Abstract
Any complex nervous system is made out of two major cell types, neurons and glial cells. A hallmark of glial cells is their pronounced ability to migrate. En route to their final destinations, glial cells are generally guided by neuronal signals. Here we show that in the developing visual system of Drosophila glial cell migration is largely controlled by glial–glial interactions and occurs independently of axonal contact. Differentiation into wrapping glia is initiated close to the morphogenetic furrow. Using single cell labeling experiments we identified six distinct glial cell types in the eye disc. The migratory glial population is separated from the wrapping glial cells by the so-called carpet cells, extraordinary large glial cells, each covering a surface area of ∼10,000 epithelial cells. Subsequent cell ablation experiments demonstrate that the carpet glia regulates glial migration in the eye disc epithelium and suggest a new model underlying glial migration and differentiation in the developing visual system.
Keywords: glia, Drosophila, development, migration, neuron-glia interaction, differentiation, eye disc
Introduction
The nervous system of complex animals comprises two cell types: neurons and glial cells whose interactions are important to sculpture a functional neural lattice. Throughout evolution glial cells have kept several characteristic features, among which is their highly motile character (Lemke, 2001; Edenfeld et al., 2005; Freeman and Doherty, 2006).
Glial cells generally leave their place of birth to migrate to positions where they control neuronal development or guarantee axonal isolation. In principle, cells can either migrate individually or can move as chains or groups of cells through the body. Peripheral glial cells derived from the neural crest move as a continuous group of cells and similarly glial cells were reported to migrate in chains in the Drosophila PNS (Gilmour et al., 2002; Aigouy et al., 2004; Lyons et al., 2005; Edenfeld et al., 2007). Likewise in the CNS, glial cells often migrate toward their final destination and on their way can be guided by the same signaling systems that are used by the navigating neuronal growth cones (Kinrade et al., 2001; Tsai and Miller, 2002; Jarjour and Kennedy, 2004).
In Drosophila, glial cell migration is also found in the developing eye. Photoreceptor neurons develop in the wake of the morphogenetic furrow that sweeps across the imaginal disc epithelium and project their axons through the optic stalk into the optic ganglia in the brain (Wolff and Ready, 1991; Freeman, 1997). As other axons in the fly PNS, the photoreceptor axons are wrapped by specific glial cells (Meinertzhagen and Hanson, 1993). These cells originate in the forming optic stalk, whose formation depends on the activity of the Focal adhesion kinase (Murakami et al., 2007) and migrate onto the larval eye disc (Choi and Benzer, 1994).
Ablation experiments show that glial cells migrate in the absence of neuronal membranes. However, ectopic photoreceptor cell neurons are able to attract glial cells over some distance (Rangarajan et al., 1999). Thus, neuronal cells are sufficient, but not necessary to regulate glial migration. In agreement with this notion are reports that document glial migration independent of axons as seen in gilgamesh, hedgehog or fear of intimacy mutants (Hummel et al., 2002; Pielage et al., 2004).
Previous work described two different glial cell types populating the eye disc (Rangarajan et al., 2001; Hummel et al., 2002). The wrapping glial cells ensheath the axons whereas surface glia surrounds the wrapping glia. A similar situation is reported for peripheral nerves where an inner wrapping layer is surrounded by subperineurial glia which is then covered by perineurial cells (Banerjee et al., 2006) (T. Stork, D. Engelen, A. Krudewig, M. Silies, R. J. Bainton, and C. Klämbt, unpublished observations). Having two glial cell layers, one in contact with neuronal membranes, one in contact only with glial membranes, the question arises how these different cell types migrate toward their final destination. Do both layers form separately or is there a common mechanism leading to the continuous formation of the two cell types? To address these questions we have revisited the migration of glial cells onto the eye disc at single cell resolution.
Materials and Methods
Immunohistochemistry and electron microscopic analyses.
Anti-Repo antibodies were obtained from the Developmental Studies Hybridoma bank (Iowa City, IA). Anti-βGal (1:1000; Cappel, Cochranville, PA), anti-phosphohistone H3 (1:1000; Upstate Biotechnology, Lake Placid, NY), anti-β-Spectrin (Hulsmeier et al., 2007), anti-GFP (1:1000; Invitrogen, Eugene, OR) and anti-HRP Cy5 (1:200; Dianova, Hamburg, Germany) antisera were used according to the manufacture's instructions. Fixation and treatment of tissues for immunohistochemistry was performed according standard procedures. For electron microscopic analyses, larval tissues were fixed in 4%PFA at 4°C overnight. After a fixation in 2% OsO4 for 1 h at room temperature and 2% uranylacetate treatment for 30 min at room temperature in the dark tissues were embedded in epon as described previously (Stollewerk et al., 1996; Stollewerk and Klämbt, 1997), but acrolein treatment was omitted. Ultrathin sections were imaged with a Zeiss EM900 with a SIS Morada digital camera. Fluorescently labeled specimens were analyzed using with a 40× oil-immersion lens on a Zeiss (Oberkochen, Germany) 510 LSM, orthogonal sections were taken using the Zeiss image browser. Cell counts were performed on confocal image stacks with sections taken at 0.8 μm distance, resolution 512 × 512 pixel; pinhole 118 μm, scan speed 8. Glial nuclei were counted using the Volocity program version 4.0. Volocity quantitation was set with an intensity range of 50–255 and a minimal volume 20 μm3. Only complete confocal stacks were counted.
DNA work.
Generation of a repoFlp transgene. A 4.4 kb large fragment of the repo promotor (Lee and Jones, 2005) was amplified by PCR and cloned into the Asp718 and NotI sites of the CaSpeR 4 vector. In contrast to (Lee and Jones, 2005), no minimal hsp70 promoter was used, but the endogenous repo TATA box. The flp coding sequence was amplified from an UASflp stock (Bloomington Stock Center, Bloomington, IN) and ligated directly 3′ to the promoter fragment, using NotI and PstI restriction sites. Further details are available on request. Germline transformation was performed according to standard procedures.
Fly strains and genetics.
All crosses were performed on standard food at 25°C unless indicated otherwise. Clonal analysis was performed using either a repoGal4 UASFlp recombinant chromosome or repoFlp transgenes in combination with different flip-out cassettes: UAS>CD2 yellow>mCD8::GFP (Wong et al., 2002), act5c>draf>τlacZ (gift from J. Skeath, Washington University in St. Louis, St. Louis, MO) act5c>CD2>Gal4, UASGFP or act5c>y>Gal4 UASlacZ (gifts from M. González-Gaitán, University of Geneva, Geneva, Switzerland). For lineage analysis and cell ablation experiments, we used c527 and Mz97 (Hummel et al., 2002) and a Gal4 driver using regulatory sequences of the moody gene (Bainton et al., 2005), UASfzr (Sigrist and Lehner, 1997) and UAShid (kindly provided by R. Lehmann, Skirball Institute, New York, NY) tubGal80ts (McGuire et al., 2003). Additional fly stocks used were w1118 and Nrx::GFP #454 (Edenfeld et al., 2006).
Results
During larval development the morphogenetic furrow sweeps across the eye disc epithelium. In its wake ommatidial units are formed by successive addition of receptor cells (Wolff and Ready, 1991; Freeman, 1997). Once receptor neurons are specified, they project their axons along the basal surface of the disc to its posterior end. Here all axonal fascicles are bundled in the optic stalk, which bridges the eye disc with the optic lobes (Fig. 1). Glial cells that originate from the optic stalk populate the posterior portion of the eye disc. They migrate onto the eye imaginal disc as photoreceptor neurons start to differentiate and send their axons to the basal side of the epithelium (Choi and Benzer, 1994) (Fig. 1C,D).
Figure 1.
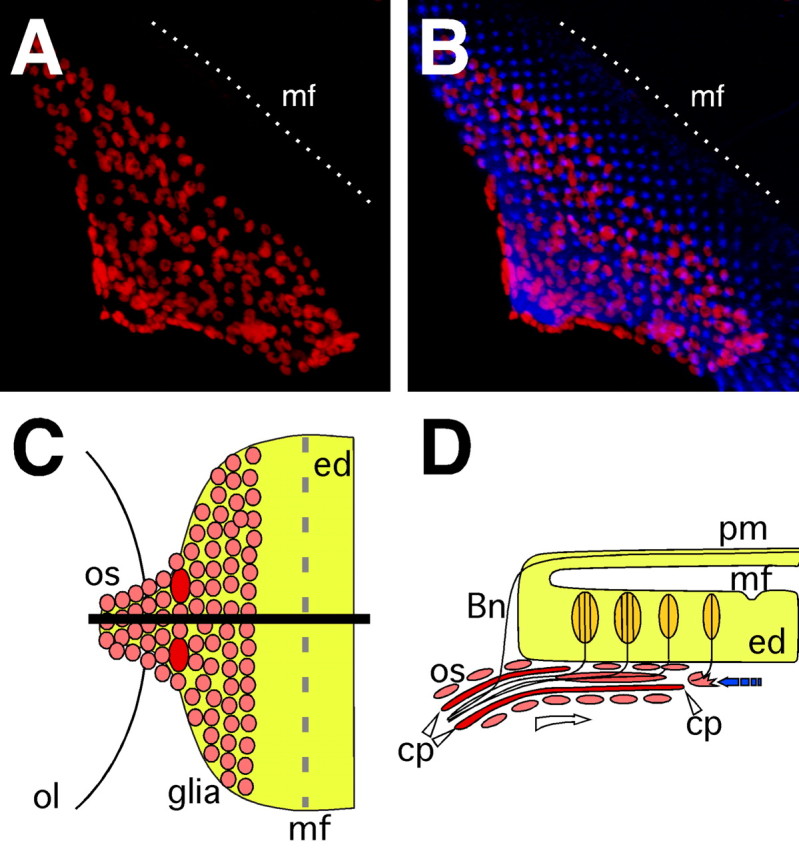
Neuron–glia interaction in the eye imaginal disc. A, B, Confocal images of dissected wild type third instar eye imaginal discs. Neurons are stained using anti-HRP antibodies (blue), glial cell nuclei are labeled using anti-Repo antibodies (red). Anterior is to the top right. In wild type eye imaginal discs migrating glial cells respect a boundary posterior to the morphogenetic furrow (mf) indicated by a white, dashed line. C, Schematic ventral view of an eye imaginal disc. Glial cells populate the eye disc from the optic stalk. The black line indicates the level of the orthogonal section shown in D. D, Glial cells migrate along the carpet cells below the eye imaginal disc (open arrow). Just posterior to the morphogenetic furrow they contact neurons, stop their migration and initiate glial differentiation back toward the CNS (filled arrow). ol, Optic lobe; os, optic stalk; cp, carpet glia; ed, eye disc; mf, morphogenetic furrow; pm, peripodial membrane; Bn, Bolwig's nerve.
Temporal and spatial pattern of gliogenesis
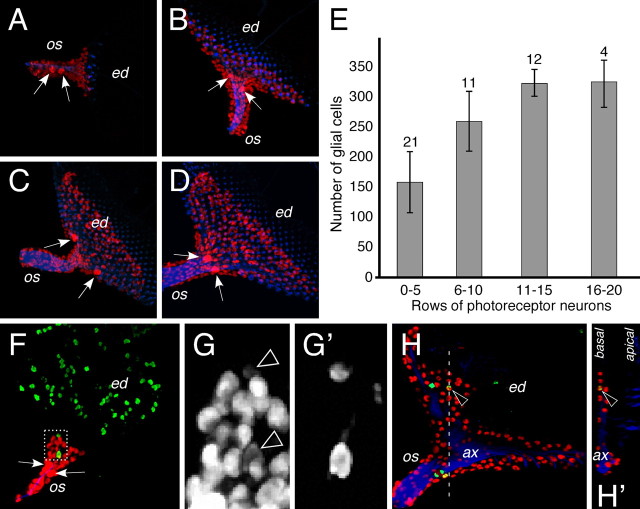
To follow glial migration we first counted the number of cells expressing the pan glial marker Repo in different developmental stages (Fig. 2, supplemental Fig. 1, available at www.jneurosci.org as supplemental material) (for details, see Materials and Methods). To control the age of the imaginal disc we also determined the number of rows with photoreceptor neurons in the eye disc. In second instar larvae an optic stalk was first observed. In this stage, which is before any neuronal differentiation in the eye disc, only six to 20 glial cells populate the stalk. When differentiation of photoreceptor neurons is initiated in the disc, ∼150 glial cells are present in the stalk. As more and more rows of photoreceptor neurons are generated in the wake of the morphogenetic furrow, glial cells populate the disc. At the end of larval development the number of glial cells increases to ∼325 cells per eye disc (Fig. 2A–E). In addition, we determined the proliferation pattern using phospho histone H3 staining (Fig. 2F–H). No pattern in the distribution of phospho histone H3 staining can be recognized suggesting that all glial cells are equally competent to divide. In general, expression of the pan glial marker Repo is reduced in dividing cells (Fig. 2G,G'). Glial proliferation occurs throughout development and is not restricted to the stalk. However, only cells located in the basal most eye disc glial layers appear to be able to divide (Fig. 2H).
Figure 2.
Formation of the eye disc glia. A–D, F–H, Larval eye imaginal disc stained for the presence of glial nuclei (Repo staining, red), neuronal membranes (HRP staining, blue) and dividing cells (phospho histone H3 staining, green). A–D, Glial cells are born in the optic stalk and migrate onto the eye imaginal disc as neuronal differentiation proceeds. Two large glial nuclei can be recognized already from early developmental stages onwards (arrows). Initially these nuclei are found in the optic stalk (os), but later are generally located at the posterior end of the eye imaginal disc (ed). Note that glial nuclei are always posterior to the differentiating neurons. E, Glial nuclei were counted using the Volocity classifier tool. The number of eye discs quantified is indicated. Discs were staged according to the number of rows of photoreceptor cells. At the end of the third instar larval stage, ∼325 glial cells can be detected in the eye imaginal disc. F, Eye imaginal disc of a second instar larvae. No neuronal differentiation can be detected in the eye imaginal disc. Dividing cells are detected after a phosphohistone H3 staining. Some glial cells divide in the stalk and generally show reduced levels of Repo expression (dashed area is shown in high magnification in (G). G, Repo expression highlights all glial nuclei. Arrowheads indicate dividing glial cells that (G′) are labeled after a phosphohistone H3 staining. H, In third instar eye imaginal discs, glial cells divide in the optic stalk (os). In addition basal glial cells located in the disc can also divide (arrowhead). H′ shows an orthogonal section at the position indicated by the white dashed line in (H. ax, Axons.
Distinct glial cell types in the eye disc
To reveal a possible morphological und functional heterogeneity among the different glial cells we generated transgenes carrying a construct that allows expression of the Flp recombinase under the control of the repo promoter (for details, see Materials and Methods). These flies as well as flies carrying a repoGal4 UASflp recombinant chromosome activate expression of the Flp recombinase specifically in all glial cells and allow labeling of individual glial cells and lineages in a flip-out approach (Struhl and Basler, 1993). Here, a constitutive promoter drives expression of a reporter gene only if an unrelated stuffer sequence is removed by an Flp/FRT-mediated recombination event (i.e., act5C>CD2>Gal4 or act5C>yellow>Gal4 together with UASCD8::GFP). Because the repo promoter confers glial expression (Campbell et al., 1994; Xiong et al., 1994; Halter et al., 1995; Jones, 2005; Lee and Jones, 2005), only glial cells are labeled. Approximately 500 eye discs were analyzed, with clone sizes ranging from single labeled glial cells to groups of >40 green fluorescent protein (GFP)-positive glial cells (Fig. 3B). Careful microscopic analysis revealed six morphologically distinct glial cell classes that are summarized in Figure 3A.
Figure 3.
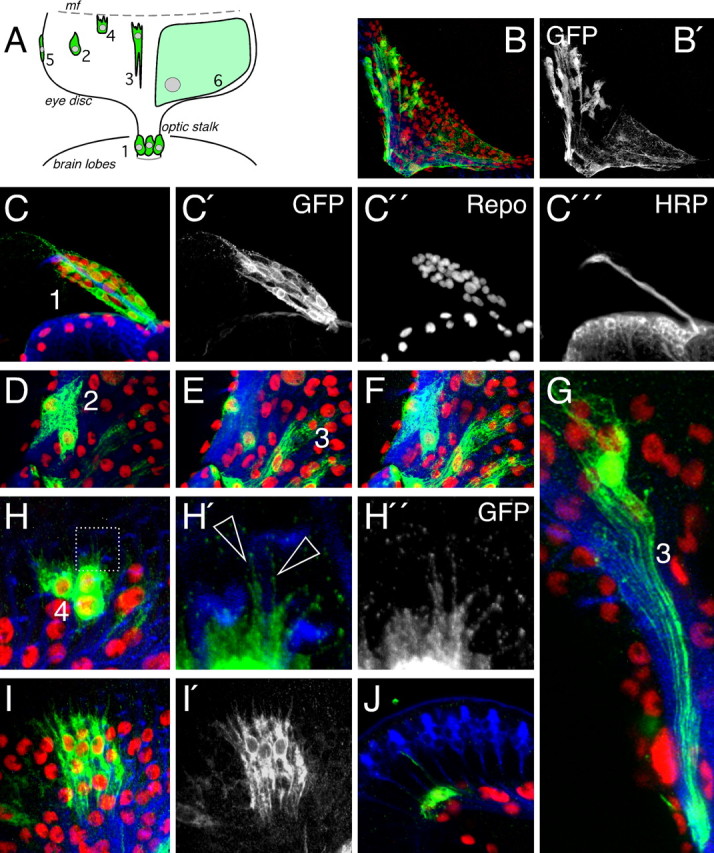
Glial cell types in the imaginal disc. Six different glial cell types are found in the eye imaginal disc. Glial cell clones were generated with repoFlp transgenic flies in a flip-out approach. See text for details. A, The position of the cell types is indicated in the schematic drawing. Anterior is up. Glial cell types 5 and 6 are shown in Figure 4. B, B′, Projection of confocal sections of an eye imaginal disc with several labeled glial cells. Glial nuclei are shown in red (Repo staining), neuronal membranes are shown in blue (HRP staining), and glial cell bodies are shown in green (anti GFP staining). Numbers correspond to the different glial cell types. C–C‴, Optic stalk of a second instar larvae, the stalk glial cells (1) display a spindle shaped morphology (C′). D, Migratory glial cell shapes (2) were only detected in the very basal plane of the eye imaginal disc. E, Confocal section taken from the same specimen at a more apical level. Differentiating glial cells (3) can only be detected close to the axonal layer (blue). F, Projection of the entire confocal stack, including images (D) and (E). G, Single cell clone of a wrapping glial cell (3). Seven wrapped fascicles can be discriminated. H, Glial cells close to the morphogenetic furrow (4) are characterized by filopodia. H′, H″, High magnification showing filopodia (arrowheads) extending along axonal membranes (blue). I, Glial cell clone of young differentiating cells. Cells extend processes in anterior and posterior directions. J, Orthogonal section through a confocal stack showing thin glial cell process along the photoreceptor neurons.
Within the optic stalk, glial cells generally adopt a spindle like appearance, forming a dense mesh of cells surrounding the photoreceptor axons (Fig. 3C). In the eye disc migratory and differentiating cells were found in different layers. In the basal most part of the eye disc, glial cells always displayed a migratory shape (Fig. 3D). The migratory cells located in the middle of the eye disc are usually spindle shaped, whereas cells toward the anterior leading edge of the glial field, which are close to the morphogenetic furrow, are characterized by numerous filopodia-like cell protrusions (Fig. 3H). This suggests that glial cells in the basal layer of the eye disc usually migrate as a chain or sheath and follow the ones at the leading edge of the glial field.
In more apical regions of the eye disc close to the axonal layer we never found glial cells with a migratory cell shape but instead cells showed prominent signs of differentiation and apparently wrap axon fascicles from the differentiating ommatidial units (Fig. 3E–G,I). Eye disc glial cells can ensheath more than one fascicle, which was also seen at the electron microscopic level (Figs. 3G, 6). Cells close to the morphogenetic furrow in general had less well established wrapping processes whereas cells in the middle or posterior domain of the eye disc showed more extensive signs of differentiation. Glial cells of the posterior domain frequently send long cellular processes that follow the axons through the optic stalk toward the brain. Wrapping glial cells can be found in the stalk where they also associate with the axons of the Bolwig's nerve which they, however, do not enwrap (Fig. 6, supplemental Fig. 2, available at www.jneurosci.org as supplemental material). In addition to wrapping of individual fascicles we frequently identified single glial processes that project along the descending photoreceptor axons into the apical region of the eye disc (Fig. 3J).
Figure 6.
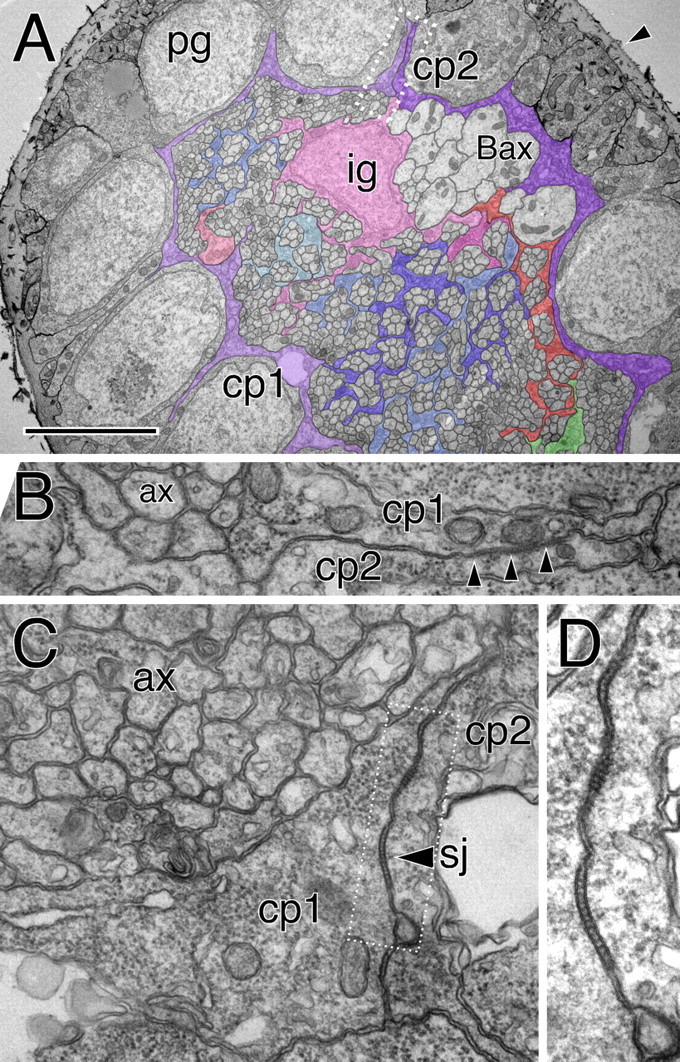
Glial cells of the optic stalk. Electron microscopic images taken from an optic stalk of a third instar wandering larvae. A, The optic stalk is surrounded by a thick neural lamella (arrowhead). The perineurial glial cells (pg) have a cuboid shape with some fine cell protrusions that extend along the axis of the stalk. Below the perineurial cells are two subperineurial cells (cp1, cp2) that are highlighted by colored shading. These cells form a continuous thin ring around the entire optic stalk. The axonal profiles of the Bolwig's organ can be recognized because of their size (Bax). One inner glial cell nucleus is seen (ig). Some glial processes that enwrap individual axons are highlighted. Here, glial processes do not surround axons at the periphery of the stalk. The boxed area is shown in higher magnification in B. Scale bar, 3.5 μm. B, High magnification of the carpet glia contact zone. Septate junctions can be recognized (arrowheads) between carpet cell one and two (cp1, cp2). Axonal profiles (ax) have not yet been engulfed by glial processes. C, D, Similar images of the septate junction area between two carpet cells (arrowhead, sj). The corresponding optic stalk is shown in supplemental Figure 3 (available at www.jneurosci.org as supplemental material).
Beside the above mentioned glial cell types, we also found several less frequent cell classes. Elongated cells that overlap adjacent glial cells in a clapboard type manner occupy the margin of the glial field. Neither in the eye imaginal disc nor in the optic stalk, where this cell type is also present, any connection to axonal membranes could be observed (Fig. 4A–C). The different glial cell types identified in the eye imaginal disc may derive from distinct progenitor pools or they originate in a sequential mode from progenitor cells that migrate on the eye disc (Fig. 1C,D).
Figure 4.
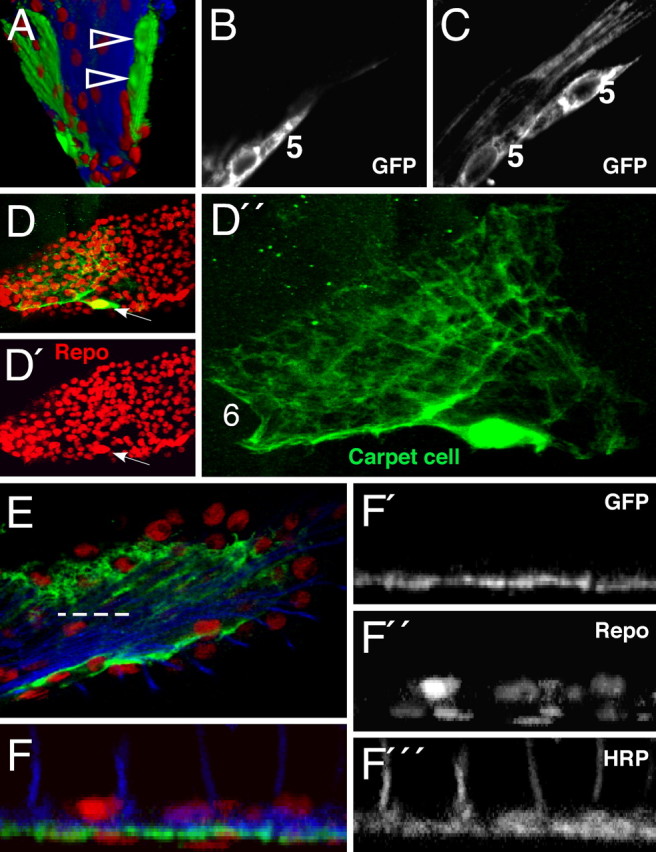
Marginal and carpet glial cells. Confocal analysis of third instar larval eye imaginal discs. Single glial cells are marked with anti-GFP antibodies (green). Glia cell nuclei are stained for Repo expression (red) and photoreceptor neurons are marked with anti-HRP antibodies (blue). A–E, Anterior is to the top. A, 3D reconstruction of an eye imaginal disc. At the margin of the eye disc, a small clone of glial cells is marked (arrowheads). B, C, Single confocal sections of this glial clone show that marginal glial cells (5) display an elongated, clapboard like shape. No contacts to axons can be detected. D, D′, Every eye disc has two large, flat carpet cells of which one is labeled in this eye disc. The carpet glial nucleus can be easily identified according to its size (arrow; see also arrows in Fig. 2). The cytoplasmic extensions of the carpet cells cover half of the eye disc. E, Triple-stained specimen showing the relation of the carpet cell to axons and other glial cells. The dashed line indicates the orthogonal section shown in F. F–F‴, Orthogonal section shows that the so-called carpet cells separate basally localized nuclei from axons (blue) and more apical located glial nuclei.
The carpet glia separates migrating and differentiating glial cell layers
An additional cell type, that we name carpet glial cells, exhibits the most striking appearance. Only two carpet cells are found in every eye disc. Their large nuclei can already be detected in an anti-Repo staining (Fig. 2A–D). When CD8::GFP expression is activated in the carpet cells we detect a fine and thin mesh of GFP-positive membranes. The carpet cells are very large and cover one half of the differentiated part of the eye disc each, which corresponds to an area of ∼10,000 epithelial cells. Orthogonal sections through z-stacks of confocal images demonstrate that the carpet cells separate the basal most localized migrating glial cells from more apical ones (Fig. 4E,F). In consequence, glial migration onto the eye disc occurs at least in part on a glial substrate.
Carpet cells obey the equator separating dorsal and ventral portion of the compound eye and never reach anterior to the differentiating photoreceptor cells. At the base of the eye imaginal disc, the carpet cells grow around the optic stalk and the posterior portion of the eye disc which they ensheath like a calla flower (supplemental Fig. 2, available at www.jneurosci.org as supplemental material). Thereby the carpet cells, which express the moody Gal4 driver, completely separate the axon area from the outer perineurial glial cell layer in which migration and cell division takes place. Carpet cells are present as soon as an optic stalk is recognized in second instar larvae (supplemental Fig. 1, available at www.jneurosci.org as supplemental material).
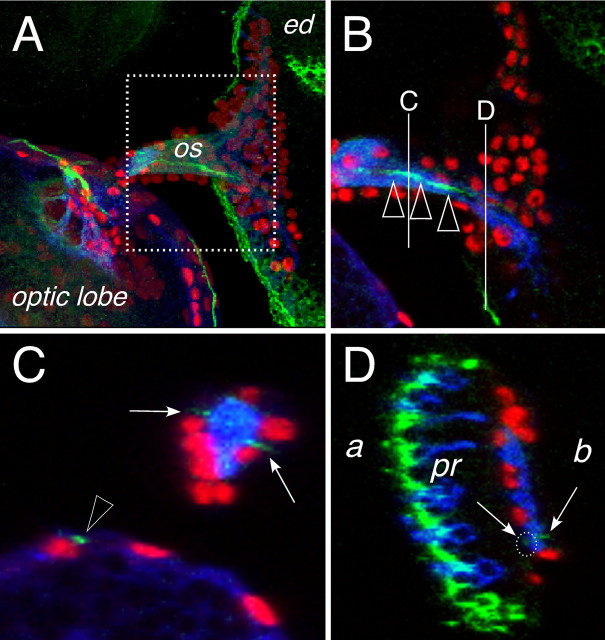
Based on the expression of moody, the carpet glia belongs to the subperineurial glia that establishes the blood–brain barrier (BBB) in insects (Carlson et al., 2000; Bainton et al., 2005; Schwabe et al., 2005) (Stork, Engelen, Krudewig, Silies, Bainton, and Klämbt, unpublished observations). To fulfill this function, the subperineurial glial cells establish septate junctions, which can be visualized by the expression of the NeurexinIV protein (Baumgartner et al., 1996; Bainton et al., 2005; Schwabe et al., 2005). NeurexinIV expression can be faithfully detected using the GFP exon trap line 454 (Edenfeld et al., 2006). Using this gene trap insertion we detected two lines of NeurexinIV expression in the optic stalk apical to the axon bundle (Fig. 5C, arrows). In the eye disc one thin line of NeurexinIV::GFP expression can be found basal to the photoreceptor axons at the later equator (Fig. 5A–D). A second, but much weaker site of NeurexinIV::GFP expression is found directly apical to the axon tracts (Fig. 5D, dotted circle). This pattern reflects the position of the carpet cell and the two expression domains are compatible with the notion that the carpet cells are septate junction forming subperineurial cells.
Figure 5.
Subperineurial cells in the eye disc. Confocal images of dissected wild type third larval instar eye imaginal discs. Neuronal membranes are shown in blue (anti-HRP staining), glial cell nuclei are labeled using anti-Repo antibodies (red), NeurexinIV::GFP expression is shown in green. Anterior is to the right. A, Overview of an eye imaginal disc attached through the optic stalk (os) to the brain lobe of a NeurexinIV::GFP-expressing larvae. The dotted area corresponds to the region shown in B. B, Single confocal section, the white lines indicate the positions of the orthogonal sections shown in C and D. The green line of NeurexinIV::GFP expression corresponds to the septate junctions formed by the carpet glia (arrowheads). C, In an orthogonal section through the optic stalk, two regions of NeurexinIV::GFP accumulation can be seen (arrows), similar NeurexinIV::GFP expressing septate junctions are seen in the optic lobe (arrowhead). D, Orthogonal section through the eye imaginal disc. Intense NeurexinIV::GFP staining can be detected on the apical part of the eye disc epithelium (a) next to the photoreceptor cells (pr). Two faint lines of GFP expression can be seen more basal (b) close to the photoreceptor axon tracts (arrows, white dotted circle), which cannot be detected in the frontal view because of the high amount of epithelial NeurexinIV expression.
Electron microscopic analysis
To further clarify whether the carpet cells belong to the subperineurial cell type we undertook an electron microscopic analysis. As already anticipated from the single cell analyses presented above, only two cell types surround the large bundle of central photoreceptor axons in the optic stalk (Fig. 6A). The outermost cell layer just below the neural lamella comprises the spindle shaped perineurial cells (Fig. 6A, pg). Between the perineurial cells and the ommatidial photoreceptor axons are two flat carpet cells (Fig. 6A, cp1, cp2). These cells do not wrap axons but form septate junctions as already suggested from the confocal analysis (Fig. 6B–D, supplemental Fig. 3, available at www.jneurosci.org as supplemental material).
The large axonal profiles of the larval photoreceptor organs, the Bolwig's organs, are found at the margin of the neuropil and provide an asymmetry to the optic stalk (Fig. 6A, Bax). The Bolwig's organ comprises 12 photoreceptor cells (Meinertzhagen and Hanson, 1993) and their axon fascicle is in contact with the carpet glia but the axons are not individually wrapped by additional glial processes (Fig. 6A). Eight to nine additional axons that presumably correspond to the two larval stemmata are closely associated with the Bolwig's nerve and degenerate during pupal stages (Tomlinson and Ready, 1987). The ommatidial fascicles are separated from the Bolwig's fascicle by an inner glial cell whose nucleus can be detected within the neuropil area (Fig. 6A, ig). We noted a gradient in glial wrapping. The peripheral fascicles are not wrapped when inner ommatidial fascicles are already wrapped as individual units (Fig. 6A, some glial cell processes are indicated by color shading).
In the eye imaginal disc, too, the carpet cells separate wrapping from nonwrapping glia (Fig. 7A). The most basal cells in the eye disc correspond to the perineurial glial cells, which belong to the migratory cell type detected by our flip out approach. Migration occurs from the stalk toward the morphogenetic furrow. In the anterior most region of the glial cell layer, the carpet cell extends about one cell posterior to the migrating glial front (Fig. 7B–D). The anterior most glial cell makes contact to the disc epithelium in which photoreceptor neurons will differentiate. This glial cell sends out fine cell protrusions between the eye disc epithelium and the covering extracellular matrix called neural lamella (Fig. 7D). Photoreceptor axons leave the eye disc epithelium and turn in a sharp angle toward the posterior of the eye disc. Sometimes, axonal profiles appear enlarged at the point where the axons leave the disc epithelium (Fig. 7E,F). As already seen in confocal analyses, thin glial cell processes project along the axonal membranes to the ommatidial unit (Figs. 3J, 7F,G). This is reminiscent to thin processes of Müller glial cells along photoreceptor neurons in the mammalian retina (Tsujikawa and Malicki, 2004). Ommatidial axons always project as fascicles. The wrapping glia generally ensheathes groups of axon fascicles and never separates individual photoreceptor axons. The axons of the Bolwig's organ are embedded within the peripodial membrane (supplemental Fig. 2, available at www.jneurosci.org as supplemental material).
Figure 7.
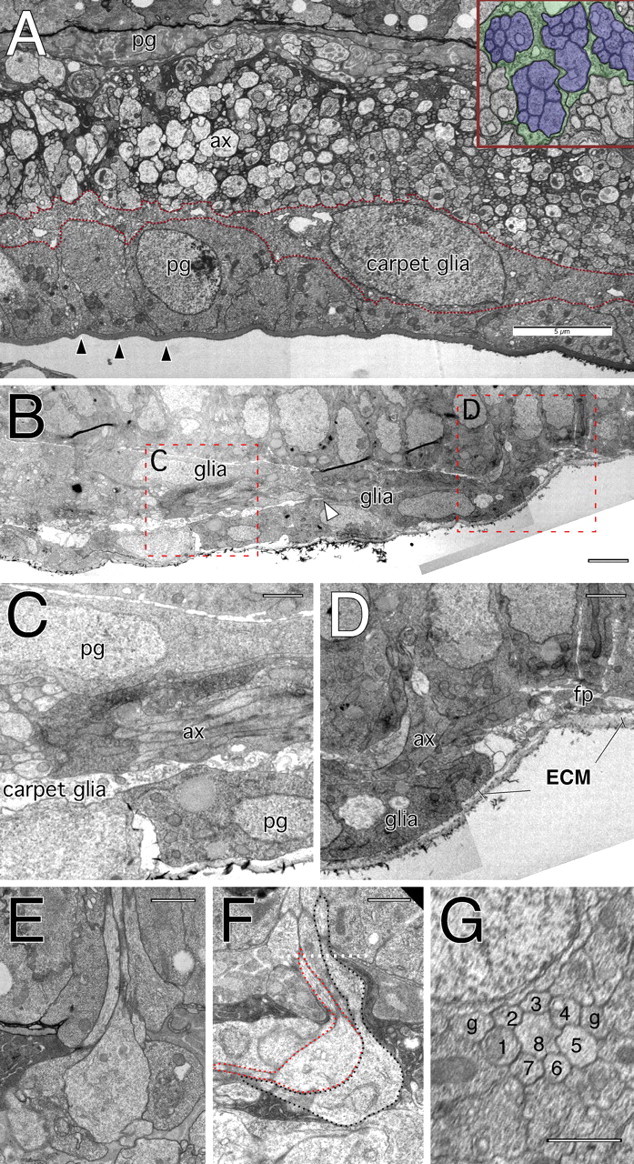
Glial cells in the eye disc. Electron microscopic images taken from an eye imaginal disc of a wandering third instar larvae. A, Cross section showing a layer of perineurial glial cells (pg) above a thick neural lamella (arrowheads). On top of the perineurium lies the carpet glia that extends thin cell processes below the axon traffic (ax). The inset shows that individual axon bundles originating from individual ommatidia are engulfed by glial processes. Many of the axons have a large diameter. B, Longitudinal section through an anterior portion of an eye imaginal disc posterior to the morphogenetic furrow. Anterior is to the right. The areas in dashed red boxes are shown in C and D. The arrowhead marks the anterior end of the carpet cell, which is slightly behind the migrating glial front. C, Three cell diameters posterior to the morphogenetic furrow, a three-layered organization can be observed below the eye imaginal disc epithelium. Most basally is the neural lamella that covers the perineurial glial cells. Above the perineurium is the carpet glia, which is a thin glial layer that prevents any axonal contact (ax) to the perineurium. Between the eye imaginal disc epithelium and the axonal layer is another layer of perineurial glial cells. D, Anterior edge of the glial field close to the morphogenetic furrow. The extracellular matrix (ECM) of the neural lamella is also found anterior to the glial cells. No processes of carpet cells can be detected and the anterior most glial cell can make contact with the axonal membranes (ax). At the anterior of the glial field an area with many filopodia processes can be seen (fp). E, F, Cross (E) and longitudinal section (F through an ommatidial fascicle projecting into the axon layer below the eye disc. Note the swollen area at the turning point. The dotted line indicates the plane of section used to take the image shown in G. G, Next to the eight axonal profiles of the different photoreceptor neurons, two glial processes can be detected. Scale bars: A, 5 μm; B, 2.5 μm; C, D, 1 μm; E–G, 1 μm.
Continuous migration and differentiation of glial cells in the eye disc
The morphological data imply a simple model underlying glial cell migration onto the eye disc (Fig. 1). Cells located at the basal side of the eye disc migrate along the carpet glia toward the morphogenetic furrow. Once the migratory cells reach the margin of the carpet cell they come in contact to epithelial and neuronal cells. After neuronal contact, glial cells start to differentiate along the axonal fascicles and grow their membranes back toward the CNS (Fig. 1).
If this sequential differentiation model is correct, ablation of the migratory glia should also affect the wrapping glia and, furthermore, early labeling of some migratory cells should lead to clones in the differentiating glia. The Gal driver line c527 is active only in the outer glial cell layer and the driver strain Mz97 is active only in the wrapping glia (Hummel et al., 2002), Figure 8A,B). Animals carrying both Gal4 drivers label all glial cells (Fig. 8C). To verify that the c527 driver is active in nonwrapping outer glial cells we used c527 UASFlp UAS>CD2 yellow>mCD8::GFP flies and generated single labeled glial cell clones (Fig. 8). Using this paradigm we detected either migratory cell types or apical perineurial cells at the apical boundary of the glial field (Fig. 8D). Likewise, Mz97-mediated Flip expression resulted only in the labeling of wrapping glial cells (Fig. 8E).
Figure 8.
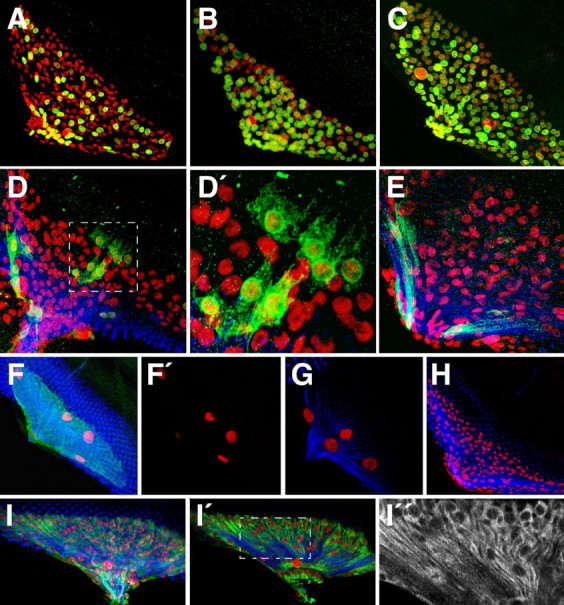
Wrapping and migrating glial cells are clonally related. Confocal images of dissected wild type third larval instar eye imaginal discs. Neurons are stained using anti-HRP antibodies (blue), glial cell nuclei are labeled using anti-Repo antibodies (red), GFP or βGal expression (green). Anterior is to the top right. A, Eye disc of a larvae with the genotype Mz97 UASlamGFP. All GFP-expressing cells also express Repo and are glial cells. Expression is found in a scattered pattern close to the axonal tracts. B, Eye disc of a larvae with the genotype: c527 UASlamGFP. C, Eye disc of a larvae with the genotype: Mz97, c527 UASlamGFP. All glial cell nuclei expressing Repo express GFP. D, Eye disc of a larvae with the genotype: c527 UASFlp UAS>CD2 y+>mCD8::GFP. Glial cells expressing the c527 driver belong to the perineurial glia type. D′, High-resolution image of D. E, Eye disc of a larvae with the genotype: Mz97 UASFlp UAS>CD2 y+>mCD8::GFP. Glial cells expressing the Mz97 driver belong to the wrapping glia type. F, F′, Expression of UASfzr and UASact5C::GFP transgenes using the panglial repoGal4 driver. Glial mitosis is efficiently blocked and only a few large glial nuclei form, which is because of endoreplication triggered by fzr expression. The few glial cells that migrate onto the eye disc show a remarkable structural plasticity. Glial cell processes (green) cover all axonal fascicles. G, A similar phenotype was observed in c527 UASfzr animals. H, A UASfzr transgene is expressed using the Mz97 Gal4 driver. No phenotypic consequences develop after fzr expression demonstrating that differentiating glial cells are postmitotic. I–I″, Eye disc of a larvae with the genotype: c527 UASFlp act>draf>τlacZ. Expression of τlacZ is activated after recombination in the c527 positive perineurial cells. τlacZ expression is maintained by the actin5c promoter and can be found in wrapping glial cells (I″) demonstrating a clonal relationship between the perineurial and the wrapping glial cell population. I shows a projection and I′ and I″ are single focal planes.
Because expression of cell death genes hid or reaper using the c527 driver resulted in early larval lethality we instead blocked mitosis of glial cells by expressing the Fizzy related (Fzr) protein (Sigrist and Lehner, 1997). Preventing mitosis in all eye disc glial cells results in a dramatic reduction in glial cell number (Fig. 8F). Similarly, expression of Fzr only in the c527-positive perineurial cell population ablated most glial cells from the eye disc, suggesting that migrating and ensheathing glia have the same origin (Fig. 8G). As Repo staining only detects glial nuclei, we visualized the entire glial cell morphology using a UASact5C::GFP construct. Surprisingly, the few remaining glial cells showed a remarkable structural plasticity and were able to extend cell processes covering all axons that developed relatively normal appearing trajectories (Fig. 8F).
Consistent with the idea that Mz97-positive wrapping glia originate from the mitotically active c527-positive glial cells, no phenotypic change was apparent when we expressed Fzr and thus blocked mitosis in the Mz97-positive wrapping glia, indicating that the wrapping glial cells are postmitotic (Fig. 8H). This also fits to the observation that dividing cells are only detected in the basal most glial cell layer (Fig. 2H). To further test whether migration of glial cells along the carpet glia results in the generation of differentiating glial cells we generated flip out clones using the c527 driver strain with UASflp and an act5C>Draf>τlacZ construct. Under this experimental condition, τlacZ expression is activated by the c527-mediated Flp expression and is then maintained because of the constitutive activity of the actin5C promoter. Thus it is possible to visualize the morphology of former c527-expressing glial cell clones even after the c527 promoter activity is turned off. Indeed cell clones generated using this paradigm clearly showed wrapping glial cells supporting the notion that glial migration and differentiation are interconnected continuous processes (Fig. 8I).
The carpet cells inhibit glial migration
A key function within this model has to be attributed to the carpet cells and we thus attempted to ablate these cells. Expression of moody is specific for the subperineurial cells (Bainton et al., 2005; Schwabe et al., 2005). In the developing eye moodyGal4 efficiently activates expression of UASlamin::GFP and UASGapGFP transgenes in the two carpet cells (Fig. 9A,B).
Figure 9.
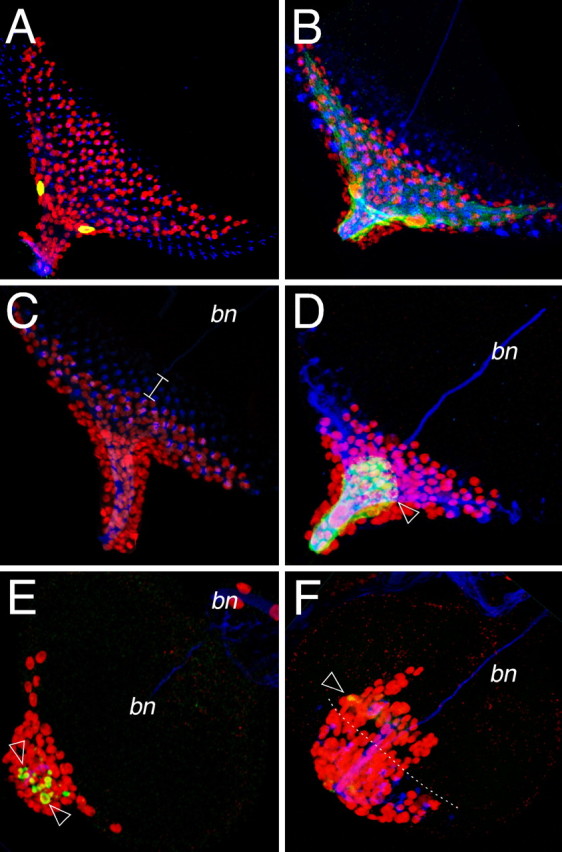
The carpet cells restrain glial cell migration. Confocal images of dissected wild type third instar eye imaginal discs. Neurons are stained using anti-HRP antibodies (blue), glial cell nuclei are labeled using anti-Repo antibodies (red), glial cell morphology is shown by GFP expression (green). Anterior is to the top right. A, Eye disc of a larvae with the genotype: moodyGal4 UASlamin::GFP. The moody driver is active only in the carpet cells. B, Eye disc of a larvae with the genotype: moodyGal4 UASGapGFP. The carpet cells can be recognized by their typical shape. C, Wild type eye imaginal disc of a third instar. Note that glial cells are posterior to the first differentiating photoreceptor cells (white line). D, After ablation of the carpet cell glial cells are found anterior to the photoreceptor cells. E, F, When only remnants of the carpet cells are left (arrowhead), the overmigration phenotype is more pronounced and glial cells enter the eye imaginal disc epithelium before photoreceptor cell differentiation.
When we used moodyGal4 to activate UAShid expression, the animals died during early larval stages presumably because we disrupted the blood brain barrier, which requires an intact subperineurial cell layer. To ablate only the moody-positive carpet cells during later developmental stages we used the Gal80ts technology (McGuire et al., 2003). Animals of the genotype moodyGal4 UASGapGFP UAShid tubGal80ts were kept at the permissive temperature for 7–10 d and then transferred to 29°C until the end of larval development. After this time regime, larvae survived until young third instar stages and their eye discs were analyzed. Ablation of the carpet cell was monitored by GFP expression. Generally, glial cells migrated further onto the imaginal disc epithelium than compared with the wild type situation. In cases were only vesicular remnants of the carpet cells could be detected we found the most pronounced overmigration phenotypes, indicating that the carpet cells are acting to retard migration of the eye disc glial cells also before the onset of photoreceptor cell differentiation in young eye imaginal discs (Fig. 9E,F).
Discussion
Here we have shown that glial migration onto the Drosophila eye imaginal disc occurs in a constant migration and differentiation mode. Unlike in many other systems, glial migration is largely regulated by glial–glial cell contact and requires the presence of two unusually large glial cells that physically separate the migrating glial population from the differentiating glial cell population.
A hallmark of all glial cells is their pronounced migratory capability. In vertebrates all PNS glial cells originate from the neural crest and follow the developing motor axon paths to eventually insulate the nerve bundles. In the CNS, oligodendrocyte progenitor cells are born in defined regions of the neural tube and migrate along specific routes to reach their final destinations in the brain (de Castro and Bribian, 2005). A negative consequence of this motility is reflected by the bad prognosis of gliomas, that are characterized by highly invasive cells that make surgical treatment impossible (Berens and Giese, 1999). Glial migration is not restricted to the vertebrate nervous system but is equally found in invertebrates (Jacob, 1984; Swales and Lane, 1985; Torrence, 1991).
During Drosophila embryonic stages, the migration of the midline glia is required for correct wrapping and patterning of the commissural axon tracts (Klambt et al., 1991; Stollewerk and Klämbt, 1997; Jacobs, 2000). The longitudinal glial cells that wrap the longitudinal connectives migrate medially from lateral regions of the ventral nerve cord toward their target axons (Jacobs et al., 1989; Kinrade et al., 2001). Glial cell migration is particularly obvious in the Drosophila PNS. As in the vertebrate nervous system most peripheral glial cells are born in the CNS and then follow axonal projections into the periphery (Sepp et al., 2000). Although motor as well as sensory axons are present, the peripheral glia of the embryo seems to particularly depend on outgrowing motor axons (M. Silies, unpublished observations).
However, migration of glia cells is not restricted to motor axons, but extensive migration of glial cells along sensory paths has been described. In the developing wing, glial cells are born in the periphery and follow the sensory neurons (Giangrande et al., 1993; Giangrande, 1994; Aigouy et al., 2004). In the developing olfactory system, glial cells are born in the periphery as well as in the CNS. The centrally born glial cells migrate along ingrowing olfactory axons toward the antennae (Sen et al., 2005). In the developing compound eye, glial cells only originate from the CNS and move toward the eye disc epithelium as photoreceptor neurons are formed in the wake of the morphogenetic furrow (Choi and Benzer, 1994). Although photoreceptor neurons appear to attract glial cells over some distance, glial migration in the eye disc can occur in the absence of photoreceptor neurons (Choi and Benzer, 1994; Rangarajan et al., 1999; Hummel et al., 2002).
The relevance of glial–glial interaction in orchestrating glial migration has been demonstrated in other systems. In the developing wing, live imaging experiments demonstrated that homotypic glial–glial cell contact regulates the extent of glia migration whereas neuron–glial interactions only affects the direction of glial migration (Aigouy et al., 2004). Similarly, spacing of zebrafish oligodendrocytes along axonal tracts requires glial–glial cell interactions (Kirby et al., 2006). Here we have shown that within the eye imaginal disc, a specialized subperineurial cell, the carpet cell restricts glial migration and might also influence glial differentiation after contact with the nerve.
The Drosophila visual system is one of the most intensively studied parts of the nervous system (Freeman, 1997; Kumar and Moses, 2001). However, analysis has focused mainly on the role of photoreceptor neurons. Although the mechanisms that pattern the eye imaginal disc and control the differentiation and targeting of photoreceptor axons are well understood, not much was known on the diversity and function of involved glial cells. Glial cells associated with the brain proliferate and form the carpet cells as well as the surrounding spindle shaped perineurial cells. These latter cells will divide throughout larval development to eventually generate the 350 glial cells found in every eye imaginal disc.
The differentiation of these glial cells is likely to require a more complex regulation to match the sequential formation of photoreceptor axons. Single-cell labeling using a repoFlp transgene demonstrated that migrating glial cell first encounter axonal projections close to the morphogenetic furrow. Thus it is ensured that all nascent axonal profiles can be contacted by glia to be equally wrapped. During this differentiation the glial cells send their processes along the axons toward the brain, thereby masking the axons from other glial cells. Consistent with this, more wrapping processes are observed in the inner optic stalk then in the peripheral regions where new axonal projections are added.
The sequential generation of glial cells in the eye also requires glial–glial cell interactions. A prime function in this process can be attributed to the carpet cells that appear to provide a physical barrier separating naive but migration competent glial cells from neuronal membranes. Ablation of the carpet cells results in an overmigration phenotype demonstrating that glial cells have a constant tendency to migrate and that they require glia–glial contacts to restrain this migratory potential. Just posterior to the morphogenetic furrow the carpet cell and the nascent photoreceptor axons induce the arrest of glial migration and the initiation of glial differentiation. Possibly, once the differentiation of wrapping glial cells takes place progression of the carpet cell toward the anterior is facilitated. This way a coordinated movement of glial cells after the morphogenetic furrow is guaranteed.
Footnotes
This work was supported by fellowships of the Cusanuswerk and the Boehringer Ingelheim Foundation (M.S.). Y.Y. acknowledges a fellowship of the graduate program “molecular cell dynamics.” This work was performed with support of the Deutsche Forschungsgemeinschaft (C.K.). We thank T. Hummel for discussion, I. Salecker for sharing EM data and discussion, and E. Naffin for help with electron microscopic analyses.
References
- Aigouy B, Van de Bor V, Boeglin M, Giangrande A. Time-lapse and cell ablation reveal the role of cell interactions in fly glia migration and proliferation. Development. 2004;131:5127–5138. doi: 10.1242/dev.01398. [DOI] [PubMed] [Google Scholar]
- Bainton RJ, Tsai LT, Schwabe T, DeSalvo M, Gaul U, Heberlein U. moody encodes two GPCRs that regulate cocaine behaviors and blood-brain barrier permeability in Drosophila. Cell. 2005;123:145–156. doi: 10.1016/j.cell.2005.07.029. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Pillai AM, Paik R, Li J, Bhat MA. Axonal ensheathment and septate junction formation in the peripheral nervous system of Drosophila. J Neurosci. 2006;26:3319–3329. doi: 10.1523/JNEUROSCI.5383-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner S, Littleton JT, Broadie K, Bhat MA, Harbecke R, Lengyel JA, Chiquet-Ehrismann R, Prokop A, Bellen HJ. A Drosophila neurexin is required for septate junction and blood-nerve barrier formation and function. Cell. 1996;87:1059–1068. doi: 10.1016/s0092-8674(00)81800-0. [DOI] [PubMed] [Google Scholar]
- Berens ME, Giese A. “… those left behind.” Biology and oncology of invasive glioma cells. Neoplasia. 1999;1:208–219. doi: 10.1038/sj.neo.7900034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell G, Goring H, Lin T, Spana E, Andersson S, Doe CQ, Tomlinson A. RK2, a glial-specific homeodomain protein required for embryonic nerve cord condensation and viability in Drosophila. Development. 1994;120:2957–2966. doi: 10.1242/dev.120.10.2957. [DOI] [PubMed] [Google Scholar]
- Carlson SD, Juang JL, Hilgers SL, Garment MB. Blood barriers of the insect. Annu Rev Entomol. 2000;45:151–174. doi: 10.1146/annurev.ento.45.1.151. [DOI] [PubMed] [Google Scholar]
- Choi KW, Benzer S. Migration of glia along photoreceptor axons in the developing Drosophila eye. Neuron. 1994;12:423–431. doi: 10.1016/0896-6273(94)90282-8. [DOI] [PubMed] [Google Scholar]
- de Castro F, Bribian A. The molecular orchestra of the migration of oligodendrocyte precursors during development. Brain Res Brain Res Rev. 2005;49:227–241. doi: 10.1016/j.brainresrev.2004.12.034. [DOI] [PubMed] [Google Scholar]
- Edenfeld G, Stork T, Klambt C. Neuron-glia interaction in the insect nervous system. Curr Opin Neurobiol. 2005;15:34–39. doi: 10.1016/j.conb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Edenfeld G, Volohonsky G, Krukkert K, Naffin E, Lammel U, Grimm A, Engelen D, Reuveny A, Volk T, Klambt C. The splicing factor crooked neck associates with the RNA-binding protein HOW to control glial cell maturation in Drosophila. Neuron. 2006;52:969–980. doi: 10.1016/j.neuron.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Edenfeld G, Altenhein B, Zierau A, Cleppien D, Krukkert K, Technau G, Klambt C. Notch and Numb are required for normal migration of peripheral glia in Drosophila. Dev Biol. 2007;301:27–37. doi: 10.1016/j.ydbio.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Freeman M. Cell determination strategies in the Drosophila eye. Development. 1997;124:261–270. doi: 10.1242/dev.124.2.261. [DOI] [PubMed] [Google Scholar]
- Freeman MR, Doherty J. Glial cell biology in Drosophila and vertebrates. Trends Neurosci. 2006;29:82–90. doi: 10.1016/j.tins.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Giangrande A. Glia in the fly wing are clonally related to epithelial cells and use the nerve as a pathway for migration. Development. 1994;120:523–534. [Google Scholar]
- Giangrande A, Murray MA, Palka J. Development and organization of glial cells in the peripheral nervous system of Drosophila melanogaster. Development. 1993;117:895–904. doi: 10.1242/dev.117.3.895. [DOI] [PubMed] [Google Scholar]
- Gilmour DT, Maischein HM, Nusslein-Volhard C. Migration and function of a glial subtype in the vertebrate peripheral nervous system. Neuron. 2002;34:577–588. doi: 10.1016/s0896-6273(02)00683-9. [DOI] [PubMed] [Google Scholar]
- Halter DA, Urban J, Rickert C, Ner SS, Ito K, Travers AA, Technau GM. The homeobox gene repo is required for the differentiation and maintenance of glia function in the embryonic nervous system of Drosophila melanogaster. Development. 1995;121:317–332. doi: 10.1242/dev.121.2.317. [DOI] [PubMed] [Google Scholar]
- Hulsmeier J, Pielage J, Rickert C, Technau GM, Klambt C, Stork T. Distinct functions of {alpha}-Spectrin and {beta}-Spectrin during axonal pathfinding. Development. 2007;134:713–722. doi: 10.1242/dev.02758. [DOI] [PubMed] [Google Scholar]
- Hummel T, Attix S, Gunning D, Zipursky SL. Temporal control of glial cell migration in the Drosophila eye requires gilgamesh, hedgehog, and eye specification genes. Neuron. 2002;33:193–203. doi: 10.1016/s0896-6273(01)00581-5. [DOI] [PubMed] [Google Scholar]
- Jacob MH. Neurogenesis in Aplysia californica resembles nervous system formation in vertebrates. J Neurosci. 1984;4:1225–1239. doi: 10.1523/JNEUROSCI.04-05-01225.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JR. The midline glia of Drosophila: a molecular genetic model for the developmental functions of glia. Prog Neurobiol. 2000;62:475–508. doi: 10.1016/s0301-0082(00)00016-2. [DOI] [PubMed] [Google Scholar]
- Jacobs JR, Hiromi Y, Patel NH, Goodman CS. Lineage, migration, and morphogenesis of longitudinal glia in the Drosophila CNS as revealed by a molecular lineage marker. Neuron. 1989;2:1625–1631. doi: 10.1016/0896-6273(89)90051-2. [DOI] [PubMed] [Google Scholar]
- Jarjour AA, Kennedy TE. Oligodendrocyte precursors on the move: mechanisms directing migration. Neuroscientist. 2004;10:99–105. doi: 10.1177/1073858403260751. [DOI] [PubMed] [Google Scholar]
- Jones BW. Transcriptional control of glial cell development in Drosophila. Dev Biol. 2005;278:265–273. doi: 10.1016/j.ydbio.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Kinrade EF, Brates T, Tear G, Hidalgo A. Roundabout signalling, cell contact and trophic support confine longitudinal glia and axons in the Drosophila CNS. Development. 2001;128:207–216. doi: 10.1242/dev.128.2.207. [DOI] [PubMed] [Google Scholar]
- Kirby BB, Takada N, Latimer AJ, Shin J, Carney TJ, Kelsh RN, Appel B. In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nat Neurosci. 2006;9:1506–1511. doi: 10.1038/nn1803. [DOI] [PubMed] [Google Scholar]
- Klambt C, Jacobs JR, Goodman CS. The midline of the Drosophila central nervous system: a model for the genetic analysis of cell fate, cell migration, and growth cone guidance. Cell. 1991;64:801–815. doi: 10.1016/0092-8674(91)90509-w. [DOI] [PubMed] [Google Scholar]
- Kumar JP, Moses K. Eye specification in Drosophila: perspectives and implications. Semin Cell Dev Biol. 2001;12:469–474. doi: 10.1006/scdb.2001.0270. [DOI] [PubMed] [Google Scholar]
- Lee BP, Jones BW. Transcriptional regulation of the Drosophila glial gene repo. Mech Dev. 2005;122:849–862. doi: 10.1016/j.mod.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Lemke G. Glial control of neuronal development. Annu Rev Neurosci. 2001;24:87–105. doi: 10.1146/annurev.neuro.24.1.87. [DOI] [PubMed] [Google Scholar]
- Lyons DA, Pogoda HM, Voas MG, Woods IG, Diamond B, Nix R, Arana N, Jacobs J, Talbot WS. erbb3 and erbb2 are essential for schwann cell migration and myelination in zebrafish. Curr Biol. 2005;15:513–524. doi: 10.1016/j.cub.2005.02.030. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, Hanson TE. The development of the optic lobe. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1993. pp. 1363–1491. [Google Scholar]
- Murakami S, Umetsu D, Maeyama Y, Sato M, Yoshida S, Tabata T. Focal adhesion kinase controls morphogenesis of the Drosophila optic stalk. Development. 2007;134:1539–1548. doi: 10.1242/dev.001529. [DOI] [PubMed] [Google Scholar]
- Pielage J, Kippert A, Zhu M, Klambt C. The Drosophila transmembrane protein Fear-of-intimacy controls glial cell migration. Dev Biol. 2004;275:245–257. doi: 10.1016/j.ydbio.2004.07.039. [DOI] [PubMed] [Google Scholar]
- Rangarajan R, Gong Q, Gaul U. Migration and function of glia in the developing Drosophila eye. Development. 1999;126:3285–3292. doi: 10.1242/dev.126.15.3285. [DOI] [PubMed] [Google Scholar]
- Rangarajan R, Courvoisier H, Gaul U. Dpp and Hedgehog mediate neuron-glia interactions in Drosophila eye development by promoting the proliferation and motility of subretinal glia. Mech Dev. 2001;108:93–103. doi: 10.1016/s0925-4773(01)00501-9. [DOI] [PubMed] [Google Scholar]
- Schwabe T, Bainton RJ, Fetter RD, Heberlein U, Gaul U. GPCR signaling is required for blood-brain barrier formation in Drosophila. Cell. 2005;123:133–144. doi: 10.1016/j.cell.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Sen A, Shetty C, Jhaveri D, Rodrigues V. Distinct types of glial cells populate the Drosophila antenna. BMC Dev Biol. 2005;5:25. doi: 10.1186/1471-213X-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepp KJ, Schulte J, Auld VJ. Developmental dynamics of peripheral glia in Drosophila melanogaster. Glia. 2000;30:122–133. doi: 10.1002/(sici)1098-1136(200004)30:2<122::aid-glia2>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Sigrist SJ, Lehner CF. Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell. 1997;90:671–681. doi: 10.1016/s0092-8674(00)80528-0. [DOI] [PubMed] [Google Scholar]
- Stollewerk A, Klämbt C. The midline glial cells are required for compartmentalisation of axon commissures in the embryonic CNS of Drosophila. Dev Genes Evol. 1997;207:401–409. doi: 10.1007/s004270050129. [DOI] [PubMed] [Google Scholar]
- Stollewerk A, Klambt C, Cantera R. Electron microscopic analysis of Drosophila midline glia during embryogenesis and larval development using beta-galactosidase expression as endogenous cell marker. Microsc Res Tech. 1996;35:294–306. doi: 10.1002/(SICI)1097-0029(19961015)35:3<294::AID-JEMT8>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Struhl G, Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- Swales LS, Lane NJ. Embryonic development of glial cells and their junctions in the locust central nervous system. J Neurosci. 1985;5:117–127. doi: 10.1523/JNEUROSCI.05-01-00117.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson A, Ready DF. Neuronal differentiation in the Drosophila ommatidium. Dev Biol. 1987;120:366–376. doi: 10.1016/0012-1606(87)90239-9. [DOI] [PubMed] [Google Scholar]
- Torrence SA. Positional cues governing cell migration in leech neurogenesis. Development. 1991;111:993–1005. doi: 10.1242/dev.111.4.993. [DOI] [PubMed] [Google Scholar]
- Tsai HH, Miller RH. Glial cell migration directed by axon guidance cues. Trends Neurosci. 2002;25:173–175. doi: 10.1016/s0166-2236(00)02096-8. [DOI] [PubMed] [Google Scholar]
- Tsujikawa M, Malicki J. Genetics of photoreceptor development and function in zebrafish. Int J Dev Biol. 2004;48:925–934. doi: 10.1387/ijdb.041890mt. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development. 1991;113:841–850. doi: 10.1242/dev.113.3.841. [DOI] [PubMed] [Google Scholar]
- Wong AM, Wang JW, Axel R. Spatial representation of the glomerular map in the Drosophila protocerebrum. Cell. 2002;109:229–241. doi: 10.1016/s0092-8674(02)00707-9. [DOI] [PubMed] [Google Scholar]
- Xiong WC, Okano H, Patel NH, Blendy JA, Montell C. repo encodes a glial-specific homeo domain protein required in the Drosophila nervous system. Genes Dev. 1994;8:981–994. doi: 10.1101/gad.8.8.981. [DOI] [PubMed] [Google Scholar]