Abstract
The basic helix-loop-helix (bHLH) transcription factor Math1 (also called Atoh1) is both necessary and sufficient for hair cell development in the mammalian cochlea (Bermingham et al., 1999; Zheng and Gao, 2000). Previous studies have demonstrated that a dynamic pattern of Math1 expression plays a key role in regulating the number and position of mechanosensory hair cells. However, the factors that regulate the temporal and spatial expression of Math1 within the cochlea are unknown. The bHLH-related inhibitors of differentiation and DNA binding (Id) proteins are known to negatively regulate many bHLH transcription factors, including Math1, in a number of different systems. Therefore, Id proteins are good candidates for regulating Math1 in the cochlea. Results from PCR and in situ hybridization indicate that Id1, Id2, and Id3 are expressed within the cochlear duct in a pattern that is consistent with a role in regulation of hair cell development. In particular, expression of Ids and Math1 overlapped in cochlear progenitor cells before cellular differentiation, but a specific downregulation of Id expression was observed in individual cells that differentiated as hair cells. In addition, progenitor cells in which the expression of Ids was maintained during the time period for hair cell differentiation were inhibited from developing as hair cells. These results indicate a key role for Ids in the regulation of expression of Math1 and hair cell differentiation in the developing cochlea.
Keywords: cochlea, transcription factor, bHLH, hearing, ear, differentiation
Introduction
The sensory epithelium of the mammalian cochlea, the organ of Corti, is a highly invariant mosaic consisting of mechanosensory hair cells and nonsensory supporting cells. The epithelium is organized as a single row of inner hair cells and three or four rows of outer hair cells separated from one another by supporting cells. The development of the organ of Corti involves the specification of a group of prosensory cells that will develop as all of the specific cell types within the structure (Kelley et al., 1995). The factors that play a role in the development of the mechanosensory hair cells and nonsensory supporting cells, are mostly unknown. However, the bHLH transcription factor Math1 has been shown to be necessary and sufficient for differentiation of both inner and outer hair cells (Bermingham et al., 1999; Zheng and Gao, 2000).
In the mouse, Math1 promoter activity and mRNAs are first detected at embryonic day 13 (E13) in the basal region of the cochlear duct in a group of cells that will develop as the sensory epithelium (Lanford et al., 2000; Woods et al., 2004). Math1 expression begins in a broad band of cells that extends from the basement membrane to the lumenal surface (Lanford et al., 2000). As development progresses, the expression pattern of Math1 extends along the length of the cochlear duct and becomes restricted to hair cells. The spatial and temporal expression of Math1 is critical for hair cell development and patterning in the organ of Corti; however, the factors that regulate this expression are unknown.
One family of regulators of bHLH transcription factors are the structurally related inhibitors of differentiation and DNA binding (Id) proteins (Benezra et al., 1990; Jen et al., 1992; Kreider et al., 1992; Sun, 1994; Desprez et al., 1995; Lister et al., 1995; Moldes et al., 1997; Yokota et al., 1999; Pan et al., 1999; Andres-Barquin et al., 2000). Id proteins contain the conserved helix-loop-helix domain that is required for dimerization with other bHLH proteins but lack the basic domain that mediates DNA binding. As a result, Ids can form heterodimers with other bHLHs, but these dimers cannot bind DNA and are inactive. Typically, to regulate transcription, tissue-specific bHLH proteins, such as Math1, must form heterodimers with ubiquitously expressed bHLHs, referred to as E-proteins (E2–2, HEB, E12, and E47) (Benezra et al., 1990; Jen et al., 1992; Norton and Atherton, 1998). Id proteins bind to and sequester E-proteins with high affinity, inhibiting the formation of functional heterodimers. As a result, Ids actively inhibit cellular differentiation (Benezra et al., 1990). The importance of Ids as regulators of differentiation was illustrated in an analysis of mice lacking both Id1 and Id3, in which tissue-specific bHLH factors, including Math1, were expressed and become functional at earlier developmental time points than in wild-type littermates, leading to premature terminal mitosis and differentiation (Lyden et al., 1999). Based on these results, we hypothesized that Ids could play a role in regulating the expression of Math1 in the developing cochlea.
Materials and Methods
Isolation of RNA and first-strand cDNA synthesis. Total RNA was isolated from the sensory epithelia of 40 cochleas using a total RNA Microprep Kit (Stratagene, La Jolla, CA). Each cochlea was dissected and treated with thermolysin to remove surrounding mesenchymal cells, as described by Montcouquiol and Kelley (2003). The reverse transcription (RT) reaction was performed using 500 ng of RNA, 10 mm deoxynucleotide triphosphates, 100 mm DTT, first-strand buffer, 40 U of RNasin RNase inhibitor (Promega, Madison, WI), 200 U of Moloney murine leukemia virus reverse transcriptase (Invitrogen, Carlsbad, CA), and 200 pmol of random hexamer primers, incubated at 37° for 2 h.
Semiquantitative RT-PCR. The PCR was performed using PCR beads (Amersham Biosciences, Little Chalfont, UK), 100 ng of cDNA, and 50 pm sense and antisense primers for each gene of interest. Id1–4 gene-specific 5′ and 3′ primers for mouse were as follows: Id1 (sense, 5′-GGTACCGTACAACCTTTCTCCAACTTC-3′; antisense, 5′-GGCTGGAGTCCATCTGGTCCCTCAGTGC-3′), Id2 (sense, 5′-CTCCAAGCTCAAGGAACTGG-3′; antisense, 5′-CGCAACCCACACAGAACTTA-3′), Id3 (sense, 5′-CTCTTGGACGACATGAACCAC-3′; antisense, 5′-AGTGAGCTCAGCTGTCTGGAT-3′), Id4 (sense, 5′-CCCAGTTAGCGAGACAGTCC-3′; antisense, 5′-TGGAATGACAAGACGAGACG-3′), and Bnr4 (sense, 5′-CTGCAACTGGGCGCAATCATCC-3′; antisense, 5′-CTTCATCAGAGTGGTCCTGGCAGTG-3′). Gene-specific amplification was performed using the following cycling conditions: one cycle of 5 min at 94°, followed by 30 cycles of 94° for 30 s, 60° for 30 s, and 72° for 1 min 30 s, followed by a final extension period of 10 min at 72°. The expected product sizes are as follows: Id1, 550 bp; Id2, 400 bp; Id3, 250 bp; Id4, 600 bp; Brn4, 300 bp; glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 500 bp.
In situ hybridization. Inner ears were dissected from mice at embryonic stages between E12 and E16, and in situ hybridization was performed as described previously (Wu and Oh, 1996).
Plasmid DNA, cochlear explant cultures, and electroporation. A plasmid vector using the pCLIG backbone (Satow et al., 2001) to direct expression of Math1 and enhanced green fluorescent protein (EGFP) as two independent transcripts (referred to as pCLIG-Math1-EGFP) was kindly provided by R. Kageyama (Kyoto University, Kyoto, Japan). The presence of the complete open reading frames for both Math1 and EGFP was confirmed by sequencing. A pCLIG-Id3-EGFP expression plasmid was constructed by inserting the full-length Id3 cDNA from a pcDNA-Id3 plasmid (kindly provided by B. Christy, University of Texas Health Sciences Center, San Antonio, TX) into the EcoRI site of the pCLIG plasmid. A pCLIG-E47-EGFP expression plasmid was constructed using PCR to generate a 2.0 kb E47 pcDNA with EcoRI ends into the EcoRI site of the pCLIG plasmid. The primers used to make the E47 cDNA were as follows: sense, 5′-CGATTCGAATTCATGAACCAGCCGCAGAGGATGGCGC-3′ and antisense, 5′-CGATTCGAATTCTCACATGTGCCCGGCGGGGTTGTG-3′.
Cochleas were dissected from mice at E13, as described by Montcouquiol and Kelley (2003). Each cochlea was transferred to a sterile Petri dish and placed in a 10 μl drop of water containing 2 μg/μl of plasmid DNA. At E13, the entire cochlear spiral is approximately one turn, and the overall shape of the structure closely resembles a flat disk with the epithelial cells that will give rise to the sensory epithelium located on one surface of the disk. For electroporation, the cochlear disk was oriented perpendicular to the surface of the Petri dish and the two electrodes were placed on opposite sides of the disk. A T820 square wave electroporator (BTX, San Diego, CA) was used for electroporation. The following parameters were used: 27 V, 30 ms duration, 9–10 pulses per cochlea. After electroporation, individual cochleas were incubated in the DNA drop for 1 min, followed by the addition of 100 μl of Fugene (Roche Diagnostics, Palo Alto, CA) and an additional 5 min incubation. At the conclusion of this incubation, cochleas were transferred to Petri dishes coated with Matrigel (BD Biosciences, San Jose, CA) and maintained in culture media with 10% fetal bovine serum for 6 d.
Immunohistochemistry. After 6 d in vitro, cochlear explants were fixed in 4% paraformaldehyde for 30 min, and immunohistochemistry was performed as described by Woods et al. (2004). The following antibodies were used: anti-EGFP (Invitrogen), anti-Myosin VIIa (kindly provided by C. Petit, Pasteur Institute, Paris, France), anti-Jagged1 (Jag1) (Santa Cruz Biotechnology, Santa Cruz, CA), anti-p75ntr (Chemicon, Temecula, CA), and anti-Prox1 (Covance, Princeton, NJ). Anti-EGFP was used because the intensity of EGFP labeling was low in some transfected cells. Significant changes in cell fate were determined using a Student's t test.
Results
Expression and localization of mRNAs for Id1–Id4 in the mouse cochlea
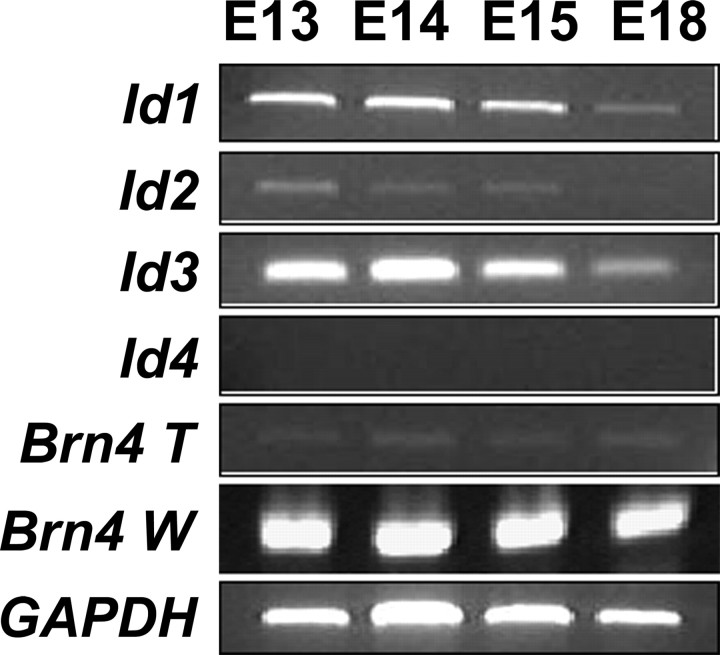
The Id family is comprised of four genes, Id1 (Benezra et al., 1990), Id2, (Sun et al., 1991), Id3 (Christy et al., 1991), and Id4 (Riechmann et al., 1994). Previous studies showed that transcripts for Id1, Id2, and Id3 are expressed in the otic vesicle of mice as early as E11.5 and in the cochlea in rats at postnatal day 1 (P1) (Jen et at., 1997; Lin et al., 2003). To determine whether Id expression occurs during the period of hair cell differentiation, transcripts for specific Id mRNAs were amplified by RT-PCR using cDNA isolated from mouse cochlear epithelia at E13, E14, E15, and E18. To confirm that the samples were not contaminated by surrounding mesenchymal cells during the epithelia isolation, transcripts for Brn4 were also amplified, because Brn4 is not expressed in the sensory epithelium but is expressed in the surrounding mesenchymal tissue (Phippard et al., 1999). Results indicated that transcripts for Id1, Id2, and Id3, but not Id4, are expressed in mouse cochlear epithelium at the time points examined (Fig. 1). The expression levels of Id1–Id3 were relatively constant at early time points but decreased at later embryonic stages (Fig. 1). All samples had very low levels of Brn4 expression, indicating minimal contamination from mesenchymal cells (Fig. 1).
Figure 1.
Expression of Id mRNAs is downregulated as the organ of Corti matures. Total RNA was isolated from cells before hair cell differentiation (E13), at the onset of hair cell differentiation (E14 and E15), and near the end of embryonic development (E18), and expression of Ids was detected by PCR. mRNAs for Id1, Id2, and Id3 are expressed at E13, E14, and E15 but are downregulated by E18. In contrast, Id4 is not expressed at any time point. Amplification of GAPDH was used to control for equal loading of cDNA in each reaction. To ensure that the cochlear samples were not contaminated by surrounding mesenchymal tissue, Brn4, a molecule that is not expressed in the developing cochlear epithelium but is expressed in adjacent mesenchyme, was amplified from cochlear epithelial samples (Brn4 T) and from samples containing both cochlear epithelium and surrounding mesenchyme (Brn4 W). Although there is a strong band for Brn4 in the Brn4 W samples at each time point, only minimal Brn4 was amplified from the Brn4 T sample, indicating minimal contamination by mesenchymal cells in the isolated epithelial samples.
To examine the cellular localization of Ids, in situ hybridizations were performed at E14, a time point near the onset of hair cell differentiation, and at E16, a time point at which differentiated hair cells can be identified based on morphology (Anniko, 1983; Lim and Anniko, 1985). Because Id4 was not detected by PCR, in situ hybridization was not done for this gene. Transcripts for Id1, Id2, and Id3 were expressed in very similar patterns in several different regions of the cochlea, including the developing cochlear duct, some mesenchymal cells, and the developing spiral ganglion at both time points (Fig. 2). Expression of all three Ids was observed throughout all three (E14) or four (E16) turns of the cochlear duct, indicating broad expression of Ids within the developing sensory epithelium (Fig. 2).
Figure 2.
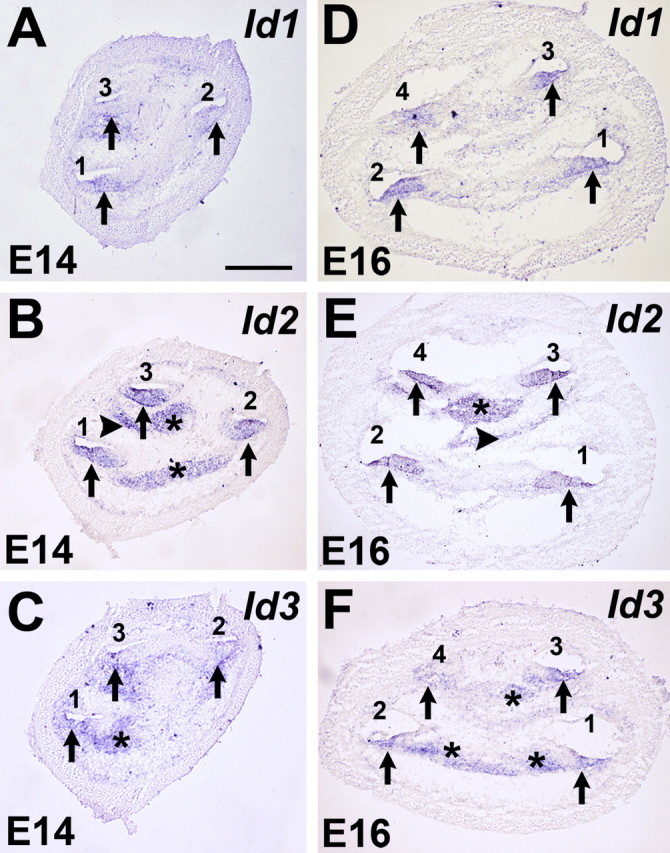
Expression of Id1, Id2, and Id3 in the developing cochlea. A–F, In situ hybridizations for Id1, Id2, and Id3 in mid-modiolar cochlear sections at E14 (A–C) and E16 (D–F). At E14, Id1 (A), Id2 (B), and Id3 (C) are broadly expressed in the floor of the cochlear duct (arrows) in each of the three cochlear turns, basal (1), middle (2), and apical (3). Expression of Id2 and Id3 was also observed in the developing spiral ganglion (B, C, asterisks), and Id2 expression was observed in some mesenchymal cells (B, arrowhead). At E16, expression of Id1 (D), Id2 (E), and Id3 (F) was still present in along the length of the cochlea duct, which has grown to four turns (numbered 1–4 as in A). Expression of Id2 and Id3 was also still present in the spiral ganglion (E and F, asterisks) and in some mesenchymal cells (E, arrowhead). Scale bar: (in A) B–F, 250 μm.
To determine the specific cellular localization of Id expression in the developing cochlea and to determine whether Id expression becomes specifically downregulated in cells that develop as hair cells, expression was examined at higher magnification within the developing sensory epithelium in cross sections from the basal turn of the cochlea at E14 and E16. In addition, because potential interactions with Math1 were of interest, transcripts for Math1 were also localized within the developing cochlea. At E14, broad expression of all three Ids was observed within the developing cochlear epithelium, including progenitor cells that will develop as the organ of Corti (Fig. 3A–C, brackets). The location of the developing prosensory region was confirmed by localization of a band of cells that expressed Math1 at E14 (Fig. 3D, bracket). By E16, individual cell types within the developing organ of Corti can be identified, although the overall level of cellular differentiation is limited (Fig. 3E). Analysis of cross sections through the basal turn of the cochlea at E16 indicated downregulation of all three Ids in developing hair cells (Fig. 3E–G), whereas Id expression was maintained in most supporting cell types at this time point. In contrast, by E16, Math1 expression is restricted to the developing inner and outer hair cells (Fig. 3I).
Figure 3.
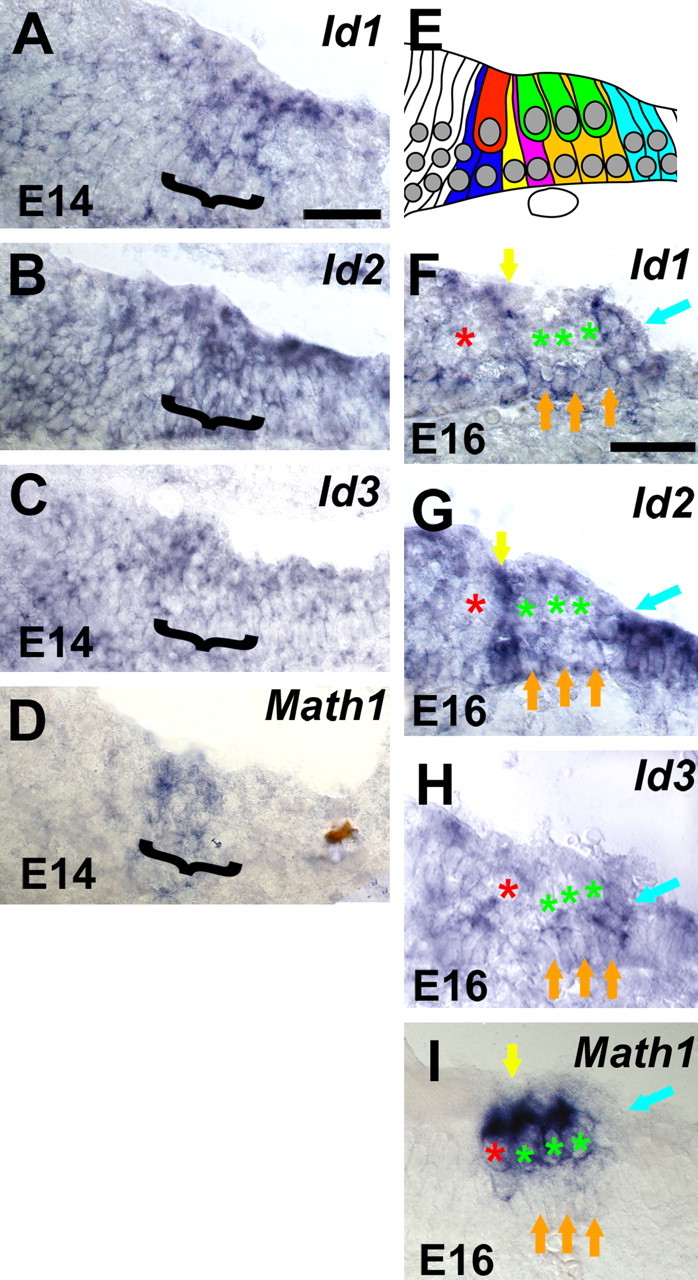
Id expression is downregulating in developing hair cells. High-magnification images of Id1, Id2, Id3, and Math 1 expression in the basal cochlear turn from mid-modiolar sections at E14 and E16. Color has been removed to provide enhanced contrast. A–C, At E14, Id1 (A), Id2 (B), and Id3 (C) are broadly expressed within the developing cochlea, including progenitor cells that will develop as the organ of Corti (bracketed region). D, At the same time point, Math 1 (bracket) is also expressed as a subset of the cells that express Ids. At this stage, Math 1 expression is an indicator of the prosensory region. E, Schematic illustration of individual cell types within the developing organ of Corti at E16. A single inner hair cell (red) and three outer hair cells (green) can be identified closer to the lumenal surface of the epithelium. Specific supporting cell types, including inner phalangeal cells (blue), inner pillar cells (yellow), outer pillar cells (magenta), Deiter's cells (orange), and Hensen's cells (blue) can also be identified based on nuclear position. At E16, expression of Id1 (F), Id2 (G), and Id3 (H) persists in supporting cell types, but there is a marked decrease in the expression of all three Ids in developing hair cells. In each panel, inner and outer hair cell nuclei are indicated with red and green asterisks. Maintained expression of Id1, Id2, and Id3 can be clearly observed in pillar cells (yellow arrows), Deiter's cells (orange arrows), and Hensen's cells (blue arrows). In contrast, by this stage, expression of Math 1 is restricted to inner (red asterisk) and outer (green asterisks) hair cells. All supporting cell types (arrows as in F) are negative for Math 1. Scale bars: (in A) B, C, 25 μm; (in E) F, G, 25 μm.
Transfection of cochlear progenitor cells
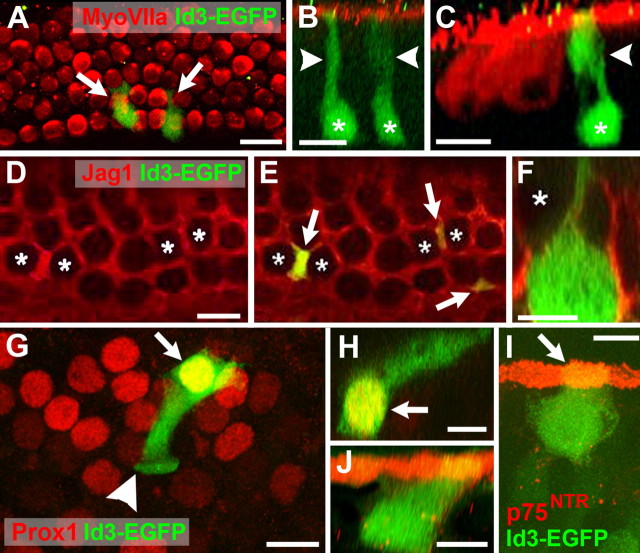
The expression data presented above strongly suggested an inhibitory role for Ids in the regulation of Math1 and hair cell differentiation. To test this hypothesis, we developed an electroporation-based protocol to transfect cochlear prosensory progenitors before determination of individual cell fates. As an initial control, cochlear explants were established from E13 animals and progenitor cells were transfected with a pCLIG-EGFP expression plasmid. After 6 d in vitro, explants were fixed and labeled with anti-EGFP to identify transfected cells and anti-Myosin VIIa to identify hair cells (Fig. 4). As reported previously (Zheng and Gao, 2000), the majority of transfected cells were located in the greater epithelial ridge (GER) cells located adjacent to the developing sensory epithelium (Fig. 4A, arrowheads). However, a number of transfected cells were consistently observed within the sensory epithelium as well (Fig. 4A, arrows). Moreover, a closer examination of individual transfected cells indicated that many had developed as hair cells based on morphology and expression of Myosin VIIa (Fig. 4B,C, arrow). To determine whether similarly transfected cells could also develop as supporting cells, additional explants were transfected with the pCLIG-EGFP vector and then labeled with markers specific for supporting cell types, including Prox1 (Bermingham-McDonogh et al., 2005) (Fig. 4D), Jagged1 (Zine et al., 2000), and p75NTR (von Bartheld et al., 1991). Examples of transfected cells that were positive for each marker were identified. (Fig. 4E,F and data not shown).
Figure 4.
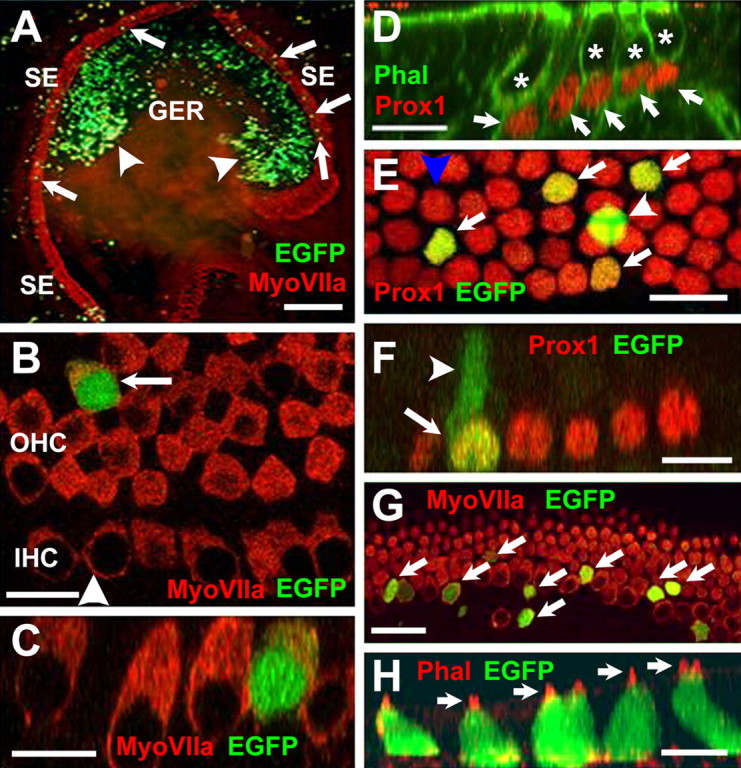
Transfection of progenitor cells in cochlear explant cultures. A, Low-magnification view of an explant culture electroporated at E13 with an EGFP expression plasmid and maintained in vitro for 6 d. Hair cells within the sensory epithelium (SE) have been labeled with anti-myosin VIIa (red), and transfected cells expressing EGFP are green. EGFP-positive cells (green) are present in the GER (arrowheads) and the SE (arrows). B, High-magnification view of the sensory epithelium in an EGFP-transfected explant. The single row in inner hair cells (IHC) and, in this case, four rows of outer hair cells (OHC) are positive for myosin VIIa (red). A single transfected cell (green) has developed as an outer hair cell (arrow). The arrowhead indicates the plane of Z-section in C. C, Confocal Z-stack cross section along the plane indicated by the arrowhead in B, illustrating hair cell morphology in the transfected cell (green). D, Prox1 is a marker for supporting cells in the organ of Corti. Confocal Z-stack of a P0 mouse cochlea. Prox1 expression is illustrated in red. Filamentous actin is labeled with phalloidin in green. Nuclei of supporting cells (arrows) are positive for Prox1 (red), whereas nuclei of hair cells (asterisks) are negative for Prox1. E, High-magnification view of the supporting cell nuclear layer in an explant transfected with EGFP. Four transfected cells (green, arrows) are also positive for Prox1. A single additional cell (arrowhead) is also transfected but appears to be located at a more lumenal position within the epithelium and is not Prox1 positive. This cell has probably developed as a hair cell. The blue arrowhead indicates plane of Z-section in F. F, Confocal Z-stack cross section through the plane indicated by the blue arrowhead in E. The single transfected Prox1-positive cell (arrow) has a morphology that is consistent with a supporting cell, including an apical projection (arrowhead) that extends to the lumenal surface. G, Overexpression of Math 1 induces development of hair cells in the sensory epithelium. Virtually all progenitor cells transfected with Math 1 (green, arrows) are positive for myosin VIIa (red), indicating that they have developed as hair cells. H, Math 1-transfected cells also develop stereociliary bundles. Confocal Z-stack cross section through a group of transfected progenitor cells (green). Stereociliary bundles on each hair cell (arrows) are visualized by phalloidin labeling of filamentous actin (red). Scale bars: A, 250 μm; B, 20 μm; C, 10 μm; D, 20 μm; E, 20 μm; F, 20 μm; G, 20 μm; H, 10 μm.
To determine whether the transfection procedure generated any bias in the determination of cell fate, the fates of transfected cells were quantified. As discussed previously, differentiation of the organ of Corti begins in a gradient that extends from the base of the cochlear spiral to the apex (Rubel, 1978). Therefore, at the time of electroporation, cells located in the apical region of the cochlea are less mature than those in the base, making the likelihood of influencing cell fate greater for cells located in the apex. Therefore, cell counts were limited to cells located in the apical 50% of the sensory epithelium of each explant. Cell fates were determined for a total of 83 cells from seven separate explants (Table 1 and supplemental Table 1, available at www.jneurosci.org as supplemental material). For the seven explants, an overall average of ∼50% of cells transfected with pCLIG-EGFP developed as hair cells (Table 1). However, an examination of the fates of the 83 individually transfected cells, regardless of the explant in which they were located, suggests a slightly higher rate of ∼54% (supplemental Table 1, available at www.jneurosci.org as supplemental material). By either method of analysis, these results indicate that approximately one-half of the progenitor cells transfected with pCLIG-EGFP develop as hair cells. These data are consistent with the approximately equivalent number of hair cells and supporting cells within the organ of Corti and indicate that progenitor cells transfected with a pCLIG-EFGP expression plasmid do not appear to be biased toward a particular cell fate within the sensory epithelium.
Table 1.
Effects of expression of EGFP, Math1, Id3, and E47 on cell fate in cochlear explants
|
cDNA |
%HC |
%Non-HC |
|---|---|---|
| EGFP (7) | 50.1 ± 18.4 | 49.9 ± 18.4 |
| Math1 (5) | 100.0 ± 0.0* | 0.0 ± 0.0* |
| Id3 (9) | 5.5 ± 12.6* | 94.5 ± 12.6* |
|
E47 (8) |
58.8 ± 16.9 |
41.2 ± 16.9 |
Expression vectors encoding full-length open reading frames for the genes listed were overexpressed in cochlear progenitor cells beginning on E13. Total number of explants analyzed for each vector are indicated in parentheses. Transfection was determined by expression of EGFP, and cell fates were determined using cell-specific markers and morphology as described in Results. Overexpression of Math1 produced a significant bias towards a hair cell fate, whereas overexpression of Id3 produced a specific bias away from the hair cell fate. Overexpression of E47 did not influence cell fate towards either cell type. Significance was determined by t test. *α = 0.01
Overexpression of Math1 in cochlear progenitor cells induces the hair cell fate
Previous results had demonstrated that transfection of Math1 into cells within the GER of postnatal rat cochlear explants resulted in the formation of ectopic hair cells (Zheng and Gao, 2000). However, the authors in that study were unable to successfully transfect cells within the sensory epithelium. Based on those results and the known effects of Math1 on hair cell formation (Bermingham et al., 1999; Woods et al., 2004), we chose to overexpress Math1 in cochlear progenitor cells to confirm that the electroporation-based transfection technique was sufficient to influence cell fate in the sensory epithelium. As expected, progenitor cells transfected with a pCLIG-Math1 expression plasmid consistently developed as hair cells based on double labeling with anti-EGFP and either the hair cell-specific antibody anti-Myosin VIIa (Fig. 4G) or phalloidin to identify the actin rich stereociliary bundles located at the lumenal surface of each hair cell (Fig. 4H). Quantification of cell fates for cells located within the sensory epithelium indicated that 100% (n = 100) of cells transfected with pCLIG-Math1 differentiated as hair cells (Table 1 and supplemental Table 1, available at www.jneurosci.org as supplemental material).
Overexpression of Id inhibits hair cell formation
To determine whether transfection of an Id protein would have a contrary effect on cell fate compared with Math1, explants were transfected with a pCLIG-Id3 expression plasmid. Because the patterns of expression for Id1, Id2, and Id3 were essentially the same within the cochlea, only a single Id, Id3, was used for transfections. Id3 was selected based on the demonstrated importance of Id1 and Id3 in neural development (Lyden et al., 1999) and the fact that it has a lower binding affinity for E47 than Id1 (Langlands et al., 1997). Therefore, overexpression of Id3 seemed less likely to have a nonspecific effect. Most of the cells transfected with the Id3 plasmid were negative for Myosin VIIa labeling (Fig. 5A), nor did these cells have stereociliary bundles as determined by labeling with phalloidin (data not shown). In addition, the morphologies of the Id3-transfected cells were consistent with supporting cells, including the extension of lumenal processes that appeared to interdigitate between adjacent hair cells (Fig. 5A–C). Also consistent with a supporting cell fate, the cell bodies of Id3-transfected cells were typically located adjacent to the basement membrane.
Figure 5.
Overexpression of Id3 inhibits hair cell differentiation but does not prevent supporting cell development in the sensory epithelium. A, Sensory epithelium in an explant transfected with Id3 (green; indicated as Id3-EGFP). Inner and outer hair cells are labeled with myosin VIIa (red). Two transfected cells (arrows) are present. Each cell is negative for myosin VIIa and has a morphology that is consistent with a supporting cell. B, Confocal Z-stack cross section of the two Id3-transfected cells in A. Each has a cell body (asterisks) located near the basement membrane and a lumenal projection (arrowheads). C, Confocal Z-stack view of a cell transfected with Id3 (green). Hair cells in the same image have been labeled with myosin VIIa in red. Note the basal position of the cell body (asterisk) and the lumenal projection (arrowhead). D, Surface view of the sensory epithelium in an explant labeled with the supporting cell marker Jagged1 (Jag1; red). Hair cells appear as black circles (asterisks), whereas interdigitating supporting cells are red. E, The same image as that in D, with three Id3-transfected cells illustrated in green (arrows). Note that each transfected cell is also positive for Jag1. F, Confocal Z-stack cross section of a cell transfected with Id3 (green). Note the large cell body located adjacent to the basement membrane and the thin extension projecting toward the lumenal surface. Also, a hair cell (asterisk) is located adjacent to the apical projection of this supporting cell, suggesting that this is a Deiter cell. G, Surface view of the sensory epithelium in an explant labeled with the supporting cell marker Prox1 (red). A single Id3-transfected cell is illustrated (green). The nucleus of this cell is positive for Prox1, making it appear yellow (arrow). The cell also extends a lumenal projection. The arrowhead indicates the plane of section for the Z-stack in H. H, Confocal Z-stack cross section of the cell in G, illustrating the lumenal projection. Note the yellow nucleus (arrow) indicating expression of Prox1. Based on the shape and position of this cell, it is most likely a third-row Deiter cell. I, A single Id3-transfected cell (green) that is also positive for p75NTR (red). This cell was located in the row of inner pillar cells, which all express p75NTR at their lumenal surfaces, giving rise to a stripe of p75NTR expression (arrow). J, Confocal Z-stack cross section of the same cell as in I. The morphology of this cell is consistent with a pillar cell. Scale bars: A, 20 μm; B–E, 10 μm; F, 5 μm; G, 10 μm; H–J, 5 μm.
Quantification of the effects of transfection with Id3 in a total of nine separate explants indicated that only ∼6% of transfected cells developed as hair cells (Table 1 and supplemental Table 1, available at www.jneurosci.org as supplemental material). This result represented a significant change in the determination of progenitor cell fate, demonstrating an important role of down-regulation of Id in the specification of progenitor cells as hair cells.
Although the morphologies of the Id3-transfected cells were consistent with a supporting cell fate, it was also possible that these cells could be arrested at the progenitor cell stage. Therefore, to determine whether these cells had developed as supporting cells, explants transfected with Id3 were double labeled with anti-EGFP and supporting cell-specific markers, including anti-Jagged1, anti-Prox1, or anti-p75NTR. Id3-transfected cells within the sensory epithelium were found to express Jag1, Prox1, and p75NTR (Fig. 5D–J), demonstrating that these cells had developed as supporting cells. These results demonstrate that, although downregulation of Ids is required for development of hair cells, Ids apparently do not inhibit supporting cell formation.
Overexpression of E47 does not inhibit Id function
Based on our results and previous studies, the loss of function of Ids should lead to either premature differentiation of cells as hair cells, an overproduction of hair cells, or both. However, targeted deletion of even two Ids, Id1 and Id3, results in embryonic lethality between E12 and E13 (Lyden et al., 1999), making it impossible to analyze the effects of deletion of even two Ids on hair cell differentiation. To overcome the limitations of functional redundancy, we overexpressed the E-protein E47. Because both Math1 and Ids form heterodimers with E47, we hypothesized that an overexpression of E47 might effectively inhibit Id function by generating a large number of Id-E47 heterodimers, thereby decreasing the level of Ids available to inhibit Math1 function. If this hypothesis was correct, an excess of E47 would result in a bias toward the hair cell fate. However, results indicated that overexpression of E47 did not change the percentage of transfected cells that developed as hair cells (Table 1 and supplemental Table 1, available at www.jneurosci.org as supplemental material).
Discussion
Id proteins regulate Math1 activity and hair cell differentiation
The generation of hair cells requires Math1 (Bermingham et al., 1999; Zheng and Gao, 2000). However, the factors that regulate Math1 activity during the development of the organ of Corti are essentially unknown. Previous results have demonstrated that the Math1 gene contains an E-box consensus binding site in its enhancer region that has been shown to play a role in autoregulation through binding of Math1/E-protein heterodimers (Helms et al., 2000), and this autoregulatory enhancer has been shown to be active in developing hair cells (Woods et al., 2004). Given that the function of Ids in various systems is to suppress binding of specific bHLH transcription factors, such as Math1, to E-proteins, it seemed likely that Ids could play a role in hair cell development through negative regulation of Math1 autoregulatory activity.
Ids can act as key regulators of binary cell fate choices
The results of numerous studies have shown that hair cells and supporting cells develop from a common progenitor within the inner ear (Kelley et al., 1995; Fekete et al., 1998; Lang and Fekete, 2001). These results suggest that one of the final cell fate choices for many progenitors in the cochlea is a binary choice between a hair cell and a supporting cell. Similarly, natural killer (NK) cells and T-cells (T) in the thymus develop from a common bipotent progenitor cell (for review, see Pear et al., 2004). Ids, and in particular Id2, play a role at the bifurcation point at which T/NK precursors must commit to either T-cells or NK lineage cells (Morrow et al., 1999; Ikawa et al., 2001). The activity of bHLH transcription factors is necessary for T-lineage differentiation; however, expression of Id2 effectively inhibits this bHLH activity, leading to commitment to the NK fate. In Id2 mutants, NK cells are greatly reduced, whereas overexpression of Ids leads to an increase in NK cell commitment (Heemskerk et al., 1997; Ikawa et al., 2001). The results presented here suggest a similar role for Ids in the choice between hair cell and supporting cell.
Two possible roles for Ids in the choice between hair cell and supporting cell should be considered. First, Ids may be indirectly involved in the commitment of hair cells through the inhibition of Math1/E-protein heterodimers. This suggestion is supported by the fact that the inhibition of differentiation by the functional inactivation of bHLH transcription factors by Ids has been reported during different developmental events including myogenesis and neurogenesis (Benezra et al., 1990; Jan and Jan, 1993; Lee, 1997; Martinsen and Bronner-Fraser, 1998). However, it is also possible that Ids play a direct role in the induction of supporting cell fates. If this were the case, expression of Ids should be sufficient to directly induce the supporting cell fate. Previous results have demonstrated that cells within the GER can be induced to develop as supporting cells (Woods et al., 2004). In the course of these studies, several hundred Id3-transfected cells were generated within the GER of cochlear explants, but none of these cells expressed any of the tested supporting cell markers (data not shown). Therefore, these results suggest that Ids indirectly regulate cell fate through inhibition of Math1.
Id gene regulation
The mechanisms that regulate the expression of Id genes in the cochlea are unknown. In particular, it is not clear how the expression of Id is downregulated in Math1-postive cells. Some studies indicate that Ids are negatively regulated by cyclin-dependent kinase 2 (Cdk2) (Hara et al., 1997). Moreover, it has been reported that Cdk2 is involved in the development of the organ of Corti (Malgrange et al., 2003). Thus, it is possible that Cdk2 could regulate Ids in the cochlea. In addition, previous data have shown that Id1–Id3 are degraded by the ubiquitin proteasome pathway (Bounpheng et al., 1999). Other studies have shown that Hes1 is able to sequester Id2, preventing dimerization (Jogi et al., 2002). However, it is unlikely that Hes1 negatively regulates Id expression, because Hes1 is only expressed in supporting cells during development of the organ of Corti. Interestingly, alternate longer splice variants have been identified for Id1 and Id3 (referred to as Id1L and Id3L). These splice variants are negative regulators of Id1 and Id3 function (Springhorn et al., 1994; Deed et al., 1996). Therefore, one possible mechanism for Id regulation in the cochlea would be a switch from expression of Id1/Id3 to Id1L/Id3L in developing hair cells. A study of the spatiotemporal pattern of Id splice variant expression could lead to valuable data regarding this possible mechanism.
Loss of Id function
The results presented here have addressed the effects of overexpression of Ids. However, it would be informative to examine the effects of loss of Id function in cochlear development. Targeted deletions have been generated for Id1, Id2, and Id3 (Yan et al., 1997; Lyden et al., 1999; Yokota et al., 1999), and the individual null mutants have been reported to have very few abnormalities and none that are associated with hearing deficit. However, the relatively subtle effects of the inactivation of a single Id gene are almost certainly the result of compensation via functional redundancy. This hypothesis is supported by the demonstration of profound developmental defects in Id1/Id3 double-mutant embryos that die at approximately E12 (Lyden et al., 1999). Because Id1, Id2, and Id3 are expressed in similar patterns within the cochlea, it seems likely that it would be necessary to generate a triple mutant to observe the full effects of loss of Id function in the ear.
In an attempt to overcome the limitations of functional redundancy, we used an Id-binding partner, the class I bHLH, E47, as a dominant negative to inhibit Id1, Id2, and Id3 in undifferentiated prosensory cells. Vinals et al. (2004) demonstrated that overexpression of E47 can sequester Id proteins, thus inhibiting Id function. However, overexpression of E47 did not have any obvious influence on progenitor cell fates. This result suggests that the absolute quantity of E47 and therefore Math1/E47 heterodimers is not limiting in this process and rather that it is the ratio of Math1/E47 to Id/E47 heterodimers within a single cell that instructs a progenitor cell to become either a hair cell (Math1/E-protein ratio is higher than the Id/E-protein ratio) or a supporting cell (Id/E-protein ratio is higher than the Math1/E-protein ratio). These findings also indicate that the role of Ids in the development of hair cells and supporting cells may extend beyond a straightforward inhibition of the formation of Math1/E-protein heterodimers.
Interestingly, previous results demonstrated that the down-regulation of the Id homolog extramacrochaetae (emc) had very subtle effects on the expression of atonal (ato), the Drosophila Math1 homolog, and photoreceptor development in Drosophila (Brown et al., 1995). However, the same study showed that the combined loss of emc and hairy, a Drosophila homolog of the HES genes, allowed ato upregulation and subsequent early differentiation of photoreceptors. This result suggests that the inhibition of Id alone may not be sufficient to influence cell fate. Rather, as will be discussed below, it may be necessary to inhibit both Id and Notch to see a loss of function phenotype.
A model for the role of Id regulation in hair cell differentiation
Based on the results presented here and in previous work, we propose the following working model for the role of Ids in the development of the organ of Corti (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). In early development, a group of cells within the cochlear duct constitute a prosensory region from which individual progenitor cells will develop as either hair cells or supporting cells. Initially, Ids are expressed throughout this group of cells. As development continues, the same cells begin to express Math1, indicating that Ids are not capable of completely inhibiting Math1 expression. These results are consistent with the demonstration that there is both Math1-dependent and Math1-independent control of Math1 transcription (Helms et al., 2000). However, in cells that express both Ids and Math1, Math1 appears incapable of upregulating its own transcription to a level sufficient to activate the downstream signaling pathway that is required for hair cell formation. It appears that, initially, there is a balance of expression levels between Id and Math1; however, as time progresses, Id expression is down-regulated in some cells, leading to increased Math1 activity and a commitment toward the hair cell fate. In contrast, in cells that maintain Id expression, Math1 activity continues to be inhibited. As individual cells begin to develop as hair cells, they upregulate expression of the Notch ligands, Jagged2 and Delta1 (Lanford et al., 1999; Morrison et al., 1999), which leads to the activation of Notch (Woods et al., 2004) and the expression of HES1 and HES5 in neighboring cells (Zheng et al., 2000; Zine et al., 2001). The simultaneous expression of Ids and HES1 or HES5 in single progenitor cells is sufficient to completely inhibit Math1 transcription, diverting these cells from a hair cell fate and leading to their formation as supporting cells through an undetermined inductive signal that is generated by adjacent hair cells (Woods et al., 2004).
Footnotes
This work was supported by funds from the Intramural Program at National Institute on Deafness and Other Communication Disorders. We thank Robert Benezra, Barbara Christy, and Ryoichiro Kageyama for providing reagents. We also thank Doris Wu and Mark Warchol for reading a previous version of this manuscript.
Correspondence should be addressed to Matthew W. Kelley, National Institute on Deafness and Other Communication Disorders/National Institutes of Health, Section on Developmental Neuroscience, 35 Convent Drive, Bethesda, MD 20892. E-mail: kelleymt@nidcd.nih.gov.
DOI:10.1523/JNEUROSCI.3859-05.2006
Copyright © 2006 Society for Neuroscience 0270-6474/06/260550-09$15.00/0
References
- Andres-Barquin PJ, Hernandez MC, Israel MA (2000) Id genes in nervous system development. Histol Histopathol 15: 603–618. [DOI] [PubMed] [Google Scholar]
- Anniko M (1983) Cytodifferentiation of cochlear hair cells. Am J Otolaryngol 4: 375–388. [DOI] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H (1990) The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 61: 49–59. [DOI] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY (1999) Math1: an essential gene for the generation of inner ear hair cells. Science 284: 1837–1841. [DOI] [PubMed] [Google Scholar]
- Bermingham-McDonogh O, Stone J, Hume C, Oesterle E (2005) Developmental expression of the homeobox transcription factor Prox1 in the murine organ of Corti. Assoc Res Otolaryngol Abstr 1353.
- Bounpheng MA, Dimas JJ, Dodds SG, Christy BA (1999) Degradation of Id proteins by the ubiquitin-proteasome pathway. FASEB J 13: 2257–2264. [PubMed] [Google Scholar]
- Brown NL, Sattler CA, Paddock SW, Carroll SB (1995) Hairy and emc negatively regulate morphogenetic furrow progression in the Drosophila eye. Cell 80: 879–887. [DOI] [PubMed] [Google Scholar]
- Christy BA, Sanders LK, Lau LF, Copeland NG, Jenkins NA, Nathans D (1991) An Id-related helix-loop-helix protein encoded by a growth factor-inducible gene. Proc Natl Acad Sci USA 88: 1815–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deed RW, Jasiok M, Norton JD (1996) Attenuated function of a variant form of the helix-loop-helix protein, Id-3, generated by an alternative splicing mechanism. FEBS Lett 393: 113–116. [DOI] [PubMed] [Google Scholar]
- Desprez PY, Hara E, Bissell MJ, Campisi J (1995) Suppression of mammary epithelial cell differentiation by the helix-loop-helix protein Id-1. Mol Cell Biol 15: 3398–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete DM, Muthukumar S, Karagogeos D (1998) Hair cells and supporting cells share a common progenitor in the avian inner ear. J Neurosci 18: 7811–7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara E, Hall M, Peters G (1997) Cdk2-dependent phosphorylation of Id2 modulates activity of E2A-related transcription factors. EMBO J 16: 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemskerk MH, Blom B, Nolan G, Stegmann AP, Bakker AQ, Weijer K, Res PC, Spits H (1997) Inhibition of T cell and promotion of natural killer cell development by the dominant negative helix loop helix factor Id3. J Exp Med 186: 1597–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms AW, Abney AL, Ben-Arie N, Zoghbi HY, Johnson JE (2000) Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development 127: 1185–1196. [DOI] [PubMed] [Google Scholar]
- Ikawa T, Fujimoto S, Kawamoto H, Katsura Y, Yokota Y (2001). Commitment to natural killer cells requires the helix-loop-helix inhibitor Id2. Proc Natl Acad Sci USA 98: 5164–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan YN, Jan LY (1993) HLH proteins, fly neurogenesis, and vertebrate myogenesis. Cell 75: 827–830. [DOI] [PubMed] [Google Scholar]
- Jen Y, Weintraub H, Benezra R (1992) Overexpression of Id protein inhibits the muscle differentiation program: in vivo association of Id with E2A proteins. Genes Dev 6: 1466–1479. [DOI] [PubMed] [Google Scholar]
- Jen Y, Manova K, Benezra R (1997) Each member of the Id gene family exhibits a unique expression pattern in mouse gastrulation and neurogenesis. Dev Dyn 208: 92–106. [DOI] [PubMed] [Google Scholar]
- Jogi A, Persson P, Grynfeld A, Pahlman S, Axelson H (2002). Modulation of basic helix-loop-helix transcription complex formation by Id proteins during neuronal differentiation. J Biol Chem 277: 9118–9126. [DOI] [PubMed] [Google Scholar]
- Kelley MW, Talreja DR, Corwin JT (1995) Replacement of hair cells after laser microbeam irradiation in cultured organs of Corti from embryonic and neonatal mice. J Neurosci 15: 3013–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreider BL, Benezra R, Rovera G, Kadesch T (1992) Inhibition of myeloid differentiation by the helix-loop-helix protein Id. Science 255: 1700–1702. [DOI] [PubMed] [Google Scholar]
- Lanford PJ, Lan Y, Jiang R, Lindsell C, Weinmaster G, Gridley T, Kelley MW (1999) Notch signalling pathway mediates hair cell development in mammalian cochlea. Nat Genet 21: 289–292. [DOI] [PubMed] [Google Scholar]
- Lanford PJ, Shailam R, Norton CR, Gridley T, Kelley MW (2000) Expression of Math1 and HES5 in the cochleae of wildtype and Jag2 mutant mice. J Assoc Res Otolaryngol 1: 161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang H, Fekete DM (2001) Lineage analysis in the chicken inner ear shows differences in clonal dispersion for epithelial, neuronal, and mesenchymal cells. Dev Biol 234: 120–137. [DOI] [PubMed] [Google Scholar]
- Langlands K, Yin X, Anand G, Prochownik EV (1997) Differential interactions of Id proteins with basic-helix-loop-helix transcription factors. J Biol Chem 272: 19785–19793. [DOI] [PubMed] [Google Scholar]
- Lee JE (1997) Basic helix-loop-helix genes in neural development. Curr Opin Neurobiol 7: 13–20. [DOI] [PubMed] [Google Scholar]
- Lim DJ, Anniko M (1985) Developmental morphology of the mouse inner ear. A scanning electron microscopic observation. Acta Otolaryngol Suppl 422: 1–69. [PubMed] [Google Scholar]
- Lin J, Ozeki M, Javel E, Zhao Z, Pan W, Schlentz E, Levine S (2003) Identification of gene expression profiles in rat ears with cDNA microarrays. Hear Res 175: 2–13. [DOI] [PubMed] [Google Scholar]
- Lister J, Forrester WC, Baron MH (1995) Inhibition of an erythroid differentiation switch by the helix-loop-helix protein Id1. J Biol Chem 270: 17939–17946. [DOI] [PubMed] [Google Scholar]
- Lyden D, Young AZ, Zagzag D, Yan W, Gerald W, O'Reilly R, Bader BL, Hynes RO, Zhuang Y, Manova K, Benezra R (1999) Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature 401: 670–677. [DOI] [PubMed] [Google Scholar]
- Malgrange B, Knockaert M, Belachew S, Nguyen L, Moonen G, Meijer L, Lefebvre PP (2003) The inhibition of cyclin-dependent kinases induces differentiation of supernumerary hair cells and Deiters' cells in the developing organ of Corti. FASEB J 17: 2136–2138. [DOI] [PubMed] [Google Scholar]
- Martinsen BJ, Bronner-Fraser M (1998) Neural crest specification regulated by the helix-loop-helix repressor Id2. Science 281: 988–991. [DOI] [PubMed] [Google Scholar]
- Moldes M, Lasnier F, Feve B, Pairault J, Djian P (1997) Id3 prevents differentiation of preadipose cells. Mol Cell Biol 17: 1796–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montcouquiol M, Kelley MW (2003) Planar and vertical signals control cellular differentiation and patterning in the mammalian cochlea. J Neurosci 23: 9469–9478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A, Hodgetts C, Gossler A, Hrabe de Angelis M, Lewis J (1999) Expression of Delta1 and Serrate1 (Jagged1) in the mouse inner ear. Mech Dev 84: 169–172. [DOI] [PubMed] [Google Scholar]
- Morrow MA, Mayer EW, Perez CA, Adlam M, Siu G (1999) Overexpression of the Helix-Loop-Helix protein Id2 blocks T cell development at multiple stages. Mol Immunol 36: 491–503. [DOI] [PubMed] [Google Scholar]
- Norton JD, Atherton GT (1998) Coupling of cell growth control and apoptosis functions of Id proteins. Mol Cell Biol 18: 2371–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Sato S, Frederick JP, Sun XH, Zhuang Y (1999) Impaired immune responses and B-cell proliferation in mice lacking the Id3 gene. Mol Cell Biol 19: 5969–5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear WS, Tu L, Stein PL (2004) Lineage choices in the developing thymus: choosing the T and NKI pathways. Curr Opin Immunol 16: 167–173. [DOI] [PubMed] [Google Scholar]
- Phippard D, Lu L, Lee D, Saunders JC, Crenshaw III EB (1999) Targeted mutagenesis of the POU-domain gene Brn4/Pou3f4 causes developmental defects in the inner ear. J Neurosci 19: 5980–5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann V, van Cruchten I, Sablitzky F (1994) The expression pattern of Id4, a novel dominant negative helix-loop-helix protein, is distinct from Id1, Id2 and Id3. Nucleic Acids Res 22: 749–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubel EW (1978) Ontogeny of structure and function in the vertebrate auditory system. In: The handbook of sensory physiology IX (Jacobson M, ed), pp 135–237. New York: Springer.
- Satow T, Bae SK, Inoue T, Inoue C, Miyoshi G, Tomita K, Bessho Y, Hashimoto N, Kageyama R (2001) The basic helix-loop-helix gene hesr2 promotes gliogenesis in mouse retina. J Neurosci 21: 1265–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springhorn JP, Singh K, Kelly RA, Smith TW (1994) Posttranscriptional regulation of Id1 activity in cardiac muscle. Alternative splicing of novel Id1 transcript permits homodimerization. J Biol Chem 269: 5132–5136. [PubMed] [Google Scholar]
- Sun XH (1994) Constitutive expression of the Id1 gene impairs mouse B cell development. Cell 79: 893–900. [DOI] [PubMed] [Google Scholar]
- Sun XH, Copeland NG, Jenkins NA, Baltimore D (1991) Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol 11: 5603–5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinals F, Reiriz J, Ambrosio S, Bartrons R, Rosa JL, Ventura F (2004) BMP-2 decreases Mash1 stability by increasing Id1 expression. EMBO J 23: 3527–3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bartheld CS, Patterson SL, Heuer JG, Wheeler EF, Bothwell M, Rubel EW (1991) Expression of nerve growth factor (NGF) receptors in the developing inner ear of chick and rat. Development 113: 455–470. [DOI] [PubMed] [Google Scholar]
- Woods C, Montcouquiol M, Kelley MW (2004) Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat Neurosci 7: 1310–1318. [DOI] [PubMed] [Google Scholar]
- Yan W, Young AZ, Soares VC, Kelley R, Benezra R, Zhuang Y (1997) High incidence of T-cell tumors in E2A-null mice and E2A/Id1 double-knockout mice. Mol Cell Biol 17: 7317–7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S, Gruss P (1999) Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature 397: 702–706. [DOI] [PubMed] [Google Scholar]
- Zheng JL, Gao WQ (2000) Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat Neurosci 3: 580–586. [DOI] [PubMed] [Google Scholar]
- Zheng JL, Shou J, Guillemot F, Kageyama R, Gao WQ (2000) Hes1 is a negative regulator of inner ear hair cell differentiation. Development 127: 4551–4560. [DOI] [PubMed] [Google Scholar]
- Zine A, Van De Water TR, de Ribaupierre F (2000) Notch signaling regulates the pattern of auditory hair cell differentiation in mammals. Development 127: 3373–3383. [DOI] [PubMed] [Google Scholar]
- Zine A, Aubert A, Qiu J, Therianos S, Guillemot F, Kageyama R, de Ribaupierre F (2001) Hes1 and Hes5 activities are required for the normal development of the hair cells in the mammalian inner ear. J Neurosci 21: 4712–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]