Abstract
Inspired by this elegant system of cellular adaptivity, we herein report the rational design of a dynamic artificial adaptive system able to sense and respond to environmental stresses in a unique sense-and-respond mode. Utilizing DNA nanotechnology, we constructed an artificial signal feedback network and anchored it to the surface membrane of a model giant membrane vesicle (GMV) protocell. Such a system would need to both senses incoming stimuli and emit a feedback response to eliminate the stimuli. To accomplish this mechanistically, our DNA-based artificial signal system, hereinafter termed DASsys, was equipped with a DNA trigger-induced DNA polymer formation and dissociation machinery. Thus, through a sequential cascade of stimulus-induced DNA strand displacement, DASsys could effectively sense and respond to incoming stimuli. Then, by eliminating the stimulus, the membrane surface would return to its initial state, realizing the formation of a cyclical feedback mechanism. Overall, our strategy opens up a route to the construction of artificial signaling system capable of maintaining homeostasis in the cellular micromilieu, and addresses important emerging challenges in bioinspired engineering.
Cells are constantly bombarded by incoming signals and cues from the outside environment. Some of these signals are adverse, e.g., invasion of foreign pathogens. Nonetheless, cells are capable of recognizing changes to the cellular micromilieu and then appropriately respond to, or otherwise dispense with, these stimuli in a manner that affords homeostasis by the ability to constantly return to prestimulus state.1–5 Mimicking this dynamic cellular adaptivity ensembles through the design and construction of artificial signaling systems based on the functional integration of artificial cell-like entities (protocells) is a major challenge that has important technological implications in bottom-up synthetic biology, bioinspired engineering and microscale engineering of intelligent machines and devices.6,7
Recently, a range of studies focused on simulating cell-like functions, such as membrane gating, gene circuit and cascade, genetic replication and cell proliferation have been reported.8–11 However, the development of protocell equipped with artificial signaling system that exhibiting cellular adaptivity with recyclable capability remains a considerable challenge. So far, colloidosomes undergoing cell-like autonomous shape oscillations and feedback-induced temporal control of breathing polymersomes, as well as beating vesicles that cause dynamic membrane deformation have been demonstrated to show the stimulation-responsive protocell behavior.12–15 Nevertheless, these studies mainly focused on morphology deformations upon activation by external or internal triggers.
To fabricate an artificial signal system into synthetic protocells, our group conceptualized the rational design of a dynamic network able to sense and respond to environmental stresses in a unique sense-and-respond mode. As a powerful molecular tool for engineering artificial molecular systems, DNA nanotechnology provides an opportunity to develop artificial dynamic networks capable of monitoring environmental changes.16 Based on dynamic DNA operations, sophisticated artificial molecular signaling systems have already been developed, including neural network computation,17 adaptive immune responsive circuits,18 logic gates19 and molecular cascades.20,21 Contributing to this array of groundbreaking discoveries, we herein report the design of a protocell with a built-in artificial signal system able to sense environmental stresses and activate cyclical feedback, thus exhibiting a simple form of cellular adaptivity.
With this in mind, we constructed a DNA-based artificial signal system (DASsys) and anchored it to the surface membrane of a model giant membrane vesicle (GMV) protocell to replicate, through biomimicry, cellular adaptivity with important implications for environmentally induced signaling and triggering of feedback networks. This process is established by dynamic DNA operations on the GMVs membrane surface and enzymes in solution (Figure 1). Through a sequential cascade of stimulus-induced DNA strand displacement reaction, the DNA-based artificial signal system, or DASsys, exhibits an active response to environmental stimulus and then eliminates the stimulus, thus setting up a cyclical feedback component with the aim of maintaining a pre-established baseline, as noted above.
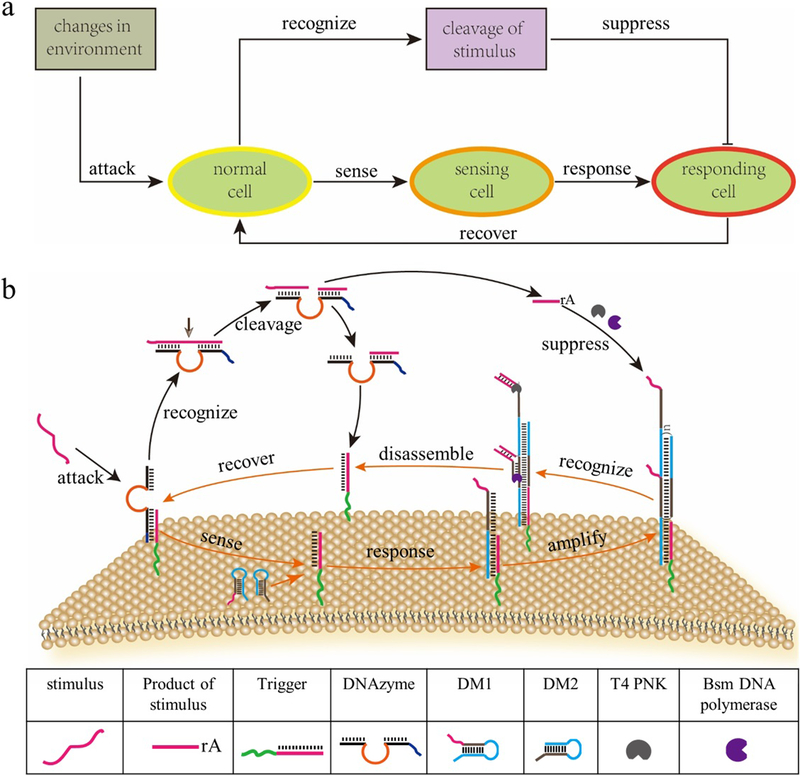
Figure 1.
Construction of DASsys model GMV protocell. (a) Design concept of DASsys-based protocell model responding to environmental stimulation. (b) Recyclable DASsys anchored on the membrane surface.
The following summarizes the reactions involved in each of the adaptivity steps, including, as noted previously, external stimulus, cell sensing and self-protection, stimulus elimination and cell recovery. These steps consisted of a network that belongs to an incoherent feedforward loop.4 Initially, ssDNA was assigned as the external stimulus. Meanwhile, cholesterol (chol)-labeled DNA trigger could spontaneously anchor on the GMV protocell’s membrane surface by hydrophobic interaction with its lipid bilayer. Afterward, DNAzyme was immobilized on the membrane surface by complementary base pairing with the DNA trigger. The binding efficiency of DNA trigger and DNAzyme was tested in solution (Figure S1), and both DNA trigger and DNAzyme hybrids could be successfully anchored on the membrane surface and used as sensors for detecting stimulus (Figure S2). The addition of the stimulus to the sensory system leads to signal transduction of DNAzyme and activation of the DNA trigger by DNA strand displacement reaction, setting the stage for sensing to take place on the GMVs membrane. Next, upon recognition of stimulus, DNA monomers (DMs) are recruited by the DNA trigger, as responding effectors, and DNA polymer is formed on the membrane surface. Upon binding with the toehold of DM1, the hairpin structure of DM1 is unfolded by chol-labeled DNA trigger. The unfolded DM1 continues to recruit DM2 through binding its toehold, followed by the formation of the DNA polymer on the surface of sensing GMVs in principle of hybridization chain reaction (HCR). Then, the stimulus is cleaved in two by DNAzyme in the present of bivalent magnesiumion, the cleaved product, binds with the toehold of DNA polymer leading to its disassembly in the presence of T4 Polynucleotide Kinase and Bsm DNA polymerase. Accordingly, the DNA polymer is reversed, and DNA trigger again binds with DNAzyme. Upon the elimination of stimulus, the membrane surface is fully recovered and able to respond to the next incoming stimulus. This DASsys was optimized and worked perfectly in Tris-HCl buffer solution (Figure S3, S4 and S5). The kinetic reaction of DNA-based artificial signal system was also characterized, as shown in Figures S6–S10. Moreover, this DNA-based artificial signaling system shows good sensitivity and precision (Figure S11, S12), due to the specific base-pairing of complementary DNA sequences.
To anchor DASsys on the membrane surface of GMVs, our model protocell, a chemically modified DNA trigger, the 3’-end of which was labeled with cholesterol and the 5’-end of which was labeled with 6-carboxyfluorescein (FAM) dye, was used. Biomimetic GMVs were prepared and used as previously reported.22 The immobilization of DNA trigger and DNAzyme on membrane surface was verified by confocal microscopy and flow cytometry (Figure S2).
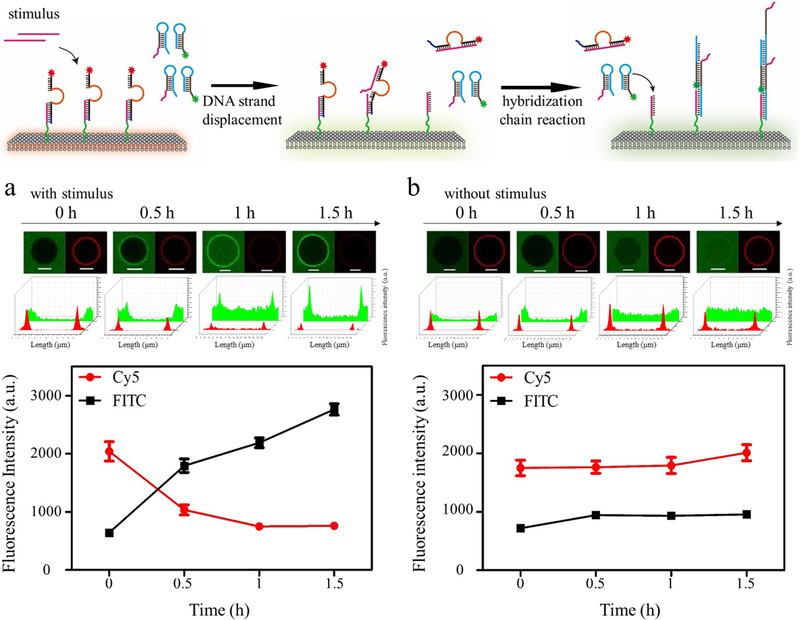
Having anchored the DNA-based sensor on the membrane surface, the signal transduction of DASsys was tested. To monitor this process, FAM-labeled DM2 and Cy5-labeled DNAzyme were used. Upon the addition of stimulus, GMVs anchored with the DNA-based sensor were observed under confocal microscope. As shown in Figure 2a, the fluorescence intensity of Cy5 on the membrane surface gradually decreased, indicating that the addition of the stimulus leads to signal transduction of DNAzyme and activation of the DNA trigger by DNA strand displacement reaction. Meanwhile, the fluorescence intensity of FAM on the membrane surface gradually increased, indicating that DMs had been recruited by the DNA trigger and that DNA polymer was formed on the membrane surface. For the control system absent stimulus, only negligible increase in green fluorescence intensity on the membrane surface was observed over a period of hours, reflecting the low background for DMs without stimulus (Figure 2b).
Figure 2.
Confocal image of GMVs modified with DNA-based sensor with (a) and without (b) the addition of stimulus. The fluorescence intensity of FAM or Cy5 was calculated. Scale bar is 5 μm.
In DASsys, recognition of stimulus by DNAzyme turns on the DNA polymer formation on the surface of GMVs, and the produced DNA polymer acts as self-protection from environmental stimulation. However, the membrane surface of GMVs will not recover to prestimulus state if the DNA polymer does not disassemble. Herein, two enzymes, T4 PNK and DNA Bsm polymerase, were used to disassemble the DNA polymer on the membrane surface. Since the 3’-end of cleaved stimulus has bound to the 2’, 3’ cyclic phosphate group (Figure S13, S14), which blocked the strand extension by DNA Bsm polymerase, T4 PNK now takes action to remove the 3’ cyclic phosphate group, allowing the cleaved stimulus product to be extended by the DNA Bsm polymerase (Figure S15).23 Thus, once the cleaved product has been separated from DNAzyme, it binds with the toehold of DNA polymer and acts as a primer for DNA extension in the presence of DNA polymerase. Subsequently, the DNA polymer immobilized on the membrane surface was disassembled (Figure S16, S17).
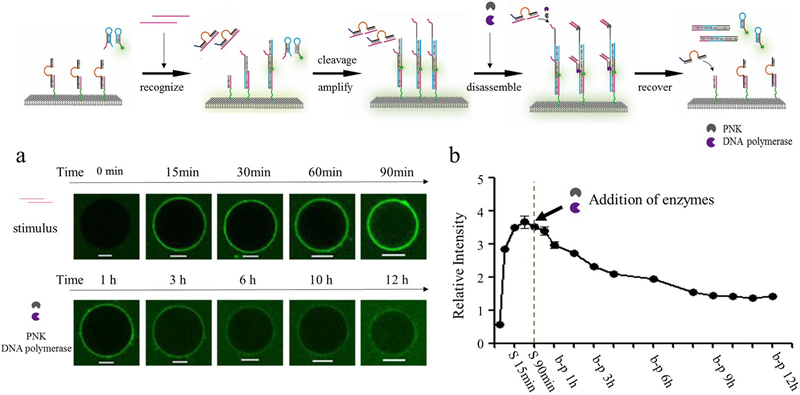
To achieve coupling of the signal transduction process with dynamic DNA assembly on GMVs, FAM-labeled DM2 was used, and the system was tested upon the addition of stimulus and enzymes consecutively. As shown in Figure 3, during the signal transduction process, including the recognition of stimulus and DNA polymer formation, fluorescence intensity on the membrane surface increases rapidly within 15 min. Upon the addition of enzymes, decreased fluorescence intensity was observed, indicating the disassembly of DNA polymer on the membrane surface. This observation shows that the signal transduction process had coupled with dynamic DNA assembly to detect external stimuli and transduce the detected signal into the output to evoke subsequent reactions, all of which represents an organism’s adaptation to environmental change (Figure S18).5,24–26
Figure 3.
Schematic representation of stimulus-induced DNA polymer assembly and disassembly after the addition of PNK and Bsm DNA polymerase. (a) Confocal images of DNA polymer anchored on membrane surface before (first panel) and after (second panel) the addition of PNK and Bsm DNA polymerase. (b) The mean fluorescence intensity of DNA polymer was calculated, and the plot shows the assembly and disassembly of DNA polymer in the presence of stimulus (S) and enzymes (b-p). Scale bar is 5 μm.
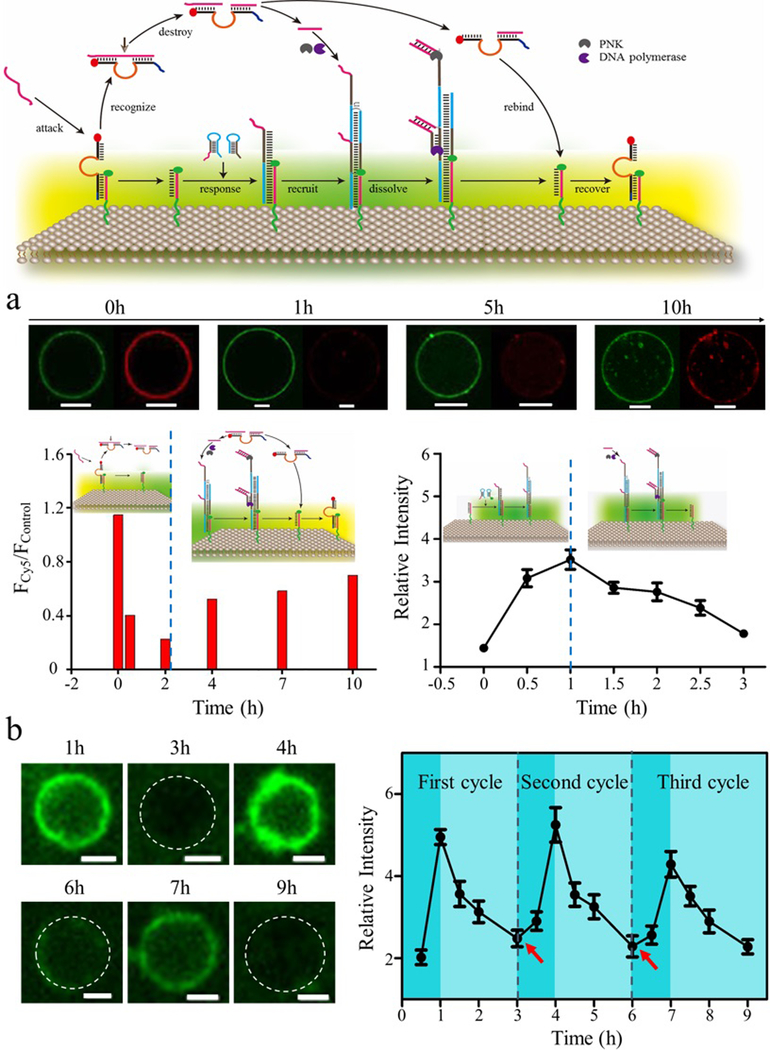
An adaptive system can both respond to an incoming stimulus and then recover to prestimulus level, enabling cells to continue sensing and responding to changes in their micromilieu.4,27,28 To verify the returning of DNAzyme on the membrane surface after the cleavage of stimulus, the fluorescence intensity of Cy5-labeled DNAzyme and FAM-labeled trigger was monitored by confocal microscopy. As shown in Figure 4a, the intact fluorescence intensity of FAM on the membrane surface indicated that the trigger was stable and could be anchored on the membrane surface during the whole process. Meanwhile, the decreased fluorescence intensity of Cy5 indicated that DNAzyme had been released from the membrane surface in the presence of stimulus. Importantly, the recovery of fluorescence intensity of Cy5 could be observed after prolonged incubation time, indicating the rebinding of DNAzyme with DNA trigger anchored on the membrane surface (Figure 4a, left).
Figure 4.
Schematic representation of recyclable DNA-based artificial signal system. (a) Confocal images of FAM-labeled trigger and Cy5-labeled DNAzyme on membrane surface at the different time period. The fluorescence intensity of Cy5 with (FCy5) or without (Fcontrol) addition of stimulus was measured, and the value of FCy5/Fcontrol was calculated. The decreased and subsequently increased fluorescence intensity of Cy5 indicated the release and rebinding of DNAzyme with trigger anchored on the membrane surface (a, left). The increased and subsequently decreased fluorescence intensity of DNA polymer indicated the assembly and disassembly of DNA polymer in response to an external stimulus (a, right). Scale bar is 5 μm. (b) Confocal images of GMVs equipped with DASsys for repeated cycling in response to external stimuli. The plot shows the assembly and disassembly of DNA polymer for repeated cycles. Arrows indicated the supplementation of stimulus and DM1. Scale bar is 2 μm.
To verify the recyclable signal system on the membrane surface, FAM-labeled DM2 was used. Upon the addition of stimulus, DNA strand displacement occurred, and the result was reflected by the increased green fluorescence intensity on the membrane surface. Meanwhile, once stimulus was cleaved into two pieces by DNAzyme, one of the cleaved pieces bound with the toehold of DNA polymer containing FAM-labeled DM2 and dissociate the fluorescence DNA polymer in the presence of DNA polymerase, as indicated by the decrease of green fluorescence intensity on the membrane surface (Figure 4a right). The signaling process is recyclable and can be switched on and off by supplementing DM1 and stimulus to the external solution, which results in the generation of reciprocal signal transduction of DASsys (Figure 4b). Significantly, the counteracting reaction was related to recovery reaction as designed, and the interactive cycle formed after recovery reaction was completed in DASsys. Thus, DASsys equipped GMVs represents a de novo synthetic approach that exhibits finely tuned continuous self-adjustment, integrated functionalities, and adaptivity to environmental stimulation.
In summary, our study reports the construction of a DNA-based signal system capable of responding to environmental stimuli, thus providing an engineering approach to induce interaction between protocell and the environment. GMVs equipped with DASsys not only sense and respond to stimuli but also can self-renew by recycling to prestimulus state, thereby permitting continuous sensing and responding in the micromilieu. The simplicity of this concept suggests that it will be broadly applicable to diverse stimuli and thus open up a wide range of applications in establishing self-protection in protocell communities and bioinspired nanotechnology.
Supplementary Material
ACKNOWLEDGMENTS
This work is supported by NSFC grants (No. 21502050, 21521063) and by NIH GM R35 127130 and NSF 1645215, Beijing National Laboratory for Molecular Sciences (BNLMS201806).
Footnotes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge on the A CS Publications website at DOI: 10.1021/jacs.8b13816.
Additional information as noted in text (PDF)
REFERENCES
- (1).Buddingh BC; van Hest JCM Artificial cells: synthetic compartments with life-like functionality and adaptivity. Acc. Chem. Res 2017, 50 (4), 769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Coates PJ; Lorimore SA; Wright EG Cell and tissue responses to genotoxic stress. J. Pathol 2005, 205 (2), 221–235. [DOI] [PubMed] [Google Scholar]
- (3).Barandiaran X; Moreno A Adaptivity from metabolism to behavior. Adapt Behav 2008, 16, 325–344. [Google Scholar]
- (4).Ma W; Trusina A; El-Samad H; Lim WA; Tang C Defining network topologies that can achieve biochemical adaptation. Cell 2009, 138 (4), 760–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Briat C; Gupta A; Khammash M Antithetic integral feedback ensures robust perfect adaptation in noisy biomolecular Networks. Cell Syst 2016, 2 (1), 15–26. [DOI] [PubMed] [Google Scholar]
- (6).Te Brinke E; Groen J; Herrmann A; Heus HA; Rivas G; Spruijt E; Huck WTS Dissipative adaptation in driven self-assembly leading to self-dividing fibrils. Nat. Nanotechnol 2018, 13 (9), 849–855. [DOI] [PubMed] [Google Scholar]
- (7).Gobbo P; Patil AJ; Li M; Harniman R; Briscoe WH; Mann S Programmed assembly of synthetic protocells into thermoresponsive prototissues. Nat. Mater 2018, 17 (12), 1145–1153. [DOI] [PubMed] [Google Scholar]
- (8).Spruijt E; Tusk SE; Bayley H DNA scaffolds support stable and uniform peptide nanopores. Nat. Nanotechnol 2018, 13 (8), 739–745. [DOI] [PubMed] [Google Scholar]
- (9).Weitz M; Kim J; Kapsner K; Winfree E; Franco E; Simmel FC Diversity in the dynamical behaviour of a compartmentalized programmable biochemical oscillator. Nat. Chem 2014, 6 (4), 295–302. [DOI] [PubMed] [Google Scholar]
- (10).Weitz M; Kim J; Kapsner K; Winfree E; Franco E; Simmel FC Diversity in the dynamical behaviour of a compartmentalized programmable biochemical oscillator. Nat. Chem 2014, 6 (4), 295–302. [DOI] [PubMed] [Google Scholar]
- (11).Kurihara K; Tamura M; Shohda K; Toyota T; Suzuki K; Sugawara T Self-reproduction of supramolecular giant vesicles combined with the amplification of encapsulated DNA. Nat. Chem 2011, 3 (10), 775–81. [DOI] [PubMed] [Google Scholar]
- (12).Che H; Cao S; van Hest JCM Feedback-induced temporal control of “breathing” polymersomes to create self-adaptive nanoreactors. J. Am. Chem. Soc 2018, 140 (16), 5356–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Litschel T; Ramm B; Maas R; Heymann M; Schwille P Beating vesicles: Encapsulated protein oscillations cause dynamic membrane deformations. Angew. Chem 2018, 130 (50), 16522–16527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Tamate R; Ueki T; Yoshida R Self-beating artificial cells: Design of cross-linked polymersomes showing self-oscillating motion. Adv. Mater 2015, 27 (5), 837–842. [DOI] [PubMed] [Google Scholar]
- (15).Tamate R; Ueki T; Yoshida R Evolved colloidosomes undergoing cell-like autonomous shape oscillations with buckling. Angew. Chem., Int. Ed 2016, 55 (17), 5179–5183. [DOI] [PubMed] [Google Scholar]
- (16).Seeman NC; Sleiman HF DNA nanotechnology. Nat. Rev. Mater 2017, 3 (1), 17068. [Google Scholar]
- (17).Qian L; Winfree E; Bruck J Neural network computation with DNA strand displacement cascades. Nature 2011, 475 (7356), 368. [DOI] [PubMed] [Google Scholar]
- (18).Han D; Wu C; You M; Zhang T; Wan S; Chen T; Qiu L; Zheng Z; Liang H; Tan W A cascade reaction network mimicking the basic functional steps of adaptive immune response. Nat. Chem 2015, 7 (10), 835–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Peng R; Zheng X; Lyu Y; Xu L; Zhang X; Ke G; Liu Q; You C; Huan S; Tan W Engineering a 3D DNA-logic gate nanomachine for bispecific recognition and computing on target cell surfaces. J. Am. Chem. Soc 2018, 140 (31), 9793–9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Xin L; Zhou C; Yang Z; Liu D Regulation of an enzyme cascade reaction by a DNA machine. Small 2013, 9 (18), 3088–3091. [DOI] [PubMed] [Google Scholar]
- (21).Chen X; Briggs N; McLain JR; Ellington AD Stacking nonenzymatic circuits for high signal gain. Proc. Natl. Acad. Sci. U. S. A 2013, 110 (14), 5386–5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Peng R; Wang H; Lyu Y; Xu L; Liu H; Kuai H; Liu Q; Tan W Facile assembly/disassembly of DNA nanostructures anchored on cell-mimicking giant vesicles. J. Am. Chem. Soc 2017, 139 (36), 12410–12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Zhao Y; Zhou L; Tang Z Cleavage-based signal amplification of RNA. Nat. Commun 2013, 4, 1493. [DOI] [PubMed] [Google Scholar]
- (24).Shoval O; Goentoro L; Hart Y; Mayo A; Sontag E; Alon U Fold-change detection and scalar symmetry of sensory input fields. Proc. Natl. Acad. Sci. U. S. A 2010, 107 (36), 15995–6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Kim J; Khetarpal I; Sen S; Murray RM Synthetic circuit for exact adaptation and fold-change detection. Nucleic Acids Res 2014, 42 (9), 6078–6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Parkinson JS; Hazelbauer GL; Falke JJ Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol 2015, 23 (5), 257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Ferrell JE Jr, Perfect and near-perfect adaptation in cell signaling. Cell Sys 2016, 2 (2), 62–67. [DOI] [PubMed] [Google Scholar]
- (28).Araujo RP; Liotta LA The topological requirements for robust perfect adaptation in networks of any size. Nat. Commun 2018, 9 (1), 1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.