Abstract
A member of regulator of G-protein signaling family, RGS9-2, is an essential modulator of signaling through neuronal dopamine and opioid G-protein-coupled receptors. Recent findings indicate that the abundance of RGS9-2 determines sensitivity of signaling in the locomotor and reward systems in the striatum. In this study we report the mechanism that sets the concentration of RGS9-2 in vivo, thus controlling G-protein signaling sensitivity in the region. We found that RGS9-2 possesses specific degradation determinants which target it for constitutive destruction by lysosomal cysteine proteases. Shielding of these determinants by the binding partner R7 binding-protein (R7BP) controls RGS9-2 expression at the posttranslational level. In addition, binding to R7BP in neurons targets RGS9-2 to the specific intracellular compartment, the postsynaptic density. Implementation of this mechanism throughout ontogenetic development ensures expression of RGS9-2/type 5 G-protein β subunit/R7BP complexes at postsynaptic sites in unison with increased signaling demands at mature synapses.
Keywords: G-protein, signal transduction, RGS proteins, protein degradation, intracellular targeting, striatum
Introduction
Heterotrimeric G-protein signaling pathways mediate a number of fundamental cellular processes including reception, proliferation, exocytosis, chemotaxis and many others. The stimuli in these systems are transmitted via molecular switches, G-proteins that oscillate between inactive GDP-bound and active GTP-bound states (Gainetdinov et al., 2004). It is well established that signaling in these pathways is critically controlled by a conserved family of proteins called regulators of G-protein signaling (RGS) which facilitate G-protein inactivation to result in faster response termination (Hollinger and Hepler, 2002).
One member of this protein family, RGS9 has recently attracted significant attention because of its essential roles in regulating vision, reward behavior and locomotion in mammals. A short splice isoform, RGS9-1, is expressed only in photoreceptors in which it sets the duration of light responses (Chen et al., 2000; Krispel et al., 2006), whereas the long splice variant, RGS9-2 is prominent in the basal ganglia, in which it modulates signaling through D2 dopamine and μ-opioid receptors to regulate reward and the coordination of movement (Rahman et al., 2003; Zachariou et al., 2003; Kovoor et al., 2005).
Accumulating evidence suggests that expression level of RGS9 critically determines signal duration in both retina and striatum. In the visual system, the absence of RGS9-1 results in delayed recovery of photoreceptors from light excitation (Chen et al., 2000) causing the disease, bradyopsia (Nishiguchi et al., 2004). Conversely, an increase in RGS9-1 levels results in faster response termination (Krispel et al., 2006). In the striatum, loss of RGS9-2 aggravates dyskinetic conditions (Kovoor et al., 2005) and hypersensitizes the reward system to stimulation with morphine and cocaine, whereas elevation in RGS9-2 expression blunts these behavioral responses (Rahman et al., 2003; Zachariou et al., 2003). Furthermore, levels of RGS9-2 were found to be specifically modulated by changes in the cascade activation induced by abused drugs (Burchett et al., 1998; Rahman et al., 2003; Zachariou et al., 2003), neurodegenerative diseases (Tekumalla et al., 2001), or sex hormones (Sharifi et al., 2004), suggesting that RGS9-2 expression modulation is an important biological parameter contributing to adaptive responses.
Given its significance, surprisingly little is known about the mechanisms involved in regulating RGS9-2 expression. RGS9-2 forms complexes with two binding partners: type 5 G-protein β subunit (Gβ5) (Snow et al., 1998; Makino et al., 1999) and R7 binding-protein (R7BP) (Drenan et al., 2005; Martemyanov et al., 2005) and both of these proteins were found to influence RGS9 stability (Kovoor et al., 2000; Chen et al., 2003; Anderson et al., 2007).
In the current study we show that the expression level of RGS9-2 in vivo is set by balancing its selective degradation by cellular cysteine proteases targeted by specific degradation determinants, and macromolecular complex formation with binding partner R7BP which shields these determinants. We demonstrate that the mechanism is involved in marked upregulation of the RGS9-2/Gβ5/R7BP complex expression in ontogeny on which it is recruited to specific signaling compartment of mature synapses, the postsynaptic density.
Materials and Methods
Antibodies and DNA constructs.
Generation of rabbit anti-R7BP NT (Martemyanov et al., 2005), rabbit anti-R9AP (Keresztes et al., 2003), sheep anti-RGS9d (Makino et al., 1999), and sheep anti-RGS9-2 CT (Martemyanov et al., 2005) has been described previously. Rabbit anti-RGS7 (7RC1) and anti-Gβ5 (SGS) were generous gifts from Dr. William Simonds (National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health). Commercial antibodies used include mouse monoclonal anti-β-actin (clone AC-15; Sigma, St. Louis, MO), anti-ubiquitin (Santa Cruz Biotechnology, Santa Cruz, CA), and PSD-95 (Abcam, Cambridge, MA). All custom generated antibodies were tested for the specificity to ensure that (1) they recognize single major band in brain extracts corresponding to size of detection subject, (2) their immunoreactivity is blocked by peptide containing specific epitope sequence and (3) antibodies efficiently performed in immunoprecipitation reactions and the identity of precipitated band was confirmed by mass-spectrometry.
RGS7, and RGS9-1 and their chimeric constructs were cloned into pcDNA3.1/V5-His-TOPO (Invitrogen, Carlsbad, CA) mammalian expression vector as detailed previously (Anderson et al., 2007). The following RGS7/RGS9 chimeras were used: #1 (RGS9 1–115, RGS7 123–469), #2 (RGS7 1–122, RGS9 116–484), #3 (RGS9 1–209, RGS7 218–469), #4 (RGS7 1–244, RGS9 210–484). Cell transfections were performed using Lipofectamine LTX reagent (Invitrogen) according to manufacturer's protocol.
Preparation of brain extracts, immunoprecipitation, and Western blotting.
Cellular lysates were prepared by homogenizing brain tissue by sonication in the IP buffer (1× PBS, 150 mm NaCl, 1% Triton X-100, protease inhibitors) followed by 10 min centrifugation at 14,000 × g. The resulting extract was used for protein concentration determination by BCA assay (Pierce, Rockford, IL). This Triton X-100 extraction procedure yielded >90% extraction efficiency of proteins under investigation and was followed for both direct immunoblot and immunoprecipitation experiments. Immunoprecipitation of RGS7 and RGS9-1 was performed using 5 μg of antibody, 10 μl of protein G beads (GE Healthcare, Little Chalfont, UK), added to the extracts. The mixtures were incubated for 1 h, washed three times with IP buffer, and proteins bound to the beads were eluted with SDS sample buffer. Samples were resolved on SDS-PAGE gel, transferred onto PVDF membrane and subjected to Western blot analysis using HRP conjugated secondary antibodies and ECL West Pico (Pierce) detection system. Signals were captured on film, scanned by densitometer and band intensities were determined using NIH ImageJ software. For quantitative analysis serial dilutions of the sample with highest concentration of antigen were loaded on the same gel and used as calibration standards for estimation of relative changes in protein levels.
Quantitative real-time reverse transcription-PCR experiments.
Striatum tissue was subjected to RNA extraction with RNA isolation kit (Stratagene, La Jolla, CA), and 0.08 μg of RNA was used to perform quantitative real-time reverse transcription (Q-RT)-PCR using SYBR Green QRT-PCR kit (Stratagene). QuantiTect primers (Qiagen, Valencia, CA) were used, which had been designed to be specific to either R7BP or RGS9 mRNA. PCR cycling and detection was performed on the 7500 Fast Real-Time PCR System, with quantification analysis by System Sequence Detection Software v1.3 (Applied Biosystems, Foster City, CA). In addition to using equal amounts of total RNA as defined by UV spectroscopy we also included internal reference controls to account for the integrity of mRNA: amplification of β-actin and GAPDH mRNAs. We found that both of these messages do not show any appreciable changes during development and consequently used β-actin as endogenous reference for normalization of both R7BP and RGS9 mRNA amplification.
Generation of R7BP knock-out mouse.
Targeting of R7BP gene was done by homologous recombination in 128 Sv/Ev mouse ES cell lines and was performed on a commercial basis at Ingenious (Stony Brook, NY).
The targeting strategy consisted in the replacement of the first two exons of R7BP gene by neomycin resistance cassette. A ∼10.8 kb region used to construct the targeting vector was first sub cloned from a positively identified BAC clone. The region was designed such that the short homology arm (SA) extends ∼2.3 kb to 3′ side of LoxP/FRT flanked Neo cassette. The long homology arm (LA) starts at the 5′ side of LoxP/FRT flanked Neo cassette and is ∼7.5kb long. The neo cassette replaces ∼5.8 kb of the gene encoding exons 1–2 (including the ATG start codon). After electroporation, G418 resistant clones were selected, confirmed for recombination by PCR and injected into blastocyst. After confirming germ-line transmission, mice were back-crossed with C57BL/6 wild-type mice for two generations. Resulting heterozygous animals were inbred to produce wild-type, heterozygote and R7BP knock-out littermates used in the experiments. R7BP knock-out mice did not show any visible abnormalities in their baseline behavior or brain morphology.
Immunogold electron microscopy.
Electron microscopic examination of immunoreactivity for RGS9-2 and R7BP in the striatum of WT and RGS9-2 KO and R7BP KO mice was performed as described previously using either preembedding or postembedding immunogold methods (Lujan et al., 1996). Briefly, brains were perfused with 4% paraformaldehyde, 0.05% glutaraldehyde and 15% (v/v) saturated picric acid in 0.1 M phosphate buffer. For preembedding immunogold labeling, brain sections (50–70 mm) were cut on a Vibratome and processed for immunocytochemical detection of RGS9-2 and/or R7BP using HRP and silver-enhanced immunogold techniques. For postembedding immunogold labeling, ultrathin sections (70–90 nm) from three Lowicryl-embedded blocks slices were cut on an Ultramicrotome and processed for immunocytochemical detection of RGS9-2 and/or R7BP.
Subcellular fractionation experiments.
Striatums from one animal were lysed in 0.5 ml of nondetergent buffer (12 mm Na2HPO4/NaH2PO4, 287 mm NaCl, 3 mm KCl, protease inhibitors, pH = 7.4), homogenized with series of needles (16, 22, 26G) and spinned down at 22,000 × g for 20 min. Supernatant was removed and pellet was resuspended in 0.5 ml of the above referenced buffer. Equal volume aliquots of supernatant and pellet were mixed with SDS sample buffer and subjected to SDS-PAGE analysis followed by Western blotting with anti-RGS9-2 antibodies. The procedure separates heavy plasma and synaptic membranes from cytosolic compartment and light microsomal membranes.
Recombinant lentiviruses and stereotaxic injections.
Recombinant lentiviruses carrying R7BP or LacZ genes were packaged and concentrated as previously described (Anderson et al., 2007). Solution containing viral particles was delivered by the means of a stereotaxic injection into the caudate–putamen regions of anesthetized mice. A total of 4 injections per brain side were performed, with injection volume of 400 nl delivered at 5 nl/s via a 30G hypodermic needle. The following stereotaxic coordinates for the injection sites were used: 0.75 mm anterior to Bregma; ±1.5 mm and ± 2.25 from midline; 2.5 mm and 3.5 mm dorsal ventral from skull surface. After injection, needle was held in place for an additional 60 s to ensure proper diffusion. Holes and wounds were closed with tissue adhesive (Vetbond; 3M, St. Paul, MN) and the animals were allowed to recover for 1 week.
Ubiquitination assays.
293FT cells were transfected with either RGS7 or RGS9-2 with or without Gβ5 and/or R7BP. Twenty hours after transfection media was supplemented with 50 μm MG132 and cells were incubated for an additional 4 h, lysed in IP buffer supplemented with 1% SDS, protease inhibitors (Complete; Roche Diagnostics, Indianapolis, IN), 50 μm MG132, and 2 mm N-ethylmaleimide (Sigma). Lysates were heated at 70°C for 10 min, diluted 10-fold with IP buffer and subjected to immunoprecipitation using RGS-specific antibodies. Immunoprecipitation eluates were resolved by SDS-PAGE and the presence of ubiquitin conjugates was detected by Western blotting with anti-ubiquitin antibodies.
Pulse-chase metabolic labeling.
Pulse-chase assays were conducted as previously described (Anderson et al., 2007). NG108–15 or 293FT cells are grown in T-25 flasks and transfected with RGS7, RGS9-1, or RGS7/RGS9 chimera in the mammalian expression construct pcDNA3.1 (Invitrogen). Gβ5 was cotransfected with RGS proteins in all experiments.
Ex vivo cultures of striatal slices.
Striatal tissue was dissected from 1 mm thick coronal brain slices and immediately placed in culturing media (DMEM, 10% FBS, penicillin/streptomycin; Invitrogen). Striatal regions from one side of the brain were treated with the indicated protease inhibitor, with the contralateral side serving as vehicle control. Striatal slices were incubated for 5 h at 37°C with or without Leupeptin, E64 (Sigma), and lactacystin (Boston Biochem, Boston, MA) and lysed in PBS, 150 mm NaCl, 1% Triton X-100. Total protein was normalized by concentration determined by BCA reagent (Pierce), and subjected to Western blot analysis.
Bioinformatics.
Disorder predictions were done using a recently developed Various Short-Long version algorithm of the Predictor of Natural Disordered Regions (PONDR-VSL1). This algorithm consists of an ensemble of logistic regression models that predict per-residue order-disorder (Obradovic et al., 2005; Peng et al., 2005). Two models are used to predict either long or short disordered regions, greater or <30 residues, and the algorithm calculates a weighted average of these predictions, in which the weights are determined by a meta-predictor that approximates the likelihood of a long disordered region within its 61-residue window. The output of the prediction is presented by a score ranging from 0 to 1 assigned to each residue which reflects its likelihood to adopt disordered conformation. The neutral probability score was set to 0.5.
Results
Expression of RGS9-2 is coinduced with R7BP in late phase of neuronal differentiation
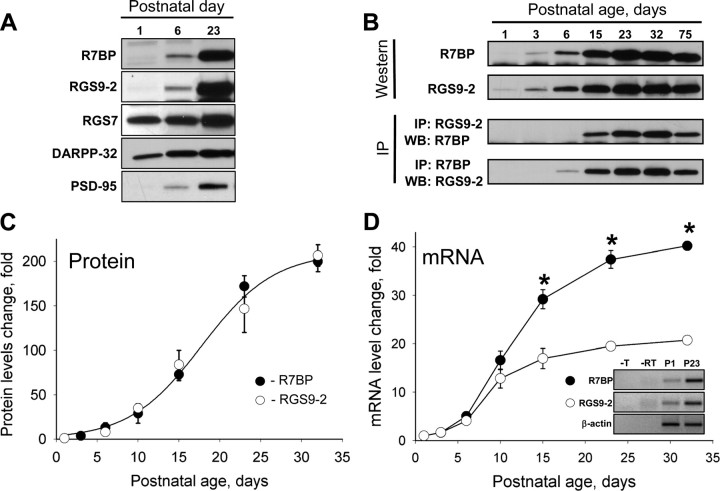
We started examining the mechanisms that set RGS9-2 expression level by analyzing the change in its protein levels during the postnatal development of the mouse striatum. Data presented in Figure 1A reveal that RGS9-2 undergoes a striking pattern of late onset expression induction. Being virtually absent in neonatal tissues until approximately day 6 of postnatal development its levels dramatically increase to reach peak levels in adult animals. Interestingly, R7BP, an interaction partner of RGS9-2 exhibited an identical level of change in expression. A similar developmental expression profile was also observed for PSD-95, a postsynaptic scaffold of mature synapses, whereas other signaling proteins such as a striatal specific regulator of dopamine signaling, DARPP-32 (Ehrlich et al., 1990) (Fig. 1A), and the close RGS9-2 homolog RGS7, exhibited only modest changes in expression levels.
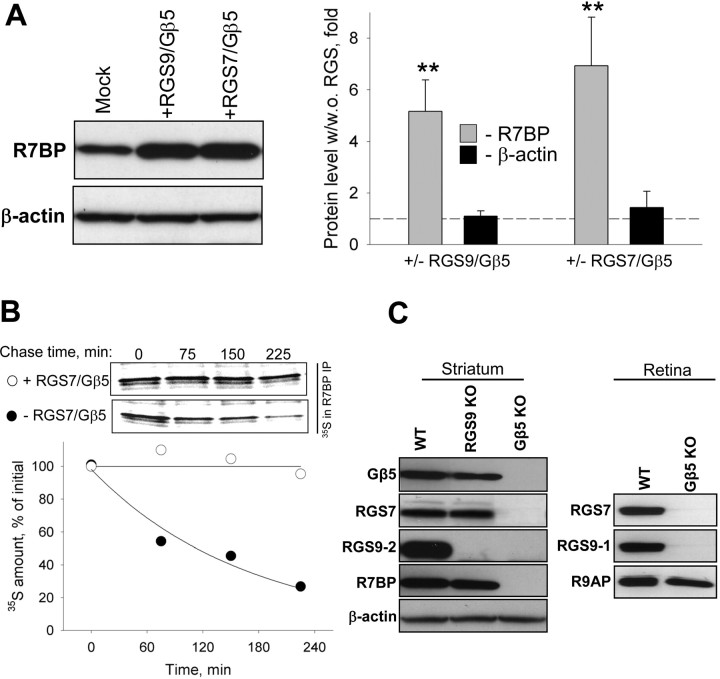
Figure 1.
Coregulation of RGS9-2 and R7BP expression during differentiation of striatal neurons. A, Western blot analysis of change in protein expression level on postnatal differentiation of striatum in Swiss-Webster mice. Striatal lysates (20 μg/lane) were analyzed for protein expression using specific antibodies. B, Time course of RGS9-2 and R7BP expression and their complex formation at the protein level. RGS9-2 and R7BP were immunoprecipitated (IP) and subjected to Western blot (WB) analysis. Immunoprecipitation efficiency in the experiments was >90%. C, Quantification of changes in levels of proteins from three separate groups analyzed as described in B. Band densities were determined by NIH ImageJ software and used to determine changes in protein levels relative to day 1 as described in Materials and Methods. D, Quantitative analysis of R7BP and RGS9-2 expression in development at mRNA level. Relative levels of RGS9-2 and R7BP mRNAs were measured by quantitative RT-PCR and normalized to the levels of β-actin mRNA amplified in parallel as an endogenous reference. Relative quantification algorithm was used in which changes in amplification threshold were normalized to sample from postnatal day 1. Data are averaged from three separate groups of animals. Inset shows gel electrophoresis of PCR products amplified from newborn (P1) or adult (P23) samples. No reverse transcriptase (-RT) or template (-T) was added in controls. *p < 0.05, statistically significant differences between relative changes in R7BP and RGS9-2 mRNA (t test).
Co immunoprecipitation experiments reveal that RGS9-2 forms a complex with R7BP as early as it becomes robustly detectable (at approximately postnatal day 6), and that the extent of the complex formation is directly proportional to the increase in the protein levels of both proteins as the brain develops (Fig. 1B). A similar pattern of expression upregulation was also observed when striatal neurons were cultured in vitro, as neurons differentiated and formed synaptic connections (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). Quantitative analysis of the protein expression kinetics indicates that both R7BP and RGS9-2 follow identical patterns of exponential increase in expression resulting in ∼200-fold increase in levels from the moment they first become detectable (Fig. 1C).
Quantitative RT-PCR analysis revealed that at the mRNA level the expression kinetics of R7BP and RGS9-2 are substantially different (Fig. 1D). The expression of both messages is very low in neonatal animals and their levels rise dramatically as animals develop. However, whereas R7BP mRNA essentially replicates exponential kinetics matching the change in its protein, the enrichment of RGS9-2 message stops much earlier. Although the abundance of RGS9-2 mRNA does not substantially change after day 10, its protein levels still increases approximately fivefold. In fact, the kinetics of RGS9-2 protein accumulation appear to follow the time course of R7BP mRNA synthesis, suggesting that the upregulation in RGS9-2 protein levels after day 10–15 of postnatal development is determined by R7BP availability.
RGS9-2 colocalizes with R7BP at postsynaptic sites of striatal neurons
The correlation of RGS9-2/R7BP late expression onset with that of major postsynaptic density organizer PSD-95, together with previously reported enrichment of both R7BP and RGS9-2 in postsynaptic densities (Song et al., 2006) has prompted us to examine their fine subcellular localization at the ultrastructural level. We have first used a postembedding immunogold labeling with specific antibodies against R7BP and RGS9-2. Although the postembedding procedure is less sensitive and does not provide perfect ultrastructural preservation, it is the only suitable method for unequivocal localization of proteins at synaptic sites (for detailed discussion, see Lujan, 2004). Data presented in Figure 2 establishes that strong immunoreactivity for both R7BP and RGS9-2 is observed at postsynaptic densities of excitatory synapses in which they are found to colocalize. Tissues processed for better ultrastructure preservation have also revealed the same colocalization pattern, although in this case antigenicity of both R7BP and RGS9-2 was greatly diminished (supplemental Fig. 2, available at www.jneurosci.org as supplemental material). To reveal the overall pattern of RGS9-2 and R7BP localization in neurons we further used a more sensitive procedure of preembedding immunogold electron microscopy (Fig. 3). This method reveals that in addition to postsynaptic densities both RGS9-2 and R7BP are also present at the extrasynaptic plasma membrane of dendritic shafts and spines with some immunoreactivity localizing to presynaptic sites. Importantly, antibody specificity was confirmed by negative control experiments with tissues isolated from the knock-out mice lacking R7BP or RGS9 (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). Quantification of the data by counting immunogold particles (2190 particles for RGS9-2 and 2163 particles for R7BP) from 3 independent labeling experiments provided the following results: 1) 88% of RGS9-2 (1752 particles) and 86% of R7BP (1691 particles) was associated with plasma membrane, whereas the remainder was found at intracellular membranes, 2) Immunoreactivity was predominantly found at postsynaptic sites (90.3% for RGS9-2 and 91.5% for R7BP), whereas only a small percentage localized presynaptically (9.7% for RGS9-2 and 8.5% for R7BP) and 3) Postsynaptic immunoreactivity was primarily found in two places: dendritic shafts (966 particles or 61% for RGS9-2 and 977 or 63% for R7BP) and spines (616 particles or 39% for RGS9-2 and 570 or 37% for R7BP).
Figure 2.
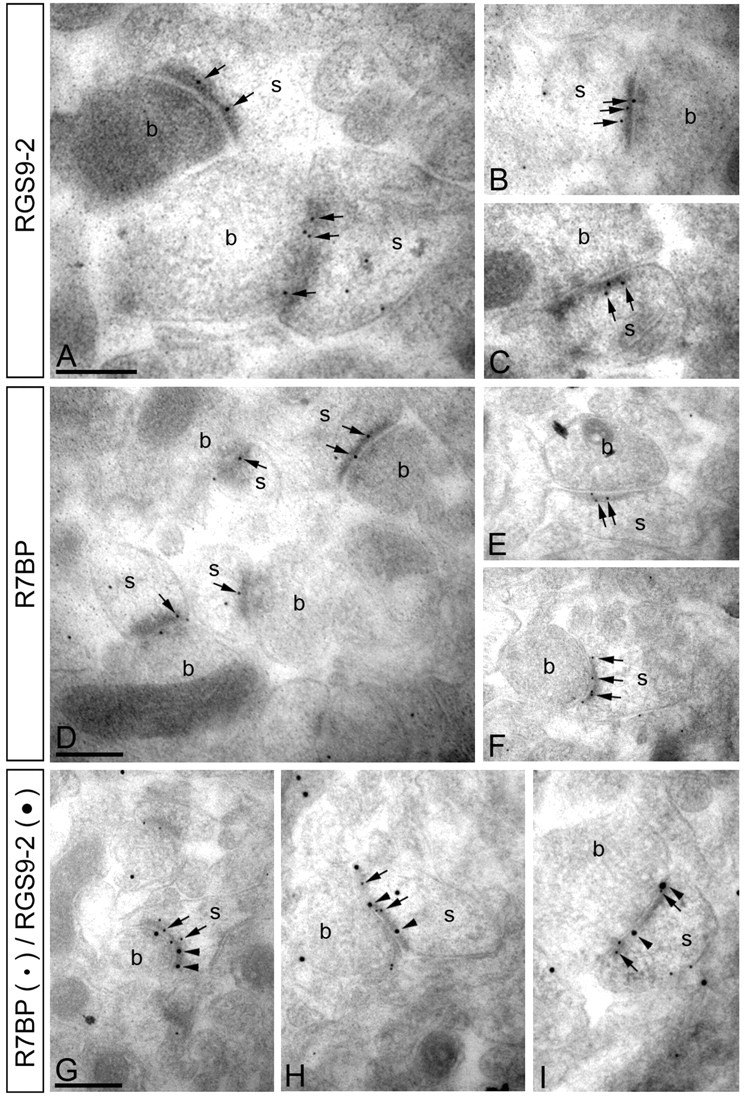
RGS9-2 and R7BP colocalize at postsynaptic densities of excitatory synapses. Electron micrographs of the striatum showing immunoreactivity for RGS9-2 and R7BP as detected using a postembedding immunogold method. A–C, Immunoparticles for RGS9-2 were found within the synaptic specialization of asymmetrical synapses (arrowheads). Gold particles were also observed at the extrasynaptic plasma membrane of spines (s) and to a lesser extent at presynaptic sites along the plasma membrane of axon terminals (b). D–F, Similarly, immunoparticles for R7BP were found within the synaptic specialization of asymmetrical synapses (arrowheads). A few immunoparticles were also observed at the extrasynaptic plasma membrane of spines (s) and at presynaptic sites along the plasma membrane of axon terminals (b). G–I, Double immunogold labeling showing colocalization of RGS9-2 (20 nm particles, arrowheads) and R7BP (10 nm particles, arrows) in individual synapses on spines (s) in the striatum. Scale bars, 0.2 μm.
Figure 3.
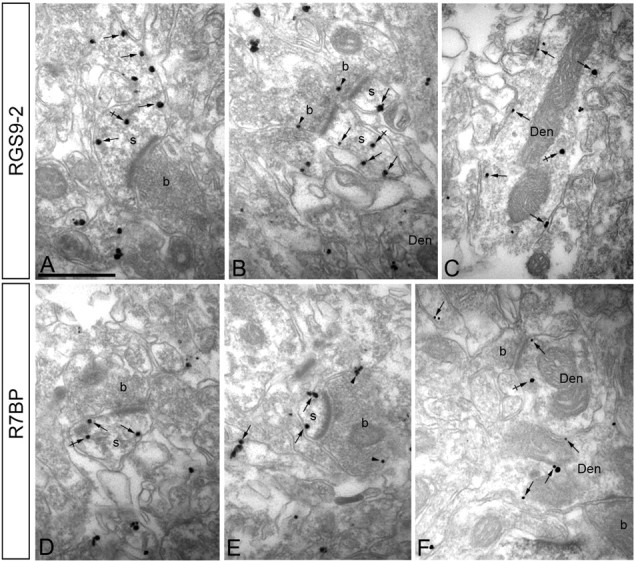
RGS9-2 and R7BP share the same subcellular localization in striatum. Electron micrographs of the striatum showing immunoreactivity for RGS9-2 (A–C) and R7BP (D–F), as detected using a preembedding immunogold method. A–C, Immunoparticles for RGS9-2 were detected associated or close (arrows) to the extrasynaptic plasma membrane of dendritic shafts (Den) and dendritic spines (s) establishing excitatory asynapses with axon terminals (b). Some immunoparticles for RGS9-2 were also detected intracellularly (crossed arrows) associated with intracellular membranes of dendritic shafts and spines. To a lesser extent, RGS9-2 immunoparticles were detected at presynaptic sites associated with the plasma membrane of axon terminals (b) (arrowheads). D–F, Immunoparticles for R7BP were detected associated or close (arrows) to the extrasynaptic plasma membrane of dendritic shafts (Den) and dendritic spines (s) establishing excitatory asynapses with axon terminals (b). Immunoparticles for R7BP were also detected intracellularly (crossed arrows) associated with intracellular membranes of dendritic shafts and spines. A few immunoparticles for R7BP were also detected at presynaptic sites associated with the plasma membrane of axon terminals (b) (arrowheads).
Knock-out of R7BP results in a selective downregulation of RGS9-2 expression and its subcellular mislocalization
To directly test the possibility that R7BP is involved in setting the expression level of RGS9-2 in vivo, we generated R7BP knock-out mice (Fig. 4A). Targeting of the R7BP locus was confirmed by PCR (Fig. 4B) and the lack of R7BP expression was confirmed at both protein (Fig. 4C) and mRNA (Fig. 4D) levels. Heterozygous animals contained approximately half of the R7BP mRNA and protein, demonstrating that R7BP protein levels are limited by the abundance of its mRNA. Strikingly, elimination of R7BP resulted in severe downregulation of RGS9-2 protein (Fig. 4C). Furthermore, reduction in R7BP levels in heterozygous mice also caused comparable reduction in the levels of RGS9-2. At the same time, both R7BP knock-outs and heterozygous animals contained normal levels of RGS9-2 mRNA comparable with that of the wild-type littermates, indicating that elimination of R7BP did not affect the synthesis of the RGS9-2 message but rather destabilized the RGS9-2 protein by changing its susceptibility to proteolysis (Anderson et al., 2007). Interestingly, knock-out of R7BP did not have any statistically significant effect on the expression levels of RGS6, RGS7 or RGS11 (data not shown), which belong to the R7 family of RGS9-like proteins and were reported to form complexes with R7BP (Drenan et al., 2005; Martemyanov et al., 2005). We have further investigated whether the observed reduction in RGS9-2 levels result from the inability of this protein to fold properly in the absence of its interaction with R7BP. This was done by monitoring RGS9-2 complex formation with its other binding partner, Gβ5, a process requiring appropriate posttranslational processing of the complex. Data presented in Figure 4E reveal that RGS9-2 efficiently coimmunoprecipitated with Gβ5 when anti-Gβ5 antibodies were used in the assay. Quantification of data from three independent experiments demonstrated that the ratio of RGS9-2 between wild-type and R7BP knock-out samples remained unchanged before and after the immunoprecipitation (8.9 ± 1.7 before IP and 9.7 ± 1.8 after the IP) indicating that the extent of RGS9-2 association with Gβ5 in R7BP knock-out is equal to that observed in wild-type mice. Because RGS9-2 complex formation with Gβ5 was previously shown to be sufficient for its functional activity (He et al., 2000; Kovoor et al., 2000; Skiba et al., 2001) we conclude that the elimination of R7BP has reduced the lifetime of RGS9-2/Gβ5 complex in the cell rather than affected its folding and/or assembly.
Figure 4.
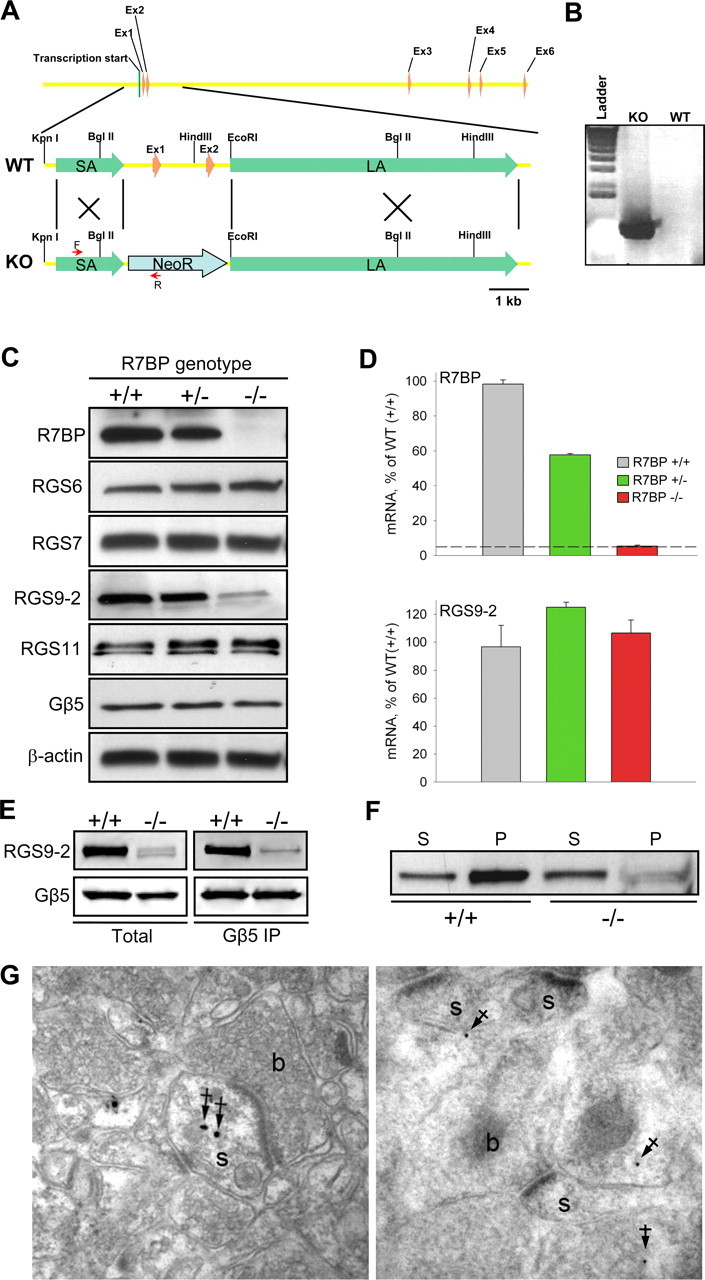
Knock-out of R7BP selectively destabilizes RGS9-2 at posttranscriptional level. A, R7BP gene structure and strategy for targeting homologous recombination. Targeting construct contained neomycin resistance cassette (NeoR) flanked by short (SA) and long (LA) homology arms. The positions of primers used to confirm the replacement are shown as red arrows. B, Confirmation of homologous recombination in genomic DNA isolated from the founder animal (KO) compared with wild-type parental strain (WT) by PCR. C, Western blot analysis of protein expression in the brains of wild-type (+/+), heterozygous (+/−) and R7BP knock-out (−/−) mice. The blot shows a representative experiment of three conducted. D, Analysis of R7BP and RGS9-2 transcript levels by quantitative RT-PCR. Total mRNA was isolated from striatal regions of adult mice was quantified by UV spectroscopy, and its equal amounts were subjected to Q-RT PCR amplification. Each experiment was conducted with samples isolated from two to three mice. Dashed line indicates the limiting level for mRNA detection. Error bars represent SEM. E, Immunoprecipitation of RGS9-2/Gβ5 complexes from wild-type and R7BP knock-out striatal tissues. Extraction and immunoprecipitation with anti-Gβ5 antibody was conducted as described in Materials and Methods. Two milligrams of total protein were used in each immunoprecipitation experiment with 14 μg of anti-Gβ5 antibody. Equal protein amounts of whole-cell extracts and volumes of the immunoprecipitation eluates were loaded in each well. The data are representative of three experiments conducted. F, Subcellular fractionation of striatal neurons into synaptic and plasma membrane containing pellet (P) and supernatant (S) containing primarily cytosol and small microsomal membranes. Striatal regions of wild-type (WT) and R7BP knock-out (R7BP KO) mice were homogenized and subjected to one step centrifugation as described in Materials and Methods. Knock-out samples were loaded at four times excess of total protein over wild-type samples. G, Preembedding (left) and postembedding (right) immunogold EM analysis of RGS9-2 localization in striatum of R7BP knock-out mice. Filled arrows indicate location of immunogold particles at intracellular sites, a predominant localization pattern of RGS9-2 in neurons of R7BP knock-outs. s, Dendritic spines; b, plasma membrane of the axon terminals.
The presence of low levels of RGS9-2/Gβ5 in R7BP knock-outs also made it possible to address the question whether R7BP is important in driving the localization of RGS9-2 in vivo. We first addressed this question by performing subcellular fractionation of brain lysates into membrane containing pellet and cytosol containing supernatant (Fig. 4F). We found that, whereas in wild-type striatal neurons most of RGS9-2 was associated with membrane fraction, in the R7BP knock-out neurons, RGS9-2 is predominantly cytoplasmic. This observation was further confirmed by immunogold labeling of RGS9-2 in the striatum of R7BP knock-out mice. Figure 4G indicate that RGS9-2 immunoreactivity was never found in postsynaptic densities, but rather was predominantly located at various intracellular sites in the spines and cell bodies. Together, these results demonstrate that in addition to being essential for maintaining high expression levels of RGS9-2, R7BP is also responsible for the localization of RGS9-2/Gβ5 complex to postsynaptic sites in native neurons.
Overexpression of R7BP in vivo leads to selective upregulation of RGS9-2 expression
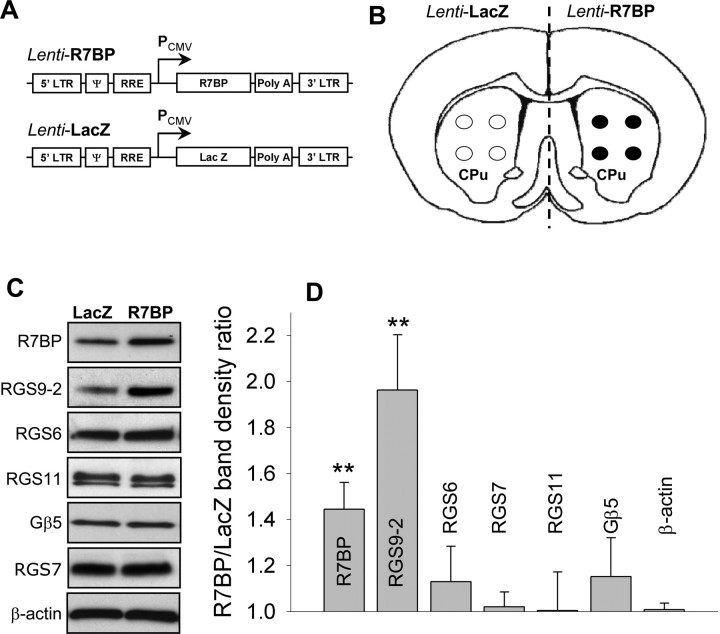
We next directly tested whether the availability of R7BP limits the expression of RGS9-2 and whether an increase in R7BP levels would be sufficient to augment RGS9-2 expression. To address this question we delivered an R7BP lentivirus (Fig. 5A) into the striatum via stereotaxic injection (Fig. 5B). Data presented in Figure 5, C and D, illustrate that we were able to achieve ∼50% overexpression of R7BP. Importantly, elevation in R7BP levels resulted in marked increase in levels of RGS9-2 but not other R7 RGS proteins. Although no statistical significance was detected, it is curious that over-expression caused larger change in RGS9-2 levels compared with R7BP (2-fold vs 1.5-fold). This is likely explained by the lower levels of endogenous RGS9-2 compared with R7BP, which is present in excess over RGS9-2 as it additionally forms complexes with other R7 family members.
Figure 5.
Lentivirus-mediated overexpression of R7BP in the striatum. A, Schematic representation of lentiviral constructs used for the expression experiments. Open reading frames for R7BP or LacZ were placed under the control of strong cytomegalovirus promoter in the context of lentiviral packaging elements (LTR, Ψ, and RRE). B, Strategy for the lentivirus mediated protein expression in vivo. Each side of the brain received four injections with either R7BP or control LacZ lentiviruses within the caudate/putamen region (CPu). C, Western blot analysis of protein expression in the CPu induced after lentiviral transduction. D, Quantification of Western blot data shown in C from three independent experiments. Band densities were determined by densitometry and used to calculate the ratio of band intensity between R7BP side to LacZ side. **p < 0.01, statistically significant changes in the levels of proteins compared with changes in β-actin expression (last bar) (t test). Error bars are SEM.
Selective degradation of RGS9-2 is mediated by its R7BP binding site
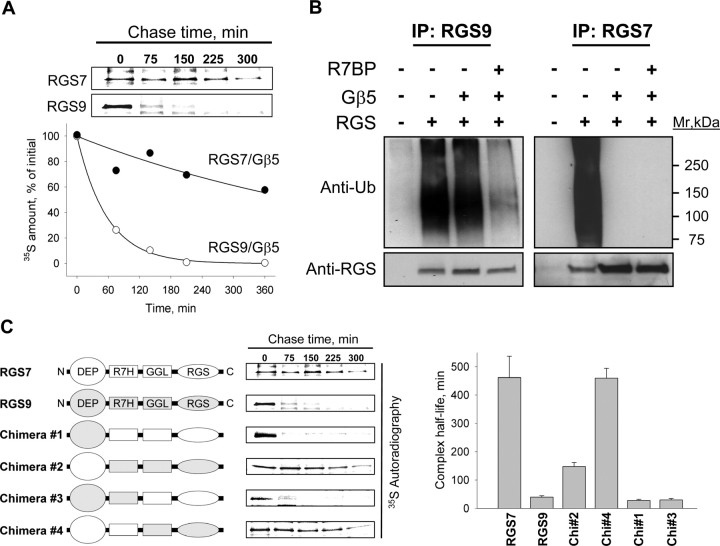
Knock-out and overexpression data (Figs. 4, 5) argue that the effect of R7BP on protein expression level is specific for RGS9-2 among all other R7 RGS protein members that form complexes with R7BP. We therefore hypothesized that the selective regulation of RGS9-2 expression by R7BP results from its higher susceptibility to degradation among the R7 family. We tested this hypothesis by comparing proteolytic degradation rates of RGS9-2 and RGS7, a representative R7 RGS protein that is not regulated in its expression by R7BP. To simplify the analysis, we used the short isoform (RGS9-1) which is the closest in its amino-acid sequence to RGS7 yet has the same degradation rate as RGS9-2 (Anderson et al., 2007). Pulse-chase metabolic labeling experiments (Fig. 6A) reveal that in contrast to an unstable RGS9-1/Gβ5 complex (half-time to degradation 39.7 ± 5 min) the complex of RGS7 with Gβ5 is ∼10 times more stable (half-time to degradation 462 ± 75 min).
Figure 6.
Selective degradation of RGS9-2/Gβ5 in the absence of R7BP is determined by N-terminal instability elements. A, Pulse-chase metabolic labeling experiments comparing degradation rates of RGS7 and RGS9-1 in 293FT cells. The blot shows the time course of 35S-Met/Cys dissipation from immunoprecipitated RGS proteins, quantified on a graph below. Approximately 25% of recombinant proteins was recovered in the immunoprecipitation eluates. B, Detection of RGS protein modification with ubiquitin in transfected 293FT cells. Top, Ubiquitination of RGS7 or RGS9-2 in the presence or absence of Gβ5 and R7BP was detected by Western blotting with anti-ubiquitin antibodies after protein immunoprecipitation (IP) with specific anti-RGS antibodies (∼25% immunoprecipitation efficiency). Bottom, Western blot detection of respective RGS protein from same immunoprecipitation fractions. C, Proteolytic stability of RGS7/RGS9 chimeras as determined by pulse-chase metabolic labeling experiments. Right panel shows quantification of protein half-life calculated from exponential fitting of degradation time course. Error bars are SEM.
Because protein modification by ubiquitination is a major cellular mechanism for selective targeting of proteins to degradation, we next checked whether RGS9-2 and RGS7 are differentially ubiquitinated. Figure 6B shows that without Gβ5, both RGS9-2 and RGS7 are heavily poly-ubiquitinated. However, coexpression with Gβ5 completely abolishes RGS7 ubiquitination. Yet Gβ5 only had a modest effect on the RGS9-2 ubiquitination, which was only significantly reduced in the presence of R7BP (Fig. 6B). Ubiquitination of RGS7 when expressed alone was reported in the past (Kim et al., 1999) and its prevention by Gβ5 observed in our study correlates very well with the requirement of the entire R7 RGS family to be associated with Gβ5 for their folding and stability (Witherow et al., 2000; Sondek and Siderovski, 2001; Chen et al., 2003). The inability of Gβ5 to fully prevent RGS9 ubiquitination and decrease its degradation rate suggests a presence of secondary instability elements in this protein that are controlled by the association with R7BP. To identify such elements, we used a chimeric mutagenesis approach exploiting the homologous nature of RGS9-1 and RGS7. We reciprocally replaced conserved domains between these proteins and analyzed the stability of the resulting chimeras in the pulse chase degradation assays in the presence of Gβ5 (Fig. 6C). Quantification of the data show that replacing the DEP domain of RGS9 with that of RGS7 partially rescues RGS9-1 instability, and an additional replacement of the R7H domain makes chimeric RGS9/Gβ5 in the absence of R7BP as stable as RGS7. Conversely, replacing either N-terminal DEP domain or DEP in combination with R7H in RGS7 dramatically destabilized this protein. This result parallels with our previous study, in which the binding site for R7BP in R7 RGS proteins was mapped to be formed by both DEP and R7H domains of R7 RGS proteins (Anderson et al., 2007). This suggests that R7BP exerts its degradation protective effects by masking the unique instability element(s) in RGS9-2 located within its R7BP binding site.
Multiple determinants at the N terminus target RGS9-2 to degradation by cysteine proteases
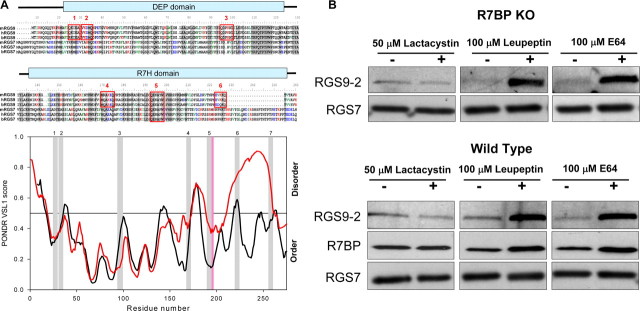
To gain insights into possible structural characteristics of the instability elements within DEP/R7H domain of RGS9-2 we performed a comparative sequence analysis between RGS7 and RGS9-2. First, RGS9-2 and RGS7 were analyzed for the presence of naturally disordered regions, predominantly unstructured segments within proteins or domains that are increasingly accepted to underlie much of the functional flexibility in proteins and serve as sites for posttranslational modifications and for association with interaction partners and ligands (Dyson and Wright, 2005; Uversky et al., 2005). We reasoned that a higher probability for the presence of these regions in RGS9-2 compared with RGS7 might account for the observed differences in their proteolytic stability, for example, by providing recognition sites for cellular degradation machinery. Analysis of disordered prediction profiles shows that both RGS7 and RGS9 have a very similar pattern of distribution for local minimums and maximums of disorder scores along the DEP/R7H domains (Fig. 7A). However, peak scores and the number of regions which exceeded the disorder threshold were significantly greater in RGS9-2.
Figure 7.
Degradation determinants and route of RGS9-2 proteolysis. A, Prediction of RGS9-2 instability elements. Top, Multiple sequence alignment of RGS9 and RGS7. Shaded areas indicate conserved regions, and red boxes mark the positions of KFERQ-like sequences. Bottom, Prediction of naturally disordered regions in N-terminal domains of RGS9 (black traces) and RGS7 (red traces) by PONDR-VSL1 algorithm. Positions of KFERQ-like motifs are marked by gray (RGS9) or pink (RGS7) bars. B, Effect of protein degradation inhibitors on RGS9-2 levels. Striatal slices from wild-type or R7BP knock-out mice were cultured ex vivo for 5 h in the presence or absence of indicated inhibitors after which protein levels were determined by Western blotting. Note that wild-type and knock-out blots were exposed for different times to reveal the extent of protein upregulation by inhibitors rather than differences in RGS9-2 levels between knock-out and wild-type samples.
The second noticeable feature that distinguishes RGS9-2 is the high occurrence of so called “KFERQ-like” motifs which are short (5–6 amino acid long) sequences that have the unusual combination of oppositely charged amino acids with hydrophobic residues and glutamates. Occurrence of these motifs in proteins is linked to their increased instability mainly by routing them to the lysosomal degradation pathway (Dice, 1990). The DEP/R7H domains of RGS9-2 contain 6 KFERQ-like sequences, whereas in contrast, only 1 such motif is present in RGS7. Superimposing positions of the KFERQ-like motifs over the disorder prediction profiles reveal that most of these sequences in RGS9 are localized to disorder peaks (Fig. 7A), whereas the only KFERQ motif present in RGS7 localized to a region of low disorder score.
Our next step was to test whether the prediction of KFERQ lysosomal degradation targeting sequences in RGS9 would be consistent with the main pathway of its degradation in cells in the absence of R7BP. Culturing striatal slices in the presence of cysteine protease inhibitors (E64 and Leupeptin), which are the major destruction enzymes of lysosomes, potently prevent RGS9-2 degradation in R7BP knock-out tissues (Fig. 7B). In contrast, the specific proteasome inhibitor lactacystin had no effect on rescuing RGS9-2 levels. Furthermore, inhibitors had similar effects on the levels of the intact RGS9-2/R7BP complex in wild-type slices suggesting that even in the presence of R7BP the regulation of RGS9-2 levels is dynamic. The effect is specific for RGS9-2/R7BP because it does not affect the levels of other R7 RGS proteins. These results suggest that the degradation of the RGS9-2/R7BP complex in native neurons is mainly controlled by cellular cysteine proteases (lysosomal pathway), rather than by proteasome.
The stability of R7BP is reciprocally controlled by its association with R7 RGS proteins
Furthermore, we observed that the inhibition of proteolysis in wild-type slices led not only to RGS9-2 upregulation but also to an increase in R7BP levels (Fig. 7B, bottom). This suggested the possibility that R7BP could also be an unstable protein whose stability might be in turn controlled by protein–protein interactions. We first tested this possibility by analyzing the stability of R7BP in transfected cells. Cotransfection with either RGS9-2/Gβ5 or RGS7/Gβ5 markedly increased the expression level of R7BP (Fig. 8A). Pulse-chase degradation experiments (Fig. 8B) revealed that like RGS9-2, R7BP is rapidly degraded (half-time to degradation is 114 ± 18 min) but is greatly stabilized when coexpressed with an R7 RGS protein. In fact, no significant degradation of R7BP/RGS7/Gβ5 complex was observed during the time course of an experiment (half-time to degradation >48 h). This observation predicts that interaction with R7 RGS proteins is essential for the stable expression of R7BP. Indeed, analysis of protein expression in Gβ5 knock-out animals which lack all four R7 RGS proteins (Chen et al., 2003), no detectable R7BP could be found in the striatum (Fig. 8C). Elimination of RGS9-2 from the striatum did not significantly affect the level of R7BP suggesting that in the absence of RGS9-2, R7BP is stabilized by association with other R7 family members which bind to this protein in the same striatal neurons. Interestingly, R7BP appears to be unique in its requirement of RGS protein association for stable expression because a homologous protein, R9AP, which serves to anchor RGS9-1 and RGS11 in the retina (Song et al., 2007) have normal expression levels in the Gβ5 knock-out.
Figure 8.
R7BP degradation is reciprocally regulated by complex formation with R7 RGS/Gβ5 proteins. A, Cotransfection with RGS7/Gβ5 and RGS9-2/Gβ5 augment R7BP expression. 293FT cells were transfected with R7BP either alone (mock) or in combination with the indicated RGS construct and Gβ5. Right shows quantification of changes in protein levels from three independent experiments. **p < 0.01, statistically significant differences in R7BP levels with and without coexpression with R7 RGS proteins (t test). B, RGS7/Gβ5 slows down the rate of R7BP proteolysis as determined by pulse-chase degradation assays. C, Effect of Gβ5 knock-out on the expression of R7BP and R9AP in striatum and retina. Tissue lysates containing equal protein concentrations were analyzed by Western blotting with the indicated antibodies.
Discussion
In this study we propose a mechanism for the regulation of the expression and localization of the key regulator of dopamine and opioid receptor signaling in the brain. In this mechanism, RGS9-2 expression level in the striatum is set by posttranslational regulation acting to rescue the protein from constitutive degradation. RGS9-2 belongs to the R7 RGS family, members of which have been found to form macromolecular complexes with two binding partners: Gβ5 and R7BP. Association with Gβ5 is required for the folding of all R7 RGS proteins which as a result, exist as constitutive RGS-Gβ5 heterodimers (Snow et al., 1999; Witherow et al., 2000; Chen et al., 2003). In turn, association of RGS9-2/Gβ5 complex with R7BP was shown to increase the proteolytic stability of the complex (Anderson et al., 2007). Here we used genetic knock-out and overexpression strategies to demonstrate that unlike Gβ5, R7BP selectively regulates the expression levels of RGS9-2 but not other R7 RGS protein complexes in vivo. We found that in the absence of R7BP, RGS9-2 is translated and forms complexes with Gβ5, but is destined to be quickly degraded by the cell's proteolytic machinery. The binding site for R7BP in RGS9-2 contains specific instability elements that target it for proteolysis by cysteine proteases. Binding of R7BP masks these degradation determinants and results in elevation of RGS9-2 expression. The instability elements are absent in other homologous R7 RGS proteins, allowing them to be stably expressed independently from R7BP association. Consequently, the expression level of the RGS9-2/Gβ5 signaling complex is determined by the availability of R7BP and is not dependent on the synthesis of RGS9-2 mRNA. Interestingly, a similar posttranslational mechanism also appears to regulate the expression of the short splice isoform, RGS9-1 whose levels in photoreceptors are determined by the R7BP homolog, R9AP (Keresztes et al., 2004; Krispel et al., 2006).
Comparative bioinformatics analysis of RGS9-2 revealed a unique combination of sequence features in the R7BP binding site, DEP/R7H domain, that confer higher susceptibility to degradation. First, this region of RGS9-2 has a high probability of local disordered regions which are increasingly viewed as hot spots of posttranslational protein modification as well as sites for association with ligands and binding partners (Dyson and Wright, 2005; Uversky et al., 2005; Radivojac et al., 2007). Second, most of the RGS9-2 regions with high disorder prediction also display unique composition of positively and negatively charged as well as hydrophobic residues in conjunction with glutamines known as KFERQ-like motifs. Although DEP/R7H domains are conserved in R7 RGS proteins, this specific combination of determinants is a unique feature of RGS9-2. Notably, KFERQ-like motifs were previously reported to target proteins to degradation mainly via the lysosomal pathway (Dice, 1990, 1992). Consistent with these reports, our data indicate that in the absence of R7BP, degradation of RGS9-2 in striatum is prevented by the inhibition of cysteine proteases of the lysosomes, but not by the specific inhibition of the proteasome. Although the present study does not unequivocally demonstrate how targeting of RGS9-2 for lysosomal degradation is accomplished, two possibilities emerged from our studies. One possibility revealed by bioinformatics analysis was the presence of disordered regions within RGS9-2 correlating with KFREQ motifs, which could serves as chaperone mediated binding sites for entry into lysosomes (Massey et al., 2006). Alternatively, another lysosomal targeting signal could be accomplished by ubiquitination within DEP/R7H disordered regions of RGS9-2, as recent data suggests that such regions could underlie ubiquitin attachment sites (Bourhis et al., 2007; Radivojac et al., 2007). Furthermore, it is becoming increasingly accepted that protein ubiquitination is involved in targeting some proteins for lysosomal degradation rather than to the proteasome (Urbe, 2005; Mukhopadhyay and Riezman, 2007). Although in this study we show that in transfected cells RGS9-2 can be ubiquitinated, the importance of this modification for targeting RGS9-2/Gβ5 degradation in striatal neurons remains to be elucidated.
The second main finding of this work is that the RGS9-2/Gβ5/R7BP complex shows striking developmental coregulation. Both RGS9-2 and R7BP are virtually absent in the brains of neonatal mice and are rapidly induced at late stages of postnatal neuronal differentiation. Similar expression time course is observed for a few postsynaptic proteins that are involved in the elaboration of the synaptic organizing characteristic of differentiated neurons such as NR2A, PSD-95, and PSD-93 (Lim et al., 1999; Sans et al., 2000). In contrast, many proteins such as DARPP-32, GluR1, SAP102 that are involved in constitutive signaling at synapses are normally expressed from early embryonic stages and exhibit rather modest upregulation (Lim et al., 1999; Sans et al., 2000). Provocatively, this suggests that the RGS9-2/R7BP complex is involved in setting signaling characteristics of the mature nervous system. Consistent with this idea, we found that both RGS9-2 and R7BP colocalize at postsynaptic densities of excitatory synapses. Importantly, this localization of the complex is mediated by R7BP, as RGS9-2 is completely absent from the postsynaptic densities of R7BP KO striatum. This result together with the importance of R7BP complex formation with RGS9-2 for its stabilization, ensures this localization pattern for RGS9-2. In addition to localizing RGS9-2, R7BP may also be important for targeting other R7 RGS proteins such as RGS6, RGS7, RGS11, with which it associates, to postsynaptic sites on nervous system development.
The described posttranslational mechanism of RGS9-2 expression and localization regulation by R7BP has several implications. First, it suggests that RGS9-2 levels could be regulated by attenuation of the lysosomal degradation pathway, perhaps, as part of the cellular response to receptor activation. Second, it suggests that changes in R7BP concentration provide a new regulatory input for modulating the sensitivity of dopamine and opioid receptor signaling set by RGS9-2. Third, the mechanism provides the specificity for the rapid alteration in concentration of one regulator among highly similar homologues. Fourth, the mechanism ensures that at the postsynaptic density RGS9-2/Gβ5 functions only in complex with R7BP. Fifth, the elimination of R7BP in Gβ5 knock-out mice indicates that R7BP is unlikely to have any independent signaling roles outside of its complex with R7 RGS proteins.
Footnotes
This work was supported by National Institutes of Health (NIH) Grants EY018139 and DA021743 (K.A.M.), MH61933 (K.W.), EY013811 (C.-K.C.), and DA 011806 (K.A.M., K.W.), the Graduate School and Academic Health Center at the University of Minnesota (K.A.M.), and Spanish Ministry of Education and Science Grant BFU-2006-01896 (R.L.). We thank Dr. William Simonds (NIH) for the generous gift of anti-RGS7 and anti-Gβ5 antibodies and Dr. Sheila Baker for critical comments on this manuscript.
References
- Anderson GR, Semenov A, Song JH, Martemyanov KA. The membrane anchor R7BP controls the proteolytic stability of the striatal specific RGS protein, RGS9-2. J Biol Chem. 2007;282:4772–4781. doi: 10.1074/jbc.M610518200. [DOI] [PubMed] [Google Scholar]
- Bourhis JM, Canard B, Longhi S. Predicting protein disorder and induced folding: from theoretical principles to practical applications. Curr Protein Pept Sci. 2007;8:135–149. doi: 10.2174/138920307780363451. [DOI] [PubMed] [Google Scholar]
- Burchett SA, Volk ML, Bannon MJ, Granneman JG. Regulators of G-protein signaling: rapid changes in mRNA abundance in response to amphetamine. J Neurochem. 1998;70:2216–2219. doi: 10.1046/j.1471-4159.1998.70052216.x. [DOI] [PubMed] [Google Scholar]
- Chen CK, Burns ME, He W, Wensel TG, Baylor DA, Simon MI. Slowed recovery of rod photoresponse in mice lacking the GTPase accelerating protein RGS9-1. Nature. 2000;403:557–560. doi: 10.1038/35000601. [DOI] [PubMed] [Google Scholar]
- Chen CK, Eversole-Cire P, Zhang HK, Mancino V, Chen YJ, He W, Wensel TG, Simon MI. Instability of GGL domain-containing RGS proteins in mice lacking the G-protein beta subunit Gβ5. Proc Natl Acad Sci USA. 2003;100:6604–6609. doi: 10.1073/pnas.0631825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- Dice JF. Selective degradation of cytosolic proteins by lysosomes. Ann NY Acad Sci. 1992;674:58–64. doi: 10.1111/j.1749-6632.1992.tb27477.x. [DOI] [PubMed] [Google Scholar]
- Drenan RM, Doupnik CA, Boyle MP, Muglia LJ, Huettner JE, Linder ME, Blumer KJ. Palmitoylation regulates plasma membrane-nuclear shuttling of R7BP, a novel membrane anchor for the RGS7 family. J Cell Biol. 2005;169:623–633. doi: 10.1083/jcb.200502007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Ehrlich ME, Rosen NL, Kurihara T, Shalaby IA, Greengard P. DARPP-32 development in the caudate nucleus is independent of afferent input from the substantia nigra. Brain Res Dev Brain Res. 1990;54:257–263. doi: 10.1016/0165-3806(90)90148-r. [DOI] [PubMed] [Google Scholar]
- Gainetdinov RR, Premont RT, Bohn LM, Lefkowitz RJ, Caron MG. Desensitization of G-protein-coupled receptors and neuronal functions. Annu Rev Neurosci. 2004;27:107–144. doi: 10.1146/annurev.neuro.27.070203.144206. [DOI] [PubMed] [Google Scholar]
- He W, Lu LS, Zhang X, El Hodiri HM, Chen CK, Slep KC, Simon MI, Jamrich M, Wensel TG. Modules in the photoreceptor RGS9-1/Gbeta5L GTPase-accelerating protein complex control effector coupling, GTPase acceleration, protein folding, and stability. J Biol Chem. 2000;275:37093–37100. doi: 10.1074/jbc.M006982200. [DOI] [PubMed] [Google Scholar]
- Hollinger S, Hepler JR. Cellular regulation of RGS proteins: Modulators and integrators of G-protein signaling. Pharmacol Rev. 2002;54:527–559. doi: 10.1124/pr.54.3.527. [DOI] [PubMed] [Google Scholar]
- Keresztes G, Mutai H, Hibino H, Hudspeth AJ, Heller S. Expression patterns of the RGS9-1 anchoring protein R9AP in the chicken and mouse suggest multiple roles in the nervous system. Mol Cell Neurosci. 2003;24:687–695. doi: 10.1016/s1044-7431(03)00231-8. [DOI] [PubMed] [Google Scholar]
- Keresztes G, Martemyanov KA, Krispel CM, Mutai H, Yoo PJ, Maison SF, Burns ME, Arshavsky VY, Heller S. Absence of the RGS9/Gbeta5 GTPase-activating complex in photoreceptors of the R9AP knockout mouse. J Biol Chem. 2004;279:1581–1584. doi: 10.1074/jbc.C300456200. [DOI] [PubMed] [Google Scholar]
- Kim E, Arnould T, Sellin L, Benzing T, Comella N, Kocher O, Tsiokas L, Sukhatme VP, Walz G. Interaction between RGS7 and polycystin. Proc Natl Acad Sci USA. 1999;96:6371–6376. doi: 10.1073/pnas.96.11.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovoor A, Chen CK, He W, Wensel TG, Simon MI, Lester HA. Co-expression of Gbeta5 enhances the function of two Ggamma subunit-like domain-containing regulators of G-protein signaling proteins. J Biol Chem. 2000;275:3397–3402. doi: 10.1074/jbc.275.5.3397. [DOI] [PubMed] [Google Scholar]
- Kovoor A, Seyffarth P, Ebert J, Barghshoon S, Chen CK, Schwarz S, Axelrod JD, Cheyette BN, Simon MI, Lester HA, Schwarz J. D2 dopamine receptors colocalize regulator of G-protein signaling 9-2 (RGS9-2) via the RGS9 DEP domain, and RGS9 knock-out mice develop dyskinesias associated with dopamine pathways. J Neurosci. 2005;25:2157–2165. doi: 10.1523/JNEUROSCI.2840-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krispel CM, Chen D, Melling N, Chen YJ, Martemyanov KA, Quillinan N, Arshavsky VY, Wensel TG, Chen CK, Burns ME. RGS expression rate-limits recovery of rod photoresponses. Neuron. 2006;51:409–416. doi: 10.1016/j.neuron.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Lim S, Naisbitt S, Yoon J, Hwang JI, Suh PG, Sheng M, Kim E. Characterization of the Shank family of synaptic proteins. Multiple genes, alternative splicing, and differential expression in brain and development. J Biol Chem. 1999;274:29510–29518. doi: 10.1074/jbc.274.41.29510. [DOI] [PubMed] [Google Scholar]
- Lujan R. Electron microscopic studies of receptor localization. In: Willars GB, Challiss RA, editors. Methods in molecular biology. Vol 259. Totowa, NJ: Humana; 2004. pp. 123–136. [DOI] [PubMed] [Google Scholar]
- Lujan R, Nusser Z, Roberts JD, Shigemoto R, Somogyi P. Perisynaptic location of metabotropic glutamate receptors mGluR1 and mGluR5 on dendrites and dendritic spines in the rat hippocampus. Eur J Neurosci. 1996;8:1488–1500. doi: 10.1111/j.1460-9568.1996.tb01611.x. [DOI] [PubMed] [Google Scholar]
- Makino ER, Handy JW, Li TS, Arshavsky VY. The GTPase activating factor for transducin in rod photoreceptors is the complex between RGS9 and type 5 G-protein beta subunit. Proc Natl Acad Sci USA. 1999;96:1947–1952. doi: 10.1073/pnas.96.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martemyanov KA, Yoo PJ, Skiba NP, Arshavsky VY. R7BP, a novel neuronal protein interacting with RGS proteins of the R7 family. J Biol Chem. 2005;280:5133–5136. doi: 10.1074/jbc.C400596200. [DOI] [PubMed] [Google Scholar]
- Massey AC, Zhang C, Cuervo AM. Chaperone-mediated autophagy in aging and disease. Curr Top Dev Biol. 2006;73:205–235. doi: 10.1016/S0070-2153(05)73007-6. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- Nishiguchi KM, Sandberg MA, Kooijman AC, Martemyanov KA, Pott JW, Hagstrom SA, Arshavsky VY, Berson EL, Dryja TP. Defects in RGS9 or its anchor protein R9AP in patients with slow photoreceptor deactivation. Nature. 2004;427:75–78. doi: 10.1038/nature02170. [DOI] [PubMed] [Google Scholar]
- Obradovic Z, Peng K, Vucetic S, Radivojac P, Dunker AK. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins. 2005;61(Suppl 7):176–182. doi: 10.1002/prot.20735. [DOI] [PubMed] [Google Scholar]
- Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK, Obradovic Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J Bioinform Comput Biol. 2005;3:35–60. doi: 10.1142/s0219720005000886. [DOI] [PubMed] [Google Scholar]
- Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman Z, Schwarz J, Gold SJ, Zachariou V, Wein MN, Choi KH, Kovoor A, Chen CK, DiLeone RJ, Schwarz SC, Selley DE, Sim-Selley LJ, Barrot M, Luedtke RR, Self D, Neve RL, Lester HA, Simon MI, Nestler EJ. RGS9 modulates dopamine signaling in the basal ganglia. Neuron. 2003;38:941–952. doi: 10.1016/s0896-6273(03)00321-0. [DOI] [PubMed] [Google Scholar]
- Sans N, Petralia RS, Wang YX, Blahos J, II, Hell JW, Wenthold RJ. A developmental change in NMDA receptor-associated proteins at hippocampal synapses. J Neurosci. 2000;20:1260–1271. doi: 10.1523/JNEUROSCI.20-03-01260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi JL, Brady DL, Koenig JI. Estrogen modulates RGS9 expression in the nucleus accumbens. NeuroReport. 2004;15:2433–2436. doi: 10.1097/00001756-200410250-00026. [DOI] [PubMed] [Google Scholar]
- Skiba NP, Martemyanov KA, Elfenbein A, Hopp JA, Bohm A, Simonds WF, Arshavsky VY. RGS9-Gbeta5 substrate selectivity in photoreceptors—opposing effects of constituent domains yield high affinity of RGS interaction with the G-protein-effector complex. J Biol Chem. 2001;276:37365–37372. doi: 10.1074/jbc.M106431200. [DOI] [PubMed] [Google Scholar]
- Snow BE, Krumins AM, Brothers GM, Lee SF, Wall MA, Chung S, Mangion J, Arya S, Gilman AG, Siderovski DP. A G-protein gamma subunit-like domain shared between RGS11 and other RGS proteins specifies binding to Gbeta5 subunits. Proc Natl Acad Sci USA. 1998;95:13307–13312. doi: 10.1073/pnas.95.22.13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow BE, Betts L, Mangion J, Sondek J, Siderovski DP. Fidelity of G-protein beta-subunit association by the G-protein gamma-subunit-like domains of RGS6, RGS7, and RGS11. Proc Natl Acad Sci USA. 1999;96:6489–6494. doi: 10.1073/pnas.96.11.6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondek J, Siderovski DP. Ggamma-like (GGL) domains: new frontiers in G-protein signaling and beta-propeller scaffolding. Biochem Pharmacol. 2001;61:1329–1337. doi: 10.1016/s0006-2952(01)00633-5. [DOI] [PubMed] [Google Scholar]
- Song JH, Waataja JJ, Martemyanov KA. Subcellular targeting of RGS9-2 is controlled by multiple molecular determinants on its membrane anchor, R7BP. J Biol Chem. 2006;281:15361–15369. doi: 10.1074/jbc.M600749200. [DOI] [PubMed] [Google Scholar]
- Song JH, Song H, Wensel TG, Sokolov M, Martemyanov KA. Localization and differential interaction of R7 RGS proteins with their membrane anchors R7BP and R9AP in neurons of vertebrate retina. Mol Cell Neurosci. 2007;35:311–319. doi: 10.1016/j.mcn.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Tekumalla PK, Calon F, Rahman Z, Birdi S, Rajput AH, Hornykiewicz O, Di Paolo T, Bedard PJ, Nestler EJ. Elevated levels of DeltaFosB and RGS9 in striatum in Parkinson's disease. Biol Psychiatry. 2001;50:813–816. doi: 10.1016/s0006-3223(01)01234-3. [DOI] [PubMed] [Google Scholar]
- Urbe S. Ubiquitin and endocytic protein sorting. Essays Biochem. 2005;41:81–98. doi: 10.1042/EB0410081. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit. 2005;18:343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- Witherow DS, Wang Q, Levay K, Cabrera JL, Chen J, Willars GB, Slepak VZ. Complexes of the G-protein subunit Gbeta5 with the regulators of G-protein signaling RGS7 and RGS9—characterization in native tissues and in transfected cells. J Biol Chem. 2000;275:24872–24880. doi: 10.1074/jbc.M001535200. [DOI] [PubMed] [Google Scholar]
- Zachariou V, Georgescu D, Sanchez N, Rahman Z, DiLeone R, Berton O, Neve RL, Sim-Selley LJ, Selley DE, Gold SJ, Nestler EJ. Essential role for RGS9 in opiate action. Proc Natl Acad Sci USA. 2003;100:13656–13661. doi: 10.1073/pnas.2232594100. [DOI] [PMC free article] [PubMed] [Google Scholar]