ABSTRACT
Marine microbes play essential roles in global energy and nutrient cycles. A primary method of determining their diversity and distribution is through sequencing of 16S ribosomal RNA genes from environmental samples. However, the perceived community composition may vary significantly based on differences in methodology, including choice of 16S variable region(s). This study investigated the influence of 16S variable region selection (V4-V5 or V6-V8) on perceived community composition and diversity for bacteria, Archaea and chloroplasts by tag-Illumina sequencing. We used 24 samples from the photic zone of the Scotian Shelf, northwest Atlantic, collected during a spring phytoplankton bloom. Taxonomic assignment and community composition varied greatly depending on the choice of variable regions while observed patterns of beta diversity were reproducible between variable regions. V4-V5 was considered the preferred variable region for future studies based on its superior recognition of Archaea, which has received little attention in bloom dynamics. The V6-V8 region captured more of the bacterial diversity, including the abundant SAR11 clades and, to a lesser extent, that of chloroplasts. However, the magnitude of difference between variable regions for bacteria and chloroplast was less than for Archaea.
Keywords: 16S rRNA, hypervariable region, marine bacterioplankton, phytoplankton, Archaea, SAR11
Choice of 16S rRNA variable region influences perceived diversity of marine bacteria, Archaea and chloroplasts, with greater impacts on alpha than beta diversity.
INTRODUCTION
Marine microbes play essential roles in global biogeochemical systems including nutrient cycling, photosynthesis and the biological pump. These contributions, coupled with their capacity for rapid growth and adaptation (Arrigo 2005), underscore the importance of increasing baseline knowledge and predictive abilities for microbial communities. Most marine microbes still lack environmentally representative cultured isolates (Epstein 2013; Rinke et al. 2013); thus, culture-independent methods are key to determining diversity, distribution and ecological roles of microbes. However, variations in the methodologies used to extract and sequence DNA from field samples as well as classifying the resulting sequence reads significantly influence the perceived community composition and subsequent conclusions on microbial diversity and ecology (Sergeant et al. 2012; Hazen, Rocha and Techtmann 2013; Cruaud et al. 2014).
Ribosomal RNA (rRNA) sequences, either 16S rRNA for Archaea, bacteria and chloroplasts or 18S rRNA for eukaryotes, are the routinely accepted standards for identifying the members of mixed microbial communities and defining the overall microbial diversity in the environment. Next-generation sequencing has opened up approaches to analyse microbial community in great details (Thompson et al. 2017). Although entire 16S rRNA gene sequences would provide the best resolution for taxonomic characterization, technological limitations on the length of sequence reads currently limit this approach to sections of the gene. Universal primers that bind to interspersed conserved regions, target-specific variable regions within the SSU rRNA genes that provide phylogenetic information on microbial taxa. The choice of ‘optimal’ variable region(s) is an ongoing debate that depends both on the sequencing technology at hand (Cruaud et al. 2014; Barb et al. 2016), the research question (Choi et al. 2017), the targeted microbial taxa (Parada, Needham and Fuhrman 2016), and the ability to compare results obtained for microbial communities collected from diverse environments by multiple investigators (Thompson et al. 2017). The level of conservation within a variable region differs between phylogenetic groups (Schmalenberger, Schwieger and Tebbe 2001; Kim, Morrison and Yu 2011). Therefore, selection of variable region(s) and the primers to amplify them may cause biases resulting in under- or overrepresentation of various taxa and subsequent inferences of community diversity and richness (Yu and Morrison 2004; Parada, Needham and Fuhrman 2016). Primer sets for both the V3-V4 and V6-V8 hypervariable regions have been used previously for studies of Arctic bacterioplankton (Comeau et al., 2011, 2012; Fadeev et al. 2018) as have V1-V2 for classifying phytoplankton from chloroplast SSU rRNA (Choi et al. 2017) and microbial communities of waste water treatment (Guo et al. 2013). The recent global ocean microbiome survey carried out in the Tara expedition used the V9 variable region in addition to extensive metagenome sequencing (Sunagawa et al. 2015). Recent improvement in the design of universal primers for the amplification of the V4-V5 region resulted in the ability to target both Archaea and bacteria within one amplicon (Guo et al. 2013; Barb et al. 2016; Parada, Needham and Fuhrman 2016; Yang, Wang and Qian 2016) supporting the acceptance of the V4-V5 region as the current standard for characterizing microbial communities from diverse environments. The improved primers more accurately represented the proportional abundance of SAR11 (Pelagibacterales) (Parada, Needham and Fuhrman 2016), typically a dominant member of marine bacterial communities (Morris et al. 2002; Giovannoni 2017).
Comparison and evaluation of variable regions have been conducted in silico, with mock communities, and with field samples (Klindworth et al. 2013; Barb et al. 2016; Parada, Needham and Fuhrman 2016; Yang, Wang and Qian 2016). The first two approaches assess the accurate assignment of sequence reads from different variable regions to known full length reference sequences, while the latter assess the congruence of results obtained from a microbial community of unknown composition. Comparison of variable regions to known full length 16S rRNA gene sequences is a first step in assessing the applicability of primer sets to diverse microbial taxonomic groups. However, analysis of field samples of unknown microbial community composition collected from diverse regional environments is an important follow up in uncovering otherwise undetected differences, or differences of greater magnitude (Parada, Needham and Fuhrman 2016).This is especially important for regions that have been undersampled with respect to microbial diversity because there are few isolated strains that can be used to build a representative mock microbial community (Zorz et al. 2019).
Blooms on the Scotian Shelf occur annually in spring and, to a lesser extent, fall due to changes in the stratification of the water column providing favourable conditions for phytoplankton in terms of access to light and nutrients (Therriault et al. 1998). Environmental microbial plankton samples from the seasonal phytoplankton bloom on the Scotian Shelf, northwest Atlantic off eastern Canada, were used to assess the similarities and differences in the characterization of the microbial community from 16S rRNA gene metabarcoding by comparing results obtained from the V4-V5 (Parada, Needham and Fuhrman 2016) and V6-V8 (Comeau, Douglas and Langille 2017) variable regions. We evaluated the resulting perceived communities to assess the differences and similarities incurred by the choice of either V4-V5 or V6-V8 variable regions on the perceived spring bloom microbial community composition of coastal North Atlantic Ocean. To these ends, amplicons of both V4-V5 and V6-V8 variable regions from 24 samples collected during the spring bloom in 2016 were sequenced and the taxonomic assignments of bacterial, Archaeal and chloroplastic 16S rRNA from both variable regions were compared.
METHODS
Sample collection
Microbial plankton samples were collected on the spring 2016 AZMP mission (HUD2016003 April 9–25) aboard the CCGS Hudson along three stations from the Halifax Line (HL, Figure S1) with samples from four depths each: HL2 (44.2664, −63.3169; 1 m, 20 m, 40 m, 80 m), HL4 (43.4813, −62.4541; 1 m, 20 m, 40 m, 60 m) and HL6 (42.8321, −61.7324; 1 m, 20 m, 50 m, 80 m). At each station water samples were collected via 12 L Niskin bottles on a CTD rosette. From each depth, 4 L of water was collected and strained (333 μm) to remove copepods and other large zooplankton. The water was then sequentially filtered through 3 μm (large/‘L’) and 0.2 μm (small/‘S’) Isopore filters (Milipore, USA) by peristaltic pump. After filtration, filters were immediately stored at −80°C.
Sample processing
DNA extraction
DNA was extracted using Qiagen's DNeasy Plant Kit (Germantown, Maryland, USA) with some modifications to manufacturer's instructions. 50 μL of lysozyme (5 mg/mL, Fisher BioReagents, Loughborough, Leicestershire, UK) was initially added to each filter sample, after which the sample was vortexed on high for 30 seconds. 400 μL of lysis buffer AP1 (from the DNeasy kit) was added to each sample followed by 45 μL of proteinase K (20 mg/mL, Fisher BioReagents, Loughborough, Leicestershire, UK). The samples were then incubated at 55°C with shaking for 1 hour. Following incubation, 4 μL of RNase A (from the DNeasy kit) were added to the samples, which were then kept on ice for 10 minutes. From here, the extraction followed manufacturer's instructions. A NanoDrop 2000 (Thermo Scientific, USA) was used to confirm DNA concentrations and purity.
Illumina miseq sequencing
Sequencing by Illimina MiSeq followed the Microbiome Amplicon Sequencing Workflow (Comeau, Douglas and Langille 2017). Samples were amplified using dual-indexing Illumina fusion primers that targeted either the 412 bp V4-V5 region of the 16S rRNA gene (515F-Y 5′-GTGYCAGCMGCCGCGGTAA and 926R 5′-CCGYCAATTYMTTTRAGTTT) (Parada, Needham and Fuhrman 2016) or the 438 bp V6-V8 region (B969F 5’-ACGCGHNRAACCTTACC and BA1406R 5’-ACGGGCRGTGWGTRCAA) (Comeau, Douglas and Langille 2017). Raw sequence files are available at the NCBI Sequence Read Archive under accession PRJNA506220.
16S rRNA sequence classification
Processing of 16S samples was conducted using QIIME1 version 1.8.0 (Caporaso et al. 2010b) following the Langille lab's workflow (Comeau, Douglas and Langille 2017, also at https://github.com/LangilleLab/microbiome_helper/wiki/16S-Bacteria-and-Archaea-Standard-Operating-Procedure). Demultiplexed, paired-end sequences were merged by PEAR version 0.9.6 (Zhang et al. 2014). Sequences less than 400 bp in length or with a quality less than 30 over 90% of bases were discarded. Chimeric sequences were removed with VSEARCH (Rognes et al. 2016). Operational Taxonomic Units (OTUs) were picked based on 97% similarity using sortmerna (Kopylova, Noe and Touzet 2012) for reference picking and sumaclust (Kopylova et al. 2016) for de novo picking. The Greengenes database version 13.8, which provides chimera-checked full-length 16S rRNA sequences (McDonald et al. 2012), was used for reference. Singletons and low-confidence OTUs that were likely due to MiSeq bleed through between runs were removed. For analysis of bacteria samples, chloroplast, mitochondrial and Archaeal sequences were removed using the function filter_taxa_from_otu_table.py. Chloroplast sequences were classified using the PhytoRef database (Decelle et al. 2015).
Rarefaction of samples were used to allow for meaningful standardization and comparison of samples (Gotelli and Colwell 2001). Bacteria sequencing depth was selected as 3500 reads to include all 24 samples. Chloroplast sequences were subset to exclude depths >40 m due to low sequence abundance at depth and rarefied to a sequencing depth of 145 reads. Few Archaea were recognised by the bacterial-specific V6-V8 primer sets and thus rarefaction was not conducted as the two variable regions could not reasonably be compared in detail.
Data analysis
All data analysis was conducted in R version 3.5.1 (R Core Team 2018) using packages as indicated. Figures were created with ggplot2 (Wickham 2009). All null hypothesis significance tests used α = 0.05. Shannon diversity was calculated using the function diversity from the package vegan (Oksanen et al. 2017).
Bacteria and chloroplast sequences from the differing variable regions were compared by proportional bar plots of taxa abundance, permutational multivariate analysis of variance (PERMAMOVA), non-metric multidimensional scaling (nMDS) and linear discriminant function analysis (DFA). These were done at the class level. PERMANOVA used the function adonis from the package vegan (Oksanen et al. ) with 1000 permutations. nMDS used the function metaMDS, also from vegan. DFA used the function lda from the package MASS (Venables and Ripley 2002). For PERMANOVA, nMDS and DFA, only taxa shared between variable regions were used. The proportional abundances of included taxa were Hellinger transformed prior to analyses as recommended for abundance data (Legendre and Gallagher 2001). nMDS and PERMANOVA used the Bray–Curtis dissimilarity on the Hellinger transformed data. Leave-one-out cross validation was conducted for both DFAs to investigate the separation of the groups and predictive abilities of the DFAs.
Numerically dominant Bacteria OTUs were investigated by category determined by variable region, size fraction, station (with HL2 and HL4 combined due to their geographic proximity and similar community composition) and depth (shallow or deep). Depth was categorised based on whether samples were above or below the mixed layer depth, which was determined using the minimum depth at which the density gradient was ≥0.01 kg/m4 and verified by visual observation (Johnson et al. 2014).
Sequences for Pelagibacteraceae OTUs 1) shared between variable regions and/or 2) the top 10 numerically dominant OTUs from each variable region were extracted. Reference 16S rRNA sequences from 21 SAR11 isolates were obtained from the Integrated Microbial Genome site of the Joint Genome institute and were used in the construction of a phylogenetic tree. OTUs and trimmed reference sequences were then aligned in Mega X (Kumar et al. 2018) using Muscle. A phylogenetic tree for each variable region was constructed based on Maximum Likelihood in Mega X with the Kimura 2-parameter model (Kimura 1980) using a gamma distribution with invariant sites, which was selected based on output of the model selection tool in Mega X by minimum Bayesian and Akaike (corrected) Information Criteria. 500 bootstraps were used to test the robustness of the models. Clades of reference sequences which consistently clustered together were trimmed to declutter the trees. Tree graphics were done in Interactive Tree of Life version 4.2.3 (Letunic and Bork 2016). Single nucleotide variants (SNVs) for the V4-V5 and V6-V8 hypervariable regions were calculated in Geneious (version 11.1.5, https://www.geneious.com), with the following parameter settings: minimum coverage of 1; minimum variant frequency of 0.1; maximum variant P-value of 10–6 (0.0001% to see variant by chance).
RESULTS
Bacteria
The number of bacterial taxa recognised and proportions of the taxa in samples differed between 16S variable regions. Significantly fewer bacterial taxa were recognised by V4-V5 than V6-V8 (paired Wilcoxon rank sum test: V = 0, P = 0.031; Figure S2, Supporting Information), thus resulting in higher ecological richness in the latter. At the level of OTU, V6-V8 recognised 2321 OTUs, of which 680 were singletons (i.e. occurred only once across all samples). In contrast, V4-V5 recognised fewer OTUs (894) but did not have any singletons.
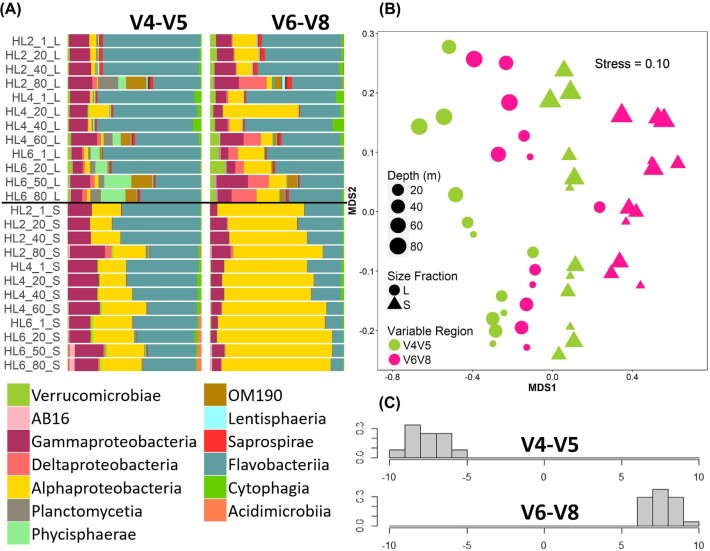
The proportional abundance of classes across the 24 samples differed significantly between variable regions (Fig. 1a) although on average, this was due to only a few classes. Examining the dominant classes, V4-V5 proportions of Flavobacteria, OM190, Phycisphaerae, and Gammaproteobacteria were greater than those in V6-V8 and the proportions of Alphaproteobacteria and Deltaproteobacteria were lower in V4-V5 than in V6-V8 (Table S1, Supporting Information). The classes that were recognised by V6-V8 and not V4-V5 (Table S1, Supporting Information) each represented at most 0.08% of the community and therefore contributed little to the proportional differences in the dominant classes. However, the Shannon diversity index was similar between variable regions (V4-V5: 4.51, V6-V8: 4.38). Of the shared classes, the proportional abundances were significantly different (PERMANOVA: F1,46 = 10.6, P < 0.001). Variable region choice thus influenced perceived community composition but had less influence on the measured Shannon diversity index within a community.
Figure 1.
Comparison of marine bacteria communities sequenced by 16S rRNA V4-V5 or V6-V8 (n = 24). (A) Proportional community class composition with partial legend of dominant classes. See Table S1 (Supporting Information) for full legend. Sample IDs are station (HL#), depth (m), size fraction (L or S). (B) Non-metric multidimensional scaling analysis of proportional Hellinger transformed class abundances using the Bray–Curtis dissimilarity matrix. (C) Linear discriminant function analysis of proportional Hellinger transformed class abundance. X-axis indicates standardised discriminant score of each sample, y-axis within-group score frequency.
The nMDS of bacteria taxa indicated clear distinction between variable regions as expected from the differences in community composition (Fig. 1b). Separation of the size fractions into two nMDSs (Figure S3) confirmed that within each size fraction the two variable regions formed distinct clusters, indicating dissimilarity. However, separation of the variable regions into two nMDSs (Figure S4) showed similar trends in beta diversity between variable regions. Variable region choice may thus not have greatly influenced perceived beta diversity in our samples.
DFA of variable regions on proportional class abundance of bacteria resulted in clear separation of variable region DFA scores (Fig. 1c). Leave-one-out cross validation was able to predict group (i.e. variable region) membership of each sample with 100% accuracy. The classes with the greatest weight in the DFA were Epsilonproteobacteria and Betaproteobacteria (Table S2, Supporting Information). The classes with the greatest weighting in the DFA were generally those with low proportional abundance. Overall, the DFA supported the results of differences in perceived community composition, as proportional abundance of classes differed enough to distinguish clearly between variable regions through DFA.
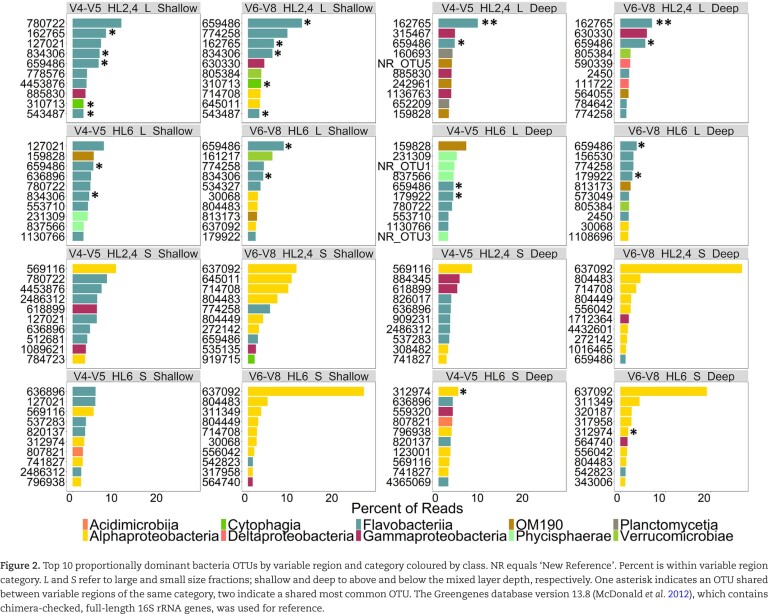
The 10 dominant OTUs by category differed both with respect to the specific OTUs and their class, especially in the small size fraction (Fig. 2). In the large size fraction, all variable region comparisons by category shared at least 2/10 OTUs and the dominant OTU belonged to the same class in all but one sample (HL6 S Shallow). In the small size fraction, the V6-V8 dominant OTUs were almost entirely Alphaproteobacteria, while the V4-V5 OTUs were more diverse. The OTU with the greatest within-category % abundance was OTU637092, belonging to the Pelagibacteraceae family (SAR11 class 1a.1), which was the dominant OTU in the V6-V8 small size fractions. However, this specific OTU was not recognised in the V4-V5 reads, possibly because of a more distributed assignment to several OTUs classified as Pelagibacteraceae.
Figure 2.
Top 10 proportionally dominant bacteria OTUs by variable region and category coloured by class. NR equals ‘New Reference’. Percent is within variable region category. L and S refer to large and small size fractions; shallow and deep to above and below the mixed layer depth, respectively. One asterisk indicates an OTU shared between variable regions of the same category, two indicate a shared most common OTU. The Greengenes database version 13.8 (McDonald et al. 2012), which contains chimera-checked, full-length 16S rRNA genes, was used for reference.
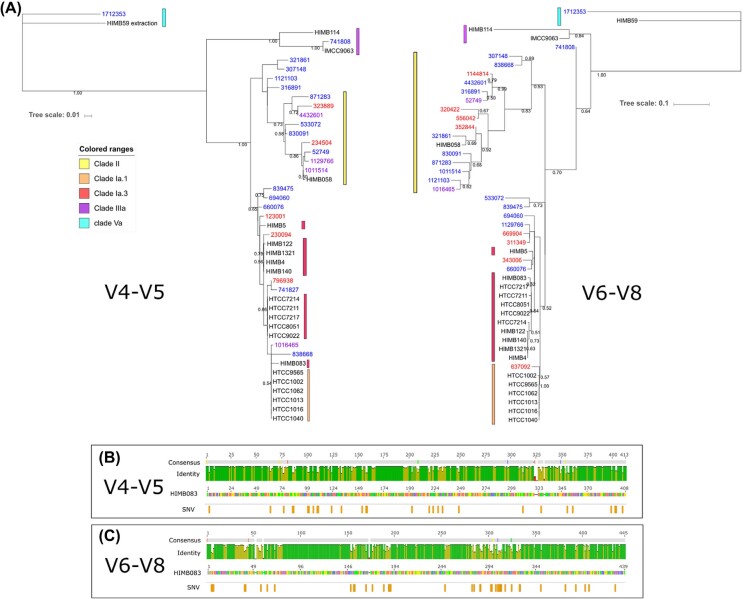
Overall, OTUs classified as Pelagibacteraceae differed between variable regions, as previously reported by Parada, Needham and Fuhrman (2016). The V4-V5 recognised 43 OTUs assigned to Pelagibacteraceae while V6-V8 recognised 413 (176 singletons), with only 18 shared Pelagibacteracea OTUs between variable regions. The proportional abundance of Pelagibacteraceae between variable regions varied greatly, with a maximum of 17% of the community by V4-V5 and 69% by V6-V8 in specific samples. To further explore the origin of this difference in OTU assignment, we constructed a phylogenetic tree with the sequences from each of the variable regions that included reference sequences from known ecotypes of Pelagibacteraceae (Fig. 3a). The longer branch length and higher bootstrap values observed in the tree generated from the V6-V8-assigned OTUs suggest that this variable region has a higher taxonomic resolution for the Pelagibacteraceae than the V4-V5 variable region. This result is corroborated by the higher number of SNVs (Kearse et al. 2012) observed in the V6-V8 (40 SNVs) variable region relative to the V4-V5 region (29 SNVs) (Fig. 3b and c).
Figure 3.
(A) Phylogenetic trees for Pelagibacteraceae by 16S variable region with 21 reference genomes representing the major clades of SAR11. Sequences used were Pelagibacteraceae OTUs shared between variable region (blue), the top 10 numerically dominant Pelagibacteraceae OTUs from each variable region (red) and reference sequences (black). Pelagibacteraceae OTUs that were both shared and top 10 are purple. Evolutionary histories were inferred using the Maximum Likelihood method based on the Kimura 2-parameter model (Kimura 1980) using a discrete Gamma distribution with invariant sites. 500 bootstraps were used; fractions on the branches indicate bootstrap values. Trees are drawn to scale, with branch lengths measured in the number of substitutions per site. The panels below the trees indicate the % identity (green = 100% while yellow is <100%) and the SNVs of 16S rRNA genes from 21 isolated SAR11 strains and OTUs used in the phylogenetic trees generated from the V4-V5 (B) and the V6-V8 (C) hypervariable regions compared to the reference strain HIMB083 that is widely distributed in the ocean (Delmont et al. 2017).
Chloroplasts
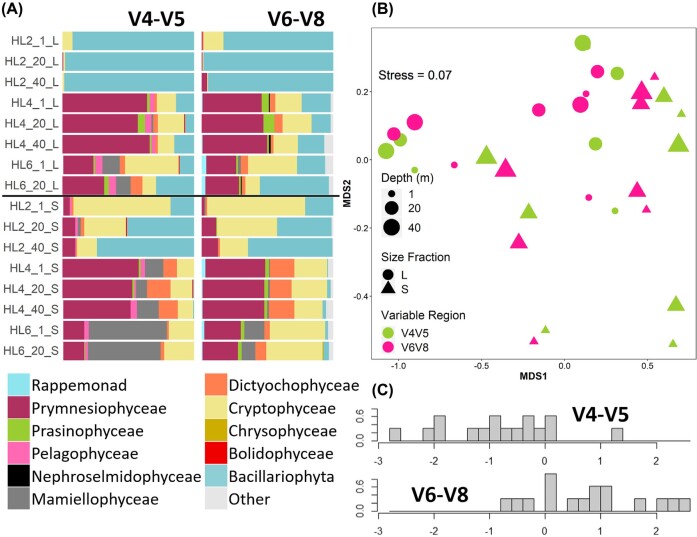
The number of chloroplast orders recognised and their proportional abundance also differed between variable regions, though to a lesser extent than bacteria (Fig. 4a; Table S3, Supporting Information). The dominant classes were Pymnesiophyceae, Mamiellophyceae, Cryptophyceae and Bacillariophyta, which were all recognised by both variable regions. Of the less abundant classes, Bolidophyceae and Pelagophyceae were recognised only by V4-V5, while Chrysophyceae and Nephroselmidophyceae were recognised only by V6-V8, as was an ‘other’ category of unrecognised sequences. As for bacteria, these excluded taxa represented small proportions of the average community across the 16 shallow samples (at most 2.63%). The classes’ proportional abundances across the samples were similar for HL2 and HL4 but differed for HL6, with relatively greater proportions of Mamiellophyceae using V4-V5 compared to V6-V8. The Shannon diversity index was similar between variable regions (V4-V5: 3.80, V6-V8: 3.91). Of the shared orders, the proportional abundances were not significantly different across all samples (PERMANOVA: F1,30 = 0.64, P = 0.501). Variable region choice thus influenced perceived community composition, primarily via recognition of rare taxa. However, the effect was less pronounced than for bacteria.
Figure 4.
Comparison of marine chloroplast communities sequenced by 16S rRNA V4-V5 or V6-V8 hypervariable regions (n = 16). (A) Proportional community class composition. See Table S3 (Supporting Information) for full legend. Sample IDs are station (HL#), depth (m), size fraction (L or S). (B) Non-metric multidimensional scaling analysis of proportional Hellinger transformed class abundances using the Bray–Curtis dissimilarity matrix. (C) Linear discriminant function analysis of proportional Hellinger transformed class abundance. X-axis indicates standardised discriminant score of each sample, y-axis within-group score frequency.
The nMDS of chloroplasts indicated little effect of variable region on beta diversity, with no clear clustering by variable region (Fig. 4b). The same result was found after separation of the size fractions into two nMDSs (Figure S5). Separation of the variable regions into two nMDSs (Figure S6) showed similar trends in beta diversity between variable regions. DFA of variable regions on proportional class abundance of chloroplasts resulted in little separation of variable region DFA scores (Fig. 4c). Leave-one-out cross validation was able to correctly predict group membership of V4-V5 samples with 67% accuracy, and of V6-V8 samples with 83% accuracy. Rappemonad had the greatest weight in the DFA followed by Prasinophyceae (Table S4, Supporting Information). The class with the least weight was Bacillariophyta. The DFA supported the results of differences in community composition, as proportional abundance of orders differed moderately as identified by the partial overlap of DFA scores. Variable region choice in this case may thus have limited influence on the perceived beta diversity.
Archaea
The V4-V5 variable region has the ability to recover sequences from Archaea, while the primers used for the V6-V8 region are specific to bacteria. This was demonstrated by the number of archaeal sequences per sample recognised by the two variable regions (Figure S7, Supporting Information), as V6-V8 found 0 sequences in 14/24 samples (mean ± SD: 3 ± 7 sequences) while V4-V5 recognised Archaea in 21/24 samples (mean ± SD: 600 ± 866 sequences). This low recognition by V6-V8 precluded further analyses as conducted with the bacteria and chloroplasts 16S. A difference between variable regions was also observed in the number of classified taxa, where V4-V5 identified more taxa at all taxonomic levels except genus (Table 1). At the phylum level, V6-V8 recognised only Crenarchaeota, while V4-V5 additionally recognised Euryarchaeota.
Table 1.
Comparison of number of classified Archaea taxa obtained by sequencing 24 samples from spring 2016 with different 16S rRNA variable regions (V4-V5 or V6-V8).
| V4-V5 | V6-V8 | |
|---|---|---|
| Phylum | 2 | 1 |
| Class | 2 | 1 |
| Order | 2 | 1 |
| Family | 3 | 1 |
| Genus | 1 | 2 |
| Distinct OTUs | 26 | 4 |
DISCUSSION
Perceived diversity of the Scotian Shelf microbial community was affected by the choice of variable region. An increased coverage of rare bacterial taxa was observed in V6-V8 and the dominant OTUs differed between the V4-V5 and V6-V8 regions. The case study of Pelagibacteraceae highlighted the differences in taxonomic assignment and proportional representation between the V4-V5 and V6-V8 variable regions. The relative abundance of Pelagibacteraceae as determined from the V6-V8 region was higher than from the V4-V5 in general, a trend that was accentuated in the deep water samples. The difference in the assignment of OTUs taxonomically related to SAR11 may be partially due to the size of the amplicons (412 nt in V4-V5 compared to 438 nt for the V6-V8 regions). In addition, we demonstrated that within the strains recovered from our study region, V6-V8 amplicon had 30% as many SNV than the V4-V5 amplicon, leading to an improved resolution for taxonomic assignment, as seen in the comparison of phylogenetic trees for the V4-V5 and V6-V8 variable regions. Notably, the V6-V8 reads identified a member of the SAR11 clade 1a.1, commonly found at high latitude, as the most abundant OTU in the deep water samples. In contrast, warm water SAR11 strains of the same clade (1a.3) were rare in the spring (Morris et al. 2002; Giovannoni 2017). This supports the view that members of the SAR11 clade 1a.1 are important members of the spring bacterial community on the Scotian Shelf (Zorz et al. 2019), while warm water members of the same clade (1a.3) were previously found in the fall months only. The fact that the V6-V8 variable region may be more effective at capturing the taxonomic diversity of the SAR11 clade than the V4V5 variable region is important given the dominance and versatility of this clade in the marine ecosystem (Delmont et al. 2017).
The targeting of important Archaeal clades by the V4-V5 primers is an advantage relative to the V6-V8 primers used here, which are specific for bacteria. Archaea have been largely ignored in bloom dynamics (Needham and Fuhrman 2016), but were detected in the V4-V5 sequence reads of almost all samples across all depth of the HL transect. Conversely, V6-V8 sequence reads detected very few Archaea sequences and did not detect the phylum Euryarchaeota. This lack of recognition was expected following previous studies (Comeau et al. 2012) and targeting of Archaeal diversity using the V6-V8 variable region requires a specific primer set that was not used here. The importance of recognition and analysis of Archaea in bloom dynamics is underscored by the high focus on bacteria and low focus on Archaea in the literature (Needham and Fuhrman 2016).
Increased analysis of perceived diversity of rare taxa of bacteria and Archaea would be beneficial to further elucidate the influence of variable region choice. This suggestion was supported by the DFA of bacteria between variable regions, as the taxa with the greatest weightings (i.e. those that differed the most) were generally low in proportional abundance. Rare or conditionally rare taxa disproportionally contribute to community dynamics (Shade et al. 2014). Given the observed superior recognition of rare bacteria classes by V6-V8, interest in rare vs dominant taxa may have a strong influence on variable region selection.
The influence of variable region choice on the community composition and beta diversity of Bacteria is consistent with results of other studies across multiple environments, as is the difference in recognition of Archaea (Cruaud et al. 2014; Barb et al. 2016; Yang, Wang and Qian 2016). These differences indicate low to moderate comparability of bacteria datasets sequenced by the different variable regions in this study. Chloroplast sequences, however, had relatively high comparability between the variable regions.
The comparison of two of the widely used variable regions in microbial community studies presented here furthers the discussion on the optimal variable region choice for marine microbes through the use of field samples compared to mock communities or in silico (Barb et al. 2016; Yang, Wang and Qian 2016). Additionally, it looked at chloroplasts 16S rRNA, while previous studies have focused on Bacteria and Archaea. Variable region comparison with field samples is important (Parada, Needham and Fuhrman 2016), as it focuses on the community of interest and the choice of variable region will depend on the study question. For example, studies on the optimal variable region in wastewater (Guo et al. 2013) may not translate well to the pelagic ocean. Our results showed that beta diversity measures (nMDS and DFA) were less affected by the choice of variable region than the alpha diversity measures (taxa recognition and community composition). This suggests that the influence of variable region choice may be minimal for studies focused on assessing the similarity of communities across an environmental gradient but will have a greater impact on studies aiming to characterise specific taxa. Given the continuous and rapid advances in sequencing technology, the selection of variable regions may become less important in the future, once full length 16S rRNA genes can be easily sequenced. However, assessing the performance of the commonly used variable regions contributes to our current and future predictive ability of the community, especially in the context of comparison to past studies.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the captain and crew of the CCGS Hudson as well as the AZMP scientific team, especially Maritimes Operational Lead Andrew Cogswell. The authors also thank Jennifer Tolman, Erin Bertrand, Brent Robicheau and Ian Luddington for useful discussion and logistical support relating to the development of this work.
FUNDING
This work was supported by the Natural Sciences and Engineering Research Council of Canada Discovery Grant program to JL; the Natural Sciences and Engineering Research Council of Canada Undergraduate Student Research Awards to CW; and the Ocean Frontier Institute.
Conflicts of Interests
None declared.
REFERENCES
- Arrigo KR. Marine microorgansisms and global nutrient cycles. Nature. 2005;437:349–55. [DOI] [PubMed] [Google Scholar]
- Barb JJ, Oler AJ, Kim HSet al. . Development of an analysis pipeline characterizing multiple hypervariable regions of 16S rRNA using mock samples. PLoS One. 2016;11:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh Jet al. . QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi CJ, Bachy C, Jaeger GSet al. . Newly discovered deep-branching marine plastid lineages are numerically rare but globally distributed. Curr Biol. 2017;27:R15–6. [DOI] [PubMed] [Google Scholar]
- Comeau AM, Douglas GM, Langille MGI. Microbiome helper: a custom and streamlined workflow for microbiome research. mSystems. 2017;2:e00127–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau AM, Harding T, Galand PEet al. . Vertical distribution of microbial communities in a perennially stratified Arctic lake with saline, anoxic bottom waters. Sci Rep. 2012;2:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau AM, Li WKW, Tremblay JÉet al. . Arctic ocean microbial community structure before and after the 2007 record sea ice minimum. PLoS One. 2011;6:e27492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruaud P, Vigneron A, Lucchetti-Miganeh Cet al. . Influence of DNA extraction method, 16S rRNA targeted hypervariable regions, and sample origin on microbial diversity detected by 454 pyrosequencing in marine chemosynthetic ecosystems. Appl Environ Microbiol. 2014;80:4626–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decelle J, Romac S, Stern RFet al. . PhytoREF: a reference database of the plastidial 16S rRNA gene of photosynthetic eukaryotes with curated taxonomy. Mol Ecol Resour. 2015;15:1435–45. [DOI] [PubMed] [Google Scholar]
- Delmont TO, Kiefl E, Kilinc Oet al. . The global biogeography of amino acid variants within a single SAR11 population is governed by natural selection. bioRxiv. 2017. https://doi.org/10.1101/170639 [Google Scholar]
- Epstein SS. The phenomenon of microbial uncultivability. Curr Opin Microbiol. 2013;16:636–42. [DOI] [PubMed] [Google Scholar]
- Fadeev E, Salter I, Schourup-kristensen Vet al. . Microbial communities in the east and west fram strait during sea ice melting season. Front Mar Sci. 2018;5:1–21.29552559 [Google Scholar]
- Giovannoni SJ. SAR11 bacteria: the most abundant plankton in the oceans. Ann Rev Mar Sci. 2017;9:231–55. [DOI] [PubMed] [Google Scholar]
- Gotelli NJ, Colwell RK. Quantifying biodiversity: procedures and pitfalls in the measurement and comparison of species richness. Ecol Lett. 2001;4:379–91. [Google Scholar]
- Guo F, Ju F, Cai Let al. . Taxonomic precision of different hypervariable regions of 16S rRNA gene and annotation methods for functional bacterial groups in biological wastewater treatment. PLoS One. 2013;8:e76185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen TC, Rocha AM, Techtmann SM. Advances in monitoring environmental microbes. Curr Opin Biotechnol. 2013;24:526–33. [DOI] [PubMed] [Google Scholar]
- Johnson C, Li W, Head Eet al. . Optical, chemical, and biological oceanographic conditions on the scotian shelf and in the eastern gulf of maine in 2013. 2014. DFO Can. Sci. Advis. Sec. Res. Doc. 2014/104 v + 49 p.
- Kearse M, Moir R, Wilson Aet al. . Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Morrison M, Yu Z. Evaluation of different partial 16S rRNA gene sequence regions for phylogenetic analysis of microbiomes. J Microbiol Methods. 2011;84:81–7. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20. [DOI] [PubMed] [Google Scholar]
- Klindworth A, Pruesse E, Schweer Tet al. . Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KopylovaE, Navas-Molina, MercierCet al. . Open-source sequence clustering methods improve the state of the art. mSystems. 2016;1::e00003–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopylova E, Noe L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–7. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li Met al. . MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P, Gallagher ED. Ecologically meaningful transformations for ordination of species data. Oecologia. 2001;129:271–80. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich Jet al. . An improved greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RM, Rappé MS, Connon SAet al. . SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420:806–10. [DOI] [PubMed] [Google Scholar]
- Needham DM, Fuhrman JA. Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nat Microbiol. 2016;1:16005. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Friendly Met al. . Vegan: community ecology package. 2017.
- Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. 2016;18:1403–14. [DOI] [PubMed] [Google Scholar]
- R Core Team . R: a language and environment for statistical computing. 2018. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
- Rinke C, Schwientek P, Sczyrba Aet al. . Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–7. [DOI] [PubMed] [Google Scholar]
- Rognes T, Flouri T, Nichols Bet al. . VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalenberger A, Schwieger F, Tebbe CC. Effect of primers hybridizing to different evolutionarily conserved regions of the small-subunit rRNA gene in PCR-based microbial community analyses and genetic profiling. Appl Environ Microbiol. 2001;67:3557–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeant MJ, Constantinidou C, Cogan Tet al. . High-throughput sequencing of 16S rRNA gene amplicons: effects of extraction procedure, primer length and annealing temperature. PLoS One. 2012;7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shade A, Jones SE, Gregory Caporaso Jet al. . Conditionally rare taxa disproportionately contribute to temporal changes in microbial diversity. MBio. 2014;5:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S, Coelho LP, Chaffron Set al. . Structure and function of the global ocean microbiome. Science2015;348:1–10. [DOI] [PubMed] [Google Scholar]
- Therriault J, Petrie B, Pepin Pet al. . Proposal for a northwest atlantic zonal monitoring program. Can Tech Rep Hydrogr Ocean Sci. 1998;194:vii+57. [Google Scholar]
- Thompson LR, Sanders JG, Mcdonald Det al. . A communal catalogue reveals Earth's multiscale microbial diversity. Nature. 2017;551:457–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables WN, Ripley BD. Modern Applied Statistics with S. 4th edn. New York: Springer, 2002. [Google Scholar]
- Wickham H, ggplot2: Elegant Graphics for Data AnaSlysis, New York: Springer, 2009. [Google Scholar]
- Yang B, Wang Y, Qian P-Y. Sensitivity and correlation of hypervariable regions in 16S rRNA genes in phylogenetic analysis. BMC Bioinformatics. 2016;17:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Morrison M. Comparisons of different hypervariable regions of rrs genes for use in fingerprinting of microbial communities by PCR-denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2004;70:4800–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Kobert K, Flouri Tet al. . PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorz J, Willis C, Comeau AMet al. . Drivers of regional bacterial community structure and diversity in the Northwest Atlantic Ocean. Front Microbiol. 2019;10:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.