Abstract
Retinal ganglion cell (RGC) axons from each eye execute a series of maneuvers as they converge on the ventral surface of the brain at the optic chiasm for sorting into the optic tracts. Heparan sulfate proteoglycans (HSPGs) are extracellular glycoproteins involved in cell-surface interactions. HSPGs exhibit massive structural diversity, conferred partly by extensive post-translational modification including differential sulfation. Here we examine the roles of HSPG sulfation in RGC axon guidance at the chiasm. We identified different axon navigation phenotypes in two heparan sulfate sulfotransferase (Hst) mutant embryos, Hs2st−/− and Hs6st1−/−, each lacking an enzyme that catalyzes a particular HSPG modification. Hs2st−/− embryos display axon disorganization at the chiasm. Hs6st1−/− embryos exhibit prolific inter-retinal innervation. We show that RGCs express Hs2st and Hs6st1 and that navigation errors made by their axons coincide with regions of high Hs2st and/or Hs6st1 expression at the chiasm. Slit proteins are expressed at particular locations in the retina and around the chiasm and are normally deployed to prevent axons entering inappropriate territories. We show that Hs2st and/or Hs6st1 expression coincides with Slit expression domains at locations where RGC axons make navigation errors in Hs2st−/− and Hs6st1−/− mutants and that Hs6st1−/− RGC axons are less sensitive to Slit2 repulsion than their wild-type counterparts in vitro. We suggest that (1) Hs2st and Hs6st1 are each deployed to generate distinct patterns of heparan sulfation on RGCs and at the optic chiasm and (2) this differential sulfation directs retinal axons through the chiasm, at least in part by modulating the response of the navigating growth cone to Slit proteins.
Keywords: Hs2st, Hs6st1, transgenic, mouse, Robo, Slit, retinal ganglion cell, optic chiasm
Introduction
During normal development, the retina generates several cell types including retinal ganglion cells (RGCs) (Cepko et al., 1996), which project axons into the optic nerve starting at embryonic day 12 (E12) in the mouse (Marcus and Mason, 1995; Marcus et al., 1995). The optic nerve joins the ventral surface of the thalamus at the optic chiasm where RGC axons are corralled into a highly defined area to generate the familiar chiasm shape. At the chiasm, RGC axons are sorted into the optic tracts and grow on toward their targets in the thalamus and superior colliculus. The topographical relationships between RGC axons and the tissues they traverse is tightly regulated throughout the developing visual pathway with axons executing a particularly intricate set of maneuvers as they approach, grow through, and exit the chiasm (Marcus and Mason, 1995; Marcus et al., 1995; Colello and Guillery, 1998; Leung et al., 2003). Here we examine the contribution made to this process by the sulfation status of heparan sulfate proteoglycans (HSPGs).
HSPGs are widely expressed in the developing brain and are important for numerous processes including RGC axon navigation. Mutant mice that lack the biosynthetic enzyme Exostosin 1 (EXT1) in developing brain (Nes-Ext1−/− mice) cannot synthesize heparan sulfate (see below) and exhibit altered brain morphology, agenesis of several commissural tracts, and an unusually prolific RGC projection into the contralateral eye (Inatani et al., 2003) (supplemental Fig. S1B, available at www.jneurosci.org as supplemental material). These and other experiments (for review, see Lee and Chien, 2004; Holt and Dickson, 2005) show that the navigating growth cone relies on HSPGs.
HSPGs are structurally diverse extracellular matrix molecules generated by the addition of heparan sulfate (HS) glycosaminoglycan (GAG) unbranched polysaccharide side chains to a core protein (for review, see Esko and Selleck, 2002) (supplemental Fig. S1A, available at www.jneurosci.org as supplemental material). There are several classes of HSPG core protein. Syndecans and Glypicans are anchored to the cell surface by a transmembrane domain or glycosylphosphatidylinositol anchor, respectively, whereas agrin and perlecan are secreted into the extracellular space. During HSPG biosynthesis, the core protein is loaded into the Golgi apparatus where repeating GAG chains of the disaccharides uronic acid and glucosamine are added by a series of polymerase enzymes including EXT1. Modifying enzymes then subject the sugar residues to epimerization, de-acetylation, and sulfation. Heparan sulfate sulfotransferase (HST) enzymes add sulfate groups to HS sugars and are named according to the sugar residue position they sulfate. Hs2st sulfates the 2-O position of uronic acid, whereas Hs3st and Hs6st enzymes sulfate the 3-O or 6-O positions of glucosamine, respectively. Animals have a battery of HSTs, so potentially produce an enormous number of differently sulfated HSPGs (for review, see Esko and Selleck, 2002; Lee and Chien, 2004). The degree to which HSPG structural diversity is exploited during development in general, and RGC axon guidance at the optic chiasm in particular, remains an open question.
Here we test the hypotheses that (1) the fine structure of HS sulfation directs RGC axons at the optic chiasm and (2) the underlying molecular mechanisms include growth cone repulsion by Slit proteins. We find that in Hs2st−/− and Hs6st1−/− mutants RGC axons make distinct errors at the chiasm coinciding with HST and Slit expression domains and that Hs6st1 is required for the avoidance of Slit2-expressing cells in vitro.
Materials and Methods
Mutant alleles and mice
The Hs2stLacZ (Hs2st−) mutant allele was obtained by insertion of a β-galactosidase (LacZ) gene trap vector into the Hs2st locus (Bullock et al., 1998) and the Hs6st1LacZ-ires-hPLAP (Hs6st1−) mutant allele by insertion of a LacZ–internal ribosomal entry site–human placental alkaline phosphatase (ires-hPLAP) gene trap vector into intron 1 of the Hs6st1 locus (Leighton et al., 2001; Mitchell et al., 2001) (see Fig. 1). We confirmed by reverse transcription-PCR (RT-PCR) that the Hs6st1 gene trap allele was unable to produce wild-type transcript (Fig. 1). The Hs2st allele was maintained on a mixed C57BL/6 × CBA pigmented background and the Hs6st1 allele on a pigmented CBA background. Hs2st mice were genotyped by PCR as described previously (McLaughlin et al., 2003). Hs6st1 mice were genotyped by LacZ staining of earclip biopsies. Embryonic homozygotes were distinguished from heterozygotes and wild types by lightly fixing the paws and incubating them at 37°C in LacZ stain (see below). Staining was detectable in homozygotes (which have two copies of Hs6st1LacZ) after ∼15 min. Staining took about twice as long to become apparent in heterozygotes (which have one copy of Hs6st1LacZ). Hs6st1+/+ animals have no LacZ so do not stain.
Figure 1.
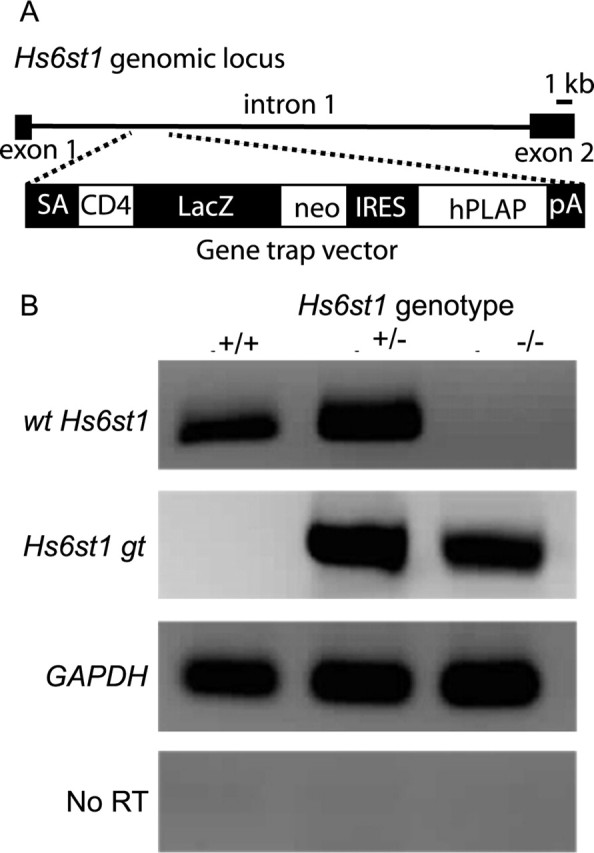
Molecular characterization of the Hs6st1 gene-trap allele. A, In mouse, the Hs6st1 gene is found on chromosome 1 and is composed of 2 exons. The gene-trap vector is composed of a splice acceptor (SA), an ORF encoding a CD4 transmembrane domain (CD4), a LacZ/neomycin (neo) fusion cassette, and an IRES and PLAP coding region followed by a polydenylation sequence (pA). B, RT-PCR analysis of Hs6st1 gene expression in wild-type, heterozygous, and homozygous mutant embryos using primers specific to the wt Hs6st1, the Hs6st1 gt, and GAPDH transcripts. Included is a no-RT control using GAPDH primers.
Molecular characterization of Hs6st1 gene-trap allele and quantification of endogenous Hs2st and Hs6st1 transcripts
cDNA synthesis.
RNA was extracted from E14.5 retina or chiasm tissue using Qiagen (Valencia, CA) RNAeasy micro kit according to the manufacturer's instructions. First-strand cDNA synthesis was performed using oligo (dT)15 primers (Promega, Madison, WI) with the Qiagen Sensiscript reverse transcriptase kit according to the manufacturer's instructions.
Characterization of the Hs6st1 gene-trap allele.
RT-PCR was performed (1) to detect the wild-type Hs6st1 (wt Hs6st1) transcript using exon-spanning primers complementary to Hs6st1 cDNA on either side of the gene-trap insertion (5′-ACGACGTGATCGTCTTCCTG-3′ and 5′-TCACACATGTGCAAGGAGGT-3′; 379 bp product), (2) to detect the Hs6st1:gene-trap (Hs6st1 gt) fusion transcript using primers complementary to Hs6st1 exon 1 and to CD4 sequences at the 5′ end of gene trap vector (5′-GACATGAAGGGCGACGAC-3′, 5′-AGTAGACTTCTGCACAGACACC-3; 500 bp product), and (3) using primers complementary to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (5′-GGGTG-TGAACCACGAGAAAT-3′ and 5′-CCTTCCACAATGCCAAAGTT-3′; 121 bp product).
Quantitative RT-PCR.
qRT-PCR was performed on cDNA from E14.5 wild-type retina (n = 3) and chiasm (n = 3) with the following primer pairs: Hs2st (5′-GCTTCCTTCTGTCTTACCTTGCTG-3′ and 5′-AGCCCACCCTTTTCTTAGTCTCC-3′; 138 bp product), Hs6st1 (5′-ACGACGTGATCGTCTTCCTG-3′ and 5′-TGGAGAAGCGAGAGAAGAGC-3′; 159 bp product), and GAPDH (5′-GGGTGTGAACCACGAGAAAT-3′ and 5′-CCTTCCACAATGCCAAAGTT-3′; 121 bp product). Quantitative RT-PCR was performed using Qiagen Quantitect SYBR green PCR kit and a DNA Engine Opticon continuous fluorescence detector (Genetic Research Instrumentation, Essex, UK). The abundance of each transcript in the original RNA sample was extrapolated from PCR reaction kinetics using Opticon software.
Tract tracing
Tract tracing was performed essentially as described by Pratt et al. (2004). Embryo heads were fixed at 4°C in 4% paraformaldehyde in PBS overnight, and 1,1′-dioctadecyl-3,3,3′,3′-tetramethyl-indocarbocyanine perchlorate (DiI) or 4-(4-dihexadecylamino)styryl)-N-methylpyridinium iodide (DiA) crystals (Invitrogen, San Diego, CA) were either (1) packed into the optic cup of one eye after removal of the lens to label axons entering or leaving the retina; in some cases, smaller focal injections were made to label a smaller number of axons or (2) placed in a line over the dorsal thalamus on one side to label axons navigating the optic tract. Heads were returned to 4% paraformaldehyde in PBS in the dark at room temperature for ∼6 weeks to allow tracers to diffuse along axons. In some cases, the brains were removed from the head, cleared in 9:1 glycerol:PBS, and imaged as whole mounts using a Zeiss (Oberkochen, Germany) Axiovert confocal LSM 510 microscope, and in other cases, heads were sectioned (200 μm) with a vibratome, cleared in 9:1 glycerol:PBS containing the nuclear counterstain TOPRO3 (1.0 μm; Invitrogen), mounted in Vectashield (Vector Laboratories, Burlingame, CA), and images acquired using an epifluorescence microscope and digital camera (Leica, Nussloch, Germany) or a TCS NT confocal microscope (Leica). For quantification of RGC projections, either (1) the total number of DiI-labeled RGCs were counted in serial vibratome sections using an epifluorescent microscope or (2) confocal optical sections sampled from serial vibratome sections were captured and DiI-labeled RGCs counted. Method 1 was used in all cases except for counting RGCs labeled by DiI injection into the contralateral thalamus when the large numbers of cells labeled were impractical to count using method 1. DiI appears orange and TOPRO3 appears red in epifluorescence images, and in confocal images, DiI appears red, DiA is green and TOPRO3 appears blue. The number of E15.5 embryos used for quantification were as follows: (1) RGC projection to the contralateral eye: Hs6st1+/+ (n = 12), Hs6st1+/− (n = 4), Hs6st1−/− (n = 7), and Hs2st−/− (n = 3); (2) RGC projection to the ipsilateral or contralateral optic tract: Hs6st1+/+ (n = 3), Hs6st1−/− (n = 3), Hs2st+/+ (n = 1), Hs2st+/− (n = 1), and Hs2st−/− (n = 1).
LacZ staining of thin frozen and thick vibratome sections
Embryonic heads were dissected and fixed overnight at 4°C in LacZ fix (4% paraformaldehyde, 0.02% NP-40, 0.01% sodium deoxycholate, 5 mm EGTA, 2 mm MgCl2 in PBS). Heads for thin frozen sections were equilibrated in 30% sucrose/PBS at 4°C, embedded in OCT (Tissue-Tek, Miles, Elkhart, IN), and sectioned (10 μm) using a cryostat. Sections were collected on poly-l-lysine-coated glass slides. Heads for thick vibratome sections were embedded in agar and sectioned (200 μm) using a vibratome. For LacZ histochemistry, sections were rinsed several times in wash buffer (2 mm MgCl2, 0.02% NP-40, 0.01% sodium deoxycholate in PBS) and transferred to LacZ stain [wash buffer supplemented with 5 mm potassium ferricyanide, 5 mm potassium ferrocyanide, and 1 mg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal)]. Frozen sections were stained for at least 20 h at 37°C and counterstained with nuclear fast red. To better visualize regions where the trapped genes were expressed strongly, we stained vibratome sections for less-prolonged periods (4 h for Hs2st+/−; 80 min for Hs6st1+/−). Stained sections were either rinsed in wash buffer and fixed in 2% gluteraldehyde and mounted in Aquamount solution (VWR Scientific, West Chester, PA) or processed for neurofilament immunohistochemistry.
Immunohistochemistry
Nkx2.2, stage-specific embryonic antigen 1, and CD44.
Standard immunohistochemistry protocols were followed. E15.5 embryo heads were fixed overnight in 4% paraformaldehyde in PBS, processed to wax, and sectioned at a thickness of 10 μm. Sections were dewaxed in xylene and rehydrated through an alcohol series. Endogenous peroxidase activity was blocked with 90% methanol:3% hydrogen peroxide. Antigens were unmasked by microwaving in 10 mm sodium citrate buffer, pH 6, before reacting overnight with the primary antibody. Nkx2.2 immunohistochemistry was performed using a 1:30 dilution of a mouse monoclonal anti-Nkx2.2 antibody (74.5A5; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) as described previously (Pratt et al., 2004) using the Envision+ (mouse) kit (K4006; DakoCytomation, High Wycombe, UK). Double-immunofluorescence immunohistochemistry was performed on wax sections blocked for 1 h at room temperature with 2% goat serum in 1% bovine serum albumin and PBS using the following primary antibodies: mouse monoclonal anti-stage-specific embryonic antigen 1 (anti-SSEA1) antibody (MC-480; Developmental Studies Hybridoma Bank) used at 1:50 and rat anti-CD44 IgG (IM7; PharMingen, San Diego, CA) used at 1:50. The primary incubation was performed overnight at 4°C. After several washes in PBS, slides were incubated in goat anti-rat Alexa Fluor 488 and goat anti-mouse Alexa Fluor 568 fluorescent secondary antibodies (A-11006, A-11004; Invitrogen), used at 1:150, for 45 min in the dark. Slides were washed with PBS, counterstained with 1:2000 TOPRO3 (Invitrogen), mounted in Mowiol, and visualized using a Leica TCSNT confocal microscope.
Neurofilament.
Tissue sections (200 μm) that had been processed for LacZ staining were incubated in blocking buffer (20% goat serum, 0.2% Triton X-100 in PBS), incubated with anti-neurofilament antibody (NA1297; 1:100 dilution in blocking buffer; Biomol, Plymouth Meeting, PA) overnight at 4°C, rinsed in 0.2% Triton X-100 in PBS, and DAB detection was performed using the Envision (rabbit) detection kit (K4010; DakoCytomation) as described by the manufacturer. After rinsing, sections were fixed in 2% gluteraldehyde and mounted in Aquamount solution (VWR Scientific)
Retinal explant/Cos7 cell cocultures
The collagen solution used in these cocultures was a mixture of rat tail and bovine collagen (BD Biosciences, Franklin Lakes, NJ) as described by Erskine et al. (2000).
Cos7 cell transfection.
Cos7 cells were transfected with a Slit2 expression construct “pSectagBhSlit2” (Erskine et al., 2000) containing a cmyc-tagged human Slit2 cDNA (hSlit2) or with an empty vector “pSecTagB” (Invitrogen) using Lipofectamine2000 (Invitrogen) according to the manufacturer's instructions. The following day, the transfected cells were trypsinized and suspended in ice-cold collagen solution at 2 × 104 cells/μl. Drops (20 μl) of this Cos7 cell/collagen suspension were placed in a tissue culture dish and allowed to set in the incubator at 37°C before addition of serum-free culture medium (Pratt et al., 2000) and overnight culture.
Retinal explant preparation.
E14.5 Hs6st1+/+ and Hs6st1−/− embryos were identified by LacZ staining paw biopsies as described above. Dissections were performed as described by Erskine et al. (2000). The eyeball was exposed, and the lens and retinal pigment epithelium teased away from the retina, which was then flattened. Explants were then cut from the peripheral third of the retina with each retina yielding ∼12 explants. Tissues were kept in ice-cold oxygenated Earle's balanced salt solution throughout the dissection. Explants were prepared from Hs6st1+/+ (n = 5) or Hs6st1−/− (n = 5) embryos.
Cocultures.
Cocultures were assembled by removing culture medium from the solidified Cos7 cell/collagen suspension and adding the retinal explants and fresh ice-cold collagen solution (13 μl). Cocultures were returned to the incubator at 37°C and the collagen allowed to set before adding serum-free culture medium and returning them to the incubator. After 40 h, the cultures were fixed in 4% paraformaldehyde/PBS and incubated in blocking solution (10% goat serum and 0.1% Triton X-100 in PBS), both for 1 h at room temperature. Cocultures were then incubated with primary antibodies [a mixture of rabbit α-neurofilament mixture (NA1297; used at 1:200; TIB MolBiol, Berlin, Germany) and mouse α-cmyc (9E10 used at 2 μg/ml; Hoffmann-La Roche, Nutley, NJ)] diluted in blocking solution overnight at 4°C followed by extensive washing with PBS and an overnight incubation at 4°C with secondary antibodies [a mixture of Alexa 488 α-rabbit (used at 1:200; Invitrogen) and Alexa 568 α-mouse (used at 1:200; Invitrogen)] diluted in blocking solution containing the DNA binding dye TOPRO3 (1 μm; Invitrogen). After extensive washing in PBS, cultures were mounted in Vectashield (Vector Laboratories) and imaged. All incubations were performed with agitation.
Imaging and quantification.
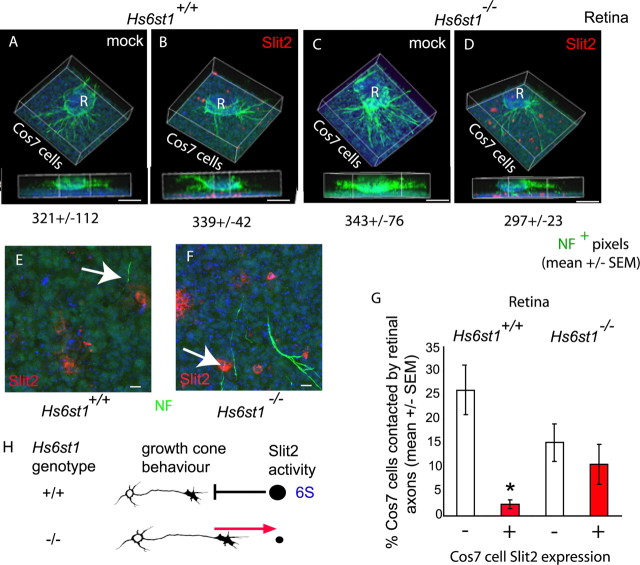
A Zeiss Axiovert confocal LSM 510 microscope was used to collect serial optical sections of each culture in the green (neurofilament+), red (hSlit2-cmyc+), and blue (TOPRO3+) channels. Zeiss and Imaris (Bitplane, Zurich, Switzerland) software were then used to (1) create a three-dimensional (3D) reconstruction of the cocultures (see Fig. 9A–D) and/or (2) reconstruct a view of the plane where RGC axons have the opportunity to contact Cos7 cells, some of which are expressing hSlit2-cmyc protein (see Fig. 9E,F). We quantified the total axon outgrowth in each culture by performing pixel counts for neurofilament immunofluorescence using 3D analysis tools on the section reconstructions. Number of cocultures used for quantification were as follows: Hs6st1+/+ with mock-transfected Cos7 cells (n = 6), Hs6st1+/+ with hSlit2-cmyc-transfected Cos7 cells (n = 8), Hs6st1−/− with mock-transfected Cos7 cells (n = 7), and Hs6st1−/− with hSlit2-cmyc-transfected Cos7 cells (n = 9).
Figure 9.
Hs6st1 is required for Slit2 avoidance by RGC axons. A–D, Collagen cultures of E14.5 Hs6st1+/+ (A, B) or Hs6st1−/− (C, D) retinal explants (R) over a bed of Cos7 cell suspension (Cos7 cells) transfected with the empty vector mock (A, C) or the hSlit2-cmyc-expressing vector Slit2 (B, D). RGC axons are immunostained for neurofilament (NF) protein (green), and hSlit2-cmyc is immunostained for cmyc (red). Nuclei are stained blue with TOPRO3. In each case, a 3D reconstruction of the culture is shown as an elevated perspective and as a side view. Neurofilament+ pixel counts (mean ± SEM) are given below each image, showing that total RGC axon outgrowth is similar under all four culture conditions. E, F, Higher magnification of the Cos7 cell layer viewed from above. E, Hs6st1+/+ RGC axons tend to avoid Slit2+ cells (the arrow points to an RGC axon that has veered away from a Slit2+ cell). F, Hs6st1−/− RGC axons frequently grow over Slit2+ Cos7 cells (the arrow points to an example). G, Histogram comparing the percentage of hSlit2-cmyc+ (red bars) and hSlit2-cmyc− (white bars) Cos7 cells contacted by Hs6st1+/+ and Hs6st1−/− RGC axons (see Results for full description of assay). The asterisk indicates a significant difference (Mann–Whitney rank–sum test, p < 0.05). H, Summary of the role of Hs6st1 in Slit2-mediated axon guidance based on these in vitro observations. The amount of Slit repulsion experienced by growth cones of each genotype is indicated by the size of the black circle. 6-O sulfated heparan (6S) allows Slit2 to exert its repulsive activity on RGC growth cones. In the absence of 6S, RGC growth cones no longer take evasive action when encountering a source of Slit2. Scale bars: A–D, 200 μm; E, F, 10 μm.
Results
The Hs6st1 gene-trap allele does not make wild-type Hs6st1 transcript
The Hs2st gene-trap allele used in this study has been shown previously to be incapable of producing wild-type Hs2st mRNA (Bullock et al., 1998) and presumed unable to produce Hs2st protein. Before commencing this study, it was necessary to characterize the Hs6st1 gene-trap allele. We prepared RNA from Hs6st1+/+ embryos and from embryos heterozygous (Hs6st1+/−) and homozygous (Hs6st1−/−) for the gene-trap insertion for analysis by RT-PCR. Previous characterization of this allele showed that the gene-trap vector was inserted into intron1 of the Hs6st1 gene as shown in Figure 1A (Leighton et al., 2001; Mitchell et al., 2001). PCR with primers spanning Hs6st1 exons 1 and 2 amplified a product of the expected size (wt Hs6st1; 379 bp) from Hs6st1+/+ and Hs6st1+/− cDNA but not from homozygous Hs6st1−/− cDNA (Fig. 1B). PCR with primers from Hs6st1 exon 1 to the CD4 sequences at the 5′ end of the gene-trap vector amplified a 500 bp Hs6st1 gt product from Hs6st1−/− and Hs6st1+/− embryos but not from homozygous Hs6st+/+ cDNA (Fig. 1B). Sequencing of this 500 bp product confirmed that it was a fusion between Hs6st1 exon1 and the 5′ end of the gene-trap vector (data not shown). The insertion of the gene-trap vector into intron 1 of the Hs6st1 gene therefore blocks production of wild-type Hs6st1 transcript and instead generates a fusion transcript between exon 1 of Hs6st1 mRNA and gene-trap vector sequences. It is therefore unlikely to produce functional Hs6st1 protein.
The structure of embryonic Hs2st−/− and Hs6st1−/− mutant brains is normal
Inatani et al. (2003) generated mutant mouse embryos that lack the major HS glycosyltransferase Ext1 throughout the developing brain (Nes-Ext1−/− mice). These animals completely lack HS throughout the developing brain and display major anatomical defects; the olfactory bulbs and cerebellum are missing, the superior colliculus is malformed, and the midline of the forebrain is extremely abnormal and is unable to support the formation of several forebrain commissures. In contrast, both Hs2st−/− and Hs6st1−/− embryos are able to produce brains that resemble the wild-type far more closely than those of the Nes-Ext1−/− embryos (Fig. 2). The olfactory bulbs are present in both of our mutants (Fig. 2D,G), and the extreme disruption of the midline of the forebrain seen in Nes-Ext1−/− embryos is not present in Hs2st−/− or Hs6st1−/− brains [compare Fig. 2E,H (present study) with Inatani et al. (2003), their Fig. 4B,D]. It has been shown previously that the Hs2st−/− cerebral cortex is undersized (McLaughlin et al., 2003), and although Hs2st−/− brains formed all the structures we looked for, they did appear somewhat smaller than in Hs2st+/+ and Hs6st1−/− brains (Fig. 2, compare D,E with A,B,G,H). We did not attempt to quantify these differences. Hs2st−/− embryos display colobomas (Bullock et al., 1998), but the histology of the retina and optic nerve were indistinguishable from those of the wild-type and Hs6st1−/− embryos (Fig. 2, compare F with C,I). Thus, differences in the navigation of RGC axons at the optic chiasm of these mutants are not secondary consequences of major anatomical defects.
Figure 2.

Embryos lacking Hs2st or Hs6st1 form a normally shaped brain. Hematoxylin and eosin-stained wax sections of heads from wild-type (A–C), Hs2st−/− (D–F), and Hs6st1−/− (G–I) E17.5 embryos. A, D, G, Sagittal sections showing the olfactory bulbs (ob), cerebral cortex (cc), thalamus (t), and superior colliculus (sc). The position of the optic chiasm (oc) is marked. B, E, H, Coronal sections through the forebrain at the point at which the optic nerves (on) have nearly reached the optic chiasm, showing the structure of the brain around the midline. C, F, I, Coronal sections through the eye showing the structure of the lens (l), retina (r), and optic nerve as it exits the eye. J, Diagram summarizing the normal development of the visual pathway. RGC axons exit the retina (r) and bundle together to form the optic nerve (on) as they navigate to the optic chiasm (oc) at the ventral midline (M), where they are sorted into the optic tracts (ot). RGC axons normally follow smooth trajectories and do not escape normal chiasm boundaries or innervate the opposite eye in large numbers. P, Posterior; A, anterior; Di, distal; Pr, proximal. Scale bars: A, D, G, 1 mm; B, C, E, F, H, I, 500 μm.
Figure 4.

The Hs6st1−/− chiasm has a normal shape but unusually large numbers of RGC axons innervate the opposite eye. All panels are horizontal sections through the chiasm (A–C, F–H) or the retina (D, E, I, J) after unilateral DiI injection into the retina. In C–E and H–J, DiA has been injected into the optic tract on the same side as the DiI-injected eye. A–E, In the wild type, relatively few RGC axons enter the contralateral optic nerve at E14.5 (A), E15.5 (C), and E17.5 (B). D, E, At E15.5, very few RGCs in the contralateral eye are retrogradely labeled with DiI. The boxed area in D is magnified in E. F–J, In the mutant, the overall shape of the chiasm closely resembles that of the wild type at E14.5 (F), E15.5 (H), and E17.5 (G). However, many more axons enter the contralateral optic nerve (con) than in the wild type at these ages. I, J, In contrast to the wild type, many RGC bodies in the contralateral retina are retrogradely labeled with DiI. The boxed area in I is magnified in J. Note that RGCs labeled with DiI from the contralateral eye are not colabeled with DiA, indicating that they do not project to the DiA injection site in the thalamus. All sections are horizontal with posterior at the top. DiI and DiA injections summarized in diagram at the bottom and indicated by red or green asterisks, respectively, in the panels. K, Histogram showing numbers of RGCs retrogradely labeled in the contralateral retina after unilateral retinal DiI injection. Significantly more retrogradely labeled RGCs were counted in the Hs6st1−/− retina then in the wild type (p = 0.006, Student's t test). Note that Hs2st−/− did not exhibit increased innervation of the opposite eye; in fact, there was a significant decrease compared with wild type (p = 0.04, Student's t test). The total numbers of DiI-labeled RGCs were counted in serial sections. All bars are means ± SEM. Student's t test for comparison with wild type, ∗p < 0.05. L, Diagram summarizing aberrant growth of RGC axons into the contralateral optic nerve of the Hs6st1−/− mutant. r, Retina; l, lens; on, optic nerve; con, optic nerve contralateral to injected retina; oc, optic chiasm; ot, optic tract; P, posterior; A, anterior; Di, distal; Pr, proximal; M, ventral midline. Scale bars: A–C, F–H, 200 μm; D, I, 100 μm; E, J, 50 μm.
RGC axons are disorganized in and around the Hs2st−/− chiasm
Unilateral DiI injections into the eye shows that in both Hs2st+/+ and Hs2st−/− embryos RGC axons exit the eye and form the optic nerve as they grow toward the optic chiasm. At the chiasm, the axons are sorted into the optic tracts. However, in the Hs2st−/− mutant, RGC axons grow up the ventral midline (Fig. 3H–J,L, arrows). In every Hs2st−/− embryo examined (n = 9), we observed abnormalities in RGC axon navigation at the chiasm that we never saw in Hs2st+/+ or Hs6st1−/− embryos. However, the size of the projection escaping the chiasm and growing up the midline varied between embryos (Fig. 3, H and I, illustrates this variability). At least some of the axons that escape from normal chiasm territory are able to rejoin the optic tracts, as shown by the fact that they can be retrogradely labeled with DiI injections into the optic tract (Fig. 3M, arrow). These escaping axons are extreme examples of a more general disorganization/defasciculation disorder at the Hs2st−/− chiasm. Smaller focal DiI injections into the retina permit visualization of axon trajectories within the chiasm (Fig. 3D,F,K,K′). In Hs2st−/− mutants, axons take erratic paths (Fig. 3K,K′), unlike the smoother more parallel formations seen in Hs2st+/+ (and Hs6st1−/−) embryos (Fig. 3D,F).
Figure 3.
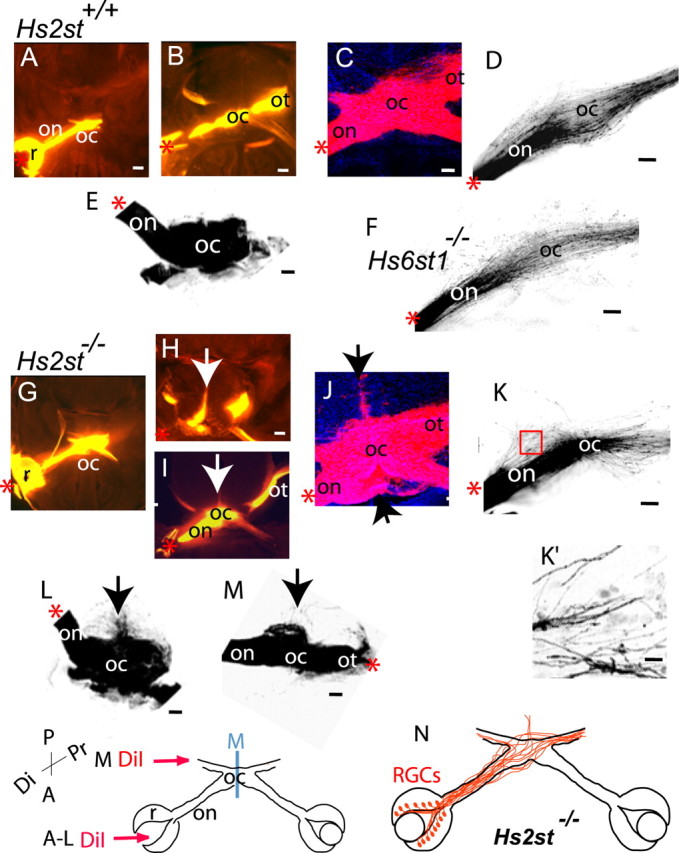
The Hs2st−/− optic chiasm is disorganized at E15.5. Images of Hs2st+/+ (A–E), Hs6st1−/− (F), and Hs2st−/− (G–M) chiasms labeled by DiI injection into the retina (A–L) or the optic tract (M) are shown. A, B, and G–I are epifluorescence images, and C–F and J–M are confocal images. The arrows in H–J, L, and M mark axons growing aberrantly outside normal chiasm territory in the mutant. A, B, and G, H are adjacent horizontal sections in which A and G are ventral to B and H. H shows a particularly extreme example of axon growth up the midline with a milder example shown in I. D, F, K, DiI labeling of the chiasm after focal DiI injection; in Hs2st+/+ (D) and Hs6st1−/− (F) embryos, RGC axons traverse the chiasm approximately parallel to one another, whereas Hs2st−/− mutant RGC axons (K) are less ordered (boxed area in K shown in K′ illustrating disordered axons). E, L, M, Three-dimensional reconstructions of DiI-labeled chiasms after DiI injection into the retina (E, L) or the optic tract (M). In each case, the underside of the reconstructed chiasm is viewed from the front. The arrows mark axons escaping up the midline in L or defasciculating at the chiasm before growing to the thalamus in M. DiI injections summarized in diagram at the bottom and indicated by red asterisks in the panels. N, Diagram summarizing aberrant RGC axon trajectories in the Hs2st−/− mutant. R, Retina; on, optic nerve; oc, optic chiasm; ot, optic tract; P, posterior; A, anterior; Di, distal; Pr, proximal; M, ventral midline. Scale bars: A, E, 400 μm; B, H, I, 200 μm; C–F, J–M, 100 μm; K′, 10 μm.
The chiasm phenotype in the Hs2st−/− mutants is very specific, and we did not find evidence that other aspects of RGC axon navigation at the chiasm were altered in these mutants (Fig. 4K). Examination of the optic tracts of Hs2st−/− embryos (n = 9) after unilateral DiI injection into the retina did not show any obvious alteration in the size of the ipsilateral projection, and we confirmed this by counting the numbers of RGCs retrogradely labeled in the ipsilateral and contralateral retina after DiI injection into the thalamus. We counted 557, 710, and 596 RGCs labeled in the contralateral retinas and 22, 17, and 17 RGCs labeled in the ipsilateral retinas of E15.5 Hs2st+/+, Hs2st+/−, and Hs2st−/− embryos, respectively (littermates, n = 1 of each genotype). We did not pursue this quantification further, so although Hs2st−/− embryos do not exhibit major alterations in the numbers of RGC axons reaching the optic tracts, we cannot exclude the possibility of a more subtle defect. Together, these results show that Hs2st activity has a specific function in maintaining the normal configuration of axons within the chiasm.
RGC axons prolifically innervate the opposite eye in Hs6st1−/− embryos
In Hs6st1−/− mutants, RGC axons make navigation errors at the optic chiasm that are completely different to those seen in Hs2st−/− mutants. Unilateral DiI injections into the eye revealed that more axons enter the contralateral optic nerve and grow toward the opposite eye in the mutant than in the wild type. At E14.5 (Fig. 4, compare A, F), more DiI-labeled axons appear in the contralateral optic nerve in the mutant than in the wild type. Very few RGC bodies in the contralateral retina are retrogradely labeled at this stage, showing that RGC axons are growing down the contralateral optic nerve but have not invaded the contralateral retina. By E15.5 (Fig. 4, compare C–E, H–J), many more of these projections continue all the way to the retina in the Hs6st1−/− mutant because large numbers of RGC bodies were retrogradely labeled in the retina contralateral to the injected eye. To quantify this misprojection, we counted the total number of DiI-labeled RGCs in serial sections of the contralateral retina and showed that four times as many RGCs project to the opposite eye in Hs6st1−/− mutants than in control embryos (Fig. 4K). Increased innervation of the opposite eye could be caused by axons branching at the chiasm and sending one branch to the thalamus and another to the contralateral eye. However, we find no evidence for this because injection of DiA into the thalamus and DiI into the eye (both on the same side) did not produce DiI/DiA double-labeled RGCs in the retina contralateral to the injections (Fig. 4J), as would have been expected if increased branching was happening.
One explanation for the increase in the number of RGC axons growing up the contralateral optic nerve instead of into the contralateral optic tract in Hs6st1−/− embryos is that they exhibit altered responsiveness to axon guidance cues in the contralateral optic tract. We therefore asked whether RGC navigation into the ipsilateral optic tract was also affected. However, we found that RGC axons are sorted into the optic tracts in normal numbers. Unilateral DiI injections into the optic tract retrogradely label similar numbers of RGC cell bodies (mean ± SEM) in the ipsilateral eye (Hs6st1+/+, 60 ± 14 vs Hs6st1−/−, 106 ± 24; Student's t test, p > 0.05). The numbers of RGC cell bodies retrogradely labeled in the retina contralateral to the thalamic DiI injection were also not significantly altered in the Hs6st−/−1 mutant (Hs6st1+/+, 72 ± 14 vs Hs6st1−/−, 63 ± 21; Student's t test, p > 0.05). Note that these contralateral RGC counts were made using the sampling method described in Materials and Methods and cannot therefore be compared directly to the ipsilateral counts. We therefore find no evidence that either the sorting of RGC axons into the optic tracts or the total number of retinal axons reaching the thalamus is altered in the Hs6st1−/− mutant.
Hs6st1 activity is therefore specifically required to prevent RGC axons growing into the contralateral optic nerve after they have crossed the midline. Hs2st mutants do not exhibit increased projection into the opposite eye; in fact, this projection was decreased in Hs2st−/− embryos (Fig. 4K). Conversely, axon trajectories within the Hs6st1−/− mutant chiasm were normal (Fig. 3F), and we did not detect axons escaping from normal chiasm territory as was seen in the Hs2st−/− mutants.
Hs2st and Hs6st1 expression by RGCs and at the optic chiasm
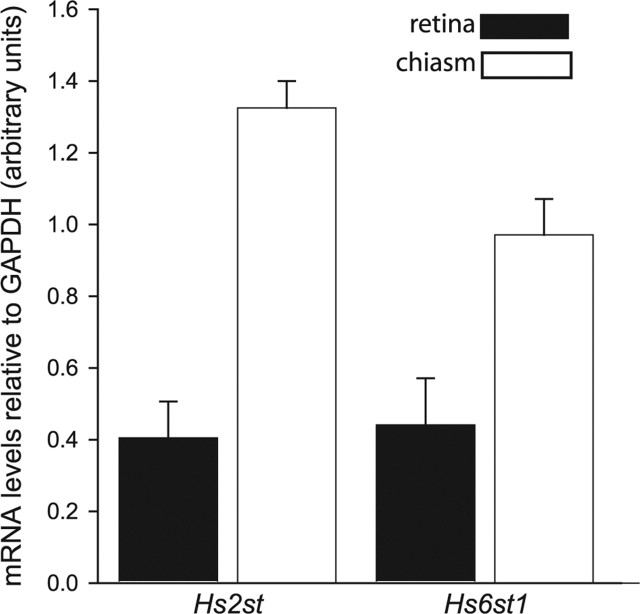
The tract-tracing experiments described above show that Hs2st and Hs6st1 play important roles in directing RGC axons at different points along their route through the optic chiasm. This raised the possibility that HS sulfated by Hs2st elicits different RGC growth cone responses to HS sulfated by Hs6st1. It is also possible that differences in the spatial arrangement of Hs2st and Hs6st1 contribute to their different roles in RGC axon guidance. First, we performed quantitative RT-PCR on RNA extracted from wild-type retina and optic chiasm. These results (normalized for expression of GAPDH mRNA) are presented in Figure 5. Both Hs2st and Hs6st1 mRNA are expressed in both the retina and in the chiasm, although in both cases, the expression in the chiasm is significantly higher. Next, we took advantage of the LacZ reporter cassette integrated into the Hs2st and Hs6st1 loci to visualize the expression of these genes in RGCs themselves and the regions they encounter as they navigate the optic chiasm. The Hs2st and Hs6st1 mutant alleles were produced by a gene-trap integration of a LacZ reporter cassette into the Hs2st locus (Bullock et al., 1998) or a LacZ-ires-hPLAP reporter cassette into the Hs6st1 locus (Leighton et al., 2001; Mitchell et al., 2001) (Fig. 1A). Histological detection of LacZ by X-gal staining thin frozen sections to completion revealed that Hs2st (Fig. 6A–D) and Hs6st1 (Fig. 7A–D) are expressed within the RGC layer of the retina and in cells surrounding RGC axons in the optic nerve and at the chiasm. Both genes are also expressed elsewhere in the brain and in non-neural tissues with variation in LacZ staining intensity between different regions. These experiments show that although Hs2st and Hs6st1 are expressed by RGCs and by the cells, their axons encounter at the chiasm there are considerable variations in levels of gene expression. To understand this in more detail, we used subsaturation LacZ staining of tissue sections to reveal sites where gene expression was particularly high. This technique relies on the fact that the rate of production of blue LacZ reaction product is proportional to the amount of LacZ enzyme present, so blue staining will appear first in places where the expression of LacZ (driven from the endogenous Hs2st or Hs6st1 promoter) is strongest.
Figure 5.
Quantitative analysis of Hs2st and Hs6st1 transcript levels in the retina and optic chiasm of E14.5 mice by qRT-PCR. Expression levels of Hs2st and Hs6st1 mRNA were each normalized to that of a ubiquitously expressed housekeeping gene, GAPDH, and expressed as mean ± SEM. Hs2st and Hs6st1 mRNAs are detectable in the retina and at the chiasm, with both transcripts present at significantly higher levels at the chiasm (Student's t test, p < 0.05).
Figure 6.
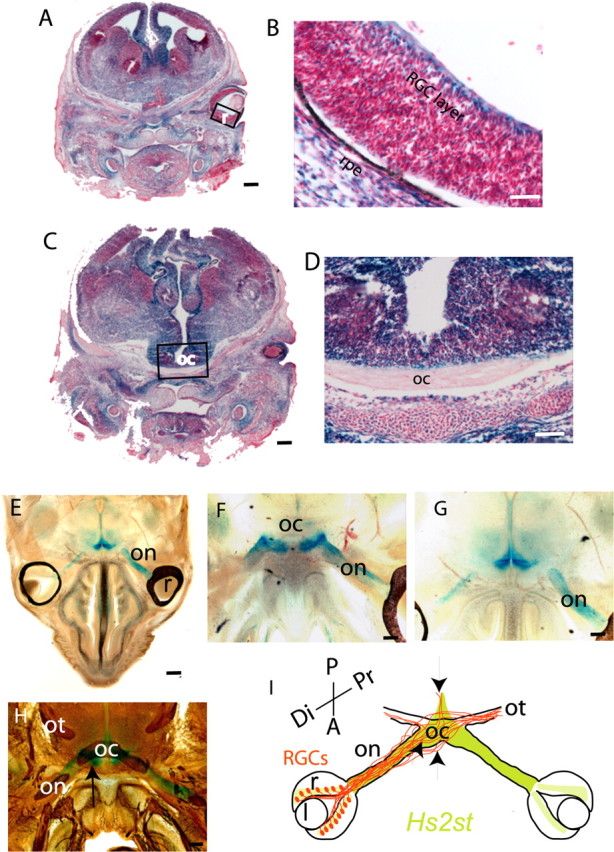
Hs2st expression in the retina and at the optic chiasm revealed by X-gal staining of E15.5 Hs2st+/LacZ sections. A–D, Thin (10 μm) coronal cryostat sections stained to completion. Hs2st is expressed widely in neural and non-neural tissues, although there is considerable variation in staining intensity between different regions. A, B, LacZ staining in the RGC layer of the retina. The boxed area in A is magnified in B. C, D, LacZ staining of cells around the optic chiasm. The boxed area in C is magnified in D. E–H, Thick (200 μm) horizontal vibratome sections through E15.5 Hs2st+/LacZ embryos in which X-gal staining was stopped before it reached completion to reveal sites of strong expression. E, Low-power view showing staining in cells surrounding the optic nerve and at the ventral midline but not in the retina. F, G, Higher magnification of consecutive sections showing staining in cells surrounding the optic nerve and at the point at which the optic nerve contacts the brain (F) and a more dorsal section showing staining dorsal and posterior to the chiasm (G). H, Section double stained with X-gal and anti-neurofilament antibody showing the relationship between neurofilament-stained RGC axons and the regions of X-gal staining. The arrow indicates coincidence between strong X-gal staining and neurofilament-labeled axons at the point at which the optic nerve joins the brain. I, Summary diagram of Hs2st expression (green). The trajectories of RGC axons (brown) at the optic chiasm in Hs2st−/− mutants is superimposed with arrows marking the coincidence between regions that normally express high levels of Hs2st and RGC axon navigation abnormalities when Hs2st function is lost (see Fig. 3). r, Retina; l, lens; on, optic nerve; oc, optic chiasm; ot, optic tract; Di, distal; Pr, proximal; A, anterior; P, posterior; rpe, retinal pigmented epithelium. Scale bars: A, C, E, 625 μm; D, F–H, 100 μm; B, 50 μm.
Figure 7.
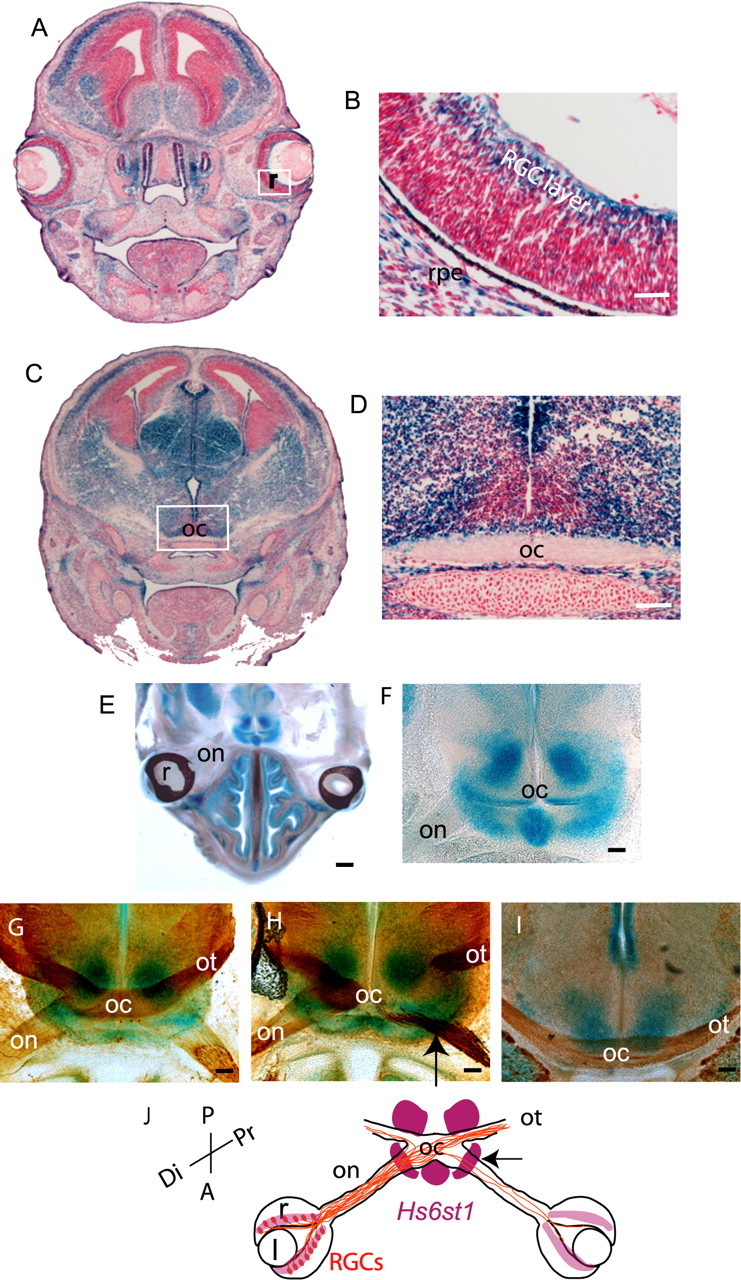
Hs6st1 expression in the retina and at the optic chiasm revealed by X-gal staining of E15.5 Hs6st1+/LacZ sections. A–D, Thin (10 μm) coronal cryostat sections stained to completion. Hs6st1 is expressed widely in neural and non-neural tissues, although there is considerable variation in staining intensity between different regions. A, B, LacZ staining in the RGC layer of the retina. The boxed area in A is magnified in B. C, D, LacZ staining of cells around the optic chiasm. The boxed area in C is magnified in D. Thick horizontal (E–H) or coronal (I) vibratome sections (200 μm) through E15.5 Hs6st1+/LacZ embryos in which X-gal staining was stopped before it reached completion to reveal sites of strong expression. E, Low-power overview showing strong staining around the chiasm but not in the retina or in cells surrounding the optic nerve. F, Higher magnification of the chiasm region. G–I, Sections double stained with X-gal and anti-neurofilament antibody. G, H, Horizontal sections through the chiasm to show the relationship between neurofilament-stained RGC axons and the regions of staining. The section in H is cut at an angle so that the left side shows RGC axons at the middle of the chiasm and the right side shows axons entering the chiasm along the optic nerve and exiting along the optic tract. The arrow indicates coincidence between strong X-gal staining and neurofilament-labeled axons at the point at which the optic nerve joins the brain. I, Coronal section through the posterior chiasm showing the lack of strong Hs6st1 expression at the midline above the chiasm. J, Summary diagram of Hs6st1 expression (purple). The trajectories of RGC axons (brown) at the optic chiasm in Hs6st1−/− mutants is superimposed with the arrow marking the coincidence between high levels of Hs6st1 expression at the point at which RGC axons misproject into the contralateral optic nerve when Hs6st1 function is lost (see Fig. 4). r, Retina; l, lens; on, optic nerve; oc, optic chiasm; ot, optic tract; Di, distal; Pr, proximal; A, anterior; P, posterior; rpe, retinal pigmented epithelium. Scale bars: A, C, E, 625 μm; D, F–I, 100 μm; B, 50 μm.
At E15.5, Hs2st is strongly expressed in cells surrounding the optic nerve, at the point at which the optic nerve joins to the brain, and around the chiasm including up the midline posterior to the chiasm (Fig. 6E–G). At E15.5, Hs6st1 is strongly expressed at the point at which the optic nerve joins the brain, at the ventral midline just anterior to the chiasm, and just posterior to the optic tract as it escapes the midline, but not at the midline itself (Fig. 7E,F). Double staining for LacZ and axonal neurofilament reveals the close relationship between RGC axons and cells expressing high levels of Hs2st (Fig. 6H) or Hs6st1 (Fig. 7G–I) at several locations along their route. The correspondence between the relative levels of Hs2st and Hs6st1 mRNA in the retina and at the chiasm detected by qRT-PCR (Fig. 5) and LacZ staining intensity (Figs. 6, 7) gives us confidence that the gene-trap LacZ alleles report accurately on endogenous gene expression.
As RGC axons navigate toward the brain, they therefore encounter environments that express differing combinations of HST activity. Both Hs2st and Hs6st1 are strongly expressed at the boundary between the optic nerve and the brain (Figs. 6H, 7H, arrows). In the region around the optic nerve and the postchiasmatic ventral midline, Hs2st alone is strongly expressed. Finally, at the prechiasmatic ventral midline and the region posterior to the optic tract, Hs6st1 alone is strongly expressed. In both Hs2st and Hs6st1 mutants, RGC axons make errors at points at which their growth cones would normally encounter these regions of high Hs2st (Fig. 6I, arrows) or Hs6st1 (Fig. 7J, arrow) expression. Note that this is not always the case because in Hs6st1−/− mutants, RGC axons make obvious errors at the optic nerve/brain boundary as they exit the chiasm but behave normally as they encounter other regions that normally express high levels of Hs6st1. It is likely that other factors are able to compensate for the loss of Hs6st1 in these regions. There are other HST genes that are expressed in the brain (Sedita et al., 2004) and it is possible that these ameliorate the effects of losing Hs2st or Hs6st1 in our mutants. Alternatively, the growth cone may be directed by several guidance cues simultaneously at a particular location, not all of which are dependent on heparan sulfation.
Key molecular chiasm markers are unaffected in Hs2st−/− and Hs6st1−/− mutants
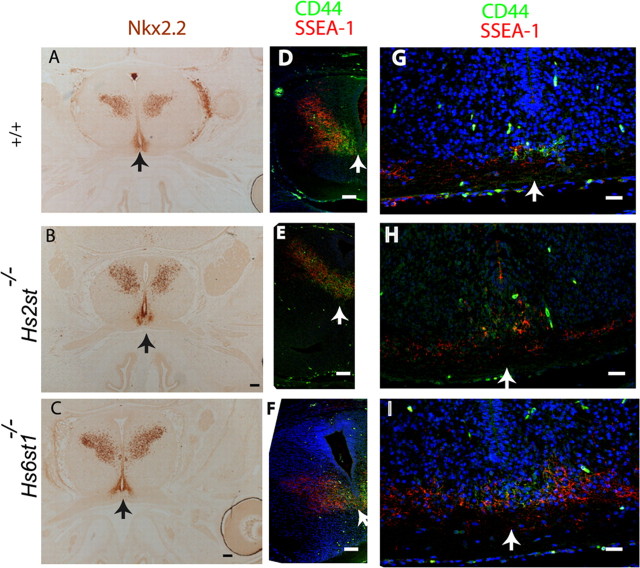
It remained a possibility that the axon guidance defects are a secondary consequence of an earlier defect in the molecular anatomy of the optic chiasm. To address this, we looked at the expression of several molecules, expressed by neurons and cells at the optic chiasm, the distribution of which is diagnostic of its normal development. One of these molecules is the transcription factor Nkx2.2, which is expressed in a domain at the midline through which RGC axons grow (Marcus et al., 1999). It is also expressed in the ventral diencephalon in a V-shape. There is no evidence for changes in Nkx2.2 expression between the wild type and mutants at E15.5 when RGC axons are navigating the optic chiasm (Fig. 8A–C). SSEA-1 is a carbohydrate moiety found on glycoproteins or glycolipids, and it has been shown to be expressed from E12 in the mouse by a population of early differentiating neurons that provide an anatomical template to guide pioneering retinal axons at the optic chiasm (Sretavan et al., 1994, 1995; Marcus and Mason, 1995). At E15.5, horizontal sections through the ventral diencephalon in wild type and Hs2st−/− and Hs6st1−/− mutants reveal that SSEA-1 is expressed prominently by a bilaterally symmetrical V-shaped population of cells and their processes in the caudal region of the ventral diencephalon (Fig. 8D–F). The tip of the “V” extends anteriorly toward the midline of the developing optic chiasm where many retinal axons navigate through at this developmental stage. In coronal sections (Fig. 8G–I), SSEA-1-positive processes grow along the retinal axons and contact the initial part of the optic tract. CD44 is a cell surface molecule expressed on the population of early differentiating chiasmatic neurons that is known to be important for chiasm formation (Sretavan et al., 1994, 1995; Marcus and Mason, 1995). CD44-positive cells occupy a more restricted area compared with SSEA-1 (Lin et al., 2005). CD44 cells and their processes are found medially but not laterally at the anterior part of the V-shaped array in wild types and mutants (Fig. 8D–I). There is some colocalization between CD44 and SSEA-1 at the midline indicated by double-labeling, but more laterally, these two molecules do not overlap. Examination of serial horizontal and coronal sections in age-matched embryos confirmed that the expression patterns of SSEA-1 and CD44 are identical to the wild-type in Hs2st−/− and Hs6st1−/− mutants. In summary, Hs2st−/− and Hs6st1−/− mutants do not show any defects in the expression of three key molecules diagnostic of proper chiasm formation, two of which are known to interact with and thought to influence the divergence of retinal axons at the optic chiasm.
Figure 8.
The expression patterns of molecules Nkx2.2, SSEA-1, and CD44 on cells around the optic chiasm are identical to the wild type in Hs2st−/− and Hs6st1−/− E15.5 embryos. The position of the midline is marked with an arrow. A–F are horizontal, anterior is down; G–I are coronal, ventral is down. A–C, Nkx2.2 immunohistochemistry on wax sections. Nkx2.2 is expressed by cells at the optic chiasm midline, posteriorly up the midline and also in the ventral diencephalon. D–I, Confocal images of cryostat sections immunostained with SSEA-1 (red) and CD44 (green) antibodies. Nuclei were counterstained with TOPRO-3 (blue). D–F, Ventral sections show one arm of the bilaterally symmetrical V-shaped SSEA-1 expression domain in the ventral diencephalon, posterior to retinal axons. CD44-positive cells and their processes are localized around the midline and overlap with SSEA-1-expressing cells at the anterior tip of the SSEA-1-expressing domain. G–I, Coronal sections through the anterior tip of the V-shaped SSEA-1 domain, just posterior to the optic chiasm. In the wild-type (G) and mutant (H, I) embryos, SSEA-1-positive processes appose retinal axons as they course over the pial surface of the ventral diencephalon. CD44 expression is confined to the midline. Scale bars: A–I, 200 μm.
Hs6st1 is needed for RGC repulsion by Slit2 in vitro
The increased growth of RGC axons into the opposite eye in Hs6st1−/− embryos phenocopies Slit1−/−; Slit2−/− embryos (Plump et al., 2002) (supplemental Fig. S1C, available at www.jneurosci.org as supplemental material), consistent with a requirement for Hs6st1 activity in Slit function. There are several lines of evidence that implicate HSPGs in the ability of RGC axons to respond to Slit (Inatani et al., 2003; Piper et al., 2006). To test the more specific hypothesis that the response to Slit is dependent on a particular heparan sulfation pattern, we compared the response of Hs6st1+/+ and Hs6st1−/− RGC axons to Cos7 cells expressing hSlit2. Retinal explants were cultured in collagen gels on a bed of a Cos7 cell/collagen gel suspension. Cos7 cells were transfected with an expression vector containing myc-tagged hSlit2 or mock transfected with an empty vector. To quantify the ability of Slit2 to repel retinal axons, we cultured retinal explants over Slit2-transfected Cos7 cells for 40 h before immunostaining the axons with α-neurofilament (Fig. 9, green fluorescence) and the Slit-expressing Cos7 cells with α-cmyc (Fig. 9, red fluorescence). Cell nuclei were stained with TOPRO3 (Fig. 9, blue fluorescence). Retinal explants project large numbers of axons, which are free to grow down into the layer of Cos7 cells; examples of these cultures are shown in Figure 9A–D with higher magnifications of axons growing in the Cos7 cell layer in Figure 9, E and F. We found no evidence that total axon outgrowth differed as a result of Hs6st1 genotype or coculture with Slit2-expressing cells because a comparison of neurofilament immunofluorescence pixel counts between the four culture conditions were very similar (Fig. 9A–D, mean ± SEM values shown below 3D reconstructions). Next, we compared the sensitivity of Hs6st1+/+ and Hs6st1−/− RGC axons to Slit2. We quantified the sensitivity of Hs6st1+/+ and Hs6st1−/− RGC axons to hSlit2 by comparing the interaction between RGC axons of each genotype and hSlit2-cmyc (Slit2+)-expressing Cos7 cells. Slit2 is normally repulsive to RGC growth cones (Erskine et al., 2000). Our aim was to test the hypothesis that Hs6st1 is required for growth cone sensitivity to Slit2. We predicted that wild-type growth cones would avoid Cos7 cells expressing Slit2 protein but mutant growth cones would be less sensitive. We compared the percentages of Slit2+ cells that were contacted by neurofilament+ RGC axons from either Hs6st1+/+ or Hs6st1−/− retinal explants. To control for a difference between Hs6st1+/+ and Hs6st1−/− cocultures that was not a direct consequence of an altered response to hSlit2, we calculated the percentage of hSlit2-cmyc− (Slit2−) cells that were contacted by neurofilament+ RGC axons in cocultures with mock-transfected Cos7 cells. The results are shown in Figure 9G. Hs6st1+/+ RGCs contacted a significantly lower percentage of Slit2+ Cos7 cells (Fig. 9G, red bar) than hSlit2-cmyc− Cos7 cells (Fig. 9G, white bar) (Mann–Whitney rank–sum test, p < 0.05), consistent with the well documented ability of Slit2 to repel RGC axons (Erskine et al., 2000). In stark contrast, Hs6st1−/− RGC axons showed no difference in the percentage of Slit2+ and Slit2− Cos7 cells they contacted (Mann–Whitney rank–sum test, p > 0.05), showing that they were insensitive to repulsion by Slit2. A significantly lower proportion of RGC axon/Slit2+ contacts were recorded in Hs6st1+/+ cultures compared with Hs6st1−/− cultures (Mann–Whitney rank–sum test, p < 0.05). We conclude that the ability of RGC growth cones to avoid Slit2-expressing cells is dependent on their ability to synthesize Hs6st1. Because HSTs are known to function in the Golgi apparatus, the most straightforward explanation for our in vitro observations is that Hs6st1 regulates the response of the growth cone to Slit2 by altering the 6-O sulfation status of HSPGs on the growth cone surface. Previous work has shown that Hs6st activity can be secreted (Habuchi et al., 1995), raising the possibility that Hs6st1 functions in the extracellular environment in our assay. Because Slit2 does not appear to be a heparan sulfate proteoglycan (Hu, 2001), it is unlikely that Hs6st1 produced by RGCs is making changes to the guidance cue itself, but we cannot rule out the possibility that Hs6st1 somehow affects the ability of Cos7 cells to present biologically active Slit2 to the RGC growth cone in vitro.
Discussion
Retinal axon navigation at the optic chiasm is controlled by genes ranging from transcription factors (for review, see Pratt and Price, 2006) to cell-surface and secreted proteins (for review, see Dickson, 2002). Manipulating a small subset of these genes produces specific alterations to chiasm organization (Deiner and Sretavan, 1999; Plump et al., 2002; Herrera et al., 2003; Williams et al., 2003). This complex genetic control may underlie variations in the details of chiasm construction between vertebrate species. HSPGs are emerging as important determinants in directing the growth cone toward its target (for review, see Lee and Chien, 2004). Genetic analysis of Caenorhabditis elegans has shown that different HS modifications are important for the navigation of particular axons, leading to the proposal of the “heparan sulfate code” hypothesis, according to which specific patterns of HS modification elicit specific axon growth behaviors (Bulow and Hobert, 2004). Additional support for this hypothesis comes from Xenopus, in which HS sulfation is required by RGC axons to navigate correctly to the tectum (Walz et al., 1997; Irie et al., 2002). The HST phenotypes reported here show that the fine structure of HSPGs modulate specific aspects of RGC axon navigation at the chiasm and provide a mechanism for fine-tuning RGC axon trajectories by altering HS sulfation patterns.
The expression pattern of Nkx2.2, a transcription factor expressed in a distinctive pattern in the ventral diencephalon (Marcus and Mason, 1995), is not altered in Hs2st−/− or Hs6st1−/− mutants, indicating that they follow the normal molecular timetable of chiasm formation. Furthermore, we failed to find alterations in the expression of SSEA-1 or CD44, cell-surface molecules known to influence the growth of retinal axons, at the chiasm of Hs2st−/− or Hs6st1−/− mutants. SSEA-1 and CD44 are expressed by an early differentiating neuronal population immediately caudal to the developing optic chiasm (Sretavan et al., 1994, 1995; Marcus and Mason, 1995; Lin et al., 2005). Contact between SSEA-1-expressing chiasm cells and the earliest growing retinal axons are thought to shape their path into the characteristic “X” shape of the optic chiasm (Sretavan et al., 1994; Marcus et al., 1995; Runko and Kaprielian, 2002). CD44 is confined to more central regions of the neuronal array than SSEA-1 and has been shown to be required for axon crossing of the midline (Sretavan et al., 1995; Lin and Chan, 2003). Different isoforms of CD44 exist and are decorated with various sulfated glycosaminoglycans (Brown et al., 1991; Piepkorn et al., 1997; Greenfield et al., 1999), and HS immunoreactivity overlaps with SSEA-1-expressing chiasm neurons (Chung et al., 2001). Hs2st and/or Hs6st1 enzymes may normally sulfate SSEA-1 and/or CD44 and influence the responsiveness of the growth cone to HSPG-dependent guidance cues such as Slit2 (Piper et al., 2006) or fibroblast growth factor (Ornitz, 2000). Improper heparan sulfation of SSEA-1 and/or CD44 could contribute to the navigation errors we observe in Hs2st−/− and/or Hs6st1−/− mutants.
The axon navigation defects we see in Hs2st−/− and Hs6st1−/− embryos colocalize with regions of high Hs2st and/or Hs6st1 expression at the chiasm. The most obvious interpretation of this is that high levels of Hs2st and/or Hs6st1 are required at these locations to supply navigation cues to RGC axons. The chronotopic and retinotopic organization of the optic nerve means that younger axons navigate along the outside of the optic nerve as it approaches and enters the chiasm (Colello and Guillery, 1998; Leung et al., 2003), so cells surrounding the optic nerve and chiasm, in which we have detected high levels of Hs2st and/or Hs6st1 expression, are well placed to direct their growth. This is a plausible mechanism because HSPGs are known to bind to axon guidance molecules including Slit and netrin and so might be involved in tethering them at the chiasm in the correct configuration to direct RGC axons. The striking coincidence of gene expression domains between Hs2st and Hs6st1 (present study) and Slit-1 and Slit-2 (Erskine et al., 2000) (summarized in supplemental Fig. S1C, available at www.jneurosci.org as supplemental material) at the chiasm suggest a functional relationship between HSTs and Slits in supplying navigation cues at the chiasm. However, it is not that simple. Hs2st and/or Hs6st1 are expressed by RGCs so may have cell-autonomous functions in the navigating growth cone. Indeed, axons projected by Hs6st1−/− RGCs in vitro are less sensitive to repulsion by Slit2 than their wild-type counterparts (present study), indicating that Hs6st1 synthesized by RGCs themselves is a factor in determining the response of their growth cones to Slit2. The HSPG Glypican-1 is expressed on RGC growth cones (Karthikeyan et al., 1994) and binds Slit2 (Ronca et al., 2001) so is a potential substrate for direct HST action on HS sulfation status on the growth cone surface. It may well be that in vivo, both RGCs and chiasm cells synthesize Hs6st1 and/or Hs2st and that the patterns of heparan sulfation on the RGC growth cones and the cells they encounter at the chiasm are both important.
Normally on their way to the chiasm, RGC axons cross the boundary between the optic nerve and the brain and the vast majority then cross the midline, rather than growing up the ipsilateral optic tract. Once RGC axons have crossed the midline, these preferences are reversed and they grow into the contralateral optic tract and avoid the optic nerve. Furthermore, the nerve/brain boundary is permissive to RGC axons as they grow toward the chiasm but nonpermissive once they have passed through it. Alterations in growth cone responsiveness to the guidance molecules netrin-1 and Slits as axons cross the midline have been well documented in both vertebrate and invertebrate systems (for review, see Dickson, 2002). The results presented here represent the first evidence that changing responses of retinal growth cones to the chiasmatic environment might be dependent on specific patterns of HS sulfation. It appears that Hs6st1 sulfation directs growth permissive interactions between RGC axons and their extracellular environment at the optic nerve/brain boundary before they have crossed the ventral midline and/or repulsive interactions after they cross. Cultured Chinese hamster ovary cells secrete Hs6st (but not Hs2st) activity into the culture medium (Habuchi et al., 1995), indicating that, in this system at least, Hs6st can be exported from the cell, whereas Hs2st never leaves the Golgi apparatus. This raises the possibility that Hs6st1 is released into the extracellular space where it modifies HS on the surface of navigating growth cones and reprograms their response to the chiasmatic environment.
How does heparan sulfation direct retinal axons at the optic chiasm? It is possible that HSPGs function directly as a receptor and/or ligand or that they modulate the interactions between other receptor/ligands. HS can interact with Robo/Slit, deleted in colorectal cancer (DCC)/netrin-1, Plexin/Semaphorin, and Eph/Ephrin navigation systems (Kantor et al., 2004; Lee and Chien, 2004), and perturbation of one or more of these could underlie the HST mutant phenotypes. Robo and its ligand Slit are important for axon guidance at the developing chiasm (Erskine et al., 2000; Plump et al., 2002). Disruption to Robo/Slit (Plump et al., 2002) or HSPG (Inatani et al., 2003; present study) function causes RGC axons to grow down the contralateral optic nerve and/or escape from the chiasm, prompting speculation that Robo/Slit and HSPG may normally cooperate to prevent these guidance errors. Gene dosage experiments in mouse (Inatani et al., 2003) and zebrafish (Lee et al., 2004) show that HSPGs and Robo/Slit cooperate to prevent RGC axons innervating the contralateral retina, and it has been demonstrated recently that Xenopus retinal axons respond to Slit2 in vitro in an HSPG-dependent manner as they navigate from the thalamus to the tectum (Piper et al., 2006). Netrin-1 is expressed around the optic nerve as it approaches the chiasm and at the point at which the optic nerve contacts the brain (Deiner and Sretavan, 1999) and therefore coincides with domains of high Hs2st and Hs6st1 expression (present study). Mice in which netrin-1/DCC signaling is disrupted display axon guidance defects at the chiasm (Deiner and Stretavan, 1999; Bertuzzi et al., 1999). Altered response to netrin-1 in the optic nerve or at the optic chiasm could therefore account for axon navigation mistakes in Hs2st or Hs6st1 mutants. Overall, the evidence available at present suggests that navigating RGC growth cones use the HS sulfation pattern to interpret Robo/Slit guidance cues, and we demonstrated directly that Hs6st1 is required for growth cone sensitivity to Slit2. However, there are other possibilities, and we note that Lee et al. (2004) identified HSPG-dependent axon guidance behaviors that were independent of Robo/Slit signaling.
In conclusion, we showed that Hs2st and Hs6st1 are expressed by the RGCs themselves and in complex patterns in cells surrounding their growth cones as they approach and navigate the optic chiasm. Mutants lacking one or other of these genes show specific axon guidance defects at the optic chiasm. We also show that the ability of RGC axons to respond to Slit2 in vitro depends on Hs6st1 synthesized by the RGC. Our results support the hypothesis that differential HS sulfation is deployed by the developing visual system to guide RGC axons into the brain.
Footnotes
This work was supported by the Medical Research Council, Biotechnology and Biological Sciences Research Council, and Wellcome Trust. We are grateful to Elizabeth Martin, Charles Prior, Duncan Mcneil, Linda Wilson (Confocal Microscope Facility), Vivian Allison, Katherine Gillies, and the Biomedical Research Resources staff for their contribution to this work. We thank Lynda Erskine and Alain Chedotal for advice on retinal culture techniques and Slit expression constructs and Valerie Wilson for supplying the Hs6st1 mutant mouse line and for helpful discussions.
References
- Bertuzzi S, Hindges R, Mui SH, O'Leary DDM, Lemke G (1999). The homeodomain protein Vax1 is required for axon guidance and major tract formation in the developing forebrain. Genes Dev 13:3092–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TA, Bouchard T, St John T, Wayner E, Carter WG (1991). Human keratinocytes express a new CD44 core protein (CD44E) as a heparan-sulfate intrinsic membrane proteoglycan with additional exons. J Cell Biol 113:207–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock SL, Fletcher JM, Beddington RS, Wilson VA (1998). Renal agenesis in mice homozygous for a gene trap mutation in the gene encoding heparan sulfate 2-sulfotransferase. Genes Dev 12:1894–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulow HE, Hobert O (2004). Differential sulphations and epimerisation define heparan sulphate specificity in nervous system development. Neuron 41:723–736. [DOI] [PubMed] [Google Scholar]
- Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D (1996). Cell fate determination in the vertebrate retina. Proc Natl Acad Sci USA 93:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KY, Leung KM, Lin L, Chan SO (2001). Heparan sulfate proteoglycan expression in the optic chiasm of mouse embryos. J Comp Neurol 436:236–247. [DOI] [PubMed] [Google Scholar]
- Colello SJ, Guillery RW (1998). The changing pattern of fibre bundles that pass through the optic chiasm of mice. Eur J Neurosci 10:3653–3663. [DOI] [PubMed] [Google Scholar]
- Deiner MS, Sretavan DW (1999). Altered midline axon pathways and ectopic neurons in the developing hypothalamus of netrin-1- and DCC-deficient mice. J Neurosci 19:9900–9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson BJ (2002). Molecular mechanisms of axon guidance. Science 298:1959–1964. [DOI] [PubMed] [Google Scholar]
- Erskine L, Williams SE, Brose K, Kidd T, Rachel RA, Goodman CS, Tessier-Lavigne M, Mason CA (2000). Retinal ganglion cell axon guidance in the mouse optic chiasm: expression and function of robos and slits. J Neurosci 20:4975–4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esko JD, Selleck JD (2002). Order out of chaos: assembly of ligand binding sites in heparin sulphate. Annu Rev Biochem 71:435–471. [DOI] [PubMed] [Google Scholar]
- Greenfield B, Wang WC, Marquardt H, Piepkorn M, Wolff EA, Aruffo A, Bennett KL (1999). Characterization of the heparan sulfate and chondroitin sulfate assembly sites in CD44. J Biol Chem 274:2511–2517. [DOI] [PubMed] [Google Scholar]
- Habuchi H, Habuchi O, Kimata K (1995). Purification and characterisation of heparan sulphate 6-sulfotransferase from the culture medium of Chinese hamster ovary cells. J Biol Chem 270:4172–4179. [DOI] [PubMed] [Google Scholar]
- Herrera E, Brown L, Aruga J, Rachel RA, Dolen G, Mikoshiba K, Brown S, Mason CA (2003). Zic2 patterns binocular vision by specifying the uncrossed retinal projection. Cell 114:545–557. [DOI] [PubMed] [Google Scholar]
- Holt CE, Dickson BJ (2005). Sugar codes for axons? Neuron 46:169–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H (2001). Cell-surface heparan sulfate is involved in the repulsive guidance activities of Slit2 protein. Nat Neurosci 47:695–701. [DOI] [PubMed] [Google Scholar]
- Inatani M, Irie F, Plump AS, Tessier-Lavigne M, Yamaguchi Y (2003). Mammalian brain morphogenesis and midline axon guidance require heparan sulphate. Science 302:1044–1046. [DOI] [PubMed] [Google Scholar]
- Irie A, Yates EA, Turnbull JE, Holt CE (2002). Specific heparan sulfate structures involved in retinal axon targeting. Development 129:61–70. [DOI] [PubMed] [Google Scholar]
- Kantor DB, Chivatakarn O, Peer KL, Oster SF, Inatani M, Hansen MJ, Flanagan JG, Yamaguchi Y, Sretevan DW, Giger RJ, Kolodkin AL (2004). Semaphorin 5A is a bifunctional axon guidance cue regulated by heparan and chondroitin sulphate proteoglycans. Neuron 44:961–975. [DOI] [PubMed] [Google Scholar]
- Karthikeyan L, Flad M, Engel M, Meyer-Puttlitz B, Margolis RU, Margolis RK (1994). Immunocytochemical and in situ hybridization studies of the heparan sulfate proteoglycan, glypican, in nervous tissue. J Cell Sci 107:3213–3222. [DOI] [PubMed] [Google Scholar]
- Lee J-S, Chien C-B (2004). When sugars guide axons: insights from heparan sulphate proteoglycan mutants. Nat Rev Genet 5:923–935. [DOI] [PubMed] [Google Scholar]
- Lee J-S, von der Hardt S, Rusch MA, Stringer SE, Stickney HL, Talbot WS, Geisler R, Nusslein-Volhard C, Selleck SB, Chien C-B, Roehl H (2004). Axon sorting in the optic tract requires HSPG synthesis by ext2 (dackel) and extl3 (boxer). Neuron 44:947–960. [DOI] [PubMed] [Google Scholar]
- Leighton PA, Mitchell KJ, Goodrich LV, Lu X, Pinson K, Scherz P, Skarnes WC, Tessier-Lavigne M (2001). Defining brain wiring patterns and mechanisms through gene trapping in mice. Nature 410:174–179. [DOI] [PubMed] [Google Scholar]
- Leung K-M, Taylor JSH, Chan S-O (2003). Enzymatic removal of chondroitin sulphates abolishes the age-related axon order in the optic tract of mouse embryos. Eur J Neurosci 17:1755–1767. [DOI] [PubMed] [Google Scholar]
- Lin L, Chan SO (2003). Perturbation of CD44 function affects chiasmatic routing of retinal axons in brain slice preparations of the mouse retinofugal pathway. Eur J Neurosci 17:2299–2312. [DOI] [PubMed] [Google Scholar]
- Lin L, Cheung AW, Chan SO (2005). Chiasmatic neurons in the ventral diencephalon of mouse embryos–changes in arrangement and heterogeneity in surface antigen expression. Brain Res Dev Brain Res 158:1–12. [DOI] [PubMed] [Google Scholar]
- Marcus RC, Mason CA (1995). The first retinal axon growth in the mouse optic chiasm: axon patterning and the cellular environment. J Neurosci 15:6389–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus RC, Blazeski R, Godement P, Mason CA (1995). Retinal axon divergence in the optic chiasm: uncrossed axons diverge from crossed axons within a midline glial specialization. J Neurosci 15:3716–3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus RC, Shimamura K, Sretavan D, Lai E, Rubenstein JL, Mason CA (1999). Domains of regulatory gene expression and the developing optic chiasm: correspondence with retinal axon paths and candidate signaling cells. J Comp Neurol 403:346–358. [PubMed] [Google Scholar]
- McLaughlin D, Karlsson F, Tian N, Pratt T, Bullock SL, Wilson VA, Price DJ, Mason JO (2003). Specific modification of heparan sulphate is required for normal cerebral cortical development. Mech Dev 120:1481–1488. [DOI] [PubMed] [Google Scholar]
- Mitchell KJ, Pinson KI, Kelly OG, Brennan J, Zupicich J, Scherz P, Leighton PA, Goodrich LV, Lu X, Avery BJ, Tate P, Dill K, Pangilinan E, Wakenight P, Tessier-Lavigne M, Skarnes WC (2001). Functional analysis of secreted and transmembrane proteins critical to mouse development. Nat Genet 28:241–249. [DOI] [PubMed] [Google Scholar]
- Ornitz DM (2000). FGFs, heparan sulfate and FGFRs: complex interactions essential for development. BioEssays 22:108–112. [DOI] [PubMed] [Google Scholar]
- Piepkorn M, Hovingh P, Bennett KL, Aruffo A, Linker A (1997). Chondroitin sulphate composition and structure in alternatively spliced CD44 fusion proteins. Biochem J 327:499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M, Anderson R, Dwivedy A, Weinl C, van Horck F, Leung KM, Cogill E, Holt C (2006). Signaling mechanisms underlying slit2-induced collapse of Xenopus retinal growth cones. Neuron 49:215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plump AS, Erskine L, Sabatier C, Brose K, Epstein CJ, Goodman CS, Mason CA, Tessier-Lavigne M (2002). Slit1 and Slit2 cooperate to prevent premature midline crossing of retinal axons in the mouse visual system. Neuron 33:219–232. [DOI] [PubMed] [Google Scholar]
- Pratt T, Price DJ (2006). Dual roles of transcription factors in forebrain morphogenesis and development of axonal pathways. In: Development and plasticity in sensory thalamus and cortex (Guido W, Molnar Z, eds) in press New York: Kluwer Academic/ Plenum.
- Pratt T, Vitalis T, Warren N, Edgar JM, Mason JO, Price DJ (2000). A role for Pax6 in the normal development of dorsal thalamus and its cortical connections. Development 127:5167–5178. [DOI] [PubMed] [Google Scholar]
- Pratt T, Tian NMM-L, Simpson TI, Mason JO, Price DJ (2004). The winged helix transcription factor Foxg1 facilitates retinal ganglion cell axon crossing of the ventral midline in the mouse. Development 2004 131:3773–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronca F, Andersen JS, Paech V, Margolis RU (2001). Characterization of Slit protein interactions with glypican-1. J Biol Chem 76:29141–29291. [DOI] [PubMed] [Google Scholar]
- Runko E, Kaprielian Z (2002). Expression of Vema in the developing mouse spinal cord and optic chiasm. J Comp Neurol 451:289–299. [DOI] [PubMed] [Google Scholar]
- Sedita J, Izvolsky K, Cardoso WV (2004). Differential expression of heparan sulfate 6-O-sulfotransferase isoforms in the mouse embryo suggests distinctive roles during organogenesis. Dev Dyn 231:782–794. [DOI] [PubMed] [Google Scholar]
- Sretavan DW, Feng L, Pure E, Reichardt LF (1994). Embryonic neurons of the developing optic chiasm express L1 and CD44, cell surface molecules with opposing effects on retinal axon growth. Neuron 12:957–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sretavan DW, Pure E, Siegel MW, Reichardt LF (1995). Disruption of retinal axon ingrowth by ablation of embryonic mouse optic chiasm neurons. Science 269:98–101. [DOI] [PubMed] [Google Scholar]
- Walz A, McFarlane S, Brickman YG, Nurcombe V, Bartlett PF, Holt CE (1997). Essential role of heparan sulfates in axon navigation and targeting in the developing visual system. Development 124:2421–2430. [DOI] [PubMed] [Google Scholar]
- Williams SE, Mann F, Erskine L, Sakurai T, Wei S, Rossi DJ, Gale NW, Holt CE, Mason CA, Henkemeyer M (2003). Ephrin-B2 and EphB1 mediate retinal axon divergence at the optic chiasm. Neuron 39:919–935. [DOI] [PubMed] [Google Scholar]