Abstract
Regulator of G-protein signaling 4 (RGS4) modulates postsynaptic signal transduction by affecting the kinetics of Gα-GTP binding. Linkage, association, and postmortem studies have implicated the gene encoding RGS4 (RGS4) as a schizophrenia susceptibility factor. Using a multimodal neuroimaging approach, we demonstrate that genetic variation in RGS4 is associated with functional activation and connectivity during working memory in the absence of overt behavioral differences, with regional gray and white matter volume and with gray matter structural connectivity in healthy human subjects. Specifically, variation at one RGS4 single nucleotide polymorphism that has been associated previously with psychosis (rs951436) impacts frontoparietal and frontotemporal blood oxygenation level-dependent response and network coupling during working memory and results in regionally specific reductions in gray and white matter structural volume in individuals carrying the A allele. These findings suggest mechanisms in brain for the association of RGS4 with risk for psychiatric illness.
Keywords: G-protein, schizophrenia, genetics, fMRI, VBM, human
Introduction
The regulator of G-protein signaling (RGS) family is a heterogeneous collection of proteins that mediate postsynaptic signal transduction in signaling pathways throughout the brain. Although diverse as a class, they share a conserved domain, the “RGS box,” which confers enzymatic activity at G-protein coupled receptors (GPCRs) (Ross and Wilkie, 2000; Hollinger and Hepler, 2002). RGS proteins accelerate the hydrolysis of GTP from Gα, thereby driving the G-protein complex into its inactive, GDP-bound state (Berman et al., 1996; Druey et al., 1996; Watson et al., 1996; Hepler et al., 1997; Huang et al., 1997). By serving as GTPase accelerating proteins at GPCRs, RGS proteins are in a position to regulate the strength and cadence of neuronal activation (Druey et al., 1996; Hollinger and Hepler, 2002).
One member of the RGS family, RGS4, is an intriguing candidate risk gene for schizophrenia. Located on chromosome 1q23.3, the RGS4 gene (Online Mendelian Inheritance in Men database entry 602516) is a 7 kb sequence encoding a 23.3 kDa protein of 205 amino acids. Across species, RGS4 is highly expressed in neocortex, striatum, and the hippocampus (Gold et al., 1997; Nomoto et al., 1997; Ingi and Aoki, 2002; Erdely et al., 2004; Larminie et al., 2004). RGS4 modulates G-protein signaling in a number of neurotransmitter systems, several of which have been linked to schizophrenia, including metabotropic glutamate (Saugstad et al., 1998; De Blasi et al., 2001), dopamine (Yan et al., 1997; Ghavami et al., 2004), and serotonin (Beyer et al., 2004; Ghavami et al., 2004) receptors. In the striatum, RGS4 regulates dopaminergic signaling in a homeostatic manner and plays a role in mediating crosstalk between D1 and D2 receptors (Geurts et al., 2002, 2003; Taymans et al., 2003, 2004). RGS4 also interacts with the ErbB3 receptor, which binds neuregulin-1, potentially important because disrupted neuregulin–Erb signaling has been implicated in the pathogenesis of schizophrenia (Stefansson et al., 2002; Harrison and Weinberger, 2004; Hashimoto et al., 2004; Law et al., 2006).
Interest in RGS4 as a candidate gene stems from a series of suggestive linkage, microarray, and association studies. A genome-wide linkage scan revealed a major schizophrenia susceptibility locus (logarithm of the odds = 6.5) at 1q21–22, a region adjacent to RGS4 (Brzustowicz et al., 2000). Using cDNA expression profile microarray probes, Mirnics et al. (2001) found significantly decreased RGS4 mRNA in the dorsolateral prefrontal cortex (DLPFC) of schizophrenic patients. Of 70 genes from 1q21–22 contained on the array, only RGS4 was differentially expressed between probands and controls (Mirnics et al., 2001). Chowdari et al. (2002) resequenced the gene and its flanking regions and identified four single nucleotide polymorphisms (SNPs) in high linkage disequilibrium (LD), three located in the 5′ domain [SNPs 1 (rs10917670), 4 (rs951436), and 7 (rs951439)] and one in intron 1 [SNP 18 (rs2661319)], that tested positive for association in three family-based samples, although the over-transmitted alleles and haplotypes differed between cohorts (Chowdari et al., 2002). This same group found that genotype at their SNP 4 (rs951436) was associated with reduced DLPFC volume in first-episode schizophrenic patients and in healthy subjects (Prasad et al., 2004). A recent meta-analysis of published and unpublished data from multiple research centers (including our own schizophrenia genetics program) suggests that RGS4 is a weak susceptibility gene for schizophrenia, with the strongest association to rs951436, although the results are not conclusive (Talkowski et al., 2006).
We used multimodal neuroimaging to investigate the impact on brain structure and function in living human subjects of allelic variation at RGS4 rs951436, the SNP most frequently identified as impacting on risk for schizophrenia and previously associated with prefrontal cortex volume. We explored the effect of this SNP on functional magnetic resonance imaging (fMRI) measures of functional activation and connectivity related to working memory and MRI-based relative gray matter (GM) and white matter (WM) volume in healthy human subjects.
Materials and Methods
Recruitment
Healthy volunteers were recruited as part of the Clinical Brain Disorders Branch Sibling Study (Protocol 95-M-0150), an ongoing examination of neurobiological abnormalities related to genetic risk for schizophrenia (Egan et al., 2000). The study was approved by the Institutional Review Board of the National Institute of Mental Health. All subjects gave written, informed consent before participation. Inclusion criteria are included as supplemental data (available at www.jneurosci.org as supplemental material).
Neural (“intermediate”) phenotypes
Samples.
We tested the hypothesized effect for RGS4 rs951436 allelic variation on several neurobiological measures related to putative intermediate phenotypes for schizophrenia in healthy individuals recruited from an ongoing study of genetic risk for schizophrenia. We analyzed fMRI data for 94 right-handed healthy Caucasian subjects of European descent (HC), matched across genotype groups for age, gender, years of education, Wechsler Adult Intelligence Scale (WAIS-IQ), and task performance (accuracy and reaction time). Structural MRI datasets were available for 106 HC subjects of European descent; these subjects were also matched across genotype groups for age, gender, years of education, and WAIS-IQ (supplemental Table 1a,b, available at www.jneurosci.org as supplemental material). Examining the impact of psychiatrically associated polymorphisms in healthy subjects has proven a useful strategy (Egan et al., 2001, 2004; Hariri et al., 2002; Callicott et al., 2005), because individual contributions of genes contributing to disease risk in a genetically complex architecture can be isolated. Moreover, by studying carefully selected normal subjects, potential disease-related confounds are avoided, and the biological effects of genetic variation in brain are more directly assayed. For 49 subjects, we had both functional and structural data, with an additional 45 subjects in our fMRI analysis and an additional 57 subjects in our structural analysis.
Function: blood oxygenation level-dependent fMRI.
We used the n-back working memory task (Callicott et al., 1999). Briefly, we used a simple block design alternating between a two-back (2B) working memory condition and a zero-back (0B) control condition. Subjects were instructed to recall stimuli (visually presented digits 1–4) seen “n” previously: two previous for 2B and the currently presented digit for 0B. Our version of the n-back task consisted of continual presentation of visual stimuli in which every number was both a probe and a target. Visual stimuli were presented via a back-projection screen, and performance (accuracy and reaction time) was recorded via the use of a button box (Callicott et al., 2003a).
For fMRI analysis, a contrast image for the 2B − 0B contrast was estimated for each subject, after preprocessing as described previously (Callicott et al., 2003a,b). To examine task-related activations and deactivations, we entered each subject's first-level contrast into a one-sample t test. To study the effects of genetic variation in RGS4, we entered the first-level contrast images into a second-level regression analysis in SPM99 (Wellcome Department of Cognitive Neurology, University College London, London, UK) with RGS4 genotype as a covariate coded 0, 1, or 2 according to the number of rs951436 “A” alleles. Regression contrasts were explicitly masked by the main effect of task (thresholded at 0.05, family-wise error correction for multiple comparisons). Post hoc, we addressed a trend-level difference in WAIS-IQ between RGS4 genotypes (∼6 IQ points) by extracting blood oxygenation level-dependent (BOLD) signal from clusters identified via correlation. BOLD values were compared across RGS4 genotypes using an analysis of covariance (ANCOVA) in SPSS (SPSS, Chicago, IL), with WAIS-IQ score as a nuisance covariate; all reported activations remained significant. To further examine the influence, if any, of this trend-level difference, we ran a multiple regression in SPM99 with RGS4 genotype and WAIS-IQ score in the model. All reported activations, with the exception of caudate, remained. Detailed descriptions of acquisition parameters and preprocessing steps are included as supplemental data (available at www.jneurosci.org as supplemental material).
Structure: voxel-based morphometry.
Our voxel-based morphometry techniques have been outlined in detail previously (Meyer-Lindenberg et al., 2006) and are included as supplemental data (available at www.jneurosci.org as supplemental material). We examined global effects of genotype on gray and white matter using a multiple regression model with RGS4 genotype as a covariate, coded 0, 1, or 2 according to the number of rs951436 “A” alleles, controlling for potential confounds including total brain volume, WAIS-IQ (full scale score), gender, and both linear and quadratic expansions of age (Buchel et al., 1996). Voxels at a significance threshold of p < 0.01, uncorrected (k > 500), are reported with reference to the Montreal Neurological Institute (MNI) standard space within SPM2 after conversion to the standard space of Talairach and Tournoux using in-house software as in previous reports (Egan et al., 2001; Callicott et al., 2005).
Connectivity.
Our methods for functional and structural connectivity were calculated as in previous reports (Pezawas et al., 2005) and are included as supplemental data (available at www.jneurosci.org as supplemental material). Seed regions for functional [left and right Brodman area (BA) 47] and structural connectivity (right BA 47) analyses were derived from the areas of maximal prefrontal activation difference in RGS4 A carriers during our working memory task.
Genotyping.
We used standard methods to extract DNA from white blood cells using the Puregene DNA purification kit (Gentra Systems, Minneapolis, MN). RGS4 genotyping was performed using the Taqman 5′-exonuclease allelic discrimination assay (Livak, 1999) obtained from Applied Biosystems (Foster City, CA) (Assay on Demand, identification number C_9619634_10).
Negative-control SNP.
We used another RGS4 SNP of similar minor allele frequency (rs1507754) that is generally regarded as nonsignificant for schizophrenia association (with the exception of Chowdari et al., 2002) as a negative-control SNP in our imaging analyses. Using the same statistical threshold [p < 0.05, false discovery rate (FDR)-corrected for multiple comparisons] as with rs951436, our analyses using the negative-control SNP rs1507754 did not find any significant differences between genotypes in any of our measures. No-call percentages were similar for both SNPs (4.3% for rs1507754 and 2.2% for rs951436), and there were no deviations from Hardy–Weinberg equilibrium for each. There were 68 subjects with both fMRI data and rs1507754 genotypes, with an overlap of 63 subjects between rs1507754 and rs951436 for fMRI analyses. There were 103 subjects with both structural MRI data and rs1507754 genotypes, with an overlap of 84 subjects between rs1507754 and rs951436 for structural MRI analyses.
Results
fMRI
A threshold of p < 0.05, false discovery rate-corrected for multiple comparisons, was applied to all imaging data. Although regional activation and volume findings do not meet this threshold, we report activation and volume differences surviving an exploratory threshold of p < 0.01, uncorrected to describe our selection of seed foci for connectivity analyses.
Task activation and deactivation
Consistent with previous reports (Callicott et al., 1999; Meyer-Lindenberg et al., 2001), we found that the two-back version of the n-back task elicits activation and deactivation in distinct networks in the human brain. Regions engaged by task performance included DLPFC, ventrolateral prefrontal cortex (VLPFC), premotor cortex, posterior parietal cortex (PPC), thalamus, basal ganglia, and cerebellum (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). We found task-related deactivation in medial prefrontal cortex, especially medial aspects of superior frontal gyrus (mPFC), hippocampal formation and parahippocampal gyrus, posterior cingulate, and superior temporal gyrus (supplemental Fig. 1, available at www.jneurosci.org as supplemental material).
RGS4 effect on BOLD response
RGS4 A allele load was associated with alterations in the neural response to working memory challenge. Regression analysis revealed a positive correlation between the number of A alleles and BOLD fMRI activation in left ventrolateral PFC [BA 47 (p < 0.001, uncorrected)]. Additionally, we found a negative correlation between A allele load and BOLD fMRI response in several regions, including posterior right inferior temporal gyrus (pITG) (BA 37), posterior right middle temporal gyrus (pMTG) (BA 39), right ventrolateral PFC (BA 47), right dorsolateral PFC (BA 46/10) (p < 0.001, uncorrected), and right caudate (p < 0.003, uncorrected.) Thus, relative to C homozygotes, A allele carriers are characterized by hypoactivation in right temporal cortex, right ventral PFC, and right caudate, in the context of hyperactivation in left ventral PFC, during working memory despite matched performance between groups. (Fig. 1, Table 1). To address the potential for type I error, we analyzed an unlinked SNP of similar minor allele frequency with few reported associations to schizophrenia diagnosis (rs1507754). We did not observe an association between allelic variation at this negative-control SNP and activation in the working memory network at the chosen threshold.
Figure 1.
Effect of RGS4 genotype on brain activation during working memory. Image depicts positive (red) and negative (green) correlations between two-back activation and the number of A alleles. Image thresholded at 0.01 uncorrected, k > 8.
Table 1.
Effect of RGS4 on working memory activation (activations thresholded at 0.01 uncorrected, k > 8)
| Location | BA | x | y | z | Cluster size (voxels) | z score | p value |
|---|---|---|---|---|---|---|---|
| Positive correlation with allele A | |||||||
| Left ventrolateral PFC | 47 | −45 | 29 | −6 | 15 | 3.43 | 0.000 |
| Left inferior parietal lobule | 40 | 55 | −35 | 40 | 8 | 2.84 | 0.002 |
| Negative correlation with allele A | |||||||
| Right inferior temporal gyrus | 37 | 51 | −58 | −2 | 39 | 3.93 | 0.000 |
| Right middle temporal gyrus | 39 | 55 | −49 | 19 | 18 | 3.52 | 0.000 |
| Right ventrolateral PFC | 47 | 34 | 14 | −11 | 18 | 3.39 | 0.000 |
| Right dorsolateral PFC | 10 | 26 | 44 | 3 | 23 | 3.10 | 0.001 |
| Right caudate | 15 | 18 | −1 | 11 | 2.78 | 0.003 |
RGS4 effect on functional connectivity
We expected that working memory connectivity associated with a putative schizophrenia risk gene would be altered in a lawful manner. Based on previous findings, we predicted increased PFC connectivity with regions that are deactivated during working memory (e.g., hippocampal formation, medial prefrontal cortex, and superior temporal gyrus), as this has been shown previously in schizophrenic subjects (Meyer-Lindenberg et al., 2005). Indeed, using the right hemisphere pITG, pMTG, VLPFC, and DLPFC foci as seed regions for functional connectivity analyses, we found a reliable pattern of disrupted connectivity associated with the A allele that is consistent with previous findings in schizophrenic patients (see Discussion). Specifically, we saw a negative correlation between RGS4 A allele load and connectivity between these foci and key nodes of the working memory network, including DLPFC and PPC (p < 0.05, corrected for multiple comparisons). Additionally, we found a positive correlation between RGS4 A allele load and connectivity between these foci and areas that are deactivated during working memory, especially mPFC, superior temporal cortex, posterior cingulate, and parahippocampal gyrus (Fig. 2, Table 2) (p < 0.05, corrected). Examining the left hemisphere ventrolateral PFC cluster as a seed region revealed the opposite pattern of A-associated connectivity. We observed a positive correlation between RGS4 A allele load and connectivity between this cluster and key working memory nodes, including DLPFC and PPC (Fig. 3, Table 2) (p < 0.05, corrected). Thus, allele A was associated with increased right ventral prefrontal connectivity with medial prefrontal cortex, superior temporal gyrus, posterior cingulate, and parahippocampal gyrus, and increased left ventral prefrontal connectivity with dorsolateral prefrontal cortex and posterior parietal cortex.
Figure 2.
Effect of RGS4 genotype on functional connectivity, right VLPFC seed. Image depicts positive correlation between the number of A alleles and working memory-related network connectivity with right VLPFC reference region. Unthresholded image; color bar represents t score values.
Table 2.
Effect of RG S4 on working memory functional connectivity (activations thresholded at t > 2.5, k > 8)
| Reference ROI | Location | BA | x | y | z | Cluster size (voxels) | z score | p value |
|---|---|---|---|---|---|---|---|---|
| Positive correlation with allele A | ||||||||
| Left ventrolateral PFC (−45, 29, −6) | Left ventrolateral PFC | 47 | −34 | 29 | −1 | 131 | 5.01 | 0.000* |
| Left dorsolateral PFC | 9 | −38 | 41 | 31 | 49 | 4.53 | 0.000* | |
| Left superior frontal gyrus | 9 | 26 | 78 | 35 | 247 | 4.43 | 0.000* | |
| Right dorsolateral PFC | 9 | 38 | 45 | 31 | 108 | 3.93 | 0.000* | |
| Right ventrolateral PFC | 47 | 38 | 21 | −1 | 113 | 3.90 | 0.000* | |
| Right supplementary motor area | 6 | 4 | 13 | 49 | 44 | 3.76 | 0.000* | |
| Left cerebellum | −45 | 46 | −33 | 36 | 3.72 | 0.000* | ||
| Right middle frontal gyrus | 6 | 40 | 2 | 50 | 88 | 3.30 | 0.000** | |
| Right ventrolateral PFC(34, 14, −11) | Right transverse temporal gyrus | 21 | 45 | −15 | −9 | 1046*** | 4.39 | 0.000* |
| Right superior temporal gyrus | 41 | 51 | −18 | 12 | % | 4.30 | 0.000* | |
| Right superior temporal gyrus | 38 | 38 | 7 | −15 | % | 4.12 | 0.000* | |
| Right parahippocampal gyrus | 38 | −1 | −10 | % | 3.91 | 0.000* | ||
| Left posterior cingulate | 30 | −19 | −50 | −8 | % | 3.88 | 0.000* | |
| Left precentral gyrus | 3 | −55 | −10 | 12 | 258*** | 4.17 | 0.000* | |
| Left superior temporal gyrus | 38 | −51 | 10 | −20 | % | 3.69 | 0.000* | |
| Left superior frontal gyrus (medial) | 8/9 | −25 | 39 | 48 | 155 | 3.75 | 0.000* | |
| Right superior frontal gyrus (medial) | 8/9 | 15 | 39 | 48 | 13 | 3.47 | 0.000* | |
| Right caudate | −11 | 19 | 16 | 49 | 3.40 | 0.000* |
*, Survives voxelwise correction for multiple comparisons; **, survives clusterwise correction for multiple comparisons; ***, peak voxel of cluster; %, peak within larger cluster above.
Figure 3.
Effect of RGS4 genotype on functional connectivity, left VLPFC seed. Image depicts positive correlation between the number of A alleles and working memory-related network connectivity with left VLPFC reference region. Unthresholded image; color bar represents t score values.
Voxel-based morphometry
RGS4 effect on brain morphology
Gray matter.
RGS4 genotype was associated with relative differences in regional gray matter volume in the human brain. Subjects carrying the RGS4 A allele showed decreased gray matter volume in right VLPFC, BA 47 (p < 0.001, uncorrected). We also noted decreased GM volume in bilateral thalamus (p < 0.001, uncorrected) and right superior temporal gyrus (p < 0.001, uncorrected) in A subjects. However, we did not observe any structural changes in left VLPFC (Fig. 4, Table 3). We did not find an association between our negative-control SNP and gray matter structure in these regions.
Figure 4.

Effect of RGS4 genotype on gray matter volume. Image depicts negative correlation between the number of A alleles and gray matter structural volume, thresholded at 0.05 uncorrected, k > 500.
Table 3.
Effect of RGS4 on gray matter (thresholded at 0.01 uncorrected, k > 500)
| Location | BA | x | y | z | Cluster size (voxels) | z score | p value |
|---|---|---|---|---|---|---|---|
| Negative correlation with allele A | |||||||
| Right superior temporal gyrus | 22 | 48 | −47 | 19 | 765 | 3.49 | 0.000 |
| Thalamus | 1 | −9 | 1 | 3751 | 3.21 | 0.001 | |
| Right ventrolateral PFC | 47 | 56 | 17 | −3 | 883 | 3.14 | 0.001 |
| Left inferior parietal lobule | 40 | −65 | −37 | 36 | 500 | 3.08 | 0.001 |
White matter.
Given our findings of decreased BOLD signal during working memory and gray matter volume in right PFC, we sought to investigate the impact of RGS4 genotype on prefrontal white matter morphology. RGS4 A carriers demonstrated significant volume decreases in right ventral prefrontal white matter (p < 0.05 corrected for multiple comparison within a right prefrontal region of interest) (Fig. 5, Table 4). We did not observe white matter volume differences in our group analysis of the negative-control SNP.
Figure 5.
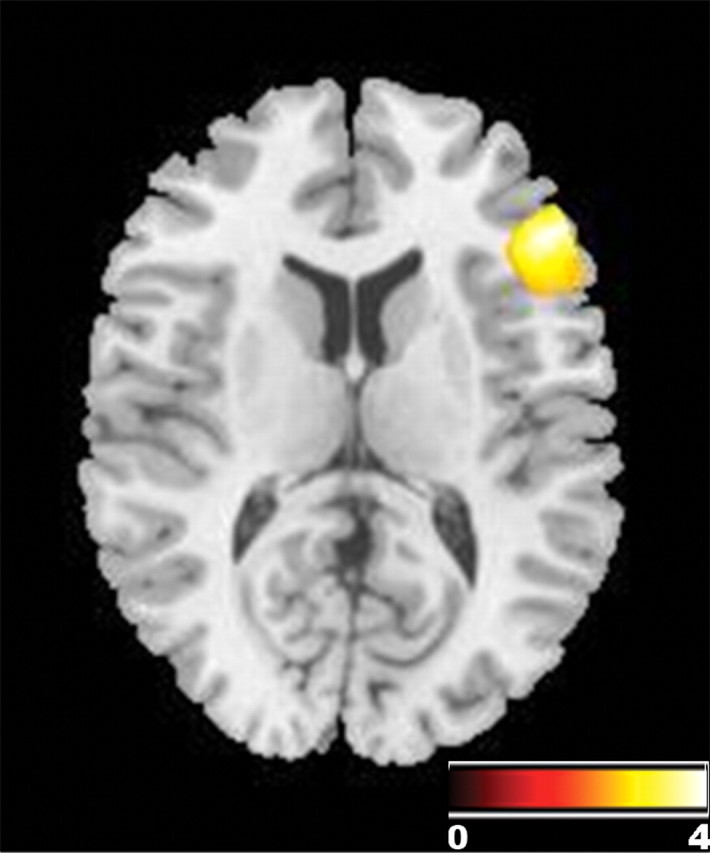
Effect of RGS4 genotype on white matter volume. Image depicts negative correlation between number of A alleles and white matter structural volume, thresholded at 0.005 uncorrected, k > 500. Rendered on single-subject T1-weighted image to visualize localization of gene effect on white matter in right ventral prefrontal region.
Table 4.
Effect of RGS4 on white matter (thresholded at 0.01 uncorrected, k > 500)
| Location | BA | x | y | z | Cluster size (voxels) | z score | p value |
|---|---|---|---|---|---|---|---|
| Negative correlation with allele A | |||||||
| Right ventral PFC | 48 | 31 | 10 | 4020 | 3.68 | 0.000* |
*, Survives voxelwise correction for multiple comparisons.
RGS4 effect on structural connectivity
Using the area of decreased GM volume in right VLPFC identified in our voxel-based morphometry analysis as a seed, we analyzed correlated changes in GM throughout the rest of the brain as a function of RGS4 genotype. Given our functional finding of increased left VLPFC BOLD, we hypothesized a correlation between right and left VLPFC GM volumes. We used a left ventral PFC seed to test our hypothesis of a region × genotype interaction for prefrontal structural connectivity and did indeed observe significantly increased connectivity between right and left VLPFC in A homozygote subjects relative to C homozygotes (p < 0.05, corrected) (Table 5). Examining the entire brain, we also found relatively increased right VLPFC–left hippocampus structural coupling in A individuals (p < 0.001, uncorrected).
Table 5.
Effect of RGS4 on structural connectivity, right ventrolateral PFC seed (thresholded at 0.01 uncorrected, k > 500)
| Location | BA | x | y | z | Cluster size (voxels) | z score | p value |
|---|---|---|---|---|---|---|---|
| Left ventrolateral PFC | 47 | −52 | 24 | 10 | 3824 | 4.17 | 0.000* |
| Left hippocampus | −29 | −9 | −13 | 1889 | 3.77 | 0.000 |
*, Survives voxelwise correction for multiple comparisons.
Functional and structural connectivity analysis using an occipital control
To test the specificity of these connectivity changes to the frontoparietal and frontotemporal networks outlined above, we examined RGS4-dependent functional and structural connectivity using a “control region” seed (a 5 mm sphere centered in occipital cortex; MNI coordinates 30, −75, 20) in which activation during working memory did not differ across RGS4 genotypes. At our threshold of p < 0.05 (FDR corrected for multiple comparisons), we did not see any differences in functional or structural connectivity related to RGS4 genotype.
Behavioral correlates of genotype-dependent changes in function and structure
To validate the relevance of our imaging findings to behavior and demonstrate that the changes evident in A carriers may be adverse and could thus be credibly claimed as contributing to disease risk, we correlated these imaging parameters with a measure of information processing speed (two-back reaction time). Reaction time during working memory and cognitive control has been shown previously to relate to BOLD activation and network connectivity, as well as individual differences in brain morphology (Rypma and D'Esposito, 1999; Posthuma et al., 2003; Haier et al., 2005; Lenartowicz and McIntosh, 2005; Liston et al., 2005). Therefore, we expected, and found, that the functional and structural alterations evident in A carriers were associated with slower information processing, i.e., increased reaction times.
Working memory BOLD
Right ventrolateral PFC BOLD and reaction time during working memory were inversely related (r = −0.22; p < 0.05) such that lower functional activation, as seen in rs951436 A subjects, predicted higher response latencies for correct trials. We found evidence for a similar relationship between reaction time and activation in the right inferior (r = −0.21; p < 0.05) and middle (r = −0.26; p < 0.05) temporal gyri.
Functional connectivity
RGS4-dependent changes in functional connectivity during working memory also correlated with reaction time. We found an inverse relationship between response latency and functional coupling between right VLPFC and left parahippocampal gyrus (r = −0.21; p < 0.05), as well as right VLPFC and left medial frontal gyrus (r = −0.21; p < 0.05).
White matter
White matter volume, which we found to be relatively decreased in rs951436 A subjects, predicted reaction time during a version of the n-back working memory task performed outside of the scanner. Right ventral prefrontal white matter volume was inversely correlated with reaction time (r = −0.22; p < 0.05).
Discussion
The present study suggests that RGS4 allelic variation may impact several MRI-based measures of functional and structural connectivity in the healthy human brain. In our sample of healthy subjects, we show that the RGS4 A allele is significantly associated with changes in the functional coupling of information processing nodes during working memory, in gray matter structural connectivity, and in regional white matter volume. Some of these alterations are qualitatively analogous, but not identical to, differences in working memory-associated BOLD response, functional connectivity, and regional gray and white matter volume demonstrated by schizophrenic patients. It is unclear exactly what these changes reflect, whether they are deleterious or compensatory, or how they may be related to the clinical association of this gene with schizophrenia. Several recent studies have linked genetic variation associated with increased risk for schizophrenia to deleterious changes in neural function and structure (Egan et al., 2001, 2004; Addington et al., 2005; Callicott et al., 2005; Cannon et al., 2005; Ohnishi et al., 2005; Gurling et al., 2006). The current finding of RGS4 A-associated alterations in brain morphology and working memory activation and connectivity in healthy subjects adds to the evidence for the impact of RGS4 on brain function and structure and may shed light on its putative role as a schizophrenia susceptibility gene.
Activation
Because these differences do not survive whole-brain correction for multiple comparisons, our finding of changes in working memory-related activation in RGS4 A allele carriers is suggestive but not compelling. However, we feel that some comment on the pattern of activation differences between genotypes may be instructive. Consistent with the long-standing observation of impaired performance on PFC-dependent tasks, neuroimaging studies have repeatedly identified abnormal prefrontal activation in schizophrenia (for review, see Callicott and Weinberger, 1999). Our finding of altered ventrolateral prefrontal engagement in A carriers accords with a number of studies in which schizophrenic patients abnormally activate this region (Barch et al., 2001; Perlstein et al., 2001; Callicott et al., 2003b; Tan et al., 2006). Ventrolateral PFC subserves several aspects of executive cognition, including inhibitory control (Liddle et al., 2001; Aron et al., 2004; Morita et al., 2004), set shifting (Nakahara et al., 2002; Shafritz et al., 2005), stimulus selection (Rushworth et al., 1997), interference suppression (Kemmotsu et al., 2005), and on-line maintenance of items in WM (Veltman et al., 2003).
In addition to differences in prefrontal activation and deactivation, RGS4 A subjects showed changes in temporal and subcortical regions that are also involved in aspects of working memory. For example, the inferior temporal area is a terminal region of the ventral visual pathway (Ungerleider and Mishkin, 1982) that is implicated in the maintenance of visual objects during working memory by single-unit recording, fMRI, and lesion studies (Miller et al., 1993; Petrides, 2000; Ranganath et al., 2004). Activation in this region may reflect a neural working memory “strategy” that is based on visuospatial, as opposed to phonological, information processing. Caudate activation in the n-back task may be specific to the manipulation of working memory representations (Lewis et al., 2004).
We observed laterality differences in WM activations and deactivations associated with the rs951436 A allele. Although there is general agreement that prefrontal cortex is a key working memory node in humans and nonhuman primates, alternate concepts of functional segregation within prefrontal cortex abound in the literature (Levy and Goldman-Rakic, 2000; Rainer and Ranganath, 2002; Wager and Smith, 2003). Along a dorsoventral axis, some groups have proposed an anatomical organization according to material type, with superior aspects of PFC associated with spatial WM and inferior regions implicated in object WM (Goldman-Rakic, 1987; Romanski, 2004). Others have suggested organization by process type, with superior PFC specialized for monitoring and manipulating information in WM and ventral PFC involved in WM rehearsal (Owen, 1997, 2000; D'Esposito et al., 1998). Along the left–right axis, there is evidence in favor of left-lateralized activations for verbal and object WM and right lateralization for spatial working memory in ventral PFC (Wager and Smith, 2003). However, our block-design version of the n-back task does not allow for separate examination of distinct working memory subcomponents, and either verbal or spatial neural strategies may be used to perform the task, limiting our ability to draw definitive conclusions from our finding of lateralized changes in prefrontal function associated with RGS4 genotype. Additional studies using more cognitively specific versions of the n-back will be key to understanding the precise neurofunctional changes associated with RGS4 variation.
Connectivity
In addition to task-related neural activation, functional connectivity, a linear measure of BOLD activity covariation between two or more regions (Ramnani et al., 2004), is emerging as a useful neuroimaging phenotype for characterizing network-level information processing dysregulation. Several studies have found aberrant functional connectivity in schizophrenic patients during the performance of cognitive tasks (Friston and Frith, 1995; Meyer-Lindenberg et al., 2001, 2005; Peled et al., 2001; Winterer et al., 2003; Foucher et al., 2005; Tan et al., 2006). For instance, Meyer-Lindenberg et al. (2001) demonstrated that schizophrenic subjects fail to disengage medial prefrontal, parahippocampal gyrus, and superior temporal gyrus during working memory; connectivity analysis identified two distributed networks involving hippocampus and inferotemporal cortex (in patients) and dorsolateral prefrontal and parietal cortex (in controls) that differentiated subjects based on disease status. Callicott et al. (2003b) replicated the finding of hippocampal overactivation during WM in poorly performing schizophrenic patients using fMRI, and a recent positron emission tomography study showed increased DLPFC–hippocampus functional connectivity in schizophrenic subjects during working memory (Meyer-Lindenberg et al., 2005). The current finding of decreased right VLPFC connectivity to DLPFC and parietal cortex and increased connectivity to medial frontal gyrus, superior temporal gyrus, and parahippocampal gyrus (areas in which schizophrenic patients activate more than healthy subjects) suggests that RGS4 rs951436 variation biases individuals with allele A toward a pattern of working memory-related functional connectivity seen previously in schizophrenic patients. That the connectivity observed in A subjects is associated with slower reaction times during WM supports our suggestion that the change in functional coupling observed in these subjects is in some way adverse or inefficient.
We have not specifically tested for a relationship between functional and structural connectivity in this dataset. This limits our ability to comment on the precise meaning of RGS4 genotype-dependent structural connectivity changes in the context of our other finding of RGS4 genotype-dependent functional connectivity changes. However, it should be noted that there is partial overlap between the patterns of functional and structural connectivity alterations evident in carriers of the RGS4 A allele, particularly with reference to right and left VLPFC connectivity and right VLPFC–hippocampus connectivity.
Morphometry
The significant correlation between white matter volume and reaction time in right ventral PFC suggests that the reductions in ventral prefrontal white matter observed in RGS4 A individuals may have some behavioral relevance. These changes parallel previous structural imaging (Paillere-Martinot et al., 2001; Suzuki et al., 2002; Ho et al., 2003; Wolkin et al., 2003; Kubicki et al., 2005; Mitelman et al., 2005) and neuropathological (Hakak et al., 2001; Hof et al., 2002; Davis et al., 2003; Flynn et al., 2003; Tkachev et al., 2003) studies, which have found reduced white matter density in schizophrenic patients. In addition, although not significant at our established threshold, rs951436 A subjects show a trend for reduced gray matter volume in several regions, including ventral PFC, superior temporal gyrus, and thalamus, highlighted in a recent meta-analysis of gray matter morphology in schizophrenic patients (Honea et al., 2005). Allelic variation in RGS4 might impact brain structure in a number of ways, because it is dynamically expressed throughout the brain during development and is thought to influence neuronal differentiation through its regulation by the paired-like homeodomain protein Phox2b (Grillet et al., 2003). In addition, RGS4 regulates signaling at receptors for other neurotransmitters (e.g., serotonin) whose roles in neurodevelopment are well established. Disrupted signaling at any of these sites during development could lead to changes in brain structure.
Limitations
A threshold of p < 0.05, corrected for multiple comparisons via FDR was applied consistently to all imaging datasets. Although white matter morphometric and connectivity findings (structural and functional) meet this threshold, regional activation and gray matter volume findings do not. Because we cannot therefore exclude the possibility that these findings resulted from type I error, we include them for the purpose of explaining the selection of regions of interest for the connectivity analyses and do not mean to imply that they are compelling in their own right.
The RGS4 SNP under investigation in this study is in high LD with the three other 5′ markers that have shown association with schizophrenia in at least one study. Therefore, its impact may represent primary effects related to one of these other SNPs or to haplotypes containing these SNPs. Likewise, there may be other currently unknown genetic variants with which it may also be in high LD. Also, given the suggestion that RGS4 SNPs are in LD with markers in other putative schizophrenia risk genes (Puri et al., 2007), we cannot rule out the possibility that the functional and structural consequences of variation in rs951436 result from the impact of another susceptibility gene in the 1q23.3 region. In other words, we cannot assert that the functional and structural changes described in this study are attributable exclusively to variation at rs951436 and not to one or more linked markers. However, the fact that there is such high LD within this narrow 5′ untranslated region suggests that we may use rs951436 as a surrogate for other linked SNPs with previous positive associations to schizophrenia and that similar results to those seen here could reasonably be expected by testing these SNPs. Importantly, testing only rs951436, the most consistently associated variant across studies and the most significant association in a recent meta-analysis (Talkowski et al., 2006), circumvents the multiple comparisons issue that would apply in testing all variants within the region of high LD in the 5′ upstream region of this gene.
Further hindering interpretation of the current data are that fact that the functional consequences of the A→C base pair substitution at rs951436 remain uncharacterized. Therefore, we are unable to offer an exact mechanism for how RGS4 variation at this or nearby loci might lead to the observed results. However, the location of this SNP, in the putative promoter region of the gene, provides a clue to likely functional outcomes after allelic variation, namely altered transcriptional efficiency of RGS4. This notion is supported by the Mirnics et al. (2001) finding of decreased RGS4 mRNA in schizophrenic PFC, although it is not clear whether altered RGS4 expression is related to genotype at this SNP. The transcriptional response of RGS4 to receptor activation has been suggested to play an important role in its ability to dynamically regulate GPCR signal transduction, especially in response to stress (Ni et al., 1999). Therefore, genetic variation affecting transcriptional response in the RGS4 gene, and by extension, expression of RGS4 protein, may lead to a dysregulated postsynaptic milieu and changes in the second-messenger pathways of RGS4-linked GPCRs, resulting in poorer stress resilience during development. This increased vulnerability to stress may play a role in the genetic association of RGS4 to schizophrenia risk.
In the current study, functional and structural changes are seen in subjects possessing an A allele. Previous positive associations to schizophrenia have been found with both allele A and C, although allele A has been most consistently associated in case-control studies (Talkowski et al., 2006). Although we cannot comment on this issue with the present data, there is some evidence for allelic heterogeneity or at the very least a complex genotype–phenotype relationship, with RGS4. Specifically, in the original association study by Chowdari et al. (2002), three independent cohorts were examined. They found significant transmission distortion in two of these but with different alleles over-transmitted in each. Notably, this group also found decreased PFC volume with allele A in the same cohort in which allele A was over-transmitted to patients (Prasad et al., 2004). The authors raise the possibility that the differing associations between the samples (and presumably, between the studies) represent independent risk factors. These variable allele findings also suggest that, if the associations are valid, this SNP is a marker monitoring different haplotypic backgrounds.
We observed a trend-level difference in WAIS-IQ score between RGS4 homozygote groups (RGS4 C/C subject mean ∼6 points higher than RGS4 A/A subject mean), potentially important considering the proposed interdependence between IQ and working memory capacity (Conway et al., 2003). However, using post hoc ANCOVA and multiple regression analyses to explicitly test for an influence of this trend on our imaging results, we found that it did not alter our findings.
Additional investigation is necessary to confirm a disease-specific impact of the changes found herein. To this end, future study of functional and structural alterations associated with rs951436 in schizophrenic subjects and their unaffected siblings is suggested; any such changes in probands should be examined with respect to clinically relevant disease markers, such as age at onset, cognitive impairment, and antipsychotic response. Additionally, analyzing the impact of RGS4 on the background of other genetic variants associated with schizophrenia (gene–gene interactions) may further elucidate the role of this gene in schizophrenia pathogenesis.
Conclusions
This study suggests a role for RGS4 variation in working memory-related activation and functional coupling and gray and white matter volume in healthy human subjects. We believe that the current findings support the notion that RGS4 variation contributes to risk for psychiatric illness.
Footnotes
This research was supported by the Intramural Research Program of the National Institute of Mental Health, National Institutes of Health. We thank Jennifer Brooke and Morgan Prust for additional analyses and technical expertise.
References
- Addington AM, Gornick M, Duckworth J, Sporn A, Gogtay N, Bobb A, Greenstein D, Lenane M, Gochman P, Baker N, Balkissoon R, Vakkalanka RK, Weinberger DR, Rapoport JL, Straub RE. GAD1 (2q31.1), which encodes glutamic acid decarboxylase (GAD67), is associated with childhood-onset schizophrenia and cortical gray matter volume loss. Mol Psychiatry. 2005;10:581–588. doi: 10.1038/sj.mp.4001599. [DOI] [PubMed] [Google Scholar]
- Aron AR, Robbins TW, Poldrack RA. Inhibition and the right inferior frontal cortex. Trends Cogn Sci. 2004;8:170–177. doi: 10.1016/j.tics.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Barch DM, Carter CS, Braver TS, Sabb FW, MacDonald A, III, Noll DC, Cohen JD. Selective deficits in prefrontal cortex function in medication-naive patients with schizophrenia. Arch Gen Psychiatry. 2001;58:280–288. doi: 10.1001/archpsyc.58.3.280. [DOI] [PubMed] [Google Scholar]
- Berman DM, Wilkie TM, Gilman AG. GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein alpha subunits. Cell. 1996;86:445–452. doi: 10.1016/s0092-8674(00)80117-8. [DOI] [PubMed] [Google Scholar]
- Beyer CE, Ghavami A, Lin Q, Sung A, Rhodes KJ, Dawson LA, Schechter LE, Young KH. Regulators of G-protein signaling 4: modulation of 5-HT(1A)-mediated neurotransmitter release in vivo. Brain Res. 2004;1022:214–220. doi: 10.1016/j.brainres.2004.06.073. [DOI] [PubMed] [Google Scholar]
- Brzustowicz LM, Hodgkinson KA, Chow EW, Honer WG, Bassett AS. Location of a major susceptibility locus for familial schizophrenia on chromosome 1q21–q22. Science. 2000;288:678–682. doi: 10.1126/science.288.5466.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchel C, Wise RJ, Mummery CJ, Poline JB, Friston KJ. Nonlinear regression in parametric activation studies. NeuroImage. 1996;4:60–66. doi: 10.1006/nimg.1996.0029. [DOI] [PubMed] [Google Scholar]
- Callicott JH, Weinberger DR. Functional brain imaging: future prospects for clinical practice. In: Weisman S, Sabshin M, Eist H, editors. Psychiatry in the new millenium. Washington, DC: American Psychiatric; 1999. pp. 119–135. [Google Scholar]
- Callicott JH, Mattay VS, Bertolino A, Finn K, Coppola R, Frank JA, Goldberg TE, Weinberger DR. Physiological characteristics of capacity constraints in working memory as revealed by functional MRI. Cereb Cortex. 1999;9:20–26. doi: 10.1093/cercor/9.1.20. [DOI] [PubMed] [Google Scholar]
- Callicott JH, Egan MF, Mattay VS, Bertolino A, Bone AD, Verchinksi B, Weinberger DR. Abnormal fMRI response of the dorsolateral prefrontal cortex in cognitively intact siblings of patients with schizophrenia. Am J Psychiatry. 2003a;160:709–719. doi: 10.1176/appi.ajp.160.4.709. [DOI] [PubMed] [Google Scholar]
- Callicott JH, Mattay VS, Verchinski BA, Marenco S, Egan MF, Weinberger DR. Complexity of prefrontal cortical dysfunction in schizophrenia: more than up or down. Am J Psychiatry. 2003b;160:2209–2215. doi: 10.1176/appi.ajp.160.12.2209. [DOI] [PubMed] [Google Scholar]
- Callicott JH, Straub RE, Pezawas L, Egan MF, Mattay VS, Hariri AR, Verchinski BA, Meyer-Lindenberg A, Balkissoon R, Kolachana B, Goldberg TE, Weinberger DR. Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc Natl Acad Sci USA. 2005;102:8627–8632. doi: 10.1073/pnas.0500515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon TD, Hennah W, van Erp TG, Thompson PM, Lonnqvist J, Huttunen M, Gasperoni T, Tuulio-Henriksson A, Pirkola T, Toga AW, Kaprio J, Mazziotta J, Peltonen L. Association of DISC1/TRAX haplotypes with schizophrenia, reduced prefrontal gray matter, and impaired short- and long-term memory. Arch Gen Psychiatry. 2005;62:1205–1213. doi: 10.1001/archpsyc.62.11.1205. [DOI] [PubMed] [Google Scholar]
- Chowdari KV, Mirnics K, Semwal P, Wood J, Lawrence E, Bhatia T, Deshpande SN, Thelma BK, Ferrell RE, Middleton FA, Devlin B, Levitt P, Lewis DA, Nimgaonkar VL. Association and linkage analyses of RGS4 polymorphisms in schizophrenia. Hum Mol Genet. 2002;11:1373–1380. doi: 10.1093/hmg/11.12.1373. [DOI] [PubMed] [Google Scholar]
- Conway AR, Kane MJ, Engle RW. Working memory capacity and its relation to general intelligence. Trends Cogn Sci. 2003;7:547–552. doi: 10.1016/j.tics.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Davis KL, Stewart DG, Friedman JI, Buchsbaum M, Harvey PD, Hof PR, Buxbaum J, Haroutunian V. White matter changes in schizophrenia: evidence for myelin-related dysfunction. Arch Gen Psychiatry. 2003;60:443–456. doi: 10.1001/archpsyc.60.5.443. [DOI] [PubMed] [Google Scholar]
- De Blasi A, Conn PJ, Pin J, Nicoletti F. Molecular determinants of metabotropic glutamate receptor signaling. Trends Pharmacol Sci. 2001;22:114–120. doi: 10.1016/s0165-6147(00)01635-7. [DOI] [PubMed] [Google Scholar]
- D'Esposito M, Aguirre GK, Zarahn E, Ballard D, Shin RK, Lease J. Functional MRI studies of spatial and nonspatial working memory. Brain Res Cogn Brain Res. 1998;7:1–13. doi: 10.1016/s0926-6410(98)00004-4. [DOI] [PubMed] [Google Scholar]
- Druey KM, Blumer KJ, Kang VH, Kehrl JH. Inhibition of G-protein-mediated MAP kinase activation by a new mammalian gene family. Nature. 1996;379:742–746. doi: 10.1038/379742a0. [DOI] [PubMed] [Google Scholar]
- Egan MF, Goldberg TE, Gscheidle T, Weirich M, Bigelow LB, Weinberger DR. Relative risk of attention deficits in siblings of patients with schizophrenia. Am J Psychiatry. 2000;157:1309–1316. doi: 10.1176/appi.ajp.157.8.1309. [DOI] [PubMed] [Google Scholar]
- Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE, Goldman D, Weinberger DR. Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci USA. 2001;98:6917–6922. doi: 10.1073/pnas.111134598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Straub RE, Goldberg TE, Yakub I, Callicott JH, Hariri AR, Mattay VS, Bertolino A, Hyde TM, Shannon-Weickert C, Akil M, Crook J, Vakkalanka RK, Balkissoon R, Gibbs RA, Kleinman JE, Weinberger DR. Variation in GRM3 affects cognition, prefrontal glutamate, and risk for schizophrenia. Proc Natl Acad Sci USA. 2004;101:12604–12609. doi: 10.1073/pnas.0405077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdely HA, Lahti RA, Lopez MB, Myers CS, Roberts RC, Tamminga CA, Vogel MW. Regional expression of RGS4 mRNA in human brain. Eur J Neurosci. 2004;19:3125–3128. doi: 10.1111/j.0953-816X.2004.03364.x. [DOI] [PubMed] [Google Scholar]
- Flynn SW, Lang DJ, Mackay AL, Goghari V, Vavasour IM, Whittall KP, Smith GN, Arango V, Mann JJ, Dwork AJ, Falkai P, Honer WG. Abnormalities of myelination in schizophrenia detected in vivo with MRI, and post-mortem with analysis of oligodendrocyte proteins. Mol Psychiatry. 2003;8:811–820. doi: 10.1038/sj.mp.4001337. [DOI] [PubMed] [Google Scholar]
- Foucher JR, Vidailhet P, Chanraud S, Gounot D, Grucker D, Pins D, Damsa C, Danion JM. Functional integration in schizophrenia: too little or too much? Preliminary results on fMRI data. NeuroImage. 2005;26:374–388. doi: 10.1016/j.neuroimage.2005.01.042. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Frith CD. Schizophrenia: a disconnection syndrome? Clin Neurosci. 1995;3:89–97. [PubMed] [Google Scholar]
- Geurts M, Hermans E, Maloteaux JM. Opposite modulation of regulators of G protein signalling-2 RGS2 and RGS4 expression by dopamine receptors in the rat striatum. Neurosci Lett. 2002;333:146–150. doi: 10.1016/s0304-3940(02)01004-2. [DOI] [PubMed] [Google Scholar]
- Geurts M, Maloteaux JM, Hermans E. Altered expression of regulators of G-protein signaling (RGS) mRNAs in the striatum of rats undergoing dopamine depletion. Biochem Pharmacol. 2003;66:1163–1170. doi: 10.1016/s0006-2952(03)00447-7. [DOI] [PubMed] [Google Scholar]
- Ghavami A, Hunt RA, Olsen MA, Zhang J, Smith DL, Kalgaonkar S, Rahman Z, Young KH. Differential effects of regulator of G protein signaling (RGS) proteins on serotonin 5-HT1A, 5-HT2A, and dopamine D2 receptor-mediated signaling and adenylyl cyclase activity. Cell Signal. 2004;16:711–721. doi: 10.1016/j.cellsig.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Gold SJ, Ni YG, Dohlman HG, Nestler EJ. Regulators of G-protein signaling (RGS) proteins: region-specific expression of nine subtypes in rat brain. J Neurosci. 1997;17:8024–8037. doi: 10.1523/JNEUROSCI.17-20-08024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman-Rakic PS. Circuitry of primate prefrontal cortex and regulation of behavior by representational memory. In: Plum F, editor. Higher functions of the brain. Bethesda, MD: American Physiological Society; 1987. pp. 373–418. [Google Scholar]
- Grillet N, Dubreuil V, Dufour HD, Brunet JF. Dynamic expression of RGS4 in the developing nervous system and regulation by the neural type-specific transcription factor Phox2b. J Neurosci. 2003;23:10613–10621. doi: 10.1523/JNEUROSCI.23-33-10613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurling HM, Critchley H, Datta SR, McQuillin A, Blaveri E, Thirumalai S, Pimm J, Krasucki R, Kalsi G, Quested D, Lawrence J, Bass N, Choudhury K, Puri V, O'Daly O, Curtis D, Blackwood D, Muir W, Malhotra AK, Buchanan RW, et al. Genetic association and brain morphology studies and the chromosome 8p22 pericentriolar material 1 (PCM1) gene in susceptibility to schizophrenia. Arch Gen Psychiatry. 2006;63:844–854. doi: 10.1001/archpsyc.63.8.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haier RJ, Jung RE, Yeo RA, Head K, Alkire MT. Structural brain variation, age, and response time. Cogn Affect Behav Neurosci. 2005;5:246–251. doi: 10.3758/cabn.5.2.246. [DOI] [PubMed] [Google Scholar]
- Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, Haroutunian V, Fienberg AA. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci USA. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri AR, Mattay VS, Tessitore A, Kolachana B, Fera F, Goldman D, Egan MF, Weinberger DR. Serotonin transporter genetic variation and the response of the human amygdala. Science. 2002;297:400–403. doi: 10.1126/science.1071829. [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry [Erratum (2005) 10:420] 2004;10:40–68. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- Hashimoto R, Straub RE, Weickert CS, Hyde TM, Kleinman JE, Weinberger DR. Expression analysis of neuregulin-1 in the dorsolateral prefrontal cortex in schizophrenia. Mol Psychiatry. 2004;9:299–307. doi: 10.1038/sj.mp.4001434. [DOI] [PubMed] [Google Scholar]
- Hepler JR, Berman DM, Gilman AG, Kozasa T. RGS4 and GAIP are GTPase-activating proteins for Gq alpha and block activation of phospholipase C beta by gamma-thio-GTP-Gq alpha. Proc Natl Acad Sci USA. 1997;94:428–432. doi: 10.1073/pnas.94.2.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho BC, Andreasen NC, Nopoulos P, Arndt S, Magnotta V, Flaum M. Progressive structural brain abnormalities and their relationship to clinical outcome: a longitudinal magnetic resonance imaging study early in schizophrenia. Arch Gen Psychiatry. 2003;60:585–594. doi: 10.1001/archpsyc.60.6.585. [DOI] [PubMed] [Google Scholar]
- Hof PR, Haroutunian V, Copland C, Davis KL, Buxbaum JD. Molecular and cellular evidence for an oligodendrocyte abnormality in schizophrenia. Neurochem Res. 2002;27:1193–1200. doi: 10.1023/a:1020981510759. [DOI] [PubMed] [Google Scholar]
- Hollinger S, Hepler JR. Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol Rev. 2002;54:527–559. doi: 10.1124/pr.54.3.527. [DOI] [PubMed] [Google Scholar]
- Honea R, Crow TJ, Passingham D, Mackay CE. Regional deficits in brain volume in schizophrenia: a meta-analysis of voxel-based morphometry studies. Am J Psychiatry. 2005;162:2233–2245. doi: 10.1176/appi.ajp.162.12.2233. [DOI] [PubMed] [Google Scholar]
- Huang C, Hepler JR, Gilman AG, Mumby SM. Attenuation of Gi- and Gq-mediated signaling by expression of RGS4 or GAIP in mammalian cells. Proc Natl Acad Sci USA. 1997;94:6159–6163. doi: 10.1073/pnas.94.12.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingi T, Aoki Y. Expression of RGS2, RGS4 and RGS7 in the developing postnatal brain. Eur J Neurosci. 2002;15:929–936. doi: 10.1046/j.1460-9568.2002.01925.x. [DOI] [PubMed] [Google Scholar]
- Kemmotsu N, Villalobos ME, Gaffrey MS, Courchesne E, Muller RA. Activity and functional connectivity of inferior frontal cortex associated with response conflict. Brain Res Cogn Brain Res. 2005;24:335–342. doi: 10.1016/j.cogbrainres.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Kubicki M, Park H, Westin CF, Nestor PG, Mulkern RV, Maier SE, Niznikiewicz M, Connor EE, Levitt JJ, Frumin M, Kikinis R, Jolesz FA, McCarley RW, Shenton ME. DTI and MTR abnormalities in schizophrenia: analysis of white matter integrity. NeuroImage. 2005;26:1109–1118. doi: 10.1016/j.neuroimage.2005.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larminie C, Murdock P, Walhin JP, Duckworth M, Blumer KJ, Scheideler MA, Garnier M. Selective expression of regulators of G-protein signaling (RGS) in the human central nervous system. Brain Res Mol Brain Res. 2004;122:24–34. doi: 10.1016/j.molbrainres.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Law AJ, Lipska BK, Weickert CS, Hyde TM, Straub RE, Hashimoto R, Harrison PJ, Kleinman JE, Weinberger DR. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5′ SNPs associated with the disease. Proc Natl Acad Sci USA. 2006;103:6747–6752. doi: 10.1073/pnas.0602002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenartowicz A, McIntosh AR. The role of anterior cingulate cortex in working memory is shaped by functional connectivity. J Cogn Neurosci. 2005;17:1026–1042. doi: 10.1162/0898929054475127. [DOI] [PubMed] [Google Scholar]
- Levy R, Goldman-Rakic PS. Segregation of working memory functions within the dorsolateral prefrontal cortex. Exp Brain Res. 2000;133:23–32. doi: 10.1007/s002210000397. [DOI] [PubMed] [Google Scholar]
- Lewis SJ, Dove A, Robbins TW, Barker RA, Owen AM. Striatal contributions to working memory: a functional magnetic resonance imaging study in humans. Eur J Neurosci. 2004;19:755–760. doi: 10.1111/j.1460-9568.2004.03108.x. [DOI] [PubMed] [Google Scholar]
- Liddle PF, Kiehl KA, Smith AM. Event-related fMRI study of response inhibition. Hum Brain Mapp. 2001;12:100–109. doi: 10.1002/1097-0193(200102)12:2<100::AID-HBM1007>3.0.CO;2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston C, Watts R, Tottenham N, Davidson MC, Niogi S, Ulug AM, Casey BJ. Frontostriatal microstructure modulates efficient recruitment of cognitive control. Cereb Cortex. 2005;16:553–560. doi: 10.1093/cercor/bhj003. [DOI] [PubMed] [Google Scholar]
- Livak KJ. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal. 1999;14:143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Poline JB, Kohn PD, Holt JL, Egan MF, Weinberger DR, Berman KF. Evidence for abnormal cortical functional connectivity during working memory in schizophrenia. Am J Psychiatry. 2001;158:1809–1817. doi: 10.1176/appi.ajp.158.11.1809. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Buckholtz JW, Kolachana B, Hariri AR, Pezawas L, Blasi G, Wabnitz A, Honea R, Verchinski B, Callicott JH, Egan M, Mattay V, Weinberger DR. From the cover: neural mechanisms of genetic risk for impulsivity and violence in humans. Proc Natl Acad Sci USA. 2006;103:6269–6274. doi: 10.1073/pnas.0511311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lindenberg AS, Olsen RK, Kohn PD, Brown T, Egan MF, Weinberger DR, Berman KF. Regionally specific disturbance of dorsolateral prefrontal-hippocampal functional connectivity in schizophrenia. Arch Gen Psychiatry. 2005;62:379–386. doi: 10.1001/archpsyc.62.4.379. [DOI] [PubMed] [Google Scholar]
- Miller EK, Li L, Desimone R. Activity of neurons in anterior inferior temporal cortex during a short-term memory task. J Neurosci. 1993;13:1460–1478. doi: 10.1523/JNEUROSCI.13-04-01460.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirnics K, Middleton FA, Stanwood GD, Lewis DA, Levitt P. Disease-specific changes in regulator of G-protein signaling 4 (RGS4) expression in schizophrenia. Mol Psychiatry. 2001;6:293–301. doi: 10.1038/sj.mp.4000866. [DOI] [PubMed] [Google Scholar]
- Mitelman SA, Buchsbaum MS, Brickman AM, Shihabuddin L. Cortical intercorrelations of frontal area volumes in schizophrenia. NeuroImage. 2005;27:753–770. doi: 10.1016/j.neuroimage.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Morita M, Nakahara K, Hayashi T. A rapid presentation event-related functional magnetic resonance imaging study of response inhibition in macaque monkeys. Neurosci Lett. 2004;356:203–206. doi: 10.1016/j.neulet.2003.11.066. [DOI] [PubMed] [Google Scholar]
- Nakahara K, Hayashi T, Konishi S, Miyashita Y. Functional MRI of macaque monkeys performing a cognitive set-shifting task. Science. 2002;295:1532–1536. doi: 10.1126/science.1067653. [DOI] [PubMed] [Google Scholar]
- Nomoto S, Adachi K, Yang LX, Hirata Y, Muraguchi S, Kiuchi K. Distribution of RGS4 mRNA in mouse brain shown by in situ hybridization. Biochem Biophys Res Commun. 1997;241:281–287. doi: 10.1006/bbrc.1997.7802. [DOI] [PubMed] [Google Scholar]
- Ohnishi T, Hashimoto R, Mori T, Nemoto K, Moriguchi Y, Iida H, Noguchi H, Nakabayashi T, Hori H, Ohmori M, Tsukue R, Anami K, Hirabayashi N, Harada S, Arima K, Saitoh O, Kunugi H. The association between the Val158Met polymorphism of the catechol-O-methyl transferase gene and morphological abnormalities of the brain in chronic schizophrenia. Brain. 2005;129:399–410. doi: 10.1093/brain/awh702. [DOI] [PubMed] [Google Scholar]
- Owen AM. The functional organization of working memory processes within human lateral frontal cortex: the contribution of functional neuroimaging. Eur J Neurosci. 1997;9:1329–1339. doi: 10.1111/j.1460-9568.1997.tb01487.x. [DOI] [PubMed] [Google Scholar]
- Owen AM. The role of the lateral frontal cortex in mnemonic processing: the contribution of functional neuroimaging. Exp Brain Res. 2000;133:33–43. doi: 10.1007/s002210000398. [DOI] [PubMed] [Google Scholar]
- Paillere-Martinot M, Caclin A, Artiges E, Poline JB, Joliot M, Mallet L, Recasens C, Attar-Levy D, Martinot JL. Cerebral gray and white matter reductions and clinical correlates in patients with early onset schizophrenia. Schizophr Res. 2001;50:19–26. doi: 10.1016/s0920-9964(00)00137-7. [DOI] [PubMed] [Google Scholar]
- Peled A, Geva AB, Kremen WS, Blankfeld HM, Esfandiarfard R, Nordahl TE. Functional connectivity and working memory in schizophrenia: an EEG study. Int J Neurosci. 2001;106:47–61. doi: 10.3109/00207450109149737. [DOI] [PubMed] [Google Scholar]
- Perlstein WM, Carter CS, Noll DC, Cohen JD. Relation of prefrontal cortex dysfunction to working memory and symptoms in schizophrenia. Am J Psychiatry. 2001;158:1105–1113. doi: 10.1176/appi.ajp.158.7.1105. [DOI] [PubMed] [Google Scholar]
- Petrides M. Dissociable roles of mid-dorsolateral prefrontal and anterior inferotemporal cortex in visual working memory. J Neurosci. 2000;20:7496–7503. doi: 10.1523/JNEUROSCI.20-19-07496.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezawas L, Meyer-Lindenberg A, Drabant EM, Verchinski BA, Munoz KE, Kolachana BS, Egan MF, Mattay VS, Hariri AR, Weinberger DR. 5-HTTLPR polymorphism impacts human cingulate-amygdala interactions: a genetic susceptibility mechanism for depression. Nat Neurosci. 2005;8:828–834. doi: 10.1038/nn1463. [DOI] [PubMed] [Google Scholar]
- Posthuma D, Baare WF, Hulshoff Pol HE, Kahn RS, Boomsma DI, De Geus EJ. Genetic correlations between brain volumes and the WAIS-III dimensions of verbal comprehension, working memory, perceptual organization, and processing speed. Twin Res. 2003;6:131–139. doi: 10.1375/136905203321536254. [DOI] [PubMed] [Google Scholar]
- Prasad KM, Chowdari KV, Nimgaonkar VL, Talkowski ME, Lewis DA, Keshavan MS. Genetic polymorphisms of the RGS4 and dorsolateral prefrontal cortex morphometry among first episode schizophrenia patients. Mol Psychiatry. 2004;10:213–219. doi: 10.1038/sj.mp.4001562. [DOI] [PubMed] [Google Scholar]
- Puri V, McQuillin A, Choudhury K, Datta S, Pimm J, Thirumalai S, Krasucki R, Lawrence J, Quested D, Bass N, Moorey H, Morgan J, Punukollu B, Kandasami G, Curtis D, Gurling H. Fine mapping by genetic association implicates the chromosome 1q23.3 gene UHMK1, encoding a serine/threonine protein kinase, as a novel schizophrenia susceptibility gene. Biol Psychiatry. 2007 doi: 10.1016/j.biopsych.2006.06.014. in press. [DOI] [PubMed] [Google Scholar]
- Rainer G, Ranganath C. Coding of objects in the prefrontal cortex in monkeys and humans. The Neuroscientist. 2002;8:6–11. doi: 10.1177/107385840200800104. [DOI] [PubMed] [Google Scholar]
- Ramnani N, Behrens TE, Penny W, Matthews PM. New approaches for exploring anatomical and functional connectivity in the human brain. Biol Psychiatry. 2004;56:613–619. doi: 10.1016/j.biopsych.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Ranganath C, DeGutis J, D'Esposito M. Category-specific modulation of inferior temporal activity during working memory encoding and maintenance. Brain Res Cogn Brain Res. 2004;20:37–45. doi: 10.1016/j.cogbrainres.2003.11.017. [DOI] [PubMed] [Google Scholar]
- Romanski LM. Domain specificity in the primate prefrontal cortex. Cogn Affect Behav Neurosci. 2004;4:421–429. doi: 10.3758/cabn.4.4.421. [DOI] [PubMed] [Google Scholar]
- Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- Rushworth MF, Nixon PD, Eacott MJ, Passingham RE. Ventral prefrontal cortex is not essential for working memory. J Neurosci. 1997;17:4829–4838. doi: 10.1523/JNEUROSCI.17-12-04829.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rypma B, D'Esposito M. The roles of prefrontal brain regions in components of working memory: effects of memory load and individual differences. Proc Natl Acad Sci USA. 1999;96:6558–6563. doi: 10.1073/pnas.96.11.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saugstad JA, Marino MJ, Folk JA, Hepler JR, Conn PJ. RGS4 inhibits signaling by group I metabotropic glutamate receptors. J Neurosci. 1998;18:905–913. doi: 10.1523/JNEUROSCI.18-03-00905.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafritz KM, Kartheiser P, Belger A. Dissociation of neural systems mediating shifts in behavioral response and cognitive set. NeuroImage. 2005;25:600–606. doi: 10.1016/j.neuroimage.2004.12.054. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, Brynjolfsson J, Gunnarsdottir S, Ivarsson O, Chou TT, Hjaltason O, Birgisdottir B, Jonsson H, Gudnadottir VG, Gudmundsdottir E, Bjornsson A, Ingvarsson B, Ingason A, Sigfusson S, Hardardottir H. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Nohara S, Hagino H, Kurokawa K, Yotsutsuji T, Kawasaki Y, Takahashi T, Matsui M, Watanabe N, Seto H, Kurachi M. Regional changes in brain gray and white matter in patients with schizophrenia demonstrated with voxel-based analysis of MRI. Schizophr Res. 2002;55:41–54. doi: 10.1016/s0920-9964(01)00224-9. [DOI] [PubMed] [Google Scholar]
- Talkowski ME, Seltman H, Bassett AS, Brzustowicz LM, Chen X, Chowdari KV, Collier DA, Cordeiro Q, Corvin AP, Deshpande SN, Egan MF, Gill M, Kendler KS, Kirov G, Heston LL, Levitt P, Lewis DA, Li T, Mirnics K, Morris DW. Evaluation of a susceptibility gene for schizophrenia: genotype based meta-analysis of RGS4 polymorphisms from thirteen independent samples. Biol Psychiatry. 2006;60:152–162. doi: 10.1016/j.biopsych.2006.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan HY, Sust S, Buckholtz JW, Mattay VS, Meyer-Lindenberg A, Egan MF, Weinberger DR, Callicott JH. Dysfunctional prefrontal regional specialization and compensation in schizophrenia. Am J Psychiatry. 2006;163:1969–1977. doi: 10.1176/ajp.2006.163.11.1969. [DOI] [PubMed] [Google Scholar]
- Taymans JM, Leysen JE, Langlois X. Striatal gene expression of RGS2 and RGS4 is specifically mediated by dopamine D1 and D2 receptors: clues for RGS2 and RGS4 functions. J Neurochem. 2003;84:1118–1127. doi: 10.1046/j.1471-4159.2003.01610.x. [DOI] [PubMed] [Google Scholar]
- Taymans JM, Kia HK, Claes R, Cruz C, Leysen J, Langlois X. Dopamine receptor-mediated regulation of RGS2 and RGS4 mRNA differentially depends on ascending dopamine projections and time. Eur J Neurosci. 2004;19:2249–2260. doi: 10.1111/j.0953-816X.2004.03336.x. [DOI] [PubMed] [Google Scholar]
- Tkachev D, Mimmack ML, Ryan MM, Wayland M, Freeman T, Jones PB, Starkey M, Webster MJ, Yolken RH, Bahn S. Oligodendrocyte dysfunction in schizophrenia and bipolar disorder. Lancet. 2003;362:798–805. doi: 10.1016/S0140-6736(03)14289-4. [DOI] [PubMed] [Google Scholar]
- Ungerleider LG, Mishkin M. Two cortical visual systems. In: Ingle DJ, editor. Analysis of visual behavior. Cambridge, MA: MIT; 1982. pp. 549–586. [Google Scholar]
- Veltman DJ, Rombouts SA, Dolan RJ. Maintenance versus manipulation in verbal working memory revisited: an fMRI study. NeuroImage. 2003;18:247–256. doi: 10.1016/s1053-8119(02)00049-6. [DOI] [PubMed] [Google Scholar]
- Wager TD, Smith EE. Neuroimaging studies of working memory: a meta-analysis. Cogn Affect Behav Neurosci. 2003;3:255–274. doi: 10.3758/cabn.3.4.255. [DOI] [PubMed] [Google Scholar]
- Watson N, Linder ME, Druey KM, Kehrl JH, Blumer KJ. RGS family members: GTPase-activating proteins for heterotrimeric G-protein alpha-subunits. Nature. 1996;383:172–175. doi: 10.1038/383172a0. [DOI] [PubMed] [Google Scholar]
- Winterer G, Coppola R, Egan MF, Goldberg TE, Weinberger DR. Functional and effective frontotemporal connectivity and genetic risk for schizophrenia. Biol Psychiatry. 2003;54:1181–1192. doi: 10.1016/s0006-3223(03)00532-8. [DOI] [PubMed] [Google Scholar]
- Wolkin A, Choi SJ, Szilagyi S, Sanfilipo M, Rotrosen JP, Lim KO. Inferior frontal white matter anisotropy and negative symptoms of schizophrenia: a diffusion tensor imaging study. Am J Psychiatry. 2003;160:572–574. doi: 10.1176/appi.ajp.160.3.572. [DOI] [PubMed] [Google Scholar]
- Yan Y, Chi PP, Bourne HR. RGS4 inhibits Gq-mediated activation of mitogen-activated protein kinase and phosphoinositide synthesis. J Biol Chem. 1997;272:11924–11927. doi: 10.1074/jbc.272.18.11924. [DOI] [PubMed] [Google Scholar]





