Abstract
Although circadian oscillation in dynamics of intracellular Ca2+ signals has been observed in both plant and animal cells, it has remained unknown whether Ca2+ signals play an in vivo role in cellular oscillation itself. To address this question, we modified the dynamics of intracellular Ca2+ signals in circadian pacemaker neurons in vivo by targeted expression of varying doses of a Ca2+ buffer protein in transgenic Drosophila melanogaster. Intracellular Ca2+ buffering in pacemaker neurons results in dose-dependent slowing of free-running behavioral rhythms, with average period >3 h longer than control at the highest dose. The rhythmic nuclear accumulation of a transcription factor known to be essential for cellular circadian oscillation is also slowed. We also determined that Ca2+ buffering interacts synergistically with genetic manipulations that interfere with either calmodulin or calmodulin-dependent protein kinase II function. These results suggest a role for intracellular Ca2+ signaling in regulating intrinsic cellular oscillation in vivo.
Keywords: Ca2+, circadian rhythm, Drosophila, calmodulin, CaMKII, transcription
Introduction
Intrinsic circadian clocks coordinate multiple aspects of organismal physiology with the 24 h rotation of the earth. In animals, autonomous cellular clocks that underlie behavioral cycles of rest and activity have been localized to particular central pacemaker neurons (Panda et al., 2002; Stanewsky, 2003). Current models of cellular oscillation in almost all organisms are based on negative transcriptional feedback loops, with transcription of “clock” genes being repressed by the proteins they encode (Hardin, 2004). Before the explosion of the molecular genetic approaches to the mechanisms of circadian clock oscillation that has led to the transcriptional feedback model, an influential theory of cellular oscillation was based on feedback between plasma membrane ionic conductances and changing intracellular ion concentrations (Njus et al., 1974). Despite many attempts to definitively falsify or support this hypothesis, however, results were ambiguous, and the genetic approaches led to the ascendance of transcriptional feedback models of cellular oscillation (for review, see Nitabach et al., 2005). More recently, studies performed in both flies and mammals have led to a revived appreciation for the potential importance of electrical and ionic signaling in cellular oscillation (Nitabach et al., 2002; Yamaguchi et al., 2003; Lundkvist et al., 2005; Maywood et al., 2006).
In addition, these and other studies have raised the possibility that negative transcriptional feedback may be neither necessary nor sufficient for circadian oscillation (Nitabach et al., 2002; Lundkvist et al., 2005; Nakajima et al., 2005; Tomita et al., 2005), leading to the proposal that circadian oscillation is an integrated cellular function that emerges out of transcriptional, signaling, and metabolic processes (Lakin-Thomas, 2006). The specific mechanisms by which these diverse cellular processes participate to give rise to cellular oscillation remain mysterious, but the key cellular signaling integrator Ca2+ is an intriguing candidate participant. Although circadian oscillation in dynamics of intracellular Ca2+ signals has been observed in both plant and animal cells (Johnson et al., 1995; Ikeda et al., 2003), it has remained unknown whether Ca2+ signals play an in vivo role in cellular oscillation itself.
Here we demonstrate that intracellular Ca2+ plays an essential role in regulating the period of free-running circadian oscillation in vivo. Expression of the vertebrate Ca2+ buffer protein parvalbumin (PV) is highly effective at modifying intracellular Ca2+ signals in transgenic fly neurons. When expressed in fly circadian pacemaker neurons, PV induces dose-dependent period lengthening of behavioral and cellular rhythms. Expression of a low dose of PV, which on its own has little effect on free-running rhythms, in the context of heterozygous point mutations in the calmodulin gene, which on their own also have little effect of free-running rhythms, results in substantial period lengthening. In addition, when this same low dose of PV is expressed with doses of a calmodulin (CaM)-dependent protein kinase II (CaMKII) inhibitory peptide that also have little effect on free-running rhythms, similar substantial period lengthening occurs. These results suggest a key role for intracellular Ca2+ signals in regulating intrinsic cellular oscillation in vivo and implicate calmodulin- and CaMKII-mediated pathways in their transduction.
Materials and Methods
Fly strains and crosses.
All crosses and behavioral experiments were performed at 25°C. Multiple independent chromosomal insertions of the upstream activating sequence (UAS)–PV transgene were obtained using standard embryo injection techniques and recombined using classical genetic methods to generate second and third chromosomes bearing two independent insertions each. Pigment dispersing factor (pdf) > 2× PV flies have pdf–galactosidase-4 (GAL4) transgene on the second chromosome and two UAS–PV transgenes on the third. pdf > 4× PV flies have pdf–GAL4 transgene on one second chromosome, two UAS–PV transgenes on the other second chromosome, and two UAS–PV transgenes on the third (the same insertions as in the pdf > 2× PV flies). pdf > 6× PV flies have pdf–GAL4 transgene on one second chromosome, two UAS–PV transgenes on the other second chromosome (the same insertions as in the pdf > 4× PV flies), and two UAS–PV transgenes on each third chromosome (the same insertions as in the pdf > 2× PV and pdf > 4× PV flies).
To generate flies expressing PV and also heterozygous for CaM mutant alleles, heterozygous Cam* mutant flies were crossed with pdf > 2× PV flies (the same transgene insertions as above). These two parental lines, as well as Cam* + pdf > 2× PV and Cam* + pdf–GAL4 (lacking UAS–PV transgenes) sibling progeny, were assayed for behavioral rhythms. In the case of ala CaMKII inhibitory peptide coexpression experiments, heat shock (hs) > ala flies were crossed to pdf > 2× PV flies (the same transgene insertions as above). The pdf > 2× PV parental line, as well as hs > ala + pdf > 2× PV and hs > ala + pdf–GAL4 (lacking UAS–PV transgenes) sibling progeny, were assayed for behavioral rhythms.
Ca2+ imaging.
The presynaptic Ib motoneuron terminal on muscle fiber 6 was backfilled with Oregon Green–BAPTA–Dextran, electrically stimulated, and imaged according to standard methods (Macleod et al., 2002), except that a cooled CCD camera (CoolSNAP HQ; Photometrics, Tucson, AZ) was used instead of an intensified CCD. This provides lower noise so that short enough exposures are possible to allow the measurement of Ca2+ signals produced by single action potentials (APs) in two-dimensional imaging mode rather than line scans.
Behavioral assays.
Free-running and diurnal rhythms of locomotor activity were assayed using an automated Trikinetics (Waltham, MA) monitoring system, and data were analyzed by Lomb–Scargle periodograms using Actimetrics (Wilmette, IL) Clocklab, each as described previously (Nitabach et al., 2006). Free-running periods of flies that shifted from long-period to short-period rhythms during the fourth week in constant darkness (DD) were excluded from the group averages depicted in Figure 4b.
Figure 4.
Parvalbumin expression in LNV circadian pacemaker neurons slows free-running behavioral rhythms. a, Representative actograms of individual flies expressing PV by combining pdf–GAL4 driver and indicated number of UAS–PV transgene insertions demonstrate dose-dependent slowing of free-running locomotor rhythms measured in DD. Nonexpressing 8× UAS–PV flies contain each of the independent transgene insertions used but no pdf–GAL4 driver. b, Flies expressing the highest doses of PV exhibit continuous lengthening of free-running period over time in DD. Flies expressing PV from four or six UAS–PV insertions exhibit longer periods than both nonexpressing flies and flies expressing PV from two UAS–PV insertions for all 4 weeks in DD (p < 0.01). Periods of flies expressing PV from six UAS–PV insertions increase over the 4 weeks in DD (p < 0.01) and are longer than those of flies expressing PV from four UAS–PV insertions during the fourth week in DD (p < 0.01). Line graph depicts mean ± SEM. c, Categorization of free-running rhythms during the fourth week in DD. Approximately 20% of flies expressing PV from six UAS–PV insertions suddenly shift to a short free-running period. (These individuals are excluded from the average periods shown in b.) The differences between control and experimental groups are statistically significant for “>24.5” (p < 0.001), “<24.5” (p < 0.001), and “<24.5 after being >24.5” (p < 0.05, χ2 test). d, Representative actogram of a pdf > 6× PV fly that shifts to a short-period rhythm. n > 30 flies per experimental condition. Multiple independent replicates reveal similar slowing and continuous lengthening of free-running period (data not shown). Statistical analysis is by repeated-measures ANOVA with Tukey–Kramer paired comparison test. e, Averaged normalized actograms of groups of flies of the indicated genotypes maintained for 3 weeks in LD, followed by release into DD for 1 week. f, Average free-running periods of flies maintained in DD for 4 weeks (measured for the 4th week; n > 12) or of flies maintained in LD for 3 weeks and then released into DD for 1 week (measured for that 1 week; n > 45). Mean ± SEM. There is no significant effect of environmental condition (i.e., maintenance in DD or maintenance in LD followed by shift to DD) on free-running period (p > 0.05; two-way ANOVA).
Anti-Myc and anti-par domain protein 1 immunocytochemistry.
Brains were dissected, stained for either anti-Myc or anti-par domain protein 1 (PDP1) immunofluorescence, and analyzed exactly as described previously (Nitabach et al., 2006). Immunofluorescence images were collected using a CCD camera mounted on a Zeiss (Oberkochen, Germany) Axioskop microscope. An average pixel value was computed for a 30 × 30 pixel region selected from each image by eye to best represent the background staining intensity in the region of tissue adjacent to the Drosophila melanogaster clock neurons (LNVs). A pixel value threshold was chosen for each image individually by eye to include pixels in the LNVs, both large and small, and to exclude background pixels. The average background pixel value for each image was then subtracted from the threshold-selected pixels of that image to yield the final threshold-selected background-subtracted images that were pseudocolored (with hotter colors representing greater pixel values) and used for quantitative analysis. Statistical analysis was performed on the integrated pixel values of the threshold-selected background-subtracted images. For statistical analysis, integrated pixel values were normalized within each day to the average absolute integrated pixel value for the time point and genotype with the highest average.
Statistics.
Significance of overall effects were determined using ANOVA, ordinary ANOVA, or repeated-measures ANOVA, as appropriate, and multiple comparisons of means were performed using either the Tukey–Kramer or Bonferroni's paired comparison tests.
Results
Functional expression of parvalbumin Ca2+ buffer protein in Drosophila neurons
To test the hypothesis that intracellular Ca2+ signals participate in cellular oscillation in vivo, we developed a novel transgenic approach in which we express the vertebrate Ca2+ buffer protein PV specifically in the ∼16–18 PDF-expressing lateral ventral pacemaker subset of LNVs, in the context of an otherwise unaffected nervous system. The LNVs are considered pacemakers of the circadian control circuit for several reasons. First, LNV ablation, pdf or pdf receptor null mutation, or electrical silencing of the LNVs each severely disrupt free-running locomotor rhythms (Renn et al., 1999; Nitabach et al., 2002; Hyun et al., 2005; Lear et al., 2005; Mertens et al., 2005). Second, cycling clock gene expression solely in the LNVs is sufficient to drive behavioral rhythms (Frisch et al., 1994). Third, genetically speeding up cellular oscillation solely in the LNVs speeds up behavioral rhythms (Stoleru et al., 2005). PV is a high-affinity Ca2+ buffer protein with slow binding kinetics that has no invertebrate homologs and has been demonstrated to be effective at buffering intracellular Ca2+ signals when heterologously expressed in cultured mammalian cells (Pusl et al., 2002). We use a modified form of PV that is fused to both a Myc epitope tag (for immunochemical detection of PV expression) and a mammalian nuclear export signal (to ensure accumulation in the cytoplasm) (Pusl et al., 2002). We tested the efficacy of the mammalian export signal to target cytoplasmic expression using anti-Myc immunocytochemistry and confirmed that PV protein accumulates both in the cytoplasm and nucleus of transgenic LNVs (Fig. 1).
Figure 1.
Parvalbumin fused to nuclear export signal localizes to both cytoplasm and nucleus of LNV pacemaker neurons. Anti-Myc and anti-PDF double immunofluorescence of adult fly brains expressing Myc-tagged PV and nonexpressing controls. PDF neuropeptide is present in the cytoplasm of the cell bodies and processes of LNVs but not in the nucleus. Colocalization of anti-PDF and anti-Myc staining demonstrates cytoplasmic localization of PV. Anti-Myc staining is also present in the LNV nuclei.
To confirm that PV is effective at modifying Ca2+ signals in vivo in transgenic fly neurons, we exploited the accessibility of the presynaptic motor neuron terminals of the larval neuromuscular junction for high signal-to-noise optical imaging of intracellular Ca2+ signals induced by action-potential-triggered depolarization (Macleod et al., 2002). Standard genetic crosses were used to generate fly larvae expressing PV pan-neuronally [using an elav (embryonic lethal, abnormal vision, Drosophila)–GAL4 driver transgene] from four independent UAS–PV transgene insertions in combination. Pan-neuronal expression of PV does not induce any lethality, reduced lifespan, or other gross behavioral impairment (data not shown). Nevertheless, PV-expressing presynaptic terminals exhibit altered intracellular Ca2+ dynamics compared with nonexpressing controls bearing multiple UAS–PV transgene insertions but no elav–GAL4 driver (Fig. 2). The transient Ca2+ increase induced by a single presynaptic AP triggered by electrical stimulation of the motor nerve is smaller in PV-expressing terminals (Fig. 2). The sustained Ca2+ increase induced by a 10 Hz train of APs is also quite different in PV-expressing terminals: the time to reach peak Ca2+ (3.7 ± 0.1 vs 2.8 ± 0.1 s; p < 0.01, t test) and the time constant of decay (Fig. 2) are each much longer in PV-expressing terminals. Interestingly, the peak Ca2+ reached during a train is somewhat higher in PV-expressing terminals (Fig. 2). This apparently paradoxical effect of Ca2+ buffer expression could be explained by some homeostatic alteration in presynaptic Ca2+ clearance mechanisms induced by long-term buffering. The fact that PV expression in fly presynaptic terminals alters the dynamics of AP-induced Ca2+ increases without abolishing either Ca2+ rises or synaptic release (as established by the viability of flies expressing PV pan-neuronally) is consistent with the modulatory effects of PV on Ca2+ signaling in the mammalian nervous system (Caillard et al., 2000; Collin et al., 2005). This indicates that PV is an effective tool for modifying the dynamics of intracellular Ca2+ signals in transgenic fly neurons without nonspecifically harming them or abolishing synaptic communication. Indeed, PV has been used in transgenic mice as a protective agent that prevents cytotoxicity and neuronal cell death in the following contexts: a transgenic model of amyotrophic lateral sclerosis, pharmacologically induced excitotoxicity, and physical injury to motoneurons (Beers et al., 2001; Van Den Bosch et al., 2002; Dekkers et al., 2004).
Figure 2.
Transgenic parvalbumin expression modifies intracellular Ca2+ signals. Dynamics of AP-induced presynaptic Ca2+ signals differ between control and PV-expressing motor neuron terminals. Ca2+ increase induced by single AP is smaller for PV-expressing terminals (p < 0.01) but decays at the same rate. Ca2+ increase induced by 10 Hz AP train is larger for PV-expressing terminals, reaches peak more slowly (see Results), and decays more slowly (p < 0.01 for all comparisons). Bar graphs depict mean ± SEM. n > 50 boutons (from at least 5 different animals) per genotype. Statistical analysis is by t test.
Ca2+ buffering in pacemaker neurons slows free-running circadian rhythms of locomotor activity
To test the hypothesis that intracellular Ca2+ signals in pacemaker neurons play an essential role in regulating the period of free-running circadian rhythms, we expressed a range of doses of PV in the LNVs using pdf–GAL4 LNV-specific driver transgene in combination with varying numbers of independent chromosomal insertions of the UAS–PV transgene. Semiquantitative anti-Myc immunocytochemistry reveals that, as expected, increasing numbers of UAS–PV transgene insertions result in increasing Myc-tagged PV accumulation in the cell bodies of the LNVs (Fig. 3). Although the anti-Myc immunofluorescence staining intensity is not significantly greater in flies with six UAS–PV transgenes than in flies with four UAS–PV transgenes, this immunocytochemical approach could be incapable of detecting incremental differences between already high levels of protein expression.
Figure 3.
Expression of increasing doses of parvalbumin by increasing number of UAS–PV transgene insertions. a, Anti-Myc immunofluorescence detection of Myc-tagged PV expressed in small and large LNV pacemaker neurons. b, c, High-magnification views of small (sLNV) and large (lLNV) LNV cell bodies and dorsomedial terminals of small LNVs, respectively. d, Normalized anti-Myc staining intensity of LNV cell bodies of flies with one copy of pdf–GAL4 driver transgene and the indicated number of UAS–PV transgenes. Anti-Myc staining of nucleus and cytoplasm analyzed as for anti-PDP1 staining in Figure 7. Mean ± SEM; n > 12 hemispheres for each genotype; p < 0.01. Statistical analysis is by one-way ANOVA with Tukey–Kramer paired comparison test.
Flies expressing PV in the LNVs from two, four, or six UAS–PV transgenes were entrained for 5 d to 12 h light/dark (LD) conditions and then released into DD for 4 weeks. The locomotor activity of individual flies was assayed using an automated infrared beam-crossing apparatus (Trikinetics). Figure 4a shows double-plotted actograms from representative individuals of the indicated genotypes. Flies with eight UAS–PV transgenes but no pdf–GAL4 driver transgene (8× UAS–PV) and flies with two UAS–PV transgenes and one pdf–GAL4 transgene (pdf > 2× PV) have free-running periods slightly shorter than 24 h, and these periods do not change over the course of 4 weeks in DD (Fig. 4a,b). In contrast, pdf > 4× PV and pdf > 6× PV flies have longer free-running periods of just under 25 h during the first week in DD, significantly longer than for 8× PV and pdf > 2× PV flies (Fig. 4a,b). Over the course of 4 weeks in DD, the free-running period of the pdf > 6× PV flies, which express the highest dose of PV, continues to lengthen to over 27 h by the fourth week, significantly longer than all of the other genotypes (Fig. 4a,b). This continuous lengthening would be consistent with either continued accumulation of PV in the LNVs or an accumulating effect of a constant dose of PV. Interestingly, ∼20% of flies expressing the highest dose of PV shift to a weak short-period rhythm during the fourth week in DD (Fig. 4c,d). Consistent with the weak short-period rhythms exhibited by pdf null mutant flies (Renn et al., 1999), this could represent the inability of non-PV-expressing clock neurons in the circadian control circuit to continue to follow the long-period rhythmicity of the PV-expressing pacemaker LNVs and consequent behavioral manifestation of intrinsic short-period cellular rhythms (Nitabach et al., 2006). Regardless of the mechanism for this period shift in a minority of PV-expressing flies, the dramatic dose-dependent behavioral effects of PV Ca2+ buffer expression in the LNV pacemaker neurons suggest a key role for intracellular LNV Ca2+ signals in setting free-running period.
The gradual continuous increase in free-running period of pdf > 6× PV flies could either be attributable to time-dependent accumulation of some cellular factor that slows cellular oscillation or, alternatively, could be a consequence of extended duration in DD in the absence of any environmental cues. To address this issue, we maintained flies in LD for 3 weeks, followed by release into DD, and compared the free-running period of these flies with flies that have been maintained in DD for 4 weeks (Fig. 4e,f). After release in to DD after 3 weeks in LD, pdf > 6× PV flies immediately begin to free run with a period of ∼27 h, whereas 8× UAS–PV flies free run with a period of ∼24 h. This suggests that the gradually increasing free-running period over extended time in DD is not the result of something unique to extended maintenance in DD but rather must be attributable to time-dependent accumulation of some cellular factor that occurs even while rhythms are entrained to a 24 h LD cycle. To address whether this accumulating factor is PV itself, we compared anti-Myc staining of pdf > 6× PV fly brains collected several days after eclosion with that of fly brains collected 32 d after eclosion (which corresponds to the end of the fourth week in DD). Contrary to what would be expected if it were continuous gradual accumulation of PV itself that leads to continuous gradual lengthening of period, we find that there is actually less normalized anti-Myc immunofluorescence in the >4-week-old flies (1.00 ± 0.11) than those just eclosed (0.68 ± 0.10) (n > 5 hemispheres per condition; p < 0.05, unpaired t test). This absence of increasing accumulation of PV itself suggests the continuous gradual alteration of some intrinsic period-determining component of the timekeeping mechanism that occurs even while entrained to a 24 h LD cycle.
The diurnal rhythms of pdf > 6× PV flies maintained in LD conditions are also different from those of control 8× UAS–PV flies (Fig. 5a). Although pdf > 6× PV flies exhibit robust “morning” and “evening” circadian anticipatory peaks of locomotor activity, their increases are phase delayed relative to those of control 8× UAS–PV flies; repeated-measures ANOVA reveals highly significant effects of genotype and of genotype × time interaction on the distribution of activity in the 6 h before both lights on and lights off (p < 0.001 for all effects). PV-expressing pdf > 6× PV flies are somewhat more active overall in LD than control 8× UAS–PV flies (0.91 ± 0.38 and 0.68 ± 0.26, respectively, mean beam crossings per minute ± SD; n > 130 flies; p < 0.001, t test), and there is a slight difference in the percentage of total activity that occurs in the dark versus the light, with pdf > 6× PV flies engaging in more of their activity in the dark than 8× UAS–PV flies (46 ± 9 and 41 ± 9, respectively, percentage of activity in dark ± SD; p < 0.001, t test). Accordingly, to confirm that the differences in the distribution of activity before the lights-on and lights-off transitions are not solely attributable to differences in total activity or apportionment of activity between day and night, we computed an anticipation phase score for each fly, defined as the percentage of activity in the 6 h period before lights-on or lights-off transition that occurs in the 3 h just before the transition. Larger anticipation phase scores reflect shifts in anticipatory activity to the 3 h period nearer to the lights-on or lights-off transition, thus indicating phase delays in anticipation. As seen in Figure 5b, PV-expressing pdf > 6× PV flies exhibit significantly larger anticipation phase scores than control 8× UAS–PV flies for both the lights-on and lights-off transitions (p < 0.001 for each, t test), indicating phase delays of activity increases for both morning and evening anticipation.
Figure 5.

Parvalbumin expression induces phase delays in morning and evening anticipatory activity in diurnal conditions. Averaged normalized relative activity profiles of flies maintained in 12 h LD conditions for 1 week. Both PV-expressing and control flies exhibit robust locomotor activity in anticipation of both lights on (morning peak) and lights off (evening peak). However, statistically significant phase delays of lights-on and lights-off anticipation occur in pdf > 6× PV flies, as measured by computing an anticipation phase score for each fly, defined as the percentage of activity in the 6 h before the lights-on (or lights-off) transition that occurs in the 3 h before the transition (p < 0.001 for both lights on and lights off, t test; n > 130 flies for each genotype).
This suggests that the differences in LD behavior do indeed represent differences in anticipation of environmental transitions and are not simply attributable to differences in the response to light and dark. The morning anticipatory peak is driven by circadian oscillation in the LNVs and requires their communication with downstream targets (Grima et al., 2004; Stoleru et al., 2004, 2005). Slowed increase in diurnal morning anticipatory activity in PV-expressing flies (Fig. 5) is therefore consistent with their slowed free-running rhythms (Fig. 4). Persistence of the morning peak in pdf > 6× PV flies establishes that PV expression does not prevent communication by the LNVs with downstream targets, including other clock neurons (Renn et al., 1999; Grima et al., 2004; Stoleru et al., 2004). Furthermore, whereas abolition of PDF signaling by LNVs leads to phase advances of evening anticipation (Renn et al., 1999; Nitabach et al., 2002), pdf > 6× PV flies exhibit phase delays of the evening activity increase, suggesting that PV-expression does not interfere with PDF signaling per se and rather influences phase, consistent with slowed free-running rhythms. Also consistent with the DD and LD behavioral effects, PV expression in the LNVs does not abolish the cyclic release of PDF from their dorsomedial terminals (Fig. 6), which has been linked to the coordination of multiple autonomous oscillators in the circadian control circuit (Lin et al., 2004; Stoleru et al., 2005; Nitabach et al., 2006). However, by the fifth day in DD, a phase shift in the rhythm of PDF accumulation is apparent, consistent with slowed free-running behavioral rhythms (Fig. 6).
Figure 6.
Robust phase-delayed PDF cycling in terminals of parvalbumin-expressing LNVs. Comparison of anti-PDF staining intensity in the dorsomedial terminals of PV-expressing and control nonexpressing LNVs on the second and fifth days in constant darkness (DD-D2 and DD-D5, respectively). Anti-PDF staining analyzed as for anti-PDP1 staining in Figure 7. Bar graph shows mean ± SEM. n > 12 hemispheres for each experimental condition. There is no significant effect of genotype on DD-D2. However, on DD-D5, there is a statistically significant effect of genotype. Control flies have similar peak levels of PDF immunoreactivity at circadian time 4 (CT4) and CT10, with lower levels at CT16 and CT22 (p < 0.05, ANOVA with Bonferroni's paired comparison test). In contrast, PV-expressing flies have similar peak levels of PDF immunoreactivity at CT4, CT10, and CT16, with a decrease only at CT22 (p < 0.05, ANOVA with Bonferroni's paired comparison test), consistent with a phase delay in PV-expressing flies. There is no significant difference in the magnitude of peak levels of PDF immunoreactivity between control and PV-expressing flies (p > 0.05, ANOVA with Bonferroni's paired comparison test).
To assess potential roles of Ca2+ signaling more broadly in the Drosophila circadian control circuit, we also expressed PV in all clock neurons, as well as in a variety of non-neuronal tissues that possess intrinsic circadian oscillators, using a timeless–GAL4 driver (tim–GAL4) (Blau and Young, 1999). We determined via immunocytochemical analysis that the highest level of PV expression is obtained in flies with two copies of the tim–GAL4 transgene and four copies of the UAS–PV transgene (2× tim > 4× PV; data not shown). PV expression in all clock neurons severely disrupts free-running rhythms of locomotor activity (Fig. 7). These severe phenotypes include arrhythmicity and an interesting behavior in which almost no beam crossings are detected for a number of consecutive days, followed by resumption of substantial numbers of beam crossings. Because the lifespan of Drosophila melanogaster starved of food is 2.5–4.5 d at 25°C (David et al., 1975), it is likely that most flies that cease, and then resume, crossing the infrared beam in the center of the monitoring tube are remaining near the food-containing end of the tube when no beam crossings occur. The average free-running period of rhythmic 2× tim > 4× PV flies on the third week in DD is 25.5 h, ∼2 h longer than control driver or UAS flies (Fig. 7) (p < 0.001, ANOVA with Tukey–Kramer paired comparison test) and nearly identical to that of pdf > 6× PV flies (Fig. 4). There was no apparent difference in the strength of rhythmicity of the rhythmic 2× tim > 4× PV flies compared with pdf > 6× PV flies. The severe phenotypes of high-level PV expression in all clock neurons are consistent with important roles for Ca2+ signaling in circadian timekeeping.
Figure 7.
Severe behavioral phenotypes induced by high-level parvalbumin expression in all clock neurons. Flies were generated homozygous for a tim–GAL4 transgene on the second chromosome and homozygous for two independent UAS–PV transgenes on the third chromosome (2× tim > 4× PV). Low-magnification (a) and high-magnification (b–d) views of brains of 2× tim > 4× PV flies subjected to anti-Myc immunofluorescence reveals high-level PV expression in all of the anatomical subgroups of clock neurons. e, Control flies homozygous for either the tim–GAL4 transgene alone (2× tim–GAL4) or the same third chromosome bearing the two independent UAS–GAL4 insertions and a second chromosome with another two independent UAS–GAL4 insertions (8× UAS–PV) exhibit robust free-running locomotor rhythms with average period of ∼23.5 h. In contrast, 2× tim > 4× PV flies expressing very high levels of PV in all clock neurons exhibit severely disrupted locomotor activity. Approximately 80% of 2× tim > 4× PV flies are arrhythmic by Lomb–Scargle periodogram analysis, and ∼30% also exhibit multiple-day gaps in their beam-crossing activity. These differences from control are highly significant by χ2 analysis (arrhythmicity, χ2 = 63.0, p < 0.0001; gaps, χ2 = 8.1, p < 0.02). The free-running period of rhythmic 2× tim > 4× PV flies, ∼25.5 h, is significantly longer than for controls (p < 0.01, one-way ANOVA, Tukey–Kramer paired comparison test). lLNV, Large LNV cell bodies; sLNV, small LNV cell bodies; LND, dorsal–lateral clock neuron; DN1–3, dorsal clock neuron groups 1–3.
Ca2+ buffering in pacemaker neurons slows free-running cellular oscillation
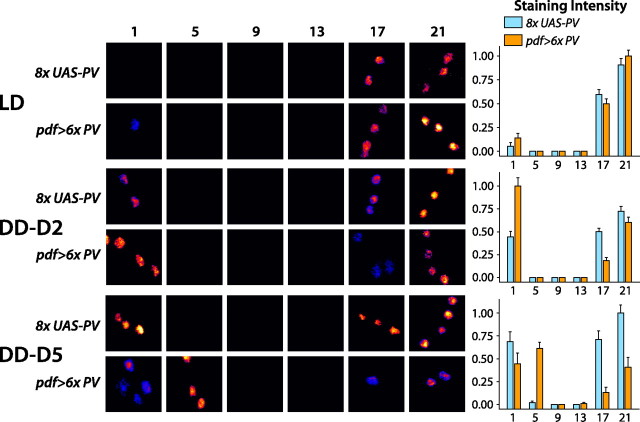
To test whether the free-running behavioral effects of PV expression result from alteration of cellular oscillation within the LNVs themselves, we directly assayed cellular oscillation using immunocytochemistry for the PDP1 transcription factor, which is known to play an essential role in circadian oscillation (Cyran et al., 2003). This protein exhibits pronounced rhythms of accumulation in the LNVs in both LD and DD (Cyran et al., 2003) and can be used as a sensitive readout of circadian oscillator phase in single cells (Nitabach et al., 2006). In environmentally entrained LD conditions, both control 8× UAS–PV and PV-expressing pdf > 6× PV LNVs exhibit robust rhythms of PDP1 accumulation with a pronounced peak at Zeitgeber time 21 (ZT21), 3 h before lights on (Fig. 8). After release into constant DD conditions, 8× UAS–PV and pdf > 6× PV LNVs both continue to exhibit robust PDP1 cycling, but the peak of PDP1 accumulation is phase delayed in pdf > 6× PV relative to 8× UAS–PV controls (Fig. 8). The peak of PDP1 accumulation that occurs near the end of subjective night of the first day in DD in 8× UAS–PV LNVs is delayed by several hours in pdf > 6× PV LNVs and thus occurs near the beginning of the following subjective morning, on the second day in DD (DD-D2) (Fig. 8). By the fifth day in DD (DD-D5), the peak of PDP1 accumulation is delayed an additional several hours in pdf > 6× PV LNVs, now occurring ∼5 h after subjective dawn (Fig. 8). Phase delays in peak PDP1 accumulation in PV-expressing LNVs are consistent with the slowing of behavioral rhythms analyzed above and indicate that the behavioral effects of LNV-specific expression of PV are a result of modification of cellular circadian oscillation.
Figure 8.
Parvalbumin expression in LNV pacemaker neurons slows free-running cellular rhythms. Anti-PDP1 clock protein immunocytochemistry reveals robust free-running cellular rhythms in PV-expressing LNVs, which are identical in magnitude and phase to nonexpressing control LNVs in 12 h LD conditions, with peak PDP1 accumulation near ZT21, 3 h before lights on. In contrast, on DD-D2, PV-expressing LNVs exhibit peak PDP1 immunoreactivity several hours later near CT1 (1 h after subjective dawn) compared with nonexpressing controls, which peak near CT21 (3 h before subjective dawn) (p < 0.01). On DD-D5, the PDP1 peak has shifted even later in PV-expressing LNVs, near CT5 (p < 0.01). Bar graphs depict mean ± SEM. n > 11 brain hemispheres per experimental condition. An independent replicate of the entire experiment revealed similar phase delays (data not shown). Statistical analysis by ANOVA with Tukey–Kramer paired comparison test.
Ca2+ buffering interacts synergistically with genetic manipulation of calmodulin and CaMKII signaling pathways
To begin to identify the downstream pathways by which intracellular Ca2+ signals influence circadian oscillation, we tested for interactions between PV-mediated Ca2+ buffering and genetic manipulations that interfere with the function of known molecular components of candidate Ca2+-sensitive pathways. Multiple Ca2+-dependent signaling events are based on initial binding of Ca2+ to CaM, a ubiquitous eukaryotic Ca2+ binding protein, followed by binding of the Ca2+/calmodulin complex to signaling enzymes such as protein kinases and phosphatases, ion channels, and other signaling molecules (for review, see Sola et al., 2001). We expressed a relatively low dose of PV from two independent transgene insertions in the LNVs of flies that were also heterozygous for a variety of CaM hypomorphic mutant alleles (Nelson et al., 1997). These heterozygous CaM mutations have little effect on free-running period on their own (Fig. 9, green circles and red triangles), similar to low-dose expression of PV in LNVs using pdf–GAL4 driver (Fig. 9, black triangles). However, for some of the CaM mutations, flies both expressing PV and heterozygous for the CaM mutation exhibit period lengthening that is significantly different from pdf > 2× PV and heterozygous CaM mutant flies (Fig. 9, blue squares, *p < 0.01, ANOVA with Tukey–Kramer paired comparison test). This synergistic effect was observed for three of four CaM point mutant alleles, Cam4c1, Cam5, and Cam7, but not for the Camn339 null or the Cam3c1 point mutant.
Figure 9.

Synergistic interaction between heterozygous calmodulin mutation and low-dose parvalbumin expression. Each line graph depicts a low dose of PV expressed from two independent UAS–PV transgenes in the context of the indicated heterozygous calmodulin mutation. Cam* (red triangles) indicates parental heterozygous calmodulin mutant flies, pdf > 2× PV (black triangles) indicates parental PV-expressing flies, Cam* + pdf-GAL4 (green circles) indicates heterozygous calmodulin mutant offspring with pdf-GAL4 driver but no UAS–PV transgenes, and Cam* + pdf > 2× PV (blue squares) indicates sibling offspring heterozygous for calmodulin mutation bearing both pdf–GAL4 and two independent UAS–PV transgenes. Asterisks indicate that offspring heterozygous for calmodulin mutation bearing both pdf–GAL4 and two independent UAS–PV transgenes have significantly longer average period than parental control heterozygous calmodulin mutant and PV-expressing flies and sibling control heterozygous calmodulin mutant offspring with pdf–GAL4 driver but no UAS–PV transgenes (p < 0.01, repeated-measures ANOVA with Tukey–Kramer paired comparison test). Minimum numbers of flies are as follows for all genotypes in experiments involving each of the calmodulin mutations: Cam3c1, n > 10; Cam4c1, n > 6; Cam5, n > 4; Cam7, n > 22; Camn339, n > 10. Number of flies are as follows for the experimental Cam* + pdf > 2× PV flies heterozygous for calmodulin mutation bearing both pdf–GAL4 and two independent UAS–PV transgenes: Cam3c1, n > 16; Cam4c1, n > 14; Cam5, n > 4; Cam7, n > 38; Camn339, n > 18.
The allele-specific synergistic effect on circadian period of combining PV-mediated Ca2+ buffering with heterozygous CaM hypomorphic alleles suggests that cytoplasmic Ca2+ signals important for circadian timekeeping are transduced, at least in part, by CaM-sensitive downstream pathways. Furthermore, this synergy provides strong support for the conclusion that PV effects on circadian rhythms are, indeed, attributable to the Ca2+ buffering capacity of PV and not attributable to some other effect of PV expression. This is because the only plausible functional convergence between the effects of PV and CaM mutation is that they both affect cellular Ca2+ signaling. The superficially paradoxical lack of synergy with the null CaM allele can be explained by the role of CaM as an abundant cellular Ca2+ buffer. Flies heterozygous for the null allele, in addition to interference with Ca2+/CaM regulation of downstream signaling pathways, also are likely to already have substantially reduced cellular Ca2+ buffering capacity and thus are less sensitive to the effects of increased Ca2+ buffering mediated by exogenously introduced PV. Consistent with this interpretation, the Cam3c1 point-mutant allele that does not interact synergistically with PV has decreased affinity for Ca2+ compared with wild type (Maune et al., 1992a,b).
Because CaMKII is an important Ca2+/CaM-sensitive target that could transduce Ca2+ signals important for circadian function, we also tested for interaction between low-dose PV expression and expression of the ala CaMKII inhibitory peptide (Griffith et al., 1993). Ubiquitous expression of ala peptide from either of two independent chromosomal insertions of a hsp70 heat-shock promoter > ala transgene (ala1 and ala2), which are constitutively active even in the absence of heat shock (Griffith et al., 1993), has little or no effect on free-running circadian period (Fig. 10, blue triangles and red circles), similarly to LNV expression of PV in pdf > 2× PV flies (Fig. 10, black squares). In contrast, flies expressing both PV and ala from the ala2 insertion exhibit period lengthening that is significantly different from that of the pdf > 2× PV and ala2 flies (Fig. 10, light blue diamonds, *p < 0.01, ANOVA with Tukey–Kramer paired comparison test). Although there is a possible trend of period lengthening by the fourth week in DD for flies expressing PV and ala from the ala1 insertion, this trend is not statistically significant (Fig. 10, green triangles). The stronger synergistic effect of ala expression from the ala2 chromosomal insertion is exactly what one would expect from the fact that ala inhibitory peptide has been demonstrated to be expressed at substantially higher levels from the ala2 insertion than from ala1 and, consequently, leads to substantially stronger behavioral phenotypes (Griffith et al., 1993). This synergy of Ca2+ buffering by PV and CaMKII inhibition by ala inhibitory peptide suggests that cytoplasmic Ca2+ signals important for circadian timekeeping are transduced, at least in part, by CaMKII-mediated phosphorylation of protein targets and also provides additional strong support for the conclusion that PV effects on circadian rhythms are, indeed, attributable to the Ca2+ buffering capacity of PV.
Figure 10.
Synergistic interaction between expression of ala CaMKII inhibitory peptide and low-dose parvalbumin expression. Flies homozygous for either the ala1 or ala2 independent chromosomal insertions of the hs > ala transgene were mated to pdf > 2× PV flies to generate flies with a single copy of the hs > ala transgene, pdf > GAL4 driver transgene, and two independent insertions of the UAS–PV transgene. pdf > 2× PV (purple diamonds) indicates parental PV-expressing flies, ala + pdf–GAL4 indicates single-copy hs > ala offspring with pdf–GAL4 driver but no UAS–PV transgenes (red and black triangles), and ala + pdf > 2× PV indicates sibling single-copy hs > ala offspring with both pdf > GAL4 driver transgene and two independent insertions of the UAS–PV transgene (blue squares and green circles). Asterisks indicate that ala2 + pdf > 2× PV flies have significantly longer free-running period than parental control pdf > 2× PV and sibling control ala2 + pdf-GAL4 flies (p < 0.01, repeated-measures ANOVA with Tukey–Kramer paired comparison test). n > 12 flies for all genotypes. n = 17 for ala2 + pdf > 2× PV experimental flies.
Discussion
Ca2+ signals play important roles in a host of cellular processes, and several studies have addressed possible roles for them in circadian oscillation. Optical imaging techniques have demonstrated the existence of circadian oscillation of intracellular Ca2+ levels in mammalian clock neurons and plant cells in vitro (Johnson et al., 1995; Colwell, 2000; Ikeda et al., 2003). The high-affinity, rapid-kinetics Ca2+ buffer BAPTA-AM damps and, ultimately, abolishes circadian rhythms of period promoter activity of cultured mammalian clock neurons in vitro without affecting their free-running period (Lundkvist et al., 2005) at concentrations that are high enough to substantially suppress Ca2+ transients and attenuate synaptic communication (Ouanounou et al., 1996). Although these studies suggest the possibility that intracellular Ca2+ signals could be important for regulating free-running cellular oscillation in vivo, this possibility has not before been tested.
Here we specifically expressed a range of doses of the high-affinity, slow-kinetics Ca2+ buffer PV in Drosophila pacemaker neurons in vivo and thereby demonstrated dramatic dose-dependent effects on cellular and behavioral rhythms (Figs. 4–8). High levels of PV do not prevent synaptic communication in either fly motoneurons or the mammalian cerebellum, only subtly alter intracellular Ca2+ dynamics, and do not interfere with cyclic PDF release (Figs. 1, 6) (Caillard et al., 2000; Collin et al., 2005). We thus propose that the effects of PV expression in the LNVs are attributable to modification of intracellular Ca2+ signals that play a direct role in autonomous cellular oscillation in vivo rather than attributable to modification of intercellular communication within the circadian control circuit. Consistent with this, a recent study performed in cultured mammalian suprachiasmatic nucleus (SCN) revealed that application of membrane-permeant Ca2+ buffer or a mixture of Ca2+ channel blockers almost immediately halts circadian transcriptional rhythms (Lundkvist et al., 2005), too quickly to likely be explained by a circuit level mechanism (Yamaguchi et al., 2003). Our studies build on these in vitro studies and generalize the suggestion of a role for Ca2+ signaling to an in vivo circadian timekeeping context. Interesting differences that remain to be explored are (1) the fact that interference with Ca2+ signals in cultured SCN does not affect free-running period and (2) the fact that the period phenotypes we observe in vivo in the fly increase their magnitude over time in either entrained or constant conditions, although PV Ca2+ buffer levels themselves do not increase.
Our studies also go beyond those performed on cultured SCN in beginning to explore the downstream Ca2+-sensitive signaling pathways through which Ca2+ signals participate in circadian timekeeping. Expression of a low dose of PV, which on its own has little effect on free-running rhythms, in the context of heterozygous point mutations in the calmodulin gene, which on their own also have little effect of free-running rhythms, results in substantial period lengthening (Fig. 9). In addition, when this same low dose of PV is expressed with doses of a CaMKII inhibitory peptide that also have little effect on free-running rhythms, similar substantial period lengthening occurs (Fig. 10). These results suggest a key role for intracellular Ca2+ signals in regulating intrinsic cellular oscillation in vivo and implicate calmodulin- and CaMKII-mediated pathways in their transduction. Interestingly, a role for CaMKII-mediated phosphorylation in regulation of the CLOCK/CYCLE core circadian transcription factor has been demonstrated recently in a reconstituted tissue culture system (Weber et al., 2006). Our studies raise a number of issues for additional investigation: the cellular sources of the Ca2+ signals important for cellular oscillation, their spatiotemporal dynamics, and the mechanisms by which Ca2+-sensitive signaling intermediates, such as Ca2+/CaM and CaMKII, couple them to circadian transcriptional feedback loops. Another good candidate for integrating Ca2+ signals into the cellular timekeeping mechanism is protein phosphatase IIA, a known Drosophila clock component that directly binds and is regulated by Ca2+ (Janssens et al., 2003; Sathyanarayanan et al., 2004). Finally, the interaction studies that revealed roles for CaM and CaMKII in Ca2+-dependent regulation of circadian timekeeping are proof-of-principle that low-dose PV expression in LNV pacemaker neurons constitutes a sensitized background suitable for large-scale forward screening for other Ca2+-sensitive cellular signaling pathways involved in circadian timekeeping.
Footnotes
M.C.H. was supported in part by a Yale Seessel Fellowship. Y.W. was supported in part by National Research Service Award from the National Institute of Neurological Disorders and Stroke (NINDS). Work in the laboratory of M.N.N. was supported in part by the Whitehall Foundation and NINDS Grants R01NS055035 and R01NS056443. Work in the laboratory of G.A.L. was supported in part by National Science Foundation Grant IOB 0543835. We thank B. Ehrlich, A. Bennett, and M. Nathanson for parvalbumin cDNA constructs and for valuable discussions and J. Blau for anti-PDP1 antiserum. We thank L. Griffith and the Bloomington Stock Center for fly stocks.
References
- Beers DR, Ho BK, Siklos L, Alexianu ME, Mosier DR, Mohamed AH, Otsuka Y, Kozovska ME, McAlhany RE, Smith RG, Appel SH. Parvalbumin overexpression alters immune-mediated increases in intracellular calcium, and delays disease onset in a transgenic model of familial amyotrophic lateral sclerosis. J Neurochem. 2001;79:499–509. doi: 10.1046/j.1471-4159.2001.00582.x. [DOI] [PubMed] [Google Scholar]
- Blau J, Young MW. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. doi: 10.1016/s0092-8674(00)81554-8. [DOI] [PubMed] [Google Scholar]
- Caillard O, Moreno H, Schwaller B, Llano I, Celio MR, Marty A. Role of the calcium-binding protein parvalbumin in short-term synaptic plasticity. Proc Natl Acad Sci USA. 2000;97:13372–13377. doi: 10.1073/pnas.230362997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin T, Chat M, Lucas MG, Moreno H, Racay P, Schwaller B, Marty A, Llano I. Developmental changes in parvalbumin regulate presynaptic Ca2+ signaling. J Neurosci. 2005;25:96–107. doi: 10.1523/JNEUROSCI.3748-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell CS. Circadian modulation of calcium levels in cells in the suprachiasmatic nucleus. Eur J Neurosci. 2000;12:571–576. doi: 10.1046/j.1460-9568.2000.00939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, Young MW, Storti RV, Blau J. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–341. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- David J, Cohet Y, Fouillet P. Resistance to starvation in insects: importance of the amount of lipid reserves in adult Drosophila melanogaster (in French) C R Acad Sci Hebd Seances Acad Sci D. 1975;280:2571–2574. [PubMed] [Google Scholar]
- Dekkers J, Bayley P, Dick JR, Schwaller B, Berchtold MW, Greensmith L. Over-expression of parvalbumin in transgenic mice rescues motoneurons from injury-induced cell death. Neuroscience. 2004;123:459–466. doi: 10.1016/j.neuroscience.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Frisch B, Hardin PE, Hamblen-Coyle MJ, Rosbash M, Hall JC. A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron. 1994;12:555–570. doi: 10.1016/0896-6273(94)90212-7. [DOI] [PubMed] [Google Scholar]
- Griffith LC, Verselis LM, Aitken KM, Kyriacou CP, Danho W, Greenspan RJ. Inhibition of calcium/calmodulin-dependent protein kinase in Drosophila disrupts behavioral plasticity. Neuron. 1993;10:501–509. doi: 10.1016/0896-6273(93)90337-q. [DOI] [PubMed] [Google Scholar]
- Grima B, Chelot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- Hardin PE. Transcription regulation within the circadian clock: the E-box and beyond. J Biol Rhythms. 2004;19:348–360. doi: 10.1177/0748730404268052. [DOI] [PubMed] [Google Scholar]
- Hyun S, Lee Y, Hong ST, Bang S, Paik D, Kang J, Shin J, Lee J, Jeon K, Hwang S, Bae E, Kim J. Drosophila GPCR Han is a receptor for the circadian clock neuropeptide PDF. Neuron. 2005;48:267–278. doi: 10.1016/j.neuron.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Sugiyama T, Wallace CS, Gompf HS, Yoshioka T, Miyawaki A, Allen CN. Circadian dynamics of cytosolic and nuclear Ca2+ in single suprachiasmatic nucleus neurons. Neuron. 2003;38:253–263. doi: 10.1016/s0896-6273(03)00164-8. [DOI] [PubMed] [Google Scholar]
- Janssens V, Jordens J, Stevens I, Van Hoof C, Martens E, De Smedt H, Engelborghs Y, Waelkens E, Goris J. Identification and functional analysis of two Ca2+-binding EF-hand motifs in the B″/PR72 subunit of protein phosphatase 2A. J Biol Chem. 2003;278:10697–10706. doi: 10.1074/jbc.M211717200. [DOI] [PubMed] [Google Scholar]
- Johnson CH, Knight MR, Kondo T, Masson P, Sedbrook J, Haley A, Trewavas A. Circadian oscillations of cytosolic and chloroplastic free calcium in plants. Science. 1995;269:1863–1865. doi: 10.1126/science.7569925. [DOI] [PubMed] [Google Scholar]
- Lakin-Thomas PL. Transcriptional feedback oscillators: maybe, maybe not. J Biol Rhythms. 2006;21:83–92. doi: 10.1177/0748730405286102. [DOI] [PubMed] [Google Scholar]
- Lear BC, Merrill CE, Lin JM, Schroeder A, Zhang L, Allada R. A G protein-coupled receptor, groom-of-PDF, is required for PDF neuron action in circadian behavior. Neuron. 2005;48:221–227. doi: 10.1016/j.neuron.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Lin Y, Stormo GD, Taghert PH. The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J Neurosci. 2004;24:7951–7957. doi: 10.1523/JNEUROSCI.2370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundkvist GB, Kwak Y, Davis EK, Tei H, Block GD. A calcium flux is required for circadian rhythm generation in mammalian pacemaker neurons. J Neurosci. 2005;25:7682–7686. doi: 10.1523/JNEUROSCI.2211-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod GT, Hegstrom-Wojtowicz M, Charlton MP, Atwood HL. Fast calcium signals in Drosophila motor neuron terminals. J Neurophysiol. 2002;88:2659–2663. doi: 10.1152/jn.00515.2002. [DOI] [PubMed] [Google Scholar]
- Maune JF, Klee CB, Beckingham K. Ca2+ binding and conformational change in two series of point mutations to the individual Ca2+-binding sites of calmodulin. J Biol Chem. 1992a;267:5286–5295. [PubMed] [Google Scholar]
- Maune JF, Beckingham K, Martin SR, Bayley PM. Circular dichroism studies on calcium binding to two series of Ca2+ binding site mutants of Drosophila melanogaster calmodulin. Biochemistry. 1992b;31:7779–7786. doi: 10.1021/bi00149a006. [DOI] [PubMed] [Google Scholar]
- Maywood ES, Reddy AB, Wong GK, O'Neill JS, O'Brien JA, McMahon DG, Harmar AJ, Okamura H, Hastings MH. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr Biol. 2006;16:599–605. doi: 10.1016/j.cub.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Mertens I, Vandingenen A, Johnson EC, Shafer OT, Li W, Trigg JS, De Loof A, Schoofs L, Taghert PH. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron. 2005;48:213–219. doi: 10.1016/j.neuron.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- Nelson HB, Heiman RG, Bolduc C, Kovalick GE, Whitley P, Stern M, Beckingham K. Calmodulin point mutations affect Drosophila development and behavior. Genetics. 1997;147:1783–1798. doi: 10.1093/genetics/147.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach MN, Blau J, Holmes TC. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell. 2002;109:485–495. doi: 10.1016/s0092-8674(02)00737-7. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Holmes TC, Blau J. Membranes, ions, and clocks: testing the njus-sulzman-hastings model of the circadian oscillator. Methods Enzymol. 2005;393:682–693. doi: 10.1016/S0076-6879(05)93036-X. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Wu Y, Sheeba V, Lemon WC, Strumbos J, Zelensky PK, White BH, Holmes TC. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J Neurosci. 2006;26:479–489. doi: 10.1523/JNEUROSCI.3915-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njus D, Sulzman FM, Hastings JW. Membrane model for the circadian clock. Nature. 1974;248:116–120. doi: 10.1038/248116a0. [DOI] [PubMed] [Google Scholar]
- Ouanounou A, Zhang L, Tymianski M, Charlton MP, Wallace MC, Carlen PL. Accumulation and extrusion of permeant Ca2+ chelators in attenuation of synaptic transmission at hippocampal CA1 neurons. Neuroscience. 1996;75:99–109. doi: 10.1016/0306-4522(96)00319-3. [DOI] [PubMed] [Google Scholar]
- Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- Pusl T, Wu JJ, Zimmerman TL, Zhang L, Ehrlich BE, Berchtold MW, Hoek JB, Karpen SJ, Nathanson MH, Bennett AM. Epidermal growth factor-mediated activation of the ETS domain transcription factor Elk-1 requires nuclear calcium. J Biol Chem. 2002;277:27517–27527. doi: 10.1074/jbc.M203002200. [DOI] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Sathyanarayanan S, Zheng X, Xiao R, Sehgal A. Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell. 2004;116:603–615. doi: 10.1016/s0092-8674(04)00128-x. [DOI] [PubMed] [Google Scholar]
- Sola C, Barron S, Tusell JM, Serratosa J. The Ca2+/calmodulin system in neuronal hyperexcitability. Int J Biochem Cell Biol. 2001;33:439–455. doi: 10.1016/s1357-2725(01)00030-9. [DOI] [PubMed] [Google Scholar]
- Stanewsky R. Genetic analysis of the circadian system in Drosophila melanogaster and mammals. J Neurobiol. 2003;54:111–147. doi: 10.1002/neu.10164. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Nawathean P, Rosbash M. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature. 2005;438:238–242. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- Tomita J, Nakajima M, Kondo T, Iwasaki H. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307:251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- Van Den Bosch L, Schwaller B, Vleminckx V, Meijers B, Stork S, Ruehlicke T, Van Houtte E, Klaassen H, Celio MR, Missiaen L, Robberecht W, Berchtold MW. Protective effect of parvalbumin on excitotoxic motor neuron death. Exp Neurol. 2002;174:150–161. doi: 10.1006/exnr.2001.7858. [DOI] [PubMed] [Google Scholar]
- Weber F, Hung HC, Maurer C, Kay SA. Second messenger and Ras/MAPK signalling pathways regulate CLOCK/CYCLE-dependent transcription. J Neurochem. 2006;98:248–257. doi: 10.1111/j.1471-4159.2006.03865.x. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]