Abstract
The kynurenine pathway is a major route of l-tryptophan catabolism producing neuroactive metabolites implicated in neurodegeneration and immune tolerance. We characterized the kynurenine pathway in human neurons and the human SK-N-SH neuroblastoma cell line and found that the kynurenine pathway enzymes were variably expressed. Picolinic carboxylase was expressed only in primary and some adult neurons but not in SK-N-SH cells. Because of this difference, SK-N-SH cells were able to produce the excitotoxin quinolinic acid, whereas human neurons produced the neuroprotectant picolinic acid. The net result of kynurenine pathway induction in human neurons is therefore predicted to result in neuroprotection, immune regulation, and tumor inhibition, whereas in SK-N-SH cells, it may result in neurotoxicity, immune tolerance, and tumor promotion. This study represents the first comprehensive characterization of the kynurenine pathway in neurons and the first description of the involvement of the kynurenine pathway as a mechanism for controlling both tumor cell neurotoxicity and persistence.
Keywords: human neurons; neuroblastoma; kynurenine pathway; indoleamine 2,3-dioxygenase; picolinic acid; quinolinic acid; neurotoxicity; tumor tolerance
Introduction
Within the CNS, >95% of l-tryptophan is catabolized through the kynurenine (KYN) pathway, resulting in the production of several neuroactive intermediates, which can be neurotoxic or neuroprotective (Fig. 1) (Stone, 1993). The NMDA receptor agonist and neurotoxin quinolinic acid (QUIN) is likely to represent the most important kynurenine pathway metabolite in terms of bioactivity (Schwarcz et al., 1983; Stone, 1993). There is good evidence that the kynurenine pathway is involved in the neurotoxicity associated with several inflammatory brain diseases (Heyes, 1996; Guillemin et al., 2005a,c; Hartai et al., 2005). In contrast to the neurotoxic activity of QUIN, kynurenic acid (KYNA) and picolinic acid (PIC), two additional kynurenine pathway products, are neuroprotective (Foster et al., 1984; Jhamandas et al., 2000). In addition to these activities, the kynurenine pathway has been found to be a key regulator of the immune response in relation to tolerance (Moffett and Namboodiri, 2003), with particular relevance to pregnancy (Munn et al., 1998) and tumor persistence. There is evidence that both tryptophan (TRP) depletion and utilization with the generation of certain kynurenine pathway products, especially QUIN, are responsible for this immune tolerance (Moffett and Namboodiri, 2003). The kynurenine pathway is also involved in certain physiological functions such as behavior, sleep, and thermoregulation (Stone, 1993; Curzon, 1996).
Figure 1.
Simplified diagram of the kynurenine pathway.
The kynurenine pathway has been shown to be fully present in macrophages and microglial cells (Guillemin et al., 2003) and partly present in astrocytes (Guillemin et al., 2001). Characterization of the kynurenine pathway in other brain cells is still unknown. To define the kynurenine pathway in human neurons, we used primary cultures of human fetal neurons, human SK-N-SH neuroblastoma cells, and normal adult brain tissue obtained at autopsy. Interferon-γ (IFN-γ) is known to be the most potent inducer for one of the first enzymes of the kynurenine pathway, indoleamine 2,3-dioxygenase (IDO) (Pemberton et al., 1997). We therefore used IFN-γ treatment to activate the neuronal kynurenine pathway enzyme expression and activity in vitro. We assessed the presence of the major kynurenine pathway enzymes by reverse transcription (RT)-PCR. In addition, we measured the concentration of important kynurenine pathway substrates and intermediates: tryptophan, kynurenine, kynurenic acid, and 3-hydroxykynurenine (3-HK), as well as the end products QUIN and PIC. We used immunocytochemistry and immunohistochemistry to study the cellular localization of some of the kynurenine pathway enzymes and metabolites. Finally, we assessed the significance of the kynurenine pathway in neurons by determining the effect of QUIN and PIC on SK-N-SH proliferation.
Materials and Methods
Human ethics approval.
These studies were approved by the Human Ethics Committees at the Universities of Sydney and New South Wales.
Reagents and chemicals.
All cell culture media and additives were from Invitrogen (Mount Waverley, Australia) unless otherwise stated. 4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI) and 1-(β-d-arabino-furanosyl)-cytosine hydrochloride (AraC) were obtained from Sigma-Aldrich (Sydney, Australia). Anti-glial fibrillary acid protein (GFAP) mouse monoclonal antibody (mAb) clone GA-5 was obtained from Novocastra (Newcastle, UK). Anti-CD68 mouse mAb clone KiM1P was generously provided by Dr. Parwaresch (University of Kiel, Kiel, Germany) and was used at a concentration of 10 μg/ml. Anti-quinolinic acid rabbit polyclonal antibody (pAb) and anti-microtubule-associated protein 2 (MAP2) mouse mAb were purchased from Millipore (Melbourne, Australia). Anti-IDO mouse mAb, anti-kynurenine hydroxylase mouse mAb, and anti-tryptophan 2,3-dioxygenase (TDO) rabbit pAb were generously provided by Profs. O. Takikawa, T. Uemura (Psychiatric Research Institute of Tokyo, Japan), and Dr. C. Miller (Johns Hopkins University, Baltimore, MD), respectively. These three antibodies were used at concentrations of 10, 10, and 2 μg/ml, respectively. Polyclonal antibodies for detection of α-amino-β-carboxymuconate-semialdehyde decarboxylase (ACMSD), quinolinate phosphoribosyltransferase (QPRT), kynurenine amino transferase-I (KAT-I), and kynureninase (KYNU) were obtained from Abnova (Taipei, Taiwan) and used at a concentration of 2 μg/ml. Alexa 488- or Alexa 594-conjugated anti-mouse IgG or anti-rabbit were purchased from Invitrogen. All commercial antibodies were used at the concentrations recommended by the manufacturer.
Cell cultures.
Human fetal brains were obtained from 14- to 18-week-old fetuses collected after therapeutic termination after informed consent. Brain cell cultures were prepared using a protocol adapted from previously described methods (Guillemin et al., 1997).
Neurons were prepared from the initial mixed brain cells as described previously (Guillemin et al., 2005b). Briefly, cells were plated in culture flasks coated with Matrigel (diluted 1/20 in Neurobasal medium) and maintained in Neurobasal medium supplemented with 2% (v/v) B-27 supplement, 2 mm Glutamax, 50 mm HEPES, 200 IU/ml penicillin G, 200 μg/ml streptomycin sulfate, and 5 mm glucose. Cultures were kept at 37°C at 5% CO2 in a humidified atmosphere. The medium was changed once to twice per week. Neurons were maintained for up to 10 weeks.
Human SK-N-SH neuroblastoma cells were obtained from American Type Culture Collection (Manassas, VA) (HTB11). The SK-N-SH cells were maintained in RPMI 1640 medium containing 10% (v/v) heat-inactivated fetal calf serum (FCS), 2 mm Glutamax, 200 IU/ml penicillin G, 200 μg/ml streptomycin sulfate, and 0.5% (w/v) glucose in 75 cm2 culture flasks (Corning, Corning, NY) and in Permanox chamber-slides (Medos, Lidcombe, Australia). Cultures were kept at 37°C at 5% CO2 in a humidified atmosphere. The medium was changed after 3 d until they reached confluence and every 2–3 d thereafter.
Human peripheral blood mononuclear cells (PBMCs) were isolated from the blood of healthy volunteers (Centre for Immunology, Sydney, Australia) using a standard Ficoll-Paque (GE Healthcare, Piscataway, NJ) density separation method as previously described (Kerr et al., 1997a). Monocyte-derived macrophages were obtained using a classic adherence method. Briefly, isolated PBMCs were plated on Falcon Primaria 24-well plates (BD, North Ryde, Australia) with RPMI medium, 10% autologous human serum, 2 mm Glutamax, 200 IU/ml penicillin G, and 200 μg/ml streptomycin sulfate. After 8 d in vitro, the human serum component of the medium was eliminated completely, and cells were maintained in AIM-V, a serum-free medium containing no detectable QUIN (Invitrogen).
Immunocytochemistry.
Brain cells were characterized as previously described (Guillemin et al., 1997). Human primary neurons and SK-N-SH cells were grown in Permanox chamber-slides for 2–3 d. After 72 h, control (untreated) and IFN-γ-treated cells were fixed with acetone/methanol 1:1 (v/v) for 20 min at −20°C. Cells were then rinsed three times with PBS, and a gentle membranous permeabilization was performed by incubation with 0.025% Triton X-100 in PBS for 10 min at 22°C. After washing, cells were incubated with 5% (v/v) nonimmune goat serum (NGS) in PBS for 45 min at 22°C, rinsed twice with PBS, and incubated for 1 h at 37°C with the specific primary mAb or pAb antibodies diluted in 5% NGS. Cells were then washed with 5% NGS solution and incubated for 1 h at 37°C with the appropriate labeled secondary antibodies (goat anti-mouse IgG or goat anti-rabbit IgG coupled to Alexa 488 or Alexa 594). Nuclear staining was performed using DAPI at 1 μg/ml for 5 min at 22°C. After several washes in PBS at 37°C, the coverslips were quickly mounted on glass slides with Fluoromount-G and were examined with an Olympus BX60 fluorescence microscope fitted with a SensiCam digital camera. Three controls were performed for each immunolabeling experiment: (1) isotypic antibody controls for mAbs and serum control for pAbs, (2) incubation with only the secondary labeled antibodies, and (3) measurement of autofluorescence of unlabeled cells.
Tissue processing and immunohistochemistry.
Blocks of the medial temporal lobe and superior frontal cortex were taken from formalin-fixed brains obtained postmortem. For this study, autopsy tissue was sampled from two neurologically normal male patients (39 and 64 years old), both with cardiac arrest as cause of death. Series of 45 μm cryosections were processed for immunohistochemistry using the avidin–biotin complex detection system as described previously (Guillemin et al., 2005c). Free-floating sections were washed in 50% ethanol for 3 × 15 min at 37°C and then incubated in 3% H2O2 for 20 min to quench endogenous peroxidase activity. Nonspecific binding sites were blocked with a solution of 1% bovine serum albumin (BSA) and 0.1% Triton X in 0.1 m Tris buffer. The tissue was then incubated for 36 h at 4°C in diluted primary antibodies to IDO, kynurenine 3-hydroxylase (KMO), KYNU, ACMSD, TDO, KAT-II, and QUIN (Fig. 1). After washing in Tris buffer, the sections were incubated for 1.5 h at room temperature in species-appropriate biotinylated secondary antibody (Vector Laboratories, Burlingame CA). After washing in Tris buffer, the sections were placed in ABCelite peroxidase reagents (1:100 in Tris buffer) for 1.5 h, washed, and transferred to the chromogen solution [DAB (brown), or SG (blue black); Vector Laboratories]. After 5 min, 25 μl of 3% H2O2 was added to the chromogen solution for color development. Sections were washed, mounted, dried, and coverslipped in Histochoice mounting medium (Amresco, Solon, OH). Selected sections were counterstained with cresyl violet for cytoarchitecture. Sections were viewed using a Leica DMLB bright-field microscope and photographed with a Jentopix Progress C14 digital camera.
Treatments of neuronal and SK-N-SH cultures.
For the characterization of the kynurenine pathway, IFN-γ (100 IU/ml) was added to 4-week-old neuronal primary cultures and SK-N-SH (1 × 106 cells/well). Messenger RNA and culture supernatants were collected after 24, 48, and 72 h.
For the proliferation assay, cultures of SK-N-SH (5 × 104 cells/well) were treated with PIC at 1 and 5 μm and QUIN at 5 μm for 24 h. Each experiment was performed in quadruplicate. Cells were trypsinized, a 200 μl aliquot of the cell suspension was mixed with 800 μl of a solution of 0.25% trypan blue (Sigma), 10 μl of this mixture was distributed inside a numeration chamber, and counting was performed in a blinded manner. Control and treated cells were counted three times. Experiments were performed in triplicate. Mean values and SEM were calculated for each treatment, and the results were plotted on a histogram. Changes in cell growth and viability were expressed as percentages and were calculated using the following formula: (number of treated cells × 100/number of cells in the untreated condition). Unpaired t tests were performed on the results obtained at 24 h. A p value of <0.05 was regarded as statistically significant.
RT-PCR detection of mRNA expression of kynurenine pathway enzymes.
The RT-PCR protocol and primer sequences were previously described (Guillemin et al., 2001). Negative controls were: (1) omission of a target template, (2) omission of reverse transcriptase, and (3) genomic DNA. Amplified products were quantified after scanning using ImageJ1.38s (NIH, Bethesda, MD). Experiments were performed in duplicate on cultures derived from five different fetal brains. Based on image analysis intensity ratios of kynurenine pathway enzyme mRNA expressed relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA, the SE was between 4 and 5%.
Gas chromatography-mass spectrometry.
Culture supernatants were assayed for QUIN as previously described (Kerr et al., 1997b). QUIN concentrations were calculated with the following formula: total concentration of QUIN (in nanomolars) detected in cell culture supernatants − concentration of QUIN (in nanomolars) present in the culture medium before addition to cells. Samples were similarly analyzed for PIC analysis using d4-picolinic acid as an internal standard. QUIN and PIC samples were analyzed by gas chromatography-mass spectrometry (GC-MS) with the spectrometer operating in electron capture negative ionization mode. Selected ions (m/z 273 for PIC and m/z 277 for d4-PIC) were then monitored (Smythe et al., 2002). The limits of quantification were <1 fmol (injected onto the column) at signal-to-noise ratios of >10:1. Experiments were performed in triplicate using supernatants from primary cultures derived from three different human fetal brains and from one SK-N-SH culture. All results are expressed as the mean ± SEM.
HPLC.
TRP, KYN, and KYNA were measured using an Agilent 1100 series HPLC system equipped with a G1329A temperature-controlled autosampler, a G1314A variable wavelength detector, a G1321A xenon flash lamp fluorescence detector, and a Zorbax 300SB C18 reversed-phase 4.6 × 250 mm column (Agilent Technologies, North Ryde, Australia) and mobile phase consisting of ammonium acetate buffer (0.1 m, pH 4.65) containing 0.02% (v/v) acetonitrile. KYN was measured by UV absorbance at 365 nm, TRP was measured using fluorescence (Ex285 nm/Em365 nm), and KYNA was measured by fluorescence (Ex254 nm/Em404 nm) after postcolumn derivatization with zinc acetate as previously described (Kapoor et al., 1994). 3-HK was measured using electrochemical detection as previously described (Heyes et al., 1988). Limits of detection for all analytes were calculated (based on the number of moles of each analyte detected in a 100 μl injection onto the HPLC column) to be the following: TRP, 65 pmol; KYN, 200 fmol; KYNA, 10 fmol; and 3-HK, 10 fmol, which is consistent with previous data (Heyes et al., 1988; Kapoor et al., 1994). Data related to cellular production of kynurenine pathway TRP metabolites have the background values (because of traces in serum and medium) for each metabolite subtracted to represent net cellular production. Each experiment was performed in triplicate using supernatants from primary cultures derived from three different human fetal brains and from one SK-N-SH culture. All results are expressed as the mean ± SEM.
Statistical analysis.
GC-MS and HPLC mean values and SEs were calculated, and the results were plotted on histograms. Unpaired t tests were performed on the results obtained at 24, 48, and 72 h for GC-MS and 72 h for HPLC. p values were generated comparing kynurenine pathway metabolite production or degradation by unstimulated cells and IFN-γ stimulated cells at the same time point. A p value of <0.05 was regarded as statistically significant.
Results
Purity of primary cultures of human fetal neurons
Consistent with our previous data (Guillemin et al., 2005b), after 7 d in culture, 90–95% of the fetal brain cells immunostained for the neuronal marker MAP2 (data not shown). The main contaminants of the neuronal cultures were astrocytes. However, growing the brain cells on Matrigel strongly limited astroglial proliferation. After 8 d in culture, 2 μm Ara-C was added for 24 h to eliminate all proliferating cells, including astrocytes and fibroblasts. Staining for CD68 indicated that <1% of the contaminating cells in the initial cultures were microglia. These monocytic cells cannot adhere to Matrigel, and the few residual microglia were rapidly removed with the regular changing of medium. Immunostaining for O4 protein (oligodendrocytes), GFAP (astrocytes), 5B5 (fibroblasts), and factor VIII (endothelial cells) did not detect any cells at either 2 weeks or 4 weeks after seeding. After 4 weeks and repeated changing of medium, purity of the neuronal cultures reached 98–99% (Fig. 2A).
Figure 2.
Immunodetection of kynurenine pathway enzymes and products in human neurons and brain. A, B, Cultures of human primary neurons (A) and of SK-N-SH cells (B) were immunostained for the enzymes IDO, TDO, KYNU, KAT-II, KMO, QPRT, and ACMSD, as well as for QUIN and MAP2 as indicated. C, Control preparations of cultures of human primary neurons: irrelevant IgG and nonimmune rabbit serum staining of human primary neurons. D, Human hippocampal sections were also immunostained for IDO, KMO, KYNU, ACMSD, TDO, KAT-II, and QUIN as indicated. Neuronal labeling for all markers was predominantly cytosolic, although occasional staining of proximal processes was seen (e.g., for KMO). Neurons of the hilus (CA4) were the most consistently labeled. Non-neuronal cells were also labeled. Punctate/vesicular labeling of probable microglial cells was widespread in all areas of cortex and hippocampus. QUIN labeling was distinctly granular, as shown in the pyramidal cells of CA1 (D).
Expression of kynurenine pathway enzymes in human fetal neurons compared with SK-N-SH neurons
Primer pairs were previously developed to identify mRNA transcripts for human IDO, TDO, KAT-I, KAT-II, KMO, KYNU, 3-hydroxyanthranilate 3,4-dioxygenase (HAAO), QPRT, and ACMSD (Guillemin et al., 2001). IFN-γ was used to induce IDO mRNA expression in primary human neurons (line 4) and SK-N-SH cells (line 2) (Fig. 3A). Neither unstimulated primary human neurons (line 1) or SK-N-SH cells (line 3) expressed IDO. IFN-γ-stimulated macrophages were used as a positive control, and a clear mRNA signal for all nine kynurenine pathway enzymes mRNA was detected as expected (line 5) (Guillemin et al., 2003). Based on image analysis of optical density (Fig. 3A), ratios of kynurenine pathway enzyme mRNA expressed relative to GAPDH mRNA were calculated (Fig. 3B); the SE was between 4 and 5% for all kynurenine pathway enzyme mRNA expression data. For all the enzymes, mRNA expression was analyzed at 24 h after application of IFN-γ or vehicle control. Time course studies (6–24 h) assessing the induction of kynurenine pathway enzyme mRNA expression in response to IFN-γ stimulation indicated that maximal mRNA expression was detected at 24 h.
Figure 3.
Detection of kynurenine pathway enzyme expression using RT-PCR. A, Ethidium bromide-stained gels showing RT-PCR for (from top to bottom) IDO, TDO, KAT-I, KAT-II, KYNU, KMO, HAAO, QPRT, ACMSD, and GAPDH expression. The first column corresponds to unstimulated SK-N-SH cells, the second to IFN-γ-stimulated SK-N-SH cells, the third to unstimulated human primary neurons, the fourth to IFN-γ-stimulated human primary neurons, and the fifth to human primary macrophages stimulated with IFN-γ and used as a positive control for kynurenine pathway enzyme expression. B, The histograms indicate the ratio of the expression of the nine kynurenine pathway enzyme mRNA relative to the GAPDH mRNA expression. Individual samples were analyzed in triplicate, and the SEM for all data was determined to be <5%.
Strong IDO expression was detected in SK-N-SH and primary neurons only after IFN-γ stimulation. TDO was expressed by both IFN-γ stimulated and unstimulated SK-N-SH and primary neurons. Interestingly, TDO expression was lower after IFN-γ stimulation when IDO was increased. Both IFN-γ stimulated and unstimulated SK-N-SH and primary neurons showed low expression for KAT-I and a very low expression for KAT-II. Indeed, the KAT-II signal was only detected in IFN-γ-stimulated SK-N-SH neurons. Expression of KYNU and KMO mRNA was present at low levels in both IFN-γ-stimulated and unstimulated SK-N-SH and primary neurons. HAAO and QPRT mRNA were clearly expressed in both SK-N-SH and primary neurons, and levels for both enzymes were slightly higher in SK-N-SH compared with primary neurons. Expression of ACMSD was not detectable in SK-N-SH neurons but was clearly present in primary neurons. ACMSD expression was higher in unstimulated compared with IFN-γ-stimulated neurons.
Immunocytochemical detection of kynurenine pathway enzymes in both IFN-γ-stimulated SK-N-SH and primary neurons (Fig. 2) was in general agreement with the RT-PCR results. In addition, we did not detect any QUIN production in primary neurons but found ACMSD expressed, whereas the reverse was found for SK-N-SH cells. In primary neurons, ACMSD labeling was mainly detected in the cytoplasm. For both SK-N-SH and primary neurons, immunostaining for IDO, TDO, KMO, KYNU, KAT-II, and QPRT was perinuclear and cytoplasmic. In primary neurons only, ACMSD labeling was mainly detected in the cytoplasm. In SK-N-SH cells, QUIN immunostaining was located in small cytoplasmic vesicles. Finally, for both SK-N-SH and primary neurons, staining for the structural protein MAP2 indicated localization throughout the cytoplasm and dendrites.
Expression of kynurenine pathway enzymes and production of kynurenine pathway metabolites in human cortex and hippocampus
Frozen 45 μm sections of superior frontal cortex and medial temporal lobe were labeled with antibodies to IDO, KMO, KYNU, QPRT, ACMSD, KAT-II, and TDO (Fig. 2D). For all enzyme labeling, there was strong staining of neurons. Astrocytes and microglia were also labeled, with identification of these cells based on cell morphology. Not all neurons were stained in each case, although there was no consistent pattern of labeling of cell types. Pyramidal neurons of CA1–4 of the hippocampus were the most consistently labeled neurons for all enzymes. KMO labeling tended to be localized to nonpyramidal neurons in the frontal cortex. Only a minority of neurons stained positive for KMO, whereas IDO and ACMSD were detected in the majority of neurons and also showed a high degree of non-neuronal staining. ACMSD immunolabeling filled the neuronal soma and occasionally extended into proximal processes. Although the factors determining the level of expression of kynurenine pathway enzymes in the intact brain remain to be clarified, the present work clearly indicates that a diverse range of neuronal types have the potential to produce these enzymes in the human brain. We previously showed that only a very low level of QUIN immunoreactivity is found in temporal neocortical and hippocampal areas in control brain tissue (Guillemin et al., 2005c) and that concentrations of QUIN in normal cerebral tissue are extremely low (<1 nm) (Guillemin et al., 2007). The present data are entirely consistent with our previous observations.
HPLC analysis of TRP, KYN, KYNA, and 3-HK secreted by SK-N-SH and primary neurons
Concentrations of TRP in Neurobasal medium and in RPMI with 10% FCS were 64.7 ± 2.5 μm and 48 ± 0.15 μm, respectively. Concentrations of KYN in Neurobasal medium and in RPMI with 10% FCS were 1.49 ± 0.24 μm and 2.11 ± 0.34 μm, respectively. KYNA and 3-HK were not detectable in the culture media. Changes in the concentration of TRP in the culture medium after 72 h (decreased in all cases) were calculated using the following formula: (TRP concentration in the cell culture supernatant − TRP concentration initially present before the medium was added to the cells). Concentrations of KYNA produced after 72 h were calculated using the following formula: (KYNA in the cell culture supernatant − KYNA initially present in the medium).
TRP was catabolized by both SK-N-SH cells and primary neuron cultures (Fig. 4A). Without stimulation, SK-N-SH and primary neurons were both able to catabolize a low but significant concentration of TRP resulting in decreases in TRP concentration of 3.43 ± 0.4 μm and 3.92 ± 1 μm, respectively. In the presence of IFN-γ, TRP loss was dramatically accelerated resulting in decreases in TRP concentration of 33.32 ± 0.4 μm and 9.31 ± 1.2 μm for SK-N-SH and primary neuron cultures, respectively. IFN-γ-stimulated SK-N-SH neurons therefore degraded 3.6-fold more TRP compared with IFN-γ-stimulated primary neurons.
Figure 4.
Quantification of kynurenine pathway products by HPLC and GC/MS. A–H, Media samples derived from cultures of SK-N-SH cells and primary neurons either untreated or treated for 24 h with 100 IU/ml IFN-γ (A–D) or at the times indicated (E–H) were analyzed by HPLC for TRP (A), KYN (B), KYNA (C), and 3-HK (D) and by using GC/MS for QUIN (E) and PIC (F, H). Statistical differences comparing the untreated and IFN-γ-treated conditions (A–D) or the change in concentration detected at the 48 or 72 h compared with the 24 h time point are indicated by the asterisks. **p < 0.02 and ***p < 0.01 (Mann–Whitney test).
Unstimulated SK-N-SH and primary neurons produced only low concentrations of KYN (0.10 ± 0.07 and 0.08 ± 0.031 μm, respectively) and did not produce detectable amounts of KYNA (Fig. 4B). IFN-γ increased SK-N-SH and primary neuron production of KYN and KYNA. Media concentrations of KYN increased to 17.66 ± 0.34 and 19.75 ± 0.37 μm for SK-N-SH cells and primary neurons, respectively, whereas media concentrations of KYNA increased to 7.16 ± 2.6 and 9.1 ± 0.49 nm for SK-N-SH and primary neurons, respectively (Fig. 4C). 3-HK secretion was detected in medium derived from unstimulated and IFN-γ-stimulated SK-N-SH cells and primary neurons (6.01 ± 2.2, 26.55 ± 0.4, 7.65 ± 0.4, and 20.25 ± 2.54 nm, respectively) (Fig. 4D).
GC/MS analysis of QUIN secreted by SK-N-SH and primary neurons
Initial QUIN concentrations in the culture media were 0.469 ± 0.056 and 0.402 ± 0.042 μm in RPMI 10% FCS and Neurobasal 2% B27, respectively. Both unstimulated and IFN-γ-stimulated primary neurons degraded QUIN (Fig. 4E). The levels of QUIN effectively catabolized were in concentrations of 0.260 ± 0.006 μm (24 h), 0.275 ± 0.06 μm (48 h), and 0.466 ± 0.084 μm (72 h) for unstimulated cultures and 0.212 ± 0.007 μm (24 h), 0.284 ± 0.013 μm (48 h), and 0.556 ± 0.039 μm (72 h) for IFN-γ-stimulated cultures. No significant differences were found in the level of QUIN catabolism when unstimulated and IFN-γ-stimulated cultures were compared. Unstimulated SK-N-SH neurons produced basal concentrations of QUIN of 0.847 ± 0.196 μm (24 h), 0.941 ± 0.07 μm (48 h), and 0.962 ± 0.121 μm (72 h). After stimulation with IFN-γ, QUIN synthesis was increased significantly to 1.1 ± 0.07 μm (24 h), 1.36 ± 0.08 μm (48 h), and 2.22 ± 0.057 μm (72 h) (Fig. 4G).
GC/MS analysis of PIC secreted by SK-N-SH and primary neurons
Initial PIC concentrations in the culture media were respectively 0.567 ± 0.31 and 0.202 ± 0.066 μm in RPMI 10% FCS and Neurobasal 2% B27, respectively. Both unstimulated and IFN-γ-stimulated primary neurons produced PIC (Fig. 4H). Primary cultures of neurons constitutively produced PIC. PIC concentrations were 0.327 ± 0.051 μm (24 h), 0.388 ± 0.13 μm (48 h), and 0.465 ± 0.1 μm (72 h) for unstimulated cultures and 0.336 ± 0.056 μm (24 h), 0.359 ± 0.011 μm (48 h), and 0.491 ± 0.09 μm (72 h) for IFN-γ-stimulated cultures, respectively. No significant differences were found in the level of PIC production between unstimulated and IFN-γ-stimulated cultures.
In SK-N-SH culture supernatants, the levels of PIC were lower than initially quantified in the culture medium (Fig. 4F). PIC concentrations were respectively −0.174 ± 0.071 μm (24 h), −0.125 ± 0.04 μm (48 h), and −0.143 ± 0.09 μm (72 h) for unstimulated cultures and −0.096 ± 0.048 μm (24 h), −0.096 ± 0.037 μm (48 h), and −0.105 ± 0.034 μm (72 h) for IFN-γ-stimulated cultures. No significant differences were found between unstimulated and IFN-γ stimulated cultures.
Effects of picolinic acid and quinolinic acid on SK-N-SH proliferation
Initially, 50 × 103 SK-N-SH cells were plated, and after 24 h without treatment (control), the cell number increased to 138.75 ± 14.7 × 103 (a 2.8-fold increase) (Fig. 5B). The number of SK-N-SH neurons present after treatment with 1 and 5 μm picolinic acid were respectively 117.5 ± 8.6 × 103 (p = 0.02) and 85.25 ± 3.9 × 103 (p = 0.0002) lower compared with the control condition (Fig. 5). The number of SK-N-SH neurons present after treatment with 5 μm quinolinic acid was 172.2 ± 18 × 103, i.e., 3.5-fold higher compared with the control (p = 0.006). Using trypan blue staining, we did not observe any significant differences in the number of dead cells between treated (7.1 × 103 for PIC 1 μm; 4.3 × 103 for PIC 5 μm; and 8.8 × 103 for QUIN 5 μm) and control cells (8.3 × 103).
Figure 5.
Effects of PIC and QUIN on SK-H-SH proliferation. A, B, Phase-contrast microscopy of SK-N-SH cell cultures (A) and histogram indicating the number of SK-N-SH cells (from left to right) in control untreated cells and cells treated with either 1 or 5 μm PIC or with 5 μm QUIN 5 for 24 h (B). p values for the comparisons with control condition were determined (Mann–Whitney test): *p < 0.05, **p < 0.01, and ***p < 0.001.
Discussion
Comparing normal and tumoral neurons, we herein reveal differences in the expression of kynurenine pathway enzymes. Primary and adult neurons expressed ACMSD and produced the neuroprotective compound PIC, whereas SK-N-SH cells did not express this enzyme. This was associated with a switch in the kynurenine pathway leading to the production of the excitotoxin QUIN. This emphasizes that caution must be exercised when using neuronal cell lines to define neuronal enzymatic pathways. More importantly, the differential pattern of neuronal kynurenine pathway expression is likely to be important in neoplastic brain diseases.
Previous studies reported IDO activity in human neoplastic cell lines (Werner-Felmayer et al., 1989; Heyes et al., 1997a). However, cell lines do not always reflect primary brain cells, and our data show that this is particularly true with the kynurenine pathway (Heyes et al., 1997a). Recently, Roy et al. (2005) showed that IDO immunoreactivity is present in rodent neurons and is upregulated by IFN-γ in neurons of the limbic cortical regions. This study also described strong IDO staining in neurons of the hippocampus and dentate gyrus, which are regions highly sensitive to excitotoxicity mediated by NMDA receptor agonists such as QUIN. However, significant species differences in the expression of the kynurenine pathway enzymes have been described (Heyes et al., 1997b), and this was one impetus for our current study, because human primary cells and tissue represent the most relevant model to study the kynurenine pathway and its relevance to human brain physiology and disease. Heyes et al. (1997a) described IDO activity in human fetal mixed brain cells but without differentiating which cell types express IDO and produce QUIN. Within the human brain, cellular localization of IDO has been found in primary astrocytes, microvascular endothelial cells, microglia, and macrophages (Heyes et al., 1996; Guillemin et al., 2001, 2005b; Schroten et al., 2001). We previously demonstrated and confirm here that primary human neurons express IDO and that the enzyme is inducible by IFN-γ (Guillemin et al., 2005b).
There are only limited published data concerning TDO expression within the brain. TDO is expressed in the cytoplasm of neurons, astrocytes, and endothelial cells but not in microglia using immunohistochemistry on sections from human frontal cortex (Miller et al., 2004, 2006). We extended these data by showing that human MAP2-positive primary neurons express TDO in their cytoplasm. We also showed for the first time that, in both primary and SK-N-SH neurons, TDO expression is decreased when IDO is activated by IFN-γ, and conversely, in the absence of IDO expression, TDO is expressed to a higher extent. This highlights a relationship between TDO and IDO expression and suggests the existence of a novel regulatory mechanism balancing the expression of the two first enzymes of the kynurenine pathway.
KAT-I, -II, and -III are responsible for synthesis of KYNA (Okuno et al., 1991; Yu et al., 2006). KAT-I immunoreactivity has been detected in astrocytes and neurons in the rat (Du et al., 1992). We found that SK-N-SH and primary neurons have a very similar and low level of expression for KAT-I and KAT-II, and both cell types produce KYNA to similar extents. KYNA is an endogenous antagonist of NMDA and α-7 nicotinic receptors resulting in neuroprotective and anticonvulsant effects (Stone, 1993). KYNA has the ability to control the vulnerability of striatal neurons to QUIN (Hilmas et al., 2001). However, ionotropic glutamate receptor ligands such as glutamate, and likely QUIN, can inhibit KYNA synthesis by rat astrocytes (Curatolo et al., 1996). Changes in KYNA concentrations have been described in several neurological disorders (Stone, 1993). Within the CNS, most KYNA is produced by astrocytes, neurons, and oligodendrocytes (Du et al., 1992; Guillemin et al., 2001; Wejksza et al., 2005). A recent study showed that KYNA is synthesized by primary cultures of rat cortical neurons (Rzeski et al., 2005) and that production, under certain conditions, can be significantly higher than in astrocytes. We confirm here that, similar to rodents, human neurons produce KYNA in μm concentrations.
Both primary neurons and SK-N-SH cells synthesize 3-HK, although the concentrations detected in conditioned medium were higher with the latter cells, paralleling the increased expression of KMO. 3-HK can promote neuronal damage by inducing oxidative stress and can also act synergistically with QUIN to potentiate its toxicity (Guidetti and Schwarcz, 1999). KYNU and KMO expression seem to be significantly expressed only in monocytic cells such as human microglia and macrophages (Guillemin et al., 2003). Interestingly, only these cell types appear to produce QUIN, and our data point toward a potential novel link between the level of expression of KYNU and KMO and the capacity to produce QUIN.
ACMSD regulates nicotinamide adenine dinucleotide biosynthesis and the generation of QUIN through the kynurenine pathway (Tanabe et al., 2002). There is an inverse relationship between ACMSD activity and QUIN production. QUIN is nonenzymatically derived from 2-aminomuconate-6-semialdehyde. Fukuoka et al. (2002) showed that ACMSD is expressed in the human brain at low levels. We found here that ACMSD is expressed by human primary and adult neurons but not by SK-N-SH cells. We previously showed that PIC is also produced by human fetal astrocytes (Guillemin et al., 2001). PIC is an endogenous neuroprotective compound within the brain (Jhamandas et al., 1990). In nanomolar concentrations, it protects against QUIN- and kainic acid-induced neurotoxicity (Vrooman et al., 1993). Unlike KYNA and other NMDA receptor antagonists, PIC can effectively block QUIN neurotoxicity but apparently without affecting the excitatory effect. The mechanism by which PIC attenuates QUIN neurotoxicity is still unclear, because PIC does not compete with QUIN at the glutamate/NMDA-binding site of the NMDA receptor. In addition, PIC can influence the immune response by increasing chemokine release (Bosco et al., 2000) and by stimulating anti-tumoral (Leuthauser et al., 1982), anti-microbial (Blasi et al., 1993), and anti-viral (Fernandez-Pol et al., 2001) activities (Fig. 6). Finally, PIC is also considered an endogenous metal chelator within the brain, which may have relevance for Alzheimer's disease (Testa et al., 1985; Melillo et al., 1996).
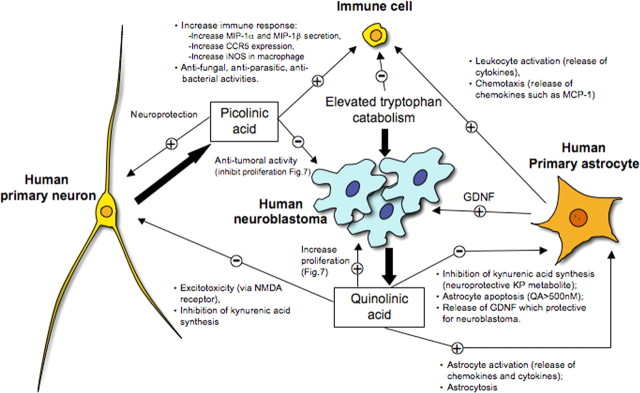
Figure 6.
Scheme indicating differences between human primary neurons and neuroblastoma cells with respect to the production of kynurenine pathway metabolites and their effects in tumor persistence and the immune response.
Several studies suggest an involvement of the kynurenine pathway in brain tumor neuropathology. Saito et al. (1993) showed that tumoral cell lines derived from human brain have the ability to convert TRP into KYN in response to IFN-γ. Other studies showed that IDO activity is present in the SK-N-SH cells and glioblastoma cells (Werner-Felmayer et al., 1989; Heyes et al., 1997a). Uyttenhove et al. (2003) showed that a majority of human tumors constitutively express IDO. Another study demonstrated that gliomas and glioneuronal tumors have an elevated TRP uptake and metabolism (Juhasz et al., 2006). This high TRP metabolism by low-grade tumors is very likely to be associated with an activation of the kynurenine pathway, which represents a mechanism regulating tumor cell growth and persistence (Juhasz et al., 2006).
The mechanism by which increased kynurenine pathway metabolism confers tumor survival is unknown (Gajewski et al., 2006). It has been shown that QUIN can directly increase proliferation rate of human glioblastoma cell line U343MG (Serio et al., 2005), and we showed here that QUIN can increase SK-N-SH neuroblastoma cell proliferation. Interestingly, QUIN also increases astroglial production of glial cell line-derived neurotrophic factor (Bresjanac and Antauer, 2000), which may induce proliferation of neuroblastoma cells and increase resistance to chemotherapeutic agents (Hansford and Marshall, 2005) (Fig. 6). However, previous studies have shown the ability of PIC to decrease tumor proliferation both in vitro (Fernandez and Johnson, 1977; Testa et al., 1985; Kovalenko et al., 1989) and in vivo (Leuthauser et al., 1982), which is in accordance with our results showing that PIC in micromolar concentrations significantly decreases SK-N-SH growth.
Recent studies suggest IDO as a target for cancer suppression in vivo (Munn and Mellor, 2004, 2007; Muller et al., 2005; Zamanakou et al., 2007). It has been shown previously that IDO inhibition induces a robust allogenic T-cell response in vitro and delays tumor growth in vivo (Friberg et al., 2002). We found that in vitro SK-N-SH produced 1–2 μm QUIN after 72 h. It is therefore possible that those concentrations of QUIN produced in vivo could reach concentrations that are cytotoxic for T-cells.
In conclusion, this study represents the first comprehensive characterization of the kynurenine pathway in human neurons. We showed that SK-N-SH neuroblastoma cells produce the excitotoxin QUIN, whereas normal primary neurons synthesize the neuroprotective molecule PIC. This differential activation of the kynurenine pathway may contribute to the persistence of neuroblastomas and might also contribute to the neurotoxicity associated with tumorogenesis (Fig. 6). These results also provide the foundation for the identification of new therapeutic strategies for cerebral tumors, because kynurenine pathway inhibitors are already available (Stone, 2000; Platten et al., 2007).
Footnotes
This work was supported by the Australian National Health and Medical Research Council (NHMRC) (fellowship and program grant), the New South Wales Department of Health, the University of New South Wales, the Rebecca L. Cooper Medical Research Foundation, the Perpetual Foundation, the Baxter Foundation, the Mason Foundation, Alzheimer Australia, Motor Neurone Disease Research Institute of Australia, and a private donation from M. Terry Gammel. We are very grateful to Prof. T. Uemura and Dr. C. Miller for providing their unique antibodies. We acknowledge the important tissue resource provided by the NHMRC Network for Brain Research into Mental Health. We thank Prof. T. W. Stone and Dr. S. Thomas for their help with the revision of this manuscript.
References
- Blasi E, Barluzzi R, Mazzolla R, Bistoni F. Differential host susceptibility to intracerebral infections with Candida albicans and Cryptococcus neoformans. Infect Immun. 1993;61:3476–3481. doi: 10.1128/iai.61.8.3476-3481.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco MC, Rapisarda A, Massazza S, Melillo G, Young H, Varesio L. The tryptophan catabolite picolinic acid selectively induces the chemokines macrophage inflammatory protein-1alpha and -1beta in macrophages. J Immunol. 2000;164:3283–3291. doi: 10.4049/jimmunol.164.6.3283. [DOI] [PubMed] [Google Scholar]
- Bresjanac M, Antauer G. Reactive astrocytes of the quinolinic acid-lesioned rat striatum express GFRalpha1 as well as GDNF in vivo. Exp Neurol. 2000;164:53–59. doi: 10.1006/exnr.2000.7416. [DOI] [PubMed] [Google Scholar]
- Curatolo L, Caccia C, Speciale C, Raimondi L, Cini M, Marconi M, Molinari A, Schwarcz R. Modulation of extracellular kynurenic acid content by excitatory amino acids in primary cultures of rat astrocytes. Adv Exp Med Biol. 1996;398:273–276. doi: 10.1007/978-1-4613-0381-7_43. [DOI] [PubMed] [Google Scholar]
- Curzon G. Brain tryptophan. Normal and disturbed control. Adv Exp Med Biol. 1996;398:27–34. [PubMed] [Google Scholar]
- Du F, Schmidt W, Okuno E, Kido R, Kohler C, Schwarcz R. Localization of kynurenine aminotransferase immunoreactivity in the rat hippocampus. J Comp Neurol. 1992;321:477–487. doi: 10.1002/cne.903210313. [DOI] [PubMed] [Google Scholar]
- Fernandez PJ, Johnson GS. Selective toxicity induced by picolinic acid in simian virus 40-transformed cells in tissue culture. Cancer Res. 1977;37:4276–4279. [PubMed] [Google Scholar]
- Fernandez-Pol JA, Klos DJ, Hamilton PD. Antiviral, cytotoxic and apoptotic activities of picolinic acid on human immunodeficiency virus-1 and human herpes simplex virus-2 infected cells. Anticancer Res. 2001;21:3773–3776. [PubMed] [Google Scholar]
- Foster AC, Vezzani A, French ED, Schwarcz R. Kynurenic acid blocks neurotoxicity and seizures induced in rats by the related brain metabolite quinolinic acid. Neurosci Lett. 1984;48:273–278. doi: 10.1016/0304-3940(84)90050-8. [DOI] [PubMed] [Google Scholar]
- Friberg M, Jennings R, Alsarraj M, Dessureault S, Cantor A, Extermann M, Mellor AL, Munn DH, Antonia SJ. Indoleamine 2,3-dioxygenase contributes to tumor cell evasion of T cell-mediated rejection. Int J Cancer. 2002;101:151–155. doi: 10.1002/ijc.10645. [DOI] [PubMed] [Google Scholar]
- Fukuoka S, Ishiguro K, Yanagihara K, Tanabe A, Egashira Y, Sanada H, Shibata K. Identification and expression of a cDNA encoding human alpha-amino-beta-carboxymuconate-epsilon-semialdehyde decarboxylase (ACMSD). A key enzyme for the tryptophan-niacine pathway and “quinolinate hypothesis”. J Biol Chem. 2002;277:35162–35167. doi: 10.1074/jbc.M200819200. [DOI] [PubMed] [Google Scholar]
- Gajewski TF, Meng Y, Harlin H. Immune suppression in the tumor microenvironment. J Immunother. 2006;29:233–240. doi: 10.1097/01.cji.0000199193.29048.56. [DOI] [PubMed] [Google Scholar]
- Guidetti P, Schwarcz R. 3-Hydroxykynurenine potentiates quinolinate but not NMDA toxicity in the rat striatum. Eur J Neurosci. 1999;11:3857–3863. doi: 10.1046/j.1460-9568.1999.00806.x. [DOI] [PubMed] [Google Scholar]
- Guillemin G, Boussin FD, Croitoru J, Franck-Duchenne M, Le Grand R, Lazarini F, Dormont D. Obtention and characterization of primary astrocyte and microglial cultures from adult monkey brains. J Neurosci Res. 1997;49:576–591. doi: 10.1002/(SICI)1097-4547(19970901)49:5<576::AID-JNR8>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Guillemin GJ, Kerr SJ, Smythe GA, Smith DG, Kapoor V, Armati PJ, Croitoru J, Brew BJ. Kynurenine pathway metabolism in human astrocytes: a paradox for neuronal protection. J Neurochem. 2001;78:1–13. doi: 10.1046/j.1471-4159.2001.00498.x. [DOI] [PubMed] [Google Scholar]
- Guillemin GJ, Smith DG, Smythe GA, Armati PJ, Brew BJ. Expression of the kynurenine pathway enzymes in human microglia and macrophages. Adv Exp Med Biol. 2003;527:105–112. doi: 10.1007/978-1-4615-0135-0_12. [DOI] [PubMed] [Google Scholar]
- Guillemin GJ, Wang L, Brew BJ. Quinolinic acid selectively induces apoptosis of human astrocytes: potential role in AIDS dementia complex. J Neuroinflammation. 2005a;2:16. doi: 10.1186/1742-2094-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemin GJ, Smythe G, Takikawa O, Brew BJ. Expression of indoleamine 2,3-dioxygenase and production of quinolinic acid by human microglia, astrocytes, and neurons. Glia. 2005b;49:15–23. doi: 10.1002/glia.20090. [DOI] [PubMed] [Google Scholar]
- Guillemin GJ, Brew BJ, Noonan CE, Takikawa O, Cullen KM. Indoleamine 2,3 dioxygenase and quinolinic acid immunoreactivity in Alzheimer's disease hippocampus. Neuropathol Appl Neurobiol. 2005c;31:395–404. doi: 10.1111/j.1365-2990.2005.00655.x. [DOI] [PubMed] [Google Scholar]
- Guillemin GJ, Brew BJ, Noonan CE, Knight TG, Smythe GA, Cullen KM. Mass spectrometric detection of quinolinic acid in microdissected Alzheimer disease plaques. International Congress Series 1304. 2007 in press. [Google Scholar]
- Hansford LM, Marshall GM. Glial cell line-derived neurotrophic factor (GDNF) family ligands reduce the sensitivity of neuroblastoma cells to pharmacologically induced cell death, growth arrest and differentiation. Neurosci Lett. 2005;389:77–82. doi: 10.1016/j.neulet.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Hartai Z, Klivenyi P, Janaky T, Penke B, Dux L, Vecsei L. Kynurenine metabolism in multiple sclerosis. Acta Neurol Scand. 2005;112:93–96. doi: 10.1111/j.1600-0404.2005.00442.x. [DOI] [PubMed] [Google Scholar]
- Heyes MP. The kynurenine pathway and neurologic disease. Therapeutic strategies. Adv Exp Med Biol. 1996;398:125–129. doi: 10.1007/978-1-4613-0381-7_20. [DOI] [PubMed] [Google Scholar]
- Heyes MP, Achim CL, Wiley CA, Major EO, Saito K, Markey SP. Human microglia convert L-tryptophan into the neurotoxin quinolinic acid. Biochem J. 1996;320:595–597. doi: 10.1042/bj3200595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyes MP, Chen CY, Major EO, Saito K. Different kynurenine pathway enzymes limit quinolinic acid formation by various human cell types. Biochem J. 1997a;326:351–356. doi: 10.1042/bj3260351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyes MP, Saito K, Chen CY, Proescholdt MG, Nowak TS, Jr, Li J, Beagles KE, Proescholdt MA, Zito MA, Kawai K, Markey SP. Species heterogeneity between gerbils and rats: quinolinate production by microglia and astrocytes and accumulations in response to ischemic brain injury and systemic immune activation. J Neurochem. 1997b;69:1519–1529. doi: 10.1046/j.1471-4159.1997.69041519.x. [DOI] [PubMed] [Google Scholar]
- Heyes MP, Saito K, Lackner A, Wiley CA, Achim CL, Markey SP. Sources of the neurotoxin quinolinic acid in the brain of HIV-1-infected patients and retrovirus-infected macaques. FASEB J. 1998;12:881–896. doi: 10.1096/fasebj.12.10.881. [DOI] [PubMed] [Google Scholar]
- Hilmas C, Pereira EF, Alkondon M, Rassoulpour A, Schwarcz R, Albuquerque EX. The brain metabolite kynurenic acid inhibits α7 nicotinic receptor activity and increases non-α7 nicotinic receptor expression: physiopathological implications. J Neurosci. 2001;21:7463–7473. doi: 10.1523/JNEUROSCI.21-19-07463.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhamandas K, Boegman RJ, Beninger RJ, Bialik M. Quinolinate-induced cortical cholinergic damage: modulation by tryptophan metabolites. Brain Res. 1990;529:185–191. doi: 10.1016/0006-8993(90)90826-w. [DOI] [PubMed] [Google Scholar]
- Jhamandas KH, Boegman RJ, Beninger RJ, Miranda AF, Lipic KA. Excitotoxicity of quinolinic acid: modulation by endogenous antagonists. Neurotox Res. 2000;2:139–155. doi: 10.1007/BF03033790. [DOI] [PubMed] [Google Scholar]
- Juhasz C, Chugani DC, Muzik O, Wu D, Sloan AE, Barger G, Watson C, Shah AK, Sood S, Ergun EL, Mangner TJ, Chakraborty PK, Kupsky WJ, Chugani HT. In vivo uptake and metabolism of alpha-[11C]methyl-L-tryptophan in human brain tumors. J Cereb Blood Flow Metab. 2006;26:345–357. doi: 10.1038/sj.jcbfm.9600199. [DOI] [PubMed] [Google Scholar]
- Kapoor V, Kapoor R, Chalmers J. Kynurenic acid, an endogenous glutamate antagonist, in SHR and WKY rats: possible role in central blood pressure regulation. Clin Exp Pharmacol Physiol. 1994;21:891–896. doi: 10.1111/j.1440-1681.1994.tb02460.x. [DOI] [PubMed] [Google Scholar]
- Kerr SJ, Armati PJ, Pemberton LA, Smythe G, Brew BJ. Kynurenine pathway inhibition with 6-chloro-D-tryptophan reduces neurotoxicity of HIV-infected macrophage supernatants (Abstract) Neurology. 1997a;48:A94. doi: 10.1212/wnl.49.6.1671. [DOI] [PubMed] [Google Scholar]
- Kerr SJ, Armati PJ, Pemberton LA, Smythe G, Tattam B, Brew BJ. Kynurenine pathway inhibition reduces neurotoxicity of HIV-1-infected macrophages. Neurology. 1997b;49:1671–1681. doi: 10.1212/wnl.49.6.1671. [DOI] [PubMed] [Google Scholar]
- Kovalenko OA, Tarasova NI, Ivannikova AG, Voronkova LN, Nikolaeva TG. [The effect of picolinic acid and iron ions on the proliferation of SPEV cells.] Tsitologiia. 1989;31:447–452. [PubMed] [Google Scholar]
- Leuthauser SW, Oberley LW, Oberley TD. Antitumor activity of picolinic acid in CBA/J mice. J Natl Cancer Inst. 1982;68:123–126. [PubMed] [Google Scholar]
- Melillo G, Bosco MC, Musso T, Varesio L. Immunobiology of picolinic acid. Adv Exp Med Biol. 1996;398:135–141. doi: 10.1007/978-1-4613-0381-7_22. [DOI] [PubMed] [Google Scholar]
- Miller CL, Llenos IC, Dulay JR, Barillo MM, Yolken RH, Weis S. Expression of the kynurenine pathway enzyme tryptophan 2,3-dioxygenase is increased in the frontal cortex of individuals with schizophrenia. Neurobiol Dis. 2004;15:618–629. doi: 10.1016/j.nbd.2003.12.015. [DOI] [PubMed] [Google Scholar]
- Miller CL, Llenos IC, Dulay JR, Weis S. Upregulation of the initiating step of the kynurenine pathway in postmortem anterior cingulate cortex from individuals with schizophrenia and bipolar disorder. Brain Res. 2006;1073–1074:25–37. doi: 10.1016/j.brainres.2005.12.056. [DOI] [PubMed] [Google Scholar]
- Moffett JR, Namboodiri MA. Tryptophan and the immune response. Immunol Cell Biol. 2003;81:247–265. doi: 10.1046/j.1440-1711.2003.t01-1-01177.x. [DOI] [PubMed] [Google Scholar]
- Muller AJ, DuHadaway JB, Donover PS, Sutanto-Ward E, Prendergast GC. Inhibition of indoleamine 2,3-dioxygenase, an immunoregulatory target of the cancer suppression gene Bin1, potentiates cancer chemotherapy. Nat Med. 2005;11:312–319. doi: 10.1038/nm1196. [DOI] [PubMed] [Google Scholar]
- Munn DH, Mellor AL. IDO and tolerance to tumors. Trends Mol Med. 2004;10:15–18. doi: 10.1016/j.molmed.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Munn DH, Mellor AL. Indoleamine 2,3-dioxygenase and tumor-induced tolerance. J Clin Invest. 2007;117:1147–1154. doi: 10.1172/JCI31178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn DH, Zhou M, Attwood JT, Bondarev I, Conway SJ, Marshall B, Brown C, Mellor AL. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science. 1998;281:1191–1193. doi: 10.1126/science.281.5380.1191. [DOI] [PubMed] [Google Scholar]
- Okuno E, Nakamura M, Schwarcz R. Two kynurenine aminotransferases in human brain. Brain Res. 1991;542:307–312. doi: 10.1016/0006-8993(91)91583-m. [DOI] [PubMed] [Google Scholar]
- Pemberton LA, Kerr SJ, Smythe G, Brew BJ. Quinolinic acid production by macrophages stimulated with IFN-gamma, TNF-alpha, and IFN-alpha. J Interferon Cytokine Res. 1997;17:589–595. doi: 10.1089/jir.1997.17.589. [DOI] [PubMed] [Google Scholar]
- Platten M, Ho PP, Steinman L. Anti-inflammatory strategies for the treatment of multiple sclerosis—tryptophan catabolites may hold the key. Drug Discov Today: Therap Strat. 2007 in press. [Google Scholar]
- Roy EJ, Takikawa O, Kranz DM, Brown AR, Thomas DL. Neuronal localization of indoleamine 2,3-dioxygenase in mice. Neurosci Lett. 2005;387:95–99. doi: 10.1016/j.neulet.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Rzeski W, Kocki T, Dybel A, Wejksza K, Zdzisinska B, Kandefer-Szerszen M, Turski WA, Okuno E, Albrecht J. Demonstration of kynurenine aminotransferases I and II and characterization of kynurenic acid synthesis in cultured cerebral cortical neurons. J Neurosci Res. 2005;80:677–682. doi: 10.1002/jnr.20505. [DOI] [PubMed] [Google Scholar]
- Saito K, Chen CY, Masana M, Crowley JS, Markey SP, Heyes MP. 4-Chloro-3-hydroxyanthranilate, 6-chlorotryptophan and norharmane attenuate quinolinic acid formation by interferon-γ-stimulated monocytes (THP-1 cells) Biochem J. 1993;291:11–14. doi: 10.1042/bj2910011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroten H, Spors B, Hucke C, Stins M, Kim KS, Adam R, Daubener W. Potential role of human brain microvascular endothelial cells in the pathogenesis of brain abscess: inhibition of Staphylococcus aureus by activation of indoleamine 2,3-dioxygenase. Neuropediatrics. 2001;32:206–210. doi: 10.1055/s-2001-17375. [DOI] [PubMed] [Google Scholar]
- Schwarcz R, Whetsell WO, Jr, Mangano RM. Quinolinic acid: an endogenous metabolite that produces axon-sparing lesions in rat brain. Science. 1983;219:316–318. doi: 10.1126/science.6849138. [DOI] [PubMed] [Google Scholar]
- Serio CD, Cozzi A, Angeli I, Doria L, Micucci I, Pellerito S, Mirone P, Masotti G, Moroni F, Tarantini F. Kynurenic acid inhibits the release of the neurotrophic fibroblast growth factor (FGF)-1 and enhances proliferation of glia cells, in vitro. Cell Mol Neurobiol. 2005;25:981–993. doi: 10.1007/s10571-005-8469-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythe GA, Braga O, Brew BJ, Grant RS, Guillemin GJ, Kerr SJ, Walker DW. Concurrent quantification of quinolinic, picolinic, and nicotinic acids using electron-capture negative-ion gas chromatography-mass spectrometry. Anal Biochem. 2002;301:21–26. doi: 10.1006/abio.2001.5490. [DOI] [PubMed] [Google Scholar]
- Stone TW. Neuropharmacology of quinolinic and kynurenic acids. Pharmacol Rev. 1993;45:309–379. [PubMed] [Google Scholar]
- Stone TW. Development and therapeutic potential of kynurenic acid and kynurenine derivatives for neuroprotection. Trends Pharmacol Sci. 2000;21:149–154. doi: 10.1016/s0165-6147(00)01451-6. [DOI] [PubMed] [Google Scholar]
- Tanabe A, Egashira Y, Fukuoka S, Shibata K, Sanada H. Expression of rat hepatic 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase is affected by a high protein diet and by streptozotocin-induced diabetes. J Nutr. 2002;132:1153–1159. doi: 10.1093/jn/132.6.1153. [DOI] [PubMed] [Google Scholar]
- Testa U, Louache F, Titeux M, Thomopoulos P, Rochant H. The iron-chelating agent picolinic acid enhances transferrin receptors expression in human erythroleukaemic cell lines. Br J Haematol. 1985;60:491–502. doi: 10.1111/j.1365-2141.1985.tb07446.x. [DOI] [PubMed] [Google Scholar]
- Uyttenhove C, Pilotte L, Theate I, Stroobant V, Colau D, Parmentier N, Boon T, Van Den Eynde BJ. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med. 2003;9:1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- Vrooman L, Jhamandas K, Boegman RJ, Beninger RJ. Picolinic acid modulates kainic acid-evoked glutamate release from the striatum in vitro. Brain Res. 1993;627:193–198. doi: 10.1016/0006-8993(93)90320-m. [DOI] [PubMed] [Google Scholar]
- Wejksza K, Rzeski W, Okuno E, Kandefer-Szerszen M, Albrecht J, Turski WA. Demonstration of kynurenine aminotransferases I and II and characterization of kynurenic acid synthesis in oligodendrocyte cell line (OLN-93) Neurochem Res. 2005;30:963–968. doi: 10.1007/s11064-005-6178-z. [DOI] [PubMed] [Google Scholar]
- Werner-Felmayer G, Werner ER, Fuchs D, Hausen A, Reibnegger G, Wachter H. Characteristics of interferon induced tryptophan metabolism in human cells in vitro. Biochim Biophys Acta. 1989;1012:140–147. doi: 10.1016/0167-4889(89)90087-6. [DOI] [PubMed] [Google Scholar]
- Yu P, Li Z, Zhang L, Tagle DA, Cai T. Characterization of kynurenine aminotransferase III, a novel member of a phylogenetically conserved KAT family. Gene. 2006;365:111–118. doi: 10.1016/j.gene.2005.09.034. [DOI] [PubMed] [Google Scholar]
- Zamanakou M, Germenis AE, Karanikas V. Tumor immune escape mediated by indoleamine 2,3-dioxygenase. Immunol Lett. 2007;111:69–75. doi: 10.1016/j.imlet.2007.06.001. [DOI] [PubMed] [Google Scholar]