Abstract
During the embryonic development of the hindbrain, movements of neuronal clusters allow the formation of mature “pools”, in particular for inferior olivary (ION) and facial motor (fMN) nuclei. The cellular mechanisms of neuron clustering remain uncharacterized. We report that the absence of the Rho–guanine exchange factor Trio, which can activate both RhoG and Rac1 in vivo, prevents the proper formation of ION and fMN subnuclei. Rac1, but not RhoG, appears to be a downstream actor in Trio-induced lamellation. In addition, we report that Cadherin-11 is expressed by a subset of neurons through the overall period of ION and fMN parcellations, and defects observed in trio mutant mice are located specifically in Cadherin-11-expressing regions. Moreover, endogenous Cadherin-11 is found in a complex with Trio when lamellation occurs. Altogether, those results establish a link between Trio activity, the subsequent Rac1 activation, and neuronal clusters organization, as well as a possible recruitment of the Cadherin-11 adhesive receptor to form a complex with Trio.
Keywords: Rho GTPases, Cadherin-11, in situ hybridization, mice, motoneurons, olivary neurons
Introduction
During development, neurons migrate from their birthplace to settle at the position they occupy in the adult. The complex architecture of the adult CNS is elaborated through a sequence of developmental events that begins with cell proliferation in the neuroepithelium and ends with synaptic stabilization of neuronal circuits. Although guidance cues involved in long-range tangential and radial migrations are getting unraveled (Barrett and Guthrie, 2001), molecular mechanisms involved in guiding a neuron to settle in a precisely defined position remain poorly characterized. The precerebellar system provides an interesting and anatomically well characterized model to study tangential migrations, and the mature organization of precerebellar nuclei, specifically with inferior olivary neurons (IONn) that form the inferior olivary nucleus (ION). During development, IONn first migrate through the submarginal stream from the dorsal neuroepithelium of the hindbrain (rhombic lips) to form the olivary primordium that is compacted in a club-shaped mass close to the floor plate at embryonic day 15 (E15) in mice (Bourrat and Sotelo, 1990). Then, within 48 h in both rat and mouse, the adult cytoarchitecture will be acquired. IONn will form three individualized lamellae in mice: the dorsal accessory olive (DAO), the principal olive (PO), and the medial accessory olive (MAO), according to Azizi and Woodward (1987). After cytoarchitectural maturation, all the olivary subnuclei are fully recognizable before birth. In the hindbrain too, the facial motoneurons (fMNn) that will form the facial motor nucleus (fMN) also show a complex organization in subnuclei. fMNn originate ventrally in the basal plate in rhombomere 4 (r4) of the hindbrain (Lumsden and Keynes, 1989). Their cell bodies first migrate tangentially from E10 in mice to reach first r5 and then r6. In r6, these neurons begin a lateral and subsequently a radial migration toward the pial surface, in which they form the fMN at approximately E14 (Studer et al., 1996). Just as IONn, fMNn also get organized in six main subnuclei: the lateral (L), dorsolateral (DL), dorsal intermediate (DI), ventral intermediate (VI), dorsomedial (DM), and ventromedial (VM) (Terashima et al., 1993). The fMNn will innervate the muscles of the second branchial arch (Gilland and Baker, 1993). Molecular events that allow the functional development of fMN subnuclei through this “second migratory period” remain poorly characterized (for review, see Wild and Zeigler, 1980). This organization appears as an intrinsic and active phenomenon, independent of their respective target, namely of the cerebellum for IONn (Wassef et al., 1992), and of facial muscles for fMNn (Terashima et al., 1993). The sequence of generation alone does not provide cues required for compartmentalizing the ION. In particular, packs of temporally related cells and those committed to form the rostral ION (MAO, PO, and DAO) undergo complex reorganizations (Bourrat and Sotelo, 1991). Nevertheless, the parcellation could result from the transient or persistent expression of a unique combination of proteins by ION (Wassef et al., 1992), including adhesion molecules (Chedotal et al., 1996).
Among adhesion molecules, Cadherins are a large family of calcium-dependent homophilic cell adhesion proteins that have been involved in various developmental processes, including morphogenesis and development of the nervous system (Saburi and McNeill, 2005). During the development of chick spinal cord, the expression of classic type II cadherins defines specific motor pools and could play a role in motor pool segregation (Price et al., 2002). In the developing mouse hindbrains, Cadherin-11 is expressed, among other structures, in the facial motor and inferior olivary nuclei (Kimura et al., 1996). In the postnatal hindbrain, Cadherin-6, Cadherin-8, and Cadherin-11 are expressed by distinct subcompartments of the ION (Suzuki et al., 1997). Recently, Cadherins have been shown to be involved in regulating the speed of neuronal migration of neurons that form specific precerebellar nuclei (Taniguchi et al., 2006). In addition, Cadherins act as signaling receptors that can activate various downstream signaling pathways, in particular Rho GTPases (Wheelock and Johnson, 2003). Rho GTPases have also emerged as key mediators of Cadherins activity (Kaibuchi et al., 1999; Braga, 2002; Charrasse et al., 2003; Wheelock and Johnson, 2003; Yap and Kovacs, 2003). Thus, increasing recent data reveal that Rho GTPases and Cadherins are linked.
Nevertheless, the biochemical link between Cadherins and Rho GTPase remained to be elucidated. The activity of Rho GTPases itself is regulated via specific GTPase activating proteins (GAPs) and guanine exchange factors (GEFs) (Etienne-Manneville and Hall, 2002), but the involvement of GTPases regulators as GAPs or GEFs in Cadherins signaling (Lanier and Gertler, 2000) remains mainly uncharacterized.
Recently, M-Cadherin has been shown to activate Rac1 through Rho–GEF Trio in myoblast cells in vitro by Charrasse et al. (2007). Trio–GEF family is a key component of intracellular signaling pathway that regulates axon guidance and neuronal migration (Bateman and Van Vactor, 2001), as well as late embryonic development of muscle, hippocampal formation, and olfactory bulb (O'Brien et al., 2000). Trio belongs to the Trio family that contains two GEF domains (Debant et al., 1996). Through its GEF-1 domain (also termed Trio-D1, the N-terminal GEF domain), Trio can activate Rac1 and RhoG, the latter subsequently activating Rac1 and cell division cycle 42 (Cdc42) (Gauthier-Rouviere et al., 1998). The role of Trio GEF-2 in the whole Trio protein has not been demonstrated, albeit the expression of the GEF-2 domain in COS-7 cells can activate RhoA (Bellanger et al., 1998).
In the present work, we investigated the role of Trio in the process of neuronal organization through clustering, which requires subtle movements of neurons to delineate a mature neuronal “pool” or nucleus. Thus, we have analyzed its role and the involvement of Rho GTPases and Cadherin-11 adhesion molecule in the final organization of ION and fMN in their ultimate domains during migration and parcellation of IONn and fMNn clusters. We report the following: (1) At birth, newborn mice deficient in the expression of trio show an abnormal organization of both ION and fMN. (2) ION and fMN properly develop in RhoG mutant mice, and the activation level of Rac1 and Cdc42 in ION and fMN is not decreased in RhoG mutant mice. (2) In contrast, Rac1 but not Cdc42 activity is decreased in trio mutant mice, which leads us to propose that the abnormal organization of both nuclei in trio mutant mice is attributable to a defective Rac1 activation. (4) Cadherin-11 is expressed in clusters of IONn and fMNn just before lamellation occurs and the cadherin-11 expression domain coincides with regions showing an impaired lamellation in the absence of trio. (5) Endogenous Cadherin-11 can form in vivo a multiproteic complex with Trio–GEF during ION and fMN organization.
Materials and Methods
Fixation.
Mouse embryos were obtained from timed mating of outbreed Swiss mice (Janvier, Le Chesnet Saint Isle, France), of heterozygous trio mutant mice (O'Brien et al., 2000), of RhoG mutant mice (Vigorito et al., 2004), and of Cadherin-11 mutant mice (provided by Prof. Takeichi, RIKEN Center for Developmental Biology, Kobe, Japan) (Manabe et al., 2000). All embryos were treated according to the recommendations of the European Union and French government. Embryonic and newborn mice were fixed with 4% paraformaldehyde solution in 0.1 m PBS, pH 7.4, and postfixed overnight at 4°C in the same fixative. The neural tubes and brains were dissected out and cryoprotected according to Bloch-Gallego et al. (1999). The frontal sections (20 μm thick) were collected using a cryostat on parallel sets of SuperFrost Plus slides and stored at −80°C until use.
Immunohistochemistry and antibodies.
Immunoreaction was performed following the methods used by Bloch-Gallego et al. (1999). Sections were incubated with the monoclonal anti-Islet-1 (1:100, 39.4D5; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) or the polyclonal anti-Peripherin (1:200; Chemicon, Temecula, CA) antibodies overnight at room temperature. These primary antibodies were revealed using secondary anti-rabbit IgG antibodies conjugated to Alexa 488 (1:400; Invitrogen, Carlsbad, CA) or cyanine 3 (1:200; Jackson ImmunoResearch, West Grove, PA). After the immunoreactions, the sections were treated with 4′,6′-diamidino-2-phenylindole (DAPI) (1 mg/ml; Vector Laboratories, Burlingame, CA).
In situ hybridization and RNA probes.
In situ hybridization (ISH) was performed on cryosections according to Bloch-Gallego et al. (1999). For the mouse Cadherin-11 subclone (Padilla et al., 1998), we linearized with HindIII (Invitrogen) enzymes and used T3 RNA polymerase (Roche, Indianapolis, IN). For the mouse Peripherin subclone, we linearized with NotI (Invitrogen) enzymes and used T7 RNA polymerase (Roche) For the mouse trio subclone, we linearized with ClaI (Invitrogen) enzymes and used T7 RNA polymerase. For the mouse RhoG subclone, we linearize with EcoRI and use T3 RNA polymerase. The mouse Brn-3.b (Brain-specific homeobox/POU domain protein 3B) Cdc42, Rac1, and neuron–glia-related cell adhesion molecule (Nr-CAM) cDNA subclones have been described previously (Backer et al., 2002).
Coimmunoprecipitation and Western blot.
Lysates were prepared from E15 and E18 ION or fMN. Freshly dissected tissues were dissociated in lysis buffer containing 50 mm Tris-HCl, pH 7.5, 120 mm NaCl, 20 mm NaF, 1 mm EDTA, 6 mm EGTA, 15 mm Natrium pyrophosphate, 1% NP-40, 0.5 mm orthovanadate, and 0.2% protease inhibiter cocktail (Sigma, St. Louis, MO), sonicated three times for 10 s on ice, and centrifuged at 16,000 × g for 10 min at 4°C. Supernatants were immunoprecipitated with anti-Trio goat monoclonal antibodies (clones C19 and D19, 1:500 dilution; Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4°C and immobilized on protein G-Sepharose gel (GE Healthcare, Little Chalfont, UK). After three washing steps with the lysis buffer, immunoprecipitated proteins were eluted by boiling with the Laemli's loading buffer, resolved on a 6 or 10% polyacrylamide gel, and blotted on to nitrocellulose membrane (GE Healthcare). After saturation in TBS/Tween 20 with 5% milk, membranes were incubated with the following antibodies: anti-Notch (1:500 dilution; Calbiochem, La Jolla, CA); anti-Abl (Abelson murine leukemia viral oncogene homolog; 1:100 dilution; Calbiochem); anti-DCC (deleted in colorectal cancer) (1:500 dilution; BD Biosciences PharMingen, San Diego, CA); anti-Cadherin-11 (1:100 dilution; Zymed Laboratories, South San Francisco, CA), anti-N-Cadherin (1:1000 dilution; Transduction Laboratories, Lexington, KY), and anti-Nr-CAM (1:3000 dilution; a kind gift from Dr. M. Grumet, Rutgers, Piscataway, NJ). Primary antibodies were revealed by incubation with HRP-conjugated anti-mouse or anti-rabbit IgG (1:25,000; Jackson ImmunoResearch). Bands were revealed with ECL+ (GE Healthcare).
To detect interaction between exogenous proteins, COS-7 cells were transfected with green fluorescent protein (GFP)–Trio and/or GFP–Cadherin-11 constructs, using JetPEI (Polyplus-transfection, New York, NY), as described by the manufacturer. Cell lysates were prepared, and immunoprecipitations were conducted as described above. Immunoprecipitated proteins were detected using a rabbit anti-GFP antibody (Invitrogen). Antibodies used for immunoprecipitation were as follows: anti-Trio goat monoclonal antibodies (C19 and D19; Santa Cruz Biotechnology), anti-Cadherin-11 mouse antibody (Zymed Laboratories), anti Brn-3.b goat monoclonal antibody (Santa Cruz Biotechnology), and anti-RhoA mouse antibody (Santa Cruz Biotechnology).
In situ detection of active Rho GTPases.
Glutathione S-transferase (GST)–Rho binding domain (RBD)–p21-activated kinase (PAK) was provided by M. A. Schwartz (University of Virginia, Charlottesville, VA) and prepared as described by Causeret et al. (2004). After fixation and cryosectioning of embryos, active Rac1 and Cdc42 activities were detected through the binding of their GST–Cdc42/Rac-interactive binding protein (CRIB)–PAK effector. At least three animals were examined for each condition tested.
Dosage of the endogenous activity of Rac1 and Cdc42 in IONn, fMNn, and whole hindbrain.
To evaluate Rac1 and Cdc42 activities in ION and fMN, E15 and E18 subdissected ION and fMN explants were lysed in 50 mm Tris, pH 7.2, 1% Triton X-100, 500 mm NaCl, 10 mm MgCl2, 1 mm PMSF, and protease inhibitors cocktail (Sigma), sonicated three times for 10 s on ice, and centrifuged at 16,000 × g for 10 min at 4°C. Cleared lysate was incubated with 100 mg of GST-fusion protein, containing CRIB domain of PAK (GST–PAK), attached to beads (Sigma) for 90 min at 4°C. The beads were then washed four times in 50 mm Tris, pH 7.2, 1% Triton X-100, 150 mm NaCl, 10 mm MgCl2, 0.1 mm PMSF, and protease inhibitors cocktail before the addition of Laemli's buffer. Samples were analyzed by Western blotting with Rac1 and Cdc42 antibodies (1:1000 and 1:400, respectively; Transduction Laboratories). Averages were analyzed, and SDs were calculated. Differences were considered as significant when p < 0.05 using Student's t test.
Results
The lamellation of both ION and fMN constitutes the ultimate step toward their mature organization. As visualized in supplemental movie 1 (available at www.jneurosci.org as supplemental material) that illustrates IONn movements from E11 to birth, IONn first compact into a club-shaped domain, and then, in a second step, they organize in subnuclei that form the characteristic lamellae, through a secondary migratory process. A similar biphase migration exists for fMNn. We analyzed here the molecular pathways that govern these cytoarchitectural maturations.
Trio-deficient mice show an abnormal cellular organization of both ION and fMN
We investigated the possible role of Trio during the IONn and fMNn mature development. Previous analysis of trio mutant mice revealed abnormal organizations in the hippocampal formation and olfactory bulb and defects in secondary myogenesis (O'Brien et al., 2000). Homozygous mutant mice for trio expression are not viable and die during late embryonic development or soon after birth. Thus, we focused our phenotypic analysis of ION and fMN organization in trio mutant mice at E15 and E17.
At E15, when the whole migrating IONn have reached the floor plate, the IONn club-shaped domain, labeled through Nr-CAM ISH (Fig. 1A–D), shows a slightly different aspect rostrally in trio mutant mice compared with wild-type (WT) mice (Fig. 1A,B). In mutants, rostral IONn come close to the floor plate and adopt an elongated aspect (Fig. 1, compare A, B). The caudal ION shows a similar aspect in wild-type and mutant mice (Fig. 1C,D). At E17, in trio mutant mice, the organization of the rostral ION is even more affected. Rostrally, IONn are absent laterally in mutant (Fig. 1F) compared with wild-type (Fig. 1E) mice. Although the DAO is formed in trio mutant mice, it presents an abnormal aspect compared with wild-type animals (Fig. 1E). The intermediate portion of the DAO is impaired. The PO shows no precise organization and no proper spreading to form both characteristic ventral and dorsal lamellae of the PO (Fig. 1F, short arrow). The aspect of the rostral MAO appears less dramatically affected than the other two lamellae (Fig. 1F, arrowhead). Similarly, when labeled through Brn-3.b ISH, at E17, the MAO, which strongly expresses Brn-3.b transcripts in the wild type (Fig. 1I, arrowhead), is properly located in trio mutant mice (Fig. 1J,K, arrowhead). The DAO is also formed, especially its most lateral part (Fig. 1J,K, large arrow), but presents a fragmented aspect. The PO is not individualized as a lamella and remains compacted (Fig. 1I–K, short arrow).
Figure 1.
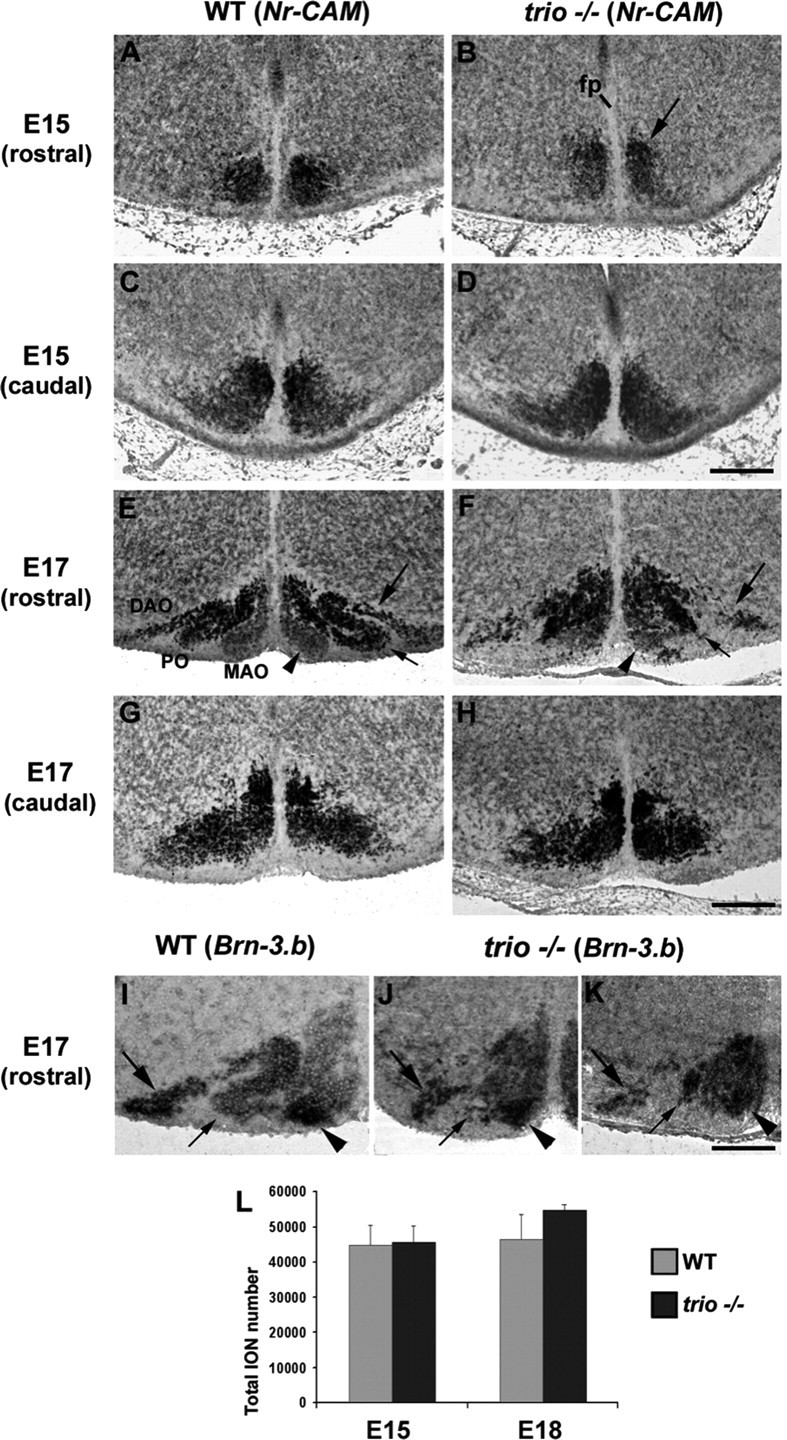
Defects of lamellation in the rostral ION in trio mutant mice. At E15, Nr-CAM transcripts allowed the visualization of the whole rostral (A, B) and caudal (C, D) ION masses in WT (A, C) and trio mutant (B, D) mice. Rostrally, ION masses were compacted closer to the floor plate (fp; B) compared with the aspect in wild type (A), but the caudal ION shows a similar aspect in wild-type (C) and trio mutant (D) mice. At E17, ION rostral lamellae were mis-developed in trio mutant mice (F), in particular the lateral DAO (large arrow in F), and the PO that still strongly synthesized Nr-CAM but remained compacted and was mis-extended laterally (short arrow in F) compared with wild type (E). The MAO was pointed with the arrowhead in F. The caudal ION showed the same aspect in both wild-type (G) and trio mutant (H) mice. Rostrally, at E17, both the MAO (arrowhead) and the most lateral part of the DAO strongly express Brn-3.b in wild-type mice (I), and those structures and expressions were maintained in mutant mice (J, K), whereas neurons in the PO remained compacted in trio−/− instead of forming an individual lamella (short arrows in I–K). The quantification of IONn (L) revealed that the number of ION at E17 was unchanged in WT (n = 45,675 ± 849) and trio mutant (46,625 ± 649) mice, with p > 0.05 (K). IONn of three wild-type and three mutant mice have been quantified. Scale bars: (in D) A–D, 140 μm; (in H) E–H, 240 μm; (in K) I–K, 160 μm.
To further determine whether the abnormal aspect of the ION at E15 and E17 was attributable to a selective loss of neurons or to a mis-organization during the phase of refinement, we quantified the total number of IONn at both E15 and E17. Nr-CAM ISH and DAPI staining (data not shown) allowed the subsequent comparison of cell density in both wild-type and trio mutant mice and showed that the cell number was neither significantly changed at E15 (n = 45,675 ± 849 and n = 46,625 ± 649 respectively; p > 0.05) nor at E17 (Fig. 1L). We concluded that the number of IONn was not changed in trio mutant mice, but their compaction and clusterization were impaired.
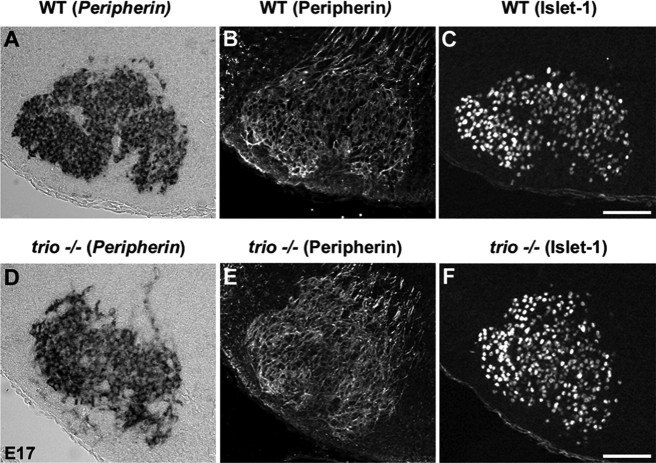
In parallel, we analyzed the aspect of the fMN (Fig. 2). At E15, the whole fMNn have correctly reached their most ventral location in trio mutant mice as revealed by anti-Peripherin and anti-Islet-1 immunostaining (data not shown), but fMNn did not properly segregate between E15 and E17 to form the various subnuclei. At E17, both Peripherin (Fig. 2A, ISH, B, IF) and Islet-1 (Fig. 2C) labelings are present in all fMN clusters. In E17 trio mutant mice, the fMNn remain poorly delimited in subnuclei after revelation of Peripherin transcripts by ISH (Fig. 2D), anti-Peripherin labeling (Fig. 2E), or Islet-1 immunostaining (Fig. 2F). The fMNn remain homogenously dispersed in the fMN.
Figure 2.
Defects in the formation of fMN subnuclei in E17 trio mutant mice. During ISH to detect Peripherin transcripts, as well as anti-Peripherin and anti-Islet-1 immunolabeling, the comparison of the structure of fMN in WT E17 embryo (A–C) and trio mutant (D–F) mice revealed that the fMN remained mis-organized in the absence of trio. Scale bars: (in C) A–C, 100 μm; (in F) D–F, 160 μm.
As in the ION, the quantification of fMNn at E17 revealed that their total number was unchanged in trio mutant mice (n = 4970 ± 390) compared with wild-type mice (n = 4580 ± 480; p > 0.05) (Fig. 2G). fMNn from four wild-type and four mutant mice have been quantified.
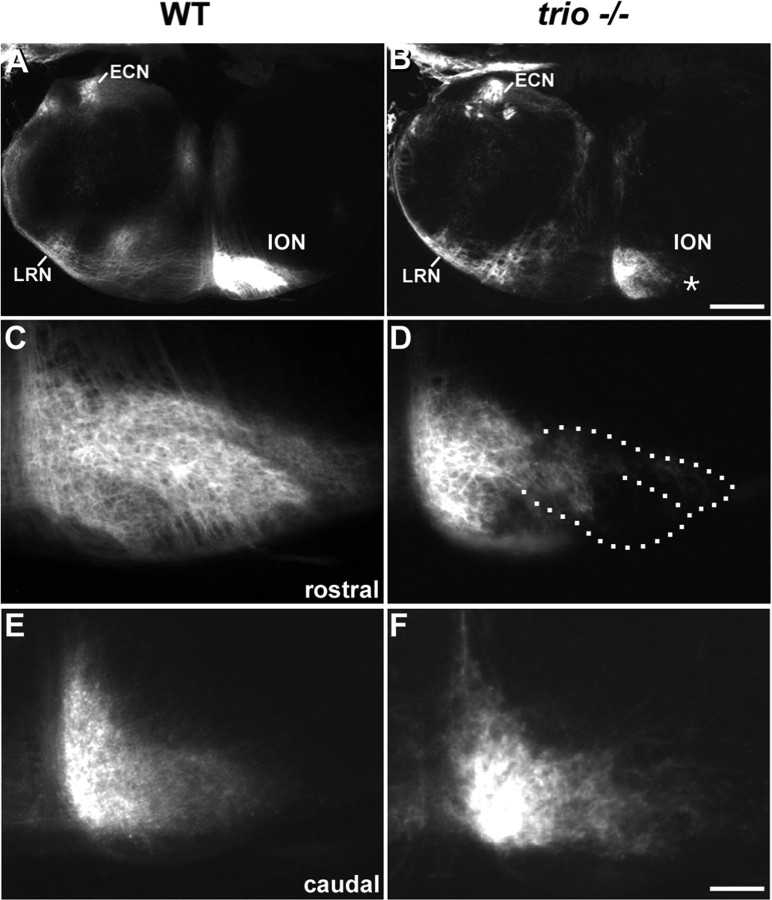
We then investigated whether the axons of mis-organized IONn could still develop in the absence of Trio. We performed unilateral insertions of DiI crystals in the cerebellum at E17 that result in a massive retrograde labeling of the ION axons. In both wild-type (Fig. 3A) and trio mutant (Fig. 3B) mice, ION are labeled contralaterally to the injection site. Although ION fibers were visualized in both wild-type (Fig. 3A) and mutant (Fig. 3B) mice, in trio mutant mice, large portions of the lateral ION (surrounded with dotted lines in Fig. 3D) do not present any retrograde labeling (Fig. 3, compare C, D), pointing out an incomplete aspect of ION masses after tracing. These labeling defects reflect the absence of IONn in the lateral PO and DAO and are in agreement with the mis-organizations of ION lamellae in the absence of Trio (Fig. 1). Interestingly, the retrograde labeling of the caudal IONn in trio mutant mice (Fig. 3F) is strictly identical to the one observed in wild-type mice (Fig. 3E). Finally, we checked that other precerebellar projections were formed correctly. Both lateral reticular nucleus (LRN) (Fig. 3A,B) and external cuneatus nucleus (ECN) (Fig. 3A,B) are retrogradely labeled ipsilaterally to the injection site and present a similar shape in both wild-type and mutant mice.
Figure 3.
Unilateral DiI injections in the cerebellum and retrograde labeling of precerebellar nuclei in both normal and trio mutant newborn mice. After the cerebellar injection in wild-type E17 mice (A and C, E at a higher magnification), the ION located contralateral to the injection site and the ipsilateral ECN and LRN were labeled (A). Crossing fibers of the olivary commissure were visualized ventrally in the olivary region. In mice mutant for the expression of trio, IONn were also labeled contralateral to the injection site (ION in B, and at a higher magnification in D, F). No labeled IONn were located in the lateral part of the ION contralateral to the injection site, in neither the lateral DAO nor the lateral aspects of the PO (black region surrounded with a dotted line in D). The aspect of the retrograde tracing in the mutant caudal ION (F) was similar to the one in the wild type (E). ECN and LRN were properly ipsilaterally retrogradely labeled (B). Scale bars: (in B) A, B, 300 μm; (in F) C–F, 108 μm.
Trio is expressed in the developing ION and fMN
We then analyzed the expression pattern of trio throughout the development of both ION and fMN. At E15, IONn masses that form the ventromedial ION show trio expression both rostrally (Fig. 4A) and caudally (data not shown). At postnatal day 0 (P0), ION expresses trio in all three lamellae (Fig. 4B). Trio transcripts are detected in fMN homogenously at E15 (data not shown), and trio synthesis is maintained at birth in all subnuclei (Fig. 4C). In conclusion, the temporal and spatial trio expression is compatible with its involvement in ION and fMN development. We tested whether kalirin, another member of the Trio family, was synthesized during the embryonic development, but no expression could be detected by ISH (data not shown).
Figure 4.
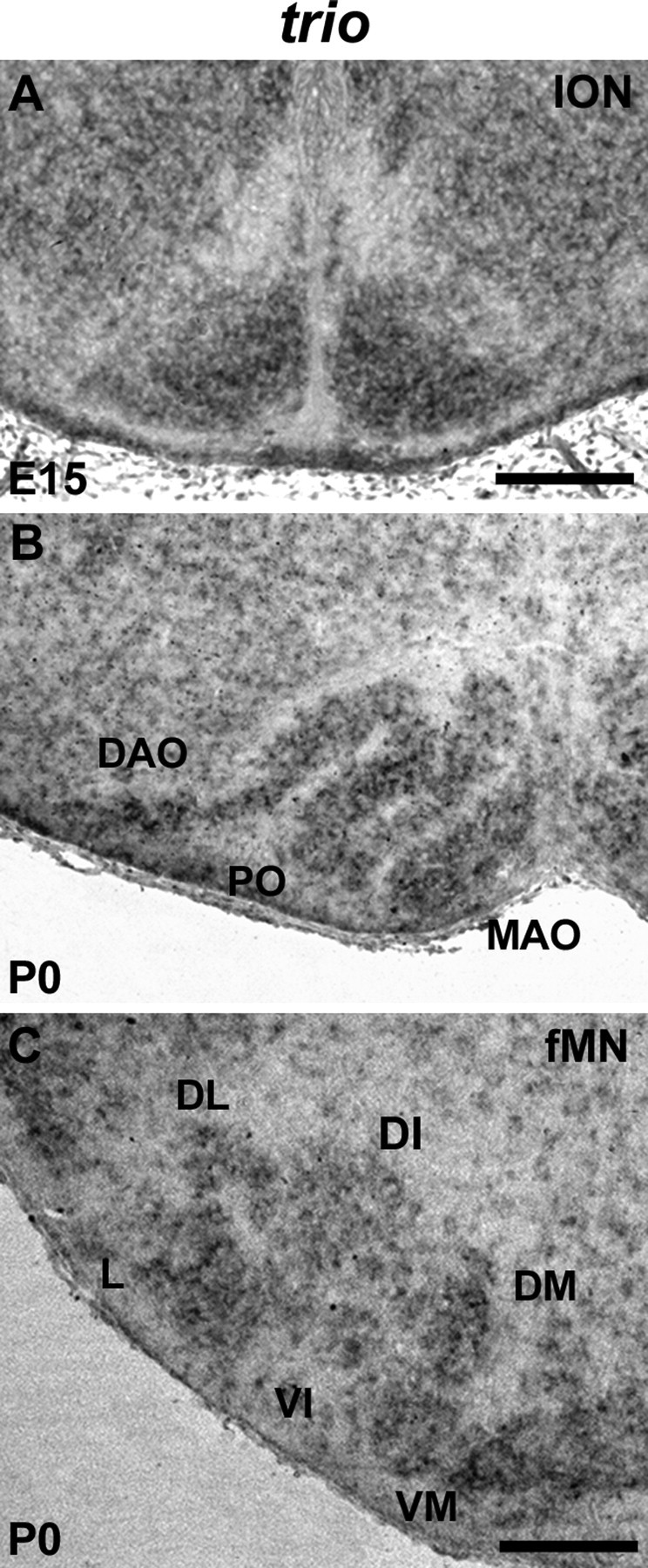
Expression of trio at E15 and P0 in ION and fMN. The ION masses showed a strong trio expression at E15 (A). At birth (B), all three ION lamellae (DAO, PO, and MAO) showed trio transcript expression. Trio was expressed in the whole fMN (C). Scale bars: A, 150 μm; (in C) B, C, 180 μm.
Expression of Rac1, Cdc42, and RhoG in ION and fMN
In vivo, Trio might activate Rac1 and/or RhoG, which in turn activates Cdc42 and Rac1 (Gauthier-Rouviere et al., 1998). Thus, we analyzed Rac1, Cdc42, and RhoG expression patterns at E15 and P0 to determine whether their expressions correlate with a physiological function during IONn and fMNn maturation.
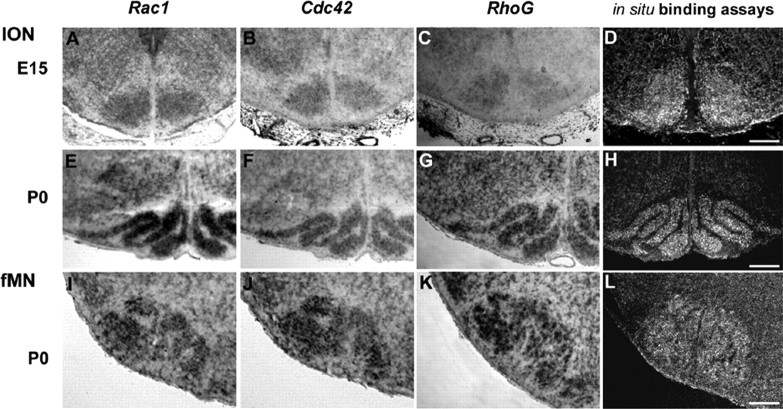
At E15, Rac1 and Cdc42 transcripts are both detected in ION (Fig. 5A,B) and fMN (data not shown). At P0, Rac1 and Cdc42 mRNA are still strongly synthesized in ION (Fig. 5E,F) and fMN (Fig. 5I,J). RhoG expression, contrary to Rac1 and Cdc42 whose transcripts are synthesized from E11 in the hindbrain (Causeret et al., 2004), only begins from E15 in both IONn and fMNn. At E15, RhoG mRNA are weakly detected in ION (Fig. 5C) and strongly in fMN (data not shown). At P0, RhoG is expressed in the whole ION lamellae (Fig. 5G), as well as in the whole fMN subnuclei (Fig. 5K).
Figure 5.
Expression of Rac1, Cdc42, and RhoG in mouse at embryonic day 15 and P0. Localization of active Rac1 and Cdc42 on sections through the hindbrain at E15 and P0. At E15, Rac1, Cdc42, and RhoG expression were located in ION masses (A–C). At P0, the whole lamellae of ION synthesized Rac1, Cdc42, and RhoG transcripts (D, E, G). In fMN, Rac1, Cdc42, and RhoG transcripts were synthesized at P0 (I–K, respectively). After incubation with RBD–Rhotekin fused to the GST and treatment with an anti-GST antibody of sections, GST–RBD–PAK-positive cells could be detected in the IONn club-shaped domain at E15 (D). A strong Rac1/Cdc42 GTPase activity was observed in the whole ION lamellae at P0 (H). Rac1/Cdc42 activity was also detected in the whole fMN (L). Scale bars: (in D) A–D, 105 μm; (in H) E–H, 215 μm; (in L) I–L, 145 μm.
Because Rho GTPases do cycle between a GDP-bound inactive state and a GTP-bound active state (Etienne-Manneville and Hall, 2002), we then assessed the activation state of those small GTPases. In situ detection of active GTPases (Causeret et al., 2004) was performed on cryosections at E15 and P0 using the GST fused to the CRIB domain of the Rac1/Cdc42 effector molecule PAK (GST–Pak CRIB), which could only bind active GTPases. At E15, active Rac/Cdc42 GTPases could be detected in the ION club-shaped domain (Fig. 5D) and in the whole fMN (data not shown). At P0, active Rac/Cdc42 GTPases are visualized on cryosections in the whole ION, with a strong binding of the effector in the medial parts of the PO (Fig. 5H). Active Rac/Cdc42 GTPases are also detected in the fMN (Fig. 5L).
Normal organization of ION and fMN in RhoG mutant mice
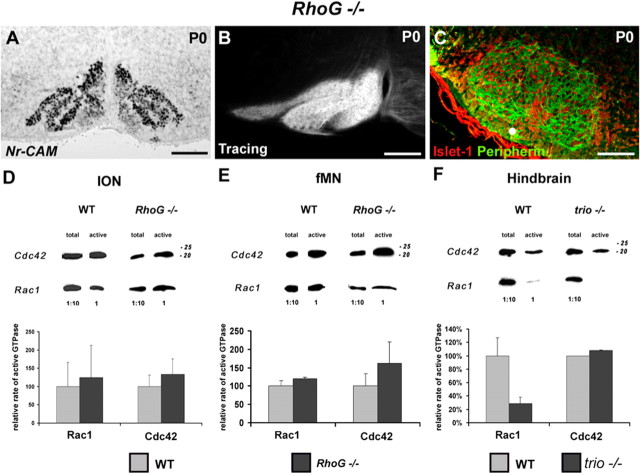
Because RhoG is one of the targets of Trio (Gauthier-Rouviere et al., 1998), we analyzed the phenotypic development of ION in RhoG mutant mice at E15 and P0. ION normally develops in the absence of RhoG, as visualized at P0 after ISH with an Nr-CAM probe (Fig. 6A). Retrograde tracing analysis confirmed that the olivocerebellar projection correctly developed and that the mature organization of the ION is preserved at P0 (Fig. 6B). In parallel, we observed the structure of the fMN at P0. The development and the aspect of fMN appear to be completely normal when compared with wild-type embryos after anti-Islet-1 and anti-Peripherin labeling (Fig. 6C).
Figure 6.
RhoG mutant mice show a normal organization and projections of IONn and a proper development of fMN. Quantification of Rho GTPases activity in E15 wild-type mice and RhoG and trio mutant mice. Nr-CAM in situ hybridization revealed a normal organization of the rostral ION in RhoG mutant mice at P0 (A). Retrograde tracing of ION allowed the visualization of the whole contralateral IO complex (B). The immunostaining with anti-Peripherin (in green) and anti-Islet-1 (in red) revealed the distinct fMN subnuclei (C). Quantification of the relative active state of Rac1 and Cdc42 after Western blot quantification in both ION (D) and fMN (E) extracts. The rate of active Rac1 and Cdc42 did not change significantly in RhoG mutant mice (p > 0.1) in either ION extracts (D) or fMN extracts (E). Western blot analysis after GST pull down was also performed on individual entire hindbrain for WT and trio mutant mice (F). The activity of Cdc42 remained unchanged in trio mutant mice, but the activity of Rac1 showed a 80% decrease. Scale bars: A, 120 μm; B, 140 μm; C, 180 μm.
Pull-down assays allow the detection of active Rac1 and Cdc42 in ION and fMN extracts from E15 wild-type and RhoG mutant mice (Fig. 6D,E). Quantification reveals an increase of 25 and 33% of the rate of active Rac1 and Cdc42, respectively, in RhoG mutant mice compared with wild-type ION (Fig. 6D). In the fMN of RhoG mutant mice, the rate of activation of both Rac1 and Cdc42 are, respectively, 19 and 62% higher than those in wild-type fMN (Fig. 6E). Thus, in RhoG mutant mice, the rate of active Rac1 and Cdc42 in ION and fMN increases during the clustering of both ION (Fig. 6D) and fMN (Fig. 6E) but not significantly when compared with activations in wild-type embryos (p > 0.05). Altogether, those results show that the disrupted activation of RhoG neither affects the rate of active Rac1 and Cdc42 at E15 nor mimics the effects of Trio depletion. Thus, RhoG is not a key component of the signaling pathway downstream of Trio in the process of ION and fMN clustering.
In E15 trio mutant mice, the relative activation state of the various Rho GTPases has also been established through GST pull-down assays using the whole hindbrain because ION and fMN dissected from a single embryo do not provide enough material for the assay. Under this condition, the ratio of active Rac1 is significantly decreased in the absence of trio (p < 0.01), being 28% of the active Rac1 ratio in wild-type hindbrains, whereas Cdc42 activity remains constant at E15 (p > 0.05), as revealed on the Western blot and the corresponding quantifications (Fig. 6F).
Altogether, these data suggest that only Rac1, but neither RhoG nor Cdc42, is involved in the signaling pathway downstream of Trio in the process of ION and fMN clustering.
Spatially restricted expression of Cadherin-11 in the developing ION and fMN
We visualized the whole club-shaped ION at E15 and the lamellated ION at birth by detection of Nr-CAM mRNA through ISH (Backer et al., 2002) (Fig. 1A–H). Rostral ION undergoes the most complex reorganizations between E15 (Fig. 1A) and E17 (Fig. 1E) and shows a highly organized aspect (Bourrat and Sotelo, 1991). We analyzed Cadherins expression, because they had been shown to control Rho GTPase activity (Charrasse et al., 2003), and the expression of specific type II cadherins defines specific motor pools and regulates their segregation (Price et al., 2002). Among Cadherins that we screened, M-Cadherin, N-Cadherin, and Cadherin-6–Cadherin-11, expression was the most striking with regard to defects observed in Trio mutant mice. At E15, Cadherin-11 transcripts are strongly synthesized in the most dorsal part of the compacted rostral IONn mass (Fig. 7A, arrow). No Cadherin-11 expression is observed in the caudal ION (Fig. 7C). In newborn mice (P0), the dorsal cap shows Cadherin-11-positive cells (Fig. 7E, arrowhead). Cadherin-11 transcripts are highly synthesized in the DAO (Fig. 7E), in the dorsal part of the PO, and the bend of the PO. The ventral part of the PO shows a lower expression (Fig. 7E, asterisk), as well as the ventral part of the MAO (Fig. 7E, short arrow). Thus, Cadherin-11 still shows a remarkable differential pattern in rostral and caudal ION and in the distinct lamellae.
Figure 7.
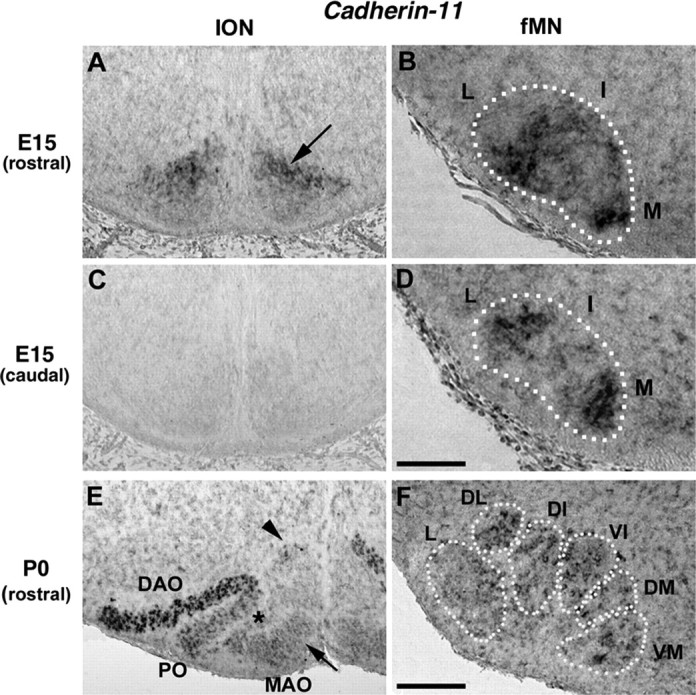
Cadherin-11 is expressed in neuronal clusters in ION and fMN at E15 and P0. At E15, Cadherin-11 was expressed in the dorsal neurons of the rostral ION masses (arrow in A). Caudally, no expression of Cadherin-11 was detected in ION masses (C). At P0 (E), Cadherin-11 was strongly synthesized in the dorsal cap (arrowhead in E) and in the DAO (E), and the intensity was much lower in the PO (asterisk) and MAO (short arrow). In the fMN, at E15 (B), the Cadherin-11 transcripts were strongly expressed in the intermediate (I) and medial (M) parts of the rostral ball-shaped fMN (B) and in the lateral (L) and medial (M) parts of the caudal fMN (D). At P0 (F), the fMN was divided into six subnuclei, and Cadherin-11 transcripts were strongly synthesized in the dorsal fMN subnuclei but lower in the lateral (L) and dorsomedial (DM) subnuclei. Scale bars: (in D) A–D, 230 μm; (in F) E, F, 150 μm.
At E15, whereas fMNn are grouped in a compacted Peripherin (data not shown), and Islet-1 (Pfaff et al., 1996), positive mass, Cadherin-11 is strongly expressed in the intermediate and medial parts of the rostral fMN and poorly expressed in its lateral portion (Fig. 7B). In the most caudal portion of the E15 fMN, Cadherin-11 is expressed in the lateral and medial portions (Fig. 7D). At P0, higher levels of Cadherin-11 transcripts are expressed in the DL and DI, in the VI, and in the VM compared with L and DM (Fig. 7F).
To summarize, the strongest Cadherin-11 synthesis is observed in neurons located in the most dorsal part of the rostral ION masses at E15 and in the most dorsolaterally located ION (DAO) at birth. Similarly, Cadherin-11 expression is located in the intermediate and medial subnuclei of the rostral fMN both at E15 and birth. Altogether, these remarkable expressions during ION and fMN development suggest that Cadherin-11 could predefine subdomains that direct neuronal clustering and localized cellular movements. It is noteworthy that neither neurons that form the LRN nor the ECN, which are two other precerebellar nuclei, express Cadherin-11. Interestingly, none of them show any defect in trio mutant mice.
Cadherin-11 interacts with Trio during ION and fMN clustering
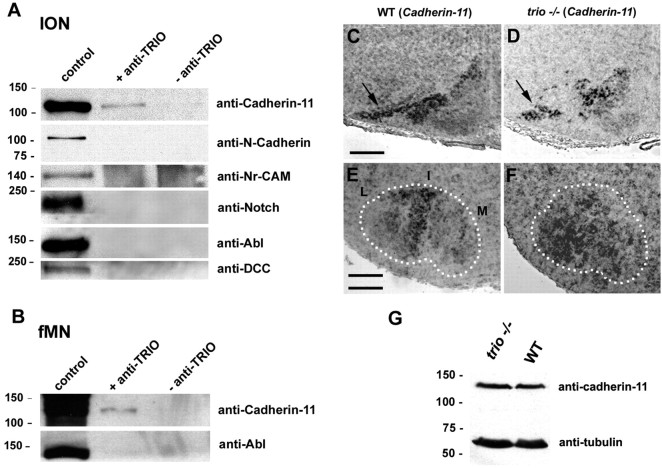
At E15, before the parcellation, we show that specific clusters express Cadherin-11 in IONn and fMNn, and, at birth, neurons that express Cadherin-11 are located in the most affected subnuclei in the absence of trio. Thus, we aimed at determining whether Trio, which can interact after COS-7 transfection with Cadherin-11 and Trio expression plasmids (supplemental Fig. 2, available at www.jneurosci.org as supplemental material), could also associate with Cadherin-11 in the developing IONn and fMNn in vivo. We prepared ION and fMN extracts at E15 that were then immunoprecipitated with an anti-Trio antibody. Complexes containing Trio were analyzed by Western blot and screened for the presence of Cadherin-11 and also Notch, Abl, and DCC, other possible partners of Trio in Drosophila (Forsthoefel et al., 2005), and N-Cadherin and Nr-CAM. Cadherin-11 was the only molecule that formed a multiproteic complex and coimmunoprecipitate with Trio at E15 in both ION and fMN (Fig. 8A,B, respectively). Thus, we unveiled an original protein complex between Cadherin-11 and Trio in vivo that could play a role in the process of lamellation.
Figure 8.
Cadherin-11 can immunoprecipitate with Trio in E15 ION and fMN extracts, and its expression is not affected in the mis-organized ION and fMN. In A, Western blot illustrating the presence of Cadherin-11, N-Cadherin, Nr-CAM, Notch, Abl, and DCC in E15 ION extracts. Cadherin-11 only could interact with Trio. Abl and Cadherin-11 were also expressed in control fMN extracts at E15, and only Cadherin-11 was immunoprecipitated with Trio (B). At E17, Cadherin-11 transcripts were expressed in the rostral ION in WT mice (C), in particular in the DAO and PO (C). Cadherin-11 remained expressed in trio mutant mice, in the mis-organized DAO at E17 (arrow in D). fMNn maintained Cadherin-11 synthesis in the absence of trio (F), but transcripts remained expressed randomly in the entire fMN but mainly expressed in the intermediate and in part of the lateral fMN subnucleus in wild-type fMN (E). Analysis of the amount of Cadherin-11 proteins in both WT and trio mutant mice by Western blot (G) reveals that the amount of Cadherin-11 is the same in WT and trio−/− hindbrains. Scale bars: (in C) C, D, 240 μm; (in E) E, F, 130 μm.
Cadherin-11 is still expressed in mis-organized ION and fMN
We analyzed the expression pattern of Cadherin-11 in trio mutant mice at E15 and E17.
In the ION, it remains expressed in the most dorsal part of the ION in E15 trio mutant mice (data not shown). At E17, Cadherin-11 is expressed in the DAO and in the dorsal PO in wild-type ION (Fig. 8C), and, in trio mutant mice, the rostral mis-developed ION still expressed Cadherin-11 (Fig. 8D, arrow) dorsally. Interestingly, the ION shows a similar aspect caudally, in which Cadherin-11 is expressed at neither E15 (Fig. 7C) nor at E17 (data not shown), in E17 trio mutant and wild-type mice (Fig. 1G,H).
In the fMN, instead of being located in the intermediate and lateral nuclei (Fig. 8E), Cadherin-11 transcripts are highly expressed but distributed in the whole mis-organized fMN (Fig. 8F).
To further analyze whether the absence of Trio could quantitatively affect Cadherin-11 expression, we performed two sets of experiments. The first one consists in counting the number of Cadherin-11-positive ION after in situ hybridization with a Cadherin-11 antisense probe combined with a DAPI staining that allows the visualization of the whole structure. We reveal that the number of Cadherin-11-positive neurons is not affected in trio knock-out (KO) (n = 1173 ± 234) compared with WT (n = 1182 ± 30) mice. Four animals of both WT and trio mutant mice were used for quantifications.
The second approach consists of a compared quantification of Cadherin-11 proteins in WT and KO hindbrains by Western blot (Fig. 8G). Indeed, only ION and fMN do express Cadherin-11 in the hindbrain at birth. After quantification, we reveal that the rate of Cadherin-11 protein is not modified in hindbrain extracts prepared from WT or trio mutant mice, whatever the presence or absence of endogenous Trio (100 arbitrary units in WT and 112 in trio mutant mice).
Thus, our data show that Cadherin-11 remains expressed at a constant proteic level in mis-organized ION and fMN in trio mutant mice, indicating that neither the rate of Cadherin-11 expression nor its spatial expression do depend on trio expression.
Discussion
The formation of the ION and fMN are carefully orchestrated processes. Despite several studies demonstrating a role of various transcription and tropic factors, as well as Rho GTPases in long-range migrations (Garel et al., 2000), the mechanisms regulating the cytoarchitectural maturation of both nuclei remain essentially unknown. Our study demonstrates in vivo that the absence of Trio impairs the organization of both ION and fMN. The ION and fMN normally develop in RhoG mutant mice. The measurement of Rho GTPases activity reveals that Rac1 activation is required for the proper organization of neuronal clusters. Cadherin-11 shows a restricted expression at E15 in both ION and fMN masses and Cadherin-11 can form a complex with Trio. The abnormal organization of clusters in trio mutant mice is localized in regions expressing Cadherin-11. We discuss these results in the context of signaling pathways affecting the mature organization of specific neuronal subpopulations.
Common molecular mechanisms that possibly cross talk and direct the clustering of hindbrain ION and fMN nuclei
So far, few factors have been proposed to direct the layering process of ION or fMN. In rig1(retinoid inducible gene 1)/Robo-3 (roundabout homolog) receptor mutant mice, ION organization is affected, but no description of fMN was reported (Marillat et al., 2004). Interestingly, reeler mice that are deficient in the extracellular signaling molecule reelin (Terashima et al., 1993) show defects in both ION and fMN organization. Time schedules for the generation, axon formation, and migration of fMNn are similar both in reeler and wild-type mice. Signaling pathways responsible for the late abnormal cytoarchitectures (i.e., lamination of the neocortex, foliation of the cerebellum, and clusterization of IONn and fMNn) through reelin are still unknown. Nevertheless, reelin could act in partnership with various receptors. Among them, apolipoprotein E receptor 2/very-low-density lipoprotein receptor (Rossel et al., 2005) and cadherin-related neuronal receptors family of protocadherins (Jossin, 2004) were recently proposed. To date, no reelin expression has been detected in ION (Schiffmann et al., 1997), and the expression domain of protocadherins in the developing hindbrain has been poorly investigated. Phenotypes resembling that of reeler are seen with mutations in mdab1 (mouse disabled 1), cyclin-dependent kinase 5 (cdk5), and p35. Mutant mice lacking p35 show lamination defects, with a more severe phenotype than trio mutant mice (Ohshima et al., 2002), and a complete lack of ION has been reported in cdk5 mutant mice (Ko et al., 2001). On the one hand, Cdk5 and p35, respectively, the catalytic and regulatory subunits of a serine/threonine kinase, could operate in a common signaling pathway with mDab and Reelin. On the other hand, it has been proposed that Cdk5 could phosphorylate Trio, increasing its GEF activity and leading to a localized activation of Rac (Xin et al., 2004). Nevertheless, its physiological relevance remains to be determined. Ohshima et al. (2002) showed that Cdk5/p35 contributes synergistically with Reelin/Dab1 to the positioning of fMNn and IONn in the developing mouse hindbrain. Thus, Trio, Reelin, Cdk5, and mDab1 could act altogether to coordinate neuronal migration and rearrangements of IONn and fMNn. In trio mutant mice, similar phenotypic defects have been retrieved in both ION and fMN, suggesting that they could use a closely related transduction pathway to get organized.
Cadherin-11 may act as a partner of Trio and Rho GTPase activation for ION and fMN to acquire their proper lamellation
It has been shown in Drosophila that the Abelson tyrosine kinase, Trio, and Enabled interact with the Netrin receptor Frazzled (Forsthoefel et al., 2005). Neither Abl nor DCC could coimmunoprecipitate in ION nor fMN extracts with Trio. It has been proposed but not demonstrated in neurons that the Trio–GEF family might interact with catenin/cadherin cell adhesion complexes (Lanier and Gertler, 2000). In myoblasts, a multiproteic complex containing M-cadherin and Trio has been detected recently (Charrasse et al., 2007). In the present report, we show that, interestingly and coincidently, in trio mutant mice, the disorganized portions of the ION are rostral and are those that express Cadherin-11. We also demonstrate that Cadherin-11 is found in a complex with Trio in ION and fMN extracts. Because Cadherin-11 is still expressed in trio mutant mice, the abnormal lamellation observed in mutant mice is not attributable to an abnormal expression of adhesion molecules in neuronal clusters but possibly to the absence of Trio recruitment that cannot further locally activate Rho GTPases and direct the proper ION and fMNn migrations. The signaling pathway involved in the IONn and fMNn movements involves Trio itself as a key regulator factor to modulate the dynamic of the actin cytoskeleton via signaling cascades involving Rho GTPases. It is interesting to underline that the association of Trio with protein partners varies according to the cell type because no association of Trio with DCC or Abl could be detected, although both proteins are present in E15 hindbrain extracts. Nevertheless, we cannot exclude that still unidentified Trio partners involved in its activation might exist. They could account for compensatory effects observed in Cadherin-11 KO mice, in which analysis could not reveal any phenotypic differences in the hindbrain organization or in the spatiotemporal expression of other cadherins (Manabe et al., 2000) (supplemental Fig. 1, available at www.jneurosci.org as supplemental material).
Rac1 is involved downstream of Trio to allow ION and fMN organization
Trio mutant mice show severe anomalies of lamellation, whereas RhoG mutant mice show a normal developmental process of the ION and fMN that acquire a normal mature aspect. The comparison between the phenotypes of trio and RhoG mutant mice leads us to favor Rac1 as being involved in ION and fMN maturation rather than Cdc42. We analyzed the activation state of Rac1 and Cdc42 in wild-type, RhoG, and Trio mutant mice. The activation of Rac1 and Cdc42 is not affected at E15 in RhoG mutant mice compared with wild-type embryos. Thus, RhoG is not involved in the signaling pathway that directs ION and fMN mature organization downstream of Trio. Because Trio can activate Cdc42 but only after RhoG activation (Gauthier-Rouviere et al., 1998), a possible Trio target that remains involved in the parcellation of both ION and fMN is Rac1 through Trio–GEF-D1. The analysis of the rate of active Rac1 shows a 72% decrease in trio mutant mice, whereas the rate of active Cdc42 remains constant in both wild-type and trio mutant mice. In conclusion, Trio affects ION and fMN cytoarchitectonic maturation through Rac1 activation. It will be interesting to analyze whether specific isoforms of Trio (McPherson et al., 2005) are expressed in various ION and fMN subregions and could regulate distinct and localized activations of Rac1 in particular. In Caenorhabditis elegans, the Rho–GEF-1 domain of UNC-73/Trio influences cell and growth cone migrations, whereas the Rho–GEF-2 domain regulates pharynx and vulva musculature, as well as synaptic neurotransmission (Steven et al., 2005). Thus, studying the expression and the role of the various Trio isoforms could bring various data about their physiological involvement. Interestingly, it has been reported that Cadherin-11 overexpression increases axon defasciculation (Marthiens et al., 2005), a phenotype similar to the one reported when the Rho/Rho kinase pathway is blocked in other cell types (Causeret et al., 2004). We propose that specific GTPases could be activated or repressed after Trio recruitment by neuronal clusters. This mechanism could be involved in micronucleokinesis and subtle migrations to organize ION and fMN nuclei.
Current model: specific regulation of distinct Rho and Rac GTPases activity in specific IONn and fMNn clusters
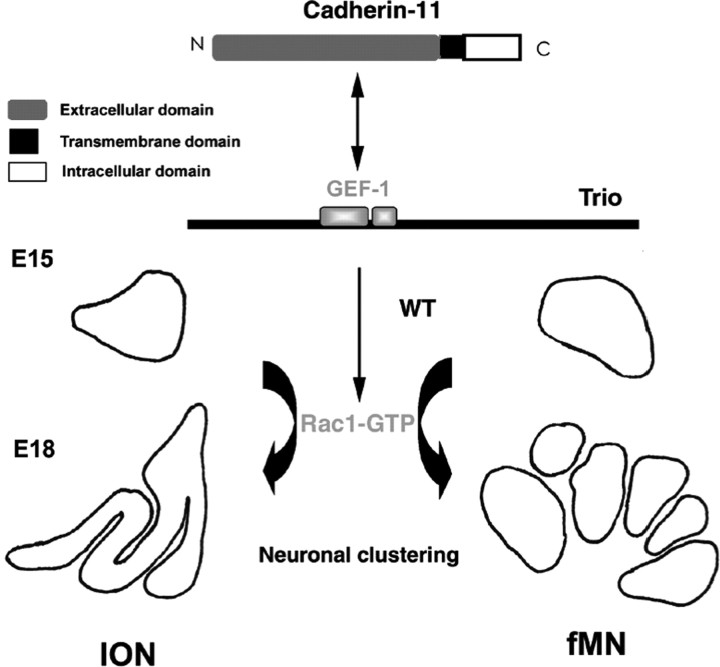
We propose that Trio could act as a molecular linker to recruit upstream and downstream partners and form a multiproteic complex to direct local rearrangement of the cytoskeleton and consequently neuronal organizations, including ION and fMN clustering in the hindbrain (model schematized in Fig. 9). It has been shown recently that several signaling pathways are activated by cadherin-mediated cell–cell contacts, particularly those involving the Rho family of small GTPases (Erasmus and Braga, 2006). Combinatorial expression of Cadherins subdivides the embryonic brain in functional subunits (Wheelock and Johnson, 2003). Various Cadherins can regulate the activity of Rho GTPases, probably through recruitment or regulation of distinct Rho–GEFs, altogether with Rho–GAPs. We propose that Cadherin-11 could be a candidate for the activation of Rac1 through Trio recruitment to allow ION and fMN lamellation. Cadherin-11 is expressed in the most dorsal ION mass, and it remains expressed in the dorsal lamellae once the IO is organized. It is interesting to underline that most defects observed in trio mutant mice concern the rostral ION, which contains Cadherin-11-expressing neurons. Because it is expressed before the settlement of IONn or fMNn to their final position, Cadherin-11 could prefigure neuronal identity and positioning of some clusters. Among the biochemical interactions of Trio with various Cadherins tested, only Cadherin-11 could be detected in a complex with Trio–GEF. Nonetheless, it remains to be demonstrated that Cadherin-11 is indeed responsible for Rac1 activation in vivo in specific clusters during the lamellation process.
Figure 9.
Molecular cascade for clustering in the hindbrain. Cadherin-11 is expressed at E15 in both ION and fMN. Cadherin-11 can interact in vivo with Trio, either directly or in a multiproteic complex with other unidentified partners. This interaction presumably activates the Rac1-specific Trio–GEF-1 in Cadherin-11-expressing clusters that results in Rac1 activation, which is required for the proper neuronal clustering to occur between E15 and P0 in both ION and fMN.
Footnotes
This work was partially funded by the Association pour la Recherche sur le Cancer (ARC 3697), the French Ministry of Research, the Association Française pour la Recherche contre les Myopathies (AFM), and Inserm. N.O. received a fellowship from AFM. We are grateful to Drs. Isabelle Le Roux and Odile De Lapeyriere for their critical reading of this manuscript and to Dr. Franck Perez for the morphing video. We thank Annie Goldman for proofreading in English.
References
- Azizi SA, Woodward DJ. Inferior olivary nuclear complex of the rat: morphology and comments on the principles of organization within the olivocerebellar system. J Comp Neurol. 1987;263:467–484. doi: 10.1002/cne.902630402. [DOI] [PubMed] [Google Scholar]
- Backer S, Sakurai T, Grumet M, Sotelo C, Bloch-Gallego E. Nr-CAM and TAG-1 are expressed in distinct populations of developing precerebellar and cerebellar neurons. Neuroscience. 2002;113:743–748. doi: 10.1016/s0306-4522(02)00221-x. [DOI] [PubMed] [Google Scholar]
- Barrett C, Guthrie S. Expression patterns of the netrin receptor UNC5H1 among developing motor neurons in the embryonic rat hindbrain. Mech Dev. 2001;106:163–166. doi: 10.1016/s0925-4773(01)00415-4. [DOI] [PubMed] [Google Scholar]
- Bateman J, Van Vactor D. The Trio family of guanine-nucleotide-exchange factors: regulators of axon guidance. J Cell Sci. 2001;114:1973–1980. doi: 10.1242/jcs.114.11.1973. [DOI] [PubMed] [Google Scholar]
- Bellanger JM, Lazaro JB, Diriong S, Fernandez A, Lamb N, Debant A. The two guanine nucleotide exchange factor domains of Trio link the Rac1 and the RhoA pathways in vivo. Oncogene. 1998;16:147–152. doi: 10.1038/sj.onc.1201532. [DOI] [PubMed] [Google Scholar]
- Bloch-Gallego E, Ezan F, Tessier-Lavigne M, Sotelo C. Floor plate and netrin-1 are involved in the migration and survival of inferior olivary neurons. J Neurosci. 1999;19:4407–4420. doi: 10.1523/JNEUROSCI.19-11-04407.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourrat F, Sotelo C. Early development of the rat precerebellar system: migratory routes, selective aggregation and neuritic differentiation of the inferior olive and lateral reticular nucleus neurons. An overview. Arch Ital Biol. 1990;128:151–170. [PubMed] [Google Scholar]
- Bourrat F, Sotelo C. Relationships between neuronal birthdates and cytoarchitecture in the rat inferior olivary complex. J Comp Neurol. 1991;313:509–521. doi: 10.1002/cne.903130311. [DOI] [PubMed] [Google Scholar]
- Braga V. Cell-cell adhesion and signalling. Curr Opin Cell Biol. 2002;14:546–556. doi: 10.1016/s0955-0674(02)00373-3. [DOI] [PubMed] [Google Scholar]
- Causeret F, Hidalgo-Sanchez M, Fort P, Backer S, Popoff MR, Gauthier-Rouviere C, Bloch-Gallego E. Distinct roles of Rac1/Cdc42 and Rho/Rock for axon outgrowth and nucleokinesis of precerebellar neurons toward netrin 1. Development. 2004;131:2841–2852. doi: 10.1242/dev.01162. [DOI] [PubMed] [Google Scholar]
- Charrasse S, Causeret M, Comunale F, Bonet-Kerrache A, Gauthier-Rouviere C. Rho GTPases and cadherin-based cell adhesion in skeletal muscle development. J Muscle Res Cell Motil. 2003;24:309–313. [PubMed] [Google Scholar]
- Charrasse S, Comunale F, Fortier M, Portales-Casamar E, Debant A, Gauthier-Rouviere C. M-Cadherin activates Rac1 GTPase through the Rho-GEF Trio during myoblast fusion. Mol Biol Cell. 2007;18:1734–1743. doi: 10.1091/mbc.E06-08-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chedotal A, Pourquie O, Ezan F, San Clemente H, Sotelo C. BEN as a presumptive target recognition molecule during the development of the olivocerebellar system. J Neurosci. 1996;16:3296–3310. doi: 10.1523/JNEUROSCI.16-10-03296.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debant A, Serra-Pages C, Seipel K, O'Brien S, Tang M, Park SH, Streuli M. The multidomain protein Trio binds the LAR transmembrane tyrosine phosphatase, contains a protein kinase domain, and has separate rac-specific and rho-specific guanine nucleotide exchange factor domains. Proc Natl Acad Sci USA. 1996;93:5466–5471. doi: 10.1073/pnas.93.11.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erasmus JC, Braga VM. Rho GTPase activation by cell-cell adhesion. Methods Enzymol. 2006;406:402–415. doi: 10.1016/S0076-6879(06)06029-0. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- Forsthoefel DJ, Liebl EC, Kolodziej PA, Seeger MA. The Abelson tyrosine kinase, the Trio GEF and Enabled interact with the Netrin receptor Frazzled in Drosophila. Development. 2005;132:1983–1994. doi: 10.1242/dev.01736. [DOI] [PubMed] [Google Scholar]
- Garel S, Garcia-Dominguez M, Charnay P. Control of the migratory pathway of facial branchiomotor neurones. Development. 2000;127:5297–5307. doi: 10.1242/dev.127.24.5297. [DOI] [PubMed] [Google Scholar]
- Gauthier-Rouviere C, Vignal E, Meriane M, Roux P, Montcourier P, Fort P. RhoG GTPase controls a pathway that independently activates Rac1 and Cdc42Hs. Mol Biol Cell. 1998;9:1379–1394. doi: 10.1091/mbc.9.6.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilland E, Baker R. Conservation of neuroepithelial and mesodermal segments in the embryonic vertebrate head. Acta Anat (Basel) 1993;148:110–123. doi: 10.1159/000147530. [DOI] [PubMed] [Google Scholar]
- Jossin Y. Neuronal migration and the role of reelin during early development of the cerebral cortex. Mol Neurobiol. 2004;30:225–251. doi: 10.1385/MN:30:3:225. [DOI] [PubMed] [Google Scholar]
- Kaibuchi K, Kuroda S, Fukata M, Nakagawa M. Regulation of cadherin-mediated cell-cell adhesion by the Rho family GTPases. Curr Opin Cell Biol. 1999;11:591–596. doi: 10.1016/s0955-0674(99)00014-9. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Matsunami H, Takeichi M. Expression of cadherin-11 delineates boundaries, neuromeres, and nuclei in the developing mouse brain. Dev Dyn. 1996;206:455–462. doi: 10.1002/(SICI)1097-0177(199608)206:4<455::AID-AJA11>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Ko J, Humbert S, Bronson RT, Takahashi S, Kulkarni AB, Li E, Tsai LH. p35 and p39 are essential for cyclin-dependent kinase 5 function during neurodevelopment. J Neurosci. 2001;21:6758–6771. doi: 10.1523/JNEUROSCI.21-17-06758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier LM, Gertler FB. From Abl to actin: Abl tyrosine kinase and associated proteins in growth cone motility. Curr Opin Neurobiol. 2000;10:80–87. doi: 10.1016/s0959-4388(99)00058-6. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Keynes R. Segmental patterns of neuronal development in the chick hindbrain. Nature. 1989;337:424–428. doi: 10.1038/337424a0. [DOI] [PubMed] [Google Scholar]
- Manabe T, Togashi H, Uchida N, Suzuki SC, Hayakawa Y, Yamamoto M, Yoda H, Miyakawa T, Takeichi M, Chisaka O. Loss of cadherin-11 adhesion receptor enhances plastic changes in hippocampal synapses modifies behavioral responses. Mol Cell Neurosci. 2000;15:534–546. doi: 10.1006/mcne.2000.0849. [DOI] [PubMed] [Google Scholar]
- Marillat V, Sabatier C, Failli V, Matsunaga E, Sotelo C, Tessier-Lavigne M, Chedotal A. The slit receptor Rig-1/Robo3 controls midline crossing by hindbrain precerebellar neurons and axons. Neuron. 2004;43:69–79. doi: 10.1016/j.neuron.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Marthiens V, Gavard J, Padilla F, Monnet C, Castellani V, Lambert M, Mege RM. A novel function for cadherin-11 in the regulation of motor axon elongation and fasciculation. Mol Cell Neurosci. 2005;28:715–726. doi: 10.1016/j.mcn.2004.12.001. [DOI] [PubMed] [Google Scholar]
- McPherson CE, Eipper BA, Mains RE. Multiple novel isoforms of Trio are expressed in the developing rat brain. Gene. 2005;347:125–135. doi: 10.1016/j.gene.2004.12.028. [DOI] [PubMed] [Google Scholar]
- O'Brien SP, Seipel K, Medley QG, Bronson R, Segal R, Streuli M. Skeletal muscle deformity and neuronal disorder in Trio exchange factor-deficient mouse embryos. Proc Natl Acad Sci USA. 2000;97:12074–12078. doi: 10.1073/pnas.97.22.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima T, Ogawa M, Takeuchi K, Takahashi S, Kulkarni AB, Mikoshiba K. Cyclin-dependent kinase 5/p35 contributes synergistically with Reelin/Dab1 to the positioning of facial branchiomotor and inferior olive neurons in the developing mouse hindbrain. J Neurosci. 2002;22:4036–4044. doi: 10.1523/JNEUROSCI.22-10-04036.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla F, Broders F, Nicolet M, Mege RM. Cadherins M, 11, and 6 expression patterns suggest complementary roles in mouse neuromuscular axis development. Mol Cell Neurosci. 1998;11:217–233. doi: 10.1006/mcne.1998.0681. [DOI] [PubMed] [Google Scholar]
- Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 1996;84:309–320. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- Price SR, De Marco Garcia NV, Ranscht B, Jessell TM. Regulation of motor neuron pool sorting by differential expression of type II cadherins. Cell. 2002;109:205–216. doi: 10.1016/s0092-8674(02)00695-5. [DOI] [PubMed] [Google Scholar]
- Rossel M, Loulier K, Feuillet C, Alonso S, Carroll P. Reelin signaling is necessary for a specific step in the migration of hindbrain efferent neurons. Development. 2005;132:1175–1185. doi: 10.1242/dev.01683. [DOI] [PubMed] [Google Scholar]
- Saburi S, McNeill H. Organising cells into tissues: new roles for cell adhesion molecules in planar cell polarity. Curr Opin Cell Biol. 2005;17:482–488. doi: 10.1016/j.ceb.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Schiffmann SN, Bernier B, Goffinet AM. Reelin mRNA expression during mouse brain development. Eur J Neurosci. 1997;9:1055–1071. doi: 10.1111/j.1460-9568.1997.tb01456.x. [DOI] [PubMed] [Google Scholar]
- Steven R, Zhang L, Culotti J, Pawson T. The UNC-73/Trio RhoGEF-2 domain is required in separate isoforms for the regulation of pharynx pumping and normal neurotransmission in C. elegans. Genes Dev. 2005;19:2016–2029. doi: 10.1101/gad.1319905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer M, Lumsden A, Ariza-McNaughton L, Bradley A, Krumlauf R. Altered segmental identity and abnormal migration of motor neurons in mice lacking Hoxb-1. Nature. 1996;384:630–634. doi: 10.1038/384630a0. [DOI] [PubMed] [Google Scholar]
- Suzuki SC, Inoue T, Kimura Y, Tanaka T, Takeichi M. Neuronal circuits are subdivided by differential expression of type-II classic cadherins in postnatal mouse brains. Mol Cell Neurosci. 1997;9:433–447. doi: 10.1006/mcne.1997.0626. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, Kawauchi D, Nishida K, Murakami F. Classic cadherins regulate tangential migration of precerebellar neurons in the caudal hindbrain. Development. 2006;133:1923–1931. doi: 10.1242/dev.02354. [DOI] [PubMed] [Google Scholar]
- Terashima T, Kishimoto Y, Ochiishi T. Musculotopic organization of the facial nucleus of the reeler mutant mouse. Brain Res. 1993;617:1–9. doi: 10.1016/0006-8993(93)90605-m. [DOI] [PubMed] [Google Scholar]
- Vigorito E, Bell S, Hebeis BJ, Reynolds H, McAdam S, Emson PC, McKenzie A, Turner M. Immunological function in mice lacking the Rac-related GTPase RhoG. Mol Cell Biol. 2004;24:719–729. doi: 10.1128/MCB.24.2.719-729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassef M, Chedotal A, Cholley B, Thomasset M, Heizmann CW, Sotelo C. Development of the olivocerebellar projection in the rat. I. Transient biochemical compartmentation of the inferior olive. J Comp Neurol. 1992;323:519–536. doi: 10.1002/cne.903230405. [DOI] [PubMed] [Google Scholar]
- Wheelock MJ, Johnson KR. Cadherins as modulators of cellular phenotype. Annu Rev Cell Dev Biol. 2003;19:207–235. doi: 10.1146/annurev.cellbio.19.011102.111135. [DOI] [PubMed] [Google Scholar]
- Wild JM, Zeigler HP. Central representation and somatotopic organization of the jaw muscles within the facial and trigeminal nuclei of the pigeon (Columba livia) J Comp Neurol. 1980;192:175–201. doi: 10.1002/cne.901920112. [DOI] [PubMed] [Google Scholar]
- Xin X, Ferraro F, Back N, Eipper BA, Mains RE. Cdk5 and Trio modulate endocrine cell exocytosis. J Cell Sci. 2004;117:4739–4748. doi: 10.1242/jcs.01333. [DOI] [PubMed] [Google Scholar]
- Yap AS, Kovacs EM. Direct cadherin-activated cell signaling: a view from the plasma membrane. J Cell Biol. 2003;160:11–16. doi: 10.1083/jcb.200208156. [DOI] [PMC free article] [PubMed] [Google Scholar]