Abstract
γ-Secretase is a multisubunit aspartyl protease complex that catalyzes intramembrane cleavage of β-amyloid precursor protein (APP), a substrate key to Alzheimer's disease pathogenesis, and of Notch, a substrate crucial for cell differentiation. How γ-secretase recognizes and selects substrates is currently barely understood. Recent data suggest that its subunit nicastrin serves as an initial substrate receptor, which might subsequently forward substrates to the active site domain located in its catalytic subunit presenilin (PS), where an additional substrate binding site has been proposed. We now used an active site domain swapping approach of PS1 with its most distant homolog, spermatogenesis defective (SPE-4), to identify sequence determinants in this region. Strikingly, when the active site domain of PS1 was exchanged with that of SPE-4, the chimeric protein, PS1/SPE-46/7, supported APP but not Notch processing. In addition, PS1/SPE-46/7 was strongly impaired in Caenorhabditis elegans Notch signaling in vivo. Mapping experiments identified a single amino acid at position x of the GxGD motif, which contains one of the two active site aspartates, to be responsible for the observed defect in Notch processing and signaling. Our data thus implicate a role of the GxGD motif in catalytic function and substrate identification of γ-secretase.
Keywords: Alzheimer's disease, amyloid β-peptide, γ-secretase, Notch, presenilin, SPE-4
Introduction
Presenilin (PS1, PS2) is the catalytic subunit of γ-secretase, an aspartyl protease complex, which mediates the regulated intramembrane cleavage of an increasing number of type I transmembrane proteins including the prototypic β-amyloid precursor protein (APP) implicated in Alzheimer's disease (AD) (De Strooper, 2003; Haass, 2004; Steiner, 2004). γ-Secretase cleavage is typically preceded by ectodomain shedding of the substrate, which removes the bulk of the extracellular domain. In the amyloidogenic pathway of APP, ectodomain cleavage by β-secretase generates a membrane-retained C-terminal fragment (CTF). The APP CTF is subsequently cleaved by γ-secretase to release the amyloid β-peptide (Aβ), which is deposited as senile plaques in the brains of patients affected with AD, and the intracellular domain (ICD) of APP (AICD) from the membrane (Haass, 2004; Steiner, 2004). γ-Secretase-dependent liberation of ICDs has been shown for many other substrates (Kopan and Ilagan, 2004) including Notch-1, a major γ-secretase substrate (De Strooper et al., 1999). When released from the membrane, the Notch-1 ICD (NICD) translocates to the nucleus where it functions as a key transcriptional regulator required for cell differentiation during development and in adulthood (Lai, 2004).
The two active site aspartates of γ-secretase are located in transmembrane domains (TMDs) 6 and 7 (Wolfe et al., 1999) of the N-terminal fragment (NTF) and CTF of PS (Thinakaran et al., 1996), which are likely derived by autoproteolysis (Wolfe et al., 1999; Edbauer et al., 2003). PSs are founding members of polytopic GxGD-type aspartyl proteases (Haass and Steiner, 2002) that include the type 4 prepilin peptidases (LaPointe and Taylor, 2000), signal peptide peptidase (SPP) (Weihofen et al., 2002), and its homologs (Krawitz et al., 2005). These protease families are characterized by a highly conserved GxGD sequence motif (Steiner et al., 2000), which includes the C-terminal active site aspartate. Homology outside this signature sequence is scarce, and the proteases carry out distinct biological functions (Haass, 2004; Steiner, 2004). Together with the catalytic subunit PS, three other integral membrane proteins, nicastrin (NCT), anterior pharynx defective (APH-1), and presenilin enhancer (PEN-2) have been identified, which are necessary and sufficient for γ-secretase complex formation and activity (Francis et al., 2002; Edbauer et al., 2003; Kimberly et al., 2003; Takasugi et al., 2003). In mammalian cells, PS1 and PS2 are the catalytic subunits of distinct γ-secretase complexes, which additionally differ in their APH-1 subunits, suggesting that γ-secretase is a heterogeneous activity (Hebert et al., 2004; Shirotani et al., 2004).
Apart from the requirement for ectodomain shedding (Struhl and Adachi, 2000), little information is available about substrate selection by γ-secretase. The C-terminal stubs that are left in the membrane after ectodomain shedding need to be recognized subsequently by γ-secretase as substrate. NCT with its large ectodomain is a candidate subunit of the γ-secretase complex for this initial substrate recognition, and indeed it has recently been shown that it serves as a γ-secretase substrate receptor (Shah et al., 2005). In addition, previous work indicated that docking of γ-secretase substrates occurs at a site very close to but different from the active site (Tian et al., 2002, 2003), and more recently it was shown that such a docking site is located in PS (Kornilova et al., 2005).
Based on these studies, we reasoned that, after initial substrate recognition by NCT, the active site domain of PS in TMDs 6 and 7 might directly contribute to subsequent substrate docking and probably a final substrate selection before cleavage by γ-secretase. We thus sought to identify sequence requirements in TMDs 6 and 7 of PS potentially involved in this process. We now have mapped a single amino acid in TMD7 at position x of the GxGD active site motif that influences APP/Notch substrate selectivity of γ-secretase. These data implicate a role of the PS active site domain in γ-secretase substrate selectivity and support the concept of a substrate-docking site close to the active site of γ-secretase in PS.
Materials and Methods
Antibodies.
The monoclonal antibody against the N terminus of PS1 (PS1N) has been described (Capell et al., 1997), and the polyclonal antibody against the C terminus of NCT (N1660) was obtained from Sigma (St. Louis, MO). Polyclonal antibody 3552 was raised to Aβ1–40 and monoclonal antibody 6E10 against Aβ1–17 was obtained from Signet Laboratories (Dedham, MA). Monoclonal antibodies against FLAG and myc tags were from Sigma (anti-FLAG, M2) and Santa Cruz Biotechnology (Santa Cruz, CA) (anti-myc, 9E10). Cleaved Notch-1 (V1744) antibody against NICD was obtained from Cell Signaling Technology (Beverly, MA).
cDNA constructs.
PS1 wild-type (wt) and mutant cDNAs were generated by PCR using appropriate primers and subcloned into the expression vector pcDNA4/HisC (Invitrogen, San Diego, CA). The suppressor/enhancer of lin-12 (sel-12) cDNA (GenBank accession number AF171064) was amplified by PCR from a Caenorhabditis elegans mixed-stage cDNA library and subcloned as a SmaI/NotI fragment under the control of a 2.8 kb sel-12 promoter fragment starting at the translational start ATG of sel-12 to generate pBY895 (Wittenburg et al., 2000). PCR-amplified cDNAs encoding homolog of presenilin (HOP-1) (amplified from the C. elegans mixed-stage cDNA library), spermatogenesis defective (SPE-4), and PS1 were subcloned into pBY895 as SmaI/NotI fragments replacing the sel-12 cDNA, thus placing them under the control of the sel-12 promoter. cDNAs encoding H6X-tagged wt PS1, PS1 L383F, and PS1/SPE-4 chimera were placed under control of the sel-12 promoter by subcloning them into pBY2019 as NcoI/XhoI fragments. All constructs were confirmed by sequencing.
Cell culture, cell lines, and cDNA transfections.
Mouse embryonic fibroblast (MEF) cells derived from PS1/2−/− mice (Herreman et al., 1999) were cultured as described previously (Steiner et al., 2002) and transiently transfected with the indicated cDNA constructs using Lipofectamine 2000 (Invitrogen) according to the instructions of the manufacturer.
Protein analysis.
PS and derivatives and NCT were analyzed by immunoblotting of cell lysates with PS1N or N1660 antibody, respectively. Processing of myc-tagged Swedish mutant APP (Wang et al., 2004) and myc-tagged F-NEXT (Okochi et al., 2002) was analyzed by immunoblotting of cell lysates with antibody 9E10. Where indicated, NICD was additionally analyzed by immunoblotting of cell lysates with Cleaved Notch-1 (V1744) antibody. To analyze secreted Aβ and F-Nβ, the medium was replaced 24 h after transfection with fresh medium and conditioned for 16 h. Aβ was analyzed from conditioned medium by combined immunoprecipitation/immunoblotting with antibodies 3552/6E10 and F-Nβ by combined immunoprecipitation with anti-FLAG M2-agarose (Sigma) and immunoblotting with antibody M2.
Immunofluorescense microscopy.
Immunofluorescense was performed as described (Wacker et al., 1997). Anti-myc antibody 9E10 was used as primary antibody and Alexa 488-labeled secondary antibody (Invitrogen, Leiden, The Netherlands) was used for detection. Fixed cells were analyzed on an Axioskop2 microscope (Zeiss, Oberkochen, Germany) equipped with a 63×/1.25 objective and standard FITC and TRITC (tetramethylrhodamine isothiocyanate) fluorescence filter sets using an Axiocam HRm camera and AxioVision software. Images were assembled and processed using Adobe Photoshop.
Transgenic lines of C. elegans and rescue assays.
To determine whether the PS constructs are able to rescue the egg-laying defect of sel-12 mutant hermaphrodites, all constructs were injected into sel-12(ar171) mutant hermaphrodites together with the coinjection marker pBY1153 (sel-12::gfp) at a concentration of 20 ng/μl, each. The egg-laying behavior of the transgenic animals was scored at 20°C as described previously (Steiner et al., 1999).
Results
A PS1-based active site domain chimera supports APP processing
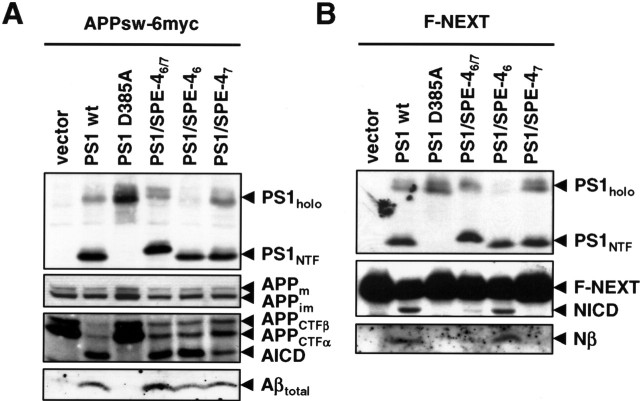
To identify sequence determinants in TMDs 6 and 7 of PS potentially involved in substrate docking and/or selection, we sought to compare the active site domain of PS1 on γ-secretase activity with that of a distant PS by swapping TMDs 6 and 7. BLAST (basic local alignment search tool) searches identified the C. elegans sperm protein SPE-4 (L'Hernault and Arduengo, 1992) as the most distant PS homolog with a low amino acid identity of 23% to PS1. Sequence comparison of PS1 with SPE-4 including the functional C. elegans PS orthologs SEL-12 and HOP-1 (Levitan and Greenwald, 1995; Westlund et al., 1999) revealed conservation of key functional residues and sequence blocks, like the critical aspartates in TMDs 6 and 7 including the GxGD active site motif and the C-terminal PALP motif (Fig. 1A). The latter motif is required for SPE-4 function (Arduengo et al., 1998) and also for PS activity in humans and Drosophila (Tomita et al., 2001; Takasugi et al., 2002; Kaether et al., 2004; Wang et al., 2004) indicating that SPE-4 functionally belongs to the PS family. Compared with PS1, SPE-4 contains a very short N terminus, a much larger cytoplasmic loop between TMDs 6 and 7, and a shorter C terminus (Fig. 1A). We then asked whether γ-secretase is functional with the related but not identical active site domain of SPE-4 and constructed the N-terminally hexahistidine-Xpress (H6X)-tagged chimeric protein PS1/SPE-46/7. In this construct, the putative TMDs 6 and 7 of PS1 are replaced with those of SPE-4 (Fig. 1B). In addition, we generated the PS1/SPE-46/7 D394A mutant in which the active site aspartate of TMD7 was changed to alanine. Constructs encoding H6X-tagged wt PS1 or inactive PS1 D385A were used as controls. We sought to assess the biological activity of these constructs in a PS-free background to avoid potential interference of endogenous wt PS in γ-secretase activity. We therefore transiently cotransfected MEF cells derived from mice deficient for PS1 and PS2 (PS−/−) (Herreman et al., 1999) with the above constructs and a construct encoding APPsw-6myc, a Swedish mutant of APP C-terminally tagged with six myc epitopes (Wang et al., 2004). As shown in Figure 2A, all four PS variants were robustly expressed. PS1/SPE-46/7 was efficiently endoproteolysed similar to wt PS1 (Fig. 2A). In contrast, PS1/SPE-46/7 D394A, like PS1 D385A, failed to undergo endoproteolysis, indicating a functional conservation of the putative SPE-4 active site domain (Fig. 2A). Both wt PS1 and PS1/SPE-46/7 underwent γ-secretase complex formation, as evident from their capability to undergo PS endoproteolysis, which occurs as final step in γ-secretase complex assembly (Kim et al., 2003; Takasugi et al., 2003). γ-Secretase complex formation was further supported by the restoration of NCT maturation (Fig. 2B), which is deficient in the absence of PS (Edbauer et al., 2002; Leem et al., 2002) and which occurs subsequently to PS endoproteolysis (Kim et al., 2003; Kimberly et al., 2003). In agreement with previous results (Nyabi et al., 2003), the recovery of NCT maturation also suggested γ-secretase complex formation (Fig. 2B) of PS1 D385A and PS1/SPE-46/7 D394A, although they were not endoproteolysed. We next investigated whether PS1/SPE-46/7 supports γ-secretase activity in APP processing using APPsw-6myc as substrate (Fig. 2C,D). Consistent with the absence of γ-secretase activity in PS−/− cells (Herreman et al., 2000; Zhang et al., 2000), APP C-terminal fragments accumulated and both AICD and Aβ failed to be generated (Fig. 2C,D). As expected, the defect in γ-secretase activity was rescued by the coexpression of wt PS1 but not by proteolytically inactive PS1 D385A. PS1/SPE-46/7, in contrast to PS1/SPE-46/7 D394A, was capable to process APPsw-6myc as judged from the robust reduction of APP CTF accumulation and the substantial generation of AICD and Aβ (Fig. 2C,D). These data show that γ-secretase can be functional with a homologous but distant GxGD-type active site domain derived from SPE-4 and that this particular active site domain is capable of APP processing.
Figure 1.
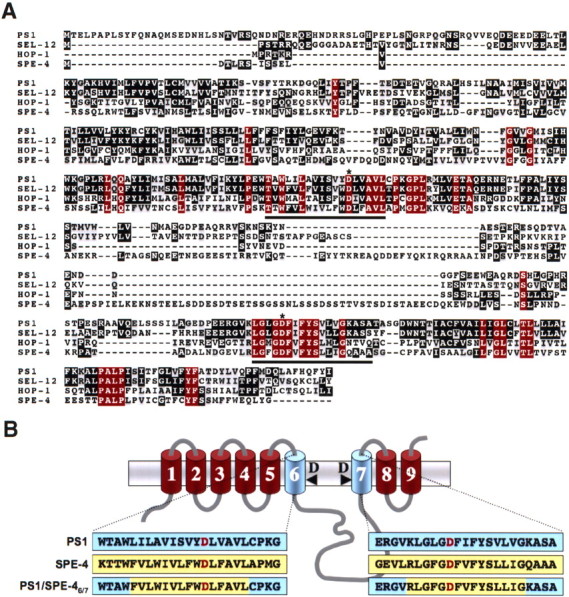
Sequence comparison of PS1 with its C. elegans homologs and schematic representation of the PS1/SPE-46/7 chimera. A, Sequence alignment of PS1 with SEL-12, HOP-1, and SPE-4 identifies the GxGD active site motif and the PALP motif as the most conserved regions between PS1 and SPE-4. The sequence alignment was generated with T-Coffee and processed with BOXSHADE. Identical amino acids residues are displayed on black or red (identical in all PSs) background and similar ones on gray background. Putative TMDs 6 and 7 comprising the active site domain are underlined. Asterisks indicate the active site aspartate residues. B, Schematic representation of PS1, SPE-4, and PS1/SPE-46/7. The primary sequences representing the putative TMDs 6 and 7 of PS1, SPE-4, and the PS1/SPE-46/7 hybrid are highlighted in blue (PS1 sequences) or yellow (SPE-4 sequences). The active site aspartate residues are displayed in red. PS is depicted according to the recent nine TMD model (Henricson et al., 2005; Laudon et al., 2005; Oh and Turner, 2005).
Figure 2.
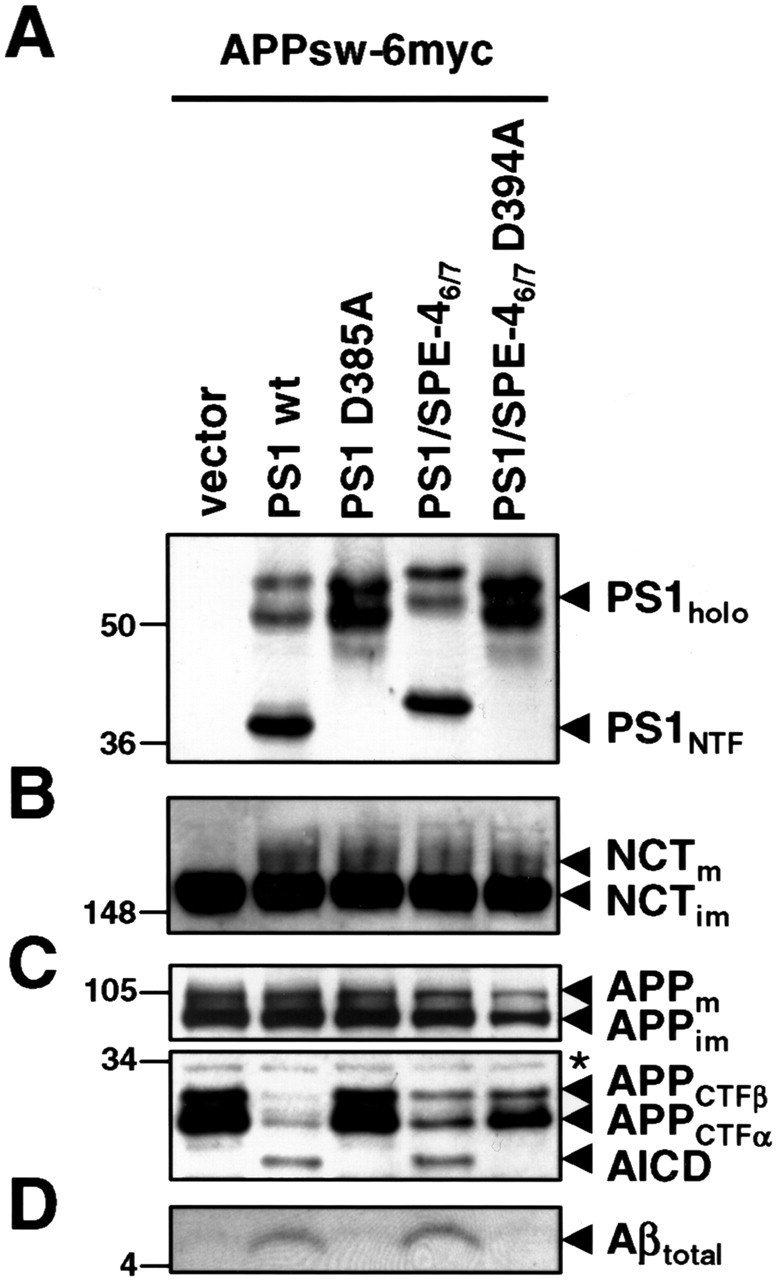
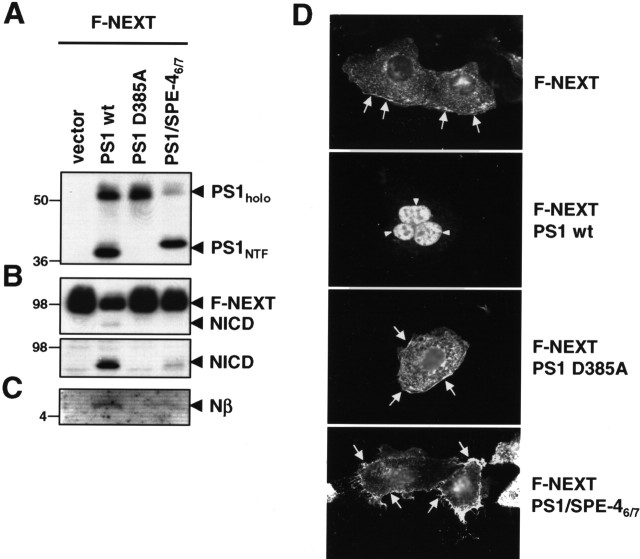
PS1/SPE-46/7 supports APP processing. A, Cell lysates of PS−/− MEF transiently cotransfected with the APPsw-6myc and the indicated H6X-tagged PS1 constructs were analyzed for PS1 expression and endoproteolysis by immunoblotting with antibody PS1N. Note that, for unknown reasons, both PS1/SPE-46/7 holoprotein and its NTF migrated slightly slower in SDS-PAGE. B, Maturation of NCT was analyzed by immunoblotting with antibody N1660. C, Full-length APP, APP CTFs, and AICD were analyzed by immunoblotting with anti-myc antibody 9E10. D, Secreted Aβ was analyzed from conditioned media by combined immunoprecipitation/immunoblotting with antibodies 3552/6E10, respectively. In A–D, molecular mass markers are shown on the left in kilodaltons. The asterisk indicates a nonspecific band.
PS1/SPE-46/7 does not process Notch
We next investigated the capability of PS1/SPE-46/7 to process Notch, a major physiological substrate of γ-secretase. We transiently cotransfected the PS−/− MEF cells with the above-described wt PS1, PS1 D385A, and PS1/SPE-46/7 constructs and a cDNA construct encoding the Notch-1-based γ-secretase substrate F-NEXT (Okochi et al., 2002). F-NEXT is a derivative of NotchΔE lacking the bulk of the ectodomain and carrying a FLAG-epitope tag at the N- and six myc-epitope tags at the C terminus (Okochi et al., 2002). Strikingly, processing of F-NEXT was strongly impaired in cells expressing PS1/SPE-46/7. Compared with wt PS1-expressing cells, F-NEXT accumulated similarly as observed in PS1 D385A-expressing cells, whereas NICD generation was strongly reduced (Fig. 3B). Likewise, the generation of F-Nβ, the secreted peptide of Notch analogous to Aβ (Okochi et al., 2002), was strongly impaired (Fig. 3C). To further corroborate these findings, immunocytochemical analysis of PS−/− MEF cells transfected with F-NEXT alone or cotransfected with the PS constructs above was performed (Fig. 3D). When PS−/− MEF cells were transfected with F-NEXT, a strong Notch staining at the plasma membrane was observed, which is consistent with an accumulation of unprocessed F-NEXT (Fig. 3B) because of the γ-secretase deficiency of these cells. Cotransfection of F-NEXT with wt PS1 restored γ-secretase activity, resulting in Notch processing as evident from the nuclear Notch staining because of the recovery of NICD formation. In contrast, cotransfection of F-NEXT with proteolytically inactive PS1 D385A did not restore γ-secretase activity as suggested from the strong plasma membrane staining of Notch. Similarly, PS−/− MEF cells transiently cotransfected with PS1/SPE-46/7 and F-NEXT revealed Notch staining at the plasma membrane. Consistent with the residual minor NICD formation (Fig. 3B), occasionally some cells showed weak nuclear staining (data not shown). These data suggest that the SPE-4-derived active site of PS1/SPE-46/7, which is able to process APP, is severely reduced in processing Notch as substrate.
Figure 3.
PS1/SPE-46/7 is defective in Notch processing. A, Cell lysates of PS−/− MEF transiently cotransfected with F-NEXT and the indicated H6X-tagged PS1 constructs were analyzed for PS expression and endoproteolysis as in Figure 2B. B, Expression and processing of F-NEXT was analyzed by immunoblotting with antibody 9E10 (top panel). NICD was additionally analyzed by immunoblotting with Cleaved Notch-1 antibody (bottom panel). C, F-Nβ levels were analyzed from conditioned media by combined immunoprecipitation/immunoblotting with anti-FLAG-M2 antibody. D, PS−/− MEF were transiently transfected with F-NEXT and the indicated H6X-tagged PS1 constructs. Forty-eight hours after transfection, cells were fixed and analyzed for Notch processing by immunofluorescence microcopy with anti-myc antibody 9E10. Expression of wt PS1, but not of PS1/SPE-46/7 or PS1 D385A, restores γ-secretase activity as indicated by nuclear Notch staining (arrowheads). Similarly as the proteolytically inactive PS1 D385A mutant, PS1/SPE-46/7 causes reduced nuclear Notch staining (arrowheads) and increased cell surface staining (arrows). In A–C, molecular mass markers are shown on the left in kilodaltons.
PS1/SPE-46/7 is deficient in Notch signaling
The above results prompted us to also analyze Notch signaling in an in vivo setting. PS1/SPE-46/7 was therefore next tested for its activity to rescue the Notch signaling deficiency caused by loss of function mutations in sel-12 (Levitan and Greenwald, 1995). These mutations cause an egg-laying defect (Egl) that can be rescued by SEL-12, HOP-1, PS1, and PS2 (Levitan et al., 1996; Baumeister et al., 1997; Li and Greenwald, 1997; Steiner et al., 1999). In contrast to wt PS1, PS1/SPE-46/7 was not able to rescue the Egl defect of the sel-12(ar171) mutant hermaphrodites (Table 1), suggesting that PS1/SPE-46/7 unlike PS1 does not support Notch signaling, consistent with the above results. We also investigated whether SPE-4 itself might be able to replace SEL-12 function. Because spe-4 expression is normally restricted to the spermatheca (L'Hernault and Arduengo, 1992; Arduengo et al., 1998), we expressed the spe-4 gene under the control of the sel-12 promoter in sel-12(ar171) mutant hermaphrodites and scored the Egl defect in the transgenic animals. Surprisingly, SPE-4, unlike PS1, SEL-12, and HOP-1, was not able to rescue the sel-12 Egl defect when expressed under the sel-12 promoter (Table 1). sel-12 mutant hermaphrodites carrying a sel-12::spe-4 extrachromosomal array never laid any eggs like the sel-12 mutants alone (Table 1) demonstrating that SPE-4 is unexpectedly unable to replace SEL-12 function.
Table 1.
Activity of different PSs to rescue the sel-12 Egl defect
| Strain | Transgenea | Genotype | Egg-laying behaviorb |
|||
|---|---|---|---|---|---|---|
| +++ | ++ | + | − | |||
| N2 | Wild type | 50 | 0 | 0 | 0 | |
| BR1129 | sel-12(ar171) | 0 | 0 | 0 | 50 | |
| BR1964 | PS1 | sel-12(ar171) | 45 | 3 | 1 | 0 |
| BR3536 | PS1/SPE-46/7 | sel-12(ar171) | 0 | 0 | 1 | 49 |
| BR3210 | PS1/SPE-46/7 | sel-12(ar171) | 0 | 0 | 2 | 48 |
| BR2364 | sel-12 | sel-12(ar171) | 48 | 2 | 0 | 0 |
| BR2993 | hop-1 | sel-12(ar171) | 50 | 0 | 0 | 1 |
| c | spe-4 | sel-12(ar171) | 0 | 0 | 0 | 34 |
| c | spe-4 | sel-12(ar171) | 0 | 0 | 0 | 27 |
| c | spe-4 | sel-12(ar171) | 0 | 0 | 0 | 16 |
aAll constructs used were untagged.
bFor each transgenic animal, the number of eggs laid were counted and were grouped into the following categories: +++, >50 eggs of progeny laid by an individual animal; ++, 15–50 eggs laid; +, 5–15 eggs laid; −, 0–5 eggs laid.
cStable lines could not be maintained because of the sterility of spe-4 transgenic animals. Instead, F1 progeny from three independent transformation experiments was scored.
Notch processing depends on the amino acid at position x of the GxGD active site motif
We next sought to define the molecular basis for the apparent substrate preference of PS1/SPE-46/7 for APP versus Notch. We first addressed the question which of the two SPE-4 TMDs was responsible for the deficiency in Notch processing and thus generated the PS1/SPE-46 and PS1/SPE-47 hybrid active site constructs in which only one TMD of PS1 is exchanged with the corresponding TMD of SPE-4. We then transiently cotransfected PS−/− MEF cells with cDNA constructs encoding the PS1/SPE-46 and PS1/SPE-47 and the APPsw-6myc or F-NEXT constructs. The PS1/SPE-46/7 construct was used for comparison in these settings. In addition, in these and the subsequent experiments, wt PS1 and PS1 D385A constructs were again included as positive and negative controls, respectively. Both PS1/SPE-46 and PS1/SPE-47 underwent γ-secretase complex formation as judged from their capability to undergo PS endoproteolysis (Fig. 4A,B) and from the NCT maturation (data not shown). Like PS1/SPE-46/7, both PS1/SPE-46 and PS1/SPE-47 were capable to process APPsw-6myc as judged from the robust reduction of APP CTF accumulation and the substantial generation of AICD and Aβ (Fig. 4A). However, in contrast to PS1/SPE-46, expression of PS1/SPE-47 strongly impaired the generation of NICD and F-Nβ from F-NEXT (Fig. 4B). Together, these results suggest that TMD7 of PS1/SPE-46/7 is responsible for the observed defect in Notch processing.
Figure 4.
Mapping of the TMD in PS1/SPE-46/7 required for Notch processing. A, PS−/− MEF were transiently cotransfected with APPsw-6myc and the indicated H6X-tagged PS1 constructs. Cell lysates were analyzed for PS expression and processing as in Figure 2A and for full-length APP, APP CTFs, and AICD as in Figure 2C. Secreted Aβ levels were analyzed from the conditioned media as in Figure 2D. B, PS−/− MEF were transiently cotransfected with F-NEXT and the indicated H6X-tagged PS1 constructs. Cell lysates were analyzed for PS expression and processing as in Figure 2A and for processing of F-NEXT by immunoblotting with 9E10 as in Figure 3B. F-Nβ levels were analyzed from the conditioned media as in Figure 3C.
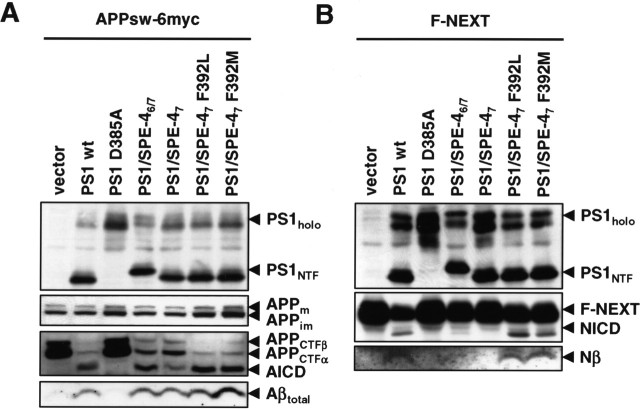
To further map the responsible sequence within the TMD7 of PS1/SPE-46/7, we made advantage of our above finding that PS1, SEL-12, and HOP-1, but not PS1/SPE-46/7 and SPE-4 was able to rescue the Notch signaling-deficient phenotype of the sel-12(ar171) mutant worms (Table 1). We reasoned that there may be differences in the conservation of crucial amino acids, which may underlie the observed rescuing activity of PS1, SEL-12, HOP-1 compared with the nonrescuing activity of PS1/SPE-46/7 and SPE-4. We thus compared the sequences of TMD7 of PS1, SEL-12, HOP-1, and SPE-4. Interestingly, we found the only potentially significant difference within their GxGD active site motifs. Although the residue x position of PS1 (L383) is rather conserved in SEL-12 (L362) and HOP-1 (M276), it is more drastically changed to a phenylalanine residue (F392) in SPE-4 (Fig. 1A,B). To address the functional significance of this amino acid change, we mutated F392 of the GFGD motif in PS1/SPE-47 to L to restore the PS1 or SEL-12 GLGD motif or to M to restore the GMGD motif of HOP-1. We then analyzed the PS1/SPE-47 F392L and PS1/SPE-47 F392M mutants using the same experimental paradigms as above. As shown in Figure 5A, when assayed in the PS−/− background, the PS1/SPE-47 F392L and PS1/SPE-47 F392M mutants underwent endoproteolysis and allowed NCT maturation (data not shown) consistent with γ-secretase complex formation. They also allowed a robust reduction of APP CTF accumulation concomitant with a notable generation of AICD and Aβ. Strikingly, both mutants allowed also the generation of substantial levels of NICD and F-Nβ, suggesting that the presence of leucine or methionine at position x within the GxGD motif is indeed crucial for the capability of PS1/SPE-47 to process Notch (Fig. 5B).
Figure 5.
Identification of phenylalanine at position x in the GxGD motif of PS1/SPE-46/7 as a critical determinant for Notch processing. A, PS−/− MEF were transiently transfected with APPsw-6myc and the indicated H6X-tagged PS1 constructs. Cell lysates were analyzed for PS expression and processing as in Figure 4A. APP processing was analyzed from cell lysates and conditioned media as in Figure 4A. B, PS−/− MEF were transiently transfected with F-NEXT and the indicated H6X-tagged PS1 constructs. Cell lysates were analyzed for PS expression and endoproteolysis and for processing of F-NEXT as in Figure 4B.
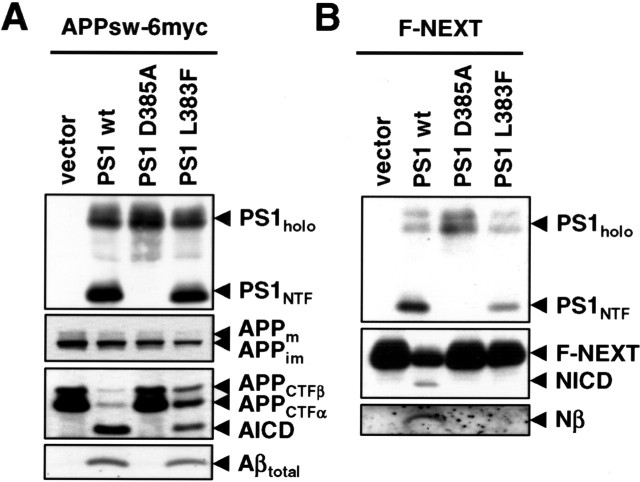
The above data suggested that the presence or absence of a phenylalanine at position x of the GxGD motif determines whether Notch can be efficiently processed or not. To obtain additional evidence for the functional importance of this residue within the GxGD motif, we next mutated L383 to F directly in PS1. We then analyzed the influence of the L383F mutation in PS1 on the processing of APP and Notch as above. The PS1 L383F mutant was endoproteolytically processed and allowed maturation of NCT (data not shown), suggesting γ-secretase complex formation. PS1 L383F also allowed robust rescue of APP CTF accumulation concomitant with a notable generation of AICD and Aβ (Fig. 6A). In sharp contrast, the PS1 L383F mutant was defective in the processing of Notch as judged from the strong reduction in NICD and F-Nβ formation (Fig. 6B). Thus, the L383 residue within the GxGD motif is also required in PS1 itself for efficient processing of Notch.
Figure 6.
Leucine 383 of PS1 is critical for Notch processing. A, PS−/− MEFs were transiently transfected with APPsw-6myc and the H6X-tagged wt PS1, PS1 D385A, or PS1 L383F constructs. Cell lysates were analyzed for PS expression and processing as in Figure 4A. APP processing was analyzed from cell lysates and conditioned media as in Figure 4A. B, PS−/− MEF were transiently transfected with F-NEXT and the H6X-tagged wt PS1, PS1 D385A, or PS1 L383F constructs. Cell lysates were analyzed for PS expression and endoproteolysis and for F-NEXT processing as in Figure 4B.
PS1 L383F is deficient in Notch signaling
To further substantiate the above findings, we examined the above constructs for their activity to rescue the sel-12 mutant deficiency in Notch signaling in vivo. The PS1/SPE-46, PS1/SPE-47, PS1/SPE-47 F392L, and PS1/SPE-47 F392M mutants and PS1 L383F were expressed under the control of the sel-12 promoter in sel-12(ar171) mutant hermaphrodites and the Egl defect in the transgenic animals was scored (Table 2). As expected from the above results, PS1/SPE-46 rescued the Egl phenotype, whereas, consistent with its defect in Notch processing, PS1/SPE-47 did not. The PS1/SPE-47 F392L and PS1/SPE-47 F392M mutants, which comprise the GLGD motif of PS1 or SEL-12 or the GMGD motif of HOP-1, respectively, showed lesser but still considerable rescuing activity. PS1 L383F, restoring the GFGD motif of SPE-4, was not able to rescue the sel-12 mutant phenotype. Thus, these data confirm in vivo that the presence of a phenylalanine residue at position x of the GxGD motif strongly impairs Notch signaling.
Table 2.
Activity of the different PS1/SPE-4 hybrid proteins to rescue the sel-12 Egl defect
| Strain | Transgenea | Genotype | Egg-laying behaviorb |
|||
|---|---|---|---|---|---|---|
| +++ | ++ | + | − | |||
| N2 | Wild type | 50 | 0 | 0 | 0 | |
| BR1129 | sel-12(ar171) | 0 | 0 | 0 | 50 | |
| BR3537 | PS1 | sel-12(ar171) | 48 | 1 | 1 | 0 |
| BR3538 | PS1/SPE-46 | sel-12(ar171) | 46 | 2 | 2 | 0 |
| BR3539 | PS1/SPE-46 | sel-12(ar171) | 48 | 0 | 2 | 0 |
| BR3540 | PS1/SPE-47 | sel-12(ar171) | 0 | 0 | 3 | 47 |
| BR3541 | PS1/SPE-47 | sel-12(ar171) | 0 | 1 | 1 | 48 |
| BR3552 | PS1/SPE-47 F392L | sel-12(ar171) | 10 | 10 | 19 | 11 |
| BR3544 | PS1/SPE-47 F392L | sel-12(ar171) | 11 | 19 | 15 | 5 |
| BR3546 | PS1/SPE-47 F392M | sel-12(ar171) | 10 | 8 | 25 | 7 |
| BR3547 | PS1/SPE-47 F392M | sel-12(ar171) | 17 | 14 | 10 | 9 |
| BR3549 | PS1 L383F | sel-12(ar171) | 0 | 1 | 4 | 45 |
| BR3550 | PS1 L383F | sel-12(ar171) | 0 | 0 | 2 | 48 |
aAll constructs used were H6X-tagged at their N termini. For all constructs, at least three independent lines were tested; only two representative lines are shown.
bFor each transgenic animal, the number of eggs laid were counted and were grouped into the following categories: +++, >50 eggs of progeny laid by an individual animal; ++, 15–50 eggs laid; +, 5–15 eggs laid; −, 0–5 eggs laid.
Discussion
In this study, using an active site domain swapping approach of PS1 with its most distant homolog SPE-4, we obtained evidence that the PS active site domain is implicated in APP/Notch substrate selectivity of γ-secretase. The active site chimera PS1/SPE-46/7 that we generated was active in γ-secretase complex formation, underwent endoproteolysis, and displayed robust γ-secretase activity in APP processing. Both endoproteolysis and γ-secretase activity were dependent on an active site aspartate. This is the first demonstration that γ-secretase can function with a related but not identical active site domain in its catalytic subunit PS. Interestingly, despite its γ-secretase activity in APP processing, PS1/SPE-46/7 was deficient in processing of Notch. In agreement with this finding, we found that, in contrast to wt PS1, PS1/SPE-46/7 failed to rescue the Notch signaling-deficient C. elegans sel-12(ar171) mutant. These data indicated that the active site domain of SPE-4 in TMDs 6 and 7 could discriminate substrates such that processing of one substrate (APP) remains possible, whereas that of another substrate (Notch) is greatly impaired. Mapping of the molecular basis for the underlying substrate discrimination between APP and Notch revealed a single amino acid difference in TMD7 as responsible site. This difference was identified as a phenylalanine at position x of the conserved GxGD motif. In the PS family of proteases, x is typically the aliphatic amino acid leucine, with the exceptions of the C. elegans homologs HOP-1 and SPE-4. Whereas a methionine is found instead of leucine in HOP-1, a rather strong amino acid exchange is present in SPE-4 with the aromatic phenylalanine. The phenylalanine is conserved in the SPE-4 homologs of the related nematodes Caenorhabditis remanei (data not shown) and Caenorhabditis briggsae, suggesting that it is an essential residue of the GxGD motif of SPE-4. We found that changing the phenylalanine to leucine or methionine to restore the GLGD or GMGD active site motifs of PS1, PS2, and SEL-12 or HOP-1, respectively, allowed recovery of Notch processing and significant activity in Notch signaling in vivo. The significance of the amino acid change could be further corroborated when we mutated the corresponding leucine in PS1 to phenylalanine. The PS1 L383F mutant allowed substantial APP processing but was strongly impaired in Notch processing and Notch signaling in vivo. This observation suggests that the residue at position x of the GxGD motif is not only of critical relevance for Notch processing in the PS1/SPE-46/7 hybrid but of general importance for Notch processing in all PSs.
The activity of PS1/SPE-46/7 in PS endoproteolysis and γ-secretase activity on APP processing also implies that SPE-4 itself has a proteolytic function. Because SPE-4 cannot functionally replace SEL-12, one may speculate that SPE-4 is a more specialized C. elegans PS protease, which may not be able to use the C. elegans Notch receptors as substrates because of the presence of the phenylalanine at position x in the GxGD motif. However, at present, we cannot exclude the possibility that the failure of SPE-4 to substitute for SEL-12 function may be attributable to a potential incapability of SPE-4 to undergo γ-secretase complex formation (as observed in mammalian cells) (data not shown). Interestingly, leucine/isoleucine–phenylalanine variations at position x in the GxGD motif are also found in and/or between the SPP/SPPL (SPP-like protease) protease families (Weihofen et al., 2002). As in PSs, these differences may have an impact on substrate selectivity. Future studies will be needed to answer these questions.
Mechanistically, the presence of a phenylalanine at position x in the GxGD motif may cause subtle differences in the conformation of TMDs 6 and 7. Although such a conformational alteration has apparently little influence on APP processing, it may affect sufficient Notch binding. Alternatively, the bulky phenylalanine may interfere with the transfer of the Notch substrate from the docking site (Esler et al., 2002), a substrate-binding site outside the active site, to the active site. The existence of a substrate binding site different from the active site had been suggested previously (Annaert et al., 2001), and strong evidence for such an exosite in γ-secretase was obtained by coisolation of APP CTFs with γ-secretase using immobilized active site-directed inhibitors (Esler et al., 2002; Beher et al., 2003). The concept of a docking site has been further supported by the noncompetitive inhibition of γ-secretase by transition-state analog inhibitors (Tian et al., 2002, 2003). The docking site was recently shown to be located in PS in very close proximity to the active site (Kornilova et al., 2005). Interestingly, the distance between the two sites has been estimated to be less than three amino acid residues in length (Kornilova et al., 2005). Our identification of a critical amino acid involved in APP/Notch substrate selection just two residues away of an active site aspartate may thus indicate that residues of the GxGD motif, in particular L383, preceding the active site aspartate in TMD7 of PS contribute to the substrate docking site of γ-secretase.
Precisely how γ-secretase recognizes and selects substrates following the obligatory step of ectodomain shedding and recognition by NCT (Shah et al., 2005) are intriguing and challenging questions. Additional studies on these questions will not only be of particular interest for targeting PS as the catalytic subunit of γ-secretase for AD treatment but should also further our understanding of intramembrane proteolysis in general.
Footnotes
This work was supported by the priority program “Cellular Mechanisms of Alzheimer's Disease” (R.B., C.H., H.S.) and the SFB596 “Molecular Mechanisms of Neurodegeneration” (C.H., H.S.) of the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie (R.B.). We thank Bart De Strooper for PS−/− MEF cells; Ralph Nixon for monoclonal antibody PS1N; Alison Goate for the APPsw-6myc construct; Steve L'Hernault for SPE-4 cDNA; Bob Barsted for the C. elegans mixed-stage cDNA library; Christine Baumeister, Alice Suelzen, and Roland Donhauser for excellent technical help; and Keiro Shirotani for critically reading this manuscript.
References
- Annaert WG, Esselens C, Baert V, Boeve C, Snellings G, Cupers P, Craessaerts K, De Strooper B (2001). Interaction with telencephalin and the amyloid precursor protein predicts a ring structure for presenilins. Neuron 32:579–589. [DOI] [PubMed] [Google Scholar]
- Arduengo PM, Appleberry OK, Chuang P, L'Hernault SW (1998). The presenilin protein family member SPE-4 localizes to an ER/Golgi derived organelle and is required for proper cytoplasmic partitioning during Caenorhabditis elegans spermatogenesis. J Cell Sci 111:3645–3654. [DOI] [PubMed] [Google Scholar]
- Baumeister R, Leimer U, Zweckbronner I, Jakubek C, Grunberg J, Haass C (1997). Human presenilin-1, but not familial Alzheimer's disease (FAD) mutants, facilitate Caenorhabditis elegans Notch signalling independently of proteolytic processing. Genes Funct 1:149–159. [DOI] [PubMed] [Google Scholar]
- Beher D, Fricker M, Nadin A, Clarke EE, Wrigley JD, Li YM, Culvenor JG, Masters CL, Harrison T, Shearman MS (2003). In vitro characterization of the presenilin-dependent γ-secretase complex using a novel affinity ligand. Biochemistry 42:8133–8142. [DOI] [PubMed] [Google Scholar]
- Capell A, Saffrich R, Olivo JC, Meyn L, Walter J, Grunberg J, Mathews P, Nixon R, Dotti C, Haass C (1997). Cellular expression and proteolytic processing of presenilin proteins is developmentally regulated during neuronal differentiation. J Neurochem 69:2432–2440. [DOI] [PubMed] [Google Scholar]
- De Strooper B (2003). Aph-1, pen-2, and nicastrin with presenilin generate an active γ-secretase complex. Neuron 38:9–12. [DOI] [PubMed] [Google Scholar]
- De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, Goate A, Kopan R (1999). A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature 398:518–522. [DOI] [PubMed] [Google Scholar]
- Edbauer D, Winkler E, Haass C, Steiner H (2002). Presenilin and nicastrin regulate each other and determine amyloid β-peptide production via complex formation. Proc Natl Acad Sci USA 99:8666–8671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbauer D, Winkler E, Regula JT, Pesold B, Steiner H, Haass C (2003). Reconstitution of γ-secretase activity. Nat Cell Biol 5:486–488. [DOI] [PubMed] [Google Scholar]
- Esler WP, Kimberly WT, Ostaszewski BL, Ye W, Diehl TS, Selkoe DJ, Wolfe MS (2002). Activity-dependent isolation of the presenilin-γ-secretase complex reveals nicastrin and a γ substrate. Proc Natl Acad Sci USA 99:2720–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R, McGrath G, Zhang J, Ruddy DA, Sym M, Apfeld J, Nicoll M, Maxwell M, Hai B, Ellis MC, Parks AL, Xu W, Li J, Gurney M, Myers RL, Himes CS, Hiebsch RD, Ruble C, Nye JS, Curtis D (2002). aph-1 and pen-2 are required for Notch pathway signaling, γ-secretase cleavage of βAPP, and presenilin protein accumulation. Dev Cell 3:85–97. [DOI] [PubMed] [Google Scholar]
- Haass C (2004). Take five-BACE and the γ-secretase quartet conduct Alzheimer's amyloid β-peptide generation. EMBO J 23:483–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Steiner H (2002). Alzheimer disease γ-secretase: a complex story of GxGD-type presenilin proteases. Trends Cell Biol 12:556–562. [DOI] [PubMed] [Google Scholar]
- Hebert SS, Serneels L, Dejaegere T, Horre K, Dabrowski M, Baert V, Annaert W, Hartmann D, De Strooper B (2004). Coordinated and widespread expression of γ-secretase in vivo: evidence for size and molecular heterogeneity. Neurobiol Dis 17:260–272. [DOI] [PubMed] [Google Scholar]
- Henricson A, Kall L, Sonnhammer EL (2005). A novel transmembrane topology of presenilin based on reconciling experimental and computational evidence. FEBS J 272:2727–2733. [DOI] [PubMed] [Google Scholar]
- Herreman A, Hartmann D, Annaert W, Saftig P, Craessaerts K, Serneels L, Umans L, Schrijvers V, Checler F, Vanderstichele H, Baekelandt V, Dressel R, Cupers P, Huylebroeck D, Zwijsen A, Van Leuven F, De Strooper B (1999). Presenilin 2 deficiency causes a mild pulmonary phenotype and no changes in amyloid precursor protein processing but enhances the embryonic lethal phenotype of presenilin 1 deficiency. Proc Natl Acad Sci USA 96:11872–11877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herreman A, Serneels L, Annaert W, Collen D, Schoonjans L, De Strooper B (2000). Total inactivation of γ-secretase activity in presenilin-deficient embryonic stem cells. Nat Cell Biol 2:461–462. [DOI] [PubMed] [Google Scholar]
- Kaether C, Capell A, Edbauer D, Winkler E, Novak B, Steiner H, Haass C (2004). The presenilin C-terminus is required for ER-retention, nicastrin-binding and γ-secretase activity. EMBO J 23:4738–4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Ikeuchi T, Yu C, Sisodia SS (2003). Regulated hyperaccumulation of presenilin-1 and the “γ-secretase” complex. Evidence for differential intramembranous processing of transmembrane substrates. J Biol Chem 278:33992–34002. [DOI] [PubMed] [Google Scholar]
- Kimberly WT, LaVoie MJ, Ostaszewski BL, Ye W, Wolfe MS, Selkoe DJ (2003). γ-Secretase is a membrane protein complex comprised of presenilin, nicastrin, Aph-1, and Pen-2. Proc Natl Acad Sci USA 100:6382–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX (2004). γ-Secretase: proteasome of the membrane? Nat Rev Mol Cell Biol 5:499–504. [DOI] [PubMed] [Google Scholar]
- Kornilova AY, Bihel F, Das C, Wolfe MS (2005). The initial substrate-binding site of γ-secretase is located on presenilin near the active site. Proc Natl Acad Sci USA 102:3230–3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawitz P, Haffner C, Fluhrer R, Steiner H, Schmid B, Haass C (2005). Differential localization and identification of a critical aspartate suggest non-redundant proteolytic functions of the presenilin homologues SPPL2b and SPPL3. J Biol Chem 280:39515–39523. [DOI] [PubMed] [Google Scholar]
- Lai EC (2004). Notch signaling: control of cell communication and cell fate. Development 131:965–973. [DOI] [PubMed] [Google Scholar]
- LaPointe CF, Taylor RK (2000). The type 4 prepilin peptidases comprise a novel family of aspartic acid proteases. J Biol Chem 275:1502–1510. [DOI] [PubMed] [Google Scholar]
- Laudon H, Hansson EM, Melen K, Bergman A, Farmery MR, Winblad B, Lendahl U, von Heijne G, Naslund J (2005). A nine-transmembrane domain topology for presenilin 1. J Biol Chem 280:35352–35360. [DOI] [PubMed] [Google Scholar]
- Leem JY, Vijayan S, Han P, Cai D, Machura M, Lopes KO, Veselits ML, Xu H, Thinakaran G (2002). Presenilin 1 is required for maturation and cell surface accumulation of nicastrin. J Biol Chem 277:19236–19240. [DOI] [PubMed] [Google Scholar]
- Levitan D, Greenwald I (1995). Facilitation of lin-12-mediated signalling by sel-12, a Caenorhabditis elegans S182 Alzheimer's disease gene. Nature 377:351–354. [DOI] [PubMed] [Google Scholar]
- Levitan D, Doyle TG, Brousseau D, Lee MK, Thinakaran G, Slunt HH, Sisodia SS, Greenwald I (1996). Assessment of normal and mutant human presenilin function in Caenorhabditis elegans. Proc Natl Acad Sci USA 93:14940–14944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Hernault SW, Arduengo PM (1992). Mutation of a putative sperm membrane protein in Caenorhabditis elegans prevents sperm differentiation but not its associated meiotic divisions. J Cell Biol 119:55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Greenwald I (1997). HOP-1, a Caenorhabditis elegans presenilin, appears to be functionally redundant with SEL-12 presenilin and to facilitate LIN-12 and GLP-1 signaling. Proc Natl Acad Sci USA 94:12204–12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyabi O, Bentahir M, Horre K, Herreman A, Gottardi-Littell N, Van Broeckhoven C, Merchiers P, Spittaels K, Annaert W, De Strooper B (2003). Presenilins mutated at Asp257 or Asp385 restore Pen-2 expression and Nicastrin glycosylation but remain catalytically inactive in the absence of wild type Presenilin. J Biol Chem 278:43430–43436. [DOI] [PubMed] [Google Scholar]
- Oh YS, Turner RJ (2005). Evidence that the COOH terminus of human presenilin 1 is located in extracytoplasmic space. Am J Physiol 289:C576–C581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okochi M, Steiner H, Fukumori A, Tanii H, Tomita T, Tanaka T, Iwatsubo T, Kudo T, Takeda M, Haass C (2002). Presenilins mediate a dual intramembraneous γ-secretase cleavage of Notch, which is required for signaling and removal of the transmembrane domain. EMBO J 21:5408–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Lee SF, Tabuchi K, Hao YH, Yu C, Laplant Q, Ball H, Dann CE III, Sudhof T, Yu G (2005). Nicastrin functions as a γ-secretase-substrate receptor. Cell 122:435–447. [DOI] [PubMed] [Google Scholar]
- Shirotani K, Edbauer D, Prokop S, Haass C, Steiner H (2004). Identification of distinct γ-secretase complexes with different APH-1 variants. J Biol Chem 279:41340–41345. [DOI] [PubMed] [Google Scholar]
- Steiner H (2004). Uncovering γ-secretase. Curr Alzheimer Res 1:175–181. [DOI] [PubMed] [Google Scholar]
- Steiner H, Duff K, Capell A, Romig H, Grim MG, Lincoln S, Hardy J, Yu X, Picciano M, Fechteler K, Citron M, Kopan R, Pesold B, Keck S, Baader M, Tomita T, Iwatsubo T, Baumeister R, Haass C (1999). A loss of function mutation of presenilin-2 interferes with amyloid β-peptide production and Notch signaling. J Biol Chem 274:28669–28673. [DOI] [PubMed] [Google Scholar]
- Steiner H, Kostka M, Romig H, Basset G, Pesold B, Hardy J, Capell A, Meyn L, Grim MG, Baumeister R, Fechteler K, Haass C (2000). Glycine 384 is required for presenilin-1 function and is conserved in polytopic bacterial aspartyl proteases. Nat Cell Biol 2:848–851. [DOI] [PubMed] [Google Scholar]
- Steiner H, Winkler E, Edbauer D, Prokop S, Basset G, Yamasaki A, Kostka M, Haass C (2002). PEN-2 is an integral component of the γ-secretase complex required for coordinated expression of presenilin and nicastrin. J Biol Chem 277:39062–39065. [DOI] [PubMed] [Google Scholar]
- Struhl G, Adachi A (2000). Requirements for presenilin-dependent cleavage of Notch and other transmembrane proteins. Mol Cell 6:625–636. [DOI] [PubMed] [Google Scholar]
- Takasugi N, Takahashi Y, Morohashi Y, Tomita T, Iwatsubo T (2002). The mechanism of γ-secretase activities through high molecular weight complex formation of presenilins is conserved in Drosophila melanogaster and mammals. J Biol Chem 277:50198–50205. [DOI] [PubMed] [Google Scholar]
- Takasugi N, Tomita T, Hayashi I, Tsuruoka M, Niimura M, Takahashi Y, Thinakaran G, Iwatsubo T (2003). The role of presenilin cofactors in the γ-secretase complex. Nature 422:438–441. [DOI] [PubMed] [Google Scholar]
- Thinakaran G, Borchelt DR, Lee MK, Slunt HH, Spitzer L, Kim G, Ratovitsky T, Davenport F, Nordstedt C, Seeger M, Hardy J, Levey AI, Gandy SE, Jenkins NA, Copeland NG, Price DL, Sisodia SS (1996). Endoproteolysis of presenilin 1 and accumulation of processed derivatives in vivo. Neuron 17:181–190. [DOI] [PubMed] [Google Scholar]
- Tian G, Sobotka-Briner CD, Zysk J, Liu X, Birr C, Sylvester MA, Edwards PD, Scott CD, Greenberg BD (2002). Linear non-competitive inhibition of solubilized human γ-secretase by pepstatin A methylester, L685458, sulfonamides, and benzodiazepines. J Biol Chem 277:31499–31505. [DOI] [PubMed] [Google Scholar]
- Tian G, Ghanekar SV, Aharony D, Shenvi AB, Jacobs RT, Liu X, Greenberg BD (2003). The mechanism of γ-secretase: multiple inhibitor binding sites for transition state analogs and small molecule inhibitors. J Biol Chem 278:28968–28975. [DOI] [PubMed] [Google Scholar]
- Tomita T, Watabiki T, Takikawa R, Morohashi Y, Takasugi N, Kopan R, De Strooper B, Iwatsubo T (2001). The first proline of PALP motif at the C terminus of presenilins is obligatory for stabilization, complex formation and γ-secretase activities of presenilins. J Biol Chem 276:33273–33281. [DOI] [PubMed] [Google Scholar]
- Wacker I, Kaether C, Kromer A, Migala A, Almers W, Gerdes HH (1997). Microtubule-dependent transport of secretory vesicles visualized in real time with a GFP-tagged secretory protein. J Cell Sci 110:1453–1463. [DOI] [PubMed] [Google Scholar]
- Wang J, Brunkan AL, Hecimovic S, Walker E, Goate A (2004). Conserved “PAL” sequence in presenilins is essential for γ-secretase activity, but not required for formation or stabilization of γ-secretase complexes. Neurobiol Dis 15:654–666. [DOI] [PubMed] [Google Scholar]
- Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B (2002). Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science 296:2215–2218. [DOI] [PubMed] [Google Scholar]
- Westlund B, Parry D, Clover R, Basson M, Johnson CD (1999). Reverse genetic analysis of Caenorhabditis elegans presenilins reveals redundant but unequal roles for sel-12 and hop-1 in Notch-pathway signaling. Proc Natl Acad Sci USA 96:2497–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenburg N, Eimer S, Lakowski B, Rohrig S, Rudolph C, Baumeister R (2000). Presenilin is required for proper morphology and function of neurons in C. elegans Nature 406:306–309. [DOI] [PubMed] [Google Scholar]
- Wolfe MS, Xia W, Ostaszewski BL, Diehl TS, Kimberly WT, Selkoe DJ (1999). Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and γ-secretase activity. Nature 398:513–517. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Nadeau P, Song W, Donoviel D, Yuan M, Bernstein A, Yankner BA (2000). Presenilins are required for γ-secretase cleavage of βAPP and transmembrane cleavage of Notch-1. Nat Cell Biol 2:463–465. [DOI] [PubMed] [Google Scholar]