Abstract
Myelin-associated inhibitors (MAIs) and chondroitin sulfate proteoglycans (CSPGs) contribute to failed regeneration after neuronal injury. MAIs and CSPGs stimulate intracellular signals including the activation of RhoA and Rho kinase to block axonal extension through targeted modifications to the cytoskeleton. RhoA and ROCK are promising targets for therapeutic intervention to promote CNS repair; however, their ubiquitous expression will limit the specificity of drugs targeted to these molecules. We have identified the cytosolic phosphoprotein CRMP4b (collapsin-response mediator protein 4b) as a protein that physically and functionally interacts with RhoA to mediate neurite outgrowth inhibition. Short interfering RNA-mediated knockdown of CRMP4 promotes neurite outgrowth on myelin substrates, indicating a critical role for CRMP4 in neurite outgrowth inhibition. Disruption of CRMP4b–RhoA binding with a competitive inhibitor attenuates neurite outgrowth inhibition on myelin and aggrecan substrates. Stimulation of neuronal growth cones with Nogo leads to colocalization of CRMP4b and RhoA at discrete regions within the actin-rich central and peripheral domains of the growth cone, indicative of a potential function in cytoskeletal rearrangements during neurite outgrowth inhibition. Together, these data indicate that a RhoA–CRMP4b complex forms in response to inhibitory challenges in the growth cone environment and regulates cytoskeletal dynamics at distinct sites necessary for axon outgrowth inhibition. Competitive inhibition of CRMP4b–RhoA binding suggests a novel, highly specific therapeutic avenue for promoting regeneration after CNS injury.
Keywords: Nogo, CRMP, TUC, glial scar, CNS regeneration, myelin inhibition, Rho GTPase
Introduction
Trauma in the adult mammalian CNS results in devastating clinical consequences as a result of the failure of injured axons to spontaneously regenerate. Chondroitin sulfate proteoglycans (CSPGs) and the myelin-associated inhibitors (MAIs) myelin-associated glycoprotein (MAG) (McKerracher et al., 1994; Mukhopadhyay et al., 1994), Nogo-A (Chen et al., 2000; GrandPre et al., 2000; Prinjha et al., 2000), and oligodendrocyte-myelin glycoprotein (Kottis et al., 2002; Wang et al., 2002) bind receptor molecules on injured axons initiating intracellular signaling cascades that block axonal regrowth (Mandemakers and Barres, 2005). In part, CSPGs and MAIs inhibit axon regeneration by disrupting Rho-GTPase-dependent cytoskeletal dynamics. Blockade of RhoA and a downstream effector, Rho kinase (ROCK), promotes axon regeneration both in vitro and in vivo (Dergham et al., 2002; Borisoff et al., 2003; Fournier et al., 2003); however, the ability of RhoA and ROCK to affect multiple physiological processes in many cell types (Riento and Ridley, 2003) highlights the need to identify novel intracellular signaling substrates of neurite outgrowth inhibition to develop more specific and potent therapeutic avenues.
In a screen to identify molecules that functionally interact with RhoA to mediate neurite outgrowth inhibition, we identified collapsin-response mediator protein 4b (CRMP4b) as a molecule that interacts with Rho GTPase in a Nogo-dependent manner. The CRMPs are a family of cytosolic phosphoproteins with five vertebrate family members (CRMP1–5) (Goshima et al., 1995; Minturn et al., 1995; Byk et al., 1996; Gaetano et al., 1997; Inatome et al., 2000). CRMP1–4 alleles each produce two transcripts, a and b, and CRMPb variants are longer N-terminal variants of the originally identified CRMPa isoforms (Yuasa-Kawada et al., 2003). Although CRMPs share significant sequence similarity with dihydropyrimidinase (DHPase), an enzyme involved in pyrimidine catabolism, no DHPase activity has been described for CRMPs (Wang and Strittmatter, 1997). Rather, a role for CRMPs in axon growth and pathfinding has been revealed. CRMPs are homologs of UNC-33, a protein that influences axon guidance and extension in Caenorhabditis elegans (Hedgecock et al., 1985; Siddiqui and Culotti, 1991). CRMP2 mediates growth cone collapse in response to the repulsive guidance cue semaphorin3A (Sema3A) (Goshima et al., 1995), and CRMP2 and CRMP4 influence neurite outgrowth (Minturn et al., 1995; Quinn et al., 1999, 2003; Yoshimura et al., 2005). Mechanistically, CRMP2 can bind to tubulin heterodimers and organizes microtubule assembly to establish axon–dendrite fate during development (Fukata et al., 2002b; Arimura et al., 2005), and CRMP4 can promote F-actin bundling (Rosslenbroich et al., 2005). Also, a role for CRMP2 in endocytosis has been described previously (Nishimura et al., 2003), and an association between CRMP4b and intersectin, an endocytic–exocytic adaptor protein, is consistent with an endocytic role for this isoform (Quinn et al., 2003). The role of the CRMPs in nervous system injury and regeneration has not been extensively studied; however, CRMP2a does have a potent neurite elongating effect in nerve regeneration in vivo (Suzuki et al., 2003). CRMP1a, CRMP2a, and CRMP5a mRNA levels increase after hypoglossal nerve injury, and CRMP4a expression is increased in regenerating adult sciatic motor neurons (Minturn et al., 1995; Suzuki et al., 2003), suggesting a more general role for CRMPs in the neuronal response to injury.
We find that CRMP4b interacts with RhoA in a Nogo-dependent manner, leading us to investigate the potential role of this complex in the inhibition of neurite outgrowth. We find that antagonism of CRMP4 or of the CRMP4b–RhoA interaction attenuates neurite outgrowth inhibition. This protein–protein interaction represents a novel, specific target for therapeutic intervention after CNS injury.
Materials and Methods
Plasmid construction.
To construct CRMP-V5 expression vectors, cDNA was amplified by PCR from rat pEGFP-CRMP constructs (generously provided by Dr. Peter McPherson, McGill University, Montreal, Quebec, Canada). The PCR product was ligated into the HindIII and XhoI sites of pcDNA 3.1 V5-His for CRMP1a, CRMP2a, and CRMP3a and into the HindIII and EcoRI sites of pcDNA 3.1 V5-His for CRMP4a. The cDNA for CRMP4b was amplified from pcDNA 3.1CRMP4bV5-His TOPO (Quinn et al., 2003) and ligated into the HindIII and EcoRI sites pcDNA 3.1 V5-His in frame with the V5-His tag. CRMP1b-V5 was constructed by amplifying the coding sequence by PCR from an expressed-sequence tag clone (IMAGE:5686818) and ligated into the EcoRI and XhoI sites of pcDNA 3.1 V5-His.
pcDNA myc-wild-type (wt) RhoA was obtained from the UMR cDNA Resource Center (http://www.cdna.org/). wt pRK5 myc-Rac1, wt pRK5 myc-Cdc42, pRK5 myc-RhoA63L, pRK5 myc-RhoAN19, pRK5 myc-RacQ61L, pRK5 myc-RacN17, pRK5 myc-Cdc42Q61L, and pRK5 myc-Cdc42N17 constructs were generously provided by Dr. Nathalie Lamarche-Vane (McGill University, Montreal, Quebec, Canada). FLAG-RhoA63L was generated by subcloning RhoA63L into the BamHI and EcoRI sites of pcDNA3FLAG. pCAG-myc-mROCK II construct was generously provided by Dr. Shuh Narumiya (Kyoto University, Kyoto, Japan).
To generate C4RIP-V5, the unique N-terminal domain of CRMP4b (residues 1–126) was introduced into the BamHI and EcoRI sites of pcDNA 3.1V5-His. pHSVC4RIP was generated by subcloning C4RIP-V5 into the HindIII and SalI sites of pHSVPrPUC. pHSVCRMP4bGFP was generated by cloning CRMP4b into the HindIII and EcoRI sites of pEGFP N2 (Clontech, Palo Alto, CA) and subsequently subcloning into the HindIII and XbaI sites of pHSVPrPUC. Chimeric CRMP4bNCRMP2-V5 was constructed by PCR by ligating residues 14–572 of CRMP2a into the EcoRI and XhoI sites of C4RIP-V5. CRMP4ΔN consists of CRMP4a with the first 12 residues replaced by a single methionine. CRMP4ΔN was amplified by PCR and ligated into the HindIII and EcoRI sites of pcDNA 3.1V5-His.
Preparation of herpes simplex viruses.
pHSVPrPUC plasmids were transfected into 2-2 Vero cells that were superinfected with 5dl 1.2 herpes simplex virus (HSV) helper virus 1 d later. Recombinant virus was amplified through three passages and stored at −80°C as described previously (Neve et al., 1997).
Preparation of recombinant proteins.
Stimulations to examine inhibitory responses were performed with alkaline phosphatase-conjugated Nogo-66 (AP-Nogo-66) purified from stably transfected HEK293 cells, Nogo-P4 peptide, or myelin. AP-Nogo-66 or AP was purified by Ni2+ affinity chromatography as described previously (Nakamura et al., 1998; Fournier et al., 2001). For all treatments, 8 nm AP-Nogo66-His or 8 nm AP was preaggregated with 100 ng/ml anti-human AP (Niederost et al., 2002). Nogo-P4 (Alpha Diagnostics, San Antonio, TX) is a 25 aa inhibitory peptide sequence (residues 31–55 of Nogo-66) sufficient to mediate the inhibitory properties of Nogo-66, a potent inhibitory component of Nogo-A (GrandPre et al., 2000). Myelin extracts were prepared from a bovine brain as described previously (Igarashi et al., 1993; Hsieh et al., 2006).
Glutathione S-transferase (GST), GST-RhoAWT, and GST-RhoA63L (construct generously provided by Dr. Keith Burridge, University of North Carolina, Chapel Hill, NC) were expressed in Escherichia coli and purified on glutathione-Sepharose as described previously (Arthur et al., 2002; Wennerberg et al., 2002). For overlay assays, RhoA was cleaved from the GST moiety via thrombin cleavage. Aggrecan was purchased from Sigma (Oakville, Ontario, Canada).
GST-RhoA pull-down assays.
PC12 cells were grown to subconfluence on collagen-coated plates in Roswell Park Memorial Institute (RPMI) medium 1640 supplemented with l-glutamine and containing 10% horse serum, 5% fetal bovine serum (FBS), and 1% penicillin/streptomycin (Invitrogen, Burlington, Ontario, Canada), after which differentiation was induced for 24 h with RPMI medium supplemented with 1% bovine serum albumin fraction V and 50 ng/ml nerve growth factor (NGF) (Millipore, Bedford, MA). GST–RhoA63L pull-downs were performed as described previously (Arthur et al., 2002). Briefly, after stimulation with AP-Nogo-66, cells were washed twice in ice-cold HEPES-buffered saline and harvested in 1 ml of ice-cold lysis buffer containing 20 mm HEPES, pH 7.3, 150 mm NaCl, 5 mm MgCl2, 1% (v/v) Triton X-100, and 1 mm phenylmethylsulfonyl fluoride (PMSF) supplemented with Complete protease inhibitors (Roche Products, Laval, Quebec, Canada). GST-precleared lysates were then mixed with 30 μg of GST-RhoA63L fusion protein bound to Sepharose beads for 1 h at 4°C. Precipitated proteins were eluted with 2× sample buffer and analyzed by SDS-PAGE on a 4–15% gradient gel, followed by silver staining.
CRMP–RhoA coimmunoprecipitation assays.
HEK293T cells were grown to subconfluence and transfected with Lipofectamine 2000 according to manufacturer instructions (Invitrogen), washed twice with ice-cold PBS, and lysed in lysis buffer A [in mm: 50 Tris, pH 7.4, 150 NaCl, 1 EDTA, 1% (v/v) Triton X-100, 1 Na3VO4, 1 NaF, 1 PMSF, and Complete protease inhibitors (Roche Diagnostics, Laval, Quebec, Canada)]. Lysates were precleared with protein A/G-agarose (Santa Cruz Biotechnology, Santa Cruz, CA) and subjected to immunoprecipitation with myc-agarose or V5-agarose (Sigma). After washing three times with ice-cold PBS, bound protein was eluted with SDS and immunoblotted with anti-Myc (9E10, 1:1000; Sigma) or anti-V5 (1:5000; Invitrogen). For time-course experiments, PC12 cells were transfected for 24 h using Lipofectamine 2000 (Invitrogen) and differentiated with 50 ng/ml NGF (Millipore) for 24 h. Cells were treated with Nogo-P4 peptide for the indicated period of time at 37°C. Cells were then lysed, and proteins were immunoprecipitated as described above.
Far Western.
Overlay of CRMP4 with RhoA was performed as described previously (McPherson et al., 1994). Briefly, HEK293T cell lysates transfected with CRMP4a-V5, CRMP4b-V5, or empty vector were subjected to immunoprecipitation with V5-agarose (Sigma). Immunoprecipitates were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and overlaid with 10 μg/ml bacterially purified RhoA for 1 h at room temperature. RhoA was detected with a rabbit anti-RhoA antibody (Santa Cruz Biotechnology).
Neurite outgrowth and growth cone collapse assays.
For neurite outgrowth assays, myelin was dried down on poly-l-lysine-coated substrates. Substrates were washed and coated with 10 μg/ml laminin for 1 h. For outgrowth on aggrecan, poly-l-lysine-coated substrates were coated with aggrecan and 10 μg/ml laminin for 2 h at 37°C. Dissociated embryonic day 13 (E13) chick dorsal root ganglion (DRG) neurons were cultured in DRG medium (F-12 medium, 10% FBS, 1% penicillin/streptomycin, 1% l-glutamine, 50 ng/ml NGF) in the presence of virus for 24 h, fixed with 4% paraformaldehyde/20% sucrose in PBS, and double stained with anti-βIII tubulin (Covance, Berkeley, CA) and anti-V5 antibody (Sigma). Neurite outgrowth lengths per cell were assessed using Image J, a public domain JAVA image processing program (http://rsb.info.nih.gov/ij/), as described previously (Fournier et al., 2003).
For growth cone collapse assays, E7 chick DRGs were cultured in DRG medium for 18 h on four-well glass chamber slides sequentially coated with poly-l-lysine and laminin as above. Sema3A-AP- or AP-conditioned medium was prepared as described previously (Takahashi et al., 1998). Explants were stimulated with Sema3A-AP- or AP-conditioned medium at indicated concentrations for 20 min and fixed with 4% paraformaldehyde, 20% sucrose, and 0.1 m NaPO4. Explants were stained with rhodamine-phalloidin and assessed for growth cone collapse as described previously (Luo et al., 1993).
CRMP-4 short interfering RNA.
For knockdown of CRMP-4a and CRMP-4b, silencer predesigned short interfering RNA (siRNA) against rat CRMP-4 was used (siRNA identification number 48833; Ambion, Austin, TX). Controls were with a scrambled siRNA (CAGCAUGGUGGUACGCUUGUAAGCA) for an ineffective CRMP4b-targeted siRNA designed with the BLOCK-IT algorithm (Invitrogen). siRNAs were validated by cotransfecting siRNAs with CRMP4-V5 in HEK293T cells for 24 h. Cell lysates were separated by SDS-PAGE and analyzed with an anti-V5 antibody. To validate siRNA efficacy in neurons, siRNA-transfected DRGs were coinfected with HSVCRMP4b–green fluorescent protein (GFP). Twenty-four hours after infection, GFP fluorescence was evaluated by fluorescence microscopy. For neurite outgrowth assays, postnatal day 5 (P5) dissociated rat DRGs were prepared and seeded on laminin substrates as described previously (Hsieh et al., 2006). After 4 h, DRGs were serum starved (in F-12 medium and 50 ng/ml NGF) and transfected with indicated siRNAs using Lipofectamine 2000. After 5 h of transfection, medium was replaced with fresh DRG medium. Twenty-four hours after transfection, DRGs were removed from the plate with EDTA and reseeded on myelin substrates. Neurons were left to grow for an additional 18 h and processed for neurite outgrowth analysis as described above.
Immunofluorescence.
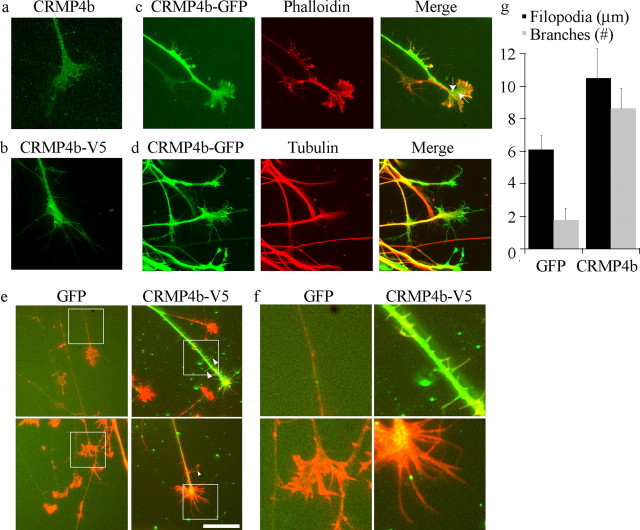
E7–E13 chick DRG explants were cultured in DRG medium on poly-l-lysine- and laminin-coated substrates for 18 h. For viral infections, recombinant viral preparations were added to the medium 1 h after plating. After 18 h, cultures were treated with myelin and fixed with 4% paraformaldehyde/20% sucrose/PBS, permeabilized in 0.2% Triton X-100, and double stained with polyclonal anti-RhoA antibody (Santa Cruz Biotechnology) and anti-CRMP4b or V5 antibody. For filopodia and branch analysis, GFP- and CRMP4bV5-infected growth cones were double stained with rhodamine-phalloidin (Invitrogen) and anti-V5 antibody. Filopodial length per growth cone was assessed using Image J, by measuring total filopodial length per growth cone averaged by the total number of filopodia per growth cone. Neurite branches were quantified by counting total branch buds per neurite.
Results
A novel Nogo-dependent interaction between CRMP4b and RhoA
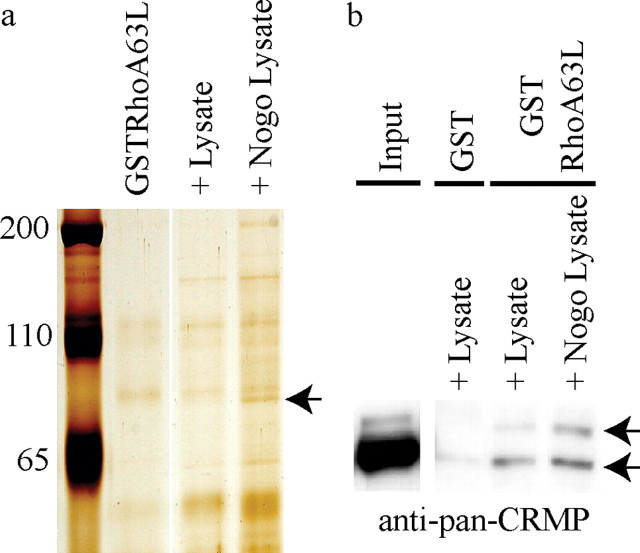
To identify molecules that functionally interact with RhoA to mediate neurite outgrowth inhibition, we screened for proteins that have enhanced affinity for a constitutively active, GTPase-deficient mutant of RhoA (RhoA63L) (Khosravi-Far et al., 1994) after treatment with Nogo-66, a potent inhibitory fragment of Nogo-A (GrandPre et al., 2000). Bacterially purified GST-RhoA63L was used as bait to precipitate proteins from PC12 cells after stimulation with AP-Nogo-66. PC12 cells were chosen for the biochemical screen based on their expression of MAI receptors (Hsieh et al., 2006) and their responsiveness to Nogo in neurite outgrowth assays (GrandPre et al., 2000) and biochemical assays evaluating RhoGTP levels (Fournier et al., 2003). Proteins interacting with GST-RhoA63L were separated by SDS-PAGE and visualized by silver staining. A 75 kDa protein with enhanced affinity for GST-RhoA63L after Nogo-66 treatment (Fig. 1a) was identified as CRMP4b by tandem mass spectrometry. The enhanced CRMP4–RhoA interaction was validated with a pan-CRMP antibody (generously provided by Dr. Peter McPherson), which recognizes the 75 kDa CRMPb isoforms and the 65 kDa CRMPa isoforms (Fig. 1b). Both 75 and 65 kDa CRMP isoforms specifically precipitate with GST-RhoA63L, whereas the interaction between the 75 kDa CRMPb isoforms and RhoA is enhanced by Nogo stimulation (Fig. 1b).
Figure 1.
A novel RhoA–CRMP4 interaction is enhanced by Nogo-66. a, GSTRhoA63L pulldown from PC12 cells stimulated with AP (+ Lysate) or AP-Nogo-66 (+ Nogo Lysate). Precipitation of a 75 kDa protein identified as CRMP4b by tandem mass spectrometry is enhanced in the AP-Nogo-66-stimulated lysate (arrow). The protein runs slightly below a nonspecific protein from the GSTRhoA63L purification, which is visible in all lanes. The GSTRhoA63L lane represents beads that were not incubated with lysates. b, GST and GSTRhoA63L pull-downs from PC12 cell lysates immunoblotted with a pan-CRMP antibody. CRMPa (65 kDa) and CRMPb (75 kDa) bands are indicated in the PC12 pull-down lane (arrows).
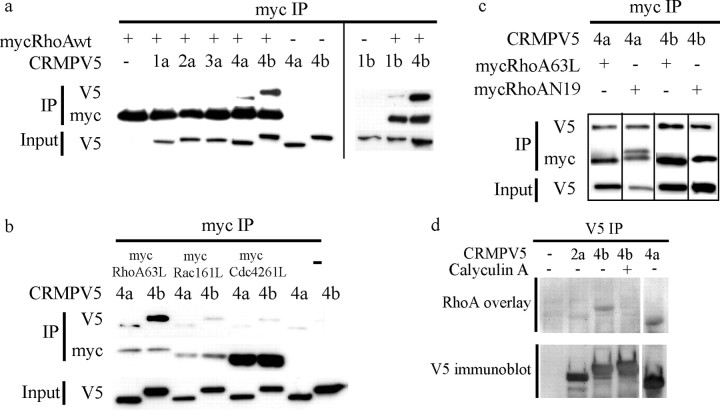
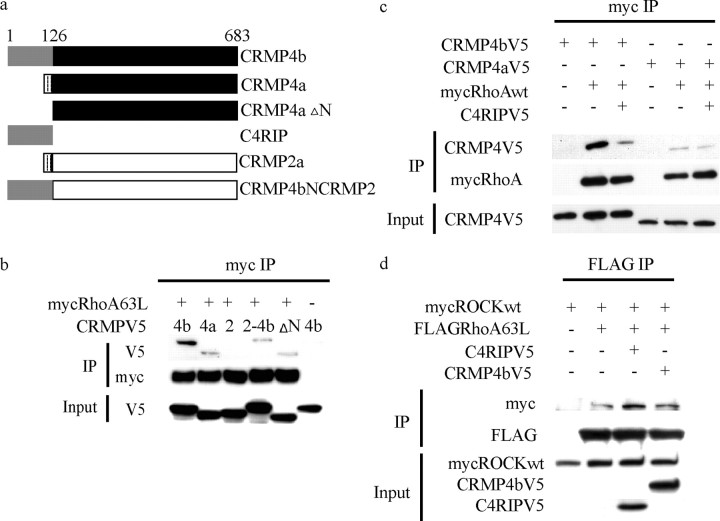
To assess the specificity of the CRMP4b–RhoA interaction, we analyzed the ability of other CRMP members to interact with RhoA. wt myc-RhoA and CRMP-V5 constructs were cotransfected in HEK293T cells. Myc-RhoA was immunoprecipitated from cell lysates, and immune complexes were analyzed for CRMP-V5. RhoA interacts preferentially with CRMP4 family members with stronger binding to CRMP4b when compared with CRMP4a (Fig. 2a). RhoA fails to bind to other CRMPa isoforms and weakly associates with CRMP1b (Fig. 2a). N-terminally tagged GFP-CRMP4b fails to associate with RhoA, suggesting that N-terminal tags interfere with this binding interaction (data not shown).
Figure 2.
The RhoA–CRMP4 interaction is highly specific, nucleotide independent, phospho dependent, and direct. a–c, HEK293T cells cotransfected with CRMP-V5 constructs and myc-tagged versions of wt and mutant Rho GTPases and subjected to myc immunoprecipitation. a, CRMP4 preferentially binds to RhoA. Data from two separate blots are separated by a vertical line. b, RhoA is the preferred binding partner for CRMP4. c, CRMP4 binding to RhoA is nucleotide independent. d, CRMP-V5 was immunoprecipitated from control or calyculin-treated HEK293T cells, separated by SDS-PAGE, and overlayed with recombinant RhoA. RhoA binds directly to CRMP4a and CRMP4b but fails to bind to phosphorylated CRMP4b. IP, Immunoprecipitation.
We then asked whether CRMP4 specifically interacts with RhoA by assessing its binding to Rac and Cdc42, two other members of the small Rho GTPases that regulate the actin cytoskeleton and positively regulate neurite outgrowth (Bishop and Hall, 2000). CRMP4a-V5 and CRMP4b-V5 were tested for binding to myc-tagged, GTP-bound active forms of RhoA (myc-RhoA63L), Rac1 (myc-Rac1Q61L), and Cdc42 (myc-Cdc42Q61L). CRMP4 binding to RhoA is markedly stronger than to Rac1 or Cdc42 (Fig. 2b). Similarly, CRMP4 binding to wt and GDP-bound forms of Rac and Cdc42 is negligible (data not shown). Together, these results demonstrate that both CRMP4a and CRMP4b specifically bind to RhoA with CRMP4b binding more strongly than CRMP4a.
CRMP4b–RhoA binding is nucleotide independent, phospho dependent, and direct
Rho GTPases cycle between an inactive GDP-bound state and an active GTP-bound state. Downstream effectors of Rho GTPases bind to Rho GTPases in the active GTP-bound state, whereas guanine nucleotide exchange factors prefer Rho GTPases in the nucleotide-free or GTP-bound state (Hall, 1994). To study the nucleotide dependence of the CRMP4–RhoA interaction, CRMP4 binding to constitutively active myc-RhoA63L or dominant-negative myc-RhoAN19 was assessed (Feig and Cooper, 1988). CRMP4a-V5 and CRMP4b-V5 interact with both myc-RhoA63L and myc-RhoAN19 (Fig. 2c), indicating that the interaction between CRMP4 and RhoA is nucleotide independent.
To assess whether the binding interaction between CRMP4 and RhoA is direct, we performed a RhoA overlay assay on CRMP-V5 immunoprecipitated from 293T cells (Fig. 2d). RhoA specifically binds to CRMP4b-V5 and CRMP4a-V5 and fails to bind to CRMP2a-V5 in the overlay assay indicating that the interaction between CRMP4b and RhoA is direct and specific.
The phospho dependence of the interaction was evaluated by stimulating CRMP4b-V5-expressing 293T cells with Calyculin A, a serine/threonine phosphatase inhibitor, before immunoprecipitating CRMP4b-V5. Calyculin-dependent CRMP4b-V5 phosphorylation is indicated by an upward mobility shift of the CRMP4b-V5 protein (Fig. 2d, V5 immunoblot). RhoA fails to bind to the phosphorylated species of CRMP4b-V5 in the overlay, demonstrating that phosphorylated CRMP4b fails to bind to RhoA.
Nogo specifically modulates the CRMP4b–RhoA interaction
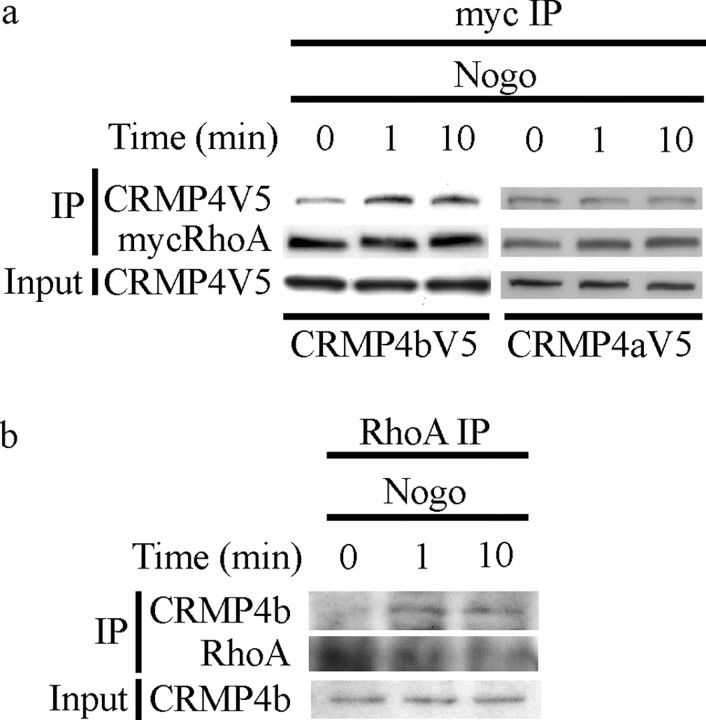
To further define the specificity and the time course of the Nogo effect on the CRMP4b–RhoA interaction, PC12 cells were cotransfected with wt myc-RhoA and CRMP4-V5 and stimulated with Nogo-P4 peptide, the minimal sequence of Nogo-66 required for Nogo-66-dependent responses (GrandPre et al., 2000). myc-RhoA was immunoprecipitated, and immune complexes were analyzed for CRMP4-V5. We detected a rapid increase in CRMP4b-V5 immunoprecipitating with myc-RhoA by 1 min after Nogo stimulation, which is maintained for 10 min (Fig. 3a). Nogo stimulation does not regulate the interaction between myc-RhoA and CRMP4a-V5 (Fig. 3a). Similar results were obtained with AP-Nogo-66 stimulation and with the active form of RhoA (data not shown), suggesting that CRMP4b is regulated independently of Nogo-dependent RhoA cycling to the GTP-bound state. The Nogo-regulated RhoA–CRMP4b interaction was also confirmed by coimmunoprecipitation of endogenous proteins in Nogo-P4-treated P8 rat cerebellar cultures, where CRMP4b and RhoA form a complex 1 and 10 min after stimulation (Fig. 3b).
Figure 3.
Nogo-P4 peptide enhances the interaction between CRMP4b and RhoA. a, b, PC12 cells transfected with myc-RhoA and CRMP4b-V5 or CRMP4a-V5; stimulated for 0, 1, or 10 min with Nogo-P4 peptide; and subjected to myc immunoprecipitation. b, P8 rat cerebellar cultures stimulated with Nogo-P4 peptide, subjected to RhoA immunoprecipitation, and analyzed for RhoA and CRMP4b. IP, Immunoprecipitation.
siRNA-mediated knockdown of CRMP4 attenuates neurite outgrowth inhibition
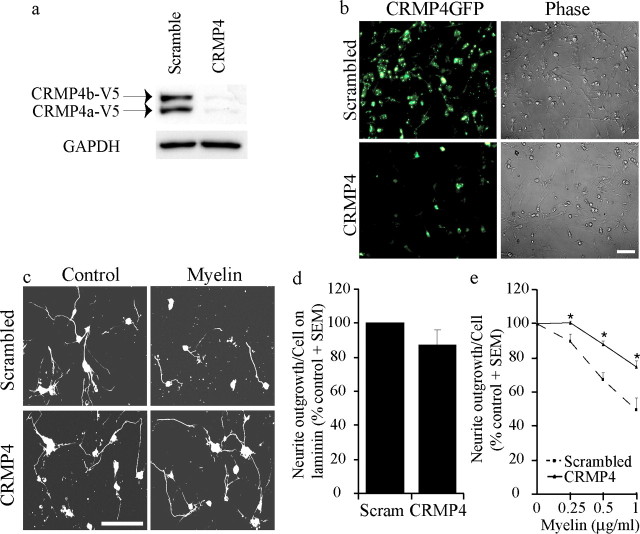
To determine whether CRMP4 function is necessary for Nogo-dependent responses, we assessed neurite outgrowth on myelin substrates in the context of CRMP4-specific siRNA. The efficacy of CRMP4-specific siRNAs was validated in transfected 293T cells. We identified one siRNA, which robustly inhibits both CRMP4a and CRMP4b expression (Fig. 4a). Lipofectamine-mediated transfection of siRNAs efficiently targets P4 rat DRG neurons, and CRMP4 siRNA introduced in this manner diminishes the expression of CRMP4b-GFP introduced by HSV-mediated infection (Fig. 4b). Rat DRGs were grown for 24 h after siRNA transfection, removed from the substrate with EDTA, and reseeded on myelin substrates for an additional 18 h. Rat DRG neuronal outgrowth is inhibited by ∼50% on a substrate coated with 1 μg/ml myelin using this protocol (Fig. 4e), a more modest inhibitory response than when dissociated DRG neurons are plated immediately on myelin substrates (see Fig. 8) (Hsieh et al., 2006). Neuronal outgrowth on the control laminin substrate is not significantly affected by the introduction of CRMP4 siRNA (Fig. 4d). However, CRMP4 siRNA-transfected P4 rat DRGs grow significantly better on myelin substrates than those transfected with scrambled siRNA (Fig. 4c,e), indicating that CRMP4 is necessary for myelin-dependent inhibition. The efficacy of the CRMP4 siRNA (Fig. 4e) may be underestimated because of the failure to transfect 100% of the DRG neurons (Fig. 4b).
Figure 4.
siRNA-mediated knockdown of CRMP4 expression promotes neurite outgrowth on myelin. a, HEK293T cells cotransfected with CRMP4-V5 and scrambled or CRMP4-targeted siRNA and analyzed by immunoblotting with anti-V5 antibody. GAPDH, Glyceraldehyde-3-phosphate dehydrogenase. b, Dissociated rat DRG neurons infected with HSVCRMP4b-GFP and transfected with scrambled or CRMP4 siRNA. Scale bar, 100 μm. c, βIII Tubulin-stained dissociated rat DRG neurons transfected with scrambled or CRMP4 siRNA and seeded on laminin (control) or myelin substrates for an 18 h neurite outgrowth assay. Scale bar, 100 μm. d, Quantitation of DRG outgrowth on control substrates with scrambled siRNA (Scram) or CRMP4 siRNA. e, Quantitation of DRG neurite outgrowth on myelin from neurons transfected with scrambled or CRMP4 siRNA. Values are normalized to baseline outgrowth on the control laminin substrate for each experiment. Determinations are from three experiments performed in duplicate. *p < 0.01 by Student's t test compared with scrambled siRNA.
Figure 8.
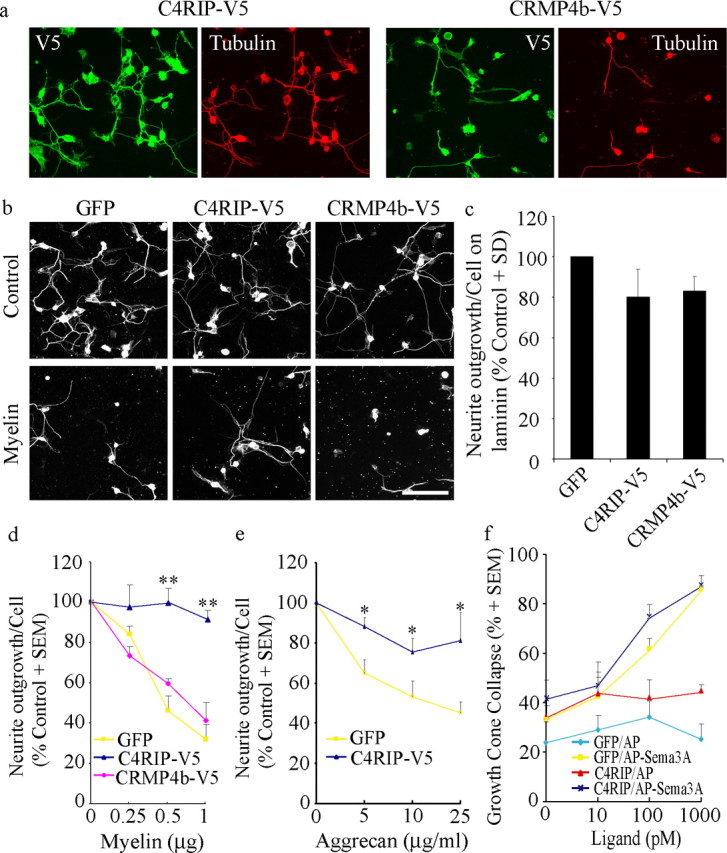
C4RIP-V5 attenuates myelin inhibition. a, E13-dissociated chick DRG neurons infected with HSV-C4RIP-V5 or HSV-CRMP4b-V5 and double stained with anti-V5 and anti-βIII tubulin antibodies to validate neuronal infection. b, E13-dissociated chick DRG neurons infected with C4RIP-V5 or CRMP4b-V5 plated on laminin (control) or myelin substrates and stained with anti-βIII tubulin to visualize neurite outgrowth. c–e, Quantitation of neurite outgrowth from GFP-, C4RIP-, or CRMP4b-infected neurons on laminin (c), myelin (d), or aggrecan (e) substrates. In c, neurite outgrowth per cell is normalized to outgrowth in GFP-infected neurons for each experiment (100%). In d and e, neurite outgrowth is normalized to growth on the laminin substrate for each dose curve (100%). Determinations are from four separate experiments. Scale bar, 100 μm. *p < 0.05 and **p < 0.01 by Student's t test compared with GFP. f, Quantitation of growth cone collapse in E8 chick DRG neurons infected with HSV-GFP or HSV-C4RIP and stimulated for 20 min with control AP ligand or AP-Sema3A ligand. Determinations are from three separate experiments performed in duplicate.
CRMP4b affects the growth cone actin cytoskeleton
CRMPs have been broadly implicated in the regulation of microtubule polymerization, actin bundling, and endocytosis, three processes that influence growth cone dynamics and neurite outgrowth (Fukata et al., 2002b; Nishimura et al., 2003; Rosslenbroich et al., 2005). To gain insight into how CRMP4b may influence neurite outgrowth inhibition, we assessed the distribution of CRMP4b in DRG growth cones. As described previously, endogenous CRMP4b has a punctate pattern within the growth cone extending throughout the central and peripheral domains (Fig. 5a)(Quinn et al., 2003). CRMP4b-V5 (Fig. 5b) and CRMP4b-GFP (Fig. 5c,d) fusion proteins also label the entire growth cone with a less distinct punctate profile, likely because of elevated cytosolic CRMP4b levels in the overexpression paradigm. C-terminal-tagged CRMP4b-GFP labels the growth cone more broadly than tubulin (Fig. 5d) and extends into the actin-rich peripheral domain (Fig. 5c) colocalizing with actin at a subset of punctae within the growth cone (Fig. 5c, arrows). Intriguingly, CRMP4b-V5 overexpression promotes the extension of filopodia from the growth cone that are, on average, 70% longer than filopodia in GFP-infected growth cones (Fig. 5e–g). This phenotype is also manifested in the DRG neurite where ectopic actin-rich branches are formed (Fig. 5e, arrowheads, f,g). A similar phenotype was promoted by CRMP4b-GFP overexpression (data not shown). On average, eight branches per neurite can be detected on CRMP4b-infected neurites compared with two branches per neurite on GFP-infected neurites (Fig. 5g). The localization of CRMP4b within the growth cone is consistent with a role in microtubule or actin dynamics. However, the filopodial and branching phenotypes promoted by CRMP4b overexpression suggest that CRMP4b may affect neuronal phenotype through an actin-based mechanism, an important observation because CRMPs have been implicated in modulating both microtubule and actin dynamics (Fukata et al., 2002a; Rosslenbroich et al., 2005). Although we cannot rule out the possibility that the fusion proteins may behave differently from native CRMP4b, both CRMP4b-V5 and CRMP4b-GFP have similar distributions to endogenous CRMP and promote similar growth cone phenotypes, suggesting that CRMP4b functionally interacts mainly with the actin cytoskeleton.
Figure 5.
CRMP4b overexpression promotes an actin-based filopodial phenotype in the neuronal growth cone and neurite. a, E13 chick DRG stained with an anti-CRMP4b antibody. b, E7 chick DRG neurons infected with HSV-CRMP4b-V5 and stained with anti-V5 antibody. c, d, E7 chick DRG neurons infected with HSV-CRMP4b-GFP and double stained with rhodamine-phalloidin (c) to label F-actin or anti-βIII tubulin antibody (d). e, E13 chick DRG neurons infected with HSV-GFP or HSV-CRMP4b-V5. GFP and CRMP4b-infected growth cones were stained with rhodamine-phalloidin (red). CRMP4b-V5 was stained with anti-V5 antibody (green). Scale bar, 10 μm. f, Magnification of boxed regions in d demonstrating enhanced neurite branching and filopodial extension in CRMP4b-V5-infected DRG neurons. g, Quantitation of number of branches per neurite and filopodial length in GFP- or CRMP4b-V5-infected DRG neurons.
To further address the potential role for CRMP4b–RhoA complexes in neurite outgrowth inhibition, the distribution of CRMP4b and RhoA during Nogo-dependent growth cone collapse was examined. Uninfected and CRMP4b-V5-infected DRG growth cones were fixed and stained for endogenous RhoA and CRMP4b or the V5 epitope tag. Before stimulation, endogenous RhoA and CRMP4b have distinct distributions within the growth cone with negligible colocalization (Fig. 6a). In unstimulated (control) growth cones, overexpressed CRMP4b-V5 also has a distinct distribution compared with endogenous RhoA (Fig. 6b). After myelin stimulation, RhoA and CRMP4b-V5 colocalize at a subset of distinct punctae within the growth cone central and peripheral domains (Fig. 6b, arrows), suggesting that a RhoA–CRMP4b complex forms in the growth cone where it may regulate actin cytoskeletal dynamics in response to inhibitory challenges.
Figure 6.
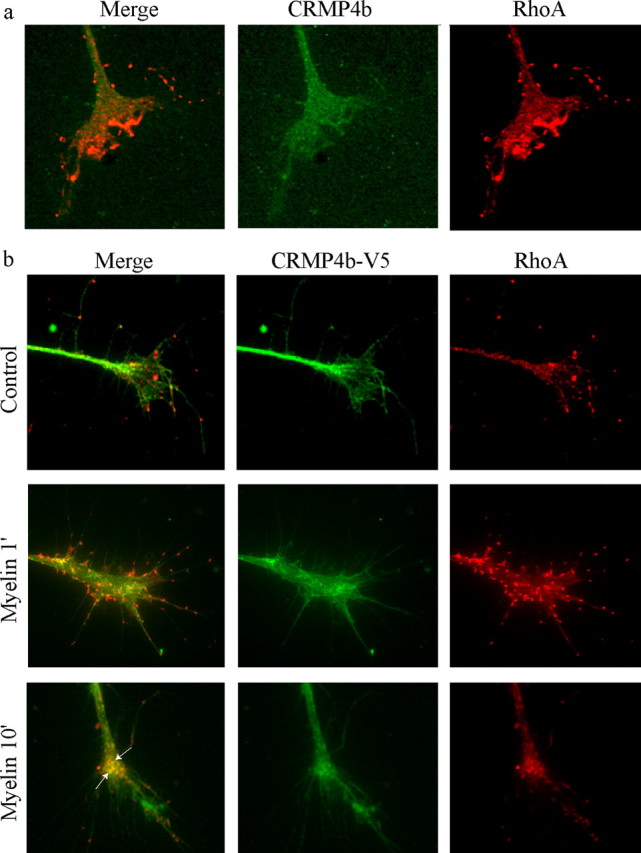
CRMP4b and RhoA colocalize at discrete punctae during myelin-dependent growth cone collapse. a, b, Immunofluorescent detection of endogenous CRMP4b and RhoA (a) or CRMP4b-V5 and RhoA (b) in control or myelin-stimulated E13 chick DRG growth cones. Arrows in the merged panels indicate areas of CRMP4b-V5-RhoA colocalization.
C4RIP-V5 attenuates CRMP4b–RhoA binding
Nogo regulation of the CRMP4b–RhoA interaction raises the possibility that CRMP4b–RhoA complex formation is critical for inhibitory signaling. The specific enhancement of CRMP4b–RhoA binding suggests that the increased affinity may be mediated by the CRMP4b N-terminal extension. We therefore assessed the ability of the CRMP4b N terminus to mediate RhoA binding. We generated a chimeric CRMP molecule consisting of the CRMP4b N terminus fused to CRMP2 (CRMP4bNCRMP2) and a CRMP4 construct consisting of the common region of CRMP4a and CRMP4b (CRMP4aΔN) (Fig. 7a). Both CRMP4bNCRMP2 and CRMP4aΔN coimmunoprecipitate with RhoA; however, both proteins bind RhoA more weakly than full-length CRMP4b (Fig. 7b). This indicates that two independent RhoA binding sites within the CRMP4b molecule are sufficient for RhoA binding and that the two sites may cooperate to mediate maximal RhoA binding; however, the N-terminal region of CRMP4b appears to be the critical site mediating Nogo-dependent recruitment to RhoA (Fig. 3).
Figure 7.
The N-terminal domain of CRMP4b is sufficient for RhoA binding and disrupts full-length CRMP4b-RhoA binding when expressed as a recombinant fusion protein. a, Schematic of CRMP constructs generated to assess CRMP4 domains involved in RhoA binding. b, HEK293T cells cotransfected with myc-RhoA63L and CRMP-V5 constructs described in a and subjected to myc immunoprecipitation. c, HEK293T cells cotransfected with myc-RhoAWT, CRMP4-V5, and C4RIP-V5 to assess the effect of C4RIP on full-length CRMP4 binding to RhoA. d, HEK293T cells cotransfected with FLAG-RhoA63L, myc-ROCK, and C4RIP-V5 to assess the effect of C4RIP on RhoA binding to ROCK. IP, Immunoprecipitation.
To specifically disrupt the Nogo-dependent interaction between full-length CRMP4b and RhoA, we generated a construct to express the unique N-terminal domain of CRMP4b (C4RIP–CRMP4b–RhoA inhibitory peptide) (Fig. 7a) fused to a V5-epitope tag. As predicted, cotransfection of C4RIP-V5 significantly diminishes RhoA binding to full-length CRMP4b without affecting binding to CRMP4a (Fig. 7c), presumably by competing for the RhoA binding sites.
The specificity of C4RIP-V5 was then evaluated by examining its effect on binding between FLAG-RhoA63L and myc-Rho kinase (myc-ROCK), a critical RhoA effector molecule for myelin inhibition. C4RIP-V5 does not diminish binding between FLAG-RhoA63L and myc-ROCK (Fig. 7d). Similarly, overexpression of CRMP4b-V5 does not affect the binding between FLAG-RhoA63L and myc-ROCK, indicating that CRMP4b does not alter the ability of RhoA to interact with this downstream effector.
C4RIP-V5 attenuates neurite outgrowth inhibition
The ability of C4RIP-V5 to attenuate CRMP4b–RhoA binding provides a valuable tool to determine whether CRMP4b–RhoA complex formation is necessary for its role in myelin inhibition. CRMP4b-V5 or C4RIP-V5 was introduced into dissociated E13 chick DRG neurons via recombinant HSV (Fig. 8a), and neurite outgrowth was assessed on myelin substrates (Fig. 8b,d). CRMP4b-V5 is not sufficient to mimic myelin inhibitory responses (Fig. 8b,c), presumably because of its failure to increase its interaction with RhoA (Fig. 6b). C4RIP significantly attenuates myelin-dependent outgrowth inhibition compared with GFP or full-length CRMP4b-infected neurons (Fig. 8b,d). Unlike previously characterized Rho and ROCK antagonists, C4RIP does not promote basal DRG outgrowth on permissive control substrates (Fig. 8b,c) (Lehmann et al., 1999; Fournier et al., 2003). DRG neurons infected with HSVC4RIP or with HSV–dominant-negative ROCK (HSV-DNROCK) (Alabed et al., 2006) are both protected from myelin inhibition; however, HSV-DNROCK promotes basal outgrowth by ∼50%.
An advantage to targeting intracellular mediators of neurite outgrowth inhibition is their potential as convergent targets, which may attenuate multiple inhibitory influences. To evaluate whether C4RIP-V5 may block additional inhibitory signals associated with CNS injury, we examined the effect of C4RIP-V5 on neurite outgrowth inhibition mediated by aggrecan, an inhibitory CSPG of the glial scar. Intriguingly, we find that C4RIP also promotes neurite outgrowth on aggrecan (Fig. 8e).
To assess the specificity of C4RIP, we tested its ability to block Sema3A-dependent growth cone collapse in E8 chick DRG neurons. Sema3A stimulation of neurons engages Rac1 GTPase and the CRMP2 isoform (Huber et al., 2003). We find that C4RIP has no effect on Sema3A-dependent growth cone collapse (Fig. 8f). Together, these results indicate that disruption of the CRMP4b–RhoA interaction protects neurons from inhibitory influences that signal through RhoA GTPase.
Discussion
Regeneration after CNS trauma is limited by the activation of intracellular pathways within the injured neuron that block axonal extension through targeted modifications to the cytoskeleton (Hsieh et al., 2006). Development of antagonists to intracellular targets of axon outgrowth inhibitors is an effective approach to circumvent the inhibitory influence of the astroglial scar and MAIs (Dergham et al., 2002; Niederost et al., 2002; Borisoff et al., 2003; Fournier et al., 2003). In this study, we have demonstrated that CRMP4b is a necessary intracellular mediator of neurite outgrowth inhibition. Our findings suggest that complex formation between CRMP4b and RhoA is critical for outgrowth inhibition and that this inhibition may be mediated through an actin-dependent phenotype. By targeting the critical CRMP4b–RhoA binding interaction, we have developed a competitive antagonist of CRMP4b–RhoA binding, which specifically promotes neurite outgrowth on inhibitory substrates, suggesting an exciting new therapeutic target for nerve repair after CNS injury.
Dynamics of the CRMP4b–RhoA interaction
We have described a specific protein interaction between RhoA and CRMP4 that does not extend to other closely related family members of the Rho GTPases nor to other CRMP family members. In the absence of Nogo stimulation, the baseline interaction between endogenous CRMP4b and RhoA is negligible in cerebellar neurons and DRG growth cones. In transfected 293T and PC12 cells, CRMP4b and RhoA do interact in the absence of Nogo; however, this interaction could be a function of protein overexpression.
Intriguingly, the RhoA–CRMP4b interaction is not dependent on the nucleotide binding state of RhoA; rather, the interaction is dependent on the phosphorylation status of CRMP4. This raises the additional possibility that the strength of the baseline interaction may vary in different cell types as a function of the complement of kinases and phosphatases.
Because the RhoA used to screen for protein interactors was purified in bacteria, it is also conceivable that dephosphorylated RhoA is a favored binding partner for CRMP4b. This raises the possibility of a novel level of RhoA regulation in response to myelin inhibitors in which the focus has primarily been on the ability of MAIs to convert RhoA to the GTP-bound form (Dergham et al., 2002; Niederost et al., 2002; Fournier et al., 2003). This idea is consistent with finding that RhoA is phosphorylated at Ser188 by protein kinase A and that RhoA phosphorylation modifies its binding to its endogenous inhibitor, Rho guanine nucleotide dissociation factor (Ellerbroek et al., 2003; Nusser et al., 2006). CRMP4 is subject to phosphorylation by glycogen synthase kinase-3β and dephosphorylation by protein phosphatase 2A (Hill et al., 2006), and it is reasonable to hypothesize that phosphorylation alters its binding properties based on similarities to CRMP2 (Uchida et al., 2005). Together this suggests a model whereby Nogo-dependent engagement of a CRMP- and/or Rho-directed phosphatase may promote formation of a CRMP4b–RhoA complex. A prediction of this model would be that CRMP4b overexpression may fail to mimic myelin inhibition in DRG neurons because its association with RhoA is not adequately enhanced in the absence of appropriate posttranslational modifications, and this is what we observe (Figs. 6, 8).
Role of N-terminal variants of the CRMPs
The recent discovery of novel N-terminal variants of the original CRMP members in rats and chicks (Quinn et al., 2003; Yuasa-Kawada et al., 2003) has revealed additional potential functions for CRMPs. The unique N-terminal extensions of the CRMPb isoforms could impart supplementary functions to each CRMP variant by mediating additional protein interactions. A reasonable possibility is that CRMP4a and CRMP4b are capable of mediating similar cytoskeletal rearrangements; however, the N-terminal region of CRMP4b is necessary for recruitment to the appropriate cytoskeletal elements within the growth cone. This is consistent with the finding that overexpression of CRMP4b but not CRMP4a leads to an increase in neurite branching (Quinn et al., 2003).
C4RIP as a therapeutic agent
Targeting CRMP4-RhoA with C4RIP is a potential avenue for therapeutic intervention. The ability of C4RIP to attenuate inhibition in response to both MAIs and CSPGs is an obvious advantage in the complex inhibitory environment after CNS injury. The enrichment of CRMP expression in the nervous system raises the possibility that additional side effects on other cell types may be limited when compared with targeting ubiquitous molecules such as RhoA or ROCK. A unique characteristic of C4RIP is its failure to affect basal neurite outgrowth. To our knowledge, this is the first example of an intracellular molecule that can be specifically ascribed to an inhibitor-dependent neurite outgrowth pathway.
The function of CRMP4b in myelin-dependent inhibition
Reasonable hypotheses for CRMP4 function in myelin inhibition include effects on microtubule dynamics, actin dynamics, and/or endocytosis. CRMP2 can bind to tubulin heterodimers and is an important organizer of microtubule assembly for establishing axon–dendrite fate during development (Fukata et al., 2002b; Arimura et al., 2005). This is partly mediated by binding to tubulin heterodimers and promoting microtubule assembly (Fukata et al., 2002b). In fact, CRMP2 undergoes ROCK-dependent phosphorylation in response to Nogo-66, and MAG and has been hypothesized to subsequently regulate microtubule dynamics (Mimura et al., 2006). Although all CRMP members can bind tubulin, CRMP4 is not a substrate for ROCK, and we were unable to detect a change in affinity between CRMP4 and tubulin in response to Nogo-66 stimulation by coimmunoprecipitation analysis in PC12 cells (data not shown). This suggests that Nogo-66 may not affect CRMP4-dependent microtubule dynamics; however, if the interaction is locally regulated within the neurite or growth cone, we may have failed to detect this by coimmunoprecipitation.
Our data suggest that a CRMP4b–RhoA complex may participate in neurite outgrowth inhibition through actin-dependent processes. An association between CRMP4b and intersectin, an endocytic–exocytic adaptor protein (Quinn et al., 2003), raises the possibility that CRMP4b could play a role in regulating endocytosis. Endocytosis during neurite inhibition may be important for regulating membrane dynamics (Fournier et al., 2000) or may target the internalization of cell-surface receptors or cell adhesion molecules enabling their temporal and spatial regulation in response to MAIs. A plausible model is one in which CRMP4b–RhoA complexes regulate localized actin rearrangements from the growth cone periphery to the central domain and subsequent endocytic events necessary for growth cone collapse and neurite withdrawal.
Footnotes
This work was supported by a grant from the Canadian Institutes of Health Research (CIHR). A.E.F. is a Tier 2 Canada Research Chair. Y.Z.A. and S.O.T. were supported by fellowships from the CIHR, and M.P. was supported by an MS Society of Canada fellowship. We thank Drs. Phil Barker and Wayne Sossin for comments and suggestions on this manuscript, Dr. Keith Burridge for RhoA constructs, Dr. Nathalie Lamarche-Vane for Rac and Cdc42 constructs, Dr. Shuh Narumiya for the ROCKII construct, and Drs. Chris Quinn and Peter McPherson for CRMP constructs and antibody.
References
- Alabed YZ, Grados-Munro E, Ferraro GB, Hsieh S, Fournier AE. Neuronal responses to myelin are mediated by ROCK. J Neurochem. 2006;96:1616–1625. doi: 10.1111/j.1471-4159.2006.03670.x. [DOI] [PubMed] [Google Scholar]
- Arimura N, Menager C, Kawano Y, Yoshimura T, Kawabata S, Hattori A, Fukata Y, Amano M, Goshima Y, Inagaki M, Morone N, Usukura J, Kaibuchi K. Phosphorylation by Rho kinase regulates CRMP-2 activity in growth cones. Mol Cell Biol. 2005;25:9973–9984. doi: 10.1128/MCB.25.22.9973-9984.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur WT, Ellerbroek SM, Der CJ, Burridge K, Wennerberg K. XPLN, a guanine nucleotide exchange factor for RhoA and RhoB, but not RhoC. J Biol Chem. 2002;277:42964–42972. doi: 10.1074/jbc.M207401200. [DOI] [PubMed] [Google Scholar]
- Bishop AL, Hall A. Rho GTPases and their effector proteins. Biochem J. 2000;348:241–255. [PMC free article] [PubMed] [Google Scholar]
- Borisoff JF, Chan CC, Hiebert GW, Oschipok L, Robertson GS, Zamboni R, Steeves JD, Tetzlaff W. Suppression of Rho-kinase activity promotes axonal growth on inhibitory CNS substrates. Mol Cell Neurosci. 2003;22:405–416. doi: 10.1016/s1044-7431(02)00032-5. [DOI] [PubMed] [Google Scholar]
- Byk T, Dobransky T, Cifuentes-Diaz C, Sobel A. Identification and molecular characterization of Unc-33-like phosphoprotein (Ulip), a putative mammalian homolog of the axonal guidance-associated unc-33 gene product. J Neurosci. 1996;16:688–701. doi: 10.1523/JNEUROSCI.16-02-00688.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MS, Huber AB, van der Haar ME, Frank M, Schnell L, Spillmann AA, Christ F, Schwab ME. Nogo-A is a myelin-associated neurite outgrowth inhibitor and an antigen for monoclonal antibody IN-1. Nature. 2000;403:434–439. doi: 10.1038/35000219. [DOI] [PubMed] [Google Scholar]
- Dergham P, Ellezam B, Essagian C, Avedissian H, Lubell WD, McKerracher L. Rho signaling pathway targeted to promote spinal cord repair. J Neurosci. 2002;22:6570–6577. doi: 10.1523/JNEUROSCI.22-15-06570.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellerbroek SM, Wennerberg K, Burridge K. Serine phosphorylation negatively regulates RhoA in vivo. J Biol Chem. 2003;278:19023–19031. doi: 10.1074/jbc.M213066200. [DOI] [PubMed] [Google Scholar]
- Feig LA, Cooper GM. Inhibition of NIH 3T3 cell proliferation by a mutant ras protein with preferential affinity for GDP. Mol Cell Biol. 1988;8:3235–3243. doi: 10.1128/mcb.8.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier AE, Nakamura F, Kawamoto S, Goshima Y, Kalb RG, Strittmatter SM. Semaphorin3A enhances endocytosis at sites of receptor-F-actin colocalization during growth cone collapse. J Cell Biol. 2000;149:411–422. doi: 10.1083/jcb.149.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier AE, GrandPre T, Strittmatter SM. Identification of a receptor mediating Nogo-66 inhibition of axonal regeneration. Nature. 2001;409:341–346. doi: 10.1038/35053072. [DOI] [PubMed] [Google Scholar]
- Fournier AE, Takizawa BT, Strittmatter SM. Rho Kinase inhibition enhances axonal regeneration in the injured CNS. J Neurosci. 2003;23:1416–1423. doi: 10.1523/JNEUROSCI.23-04-01416.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata Y, Itoh TJ, Kimura T, Menager C, Nishimura T, Shiromizu T, Watanabe H, Inagaki N, Iwamatsu A, Hotani H, Kaibuchi K. CRMP-2 binds to tubulin heterodimers to promote microtubule assembly. Nat Cell Biol. 2002a;4:583–591. doi: 10.1038/ncb825. [DOI] [PubMed] [Google Scholar]
- Gaetano C, Matsuo T, Thiele CJ. Identification and characterization of a retinoic acid-regulated human homologue of the unc-33-like phosphoprotein gene (hUlip) from neuroblastoma cells. J Biol Chem. 1997;272:12195–12201. doi: 10.1074/jbc.272.18.12195. [DOI] [PubMed] [Google Scholar]
- Goshima Y, Nakamura F, Strittmatter P, Strittmatter SM. Collapsin-induced growth cone collapse mediated by an intracellular protein related to UNC-33. Nature. 1995;376:509–514. doi: 10.1038/376509a0. [DOI] [PubMed] [Google Scholar]
- GrandPre T, Nakamura F, Vartanian T, Strittmatter SM. Identification of the Nogo inhibitor of axon regeneration as a Reticulon protein. Nature. 2000;403:439–444. doi: 10.1038/35000226. [DOI] [PubMed] [Google Scholar]
- Hall A. Small GTP-binding proteins and the regulation of the actin cytoskeleton. Annu Rev Cell Biol. 1994;10:31–54. doi: 10.1146/annurev.cb.10.110194.000335. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, Culotti JG, Thomson JN, Perkins LA. Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev Biol. 1985;111:158–170. doi: 10.1016/0012-1606(85)90443-9. [DOI] [PubMed] [Google Scholar]
- Hill JJ, Callaghan DA, Ding W, Kelly JF, Chakravarthy BR. Identification of okadaic acid-induced phosphorylation events by a mass spectrometry approach. Biochem Biophys Res Commun. 2006;342:791–799. doi: 10.1016/j.bbrc.2006.02.029. [DOI] [PubMed] [Google Scholar]
- Hsieh S, Ferraro GB, Fournier AE. Myelin-associated inhibitors regulate cofilin phosphorylation and neuronal inhibition through LIM kinase and Slingshot phosphatase. J Neurosci. 2006;26:1006–1015. doi: 10.1523/JNEUROSCI.2806-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A, Kolodkin A, Ginty D, Cloutier J-F. Signaling at the growth cone: ligand-receptor complexes and the control of axon growth and guidance. Annu Rev Neurosci. 2003;26:509–563. doi: 10.1146/annurev.neuro.26.010302.081139. [DOI] [PubMed] [Google Scholar]
- Igarashi M, Strittmatter SM, Vartanian T, Fishman MC. Mediation by G proteins of signals that cause collapse of growth cones. Science. 1993;259:77–79. doi: 10.1126/science.8418498. [DOI] [PubMed] [Google Scholar]
- Inatome R, Tsujimura T, Hitomi T, Mitsui N, Hermann P, Kuroda S, Yamamura H, Yanagi S. Identification of CRAM, a novel unc-33 gene family protein that associates with CRMP3 and protein-tyrosine kinase(s) in the developing rat brain. J Biol Chem. 2000;275:27291–27302. doi: 10.1074/jbc.M910126199. [DOI] [PubMed] [Google Scholar]
- Khosravi-Far R, Chrzanowska-Wodnicka M, Solski PA, Eva A, Burridge K, Der CJ. Dbl and Vav mediate transformation via mitogen-activated protein kinase pathways that are distinct from those activated by oncogenic Ras. Mol Cell Biol. 1994;14:6848–6857. doi: 10.1128/mcb.14.10.6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottis V, Thibault P, Mikol D, Xiao ZC, Zhang R, Dergham P, Braun PE. Oligodendrocyte-myelin glycoprotein (OMgp) is an inhibitor of neurite outgrowth. J Neurochem. 2002;82:1566–1569. doi: 10.1046/j.1471-4159.2002.01146.x. [DOI] [PubMed] [Google Scholar]
- Lehmann M, Fournier A, Selles-Navarro I, Dergham P, Sebok A, Leclerc N, Tigyi G, McKerracher L. Inactivation of Rho signaling pathway promotes CNS axon regeneration. J Neurosci. 1999;19:7537–7547. doi: 10.1523/JNEUROSCI.19-17-07537.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Raible D, Raper JA. Collapsin: a protein in brain that induces the collapse and paralysis of neuronal growth cones. Cell. 1993;75:217–227. doi: 10.1016/0092-8674(93)80064-l. [DOI] [PubMed] [Google Scholar]
- Mandemakers WJ, Barres BA. Axon regeneration: it's getting crowded at the gates of TROY. Curr Biol. 2005;15:R302–R305. doi: 10.1016/j.cub.2005.04.002. [DOI] [PubMed] [Google Scholar]
- McKerracher L, David S, Jackson DL, Kottis V, Dunn RJ, Braun PE. Identification of myelin-associated glycoprotein as a major myelin-derived inhibitor of neurite growth. Neuron. 1994;13:805–811. doi: 10.1016/0896-6273(94)90247-x. [DOI] [PubMed] [Google Scholar]
- McPherson PS, Czernik AJ, Chilcote TJ, Onofri F, Benfenati F, Greengard P, Schlessinger J, De Camilli P. Interaction of Grb2 via its Src homology 3 domains with synaptic proteins including synapsin I. Proc Natl Acad Sci USA. 1994;91:6486–6490. doi: 10.1073/pnas.91.14.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura F, Yamagishi S, Arimura N, Fujitani M, Kubo T, Kaibuchi K, Yamashita T. Myelin-associated glycoprotein inhibits microtubule assembly by a rho-kinase-dependent mechanism. J Biol Chem. 2006;281:15970–15979. doi: 10.1074/jbc.M510934200. [DOI] [PubMed] [Google Scholar]
- Minturn JE, Fryer HJ, Geschwind DH, Hockfield S. TOAD-64, a gene expressed early in neuronal differentiation in the rat, is related to unc-33, a C. elegans gene involved in axon outgrowth. J Neurosci. 1995;15:6757–6766. doi: 10.1523/JNEUROSCI.15-10-06757.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay G, Doherty P, Walsh FS, Crocker PR, Filbin MT. A novel role for myelin-associated glycoprotein as an inhibitor of axonal regeneration. Neuron. 1994;13:757–767. doi: 10.1016/0896-6273(94)90042-6. [DOI] [PubMed] [Google Scholar]
- Nakamura F, Tanaka M, Takahashi T, Kalb RG, Strittmatter SM. Neuropilin-1 extracellular domains mediate semaphorin D/III-induced growth cone collapse. Neuron. 1998;21:1093–1100. doi: 10.1016/s0896-6273(00)80626-1. [DOI] [PubMed] [Google Scholar]
- Neve RL, Howe JR, Hong S, Kalb RG. Introduction of the glutamate receptor subunit 1 into motor neurons in vitro and in vivo using a recombinant herpes simplex virus. Neuroscience. 1997;79:435–447. doi: 10.1016/s0306-4522(96)00645-8. [DOI] [PubMed] [Google Scholar]
- Niederost B, Oertle T, Fritsche J, McKinney RA, Bandtlow CE. Nogo-A and myelin-associated glycoprotein mediate neurite growth inhibition by antagonistic regulation of RhoA and Rac1. J Neurosci. 2002;22:10368–10376. doi: 10.1523/JNEUROSCI.22-23-10368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Fukata Y, Kato K, Yamaguchi T, Matsuura Y, Kamiguchi H, Kaibuchi K. CRMP-2 regulates polarized Numb-mediated endocytosis for axon growth. Nat Cell Biol. 2003;5:819–826. doi: 10.1038/ncb1039. [DOI] [PubMed] [Google Scholar]
- Nusser N, Gosmanova E, Makarova N, Fujiwara Y, Yang L, Guo F, Luo Y, Zheng Y, Tigyi G. Serine phosphorylation differentially affects RhoA binding to effectors: implications to NGF-induced neurite outgrowth. Cell Signal. 2006;18:704–714. doi: 10.1016/j.cellsig.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Prinjha R, Moore SE, Vinson M, Blake S, Morrow R, Christie G, Michalovich D, Simmons DL, Walsh FS. Inhibitor of neurite outgrowth in humans. Nature. 2000;403:383–384. doi: 10.1038/35000287. [DOI] [PubMed] [Google Scholar]
- Quinn CC, Gray GE, Hockfield S. A family of proteins implicated in axon guidance and outgrowth. J Neurobiol. 1999;41:158–164. [PubMed] [Google Scholar]
- Quinn CC, Chen E, Kinjo TG, Kelly G, Bell AW, Elliott RC, McPherson PS, Hockfield S. TUC-4b, a novel TUC family variant, regulates neurite outgrowth and associates with vesicles in the growth cone. J Neurosci. 2003;23:2815–2823. doi: 10.1523/JNEUROSCI.23-07-02815.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riento K, Ridley AJ. Rocks: multifunctional kinases in cell behaviour. Nat Rev Mol Cell Biol. 2003;4:446–456. doi: 10.1038/nrm1128. [DOI] [PubMed] [Google Scholar]
- Rosslenbroich V, Dai L, Baader SL, Noegel AA, Gieselmann V, Kappler J. Collapsin response mediator protein-4 regulates F-actin bundling. Exp Cell Res. 2005;310:434–444. doi: 10.1016/j.yexcr.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Siddiqui SS, Culotti JG. Examination of neurons in wild type and mutants of Caenorhabditis elegans using antibodies to horseradish peroxidase. J Neurogenet. 1991;7:193–211. doi: 10.3109/01677069109167433. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Nakagomi S, Namikawa K, Kiryu-Seo S, Inagaki N, Kaibuchi K, Aizawa H, Kikuchi K, Kiyama H. Collapsin response mediator protein-2 accelerates axon regeneration of nerve-injured motor neurons of rat. J Neurochem. 2003;86:1042–1050. doi: 10.1046/j.1471-4159.2003.01920.x. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Nakamura F, Jin Z, Kalb RG, Strittmatter SM. Semaphorins A and E act as antagonists of neuropilin-1 and agonists of neuropilin-2 receptors. Nat Neurosci. 1998;1:487–493. doi: 10.1038/2203. [DOI] [PubMed] [Google Scholar]
- Uchida Y, Ohshima T, Sasaki Y, Suzuki H, Yanai S, Yamashita N, Nakamura F, Takei K, Ihara Y, Mikoshiba K, Kolattukudy P, Honnorat J, Goshima Y. Semaphorin3A signalling is mediated via sequential Cdk5 and GSK3beta phosphorylation of CRMP2: implication of common phosphorylating mechanism underlying axon guidance and Alzheimer's disease. Genes Cells. 2005;10:165–179. doi: 10.1111/j.1365-2443.2005.00827.x. [DOI] [PubMed] [Google Scholar]
- Wang KC, Koprivica V, Kim JA, Sivasankaran R, Guo Y, Neve RL, He Z. Oligodendrocyte-myelin glycoprotein is a Nogo receptor ligand that inhibits neurite outgrowth. Nature. 2002;417:941–944. doi: 10.1038/nature00867. [DOI] [PubMed] [Google Scholar]
- Wang LH, Strittmatter SM. Brain CRMP forms heterotetramers similar to liver dihydropyrimidinase. J Neurochem. 1997;69:2261–2269. doi: 10.1046/j.1471-4159.1997.69062261.x. [DOI] [PubMed] [Google Scholar]
- Wennerberg K, Ellerbroek SM, Liu RY, Karnoub AE, Burridge K, Der CJ. RhoG signals in parallel with Rac1 and Cdc42. J Biol Chem. 2002;277:47810–47817. doi: 10.1074/jbc.M203816200. [DOI] [PubMed] [Google Scholar]
- Yoshimura T, Kawano Y, Arimura N, Kawabata S, Kikuchi A, Kaibuchi K. GSK-3beta regulates phosphorylation of CRMP-2 and neuronal polarity. Cell. 2005;120:137–149. doi: 10.1016/j.cell.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Yuasa-Kawada J, Suzuki R, Kano F, Ohkawara T, Murata M, Noda M. Axonal morphogenesis controlled by antagonistic roles of two CRMP subtypes in microtubule organization. Eur J Neurosci. 2003;17:2329–2343. doi: 10.1046/j.1460-9568.2003.02664.x. [DOI] [PubMed] [Google Scholar]