Tof-2 mass spectrometer (Waters, Milford, MA). Typical gradient conditions used to elute peptides from the column were 0–50% B (where A = 98:2 H2O/acetonitrile incorporating 0.3% formic acid, and B = 9:1 H2O/acetonitrile incorporating 0.3% formic acid) in 10 min, 50–100% B in 2 min, 100% B for 2 min, and 100–2% B in 30 s. The Q-Tof-2 electrospray tandem mass spectrometer was operated in a data-dependant acquisition mode such that continuous cycles of one MS survey scan followed by product ion MS/MS scans of the four most abundant ions in each survey scan were recorded. MS survey and product ion spectra integration times were 1 s. Collision offset was determined automatically based on precursor ion mass and charge state. Product ion data were searched against the Mascot protein database.
In vitro kinase assay.
CHO-K1 cells stably expressing ErbB4 or ErbB4/Fyn–V5 were treated with 7.2 nm NRG1α2 and lysed in 500 μl PTK extraction buffer (SignaTECT Protein Tyrosine Kinase Assay System; Promega, Madison, WI) containing Complete Protease Inhibitor Cocktail without EDTA (Roche) and Phosphatase Inhibitor Cocktail Set II, as well as 25 U of Benzonase Nuclease. ErbB4 and Fyn–V5 were immunoprecipitated from cell lysates using α-ErbB4 or α-V5 antibodies, respectively. In vitro kinase assays with the immunoprecipitates were performed using a SignaTECT Protein Tyrosine Kinase Assay System (Promega) according to the instructions of the manufacturers. Substrate 1 was used for ErbB4 and substrate 2 for Fyn.
Luciferase reporter assay.
CHO-K1 cells carrying pSRE-Luc (PathDetect; Stratagene) and stably expressing ErbB4 or ErbB4/Fyn–V5 were plated in serum-free media (96-well format) and treated for 4 h with 0–57.6 nm NRG1α2. Assay was performed using the Dual-Luciferase Reporter Assay System kit (Promega) according to the instructions of the manufacturer. Plates were read with a TR 717 Microplate Luminometer (Applied Biosystems, Foster City, CA). In this assay, the NRG1α2 EGF domain protein had an EC50 of 0.7–1.0 nm, whereas the ECD of NRG1β1 had an EC50 of 0.7 nm (see Fig. 2 and data not shown).
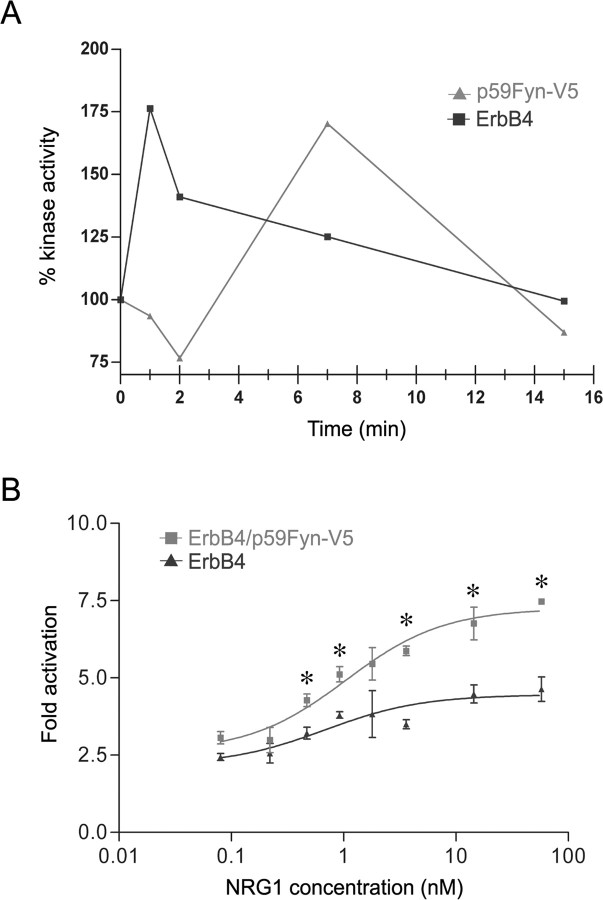
Figure 2.
NRG1 regulation of the Fyn kinase activity. ErbB4 and Fyn kinase activities were increased in CHO-K1 cells expressing ErbB4 (■) or coexpressing ErbB4 and Fyn–V5 (▴) in response to treatment with 7.2 nm NRG1. A, Fyn kinase activity was measured in the presence of ErbB4 using a substrate with higher affinity for Fyn. Maximum ErbB4 activity was observed within 1 min of stimulation (■), whereas Fyn kinase activity peaked at ∼7 min (▴) in response to NRG1 (n > 3). B, Fyn potentiates NRG1-induced signal transduction activity as measured with an MAPK pathway reporter gene. CHO-K1 cells carrying the pSRE–Luc plasmid and stably expressing either ErbB4 or coexpressing ErbB4 and Fyn–V5 were activated with varying concentrations of NRG1 for 4 h at 37°C and subsequently assayed for luciferase activity. Induction of luciferase activity was increased by approximately twofold (*p < 0.05 after applying the Bonferroni's correction for multiple t tests), whereas the EC50 for NRG1 was unchanged (0.7 vs 1.0 nm, respectively) (n = 3).
Knock-out mice.
NRG1(ΔTM)+/− and ErbB4+/− mutant mice were obtained from Prof. Richard Harvey (The Victor Chang Cardiac Research Institute, Sydney, New South Wales, Australia) and from Prof. Greg Lemke (The Salk Institute for Biological Studies, La Jolla, CA), respectively, as acknowledged previously (Stefansson et al., 2002). The NRG1(ΔEGF)+/− mice (Erickson et al., 1997) were obtained from Genentech (South San Francisco, CA). The Fyn−/− mutant mice were obtained from The Jackson Laboratory (Bar Harbor, ME). All lines were maintained by mating to wild-type C57BL/6. Mice were housed under a standard 12 h light/dark cycle with access to food and water ad libitum. All animal work was approved by the respective Institutional Animal Care and Use Committees and conducted in accordance with appropriate national regulations concerning animal welfare.
Clozapine treatment.
Clozapine was administered by intraperitoneal injection as described previously (Stefansson et al., 2002).
Slice preparation.
NRG1(ΔEGF)+/− mice (4–16 weeks of age) and age-matched controls were used for all electrophysiology experiments. Experiments and subsequent analysis were performed by investigators blinded to the genotype of the mice. Slices were prepared as described previously (Misner and Sullivan, 1999). Briefly, mice were anesthetized with halothane, the brains were removed, and coronal slices (350 μm) were cut in ice-cold solution (in mm: 120 NaCl, 3.5 KCl, 0.7 CaCl2, 4.0 MgCl2, 1.25 NaH2PO4, 26 NaHCO3, and 10 glucose) bubbled with 95% O2/5% CO2. The hippocampus was dissected and placed in a chamber containing recording buffer (in mm: 120 NaCl, 3.5 KCl, 2.6 CaCl2, 1.3 MgCl2, 1.25 NaH2PO4, 26 NaHCO3, and 10 glucose) perfused with 95% O2/5% CO2. Slices were incubated at room temperature at least 1 h before recording.
Electrophysiology.
All experiments were performed as described previously (Misner and Sullivan, 1999). Individual slices were placed in a submerged recording chamber and were perfused with recording solution at a rate of 1–2 ml/min. Schaffer collateral–commissural fibers were stimulated every 20 s unless otherwise noted. Stimulus intensity was adjusted to evoke 30–40% maximal stimulation, and the initial slopes of field EPSPs were measured for field potential recordings. Data are expressed as mean ± SEM.
Paired-pulse facilitation (PPF) was studied by applying pairs of stimuli at varying interpulse intervals (20–200 ms). The slope of the response to the second pulse (P2) was averaged over 5–10 trials and divided by the averaged slope or amplitude of the response to the first pulse (P1) to give a ratio (P2/P1).
Population spikes were recorded in the CA1 pyramidal cell layer, and the amplitude of the spike from the positive to negative peak was measured. Paired-pulse inhibition was studied by applying pairs of stimuli at an interpulse interval of 10 ms. The paired-pulse ratio (P2/P1) was calculated as described for facilitation experiments.
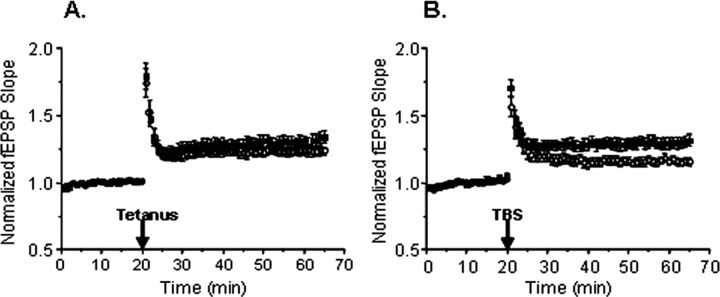
Long-term potentiation (LTP) was induced by either tetanic or theta-burst stimulation. The responses were divided by the initial slope of the first field potential to normalize the data. Tetanic stimulation consisted of five trains of 100 Hz stimulation lasting 200 ms at an intertrain interval of 10 s. Synaptic fatigue was studied by measuring the synaptic responses during the first high-frequency train. Theta-burst stimulation consisted of 10 trains of four pulses delivered at a frequency of 100 Hz separated by 200 ms.
Recombinant NRG1 comprising the entire extracellular domain of NRG1β1 was applied to some slices at concentrations between 0.1 and 10.0 nm after recording of a stable baseline. Control slices were treated with vehicle (0.01% BSA).
Results
Fyn is an ErbB4 binding protein
To characterize components of the NRG1 signaling pathway downstream of ErbB4, the intracellular domain of ErbB4 (ErbB4i) was used as a bait in a yeast two-hybrid interaction screen. This bait is capable of autophosphorylation on tyrosine in yeast cells. Tyrosine phosphorylation is absent on ErbB4 baits in which mutations were introduced either in the ATP binding site, K751A, or the catalytic site, D843A (data not shown). Screening of a human hippocampus cDNA library yielded a partial cDNA clone comprising the Src homology 2 (SH2) and SH3 domains of Fyn (amino acids 60–252, RefSeq accession number NM_002037). No Fyn clones were isolated when the same cDNA library was screened using ErbB4 K751A as bait, nor did the ErbB4 K751A or D843A bait interact with Fyn in the yeast two-hybrid growth assay (Fig. 1A). These results suggest that Fyn and ErbB4 can interact without the need of an adapter protein such as PSD-95, that ErbB4-Fyn interaction is dependent on tyrosine phosphorylation of ErbB4, and that the binding likely involves the Fyn SH2 domain.
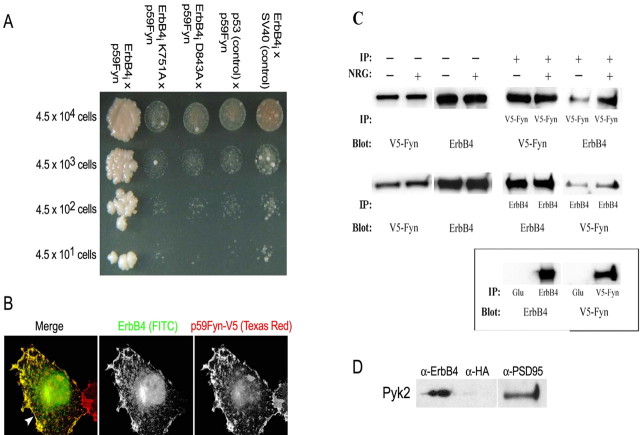
Figure 1.
Fyn and Pyk2 interact with ErbB4. Fyn and ErbB4 interact in a yeast two-hybrid growth assay. A, The intracellular domain of ErbB4 physically interacts with a partial cDNA clone comprising the SH2 and SH3 domains of Fyn (ErbB4i; lane 1) but not with ErbB4 baits containing mutations that inactivate kinase activity (ErbB4i K751A and ErbB4i D843A; lanes 2, 3) or with control bait or prey plasmids (lanes 4, 5) (n > 3). B, Full-length ErbB4 (green) and Fyn–V5 (red) colocalize at the cytoplasmic membrane (arrow) when transiently expressed in COS7 cells and visualized by deconvolution microscopy (n > 3) and magnification at 1000×. C, Full-length Fyn–V5 and ErbB4 were stably coexpressed in CHO-K1 cells. Fyn–V5 coimmunoprecipitates with anti-ErbB4 antibodies, as shown by Western blot using anti-V5 antibodies for detection. Stimulation with 7.2 nm NRG1 for 10 min enhanced the interaction (top). ErbB4 is also coimmunoprecipitated with V5 antibody as shown by Western blot using anti-ErbB4 antibodies for detection, and the interaction is enhanced by NRG1 stimulation of the cells as before (middle). No interaction between the two proteins was detected using the control antibody Glu (bottom). D, Pyk2 coimmunoprecipitates with ErbB4 (n = 3) and PSD-95 (n = 2) from mouse brain hippocampus (lanes 1 and 3, respectively) as shown by Western blot with detection using anti-Pyk2. No coimmunoprecipitation was observed with control antibodies (anti-HA; lane 2). IP, Immunoprecipitation.
To determine whether these proteins interact in the context of a mammalian cell, full-length ErbB4 and Fyn–V5 were transiently coexpressed in COS7 cells. As shown in Figure 1B, these proteins colocalized to the plasma membrane. This colocalization was independent of ErbB4 receptor activation by NRG1, perhaps because of high basal levels of ErbB4 tyrosine phosphorylation in the transfected cells (data not shown).
To demonstrate that ErbB4 and Fyn exist in a complex, we performed coimmunoprecipitations from CHO-K1 cells stably expressing both proteins. Complexes containing ErbB4 were immunoprecipitated using anti-V5 (recognizing Fyn) but not by control anti-Glu antibodies (Fig. 1C). Fivefold more ErbB4 was coimmunoprecipitated with Fyn from cells stimulated with NRG1, suggesting that activation of the receptor facilitates the recruitment of Fyn into the ErbB4 protein complex. In addition, immunoprecipitation of ErbB4 coprecipitates Fyn, further confirming the association of these two proteins (Fig. 1C). These results demonstrate that ErbB4 and Fyn can exist in a protein complex in mammalian cells and that the formation of the complex is facilitated by ErbB4 activation and consequent tyrosine phosphorylation.
ErbB4 binding partners in brain
ErbB4 has been shown to be a component of the postsynaptic density in brain, a multiprotein complex containing various protein kinases, including Fyn and other SFKs, the NMDAR, and scaffolding proteins such as PSD-95 (Husi et al., 2000). A specific interaction of PSD-95 and ErbB4 has been demonstrated previously in brain (Garcia et al., 2000; Huang et al., 2000). To further characterize its binding partners in brain, ErbB4 was immunoprecipitated from mouse hippocampal lysates, and associated proteins were analyzed using in-gel tryptic digestion and mass fragmentation. One associated protein contained a peptide with the amino acid sequence NH2-EIISEVQR-CO2H, a perfect match to DDEF2 (development and differentiation enhancing factor 2), a Pyk2 binding protein and an SFK substrate. This suggested that Pyk2, a nonreceptor SFK that, like Fyn, has been implicated in the modulation of NMDAR function (Huang et al., 2001), also associates with ErbB4. This was confirmed by anti-Pyk2 immunoblotting of anti-ErbB4 coimmunoprecipitates (Fig. 1D). Pyk2 was also coimmunoprecipitated from mouse hippocampus using antibodies directed against PSD-95, confirming a previous observation that PSD-95 and Pyk2 are components of the multiprotein postsynaptic density complex (Husi et al., 2000). No coimmunoprecipitation of Pyk2 was observed when anti-HA antibodies were used for immunoprecipitation (Fig. 1D, middle lane).
NRG1 regulation of Fyn kinase
The finding that Fyn and ErbB4 are found in the same protein complex suggests that Fyn may be a downstream effector of NRG1. The initial step of NRG1 signaling is activation of ErbB4 kinase and autophosphorylation on tyrosine residues. An increase in ErbB4 tyrosine phosphorylation and associated kinase activity was observed when CHO-K1 cells expressing ErbB4 were exposed to NRG1. After immunoprecipitation of ErbB4, kinase activity was measured using a peptide substrate. Maximum ErbB4 kinase activity was detected within 1 min of NRG1 exposure and returned to the prestimulation level by 15 min (Fig. 2A).
To determine whether Fyn was activated by NRG1 signaling, we determined whether NRG1 binding to ErbB4 leads to a change in Fyn phosphorylation. Fyn kinase is inhibited in the basal state through an intramolecular SH2 interaction with phosphorylated Y531 and can be activated directly by a second kinase through phosphorylation on Y420 (Salter and Kalia, 2004). Alternatively, Fyn can be activated by dephosphorylation of Y531 or through displacement of the SH2 domain by a second tyrosine-phosphorylated binding partner, thereby allowing autophosphorylation of Y420. NRG1 treatment of HEK293 cells stably expressing ErbB4 and Fyn resulted in 5.4-fold increase in tyrosine phosphorylation of ErbB4 and activation of the downstream effector Erk1/2 (p44/42) as evident by elevation of the T202/Y204 phosphorylation (Fig. 3A). To monitor the effect of NRG1 stimulation on Fyn, the changes in phosphorylation levels on Y531 and Y420 were evaluated. Densiometric scanning of the phosphorylation bands were made, and a ratio was obtained by normalizing to the total Fyn–V5 levels. This revealed a slight decrease in PY531phosphorylation (0.8×) after NRG1 stimulation. However, NRG1 stimulation leads to a 2.5-fold increase in PY420 phosphorylation. This NRG1-stimulated increase in Fyn Y420 phosphorylation could be attributable to stimulation of Fyn autophosphorylation, to phosphorylation of Fyn by ErbB4, or to phosphorylation of Fyn by an ErbB4-activated tyrosine kinase other than Fyn.
Figure 3.
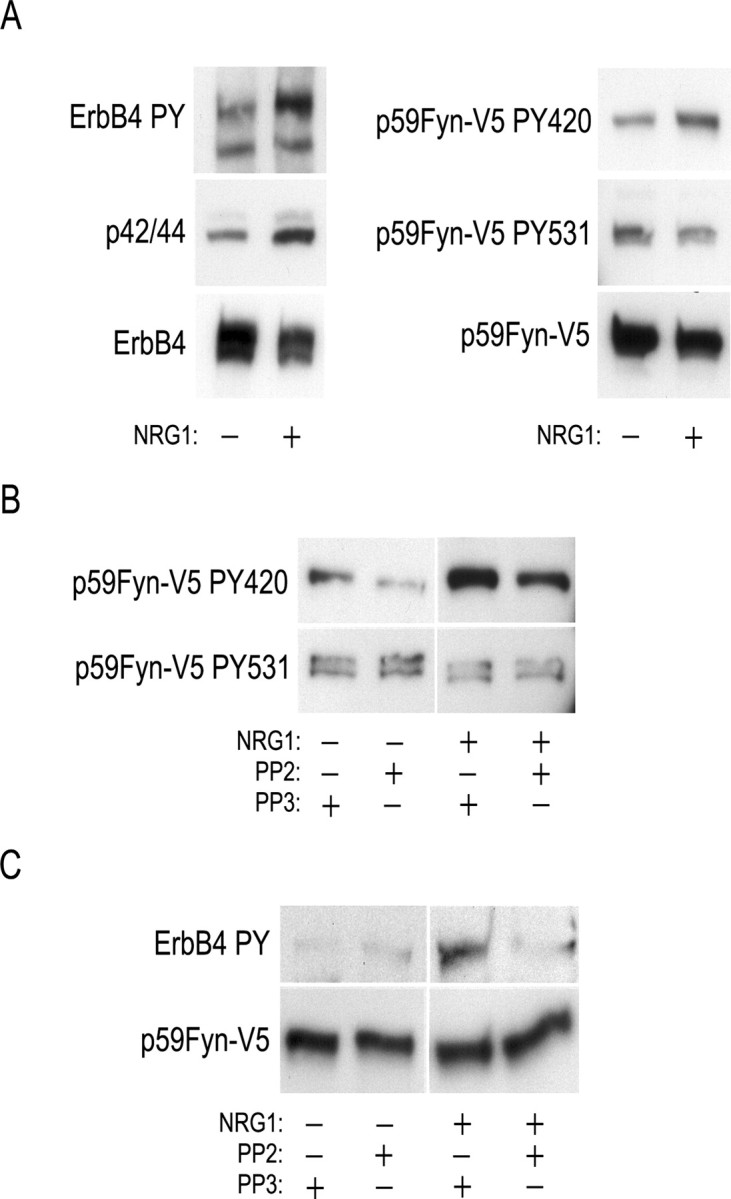
NRG1 activation of ErbB4 regulates Fyn phosphorylation in HEK293 cells coexpressing ErbB4 and Fyn–V5. A, NRG1 stimulation (7.2 nm for 5 min) resulted in phosphorylation of ErbB4 on tyrosine residues and activation of MAPK p44/42 (left top and middle panel, respectively) as shown by Western blot. NRG1 stimulation increased phosphorylation of Fyn on Y420, but had only minor affect on Y531 phosphorylation (right top and middle, respectively) (n = 3). B, NRG1-induced phosphorylation of Fyn Y420 was reduced, but not eliminated, after preincubation of the cells with the Src-family kinase inhibitor PP2 at 25 nm for 16 h (top). The control compound PP3 had no effect. Fyn Y531 phosphorylation was unaffected by NRG1 signaling or the presence of PP2 or PP3 (n = 3). C, NRG1-induced tyrosine phosphorylation of ErbB4 was impaired in the presence of PP2. However, the ErbB4 phosphorylation without NRG1 stimulation was unaffected by PP2 (n = 3).
These alternatives could be distinguished by using PP2, a relatively selective SFK inhibitor, to block Fyn activity, whereas ErbB4 signaling was stimulated with NRG1 (Fig. 3B). Fyn inhibition is highly sensitive to PP2 with an IC50 of 5 nm. We used as a control PP3, a protein kinase inhibitor related to PP2 that has no effect on Fyn or other SFKs. In HEK293 cells coexpressing ErbB4 and Fyn, Fyn was phosphorylated at a low level on Y420 when treated with PP3. This could be reduced by preincubation for 16 h with 25 nm PP2, suggesting that Fyn Y420 phosphorylation was attributable to autophosphorylation or phosphorylation by another SFK. After NRG1 stimulation, an increase in Fyn Y420 phosphorylation was seen, even under conditions in which PP2 was used to inhibit Fyn autophosphorylation. The control compound PP3 had no effect on Fyn phosphorylation. Thus, ErbB4 or an ErbB4-activated tyrosine kinase can phosphorylate Fyn on Y420 in response to NRG1 stimulation and activate this kinase directly.
To test whether the observed increase in Fyn Y420 phosphorylation was associated with enhanced catalytic activity, we measured Fyn kinase activity after NRG1 stimulation (Fig. 2A). Fyn kinase activity was increased within 5 min of NRG1 stimulation, reached maximum activity after 7 min, and returned to prestimulation levels within 15 min. These results confirm that, in mammalian cells, Fyn kinase is activated in response to NRG1 signaling through ErbB4 either directly or indirectly through Y420 phosphorylation.
Fyn potentiates ErbB4-mediated NRG1 signaling
We observed greater tyrosine phosphorylation of ErbB4 in the presence of Fyn. This suggested that Fyn might amplify NRG1 signaling by increasing ErbB4 tyrosine phosphorylation. To test this possibility, we examined NRG1-induced phosphorylation of ErbB4 under conditions in which Fyn and related Src-family kinases were inhibited by PP2. PP2 greatly reduced NRG1-stimulated ErbB4 tyrosine phosphorylation (Fig. 3C), whereas the control compound PP3 had no effect. However, no difference in ErbB4 phosphorylation was observed after PP2 or PP3 incubation in the absence of NRG1, nor on NRG1-induced tyrosine phosphorylation in HEK293 cells expressing only ErbB4 (data not shown). Our results suggest that Fyn is activated in response to NRG1 signaling and that activated Fyn contributes to ErbB4 phosphorylation on tyrosine residues. This could conceivably result in the creation of additional SH2 domain docking sites, thus leading to further activation of Fyn as part of a positive feedback mechanism involving NRG1, ErbB4, and SFKs.
Because Fyn kinase activity appears to contribute to NRG1-induced tyrosine phosphorylation of ErbB4, we speculated that NRG1-dependent signal transduction activity would be enhanced in cells overexpressing Fyn kinase. To test this hypothesis, we generated a CHO-K1 line expressing only ErbB4 and a subclone coexpressing ErbB4 and Fyn (Fig. 2B). The parental ErbB4-expressing cell line and the subclone coexpressing ErbB4 and Fyn continued to express similar levels of ErbB4, as shown by Western blot. Both lines contained a pSRE–Luc reporter gene sensitive to MAPK pathway activation. Expression of the luciferase reporter gene by NRG1 was increased twofold in cells coexpressing ErbB4 and Fyn compared with cells expressing only ErbB4 (p < 0.05). No significant differences in baseline luciferase activity or in the EC50 for NRG1 stimulation of reporter gene expression (∼1 nm) were found between the two cell lines. Thus, our results suggest that Fyn is a downstream amplifier of NRG1 signaling through ErbB4.
NRG1 signaling modulates NR2B tyrosine phosphorylation in retinoic acid differentiated human BE(2)-M17 neuroblastoma cells
Fyn and Pyk2 have been implicated in the phosphorylation of the NR2A and NR2B subunits of the NMDAR (Tezuka et al., 1999; Nakazawa et al., 2001; Heidinger et al., 2002). It also has been reported that NR2B Y1472 phosphorylation is increased after induction of LTP in the hippocampal CA1 region (Nakazawa et al., 2001) and that Pyk2 is required for LTP induction (Yang et al., 2003). As described above, we find that the SFKs Fyn and Pyk2 both associate with ErbB4. We therefore tested whether NRG1 signaling, through activation of SFKs, results in changes in the phosphorylation state of regulatory tyrosine residues on NR2B.
For convenience, we conducted these experiments using BE(2)-M17 human neuroblastoma cells. These cells have a neuron-like appearance after differentiation for 72 h in the presence of 10 μm RA (Hanada et al., 1993), and we found by Western blot that BE(2)-M17 express ErbB4, Fyn, Src, Pyk2, PSD-95, and NR1 (Fig. 4 and data not shown). We further found that RA differentiation induces NR2B but not NR2A, allowing the cells to express an NMDAR complex containing NR1 and NR2B (Fig. 4 and data not shown). After RA stimulation, BE(2)-M17 cells become sensitive to glutamate-induced excitotoxicity, implying that NR1 and NR2B assemble into a functional complex (data not shown).
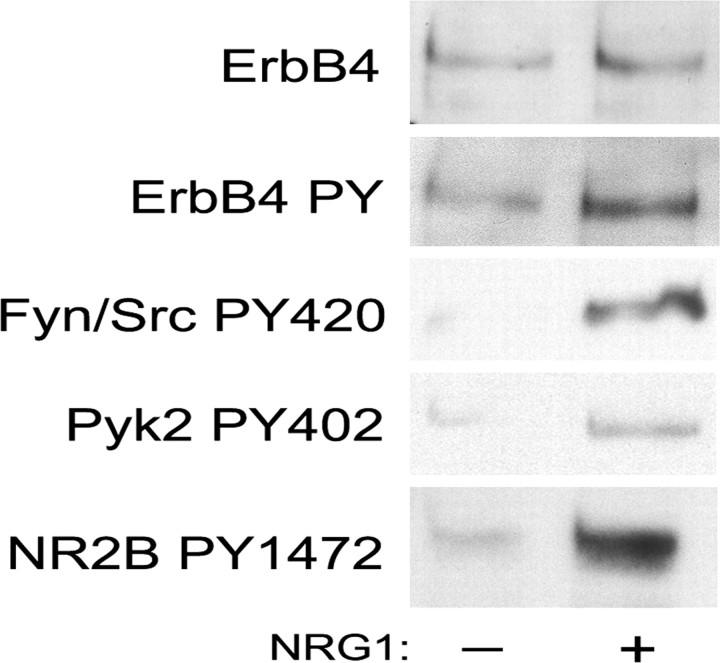
Figure 4.
NRG1 modulates tyrosine phosphorylation on the NMDAR NR2B subunit in human neuroblastoma cells. Human BE(2)-M17 neuroblastoma cells were differentiated with retinoic acid and stimulated with NRG1α2 (14.4 nm for 10 min). NRG1 stimulation resulted in tyrosine phosphorylation of ErbB4 (second panel), Fyn/Src on Y420 (third panel), Pyk2 on Y402 (fourth panel), and NR2B on Y1472 (bottom panel) as shown by Western blot. Total ErbB4 provided a control for protein loading (top panel) (n > 3).
Retinoic acid-differentiated BE(2)-M17 cells thus provided a model system in which to examine the coupling of NRG1 activation of ErbB4 to NMDAR phosphorylation. When differentiated BE(2)-M17 cells were stimulated with NRG1 for 10 min, we found increased ErbB4 tyrosine phosphorylation (Fig. 4), increased Fyn/Src Y420 phosphorylation (indicative of increased SFK activity), and increased Pyk2 Y402 phosphorylation. Pyk2 Y402 is an autophosphorylation site involved in SFK recruitment (Dikic et al., 1996). Like Fyn, Pyk2 also has been implicated in the modulation of NMDAR function (Huang et al., 2001). Pyk2 might be phosphorylated directly by ErbB4 or Fyn or indirectly through MAPK pathway activation attributable to NRG1 stimulation. Importantly, we observed a coincident increase in NR2B Y1472 phosphorylation that is associated with Fyn and Pyk2 activation. Thus, NRG1 signaling through ErbB4 modulates NR2B tyrosine phosphorylation in neuroblastoma cells. We lack the reagents to distinguish between Fyn and Pyk2 as proximal effectors in BE(2)-M17 cells. Furthermore, because we do not have information about the expression of other ErbB family members (ErbB1, EebB2, and ErbB3) in the BE(2)-M17, we cannot exclude the possibility that other family members might contribute to the phosphorylation of NR2B in these cells.
NR2B is hypophosphorylated in NRG1(ΔTM)+/− and ErbB4+/− mutant mice
Having shown that NRG1 influences NR2B phosphorylation in a cellular model, we hypothesized that the abnormal behavioral phenotype seen in NRG1+/− and ErbB4+/− mutant mice (Stefansson et al., 2002) might be related to altered NR2B Y1472 phosphorylation. Indeed, we found that Y1472 was hypophosphorylated in hippocampal lysates from 9-month-old NRG1(ΔTM)+/− or ErbB4+/− mutant mice compared with age- and sex-matched controls (Fig. 5A). There was no apparent difference in total NR2B protein between the mutant and control mice. As reported by others, NR2B Y1472 phosphorylation was reduced in Fyn−/− null mice (Nakazawa et al., 2001). Interestingly, Y1472 phosphorylation in both NRG1(ΔTM)+/− and ErbB4+/− mice is reduced to the level seen in Fyn−/− null mice, suggesting that NRG1 is the major regulator of Fyn-mediated NR2B tyrosine phosphorylation in the hippocampus. Fyn/Src Y420 phosphorylation was also reduced in the mutant mice compared with the controls, suggesting attenuated SFK activity in an environment of altered NRG1 signaling. Although the anti-Fyn/Src PY420 antibody reagent used for Western blotting does not distinguish between Fyn and Src, the signal is virtually absent in Fyn−/− null mice, suggesting that Fyn is the predominant kinase in the hippocampal lysate. Although we could not distinguish between Fyn and Pyk2 as effectors of NR2B Y1472 phosphorylation in BE(2)-M17 cells, the results presented in Figure 5 indicate that Fyn plays a dominant role in the hippocampus. Our results suggest that, in animals deficient in components of the NRG1 signaling pathway, altered regulation of NR2B Y1472 phosphorylation may cause defects in NMDAR function.
Figure 5.
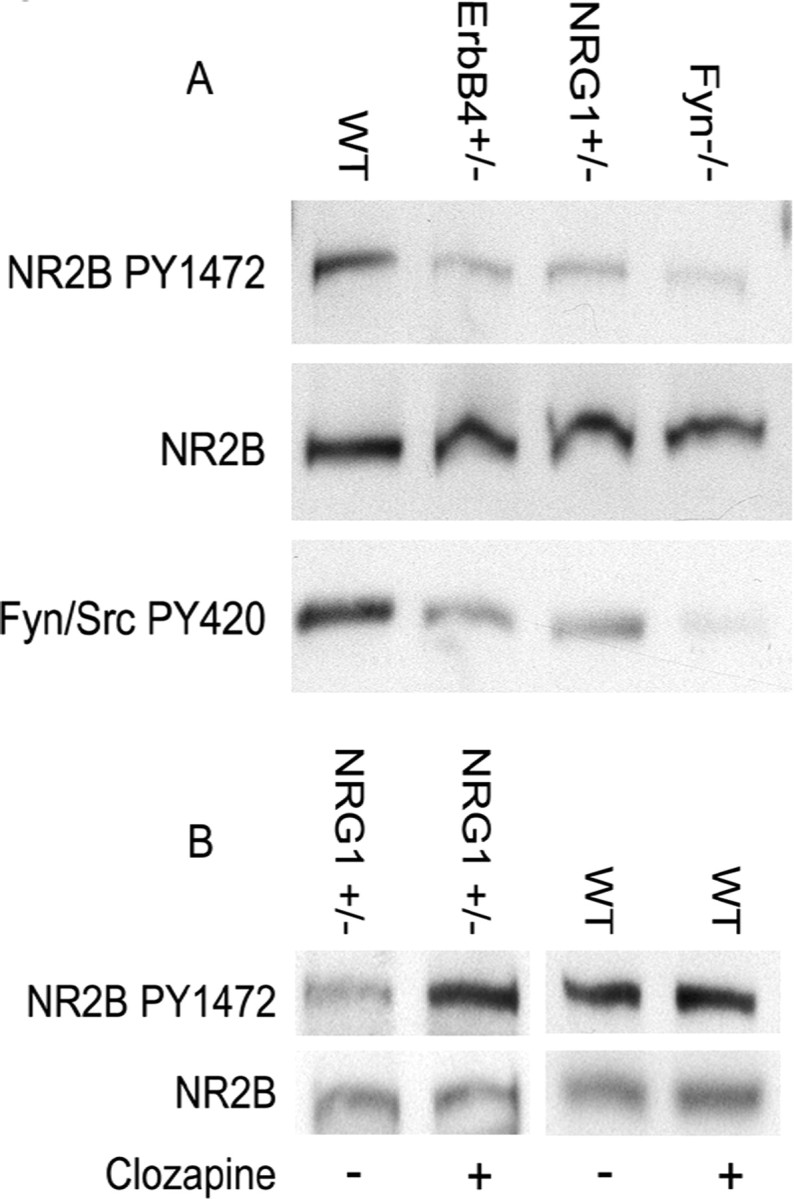
NMDAR is hypophosphorylated in NRG1(ΔTM)+/− and ErbB4+/− mutant mice, and hypophosphorylation is reversed by clozapine. A, NR2B Y1472 was hypophosphorylated in hippocampal lysates from NRG1(ΔTM)+/− and ErbB4+/− mutant mice (top, lanes 2 and 3, respectively) compared with age- and sex-matched wild-type (WT) C57BL/6 mice as shown by Western blot. Fyn/Src Y420 phosphorylation also was reduced in ErbB4+/− and NRG1(ΔTM)+/− mutant mice (bottom, lanes 2 and 3, respectively). Hippocampal lysates from Fyn−/− mice served as a control for NR2B Y1472 hypophosphorylation, as well as for Fyn/Src Y420 phosphorylation (fourth lane, top and bottom panels, respectively). Data are representative of five to six animals in at least three independent experiments; 25 μg of total protein was loaded per lane. B, Clozapine reverses NR2B hypophosphorylation in NRG1(ΔTM)+/− mice. NR2B Y1472 phosphorylation (top) was increased 2.5- to 3-fold in NRG1(ΔTM)+/− mice when normalized against total loading of NR2B (bottom). The same dose of clozapine had no effect on NR2B Y1472 phosphorylation (top) when administered to age- and sex-matched wild-type mice. Data representative of two to three animals in at least two independent experiments; 20 μg of total protein was loaded per lane.
NR2B hypophosphorylation in NRG1(ΔTM)+/− mutant mice is reversed by clozapine
We reported previously that the behavioral abnormality of NRG1(ΔTM)+/− mutant mice in the open-field test was reversible within 25 min of treatment with clozapine at a intraperitoneal dose of 1 mg/kg (Stefansson et al., 2002). This dose had no obvious effect on wild-type controls, although higher doses were sedating. Therefore, we administered this same dose of clozapine to wild-type and NRG1+/− mice and prepared hippocampal lysates 25 min later. NR2B Y1472 phosphorylation was increased 2.5- to 3-fold when normalized to total NR2B (Fig. 5B, compare lanes 1, 2). NR2B Y1472 phosphorylation in clozapine-treated NRG1(ΔTM)+/− mice was indistinguishable from wild-type control lysates, regardless of whether the wild-type animals received clozapine (Fig. 5B, lanes 2–4). Thus, a dose of clozapine that reverses the abnormal behavior in NRG1(ΔTM)+/− mice restores NR2B Y1472 phosphorylation to normal levels.
Paired-pulse facilitation is enhanced in NRG1(ΔEGF)+/− mice
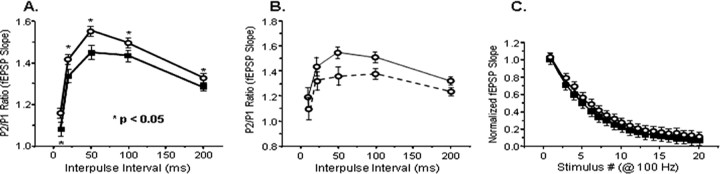
Postsynaptic excitatory responses and short-term plasticity were examined to determine the effects of NRG1 gene deletion on synaptic transmission. In these experiments, a second NRG1 knock-out line, carrying a deletion of the entire EGF domain, was used (Erickson et al., 1997). To the degree that we have examined them, these mice exhibit the same phenotype as the NRG1(ΔTM)+/− line, including homozygous lethality, hyperactivity in the open-field assay, and decreased NMDAR expression (Gerlai et al., 2000 and data not shown). Basal synaptic transmission, as measured by postsynaptic responses to increasing stimulation intensities, was not altered in NRG1(ΔEGF)+/− mice (data not shown). PPF, a form of short-term plasticity believed to reflect presynaptic function (Zucker, 1989), was measured in both wild-type and heterozygous slices (Fig. 6A). Over all interpulse intervals tested (10–200 ms), PPF was enhanced in slices from NRG1(ΔEGF)+/− mice compared with wild type (p < 0.05, two-sample t test). Moreover, these effects were reversed by acute treatment (30 min) with 1 nm NRG1β1 (Fig. 6B) (p < 0.05, one-sample t test). The rate of synaptic fatigue during high-frequency stimulation, an additional measure of presynaptic function, was similar between wild-type and heterozygous slices (Fig. 6C). Finally, paired-pulse inhibition of the population spike, a measure of inhibitory transmission mediated by GABAA receptors, was tested by evoking two population spikes 10 ms apart in the CA1 pyramidal cell layer and expressing the magnitude of the second spike as a percentage of the first spike. Paired-pulse inhibition was decreased in NRG1(ΔEGF)+/− slices (64% compared with 52%) but was not statistically significant from wild type (p > 0.05, two-sample t test; data not shown). Together, these results suggest that NRG1 gene deletion selectively and reversibly alters paired-pulse facilitation, without effects on additional measures of basal transmission.
Figure 6.
Effects of NRG-1 gene deletion on basal synaptic transmission. A, PPF was measured by applying two closely spaced stimuli, and the ratio of the second response to the first response was calculated. PPF was enhanced in NRG1(ΔEGF)+/− slices (open circles; n = 67) compared with wild-type slices (filled squares; n = 76). B, PPF was measured in untreated NRG1(ΔEGF)+/− slices (solid line; n = 18), then slices were treated for 30 min with 1 nm NRG1β1, and facilitation was measured again (dotted line). C, The rate of synaptic fatigue during a 100 Hz stimulus train was fit by a single-exponential function. The resulting rates were similar between wild-type (filled squares; 5.5 stimuli; n = 24) and NRG1+/− (open circles; 6.2 stimuli; n = 17) slices.
Theta burst-induced LTP is impaired in NRG1(ΔEGF)+/− mice
Because previous studies demonstrated that bath application of NRG1 to hippocampal slices from wild type rats impaired tetanus-induced LTP (Huang et al., 2000), we next examined the effects of NRG1 gene deletion on LTP induced by high-frequency stimulation (Fig. 7A). Although the mean potentiation 45 min after tetanus was lower in NRG1(ΔEGF)+/− slices compared with wild-type slices (123.9 ± 3.4 vs 131.8 ± 4.7%), this was not statistically different (p > 0.1, two-sample t test), indicating that LTP induced by tetanic stimulation was not altered in NRG1(ΔEGF)+/− slices. Interestingly, LTP induced by theta-burst stimulation (Fig. 7B), which was also reversed by NRG1 in mice (Kwon et al., 2005), was impaired in NRG1(ΔEGF)+/− slices compared with controls (116.0 ± 2.8 vs 130.4 ± 4.3%; p < 0.05, two-sample t test). These results suggest that NRG1 may differentially regulate synaptic plasticity, depending on the type of stimulus used.
Figure 7.
Effects of NRG1 gene deletion on LTP. A, LTP was induced by five trains of 100 Hz stimuli lasting 200 ms, with each train separated by 10 s. Data are normalized to baseline values preceding LTP induction, and mean potentiation was measured 45 min after tetanus. LTP was not significantly different between wild-type (filled squares; n = 24) and NRG1(ΔEGF)+/− (open circles; n = 17) slices. B, LTP was induced by theta-burst stimulation (TBS) consisting of 10 sets of four stimuli given at 100 Hz, with each set separated by 200 ms. Theta-burst-induced LTP was significantly diminished in NRG1(ΔEGF)+/− slices (open circles; n = 13) compared with wild-type slices (filled squares; n = 13).
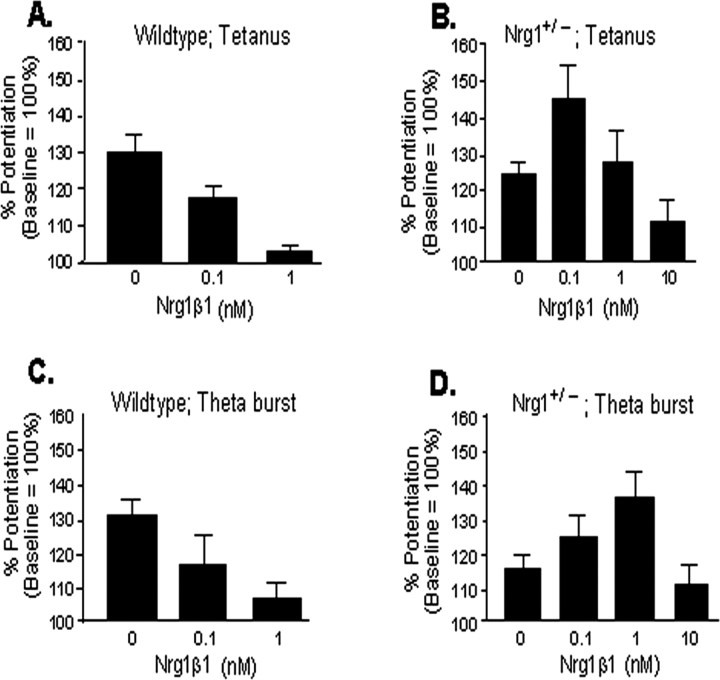
Acute NRG1 treatment has differential effects on LTP in NRG1(ΔEGF)+/− mice
Finally, we examined the effects of acute application of the soluble extracellular domain of NRG1β1 on LTP induced by both high-frequency and theta-burst stimulation in wild-type and NRG1(ΔEGF)+/− slices. Bath application of NRG1β1 for 30 min had no effect on basal synaptic strength at any concentration tested, in agreement with previous reports (data not shown). Thirty minute pretreatment with NRG1β1 dose dependently inhibited tetanus-induced LTP of wild-type slices (Fig. 8A), with maximal inhibition at 1 nm (130.1 ± 4.6% for untreated slices vs 102.3 ± 2.1% for treated slices). Similarly, NRG1β1 pretreatment inhibited LTP induced by theta-burst stimulation (Fig. 8C), with complete inhibition at 1 nm (130.4 ± 4.2 vs 106.4 ± 4.6%). Surprisingly, acute treatment with NRG1β1 had very different results in NRG1(ΔEGF)+/− slices. Compared with untreated slices, tetanus-induced LTP was enhanced in the presence of 0.1 nm NRG1β1 (123.8 ± 3.3% for untreated vs 144.9 ± 8.8% for 0.1 nm NRG1β1 treated) (Fig. 8B). This effect was reversed when higher concentrations were applied (128.1 ± 10.8% at 1 nm and 112.1 ± 4.3% at 10 nm). Finally, the impairment of theta-burst-induced LTP observed in NRG1(ΔEGF)+/− slices was reversed by acute treatment of lower concentrations of NRG1β1 but not at the highest concentration tested (Fig. 8D). The maximal effect was observed at 1 nm (116.0 ± 2.6% for untreated vs 135.5 ± 7.5% for treated), the same concentration that reversed the effects on paired-pulse facilitation. NRG1β1 at 0.1 nm slightly enhanced (124.9 ± 4.6%) whereas 10 nm slightly inhibited (111.0 ± 4.8%) LTP induced by theta-burst stimulation. These results suggest that LTP can be impaired by either too little or too much NRG1β1 and that an optimal concentration range of NRG1 exists for synaptic plasticity.
Figure 8.
Effects of exogenously applied NRG1 on LTP in wild-type and NRG1(ΔEGF)+/− slices. A, NRG1β1 was applied to wild-type slices at varying concentrations for 30 min after a stable baseline of at least 10 min was achieved. LTP was induced as described previously by a tetanic train. NRG1β1 inhibited tetanus-induced LTP in wild-type slices, ∼50% at 0.1 nm (n = 18), and completely ablated it at 1 nm (n = 8). B, NRG1β1 enhanced tetanus-induced LTP at low concentrations (0.1 nm; n = 9) in NRG1(ΔEGF)+/− slices but inhibits LTP at higher concentrations of 1 nm (n = 8) and 10 nm (n = 5). C, NRG1β1 inhibited theta-burst-induced LTP in wild-type slices, ∼50% at 0.1 nm (n = 4) and completely at 1 nm (n = 4). D, Acute treatment with NRG1β1 dose-dependently reversed deficits in theta-burst-induced LTP in NRG1(ΔEGF)+/− slices. The deficit was partially ameliorated with 0.1 nm (n = 11) and 10 nm (n = 5) NRG1β1 and was maximally reversed with 1 nm NRG1β1 (n = 15).
Discussion
In this study, we show that two nonreceptor SFKs, Fyn and Pyk2, are associated with ErbB4, the predominant neuronal receptor for NRG1, and that NRG1 stimulation results in activation of Fyn kinase, as well as phosphorylation of Pyk2 on Y402, a site associated with SFK recruitment. Both kinases have been implicated previously in the regulation of NMDAR function through tyrosine phosphorylation of the NR2A and NR2B subunits (Tezuka et al., 1999; Huang et al., 2001; Nakazawa et al., 2001). Activation of SFKs increases NMDAR channel open probability and mean open time (Yu et al., 1997). The NMDAR plays a key role in the synaptic plasticity underlying induction of hippocampal LTP and long-term depression, and this is dependent on NMDAR subunit composition (Liu et al., 2004). Fyn has been implicated in vivo in the regulation of induction of LTP in mouse hippocampus (Grant et al., 1992; Kojima et al., 1997; Nakazawa et al., 2001).
NMDAR phosphorylation by Fyn and Pyk2, in addition to our own data identifying NRG1 as a gene playing a role in the pathogenesis of schizophrenia (Stefansson et al., 2002), prompted us to test for NRG1-dependent phosphorylation of an important regulatory site on NR2B, Y1472. Phosphorylation of brain NR2B Y1472 has been demonstrated previously to increase with induction of LTP (Nakazawa et al., 2001). Here we show that NRG1 signaling results in phosphorylation of NR2B Y1472 in BE(2)-M17 human neuroblastoma cells. Furthermore, NR2B Y1472 was found to be hypophosphorylated in NRG1+/− and ErbB4+/− mutant mice, and synaptic plasticity in the NRG1+/− was altered with impaired theta-burst LTP and enhanced LTP in response to NRG1. Both the NRG1+/− and ErbB4+/− strains exhibit modest behavioral abnormalities (Stefansson et al., 2002), and we showed previously that the hyperactivity of NRG1+/− mutant mice in the open-field test was reversible by the atypical antipsychotic drug clozapine (Stefansson et al., 2002). Although the clinical efficacy of clozapine is attributable in part to its combined activity on numerous neurotransmitter receptors (Miyamoto et al., 2005), we show here that it can also reverse the deficient NR2B Y1472 phosphorylation seen in untreated NRG1+/− mice. Thus, reversal of the behavioral abnormality is associated with restoration of NR2B Y1472 phosphorylation. Interestingly, clozapine has also been shown to facilitate potentiation of synaptic transmission through increased, NMDAR-mediated, EPSCs (Gemperle et al., 2003).
Together, these data suggest to us that NRG1-associated susceptibility to schizophrenia is at least partly associated with hypofunction of NRG1 signaling through ErbB4, Fyn, and other associated kinases such as Pyk2, that phosphorylate regulatory sites on NMDAR subunits, resulting in abnormal modulation of excitatory glutamatergic neurotransmission.
Modulation of LTP by NRG1 in normal rodents has been investigated by several groups (Huang et al., 2000; Gu et al., 2005; Kwon et al., 2005). All of these investigators found that NRG1 inhibited tetanic LTP in hippocampal slices. In agreement with these studies, NRG1 dose dependently inhibited both tetanus and theta-burst-induced LTP in wild-type slices. Comparing NRG1+/− slices and wild-type controls, we do not find any difference in tetanus-induced LTP but do find a deficit in theta-burst LTP. The deficits in theta-burst LTP may be in part attributable to decreased release probability in NRG1+/− slices indicated by the enhancement of paired-pulse facilitation, such that neurotransmitter release during theta-burst stimulation was not sufficient to induce robust LTP. Acute NRG1 treatment dose dependently increased theta-burst LTP, fully reversing the deficit at 1 nm. Furthermore, NRG treatment decreased paired-pulse facilitation within the same slices after 20 min (data not shown), suggesting an increase in release probability similar to wild-type controls. In NRG1+/− slices, NRG1 dramatically increases tetanus-induced LTP, demonstrating a clear and significant difference in synaptic function between wild-type and NRG1+/− animals. Results in the NRG1+/− mice are consistent with the hypothesis that hypofunction in the NRG1 signaling pathway causes a decrement in LTP that can be reversed by restoration of the NRG1/ErbB4 signaling pathway.
By providing a biochemical mechanism linking NRG1 signaling, through activation of Fyn and Pyk2, to the NMDAR complex, we are able to incorporate a number of recently published schizophrenia susceptibility genes into a unified model for this disease. In this model, NRG1 signaling, through Fyn kinase, leads to increased NMDAR activation. NMDAR-mediated calcium influx, but not calcium release from other sources, has been shown to activate the Ca2+-dependent phosphatase calcineurin (PPP3CC). The gene encoding PPP3CC has been associated previously with schizophrenia (Gerber et al., 2003). In a negative feedback loop, a calcium-induced increase in calcineurin phosphatase activity leads to inactivation of Fyn via activation of STEP [striatum enriched protein tyrosine phosphatase (PTPN5)] (Nguyen et al., 2002; Paul et al., 2003). STEP depresses NMDAR activity in CA1 hippocampal neurons, and its overexpression blocks induction of NMDAR-dependent LTP, whereas inhibition of STEP enhances NMDAR-mediated, excitatory neurotransmission (Pelkey et al., 2002). Functional activation of the NMDAR requires the binding of a coagonist, either glycine or d-serine. d-Serine levels in brain are regulated by d-amino acid oxidase (DAAO). DAAO, as well as its activator G72, have also been implicated as genetic risk factors for schizophrenia (Chumakov et al., 2002). Our results demonstrating that attenuation of NRG1 signaling leads to changes in NMDAR-mediated neurotransmission is consistent with genetic data in schizophrenia that implicates several genes involved in glutamatergic signaling. The basis for disease susceptibility may be altered adaptability of the brain to experience and the changes in synaptic efficacy that underlie such adaptation, attributable to misregulation of multiple pathways impacting on the modulation of the NMDAR complex. The NRG1+/− mutant mice provide a novel genetic model of schizophrenia in which a biochemical deficit correlated to NMDAR channel modulation can be corrected by an atypical antipsychotic drug. Thus, these data, together with work of many others, suggest that restoration of NMDAR modulation may restore normal brain function in schizophrenia.
Footnotes
We thank Thora Dagbjartsdottir, Palmi Atlason, Thorhildur Gudsteinsdottir, Jon Mar Bjornsson, and Gudmundur Sigthorsson for assistance with animal work and Rick Salazar for generating the CHO-ErbB4/SRE-luc cell line.
References
- Aston C, Jiang L, Sokolov BP. Microarray analysis of postmortem temporal cortex from patients with schizophrenia. J Neurosci Res. 2004;15:858–866. doi: 10.1002/jnr.20208. [DOI] [PubMed] [Google Scholar]
- Buonanno A, Fischbach GD. Neuregulin and ErbB receptor signaling pathways in the nervous system. Curr Opin Neurobiol. 2001;11:287–296. doi: 10.1016/s0959-4388(00)00210-5. [DOI] [PubMed] [Google Scholar]
- Chen S, Velardez MO, Warot X, Yu ZX, Miller SJ, Cros D, Corfas G. Neuregulin 1–erbB signaling is necessary for normal myelination and sensory function. J Neurosci. 2006;26:3079–3086. doi: 10.1523/JNEUROSCI.3785-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumakov I, Blumenfeld M, Guerassimenko O, Cavarec L, Palicio M, Abderrahim H, Bougueleret L, Barry C, Tanaka H, La Rosa P, Puech A, Tahri N, Cohen-Akenine A, Delabrosse S, Lissarrague S, Picard FP, Maurice K, Essioux L, Millasseau P, Grel P, et al. Genetic and physiological data implicating the new human gene G72 and the gene for d-amino acid oxidase in schizophrenia. Proc Natl Acad Sci USA. 2002;99:13675–13680. doi: 10.1073/pnas.182412499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corfas G, Roy K, Buxbaum JD. Neuregulin 1-erbB signaling and the molecular/cellular basis of schizophrenia. Nat Neurosci. 2004;7:575–580. doi: 10.1038/nn1258. [DOI] [PubMed] [Google Scholar]
- Coyle JT, Tsai G, Goff D. Converging evidence of NMDA receptor hypofunction in the pathophysiology of schizophrenia. Ann NY Acad Sci. 2003;1003:318–327. doi: 10.1196/annals.1300.020. [DOI] [PubMed] [Google Scholar]
- Dikic I, Tokiwa G, Lev S, Courtneidge SA, Schlessinger J. A role for Pyk2 and Src in linking G-protein-coupled receptors with MAP kinase activation. Nature. 1996;383:547–550. doi: 10.1038/383547a0. [DOI] [PubMed] [Google Scholar]
- Edmondson RD, Vondriska TM, Biederman KJ, Zhang J, Jones RC, Zheng Y, Allen DL, Xiu JX, Cardwell EM, Pisano MR, Ping P. Protein kinase C epsilon signaling complexes include metabolism- and transcription/translation-related proteins: complimentary separation techniques with LC/MS/MS. Mol Cell Proteomics. 2002;1:421–433. doi: 10.1074/mcp.m100036-mcp200. [DOI] [PubMed] [Google Scholar]
- Erickson SL, O'Shea KS, Ghaboosi N, Loverro L, Frantz G, Bauer M, Lu L, Moore MW. ErbB3 is required for normal cerebellar and cardiac development: a comparison with ErbB2- and heregulin-deficient mice. Development. 1997;124:4999–5011. doi: 10.1242/dev.124.24.4999. [DOI] [PubMed] [Google Scholar]
- Falls DL. Neuregulins: functions, forms, and signaling strategies. Exp Cell Res. 2003;284:14–30. doi: 10.1016/s0014-4827(02)00102-7. [DOI] [PubMed] [Google Scholar]
- Freedman R. Schizophrenia. N Engl J Med. 2003;349:1738–1749. doi: 10.1056/NEJMra035458. [DOI] [PubMed] [Google Scholar]
- Garcia RA, Vasudevan K, Buonanno A. The neuregulin receptor ErbB-4 interacts with PDZ-containing proteins at neuronal synapses. Proc Natl Acad Sci USA. 2000;97:3596–3601. doi: 10.1073/pnas.070042497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemperle AY, Enz A, Pozza MF, Luthi A, Olpe HR. Effects of clozapine, haloperidol and iloperidone on neurotransmission and synaptic plasticity in prefrontal cortex and their accumulation in brain tissue: an in vitro study. Neuroscience. 2003;117:681–695. doi: 10.1016/s0306-4522(02)00769-8. [DOI] [PubMed] [Google Scholar]
- Gerber DJ, Hall D, Miyakawa T, Demars S, Gogos JA, Karayiorgou M, Tonegawa S. Evidence for association of schizophrenia with genetic variation in the 8p21.3 gene, PPP3CC, encoding the calcineurin gamma subunit. Proc Natl Acad Sci USA. 2003;100:8993–8998. doi: 10.1073/pnas.1432927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlai R, Pisacane P, Erickson S. Heregulin, but not ErbB2 or ErbB3, heterozygous mutant mice exhibit hyperactivity in multiple behavioral tasks. Behav Brain Res. 2000;109:219–227. doi: 10.1016/s0166-4328(99)00175-8. [DOI] [PubMed] [Google Scholar]
- Grant SG, O'Dell TJ, Karl KA, Stein PL, Soriano P, Kandel ER. Impaired long-term potentiation, spatial learning, and hippocampal development in fyn mutant mice. Science. 1992;258:1903–1910. doi: 10.1126/science.1361685. [DOI] [PubMed] [Google Scholar]
- Gu Z, Jiang Q, Fu AKY, Ip NY, Yan Z. Regulation of NMDA receptors by neuregulin signaling in prefrontal cortex. J Neurosci. 2005;25:4974–4984. doi: 10.1523/JNEUROSCI.1086-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, Haroutunian V, Fienberg AA. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci USA. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada M, Krajewski S, Tanaka S, Cazals-Hatem D, Spengler BA, Ross RA, Biedler JL, Reed JC. Regulation of Bcl-2 oncoprotein levels with differentiation of human neuroblastoma cells. Cancer Res. 1993;53:4978–4986. [PubMed] [Google Scholar]
- Harrison PJ, Owen MJ. Genes for schizophrenia? Recent findings and their pathophysiological implications. Lancet. 2003;361:417–419. doi: 10.1016/S0140-6736(03)12379-3. [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- Hasimoto R, Straub RE, Weickert CS, Hyde TM, Kleinman JE, Weinberger DR. Expression analysis of neuregulin-1 in the dorsolateral prefrontal cortex in schizophrenia. Mol Psychiatry. 2004;9:299–307. doi: 10.1038/sj.mp.4001434. [DOI] [PubMed] [Google Scholar]
- Heidinger V, Manzerra P, Wang XQ, Strasser U, Yu SP, Choi DW, Behrens MM. Metabotropic glutamate receptor 1-induced upregulation of NMDA receptor current: mediation through the Pyk2/Src-family kinase pathway in cortical neurons. J Neurosci. 2002;22:5452–5461. doi: 10.1523/JNEUROSCI.22-13-05452.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Lu W, Ali DW, Pelkey KA, Pitcher GM, Lu YM, Aoto H, Roder JC, Sasaki T, Salter MW, MacDonald JF. CAKbeta/Pyk2 kinase is a signaling link for induction of long-term potentiation in CA1 hippocampus. Neuron. 2001;29:485–496. doi: 10.1016/s0896-6273(01)00220-3. [DOI] [PubMed] [Google Scholar]
- Huang YZ, Won S, Ali DW, Wang Q, Tanowitz M, Du QS, Pelkey KA, Yang DJ, Xiong WC, Salter MW, Mei L. Regulation of neuregulin signaling by PSD-95 interacting with ErbB4 at CNS synapses. Neuron. 2000;26:443–455. doi: 10.1016/s0896-6273(00)81176-9. [DOI] [PubMed] [Google Scholar]
- Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci. 2000;3:661–669. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- Kojima N, Wang J, Mansuy IM, Grant SG, Mayford M, Kandel ER. Rescuing impairment of long-term potentiation in fyn-deficient mice by introducing Fyn transgene. Proc Natl Acad Sci USA. 1997;94:4761–4765. doi: 10.1073/pnas.94.9.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konradi C, Heckers S. Molecular aspects of glutamate dysregulation: implications for schizophrenia and its treatment. Pharmacol Ther. 2003;97:153–179. doi: 10.1016/s0163-7258(02)00328-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon OB, Longart M, Vullhorst D, Hoffman DA, Buonanno A. Neuregulin-1 reverses long-term potentiation at CA1 hippocampal synapses. J Neurosci. 2005;25:9378–9383. doi: 10.1523/JNEUROSCI.2100-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law AJ, Lipska BK, Weickert CS, Hyde TM, Straub RE, Hashimoto R, Harrison PJ, Kleinman JE, Weinberger DR. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5′ SNPs associated with the disease. Proc Natl Acad Sci USA. 2006;103:6747–6752. doi: 10.1073/pnas.0602002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Wong TP, Pozza MF, Lingenhoehl K, Wang Y, Sheng M, Auberson YP, Wang YT. Role of NMDA receptor subtypes in governing the direction of hippocampal synaptic plasticity. Science. 2004;304:1021–1024. doi: 10.1126/science.1096615. [DOI] [PubMed] [Google Scholar]
- Misner DL, Sullivan JM. Mechanism of cannabinoid effects on long-term potentiation and depression in hippocampal CA1 neurons. J Neurosci. 1999;19:6795–6805. doi: 10.1523/JNEUROSCI.19-16-06795.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S, Duncan GE, Marx CE, Lieberman JA. Treatments for schizophrenia: a critical review of pharmacology and mechanisms of action of antipsychotic drugs. Mol Psychiatry. 2005;10:79–104. doi: 10.1038/sj.mp.4001556. [DOI] [PubMed] [Google Scholar]
- Moghaddam B. Bringing order to the glutamate chaos in schizophrenia. Neuron. 2003;40:881–884. doi: 10.1016/s0896-6273(03)00757-8. [DOI] [PubMed] [Google Scholar]
- Nakazawa T, Komai S, Tezuka T, Hisatsune C, Umemori H, Semba K, Mishina M, Manabe T. Characterization of Fyn-mediated tyrosine phosphorylation sites on GluR epsilon 2 (NR2B) subunit of the N-methyl-d-aspartate receptor. J Biol Chem. 2001;276:693–699. doi: 10.1074/jbc.M008085200. [DOI] [PubMed] [Google Scholar]
- Nguyen TH, Liu J, Lombroso PJ. Striatal enriched phosphatase 61 dephosphorylates Fyn at phosphotyrosine 420. J Biol Chem. 2002;277:24274–24279. doi: 10.1074/jbc.M111683200. [DOI] [PubMed] [Google Scholar]
- Ninan I, Jasrdemark KE, Liang X, Wang RY. Calcium/calmodulin-dependent kinase II is involved in the facilitating effect of clozapine on NMDA and electrically evoked responses in the medial prefrontal cortical pyramidal cells. Synapse. 2003;47:285–294. doi: 10.1002/syn.10175. [DOI] [PubMed] [Google Scholar]
- Norton N, Moskvina V, Morris DW, Bray NJ, Zammit S, Williams NM, Williams HJ, Preece AC, Dwyer S, Wilkinson JC, Spurlock G, Kirov G, Buckland P, Waddington JL, Gill M, Corvin AP, Owen MJ, O'Donovan MC. Evidence that interaction between neuregulin 1 and its receptor erbB4 increases susceptibility to schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2006;141:96–101. doi: 10.1002/ajmg.b.30236. [DOI] [PubMed] [Google Scholar]
- Ozaki M, Sasner M, Yano R, Lu HS, Buonanno Neuregulin-β induces expression of an NMDA-receptor subunit. Nature. 1997;390:691–694. doi: 10.1038/37795. [DOI] [PubMed] [Google Scholar]
- Paul S, Nairn AC, Wang P, Lombroso PJ. NMDA-mediated activation of the tyrosine phosphatase STEP regulates the duration of ERK signaling. Nat Neurosci. 2003;6:34–42. doi: 10.1038/nn989. [DOI] [PubMed] [Google Scholar]
- Pelkey KA, Askalan R, Paul S, Kalia LV, Nguyen TH, Pitcher GM, Salter MW, Lombroso PJ. Tyrosine phosphatase STEP is a tonic brake on induction of long-term potentiation. Neuron. 2002;34:127–138. doi: 10.1016/s0896-6273(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Rosenfeld J, Capdevielle J, Guillemot JC, Ferrara P. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- Roysommuti S, Carroll SL, Wyss JM. Neuregulin-1β modulates in vivo entorhinal-hippocampal synaptic transmission in adult rats. Neuroscience. 2003;121:779–785. doi: 10.1016/s0306-4522(03)00503-7. [DOI] [PubMed] [Google Scholar]
- Salter MW, Kalia LV. Src kinases: a hub for NMDA receptor regulation. Nat Rev Neurosci. 2004;5:317–328. doi: 10.1038/nrn1368. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, Brynjolfsson J, Gunnarsdottir S, Ivarsson O, Chou TT, Hjaltason O, Birgisdottir B, Jonsson H, Gudnadottir VG, Gudmundsdottir E, Bjornsson A, Ingvarsson B, Ingason A, Sigfusson S, Hardardottir H, et al. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Task Force on DSM-IV. Diagnostic and statistical manual of mental disorders DSM-IV-TR. Washington, DC: American Psychiatric Publishing; 2000. [Google Scholar]
- Tezuka T, Umemori H, Akiyama T, Nakanishi S, Yamamoto T. PSD-95 promotes Fyn-mediated tyrosine phosphorylation of the N-methyl-d-aspartate receptor subunit NR2A. Proc Natl Acad Sci USA. 1999;96:435–440. doi: 10.1073/pnas.96.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YT, Salter MW. Regulation of NMDA receptors by tyrosine kinases and phosphatases. Nature. 1994;369:233–235. doi: 10.1038/369233a0. [DOI] [PubMed] [Google Scholar]
- Wittmann M, Marino MJ, Henze DA, Seabrook GR, Conn PJ. Clozapine potentiation of N-methyl-d-aspartate receptor current in the nucleus accumbens: role of NR2B and protein kinase A/Src kinases. J Pharmacol Exp Ther. 2005;313:594–603. doi: 10.1124/jpet.104.080200. [DOI] [PubMed] [Google Scholar]
- Yang YC, Ma YL, Chen SK, Wang CW, Lee EH. Focal adhesion kinase is required, but not sufficient, for the induction of long-term potentiation in dentate gyrus neurons in vivo. J Neurosci. 2003;23:4072–4080. doi: 10.1523/JNEUROSCI.23-10-04072.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XM, Askalan R, Keil GJ, Salter MW. NMDA channel regulation by channel-associated protein tyrosine kinase Src. Science. 1997;275:674–678. doi: 10.1126/science.275.5300.674. [DOI] [PubMed] [Google Scholar]
- Zucker RS. Short-term synaptic plasticity. Annu Rev Neurosci. 1989;12:13–31. doi: 10.1146/annurev.ne.12.030189.000305. [DOI] [PubMed] [Google Scholar]