Abstract
MAM (meprin/A5 protein/receptor protein tyrosine phosphatase mu) domain glycosylphosphatidylinositol anchor 1 (MDGA1), a unique cell surface glycoprotein, is similar to Ig-containing cell adhesion molecules that influence neuronal migration and process outgrowth. We show in postnatal mice that MDGA1 is expressed by layer 2/3 neurons throughout the neocortex. During development, MDGA1 is expressed in patterns consistent with its expression by migrating layer 2/3 neurons, suggesting a role for MDGA1 in controlling their migration and settling in the superficial cortical plate. To test this hypothesis, we performed loss-of-function studies using RNA interference (RNAi) targeting different sequences of mouse MDGA1. RNAi or empty vectors were coelectroporated with an enhanced green fluorescent protein reporter in utero into the lateral ventricle at embryonic day 15.5 to transfect progenitors of superficial layer neurons; the distributions of transfected neurons were analyzed late on postnatal day 0. We found a direct correlation between effectiveness of an RNAi in suppressing MDGA1 expression and disrupting migration of superficial layer neurons. An RNAi with no effect on MDGA1 expression has no effect on the migration. In contrast, an RNAi that suppresses MDGA1 expression also blocks proper migration of transfected superficial layer neurons, with essentially all transfected cells found deep in the cortical plate or beneath it. This migration defect is rescued by cotransfection of a rat MDGA1 expression construct along with the effective RNAi, confirming that the RNAi effect is specific to diminishing mouse MDGA1 expression. RNAi transfections of deep layer neurons that do not express MDGA1 do not significantly affect their migration. We conclude that MDGA1 acts cell autonomously to control the migration of MDGA1-expressing superficial layer cortical neurons.
Keywords: cell adhesion molecules, cortical development, cortical lamination, Ig superfamily, in utero electroporation, RNAi
Introduction
The mammalian neocortex is distinguished from other regions of the cerebral cortex by its six major layers. Each layer is comprised of a heterogeneous population of neurons broadly classified into two general types: glutamatergic, including all projection neurons, and GABAergic interneurons. Most GABAergic interneurons are generated within the medial and caudal ganglionic eminences and migrate tangentially in the intermediate zone (IZ) or marginal zone (MZ) until they reach their cortical destination, whereupon they turn and migrate radially into the cortical plate (CP) (Marin and Rubenstein, 2003; Kriegstein and Noctor, 2004). Glutamatergic neurons are generated by progenitors within the ventricular zone (VZ) and subventricular zone (SVZ) of dorsal telencephalon and migrate radially along processes of radial glia into the overlying CP in an “inside-out” pattern [i.e., early-born neurons form deeper layers and later-born neurons migrate past them to form more superficial layers (Rakic, 1972; Kriegstein and Noctor, 2004)].
Molecules required for radial migration and inside-out cortical lamination include reelin, a protein secreted by Cajal–Retzius neurons of the MZ, its receptors, the very low-density lipoprotein receptor/apolipoprotein E receptor type 2, and downstream components of this signaling pathway that influence migration in part through regulating cytoskeletal proteins, including doublecortin (Bielas et al., 2004; Tsai and Gleeson, 2005). The POU domain transcription factors Brn-1 and Brn-2 also have roles in patterning of superficial layers (McEvilly et al., 2002; Sugitani et al., 2002). Cell adhesion molecules (CAMs) play important roles in the interactions of migrating neurons with glial processes (Zheng et al., 1996; Anton et al., 1999). Two CAMs involved in neuronal migration are astrotactin (Adams et al., 2002) and the integrin α3β1, which to date is the only CAM reported to control cortical radial migration (Anton et al., 1999; Dulabon et al., 2000).
Here, we report a role in cortical radial migration for a unique Ig superfamily protein, meprin/A5 protein/receptor protein tyrosine phosphatase mu (MAM) domain glycosylphosphatidylinositol anchor 1 (MDGA1), that we originally cloned in the rat (Litwack et al., 2004). MDGA1, an IgCAM anchored to the extracellular surface of the cell membrane by a GPI-linkage, contains six Ig domains, a fibronectin III domain, and uniquely for IgCAMs, a MAM domain (Litwack et al., 2004). MDGA1 is structurally similar to other IgCAMs, such as the L1 family and axonin 1, which have roles in cell adhesion, migration, and process outgrowth (Walsh and Doherty, 1997; Panicker et al., 2003). We cloned full-length mouse MDGA1 and show that it is expressed in neocortex by layer 2/3 neurons throughout their development. We hypothesize that MDGA1 is involved in controlling the migration and lamination of layer 2/3 neurons. We address this issue using RNA interference (RNAi) combined with in utero electroporation in mice, an approach recently used to reveal a function for doublecortin in radial migration of cortical neurons (Bai et al., 2003).
Materials and Methods
Animals.
C57BL/6 mice were used to examine MDGA1 expression and to identify its cDNA sequence. Timed pregnant ICR mice were used for in utero electroporation. The morning of the vaginal plug is embryonic day 0.5 (E0.5), and the first postnatal day (P) is P0. Experiments were done following institutional guidelines and approved protocols.
Microscopy.
Digital images were obtained with a Zeiss (Thornwood, NY) LCM510 confocal microscope or a Retiga digital camera on a Nikon (Tokyo, Japan) Microphot microscope with OpenLab software (Improvision, Conventry, UK).
In situ hybridization.
Radioactive in situ hybridization of MDGA1 was done on wild-type C57BL/6 mouse embryos and neonates from E9.5 to P7 as described previously (Litwack et al., 2004).
Vectors and in utero electroporation.
Four different targeted regions that only match with MDGA1 cDNA sequence were selected to make H1 promoter-based RNAi vectors (pSUPER) (Brummelkamp et al., 2002). The four 19-nucleotide target sequences are as follows: M1, GGAGGATAACATCAGCGAG; M2, GTCTATCCGCGTGGACGTG; M5, CATTTCCTCAGAGACAGTA; M6, CGTACGACCCCGGGAGGCT. To evaluate their efficiency, each RNAi vector, or as a control an empty pSUPER (‘E1′) vector, was cotransfected with a Myc-tagged MDGA1 expressing vector in pcDNA3.1 (−)/Myc-His B (Invitrogen, San Diego, CA) (RNAi, 4.0 μg; MDGA1-Myc, 1.0 μg) into COS-7 cells using PolyFect Transfection Reagent (Qiagen, Hilden, Germany). After 24 h, expression of MDGA1-Myc protein was visualized with anti-Myc mouse monoclonal antibody (9E10) and a fluorescent secondary. Quantification of labeling was done using NIH Image and Adobe Photoshop (Adobe Systems, San Jose, CA) as described previously (Pak et al., 2004). To determine relative MDGA1 protein levels, total pixel intensity of fluorescence for each RNAi vector was expressed as a percentage of that obtained with the E1 control vector (normalized as 100%). The number of MDGA1-expressing cells was also counted and indicated as a corrected absolute number. Immunolabeled cells were marked using NIH Image. Counts were done with thresholds set at four different pixel intensity levels (>50, >100, >150, >200; software maximum is 256). Experiments were done five times using the same conditions, normalized between each experiment.
Plasmids used for in utero electroporation were prepared using the EndoFree Plasmid kit (Qiagen). To fluorescently label cells transfected in vivo with RNAi, we cotransfected a pCAG-enhanced green fluorescent protein (eGFP) vector (Saito and Nakatsuji, 2001). One to 2 μl of plasmid (molecular ratio for pCAG-GFP and RNAi vectors, 1:3; final concentration of mixed DNA, 1.5 μg/μl) with Fast Green (0.01%; Sigma, St. Louis, MO) in PBS were microinjected using fine-tipped glass capillaries into the right lateral ventricle of each embryo. For the “rescue” experiments, the molecular ratio for the M1 RNAi, pCMV empty vector or pCMV-rMDGA1, and pCAG-GFP is 3:2:1, and the final concentration of mixed DNA is 1.5 μg/μl. Electroporation was done by five pulses at 30 V discharge with a duration of 50 ms at 950 ms intervals. Brains were fixed and coronally sectioned either 24 h later to confirm efficiency and restriction of electroporations or late on P0 for analyses.
Results
A single full-length mouse MDGA1 cDNA was isolated by reverse transcription-PCR from a cDNA library of P7 mouse basilar pons. Homologies in the coding region of cDNA and protein sequences (supplemental Fig. S1, available at www.jneurosci.org as supplemental material) are 94 and 93% between mouse and rat, respectively, and 89 and 92% between mouse and human, respectively.
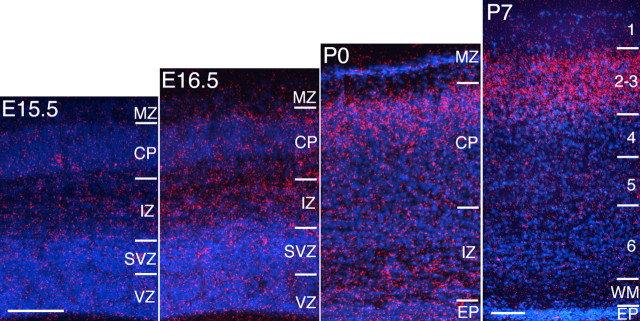
In situ hybridization reveals a weak transient expression of MDGA1 early in cortical development, from E9.5 to E13.5, limited to the MZ/preplate (data not shown). At E15.5 and later, MDGA1 expression is no longer detected in the MZ (Fig. 1). Signal above background is not detected at any age in the VZ/SVZ. At E16.5, modest expression is evident in the IZ. By P0, a band of MDGA1 expression is clearly evident in the superficial CP (the dense CP that will differentiate into layers 2/3); in addition, scattered cells deeper in the CP and IZ express MDGA1. By P7, expression of MDGA1 is limited to layers 2/3 throughout most of the cortex (Fig. 1) but is not detected in adults (data not shown). Layer 2/3 neurons are generated from E15 through E17, with peak generation on E16 (Takahashi et al., 1999); a substantial proportion reach the superficial CP by birth (Bayer and Altman, 1991). Thus, these data indicate that MDGA1 is expressed by layer 2/3 neurons during their migration and settling in the CP.
Figure 1.
Laminar expression of MDGA1 in the developing cortex. In situ hybridization of MDGA1 expression in the developing mouse cortex is shown. At E16.5, MDGA1 is modestly expressed in the IZ. At P0, expression is robust in the CP, and dense clusters of silver grains (red) deeper in the CP and IZ suggest MDGA1 expression by migrating neurons. By P7, robust expression is localized to layers 2/3. EP, Ependymal layer; WM, white matter. Scale bars, 100 μm; bar in E15.5 is E15.5 to P0.
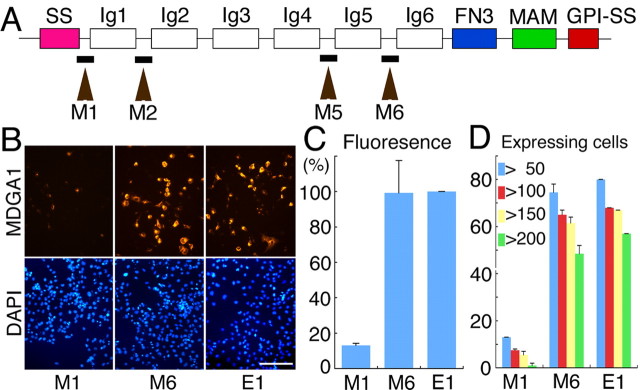
To investigate a role for MDGA1 in the migration of superficial layer neurons, we adopted small interference RNA (RNAi) to suppress MDGA1 expression using a pSUPER vector to produce short hairpin RNAs (Brummelkamp et al., 2002) by in utero electroporation in mice. Four RNAi vectors were made with unique nucleotide sequences of inter-Ig domains specific to mouse MDGA1 (Fig. 2A). The RNAi vectors vary in their effectiveness in suppressing MDGA1 protein expression, shown by cotransfecting each vector and a myc-tagged MDGA1 expression construct into COS-7 cells (Fig. 2B). Quantitation of relative levels of MDGA1 protein or number of MDGA1-expressing cells revealed by fluorescence immunostaining for the myc-tag shows that the M1 RNAi vector is very effective in suppressing MDGA1 expression, whereas the M6 RNAi vector has no detectable effect and is indistinguishable from the empty pSUPER (‘E1′) vector (Fig. 2C,D).
Figure 2.
Targeted sequences for RNAi vectors differentially suppress MDGA1 protein expression in vitro. A, MDGA1 domain structure and selected target regions to make RNAi vectors; the four target regions (M1, M2, M5, M6) were 19 bp sequences located between the Ig domains. SS, Signal peptide sequence; FN3, fibronectin type 3 repeat; MAM, MAM domain; GPI, glycosylphosphatidylinositol anchor. B, COS-7 cells cotransfected with a pcDNA3.1 vector containing MDGA1 cDNA tagged with a Myc epitope and an RNAi pSUPER vector or en empty pSUPER vector (E1); RNAi vector name (M1, M6) denotes sequence location shown in A. Shown is immunofluorescence using a Myc antibody 24 h after transfection; the secondary antibody was conjugated to Alexa568 (orange). Cultures are DAPI stained (blue). Scale bar, 100 μm C, Histogram of fluorescence intensity of MDGA1-Myc protein. D, Histogram of the number of MDGA1-positive cells with a total pixel intensity above the indicated thresholds. Quantitative data for M2 is statistically indistinguishable from that shown in C and D for M1 and M5 from that shown for M6.
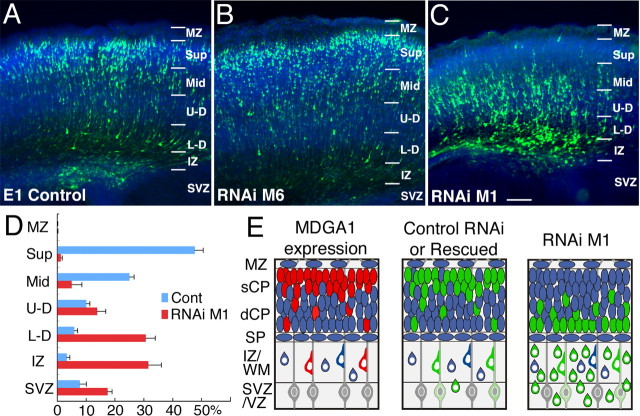
For our in vivo studies, we used the M1 RNAi vector to suppress MDGA1 expression, and both the E1 empty vector and the M6 RNAi vector as controls for potential nonspecific effects of RNAi and vector expression. We performed in utero electroporations of the vectors in E15.5 embryos; an eGFP vector was coelectroporated to mark cells transfected with the RNAi or E1 vectors (Fig. 3A). Because of the timing of these transfections and the dynamics of transgene expression, the majority of the GFP-marked cells should be layer 2/3 neurons, although a very small proportion of layer 4 neurons are generated after E15.5 and therefore could be transfected (Takahashi et al., 1999). Embryos examined 1 d later show discrete transfection domains of GFP-labeled cells within the VZ and SVZ and restricted to dorsal telencephalon (data not shown). To determine the effect of the transfections on neuronal migration, electroporated mice were perfused late on P0, when a substantial proportion of layer 2/3 neurons have reached the superficial CP; only healthy appearing nursing pups were fixed. Twenty-three of the 26 control transfected brains (coelectroporated with GFP, and either M6 RNAi or E1 vectors) and 31 of the 33 M1/GFP cotransfected brains had cortical GFP signal visible in whole mounts fixed at P0.
Figure 3.
Transfection of M1 RNAi to suppress MDGA1 selectively results in aberrant deep distribution of superficial layer neurons. Distribution of GFP-labeled neurons late on P0 following cotransfections on E15.5 with a GFP and E1 vector (A), GFP and M6 RNAi vector (B), and GFP and M1 RNAi vector (C). D, Five M1 cases and five control cases (4 E1 and 1 M6) were quantitatively analyzed. The cortical wall was subdivided as indicated, and numbers of GFP-labeled transfected cells in each layer were counted and expressed as a percentage of the total. Statistical analyses comparing the number of labeled cells in each layer between control and M1 transfected cases (ANOVA, Bonferroni’s test) are shown. Statistical difference is seen for each set of layers (p < 0.01) except MZ and U-D layers. Scale bars, 100 μm. E, Schematics of late P0 cortex showing an expression pattern of MDGA1 and distribution of GFP-labeled neurons transfected with M1 RNAi or control (M6 RNAi, E1) vectors at E15.5. Expression of MDGA1 is similar to distribution of GFP-labeled neurons transfected with control vectors or in rescue experiments (cotransfections with M1 RNAi and rat MDGA1 cDNA vectors). However, distribution of GFP-labeled neurons transfected with an M1 RNAi vector is aberrantly displaced deep. sCP, Superficial cortical plate; dCP, deep cortical plate; Sup, superficial CP; Mid, middle CP; U-D, upper-deep CP; L-D, lower-deep CP; others are as in Figure 1.
Brains with similar transfection domains were sectioned: these included 10 control transfected brains (five each of M6 and E1 vectors) and 12 M1 transfected brains. No morphological defects are evident in these transfected brains, and cortical architecture appears normal in 4′,6′-diamidino-2-phenylindole (DAPI)-stained sections. Essentially all of the GFP-labeled cells have a pyramidal-like morphology characteristic of layer 2/3 neurons and immunostain for the neuronal marker TuJ1 (class III β-tubulin) (supplemental Fig. S2, available at www.jneurosci.org as supplemental material); however, their laminar distribution differs substantially between brains electroporated with the control vectors (M6 RNAi or E1 empty) versus the M1 RNAi vector. In brains transfected with either the E1 (Fig. 3A) or M6 vector (Fig. 3B), the majority of GFP-labeled cells are in the superficial layers in a distribution that closely resembles the distribution of MDGA1-expressing cells in nontransfected P0 cortex (Fig. 1). In contrast, in brains transfected with the M1 vector, the majority of GFP-labeled cells are deep in the CP or beneath it in the IZ or deeper (Fig. 3C).
The laminar (i.e., radial) distributions of GFP-labeled cells were quantified at late P0 from sections of electroporated brains with very similar transfection domains (five brains electroporated with the M1 RNAi vector and five with control vectors: one M6 RNAi and four E1 empty vectors). In the controls, the mean number of GFP-labeled cells is 370 ± 29 (total of 1851; range, 270–448); the M1 cases have a mean of 268 ± 30 labeled cells (total of 1342; range, 196–364).
In control transfected brains, 74% of GFP-labeled cells are in the upper half of the CP, with 49% in the superficial most quarter of the CP (the dense CP that will become layers 2/3) (Fig. 3D). In contrast, in M1 transfected brains, only 6% of GFP-labeled cells are in the upper half of the CP, with virtually no labeled cells (1%) in the superficial most quarter of the CP; 80% of all labeled cells are in the deepest one-quarter of the CP or beneath it (Fig. 3D). The difference in the distribution of GFP-labeled cells between control and M1 transfected brains is statistically significant for each “layer,” with the exception of the MZ, which contains virtually no labeled cells, and the “upper-deep” layer of the CP, which is the layer where the transition occurs in the superficial versus deep bias in the distribution of transfected cells between control and M1 electroporated brains (Fig. 3D). Thus, transfection with the M1 RNAi vector in migrating layer 2/3 neurons inverts their distribution from superficial to deep (Fig. 3D).
As described in Figure 2, the M6 vector has no detectable effect on expression of MDGA1 in vitro, whereas the M1 vector substantially suppresses it (Fig. 2). We also analyzed the effect of these RNAi vectors on MDGA1 expression in vivo and obtained similar findings; the M6 vector has no detectable effect on MDGA1 expression (n ; 3) (Fig. 4A,C), whereas MDGA1 expression is diminished in correlation with domains of M1 transfections (n ; 2) (Fig. 4B,D). Thus, both the in vitro and in vivo data indicate that the M1 RNAi vector selectively diminishes MDGA1 expression, whereas the M6 RNAi vector does not.
Figure 4.
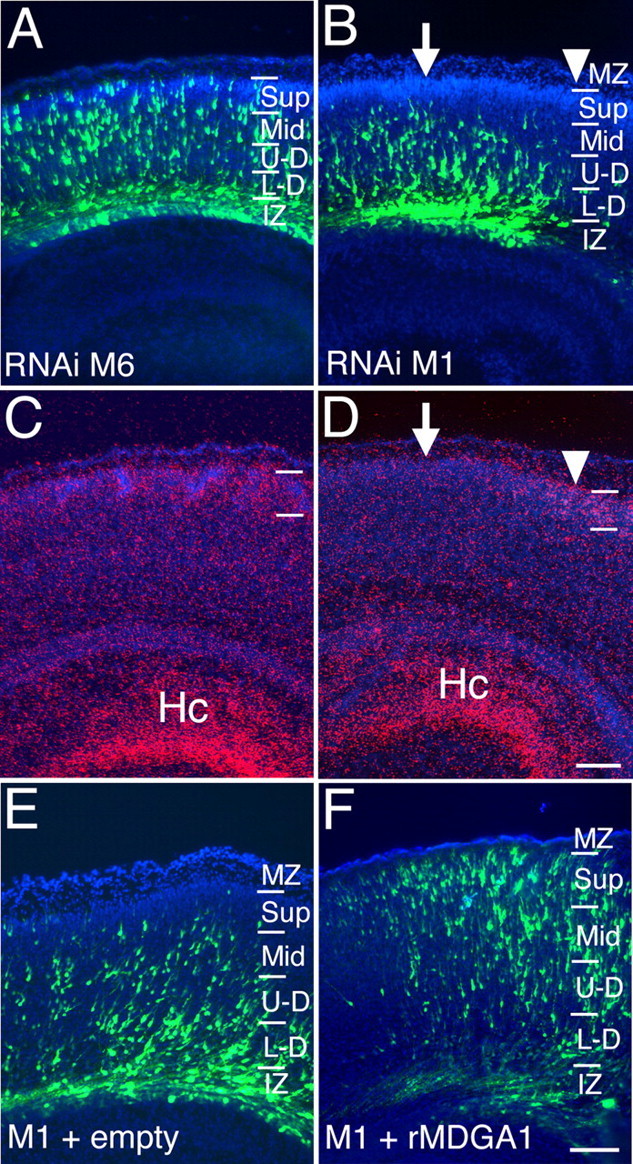
Differential suppression of MDGA1 expression in vivo by mouse RNAi vectors with rescue of RNAi-induced migration defect by cotransfection of rat MDGA1. A–D, Distribution of GFP-labeled neurons late on P0 following cotransfection with GFP and M6 (A) or GFP and M1 RNAi vectors at E15.5 (B). In situ hybridization using S35-labeled riboprobes for MDGA1 (C, D) on adjacent sections of A and B, respectively. M6 RNAi transfection has no detectable effect on MDGA1 expression. However, MDGA1 expression is diminished in superficial layers above the domain of M1 RNAi transfection (arrow) but not adjacent to it (arrowhead). E, F, Distribution of radially migrating neurons cotransfected with M1 RNAi plus empty pCMV plus GFP vectors (control) (E) and M1 RNAi plus pCMV rat-MDGA1 (rMDGA1) plus GFP vectors (F). M1 RNAi target sequence (GGAGGATAACATCAGCGAG) and rat MDGA1 cDNA (-GGAGGATAATATCAGCGAG-) have a one nucleotide mismatch (bolded). In control cases (E), most GFP-labeled cells are deep, and virtually none are in the superficial CP. As shown in F, cotransfection with rat MDGA1 expression vector essentially corrects the migration defect and shifts distribution of GFP-labeled cells compared with D, with many in superficial CP. Scale bars, 100 μm. Hc, Hippocampal formation; other abbreviations are as in Figure 1.
To corroborate further the specificity of the M1 RNAi vector for selectively suppressing MDGA1, resulting in aberrant migration of MDGA1 expressing layer 2/3 neurons, we did two complementary experiments. In the first set, we performed a “rescue” experiment in which we repeated the same M1/GFP transfections at E15.5 but cotransfected with either a rat MDGA1 cDNA expression construct (n ; 2) or as a control with an empty expression vector (n ; 2). Importantly, the M1 vector against mouse MDGA1 has only a one nucleotide mismatch with the same sequence stretch in the rat MDGA1 (Fig. 4F). When analyzed late on P0, the distribution of GFP-labeled cells in cases cotransfected with the M1 RNAi and control expression vectors are qualitatively the same as in cases transfected with only the M1 RNAi vector (i.e., the cells exhibit a migration defect and are distributed deep to their normal positions) (Fig. 4E). In contrast, in cases cotransfected with the M1 RNAi and rat MDGA1 expression vectors, a substantial proportion of the GFP marked cells are positioned superficially in the CP (Fig. 4F). Thus, overexpression of rat MDGA1 in cells transfected with the M1 RNAi mostly rescues the migration defect. In the second set of experiments, we show that transfections at E12.5 of the M1 RNAi vector into deep layer neurons that do not express MDGA1 does not disrupt their migration (supplemental Fig. S3, available at www.jneurosci.org as supplemental material). Together, these findings demonstrate the specificity of the M1 RNAi for mouse MDGA1 and indicate that the defective migration of layer 2/3 neurons transfected with M1 RNAi is specific to their diminished expression of MDGA1.
Discussion
We show that MDGA1, a unique IgCAM, is expressed in mouse neocortex by layer 2/3 neurons during their migration and settling superficially in the CP. We go on to show, using RNAi and in utero electroporation, that suppression of MDGA1 expression in migrating layer 2/3 neurons disrupts their migration, and over the period of our analyses prevents the transfected neurons from reaching the superficial CP (Fig. 3E).
To generate the RNAi vectors, we chose stretches of cDNA between the Ig domains, because they have the least homology to other genes. These RNAi vectors have varying effectiveness in suppressing MDGA1 expression. Importantly, we found a direct correlation between the effectiveness of an RNAi vector in suppressing MDGA1 expression, both in vitro and in vivo, and its ability to disrupt in vivo the migration of layer 2/3 neurons: the M1 RNAi vector blocks MDGA1 expression and also blocks proper migration of layer 2/3 neurons, whereas the M6 RNAi vector has no evident effect on MDGA1 expression nor on migration.
These findings argue for the specificity of the effect of the M1 RNAi vector, which we also directly demonstrated by rescuing the M1 RNAi induced migration defect by cotransfecting a rat MDGA1 expression vector with the M1 RNAi vector (Fig. 3E). The mouse MDGA1 sequence against which the M1 RNAi is directed has only a single nucleotide difference from the equivalent sequence of rat MDGA1, which strongly argues for the specificity of the M1 RNAi for mouse MDGA1. Other findings also strongly imply that the observed migration defect is caused by the suppression of MDGA1 in the migrating neurons themselves. First, MDGA1 is expressed by layer 2/3 neurons as they are migrating and forming the superficial layer of the CP but is not expressed by VZ/SVZ cells, and therefore not by radial glia that guide migrating neurons to the CP (Rakic, 1972). In addition, at the developmental stages at which we have performed the in utero electroporations, MDGA1 is not expressed by cells in the MZ and therefore not by Cajal–Retzius neurons that express reelin, a protein required for proper radial migration of cortical neurons and CP lamination (D’Arcangelo et al., 1995). Essentially no cells in the MZ are transfected and therefore could not be influenced by the vectors. Additionally, transfections done with the E1 control and M1 RNAi vectors at E12.5, coincident with the generation of deep layer neurons that do not express MDGA1, do not appear to affect the migration of the transfected neurons (supplemental Fig. S3, available at www.jneurosci.org as supplemental material). Therefore, we conclude that the aberrant migration of superficial layer neurons transfected with the M1 RNAi vector is caused by a cell autonomous effect of the suppression of MDGA1 protein.
Our findings indicate a critical role of MDGA1 in the proper migration of superficial layer cortical neurons that normally express it. RNAi suppression of MDGA1 expression results in a failure of superficial layer neurons to reach even the upper half of the CP, and essentially all transfected neurons are very deep in the CP or beneath it (Fig. 3E). This aberrant deep distribution of superficial layer neurons could be attributable to a dramatic slowing in their migration or to a defect in their final laminar distribution. A related issue is whether MDGA1 is required for the formation of layer 2/3. However, these issues are difficult to address using in utero electroporation of RNAi vectors because of the transient suppression of expression and the difficulty in transfecting a sufficient proportion of layer 2/3 neurons.
MDGA1 is also expressed by a select number of other neuronal populations as they migrate, for example basilar pontine neurons and D1 spinal interneurons (Litwack et al., 2004), and by inference might be involved in controlling their tangential migration. Our findings minimize the risk and provide a justification for making the investments to generate additional reagents and genetically engineered mice necessary to advance the work presented here and to explore more broadly the role of MDGA1 in neuronal migration and other aspects of neural development.
Footnotes
This work was supported by the National Institutes of Health (D.D.M.O.) and a fellowship from Uehara Memorial Foundation (A.T.). We thank David Litwack for early input and Carlos Garcia for comments. None of the authors has a financial interest related to this work.
References
- Adams NC, Tomoda T, Cooper M, Dietz G, Hatten ME (2002). Mice that lack astrotactin have slowed neuronal migration. Development 129:965–972. [DOI] [PubMed] [Google Scholar]
- Anton ES, Kreidberg JA, Rakic P (1999). Distinct functions of alpha3 and alpha(v) integrin receptors in neuronal migration and laminar organization of the cerebral cortex. Neuron 22:277–289. [DOI] [PubMed] [Google Scholar]
- Bai J, Ramos RL, Ackman JB, Thomas AM, Lee RV, LoTurco JJ (2003). RNAi reveals doublecortin is required for radial migration in rat neocortex. Nat Neurosci 6:1277–1283. [DOI] [PubMed] [Google Scholar]
- Bayer S, Altman J (1991). In: Neocortical development New York: Raven.
- Bielas S, Higginbotham H, Koizumi H, Tanaka T, Gleeson JG (2004). Cortical neuronal migration mutants suggest separate but intersecting pathways. Annu Rev Cell Dev Biol 20:593–618. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R (2002). A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550–553. [DOI] [PubMed] [Google Scholar]
- D’Arcangelo G, Miao GG, Chen SC, Soares HD, Morgan JI, Curran T (1995). A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature 374:719–723. [DOI] [PubMed] [Google Scholar]
- Dulabon L, Olson EC, Taglienti MG, Eisenhuth S, McGrath B, Walsh CA, Kreidberg JA, Anton ES (2000). Reelin binds alpha3beta1 integrin and inhibits neuronal migration. Neuron 27:33–44. [DOI] [PubMed] [Google Scholar]
- Kriegstein AR, Noctor SC (2004). Patterns of neuronal migration in the embryonic cortex. Trends Neurosci 27:392–399. [DOI] [PubMed] [Google Scholar]
- Litwack ED, Babey R, Buser R, Gesemann M, O’Leary DDM (2004). Identification and characterization of two novel brain-derived immunoglobulin superfamily members with a unique structural organization. Mol Cell Neurosci 25:263–274. [DOI] [PubMed] [Google Scholar]
- Marin O, Rubenstein JL (2003). Cell migration in the forebrain. Annu Rev Neurosci 26:441–483. [DOI] [PubMed] [Google Scholar]
- McEvilly RJ, de Diaz MO, Schonemann MD, Hooshmand F, Rosenfeld MG (2002). Transcriptional regulation of cortical neuron migration by POU domain factors. Science 295:1528–1532. [DOI] [PubMed] [Google Scholar]
- Pak W, Hindges R, Lim YS, Pfaff SL, O’Leary DDM (2004). Magnitude of binocular vision controlled by islet-2 repression of a genetic program that specifies laterality of retinal axon pathfinding. Cell 119:567–578. [DOI] [PubMed] [Google Scholar]
- Panicker AK, Buhusi M, Thelen K, Maness PF (2003). Cellular signalling mechanisms of neural cell adhesion molecules. Front Biosci 8:d900–d911. [DOI] [PubMed] [Google Scholar]
- Rakic P (1972). Mode of cell migration to the superficial layers of fetal monkey neocortex. J Comp Neurol 145:61–83. [DOI] [PubMed] [Google Scholar]
- Saito T, Nakatsuji N (2001). Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev Biol 240:237–246. [DOI] [PubMed] [Google Scholar]
- Sugitani Y, Nakai S, Minowa O, Nishi M, Jishage K, Kawano H, Mori K, Ogawa M, Noda T (2002). Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev 16:1760–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Goto T, Miyama S, Nowakowski RS, Caviness VS Jr (1999). Sequence of neuron origin and neocortical laminar fate: relation to cell cycle of origin in the developing murine cerebral wall. J Neurosci 19:10357–10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai LH, Gleeson JG (2005). Nucleokinesis in neuronal migration. Neuron 46:383–388. [DOI] [PubMed] [Google Scholar]
- Walsh FS, Doherty P (1997). Neural cell adhesion molecules of the immunoglobulin superfamily: role in axon growth and guidance. Annu Rev Cell Dev Biol 13:425–456. [DOI] [PubMed] [Google Scholar]
- Zheng C, Heintz N, Hatten ME (1996). CNS gene encoding astrotactin, which supports neuronal migration along glial fibers. Science 272:417–419. [DOI] [PubMed] [Google Scholar]