Abstract
We investigated the role of 3D genome architecture in instructing functional properties of glioblastoma stem cells (GSCs) by generating sub-5-kb resolution 3D genome maps by in situ Hi-C. Contact maps at sub-5-kb resolution allow identification of individual DNA loops, domain organization, and large-scale genome compartmentalization. We observed differences in looping architectures among GSCs from different patients, suggesting that 3D genome architecture is a further layer of inter-patient heterogeneity for glioblastoma. Integration of DNA contact maps with chromatin and transcriptional profiles identified specific mechanisms of gene regulation, including the convergence of multiple super enhancers to individual stemness genes within individual cells. We show that the number of loops contacting a gene correlates with elevated transcription. These results indicate that stemness genes are hubs of interaction between multiple regulatory regions, likely to ensure their sustained expression. Regions of open chromatin common among the GSCs tested were poised for expression of immune-related genes, including CD276. We demonstrate that this gene is co-expressed with stemness genes in GSCs and that CD276 can be targeted with an antibody-drug conjugate to eliminate self-renewing cells. Our results demonstrate that integrated structural genomics data sets can be employed to rationally identify therapeutic vulnerabilities in self-renewing cells.
Glioblastoma (GBM) is the most common malignant brain tumor in adults, and current treatments are mostly palliative (Stupp et al. 2005). Development of rationally designed new treatments for this malignancy has been hampered by inter-tumoral and intra-tumoral heterogeneity (Verhaak et al. 2010; Brennan et al. 2013). Recent work has shown that intra-tumoral heterogeneity can be observed at the genomic, epigenomic, and functional level. Genomic heterogeneity is manifested by the co-occurrence of multiple subclonal compartments in primary tumors and their dynamic selection at recurrence (Snuderl et al. 2011; Szerlip et al. 2012; Sottoriva et al. 2013; Johnson et al. 2014; Meyer et al. 2015). Functional heterogeneity is reflected by the nonequipotency of GBM cells: Only a subpopulation of cells with self-renewal properties contributes to sustained tumor growth.
These self-renewing cells can engraft in transplantation experiments in immunocompromised mice, and they contribute to genetic tumor evolution (Galli et al. 2004; Singh et al. 2004; Lan et al. 2017). We refer to these cells as cancer stem cells, or GBM stem cells (GSCs). GSCs are resistant to standard-of-care therapy and are thought to be responsible for tumor recurrence (Bao et al. 2006; Eramo et al. 2006). We and others have shown that chromatin (Heddleston et al. 2012; Gallo et al. 2013, 2015) and transcriptional profiles (Patel et al. 2014; Suvà et al. 2014) differ between GSCs and non-GSCs, suggesting that intra-tumoral epigenetic heterogeneity may reflect functional differences between populations of stem-like and more differentiated cells. It is therefore possible that specific epigenetic and chromatin states are pivotal in maintaining a functional hierarchy in GBM, similar to their roles in preserving hierarchical systems required for normal tissue homeostasis (Abdouh et al. 2009; Suvà et al. 2009; Heddleston et al. 2012; Gallo et al. 2013; Kim et al. 2013; Jin et al. 2017; Miller et al. 2017). However, it is not currently known how chromatin states and cancer-associated genetic lesions interact to shape overall three-dimensional (3D) genome architecture in GSCs.
3D genome organization refers to the stereotypical folding of each chromosome in three-dimensional space. Hi-C sequencing (Lieberman-Aiden et al. 2009) allows characterization of genome architecture by measuring the frequency of physical interactions between genomic regions. Throughout this manuscript, we use the following terms to describe different features of genome architecture as inferred from Hi-C data: “Interactions” refer broadly to any above-background frequency of DNA contacts; “loops” refer to discrete points of high contact frequency between nonadjacent genomic regions; and “domains” refer to contiguous genomic regions involved in self-interactions that occur at higher frequencies than with neighboring regions. Loops frequently occur at the edges of a domain, are usually nucleated by the presence of antiparallel binding sites for CCCTC-binding factor (CTCF) (Rao et al. 2014), and often serve to connect regulatory regions, such as enhancers and superenhancers (SEs) (Whyte et al. 2013), to their target genes. Genomes can be further segregated into type A and type B compartments, which broadly correspond to open and closed chromatin, respectively (Lieberman-Aiden et al. 2009; Dixon et al. 2012; Stevens et al. 2017). Integration of information on loops, domains, and compartment organization is required to achieve precise understanding of genome organization for a specific cell type.
Recent work has shown that disruption of genome architecture may contribute to the etiology of developmental disorders (Lupiáñez et al. 2015) and cancer (Flavahan et al. 2016) through dysregulation of gene expression programs. The global impact of genome architecture on transcriptional programs in gliomas and other brain tumors has been difficult to assess because of the lack of high-resolution 3D genome maps for these cancers. We hypothesized that differences in 3D genome architecture could explain transcriptional heterogeneity between patients with GBM. We have therefore generated sub-5-kb resolution Hi-C contact maps from patient-derived primary GSC cultures. Integration of Hi-C, chromatin immunoprecipitation (ChIP-seq), and transcriptomics (RNA-seq) data sets for each sample generated a comprehensive view of transcriptional control mechanisms in GSCs. We provide proof-of-principle that 3D genome information can be used to identify new potential therapeutic targets to curb the self-renewal properties of GSCs.
Results
Genes contacted by DNA loops in GSCs exhibit elevated expression
To characterize 3D genome architecture in GBM, we generated sub-5-kb resolution DNA contact maps using in situ Hi-C (Rao et al. 2014) from low-passage GSC cultures from three patients diagnosed with GBM (G523, G567, and G583) (patient metadata presented in Supplemental Table S1). Two biological replicates were performed for each culture. Because inter-replicate contact map Pearson correlation values exceeded 0.9, biological replicates were merged to maximize contact map resolution for each culture (Supplemental Fig. S1A; Supplemental Table S2; see Methods). We annotated loops, domains, and compartment membership genome-wide for each sample (Supplemental Fig. S1B–F; Supplemental Tables S3–S5). Although two-thirds of genes (16,466 of 24,261 annotated) were not involved in loop formation in any sample, 7795 genes were engaged by at least one loop and 3111 genes were engaged with at least two loops in any one GSC culture (Supplemental File S1). Subsequently, we integrated Hi-C and RNA-seq data for these cultures to investigate the impact of loop formation on transcriptional profiles (for RNA-seq summary statistics, see Supplemental Table S6). We found that genes contacted by a single loop were expressed at significantly higher levels than those that were not part of any loop. Genes contacted by two or more loops were expressed at significantly higher levels than genes contacted by a single loop (Supplemental Fig. S1G). This finding suggested that gene expression can be enhanced by the convergence of multiple regulatory regions.
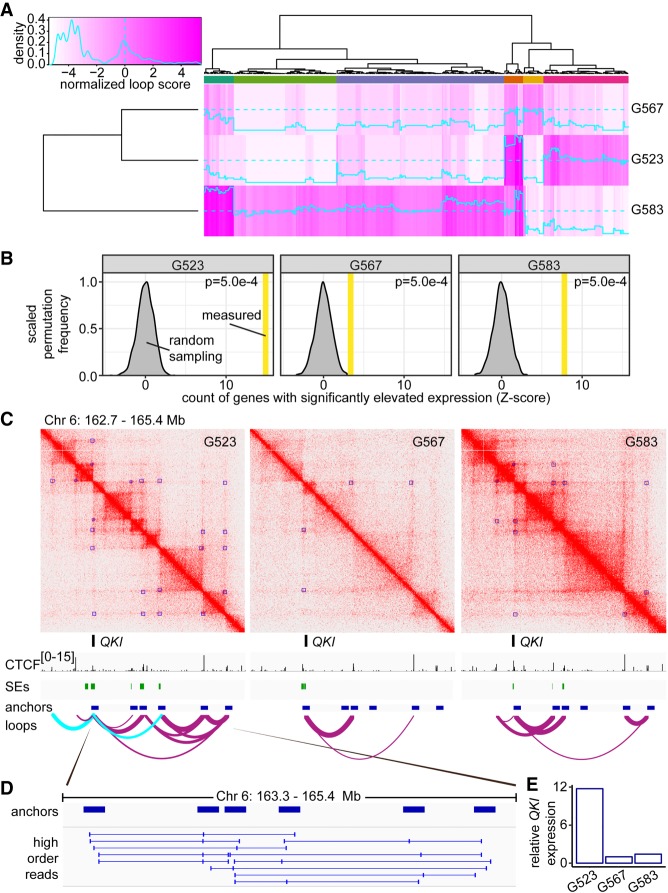
Culture-specific loops drive differential expression between GSCs
Overall, loop calls between GSC cultures were highly correlated, with pairwise Spearman's correlation scores ranging from 0.79–0.82 between cultures (Supplemental Fig. S1C). However, despite broad similarity, there remained many differences in loop presence and loop strength between cultures (Fig. 1A). To better understand the interplay between genome architecture and chromatin marks in achieving GSC culture-specific gene expression profiles, we generated ChIP-seq data sets for CTCF and the open chromatin mark H3K27ac (see Supplemental Fig. S1H–M for quality control). ChIP-seq, SE calls, and Hi-C data were then integrated for each GSC culture. Genes that contacted an SE via a loop had significantly elevated expression (Supplemental Fig. S1N). To examine whether differential looping impacted transcriptional output, we identified culture-specific loops and tested for inter-culture differential expression of genes at the anchors of these loops using RNA-seq data sets. Permutation analysis indicated that culture-specific loops were enriched for genes with elevated expression in that culture as compared to the others (Fig. 1B). These results suggest that 3D genome architecture may contribute to transcriptional heterogeneity in GBM cells.
Figure 1.
Culture-specific loops promote gene expression in GSCs. (A) Heat map of loop scores for loops in the top 2% of variance between cultures. Top multicolored bar indicates clusters of loops with shared patterns of differential representation between the cultures. Loops called by HiCCUPS as 5-kb–100-kb resolution merged loops throughout this figure. (B) Enrichment for genes with elevated expression at culture-specific loops. Gray curve: frequency of detecting significantly elevated expression determined by 2000 permutations of randomly sampled expression values from genes with nonunique loops. Vertical bar: measured number of differentially expressed genes found overlapping culture-specific loops, expressed as Z-score. (C) Hi-C contact maps for G523, G567, and G583 surrounding the QKI locus displayed at 5-kb resolution. Green track: superenhancers called using ROSE. Purple arc tracks: loops identified by HiCCUPS (union of 5-, 10-, and 25-kb calls) with thickness proportional to loop score. Cyan arc tracks: culture-specific loops to QKI present in G523. (D) Chimeric reads derived from the same DNA fragment that aligns to more than two loop anchors. (E) Expression of QKI in G523, G567, and G583 was determined by RNA-seq. Y-axis represents read counts normalized to G567 to give fold enrichment values.
A representative example of culture-specific loops affecting transcription is offered by the QKI locus, which differentially engages SEs in each of the three GSC cultures we profiled. The number of SEs looped to QKI in G523, G567, and G583 was three, zero, and two, respectively (Fig. 1C, green track). G523 had two culture-specific loops contacting the QKI gene (Fig. 1C, cyan arc tracks). Additionally, the QKI locus exhibited many transitive loops—multiple loops that share common anchor loci, potentially allowing a higher-order, multiloop hub to exist within an individual cell (Rao et al. 2017). To investigate the possibility that QKI could be regulated by a transitive loop hub, we reanalyzed Hi-C library sequence alignments near these loop anchors for evidence of split alignments corresponding to more than two DNA regions within the same sequenced molecule. This analysis revealed interactions between triplets of loop anchors in different combinations (Fig. 1D), supporting the notion that a multiloop hub places QKI in direct contact with multiple control regions concurrently within individual cells. Consistent with the observation of multiple, culture-specific loops, QKI expression was nearly 12 times higher in culture G523 than G567 and G583 (Fig. 1E). Together, these results demonstrate that culture-specific loops are associated with the transcriptional differences evident between GSC cultures.
Genomic structural variants result in new SE-promoter interactions
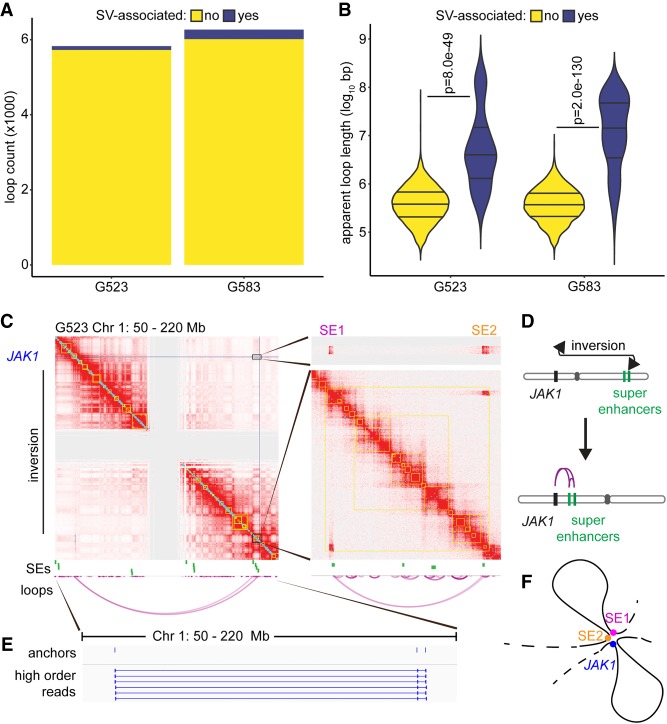
A wide range of loop lengths was observed within each GSC culture. Over 80% of the loops detected were <1 Mb in length (Supplemental Fig. S1B), which corresponds with the average length of contact domains in human cells (Dixon et al. 2012; Nora et al. 2012). However, we also observed many putative long-range contacts between genomic regions over 1 Mb apart, including rare loops spanning an apparent distance >100 Mb (Supplemental Fig. S1B; Supplemental Table S3). We hypothesized that these unusually long loop lengths could be explained by structural variants (SVs) in chromosomes, such as deletions or inversions. Upon alignment of Hi-C reads to the unrearranged genome, large SVs would cause DNA interactions to appear at much greater distances than their true molecular distance following rearrangement in the GSC genomes. We therefore used Manta (Chen et al. 2016) to annotate SVs using whole-genome sequences available for cultures G523 and G583 (Supplemental Files S2, S3) and intersected SV loci with loop anchors to identify SV-associated loops (Supplemental Files S4, S5). Although SV-associated loops represented <5% of loop calls for each culture (Fig. 2A), the median apparent length of these loops was over an order of magnitude longer than loops without nearby SVs (Fig. 2B). The percentages of SV-associated loops reported here should be considered to be the lower bound of the true prevalence; numerous inter-chromosomal SVs were evident in the GSC contact maps, but inter-chromosomal loops could not be assessed using HiCCUPS as this method is currently limited to identifying only intra-chromosomal loops.
Figure 2.
Genomic rearrangements cause differential superenhancer interactions in GSCs. (A) Number of loops associated with local SVs. Loops called by HiCCUPS as 5-kb–100-kb resolution merged loops throughout this figure. (B) Loop length separated by SV status. SV-associated loops tend to connect genomic loci separated by a much larger apparent distance, although this is unlikely the true molecular distance following chromosomal rearrangement. P-values calculated by Wilcoxon rank-sum test. (C) Hi-C contact maps assuming a standard chromosomal order indicate the formation of a ∼140-Mb loop connecting JAK1 to two superenhancers at the other end of Chromosome 1. The central gray region lacks signal throughout due to repetitive pericentromeric regions with ambiguous sequence alignments. Contact maps displayed at 250-kb resolution for the left panel and 5-kb resolution for the right panels. Loops represent the union of 5-, 10-, and 25-kb HiCCUPS calls. (D) Schematic indicating how a large inversion brings the superenhancers and JAK1 in close proximity. (E) Chimeric reads aligning to JAK1 and both superenhancers. Additional higher-order reads were detected, but not all are displayed due to redundancy. (F) Diagrammatic representation of the convergence of two SEs (SE1 and SE2) to the JAK1 locus in G523.
For a representative example of the effects of large SVs on 3D genome architecture and transcriptional output, we examined the JAK1 locus in culture G523. A large inversion spanning 140 Mb, as well as other large associated deletions, shifted two SEs normally located on the q-arm of Chromosome 1 close to JAK1 on the p-arm (Fig. 2C,D). Hi-C data showed clear physical interactions between the two SEs (SE1 and SE2) and JAK1, as illustrated by the heat maps and the loop arc plots (Fig. 2C). These interactions were not identified in the other two GSC cultures, which did not carry the Chromosome 1 inversion. Our computational analysis found high-order reads mapping to SE1, SE2, and JAK1 (Fig. 2E), providing evidence that these three elements form a contact hub in at least a subpopulation of cells (Fig. 2F). This example highlights how a gene with known roles in GBM (McFarland et al. 2011) and other cancer types (Thomas et al. 2015) can be regulated by nonstandard SEs as a result of structural variation.
Genes connected to multiple loops are enriched for stemness and cancer genes
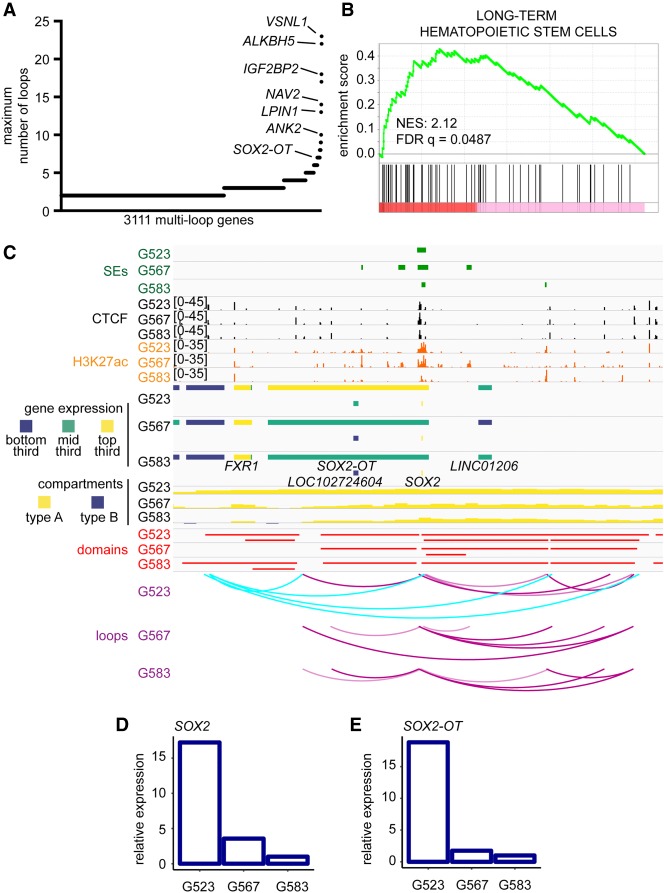
Following the observation that transcription is elevated at genes possessing multiple loops (Supplemental Fig. S1G), we hypothesized that genes with extensive looping would be enriched for genes promoting maintenance of self-renewal, a key cancer stem cell property (Al-Hajj and Clarke 2004; Laks et al. 2009). First, we annotated the number of loops contacting each gene in our GSC cultures and identified 3111 genes that were targeted by at least two loops in any one GSC culture (Supplemental File S1). Certain genes displayed extreme interactivity, with the most highly looped gene in our cultures being VSNL1, targeted by 23 loops (Fig. 3A). We validated these findings by using C3D which identifies 3D interactions between cis-regulatory elements based on correlations in chromatin accessibility between loci in different samples (Supplemental Fig. S2A; Mehdi et al. 2019).
Figure 3.
Interplay of 3D genome organization and chromatin features in transcriptional control of stemness genes in GBM. (A) Genes ranked by the number of loops they contact. Only genes with at least two loops are displayed. Loops called by HiCCUPS as 5-kb–100-kb resolution merged loops throughout this figure. (B) Gene set enrichment analysis based on gene ranking in A. (C) Integration of Genome Browser tracks for ROSE superenhancer calls, CTCF ChIP-seq, H3K27ac ChIP-seq, RNA-seq, compartments (50-kb), domains (10-kb), and loops (union of 5-, 10-, and 25-kb calls) determined by Hi-C at the SOX2 locus. Cyan arc tracks indicate a hub of culture-specific loops in G523. (D,E) Relative expression of SOX2 and SOX2-OT as determined by RNA-seq.
Of the genes with the highest numbers of loops, many had known associations with brain tumor and stem cell biology (summarized in Supplemental Table S7). Accordingly, we ranked genes based on the maximum number of loops they were associated with in any one GSC culture and performed gene set enrichment analysis (GSEA) (Fig. 3B; Subramanian et al. 2005). We found significant positive enrichment of genes involved in regulation of insulin secretion (normalized enrichment score [NES]: 2.19; q = 0.0886) (Supplemental Fig. S2B), metastasis (NES: 2.16; q = 0.0530) (Supplemental Fig. S2C), and long-term hematopoietic stem cells (Ivanova et al. 2002) (NES: 2.12; q = 0.0487) (Fig. 3B). Overall, our data show that highly connected gene sets in GSCs are significantly enriched for genes with established roles in cancer and in regulation of stemness properties (Supplemental Fig. S2A).
Integration of Hi-C and chromatin data allows dissection of mechanisms of transcriptional regulation
Expression of stemness genes is important for maintenance of GSC self-renewal. Some of these stemness factors are expressed at different levels in GSCs from different patients. Therefore, we assessed transcriptional differences at established stemness genes and the underlying differences in chromatin marks and genome architecture at such loci.
The locus surrounding SOX2 exhibits similar compartment and domain organization between GSC samples. All three cultures indicate strong H3K27ac signal and identify the region overlapping SOX2 as an SE locus. Distinctively, the culture G523 possesses several culture-specific loops emanating from an upstream region (Fig. 3C). High-order reads indicate that SOX2 simultaneously is engaged in formation of multiple loops in G523 (Supplemental Fig. S3A). Accordingly, SOX2 expression is found to be much higher in G523 compared to the other cultures (Fig. 3D). Sharing a similar genomic context, the overlapping noncoding RNA SOX2-OT is also found to be elevated in G523 (Fig. 3E). Similar observations were made for ASCL1, which was shown to be important for the maintenance of self-renewal and tumorigenic properties of GSCs (Supplemental Fig. S3B,C; Rheinbay et al. 2013; Suvà et al. 2014). These findings suggest that genome architecture and chromatin states are not independently sufficient to achieve specific transcriptional outputs.
Differential genome compartmentalization between GSC cultures affects transcriptional output
Despite many transcriptional differences between GSC cultures being associated with differential loops, the differential compartmentalization observed at the ASCL1 locus prompted us to assess the influence of compartmentalization on transcription genome-wide. Type A/B compartment status was determined for 93% of the genome in at least one culture (2.86 Gbp of 3.09 Gbp), with similar numbers and sizes of compartments identified for each culture (Supplemental Table S5). The correlation in compartment membership between GSCs ranged from 0.57 to 0.73 (Spearman's correlation) (Supplemental Fig. S1E), with only 53% of compartment assignment (1.52 Gbp of 2.86 Gbp defined in any culture) being homogenously called as type A or type B in all three cultures. This suggests a higher degree of dissimilarity at the compartment level than at the loop level of genome architecture (Supplemental Fig. S1C). As expected, genes in type A compartments exhibited elevated expression, while genes in type B compartments were generally repressed (Supplemental Fig. S4A,B). The number of differentially expressed genes was significantly increased in culture-specific compartments (Supplemental Fig. S4C).
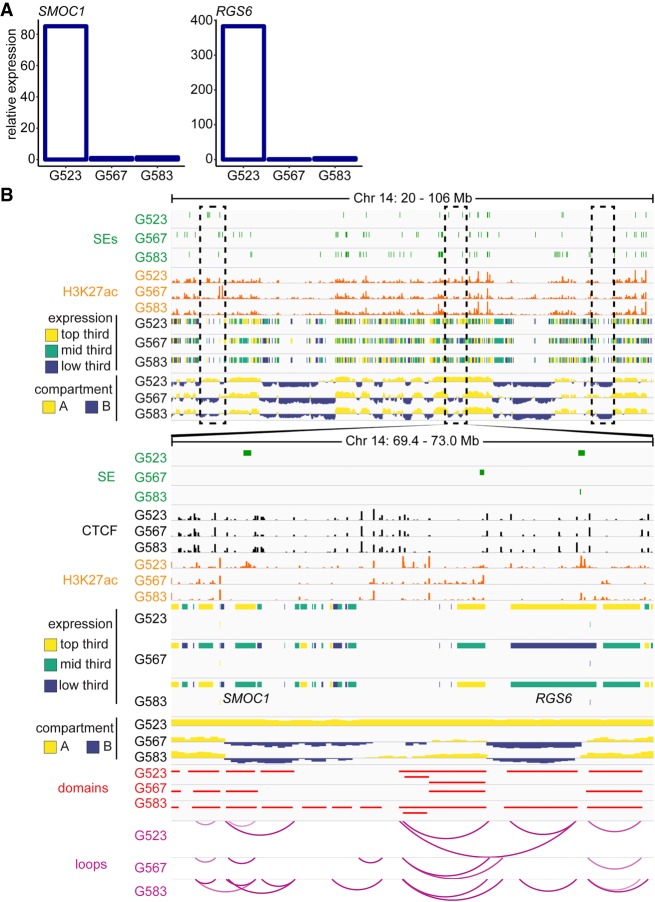
To examine the effects of differential compartmentalization on gene regulation, we present the region of Chromosome 14 harboring SMOC1 and RGS6. Expression was ∼80× and ∼400× higher in G523 as compared to the other cultures for SMOC1 and RGS6, respectively (Fig. 4A). Differences in loops and domain structures between cultures are subtle in this region, however; only culture G523 exhibits type A compartmentalization spanning these genes (Fig. 4B). Of note, H3K27ac signal was present at the RGS6 locus in both G523 and G583, yet G583 did not exhibit the same elevated transcription. These data highlight differential compartmentalization as an important source of transcriptional heterogeneity between GSC cultures.
Figure 4.
Culture-specific compartmentalization of SMOC1 and RGS6. (A) Relative expression of SMOC1 and RGS6. (B) Example of culture-specific compartmentalization in GSCs. Compartmentalization called at 50-kb, domains at 10-kb, and loops as the union of 5-, 10-, and 25-kb calls.
Using Hi-C data to identify potential therapeutic targets
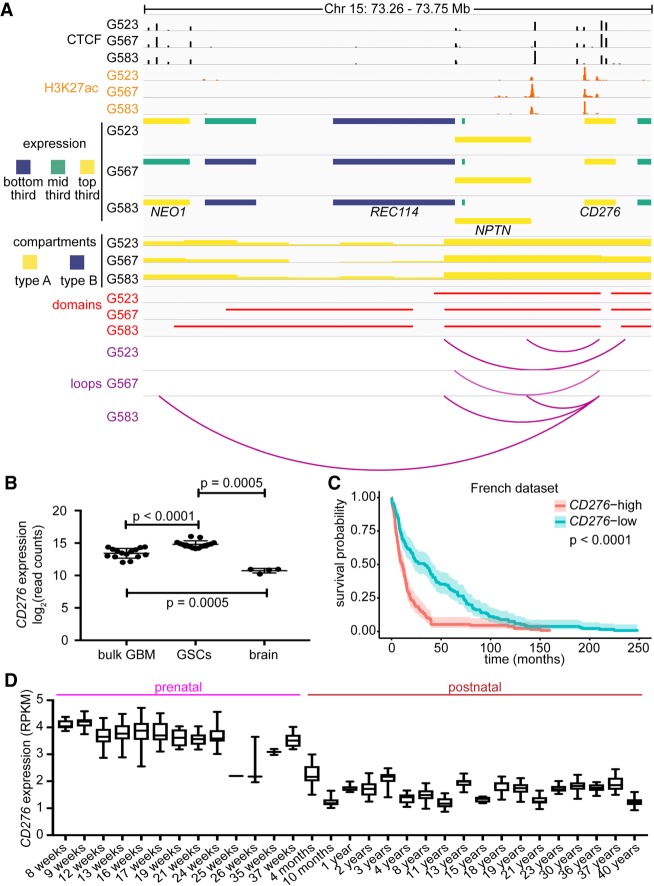
Having established that regions of differential genome architecture can account for culture-specific differences in transcription, we next sought to determine if shared elements of genome architecture between GSC cultures could identify genes important for the key stemness property of self-renewal. To identify potential therapeutic targets in GSCs, we looked for genes that met the following criteria: (1) type A compartment status in all GSC cultures; (2) share a common loop among all GSC cultures; (3) expressed >10 FPKM in all GSC cultures; (4) encode cell surface proteins as determined by Gene Ontology (GO) Term 0009986; and (5) potentially targetable with current or experimental compounds. The gene CD276 (also known as B7-H3) met all these criteria. In all three GSC cultures, the CD276 locus was contacted by a CTCF-delimited loop ∼140 kb upstream of the 5′ ends of genes NPTN and REC114 (Fig. 5A). Additional loops connect CD276 to a putative upstream enhancer element in G523 and G583, while G583 possesses a longer, culture-specific loop to CTCF binding sites within the gene body of NEO1. Therefore, although all three GSC cultures have high expression of CD276 (top third of expression genome-wide) (Fig. 5A) and similar H3K27ac signals, its transcriptional regulation may be further tuned by differential genome architecture. This is reflected by different transcriptional outputs for CD276 among the three GSC cultures (Supplemental Fig. S5A).
Figure 5.
CD276 is an immune gene with elevated expression in GSCs. (A) 3D genome and chromatin landscape at the CD276 locus. This panel integrates CTCF ChIP-seq, H3K27ac ChIP-seq, RNA-seq, compartments (50-kb), domains (10-kb), and loops (union of 5-, 10-, and 25-kb calls) determined by Hi-C for three patient-derived GSC cultures (G523, G567, and G583). (B) Expression of CD276 was determined by RNA-seq in bulk GBM samples (n = 76), GSCs (n = 76), and non-neoplastic brain tissue (n = 4). P-values were calculated with the Mann–Whitney U statistical test. (C) Survival of glioma patients stratified by CD276 expression in the French data set. Median gene expression was used to stratify patients. P-value was derived with log-rank statistics. Shading around curve indicates 95% confidence interval. (D) Patterns of expression of CD276 in the prenatal and postnatal human brain. Data were extracted from BrainSpan.
CD276 is a putative immune checkpoint regulator (Lee et al. 2017) and is thought to be a repressor of T cell activation, preventing release of IFNG by T cells and impairing CD4+ cell proliferation (for review, see Picarda et al. 2016). However, its exact role in immunomodulation is still controversial (Hofmeyer et al. 2008). CD276 is aberrantly expressed in a number of solid tumors and is detected on both tumor cells and in the tumor vasculature (Seaman et al. 2017). Indeed, CD276 exhibits elevated expression in GBM relative to lower grade astrocytomas, promotes tumor infiltration, and allows cells to resist lysis by NK cells (Lemke et al. 2012). We found that CD276 expression was elevated in GSC cultures (n = 76) compared to bulk GBM (n = 76; P < 0.001) or non-neoplastic brain tissue (n = 4; P = 0.0005) (Fig. 5B).
We next asked whether transcription of CD276 is associated with a previously published 16-gene GBM self-renewal signature (Suvà et al. 2014). We calculated Pearson correlation scores for CD276, the 16-gene signature, and for a selected subset of immune genes that we found to be highly expressed in GSCs, using bulk RNA-seq data from a collection of patient-derived GSC cultures (n = 76). We found that, although CD276 expression was most strongly correlated with other immune modulators (IFNGR1, IFNGR2, TNFRSF1A, and TNFSRF1B), it was also positively correlated with a cluster of self-renewal genes, including VAX2, SOX21, and CITED1 (Supplemental Fig. S5B). This association with the stemness signature was unique to CD276, as the other immune genes found were anticorrelated with the stemness signatures. Further meta-analysis of published data sets showed that expression of CD276 stratifies adult glioma patients, with high CD276 expression being a negative prognostic factor (Fig. 5C; Supplemental S5C,D,E). Transcription data from the BrainSpan atlas (Miller et al. 2014) reveals that CD276 expression in the human brain is higher during the prenatal period (average RPKM 3.74) than the postnatal years (average RPKM 1.72; two-tailed t-test P = 1.04 × 10−211) (Fig. 5D). This immune-related gene might therefore be associated with a primordial developmental program, providing a rationale for its association with the expression of self-renewal genes in GSCs.
Targeting CD276 reduces self-renewal properties of GSCs
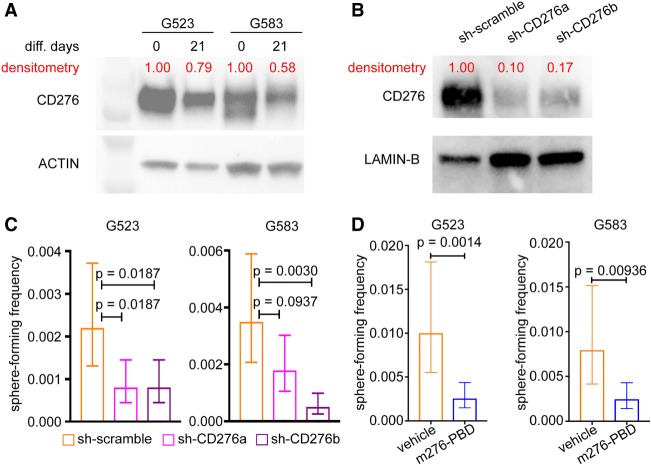
Considering the co-expression of CD276 and stemness genes in GSCs, we further explored a possible role of CD276 in the self-renewal program of GSCs. To assess CD276 levels in cells with reduced self-renewal capacity, we induced loss of self-renewal using an established growth factor withdrawal protocol (Pollard et al. 2009; Gallo et al. 2015). Growth factor withdrawal resulted in decreased protein levels for CD276 in G523 and G583 (Fig. 6A), although it had no effect on G567 (Supplemental Fig. S6A). In a complementary experiment, we hypothesized that reducing levels of CD276 could cause decreased GSC self-renewal. Accordingly, we knocked down CD276 using two inducible shRNA constructs (sh-CD276a and sh-CD276b) (Fig. 6B). The effects of CD276 knockdown on self-renewal were assessed with in vitro limiting dilution assays (LDAs). For both G523 and G583, knockdown of CD276 caused significant reduction of self-renewal (Fig. 6C). These data functionally validate a role of CD276 in maintaining the self-renewing state of GSCs.
Figure 6.
CD276 as a potential therapeutic target. (A,B) Western blots comparing CD276 levels between cultures exposed to (A) growth factor withdrawal, or (B) shRNA constructs targeting CD276. Densitometry was performed to normalize CD276 signals to their respective loading controls. (C) Limiting dilution analysis for G523 and G583 transfected with either scramble (control) or shRNA constructs targeting CD276 (sh-CD276a and sh-CD276b). Data show mean sphere-forming frequency. Error bars correspond to 95% confidence interval. P-values were determined with ELDA. (D) Limiting dilution analysis for G523 and G583 treated with m276-PBD or vehicle control. Data show mean sphere-forming frequency. Error bars correspond to 95% confidence interval. P-values were determined with ELDA.
Subsequently, we explored tools that allow targeting of CD276+ cells and have translational potential. Therapeutics against CD276, including a bispecific CD3-CD276 antibody (ClinicalTrials.gov identifier NCT02628535), are active agents used in clinical trials. We explored an alternative strategy, using an anti-CD276 monoclonal antibody conjugated with pyrrolobenzodiazepine (PBD) to selectively eradicate CD276+ cells. This m276-PBD antibody-drug conjugate (ADC) was recently shown to be selective in delivering the PBD warhead to CD276+ cells in tumors and vasculature and to reduce tumor burden in several xenograft and mouse models of solid tumors (Seaman et al. 2017). We performed LDAs by administering IC10 concentrations of m276-PBD (Supplemental Fig. S6B; see Methods) to G523 and G583 to assess the effects of the ADC on their self-renewal. For both G523 and G583, m276-PBD effectively curbed self-renewal (Fig. 6D). Together, these data strongly suggest that targeting CD276+ cells might be a promising strategy to curb cancer stem cell properties in adult gliomas.
Discussion
We report that inter-sample 3D genome heterogeneity is a molecular feature of GBM. Generation of sub-5-kb Hi-C contact maps allowed us to call loops connecting regulatory regions to their target genes. Together with the other genomic data sets we generated, these Hi-C maps will be an important resource for the brain tumor field and for future studies on the relationships between 3D genome, epigenetic factors, and transcriptional regulation.
Our Hi-C maps highlighted that GSC cultures exhibit clear differences in DNA looping and compartmentalization. The total number of looping interactions for a specific gene was correlated with increased expression, and culture-specific loops had a strong effect on potentiating transcriptional output. Some of these culture-specific loops engage SEs that are not available in other cultures and might reflect mechanisms of epigenetic evolution of the cancer cells. Our results point to an important contribution of genome architecture to transcriptional heterogeneity in GBM.
Using primary surgical resections, we previously reported that the key self-renewal factors like SOX2 are widely expressed in GBM cells, with between 5% and 50% of cells being SOX2+ in different samples (Gallo et al. 2013). This result is in apparent contradiction with functional studies that showed that cancer stem cells are rare in this tumor type (Singh et al. 2004). However, our data show that numerous stemness genes tend to be co-expressed in GSCs. We propose that coregulation of many stemness genes is achieved through the combined action of epigenetic inputs and transcription factor occupancy to achieve a consensus 3D genome state conducive to GSC self-renewal, with the expectation that perturbation of this state could destabilize transcriptional programs of self-renewal, with consequent loss of stemness properties. Considering that GSCs are thought to nucleate tumor relapse, identification of methods to induce loss of self-renewal properties is a key goal of GSC research and could allow relapse to be delayed or prevented in patients.
By identifying 3D genome features common among GSC cultures, we aimed at identifying new members of the self-renewal signature. By integrating these findings with RNA-seq data sets for 76 patient-derived primary GSC cultures, we found that CD276 is expressed at higher levels in GSCs than in their matched bulk GBM samples or in non-neoplastic brain. Recent preclinical studies showed that an m276-PBD ADC was effective at drastically reducing or even abrogating tumor growth in several cancer models (Seaman et al. 2017). Here, we show that m276-PBD can be used to target CD276+ GSCs to curb their self-renewal potential. Of note, m276-PBD kills cells in a cell cycle-independent manner. This is important, because self-renewing cells in vivo have been shown to be relatively slow-cycling (Lan et al. 2017), partially explaining their resistance to radiotherapy and the alkylating agent temozolomide that are currently used to treat GBM patients. During the review process, another group published data in support of a role for CD276 in a stemness circuitry in GBM (Zhang et al. 2019). We propose that adding m276-PBD to the current standard of care might result in the eradication of self-renewing cells and slow the emergence of recurrences. Besides m276-PBD, other strategies have been developed to target CD276+ cells in cancer. One of them, a CD3-CD276 bispecific antibody, is currently in clinical trials for several solid malignancies, including neuroblastoma and head and neck cancer. Adding GBM patients to these clinical trials could be beneficial for patients. Additionally, chimeric antigen receptor (CAR) T cells could be used to target CD276, exploiting the several-fold higher expression of CD276 in GSCs compared to non-neoplastic brain. The identification of CD276 as a putative new therapeutic target in GBM provides proof-of-principle that structural genomics and other genomic data sets can be successfully integrated to delineate self-renewal states and their vulnerabilities.
Methods
GSC cell culture
Use of all specimens and derivation of primary cultures was approved by the Health Research Ethics Board of Alberta and the research ethics board of the Hospital for Sick Children (Toronto, ON). Sample acquisition and derivation of GSC primary cultures G523, G567, and G583 were performed using previously described methods (Pollard et al. 2009). Primary glioblastoma cultures were grown in NeuroCult NS-A Basal Medium and Proliferation Supplement (StemCell Technologies, #05751), supplemented with 20 μg/mL rhEGF (Peprotech, AF-100-15), 10 μg/mL bFGF (StemCell Technologies, #78003), and 2 μg/mL heparin (StemCell Technologies, #07980). Cultures were grown adherently on Corning Primaria dishes coated with poly-L-ornithine (Sigma-Aldrich, P4957) and laminin (Sigma-Aldrich, L2020), under standard temperature, oxygen, and humidity conditions. All cultures were used within the first 20 passages of generation. Adherent cells were disassociated with Accutase (Stemcell Technologies, #07920) and plated onto fresh, coated plates when confluence reached 80%–90%. Viability and cell numbers were determined using Countess II (Thermo Fisher Scientific, AMQAX1000).
Loss of self-renewal assays
Induced loss of self-renewal in GSCs was performed according to established protocols (Pollard et al. 2009; Gallo et al. 2015). Briefly, 24 h after replating, the media was replaced with NS-A media lacking EGF and FGF. After 1 wk, the media was replaced with differentiation media consisting of half-volume NS-A media (without NeuroCult supplement), half-volume Neurobasal media (Thermo Fisher Scientific, #21103049), 2× B27 supplement (Thermo Fisher Scientific, #17504-044), and 0.5× N2 supplement (Thermo Fisher Scientific, #17502001). After 2 wk in this media, cells were collected for analysis.
Knockdown of CD276 with short hairpin RNA
Targeted gene silencing was performed with shRNA constructs targeting CD276 (Dharmacon) (see Supplemental Methods for construct details). Nucleofection was performed using the Mouse Neural Stem Cell Nucleofector Kit (Lonza, VPG-1004) and Amaxa Biosystems Nucleofector I, program A-33. For each nucleofection, 6 μg of shRNA plasmid was applied to 2 × 106 cells. Following nucleofection, cells were allowed to recover for 24 h in media with 1× antibiotic-antifungal. Expression was then induced with 1 µg/mL doxycycline for 24 h. Selection for transfected cells was performed by adding 1.5 µg/mL of puromycin to media for 48 h, after which cells were harvested for western blot and/or limiting dilution assays.
Dose-response curves for m276-PBD
Cells were plated at 3000 cells/well in NS media into laminin–poly-L-ornithine-coated 96-well-plates (Corning). The antibody-drug conjugate, m276-PBD, was tested at different concentrations (40 nM–1.5 nM range) (see Supplemental Fig. S6B) in PBS. Each concentration was applied to six wells for each culture; PBS-only was used as a control. Cell viability was assessed on day 4. alamarBlue (Thermo Fisher Scientific, Cat# DAL1025) was added and incubated at 37°C in the dark for 4 h. Fluorescence was measured on the Spectramax spectrophotometer and normalized to the PBS control.
Limiting dilution assays
Cells were plated on uncoated 96-well plates in a twofold dilution series spanning from 2000 down to 4 cells per well in NeuroCult NS-A media (StemCell Technologies, #05751). A total of six replicates were plated for each concentration. Sphere formation frequency was estimated using ELDA (Hu and Smyth 2009).
For m276-PBD: Drug was diluted to 2× the LD10 of the culture (LD10 for G523: 5 nM; LD10 for G583: 0.2 nM) in NS-A media. For treated wells, the compound was applied to a final concentration of 1× LD10, while an equivalent amount of PBS was applied to the control plate. Sphere formation was scored on day 4.
For CD276 knockdown: Cells transfected with either sh-CD276 constructs or scramble shRNA control (see “Knockdown of CD276 with short hairpin RNA” above) were used in LDAs as described above. Sphere formation was scored on day 7.
Western blots
Protein concentration was determined using a DC (detergent-compatible) protein assay (Bio-Rad, #5000112). Samples were prepared in 20 μL at 15 μg/μL in Laemmli loading buffer. Samples were run on 7.5% Mini-PROTEAN gels (Bio-Rad, #4568025). Primary antibodies used: anti-CD276 (Cell Signaling, #14508S, Lot# 1) at 1:1000; mAb Anti-β-Actin (Sigma-Aldrich, A5441, Lot# 127M4866V, Clone AC-15) at 1:1000; anti-LAMIN B1 antibody (Abcam, #16048, Lot# GR3197369-1) at 1:10,000. Secondary antibodies used: goat anti-rabbit IgG H&L (HRP) (Abcam, #6721, Lot# GR3192725-6) at 1:20,000; goat anti-mouse IgG H&L (HRP) (Abcam, #6789, Lot# GR242773-5) at 1:5000. Densitometry was performed with ImageJ (https://imagej.nih.gov) (Schneider et al. 2012).
Hi-C library generation and sequencing
Hi-C libraries were generated by an in situ Hi-C library protocol (Rao et al. 2014) with minor modifications. Cellular inputs consisted of 2.5 million cells from GSC cultures that had been grown to passage 11–15. See Supplemental Methods for complete library generation protocol.
For quality control of our Hi-C libraries, we generated ∼20 million reads per library with a NextSeq 500 (Illumina) at the Centre for Health Genomics and Informatics (University of Calgary) and assessed library quality using HiCUP (Wingett et al. 2015). Final sequencing was performed on an Illumina HiSeq 2500 at the Princess Margaret Cancer Centre (Toronto, ON). Analysis of Hi-C libraries was performed using Juicer and the associated Juicer Tools (Durand et al. 2016). Loop calls were generated using HiCCUPS and represent the union of calls made at 5-, 10-, and 25-kb resolution. Domains were called using Arrowhead at each resolution of 5-, 10-, 25-, 50-, and 100-kb, with only 10-kb domain calls presented in the figures. Compartment calls were generated from principal components of the Pearson correlation matrix of 50-kb contact maps. See Supplemental Methods for all parameters used.
Enrichment of genomic features
Enriched gene sets were identified using GSEA (Subramanian et al. 2005). Statistical significance of overlap between differentially expressed genes, loops, compartments, methylation probes, CTCF sites, and structural variants was calculated by permutation testing wherein the observed intersection between two features was compared to the overlap measured by iterative subsampling of a larger background data set. Permutation calculations and plot generation were performed using R 3.5.3 (R Core Team 2019).
Evidence of higher-order reads
To identify sequencing reads supporting higher-order loops, all reads proximate to loop anchors of interest were extracted from the “abnormal.sam” alignment file produced by Juicer. This file contains alignments for reads that failed to be incorporated in the Hi-C contact map, including those that may have ambiguously aligned to more than two genomic positions. Reads were identified as potential higher-order reads if they had three alignments on the same chromosome with each separated by at least 100 kb. Interactions of more than three positions were not detected from a single fragment likely due to rarity of these fragments within the libraries as well limitations of sequencing depth and length.
Chromatin immunoprecipitation with highly parallel sequencing (ChIP-seq)
ChIP-seq was done as previously described (Magnani et al. 2013; Gallo et al. 2015) with some modifications. Antibodies used were anti-CTCF (Cell Signaling, #3418, Lot #1) and anti-H3K27ac (Active Motif, #39133, Lot# 31814008). ChIP-seq peaks were called using MACS2 callpeak (Zhang et al. 2008) on filtered alignments from BWA-MEM (Li and Durbin 2009). Superenhancer regions were then identified using ROSE (Lovén et al. 2013; Whyte et al. 2013) on our H3K27ac data. See Supplemental Methods for the complete library generation protocol and analysis parameters.
C3D Analysis
C3D (Mehdi et al. 2019) was run using signal from individual replicate bedGraph tracks for H3K27ac ChIP, open regions as the union of H3K27ac peak calls for all three GSC cultures, and interaction anchors as gene bodies ±5 kb. An interaction window of 5 Mb around gene bodies was considered, and only interactions with q-value ≤0.1 were maintained.
Stem cell and immune gene clustering
Log normalized read counts from RNA-sequencing data were analyzed using R 3.4.3 (R Core Team 2017). A Pearson correlation was computed between all genes, and hierarchical clustering was applied to the result to cluster genes with similar expression patterns. Results were plotted using heatmap.2 in the gplots package (https://cran.r-project.org/web/packages/gplots/index.html).
Bulk RNA-seq data sets
RNA from tissue and cells was extracted using a Qiagen AllPrep DNA/RNA/miRNA Universal kit (catalog # 80224). Strand-specific RNA-seq (ssRNA-seq) libraries were constructed from total RNA samples using plate-based protocols. Libraries were sequenced at 75-bp PET using V4 chemistry on a HiSeq 2500 instrument (Illumina) at the Genome Sciences Centre (Vancouver, BC). Reads were aligned with the STAR aligner (Dobin et al. 2013) v2.4.2a to hg38 human reference genome (from iGenome). The R Bioconductor DESeq2 package (Love et al. 2014) was used for normalization and vst transformation of the gene expression matrix.
Structural variant analysis
WGS sequencing (150-bp paired-read) for GSC culture and matched blood samples was generated using a HiSeq X Illumina sequencer. WGS data were aligned with BWA to the hg38 human reference genome (iGenome) and further processed according to the GATK (McKenna et al. 2010) best practice pipeline. The WGS data set has a coverage of, on average, 60×.
Manta was run on the processed BAM files using the default parameters with the matched blood WGS data as a reference to detect somatic SV in the GSC lines. Loop anchors overlapping predicted SVs were then identified genome-wide using BEDTools (Quinlan and Hall 2010) intersect. Additional loops were identified as SV-associated upon manual review of loop calls and Hi-C contacts.
Data access
Raw Hi-C sequencing data generated in this study have been submitted to the European Genome-phenome Archive (EGA; https://www.ebi.ac.uk/ega/) under accession number EGAS00001003493. Processed Hi-C data generated in this study have been submitted to the WashU Epigenome Browser (http://epigenomegateway.wustl.edu/browser/; Li et al. 2019) and can be visualized at the URL http://epigenomegateway.wustl.edu/browser/?genome=hg38&position=chr3:179800000-182700000&hub=https://wangftp.wustl.edu/hubs/johnston_gallo/johnston_gallo_datahub.json. All processed data are available for download at https://wangftp.wustl.edu/hubs/johnston_gallo/. Raw ChIP-seq data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE121601. Bulk RNA-seq data generated in this study have been submitted to the European Genome-phenome Archive (EGA; https://ega-archive.org/), which is hosted by the EBI and the CGR, under accession number EGAS00001003700.
Supplementary Material
Acknowledgments
Funding for this work was provided by: an operating grant from the Alliance for Cancer Gene Therapy to M.G.; the Canadian Institutes of Health Research (CIHR) early career award (Institute of Cancer Research) to M.G. (ICT-156651); a Cancer Research Society Scholarship for the Next Generation of Scientists grant to M.G.; a discovery grant from the Natural Sciences and Engineering Research Council of Canada (NSERC) to M.G.; and a Clark Smith Scholarship to M.J.J. Research was supported by SU2C Canada Cancer Stem Cell Dream Team Research Funding (SU2C-AACR-DT-19-15) provided by the Government of Canada through Genome Canada and the Canadian Institute of Health Research, with supplemental support from the Ontario Institute for Cancer Research, through funding provided by the Government of Ontario. Stand Up To Cancer Canada is a Canadian Registered Charity (Reg. # 80550 6730 RR0001).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.246520.118.
References
- Abdouh M, Facchino S, Chatoo W, Balasingam V, Ferreira J, Bernier G. 2009. BMI1 sustains human glioblastoma multiforme stem cell renewal. J Neurosci 29: 8884–8896. 10.1523/JNEUROSCI.0968-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hajj M, Clarke MF. 2004. Self-renewal and solid tumor stem cells. Oncogene 23: 7274–7282. 10.1038/sj.onc.1207947 [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, Hjelmeland AB, Rich JN, Dewhirst MW, Shi Q, Hao Y, McLendon RE, Bigner DD. 2006. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444: 756–760. 10.1038/nature05236 [DOI] [PubMed] [Google Scholar]
- Brennan CW, Verhaak RGW, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al. 2013. The somatic genomic landscape of glioblastoma. Cell 155: 462–477. 10.1016/j.cell.2013.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, Cox AJ, Kruglyak S, Saunders CT. 2016. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32: 1220–1222. 10.1093/bioinformatics/btv710 [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, Rao SSP, Huntley MH, Lander ES, Aiden EL. 2016. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst 3: 95–98. 10.1016/j.cels.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eramo A, Ricci-Vitiani L, Zeuner A, Pallini R, Lotti F, Sette G, Pilozzi E, Larocca LM, Peschle C, De Maria R. 2006. Chemotherapy resistance of glioblastoma stem cells. Cell Death Differ 13: 1238–1241. 10.1038/sj.cdd.4401872 [DOI] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suvà ML, Bernstein BE. 2016. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529: 110–114. 10.1038/nature16490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A. 2004. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 64: 7011–7021. 10.1158/0008-5472.CAN-04-1364 [DOI] [PubMed] [Google Scholar]
- Gallo M, Ho J, Coutinho F, Vanner R, Lee L, Head R, Ling E, Clarke I, Dirks P. 2013. A tumorigenic MLL-homeobox network in human glioblastoma stem cells. Cancer Res 73: 417–427. 10.1158/0008-5472.CAN-12-1881 [DOI] [PubMed] [Google Scholar]
- Gallo M, Coutinho FJ, Vanner RJ, Gayden T, Mack SC, Murison A, Remke M, Li R, Takayama N, Desai K, et al. 2015. MLL5 orchestrates a cancer self-renewal state by repressing the histone variant H3.3 and globally reorganizing chromatin. Cancer Cell 28: 715–729. 10.1016/j.ccell.2015.10.005 [DOI] [PubMed] [Google Scholar]
- Heddleston JM, Wu Q, Rivera M, Minhas S, Lathia JD, Sloan AE, Iliopoulos O, Hjelmeland AB, Rich JN. 2012. Hypoxia-induced mixed-lineage leukemia 1 regulates glioma stem cell tumorigenic potential. Cell Death Differ 19: 428–439. 10.1038/cdd.2011.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmeyer KA, Ray A, Zang X. 2008. The contrasting role of B7-H3. Proc Natl Acad Sci 105: 10277–10278. 10.1073/pnas.0805458105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Smyth GK. 2009. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 347: 70–78. 10.1016/j.jim.2009.06.008 [DOI] [PubMed] [Google Scholar]
- Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. 2002. A stem cell molecular signature. Science 298: 601–604. 10.1126/science.1073823 [DOI] [PubMed] [Google Scholar]
- Jin X, Kim LJY, Wu Q, Wallace LC, Prager BC, Sanvoranart T, Gimple RC, Wang X, Mack SC, Miller TE, et al. 2017. Targeting glioma stem cells through combined BMI1 and EZH2 inhibition. Nat Med 23: 1352–1361. 10.1038/nm.4415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, Fouse SD, Yamamoto S, Ueda H, Tatsuno K, et al. 2014. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 343: 189–193. 10.1126/science.1239947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, Oh YT, Kim H, Rheey J, Nakano I, et al. 2013. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell 23: 839–852. 10.1016/j.ccr.2013.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laks DR, Masterman-Smith M, Visnyei K, Angenieux B, Orozco NM, Foran I, Yong WH, Vinters H V, Liau LM, Lazareff JA, et al. 2009. Neurosphere formation is an independent predictor of clinical outcome in malignant glioma. Stem Cells 27: 980–987. 10.1002/stem.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan X, Jörg DJ, Cavalli FMG, Richards LM, Nguyen L V, Vanner RJ, Guilhamon P, Lee L, Kushida MM, Pellacani D, et al. 2017. Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature 549: 227–232. 10.1038/nature23666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Martin-Orozco N, Zheng P, Li J, Zhang P, Tan H, Park HJ, Jeong M, Chang SH, Kim BS, et al. 2017. Inhibition of the B7-H3 immune checkpoint limits tumor growth by enhancing cytotoxic lymphocyte function. Cell Res 27: 1034–1045. 10.1038/cr.2017.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke D, Pfenning PN, Sahm F, Klein AC, Kempf T, Warnken U, Schnölzer M, Tudoran R, Weller M, Platten M, et al. 2012. Costimulatory protein 4IgB7H3 drives the malignant phenotype of glioblastoma by mediating immune escape and invasiveness. Clin Cancer Res 18: 105–117. 10.1158/1078-0432.CCR-11-0880 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Hsu S, Purushotham D, Sears RL, Wang T. 2019. WashU Epigenome Browser update 2019. Nucleic Acids Res 47: W158–W165. 10.1093/nar/gkz348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, Van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153: 320–334. 10.1016/j.cell.2013.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161: 1012–1025. 10.1016/j.cell.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani L, Stoeck A, Zhang X, Lanczky A, Mirabella AC, Wang T-L, Gyorffy B, Lupien M. 2013. Genome-wide reprogramming of the chromatin landscape underlies endocrine therapy resistance in breast cancer. Proc Natl Acad Sci 110: E1490–E1499. 10.1073/pnas.1219992110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland BC, Ma J-Y, Langford CP, Gillespie GY, Yu H, Zheng Y, Nozell SE, Huszar D, Benveniste EN. 2011. Therapeutic potential of AZD1480 for the treatment of human glioblastoma. Mol Cancer Ther 10: 2384–2393. 10.1158/1535-7163.MCT-11-0480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehdi T, Bailey SD, Guilhamon P, Lupien M. 2019. C3D: a tool to predict 3D genomic interactions between cis-regulatory elements. Bioinformatics 35: 877–879. 10.1093/bioinformatics/bty717 [DOI] [PubMed] [Google Scholar]
- Meyer M, Reimand J, Lan X, Head R, Zhu X, Kushida M, Bayani J, Pressey JC, Lionel AC, Clarke ID, et al. 2015. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc Natl Acad Sci 112: 851–856. 10.1073/pnas.1320611111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. 2014. Transcriptional landscape of the prenatal human brain. Nature 508: 199–206. 10.1038/nature13185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller TE, Liau BB, Wallace LC, Morton AR, Xie Q, Dixit D, Factor DC, Kim LJY, Morrow JJ, Wu Q, et al. 2017. Transcription elongation factors represent in vivo cancer dependencies in glioblastoma. Nature 547: 355–359. 10.1038/nature23000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, Van Berkum NL, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. 10.1038/nature11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed B V, Curry WT, Martuza RL, et al. 2014. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344: 1396–1401. 10.1126/science.1254257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picarda E, Ohaegbulam KC, Zang X. 2016. Molecular pathways: targeting B7-H3 (CD276) for human cancer immunotherapy. Clin Cancer Res 22: 3425–3431. 10.1158/1078-0432.CCR-15-2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard SM, Yoshikawa K, Clarke ID, Danovi D, Stricker S, Russell R, Bayani J, Head R, Lee M, Bernstein M, et al. 2009. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell 4: 568–580. 10.1016/j.stem.2009.03.014 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2017. R: a language and environment for statistical computing. R; Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- R Core Team. 2019. R: a language and environment for statistical computing. R; Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huang SC, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon KR, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, et al. 2017. Cohesin loss eliminates all loop domains. Cell 171: 305–320.e24. 10.1016/j.cell.2017.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinbay E, Suvà ML, Gillespie SM, Wakimoto H, Patel AP, Shahid M, Oksuz O, Rabkin SD, Martuza RL, Rivera MN, et al. 2013. An aberrant transcription factor network essential for Wnt signaling and stem cell maintenance in glioblastoma. Cell Rep 3: 1567–1579. 10.1016/j.celrep.2013.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman S, Zhu Z, Saha S, Zhang XM, Yang MY, Hilton MB, Morris K, Szot C, Morris H, Swing DA, et al. 2017. Eradication of tumors through simultaneous ablation of CD276/B7-H3-positive tumor cells and tumor vasculature. Cancer Cell 31: 501–515.e8. 10.1016/j.ccell.2017.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. 2004. Identification of human brain tumour initiating cells. Nature 432: 396–401. 10.1038/nature03128 [DOI] [PubMed] [Google Scholar]
- Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, Akhavanfard S, Cahill DP, Aldape KD, Betensky RA, et al. 2011. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell 20: 810–817. 10.1016/j.ccr.2011.11.005 [DOI] [PubMed] [Google Scholar]
- Sottoriva A, Spiteri I, Piccirillo SGM, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavaré S. 2013. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci 110: 4009–4014. 10.1073/pnas.1219747110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens TJ, Lando D, Basu S, Atkinson LP, Cao Y, Lee SF, Leeb M, Wohlfahrt KJ, Boucher W, O'Shaughnessy-Kirwan A, et al. 2017. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544: 59–64. 10.1038/nature21429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp R, Mason W, van den Bent MJ, Weller M, Fisher BM, Taphoorn MJB, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. 2005. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352: 987–996. 10.1056/NEJMoa043330 [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvà ML, Riggi N, Janiszewska M, Radovanovic I, Provero P, Stehle JC, Baumer K, Le Bitoux MA, Marino D, Cironi L, et al. 2009. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res 69: 9211–9218. 10.1158/0008-5472.CAN-09-1622 [DOI] [PubMed] [Google Scholar]
- Suvà ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD, Riggi N, Chi AS, Cahill DP, Nahed B V, et al. 2014. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 157: 580–594. 10.1016/j.cell.2014.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y, Ozawa T, Holland EC, Huse JT, Jhanwar S, et al. 2012. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci 109: 3041–3046. 10.1073/pnas.1114033109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SJ, Snowden JA, Zeidler MP, Danson SJ. 2015. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br J Cancer 113: 365–371. 10.1038/bjc.2015.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. 2010. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17: 98–110. 10.1016/j.ccr.2009.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153: 307–319. 10.1016/j.cell.2013.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingett S, Ewels P, Furlan-Magaril M, Nagano T, Schoenfelder S, Fraser P, Andrews S. 2015. HiCUP: pipeline for mapping and processing Hi-C data. F1000Res 4: 1310 10.12688/f1000research.7334.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wang J, Marzese DM, Wang X, Yang Z, Li C, Zhang H, Zhang J, Chen CC, Kelly DF, et al. 2019. B7H3 regulates differentiation and serves as a potential biomarker and theranostic target for human glioblastoma. Lab Invest 10.1038/s41374-019-0238-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.