Abstract
Nuclear lamins are intermediate filament proteins that polymerize to form the nuclear lamina on the inner aspect of the inner nuclear membrane. Long known to be essential for maintaining nuclear structure and disassembling/reassembling during mitosis in metazoans, research over the past dozen years has shown that mutations in genes encoding nuclear lamins, particularly LMNA encoding the A-type lamins, cause a broad range of diverse diseases, often referred to as laminopathies. Lamins are expressed in all mammalian somatic cells but mutations in their genes lead to relatively tissue-selective disease phenotypes in most cases. While mutations causing laminopathies have been shown to produce abnormalities in nuclear morphology, how these disease-causing mutations or resultant alterations in nuclear structure lead to pathology is only starting to be understood. Despite the incomplete understanding of pathogenic mechanisms underlying the laminopathies, basic research in cellular and small animal models has produced promising leads for treatments of these rare diseases.
Keywords: lamin, nuclear envelope, laminopathy, progeria, cardiomyopathy
Nuclear lamins
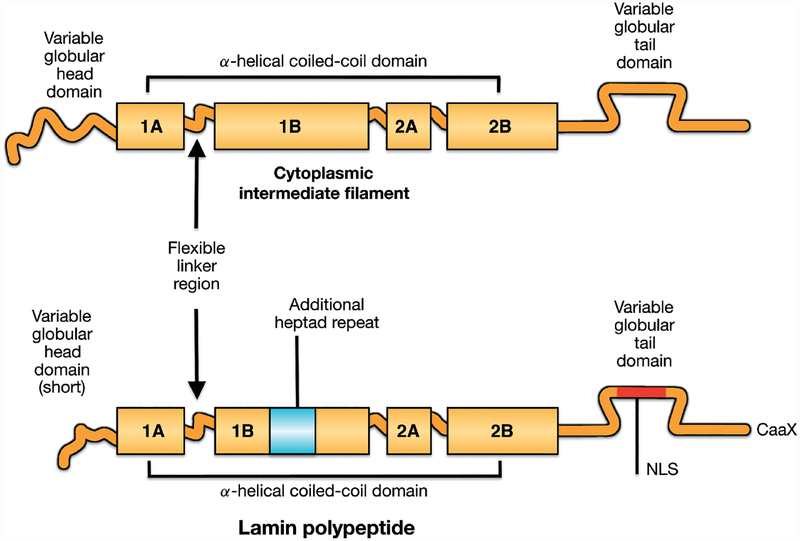
Lamins are type V intermediate filament proteins. They are located primarily at the inner aspect of the inner nuclear membrane, where they polymerize to form a higher-ordered structure called the nuclear lamina. Lamins appear to be expressed in all or most metazoans but are absent from yeast, other unicellular organisms and plants [1]. Vertebrate lamins are similar to their cytoplasmic intermediate filament homologues but have several distinguishing features (Figure 1). Like all intermediate filament proteins, lamins contain a highly conserved α-helical coiled-coil rod domain, composed of heptad repeats of amino acids flanked by variable amino-terminal head and carboxyl-terminal tail domains. However, the rod domains of lamins have 42 additional amino acids (six heptad repeats) within coil 1B compared to vertebrate cytoplasmic intermediate filament proteins [2]. Lamins also have shorter head domains than most cytoplasmic intermediate filament proteins. The tail domains of lamins contain a nuclear localization signal, which is necessary for their active nuclear transport after synthesis [3]. The tail domains also contain an immunoglobulin-like fold motif near the nuclear localization signal [4,5]. Most lamins (in mammals, all except lamin C and lamin C2) contain a motif of amino acid sequence cysteine–aliphatic–aliphatic–any (CaaX) at their carboxyl-termini, which initiates a series of enzymatic reactions that lead to farnesylation and carboxymethylation of the cysteine and endoproteolytic cleavage of–aaX [6]. In contrast to cytoplasmic intermediate filament proteins, lamins also contain consensus sites for mitotically active protein kinases, the phosphorylation of which directs the disassembly of the nuclear lamina during mitosis [7].
Figure 1.
Schematic diagrams showing vertebrate intermediate filament protein structures and differences between cytoplasmic intermediate filament proteins and nuclear lamins. All intermediate filament proteins, including lamins, have a conserved domain structure consisting a central α-helical coiled-coil rod domain consisting of four coiled coils (1A, 1B, 2A, 2B), based on heptad repeats, interrupted by flexible linker domains and a variable globular tail domain. Compared to vertebrate cytoplasmic intermediate filament proteins, lamins contain six additional heptad repeats (42 amino acids) in coil 1B, a nuclear localization signal (NLS) near an immunoglobulin-like fold domain in the carboxyl-terminal tail and, in most lamins, a CaaX motif at the carboxyl-terminus. Reproduced with permission from Hutchison CJ, Worman HJ. A-type lamins: guardians of the soma? Nat Cell Biol 2004; 6: 1062–1067 [1].
Lamins are generally divided into A-type and B-type, depending upon their structural similarities and isoelectric points. In mammals, three genes encode lamins (Table 1). In humans, LMNB1 on chromosome 5 encodes lamin B1, which is expressed in all or most somatic cells [8]. LMNB2 on human chromosome 19 encodes lamin B2, also expressed in all or most somatic cells, and lamin B3, a germ cell-specific isoform that arises by alternative RNA splicing [9,10]. LMNA on human chromosome 1 encodes the A-type lamins, with lamin A and lamin C being the major isoforms arising by alternative RNA splicing that are expressed in most terminally differentiated cells [11]. A-type lamins are lacking from undifferentiated cells, such as early embryos and lymphoblasts [12–15]. LMNA also encodes a germ cell-specific isoform, lamin C2, and a poorly studied isoform, lamin A 10 [16,17].
Table 1.
Human nuclear lamins
| Protein | Expression | Gene |
|---|---|---|
| Lamin A | Most differentiated somatic cells | LMNA |
| Lamin C | Most differentiated somatic cells | LMNA |
| Lamin C2 | Germ cells | LMNA |
| Lamin AΔ10 | Unclear | LMNA |
| Lamin B1 | Most or all somatic cells | LMNB1 |
| Lamin B2 | Most or all somatic cells | LMNB2 |
| Lamin B3 | Germ cells | LMNB2 |
Lamins A and C are identical for their first 566 amino acids and differ in their carboxyl-terminal tail domains [18,19]. Lamin A is synthesized as a precursor, prelamin A, which contains a CaaX motif at its amino-terminus. After farnesylation (catalysed by protein farnesyltransferase), carboxymethylation (catalysed by isoprenylcysteine carboxylmethyltransferase) and–aaX cleavage (catalysed by the endoproteases RCE1 and ZMPSTE24), prelamin A is recognized by ZMPSTE24, which cleaves it 15 amino acids from the farnesylated cysteine [6,20]. Hence, in contrast to the B-type lamins, lamins A and C are not farnesylated when assembled in the nuclear lamina.
Many different cellular functions have been attributed to the nuclear lamina [21]. Most cell biologists would agree that one function is to provide structural support to the nucleus, perhaps also regulating the spacing of nuclear pore complexes. A role for the nuclear lamina in regulating chromatin organization and gene expression has been widely hypothesized and repositioning of genes to the nuclear lamina leads to transcriptional repression [22]. However, the exact physiological role of lamins in gene expression remains unknown. Some research suggests that B-type lamins function in fundamental biological processes, including DNA replication and mitotic spindle pole formation [23–26]. However, mice deficient in lamin B1 or lamin B2 survive embryonic development, with the most predominant abnormalities in lamin B2-null mice involving brain development, and mouse keratinocytes genetically engineered to be deficient in all B-type lamins proliferate normally [27–29]. Cells without A-type lamins also proliferate and Lmna knock-out mice survive beyond birth, eventually dying from cardiac and skeletal muscle abnormalities [30].
Laminopathies
In 1999, Bonne et al [31] reported that mutations in LMNA encoding A-type lamins cause autosomal-dominant Emery–Dreifuss muscular dystrophy. This opened the floodgates to discoveries over the next decade that mutations in the same LMNA gene cause more than a dozen previously defined rare clinical disorders called laminopathies (Table 2). As a result, research in clinical genetics has changed the way cell biologists view the nuclear lamins and nuclear lamina.
Table 2.
Mutations in LMNA cause rare clinical disorders often called laminopathies
| Autosomal Emery-Dreifuss muscular dystrophy |
| Cardiomyopathy dilated 1A |
| Limb girdle muscular dystrophy type 1B |
| Congenital muscular dystrophy |
| ‘Heart-hand’ syndrome |
| Dunnigan-type familial partial lipodystrophy |
| Lipoatrophy with diabetes and other features of insulin resistance |
| Mandibuloacral dysplasia |
| Charcot-Marie-Tooth disorder type 2B1 |
| Hutchinson-Gilford progeria syndrome |
| Atypical Werner syndrome |
| Restrictive dermopathy |
| Variant progeroid disorders |
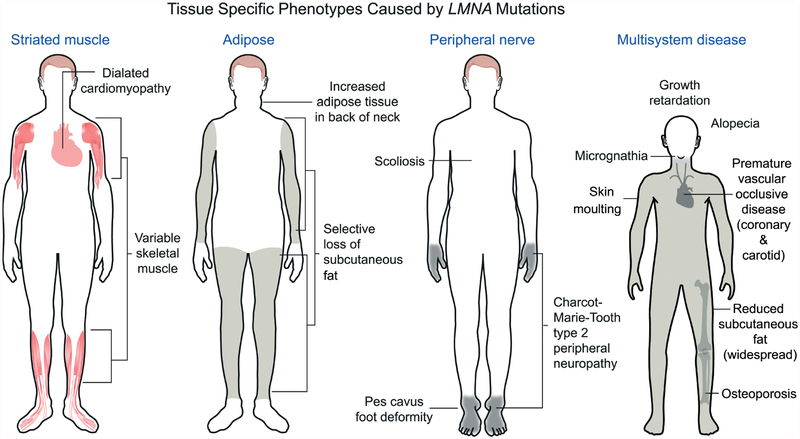
A careful look at the clinical disorders caused by LMNA mutations shows that they can be grouped into those that affect either striated muscle, adipose tissue (with consequent metabolic abnormalities), peripheral nerve or multiple systems with signs of accelerated ageing (Figure 2). While there is some overlap between affected organs and tissues, this classification is for the most part robust.
Figure 2.
Different LMNA mutations cause diseases that affect striated muscle, adipose, peripheral nerve or multiple systems with features of accelerated ageing. Most LMNA mutations are autosomal-dominant and cause dilated cardiomyopathy with variable skeletal muscle involvement. This includes the classical Emery–Dreifuss muscular dystrophy phenotype, as shown in the diagram, with scapulohumeral–peroneal distribution of skeletal muscle involvement, concurrent tendon contractures and dilated cardiomyopathy. Autosomal-dominant missense mutations in LMNA, the large majority of which cause a change in the surface charge of the immunoglobulin-like fold of lamin A and lamin C, cause Dunnigan-type familial partial lipodystrophy, with selective loss of subcutaneous fat from the extremities, fat accumulation in the neck and face and insulin resistance and diabetes mellitus. An autosomal recessive LMNA mutation that leads to an arginine-to-cysteine amino acid substitution at residue 298 causes a Charcot–Marie–Tooth type 2 peripheral neuropathy, characterized clinically by a stocking–glove sensory neuropathy, resultant pes cavus foot deformity and other variable features, such as scoliosis. The sporadic cytosine to thymine transversion in codon 608 of exon 11 of LMNA causes Hutchinson–Gilford progeria syndrome, which has features of accelerated ageing, such as sclerotic skin, joint contractures, micrognathia, alopecia, fingertip tufting, distal-joint abnormalities, growth impairment and vascular abnormalities, generally leading to death during the second decade due to myocardial infarction or stroke. Other LMNA mutations can also cause progeroid syndromes with similar features; a recessive LMNA mutation causing an arginine-to-histidine amino acid substitution at residue 527 in the immunoglobulin-like fold causes mandibuloacral dysplasia, a disorder with a combination of progeroid features and partial lipodystrophy. Reproduced with permission from: Dauer WT, Worman HJ. The nuclear envelope as a signalling node in development and disease. Dev Cell 2009; 17: 626–638 [65].
The first-described laminopathies were diseases of striated muscle [31]. Autosomal-dominant Emery–Dreifuss muscular dystrophy is characterized by early joint contractures, primarily involving the elbows, ankles and neck, followed by progressive muscle weakness and wasting, beginning in the upper arms and lower legs and progressing to muscles in the shoulders and hips [32]. The life-threatening feature of the disease, however, is dilated cardiomyopathy with early conduction system abnormalities. The same LMNA mutations that cause Emery–Dreifuss muscular dystrophy can also cause dilated cardiomyopathy, with minimal to no skeletal muscle disease or a limb-girdle muscular dystrophy with dilated cardiomyopathy [33,34]. Hence, Emery–Dreifuss muscular dystrophy, isolated dilated cardiomyopathy and limb-girdle muscular dystrophy 1B are actually a spectrum of overlapping clinical phenotypes caused by LMNA mutations, with dilated cardiomyopathy as the unifying feature [35]. LMNA mutations have also been associated with cases of congenital muscular dystrophy with heart involvement and ‘heart–hand syndrome’, in which cardiomyopathy is associated with congenital limb abnormalities [36,37]. LMNA mutations that cause striated muscle diseases generally lead to amino acid substitutions throughout lamins A and C, small in-frame deletions, RNA splicing defects or haploinsufficiency of A-type lamins. The Emery–Dreifuss muscular dystrophy phenotype can also be inherited in an X-linked manner. This occurs as a result of mutations in the EMD gene, which encodes an integral membrane protein called emerin [38]. Emerin is localized to the inner nuclear membrane and in most cases of X-linked Emery–Dreifuss muscular dystrophy is lacking from the nuclear envelope [39,40]. Emerin and A-type lamins bind to each other and A-type lamins are essential for retaining emerin in the inner nuclear membrane [30,41]. However, the functional significance of the lamin–emerin interaction and why mutations in the genes encoding these different proteins can cause the same striated muscle disease phenotype remain unknown.
In 2000, several groups showed that mutation in LMNA cause disease of adipose tissue, specifically Dunnigan-type familial partial lipodystrophy [42–44]. Dunnigan-type familial partial lipodystrophy is autosomal-dominantly inherited and presents with loss of adipose tissue from the extremities around the onset of puberty, with consequent insulin resistance, diabetes mellitus, hypertriglyceridaemia and complications such as hepatic steatosis [42–46]. Approximately 85–90% of mutations that cause this disease are in exon 8 of LMNA and lead to amino acid substitutions that change the surface charge of the immunoglobulin-like fold domain in the carboxyl-terminal tail of lamins A and C [4,5]. Homozygous missense mutations, most causing an arginine-to-methionine substitution, within the immunoglobulin-like fold domain cause mandibuloacral dysplasia, a syndrome with partial lipodystrophy and congenital abnormalities mostly affecting the skeleton [47]. In contrast, amino acids substitutions in the immunoglobulin-like fold that occur in striated muscle diseases are predicted to lead to significant overall disruption of its tertiary structure [4,5]. Various other mutations in LMNA have been reported in subjects with abnormalities in adipose tissue, insulin sensitivity and fat metabolism but without the typical Dunnigan-type familial partial lipodystrophy phenotype [48–50].
A LMNA mutation leading to a homozygous arginine-to-cysteine amino acid substitution at position 298 in the rod domain of A-type lamins has been reported to cause an autosomal recessive peripheral neuropathy, specifically Charcot–Marie–Tooth type 2 disorder type 2B1 [51]. Affected subjects suffer from an axonal peripheral neuropathy with variability in the age of onset and the course of the disease [52]. Sciatic nerves of Lmna-null mice have a reduction of axon density, axonal enlargement and non-myelinated axons similar to phenotypes of human peripheral axonal neuropathies [51]. Heterozygous deletion of the LMNA initiator codon has been reported to cause a phenotype with features of both autosomal-dominant Emery–Dreifuss muscular dystrophy and peripheral neuropathy [53]. Hence, peripheral neuropathy appears to result from some type of selective loss of some function or property of A-type lamins.
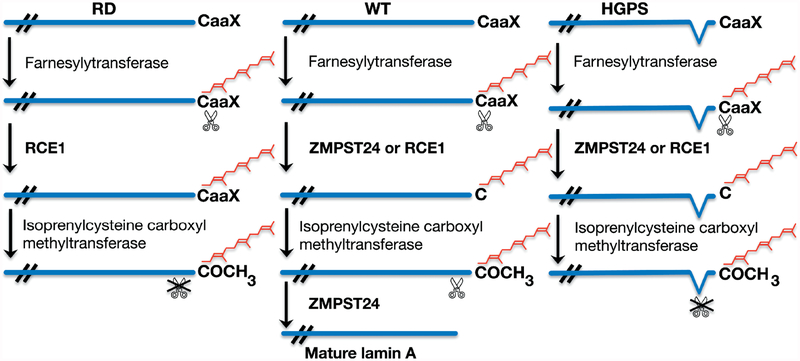
The other group of diseases resulting from LMNA mutations has defects in multiple organ systems with signs of accelerated ageing. Individuals with mandibuloacral dysplasia caused by LMNA mutation have partial lipodystrophy but also mandibular and clavicular hypoplasia, acroosteolysis of the distal fingers, delayed closure of the cranial suture, joint contractures, short adult stature and growth retardation [37]. Several of these phenotypic abnormalities overlap those present in progerias, disorders of accelerated ageing [54]. Hutchinson–Gilford progeria syndrome is a sporadic, autosomal-dominant syndrome characterized by features of accelerated ageing, including sclerotic skin, joint contractures, micrognathia, alopecia, fingertip tufting, distal-joint abnormalities, growth impairment and vascular abnormalities, generally leading to death during the second decade due to myocardial infarction or stroke [55]. This syndrome is caused by a sporadic cytosine-to-thymine transversion in codon 608 of exon 11 of LMNA [56,57]. It generates a RNA a splice donor site that leads to expression of a prelamin A variant with 50 amino acids deleted from the carboxyl-terminal tail region, including the ZMPSTE24 endoprotease site. Homozygous mutation in the gene encoding ZMPSTE24 also results in a progeriod syndrome, neonatal lethal restrictive dermopathy [58,59]. In Hutchinson–Gilford progeria syndrome and restrictive dermopathy, the genetic defects respectively result in expression of either a prelamin A variant or unprocessed prelamin A, both of which retain a farnesylated and carboxymethylated carboxyl-terminal cysteine (Figure 3). Expression of these abnormally farnesylated proteins appears to be important in the pathophysiology of the progeroid phenotypes (see below); however, point mutations in LMNA that do not lead to abnormally farnesylated protein variants have also been associated with atypical progeroid syndromes [60–62].
Figure 3.
Processing of prelamin A in wild-type (WT) cells occurs in a series of sequential enzymatic reactions that lead to farnesylation, endoproteolytic cleavage of–aaX, carboxymethylation and a second endoproteolytic cleavage catalysed by ZMSTE24. In restrictive dermopathy (RD), there is no ZMPSTE24 activity, which results in accumulation of farnesylated, carboxymethylated prelamin A. In Hutchinson–Gilford progeria syndrome (HGPS), the second site for cleavage catalysed by ZMPSTE24 is deleted, which leads to accumulation of progerin, a truncated variant of farnesylated, carboxymethylated prelamin A. Reproduced with permission from: Worman HJ, Östlund C, Wang Y. Diseases of the nuclear envelope. Cold Spring Harb Perspect Biol 2010; 2: a000760 [66].
While mutations in LMNA cause diverse disease phenotypes, fewer disease-causing mutations in genes encoding B-type lamins have been reported. Duplication of LMNB1, which leads to an increase in lamin B1 expression, causes adult-onset autosomal-dominant leukodystrophy, a slowly progressive neurological disorder characterized by symmetrical widespread myelin loss in the central nervous system [63]. Mutations in LMNB2 have been reported to lead to susceptibility to acquired partial lipodystrophy [64]. In mice, homozygous Lmnb1 and Lmnb2 deletions both cause neonatal lethality, with Lmnb2-null mice having abnormal development of the cerebral cortex and cerebellum [27,28]. Mutations in genes encoding several lamin-associated proteins of the nuclear envelope cause a variety of diseases, also sometimes referred to as laminopathies (reviewed previously in [65,66]).
Pathophysiology and potential treatments
Research on the genetic mutations causing laminopathies took off in the first decade of the twenty-first century, but understanding of disease pathogenesis lagged. Research on pathophysiology using cellular and animal models subsequently began to catch up. Some of this research has already led to early stage clinical trials for these rare diseases.
The first cellular pathophysiological observation in laminopathies was that mutations in LMNA generally lead to abnormal nuclear morphology. When A-type lamins are absent from cells that normally express them, the nuclei are irregular in shape with herniations of the nuclear envelope; in addition, nuclear pore complexes cluster, B-type lamins are partly mislocalized and emerin redistributes from the inner nuclear membrane to the bulk endoplasmic reticulum (reviewed previously in [21,65–69]). Cells expressing disease-causing A-type lamin variants have lobulations or blebbing of the nuclear envelope, honeycombing of the lamina, increased nuclear surface area, thickening of the nuclear lamina, aberrant intranuclear foci of lamins, loss of peripheral heterochromatin and aberrant clustering of nuclear pore complexes [21,65–69]. These morphological alterations depend upon cell type, lamin protein sequence, protein expression levels in transfected cells and culture conditions. Along with altered nuclear morphology, deficiencies of A- and B-type lamins and expression of certain lamin A variants lead to abnormalities in nuclear mechanics and cellular mechanotransduction [70–74]. Lack of A-type lamins and expression of variants in striated muscle diseases also cause abnormal positioning of nuclei in migrating fibroblasts, which likely results from defective connections between the nucleus and cytoplasm [74,75].
Although alterations in nuclear morphology have been extensively examined in transfected cultured cells, fibroblasts from affected human subjects, fibroblasts from mouse models of the diseases and in situ in tissues, the direct connection to disease pathophysiology remains unknown. Nonetheless, studies from mouse models of Hutchinson–Gilford progeria syndrome and restrictive dermopathy show a correlation between abnormal nuclear morphology and disease phenotypes in animals. Mice with the gene encoding the prelamin A endoprotease ZMPSTE24 deleted have a progeroid phenotype and accumulate farnesylated prelamin A [76,77]. Both genetic reduction of prelamin A and treatment with a protein farnesyltransferase inhibitor that blocks prelamin A farnesylation lead to improvement in the disease phenotype in these mice [78,79]. These original results have been confirmed by treating ZMPSTE24-deficient mice with statins and aminobisphosphonates, which inhibit prenylation of proteins [80]. Similarly, treatment with protein farnesyltransferase inhibitors improves abnormal phenotypes and prolongs survival in mouse models of Hutchinson–Gilford progeria syndrome that express progerin [81–83]. Expression of progerin variants that cannot be farnesylated because of mutations in the CaaX motif, depending on the sequence alteration to CaaX, either reduces the severity of or abolishes the progeroid phenotype in knock-in mice [84,85]. These studies have established that farnesyl modification of the prelamin A variants is clearly involved in disease pathogenesis in Hutchinson–Gilford progeria syndrome and restrictive dermopathy. In addition to ameliorating progeroid phenotypes in animals, protein fanesyltransferase inhibitor treatment of cultured fibroblasts from human subjects with Hutchinson–Gilford progeria syndrome and restrictive dermopathy, animal modes of these diseases and transfected cells expressing progerin leads to significant improvements in nuclear morphology [86–90]. Hence, there is a correlation between improvement in abnormal nuclear morphology and improvement in whole animal phenotypes when farnesylation of the pathogenic prelamin A variants is blocked. This basic cell biology and small animal research has led to clinical trials of protein prenylation inhibitors in human subjects with Hutchinson–Gilford progeria syndrome [91]. However, abnormal nuclear morphology per se does not appear to be the direct cause of tissue or organ dysfunction, as progerin expression, ZMPSTE24 depletion and A-type lamin depletion can induce significant alterations in nuclear shape in tissues and organs without pathology [30,80,92].
While expression of farnesylated prelamin A variants are at least partly responsible for the progeroid phenotypes in Hutchinson–Gilford progeria syndrome and restrictive dermopathy, the downstream pathways affected by these abnormal proteins or the resultant alterations in nuclear morphology are less well established. Accumulation of progerin and farnesylated prelamin A has been correlated with defective DNA repair mechanisms and genomic instability [93–95]. Progerin binds to DNA-dependent protein kinase catalytic subunit, which functions in genomic stability [96]. Expression of progerin also leads to telomere dysfunction and induction of senescence [97–99].
Abnormalities in signalling pathways appear to be perturbed in cells expressing progerin or farnesylated prelamin A. Progerin activates the Notch signalling pathway [100]. Defective canonical Wnt signalling occurs in cells from mouse models of Hutchinson–Gilford progeria syndrome and affected human subjects [101]. Deficiency of ZMPSTE24 similarly causes alterations in Wnt signalling [102]. The Notch and Wnt signalling pathways are important in controlling stem cell proliferation and differentiation, hence stem cell dysfunction may be a pathogenic factor in progerias caused by alteration in A-type lamins [100–102]. Rapamycin has also been shown to improve abnormal phenotypes in cells from subjects with Hutchinson–Gilford progeria syndrome, suggesting that the mTOR signalling axis may be involved in pathophysiology [103].
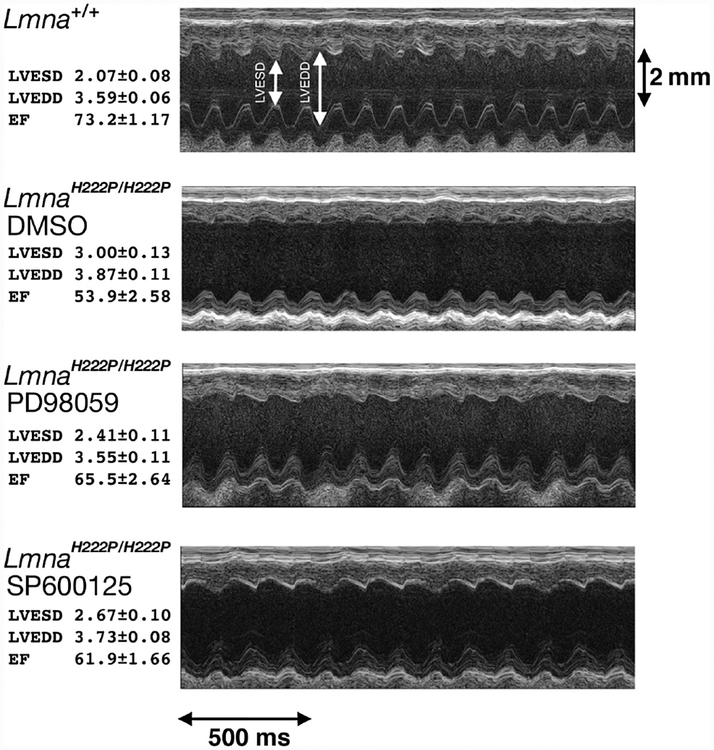
The nuclear envelope may generally function in integrating and regulating different signal transduction pathways [65]. Abnormalities in signal transduction appear to underlie the pathophysiology of cardiomyopathy caused by LMNA mutations. Hearts of a knock-in mouse model of autosomal Emery–Dreifuss muscular dystrophy have perturbations in several signal transduction pathways, including increased mitogen-activated protein kinase signalling [104]. Similar alterations in mitogen-activated protein kinase signalling occur in the hearts of mice lacking emerin, which is not expressed in most cases of human X-linked Emery–Dreifuss muscular dystrophy [105]. Pharmacological blockade of signalling in the extracellular signal-regulated kinase and c-Jun N-terminal kinase branches of the mitogen-activated protein kinase cascade in mice with an Lmna mutation prevent left ventricular dilatation and deterioration in cardiac contractility, if administered prior to the onset of detectable heart disease [106,107]. Administration of these drugs after the mice have developed deterioration in heart function improves cardiac ejection fraction and blunts further increases in left ventricular dilatation (Figure 4) [108]. Treatment also decreases cardiac fibrosis, an end-stage and irreversible feature of cardiomyopathy caused by LMNA mutation [107,108]. While it remains unclear how alterations in A-type lamins lead to activation of mitogen-activated protein kinase signalling in the heart, these studies clearly show that the abnormal activation involved is the pathophysiology of dilated cardiomyopathy caused by LMNA mutation. Inhibitors of extracellular signal-regulated kinase signalling are currently in human clinical trials for other indications and could potentially be tested in human subjects with LMNA cardiomyopathy.
Figure 4.
Representative transthoracic M-mode echocardiograms taken from wild-type mice (Lmna+/+), Lmna mutant mice that develop cardiomyopathy receiving placebo (LmnaH222P/H222P DMSO), Lmna mutant mice treated with an inhibitor of extracellular signal-regulated kinase signalling (LmnaH222P/H222P PD98059) and Lmna mutant mice treated with an inhibitor of c-Jun N-terminal kinase signalling (LmnaH222P/H222P SP600125). Left ventricular end systolic diameter (LVESD) and left ventricular end diastolic diameter (LVEDD) are indicated in the top echocardiographic tracing. At left, means ± standard errors for LVESD, LVEDD and the cardiac ejection fraction (EF), a measure of cardiac contractility, are given for mice in each group. Both PD98059 and SP600125 significantly improve the ejection fraction compared to placebo. This figure is based on Figure 3 and data are used with permission from: Wu W, Muchir A, Shan J, et al. Mitogen-activated protein kinase inhibitors improve heart function and prevent fibrosis in cardiomyopathy caused by mutation in lamin A/C gene. Circulation 2011; 123: 53–61 [108].
Conclusions
Mutations in genes encoding nuclear lamins, particularly LMNA encoding the A-type lamins, cause a range of phenotypically diverse diseases. The phenotypes and genetic abnormalities of these disorders have been extensively described. A significant amount of current research is aimed at deciphering pathogenic mechanisms and some has connected mutations in LMNA to posttranslational protein modifications and alterations in cell signalling pathways that can be connected to pathophysiological processes and targeted by small molecule therapeutics. While laminopathies are rare diseases, the disease phenotypes that result from mutations in genes encoding lamins are fairly common, such as dilated cardiomyopathy, insulin resistance and even ageing. In this regard, research on these fascinating rare diseases should provide insights into common disorders.
Acknowledgment
This study was supported by grants from the National Institutes of Health (Grant Nos RO1AR048997, RO1NS059352 and RO1HD070713), the Muscular Dystrophy Association (Grant No. MDA172222) and the New York City Partnership Foundation Inc.
Footnotes
Conflict of interest: The author is an inventor on a pending PCT patent application on methods for treating and/or preventing cardiomyopathies by ERK and JNK inhibition, filed by the Trustees of Columbia University in the City of New York.
References
- 1.Melcer S, Gruenbaum Y, Krohne G. Invertebrate lamins. Exp Cell Res 2007; 313: 2157–2166. [DOI] [PubMed] [Google Scholar]
- 2.Hutchison CJ, Worman HJ. A-type lamins: guardians of the soma? Nat Cell Biol 2004; 6: 1062–1067. [DOI] [PubMed] [Google Scholar]
- 3.Frangioni J, Neel B Use of a general purpose mammalian expression vector for studying intracellular protein targeting: identification of critical residues in the nuclear lamin A/C nuclear localisation sequence. J Cell Sci 1993; 105: 481–488. [DOI] [PubMed] [Google Scholar]
- 4.Dhe-Paganon S, Werner ED, Chi YI, et al. Structure of the globular tail of nuclear lamin. J Biol Chem 2002; 277: 17381–17384. [DOI] [PubMed] [Google Scholar]
- 5.Krimm I, Östlund C, Gilquin B, et al. The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy. Structure 2002; 10: 811–823. [DOI] [PubMed] [Google Scholar]
- 6.Rusiñol AE, Sinensky MS. Farnesylated lamins, progeroid syndromes and farnesyl transferase inhibitors. J Cell Sci 2006; 119: 3265–3272. [DOI] [PubMed] [Google Scholar]
- 7.Nigg EA. Assembly and cell cycle dynamics of the nuclear lamina. Semin Cell Biol 1992; 3: 245–253. [DOI] [PubMed] [Google Scholar]
- 8.Lin F, Worman HJ. Structural organization of the human gene (LMNB1) encoding nuclear lamin B1. Genomics 1995; 27: 230–236. [DOI] [PubMed] [Google Scholar]
- 9.Biamonti G, Giacca M, Perini G, et al. The gene for a novel human lamin maps at a highly transcribed locus of chromosome 19 which replicates at the onset of S-phase. Mol Cell Biol 1992; 12: 3499–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furukawa K, Hotta Y. cDNA cloning of a germ cell specific lamin B3 from mouse spermatocytes and analysis of its function by ectopic expression in somatic cells. EMBO J 1993; 12: 97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin F, Worman HJ. Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J Biol Chem 1993; 268: 16321–16326. [PubMed] [Google Scholar]
- 12.Stewart C, Burke B. Teratocarcinoma stem cells and early mouse embryos contain only a single major lamin polypeptide closely resembling lamin B. Cell 1987; 51: 383–392. [DOI] [PubMed] [Google Scholar]
- 13.Guilly MN, Bensussan A, Bourge JF, et al. A human T lymphoblastic cell line lacks lamins A and C. EMBO J 1987; 6: 3795–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Worman HJ, Lazaridis I, Georgatos SD. Nuclear lamina heterogeneity in mammalian cells. Differential expression of the major lamins and variations in lamin B phosphorylation. J Biol Chem 1988; 263: 12135–12141. [PubMed] [Google Scholar]
- 15.Röber RA, Weber K, Osborn M. Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animal: a developmental study. Development 1989; 105: 365–378. [DOI] [PubMed] [Google Scholar]
- 16.Furukawa K, Inagaki H, Hotta Y. Identification and cloning of an mRNA coding for a germ cell-specific A-type lamin in mice. Exp Cell Res 1994; 212: 426–430. [DOI] [PubMed] [Google Scholar]
- 17.Machiels BM, Zorenc AH, Endert JM, et al. An alternative splicing product of the lamin A/C gene lacks exon 10. J Biol Chem 1996; 271: 9249–9253. [DOI] [PubMed] [Google Scholar]
- 18.Fisher DZ, Chaudhary N, Blobel G. cDNA sequencing of nuclear lamins A and C reveals primary and secondary structural homology to intermediate filament proteins. Proc Natl Acad Sci USA 1986; 83: 6450–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKeon FD, Kirschner MW, Caput D. Homologies in both primary and secondary structure between nuclear envelope and intermediate filament proteins. Nature 1986; 319: 463–468. [DOI] [PubMed] [Google Scholar]
- 20.Davies BS, Fong LG, Yang SH, et al. The posttranslational processing of prelamin A and disease. Annu Rev Genomics Hum Genet 2009; 10: 153–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dechat T, Pfleghaar K, Sengupta K, et al. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev 2008; 22: 832–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reddy KL, Zullo JM, Bertolino E, et al. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 2008; 452: 243–247. [DOI] [PubMed] [Google Scholar]
- 23.Spann TP, Moir RD, Goldman AE, et al. Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J Cell Biol 1997; 136: 1201–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellis DJ, Jenkins H, Whitfield WG, et al. GST–lamin fusion proteins act as dominant negative mutants in Xenopus egg extract and reveal the function of the lamina in DNA replication. J Cell Sci 1997; 110: 2507–2518. [DOI] [PubMed] [Google Scholar]
- 25.Moir RD, Spann TP, Herrmann H, et al. Disruption of nuclear lamin organization blocks the elongation phase of DNA replication. J Cell Biol 2000; 149: 1179–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai MY, Wang S, Heidinger JM, et al. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science 2006; 311: 1887–1893. [DOI] [PubMed] [Google Scholar]
- 27.Vergnes L, Péterfy M, Bergo MO, et al. Lamin B1 is required for mouse development and nuclear integrity. Proc Natl Acad Sci USA 2004; 101: 10428–10433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coffinier C, Chang SY, Nobumori C, et al. Abnormal development of the cerebral cortex and cerebellum in the setting of lamin B2 deficiency. Proc Natl Acad Sci USA 2010; 107: 5076–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang SH, Chang SY, Yin L, et al. An absence of both lamin B1 and lamin B2 in keratinocytes has no effect on cell proliferation or the development of skin and hair. Hum Mol Genet 2011; 20: 3537–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sullivan T, Escalante-Alcalde D, Bhatt H, et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol 1999; 147: 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonne G, Di Barletta MR, Varnous S, et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery–Dreifuss muscular dystrophy. Nat Genet 1999; 21: 285–288. [DOI] [PubMed] [Google Scholar]
- 32.Muchir A, Worman HJ. Emery–Dreifuss muscular dystrophy. Curr Neurol Neurosci Rep 2007; 7: 78–83. [DOI] [PubMed] [Google Scholar]
- 33.Fatkin D, MacRae C, Sasaki T, et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N Engl J Med 1999; 341: 1715–1724. [DOI] [PubMed] [Google Scholar]
- 34.Muchir A, Bonne G, van der Kooi AJ, et al. Identification of mutations in the gene encoding lamins A/C in autosomal dominant limb girdle muscular dystrophy with atrioventricular conduction disturbances (LGMD1B). Hum Mol Genet 2000; 9: 1453–1459. [DOI] [PubMed] [Google Scholar]
- 35.Lu JT, Muchir A, Nagy PL, et al. LMNA cardiomyopathy: cell biology and genetics meet clinical medicine. Dis Model Mech 2011; 4: 562–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quijano-Roy S, Mbieleu B, Bönnemann CG, et al. De novo LMNA mutations cause a new form of congenital muscular dystrophy. Ann Neurol 2008; 64: 177–186. [DOI] [PubMed] [Google Scholar]
- 37.Renou L, Stora S, Yaou RB, et al. Heart–hand syndrome of Slovenian type: a new kind of laminopathy. J Med Genet 2008; 45: 666–671. [DOI] [PubMed] [Google Scholar]
- 38.Bione S, Maestrini E, Rivella S, et al. Identification of a novel X-linked gene responsible for Emery–Dreifuss muscular dystrophy. Nat Genet 1994; 8: 323–327. [DOI] [PubMed] [Google Scholar]
- 39.Nagano A, Koga R, Ogawa M, et al. Emerin deficiency at the nuclear membrane in patients with Emery–Dreifuss muscular dystrophy. Nat Genet 1996; 12: 254–259. [DOI] [PubMed] [Google Scholar]
- 40.Manilal S, Nguyen TM, Sewry CA, et al. The Emery–Dreifuss muscular dystrophy protein, emerin, is a nuclear membrane protein. Hum Mol Genet 1996; 5: 801–808. [DOI] [PubMed] [Google Scholar]
- 41.Clements L, Manilal S, Love DR, et al. Direct interaction between emerin and lamin A. Biochem Biophys Res Commun 2000; 267: 709–714. [DOI] [PubMed] [Google Scholar]
- 42.Cao H, Hegele RA. Nuclear lamin A/C R482Q mutation in canadian kindreds with Dunnigan-type familial partial lipodystrophy. Hum Mol Genet 2000; 9: 109–112. [DOI] [PubMed] [Google Scholar]
- 43.Shackleton S, Lloyd DJ, Jackson SN, et al. LMNA, encoding lamin A/C, is mutated in partial lipodystrophy. Nat Genet 2000; 24: 153–156. [DOI] [PubMed] [Google Scholar]
- 44.Speckman RA, Garg A, Du F, et al. Mutational and haplotype analyses of families with familial partial lipodystrophy (Dunnigan variety) reveal recurrent missense mutations in the globular C-terminal domain of lamin A/C. Am J Hum Genet 2000; 66: 1192–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vigouroux C, Magré J, Vantyghem MC, et al. Lamin A/C gene: sex-determined expression of mutations in Dunnigan-type familial partial lipodystrophy and absence of coding mutations in congenital and acquired generalized lipoatrophy. Diabetes 2000; 49: 1958–1962. [DOI] [PubMed] [Google Scholar]
- 46.Lüdtke A, Genschel J, Brabant G, et al. Hepatic steatosis in Dunnigan-type familial partial lipodystrophy. Am J Gastroenterol 2005; 100: 2218–2224. [DOI] [PubMed] [Google Scholar]
- 47.Novelli G, Muchir A, Sangiuolo F, et al. Mandibuloacral dysplasia is caused by a mutation in LMNA-encoding lamin A/C. Am J Hum Genet 2002; 71: 426–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caux F, Dubosclard E, Lascols O, et al. A new clinical condition linked to a novel mutation in lamins A and C with generalized lipoatrophy, insulin-resistant diabetes, disseminated leukomelanodermic papules, liver steatosis, and cardiomyopathy. J Clin Endocrinol Metab 2003; 88: 1006–1013. [DOI] [PubMed] [Google Scholar]
- 49.Decaudain A, Vantyghem MC, Guerci B, et al. New metabolic phenotypes in laminopathies: LMNA mutations in patients with severe metabolic syndrome. J Clin Endocrinol Metab 2007; 92: 4835–4844. [DOI] [PubMed] [Google Scholar]
- 50.Dutour A, Roll P, Gaborit B, et al. High prevalence of laminopathies among patients with metabolic syndrome. Hum Mol Genet 2011; 20: 3779–3786. [DOI] [PubMed] [Google Scholar]
- 51.De Sandre-Giovannoli A, Chaouch M, Kozlov S, et al. Homozygous defects in LMNA, encoding lamin A/C nuclear-envelope proteins, cause autosomal recessive axonal neuropathy in human (Charcot–Marie–Tooth disorder type 2) and mouse. Am J Hum Genet 2002; 70: 726–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tazir M, Azzedine H, Assami S, et al. Phenotypic variability in autosomal recessive axonal Charcot–Marie–Tooth disease due to the R298C mutation in lamin A/C. Brain 2004; 127: 154–163. [DOI] [PubMed] [Google Scholar]
- 53.Walter MC, Witt TN, Weigel BS, et al. Deletion of the LMNA initiator codon leading to a neurogenic variant of autosomal dominant Emery–Dreifuss muscular dystrophy. Neuromuscul Disord 2005; 15: 40–44. [DOI] [PubMed] [Google Scholar]
- 54.Agarwal AK, Kazachkova I, Ten S, et al. Severe mandibuloacral dysplasia-associated lipodystrophy and progeria in a young girl with a novel homozygous Arg527Cys LMNA mutation. J Clin Endocrinol Metab 2008; 93: 4617–4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Merideth MA, Gordon LB, Clauss S, et al. Phenotype and course of Hutchinson–Gilford progeria syndrome. N Engl J Med 2008; 358: 592–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eriksson M, Brown WT, Gordon LB, et al. Recurrent de novo point mutations in lamin A cause Hutchinson–Gilford progeria syndrome. Nature 2003; 423: 293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Sandre-Giovannoli A, Bernard R, Cau P, et al. Lamin A truncation in Hutchinson–Gilford progeria. Science 2003; 300: 2055. [DOI] [PubMed] [Google Scholar]
- 58.Navarro CL, Cadiñanos J, De Sandre-Giovannoli A, et al. Loss of ZMPSTE24 (FACE-1) causes autosomal recessive restrictive dermopathy and accumulation of lamin A precursors. Hum Mol Genet 2005; 14: 1503–1513. [DOI] [PubMed] [Google Scholar]
- 59.Moulson CL, Go G, Gardner JM, et al. Homozygous and compound heterozygous mutations in ZMPSTE24 cause the laminopathy restrictive dermopathy. J Invest Dermatol 2005; 125: 913–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen L, Lee L, Kudlow BA, et al. LMNA mutations in atypical Werner’s syndrome. Lancet 2003; 362: 440–445. [DOI] [PubMed] [Google Scholar]
- 61.Verstraeten VL, Broers JL, van Steensel MA, et al. Compound heterozygosity for mutations in LMNA causes a progeria syndrome without prelamin A accumulation. Hum Mol Genet 2006; 15: 2509–2522. [DOI] [PubMed] [Google Scholar]
- 62.Garg A, Subramanyam L, Agarwal AK, et al. Atypical progeroid syndrome due to heterozygous missense LMNA mutations. J Clin Endocrinol Metab 2009; 94: 4971–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Padiath QS, Saigoh K, Schiffmann R, et al. Lamin B1 duplications cause autosomal dominant leukodystrophy. Nat Genet 2006; 38: 1114–1123. [DOI] [PubMed] [Google Scholar]
- 64.Hegele RA, Cao H, Liu DM, et al. Sequencing of the reannotated LMNB2 gene reveals novel mutations in patients with acquired partial lipodystrophy. Am J Hum Genet 2006; 79: 383–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dauer WT, Worman HJ. The nuclear envelope as a signaling node in development and disease. Dev Cell 2009; 17: 626–638. [DOI] [PubMed] [Google Scholar]
- 66.Worman HJ, Östlund C, Wang Y. Diseases of the nuclear envelope. Cold Spring Harb Perspect Biol 2010; 2: a000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Young SG, Fong LG, Michaelis S Prelamin A, Zmpste24, misshapen cell nuclei, and progeria—new evidence suggesting that protein farnesylation could be important for disease pathogenesis. J Lipid Res 2005; 46: 2531–2558. [DOI] [PubMed] [Google Scholar]
- 68.Worman HJ, Bonne G. ‘Laminopathies’: a wide spectrum of human diseases. Exp Cell Res 2007; 313: 2121–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Worman HJ, Fong LG, Muchir A, et al. Laminopathies and the long strange trip from basic cell biology to therapy. J Clin Invest 2009; 119: 1825–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lammerding J, Schulze PC, Takahashi T, et al. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest 2004; 113: 370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ji JY, Lee RT, Vergnes L, et al. Cell nuclei spin in the absence of lamin b1. J Biol Chem 2007; 282: 20015–20026. [DOI] [PubMed] [Google Scholar]
- 72.Lee JS, Hale CM, Panorchan P, et al. Nuclear lamin A/C deficiency induces defects in cell mechanics, polarization, and migration. Biophys J 2007; 93: 2542–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Verstraeten VL, Ji JY, Cummings KS, et al. Increased mechanosensitivity and nuclear stiffness in Hutchinson–Gilford progeria cells: effects of farnesyltransferase inhibitors. Aging Cell 2008; 7: 383–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hale CM, Shrestha AL, Khatau SB, et al. Dysfunctional connections between the nucleus and the actin and microtubule networks in laminopathic models. Biophys J 2008; 95: 5462–5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Folker ES, Östlund C, Luxton GW, et al. Lamin A variants that cause striated muscle disease are defective in anchoring trans-membrane actin-associated nuclear lines for nuclear movement. Proc Natl Acad Sci USA 2011; 108: 131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bergo MO, Gavino B, Ross J, et al. Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness, and a prelamin A processing defect. Proc Natl Acad Sci USA 2002; 99: 13049–13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pendás AM, Zhou Z, Cadiñanos J, et al. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase-deficient mice. Nat Genet 2002; 31: 94–99. [DOI] [PubMed] [Google Scholar]
- 78.Fong LG, Ng JK, Meta M, et al. Heterozygosity for Lmna deficiency eliminates the progeria-like phenotypes in Zmpste24-deficient mice. Proc Natl Acad Sci USA 2004; 101: 18111–18116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fong LG, Frost D, Meta M, et al. A protein farnesyltransferase inhibitor ameliorates disease in a mouse model of progeria. Science 2006; 311: 1621–1623. [DOI] [PubMed] [Google Scholar]
- 80.Varela I, Pereira S, Ugalde AP, et al. Combined treatment with statins and aminobisphosphonates extends longevity in a mouse model of human premature aging. Nat Med 2008; 14: 767–772. [DOI] [PubMed] [Google Scholar]
- 81.Yang SH, Meta M, Qiao X, et al. A farnesyltransferase inhibitor improves disease phenotypes in mice with a Hutchinson–Gilford progeria syndrome mutation. J Clin Invest 2006; 116: 2115–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang SH, Qiao X, Fong LG, et al. Treatment with a farnesyltransferase inhibitor improves survival in mice with a Hutchinson–Gilford progeria syndrome mutation. Biochim Biophys Acta 2008; 1781: 36–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Capell BC, Olive M, Erdos MR, et al. A farnesyltransferase inhibitor prevents both the onset and late progression of cardiovascular disease in a progeria mouse model. Proc Natl Acad Sci USA 2008; 105: 15902–15907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang SH, Andres DA, Spielmann HP, et al. Progerin elicits disease phenotypes of progeria in mice whether or not it is farnesylated. J Clin Invest 2008; 118: 3291–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang SH, Chang SY, Ren S, et al. Absence of progeria-like disease phenotypes in knock-in mice expressing a non-farnesylated version of progerin. Hum Mol Genet 2011; 20: 436–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang SH, Bergo MO, Toth JI, et al. Blocking protein farnesyltransferase improves nuclear blebbing in mouse fibroblasts with a targeted Hutchinson–Gilford progeria syndrome mutation. Proc Natl Acad Sci USA 2005; 102: 10291–10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Toth JI, Yang SH, Qiao X, et al. Blocking protein farnesyltransferase improves nuclear shape in fibroblasts from humans with progeroid syndromes. Proc Natl Acad Sci USA 2005; 102: 12873–12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Capell BC, Erdos MR, Madigan JP, et al. Inhibiting farnesylation of progerin prevents the characteristic nuclear blebbing of Hutchinson–Gilford progeria syndrome. Proc Natl Acad Sci USA 2005; 102: 12879–12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mallampalli MP, Huyer G, Bendale P, et al. Inhibiting farnesylation reverses the nuclear morphology defect in a HeLa cell model for Hutchinson–Gilford progeria syndrome. Proc Natl Acad Sci USA 2005; 102: 14416–14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Glynn MW, Glover TW. Incomplete processing of mutant lamin A in Hutchinson–Gilford progeria leads to nuclear abnormalities, which are reversed by farnesyltransferase inhibition. Hum Mol Genet 2005; 14: 2959–2969. [DOI] [PubMed] [Google Scholar]
- 91.Gordon LB, Harling-Berg CJ, Rothman FG. Highlights of the 2007 Progeria Research Foundation scientific workshop: progress in translational science. J Gerontol A Biol Sci Med Sci 2008; 63: 777–787. [DOI] [PubMed] [Google Scholar]
- 92.Wang Y, Panteleyev AA, Owens DM, et al. Epidermal expression of the truncated prelamin A causing Hutchinson–Gilford progeria syndrome: effects on keratinocytes, hair and skin. Hum Mol Genet 2008; 17: 2357–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu B, Wang J, Chan KM, et al. Genomic instability in laminopathy-based premature aging. Nat Med 2005; 11: 780–785. [DOI] [PubMed] [Google Scholar]
- 94.Liu Y, Wang Y, Rusinol AE, et al. Involvement of xeroderma pigmentosum group A (XPA) in progeria arising from defective maturation of prelamin A. FASEB J 2008; 22: 603–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Constantinescu D, Csoka AB, Navara CS, et al. Defective DSB repair correlates with abnormal nuclear morphology and is improved with FTI treatment in Hutchinson–Gilford progeria syndrome fibroblasts. Exp Cell Res 2010; 316: 2747–2759. [DOI] [PubMed] [Google Scholar]
- 96.Liu GH, Barkho BZ, Ruiz S, et al. Recapitulation of premature ageing with iPSCs from Hutchinson–Gilford progeria syndrome. Nature 2011; 472: 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kudlow BA, Stanfel MN, Burtner CR, et al. Suppression of proliferative defects associated with processing-defective lamin A mutants by hTERT or inactivation of p53. Mol Biol Cell 2008; 19: 5238–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Benson EK, Lee SW, Aaronson SA. Role of progerin-induced telomere dysfunction in HGPS premature cellular senescence. J Cell Sci 2010; 123: 2605–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cao K, Blair CD, Faddah DA, et al. Progerin and telomere dysfunction collaborate to trigger cellular senescence in normal human fibroblasts. J Clin Invest 2011; 121: 2833–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Scaffidi P, Misteli T. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat Cell Biol 2008; 10: 452–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hernandez L, Roux KJ, Wong ES, et al. Functional coupling between the extracellular matrix and nuclear lamina by Wnt signaling in progeria. Dev Cell 2010; 19: 413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Espada J, Varela I, Flores I, et al. Nuclear envelope defects cause stem cell dysfunction in premature-aging mice. J Cell Biol 2008; 181: 9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cao K, Graziotto JJ, Blair CD, et al. Rapamycin reverses cellular phenotypes and enhances mutant protein clearance in Hutchinson–Gilford progeria syndrome cells. Sci Transl Med 2011; 3: 89ra58. [DOI] [PubMed] [Google Scholar]
- 104.Muchir A, Pavlidis P, Decostre V, et al. Activation of MAPK pathways links LMNA mutations to cardiomyopathy in Emery–Dreifuss muscular dystrophy. J Clin Invest 2007; 117: 1282–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Muchir A, Pavlidis P, Bonne G, et al. Activation of MAPK in hearts of EMD null mice: similarities between mouse models of X-linked and autosomal dominant Emery–Dreifuss muscular dystrophy. Hum Mol Genet 2007; 16: 1884–1895. [DOI] [PubMed] [Google Scholar]
- 106.Muchir A, Shan J, Bonne G, et al. Inhibition of extracellular signal-regulated kinase signaling to prevent cardiomyopathy caused by mutation in the gene encoding A-type lamins. Hum Mol Genet 2009; 18: 241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu W, Shan J, Bonne G, et al. Pharmacological inhibition of c-Jun N-terminal kinase signaling prevents cardiomyopathy caused by mutation in LMNA gene. Biochim Biophys Acta 2010; 1802: 632–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wu W, Muchir A, Shan J, et al. Mitogen-activated protein kinase inhibitors improve heart function and prevent fibrosis in cardiomyopathy caused by mutation in lamin A/C gene. Circulation 2011; 123: 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]