Abstract
NR3A is expressed widely in the developing CNS of mammals. Coassembly of NR3A with NR1 and NR2 modifies NMDA receptor-mediated responses, reducing calcium permeability and single-channel conductance. The ligand binding properties of NR3A are unknown but shape the role NR3A plays when incorporated into NMDA receptors. Here, a soluble NR3A ligand binding domain (NR3A S1S2) was constructed based on amino acid sequence alignments with other glutamate receptor ion channels and is expressed in Escherichia coli. After purification by affinity, gel filtration, and ion exchange chromatography, NR3A S1S2 behaves as a monomer even at a concentration of 20 mg/ml, as determined by size-exclusion chromatography and dynamic light scattering. NR3A S1S2 has very high affinity for glycine with an apparent dissociation constant (Kd) of 40 nm, 650-fold less than the Kd for NR1. Glutamate, which binds to NR2 subunits, also binds to NR3A, but with very low affinity (Kd = 9.6 mm); in contrast, binding of glutamate to NR1 was not detectable even at a 300 mm concentration. The antagonist binding profiles of NR3A and NR1 also show striking differences. 6-Cyano-2,3-dihydroxy-7-nitro-quinoxaline (CNQX), and its analog CGP78608, bind to NR3A S1S2 with low micromolar affinity, whereas for NR1, the affinity of CGP78608 increases 1000-fold compared with CNQX. Other high-affinity NR1 antagonists also show very weak binding to NR3A. Proteolysis protection experiments reveal that CNQX and CGP78608 bind to and stabilize domain 1 of NR3A S1S2 but increase proteolysis of domain 2, indicating that they produce conformational changes distinct from those induced by glycine and d-serine.
Keywords: glutamate receptor, NMDA, gating, antagonist, agonist, proteolysis
Introduction
NMDA receptor ion channels have attracted substantial interest because of their unique roles in synaptic plasticity and memory formation (Nakanishi, 1992; Collingridge and Bliss, 1995). Conventional NMDA receptors are composed of obligate NR1 subunits that bind the coagonist glycine and of NR2A to NR2D subunits that bind glutamate (Monyer et al., 1992; Furukawa et al., 2005). Discrepancies between the properties of native NMDA receptors and those expressed from cloned NR1 and NR2 subunits led to identification of NR3A and NR3B (Ciabarra et al., 1995; Sucher et al., 1995; Nishi et al., 2001; Chatterton et al., 2002).
The physiological roles of NR3A and NR3B, which share only 24–29% amino acid sequence identity with NR1 and NR2, have yet to be fully established. When coexpressed with NR1 and NR2, NR3A and NR3B act as modulators of ion flux, reducing single-channel conductance and permeability to Ca2+ (Das et al., 1998; Sasaki et al., 2002; Matsuda et al., 2003). In NR3A−/− mice, spine density, and the amplitude of NMDA receptor currents, increase compared with wild type (Das et al., 1998). In mammalian cell lines and neurons, NR1 is obligatory for the trafficking of NR3A and NR3B to the plasma membrane, but ion channels formed by coassembly of NR1 and NR3A in the absence of NR2 are not functional (Perez-Otano et al., 2001; Matsuda et al., 2003). However, in oocytes, coexpression of NR1 with NR3A or NR3B forms a novel excitatory glycine receptor (Chatterton et al., 2002). Whether such an excitatory glycine receptor exists in vivo is controversial. Glycine triggers neuronal bursting in electrophysiological experiments on cerebrocortical neurons (Chatterton et al., 2002). However, a similar response occurs in cultures from NR1−/− mice, indicating that NMDA receptors and thus NR3 subunits are unlikely to be involved (Matsuda et al., 2003).
Surprisingly, the ligand binding properties of the NR3 family are unknown, and it has been proposed that NR3A is a regulatory subunit that does not bind agonists (Villmann et al., 1999), that NR3A and NR3B are glycine-binding subunits (Chatterton et al., 2002), and that NR3B is likely to bind glutamate (Nishi et al., 2001). The ligand binding domain for many subtypes of glutamate receptor ion channels (iGluRs) can be isolated by genetic manipulation and expressed as soluble proteins in Escherichia coli; this has been achieved for the AMPA receptor GluR2 subunit (Chen and Gouaux, 1997), the kainate receptor GluR5 and GluR6 subunits (Mayer, 2005a; Nanao et al., 2005; Naur et al., 2005), and the NMDA receptor NR1 and NR2A subunits (Furukawa and Gouaux, 2003; Furukawa et al., 2005). These studies established that selectivity and affinity of the isolated ligand binding cores faithfully replicates that of intact membrane-bound receptors. Here, we report synthesis of the NR3A subunit ligand binding domain based on amino acid sequence alignments between NR3A, NR1, and NR2A. We find that NR3A is a high-affinity glycine binding subunit, with a unique selectivity profile strikingly different from that for NR1. Glutamate also binds to NR3A, but at concentrations too low to be of physiological significance.
Materials and Methods
Protein expression and purification.
Based on amino acid sequence alignments with the NR1 and NR2A ligand binding domains, the crystal structures of which have been solved (Furukawa and Gouaux, 2003; Furukawa et al., 2005), two peptides, N511-R660 and E776-K915, were isolated by PCR from the cDNA for full-length NR3A and joined by a GT dipeptide linker. The resulting NR3A S1S2 construct was then inserted into a modified version of the vector pET22b(+) (Novagen, Madison, WI), with an N-terminal His8SSGLVPRGS affinity tag and thrombin cleavage site at its N terminus. The coding region of the expression plasmid was confirmed by DNA sequencing. NR3A S1S2 was expressed as a recombinant protein in OrigamiB (DE3) Escherichia coli (Novagen). Cultures were grown in terrific broth medium supplemented with 100 μg/ml ampicillin, 15 μg/ml kanamycin, and 12.5 μg/ml tetracycline, to an OD600 value of 2.5–3.0 at 37°C; the cultures were then cooled to 17°C by immersion in ice and induced with 100 μm isopropyl-β-d-thiogalactopyranoside (IPTG); the cultures were grown overnight and typically reached an OD600 value of 4.5. Although NR3A S1S2 formed inclusion bodies, we were able to capture a soluble protein, present at ∼2 mg per liter of cells, using affinity chromatography.
The lysate from 12 L cultures was centrifuged at 40,000 rpm in a Type 45 Ti rotor, and the supernatant was loaded onto a 5 ml Ni-NTA affinity column and eluted with a step from 50 to 200 mm imidazole. Gel filtration size exclusion chromatography (SEC) with a Superose 12 XK26/70 column was used to remove aggregated protein; the eluate was digested at room temperature with thrombin at a 1:400 ratio for 1 h and further purified by SP Sepharose ion-exchange chromatography. The main peak was eluted at 180 mm NaCl and analyzed by SDS-PAGE, SEC, dynamic light scattering (DynaPro; Wyatt Technology, Santa Barbara, CA), and electrospray ionization (ESI) time of flight mass spectrometry. All chromatography buffers contained 1–2 mm glycine; the columns and chromatography media were from Amersham Biosciences (Piscataway, NJ). Purified NR3A S1S2 was frozen by flash cooling in liquid nitrogen at 10 mg/ml and stored at −80°C. NR1 S1S2 was prepared following published procedures (Furukawa and Gouaux, 2003).
Proteolysis protection assays.
Apo protein was prepared by exhaustive dialysis (>1015-fold dilution) against a glycine-free buffer containing 10 mm HEPES, 20 mm NaCl, and 1 mm EDTA, pH 7.0. The protein was then diluted to 1 mg/ml with buffer containing 10 mm HEPES, pH 7.4, 100 mm NaCl, 1 mm EDTA, and 10 mm CaCl2. The apo protein was preincubated with 1 mm concentrations of selected NR1 ligands (Tocris Cookson, Ellisville, MO; and Sigma, St. Louis, MO) for 15 min at an ambient temperature before the addition of trypsin at a 1:20 (w/w) ratio. The reactions were incubated at 25°C for 30 min and stopped by adding 1 mm 4-(2-aminoethyl)-benzenesulfonylfluoride (PEFA), followed by 25 mm EDTA. Analytical SEC of digestion reactions with 2 mm CNQX and NR3A S1S2 at 7 mg/ml was performed on a Superose 12 HR 10/30 column equilibrated with 10 mm HEPES, pH 7.0, 250 mm NaCl, 1 mm EDTA and 1 mm PEFA; the same buffer with 10 mm DTT was used for SEC under reducing conditions. The sequence of the peptide fragments was determined by a combination of matrix assisted laser-desorption ionization time of flight (MALDI-TOF) mass spectral analysis of the proteolysis reactions and N-terminal Edman sequencing, after SDS-PAGE and transfer to polyvinylidene difluoride membrane.
Radiolabel ligand binding and displacement assays.
Once it had been determined that glycine, but not glutamate or the high-affinity NR1 antagonist MDL105519, showed strong protection in proteolysis assays, the Kd value for binding of glycine to NR3A S1S2 was determined by a saturation binding assay in ligand binding buffer containing 10 mm HEPES, pH 7.2, 150 mm NaCl, 0.2 mm EDTA, 10% glycerol, 10 nm NR3A S1S2, and 60 Ci/mmol [3H]glycine (PerkinElmer Life Sciences, Norwalk, CT) varying from 4.7 to 300 nm. Displacement assays were performed as described previously (Furukawa and Gouaux, 2003; Mayer, 2005a), using exhaustively dialyzed apo protein and either 41.8 nm [3H]glycine for NR3A S1S2 or 11.3 nm [3H]MDL105519 (71 Ci/mmol; Perkin-Elmer Life Sciences) for NR1 S1S2. Kd values are reported as mean ± SEM calculated from single binding site fits to three to six displacement curves per ligand, corrected for the binding of [3H]glycine or [3H]MDL105519, using experimentally determined Kd values of 40.4 nm for NR3A S1S2-glycine and 5.86 nm for NR1 S1S2-MDL105519.
Results
Our initial experimental step was to purify to homogeneity and verify the identity of the NR3A S1S2 construct (Fig. 1). The ESI mass spectrum for NR3A S1S2 gave an experimental molecular weight (MW) of 32,895 Da, 4 Da less than the theoretical mass calculated from the amino acid sequence. The mass difference most likely results from the loss of four hydrogen atoms by formation of disulfide bonds between Cys543–Cys575 and Cys859–Cys913, which are conserved in the NR3 and NR1 subunits and for which disulfide bonds are observed in NR1 crystal structures (Furukawa and Gouaux, 2003; Inanobe et al., 2005). As shown in Figure 1B, NR3A S1S2 migrated as a monomer of an apparent MW of 33.5 kDa during SEC, and when concentrated to 6 mg/ml, it gave an apparent MW of 37.71 ± 0.95 kDa, a polydispersity of 17.7 ± 3.7%, in a dynamic light scattering assay (Fig. 1C); similar results were obtained at a concentration of 20 mg/ml. This behavior is similar to that observed for wild-type NR1 S1S2 (Furukawa et al., 2005) and indicates that NR3 does not form stable dimers at a protein concentration of ∼500 μm.
Figure 1.
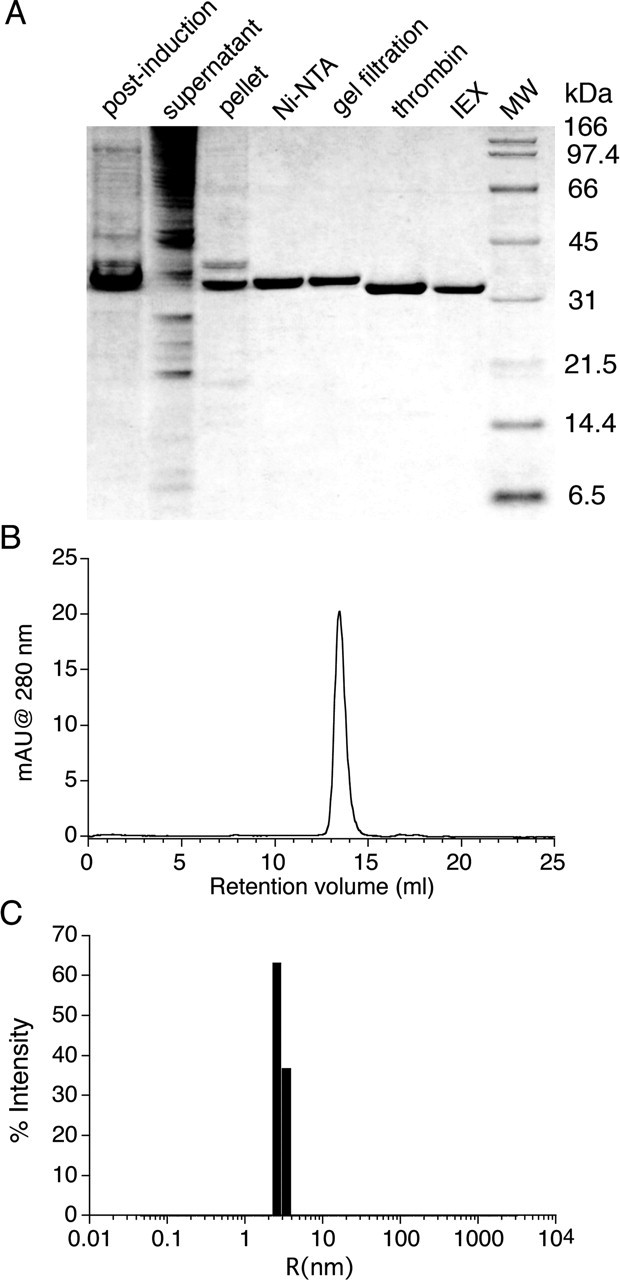
Purification and characterization of soluble NR3A S1S2. A, SDS-PAGE analysis at different stages of purification. post-induction, Whole-cell lysate after overnight induction with 100 μm IPTG; supernatant, lysate after centrifugation, before loading onto the Ni-NTA column; pellet, lysate after centrifugation; Ni-NTA, His-tagged NR3A S1S2 protein eluted from the Ni-NTA column at 200 mm imidazole; gel filtration, eluate from the Superose 12 XK26/70 column; thrombin, NR3A S1S2 digestion product after removal of the His tag; IEX, eluate from the SP Sepharose ion-exchange column. B, Superose 12 HR 10/30 SEC profile of 50 μg of NR3A S1S2 at 0.5 mg/ml; the peak eluted at 13.46 ml, corresponding to an apparent MW of 33.5 kDa. mAU, milli absorbance unit. C, Dynamic light scattering analysis of NR3A S1S2 at 6 mg/ml reveals a monodisperse profile (polydispersity, 18%) of radius 2.8 nm, corresponding to an apparent MW of 37.7 kDa.
Trypsin digestion of apo- and ligand-bound NR3A S1S2
Proteolysis protection assays provide a convenient method to rapidly screen proteins for ligand-induced conformational changes and to identify candidate agonists and antagonists for the ligand binding domains of neurotransmitter receptors (Chen et al., 1998). As shown in Figure 2A, glycine (1 mm) protected NR3A S1S2 from digestion by trypsin, whereas the apo protein was rapidly hydrolyzed. Similar protection against proteolysis occurs when the AMPA receptor GluR2 S1S2 construct is incubated with trypsin in the presence of 1 mm glutamate (Chen et al., 1998). The result of this experiment suggests that NR3A binds glycine with high affinity. Subsequent proteolysis protection experiments were used to screen a broad range of agonists and antagonists that are known to bind to the glycine recognition site on the NMDA receptor NR1 subunit. These experiments revealed that, in addition to glycine and d-serine, the NR1 partial agonists 1-aminocyclopropane-1-carboxylic acid (ACPC) and 1-aminocyclobutane-1-carboxylic acid (ACBC) also showed proteolysis protection, although the effect was weaker than for glycine or d-serine (Fig. 2B); strikingly, the digestion profiles of the peptide fragments produced for glycine- and d-serine-bound NR3A are virtually indistinguishable. Other NR1 partial agonists, l-serine and d-cycloserine (DCS), showed little or no protection, suggesting that if they bind to NR3A, their affinity is much lower than that for glycine and d-serine. The NMDA receptor NR2 subunit agonists l-glutamate and NMDA also show no protective activity, indicating that if NR3A binds glutamate, then the affinity must be much lower than for neutral amino acids.
Figure 2.
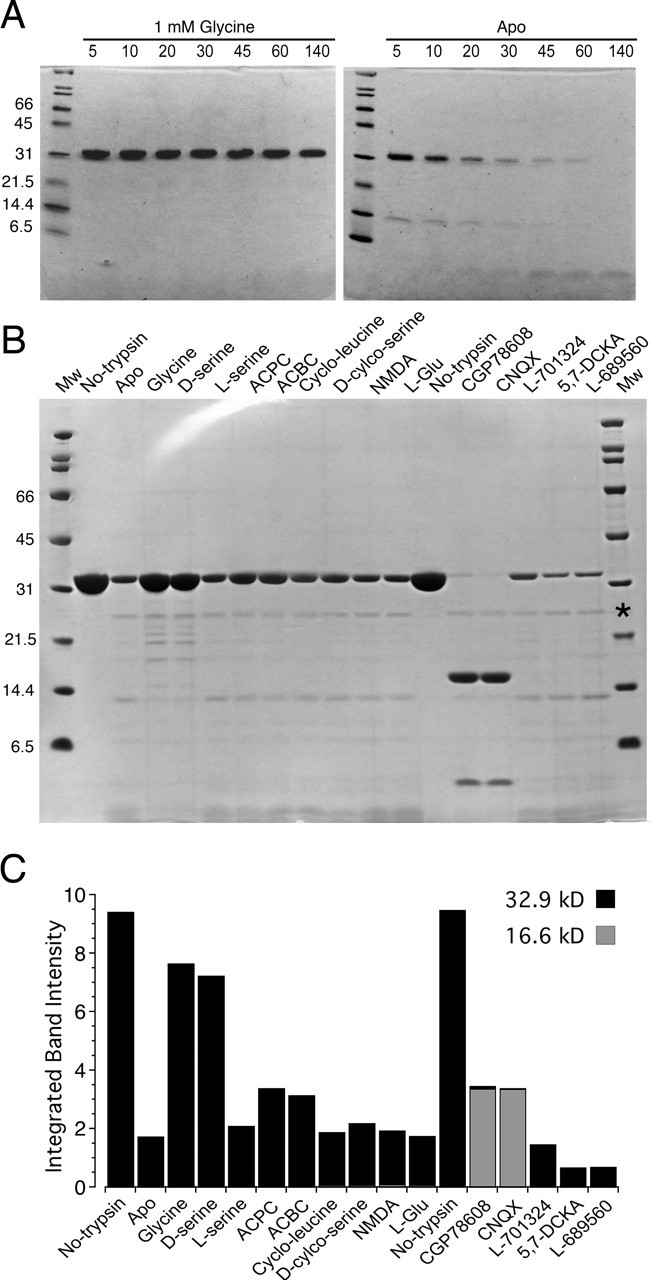
Proteolysis protection assays for NR3A S1S2. A, SDS-PAGE analysis of the digestion of NR3A S1S2 by trypsin at a ratio of 20:1 in the presence of 1 mm glycine (left) and for apo protein (right); the numbers above each lane indicate the cumulative digestion time in minutes. B, SDS-PAGE analysis of the digestion by trypsin of 8 μg of NR3A S1S2 per lane, for 30 min at 25°C, at a ratio of 20:1 in the presence of the indicated ligands at a concentration of 1 mm. The asterisk in the MW marker lane indicates the position at which trypsin migrates, which is visible as a faint band for all lanes, except the two controls labeled No-trypsin. C, Densitometric analysis of the digestion results shown in B for the 32.9 kDa band (black bar) corresponding to full-length NR3A S1S2 and the 16.6 kDa fragment (gray bar). L-Glu, l-Glutamate.
We also screened a series of eight NR1 subunit competitive antagonists that showed more complex proteolysis patterns than observed for NR1 agonists; three types of behavior were observed. The most intriguing results came from two quinoxalinediones, 6-cyano-2,3-dihydroxy-7-nitro-quinoxaline (CNQX) and [(1S)-1-[[(7-bromo-1,2,3,4-tetrahydro-2,3-dioxo-5-quinoxalinyl)methyl]amino]ethyl]phosphonic acid hydrochloride (CGP78608), both of which increased the extent of digestion of full-length NR3A S1S2 compared with the result for apo protein, but which, in addition, caused the appearance of a prominent peptide fragment of an apparent MW of 16.6 kDa on SDS-PAGE (Fig. 2B). A second pattern was observed for the quinolines 7-chlorokynurenic acid (7-CKA), 5,7-dichlorokynurenic acid (5,7-DCKA), and trans-2-carboxy-5,7-dichloro-4-phenylaminocarbonylamino-1,2,3,4-tetrahydroquinoline (L689560), which also enhanced proteolysis, but without stabilizing the 16.6 kDa MW peptide. The third pattern was observed for kynurenic acid, cycloleucine, and 7-chloro-4-hydroxy-3-(3-phenoxy)phenyl-2(1H)-quinoline (L701324), for which the proteolysis profile was similar to that for apo protein. Densitometric analysis gave the following order of protection, calculated as the ratio of the density of a 32.9 kDa band compared with the value when trypsin was omitted from the reaction (in %): 81 glycine, 76 d-serine, 35 ACPC, 33 ACBC, 22 DCS, 22 l-serine, 20 NMDA, 19 cycloleucine, 18 apo protein, 18 l-glutamate, 15 L701324, 15 kynurenic acid, 7 L689560, 8 7-CKA, 7 5,7-DCKA, 1 CGP78608, and 0.3 CNQX (Fig. 2C). The density for the 16.6 kDa fragment was <2% for all other conditions compared with the results for CGP78608 and CNQX.
CNQX and CGP78608 protect domain 1 from hydrolysis
The proteolysis results for CNQX and CGP78608 suggest that when these NR1 antagonists bind to the ligand recognition site of NR3A they induce a conformational change distinct from both the apo state and that produced by glycine and d-serine. The antagonist-induced conformation exposes proteolysis sites that are either occluded or less frequently accessible in the apo- and glycine-bound states, increasing sensitivity to proteolysis by trypsin. At the same time, these antagonists induce strong protection of a 16.6 kDa peptide, named fragment A, that is almost 50% of the mass of the NR3A S1S2 construct, together with a 6 kDa peptide named fragment B. Fragment A is too small to be domain 1 and too large to be domain 2, for which the mass values are 20.2 and 12.7 kDa, respectively, calculated by assigning Phe649 and Tyr873 as the boundary of domain2, with domain 1 constituting the remainder of the NR3A S1S2 construct, based on alignment with the crystal structure of the NR1 ligand binding core (Furukawa and Gouaux, 2003). Likewise, the composition of fragment B is not immediately obvious from its mass alone.
Surprisingly, when the CNQX- or CGP78608-protected proteolysis reactions were analyzed by gel filtration chromatography under oxidizing conditions, we observed a complex elution profile with two high molecular peaks, at 13.0 and 13.9 ml, corresponding to apparent MWs of 42.5 and 26.7 kDa, respectively, followed by multiple small peaks corresponding to low molecular proteolytic fragments (Fig. 3A). When intact NR3A S1S2 was run on the same column, it eluted at 13.5 ml, corresponding to an apparent MW of 33.5 kDa. The eluate from the gel filtration column was analyzed by reducing SDS-PAGE, which revealed that the fractions eluting at 13.0 and 13.9 ml both contained fragments A and B, in approximately stoichiometric quantities (Fig. 3B). This is a surprising result given the nearly threefold difference in mass of fragments A and B and suggests they are part of a stable complex that is resistant to further proteolysis in the presence of CNQX and CGP78608. When the same fractions from the gel filtration column were analyzed by nonreducing SDS-PAGE, we observed an additional peptide of intermediate MW named fragment C, which coeluted at 13.0 ml with fragment A, whereas the composition of the peak eluting at 13.9 ml was unchanged (Fig. 3B). We then ran the gel filtration column under reducing conditions, with 10 mm DTT added to the elution buffer, and observed that the peak at 13.0 ml was absent, whereas the main peak again eluted at 13.9 ml (Fig. 3A). Analysis by SDS-PAGE revealed that the peak eluting at 13.9 ml again contained fragments A and B in stoichiometric amounts, indicating that the complex is also stable in the absence of disulfide bond formation.
Figure 3.
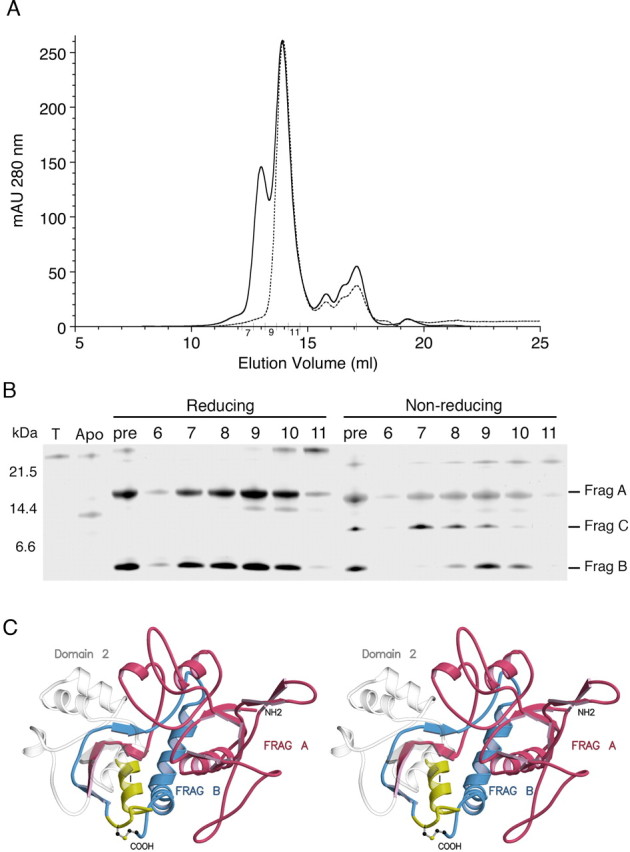
CNQX protects domain 1 from proteolysis. A, Superose 12 HR 10/30 SEC profile of 1.4 mg of NR3A S1S2 at 7 mg/ml digested with trypsin at a ratio of 20:1 for 30 min in the presence of 2 mm CNQX (solid line); the column was run again, with 10 mm DTT added to the elution buffer, for a sample treated with 10 mm DTT after the digestion reaction had been stopped with PEFA and EDTA (dashed line). B, SDS-PAGE analysis of a silver-stained gel for fractions from the SEC run under oxidizing conditions (chromatogram drawn with a solid line). T, Samples with trypsin but no NR3A S1S2; Apo, NR3A S1S2 digested under identical conditions but without CNQX; pre, NR3A S1S2 digested in the presence of CNQX, before gel filtration; lanes 6–11, fractions 6–11 from the SEC column that were either treated with BME before SDS-PAGE (Reducing) or run without BME (Non-reducing); Frag, fragment. C, Stereoview of a homology model of the NR3A S1S2 construct colored to indicate the location of fragments (FRAG) A, B, and C; the remainder of the protein is drawn as a transparent object to indicate cleavage of domain 2 into small peptides that were not resolved by SDS-PAGE.
We then used a combination of amino acid sequencing and mass spectrometry to deduce the composition of fragments A, B, and C and mapped these to a homology model based on the crystal structure of the NR1 subunit. N-terminal Edman sequencing identified the start of fragments A and B as GSNKLHLRVVT and LLTVGKPFAI, respectively. Analysis by MALDI-TOF of CNQX-protected proteolytic fragments under reducing conditions gave m/z values of 16,625 Da for fragment A and 6047 Da for fragment B. In combination with the results of N-terminal sequencing, we deduced that fragment A corresponds to the N terminus of the NR3A S1S2 construct, from N511 to R658, with the first two amino acid vector-encoded residues that precede the start of domain 1. In a homology model based on the crystal structure of the NR1 S1S2 construct, fragment A terminates in domain 2 immediately before helix I (Fig. 3C). Fragment B starts at L861, immediately after the end of helix I, and ends at K915, the C terminus of the NR3A S1S2 construct. Mass spectral analysis of the CNQX-protected proteolysis reactions prepared using oxidizing conditions showed the appearance of a peak of m/z value 7601, whereas the 6047 Da peak was reduced in amplitude. This matches results obtained by SDS-PAGE under nonreducing conditions. We explain the formation of fragment C as a result of a disulfide bond between Cys859 and Cys913, which connects fragment B to a small peptide starting at A847 and ending at K860, for which the theoretical MW is 1555 (Fig. 3C). The small peptide corresponds to helix I in domain 2. Under oxidizing conditions, the predicted mass of fragment B, linked via a disulfide bond to helix I, is 7600 Da, nearly identical to the experimentally observed peak of m/z 7601. The coelution of fragments A and B or fragments A and C, during gel filtration, indicates that when CNQX and CGP78608 bind to NR3A, they form a complex that protects the whole of domain 1, the interdomain β strands that link domains 1 and 2, and helix I in domain 2 from proteolysis by trypsin. This is the first biochemical demonstration that domain 1 of the glutamate ligand binding core forms a stable entity in solution that can be isolated from domain 2. This reinforces the interpretation of crystallographic and kinetic experiments on AMPA receptor ligand binding cores, which suggested that ligands first bind to domain 1, in which the ligand binding site exists as a preorganized assembly, and then trigger allosteric transitions to a conformational state supporting interactions with domain 2 (Abele et al., 2000; Armstrong and Gouaux, 2000). The higher apparent MW for the CNQX-protected proteolytic fragments observed during gel filtration, under either oxidizing or reducing conditions (Fig. 3A), compared with the results obtained by SDS-PAGE and mass spectrometry, most likely represents a change in shape of the peptide fragments compared with native NR3A S1S2, probably as a result of unwinding of helix I.
Agonist binding properties of NR3A S1S2
To determine the affinity of the NR3A subunit ligand binding core for glycine, a saturation binding assay was performed using [3H]glycine over the concentration range 4.7–300 nm (Fig. 4A). Analysis of this experiment gave a Kd value of 40.4 nm, ∼650-fold less than for the published value for the NR1 subunit (Table 1). We then performed displacement assays using [3H]glycine to measure the affinity of additional ligands for NR3A. d-Serine, which acts as an agonist at recombinant NMDA receptors generated by coexpression of NR1 and NR2 subunits (Wafford et al., 1995), inhibits the activation by glycine of NMDA receptors generated by coexpression of NR1 and NR3 subunits in Xenopus oocytes (Chatterton et al., 2002). Like glycine, d-serine also binds to NR3A S1S2 with high affinity, Kd = 643 nm, a value only 16-fold greater than the Kd of NR3A for glycine and 11-fold less than the Kd of d-serine for NR1. The binding of serine is stereo selective; the 158 μm Kd for the l-isomer is 240-fold greater than the Kd for d-serine and nearly 4000-fold larger than the value for glycine. The ratio of the affinity of the isomers of serine for NR3A is quantitatively similar to that for NR1 (Furukawa and Gouaux, 2003). The high affinity of glycine and d-serine but not of l-serine for NR3A are consistent with the proteolysis data shown in Figure 2, which reveals that both ligands produce an agonist-like profile distinct from that for NR1 antagonists.
Figure 4.
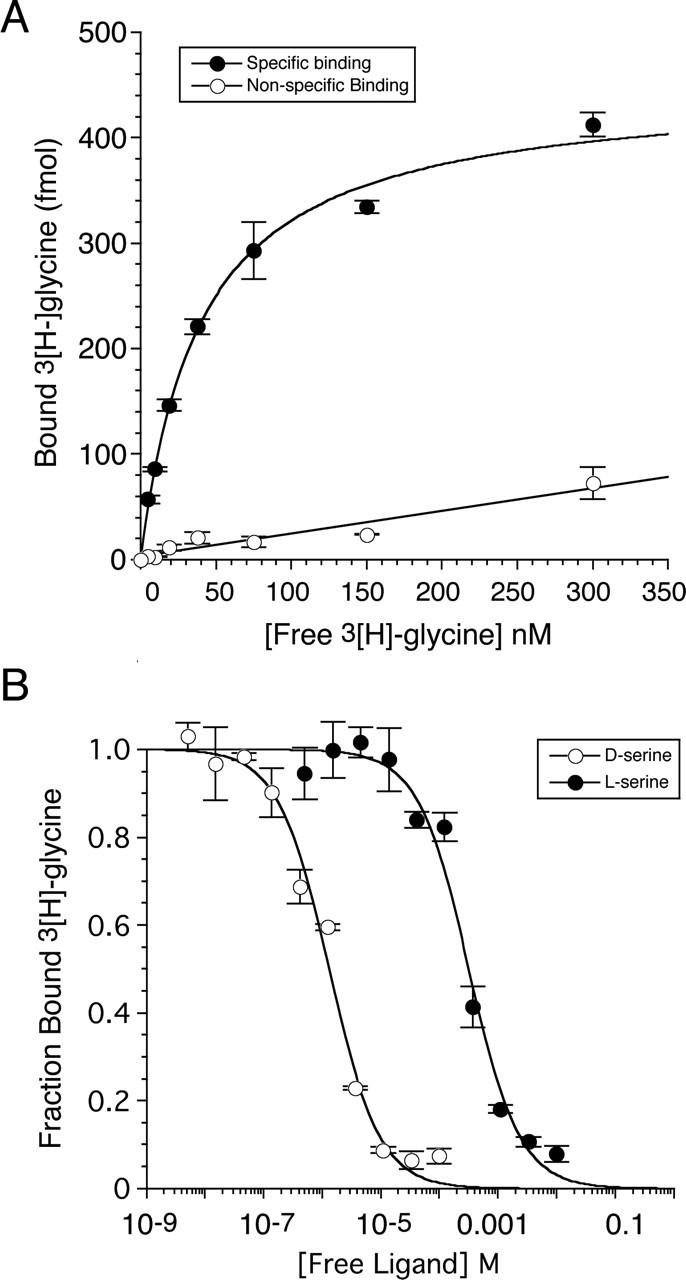
Agonist binding assays for NR3A S1S2. A, Saturation binding analysis with [3H]glycine; nonspecific binding was determined in the presence of 100 mm glycine. The fitted curve is a single binding site isotherm with a Kd of 40.4 nm. B, Displacement analysis for d-serine and l-serine; the fitted curves are for a single binding site model with competition between the cold ligands and 41.8 nm [3H]glycine. The data points show the mean ± SEM for three observations per experiment.
Table 1.
Ligand affinity for NR3A S1S2 and NR1 S1S2
| NR3A S1S2 | NR1 S1S2 | NR1/NR3A ratio | |
|---|---|---|---|
| NR1 agonists | |||
| Glycine | 40.4 ± 3.7 nm | 26.4 μma | 653 |
| d-Serine | 643 ± 47 nm | 7.02 μma | 10.9 |
| l-Serine | 158 ± 17 μm | 2.3 mma | 14.6 |
| DCS | 277 ± 8 μm | 241 μma | 0.87 |
| ACPC | 97.7 ± 5.5 μm | 4.8 μmb | 0.049 |
| ACBC | 14.4 ± 1.2 μm | 830 μmb | 57.6 |
| NR2 agonists | |||
| l-Glutamate | 9.6 ± 1.7 mm | ≫300 mmc | ≫30 |
| NMDA | >10 mm | ND | ND |
| NR1 antagonists | |||
| CNQX | 2.51 ± 0.35 μm | 6.33 ± 1.64 μm | 2.52 |
| CGP78608 | 5.52 ± 0.13 μm | 6.40 ± 0.89 nm | 1.2 × 10−3 |
| L689560 | 305 ± 41 μm | 29.3 ± 1.4 nm | 9.6 × 10−5 |
| Kynurenic acid | 14.7 ± 7.7 mm | 53 ± 29 μm | 3.6 × 10−3 |
| 7-CKA | 620 ± 115 μm | 2.6 ± 0.3 μm | 4.2 × 10−3 |
| 5,7-DCKA | 647 ± 61 μm | 0.54 μma | 8.4 × 10−4 |
| L701324 | >100 μmd | 941 ± 151 nm | < 9.4 × 10−3 |
| Cycloleucine | >100 mm | 15.3 mmb | <0.15 |
Kd values are mean ± SEM for three to six observations per ligand. ND, Not determined.
aWhere indicated, values for NR1 were taken from Furukawa and Gouaux (2003).
bWhere indicated, values for NR1 were taken from Inanobe et al. (2005).
cl-Glutamate solubility was <2.5 m in LBB.
dL701324 solubility was <3.6 × 10−4 m in ligand binding buffer.
Analysis of the affinity of NR1 subunit partial agonists for NR3A reveals substantial differences in the ligand binding properties of the two receptor subtypes despite their common activation by glycine. The rank order of affinity for NR3A is ACBC (Kd = 14.4 μm) > ACPC (Kd = 97.7 μm) > DCS (Kd = 277 μm), whereas for NR1, the rank order is ACPC (Ki = 4.8 μm), > DCS (Ki = 241 μm) > ACBC (Ki = 830 μm). NR1 shows absolute selectivity for glycine over glutamate, which the crystal structure of the NR1 ligand binding core reveals to result from steric occlusion of the glutamate ligand γ carboxyl group by the side chain of Trp731 (Furukawa and Gouaux, 2003). In NR3A and NR3B, the smaller Met residue replaces Trp731, and thus it was possible that NR3A and NR3B could bind glutamate as has been proposed previously (Nishi et al., 2001). To test this, displacement assays were performed using l-glutamate and NMDA. NR3A was observed to show low affinity binding for l-glutamate, with a Kd of 9.55 mm; for NMDA, no displacement was observed at a concentration of 1 mm. Binding with such low affinity is unusual but not without precedent, and for the NR1 subunit cycloleucine (Ki = 15.3 mm) binds with comparably low affinity (Inanobe et al., 2005). As a control to exclude an artifact resulting from contamination by glycine of the glutamate solutions prepared for this assay, we repeated the experiment using NR1 S1S2 and 3[H]MDL105519 to measure ligand binding to NR1 subunits as described previously (Furukawa and Gouaux, 2003). At a concentration of 300 mm, l-glutamate did not produce any detectable displacement of [3H]MDL105519 for binding assays with NR1, whereas for NR3A, the agonist binding site was nearly saturated at this concentration of glutamate (Fig. 5A).
Figure 5.
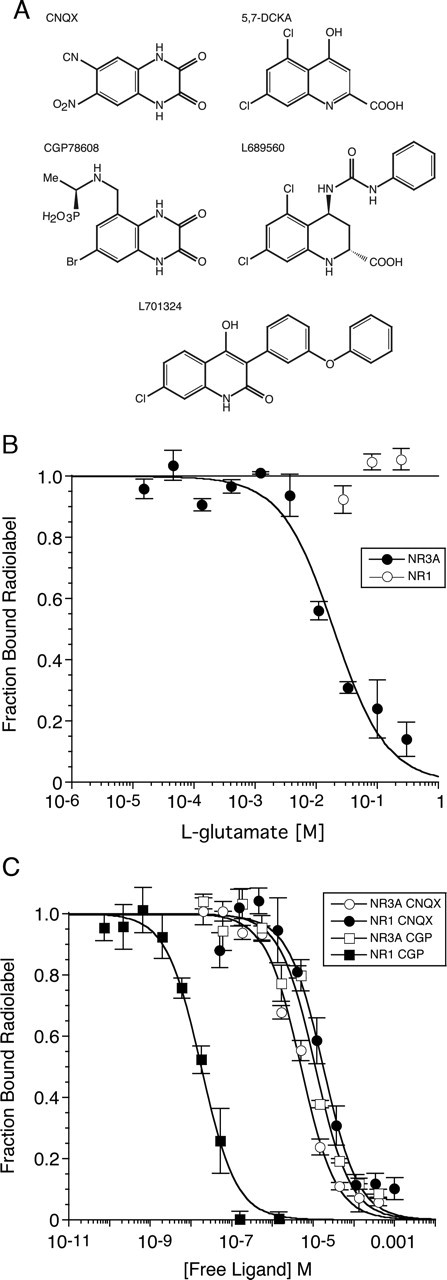
NR3A and NR1 show different ligand selectivity. A, Chemical structures of NR1 antagonists tested for activity at NR3A using proteolysis protection and [3H]glycine displacement assays. B, Displacement curves for glutamate binding to NR3A and NR1 S1S2 constructs, using 41.8 nm [3H]glycine for NR3A and 11.3 nm [3H]MDL105519 for NR1. For NR3A, the fitted curve is a single binding site model with competition between glutamate and [3H]glycine and a Kd of 9.5 mm for glutamate. C, Displacement curves for CNQX and CGP 78608 performed as described above; the fitted curves give Kd values of 2.4 and 6.1 μm for binding of CNQX to NR3A and NR1, respectively, and 5.5 μm and 6.1 nm for the binding of CGP78608 (CGP). The data points show the mean ± SEM for three observations per experiment.
Antagonist binding properties of NR3A S1S2
Numerous competitive antagonists have been developed for NR1, but it is not known whether any of these bind to NR3 subunits. We tested both the first-generation glycine site antagonists CNQX and kynurenic acid, as well as the more potent compounds CGP78608, 5,7-DCKA, L689560, and L701324 (Fig. 5). Cycloleucine, which extends the series ACPC and ACBC and causes a switch from partial agonist to antagonist activity at NR1 (Inanobe et al., 2005), was also tested for binding to NR3A. The quinoxalinedione CNQX shows little discrimination between NR3A and NR1, and its affinity for NR3A (Kd = 2.51 μm) is only 2.5-fold higher than for NR1 (Table 1). The CNQX derivative CGP78608, however, shows strong selectivity for NR1, because of a 1000-fold increase in affinity compared with CNQX, with essentially no change in affinity for NR3A (Table 1). The quinoline derivatives 5,7-DCKA, L689560, and L701324 were all strongly selective for NR1, to which they bind with affinities in the nanomolar range, whereas for NR3A, the Kd values were in the 100 μm range, giving selectivity ratios of 102–104 for NR1/NR3A (Table 1). The parent compound kynurenic acid bound with very low affinity to NR3A (Kd = 14.7 mm); the Kd decreased 25-fold for the 7-chloro derivative, with no further change for 5,7-DCKA (Table 1). In contrast, for NR1, the Kd for 7-CKA decreased 20-fold compared with the parent compound, with an additional fivefold reduction for 5,7-DCKA.
Discussion
We have determined the ligand binding properties of the NMDA receptor NR3A subunit by isolating its ligand binding core and expressing it as a highly purified water-soluble protein. We took this approach because the results of a traditional analysis, using binding experiments or functional assays on native or recombinant NMDA receptors, would be very difficult to evaluate for two reasons. First, NR3 subunits do not assemble to form functional receptors in the absence of other NMDA receptor subunits. In mammalian cells, NR3 must be coexpressed with both NR1 and NR2 subunits to generate functional ion channels (Perez-Otano et al., 2001; Matsuda et al., 2003). This makes it impossible to separate the contributions from each receptor subtype and thus obtain quantitative estimates of ligand affinity for NR3 in isolation from NR1. A similar problem occurs when NR1 and NR3 are coexpressed in Xenopus oocytes (Chatterton et al., 2002). Second, it is well known that NMDA receptors exhibit very complex allosteric interactions, such that the affinity for glycine binding varies over a 10-fold range, depending on which of the four NR2 subunits coassemble with NR1 (Monyer et al., 1992; Laurie and Seeburg, 1994; Priestley et al., 1995). Currently, it is impossible to determine whether coassembly with other NMDA receptor subtypes exerts an allosteric influence on NR3 subunits. Also, it is not possible to determine whether the affinity of NR1 and NR3 for glycine changes in triheteromeric receptor assemblies, as occurs when NR1 and NR2 are coexpressed. In light of this, knowledge of the ligand binding properties of NR3 subunits in isolation provides an important reference point from which to begin to address their functional role in vivo.
Agonist binding properties of NR3A
Electrophysiological assays suggest that glycine binds to both the NR1 and NR3 subunits when these are coexpressed in Xenopus oocytes (Chatterton et al., 2002). For both NR3A and NR3B, desensitization precluded the accurate measurement of concentration response curves, but it is clear that when coexpressed with NR1, the receptors generated by coassembly with NR3A are more sensitive to glycine than those generated by coassembly of NR1 with NR2 (Chatterton et al., 2002). Although these experiments are consistent with binding of glycine to NR3, they do not exclude an allosteric influence of NR3 on NR1, which could occur even if NR3 did not bind glycine. Our experiments with the isolated ligand binding cores of NR3A and NR1 reveal that the affinity of NR3A for glycine, Kd = 40 nm, is 650 times that for NR1, suggesting that in vivo NR3A subunits are likely to be saturated by glycine or, as discussed below, d-serine. In electrophysiological experiments with Xenopus oocytes, d-serine acts as an antagonist of responses to glycine when NR3 subunits are coexpressed with NR1 (Chatterton et al., 2002). This was a surprising result given that d-serine is an established glycine site agonist for native NMDA receptors (Kleckner and Dingledine, 1988), and indeed d-serine may even be the main endogenous ligand for NR1 subunits (Wolosker et al., 1999; Shleper et al., 2005). In proteolysis protection assays, we find that d-serine produces a digestion profile identical to that produced by glycine and distinct from that produced by the NR1 glycine site competitive antagonists CNQX and CGP78608. Because d-serine, CNQX, and CGP78608 all bind to NR3A with approximately similar affinities, with Kd values ranging from 0.6 to 5 μm, it seems likely that d-serine acts as an agonist at NR3A and perhaps produces functional antagonism as a result of rapid desensitization. Such a mechanism would not be without precedent and was described for the high-affinity kainate receptor agonist (2S,4R)-4-methyl glutamate (Zhou et al., 1997). The results of proteolysis protection assays for other NR1 subunit agonists and partial agonists, which are active in binding assays with NR3A, were not informative with respect to their digestion profile, because of the weak protection observed for l-serine, DCS, ACPC, and ACBC at the concentrations tested.
Antagonist binding properties of NR3A
Of the eight NR1 subunit antagonists tested for binding to NR3A, only CNQX and CGP78608 showed activity, with a low micromolar Kd. CNQX shows very little discrimination between NR1 and NR3, with Kd values differing only 2.5-fold. All other NR1 antagonists exhibit strong selectivity for NR1, with Kd values 103 to 104 times larger for NR3A. This suggests that when used at appropriate concentrations, it should be possible to selectively block the activation of NR1 subunits in heteromeric assemblies containing both NR1 and NR3 subunits; the best ligands for this purpose would be L689560 and 5,7-DCKA. A complication of such experiments arises from the 650-fold higher affinity of glycine for NR3A compared with NR1, which will offset the difference in affinity for antagonists for responses measured at equilibrium (Neyton and Paoletti, 2006). However, for brief applications of agonists or during synaptic activation of NMDA receptors, this complication is largely avoided because of the slow rate of dissociation that is typical for high-affinity NMDA receptor competitive antagonists (Benveniste and Mayer, 1991). An interesting result that arises from comparison of competitive antagonist affinity for NR1 versus NR3A subunits is that the second- and third-generation NR1 subunit antagonists have substantially increased affinity for NR1 but little change in affinity for NR3. Thus, compared with CNQX, the quinoxalinedione derivative CGP78608 binds with 1000-fold higher affinity to NR1 but 2-fold lower affinity to NR3A. Likewise, 5,7-DCKA binds to NR1 with a fivefold higher affinity than 5-chlorokynurenic acid, whereas for NR3A, these compounds have equal affinity. These results suggest that the ligand binding profiles of NR1 and NR3A are substantially different and that it should be possible to develop antagonists that bind selectively to NR3A and perhaps to NR3B.
Mechanisms underlying the different binding properties of NR3A and NR1
The isolation of the ligand binding core of NR3A paves the way for a structural analysis by x-ray crystallography. Although we have obtained crystals of a glycine complex, these are small, suffer radiation damage, and have low crystallographic symmetry. However, amino acid sequence alignments with the NR1 subunit, for which a crystal structure has been solved, and homology models based on this already point to some key differences between NR1 and NR3 subunits. The ligand binding cavity of the NR1 subunit glycine complex (volume, 56 Å3) is four to six times smaller than that of the glutamate-bound complexes of other iGluR subunits (Mayer, 2005b). Conformational rearrangements involving the Val689 and Trp731 side chains increase the volume of the NR1 cavity to 72 and 116 Å3 in the ACPC and ACBC complexes (Inanobe et al., 2005). In NR3A, these side chains are replaced by alanine and methionine, respectively. The substitution of these smaller residues will increase the volume of the ligand binding cavity in the NR3 subunits, and it is likely that this removes steric constraints that prevent binding of glutamate to NR1 but not NR3A.
The partial agonists ACPC and ACBC make important contacts with the side chain of Val689 in NR1 (Inanobe et al., 2005), and the substitution of Val689 by Ala in NR3A as well as the replacement of Trp731 by Met are likely to play a key role in the different affinities of NR1 and NR3A for ACPC, ACBC, and cycloleucine. Strikingly, the affinity of ACBC for NR3A is nearly 60-fold greater than for NR1, and the rank order of affinity for these ligands differs: ACBC > ACPC > cycloleucine for NR3 versus ACPC > ACBC > cycloleucine for NR1. This suggests that in NR3A, the ligand binding cavity has been altered so that the four carbon atom carbocyclic ring of ACBC fits the binding cavity of NR3A more precisely than the smaller and larger carbocyclic rings of ACPC and cycloleucine. Likewise, the van der Waals contact with the side chain of Trp731 made by the 5-halogen atom of the NR1 antagonist 5,7-DCKA is likely to be lost in NR3 subunits because of the substitution of Met at the equivalent position. Finally, we note that replacement of Phe754 by Ala in NR3A removes an important interaction between the hinge region linking domains 1 and 2 in NR1 with helix F (Inanobe et al., 2005).
In the absence of structural data, it is less easy to explain why NR3A binds glycine with 650-fold higher affinity than NR1, because the key residues that bind glycine are conserved in the two subunits. Likely mechanisms include differences in contacts between domains 1 and 2 that stabilize the closed-cleft glycine-bound conformation more effectively in the NR3 complex. Solvent-mediated hydrogen bond networks in the expanded cavity of the NR3 ligand binding core also likely play a role.
In summary, we show that the agonist and antagonist binding properties of NR3A have a unique profile distinct from that for NR1. The NR3A subunit is widely expressed in the embryonic and early postnatal nervous system. Its high affinity for glycine and d-serine suggests that it will be saturated at ambient amino acid concentrations in vivo. The functional consequences of this for NMDA receptor function in the triheteromeric NMDA receptor assemblies that likely exist in vivo have yet to be established, but the information provided here will be useful in resolving this issue.
Footnotes
This work was supported by the intramural research program of the National Institute of Child Health and Human Development, National Institutes of Health (NIH), Department of Health and Human Services. We thank Drs. Stuart Lipton and Dongxian Zhang for the gift of plasmids encoding the cDNAs for NR3A and NR3B, and Dr. H. Furukawa for sharing the purification protocol for NR1S2S2. Mass spectral analysis and Edman sequencing was performed by Howard Jaffe [Protein/Peptide Sequencing Facility, National Institute of Neurological Disorders and Stroke (NINDS), NIH]. Nucleic acid sequencing was performed by the NINDS DNA sequencing facility.
References
- Abele R, Keinanen K, Madden DR (2000). Agonist-induced isomerization in a glutamate receptor ligand-binding domain. A kinetic and mutagenetic analysis. J Biol Chem 275:21355–21363. [DOI] [PubMed] [Google Scholar]
- Armstrong N, Gouaux E (2000). Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron 28:165–181. [DOI] [PubMed] [Google Scholar]
- Benveniste M, Mayer ML (1991). Structure-activity analysis of binding kinetics for NMDA receptor competitive antagonists: the influence of conformational restriction. Br J Pharmacol 104:207–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterton JE, Awobuluyi M, Premkumar LS, Takahashi H, Talantova M, Shin Y, Cui J, Tu S, Sevarino KA, Nakanishi N, Tong G, Lipton SA, Zhang D (2002). Excitatory glycine receptors containing the NR3 family of NMDA receptor subunits. Nature 415:793–798. [DOI] [PubMed] [Google Scholar]
- Chen GQ, Gouaux E (1997). Overexpression of a glutamate receptor (GluR2) ligand binding domain in Escherichia coli: application of a novel protein folding screen. Proc Natl Acad Sci USA 94:13431–13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GQ, Sun Y, Jin R, Gouaux E (1998). Probing the ligand binding domain of the GluR2 receptor by proteolysis and deletion mutagenesis defines domain boundaries and yields a crystallizable construct. Protein Sci 7:2623–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciabarra AM, Sullivan JM, Gahn LG, Pecht G, Heinemann S, Sevarino KA (1995). Cloning and characterization of chi-1: a developmentally regulated member of a novel class of the ionotropic glutamate receptor family. J Neurosci 15:6498–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingridge GL, Bliss TV (1995). Memories of NMDA receptors and LTP. Trends Neurosci 18:54–56. [PubMed] [Google Scholar]
- Das S, Sasaki YF, Rothe T, Premkumar LS, Takasu M, Crandall JE, Dikkes P, Conner DA, Rayudu PV, Cheung W, Chen HS, Lipton SA, Nakanishi N (1998). Increased NMDA current and spine density in mice lacking the NMDA receptor subunit NR3A. Nature 393:377–381. [DOI] [PubMed] [Google Scholar]
- Furukawa H, Gouaux E (2003). Mechanisms of activation, inhibition and specificity: crystal structures of NR1 ligand-binding core. EMBO J 22:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H, Singh SK, Mancusso R, Gouaux E (2005). Subunit arrangement and function in NMDA receptors. Nature 438:185–192. [DOI] [PubMed] [Google Scholar]
- Inanobe A, Furukawa H, Gouaux E (2005). Mechanism of partial agonist action at the NR1 subunit of NMDA receptors. Neuron 47:71–84. [DOI] [PubMed] [Google Scholar]
- Kleckner NW, Dingledine R (1988). Requirement for glycine in activation of NMDA-receptors expressed in Xenopus oocytes. Science 241:835–837. [DOI] [PubMed] [Google Scholar]
- Laurie DJ, Seeburg PH (1994). Ligand affinities at recombinant N-methyl-d-aspartate receptors depend on subunit composition. Eur J Pharmacol 268:335–345. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Fletcher M, Kamiya Y, Yuzaki M (2003). Specific assembly with the NMDA receptor 3B subunit controls surface expression and calcium permeability of NMDA receptors. J Neurosci 23:10064–10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML (2005a). Crystal structures of the GluR5 and GluR6 ligand binding cores: molecular mechanisms underlying kainate receptor selectivity. Neuron 45:539–552. [DOI] [PubMed] [Google Scholar]
- Mayer ML (2005b). Glutamate receptor ion channels. Curr Opin Neurobiol 15:282–288. [DOI] [PubMed] [Google Scholar]
- Monyer H, Sprengel R, Schoepfer R, Herb A, Higuchi M, Lomeli H, Burnashev N, Sakmann B, Seeburg PH (1992). Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science 256:1217–1221. [DOI] [PubMed] [Google Scholar]
- Nakanishi S (1992). Molecular diversity of glutamate receptors and implications for brain function. Science 258:597–603. [DOI] [PubMed] [Google Scholar]
- Nanao MH, Green T, Stern-Bach Y, Heinemann SF, Choe S (2005). Structure of the kainate receptor subunit GluR6 agonist-binding domain complexed with domoic acid. Proc Natl Acad Sci USA 1708–1713. [DOI] [PMC free article] [PubMed]
- Naur P, Vestergaard B, Skov LK, Egebjerg J, Gajhede M, Kastrup JS (2005). Crystal structure of the kainate receptor GluR5 ligand-binding core in complex with (S)-glutamate. FEBS Lett 579:1154–1160. [DOI] [PubMed] [Google Scholar]
- Neyton J, Paoletti P (2006). Relating NMDA receptor function to receptor subunit composition: limitations of the pharmacological approach. J Neurosci 26:1331–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi M, Hinds H, Lu HP, Kawata M, Hayashi Y (2001). Motoneuron-specific expression of NR3B, a novel NMDA-type glutamate receptor subunit that works in a dominant-negative manner. J Neurosci 21:RC185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Otano I, Schulteis CT, Contractor A, Lipton SA, Trimmer JS, Sucher NJ, Heinemann SF (2001). Assembly with the NR1 subunit is required for surface expression of NR3A-containing NMDA receptors. J Neurosci 21:1228–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priestley T, Laughton P, Myers J, Le Bourdelles B, Kerby J, Whiting PJ (1995). Pharmacological properties of recombinant human N-methyl-d-aspartate receptors comprising NR1a/NR2A and NR1a/NR2B subunit assemblies expressed in permanently transfected mouse fibroblast cells. Mol Pharmacol 48:841–848. [PubMed] [Google Scholar]
- Sasaki YF, Rothe T, Premkumar LS, Das S, Cui J, Talantova MV, Wong HK, Gong X, Chan SF, Zhang D, Nakanishi N, Sucher NJ, Lipton SA (2002). Characterization and comparison of the NR3A subunit of the NMDA receptor in recombinant systems and primary cortical neurons. J Neurophysiol 87:2052–2063. [DOI] [PubMed] [Google Scholar]
- Shleper M, Kartvelishvily E, Wolosker H (2005). d-Serine is the dominant endogenous coagonist for NMDA receptor neurotoxicity in organotypic hippocampal slices. J Neurosci 25:9413–9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucher NJ, Akbarian S, Chi CL, Leclerc CL, Awobuluyi M, Deitcher DL, Wu MK, Yuan JP, Jones EG, Lipton SA (1995). Developmental and regional expression pattern of a novel NMDA receptor-like subunit (NMDAR-L) in the rodent brain. J Neurosci 15:6509–6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villmann C, Strutz N, Morth T, Hollmann M (1999). Investigation by ion channel domain transplantation of rat glutamate receptor subunits, orphan receptors and a putative NMDA receptor subunit. Eur J Neurosci 11:1765–1778. [DOI] [PubMed] [Google Scholar]
- Wafford KA, Kathoria M, Bain CJ, Marshall G, Le Bourdelles B, Kemp JA, Whiting PJ (1995). Identification of amino acids in the N-methyl-d-aspartate receptor NR1 subunit that contribute to the glycine binding site. Mol Pharmacol 47:374–380. [PubMed] [Google Scholar]
- Wolosker H, Blackshaw S, Snyder SH (1999). Serine racemase: a glial enzyme synthesizing d-serine to regulate glutamate-N-methyl-d-aspartate neurotransmission. Proc Natl Acad Sci USA 96:13409–13414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou LM, Gu ZQ, Costa AM, Yamada KA, Mansson PE, Giordano T, Skolnick P, Jones KA (1997). (2S,4R)-4-methylglutamic acid (SYM 2081): a selective, high-affinity ligand for kainate receptors. J Pharmacol Exp Ther 280:422–427. [PubMed] [Google Scholar]


