Abstract
Human bitter taste is mediated by ∼25 members of the human TAS2 receptor (hTAS2R) gene family. The hTAS2R genes are expressed in taste buds of gustatory papillae on the tongue surface. Because many naturally occurring bitter compounds are toxic, bitter taste receptors are believed to serve as warning sensors against the ingestion of toxic food compounds. An important question is whether bitter taste receptor cells are a homogeneous, broadly tuned population of cells, which uniformly express all bitter taste receptor genes, or not. Gene expression analyses in rodents demonstrated an essentially overlapping expression of TAS2R genes indicating a broad tuning, whereas functional in vivo analyses suggest a narrow tuning. The present study demonstrates the expression of all 25 human TAS2R genes in taste receptor cells of human circumvallate papillae. As shown by in situ hybridization experiments, the expression of hTAS2R genes differs in both the apparent level of expression and the number of taste receptor cells expressing these genes, suggesting a heterogeneous bitter taste receptor cell population. Differences in gene expression levels were verified by quantitative reverse transcription-PCR experiments for a subset of hTAS2R genes. Direct evidence for the heterogeneity of bitter taste receptor cells is provided by dual-labeling in situ hybridizations with selected pairs of hTAS2R gene-specific probes. Functional coexpression experiments in heterologous cells show competition among hTAS2Rs, indicating a possible biological reason for the observed expression pattern. From the data, we conclude that human bitter taste receptor cells are tuned to detect a limited subset of bitter stimuli.
Keywords: human bitter taste, gene expression, hTAS2R genes, heterologous expression, in situ hybridization, taste perception
Introduction
Of the five basic taste qualities of vertebrates, bitter taste is most complex. An enormous number of structurally diverse bitter tastants are detected by ∼25 receptors belonging to the human TAS2 receptor (hTAS2R) gene family (Meyerhof, 2005). In general, it is believed that TAS2Rs serve as warning sensors against the ingestion of toxic food compounds, although the correlation between bitterness and toxicity can vary (Glendinning, 1994). Morphologically, bitter taste receptor cells (TRCs) are located in gustatory papillae on the tongue in which they are organized, together with TRCs specific for the other taste qualities, in groups of 60–100 cells, the taste buds. Additionally, taste buds are found on the soft palate, larynx, and pharynx (Miller, 1995). Aside from the taste system, TAS2R genes are expressed in solitary chemosensory cells (Sbarbati et al., 1999) within the respiratory (Finger et al., 2003) and digestive tract (Wu et al., 2002), raising the intriguing possibility that not all TAS2R genes may have gustatory functions.
Behavioral and psychophysical studies performed in several species, including humans, did not provide a clear answer to the question of whether vertebrates can discriminate between bitter compounds. Whereas some studies indicate that bitter taste processing is, depending on the substances tested, not uniform (McBurney et al., 1972; Whitney and Harder, 1994; Dahl et al., 1997; Delwiche et al., 2001; Frank et al., 2004), other studies concluded that purely bitter tasting compounds cannot be distinguished (Spector and Kopka, 2002).
A non-uniform expression of TAS2R genes in TRCs would be a prerequisite for an animal's ability to discriminate between bitter stimuli at the cellular level. Several studies investigated the coexpression of TAS2Rs in rodent TRCs. It is undisputed that rodent TAS2R genes exhibit an overlapping expression pattern in TRCs as investigated by in situ hybridizations, but the degree of overlap, however, is still a matter of debate (Adler et al., 2000; Matsunami et al., 2000). A genetic rescue strategy using TAS2R promoter sequences guiding the expression of phospholipase Cβ2 (PLCβ2), a critical component of bitter taste signal transduction, in bitter taste-deficient PLCβ2−/− mice restores normal bitter taste to all compounds tested, demonstrating coexpression of PLCβ2 with endogenous TAS2Rs in a sufficient number of TRCs (Mueller et al., 2005). In vivo experiments, however, demonstrated that most cells responded to only one or two of five bitter stimuli presented, indicating functional heterogeneity of the TRC population (Caicedo and Roper, 2001).
In the present study, we provide a comprehensive analysis of the expression of all 25 hTAS2Rs in gustatory papillae. Expression of hTAS2Rs was monitored on a cellular level. Because the in situ hybridization data revealed that the population of bitter TRCs in human circumvallate papillae (CV) is not uniform, we performed dual-label in situ hybridization of hTAS2R pairs to substantiate this finding. Quantitative PCR of selected TAS2R cDNAs was used to correlate apparent signal levels and expression frequencies obtained by in situ hybridization reactions with corresponding cDNA amounts. Finally, functional experiments were done elucidating potential consequences for human bitter taste arising from the observed expression characteristics.
Materials and Methods
Reverse transcription-PCR.
Biopsy materials of human CV and nonchemosensory filiform papillae were subjected to RNA extractions using TRIzol reagent (Invitrogen, Karlsruhe, Germany). For removal of contaminating genomic DNA, RNAs were subjected to DNaseI (Invitrogen) digestion for 30 min at room temperature according to the instructions of the manufacturer. cDNA synthesis was done using SuperScript II reverse transcriptase and random hexamers (Invitrogen) following the recommendations of the manufacturer. Identical reactions omitting reverse transcriptase were performed to generate negative control templates. cDNA corresponding to 7–9 ng of reverse-transcribed RNA was used for each bitter taste receptor amplification reaction. For each receptor, an amplification of cDNA over 40 cycles and a negative control reaction was performed using TaqDNA-polymerase and standard PCR conditions. Oligonucleotide sequences are given in Table 1. Amplification of hTAS2R cDNA was visualized by agarose gel electrophoresis. Because the eight receptors hTAS2R43, -44, -45, -46, -47, -48, -49, and -50 are closely related to each other, exhibiting nucleotide sequence identities between 80 and >90%, cross-amplification was carefully controlled. Amplification of equimolar mixtures of the cloned receptor cDNAs of hTAS2R43–hTAS2R50 with the corresponding primer pairs resulted in amplicons of the expected size, whereas the primer pairs failed to amplify from identical mixtures containing only the seven related but excluding the specific receptor cDNA.
Table 1.
List of oligonucleotides used for RT-PCR analyses of human circumvallate papillae cDNA
| Oligonucleotide | Sequence (5′ to 3′) | Ann. | Amplicon size |
|---|---|---|---|
| hTAS2R1_for | TGTGGTGGTGAATGGCATTG | 62°C | 790 bp |
| hTAS2R1_rev | CAGCACTTACTGTGGAGGAGGAAC | ||
| hTAS2R3_for | ACACATGATTCAGGGATAATAATGCAAA | 60°C | 697 bp |
| hTAS2R3_rev | TTAGCCATCTTGGTTTTTGGTAGGAAATT | ||
| hTAS2R4_for | TACAGTGGTCAATTGCAAAACTTGG | 62°C | 726 bp |
| hTAS2R4_rev | AATGTCCTGGAGAGTAAAGGGTGG | ||
| hTAS2R5_for | TGGTCCTCATATAACCTCATTATCCTGG | 63°C | 668 bp |
| hTAS2R5_rev | CTGCCATGAGTGTCTCCCA | ||
| hTAS2R7_for | TGTTTTATATTGGTGCTATATCCAGATGTCTATGC | 64°C | 664 bp |
| hTAS2R7_rev | GGATAAATGAATGACTTGAGGGGTAGATTAGAG | ||
| hTAS2R8_for | CAATTTAGTTATCGCCAGAATTTGTTTGATC | 64°C | 722 bp |
| hTAS2R8_rev | TTATTTAAAACAATTAAAATAAGTGAGTGACCCAAGG | ||
| hTAS2R9_for | TGAATTGACCATAGGGATTTGGG | 62°C | 807 bp |
| hTAS2R9_rev | ATAATTAGAATGAATGAATGGCTTGATGG | ||
| hTAS2R10_for | GACTTGTAAACTGCATTGACTGTGCC | 62°C | 784 bp |
| hTAS2R10_rev | AAAGAGGCTTGCTTTAGCTTGCTG | ||
| hTAS2R13_for | GGGTCAGTAAAAGAGAGCTGTCCTC | 61°C | 743 bp |
| hTAS2R13_rev | ATCAGAAGAAAGGAGTGGCTTGAAG | ||
| hTAS2R14_for | GCTTTGGCAATCTCTCGAATTAGC | 60°C | 771 bp |
| hTAS2R14_rev | CTCTAAATTCTTTGTGACCTGAGGGC | ||
| hTAS2R16_for | CCTGGGAATTTTTTAATATCCTTACATTCTGGT | 63°C | 449 bp |
| hTAS2R16_rev | GAAGCGCGCTTTCATGCTT | ||
| hTAS2R38_for | ACAGTGATTGTGTGCTGCTG | 58°C | 766 bp |
| hTAS2R38_rev | GCTCTCCTCAACTTGGCATT | ||
| hTAS2R39_for | TGTCGCCATTTCTCATCACCTTA | 62°C | 849 bp |
| hTAS2R39_rev | ATTGAGTGGCTGGCAGGGTAG | ||
| hTAS2R40_for | AGAGTGCATCACTGGCATCCTT | 60°C | 685 bp |
| hTAS2R40_rev | GAGGATGAGAAAGTAGCTGGTGGC | ||
| hTAS2R41_for | GGTTGCTGCCCTTGGATATGA | 63°C | 740 bp |
| hTAS2R41_rev | TGAAGATGAGGATGAAGGGATGG | ||
| hTAS2R42_for | ATGGCCACCGAATTGGACA | 62°C | 850 bp |
| hTAS2R42_rev | GCTTGCTGTTTCCCAGAATGAG | ||
| hTAS2R43_for | GCTAATGGCTTCATAGCAC | 52°C | 589 bp |
| hTAS2R43_rev | GATCTTGAGATCCTTTACCAC | ||
| hTAS2R44_for | CATTGGTAAATTCCATTGAGC | 53°C | 653 bp |
| hTAS2R44_rev | GATATCATTATGGACAGAAAGTAAAC | ||
| hTAS2R45_for | CTCCTTTGCTGACCAAATTGTC | 58°C | 710 bp |
| hTAS2R45_rev | GAACGGGTGGGCTGAAGAAC | ||
| hTAS2R46_for | GAGTTGAATCCAGCTTTTAAC | 52°C | 607 bp |
| hTAS2R46_rev | ATAGCTGAATGCAATAGCTTC | ||
| hTAS2R47_for | GGTGTTATTACTACATTGGTATGCAACTC | 59°C | 604 bp |
| hTAS2R47_rev | AAGACAGGTTGCTTTTCCAGC | ||
| hTAS2R48_for | GGTTTACTCTGGGTCATGTTATTC | 56°C | 607 bp |
| hTAS2R48_rev | TTTGCTCTGCTGTGTCCTAAG | ||
| hTAS2R49_for | GCACTGATAAATTTCATTGCCTGG | 62°C | 786 bp |
| hTAS2R49_rev | TTGTTCCCCCAAATCAGAATGAAT | ||
| hTAS2R50_for | GGTAAATTTCATTGACTGGGTGAAGAG | 62°C | 710 bp |
| hTAS2R50_rev | CCTTGCTAACCATGACAACTGGG | ||
| hTAS2R60_for | CAGGCAATGGCTTCATCACTG | 63°C | 748 bp |
| hTAS2R60_rev | TCCCACACCCAGAATTTAAAGTCC |
The oligonucleotide sequences, the annealing temperatures used for amplification reactions (Ann.), and the corresponding amplicon sizes are given. for, Forward; rev, reverse.
Quantitative RT-PCR.
For cDNA preparation from human CV, see the previous section. Gene-specific primers and TaqMan probes were used to amplify hTAS2R1, hTAS2R16, and hTAS2R38 (hTAS2R1_forward, 5′-TCGGCGTTTTCTATTGTGCCAAGG-3′; hTAS2R1_reverse, 5′-CATATAGCAGAGACCCCAGGATCATCC-3′; TaqMan probe, 5′-TGCCAGCGTCCGTCACCCACTCTTCATC-3′; hTAS2R16_forward, 5′-GGTCACTGCAATCCAAGCATGAAAGC-3′; hTAS2R16_reverse, 5′-AGCTTCCCAGACCCATAACCAACATC-3′; TaqMan probe, 5′-CGCTTCACTGCCCTGAGGTCCCTTGCCGTC-3′; hTAS2R38_forward, 5′-AAATAGGGGTGATGGTTTGTGTTGGG-3′; hTAS2R38_reverse, 5′-TCAGCACAGTGTCCGGGAATCTG-3′; TaqMan probe, 5′-CTTGTGGTCGGCTCTTACCTTCAGGCTGCTCT-3′). For quantification analyses, the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH), for which primers and probe were designed and synthesized by the Applied Biosystem (Foster City, CA) TaqMan Gene Expression Assay customer service, served as internal control. Quantitative real-time PCR analysis was performed on a TaqMan ABI Prism 7300 Real Time PCR System (Applied Biosystems, Darmstadt, Germany). For quantitative RT (qRT)-PCR analysis of hTAS2R1 and hTAS2R38, both forward and reverse primers were used at a concentration of 0.3 μm, whereas the primer pair of TAS2R16 was used at a concentration of 0.6 μm. In all cases, the probes were used at a final concentration of 0.1 μm. For the PCR reaction, 100 ng of cDNA template was incubated with iTaq Supermix with ROX containing dNTPs with dUTP and iTaq DNA polymerase (P/N 170-8854; Bio-Rad, Hercules, CA) in a final volume of 25 μl. Cycling parameters were as follows: 95°C for 3 min for initial denaturation, followed by 40 cycles of 15 s at 95°C, 45 s at 60°C for hTAS2R16 and hTAS2R38, and 15 s at 95°C, 45 s at 58°C for hTAS2R1, respectively. Each cDNA (+RT) and RNA (−RT) sample was tested in triplicate, and mean threshold cycle values were reported. Furthermore, for each reaction, a sample without template was included as a negative control. Raw data were acquired and processed with the Applied Biosystem 7300 System software (Applied Biosystems) and further analyzed with Microsoft (Seattle, WA) Excel.
Preparation of probes for in situ hybridization.
The open reading frames of all 25 hTAS2Rs were cloned into a vector cassette adding an N-terminal rat somatostatin receptor subtype 3 (sst3) tag and a C-terminally located herpes simplex virus glycoprotein D epitope (HSV) originally described by Bufe et al. (2002). PCR reactions using Pfu DNA-polymerase and primers specific for sequences of the sst3 and HSV tags were used to generate template DNA and to add T3- and T7-RNA-polymerase promoter sequences for subsequent in vitro transcriptions. Some hTAS2R cDNAs were cloned into the transcription vector pBluescriptKS+ (Stratagene, Heidelberg, Germany) and linearized with appropriate restriction endonucleases before in vitro transcription. In vitro transcriptions in the presence of either digoxigenin- or fluorescein-labeled ribonucleotides (Roche Applied Science, Mannheim, Germany) to generate sense and antisense riboprobes were done using standard procedures (Behrens et al., 2000). For single-label in situ hybridizations, all probes were hybridized to template plasmid DNA spotted onto nitrocellulose membranes. After color development, densitometry was performed to adjust probes to similar detection efficiencies. Additionally, for the eight highly homologous receptors hTAS2R43–hTAS2R50, the extent of cross-hybridization was determined by dot-blot analyses. For this experiment, identical amounts of all eight receptors were spotted onto membranes and subjected to hybridization experiments with the corresponding antisense riboprobes under the conditions used for the single-label in situ hybridizations. The probes for the receptors hTAS2R48 and hTAS2R49, which are <90% homologous to each other, detected only the cDNAs of the corresponding receptors. All other riboprobes detected the corresponding homologous receptor cDNAs best. Hybridization with a riboprobe specific for hTAS2R50 resulted in a very faint signal with hTAS2R48 cDNA. Additionally, some degree of hybridization across those receptors showing >90% sequence identity, hTAS2R43–hTAS2R47, was observed. hTAS2R47 barely cross-reacts with hTAS2R46 > hTAS2R44. hTAS2R46 moderately cross-reacts with hTAS2R47 and barely with hTAS2R44 > hTAS2R43. hTAS2R45 weakly cross-reacts with hTAS2R47, hTAS2R43, and hTAS2R44. hTAS2R44 moderately cross-reacts with hTAS2R47 and hTAS2R43 and barely with hTAS2R46 > hTAS2R45. hTAS2R43 barely cross-reacts with hTAS2R46 > hTAS2R45 > hTAS2R44.
In situ hybridization.
Sections of 10 μm thickness of human CV were cut with a cryostat (Microm, Walldorf, Germany), thaw-mounted onto positively charged glass slides (Menzel, Braunschweig, Germany), and stored at −80°C. For in situ hybridization, sections were fixed with 4% paraformaldehyde in PBS (in mm: 20 NaH2PO4, 80 Na2HPO4, and 65 NaCl, pH 7.4) containing 1 mm MgCl2 and adjusted to pH 7.2, rinsed with PBS, and incubated with 0.2 m HCl for 10 min and with 1% Triton X-100 in PBS for 2 min. After a brief rinse with PBS, sections were acetylated in 0.1 m triethanolamine, pH 8.0, by dropwise addition of acetic anhydride to a final concentration of 0.25% for 10 min. Prehybridization of the sections was done for 5 h at 50°C in prehybridization solution (50% formamide, 0.75 m NaCl, 25 mm PIPES, 25 mm EDTA, 5× Denhardt's reagent, 0.2% SDS, 250 μg/ml Escherichia coli tRNA, and 250 μg/ml salmon testis DNA, pH 6.8). For hybridization of the tissue, prehybridization buffer containing 10% (w/v) dextran sulfate and 5% (v/v) of the sensitivity-matched riboprobes was incubated for 10 min at 85°C before it was applied onto the sections. Hybridization was allowed to run overnight at 50°C in a chamber humidified with 50% formamide. Next, sections were washed three times with 2× SSC at room temperature, incubated in RNase buffer (2 μg/ml RNase A, 0.5 m NaCl, 10 mm Tris-HCl, and 1 mm EDTA, pH 7.5) for 30 min at 37°C. After one 15 min incubation in 0.4× SSC at room temperature and two washing steps in 0.4× SSC at 50°C for 30 min each, the slides were equilibrated in buffer 1 (0.1 m maleic acid and 0.15 m NaCl, pH 7.5) for 5 min at room temperature. Blocking of the tissue was done using 1% blocking reagent (Roche Applied Science) in buffer 1 for 1 h at room temperature before the anti-digoxigenin antibody coupled to alkaline phosphatase (Roche Applied Science) was applied at a dilution of 1:750 in the same buffer and left on the sections for another hour at room temperature. Unbound antibodies were removed by two subsequent washes with buffer 1 at room temperature for 30 min each. After equilibration in buffer 3 (0.1 m Tris-HCl, 0.1 m NaCl, and 50 mm MgCl2, pH 9.5) for 5 min at room temperature, color substrate [0.175 mg/ml 5-bromo-4-chlor-indolyl-phosphate (BCIP) and 0.25 mg/ml nitroblue-tetrazolium-chloride (NBT) in buffer 3] was applied and left on the slides placed in darkness overnight at room temperature. Finally, the reaction was stopped by 5 min incubation in TE (Tris-EDTA) buffer. The slides were mounted with glass coverslips and mounting medium, before microphotographs were taken using a microscope (Axioplan; Zeiss, Goettingen, Germany) connected to a CCD camera (RT slider; Diagnostic Instruments, Sterling Heights, MI).
For quantification, all stained and unstained intragemmal cells on the sections were counted independently by three individuals. The percentage of positive cells was calculated. The mean values and SDs were calculated using Microsoft Excel and summarized in Table 3.
Table 3.
Quantification of taste receptor cells stained by in situ hybridizations with individual hTAS2R probes
| Receptor | Positive intragemmal cells (%) |
|---|---|
| hTAS2R1 | 7.8 ± 1.3 |
| hTAS2R3 | 1.5 ± 0.1 |
| hTAS2R4 | 0.9 ± 0.2 |
| hTAS2R5 | 0.7 ± 0.1 |
| hTAS2R7 | 0.7 ± 0.1 |
| hTAS2R8 | 7.4 ± 0.6 |
| hTAS2R9 | 0.7 ± 0.2 |
| hTAS2R10 | 4.5 ± 1.1 |
| hTAS2R13 | 4.4 ± 1.1 |
| hTAS2R14 | 5.4 ± 1.7 |
| hTAS2R16 | 3.0 ± 0.6 |
| hTAS2R38 | 10.7 ± 4.1 |
| hTAS2R39 | 4.2 ± 1.1 |
| hTAS2R40 | 0.9 ± 0.3 |
| hTAS2R41 | 1.4 ± 0.1 |
| hTAS2R42 | 6.9 ± 1.1 |
| hTAS2R43 | 6.5 ± 1.8 |
| hTAS2R44 | 9.0 ± 2.1 |
| hTAS2R45 | 7.4 ± 0.8 |
| hTAS2R46 | 9.3 ± 2.8 |
| hTAS2R47 | 6.3 ± 3.8 |
| hTAS2R48 | 4.3 ± 1.0 |
| hTAS2R49 | 2.6 ± 0.5 |
| hTAS2R50 | 5.6 ± 1.2 |
| hTAS2R60 | 6.4 ± 0.9 |
| h α-Gustducin | 30 ± 0.9 |
All 25 human bitter taste receptors and human α -gustducin are listed together with their corresponding expression frequencies. All cells within taste buds were counted by three persons independently. The fraction of positive cells versus the total number of intragemmal cells is expressed as percentage.
Two-color fluorescence in situ hybridization.
Pretreatment of sections, hybridization, and stringency washes were mainly done as described for the single-detection procedure. During the prehybridization and hybridization a formamide concentration of 33% was maintained instead of 50% formamide used for the single-detection in situ hybridization. Accordingly, the final two stringency washing steps were done at 40°C (instead of 50°C) in 0.4× SSC for 30 min each. Next, the sections were equilibrated in TNT-buffer (0.15 m NaCl, 0.1 m Tris-HCl, and 0.05% Tween 20, pH 7.5) for 5 min at room temperature and then blocked with TNB-buffer [0.5% TSA blocking reagent (PerkinElmer, Rodgau-Juegesheim, Germany) in TNT buffer] for 1 h at room temperature. Incubation with antibodies [1:500 anti-fluorescein coupled to alkaline phosphatase and 1:750 anti-digoxigenin coupled to horseradish peroxidase (Roche Applied Science, Mannheim, Germany)] was done in TNB buffer for 30 min at room temperature. After washing three times for 5 min each with TNT buffer, fluorescein–tyramide 1:50 diluted in diluent (PerkinElmer) was applied onto the slides. The horseradish peroxidase reaction was allowed to run for 10 min at room temperature in darkness. After three washes with TNT buffer (5 min each at room temperature), the tissue was equilibrated by an incubation in 0.1 m Tris-HCl, pH 8.2 (5 min at room temperature), before color substrate [0.25 mg/ml naphthol, 1 mg/ml fast red, and 0.2 mg/ml levamisole (Roche Applied Science) in 0.1 m Tris-HCl, pH 8.2] was applied and left on the slides (kept in the dark) overnight at room temperature. The reaction was stopped by incubation with TE buffer (5 min, room temperature), and sections were mounted with glass coverslips using a water-based mounting medium. Images were taken by confocal laser scanning microscopy (TCS SP2; Leica, Bensheim, Germany). The entire deepness of the tissue was sequentially scanned with the excitation wavelengths 488 nm (fluorescein) and 543 nm (fast red), respectively. Emissions of the fluorochromes were detected between 500 and 530 nm (fluorescein) and 570 and 640 nm (fast red), respectively. The acquired image stack was flattened using the averaging function of the Leica confocal software.
Functional expression of hTAS2R cDNAs.
Functional expression of bitter taste receptors in human embryonic kidney HEK 293T–Gα16gust44 cells and subsequent calcium imaging analyses were done as described previously (Behrens et al., 2004). Briefly, the receptor cDNAs were all contained within the identical expression cassette as described previously (Bufe et al., 2002) adding a sst3 export tag to the N terminus of the receptor and a herpes simplex virus glycoprotein D epitope to its C terminus. hTAS2R16 and hTAS2R38, respectively, were used as reference bitter taste receptors, because their agonists salicin (hTAS2R16) (Bufe et al., 2002) and propylthiocarbamide (PTC) (hTAS2R38) (our unpublished observations) (Kim et al., 2003) are highly selective for the corresponding receptors, thus excluding cross-activation phenomena. The receptor mixtures were composed according to Table 2, transiently transfected into HEK 293T–Gα16gust44 cells using Lipofectamine 2000 (Invitrogen), incubated for 24 h, and stimulated with different concentrations of their selective agonists [30 mm, 10 mm, 3 mm, 1 mm, 300 μm, 100 μm, and 30 μm salicin for hTAS2R16; 30 μm, 10 μm, 3 μm, 1 μm, 300 nm, 100 nm, and 30 nm PTC for hTAS2R38 (PAV taster variant)]. Increases in intracellular calcium levels were visualized by Fluo-4-AM (Invitrogen) using a fluorometric imaging plate reader (FLIPR; Molecular Devices, Sunnyvale, CA). Calcium responses of at least three independent experiments performed in triplicates were baseline corrected and expressed as ΔF/F. Dose–response curves were calculated using SigmaPlot, and statistical analysis (one-way ANOVA with Tukey's post hoc test) was done with the GraphPad Software (San Diego, CA) Prism 3 software package. To control for effects that are not specifically related to hTAS2Rs, an unrelated G-protein-coupled receptor, the rat sst3 receptor, was used instead of the empty cassette vector in two independent experiments.
Table 2.
Pools of receptor expression constructs used for transient transfections of HEK 293T cells
| Reference receptor | Reference receptor only | Mixture of 3 | Mixture of 6 | Mixture of 12 | Mixture of 17 | Mixture of 25 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hTAS2R16 | 1p | hTAS2R16 | 1p | hTAS2R16 | 3p | Mix of 3 | 6p | Mix of 6 | 12p | Mix of 12 | 17p | Mix of 17 |
| 24p | Vector | hTAS2R14 | hTAS2R10 | 1p | hTAS2R1 | 1p | hTAS2R9 | 1p | hTAS2R3 | |||
| hTAS2R38 | hTAS2R44 | hTAS2R4 | hTAS2R39 | hTAS2R5 | ||||||||
| or | 22p | Vector | hTAS2R46 | hTAS2R8 | hTAS2R42 | hTAS2R7 | ||||||
| 1p | hTAS2R16 | 19p | Vector | hTAS2R13 | hTAS2R50 | hTAS2R40 | ||||||
| 24p | rsst3 | hTAS2R47 | hTAS2R60 | hTAS2R41 | ||||||||
| hTAS2R48 | 8p | Vector | hTAS2R43 | |||||||||
| 13p | Vector | hTAS2R45 | ||||||||||
| hTAS2R49 | ||||||||||||
| hTAS2R38 | 1p | hTAS2R38 | 1p | hTAS2R16 | 3p | Mix of 3 | 6p | Mix of 6 | 12p | Mix of 12 | 17p | Mix of 17 |
| 24p | Vector | hTAS2R14 | hTAS2R10 | 1p | hTAS2R1 | 1p | hTAS2R9 | 1p | hTAS2R3 | |||
| hTAS2R38 | hTAS2R44 | hTAS2R4 | hTAS2R39 | hTAS2R5 | ||||||||
| or | 22p | Vector | hTAS2R46 | hTAS2R8 | hTAS2R42 | hTAS2R7 | ||||||
| 1p | hTAS2R38 | 19p | Vector | hTAS2R13 | hTAS2R50 | hTAS2R40 | ||||||
| 24p | rsst3 | hTAS2R47 | hTAS2R60 | hTAS2R41 | ||||||||
| hTAS2R48 | 8p | Vector | hTAS2R43 | |||||||||
| 13p | Vector | hTAS2R45 | ||||||||||
| hTAS2R49 | ||||||||||||
To investigate competition between coexpressed hTAS2Rs, mixtures of expression constructs containing always 1 part (p) of the reference receptors hTAS2R16 or hTAS2R38 were used. The indicated constructs were added to fill up to 25 parts. Vector refers to the empty expression vector, pcDNA5FRT.
Results
To investigate whether all human TAS2R genes are expressed in CV, we performed RT-PCR experiments specific for all 25 bitter receptor cDNAs (Fig. 1 A). Although the intensity of PCR fragments varies considerably between the 25 hTAS2R cDNAs, all bitter taste receptors were detected. A selection of 10 hTAS2Rs, including receptors that are only weakly detected by in situ hybridization (hTAS2R3, -4, -5, -7, -8, -40, and -41) (compare with Fig. 2 A), and three previously published hTAS2Rs (hTAS2R14, -16, and -38) could not be detected by amplification of cDNA from human nonchemosensory filiform papillae, demonstrating taste tissue-specific expression (Fig. 1 B).
Figure 1.
RT-PCR of hTAS2R1–hTAS2R60 from human circumvallate papillae and nonchemosensory filiform papillae. Random primed cDNA was synthesized from DNaseI digested total RNA of human circumvallate papillae and filiform papillae by reverse transcription. For negative controls, reverse transcriptase was omitted from the reactions. A, Agarose gel electrophoresis of PCR reactions of human circumvallate papillae specific for hTAS2R1, -3, -4, -5, -7, -8, -9, -10, -13, -14, -16, -38, and -39 obtained by reverse transcription (+) and RT controls (−) are shown in the top. Reaction products specific for hTAS2R40, -41, -42, -43, -44, -45, -46, -47, -48, -49, -50, and -60 are shown in the bottom. B, RT-PCR analysis of human filiform papillae for GAPDH as positive control (Co) and hTAS2R3, -4, -5, -7, -8, -14, -16, -38, -40, and -41. M, Molecular weight standard (φX174/HaeIII).
Figure 2.
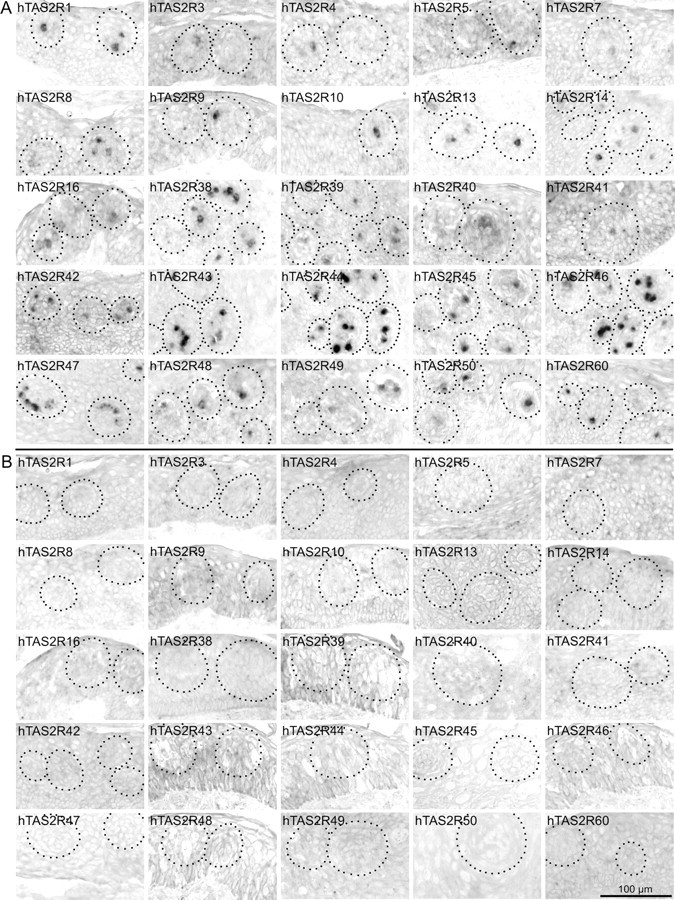
Human TAS2R gene expression in human circumvallate papillae. A, Ten micrometer cross-sections of human circumvallate papillae were hybridized with individual digoxigenin-labeled antisense riboprobes specific for all 25 hTAS2R genes (A). Standard colorimetry using NBT and BCIP was performed, resulting in red-colored precipitates within cells expressing the corresponding hTAS2R gene. The areas containing taste buds are indicated by dotted lines. Individual panels are labeled according to the hTAS2R-specific probes used for hybridization. Note that individual cells exclusively within taste buds are labeled. B same as in A, probes corresponding to the sense strand of the individual hTAS2R genes were used for negative controls. Scale bar, 100 μm.
Individual in situ hybridizations of cross-sections through human CV were performed to detect hTAS2R gene expression at the cellular level. mRNAs of all 25 hTAS2R genes were detected within individual, stained cells that are located in taste buds (Fig. 2 A). Both the number of stained cells per bud and the signal intensity observed for the individual probes varies substantially, indicating differential expression of bitter taste receptor genes within human CV. Whereas for most receptor mRNAs strong signals were readily obtained, hTAS2R3, -7, -9, -16, and -41 initially showed only faint or no signals, although all probes have been adjusted to similar apparent detection sensitivities. By doubling the concentration of probes and reducing the incubation time of the RNase A digestion step during the washing procedure, robust signals were obtained for hTAS2R3, -9, and -16. Even after these modifications of the in situ hybridization procedure, hTAS2R7 and -41 specific signals remained faint. Specificities of the reactions are demonstrated by the absence of signals if using sense probes (Fig. 2 B). In addition to differences in the staining intensity, the number of positive cells varied between the different hTAS2Rs. Counting of positive cells within taste buds allowed dividing the hTAS2Rs roughly into three groups (Table 3). Twelve hTAS2Rs were quite abundantly expressed, showing labeling of >5% of all intragemmal cells per section on average (hTAS2R1, -8, -14, -38, -42, -43, -44, -45, -46, -47, -50, and -60). Six receptors are moderately expressed with frequencies between >1.5 and <5% (hTAS2R10, -13, -16, -39, -48, and -49). Seven receptors show expression in only ∼1% of the cells (hTAS2R3, -4, -5, -7, -9, -40, and -41). None of the receptors is expressed with a frequency comparable with human α-gustducin (30%; data not shown).
In situ hybridizations performed with a total number of nine human fungiform papillae of three different individuals using either single hTAS2R probes (e.g., hTAS2R14 and -38), a mixture of three probes (hTAS2R1, -14, and -38), or a mixture of five probes (hTAS2R1, -10, -14, -38, and -46) did not reveal positive cells (data not shown).
To validate whether a single receptor probe labels only a subset of all bitter TRCs or if different cell numbers might arise from fluctuations in the number of bitter TRCs per section, we performed dual-label in situ hybridization using a digoxigenin-labeled probe specific for human α-gustducin in combination with a fluorescein-labeled probe for hTAS2R14 (Fig. 3). Of the 31 gustducin-positive cells, 19% (six cells) are also hTAS2R14 positive. Because only 30% of the intragemmal cells are gustducin positive (Table 3), 19% hTAS2R14/gustducin-positive cells extrapolated onto all intragemmal cells would result in 5.7%, thus matching closely our results obtained by the single-label in situ hybridizations (5.4 ± 1.7% for hTAS2R14).
Figure 3.
Human TAS2R mRNA is present in a subset of α-gustducin-expressing cells. Dual-color in situ hybridization was done using a digoxigenin-labeled antisense riboprobe specific for human α-gustducin and a fluorescein-labeled antisense riboprobe specific for hTAS2R14. For the detection of hybridization signals, an anti-digoxigenin antibody coupled to horseradish peroxidase was used in combination with fluorescein–tyramide solution, resulting in a green fluorescent color precipitate specific for α-gustducin (A). The fluorescein-labeled hTAS2R14 probe was detected with an anti-fluorescein antibody coupled to alkaline phosphatase in combination with fast red solution, leading to a red fluorescent color precipitate specific for hTAS2R14 (B). C, Overlay of A and B. D same as C superimposed on a topographical black and white image of the scanned specimen. Pictures were taken by confocal laser scanning microscopy (Leica TCS SP2) of the entire depth of the tissue. The resulting z-stack was flattened using the averaging function of the Leica confocal software. Cells expressing α-gustducin and hTAS2R14 are labeled with arrows (C). Scale bar, 40 μm.
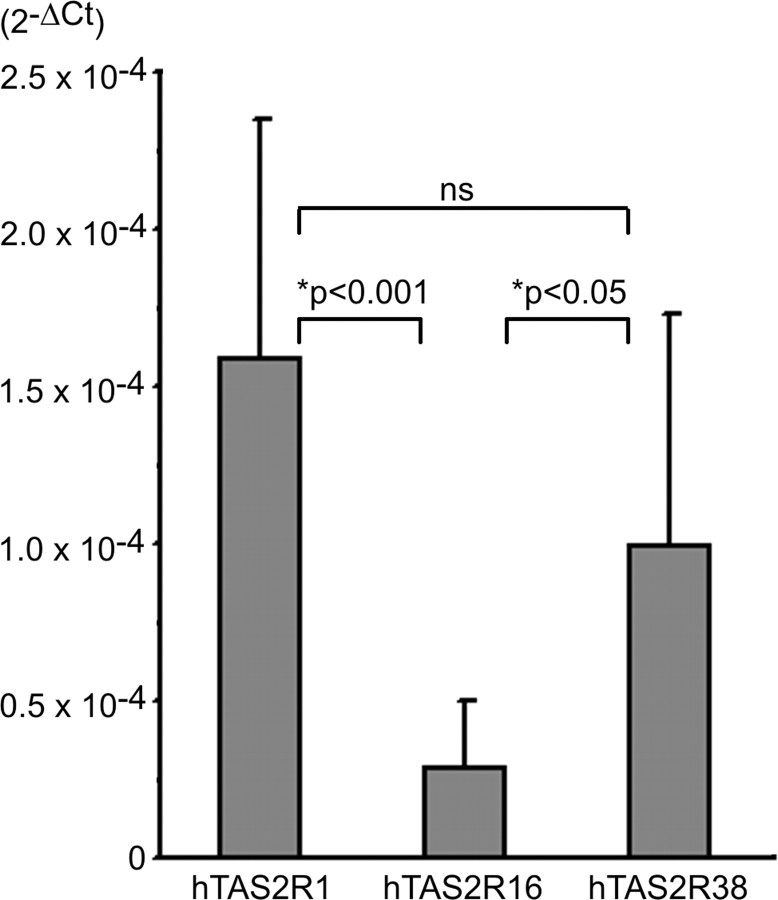
Single receptor in situ hybridizations and cellular quantification revealed that receptors differ in both their apparent expression levels and their expression frequencies. To verify these results and to determine the total amounts of mRNAs present in human gustatory tissue, we performed quantitative PCR analyzes for three hTAS2Rs differing in their expression using total RNA extracted from human CV (Fig. 4). Interestingly, hTAS2R1 mRNA, which appears to be expressed at an apparently similar level but with slightly lower frequency when compared with hTAS2R38 (compare with Fig. 2 A) showed the strongest overall expression. The amount of hTAS2R38 mRNA is only 62% of the hTAS2R1 mRNA, although the difference is not statistically significant. The lowest amount was detected for hTAS2R16 mRNA. This result verifies our observation that hTAS2R16 signals were not detectable using our standard protocol for single-label in situ hybridizations.
Figure 4.
Quantification of hTAS2R mRNAs in human circumvallate papillae. Random primed cDNA synthesized from human circumvallate papillae was used as template for quantitative PCR specific for hTAS2R1, -16, -and -38. The amount of each hTAS2R cDNA was standardized to the housekeeping gene GAPDH and expressed as 2-ΔCt (y-axis).
Unlike a previous report on bitter taste receptor expression in rats, which indicates that most, if not all, TAS2Rs are coexpressed suggesting a very homogeneous population of bitter TRCs in this organism (Adler et al., 2000), the human bitter TRCs appear less uniform. We therefore performed in situ hybridizations using mixtures of probes specific for two pairs of receptors displaying moderate expression frequencies (Fig. 5). Hybridization of adjacent sections with single probes revealed a similar number of positive taste receptor cells for hTAS2R14 (Fig. 5 A,D) and hTAS2R50 (Fig. 5 B,E) with 23 and 25 cells, respectively. However, hybridization with a mixture of both probes (Fig. 5 C,F) resulted not only in stronger signals indicative of overlapping expression but also in the staining of more cells (38), which indicates that some TRCs express only one of the two receptors. In situ hybridizations with hTAS2R13 (Fig. 5 G), hTAS2R47 (Fig. 5 H), and a mixture of hTAS2R13 and hTAS2R47 probes (Fig. 5 I) reveal similar results.
Figure 5.

In situ hybridizations of human circumvallate papillae using mixed probes specific for pairs of hTAS2Rs. Cross-sections of human circumvallate papillae were hybridized with digoxigenin-labeled antisense probes specific for hTAS2R14 (A, D), hTAS2R50 (B, E), or a mixture of both probes (C, F). Standard colorimetry using NBT and BCIP was performed, resulting in red-colored precipitates within cells expressing the corresponding hTAS2R gene. Note that the staining intensity of a subpopulation of cells and the number of positive cells increased when a mixture of hTAS2R14 and hTAS2R50 probes was used for hybridization. G–I, In situ hybridizations using probes specific for hTAS2R13 (G), hTAS2R47 (H), and a mixture of hTAS2R13 and hTAS2R47 probes (I). Scale bars: A–C, 250 μm; D–I, 100 μm.
Direct evidence for the degree of cellular coexpression of bitter taste receptor genes was obtained by dual-label in situ hybridizations using differently labeled pairs of hTAS2R-specific probes (Fig. 6). Although cells showing an overlapping expression of the pairs of genes under investigation are frequently found (hTAS2R1 and hTAS2R10, hTAS2R14 and hTAS2R1, hTAS2R10 and hTAS2R14, hTAS2R14 and hTAS2R38, hTAS2R39 and hTAS2R1, hTAS2R38 and hTAS2R1), many cells only express one of the two receptors (hTAS2R1 or hTAS2R10, hTAS2R10 or hTAS2R14, hTAS2R14 or hTAS2R38, hTAS2R39 or hTAS2R1, hTAS2R38 or hTAS2R1).
Figure 6.
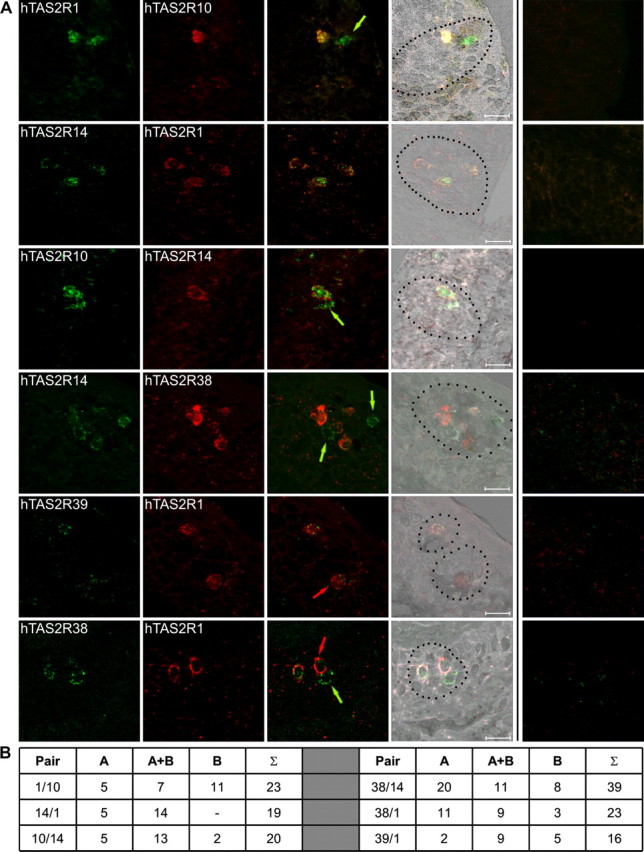
Dual-color in situ hybridizations of selected pairs of bitter receptor mRNAs. A, Dual-color in situ hybridizations were done using digoxigenin-labeled antisense riboprobes and fluorescein-labeled antisense riboprobes for the simultaneous detection of pairs of hTAS2R mRNAs. For the detection of hybridization signals, an anti-digoxigenin antibody coupled to horseradish peroxidase was used in combination with fluorescein–tyramide solution, resulting in a green fluorescent color precipitate shown in the first row. The fluorescein-labeled probes were detected with an anti-fluorescein antibody coupled to alkaline phosphatase in combination with fast red solution, leading to a red fluorescent color precipitate shown in the second row. Overlays of green and red channels are shown in the third row. Overlays superimposed onto topographical black and white images are depicted in the fourth row. Overlays of the green and red channels obtained after in situ hybridizations with the corresponding sense probes are depicted in the fifth row for negative controls. Pairs of receptors used for dual-color in situ hybridizations were as follows (from top to bottom): hTAS2R1 (green) and hTAS2R10 (red); hTAS2R14 (green) and hTAS2R1 (red); hTAS2R10 (green) and hTAS2R14 (red); hTAS2R14 (green) and hTAS2R38 (red); hTAS2R39 (green) and hTAS2R1 (red); hTAS2R38 (green) and hTAS2R1 (red). Pictures were taken by confocal laser scanning microscopy (Leica TCS SP2) of the entire depth of the tissue. The resulting z-stack was flattened using the averaging function of the Leica confocal software. Cells expressing only a single receptor are labeled with arrows in the corresponding color. Scale bars, 20 μm. B, Quantification of the number of cells expressing receptor A, receptor A together with receptor B (A+B), or only receptor B for each pair of receptors used for dual-label in situ hybridizations.
To evaluate possible physiological consequences of hTAS2R coexpression at variable levels of complexity, we cotransfected different mixtures of 1, 3, 6, 12, 17, and 25 hTAS2R expression constructs into HEK 293T–Gα16gust44 cells and challenged the cells with bitter stimuli selective for a single receptor present at constant amounts in these mixtures while monitoring dose–response relationships. When cells transfected with hTAS2R16 DNA were stimulated with different concentrations of salicin, a dose–response relationship comparable with published results was observed (Fig. 7, top). The calculated EC50 of 2.0 ± 0.1 mm salicin, although obtained with an 1- to 25-fold diluted construct, closely resembled the EC50 of 1.4 ± 0.2 mm for salicin published previously (Bufe et al., 2002). The cotransfection with two additional receptors resulted in a decreased signal amplitude. Although the efficacy of hTAS2R16 in a pool of receptors is only approximately one-third (at salicin concentrations of 3 and 10 mm) if compared with hTAS2R16 transfected alone, the potency with an EC50 of 1.8 ± 0.3 mm salicin remained essentially unchanged. Transfecting a mixture of six hTAS2Rs including hTAS2R16 reduced signal amplitudes further (EC50 of 2.8 ± 0.2 mm salicin). Cotransfection with 11 additional receptors neither changed EC50 (2.3 ± 0.3 mm salicin) significantly nor reduced signal amplitudes further. The presence of 17 and 24 additional receptors left only ∼10% of the originally observed response amplitudes, whereas EC50 values, with 2.1 mm (extrapolated) for 17 additional receptors and with 2.2 ± 0.8 mm for 24 additional receptors, respectively, remained nearly constant. Although cotransfection with a 24-fold excess of the unrelated receptor, rat sst3, appeared slightly less potent compared with coexpression of all hTAS2Rs, the results were not statistically different. A similar experiment using hTAS2R38 as reference receptor gave comparable results (Fig. 7, bottom). In this case, an EC50 value of 2.6 ± 0.1 μm for PTC of the 1:25 diluted hTAS2R38 [PAV-taster variant (Bufe et al., 2005)] was determined. This time, coexpression of hTAS2R38 with two other bitter taste receptors only reduced signal amplitudes to ∼70% of the originally observed values. Again, the receptor potency was not significantly changed (EC50 of 2.1 ± 0.4 μm PTC). The signal amplitudes decreased even further if more hTAS2R cDNA constructs were cotransfected, thus paralleling the results obtained with hTAS2R16 as reference receptor. Also in case of the hTAS2R38, similar competition effects were observed for cotransfected hTAS2Rs and rat sst3.
Figure 7.
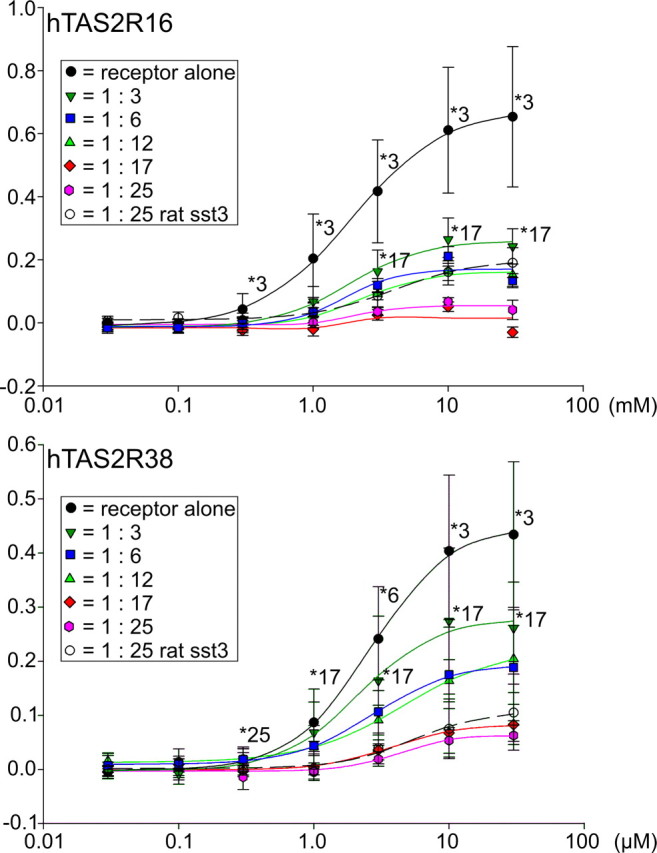
Functional coexpression of hTAS2R constructs in HEK 293T cells. Constant amounts of hTAS2R16 and hTAS2R38, respectively, were transiently transfected into HEK 293T cells stably expressing the chimeric G-protein subunit Gα16gust44. hTAS2R16 and hTAS2R38 constructs were transfected alone, together with two additional hTAS2R constructs (1:3), together with five additional hTAS2R constructs (1:6), together with 11 additional hTAS2R constructs (1:12), together with 16 additional hTAS2R constructs (1:17), together with 24 additional hTAS2R constructs (1:25), or together with the unrelated rat sst3 receptor (1:25 rat sst3). The empty expression vector used for the cloning of the hTAS2R constructs was used to supplement for resulting variations in cDNA amounts between plasmid mixtures for transfection. Cells were loaded with the calcium-sensitive dye Fluo-4-AM and challenged with different concentrations of the specific agonists salicin (hTAS2R16) and phenylthiocarbamide (hTAS2R38). Changes in fluorescence were monitored, baseline subtracted, and plotted as ΔF/F (y-axis). Dose–response curves were calculated using SigmaPlot 2000. For statistical analyses, the GraphPad Prism software package was used. Asterisks and numbers indicated statistical significance of at least p < 0.01 and refer to the closest dataset for which significance has been observed.
Discussion
In the present study, we elucidated whether all hTAS2R genes are expressed in gustatory tissue and, hence, whether the entire hTAS2R gene family might serve as taste receptors. Previous studies demonstrated that all functionally analyzed TAS2Rs respond to substances known to taste bitter to humans [hTAS2R4, denatonium benzoate (Chandrashekar et al., 2000); hTAS2R7, strychnine (Sainz et al., 2007); hTAS2R10, strychnine (Bufe et al., 2002); hTAS2R14, picrotoxin (Behrens et al., 2004); hTAS2R16, salicin (Bufe et al., 2002); hTAS2R38, 6-n-propyl-2-thiouracil (Kim et al., 2003; Bufe et al., 2005); hTAS2R43/44, aristolochic acid (Kuhn et al., 2004); hTAS2R47, 6-nitrosaccharin (Pronin et al., 2004); only single compounds listed for each receptor] or aversive to rodents [mouse T2R5, cycloheximide; mouse T2R8, denatonium benzoate (Chandrashekar et al., 2000); rat T2R9, cycloheximide (Bufe et al., 2002)]. These findings support the conclusion that most likely all TAS2Rs are indeed bitter sensors. However, for humans, only nine hTAS2Rs are analyzed for function and five hTAS2R genes for gustatory expression, respectively (Chandrashekar et al., 2000; Bufe et al., 2002, 2005; Behrens et al., 2004; Kuhn et al., 2004; Pronin et al., 2004), leaving the majority of hTAS2Rs uncharacterized. Reports on the extragustatory expression of TAS2R genes in respiratory epithelia (Finger et al., 2003), gastrointestinal tissues and cells derived thereof (Wu et al., 2002, 2005), and testes (Matsunami et al., 2000; Behrens et al., 2006) indicate additional nongustatory functions for TAS2Rs, making it necessary to confirm for every single TAS2R its potential role in taste sensation.
By RT-PCR analyses and in situ hybridizations, we now detected mRNAs of the full complement of hTAS2R genes in TRCs, thereby confirming a possible gustatory function for the entire gene family. Our attempts to detect hTAS2R gene expression in a limited number of fungiform papillae failed. In view of the fact that humans clearly detect bitter stimuli at the tip of the tongue in which fungiform papillae are located (Collings, 1974), the most likely explanation for this negative result is that only a subset of human fungiform taste buds might express hTAS2Rs, thus paralleling observations in rats in which only ∼10% of fungiform taste buds expressed TAS2R genes (Adler et al., 2000).
Another major goal of our analyses was to determine whether the human bitter TRC population is homogenous or heterogenous. The latter could provide a cellular basis for a potential discrimination of bitter stimuli. Currently, it is still controversial whether the mammalian bitter TRC population is uniform, expressing all functional TAS2R genes within each bitter TRC, or not. From gene expression analyses in the rat gustatory system, it was suggested that the expression patterns of TAS2Rs primarily overlap (Adler et al., 2000). An extensive coexpression of TAS2R genes has also been concluded from the observation that bitter taste deficient mice lacking the PLCβ2 gene can be genetically rescued to taste bitter compounds by the reintroduction of PLCβ2 under the control of TAS2R gene promoters. This finding demonstrated that each of the three different TAS2R promoters tested restores PLCβ2 signaling in bitter TRCs expressing the receptors for the selected eight bitter agonists (Mueller et al., 2005). Gene expression analyses of mouse gustatory tissues suggested a more limited coexpression of TAS2Rs (Matsunami et al., 2000). Whereas gene expression analyses and genetic-rescue experiments suggest a broadly tuned rather homogeneous bitter TRC population in rodents, physiological experiments demonstrate that most bitter TRCs only respond to one or two of five bitter stimuli, thus reacting functionally dissimilar (Caicedo and Roper, 2001).
Our results obtained by in situ hybridizations show that hTAS2R gene expression differs in both the apparent expression level and the average number of TRCs (Fig. 2 A, Table 3). This observation is further supported by qRT-PCR analyses of selected hTAS2Rs demonstrating that also the total amounts of hTAS2R mRNAs differ (Fig. 4). Because hTAS2R-specific antibodies are not available, it remains to be determined whether the different mRNA levels lead to functionally relevant differences in hTAS2R protein amounts. For the five highly homologous hTAS2Rs, hTAS2R43–hTAS2R47, limited cross-hybridizations on control blots were evident (see Material and Methods). Therefore, the observed expression frequencies may be slightly overestimated for these receptors. However, staining of directly adjacent sections (Fig. 2, e.g., hTAS2R43 and hTAS2R44) showing different patterns of hybridization signals for closely related hTAS2Rs, provide no indication of cross-hybridization in the in situ hybridization experiments. Nevertheless, an overestimation of the number of labeled cells per probe attributable to possible cross-hybridization would further support the existence of a heterogenous bitter TRC population.
Most strikingly, our dual-label in situ hybridizations (Fig. 6) clearly show that the human bitter TRC population is heterogeneous. Because our data point to a limited number of hTAS2R genes expressed on average in each TRC, the question arises how many receptors may be present per cell. To answer this question, we first need to determine the proportion of bitter responsive intragemmal cells. This value cannot exceed 30%, because bitter TRCs are a fraction of α-gustducin-positive cells, which we determined to be 30% of the intragemmal cells. Most likely, the true value is clearly smaller because α-gustducin-positive cells also include umami and sweet responsive cells (Wong et al., 1996; He et al., 2004). The value cannot fall below 11%, which corresponds to the proportion of intragemmal cells expressing the most frequent TAS2R, hTAS2R38. Next, we determine the probability of a bitter TRC to express a given TAS2R. Human TAS2R1 for example is present in ∼7.8% of the intragemmal cells or, based on above estimations, in 26–71% of the bitter TRCs. In other words, the probability of a bitter TRC to express hTAS2R1 is between 0.26 and 0.71. Performing the same calculations for all 25 hTAS2Rs and summing up the individual probabilities results in the average probability of 3.95–10.77 for a bitter TRC to express TAS2Rs. This means that no more than 4–11 TAS2Rs are coexpressed on average in a single cell. This unexpectedly low number of coexpressed TAS2Rs explains the observation by Caicedo and Roper (2001) showing that most TRCs responded to only one or two of five bitter stimuli tested.
If all TAS2R genes are coordinately regulated, then every hTAS2R could be coexpressed with any other hTAS2R in some but not all bitter TRCs. This conjecture does not contradict the observation of Mueller et al. (2005) who rescued PLCβ2 gene-targeted mice for tasting bitter substances by transgenic PLCβ2 expression driven by different TAS2R gene promoters. If, however, TAS2R gene expression is not coordinately regulated, we might expect that some TAS2R combinations are not realized in TRCs. Only a future detailed investigation of TAS2R gene expression could clarify this point.
Our results still have another inevitable implication. The number of possibilities to combine the estimated maximal 11 of 25 TAS2Rs in a TRC, which is defined by the algorithm 25! ÷ (25 − 11)! ÷ 11!, is ∼4.5 × 106. This value exceeds the number of vallate taste buds and TRCs. Estimations range from 4–18 circumvallate papillae per tongue, 234–400 taste buds per papilla and 50–120 cells per bud (Miller, 1995). This means that no two vallate TRCs or buds may be identical with regard to their TAS2R repertoire. This heterogeneity may well be maintained at the level of ganglion cells. A study on the innervation pattern of fungiform taste buds demonstrated that, on average, individual taste buds are contacted by four to five ganglion cells (Zaidi and Whitehead, 2006). This raises the intriguing possibility that, on average, only one ganglion cell per taste modality per taste bud exists. This assumption is also supported by the observation that different sensitivities for a collection of bitter compounds are evident in individual afferent fibers of glossopharyngeal and chorda tympani nerves (Dahl et al., 1997). Consequently, single neurons in the rat nucleus of the solitary tract, the first relay station for taste information within the CNS, differentially respond to bitter stimuli (Geran and Travers, 2006). Clearly, the ultimate answer has to come from detailed psychophysical or behavioral experiments because such data represent the integrated output of neural processing. So far, these studies have provided controversial results about discrimination among bitter stimuli in humans and animals (Yokomukai et al., 1993; Delwiche et al., 2001; Spector and Kopka, 2002; Keast et al., 2003; Brasser et al., 2005).
To investigate how coexpression of multiple receptors in a population of cells might affect receptor responsiveness, we expressed different cDNA mixtures in heterologous HEK 293 cells (Fig. 7). This experiment demonstrated that hTAS2Rs compete with each other for common cellular components. This resulted in reduced signal amplitudes already when using receptor mixtures of limited complexity. The amplitudes decreased further with increasing complexities of receptor mixtures. Because cotransfection of the unrelated receptor rat sst3 resulted in similar decreases in signal amplitudes, it appears likely that competition might occur during all phases of the life of a receptor, from biosynthesis to signaling and ultimately its turnover. Similar competition effects could also play a role in vivo, suggesting that a smaller number of coexpressed hTAS2R genes might be beneficial for the signal intensity generated by the entire population of bitter TRC.
Footnotes
This work was supported by a grant from the German Science Foundation (Deutsche Forschungsgemeinschaft Grant Me 1024/2-1/2). We thank Ulrike Lerner for her excellent technical assistance and Stefanie Manthey for her help with the quantification of bitter taste receptor cells.
References
- Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJ, Zuker CS. A novel family of mammalian taste receptors. Cell. 2000;100:693–702. doi: 10.1016/s0092-8674(00)80705-9. [DOI] [PubMed] [Google Scholar]
- Behrens M, Venkatraman G, Gronostajski RM, Reed RR, Margolis FL. NFI in the development of the olfactory neuroepithelium and the regulation of olfactory marker protein gene expression. Eur J Neurosci. 2000;12:1372–1384. doi: 10.1046/j.1460-9568.2000.00032.x. [DOI] [PubMed] [Google Scholar]
- Behrens M, Brockhoff A, Kuhn C, Bufe B, Winnig M, Meyerhof W. The human taste receptor hTAS2R14 responds to a variety of different bitter compounds. Biochem Biophys Res Commun. 2004;319:479–485. doi: 10.1016/j.bbrc.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Behrens M, Bartelt J, Reichling C, Winnig M, Kuhn C, Meyerhof W. Members of RTP and REEP gene families influence functional bitter taste receptor expression. J Biol Chem. 2006;281:20650–20659. doi: 10.1074/jbc.M513637200. [DOI] [PubMed] [Google Scholar]
- Brasser SM, Mozhui K, Smith DV. Differential covariation in taste responsiveness to bitter stimuli in rats. Chem Senses. 2005;30:793–799. doi: 10.1093/chemse/bji071. [DOI] [PubMed] [Google Scholar]
- Bufe B, Hofmann T, Krautwurst D, Raguse JD, Meyerhof W. The human TAS2R16 receptor mediates bitter taste in response to beta-glucopyranosides. Nat Genet. 2002;32:397–401. doi: 10.1038/ng1014. [DOI] [PubMed] [Google Scholar]
- Bufe B, Breslin PA, Kuhn C, Reed DR, Tharp CD, Slack JP, Kim UK, Drayna D, Meyerhof W. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr Biol. 2005;15:322–327. doi: 10.1016/j.cub.2005.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo A, Roper SD. Taste receptor cells that discriminate between bitter stimuli. Science. 2001;291:1557–1560. doi: 10.1126/science.291.5508.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zuker CS, Ryba NJ. T2Rs function as bitter taste receptors. Cell. 2000;100:703–711. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- Collings VB. Human taste responses as a function of locus of stimulation on the tongue and soft palate. Percept Psychophys. 1974;16:169–174. [Google Scholar]
- Dahl M, Erickson RP, Simon SA. Neural responses to bitter compounds in rats. Brain Res. 1997;756:22–34. doi: 10.1016/s0006-8993(97)00131-5. [DOI] [PubMed] [Google Scholar]
- Delwiche JF, Buletic Z, Breslin PA. Covariation in individuals' sensitivities to bitter compounds: evidence supporting multiple receptor/transduction mechanisms. Percept Psychophys. 2001;63:761–776. doi: 10.3758/bf03194436. [DOI] [PubMed] [Google Scholar]
- Finger TE, Bottger B, Hansen A, Anderson KT, Alimohammadi H, Silver WL. Solitary chemoreceptor cells in the nasal cavity serve as sentinels of respiration. Proc Natl Acad Sci USA. 2003;100:8981–8986. doi: 10.1073/pnas.1531172100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank ME, Bouverat BP, MacKinnon BI, Hettinger TP. The distinctiveness of ionic and nonionic bitter stimuli. Physiol Behav. 2004;80:421–431. doi: 10.1016/j.physbeh.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Geran LC, Travers SP. Single neurons in the nucleus of the solitary tract respond selectively to bitter taste stimuli. J Neurophysiol. 2006;96:2513–2527. doi: 10.1152/jn.00607.2006. [DOI] [PubMed] [Google Scholar]
- Glendinning JI. Is the bitter rejection response always adaptive? Physiol Behav. 1994;56:1217–1227. doi: 10.1016/0031-9384(94)90369-7. [DOI] [PubMed] [Google Scholar]
- He W, Yasumatsu K, Varadarajan V, Yamada A, Lem J, Ninomiya Y, Margolskee RF, Damak S. Umami taste responses are mediated by α-transducin and α-gustducin. J Neurosci. 2004;24:7674–7680. doi: 10.1523/JNEUROSCI.2441-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keast RS, Bournazel MM, Breslin PA. A psychophysical investigation of binary bitter-compound interactions. Chem Senses. 2003;28:301–313. doi: 10.1093/chemse/28.4.301. [DOI] [PubMed] [Google Scholar]
- Kim UK, Jorgenson E, Coon H, Leppert M, Risch N, Drayna D. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 2003;299:1221–1225. doi: 10.1126/science.1080190. [DOI] [PubMed] [Google Scholar]
- Kuhn C, Bufe B, Winnig M, Hofmann T, Frank O, Behrens M, Lewtschenko T, Slack JP, Ward CD, Meyerhof W. Bitter taste receptors for saccharin and acesulfame K. J Neurosci. 2004;24:10260–10265. doi: 10.1523/JNEUROSCI.1225-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami H, Montmayeur JP, Buck LB. A family of candidate taste receptors in human and mouse. Nature. 2000;404:601–604. doi: 10.1038/35007072. [DOI] [PubMed] [Google Scholar]
- McBurney DH, Smith DV, Shick TR. Gustatory cross adaptation: sourness and bitterness. Percept Psychophys. 1972;11:228–232. [Google Scholar]
- Meyerhof W. Elucidation of mammalian bitter taste. Rev Physiol Biochem Pharmacol. 2005;154:37–72. doi: 10.1007/s10254-005-0041-0. [DOI] [PubMed] [Google Scholar]
- Miller IJ Jr, editor. Anatomy of the peripheral taste system. New York: Dekker; 1995. [Google Scholar]
- Mueller KL, Hoon MA, Erlenbach I, Chandrashekar J, Zuker CS, Ryba NJ. The receptors and coding logic for bitter taste. Nature. 2005;434:225–229. doi: 10.1038/nature03352. [DOI] [PubMed] [Google Scholar]
- Pronin AN, Tang H, Connor J, Keung W. Identification of ligands for two human bitter T2R receptors. Chem Senses. 2004;29:583–593. doi: 10.1093/chemse/bjh064. [DOI] [PubMed] [Google Scholar]
- Sainz E, Cavenagh MM, Gutierrez J, Battey JF, Northup JK, Sullivan SL. Functional characterization of human bitter taste receptors. Biochem J. 2007;403:537–543. doi: 10.1042/BJ20061744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbarbati A, Crescimanno C, Bernardi P, Osculati F. Alpha-gustducin-immunoreactive solitary chemosensory cells in the developing chemoreceptorial epithelium of the rat vallate papilla. Chem Senses. 1999;24:469–472. doi: 10.1093/chemse/24.5.469. [DOI] [PubMed] [Google Scholar]
- Spector AC, Kopka SL. Rats fail to discriminate quinine from denatonium: implications for the neural coding of bitter-tasting compounds. J Neurosci. 2002;22:1937–1941. doi: 10.1523/JNEUROSCI.22-05-01937.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney G, Harder DB. Genetics of bitter perception in mice. Physiol Behav. 1994;56:1141–1147. doi: 10.1016/0031-9384(94)90358-1. [DOI] [PubMed] [Google Scholar]
- Wong GT, Gannon KS, Margolskee RF. Transduction of bitter and sweet taste by gustducin. Nature. 1996;381:796–800. doi: 10.1038/381796a0. [DOI] [PubMed] [Google Scholar]
- Wu SV, Rozengurt N, Yang M, Young SH, Sinnett-Smith J, Rozengurt E. Expression of bitter taste receptors of the T2R family in the gastrointestinal tract and enteroendocrine STC-1 cells. Proc Natl Acad Sci USA. 2002;99:2392–2397. doi: 10.1073/pnas.042617699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SV, Chen MC, Rozengurt E. Genomic organization, expression, and function of bitter taste receptors (T2R) in mouse and rat. Physiol Genomics. 2005;22:139–149. doi: 10.1152/physiolgenomics.00030.2005. [DOI] [PubMed] [Google Scholar]
- Yokomukai Y, Cowart BJ, Beauchamp GK. Individual differences in sensitivity to bitter-tasting substances. Chem Senses. 1993;18:669–681. [Google Scholar]
- Zaidi FN, Whitehead MC. Discrete innervation of murine taste buds by peripheral taste neurons. J Neurosci. 2006;26:8243–8253. doi: 10.1523/JNEUROSCI.5142-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]