Abstract
Palmitoylation affects the trafficking, stability, aggregation, and/or functional activity of a substantial number of neuronal proteins. We identified mutations in dHIP14, the Drosophila homolog of the human palmitoyl transferase, Huntingtin-interacting protein 14 (HIP14). HIP14 was previously reported to localize primarily to Golgi and to palmitoylate the neuronal proteins synaptosome-associated protein 25 (SNAP-25), PSD-95 (postsynaptic density-95), GAD65, Synaptotagmin, and Huntingtin in mammalian neurons. We find dHIP14 to be an essential maternal effect gene required for photoreceptor synaptic transmission and for proper in vivo expression of the palmitoylated presynaptic proteins SNAP-25 and cysteine string protein. In non-neuronal cells in the fly, dHIP14 protein is found in Golgi. However, in fly neurons, we find dHIP14 primarily in presynaptic terminals, something we also observe with HIP14. In mammalian neurons, we also find a significant fraction of HIP14 colocalizing with a synaptic vesicle marker. Based on localization of the palmitoyl transferase HIP14 within the presynaptic nerve terminal, we propose palmitoylation as a possible mechanism that may be operating to rapidly regulate synaptic efficacy.
Keywords: Huntingtin, plasticity, palmitoylation, Drosophila, photoreceptor, membrane trafficking
Introduction
The importance of palmitoylation in diverse biological processes including pattern formation, immune system signaling, sensory transduction, cell growth, and especially neuronal development and physiology has become increasingly apparent in recent years (for review, see Huang and El-Husseini, 2005; Resh, 2006; Linder and Deschenes, 2007). Aberrant palmitoylation has also been implicated in human diseases including cardiovascular disease (Ferri et al., 2005), lung disease (ten Brinke et al., 2002), cancer, (Ducker et al., 2004; Zhou et al., 2004), and neurodegenerative disease (Yanai et al., 2006). Palmitoylation involves the attachment of the 16 carbon saturated fatty acid palmitate to specific cysteines via a thioester linkage. Unlike other forms of acylation, such as prenylation or myristoylation, S-palmitoylation is reversible, making palmitoylation a potentially dynamic posttranslational mechanism for regulating protein trafficking, stability, aggregation, and functional activity.
In a prominent example of the modulatory activity of palmitoylation on neuronal function, postsynaptic density-95 (PSD-95) was demonstrated to regulate synaptic strength via activity-dependent palmitate cycling in hippocampal neurons (El-Husseini et al., 2002). Trafficking and/or functional activity of other synaptic proteins including AMPA receptors (Hayashi et al., 2005), the GABAA receptor (Keller et al., 2004), the β2A subunit of Ca+2 channels (Hurley et al., 2000), and the α7 nicotinic acetylcholine receptor (Drisdel et al., 2004) has also been demonstrated in cultured neurons to depend on palmitoylation. Because palmitoylation is thought to occur in the endoplasmic reticulum (ER) and/or Golgi, the timescale on which palmitoylation is expected to exert functional effects on synapses is large, with a delay of minutes to hours for transport to terminals.
The best characterized mammalian palmitoyl transferase, Huntingtin-interacting protein 14 (HIP14), was originally identified based on its ability to interact with Huntingtin, the causative agent of Huntingtin's disease (Singaraja et al., 2002). HIP14 has since been shown to directly palmitoylate Huntingtin at cysteine 214, and the palmitoylation state of Huntingtin was correlated with the tendency of polyglutamine-expanded Huntingtin to form the aggregates associated with the disease (Yanai et al., 2006). Furthermore, HIP14 shows a differential binding affinity for polyglutamine expanded versus normal Huntingtin (Singaraja et al., 2002). These observations suggest palmitoylation may play a critical role in the pathology of Huntington's disease.
Here, we report the characterization of a Drosophila HIP14 (dHIP14) mutant. dHIP14 is an essential maternal effect gene that when mutant results in lethality at either the first instar (maternal contribution absent) or pharate adult (maternal contribution present) developmental stages. Characterization of dHIP14 protein expression revealed ubiquitous expression in all tissues examined with subcellular localization to Golgi in non-neuronal cells and preferential localization to presynaptic terminals in neurons. Like dHIP14, human HIP14 also localizes to presynaptic terminals in Drosophila neurons. In cultured mammalian hippocampal neurons, there is also partial colocalization of human HIP14 with synaptic vesicle markers. The palmitoylated proteins synaptosome-associated protein 25 (SNAP-25) and cysteine string protein (CSP) were mislocalized and/or exhibited reduced expression in dHIP14 mutants, indicating they are potential in vivo palmitoylation substrates of dHIP14. Our results suggest dHIP14 may regulate synapse function within the presynaptic terminal via palmitoylation.
Materials and Methods
Fly stocks.
The following stocks were obtained from the indicated sources: P{EPgy2}CG6017EY09853, P{Rh1-GAL4}2, and P{UAS-FLP1.D}JD1 (Bloomington Drosophila Stock Center, Bloomington, IN); P{nos-GAL4} (Ruth Lehmann, Skirball Institute, New York, NY) (Van Doren et al., 1998); l(3)72Ad alleles (Jim Kennison, National Institutes of Health, Bethesda, MD) (Brizuela et al., 1994) and Dan Kalderon (Columbia University, New York, NY) (Melendez et al., 1995); and cn bw sp chromosome (Gerry Rubin Laboratory, Ashburn, VA). The P{nos-GAL4}, P{UAS-FLP.D}JD1 chromosome (2) was generated by meiotic recombination.
Mutagenesis.
Mutagenesis and screening for photoreceptor synaptic transmission defective mutants was performed as described previously (Babcock et al., 2003) except the strains used were yw; P{w+}FRT79isogenized and yw; P{w+}FRT79 GMR-hid CL3L P{ry+} ey-FLP/TM6,y+.
Electroretinograms.
Electroretinograms (ERGs) were performed by giving 1 s pulses of light to dark adapted flies in which a sharp electrode filled with 3 m KCl was placed in the thorax and a reference electrode filled with 85 mm NaCl was placed on the surface of the eye.
Molecular biology.
The pCasPeR4 dHIP14YFP genomic rescue/expression construct was generated from genomic DNA obtained from BACR18K24 as a 6.7 kb KpnI fragment cloned into the KpnI site of pBluescript (pBS). A NotI restriction site was introduced by PCR to replace the dHIP14 TAA stop codon, followed by insertion of a ∼800 bp NotI flanked yellow fluorescent protein (YFP) encoding PCR fragment into the NotI site to create pBS dHIP14YFP. The 5.6 kb KpnI/BglII fragment from pBS dHIP14YFP was then cloned into the KpnI/BamHI sites of pCasPeR4 to generate pCasPeR4 dHIP14YFP. Four repeats of a linker encoding the amino acids GGS were inserted amino-terminal to the coding sequence of YFP when the initial version of pCasPeR 4 dHIP14YFP without the linker was found not to be fluorescent nor detectable with anti-green fluorescent protein (GFP) antibodies. pUAST-dHIP14TOM was generated by first inserting a 2.3 kb EcoRI fragment containing the dHIP14 ORF from LD10758 into the EcoRI site of pUAST to create pUAST-dHIP14. Next, a ∼1.5 kb NotI flanked tdTomato (Shaner et al., 2004) PCR fragment was inserted into the NotI site of pCasPeR4 dHIP14YFP to create pCasper4 dHIP14TOM. Finally, a 5.2 kb SfiI/BglII fragment from pCasPeR4 dHIP14TOM was cloned into the SfiI/BglII sites of pUAST-dHIP14 to create pUAST-dHIP14TOM. The 4× GGS linker was also included at the amino terminus of tdTomato. pUAST-HIP14GFP was generated by inserting a 2.8 kb EcoRI-partial/NotI fragment from pClneo-HIP14GFP (Singaraja et al., 2002) into the EcoRI/NotI sites of pUAST to create pUAST-HIP14GFP.
Immunohistochemistry.
First and third instar larvae were filleted in HL3 (Stewart et al., 1994) and attached to Sylgard-coated slides using Nexaband glue (first instar) or minutien pins (third instar), fixed in freshly prepared 4% paraformaldehyde in PBS for 10 min at room temperature (RT), blocked for 1 h in PBS plus 0.5% Triton X-100 plus 5% normal goat serum (PBSTNGS) at RT, incubated with primary antibody overnight (O/N) at 4°C, washed three times 5 min in PBS, incubated with secondary antibody 1–2 h at RT, washed three times 5 min in PBS, before confocal microscopy using an Achroplan 20×/0.5 or 40×/0.8 water immersion lens, or Plan Apochromat 63×/1.4 oil lens, using a Zeiss 510 Meta (Zeiss, Oberkochen, Germany) camera and acquisition software. First instar larvae shown in Figure 3 were fixed for 3 min, and primary antibody incubation was for 1 h at RT. For fly head cryostat sections, flies were decapitated, their proboscis and air sacs were removed, and head capsules were fixed in freshly prepared 4% paraformaldehyde for 3 h at 4°C, incubated in sucrose in PBS (5%, 10 min; 12.5%, 10 min; 25% O/N), placed in OCT, frozen in liquid N2, cut into 10 μm cryostat sections, and air dried for >30 min. Slides were processed for immunolabeling as described above. Primary rat hippocampal neuron cultures were plated on coverslips that were fixed for 10 min in freshly prepared 4% paraformaldehyde and processed as above. Primary antibodies and dilutions were as follows: mouse anti-Chaoptin (24B10), 1:50; mouse anti-Synapsin (3C11), 1:50; mouse anti-Dlg (4F3), 1:50; mouse anti-CSP (6D6), 1:50; mouse anti-Bruchpilot (nc82), 1:100; mouse anti-SV2, 1:50 (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA); rabbit anti-Synaptotagmin (Littleton et al., 1993), 1:2000; rabbit anti-SNAP-25 (Rao et al., 2001), 1:200; mouse anti-Golgi (7H6D7C2) (Stanley et al., 1997) (345867; Calbiochem, La Jolla, CA), 1:500; rabbit anti-DsRed (recognizes tdTomato) (632496; Clontech, Mountain View, CA), 1:1000; rabbit anti-GFP-Alexa 488 (recognizes YFP) (Invitrogen, San Diego, CA; A-21311), 1:250; rat anti-neuronal synaptobrevin (Wu et al., 1999), 1:250. Secondary antibodies were as follows: goat anti-mouse Cy3 and goat anti-rabbit Cy3 (Jackson ImmunoResearch, West Grove, PA), and goat anti-mouse Alexa 488 and goat anti-rat Alexa 647 (Invitrogen). All secondary antibodies were prepared per the manufacturer's instructions and used at 1:400. For the confocal imaging in Figure 6, settings were established on control animals and maintained on dHIP14ex11 mutants.
Figure 3.
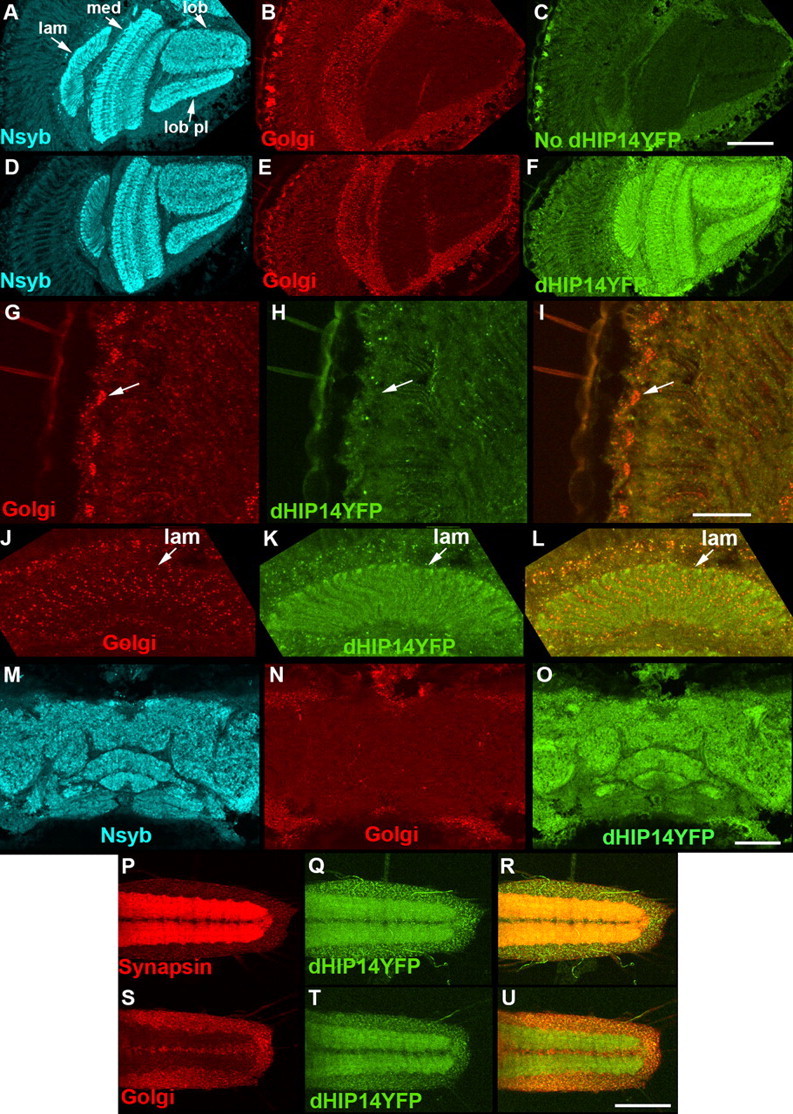
dHIP14YFP localizes to synapses in optic lobe, central brain, and larval ventral nerve cord neurons. A–F, Cryostat sections of the optic lobes of a control fly (No dHIP14YFP) (A–C) and a dHIP14YFP fly (D–F) triple-labeled with the synaptic vesicle marker neuronal-synaptobrevin (N-syb) (A, D), a Golgi marker (B, E), and anti-YFP (C, F). dHIP14YFP distributes to the synaptic neuropil of the optic lobe in a pattern very similar to the synaptic vesicle marker N-syb and clearly distinct from the Golgi marker. Background levels of fluorescence for the anti-YFP antibody can be observed in C. lam, Lamina; med, medula; lob, lobula; lob pl, lobula plate. Scale bar, 50 μm. G–L, High-resolution images of cryostat sections of the cell body (G–I) and lamina (J–L) regions of photoreceptors of a dHIP14YFP fly double-immunolabeled with a Golgi marker (G, J), anti-YFP (H, K), and the overlay (I, L). The arrow (G–I) points to the same cell body in each. dHIP14YFP is not detected in either the photoreceptor or presumptive lamina cell body Golgi, but is present in non-neuronal cell bodies adjacent to the lamina. Scale bar, 20 μm. M–O, Cryostat sections of the central brain of a dHIP14YFP fly triple-labeled for the synaptic vesicle marker N-syb (M), a Golgi marker (N), and anti-YFP (O). Similar to the optic lobe, the vast majority of dHIP14YFP localizes in the pattern of the synaptic vesicle marker, but note the overlap with the Golgi marker in adjacent non-neuronal regions. Scale bar, 50 μm. P–R, First instar ventral nerve cord of a dHIP14YFP larvae (VNC) labeled with Synapsin (P), direct dHIP14YFP fluorescence (Q), and the overlay (R). The majority of dHIP14YFP colocalizes with the synaptic vesicle marker Synapsin in the synaptic neuropil region of the VNC. S–U, First instar VNC of a dHIP14YFP larvae labeled with a Golgi marker (S), direct dHIP14YFP fluorescence (T), and the overlay (U). The Golgi marker is restricted to the cell body region of the VNC, whereas a portion of dHIP14YFP is present in the cell body, but most distributes to the synaptic neuropil. Scale bar, 50 μm. Genotypes are as follows: yw; cn bw sp/cn bw sp (A–C), yw; cn bw sp/cn bw sp; dHIP14ex11, dHIP14YFP/dHIP14ex11, dHIP14YFP (D–O), yw; dHIP14ex11, dHIP14YFP/dHIP14ex11, dHIP14YFP (P–U). lam, Lamina; med, medulla; lob, lobula; lob pl, lobula plate.
Figure 6.
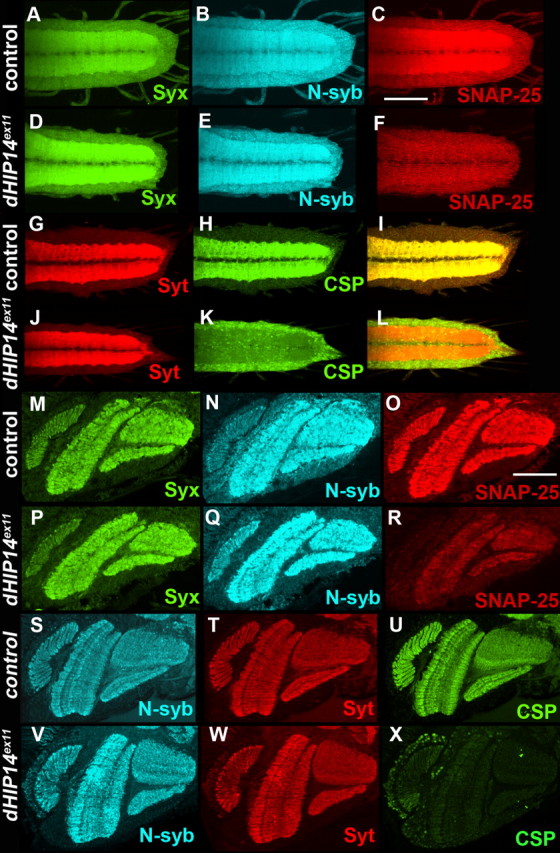
Subcellular localization and abundance of SNAP-25 and CSP are aberrant in dHIP14 mutants. A–F, First instar larval ventral nerve cords of control (A–C) and a dHIP14 mutant (D–F) triple-labeled for Syntaxin (A, D), neuronal-synaptobrevin (B, E), and SNAP-25 (C, F). Syntaxin and N-syb expression are not affected in the dHIP14 mutant, but SNAP-25 is significantly reduced and more diffuse. G–L, First instar larval ventral nerve cords of control (G–I) and dHIP14 mutant (J–L) double-labeled for the synaptic vesicle marker Synaptotagmin (G and J), cysteine string protein (H, K), and the overlay (I, L). Synaptotagmin localization to the synaptic neuropil is not affected in the dHIP14 mutant, but CSP is mislocalized to the cell body region of VNC neurons. Scale bar, 50 μm. M–R, Cryostat sections of optic lobes of control (M–R) and a dHIP14 mutant triple-labeled for Syntaxin (M, P), N-syb (N, Q), and SNAP-25 (O, R). Syntaxin and N-syb expression are unaffected in the dHIP14 mutant, whereas SNAP-25 shows a severe reduction. S–X, Cryostat sections of optic lobes of control (S–U) and a dHIP14 mutant (V–X) triple-labeled for N-syb (S, V), Synaptotagmin (T, W), and CSP (U, X). N-syb and Synaptotagmin expression are not affected in the dHIP14 mutant, whereas CSP is almost completely absent. Scale bar, 50 μm. Genotypes are as follows: yw; nos-GAL4, UAS-FLP or +/+; FRT79/FRT79 from yw; nos-GAL4, UAS-FLP/+; FRT79 ovoD 3L/FRT79 females (A–C, G–I), yw; nos-GAL4, UAS-FLP or +/+; FRT79 dHIP14ex11/FRT79 dHIP14ex11 from yw; nos-GAL4, UAS-FLP/+; FRT79 ovoD 3L/FRT79 dHIP14ex11 females (D–F, J–L), yw; cn bw sp/cn bw sp; FRT79/FRT79 (M–O, S–U), yw; cn bw sp/cn bw sp; FRT79 dHIP14ex11/FRT79 dHIP14ex11 (P–R, V–X).
Cell culture.
Hippocampal neuron cultures from embryonic day 18 rats were established. After 3 d in vitro, cultures were infected with lentivirus expressing HIP14GFP. Ten days later, coverslips containing the neurons were fixed in freshly prepared 4% paraformaldehyde for 10 min and processed for immunostaining as described above.
Results
Identification of dHIP14 mutants
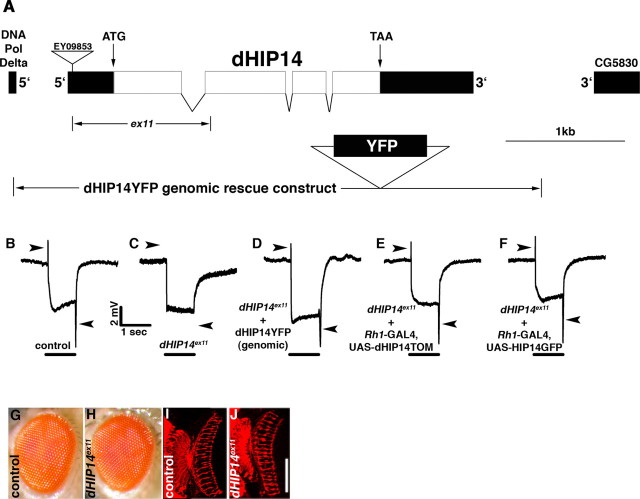
To identify previously unknown genes required for synapse function, a forward genetic screen was performed using a genetic technique that generates flies with exclusively homozygous mutant eyes (Stowers and Schwarz, 1999). In these flies, all other cell types, except for the eye, are heterozygous, and thus functionally wild type, making it possible to recover mutations in essential synaptic genes, because the fly eye is not required for viability or fertility. After ethylmethylsulfonate mutagenesis of their male parents, F1 progeny with exclusively homozygous mutant photoreceptors were screened for aberrant phototaxis behavior. Single male nonphototactic F1 flies were individually mated to appropriate females (see Materials and Methods) to generate F2 flies with homozygous mutant photoreceptors. These flies were screened by ERG measurements. Because fly ERGs have distinguishable phototransduction and synaptic components, this analysis enabled a determination of whether the nonphototactic behavior was attributable to mutations in genes required for phototransduction or synaptic transmission. Two independent noncomplementing alleles were identified. Deletion mapping using the Bloomington 3L deficiency kit narrowed the location of these alleles to the 72AB region that is uncovered by Df(3L)th102. Additional deletion mapping and complementation testing with existing alleles in the region determined that both mutants were allelic to the previously identified l(3)72Ad complementation group (Brizuela et al., 1994; Melendez et al., 1995). The l(3)72Ad complementation group mapped to a region containing six candidate genes, three of which had associated nonlethal P-elements. Each of these were used to generate excision alleles that were subsequently complementation tested for lethality against the l(3)72Ad alleles recovered in the screen. One of these nonlethal P-elements, EY09853, produced two noncomplementing excision alleles. Molecular analysis of these two excision alleles showed that the lesions were entirely contained within the predicted CG6017 gene, thus demonstrating that this is the gene corresponding to l(3)72Ad (hereafter referred to as dHIP14 based on sequence homology). The larger of the two lesions, dHIP14ex11, removed the translation start codon and the first 203 amino acids of the predicted CG6017 ORF. The location of EY09853 and the dHIP14ex11 lesion is indicated in Figure 1A.
Figure 1.
dHIP14 is required for photoreceptor synaptic transmission, but not photoreceptor development. A, Molecular and genetic map of the dHIP14 genomic region. The locations of the four exons of the dHIP14 gene including the translation start and stop codons are shown along with the terminal exons from the adjacent DNA Pol Delta and CG5830 genes. The position of the EY09853 P-element in the 5′-untranslated region used to generate the ex11 excision allele as well as the extent of dHIP14ex11 deletion is indicated. The genomic region included in the dHIP14YFP genomic rescue/expression construct is shown at the bottom. B–F, ERG recordings. B, The “on” and “off” transients corresponding to the synaptic components of the ERG (indicated with arrowheads) are present in the isoparental control. C, The synaptic components of the ERG are absent in flies with homozygous dHIP14ex11 mutant photoreceptors (arrowheads point to where the synaptic components of the ERG should be). D, The dHIP14YFP genomic rescue construct restores the synaptic components of the ERG in dHIP14ex11 mutants. E, Expression of dHIP14TOM in photoreceptors restores the synaptic components of the ERG in flies with homozygous dHIP14ex11 mutant photoreceptors. F, Expression of human HIP14GFP in photoreceptors restores the synaptic components of the ERG in flies with homozygous dHIP14ex11 mutant photoreceptors. The bars indicate the presence of the light stimulus. G, H, External surface of isoparental control (G) and dHIP14ex11 mutant eyes are indistinguishable. I, J, Cryostat sections of homozygous photoreceptor clones from isoparental control (I) and dHIP14ex11 (J) flies immunolabeled with the photoreceptor axonal marker mAb24B10. dHIP14ex11 photoreceptor axon targeting is essentially indistinguishable from the control. Scale bar, 50 μm. Genotypes are as follows: yw; FRT79 GMR-hid CL3L ey-FLP/FRT79 (B, I), yw; FRT79 GMR-hid CL3L ey-FLP/FRT79 dHIP14ex11 (C, J), yw; cn bw sp/cn bw sp; dHIP14ex11, dHIP14YFP/dHIP14ex11, dHIP14YFP (D), yw; Rh1-GAL4/UAS-dHIP14TOM; FRT79 GMR-hid CL3L ey-FLP/FRT79 dHIP14ex11 (E), yw; Rh1-GAL4/UAS-HIP14GFP; FRT79 GMR-hid CL3L ey-FLP/FRT79 dHIP14ex11.
ERGs from flies of a control and those with dHIP14ex11 homozygous mutant photoreceptors are shown in Figure 1, B and C. The synaptic components of the ERG, the so-called “on” and “off” transients, are upward and downward spikes, respectively, associated with the initiation and cessation of the light stimulus (Fig. 1B,C, arrowheads) (Heisenberg 1971). Although the slow phototransduction component is essentially normal, the synaptic components are absent from the dHIP14ex11 ERG indicating a specificity of the dHIP14ex11 mutation for synaptic transmission. In addition to having normal phototransduction, dHIP14ex11 eyes appear to have normal external morphology and photoreceptor axon development (Fig. 1G–J). These data suggest the defect in the dHIP14ex11 mutant is physiological, although we cannot rule out that subtle morphological or developmental defects could yet be revealed on more detailed analysis.
dHIP14 is an essential maternal effect gene
Lethality occurs in dHIP14ex11/dHIP14ex11 (homozygous) as well as dHIP14ex11/deletion (hemizygous) mutants at the pharate adult stage of development. These observations establish dHIP14 as an essential gene whose function is not restricted to photoreceptors. They also suggest that dHIP14ex11 is a strong loss of function, or a null, allele because there are no apparent phenotypic differences between dHIP14ex11 homozygotes and hemizygotes. Throughout larval development, dHIP14ex11 mutants exhibit normal and vigorous foraging behavior with no discernible developmental delay. When dissected out of their pupal cases on the day their heterozygous siblings in the same vial are eclosing, dHIP14ex11 homozygous or hemizygous mutants appear fully developed and are clearly alive, but are weak, uncoordinated, and cannot stand upright. ERG assays on such pharate adults are indistinguishable from flies with homozygous dHIP14ex11 mutant photoreceptors (data not shown). However, when the maternal component of dHIP14 is removed by using dHIP14ex11 germline clone female mothers, dHIP14ex11 homozygous mutants exhibit a large shift in lethal phase from pharate adult to first instar larva. Maternal and zygotic dHIP14ex11 first instar larvae eclose without a developmental delay from their egg cases, have a normal external appearance, but are sluggish and feed very little. They do, however, exhibit a robust withdrawal response when poked with a sharp wire, suggesting that a general lack of synaptic transmission is not the cause for the observed lethality and that different neurons may have differential requirements for dHIP14. Interestingly, the lethality of dHIP14ex11 homozygotes was rescued by transgenic expression of dHIP14 exclusively in the nervous system using the pan-neuronal driver elav-GAL4, indicating the essential function of dHIP14 is restricted to the nervous system.
dHIP14 is the Drosophila homolog of the palmitoyl transferase HIP14
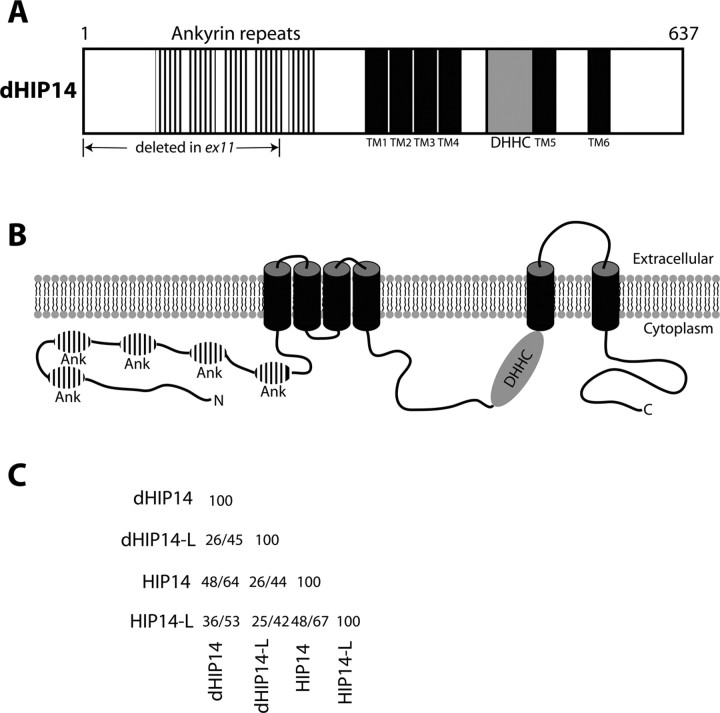
Blast searches using the 637 aa dHIP14 predicted protein revealed strong identity (48%) to human HIP14. HIP14 was identified in a yeast two-hybrid screen for proteins that interact with the Huntingtin neurodegenerative disease protein (Singaraja et al., 2002). The homology domains of this protein include five ankyrin repeats in the amino half of the protein followed by six predicted transmembrane domains (Fig. 2A,B). HIP14 has been demonstrated to be a palmitoyl transferase whose substrates include numerous neuronal proteins including SNAP-25, PSD-95, GAD65, Synaptotagmin, and Huntingtin (Huang et al., 2004; Yanai et al., 2006). A signature domain of palmitoyl transferases is a ∼50 aa region containing the sequence DHHC that is believed to form the active site. This DHHC sequence is present in dHIP14 just before the fifth predicted transmembrane domain (Fig. 2A,B, shaded in gray). The predicted membrane topology of dHIP14 is shown in Figure 2B. Note that the ankyrin repeats (which are protein–protein interaction domains that may function in substrate recognition) and the DHHC active site region are both predicted to reside on the cytoplasmic face of the lipid bilayer. Searches of the Drosophila protein database reveal one other dHIP14 ortholog (dHIP14-like) with homology throughout the length of the protein (26% identity). This ortholog is also present in humans and a table of percent identity/similarity of both the human and Drosophila homologs of HIP14 and HIP14-like is shown (Fig. 2C).
Figure 2.
Predicted domains and topology of dHIP14 protein. A, Schematic diagram of the predicted dHIP14 protein. Five ankyrin repeats are predicted in the amino terminal half of the protein followed by six predicted transmembrane domains. The presumptive active site of dHIP14 is the ∼50 aa DHHC region shaded in gray. B, Predicted topology of the dHIP14 protein in a lipid bilayer. The five ankyrin repeats and the DHHC region are both located on the cytoplasmic face of the bilayer. C, Percent identity/similarity table of Drosophila and human homologs of HIP14 and HIP14-like.
dHIP14 localizes to presynaptic terminals
As a first step in gaining insight into the mechanism by which loss of dHIP14 results in a synaptic transmission phenotype, we determined the cellular and subcellular localization of the dHIP14 protein. The approach we took was to generate transgenic flies containing a dHIP14 genomic rescue construct with a yellow fluorescent protein tag (dHIP14YFP). This construct contains 4.9 kb of genomic DNA extending from ∼500 bp upstream of the dHIP14 transcription start site to ∼600 bp downstream of the transcription stop site (Fig. 1A). In addition, dHIP14YFP contains a ∼800 bp fragment encoding YFP that is inserted in-frame at the carboxyl terminus of the predicted dHIP14 ORF. If dHIP14YFP faithfully recapitulates endogenous dHIP14 expression and function, then it should rescue both the lethality and the ERG phenotype of dHIP14 mutants. Indeed, homozygous dHIP14ex11 animals from both dHIP14ex11 germline clone and dHIP14ex11 heterozygous mothers are fully rescued for lethality by dHIP14YFP (data not shown), as is the ERG phenotype of flies with homozygous dHIP14ex11 mutant photoreceptors (Fig. 1D). These results show the dHIP14 maternal effect can be rescued by zygotic dHIP14 expression. In addition, these results indicate dHIP14YFP contains all necessary regulatory and protein coding sequences for dHIP14 function and there is thus every reason to expect dHIP14YFP expression mirrors that of endogenous dHIP14.
To assess dHIP14YFP expression, YFP was used as an epitope tag in immunohistochemistry experiments. Background levels of fluorescence of a control fly that does not contain dHIP14YFP are shown in Figure 3C in cryostat sections of the optic lobe also labeled for the synaptic vesicle-specific marker neuronal synaptobrevin (N-syb) (Fig. 3A) and a Golgi-specific marker (Fig. 3B). Similarly labeled cryostat sections of dHIP14YFP flies showed strong YFP labeling that overlapped with N-syb in the optic lobe, and that was not present in the control (Fig. 3D–F), indicating that dHIP14YFP localizes to the neuropil, including the lamina and medulla where photoreceptors make their synaptic connections to second-order neurons. This dHIP14YFP expression is clearly distinct from the Golgi expression pattern and was striking given the conclusion of previous studies in rodents that HIP14 localized primarily to the Golgi, with some expression in cytoplasmic vesicles but no mention of synaptic expression (Singaraja et al., 2002; Huang et al., 2004). Whereas the most concentrated localization of dHIP14YFP in the optic lobe overlapped with that of the synaptic vesicle marker, in non-neuronal cells bordering the synaptic neuropil, dHIP14YFP expression coincided with that of the Golgi marker.
To further examine dHIP14YFP localization, we used high-magnification imaging to assess expression in the photoreceptor cell body and lamina regions. High-resolution images of the cell body region of the photoreceptors show labeling with the Golgi marker, but dHIP14YFP expression is not detectable (Fig. 3G–I, arrow indicates a single photoreceptor cell body). Thus, in photoreceptor neurons, dHIP14YFP localizes primarily, if not exclusively, to presynaptic terminals. High-resolution views of the presumptive lamina cell bodies reveals a similar pattern (Fig. 3J–L) in that the punctate labeling of the Golgi marker is not observed for dHIP14YFP. Once again, outside the neuronal layer there is overlap between Golgi and dHIP14YFP. The expression of dHIP14YFP in the adult Drosophila brain is similar to that of the optic lobe. The vast majority of dHIP14YFP expression is coincident with the synaptic vesicle marker N-syb, with colocalization with the Golgi marker observed in non-neuronal cells (Fig. 3M–O). To determine whether the localization of dHIP14YFP to neuronal processes and away from Golgi is generalizable to other stages of development, we also assessed dHIP14YFP expression in the larval CNS. As in the adult CNS, the bulk of dHIP14YFP colocalized with the synaptic vesicle marker (Fig. 3P–R), with lesser amounts of dHIP14YFP coincident with the Golgi marker outside the synaptic neuropil (Fig. 3S–U).
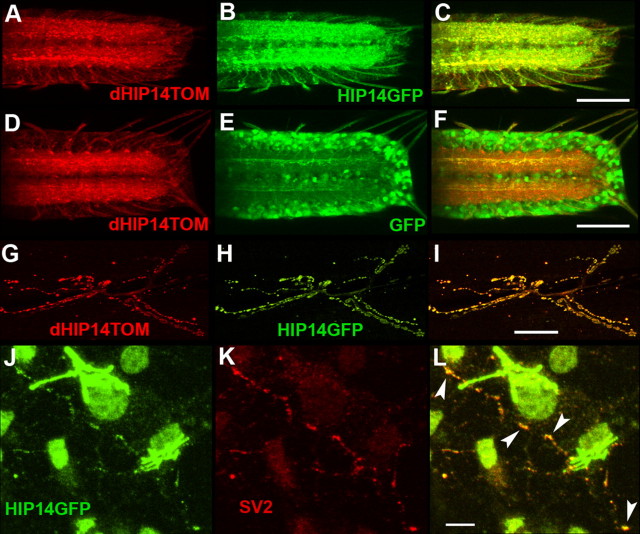
We also assessed dHIP14YFP expression in non-neuronal tissues and the larval neuromuscular junction. dHIP14YFP appeared to be ubiquitously expressed in all tissues examined. In non-neuronal tissues, it colocalized precisely with the Golgi marker. As an example, a leg imaginal disc is shown in Figure 4A–C. At the neuromuscular junction (NMJ), we also observed dHIP14YFP colocalizing with synaptic vesicle markers with dHIP14YFP expression in the surrounding muscle colocalizing with the Golgi marker (Fig. 4G–I) (background levels of anti-YFP labeling observed at the NMJ of a non-dHIP14YFP fly are shown in Fig. 4F). Analysis of the expression of dHIP14tdTOM, a genomic construct identical to dHIP14YFP except for the substitution of the fluorescent protein tdTomato for YFP, in the larval ventral nerve cord (VNC) and NMJ showed indistinguishable synaptic localization as that of dHIP14YFP (data not shown), indicating the synaptic localization of dHIP14 is independent of the fluorescent tag.
Figure 4.
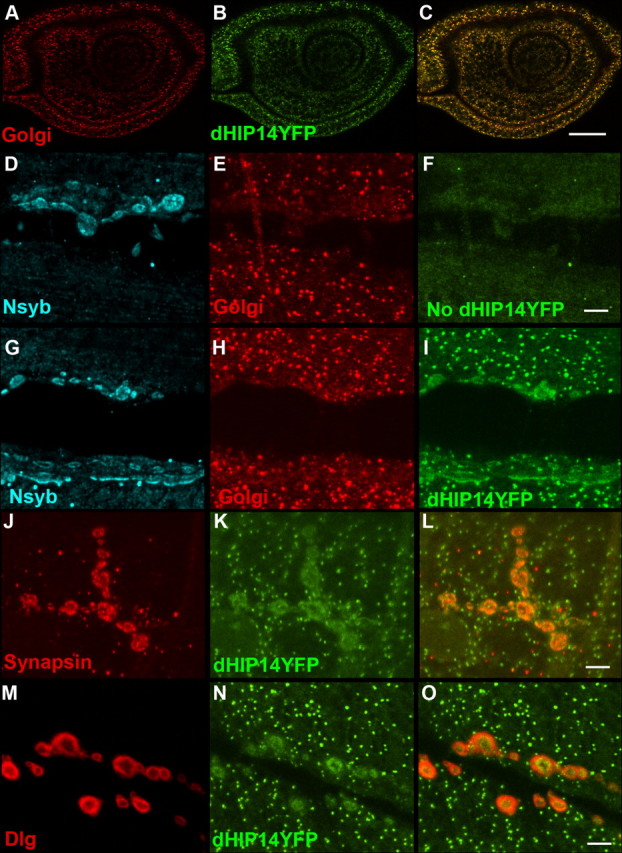
dHIP14YFP localizes to presynaptic terminals of motor neurons but Golgi in non-neuronal tissues. A–C, Leg imaginal disc of a dHIP14YFP third instar larvae labeled with a Golgi marker (A), anti-YFP (B), and the overlay (C). In non-neuronal tissues such as the leg imaginal disc, dHIP14YFP expression is indistinguishable from the Golgi marker. Scale bar, 25 μm. D–I, Neuromuscular junction of a control (No dHIP14YFP) and a dHIP14YFP third instar larvae triple-labeled with the synaptic vesicle marker N-syb (D, G), a Golgi marker (E, H), and anti-YFP (F, I). dHIP14YFP colocalizes with N-syb in the presynaptic terminal of motor neurons and with the Golgi marker in the surrounding muscle. Background levels of fluorescence for the anti-YFP antibody can be observed in F. Scale bar, 5 μm. J–L, Neuromuscular junction of a dHIP14YFP third instar larvae double-labeled with the presynaptic marker Synapsin (J), anti-YFP (K), and the overlay (L). dHIP14YFP and Synapsin expression are tightly correlated. Scale bar, 5 μm. M–O, Neuromuscular junction of a dHIP14YFP third instar larvae doubled with the postsynaptic marker Dlg (M), anti-YFP (N), and the overlay (O). Haloes of Dlg surround the more restricted dHIP14YFP expression in the overlay suggesting dHIP14YFP is not localizing postsynaptically at the NMJ. Scale bar, 5 μm. Genotypes are as follows: yw; dHIP14ex11, dHIP14YFP/dHIP14ex11, dHIP14YFP (A–C, G–O), yw; FRT79/FRT79 (D–F).
To determine whether the overlap of dHIP14YFP and N-syb was in the motor axons or at postsynaptic sites in the muscle, labeling was compared with both presynaptic and postsynaptic markers. As shown in Figure 4, J–L, dHIP14YFP coincides with the synaptic vesicle marker Synapsin in the neuromuscular presynaptic terminal. In contrast, a comparison of dHIP14YFP localization with that of the postsynaptic marker Discs large (Dlg), the Drosophila homolog of mammalian PSD-95 (Fig. 4M–O), reveals halos of Dlg surrounding smaller regions of dHIP14YFP, suggesting that dHIP14YFP localizes in the presynaptic terminal. In support of this conclusion, dHIP14TOM (dHIP14 tagged with the tomato fluorescent protein) (see below) that was ectopically expressed specifically in the muscle (data not shown) showed no preferential accumulation at the synapse. These results suggest that dHIP14 localizes presynaptically and not postsynaptically. Within the presynaptic terminal, dHIP14YFP is presumably membrane-associated because it has six predicted transmembrane domains. Close comparison of the localization of dHIP14YFP with the synaptic vesicle markers N-syb (Fig. 4G,I) and Synapsin (Fig. 4J,L) reveal subtle differences. Our interpretation is that dHIP14YFP appears more contiguous in regions in which the synaptic vesicle markers are punctate and thus that it is more likely that dHIP14YFP is associated with the plasma membrane. However, localization with synaptic vesicles cannot be ruled out based on our analysis.
Human HIP14 localizes to presynaptic terminals
To investigate further the subcellular localization of the protein, the Drosophila and human homologs of HIP14 were transgenically expressed in specific fly tissues using the GAL4/UAS system. To distinguish them from one another, Drosophila HIP14 was tagged with the tomato fluorescent protein (dHIP14TOM), whereas human HIP14 was tagged with GFP (HIP14GFP). To assess function of these proteins, we tested their ability to functionally rescue for lack of dHIP14 in the mutant flies. ERGs were performed on flies with homozygous dHIP14ex11 mutant photoreceptors in which either dHIP14 or hHIP14 were expressed using the photoreceptor-specific rhodopsin 1 promoter. As shown in Figure 1, E and F, both transgenes equally and fully restore photoreceptor synaptic transmission. The ability to functionally restore synaptic transmission in animals that have lost dHIP14 suggests that the fluorescent protein-tagged versions of the Drosophila and human homologs of HIP14 are targeted to the native location and carry out the native function of endogenous dHIP14. To determine directly the subcellular distribution of dHIP14TOM and HIP14GFP in neurons, the two transgenes were expressed simultaneously in the same animals in the larval nervous system using the panneuronal driver elav-GAL4. As shown in Figure 5, A–C, dHIP14TOM and HIP14GFP overlap and localize predominantly to the synaptic neuropil of the VNC in a pattern similar to that of the genomic rescue construct dHIP14YFP (Fig. 3P–U). As a control, dHIP14TOM was also simultaneously coexpressed using the elav-GAL4 driver with GFP, which was not fused to any other protein and so was free to diffuse in the cytoplasm (Fig. 5D–F). Whereas dHIP14TOM localizes predominantly to the synaptic neuropil, GFP remains mostly in the cell body region of ventral nerve cord neurons with little diffusing to the presynaptic terminal. Thus, the localization of HIP14GFP to the synaptic neuropil is dependent on an intrinsic property of HIP14. We also assessed the subcellular localization of dHIP14TOM and HIP14GFP at the NMJ in the same animals. The results at the NMJ are similar to those in the VNC. dHIP14TOM and HIP14GFP colocalize in the presynaptic terminals of neuromuscular synapses in an almost indistinguishable pattern (Fig. 5G–I). Ectopic expression of YFP-dHIP14, a dHIP14 fusion with YFP at the amino terminus, using the elav-GAL4 driver showed synaptic localization at the larval VNC and NMJ not distinguishable from dHIP14TOM (data not shown), indicating the synaptic localization of dHIP14 is independent of both fluorescent protein tag and fusion terminus. We conclude that in Drosophila HIP14 and its Drosophila counterpart are targeted to presynaptic nerve terminals in vivo.
Figure 5.
Human HIP14GFP localizes to synapses in Drosophila VNC and motor neurons, and in cultured rat hippocampal neurons. A–C, Subcellular localization in the third instar larval VNC of simultaneously expressed dHIP14TOM (A), HIP14GFP (B), and the overlay (C). dHIP14TOM and HIP14GFP localize almost indistinguishably to the synaptic neuropil of the VNC. Scale bar, 50 μm. D, E, Subcellular localization in the third instar larval ventral nerve cord of simultaneously expressed dHIP14TOM (D), GFP (control, unfused) (E), and the overlay. When not fused to HIP14, GFP localizes to the cell bodies of VNC neurons, distinct from the VNC synaptic neuropil localization of dHIP14TOM. Human HIP14GFP localization to the VNC synaptic neuropil is thus dependent on an intrinsic property of HIP14. Scale bar, 50 μm. G–I, Subcellular localization in third instar neuromuscular junction of simultaneously expressed dHIP14TOM (G), HIP14GFP (H), and the overlay (I). Scale bar, 25 μm. J–L, Subcellular localization in cultured rat hippocampal neurons of lentivirus-expressed HIP14GFP (J), the synaptic vesicle marker SV2 (K), and the overlay (L). The majority of HIP14GFP localizes to the presumptive Golgi, but a significant fraction colocalizes in a punctate pattern with the synaptic vesicle marker SV2. Scale bar, 10 μm. Genotypes are as follows: yw; UAS-dHIP14TOM/UAS-HIP14GFP; elav-GAL4/+ (A–C, G–I), yw; UAS-dHIP14TOM/UAS-GFP; elav-GAL4/+ (D–F).
Given our finding that human HIP14GFP localizes presynaptically in Drosophila, we decided to assess HIP14GFP targeting in cultured mammalian neurons. We expressed HIP14GFP in cultured rat hippocampal neurons using a lentiviral expression system. As previously observed (Singaraja et al., 2002; Huang et al., 2004), HIP14GFP predominantly localizes to cell bodies, at locations known to correspond to Golgi. However, HIP14GFP was also found to colocalize with the synaptic vesicle marker SV2 in fine processes (Fig. 5J–L) (overlap indicated by arrowheads in Fig. 5L). This demonstrates HIP14 is also a presynaptic protein in mammals.
SNAP-25 and CSP expression are altered in dHIP14 mutants
Previous studies have established HIP14 as a palmitoyl transferase and a number of in situ and in vitro HIP14 substrates have been identified including SNAP-25, PSD-95, GAD65, Synaptotagmin, and Huntingtin (Huang et al., 2004; Yanai et al., 2006). Because palmitoylation is thought to be necessary for the proper trafficking and subcellular localization of some palmitoylated proteins, we assayed the expression of a number of neuronal proteins in Drosophila dHIP14ex11 mutants in the hope of identifying potential in vivo dHIP14 substrates. In particular, we analyzed the distribution of several proteins known to be palmitoylated, including Synaptotagmin, N-syb, SNAP-25, Dlg, and cysteine string protein, as well as several proteins not believed to be palmitoylated including Syntaxin, Synapsin, and Bruchpilot. We analyzed the distribution of these proteins in the ventral nerve cord of first instar larval maternally and zygotically deficient for dHIP14. All of the proteins exhibited expression levels and spatial distributions not noticeably different from controls except for SNAP-25 and CSP. SNAP-25 expression was clearly reduced and more diffuse in the dHIP14 mutant VNC, compared with the control (Fig. 6, compare C, F). Syntaxin and N-syb expression in these animals was indistinguishable between the dHIP14ex11 mutant and control (Fig. 6; for Syntaxin, compare A, D; for N-syb, compare B, E). CSP was even more severely mislocalized than SNAP-25, because it remained in the cell bodies of the larval VNC neurons and did not appear in the synaptic neuropil (Fig. 6, compare H, K). Synaptotagmin expression in these same animals did not appear to be affected (Fig. 6, compare G, J).
Expression patterns of SNAP-25 and CSP were also aberrant in the optic lobe of dHIP14ex11/dHIP14ex11 pharate adults. Control and dHIP14ex11 pharate adults were dissected out of their pupal cases at the same time their siblings were eclosing and were alive at the time of head dissection as evidenced by leg and proboscis movement. As in the larval VNC, the localization of Syntaxin, N-syb, and Synaptotagmin were similar between dHIP14ex11 mutants and control (Fig. 6; for Syntaxin, compare M, P; for N-syb, compare N with Q, S, V; for Synaptotagmin, compare T, W). In contrast, optic lobe expression of both SNAP-25 and CSP was severely reduced in the dHIP14ex11 mutant (Fig. 6; for SNAP-25, compare O, R; for CSP, compare U, X). In summary, the abundance and subcellular distribution of the palmitoylated proteins SNAP-25 and CSP are selectively affected in the neurons of the dHIP14 mutant, whereas the expression of several other palmitoylated and nonpalmitoylated presynaptic proteins is not. SNAP-25 and CSP are thus potential palmitoylation substrates of dHIP14 in vivo.
Discussion
We report the identification of mutations in dHIP14, the Drosophila homolog of the human Huntingtin-interacting protein 14 palmitoyl transferase. To our knowledge, this is the first report of mutations in a DHHC palmitoyl transferase in any species with a nervous system. Our initial characterization of dHIP14 mutants determined that dHIP14 is required in photoreceptors for synapse function and in other neurons for organismal viability. dHIP14 protein was found to be expressed in all tissues examined. In non-neuronal tissues it localizes to the Golgi, whereas in neurons it localizes predominantly to presynaptic nerve terminals. Human HIP14 was also shown to localize to presynaptic terminals in flies and in cultured mammalian neurons. Aberrant neuronal expression of the palmitoylated proteins SNAP-25 and CSP was observed in dHIP14 mutants, making these presynaptic proteins candidate in vivo palmitoylation substrates of dHIP14 in neurons and providing a likely explanation for the photoreceptor synaptic transmission defect and lethality of the dHIP14 mutant.
dHIP14 localization to presynaptic terminals
We observed expression of dHIP14 in all tissues examined using the genomic rescue/expression construct dHIP14YFP. In non-neuronal tissues, such as the larval imaginal discs and muscles, we find dHIP14YFP localizing precisely with a Golgi marker. In contrast, in neurons dHIP14YFP localizes to synapses in the adult optic lobe, adult central brain, larval ventral nerve cord, and the larval NMJ. Furthermore, our NMJ colocalization of dHIP14YFP with presynaptic, but not postsynaptic, markers at the NMJ suggests that dHIP14YFP is present in the presynaptic compartment. This is consistent with the genetic results, mentioned above, in which the defective ERG of flies with homozygous dHIP14 mutant photoreceptors is a consequence of a presynaptic defect in the photoreceptor cell. Additionally, in certain neurons, including photoreceptor and presumptive lamina neurons, although we observed high levels of dHIP14 at synapses, it was absent from Golgi, or at least not within our detection limit. Our observation of dHIP14 localization to synapses was striking in light of previous studies of mammalian HIP14 expression in which Golgi and cytoplasmic vesicle, but not synapse, localization was reported (Singaraja et al., 2002; Huang et al., 2004). Such a divergence between Drosophila and mammalian homologs as closely related as dHIP14/HIP14 (48% identity) in subcellular localization is rare, if not unknown. To investigate this apparent difference, we expressed human HIP14 in Drosophila neurons and found that human HIP14 localized to synapses in a pattern indistinguishable from dHIP14 and also that it rescued the ERG phenotype as effectively as dHIP14 when expressed in photoreceptors. This indicates that, in Drosophila in vivo, dHIP14 and its human homolog HIP14 are interchangeable and function in presynaptic nerve terminals. Moreover, in cultured rat hippocampal neurons, we observed that, in addition to HIP14 in the cell body, a significant fraction of HIP14 colocalized with a synaptic vesicle marker in fine processes. This indicates that, as in Drosophila, HIP14 is present in the presynaptic compartment.
dHIP14 palmitoylation substrates
In an effort to identify potential in vivo dHIP14 palmitoylation substrates, we assessed the expression of a number of neuronal proteins in dHIP14 mutant animals. Only SNAP-25 and CSP were aberrantly expressed in the dHIP14 mutant. SNAP-25 is a plasma membrane-associated SNARE (SNAP receptor) protein that binds to Syntaxin and N-syb to form the synaptic core complex, which is believed to mediate docking and fusion of synaptic vesicles with the plasma membrane (Sorensen, 2005). CSP is a synaptic vesicle-associated protein that complexes with Hsc70 and SGT to form a chaperone complex important for the proper folding of proteins that are critical for synaptic vesicle exocytosis (Tobaben et al., 2001). Both SNAP-25 and CSP lack integral transmembrane domains and exhibit palmitoylation-dependent membrane targeting (Gonzalo and Linder, 1998; Chamberlain and Burgoyne, 1998). SNAP-25 has been shown to be directly palmitoylated by HIP14 in vitro, to colocalize with HIP14 in puncta on tubulovesicular structures in cultured hippocampal neurons, and to exhibit enhanced palmitoylation on ectopic expression of HIP14 (Huang et al., 2004). Our results showing aberrant SNAP-25 expression in dHIP14 mutants suggests this enzyme/substrate pairing is likely to be conserved in Drosophila.
Although CSP has been known to be palmitoylated on its cysteine string for some time (Gundersen et al., 1994), its palmitoyl transferase has not been identified. The simplest explanation for the aberrant localization of CSP in the dHIP14 mutant is that dHIP14 directly palmitoylates CSP and that the proper expression of CSP is palmitoylation dependent. However, another possibility that cannot presently be excluded is that one or more as-yet-unidentified proteins that CSP depends on for localization/stabilization is palmitoylated by dHIP14 and that CSP expression is affected indirectly by the failure of dHIP14 to palmitoylate another protein(s). Biochemical experiments will ultimately be needed to distinguish between these possibilities.
Previous studies of human HIP14 identified Synaptotagmin (Huang et al., 2004) and differed on PSD-95 (Dlg in Drosophila) (Fukata et al., 2004; Huang et al., 2004) as other HIP14 palmitoylation substrates. However, we detected no differences in expression of the Drosophila homologs of these proteins in the dHIP14 mutant, suggesting either that they do not depend on palmitoylation for their localization or that other palmitoyl transferase(s) recognize them as substrates.
dHIP14 phenocopy of SNAP-25 and CSP mutants
Given the dramatic effects on SNAP-25 and CSP localization and expression levels, the dHIP14 mutant phenotype might be expected to phenocopy mutations in these genes. This seems to apply somewhat to SNAP-25, but less so for CSP. The same phenotypes of pharate adult lethality and a synaptic transmission defective ERG of zygotically deficient dHIP14ex11 are highly reminiscent of those observed for SNAP-25 mutants (Vilinsky et al., 2002). Thus, the lethality associated with dHIP14 mutants may be attributable to a lack of SNAP-25 function that results from failure of dHIP14 to palmitoylate SNAP-25. The correlation of phenotypes between zygotically and maternally deficient dHIP14ex11 mutants and CSP mutants is not as similar. Whereas such dHIP14ex11 mutants die as first instar larvae, CSP mutants are lethal as early as late embryogenesis (Zinsmaier et al., 1994). Thus, because dHIP14 maternal and zygotic mutants exhibit a later lethal phase than CSP mutants, CSP may retain some residual function in the absence of palmitoylation. Alternatively, partial redundancy of CSP palmitoylation by dHIP14-L, or other palmitoyl transferase(s), could explain the later dHIP14 lethal phase. Yet another possibility is the failure of dHIP14 to palmitoylate an as-yet-unidentified palmitoylation substrate with an essential palmitoylation-dependent function during first instar.
Implications of palmitoylation within the presynaptic nerve terminal
The implications of the presynaptic terminal localization of dHIP14 are potentially significant. If, as believed previously, palmitoylation of presynaptic neuronal proteins occurred primarily in the Golgi, and to a lesser extent at other subcellular locations outside the presynaptic terminal, the effects of palmitoylation on synaptic function would only be realized on a slow timescale, after a lag of many minutes for axonal transport of newly palmitoylated proteins to the presynaptic terminal. However, the presence of dHIP14 in the presynaptic terminal opens the possibility that palmitoylation occurs within the terminal near the active zone in the plasma membrane (where SNAP-25 localizes) and/or on synaptic vesicles (where CSP localizes). This would mean that palmitoylation, not unlike phosphorylation, could modify protein activity and synaptic transmission on a much shorter timescale (milliseconds to seconds). This possibility seems even more plausible given the observation that palmitoyl-protein thioesterases, the enzymes antagonistic to palmitoyl transferases, which cleave palmitate from palmitoylated proteins, have been observed to colocalize with synaptic vesicle markers in axons (Heinonen et al., 2000; Ahtiainen et al., 2003). Thus, the activity of presynaptic proteins that mediate synaptic transmission could be modulated by a palmitoylation/depalmitoylation cycle on a fast timescale that corresponds to the changes in synaptic efficacy observed in short-term synaptic plasticity.
Studies of palmitate turnover on CSP, however, revealed that CSP palmitoylation is uncharacteristically stable and no cycle of palmitoylation/depalmitoylation was observed (Gundersen et al., 1996; van de Goor and Kelly, 1996). CSP is thus not likely to mediate short-term changes in synaptic function via palmitoylation. SNAP-25, on the other hand, is a much better candidate for mediating short-term effects on synapse function via palmitoylation. The half-life of palmitoylation of SNAP-25 has been measured at 3 h (Lane and Liu, 1997), and thus a palmitoylation/depalmitoylation cycle for SNAP-25 exists and could be a mechanism for altering synapse function in the short term. Whereas no expression differences were found for synaptotagmin, neuronal-synaptobrevin, or Dlg/PSD-95 in the dHIP14 mutant, these palmitoylated proteins as well as other as-yet-unidentified palmitoylation substrates of dHIP14 also have the potential to mediate short-term changes in synapse function. If, as we suppose, a palmitoylation cycle does occur within the presynaptic terminal, understanding palmitoylation-dependent changes in synapse function will include determining what and how intracellular and extracellular signals regulate palmitoyl transferase activity. Given palmitoylation functions in signaling pathways in non-neuronal cells, it may be that a subset of DHHC palmitoyl transferases also localize outside the ER/Golgi in non-neuronal cells and that palmitoylation also acts more broadly as a short timescale signaling mechanism.
Footnotes
This work was supported by National Institutes of Health Grants K01DA19458 (R.S.S.) and R01MH60711 (E.Y.I.). We thank Sophie Pautot for preparation of lentivirus and culturing of hippocampal neurons. We thank Douglas Guarnieri, Kristin Scott, and members of the Isacoff Laboratory for critical reading of this manuscript and Corey Goodman for support during the mutagenesis stage of this project. Data for this study were acquired and analyzed at the CRL Molecular Imaging Center at the University of California, Berkeley.
References
- Ahtiainen L, Van Diggelen OP, Jalanko A, Kopra O. Palmitoyl protein thioesterase 1 is targeted to the axons in neurons. J Comp Neurol. 2003;455:368–377. doi: 10.1002/cne.10492. [DOI] [PubMed] [Google Scholar]
- Babcock MC, Stowers RS, Leither J, Goodman CS, Pallanck LJ. A genetic screen for synaptic transmission mutants mapping to the right arm of chromosome 3 in Drosophila. Genetics. 2003;165:171–183. doi: 10.1093/genetics/165.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizuela BJ, Elfring L, Ballard J, Tamkun JW, Kennison JA. Genetic analysis of the brahma gene of Drosophila melanogaster and polytene chromosome subdivisions 72AB. Genetics. 1994;137:803–813. doi: 10.1093/genetics/137.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain LH, Burgoyne RD. The cysteine-string domain of the secretory vesicle cysteing-string protein is required for membrane targeting. Biochem J. 1998;335:205–209. doi: 10.1042/bj3350205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drisdel RC, Manzana E, Green WN. The role of palmitoylation in functional expression of nicotinic α7 receptors. J Neurosci. 2004;24:10502–10510. doi: 10.1523/JNEUROSCI.3315-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducker CE, Stettler EM, French KJ, Upson JJ, Smith CD. Huntingtin interacting protein 14 is an oncogenic human protein: palmitoyl acyltransferase. Oncogene. 2004;23:9230–9237. doi: 10.1038/sj.onc.1208171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Husseini AE, Schnell, Dakoji S, Sweeney N, Zhou Q, Prange O, Gauthier-Campbell C, Aguilera-Moreno A, Nicoll RA, Bredt DS. Synaptic strength regulated by palmitate cycling on PSD-95. Cell. 2002;108:849–863. doi: 10.1016/s0092-8674(02)00683-9. [DOI] [PubMed] [Google Scholar]
- Ferri N, Paoletti R, Corsini A. Lipid-modified proteins as biomarkers for cardiovascular disease: a review. Biomarkers. 2005;10:219–237. doi: 10.1080/13547500500216660. [DOI] [PubMed] [Google Scholar]
- Fukata M, Fukata Y, Adesnik H, Nicoll RA, Bredt DS. Identification of PSD-95 palmitoylating enzymes. Neuron. 2004;44:987–996. doi: 10.1016/j.neuron.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Gonzalo S, Linder ME. SNAP-25 palmitoylation and plasma membrane targeting require a functional secretory pathway. Mol Biol Cell. 1998;9:585–597. doi: 10.1091/mbc.9.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen CB, Mastrogiacomo A, Faull K, Umbach JA. Extensive lipidation of a Torpedo cysteine string protein. J Biol Chem. 1994;269:19197–19199. [PubMed] [Google Scholar]
- Gundersen CB, Umbach JA, Mastrogiacomo A. Cysteine-string proteins: a cycle of acylation and deacylation. Life Sci. 1996;58:2037–2040. doi: 10.1016/0024-3205(96)00195-6. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Rumbaugh G, Huganir RL. Differential regulation of AMAP receptor subunit trafficking by palmitoylation of two distinct sites. Neuron. 2005;47:709–723. doi: 10.1016/j.neuron.2005.06.035. [DOI] [PubMed] [Google Scholar]
- Heinonen O, Kyttala A, Lehmus E, Paunio T, Peltonen L, Jalanko A. Expression of palmitoyl-protein thioesterase in neurons. Mol Gen Metab. 2000;69:123–129. doi: 10.1006/mgme.2000.2961. [DOI] [PubMed] [Google Scholar]
- Heisenberg M. Separation of receptor and lamina potentials in the electroretinogram of normal and mutant Drosophila. J Exp Biol. 1971;55:85–100. doi: 10.1242/jeb.55.1.85. [DOI] [PubMed] [Google Scholar]
- Huang K, El-Husseini A. Modulation of protein trafficking and function by palmitoylation. Curr Opin Neurobiol. 2005;15:527–535. doi: 10.1016/j.conb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Huang K, Yanai A, Kang R, Arstikaitis P, Singaraja RR, Metzler M, Mullard A, Haigh B, Gauthier-Campbell C, Gutekunst CA, Hayden MR, El-Husseini A. Huntingtin-interacting protein HIP14 is a palmitoyl transferase involved in palmitoylation and trafficking of multiple neuronal proteins. Neuron. 2004;44:977–986. doi: 10.1016/j.neuron.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Cahill AL, Currie KP, Fox AP. The role of dynamic palmitoylation in Ca2+ inactivation. Proc Natl Acad Sci USA. 2000;97:9293–9398. doi: 10.1073/pnas.160589697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller CA, Yuan X, Panzanelli P, Martin ML, Alldred M, Sassoe-Pognetto M, Luscher B. The γ2 subunit of GABAA receptors is a substrate for palmitoylation by GODZ. J Neurosci. 2004;24:5881–5891. doi: 10.1523/JNEUROSCI.1037-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane SR, Liu Y. Characterization of the palmitoylation domain of SNAP-25. J Neurochem. 1997;69:1864–1869. doi: 10.1046/j.1471-4159.1997.69051864.x. [DOI] [PubMed] [Google Scholar]
- Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- Littleton JT, Bellen HJ, Perin MS. Expression of synaptotagmin in Drosophila reveals transport and localization of synaptic vesicles to the synapse. Development. 1993;118:1077–1088. doi: 10.1242/dev.118.4.1077. [DOI] [PubMed] [Google Scholar]
- Melendez A, Li W, Kalderon D. Activity, expression and function of a second Drosophila protein kinase A catalytic subunit gene. Genetics. 1995;141:1507–1520. doi: 10.1093/genetics/141.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Stewart BA, Rivlin PK, Vilinsky I, Watson BO, Lang C, Boulianne G, Salpeter MM, Deitcher DL. Two distinct effects on neurotransmission in a temperature-sensitive SNAP-25 mutant. EMBO J. 2001;20:6761–6771. doi: 10.1093/emboj/20.23.6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resh MD. Palmitoylation of ligands, receptors, and intracellular signaling molecules. Sci STKE. 2006;359:re14. doi: 10.1126/stke.3592006re14. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. Red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Singaraja RR, Hadano S, Metzler M, Givan S, Wellington CL, Warby S, Yanai A, Gutekunst CA, Leavitt BR, Yi H, Fichter K, Gan L, McCutcheon K, Chopra V, Michel J, Hersch SM, Ikeda JE, Hayden MR. Hum Mol Genet. 2002;11:2815–2828. doi: 10.1093/hmg/11.23.2815. [DOI] [PubMed] [Google Scholar]
- Sorensen JB. SNARE complexes prepare for membrane fusion. Trends Neurosci. 2005;28:453–455. doi: 10.1016/j.tins.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Stanley H, Botas J, Malhotra V. The mechanism of Golgi segregation during mitosis is cell type-specific. Proc Natl Acad Sci USA. 1997;23:14467–14470. doi: 10.1073/pnas.94.26.14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart BA, Atwood HL, Renger JJ, Wang J, Wu CF. Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solutions. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1994;175:179–191. doi: 10.1007/BF00215114. [DOI] [PubMed] [Google Scholar]
- Stowers RS, Schwarz TL. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics. 1999;152:1631–1639. doi: 10.1093/genetics/152.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Brinke A, van Golde LM, Batenburg JJ. Palmitoylation and processing of the lipopeptide surfactant protein C. Biochem Biophys Acta. 2002;1583:253–265. doi: 10.1016/s1388-1981(02)00248-2. [DOI] [PubMed] [Google Scholar]
- Tobaben S, Thakur R, Fernandez-Chacon R, Sudhor TC, Rettig J, Stahl B. A trimeric protein complex functions as a synaptic chaperone machine. Neuron. 2001;31:987–999. doi: 10.1016/s0896-6273(01)00427-5. [DOI] [PubMed] [Google Scholar]
- van de Goor J, Kelly RB. Association of Drosophila cysteine string proteins with membranes. FEBS Lett. 1996;380:251–256. doi: 10.1016/0014-5793(96)00026-9. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Williamson AL, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol. 1998;8:243–246. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- Vilinsky I, Stewart BA, Drummond J, Robinsin I, Deitcher DL. A Drosophila SNAP-25 null mutant reveals context-dependent redundancy with SNAP-24 in neurotransmission. Genetics. 2002;162:259–271. doi: 10.1093/genetics/162.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MN, Fergestad T, Lloyd TE, He Y, Broadie K, Bellen HJ. Syntaxin 1A interacts with multiple exocytotic proteins to regulate neurotransmitter release in vivo. Neuron. 1999;23:593–605. doi: 10.1016/s0896-6273(00)80811-9. [DOI] [PubMed] [Google Scholar]
- Yanai A, Huang K, Kang R, Singaraja RR, Arstikaitis P, Gan L, Orban PC, Mullard A, Cowan CM, Raymond LA, Drisdel RC, Green WN, Ravikumar B, Rubinsztein DC, El-Husseini A, Hayden MR. Palmitoylation of huntingtin by HIP14 is essential for its trafficking and function. Nat Neurosci. 2006;9:824–831. doi: 10.1038/nn1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Liu L, Reddivari M, Zhang XA. The palmitoylation of metastasis suppressor KAI1/CD82 is important for its motility- and invasiveness-inhibitory activity. Cancer Res. 2004;64:7455–7463. doi: 10.1158/0008-5472.CAN-04-1574. [DOI] [PubMed] [Google Scholar]
- Zinsmaier KE, Eberle KK, Buchner E, Walter N, Benzer S. Paralysis and early death in cysteine string protein mutants of Drosophila. Science. 1994;263:977–980. doi: 10.1126/science.8310297. [DOI] [PubMed] [Google Scholar]