Abstract
Rheumatic diseases have complex aetiologies that are not fully understood, which makes the study of pathogenic mechanisms in these diseases a challenge for researchers. Next-generation sequencing (NGS) and related omics technologies, such as transcriptomics, epigenomics and genomics, provide an unprecedented genome-wide view of gene expression, environmentally responsive epigenetic changes and genetic variation. The integrated application of NGS technologies to samples from carefully phenotyped clinical cohorts of patients has the potential to solve remaining mysteries in the pathogenesis of several rheumatic diseases, to identify new therapeutic targets and to underpin a precision medicine approach to the diagnosis and treatment of rheumatic diseases. This Review provides an overview of the NGS technologies available, showcases important advances in rheumatic disease research already powered by these technologies and highlights NGS approaches that hold particular promise for generating new insights and advancing the field.
Technical advances and the decreasing cost of next-generation sequencing (NGS) technologies have led to the development and rapid application of methods to assess gene expression, chromatin accessibility and structure, DNA methylation and DNA sequences at the genome-wide level. Subsequent advances have enabled these technologies to be scaled for the analysis of single cells. These approaches have been extensively applied to the study of fundamental biological processes, including immune and autoimmune responses1,2. In diseases such as cancer, which have a strong genetic component, these technologies have already had a transformative effect on our understanding of pathogenesis and in the development of precision medicine therapeutic approaches. In rheumatic diseases, analysis of genome-wide association studies (GWAS) has revealed many common allelic variants that are associated with small increases in disease risk; however, the majority of disease heritability has yet to be accounted for, and non-genetic factors, such as environmental factors, have a large effect on disease risk and progression.
The challenge in applying NGS technologies to gain insight into complex human conditions, such as rheumatic diseases, lies in appreciating what questions can be meaningfully answered with these tools (BOX 1), including how to sift through the noise within the large data sets and effectively deduce the causal mechanisms of disease aetiology and pathogenesis. Given the rapid development of NGS technologies and the complexity of clinical data sets, collaborative teams of rheumatologists, molecular scientists and computational experts will be required to effectively couple NGS high-throughput genomic information with clinical information that can be used to elucidate the fundamentals of rheumatic disease pathology.
Box 1 |. Addressing challenges in rheumatic disease research using omics technologies.
Researchers working on rheumatic diseases are faced with several challenges, some of which can be addressed using omics technologies. Details of five such challenges are provided here.
Disease heterogeneity confounds precise diagnosis and results in variable responses to therapy
The application and integration of omics technologies for patients with similar clinical syndromes who respond differently to therapy might provide objective molecular data to stratify these patients into subsets on the basis of distinct underlying pathogenic mechanisms. This stratification could facilitate a precision medicine approach to treatment and might also provide molecular data to guide the interpretation of clinical trial results in which subgroups of patients respond differentially to a therapy.
The relative importance of genetics versus environment in rheumatic diseases is unclear
Environmental influences are difficult to measure, but as many epigenetic features are readily modifiable by environmental cues and can be stably maintained over time, epigenomic technologies might enable the assessment of environmental factors in pathogenesis.
Knowledge of the molecular drivers of pathogenesis is incomplete
Open-ended unbiased omics approaches could facilitate the discovery of novel pathogenic molecules and disease pathways.
The precise cellular drivers of pathogenesis are not well understood
Single-cell transcriptomics provides an unprecedented view of known and novel cellular phenotypes associated with disease states, which enables more precise identification and targeting of pathogenic cell types.
The mechanisms responsible for variability in responses to therapy are not well understood
Analysis of drug responses in vivo and in ex vivo organoid cultures using omics approaches could help to identify the basis for drug sensitivity and epigenetic mechanisms of drug resistance. Single-cell analysis of drug responses can provide insights into the mechanisms of action in different pathogenic cell populations and provide a rational basis for the use of combination therapies.
In this Review, we discuss three main omics NGS approaches — transcriptomics, epigenomics and genome sequencing — that are being applied to the study of rheumatic diseases (FIG. 1). These techniques have the potential to provide insight into the interaction of environmental and genetic factors in the pathogenesis of rheumatic diseases. We also discuss how whole-exome and whole-genome sequencing can reveal so-called missing heritability by uncovering rare mutations and can help researchers to identify somatic mutations in haematopoietic progenitors that promote autoimmunity and inflammation. The novel cell types and molecular pathways involved in the pathogenesis of rheumatic diseases that these omics approaches could potentially expose might lead to new therapeutic targets and strategies.
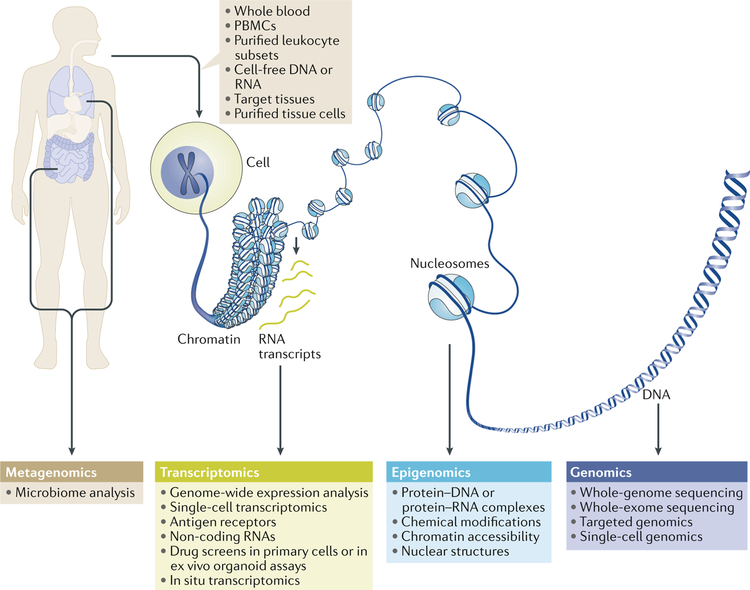
Fig. 1 |. Overview of next-generation sequencing applications.
Large amounts of transcriptomic, epigenomic, genomic and metagenomic sequencing data can be generated from an individual. Sources of nucleic acids for sequencing include cells obtained from blood or tissues and cell-free DNA or RNA. Microbiome analysis (metagenomics) can be performed on microorganisms from various tissues including the gut, skin and urogenital tract. Linking strong hypotheses with the most informative and robust techniques in carefully characterized cohorts of patients holds promise for potentially groundbreaking discoveries in rheumatic diseases. PBMCs, peripheral blood mononuclear cells.
Transcriptomics
The term transcriptomics encompasses several technologies that can be used for the high-throughput identification of RNA species. Genome-wide gene expression profiling using RNA sequencing (RNA-seq) has already been widely used to study rheumatic diseases, although its utility in the clinic awaits the identification of robust patterns of gene expression that meaningfully inform treatment decisions3. Whereas DNA sequencing can be used to identify the mutations that cause specific cancers and thus aid diagnosis and inform treatment options, the complex genetics and important role of environmental factors in rheumatic diseases render a particular need for approaches (including functional genomic assays such as RNA-seq) that measure facets of the active genome. By using such assays, researchers might uncover much-needed objective measures of flare episodes and treatment response profiles that could aid rheumatic disease stratification3.
Transcriptome analysis across all cells and tissues of the human body is orders of magnitude more complex than sequencing an individual’s genome, as each cell in the body possesses a unique transcriptional profile that can dynamically change in response to many triggers, such as diurnal cycles, ageing, food intake, exposure to microorganisms, exposure to inhalants and genetics. The extensive variability in clinical manifestations and the chronic nature of rheumatic diseases further complicate the interpretation of transcriptomic data sets4. Thus, as with the human genome and GWAS projects, large-scale consortia will undoubtedly be instrumental in generating meaningful transcriptomic data sets for rheumatic diseases. Rheumatologists should be encouraged to join such efforts and to communicate closely with the computational scientists who process these data sets using emerging technologies such as artificial intelligence. Thoughtfully and meticulously collected clinical information and continuous consultation between scientists and physicians who understand the nuances of clinical rheumatology will be instrumental to the successful interpretation of such data.
To capture meaningful transcriptomic patterns amidst this complexity, it will be important for some research projects to focus on specific cell subsets in a particular cohort of patients while others look broadly by conducting meta-analyses across thousands of independent transcriptomics studies. Complementing transcriptomics data with other high-dimensional technologies such as mass cytometry and high-throughput molecular imaging techniques will also be important. Technical challenges related to RNA integrity and quantity, which are a particular challenge when working with clinical samples, have been addressed by standardized viable preservation and dissociation protocols for tissues commonly affected in rheumatic diseases, such as synovium, kidney, skin and urine5–7. Together with improved protocols for robust low-input sequencing, the ability to analyse low-quality RNA holds great value, for example, by enabling analysis of gene expression in formalin-fixed paraffin-embedded samples8.
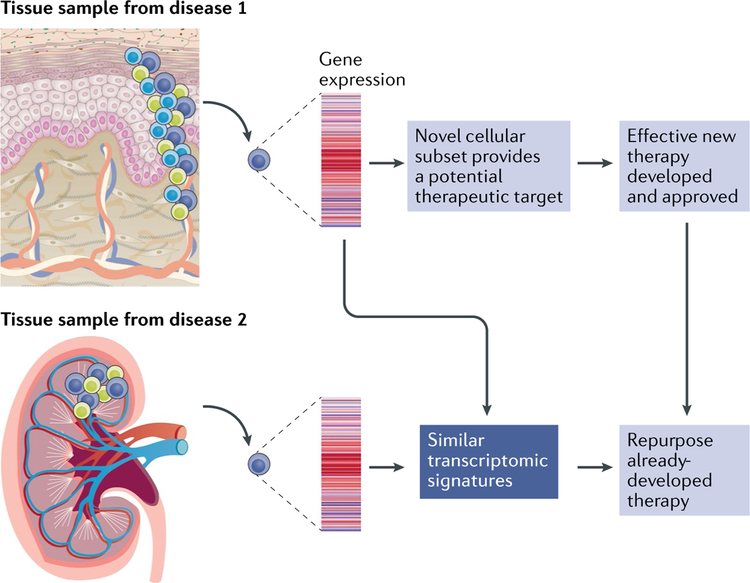
As with any discovery project that aims to implicate objective molecular patterns in human disease, deducing which patterns are causing, rather than reacting to, the pathology is challenging. To address causality in the human system, ex vivo organoid assays using primary cells from patients combined with genetics and disease-relevant pharmacologic perturbations will be a valuable approach. Such assays can then inform animal models that are used to dissect disease mechanisms relevant to the human patient. This approach is not only applicable to transcriptomic studies but also represents a valuable overall strategy for various omics, as well as hypothesis-driven approaches. With clear pathological pathways identified in various rheumatic diseases, the potential identification of pathological pathways shared between cell subsets in disparate rheumatic conditions could lead to the immediate and effective repurposing of approved medications for patients with rheumatic diseases (FIG. 2).
Fig. 2 |. Using transcriptomics to gain insight into rheumatic diseases and develop new therapeutics.
Single-cell transcriptomic techniques have already provided initial findings of novel cell types and activation states in tissues affected by rheumatic diseases. Further advances could include identification of an immune cell that might share a similar transcriptomic profile in distinct tissues and diseases, such as the kidney in lupus nephritis and the skin in systemic sclerosis, thereby providing support for evidence-based therapeutic targeting of the same pathway in distinct clinical conditions. Linking these findings with the most appropriate therapeutics will be the important next step, which could involve repurposing medications developed for other diseases that share similar molecular transcriptomic patterns.
Genome-wide gene expression analyses
Although microarray assays first introduced the power of profiling thousands of unique transcripts to rheumatic disease research9,10, NGS provides richer molecular information and greater scalability by capturing the expression of all known (and as yet unknown) genes, non-coding RNAs and alternatively spliced transcripts. The standard RNA-seq protocol involves generating a cDNA library that is sequenced as short fragments or reads, which are then computationally assigned to a gene by aligning their sequences to a reference genome and are ultimately tallied for abundance. The gene expression levels from samples are then compared to create lists of differentially expressed genes (DEGs)11, which can be further examined using pathway and network analysis programmes. Pathway databases computationally score lists of genes and their relative abundance against previously defined gene sets known to be involved in cellular pathways or physiological responses. Computational programmes have even begun to address how gene expression profiles relate to spatial organization of cells and tissues in vivo, and emerging new techniques have begun to be used to directly assay transcriptional signatures in situ, promising to provide insights into the intercellular networks that drive tissue function and physiological systems12.
Bulk gene expression analyses.
Transcriptomic analyses of mixed cell populations or whole tissue preparations are relatively easy to generate, even in clinical settings. Such work has provided evidence for specific gene signatures in neutrophils that are associated with cardiovascular disease in patients with systemic lupus erythematosus (SLE)13 and for plasma cell-related genes as robust markers of rheumatoid arthritis (RA) in synovial tissue14,15. However, the sequencing of complex mixtures of cells (synovial tissue samples or peripheral blood mononuclear cells) has been less effective at revealing robust gene expression signatures in patients with clear clinical differences or with different responses to treatment16,17. To increase the chance of detecting meaningful gene expression changes, different cell types have been purified from patient samples by fluorescent-activated cell sorting or other means and then analysed as distinct populations of cells18. For example, in a 2018 study, a sepsis-like signature was identified in neutrophils from patients with systemic juvenile idiopathic arthritis (sJIA) that decreased upon treatment with an IL-1 receptor antagonist19. In another study, macrophages were purified from synovial tissue samples from patients with RA or osteoarthritis (OA), and numerous gene modules were identified that correlated with disease activity and therapy20. The drawbacks of sequencing whole tissue fragments and mixed cell populations include difficulties in detecting changes in rare cell types, understanding which cell type is responsible for the detected changes in gene expression and knowing the extent to which changes in gene expression are attributable to differences in the composition of mixtures of cells. The latter limitation can be partially overcome by predictive deconvolution algorithms, and more definitively by cell sorting, but has also provided motivation for the development of alternative approaches such as single-cell sequencing.
Single-cell gene expression analyses.
Single-cell transcriptomics is an accessible, cutting-edge NGS approach, thanks in part to the development of technologies to capture and interrogate gene expression in single cells and to the open-access mindset of the bioinformatics community. The capacity to identify the transcriptomic profiles of thousands of individual cells in a sample provides the opportunity to tease apart how specific cell subtypes contribute to the expression patterns observed in mixed cell population transcriptomics. This approach also offers the potential to understand the variability in purified populations (which can be computationally interpreted for signs of directional differentiation among the cells) and, importantly, enables the discovery and confident classification of extremely rare cell types using high-dimensional data. The comprehensive and unbiased description of all cell types extracted from a given sample is possible with no a priori knowledge of the cellular composition of tissues; however, assigning an identity to a cell cluster defined by gene expression usually requires imputation on the basis of known profiles of cell type-specific gene expression. Another limitation of single-cell sequencing relates to the depth of sequencing possible — currently available technologies capture expression of only a few thousand of the most highly expressed genes per cell, which can make it difficult to segregate related cell types or to identify pathways or phenotypes comprising genes expressed at low levels. This limitation can be addressed by combining single-cell sequencing with bulk sequencing of sorted cell populations or with analysis of surface markers using DNA-barcoded antibodies, thereby providing a greater depth of coverage and potential insight into phenotype.
Outstanding reviews from 2018 have broadly highlighted single-cell RNA-seq in immunological studies21,22, and the accompanying Review by Cheung et al. provides a further focus on single-cell sequencing in rheumatic disease research23. However, it is worth mentioning the results of two studies24,25 that used single-cell RNA-seq on synovial tissue from patients with RA to reveal multiple new cell types and activation states in the primary tissue affected in this disease. In these studies24,25, three fibroblast subsets (one lining and two sublining) were defined by their transcriptional profiles, which could represent non-immune-system-related drug targets. An extensive enrichment of programmed cell death protein 1+ peripheral T follicular helper cells was also confirmed in RA synovium25, which had previously been identified by mass cytometry26.
Multidimensional analyses of gene expression data.
Transcriptomic profiling of patients with rheumatic diseases has been implemented in a large numbers of studies, and over 4,000 gene expression profiles have been deposited in public databases such as Gene Expression Omnibus. Sophisticated mathematical algorithms designed to coalesce data from these vast global studies could be used to generate insights into clinically relevant parameters such as responses to treatment27 in population-wide gene expression patterns in patients with rheumatic diseases, as has already been done for inflammatory bowel disease and sepsis28,29. Several large-scale consortia have already been established to aid the discovery and sharing of omics data relevant to rheumatic diseases. The Accelerating Medicine Partnership (AMP) consortium was created to identify promising biological targets directly from human tissues affected by RA and SLE using transcriptomics (sorted cell populations and single-cell analysis) and mass cytometry6,30,31. During development of the pipelines for high-dimensional analyses of samples from cohorts of patients undergoing different drug treatments, the team of rheumatologists, scientists and bioinformaticians in the AMP has discovered disease-associated immune and stromal cells in various activation states in synovium from patients with RA and kidney tissue from patients with lupus nephritis, critically reshaping our view of the pathological pathways involved in these rheumatic diseases6,30,31. Primary data from the AMP RA and SLE network are available through ImmPort. Other consortia such as PRECISESADS, The LifeTime Initiative, the Human Cell Atlas and the International Human Epigenome Consortium will also be instrumental in laying the foundation for investigation of the tissues and cell types relevant to autoimmune diseases.
Targeting specific RNA species
The basic RNA-seq protocol can be modified to capture specific RNA subtypes, including mRNA (by poly-A selection or ribosomal RNA depletion), nascent transcripts, nuclear RNA, chromatin-associated RNA and ribosome-associated RNA. Paired-end sequencing enables the genome-wide identification of alternatively spliced RNA isoforms that can generate multiple novel gene products within each cell. Sequencing can also be targeted to a specific type of gene, such as antigen receptor gene products, or to non-coding RNA species.
Molecules with some of the strongest associations with the rheumatic diseases are HLA haplotypes and autoantibodies; a large number of transcriptomic studies in rheumatic diseases have therefore used T cell receptor (TCR) and B cell receptor (BCR) sequencing. Whereas TCR and BCR sequences can be deduced from DNA sequencing, sequencing of antigen receptor recombination products is often performed using RNA as the starting template. For example, a 2018 study of TCR diversity in RA that used RNA-seq of the TCR β chains showed considerable overlap of dominant TCR clones within synovial compartments and between affected joints, implicating common and potentially therapeutically targetable lymphocytes in the pathogenesis of RA32. In another study, this technology was used to provide evidence that, in the blood of patients with RA, HLA-DRB1 shared epitope alleles associate with a reduced diversity of the TCR repertoire33.
Regarding BCRs and antibodies, a single-cell sequencing study showed that circulating IgA+ plasma-blasts (present at high concentrations in individuals at high risk of developing RA) had shared antibody repertoires with IgG+ plasmablasts, which the authors suggest implicates a mucosal immune reaction in the onset of RA34. In another study, NGS was used with blood from patients with SLE to identify somatic mutations in anti-nuclear antibody sequences that increased the affinity of the antibodies for DNA35. Several groups have integrated antigen–tetramer isolation techniques with single-cell BCR sequencing to define continuous affinity maturation of anti-citrullinated protein antibody (ACPA)-producing and rheumatoid factor-producing plasmablasts, linking these findings to ACPA epitope spreading and inherent phenotypic differences between rheumatoid factor-producing and ACPA-producing plasmablasts36–38. Furthermore, in a study on primary Sjögren syndrome (pSS), researchers combined proteomic mass spectrometry of rheumatoid factors with immunoglobulin heavy chain repertoire sequencing to identify antibody clonotypes that then served as biomarkers for the severity and course of cryoglobulin-associated disease39. Emerging techniques that link antigen receptor sequencing with high-throughput antigen binding assays and single-cell transcriptomics promise to increase our understanding of autoantigens and their cognate antibodies, as well as TCRs and the cells that produce them, in rheumatic diseases40.
Epigenomics
Epigenetic control of the genome occurs at many levels, including the covalent modification of DNA-associated proteins such as histones, the direct chemical modification of DNA and changes in chromatin accessibility and higher order structure41,42. The term epigenome, also known as the epigenomic landscape, refers to the genome-wide pattern of chromatin accessibility, DNA modification, transcription factor binding and chromatin-modifying enzyme binding that is unique to each cell type. Such epigenetic patterns are plastic during inflammatory processes, and epigenetic regulation is thought to be important in inflammation and in disease states43–45; thus, it is important to understand how epigenetic changes can contribute to rheumatic diseases. Insights into cellular epigenetic landscapes can be gained through the use of high-throughput, genome-wide methods to analyse chromatin accessibility, histone modification, transcription factor occupancy and DNA methylation45,46. Such methods include bisulfite sequencing (BS-seq), which measures DNA methylation, and DNase I hypersensitive sites sequencing (DNase-seq), formaldehyde-assisted isolation of regulatory elements with sequencing (FAIRE-seq) and assay for transposase-accessible chromatin using sequencing (ATAC-seq), which reveal nucleosome-depleted regions known as open chromatin. By contrast, chromatin immunoprecipitation sequencing (ChIP–seq) uses antibodies specific for histone marks or transcription factors to reveal genome-wide patterns of histone modifications, which can promote, repress or stabilize gene expression, and transcription factor occupancy.
Studying histone modification
Genome-wide epigenetic analysis of histone modifications and chromatin accessibility in patients with rheumatic diseases is still at an early stage, although epigenetic changes in disease-relevant genes have been detected in some rheumatic diseases47. For example, gene enhancers are altered in monocytes from patients with SLE compared with monocytes from healthy individuals48, as are histone H3 trimethylation at lysine 4 (H3K4me3) patterns, particularly at elements associated with interferon signalling49. Changes also exist in chromatin accessibility at B cell activation genes in B cells from patients with SLE undergoing flare50. Profiling of synovial fibroblasts from patients with RA for various histone modifications (such as H3K4me3, histone H3 acetylation at lysine 27 (H3K27ac), histone H3 monomethylation at lysine 4 (H3K4me1) and histone H3 trimethylation at lysine 36 (H3K36me3)) and for open chromatin has revealed differences to synovial fibroblasts from patients with OA, as well as an association of epigenomic marks with active enhancers and promoters of immune-related genes51. Although such studies represent an important step forward for the use of epigenomic analysis for rheumatic diseases, considerable challenges remain in demonstrating a causal link between epigenetic marks and gene expression patterns that define cell activation states and phenotypes.
An important epigenetic concept to be considered in the dysregulated inflammatory responses in rheumatic diseases is the establishment of memory-like cellular phenotypes that can either promote or inhibit the responses of innate immune cells to environmental stimuli52. Specifically, the exposure of innate immune cells to certain triggers such as microbial products results in epigenetic imprinting that manifests as either a state of immunological tolerance53 or of heightened responsiveness, termed trained immunity54. Tolerant macrophages have less-accessible chromatin and fewer active histone marks (such as H3K4me3 and H3K27ac) at the promoters and enhancers of inflammation-related genes55,56, as well as a defect in Toll-like receptor (TLR) signalling53 compared with naive unstimulated macrophages. By contrast, trained monocytes gain active histone marks at distal regulatory elements; upon re-stimulation, these genomic regions are readily acetylated and rapidly and robustly transcribed54.
Both tolerance and trained immunity represent clinically relevant functional states, and the development of innate memory is relevant to inflammation52,53. Pertinent to autoimmunity, microbiome-derived products can differentially affect the tolerization of innate immune cells and the emergence of autoimmune diseases such as diabetes57, and exposure to type I interferons can abolish macrophage tolerance in a chromatin-dependent manner55,58. Trained immunity can also counteract tolerance-related epigenetic changes56 and thereby contribute to the nonspecific protective effects of vaccination and affect susceptibility to subsequent infections. A 2018 study59 showed that training or tolerization of innate immune cells in the periphery led to the stable epigenetic reprogramming of microglia in the brain, which in turn affected the development of disease in mouse models of Alzheimer disease and stroke. The results of this study59 support the concept that environmental stimuli at distal sites can affect inflammatory responses in distinct tissues, thereby contributing to pathogenesis. It will be important to determine whether defects in establishing tolerance and trained immunity contribute to rheumatic diseases. An additional possibility is that training-induced epigenetic changes that stabilize inflammation-related gene expression might contribute to resistance to therapies that target upstream signalling events. Overall, tolerance and trained immunity could be representative examples of a pervasive phenomenon of epigenetic conditioning by environmental challenges, which might control several fundamental features of rheumatic disease pathology.
Studying the chromatin landscape
Numerous studies have shown that the majority of disease-associated allelic variants fall outside of protein-coding sequences and instead lie in cis-regulatory regions such as gene enhancers60–64, including common autoimmune disease-associated polymorphisms65. SLE-associated polymorphisms in HLA-DR, HLA-DQ and BANK1 loci have been implicated as expression quantitative trait loci (eQTLs) that control gene expression66,67. As few disease-associated eQTLs have been mapped relative to the large numbers of disease-associated single nucleotide variants discovered, it is challenging to assign distal enhancers that harbour disease-associated allelic variants to their target genes and to determine causal relationships between these variants, gene expression and disease states. Advances that have begun to address causal links between the chromatin landscape and functional gene expression include the development of techniques to study chromatin conformation and looping42 and CRISPR–Cas9-mediated genome editing68. Techniques that map chromatin interactions such as Hi-C42 have been combined with ChIP to map chromatin contact points that associate with specific proteins69. In this process, known as HiChIP, interacting DNA fragments are covalently linked before ChIP analysis is performed to detect particular histone modifications or chromatin-associated proteins. The detection of enhancer–promoter interactions enables the identification of target genes for a particular enhancer. HiChIP has so far been used to generate high-resolution maps of enhancer–promoter contacts in primary human T cells and to map the target genes of enhancers that harbour autoimmune disease-associated allelic variants69. This approach will provide a valuable framework for mapping distal cis-regulatory regions and disease-associated variants for target genes in rheumatic diseases.
Genome editing technologies such as the CRISPR–Cas9 system enable manipulation of the epigenome to identify the effect of epigenetic patterns on cellular function68. This approach has been used to support the auto-immune disease-relevant importance of an enhancer in the TNFAIP3 locus70. Nuclease-deficient Cas9 fused with functional domains of the epigenetic regulators KRAB, TET1 and p300 has also been used to target these proteins to specific regulatory regions to determine the role of particular epigenetic patterns in gene expression and cell function71. Overall, the CRISPR–Cas9 system could be used to help identify which genes are regulated by disease-associated enhancers and which epigenetic mechanisms regulate gene expression in rheumatic diseases. Such epigenetic mechanisms could potentially be therapeutically targeted to treat autoimmune and inflammatory diseases.
Studying DNA methylation
DNA methylation involves the addition of a methyl group to a cytosine residue by DNA methyltransferases (DNMTs), most commonly at cytosine–guanine (CpG) dinucleotide locations (known as CpG islands)72. Typically, DNA methylation in promoter CpG islands blocks transcription and promotes gene silencing73,74, whereas variable effects occur when CpG islands in gene bodies, introns or enhancers are methylated75. DNA methylation has been extensively analysed in an attempt to uncover the underlying epigenetic mechanisms that control rheumatic diseases. A typical workflow involves identifying differentially methylated regions (DMRs) and associating them with genes. These genes are then annotated with known pathways and biological activities that can either confirm known functions or reveal unexpected functions of relevance for the disease being studied51. Transcription factor motif enrichment analysis can be included as part of the workflow to enable the discovery of motifs that potentially characterize DMRs. The ultimate goal of DNA methylation studies in rheumatic diseases is to compare epigenetic signatures across diseases and disease stages and to identify candidate disease biomarkers.
NGS technologies are aiding epigenomics research by enabling capture of the distribution of DNA methylation across the genome76,77. The best-established NGS-based experimental protocols for detecting methylated cytosines across the genome are BS-seq and whole-genome bisulfite sequencing (WGBS)78–80. A variation of these techniques is reduced representation bisulfite sequencing (RRBS)81, in which only a CpG-rich fraction of the genome is sequenced. RRBS typically ensures isolation of ~85% of CpG islands in the human genome82 and is the method of choice when the high costs of WGBS would compromise sequencing coverage. NGS and array platforms for DNA methylation studies have been reviewed eleswhere82.
Before NGS technologies were available, array-based profiling approaches were often used for genome-wide DNA methylation studies, such as the Illumina Infinium HumanMethylation450K BeadChIP and its successor, the MethylationEPIC kit82,83. Chip-based DNA methylation studies showed that methylation patterns are distinct in cultured synovial fibroblasts from patients with RA compared with those from patients with OA and from a control cell line83. In a follow-up study, differentially methylated genes were identified that participate in RA-related pathways and a DNA methylation signature was confirmed in synovial fibroblasts from patients with RA that remained unaltered across passages and between replicates84. Two more studies addressed the question of whether DNA methylation patterns change as a result of disease progression and duration85 or differ between joints51. DNA methylation patterns in synovial fibroblasts from patients with early RA differed from those from patients with longstanding RA, suggesting that DNA methylation changes occur early in the disease process and change over time85. Different joints also seemed to have distinct methylation signatures51. An integrated transcriptomic and epigenomic analysis of synovial fibroblasts obtained from different anatomical sites showed joint-specific DNA methylation, chromatin marks and transcription that was associated with differential expression of homeobox genes, which confer positional memory along the body axis86. Thus, joint-specific epigenomic features can contribute to distinct synovial fibroblast functions and help explain the differential involvement of various joints in RA.
Synovial fibroblasts have also been compared at the epigenomic level with peripheral blood cells from patients with RA, resulting in an RA-associated methylation signature of hypermethylated loci that is similar in blood cells and synovial fibroblasts87; if this blood cell methylation pattern is found to differ substantially from healthy individuals, it could potentially function as an RA biomarker. Along the same lines, distinct DNA methylation profiles in peripheral blood cell subpopulations were associated with different clinical outcomes in patients with RA, suggesting that specific DNA methylation changes might serve as predictors of disease progression and severity88. In SLE, genome-wide DNA methylation studies have revealed DMRs that are associated with the production of clinically relevant SLE-related autoantibodies89 and with nephritis90, findings that suggest potential relationships between DNA methylation patterns and components of the clinical spectrum of SLE. Additionally, a genome-wide study of salivary gland tissue from 28 women with pSS identified a hypomethylated DMR signature associated with inflammation-related genes91.
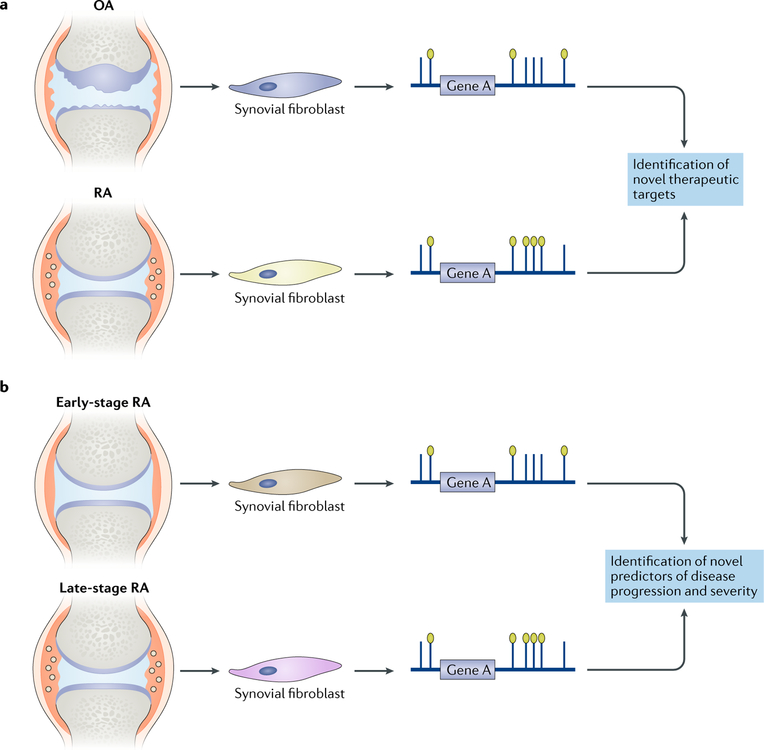
Notably, a 2018 genome-wide, multi-omics study to characterize the epigenomic landscape of synovial fibroblasts from patients with RA demonstrated the potential of NGS to identify unanticipated therapeutic targets51. The integrative use of WGBS with other NGS technologies to study RNA expression, patterns of open chromatin and histone modifications unexpectedly revealed ‘Huntington disease signalling’ as the most enriched biological pathway in synovial fibroblasts from patients with RA compared with those from patients with OA. Further analysis of the genes in this pathway suggested that HIP1, a gene that is particularly prominent in this pathway and has been previously associated with cell movement in cancer, has a role in the invasion of synovial fibroblasts into cartilage. This study51 represents an example of how a genome-wide and unbiased multidimensional view of synovial fibroblasts from patients with RA at the epigenomic level can lead to the discovery of novel potential therapeutic targets (FIG. 3).
Fig. 3 |. Using epigenomics to gain insight into rheumatic diseases.
a | Epigenomic analysis of synovial fibroblasts has already provided insights into mechanisms that distinguish cells from different anatomic locations, and DNA methylation analysis has identified differences between synovial fibroblasts in osteoarthritis (OA) and rheumatoid arthritis (RA) that might lead to the identification of novel therapeutic targets. b | Differences in DNA methylation have been detected between synovial fibroblasts in early-stage and late-stage RA and might be important for prognosis. As epigenomic changes are often stable, they can confer transcriptional memory and reflect environmental influences on pathogenesis.
Genome sequencing
The advent of NGS technologies (BOX 2) has revolutionized the discovery of pathogenic genetic variants, such as variants in rare inherited diseases, de novo variants in genetic diseases with sporadic onset and somatic mosaicism as a cause of disease. Before NGS technologies were available, the identification of disease-causing genes required the laborious process of positional cloning, linkage analysis92 and Sanger sequencing and often took years to uncover an underlying causative gene. Such was the discovery of mutations in the MEFV gene as the genetic cause of familial Mediterranean fever93. More than 20 years later, NGS of DNA has eclipsed these methods and improved the scope and speed of mutation discovery; diagnostic exome sequencing is now a commercial commodity costing less than US$1,000 per sample and taking just a few weeks to return results.
Box 2 |. Next-generation sequencing technology.
The term next-generation places the current technologies in contrast with the preceding Sanger and Maxam-Gilbert sequencing methods. Next-generation sequencing (NGS) technologies can perform unbiased sequencing of ~104–108 nucleotide stretches in parallel and hence are often referred to as massively parallel, high-throughput or high-dimensional sequencing. The most widely used technology from Illumina uses fragmented DNA or cDNA, and the transcribed product of each fragment is referred to as a read. Several NGS platforms use optical detection of light-emitting tags on nucleotides as a readout, which are added as single strands and transcribed by a polymerase. Other technologies use ionic disturbances as nucleotides are added as a readout, whereas Nanopore stands alone as a novel technology in which long stretches of DNA or RNA are funnelled through a biological pore while the electrical output from each nucleotide is measured. Details of some of the most commonly used NGS platforms are provided in the table below.
| NGS platform | Input | Readout | Advantages and/or disadvantages |
|---|---|---|---|
| Illumina | • Short fragments of DNA • cDNA |
Polymerase with an optical readout | • Low error rate (2%) • Widely used |
| Ion Torrent | • Short fragments of DNA • cDNA |
Ionic disturbance readout | • Fast readout • Low error rate (2%) |
| PacBio | • Long fragments of DNA • RNA |
Polymerase with an optical readout | • Low error rate (<1%) on multiple passes • Real time • Fast |
| Nanopore | • Long fragments of DNA • RNA |
Protein pore with an electrical readout | • Real time • Fast • High error rate (13%) |
| Chromium 10×: linked reads | • Long fragments of DNA • RNA |
Polymerase with an optical readout | • Uses molecular barcodes to tag reads that come from the same long DNA fragments • Uses Illumina-based short fragment sequencing |
Exome sequencing of affected individuals and their relatives provides an efficient and unbiased method to identify pathogenic mutations by screening for coding variants. A typical workflow involves sequencing the DNA of affected and unaffected individuals in a family and then using various inheritance models, such as autosomal dominant, recessive and X-linked, to filter the results for mutations correlating with disease status. Large families help to facilitate the identification of pathogenic variants, but even the sequencing of trios consisting of two parents and a child can lead to the identification of mutations94, particularly in early-onset severe disease in which there is a de novo pathogenic mutation in the affected child. A combined sequencing and family study approach led to the discovery of haploinsufficiency of TNF-induced protein 3 (also known as A20) as a result of loss-of-function mutations in TNFAIP3, which caused an autosomal dominant earlyonset inflammatory syndrome resembling Behçet syndrome95. The TNFAIP3 mutations identified in the first two families in this study95 were discovered by sequencing DNA from affected and unaffected family members. Comparison of candidate rare variants between the two families yielded TNFAIP3 as the only commonly mutated gene. Subsequently, targeted sequencing of TNFAIP3 exons in similarly affected individuals identified three additional families with loss-of-function mutations95.
As whole-genome sequencing becomes comparable in cost to exome sequencing, its use as a discovery tool in Mendelian disease is becoming more feasible. Although the sequencing depth is lower (typically 30×coverage for whole-genome sequencing versus 50–100× coverage for exome sequencing), coverage is more uniform across genomic regions in whole-genome sequencing owing to the absence of hybridization-based selection of specific sequences, making possible the detection of mutations in regulatory regions and non-coding RNA, as well as structural variations96. In a survey of Mendelian disease-causing variants curated from the Online Mendelian Inheritance in Man (OMIM) and ClinVar databases, 45% were found to be non-coding97, suggesting that whole-genome sequencing has the potential to identify disease-causing variants outside of the exome. The importance of non-coding variation is emerging in both Mendelian and complex immune-mediated disorders, such as the 2018 discovery of a structural variant in the 5ʹ-untranslated region of RAB27A that is associated with late-onset haemophagocytic lymphohistiocytosis98.
Whereas the underlying cause of rare diseases can often be pinpointed to a single gene, the genetic causes of complex autoimmune disorders such as SLE remain elusive. Novel technologies have helped to advance knowledge in this field, for example, in the investigation of rare variants that might be important in SLE susceptibility. Mutations in TREX1 that occur in patients with the rare, early-onset inflammatory disease Aicardi–Goutieres syndrome (which resembles SLE in certain features) have also been found by exome sequencing in ~0.5% of patients with SLE99. Similarly, targeted sequencing studies that focused on loci previously identified by GWAS have aided our understanding of the connection between associated genes and disease by capturing rare variants that were undetected by GWAS. Several rare variants associated with inflammatory bowel disease have been identified in this way100,101.
Somatic mosaic mutations are a class of pathogenic mutations that can be missed by standard exome and genome sequencing and that are only now being detected. Somatic mutations are not present in the germ line and instead arise in tissue cells, including stem and progenitor cells, after birth and as such are present in only a subset of cells, resulting in a mosaic phenotype in adult organisms. A subset of patients with symptoms that are indistinguishable from those of Mendelian auto-inflammatory diseases, such as TNF receptor-associated periodic syndrome (TRAPS), neonatal-onset multisystem inflammatory disease (NOMID) and autoimmune lymphoproliferative syndrome (ALPS), lack a germline genetic diagnosis. For some of these patients, somatic mosaicism of the pathogenic gene can cause a similar or milder phenotype that mimics the germline disorder. For example, in patients with cryopyrinopathies related to mutations in NLRP3, embryonic and haematopoietic lineage-restricted somatic mutations in NLRP3 have been identified that have variant allele frequencies as low as 5%102–104. Somatic mutations in TNFRSF6 (also known as FAS) that are restricted to lymphocyteshave also been identified in patients with ALPS105, which is commonly caused by dominant negative germline mutations in this gene. Interest in the role of somaticmutations in complex autoimmune diseases is also increasing; a 2017 study showed the presence of somatic mutations in various immune-related genes in clonally expanded cytotoxic T cells from patients with RA106. However, further work is needed to understand the functionalimplications of such mutations, and similar studies are underway in other autoimmune diseases.
Previously, somatic mutations were identified by the labour-intensive process of subcloning and Sanger sequencing, but somatic mimics of germline syndromes have now been discovered as a result of targeted sequencing or exome sequencing studies, which have identified such mutations in TNFR1 (also known as TNFRSF1A) and NLRC4, thus expanding the spectrum of mosaic mutations known to cause autoinflammatory diseases107,108. Exome sequencing enables the detection of novel somatic mutations when paired samples of affected and unaffected tissues are used, but the current limit of detection is an allele frequency of ~5%. Targeted sequencing technologies such as multiplexed PCR amplicon sequencing and hybridization-based enrichment of selected genes109 enable sequencing at a high depth, which lowers the limit of detection. Careful selection of the cell types in the input sample is crucial to enable detection of mutations in the target cell population, depending on its abundance. Single-cell sequencing of DNA and RNA could also aid the identification of somatic mutations in rare populations of cells.
Future perspectives
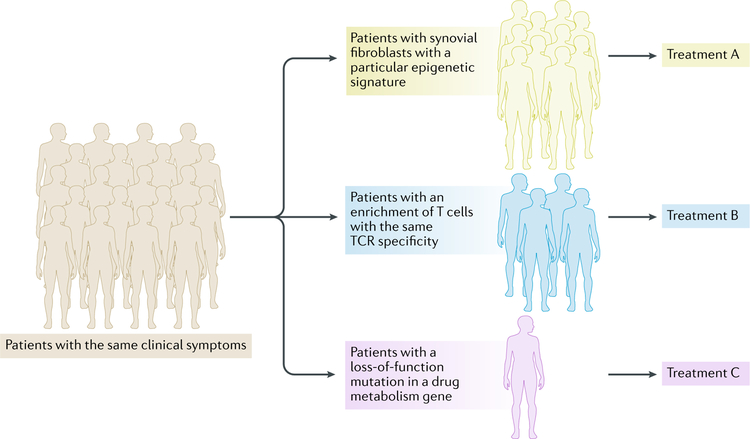
Rheumatologists have long appreciated that patients with similar clinical syndromes and the same diagnosis can fall into distinct subsets. Precision medicine relies on the idea that accurately dividing patients into subsets according to pathogenic mechanisms and molecular contributors to disease will enable the selection of personalized, and therefore more effective, targeted therapies. The NGS approaches described in this Review expand our ability to characterize individual patients beyond current efforts that mainly rely on careful clinical phenotyping, blood tests and imaging. The application of NGS technology to precision medicine has already been successful in oncology, in which the sequencing of tumour DNA has identified causal mutations that can be therapeutically targeted110. This approach is more challenging in rheumatic diseases, in which the genetics are complex and it seems unlikely that mutations in individual genes are predominantly causing pathogenesis (unless somatic mutation is more common than currently appreciated). One emerging approach is the use of transcriptomics to identify gene expression signatures, molecular pathways or pathogenic cell types that are associated with, and possibly contribute to the pathogenesis of, disease in individual patients (FIG. 4). The use of epigenomics could also lead to the identification of disease pathways that are either activated by environmental factors or related to allelic variation in gene regulatory elements. The identification of stable and persistent chromatin or DNA methylation changes can also yield insight into why some patients are resistant to therapies that target upstream signalling pathways. It is our hope that the precise phenotyping of patients and identification of pathogenic pathways using NGS approaches will lead to personalized therapeutic strategies.
Fig. 4 |. Next-generation sequencing for precision medicine.
Objective molecular data generated using next-generation sequencing approaches can aid in the stratification of patients with similar or shared clinical features. Thus, patients could be stratified on the basis of epigenomic signatures, transcriptomic profiles or genetically determined gene expression differences. This information could inform clinicians as to the drugs that are most likely to target these patterns in subsets of patients, provide a rationale for combination therapies and/or provide insights for disease aetiology. TCR, T cell receptor.
In the future, it will be important to use NGS technologies in an integrated manner to gain novel and deeper insights into rheumatic diseases (BOX 3). For example, it has become clear that crosstalk between genes and the environment can be deciphered at the molecular level by analysing epigenetic changes and mechanisms. Thus, it should be possible to use epigenomic analysis to understand the molecular mechanisms that integrate the effects of nature and nurture in rheumatic disease pathogenesis and progression. Another fertile area for future research is integrated transcriptomic and epigenomic analysis of single cells, which will enable the deconvolution and identification of signals, pathways, cell types and pathogenic mechanisms that are not detected in the analysis of mixed cell populations or whole tissues. It will be important to extend transcriptomic analysis to various types of non-coding RNAs, which can have stable epigenetic effects and shape immune cell function. The expansion of whole-exome sequencing and whole-genome sequencing efforts will also help to resolve the important question of the magnitude of the contribution of rare variants or even mutations to disease, thus bringing greater clarity to the genetics of rheumatic diseases and possibly providing an explanation for missing heritability.
Box 3 |. The integration of omics technologies.
The integration of data obtained using distinct omics technologies is computationally challenging but can help overcome the weaknesses in individual approaches and provide deeper mechanistic insight into the pathogenesis of rheumatic diseases.
Integration of bulk sequencing data and single-cell sequencing data
Single-cell sequencing requires no a priori knowledge of cell populations within a tissue and provides insights into the heterogeneity of cell populations and subpopulations and the relationships between different types of cells but is limited to the detection of highly expressed genes. Greater insight into cell phenotypes and disease pathways can be obtained by integrating single-cell sequencing with deep sequencing of sorted cell populations.
Integration of mRNa data with protein expression data
mRNA expression is not necessarily reflected at the protein level. This limitation can be overcome by integrating RNA sequencing with high-dimensional protein expression data, such as those obtained by mass cytometry. Technologies such as RNA expression and protein sequencing (REAP-seq) and cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) incorporate cell surface protein-specific oligonucleotide-coupled antibodies into single-cell sequencing approaches to simultaneously obtain mRNA and protein expression data.
Integration of epigenomic data and transcriptomic data
Analysis of correlations between epigenomic changes and gene expression data can provide support for causal relationships between epigenetic mechanisms and gene expression. For example, an increased occupancy of a transcription factor at a regulatory region of a particular gene, as detected by chromatin immunoprecipitation sequencing (ChIP–seq), that associates with an increase in mRNA expression of the gene would suggest that the transcription factor might directly increase the expression of the gene.
Integration of DNa sequencing data with epigenomic data and transcriptomic data
The integration of genomic, transcriptomic and epigenomic data provides insights into how disease-associated mutations and allelic variants affect the epigenome and can lead to the identification of causal variants that are linked with pathogenic gene expression.
Conclusions
In conclusion, NGS technologies have rapidly advanced and are being used by teams of rheumatologists, molecular scientists and computational biologists to address important outstanding questions in rheumatology about disease aetiology, pathogenesis and prognosis, including the role of genes versus the environment and the variability in disease phenotypes, the course of disease over time and responses to therapy. One, as yet underappreciated, potential explanation for the variability in disease phenotypes and severity, especially in patients with lateonset disease, is somatic mutation in haematopoietic progenitor cells that undergo clonal expansion with age or in response to environmental challenges or antigens; advances that increase the sensitivity of somatic mutation detection will clarify whether somatic mosaicism is an important mechanism of disease. The knowledge gained from the integrated application of NGS technologies, coupled with careful and detailed analysis of individual patients, will enable the development of a precision medicine approach that links therapies to molecular contributors to disease and that provides insight into combination therapies that can induce remission. Omics studies are likely to shed light on additional important questions such as sex bias in autoimmune disease. NGS approaches can help researchers to address the remaining roadblocks to a mechanistic understanding of complex rheumatic diseases, which will undoubtedly reveal new pathogenic pathways and provide novel therapeutic targets.
Key points.
Next-generation sequencing (NGS) technologies have the potential to provide insight into the interaction between environmental factors and genetics in the pathogenesis of rheumatic diseases.
Transcriptomic studies have revealed disease-related pathways and novel pathogenic cell types in rheumatic diseases.
Epigenomic studies have revealed memory-related phenomena that might help to explain the chronicity of disease and have linked enhancers harbouring disease-associated allelic variants with target genes.
Whole-genome sequencing and exome sequencing have revealed causal mutations in rare Mendelian autoinflammatory diseases.
NGS approaches will substantially contribute to the application of precision medicine in rheumatology.
Acknowledgements
The work of L.T.D., L.B.I. and K.-H.P.-M. was supported by grants from the US National Institutes of Health (NIH).
Footnotes
Competing interests
R.M.S. declares that he is an employee of Novartis. The other authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reviewer information
Nature Reviews Rheumatology thanks P. Gaffney and the other anonymous reviewers for their contribution to the peer review of this work.
Related links
AMP RA and SLE network: https://amp-ralupus.stanford.edu/about/ra-lupus-amp-project/
Gene Expression Omnibus: https://www.ncbi.nlm.nih.gov/geo/
Human Cell Atlas: https://www.humancellatlas.org/
ImmPort: https://www.immport.org/home/
International Human Epigenome Consortium: http://ihec-epigenomes.org/welcome/
PRECISESADS: http://www.precisesads.eu/
The LifeTime initiative: https://lifetime-fetflagship.eu/
References
- 1.Banchereau R, Cepika AM, Banchereau J & Pascual V Understanding human autoimmunity and autoinflammation through transcriptomics. Annu. Rev. Immunol 35, 337–370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ermann J, Rao DA, Teslovich NC, Brenner MB & Raychaudhuri S Immune cell profiling to guide therapeutic decisions in rheumatic diseases. Nat. Rev. Rheumatol 11, 541–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byron SA, Van Keuren-Jensen KR, Engelthaler DM, Carpten JD & Craig DW Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat. Rev. Genet 17, 257–271 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis MM & Brodin P Rebooting human immunology. Annu. Rev. Immunol 36, 843–864 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donlin LT et al. Methods for high-dimensional analysis of cells dissociated from cryopreserved synovial tissue. Arthritis Res. Ther 20, 139 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Der E et al. Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat. Immunol (in the press). [DOI] [PMC free article] [PubMed]
- 7.Rao DA et al. A protocol for single-cell transcriptomics from cryopreserved renal tissue and urine for the Accelerating Medicine Partnership (AMP) RA/SLE network. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/275859v1 (2018).
- 8.Eikrem O et al. Transcriptome sequencing (RNAseq) enables utilization of formalin-fixed, paraffinembedded biopsies with clear cell renal cell carcinoma for exploration of disease biology and biomarker development. PLOS ONE 11, e0149743 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banchereau R et al. Personalized immunomonitoring uncovers molecular networks that stratify lupus patients. Cell 165, 1548–1550 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Dennis G Jr. et al. Synovial phenotypes in rheumatoid arthritis correlate with response to biologic therapeutics. Arthritis Res. Ther 16, R90 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa-Silva J, Domingues D & Lopes FM RNA-Seq differential expression analysis: An extended review and a software tool. PLOS ONE 12, e0190152 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlucci PM et al. Neutrophil subsets and their gene signature associate with vascular inflammation and coronary atherosclerosis in lupus. JCI Insight 3, 99276 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole S et al. Integrative analysis reveals CD38 as a therapeutic target for plasma cell-rich pre-disease and established rheumatoid arthritis and systemic lupus erythematosus. Arthritis Res. Ther 20, 85 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orange DE et al. Identification of three rheumatoid arthritis disease subtypes by machine learning integration of synovial histologic features and RNA sequencing data. Arthritis Rheumatol 70, 690–701 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walsh AM et al. Triple DMARD treatment in early rheumatoid arthritis modulates synovial T cell activation and plasmablast/plasma cell differentiation pathways. PLOS ONE 12, e0183928 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuppen BVJ et al. RNA sequencing to predict response to TNF-alpha inhibitors reveals possible mechanism for nonresponse in smokers. Expert Rev. Clin. Immunol 14, 623–633 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Teitsma XM et al. Identification of differential co-expressed gene networks in early rheumatoid arthritis achieving sustained drug-free remission after treatment with a tocilizumab-based or methotrexate-based strategy. Arthritis Res. Ther 19, 170 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ter Haar NM et al. Reversal of sepsis-like features of neutrophils by interleukin-1 blockade in patients with systemic-onset juvenile idiopathic arthritis. Arthritis Rheumatol 70, 943–956 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Mandelin AM 2nd et al. Transcriptional profiling of synovial macrophages using minimally invasive ultrasound-guided synovial biopsies in rheumatoid arthritis. Arthritis Rheumatol 70, 841–854 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giladi A & Amit I Single-cell genomics: a stepping stone for future immunology discoveries. Cell 172, 14–21 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Landhuis E Single-cell approaches to immune profiling. Nature 557, 595–597 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Cheung P, Khatri P, Utz PJ & Kuo AJ Single-cell technologies — studying rheumatic diseases one cell at a time. Nat. Rev. Rheumatol 10.1038/s41584-019-0220-z (2019). [DOI] [PMC free article] [PubMed]
- 24.Mizoguchi F et al. Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat. Commun 9, 789 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephenson W et al. Single-cell RNA-seq of rheumatoid arthritis synovial tissue using low-cost microfluidic instrumentation. Nat. Commun 9, 791 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao DA et al. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature 542, 110–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim TH, Choi SJ, Lee YH, Song GG & Ji JD Gene expression profile predicting the response to anti-TNF treatment in patients with rheumatoid arthritis; analysis of GEO datasets. Joint Bone Spine 81, 325–330 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Gaujoux R et al. Cell-centred meta-analysis reveals baseline predictors of anti-TNFα non-response in biopsy and blood of patients with IBD. Gut 68, 604–614 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sweeney TE et al. Unsupervised analysis of transcriptomics in bacterial sepsis across multiple datasets reveals three robust clusters. Crit. Care Med 46, 915–925 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang F et al. Defining inflammatory cell states in rheumatoid arthritis joint tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol (in the press). [DOI] [PMC free article] [PubMed]
- 31.Arazi A, R. D. et al. The immune cell landscape in kidneys of lupus nephritis patients. Nat. Immunol (in the press). [DOI] [PMC free article] [PubMed]
- 32.Musters A et al. In rheumatoid arthritis, synovitis at different inflammatory sites is dominated by shared but patient-specific T cell clones. J. Immunol 201, 417–422 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Sakurai K et al. HLA-DRB1 shared epitope alleles and disease activity are correlated with reduced T cell receptor repertoire diversity in CD4+T cells in rheumatoid arthritis. J. Rheumatol 45, 905–914 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Kinslow JD et al. Elevated IgA plasmablast levels in subjects at risk of developing rheumatoid arthritis. Arthritis Rheumatol 68, 2372–2383 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakakibara S et al. Clonal evolution and antigen recognition of anti-nuclear antibodies in acute systemic lupus erythematosus. Sci. Rep 7, 16428 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu DR et al. T cell-dependent affinity maturation and innate immune pathways differentially drive autoreactive B cell responses in rheumatoid arthritis. Arthritis Rheumatol 70, 1732–1744 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elliott SE et al. Affinity maturation drives epitope spreading and generation of proinflammatory anti-citrullinated protein antibodies in rheumatoid arthritis. Arthritis Rheumatol 70, 1946–1958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Titcombe PJ et al. Pathogenic citrulline-multispecific B cell receptor clades in rheumatoid arthritis. Arthritis Rheumatol 70, 1933–1945 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang JJ et al. Molecular profiling and clonal tracking of secreted rheumatoid factors in primary Sjogren’s syndrome. Arthritis Rheumatol 70, 1617–1625 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Gee MH et al. Antigen identification for orphan T cell receptors expressed on tumor-infiltrating lymphocytes. Cell 172, 549–563 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allis CD & Jenuwein T The molecular hallmarks of epigenetic control. Nat. Rev. Genet 17, 487–500 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Rivera CM & Ren B Mapping human epigenomes. Cell 155, 39–55 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alvarez-Errico D, Vento-Tormo R, Sieweke M & Ballestar E Epigenetic control of myeloid cell differentiation, identity and function. Nat. Rev. Immunol 15, 7–17 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Smale ST, Tarakhovsky A & Natoli G Chromatin contributions to the regulation of innate immunity. Annu. Rev. Immunol 32, 489–511 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Ivashkiv LB & Park SH Epigenetic regulation of myeloid cells. Microbiol. Spectr 10.1128/microbiolspec.MCHD-0010-2015 (2016). [DOI] [PMC free article] [PubMed]
- 46.Wang KC & Chang HY Epigenomics: technologies and applications. Circ. Res 122, 1191–1199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ballestar E & Li T New insights into the epigenetics of inflammatory rheumatic diseases. Nat. Rev. Rheumatol 13, 593–605 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Shi L et al. Monocyte enhancers are highly altered in systemic lupus erythematosus. Epigenomics 7, 921–935 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Z et al. H3K4 tri-methylation breadth at transcription start sites impacts the transcriptome of systemic lupus erythematosus. Clin. Epigenet 8, 14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scharer CD et al. ATAC-seq on biobanked specimens defines a unique chromatin accessibility structure in naive SLE B cells. Sci. Rep 6, 27030 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ai R et al. Comprehensive epigenetic landscape of rheumatoid arthritis fibroblast-like synoviocytes. Nat. Commun 9, 1921 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Netea MG, Latz E, Mills KH & O’Neill LA Innate immune memory: a paradigm shift in understanding host defense. Nat. Immunol 16, 675–679 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Biswas SK & Lopez-Collazo E Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol 30, 475–487 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Saeed S et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 345, 1251086 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park SH et al. Type I interferons and the cytokine TNF cooperatively reprogram the macrophage epigenome to promote inflammatory activation. Nat. Immunol 18, 1104–1116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Novakovic B et al. Beta-glucan reverses the epigenetic state of LPS-induced immunological tolerance. Cell 167, 1354–1368 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vatanen T et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 165, 842–853 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi L et al. Endotoxin tolerance in monocytes can be mitigated by alpha2-interferon. J. Leukoc. Biol 98, 651–659 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wendeln AC et al. Innate immune memory in the brain shapes neurological disease hallmarks. Nature 556, 332–338 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanyal A, Lajoie BR, Jain G & Dekker J The long-range interaction landscape of gene promoters. Nature 489, 109–113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ye CJ et al. Intersection of population variation and autoimmunity genetics in human T cell activation. Science 345, 1254665 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raj T et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science 344, 519–523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee MN et al. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science 343, 1246980 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Farh KK et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Raj P et al. Regulatory polymorphisms modulate the expression of HLA class II molecules and promote autoimmunity. eLife 5, e12089 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martinez-Bueno M et al. Trans-ethnic mapping of BANK1 identifies two independent SLE-risk linkage groups enriched for co-transcriptional splicing marks. Int. J. Mol. Sci 19, E2331 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pulecio J, Verma N, Mejia-Ramirez E, Huangfu D & Raya A CRISPR/Cas9-based engineering of the epigenome. Cell Stem Cell 21, 431–447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mumbach MR et al. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat. Genet 49, 1602–1612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sokhi UK et al. Dissection and function of autoimmunity-associated TNFAIP3 (A20) gene enhancers in humanized mouse models. Nat. Commun 9, 658 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liao HK et al. In vivo target gene activation via CRISPR/Cas9-mediated trans-epigenetic modulation. Cell 171, 1495–1507 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Robertson KD DNA methylation and human disease. Nat. Rev. Genet 6, 597–610 (2005). [DOI] [PubMed] [Google Scholar]
- 73.Doody KM, Bottini N & Firestein GS Epigenetic alterations in rheumatoid arthritis fibroblast-like synoviocytes. Epigenomics 9, 479–492 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hammaker D & Firestein GS Epigenetics of inflammatory arthritis. Curr. Opin. Rheumatol 30, 188–196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jin B, Li Y & Robertson KD DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer 2, 607–617 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mardis ER Next-generation DNA sequencing methods. Annu. Rev. Genomics Hum. Genet 9, 387–402 (2008). [DOI] [PubMed] [Google Scholar]
- 77.Shendure J & Ji H Next-generation DNA sequencing. Nat. Biotechnol 26, 1135–1145 (2008). [DOI] [PubMed] [Google Scholar]
- 78.Feil R, Charlton J, Bird AP, Walter J & Reik W Methylation analysis on individual chromosomes: improved protocol for bisulphite genomic sequencing. Nucleic Acids Res 22, 695–696 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lister R et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reinders J & Paszkowski J Bisulfite methylation profiling of large genomes. Epigenomics 2, 209–220 (2010). [DOI] [PubMed] [Google Scholar]
- 81.Meissner A et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res 33, 5868–5877 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kurdyukov S & Bullock M DNA methylation analysis: choosing the right method. Biology 5, E3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nakano K, Whitaker JW, Boyle DL, Wang W & Firestein GS DNA methylome signature in rheumatoid arthritis. Ann. Rheum. Dis 72, 110–117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Whitaker JW et al. An imprinted rheumatoid arthritis methylome signature reflects pathogenic phenotype. Genome Med 5, 40 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ai R et al. DNA methylome signature in synoviocytes from patients with early rheumatoid arthritis compared to synoviocytes from patients with longstanding rheumatoid arthritis. Arthritis Rheumatol 67, 1978–1980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Frank-Bertoncelj M et al. Epigenetically-driven anatomical diversity of synovial fibroblasts guides joint-specific fibroblast functions. Nat. Commun 8, 14852 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rhead B et al. Rheumatoid arthritis naive T cells share hypermethylation sites with synoviocytes. Arthritis Rheumatol 69, 550–559 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mok A et al. Hypomethylation of CYP2E1 and DUSP22 promoters associated with disease activity and erosive disease among rheumatoid arthritis patients. Arthritis Rheumatol 70, 528–536 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chung SA et al. Genome-wide assessment of differential DNA methylation associated with autoantibody production in systemic lupus erythematosus. PLOS ONE 10, e0129813 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mok A et al. Genome-wide profiling identifies associations between lupus nephritis and differential methylation of genes regulating tissue hypoxia and type 1 interferon responses. Lupus Sci. Med 3, e000183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cole MB et al. Epigenetic signatures of salivary gland inflammation in Sjogren’s syndrome. Arthritis Rheumatol 68, 2936–2944 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Puliti A, Caridi G, Ravazzolo R & Ghiggeri GM Teaching molecular genetics: chapter 4—positional cloning of genetic disorders. Pediatr. Nephrol 22, 2023–2029 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.The International FMF Consortium. Ancient missense mutations in a new member of the RoRet gene family are likely to cause familial Mediterranean fever. Cell 90, 797–807 (1997). [DOI] [PubMed] [Google Scholar]
- 94.Zhu X et al. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet. Med 17, 774 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhou Q et al. Loss-of-function mutations in TNFAIP3 leading to A20 haploinsufficiency cause an early onset autoinflammatory syndrome. Nat. Genet 48, 67–73 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zappala Z & Montgomery SB Non-coding loss-of-function variation in human genomes. Hum. Hered 81, 78–87 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ma M et al. Disease-associated variants in different categories of disease located in distinct regulatory elements. BMC Genomics 16, S3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tesi B et al. A RAB27A 5ʹ untranslated region structural variant associated with late-onset hemophagocytic lymphohistiocytosis and normal pigmentation. J. Allergy Clin. Immunol 142, 317–321 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Namjou B et al. Evaluation of the TREX1 gene in a large multi-ancestral lupus cohort. Genes Immun 12, 270–279 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Beaudoin M et al. Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLOS Genet 9, e1003723 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cardinale CJ et al. Targeted resequencing identifies defective variants of decoy receptor 3 in pediatric-onset inflammatory bowel disease. Genes Immun 14, 447 (2013). [DOI] [PubMed] [Google Scholar]
- 102.Nakagawa K et al. Somatic NLRP3 mosaicism in Muckle-Wells syndrome. A genetic mechanism shared by different phenotypes of cryopyrin-associated periodic syndromes. Ann. Rheumat. Dis 74, 603–610 (2015). [DOI] [PubMed] [Google Scholar]
- 103.Tanaka N et al. High incidence of NLRP3 somatic mosaicism in patients with chronic infantile neurologic, cutaneous, articular syndrome: results of an international multicenter collaborative study. Arthritis Rheum 63, 3625–3632 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhou Q et al. Cryopyrin-associated periodic syndrome caused by a myeloid-restricted somatic NLRP3 mutation. Arthritis Rheumatol 67, 2482–2486 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Holzelova E et al. Autoimmune lymphoproliferative syndrome with somatic Fas mutations. N. Engl. J. Med 351, 1409–1418 (2004). [DOI] [PubMed] [Google Scholar]
- 106.Savola P et al. Somatic mutations in clonally expanded cytotoxic T lymphocytes in patients with newly diagnosed rheumatoid arthritis. Nat. Commun 8, 15869 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rowczenio DM et al. Late-onset cryopyrin-associated periodic syndromes caused by somatic NLRP3 mosaicism—UK single center experience. Front. Immunol 8, 1410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yuri K et al. Identification of a high-frequency somatic NLRC4 mutation as a cause of autoinflammation by pluripotent cell–based phenotype dissection. Arthritis Rheumatol 69, 447–459 (2017). [DOI] [PubMed] [Google Scholar]
- 109.Chung J et al. The minimal amount of starting DNA for Agilent’s hybrid capture-based targeted massively parallel sequencing. Sci. Rep 6, 26732 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Grossman RL et al. Toward a shared vision for cancer genomic data. N. Engl. J. Med 375, 1109–1112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]