Abstract
Background
Cecal intubation time is an important component for quality colonoscopy. Cecum is the turning point that determines the insertion and withdrawal phase of the colonoscope. For this reason, obtaining information related with location of the cecum in the endoscopic procedure is very useful. Also, it is necessary to detect the direction of colonoscope’s movement and time-location of the cecum.
Methods
In order to analysis the direction of scope’s movement, the Horn–Schunck algorithm was used to compute the pixel’s motion change between consecutive frames. Horn–Schunk-algorithm applied images were trained and tested through convolutional neural network deep learning methods, and classified to the insertion, withdrawal and stop movements. Based on the scope’s movement, the graph was drawn with a value of +1 for insertion, −1 for withdrawal, and 0 for stop. We regarded the turning point as a cecum candidate point when the total graph area sum in a certain section recorded the lowest.
Results
A total of 328,927 frame images were obtained from 112 patients. The overall accuracy, drawn from 5-fold cross-validation, was 95.6%. When the value of “t” was 30 s, accuracy of cecum discovery was 96.7%. In order to increase visibility, the movement of the scope was added to summary report of colonoscopy video. Insertion, withdrawal, and stop movements were mapped to each color and expressed with various scale. As the scale increased, the distinction between the insertion phase and the withdrawal phase became clearer.
Conclusion
Information obtained in this study can be utilized as metadata for proficiency assessment. Since insertion and withdrawal are technically different movements, data of scope’s movement and phase can be quantified and utilized to express pattern unique to the colonoscopist and to assess proficiency. Also, we hope that the findings of this study can contribute to the informatics field of medical records so that medical charts can be transmitted graphically and effectively in the field of colonoscopy.
Keywords: Colonoscopy, Cecum, Cecal-location, Summary report, CNN, Deep learning
Introduction
A polyp is an abnormal tissue growth and is commonly found in the intestine (Haggar & Boushey, 2009). Since all colon and rectal cancers arise from a polyp, it is crucial to detect polyps in the early stage and treat them before they progress to being cancerous (Leslie et al., 2002). Colonoscopy is the most commonly used method to detect polyps and most available method (Nishihara et al., 2013; Rex, 2002). For this reason, the demand for colonoscopy continues to increase (Quintero et al., 2012).
Cecal intubation time (CIT) provides various data, which may be important indicators (Marshall & Barthel, 1993; Bernstein et al., 2005). In addition, when performing colonoscopy, the cecum is the turning point that determines the insertion phase and withdrawal phase of the colonoscope, that is, the gastroenterologist inserts the colonoscope close to the appendix and withdraws it from the cecum (Fatima et al., 2008). However, there are individual (i.e., patient and gastroenterologist) differences in the sequence and process of advancing the colonoscopy (Spier et al., 2010; Rex, 2001; Saifuddin et al., 2000). Thus, obtaining information about the time-location of the cecum in the colonoscopic procedure is very useful.
Information about the time-location of the cecum can also be helpful when checking colonoscopy videos. Because analyzing a video requires much time and concentration, it places a great burden on the physician, especially when the doctor re-watches the video or shares the video due to change of doctor or hospital (Terada, 2015; Hu et al., 2016). Knowing the time-location of the cecum can help physicians to distinguish the insertion phase and the withdrawal phase of the colonoscope (Fatima et al., 2008), which can help reduce the burden on video observation. In addition, because detailed examination was undertaken mostly during the withdrawal phase of the scope after reaching the cecum, knowing the time-location of the cecum and distinction between the insertion phase and withdrawal phase of the colonoscope is important (Barclay et al., 2006; Barclay, Vicari & Greenlaw, 2008; Moritz et al., 2012). Furthermore, the CIT and withdrawal phase can also be useful as metadata (Taber & Romagnuolo, 2010; Lee et al., 2009). The location of other anatomic sites such as the T-colon and S-colon can be inferred on the assumption that the location of the cecum is known (Cherian & Singh, 2004).
Moreover, the movement data of the colonoscope can be utilized for proficiency assessment (Lee et al., 2009; Snyder et al., 2010). Since insertion and withdrawal are technically different movements, the pattern, combination, and repetition of insertion/withdrawal/stop can be utilized to express individual features of the colonoscopist and to assess proficiency (Marshall, 1995; Benson et al., 2010; Anderson et al., 2001). Thus, it is necessary to detect the direction of the scope’s movement and time-location information of the cecum.
Recently, with advances in computer technology and equipment, gastroenterologists do not need to record these data anymore (Denny et al., 2010; Leiman et al., 2016). In various medical fields, systems that automatically record medical reports are being developed (Münzer, Schoeffmann & Böszörmenyi, 2018; Yuan, Li & Meng, 2016; Greenhalgh et al., 2010; Taira, Soderland & Jakobovits, 2001). In our previous study, we developed a useful system that automatically extracts meaningful information (namely, bleeding, polypectomy, tool, residue, thin wrinkle, folded wrinkle) from colonoscopy videos using support vector machine (SVM) and provides such information on the summary report with color-coded timeline visualization (Cho et al., 2018).
Horn–Schunk algorithms are the most popular differential algorithms that have been used for many applications and have been referenced for many performance evaluation models (Meinhardt-Llopis, Pérez & Kondermann, 2013). The Horn–Schunk algorithm is a technique used to identify the image velocity or motion vector based on spatial temporal gradient technique that computes the image velocity from spatiotemporal derivatives of the image intensity (Gong & Bansmer, 2015; Horn & Schunck, 1981). Through this algorithm, we can compute the pixel’s motion change between consecutive frames (Bruhn, Weickert & Schnörr, 2005). In other words, it is possible to know the movement direction of the pixels in the current frame relative to the previous frame; thus, it is possible to determine the direction in which the current frame is moving.
Therefore, in this study, we aimed to provide more useful information to physicians by recording the direction of the scope’s movement and time-location of the cecum using Horn–Schunk algorithm by applying convolutional neural network (CNN).
Material & Methods
This study was approved by the institutional review board of the Seoul National University Hospital (IRB No. 1509-062-703), and it was conducted in accordance with the Declaration of Helsinki. Informed consent was obtained from all participants before any study-related procedures were performed. This prospective, single-center trial enrolled patients aged 19 to 75 years who underwent colonoscopy for screening, surveillance, or therapy such as polypectomy at Seoul National University Hospital, a tertiary referral center in Korea, from August 2016 to December 2016.
Acquisition of colonoscopy video
In this study, colonoscopy was performed using a high-resolution colonoscopy device (CV260SL, Olympus, Tokyo, Japan). Colonoscopy videos were acquired using a video capture card (SkyCaputre U6T; Skydigital, Seoul, Korea), after signal branching from the CV260SL.
The video was converted to an MP4 format to avoid alteration of the resolution, and the resolution was 1920*1080, 30fps. Videos were acquired from 112 patients, and the play time was about 20-40 min. The colonoscopy video was decomposed into frames. A frame was extracted as a PNG file per 0.5 s using Virtualdub software. All frames were cropped to 850*750 pixels to extract only the colonoscopy area, excluding patient information and the settings.
Classification of informative frames
As the play time of the colonoscopy videos was long and had much noise, it was inefficient to use the whole video (Vilariño et al., 2007). Thus, frames were extracted to select only meaningful information or applied to image processing. In the colonoscopy video, the frame that has meaningful information is called informative frame, and the other is called non-informative frame (Vilariño et al., 2007). Since non-informative frames are frames that do not have meaningful information in the image itself, they have had a negative effect on training (Ballesteros et al., 2016). In this study, only informative frames were used. The classification accuracy of the informative frame used in this study was 99.4%.
Horn–Schunk algorithm
As the Horn–Schunk algorithm can only estimate small motions and compares the current frame with the previous frame to calculate the motion vector, the time interval between the two frames has a significant effect on the outcome (Meinhardt-Llopis, Pérez & Kondermann, 2013; Horn & Schunck, 1981). If the interval is too large, the calculation error will increase beyond the small motion, and if the interval is too small, the movement of the scope will not be considered sufficiently. Considering this limitation, we extracted the frame per 0.5 s.
Because the image inside the large intestine had varied shapes and colors depending on the conditions of position, health status, and patient’s condition, it is advantageous to remove other information from the frame image for training, leaving only information about the position using Horn-Schunck algorithm. After Horn-Schunck algorithm application, we acquired the image shown in Fig. 1 by expressing the direction change of the pixels inside the frame in color. Since all elements inside the frame do not move in the same direction consistently, various colors appear mixed, as shown in Fig. 1. To understand the pattern of colors according to these positions, we used CNN deep learning in the next step.
Figure 1. Horn and Schunk algorithm application.
(A) Original image frame, (B) Horn and Schunk algorithm applied image, (C) motion vector and its color expression.
Three-direction classification of informative frame
In this paper, Horn-Schunck algorithm-applied color images were classified into three types: insertion, withdrawal, and stop. Therefore, frames from colonoscopy images of 112 patients were classified as insert, withdrawal, and stop as the standard for deep training set. To make a standard setting, a colonoscopy video was reproduced at a speed of 0.7 times, and a user observing it pressed the direction key of the keyboard in real time. The up, down, and space buttons were matched to insert, withdrawal, and stop, respectively, and the frames were classified based on the input keyboard values. Five gastroenterologist with more than 5 years experience participated in this work, and the values selected by more than three were used as the standard (Fallah & Niakan Kalhori, 2017). An overview of the entire process is shown in Fig. 2.
Figure 2. Overview and process of the proposed system.
After extracting the frame from the video, only the informative frame is extracted by the SVM. In the informative frame, the motion vector is expressed in color by Horn and Schunk algorihm. Through the CNN deep learning method, these color images were classified into three types: insertion, withdrawal, and stop.
CNN machine learning for three-direction classification
CNN deep learning was applied based on the standard created above (Le, Ho & Ou, 2017; Ho & Ou, 2018; Taju et al., 2018). Horn-Schunck algorithm-applied color images were used as training data and test data. Unlike previous research using SVM, we used CNN, a deep learning technique, because it was difficult to select features from the Horn-Schunck algorithm-applied color image (Rajaraman et al., 2018).
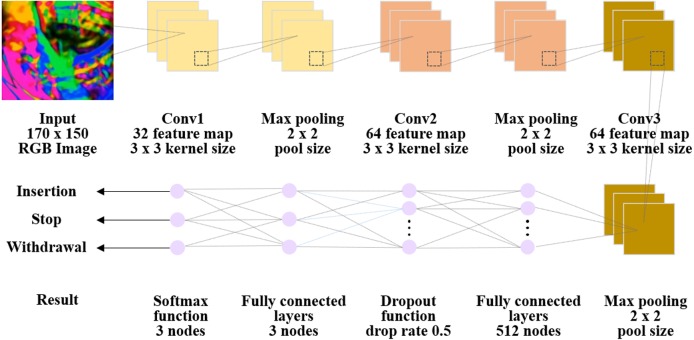
The CNN structure and training procedure are described in Fig. 3. The network architecture consisted of an input stage, a feature extraction stage with three convolutional layers, and an output classification stage (Nagasawa et al., 2018; Jang et al., 2018). The input stage received the Horn-Schunck algorithm applied color image which was converted to 170*150 pixels. The feature extraction stage that followed a triple ternary convolution block structure was composed of convolution, pooling, and activation functions. The rectified linear unit (ReLU) activation function and max polling layer were placed after each convolutional layer. The classification stage included fully connected layers and a dropout function (drop rate of 0.5) and provided an output from a Softmax function. Fully connected layer were classified into three classes using the Softmax function.
Figure 3. Overall architecture of the convolutional neural network (CNN) deep learning model.
All image were reduced to 170 × 150 and were input into the model. Next, it was passed through all convolution layers and the entire binding layer, and it was classified into three classes.
The learning was carried out with batch size 128 (set by experimental trials) images. The model is optimized for hyper-parameters by a randomized grid search method (Bergstra & Bengio, 2012). We initialized search ranges to be [1e–7 1e–2], [0.8 0.99], and [1e–10 1e–2] for the learning rate, SGD, and L2-regularization parameters, respectively.
Finally, the trained network was validated using the 5-fold cross-validation method (Ou, 2016; Ou, 2017; Le, Sandag & Ou, 2018). Five groups of data were established such that four groups were used for training and the remaining groups were used for validation. Each group was designed to participate in validation in turn. The performance was measured using the mean of the validation results for each group.
The model was trained and tested on a Windows 10 Pro system with Intel i9-9900K CPU 3.6-GHz processor, 2TB SDD, 64 GB RAM, GeForce RTX 2070 Gaming OC D6 8GB.
Cecum time-location calculation
To determine the location of the cecum, we analyzed the direction of the scope’s movement. Based on the results of the analysis, the graph was drawn with a value of +1 for insertion, −1 for withdrawal, and 0 for stop. Because the cecum is a large turning point that distinguishes the insertion phase from the withdrawal phase, the turning point of transition from insertion to withdrawal is the candidate for the cecum. However, some scopic insertion-withdrawal repeat movements often occur during colonoscopy, and turning points can be found very often. Therefore, in this study, the scope movement for a certain range was analyzed to find the real turning point. All points that transitioned from insertion to withdrawal were recorded. These turning points were referred to as a cecum candidate point.
We set a certain range “t” and calculated the sum of the graph area around the turning points. Since the value is +1 during insertion, the area of the graph will be positive, and when the value is −1 during withdrawal, the area of the graph will be negative. At the turning point with a certain range “t,” when the total sum of the graph areas is the minimum value, that turning point is regarded as the cecum (Fig. 4). If the value “t” is too small, light repetitive movements of insertion-withdrawal may have a minimum area value, so “t” is set to a sufficiently large value. For several candidate points, the graph area was calculated based on range “t.” The range “t” was set to 10, 20, and 30 s.
Figure 4. The movement of the scope.
(A) Original curve graph of insertion and withdrawal movement, (B) graph area calculation for cecal position prediction, (C) cecum candidate point.
Focus group interview (FGI) for the proposed system
In this study, we proposed the system that can analyze the colonoscopy video and provide meaningful direction information of endoscope’s movement and time-location information of the cecum via visualized summary report. To ask for comments on the system and reflect gastroenterologists’ requirements, we had implemented a FGI. Seven gastroenterologists with more than 5 years experience participated in this FGI, and it was conducted in the face-to-face meeting. The comments on necessity referred in this research were selected by FGI, and ideas about visualization were also recruited and validated. This FGI simply asked positive and negative opinions, and the results are as follows. All seven doctors determined that this system would positively benefit physicians and patients. Six doctors agreed that a visualized summary report based on this system would be helpful for medical records management and data sharing. Five doctors agreed that the results could be used for proficiency testing.
Results
In this study, we trained and tested the CNN models using 328,927 frames from 112 colonoscopy videos. To facilitate physician’s diagnosis during colonoscopy, we recruited most patients receiving colonoscopy for the second time.
Classification according to direction of scope’s movement
Since the features of Horn–Schunk-algorithm applied images were difficult to specify, we applied the CNN deep learning method to the images for high accuracy. The overall accuracy, drawn from 5-fold cross-validation, was 95.6%. Table 1 shows the classification results in confusion matrix form. Each cell in the Table 1 represents the number of samples classified by the trained CNN algorithm. The vertical title indicates the actual class (input class of the algorithm), whereas the horizontal title indicates the classified results (output class of the algorithm). Performance comparisons according to the indicators of recall, precision, and f1 score for each class are presented in Table 2. These numbers can be used for alternative algorithm assessments (Kainz, Pfeiffer & Urschler, 2017).
Table 1. The confusion matrix of classification results using the 5-fold cross-validation of the CNN algorithm.
| Target class | Sum by row | ||||
|---|---|---|---|---|---|
| Insertion | Withdrawal | Stop | |||
| Output class | Insertion | 116,437 | 1,533 | 1,379 | 119,349 |
| Withdrawal | 1,586 | 107,336 | 1,757 | 110,679 | |
| Stop | 3,484 | 4,603 | 90,812 | 98,899 | |
| Sum by column | 121,507 | 113,472 | 93,948 | 328,927 | |
Table 2. Performance indicators of the CNN and other algorithms for the three individual classes.
| Class/Indicators | Insertion | Withdrawal | Stop | |
|---|---|---|---|---|
| CNN | Recall | 0.958 | 0.946 | 0.967 |
| Precision | 0.976 | 0.969 | 0.918 | |
| F1 score | 0.967 | 0.958 | 0.942 | |
| VGG-16 | Recall | 0.95 | 0.916 | 0.964 |
| Precision | 0.953 | 0.959 | 0.911 | |
| F1 score | 0.952 | 0.937 | 0.937 | |
| LeNET | Recall | 0.907 | 0.833 | 0.872 |
| Precision | 0.874 | 0.909 | 0.83 | |
| F1 score | 0.89 | 0.87 | 0.85 | |
| SVM | Recall | 0.844 | 0.775 | 0.799 |
| Precision | 0.804 | 0.832 | 0.785 | |
| F1 score | 0.823 | 0.802 | 0.792 | |
Cecum time-location calculation
We regarded the turning point as a cecum candidate point when the total area sum in a certain section recorded the lowest. We checked the existence of the actual cecum within a certain interval around the cecum candidate point. The accuracy according to “t” values is shown in Fig. 5 and Table 3. The position of the cecum recorded by three clinicians was compared with the results of the proposed system.
Figure 5. Accuracy and number of cases when cecum is detected according to “t” values ranging from 10 s to 30 s.
If “t” is less than 10 s, light repetitive insertion-withdrawal movements may have a minimum area value, so “t” is set to a sufficiently large value. Considering this, we analyzed the results when t values were 10, 20, and 30 s.
Table 3. Accuracy of cecum discovery according to the set certain range “t”.
| t = 10 | t = 20 | t = 30 | |
|---|---|---|---|
| Accuracy | 83.9% | 94.6% | 96.4% |
Summary reports visualized in the timeline
The movement of the scope, shown in Fig. 4 as a graph, was changed to a color code to increase visibility. Insertion, withdrawal, and stop were mapped to each color and expressed in 1 s, as shown in Fig. 6A. Again, the scale was changed to a 10 s scale, 30 s scale to increase visibility (Figs. 6B, 6C). One 10 s square represents 10 s. One 30 s square represents 30 s.
Figure 6. Insertion, withdrawal, and stop were mapped to each color and expressed with various scale.
(A) 1 second per square, (B) 10 seconds per square, (C) 30 seconds per square, (D) recording the direction of scope’s movement and cecum time-location information on SRCV.
At the 1 s scale, the result was too microscopic and not suitable for intuitive observation. However, since the outlier belonging to a small number was removed from the 10 s scale, the movement of the scope and the position of the cecum could be intuitively grasped. In addition, as the scale increased, the distinction between the insertion phase and the withdrawal phase became clearer.
Discussion
In this study, we developed and verified a system that can detect the direction of scope’s movement from colonoscopy video using Horn–Schunk algorithm by applying CNN deep learning methods. The motion change of the video is extracted through the Horn–Schunk algorithm for calculating optical flow. Through this algorithm, we can compute the pixel’s motion change between consecutive frames, and the direction of scope’s movement was trained and determined through the CNN. The extracted information about the direction of scope’s movement was visualized and added to the summary report of colonoscopy video (SRCV). In addition, the time-location of the cecum was calculated based on the results of direction of the scope’s movement and included in the SRCV. This proposed system can assist physician’s observation of the colonoscopy video and provide helpful information.
The videos used in this research were favorable for image acquisition and image processing because skilled physician performed the colonoscopy. This may have resulted in lowering the false-negative results when the system was applied to the video. In a previous study, we extracted 1 frame of images in 0.3 s from a colonoscopy video. The higher the extraction frequency, the more frames can be extracted, so more images can be used in the system. However, in applying the Horn-Schunck algorithm in this study, we extracted one frame at 0.5 s. Note that the Horn-Schunck algorithm can only calculate motion vectors for small changes (Meinhardt-Llopis, Pérez & Kondermann, 2013; Horn & Schunck, 1981). Therefore, if the interval between frames is too large, Horn-Schunck algorithm is not applicable. However, to calculate the motion vector based on the difference between the previous frame and the current frame, there should be a proper change between the two frames. In our colonoscopy video, a 0.5 s period yielded optimal results. As the standard setting, observing the colonoscopy video at a speed of 0.7 times to avoid misreading directions, participation of five fellow doctors, and selecting values more than three made the accuracy of the gold standard more reliable (Cho et al., 2016; Fallah & Niakan Kalhori, 2017; Cho et al., 2017)
In previous studies, we have classified informative frames and non-informative frames from the colonoscopy video with high accuracy (Cho et al., 2018). The SVM was used as it is suitable for image classification (Park, Jang & Yoo, 2016). The criteria for classification of informative frame and non-informative frame were as follows: presence of noise such as color separation phenomenon, blur caused by motion, and non-observable screen such as excessive darkness, brightness, and enlarged screen (Ballesteros et al., 2016). The mean, variance, skewness, correlation, contrast, energy of Laplacian, and energy of gradient values were acquired from the decomposed frames, and these values were used as features of the SVM model to classify the informative and non-informative frames (Sonka, Hlavac & Boyle, 2014; Hua, Fu-Long & Li-Cheng, 2006). SVM modeling was performed with the 5-fold cross-validation method.
We did not consider real-time use in this study. This is because the movement of the scope is meaningful when analyzed with the whole video. When the insertion and withdrawal movements of the scope throughout the video are analyzed, a clear distinction between the insertion phase and the withdrawal phase can be obtained. As a result, the position of the cecum can be accurately calculated.
In the methods section, we used the certain range value “t” to find the real cecum point accurately. When analyzing 112 patient images, we found numerous light insertion-withdrawal repeat movements. With the exception of polypectomy, the scopes frequently repeat insertion-withdrawal movements to find the polyps (Anderson et al., 2016). In addition, withdrawal movement was often accompanied by smooth insertion (Simmons et al., 2006). Most repetitive movements did not exceed 10 s. In other words, if “t” is less than 10 s, light repetitive insertion-withdrawal movements may have a minimum area value, so “t” is set to a sufficiently large value. Considering this, we analyzed the results when t values were 10, 20, and 30 s.
In this study, we used the CNN deep learning method to classify the movement of the scope. The images used for deep learning input were the Horn-Schunck algorithm-applied images, and the motion vector was expressed in color. In the images, the layer did not have to be deep because the directions were classified according to the color distribution only. The CNN architecture used in this study was developed with reference to LeNET and VGG, and the layer structure was experimentally optimized. The trained network was validated using the 5-fold cross-validation method, and overfitting did not occur. As a limitation of this study, we did not test data from outside the hospital. In a future study, we plan to conduct research and testing with independent data from other hospitals.
During colonoscopy, the CIT was recorded by a physician. However, as the technology is under development today, the trend is to automatically store the data generated during the medical procedure, and the information generated during colonoscopy also needs to be automatically analyzed and stored (Münzer, Schoeffmann & Böszörmenyi, 2018; Yuan, Li & Meng, 2016; Greenhalgh et al., 2010; Taira, Soderland & Jakobovits, 2001; Terada, 2015). In our previous study, we have already developed a system that automatically analyzes and records colonoscopy videos, and through the system, the video was classified by types and visualized as summary report to communicate meaning effectively (Cho et al., 2018). In addition to the previous research, it will be possible to provide more useful information to physicians via recording the direction of scope’s movement and cecum time-location information (Fig. 6D).
In this study, only the insertion, withdrawal, and stop phases were analyzed and determined. Through FGI, gastroenterologists have expressed the need to identify and visualize the anatomical structure of the large intestine. In a future study, if left and right movements can be recognized, the direction of the colonoscope can be recorded in more detail. Based on this, it is expected that the anatomical structure of the large intestine can be deduced and the anatomical position with time can be indicated.
Conclusions
Information obtained in this study can be utilized as metadata for proficiency assessment. Since insertion and withdrawal are technically different movements, data of scope’s movement and phase can be quantified and utilized to express pattern unique to the colonoscopist and to assess proficiency (Taber & Romagnuolo, 2010; Snyder et al., 2010; Benson et al., 2010). When viewed with reference to the SRCV, we thought that the colonoscopist might have limited proficiency if there were repetitive insertion-withdrawal movements or no movements at all, which was not related to polyps, bleeding, or polypectomy.
With the proposed system, we believe that if the current handwritten medical records can be automatically summarized together with more detailed, graphical information, it will be useful for physicians and patients and will improve medical services (Ahn, Choi & Kim, 2016; Denny et al., 2010). The results of this study may contribute to improving the medical record after colonoscopy has finished. We hope that the findings of this study can contribute to the informatics field of medical records so that medical charts can be transmitted graphically and effectively in the field of colonoscopy.
Supplemental Information
Original frame image (raw data) was extracted from colonoscopy video.
In this study, colonoscopy was performed using a high-resolution colonoscopy device (CV260SL, Olympus, Tokyo, Japan). Colonoscopy videos were acquired using a video capture card (SkyCaputre U6T, Skydigital, Yongsan, Korea), after signal branching from the CV260SL. The video was converted to an MP4 format to avoid alteration of the resolution, and the resolution was 1920*1080, 30fps. The colonoscopy video was decomposed into frames. A frame was extracted as a PNG file per 0.5 s using Virtualdub software.
Original frame image (raw data) was extracted from colonoscopy video.
In this study, colonoscopy was performed using a high-resolution colonoscopy device (CV260SL, Olympus, Tokyo, Japan). Colonoscopy videos were acquired using a video capture card (SkyCaputre U6T, Skydigital, Yongsan, Korea), after signal branching from the CV260SL. The video was converted to an MP4 format to avoid alteration of the resolution, and the resolution was 1920*1080, 30fps. The colonoscopy video was decomposed into frames. A frame was extracted as a PNG file per 0.5 s using Virtualdub software.
Funding Statement
This study was supported by the National Research Foundation of Korea (NRF) funded by the Korean Government (2018M1A3A3A02065779 to Sungwan Kim), and MSIT (Ministry of Science and ICT), Korea, under the ITRC (Information Technology Research Center) support program (IITP-2019-2018-0-01833) supervised by the IITP (Institute for Information & communications Technology Promotion). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Hyoun-Joong Kong, Email: gongcop@cnu.ac.kr.
Sungwan Kim, Email: sungwan@snu.ac.kr.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Minwoo Cho conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Jee Hyun Kim and Kyoung Sup Hong performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, approved the final draft.
Joo Sung Kim analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Hyoun-Joong Kong and Sungwan Kim conceived and designed the experiments, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Human Ethics
The following information was supplied relating to ethical approvals (i.e., approving body and any reference numbers):
This study was approved by the institutional review board of the Seoul National University Hospital (IRB No. 1509-062-703), and it was conducted in accordance with the Declaration of Helsinki.
Data Availability
The following information was supplied regarding data availability:
The raw data is available at Figshare: Cho, Minwoo (2019): Colonoscopy images. figshare. Dataset. https://doi.org/10.6084/m9.figshare.7937336.v1.
References
- Ahn, Choi & Kim (2016).Ahn M, Choi M, Kim Y. Factors associated with the timeliness of electronic nursing documentation. Healthcare Informatics Research. 2016;22(4):270–276. doi: 10.4258/hir.2016.22.4.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson et al. (2001).Anderson JC, Messina CR, Cohn W, Gottfried E, Ingber S, Bernstein G, Coman E, Polito J. Factors predictive of difficult colonoscopy. Gastrointestinal Endoscopy. 2001;54(5):558–562. doi: 10.1067/mge.2001.118950. [DOI] [PubMed] [Google Scholar]
- Anderson et al. (2016).Anderson BW, Smyrk TC, Anderson KS, Mahoney DW, Devens ME, Sweetser SR, Kisiel JB, Ahlquist DA. Endoscopic overestimation of colorectal polyp size. Gastrointestinal Endoscopy. 2016;83(1):201–208. doi: 10.1016/j.gie.2015.06.058. [DOI] [PubMed] [Google Scholar]
- Ballesteros et al. (2016).Ballesteros C, Trujillo M, Mazo C, Chaves D, Hoyos J. Iberoamerican congress on pattern recognition. Springer; Cham: 2016. Automatic classification of non-informative frames in colonoscopy videos using texture analysis; pp. 401–408. [Google Scholar]
- Barclay et al. (2006).Barclay RL, Vicari JJ, Doughty AS, Johanson JF, Greenlaw RL. Colonoscopic withdrawal times and adenoma detection during screening colonoscopy. New England Journal of Medicine. 2006;355(24):2533–2541. doi: 10.1056/NEJMoa055498. [DOI] [PubMed] [Google Scholar]
- Barclay, Vicari & Greenlaw (2008).Barclay RL, Vicari JJ, Greenlaw RL. Effect of a time-dependent colonoscopic withdrawal protocol on adenoma detection during screening colonoscopy. Clinical Gastroenterology and Hepatology. 2008;6(10):1091–1098. doi: 10.1016/j.cgh.2008.04.018. [DOI] [PubMed] [Google Scholar]
- Benson et al. (2010).Benson ME, Reichelderfer M, Said A, Gaumnitz EA, Pfau PR. Variation in colonoscopic technique and adenoma detection rates at an academic gastroenterology unit. Digestive Diseases and Sciences. 2010;55(1):166–171. doi: 10.1007/s10620-008-0703-2. [DOI] [PubMed] [Google Scholar]
- Bergstra & Bengio (2012).Bergstra J, Bengio Y. Random search for hyper-parameter optimization. Journal of Machine Learning Research. 2012;13(Feb):281–305. [Google Scholar]
- Bernstein et al. (2005).Bernstein C, Thorn M, Monsees K, Spell R, O’Connor JB. A prospective study of factors that determine cecal intubation time at colonoscopy. Gastrointestinal Endoscopy. 2005;61(1):72–75. doi: 10.1016/S0016-5107(04)02461-7. [DOI] [PubMed] [Google Scholar]
- Bruhn, Weickert & Schnörr (2005).Bruhn A, Weickert J, Schnörr C. Lucas/Kanade meets Horn/Schunck: combining local and global optic flow methods. International Journal of Computer Vision. 2005;61(3):211–231. [Google Scholar]
- Cherian & Singh (2004).Cherian S, Singh P. Is routine ileoscopy useful? An observational study of procedure times, diagnostic yield, and learning curve. The American Journal of Gastroenterology. 2004;99(12):2324–2329. doi: 10.1111/j.1572-0241.2004.40730.x. [DOI] [PubMed] [Google Scholar]
- Cho et al. (2018).Cho M, Kim JH, Kong HJ, Hong KS, Kim S. A novel summary report of colonoscopy: timeline visualization providing meaningful colonoscopy video information. International Journal of Colorectal Disease. 2018;33(5):549–559. doi: 10.1007/s00384-018-2980-3. [DOI] [PubMed] [Google Scholar]
- Cho et al. (2017).Cho M, Lee DH, Doh EJ, Kim Y, Chung JH, Kim HC, Kim S. Development and clinical validation of a novel photography-based skin erythema evaluation system: a comparison with the calculated consensus of dermatologists. International Journal of Cosmetic Science. 2017;39(4):426–434. doi: 10.1111/ics.12393. [DOI] [PubMed] [Google Scholar]
- Cho et al. (2016).Cho M, Lee DH, Kim Y, Koh W, Chung JH, Kim HC, Kim S. Development and clinical validation of a novel photography-based skin pigmentation evaluation system: a comparison with the calculated consensus of dermatologists. International Journal of Cosmetic Science. 2016;38(4):399–408. doi: 10.1111/ics.12303. [DOI] [PubMed] [Google Scholar]
- Denny et al. (2010).Denny JC, Peterson JF, Choma NN, Xu H, Miller RA, Bastarache L, Peterson NB. Extracting timing and status descriptors for colonoscopy testing from electronic medical records. Journal of the American Medical Informatics Association. 2010;17(4):383–388. doi: 10.1136/jamia.2010.004804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallah & Niakan Kalhori (2017).Fallah M, Niakan Kalhori SR. Systematic review of data mining applications in patient-centered mobile-based information systems. Healthcare Informatics Research. 2017;23(4):262–270. doi: 10.4258/hir.2017.23.4.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatima et al. (2008).Fatima H, Rex DK, Rothstein R, Rahmani E, Nehme O, Dewitt J, Helper D, Toor A, Bensen S. Cecal insertion and withdrawal times with wide-angle versus standard colonoscopes: a randomized controlled trial. Clinical Gastroenterology and Hepatology. 2008;6(1):109–114. doi: 10.1016/j.cgh.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Gong & Bansmer (2015).Gong X, Bansmer S. Horn–Schunck optical flow applied to deformation measurement of a birdlike airfoil. Chinese Journal of Aeronautics. 2015;28(5):1305–1315. doi: 10.1016/j.cja.2015.07.005. [DOI] [Google Scholar]
- Greenhalgh et al. (2010).Greenhalgh T, Stramer K, Bratan T, Byrne E, Russell J, Potts HW. Adoption and non-adoption of a shared electronic summary record in England: a mixed-method case study. BMJ. 2010;340 doi: 10.1136/bmj.c3111. Article c3111. [DOI] [PubMed] [Google Scholar]
- Haggar & Boushey (2009).Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clinics in Colon and Rectal Surgery. 2009;22:191–197. doi: 10.1055/s-0029-1242458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho & Ou (2018).Ho QT, Ou YY. Classifying the molecular functions of Rab GTPases in membrane trafficking using deep convolutional neural networks. Analytical Biochemistry. 2018;555:33–41. doi: 10.1016/j.ab.2018.06.011. [DOI] [PubMed] [Google Scholar]
- Horn & Schunck (1981).Horn BK, Schunck BG. Determining optical flow. Artificial Intelligence. 1981;17(1–3):185–203. doi: 10.1016/0004-3702(81)90024-2. [DOI] [Google Scholar]
- Hu et al. (2016).Hu E, Sakanashi H, Nosato H, Takahashi E, Suzuki Y, Takeuchi K, Aoki H, Murakawa M. Bleeding and tumor detection for capsule endoscopy images using improved geometric feature. Journal of Medical and Biological Engineering. 2016;36(3):344–356. doi: 10.1007/s40846-016-0138-8. [DOI] [Google Scholar]
- Hua, Fu-Long & Li-Cheng (2006).Hua BO, Fu-Long MA, Li-Cheng J. Research on computation of GLCM of image texture. Acta Electronica Sinica. 2006;1(1):155–158. [Google Scholar]
- Jang et al. (2018).Jang Y, Kim S, Kim K, Lee D. Deep learning-based classification with improved time resolution for physical activities of children. PeerJ. 2018;6:e5764. doi: 10.7717/peerj.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainz, Pfeiffer & Urschler (2017).Kainz P, Pfeiffer M, Urschler M. Segmentation and classification of colon glands with deep convolutional neural networks and total variation regularization. PeerJ. 2017;5:e3874. doi: 10.7717/peerj.3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, Ho & Ou (2017).Le NQK, Ho QT, Ou YY. Incorporating deep learning with convolutional neural networks and position specific scoring matrices for identifying electron transport proteins. Journal of Computational Chemistry. 2017;38(23):2000–2006. doi: 10.1002/jcc.24842. [DOI] [PubMed] [Google Scholar]
- Le, Sandag & Ou (2018).Le NQK, Sandag GA, Ou YY. Incorporating post translational modification information for enhancing the predictive performance of membrane transport proteins. Computational Biology and Chemistry. 2018;77:251–260. doi: 10.1016/j.compbiolchem.2018.10.010. [DOI] [PubMed] [Google Scholar]
- Lee et al. (2009).Lee HL, Eun CS, Lee OY, Jeon YC, Han DS, Sohn JH, Yoon BC, Choi HS, Hahm JS, Lee MH, Lee DH, Moon W, Kim SY. Significance of colonoscope length in cecal insertion time. Gastrointestinal Endoscopy. 2009;69(3):503–508. doi: 10.1016/j.gie.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Leiman et al. (2016).Leiman DA, Metz DC, Ginsberg GG, Howell JT, Mehta SJ, Ahmad NA. A novel electronic medical record–based workflow to measure and report colonoscopy quality measures. Clinical Gastroenterology and Hepatology. 2016;14(3):333–337. doi: 10.1016/j.cgh.2015.12.001. [DOI] [PubMed] [Google Scholar]
- Leslie et al. (2002).Leslie A, Carey FA, Pratt NR, Steele RJC. The colorectal adenoma–carcinoma sequence. British Journal of Surgery. 2002;89(7):845–860. doi: 10.1046/j.1365-2168.2002.02120.x. [DOI] [PubMed] [Google Scholar]
- Marshall (1995).Marshall JB. Technical proficiency of trainees performing colonoscopy: a learning curve. Gastrointestinal Endoscopy. 1995;42(4):287–291. doi: 10.1016/S0016-5107(95)70123-0. [DOI] [PubMed] [Google Scholar]
- Marshall & Barthel (1993).Marshall JB, Barthel JS. The frequency of total colonoscopy and terminal ileal intubation in the 1990s. Gastrointestinal Endoscopy. 1993;39(4):518–520. doi: 10.1016/S0016-5107(93)70162-5. [DOI] [PubMed] [Google Scholar]
- Meinhardt-Llopis, Pérez & Kondermann (2013).Meinhardt-Llopis E, Pérez JS, Kondermann D. Horn-schunck optical flow with a multi-scale strategy. Image Processing on Line. 2013;2013:151–172. [Google Scholar]
- Moritz et al. (2012).Moritz V, Bretthauer M, Ruud HK, Glomsaker T, De Lange T, Sandvei P, Huppertz-Hauss G, Kjellevold Ø, Hoff G. Withdrawal time as a quality indicator for colonoscopy–a nationwide analysis. Endoscopy. 2012;44(05):476–481. doi: 10.1055/s-0032-1306898. [DOI] [PubMed] [Google Scholar]
- Münzer, Schoeffmann & Böszörmenyi (2018).Münzer B, Schoeffmann K, Böszörmenyi L. Content-based processing and analysis of endoscopic images and videos: a survey. Multimedia Tools and Applications. 2018;77(1):1323–1362. doi: 10.1007/s11042-016-4219-z. [DOI] [Google Scholar]
- Nagasawa et al. (2018).Nagasawa T, Tabuchi H, Masumoto H, Enno H, Niki M, Ohsugi H, Mitamura Y. Accuracy of deep learning, a machine learning technology, using ultra-wide-field fundus ophthalmoscopy for detecting idiopathic macular holes. PeerJ. 2018;6:e5696. doi: 10.7717/peerj.5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara et al. (2013).Nishihara R, Wu K, Lochhead P, Morikawa T, Liao X, Qian ZR, Inamura K, Kim SA, Kuchiba A, Yamauchi M, Imamura Y, Willett WC, Rosner BA, Fuchs CS, Giovannucci E, Ogino S, Chan AT. Long-term colorectal-cancer incidence and mortality after lower endoscopy. New England Journal of Medicine. 2013;369(12):1095–1105. doi: 10.1056/NEJMoa1301969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou (2016).Ou YY. Prediction of FAD binding sites in electron transport proteins according to efficient radial basis function networks and significant amino acid pairs. BMC Bioinformatics. 2016;17(1):298. doi: 10.1186/s12859-016-1163-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou (2017).Ou YY. Identifying the molecular functions of electron transport proteins using radial basis function networks and biochemical properties. Journal of Molecular Graphics and Modelling. 2017;73:166–178. doi: 10.1016/j.jmgm.2017.01.003. [DOI] [PubMed] [Google Scholar]
- Park, Jang & Yoo (2016).Park BE, Jang WS, Yoo SK. Texture analysis of supraspinatus ultrasound image for computer aided diagnostic system. Healthcare Informatics Research. 2016;22(4):299–304. doi: 10.4258/hir.2016.22.4.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero et al. (2012).Quintero E, Hassan C, Senore C, Saito Y. Progress and challenges in colorectal cancer screening. Gastroenterology Research and Practice. 2012;2012 doi: 10.1155/2012/846985. Article 846985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaraman et al. (2018).Rajaraman S, Antani SK, Poostchi M, Silamut K, Hossain MA, Maude RJ, Jaeger S, Thoma GR. Pre-trained convolutional neural networks as feature extractors toward improved malaria parasite detection in thin blood smear images. PeerJ. 2018;6:e4568. doi: 10.7717/peerj.4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex (2001).Rex DK. Effect of variable stiffness colonoscopes on cecal intubation times for routine colonoscopy by an experienced examiner in sedated patients. Endoscopy. 2001;33(01):60–64. doi: 10.1055/s-2001-11179. [DOI] [PubMed] [Google Scholar]
- Rex (2002).Rex DK. Rationale for colonoscopy screening and estimated effectiveness in clinical practice. Gastrointestinal Endoscopy Clinics of North America. 2002;12(1):65–75. doi: 10.1016/S1052-5157(03)00058-8. [DOI] [PubMed] [Google Scholar]
- Saifuddin et al. (2000).Saifuddin T, Trivedi M, King PD, Madsen R, Marshall JB. Usefulness of a pediatric colonoscope for colonoscopy in adults. Gastrointestinal Endoscopy. 2000;51(3):314–317. doi: 10.1016/S0016-5107(00)70361-0. [DOI] [PubMed] [Google Scholar]
- Simmons et al. (2006).Simmons DT, Harewood GC, Baron TH, Petersen BT, Wang KK, Boyd-Enders F, Ott BJ. Impact of endoscopist withdrawal speed on polyp yield: implications for optimal colonoscopy withdrawal time. Alimentary Pharmacology & Therapeutics. 2006;24(6):965–971. doi: 10.1111/j.1365-2036.2006.03080.x. [DOI] [PubMed] [Google Scholar]
- Snyder et al. (2010).Snyder CW, Vandromme MJ, Tyra SL, Hawn MT. Retention of colonoscopy skills after virtual reality simulator training by independent and proctored methods. The American Surgeon. 2010;76(7):743–746. [PubMed] [Google Scholar]
- Sonka, Hlavac & Boyle (2014).Sonka M, Hlavac V, Boyle R. Image processing, analysis, and machine vision. Boston: Cengage Learning; 2014. [Google Scholar]
- Spier et al. (2010).Spier BJ, Benson M, Pfau PR, Nelligan G, Lucey MR, Gaumnitz EA. Colonoscopy training in gastroenterology fellowships: determining competence. Gastrointestinal Endoscopy. 2010;71(2):319–324. doi: 10.1016/j.gie.2009.05.012. [DOI] [PubMed] [Google Scholar]
- Taber & Romagnuolo (2010).Taber A, Romagnuolo J. Effect of simply recording colonoscopy withdrawal time on polyp and adenoma detection rates. Gastrointestinal Endoscopy. 2010;71(4):782–786. doi: 10.1016/j.gie.2009.12.008. [DOI] [PubMed] [Google Scholar]
- Taira, Soderland & Jakobovits (2001).Taira RK, Soderland SG, Jakobovits RM. Automatic structuring of radiology free-text reports. Radiographics. 2001;21(1):237–245. doi: 10.1148/radiographics.21.1.g01ja18237. [DOI] [PubMed] [Google Scholar]
- Taju et al. (2018).Taju SW, Nguyen TTD, Le NQK, Kusuma RMI, Ou YY. DeepEfflux: a 2D convolutional neural network model for identifying families of efflux proteins in transporters. Bioinformatics. 2018;34(18):3111–3117. doi: 10.1093/bioinformatics/bty302. [DOI] [PubMed] [Google Scholar]
- Terada (2015).Terada T. Histopathological study using computer database of 10 000 consecutive gastric specimens:(1) benign conditions. Gastroenterology Report. 2015;3(3):238–242. doi: 10.1093/gastro/gou093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilariño et al. (2007).Vilariño F, Lacey G, Zhou J, Mulcahy H, Patchett S. Iberian conference on pattern recognition and image analysis. Springer; Berlin, Heidelberg: 2007. Automatic labeling of colonoscopy video for cancer detection; pp. 290–297. [Google Scholar]
- Yuan, Li & Meng (2016).Yuan Y, Li B, Meng MQH. Improved bag of feature for automatic polyp detection in wireless capsule endoscopy images. IEEE Transactions on Automation Science and Engineering. 2016;13(2):529–535. doi: 10.1109/TASE.2015.2395429. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Original frame image (raw data) was extracted from colonoscopy video.
In this study, colonoscopy was performed using a high-resolution colonoscopy device (CV260SL, Olympus, Tokyo, Japan). Colonoscopy videos were acquired using a video capture card (SkyCaputre U6T, Skydigital, Yongsan, Korea), after signal branching from the CV260SL. The video was converted to an MP4 format to avoid alteration of the resolution, and the resolution was 1920*1080, 30fps. The colonoscopy video was decomposed into frames. A frame was extracted as a PNG file per 0.5 s using Virtualdub software.
Original frame image (raw data) was extracted from colonoscopy video.
In this study, colonoscopy was performed using a high-resolution colonoscopy device (CV260SL, Olympus, Tokyo, Japan). Colonoscopy videos were acquired using a video capture card (SkyCaputre U6T, Skydigital, Yongsan, Korea), after signal branching from the CV260SL. The video was converted to an MP4 format to avoid alteration of the resolution, and the resolution was 1920*1080, 30fps. The colonoscopy video was decomposed into frames. A frame was extracted as a PNG file per 0.5 s using Virtualdub software.
Data Availability Statement
The following information was supplied regarding data availability:
The raw data is available at Figshare: Cho, Minwoo (2019): Colonoscopy images. figshare. Dataset. https://doi.org/10.6084/m9.figshare.7937336.v1.