Abstract
In contrast to classical transmitters, the detailed structures and cellular and synaptic actions of neuropeptides are less well described. Peptide mass profiling of single identified neurons of the mollusc Lymnaea stagnalis indicated the presence of 17 abundant neuropeptides in the cardiorespiratory neuron, visceral dorsal 1 (VD1), and a subset of 14 peptides in its electrically coupled counterpart, right parietal dorsal 2. Altogether, based on this and previous work, we showed that the high number of peptides arises from the expression and processing of four distinct peptide precursor proteins, including a novel one. Second, we established a variety of posttranslational modifications of the generated peptides, including phosphorylation, disulphide linkage, glycosylation, hydroxylation, N-terminal pyroglutamylation, and C-terminal amidation. Specific synapses between VD1 and its muscle targets were formed, and their synaptic physiology was investigated. Whole-cell voltage-clamp analysis of dissociated heart muscle cells revealed, as tested for a selection of representative family members and their modifications, that the peptides of VD1 exhibit convergent activation of a high-voltage-activated Ca current. Moreover, the differentially glycosylated and hydroxylated α2 peptides were more potent than the unmodified α2 peptide in enhancing these currents. Together, this study is the first to demonstrate that single neurons exhibit such a complex pattern of peptide gene expression, precursor processing, and differential peptide modifications along with a remarkable degree of convergence of neuromodulatory actions. This study thus underscores the importance of a detailed mass spectrometric analysis of neuronal peptide content and peptide modifications related to neuromodulatory function.
Keywords: mollusk, MALDI-TOF mass spectrometry, glycopeptide, posttranslational modification, HVA calcium channel, neuromodulation
Introduction
Neuropeptides play many important roles in the generation of behavior and the regulation of physiological processes in both vertebrates and invertebrates. Often, multiple neuropeptides are coexpressed in single cells, and their combinatorial action through divergent as well as convergent pathways enhance the information processing capacity of neurons in the nervous system (Kupfermann, 1991; Peaker, 1992; Zupanc, 1996; Brezina and Weiss, 1997; Hokfelt et al., 2000; Kupfermann and Weiss, 2001; Nusbaum et al., 2001; Merighi, 2002). This emphasizes the importance of detailed knowledge of the full complement of the chemical diversity of the coexpressed neuropeptides in relation to their possible action as cotransmitters in neuronal communication.
In principle, peptide diversity can be generated via several mechanisms, namely (1) cell-specific coexpression of distinct peptide precursor genes (Cropper et al., 1987; Church and Lloyd, 1991; Hekimi et al., 1991; Vilim et al., 1996; Jimenez et al., 1998; Wood et al., 2000), (2) alternative splicing of a pre-mRNA encoding different precursors (Saunders et al., 1992; Bogerd et al., 1993), (3) alternative proteolytic processing of a precursor protein (Eipper and Mains, 1980) and (4) posttranslational modifications of the peptides (Loh, 1992; Dockray et al., 1996). In all instances, a peptidomics approach based on mass spectrometry has proven to be superior to other classic biochemical approaches for defining peptide diversity at the level of single cells and small biopsies (Jimenez and Burlingame, 1998; Li et al., 2000; Schrader and Schulz-Knappe, 2001). Although studies using this methodology have revealed that the coexistence of multiple neuropeptides in single (invertebrate) neurons is a general phenomenon, the physiological significance of the full chemical diversity of peptides, including that generated by posttranslational modifications, has not been determined.
The identifiable neuron visceral dorsal 1 (VD1) and the electrically coupled right parietal dorsal 2 (RPD2) in the brain of the mollusc Lymnaea stagnalis provide a convenient model preparation in which to examine the generation of peptide diversity in the context of biological function, namely, the modulation of cardiorespiratory behavior (Janse et al., 1985; Kerkhoven et al., 1991). Previous studies have identified three peptides in VD1 and RPD2, of which two were shown to possess cardioexcitatory effects (Bogerd et al., 1994). Subsequent mass spectrometric analysis of single VD1 and RPD2 neurons (Jimenez et al., 1994, 1998) revealed overlapping peptide profiles and the presence of a total of 17 abundant mass peaks, representing putative peptides. This finding suggested that the peptide phenotype of VD1 and RPD2 is far more complex than reported previously (Bogerd et al., 1994) because of the expression of hitherto unknown neuropeptides and/or posttranslational processing.
To begin understanding the significance of neuropeptide diversity in VD1 and RPD2 and its functional correlate on target cells, we first performed a peptidomics approach to characterize the unidentified neuropeptides present in VD1 and RPD2, including their posttranslational modifications. Subsequent functional analysis of VD1 and RPD2 peptides on its target cells, i.e., heart muscle cells, showed that peptides derived from distinct precursor proteins and differentially modified forms exhibit convergent yet differential activation of the same calcium channel current in heart muscle fibers.
Materials and Methods
Animals
Adult specimens of L. stagnalis (shell height, 30–35 mm) bred in the laboratory under standard conditions were used. The animals were kept at 20°C and a 12 h light/dark cycle and were fed lettuce ad libitum.
Extraction and purification of peptides
Dissected VD1 and RPD2, which shares the same set of peptides as VD1 with the exception of the small cardioactive peptides (SCPs) (Jimenez et al., 1998), were collected on solid carbon dioxide and stored at –20°C. Pooled neurons (total, 800) were extracted by boiling in 0.1 m acetic acid for 8 min and centrifuged for 10 min at 4°C, and the supernatant was fractionated by HPLC using a Nucleosil C18 column (5 μm; 250 × 2.1 mm) with an increasing concentration of acetonitrile in 7 mm trifluoroacetic acid (TFA). The flow rate was 300 μl/min, and 1 min fractions were collected. Each fraction was screened by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) to search for the molecular ion species that were detected in the single VD1 mass spectrum. In some cases, fractions that contained the molecules of interest were further separated using the same Nucleosil column with increasing acetonitrile concentration in 0.05% HCl.
Mass spectrometry
For MALDI-TOF-MS, an aliquot (0.5 μl) of each sample (HPLC fraction or enzymatic digest) was mixed with 1 μl of matrix (10 mg 2,5-dihydroxy-benzoic acid and 1 mg 5-methoxy-2-benzoic acid dissolved in 1 ml of 7.5 mm trifluoroacetic acid in 30% acetonitrile). After drying of the sample, the target was placed into a laboratory-built laser desorption reflectron time-of-flight mass spectrometer equipped with a pulsed nitrogen laser (337 nm; pulse width, 3 ns) for analysis, as described previously (Jimenez et al., 1998). The spectra were externally calibrated using a peptide mixture, yielding an accuracy of 0.05%.
To obtain peptide sequence information, post-source decay mass spectra were recorded on a VG Tofspec SE time-of-flight mass spectrometer (Micromass, Manchester, UK) equipped with a nitrogen laser operated in reflectron mode, as described previously (Matsui et al., 1997). Samples were prepared as described above.
Electrospray MS and fragmentation spectra were recorded using a triple–quadrupole instrument (API300) from MDS-Sciex (Concord, Ontario, Canada) with a standard ion spray source that was infused at 5 μl/min. The collisionally induced dissociation gas used was nitrogen, and the collision energy was 35–50 eV, 0.2 Da step size, and 5–10 s total scan time.
Amino acid sequence analysis
Amino acid sequencing was performed by automated Edman degradation in a model 473 pulse liquid phase sequencer (Applied Biosystems, Foster City, CA), using the sequencing program recommended by the company. The sequence data were also used for quantification of the amount of peptides purified.
Enzymatic digestions
Purified peptide I was incubated with 1 μg of endo-lys-c (Sigma, St. Louis, MO) in 100 μl of 50 mm ammonium acetate buffer containing 1 mm dithiothreitol for 18 h at 20°C. Purified peptide K and α2 peptides were incubated with 1 μg of trypsin (Promega, Madison, WI) in 100 μlof 50 mm ammonium carbonate buffer for 18 h at 20°C. Dephosphorylation of peptide K was performed by incubation with calf intestine alkaline phosphatase (Promega) in 100 μl of 50 mm ammonium carbonate buffer containing 1 mm magnesium chloride for 30 min at 30°C. Digested peptides were directly analyzed by MALDI-TOF-MS and in some cases purified by reversed-phase HPLC using trifluoroacetic acid as counterion.
Cloning and sequencing of pro-I cDNA, Northern blotting, and in situ hybridization
To obtain the cDNA sequence of the precursor of peptide I (pro-I) first two degenerate nested antisense primers oligo VD cells (OLVD2) [5′-GGAATTCGGIACNGGNAC(GA)AA(TC)TCNC(TG)(GA)TC-3′] and OLVD3 [5′-GGAATTCGIAC(GA)AA(TC)TCNC(TG)(GA)TC(TC)TC(GA)TANGC-3′] (Isogen Bioscience, Ijsselstein, The Netherlands) containing 5′-EcoRI restriction sites, designed on the partial peptide I sequence KAYEDREFVPVVPK were used. Then, two specific nested sense primers, I-peptide query sense (IQS3) (5′-GGGAGCTCAGTTATATCCAGACTGCAG-3′) and IQS4 (5′-GGGATCCAGACTGCAGAACGGAG-3′) (Isogen Bioscience) containing 5′-BamHI restriction sites, designed on the 5′ end of the obtained 5′ rapid amplification of cDNA ends (RACE) I-clone, were used. The specific peptide I primers were used in combination with an anchored dT primer [adaptor oligo A (ADA)-(dT20)] and its respective adaptor primers ADA-I and ADA-II (Tensen et al., 1994) in 5′ RACE and 3′ RACE PCR on hexanucleotide primed cDNA from single RPD2 neurons as described previously (Spijker et al., 1999) with the following modifications. Three individual RPD2s were isolated using a fine glass pipette (tip diameter, ±100 μm) and transferred to a glass plate. After removal of superfluous liquid, 11 μl of water containing 400 pmol of hexanucleotides was added and mixed, and the sample was heated and processed for cDNA synthesis. After incubation, the cDNA was phenolized and precipitated. After dATP tailing, each cDNA synthesis was used for a first, a second, and a third round of amplification using 50 pmol of I-primer (OLVD2 or IQS3) in combination with ADA-(dT20), 50 pmol of I-primer (OLVD2 or IQS3) in combination with 50 pmol of ADA-I, and 50 pmol of nested I-primer (OLVD3 or IQS4) in combination with 50 pmol of nested adaptor primer ADA-II. Amplified cDNA of 5′ and 3′ RACE was digested with EcoRI/SacI or with BamHI/SacI and separated on an agarose gel. Fragments were cloned and sequenced in both orientations according to the dideoxy chain termination method (Sanger et al., 1977), using T7 DNA polymerase.
Total RNA from visceral and parietal ganglia was isolated (Chomczynski and Sacchi, 1987), and ∼10 μg of RNA was glyoxylated, fractionated on a 1.8% agarose gel, and transferred to charged nylon membranes. A peptide-I-specific 3′ RACE clone (nucleotides 79–633) was used to make a randomly primed probe, labeled with [μ-32P]dATP (specific activity >109 cpm/μg) by primer extension. The membrane was hybridized in 6× SSC, 0.2% SDS, 5× Denhardt's solution, 10 μg/ml tRNA, and 10 μg/ml herring sperm DNA, at 65°C for 16 h, subsequently washed in 0.5× SSC and 0.2% SDS at 65°C for 45 min, and then autoradiographed.
The cellular localization of mRNA encoding peptide I was studied in sections of the visceral and right parietal ganglia of L. stagnalis by in situ hybridization using a digoxigenin–dUTP-labeled cRNA. Parallel sections were used to confirm the simultaneous presence of α1 peptide in VD1 and RPD2 using immunocytochemistry. Tissue was fixed in 1% paraformaldehyde and 1% acetic acid (Smit et al., 1996). cRNA probes were synthesized from 100 ng of a PCR fragment of the M13 vector containing a partial I-peptide cDNA (nucleotides 1–260), including the T3 and T7 RNA polymerase promotor. Hybridization, washing, and visualization were done as described by Smit et al. (1996).
Cardiac muscle dissociation and neuron isolation
Muscle cells were dissociated from dissected atria and ventricles under sterile conditions using 0.2% dispase/collagenase in defined medium (Ridgway et al., 1991) (one auricle or ventricle per ml) for 4 h at 30°C while gently shaking. The dissociated cells were subsequently washed three times with defined medium and sedimented at low speed, before plating. Dissociated muscle fibers were kept in defined medium in Costar (Cambridge, MA) dishes at 20°C for 1–4 d (Lee et al., 2002). All neuronal culture procedures were performed as described previously (Syed et al., 1990). In brief, VD1 and right pedal dorsal 1 (RPeD1) were mechanically isolated under sterile conditions from the brain by applying gentle suction through a fire-polished pipette. Isolated cells were plated directly on poly-l-lysine-coated dishes containing brain-conditioned medium. After 24 h in culture, muscle cells were plated on top of neurites of VD1, and synapses were allowed to form during 24 h.
Electrophysiology
Simultaneous intracellular recordings from cocultured cells. For intracellular recordings, conventional electrophysiological techniques were used as described previously (Syed et al., 1990). Glass microelectrodes (resistance of 20–40 MΩ) were filled with a saturated solution of potassium sulfate (K2SO4), and neurons were impaled using Narishige (Tokyo, Japan) micromanipulators (models M202 and M204). The electrophysiological signals were amplified (amplifier model IR-283; Neurodata Instruments, New York, NY), displayed on a digital storage oscilloscope (Philips PM 3394), and recorded on a Gould Instruments (Valley View, OH) chart recorder (model TA240S).
Voltage-clamp recordings. Dissociated ventricle and auricle cells were bathed in HEPES-buffered saline (HBS) (see above). To record calcium currents, we used Ba2+ as charge carrier. HBS was washed out and replaced under continuous perfusion by a solution containing the following (in mm): 40 NaCl, 2 BaCl2, 10 HEPES, and 2 4-aminopyridine, pH 7.8 adjusted with tetraethylammonium (TEA)-OH. The internal pipette solution contained the following (in mm): 5 TEA-Cl, 29 CsCl, 2.3 CaCl2,10 HEPES, 11 EGTA, 2 ATP-Mg, and 0.1 GTP-Tris, pH 7.4 adjusted with CsOH. The calculated free calcium concentration was 10–8 m. Peptides were administered by micropressure ejection or Y-tube application. The Y-tube system used a pipette that was held under negative pressure and continuously perfused with a solution containing the relevant peptide. The tip of this pipette (diameter of ∼20 μm) was placed near the recorded cell. By switching on an electrically operated valve, the negative pressure was interrupted and a gentle flow of peptide solution, driven by gravity, was perfused over the cell. In this way, peptide application was accomplished within 500 ms. Whole-cell voltage-clamp experiments were performed with a List Electronics (Darmstadt, Germany) EPC 7 or an Axopatch 2B amplifier (Axon Instruments, Union City, CA), using a 0.1 GΩ feedback resistance. Pipettes (2–6 MΩ) were pulled on a Flaming/Brown P-87 (Sutter Instruments, Novato, CA) horizontal microelectrode puller from Clarke Electromedical Instruments (Pangbourne, UK) GC-150 glass. In the cell-attached configuration (seal resistance of ∼1 GΩ or more), the fast capacitive transients of the patch pipette were compensated. After disruption of the patch membrane, series resistance (≤7 MΩ) and cell capacitance (≤100 pF) were compensated. In the whole-cell mode, we waited ∼15 min before measuring barium currents, to equilibrate the interior of the cell with the pipette solution. Data acquisition was controlled by a CED 1401 analog-to-digital converter (Cambridge Electronics Design, Cambridge, UK) connected to an Intel 80486-based computer, run with voltage-clamp software developed in our laboratory. The current recordings were filtered at 1–5 kHz, sampled at >2 kHz, and stored on-line. This system allowed simultaneous application of voltage steps, acquisition of current recordings, and timed application of peptides.
Data analysis. The data points of the dose–response curve were fitted by a four-parameter Hill function (r = 0.9908) using Sigma Plot 2000 software (SPSS, Chicago, IL). A one-way ANOVA in conjunction with a post hoc Tukey–Kramer test was used to assess the significance of dose dependence. A Student's t test was used (p < 0.05) for comparison of the effect of the differentially modified α2 peptides.
Results
Purification and characterization of VD1 peptides
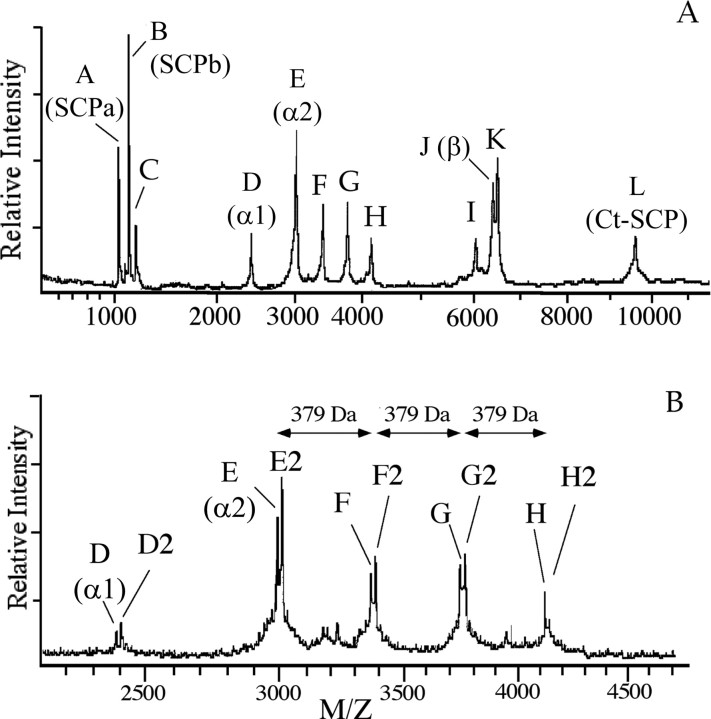
Profiling of single VD1 and RPD2 neurons by MALDI-TOF-MS revealed a multitude of major molecular ions that are named molecules A–L (Fig. 1A) (Jimenez et al., 1998). Closer examination of the mass region containing molecules D–H (Fig. 1B) revealed that mass peaks D–H are each accompanied by an extra peak at a mass increment of 16 Da, which are named accordingly as molecules D2, E2, F2, G2, and H2 (Fig. 1B). Previous studies have identified some of the VD1 and RPD2 molecules as bioactive peptides; peptides A, B, and L are uniquely expressed in VD1 and represent the small cardioactive peptides A, B, and the C-terminal peptide, respectively, that are all derived from the small cardioactive peptide precursor (Jimenez et al., 1998). Peptides D, E, and J correspond to the α1, α2, and β peptides, respectively, derived from the alternatively spliced α1 and α2 precursors expressed in both VD1 and RPD2 (Bogerd et al., 1994), with the β peptide contained in both precursors. The remaining mass peaks, i.e., molecules C, D2, E2, F, F2, G, G2, H, H2, I, and K, are present in both VD1 and RPD2 (Jimenez et al., 1998) and do not correspond to previously characterized peptides, suggesting that they may represent novel and/or posttranslationally modified peptides. Moreover, these peptides are present in all single-cell VD1 spectra analyzed to date (up to a few hundred), indicating the robustness of their expression. To gain additional insight into these molecules, we purified them from 800 individually dissected VD1 and RPD2, using a C18 column with increasing acetonitrile concentration in TFA (Fig. 2). Screening of each HPLC fraction with MALDI-TOF-MS allowed us to localize the fractions with the molecules of interest (Figs. 1, 2), which were then subjected to additional purification using reversed-phase HPLC (if necessary), structural characterization, and functional studies. Table 1 contains a summary of the mass spectrometry and sequencing data of the peptides identified in this study.
Figure 1.
MALDI mass spectra of a single VD1 neuron. The mass spectrum of VD1 (A) and an x-axis expansion displaying the mass range of m/z of 2000–4200 (B) show the presence of multiple molecular ions labeled A–L. Molecular ions D2–H2 occur at a 16 Da mass increment from D–H, respectively. Molecular ions D, E, and J correspond to peptides encoded by the α peptide precursors (Bogerd et al., 1994) and molecular ions A, B, and L to peptides derived from the SCP precursor (Jimenez et al., 1998). Molecular ions C, D2, E2, F and F2, G and G2, H and H2, I, and K represent hitherto unidentified putative peptides. x-Axis, mass-to-charge ratio; y-axis, ion intensity in arbitrary units.
Figure 2.
Purification of peptides from neurons VD1 and RPD2 using reverse-phase HPLC. Peptides were extracted from 800 VD1 and RPD2 neurons and resolved on a C18 column using an acetonitrile gradient in 7.5 mm TFA as a counter ion. Fractions eluting at 23,42,43,45,48,51, 53, 54, 55, 67, and 113 min contained molecules with masses that corresponded to molecules C, H2, H, G2, G, F2, F, D2, E2, I, and K, respectively, of the VD1 mass spectra in Figure 1. The elution positions of peptides F, G, and H as well as those of the previously identified α1, α2, and β peptides (Bogerd et al., 1994) and SCPs (Jimenez et al., 1998) are indicated. In some cases, to obtain pure peptide, it was necessary to further resolve peptides on a second gradient using HCl as a counter ion. The dashed line indicates the gradient of acetonitrile.
Table 1.
Summary of the molecular weight, sequence, and peptide mapping data obtained for the novel VD1 peptides of the Lymnaea brain (compare with Fig. 1)
|
Mass peak |
Measured MH+ VD1 |
Measured MH+ HPLC |
Peptide name |
Calculated unmodified MH+ |
Mass difference (Da) |
Modification |
|---|---|---|---|---|---|---|
| C | 1160.0 | 1160.1 | DRRLNGDTVN | 1159.9 | 0 | |
| D2 | 2418.1a | 2417.0a | α1 | 24001.7 | 16 | Hydroxylation |
| E2 | 3013.2a | 3012.7a | α2 | 2996.4 | 16 | Hydroxylation |
| F | 3375.8a | 3375.7a | α2 | 2996.4 | 1×379 | Monoglycosylation |
| F2 | 3392.4a | 3391.8a | α2 | 2996.4 | 395 | Monoglycosylation and hydroxylation |
| G | 3755.0a | 3754.8a | α2 | 2996.4 | 759 (2× 379) | Diglycosylation |
| G2 | 3771.4a | 3770.8a | a2 | 2996.4 | 775 | Diglycosylation and hydroxylation |
| H | 4134.8a | 4133.6a | α2 | 2996.4 | 1138 (3× 379) | Triglycosylation |
| H2 | 4151.3a | 4149.5a | α2 | 2996.4 | 1154 | Triglycosylation and hydroxylation |
| I | 6020.2 | 6019.1 | LyCCAP | 6027.1 | 6 | 3× disulphide bridge |
| K |
6455.4 |
6457.0 |
B |
6375.9 |
80 |
Phosphorylation |
The molecular weight of each calculated and measured protonated molecular ion (MH+) is listed, as well as the approximate mass difference from the calculated unmodified mass. The mass accuracy was 0.01-0.05%. The peptide names were assigned based on Edman degradation data. Monoglycosylation, diglycosylation, and triglycosylation indicates modification of one, two, or three Thr residues, respectively, in the N terminus of the α2 peptide with the 379 Da glycomoiety (HexNAc—hexuronic acid). For details of the amino acid sequence and modifications of α2 and β peptides, see Figures 3 and 4, respectively. For the cDNA precursor and deduced amino acid sequence of peptide I/LyCCAP, see Figure 5. Peptides A, B, D, E, J, and L are not listed because they correspond to the previously identified SCP precursor peptides (Jimenez et al., 1998) and unmodified α1, α2, and β peptide (Bogerd et al., 1994).
All α peptides (D2, E2, F, F2, G, G2, H, and H2) also contain a disulphide bridge.
MALDI-TOF-MS screening of the HPLC fractions revealed that molecule C [mass-to-charge ratio (m/z) of 1160.1] (Table 1) eluted in fraction 23, and the structure was established by Edman degradation as DRRLNGDTVN. This peptide corresponds to residues 38–47 of the N-terminal of the α peptide prohormones (Bogerd et al., 1993). Peptide material in fractions eluting at 42, 43, 45, 48, 51, 53, 54, and 55 min (Fig. 2) contained molecular ions corresponding to molecules H2 (m/z of 4149.5), H (m/z of 4133.6), G2 (m/z of 3770.8), G (m/z of 3754.8), F2 (m/z of 3391.8), F (m/z of 3375.7), D2 (m/z of 2417.0), and E2 (m/z of 3012.7), respectively, as those detected in the single-cell mass spectrum of VD1 (Fig. 1, Table 1). Some of these peptides were further resolved using reversed-phase HPLC with HCl as a counterion (data not shown). Molecule D2, which occurs at a mass increment of 16 Da to α1 peptide, yielded an α1 peptide sequence (DMYEGLAGRCQHHPRNCPGFN), indicating a modification (see below). Molecules F, G, and H are 379 Da (molecule F), two times 379 Da (molecule G), and three times 379 Da (molecule H) heavier than α2 peptide, suggesting that they represent multiple posttranslationally modified forms of this peptide. Indeed, Edman sequencing of purified molecules F, G, and H all yielded the same amino acid sequence, namely that of the α2 peptide (DMVTTTRIGTGGLAGRCQHHPRNCPGFN). In addition, F2, G2, and H2, which differ each by 16 Da to F, G, and H, respectively, also yielded an α2 peptide sequence.
To locate the site(s) of the posttranslational modification(s), ∼10% of the purified peptides F, G, and H and their +16 Da variants were pooled together and digested with trypsin. The remainder was kept for bioassay experiments. For comparison, synthetic α2 peptide was also digested with trypsin (Fig. 3A). As expected, trypsin cleavage of synthetic α2 peptide yields three specific peptides (at m/z of 802.1, 825.1, and 1426.3) that cover the full peptide sequence (Fig. 3A, right, B). The molecular ion at m/z of 1426.3 represents tryptic peptides 3 and 4, which are still connected via a disulphide bridge. After trypsin cleavage of the pool of modified α2 peptides F, F2, G, G2, H, and H2, three specific peptides were detected, namely those corresponding to tryptic peptides 2, disulphide-linked peptides 3 + 4, and disulphide-linked peptides 3 + 4(+16 Da) (see below) (Fig. 3A, left). Tryptic peptide 1 (m/z of 825.1) was absent from the spectrum, suggesting that the site of the 379 Da modification resides in the N-terminal part of the α2 peptide. However, the expected posttranslationally modified tryptic peptide 1 was not detected at mass increments of 379 Da of the native peptide. Therefore, under the present MS conditions, the structure of modified tryptic peptide 1 may be unfavorable for the desorption/ionization process and so cannot be effectively detected and further analyzed. Alternatively, to obtain structural information on the 379 Da modification, we subjected the intact variant α2 peptides to low-energy collisions on a triple–quadrupole instrument. Figure 3C shows the MS/MS spectrum of peptide F2. The low-mass region contains numerous carbohydrate-related fragments, indicating the presence of N-acetyl hexosamine (HexNac) (m/z of 204.0) and HexNac-related ions (m/z of 186.0, 168, 144.3, 137.8, and 126.5). The ions at m/z of 186 and 168 may be explained as water losses. The ion at 144 most likely arises from protonated HexNac by a formal loss of acetic acid, i.e., loss of water and additional loss of the amide-linked acetyl group. The ion at 126 would then correspond to loss of acetyl and loss of two water molecules. The same ions were detected in the low-mass regions of G2 and H2 (data not shown).
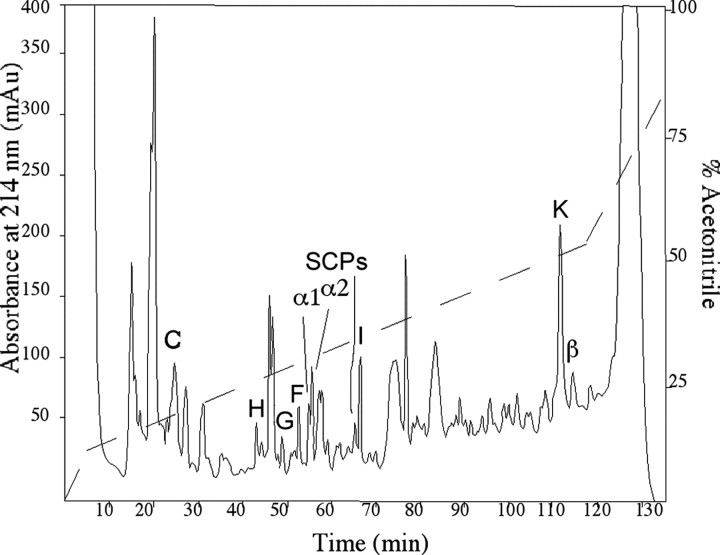
Figure 3.
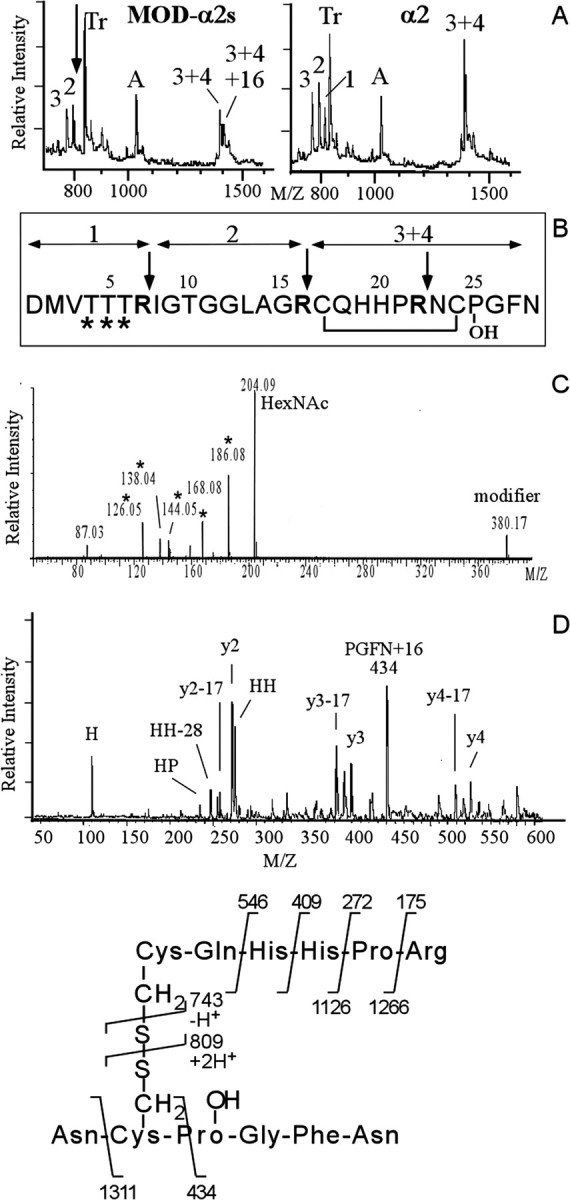
Structural characterization of the modifiedα2 peptides reveals multiple differentially glycosylated forms of the α2 peptide that in addition contains a HyP residue. A, MALDI mass spectra of a tryptic digest of the pooled modifiedα2 peptides F, F2, G, G2, H, and H2 (left) and synthetic α2 peptide (right). The arrow in A indicates the location of α2 tryptic peptide 1 that is expected to be modified (see B) and that has disappeared in the spectrum of the digested modified α2 peptides. Numbers indicate expected α2-specific tryptic peptides. Tr, Autoproteolytic fragments of trypsin. A, A-specific ion. B, Primary sequence of the α2 peptide. Trypsin cleaves after basic residues (indicated by the arrows), yielding four tryptic peptides. Fragments 3 and 4 are held together by a disulphide bridge. The sites predicted to be glycosylated are indicated by an asterisk. C, Electrospray MS/MS spectrum (the lower mass range) of purifiedα2 variant peptide F reveals glycosylation of F. Note the abundance of HexNac (at m/z of 204) and HexNac-related ions (m/z of 186, 168, 144, and 126). D, Identification of the 16 Da moiety on the modified α2 peptides as hydroxy-proline. Post-source decay spectrum (the lower mass range) of the trypsin digest fragment (3 + 4)+16 Da selected from the digest mixture of the pool of modifiedα2 peptides in A. The 16 Da mass addition is located to the four C-terminal residues PGFN, indicating a hydroxy-proline at position 25 instead of a proline residue. See E for the product assignments. Other fragment ions of the y- and a-series are indicated.
The spectrum of the modified α2 peptides digested with trypsin also indicated that the site of the 16 Da modification on α2 peptides F2, G2, and H2 is at the C terminal of the peptides. This can be deduced from the presence of an additional peak with a mass corresponding to that of the disulphide-linked tryptic peptides 3 and 4, +16 Da (Fig. 3A, left). To further characterize this fragment, we subjected it to post-source decay analysis (Fig. 3D), which yielded y ions at m/z of 175 (y1), 272 (y2), 409 (y3), and 546 (y4). Cleavage at the disulphide bond yielded the fragment ions at m/z of 809 and 743. The 16 Da mass increment was located on the C-terminal fragment PGFN (at m/z of 435). No additional sequence information was observed on the PGFN moiety. Among these four amino acid residues, a modification of 16 Da is most likely located on the proline residue, because it corresponds to the commonly occurring hydroxy-proline residue. Furthermore, during Edman sequencing, an extra peak in addition to proline was also observed, which eluted in a position expected for hydroxy-proline (data not shown).
In summary, the above sequencing and peptide mapping data of the α peptide family members identified peptide E2 as hydroxy-proline-containing α2 peptide (HyP-α2 peptide), peptide F as monoglycosylated α2 peptide, peptide F2 as monoglycosylated HyP-α2 peptide, peptide G as diglycosylated α2 peptide, peptide G2 as diglycosylated HyP-α2 peptide, peptide H as triglycosylated α2 peptide, peptide H2 as triglycosylated HyP-α2 peptide, and peptide D2 tentatively as HyP-α1 peptide.
Edman degradation of the molecule of 6455.5 Da in fraction 113 (Fig. 2) corresponding to molecule K in the VD1 spectrum (Fig. 1) revealed the N-terminal sequence of the β peptide (GSPYEPSLKDSFADSLTGKDQI...). This suggests that peptide K and the β peptide have the same peptide backbone and differ by a posttranslational modification of 80 Da, presumably the phosphorylation of a single amino acid residue. To confirm this assumption, peptide K was digested with alkaline phosphatase, which removed the phosphate group from the peptide, resulting in a reduced mass of peptide K by 80 Da (Fig. 4A). Using software to predict phosphorylation sites (Netphos 2.0; http://www.cbs.dtu.dk/services/NetPhos/) (Blom et al., 1999), the β peptide sequence contains eight predicted phosphorylation sites (Fig. 4B). To locate the site of phosphorylation, peptide K was digested with trypsin, and the tryptic fragments were detected by mass spectrometry. Figure 4C shows that the C-terminal tryptic peptide is detected at a mass increment of 80 Da, suggesting that either the Thr at position 50 (Netphos score of 0.623) or the Ser at position 53 (Netphos score of 0.992) (Fig. 4B) is phosphorylated, with the latter being more likely.
Figure 4.
Phosphatase treatment and tryptic peptide mapping of purified peptide K demonstrates phosphorylation of the C terminal of the β peptide. A, MALDI mass analysis of peptide K before (left) and after (right) dephosphorylation) using calf-intestine alkaline phosphatase (CIAP) shows removal of the 80 Da, phosphate moiety. B, Primary sequence of the β peptide. Trypsin cleaves after basic residues (indicated by the arrows), yielding five digest fragments, containing residues 1–9, 10–19, 20–35, 36–43, and 44–57, in case of a complete digestion. The putative phosphorylation sites are indicated with an asterisk. C, MALDI mass analysis of a tryptic digest of the phosphorylated β peptide (purified peptide K). Numbers indicate the residues of the trypsin fragments. Note the 80 Da mass addition to the partially cleaved C-terminal fragment 36–57. Unidentified molecular ions are labeled with a question mark.
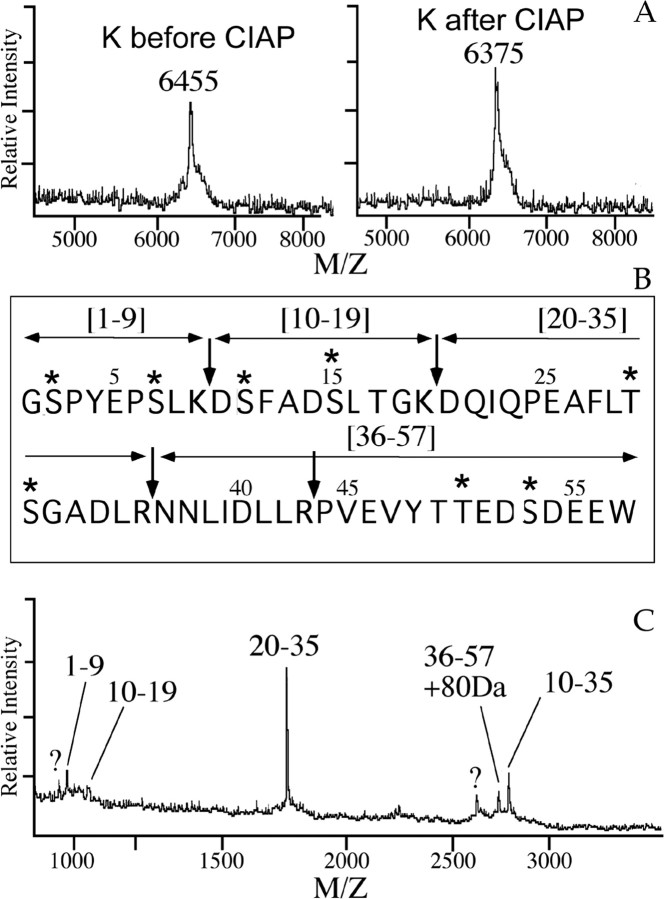
Molecule I at m/z of 6020.2 in the single-cell spectrum of VD1 (Fig. 1A) was located in fraction 67 (m/z of 6019.1) of the HPLC fractions (Fig. 2). Edman sequencing revealed the N-terminal sequence of this peptide as GPIGAKKF, which is a unique sequence not present in the National Center for Biotechnology Information (NCBI) database. To obtain internal sequence information, the purified molecule I was digested with endo-lys-c, and the fragments were purified by a C18 column (Fig. 5A). Amino acid sequence analysis of the main fragment at m/z of 1451.2 Da in fraction 36 yielded the sequence AYEDREFVPVPK, which is also not present in the NCBI database. The obtained amino acid sequences indicate that peptide I is a novel peptide. These sequences were subsequently used to synthesize degenerate oligonucleotides for a PCR experiment to characterize the neuropeptide precursor dubbed the peptide I precursor.
Figure 5.
Peptide mapping of purified molecule I, PCR strategy, and cDNA encoding peptide I. A, Purification of the endo-lys-c digested peptides of molecule I by reverse-phase HPLC. The main UV absorbing peak indicated contained the digest fragment of 1451.2 Da, which was subjected to amino acid sequence analysis. The dashed line indicates the gradient of acetonitrile. Inset in A, MALDI mass spectrum of the endo-lys-c digest of molecule I before purification. B, Schematic representation of the sequence strategy to obtain the cDNA of the peptide I. Both 5′ and 3′ RACE clones were sequenced in both orientations. Scale bar, 500 nt. C, Northern blot analysis of visceral and parietal ganglia shows a single band of 1.8 kb when hybridized to a peptide I-specific random labeled [γ-32P]dATP probe. The transcript size markers (yeast ribosomal RNAs) 26S (3400 bases) and 18S (1800 bases) are indicated. D, Nucleotide sequence and deduced amino acid sequence of peptide I cDNA. The number of nucleotides is indicated at the end of each line. Amino acid sequence number of peptide I starts at the predicted N-terminal residue (vertical arrow) and is indicated above the sequence. The consensus for polyadenylation is shown in bold.
Together, the peptide purification and characterization data explained all of the major unidentified molecular ions from the VD1 single-cell spectrum and revealed both novel peptide expression as well as extensive posttranslational modifications of previously identified peptides.
Characterization of the novel peptide I precursor and cellular localization of the transcript
To elucidate the structure of the precursor encoding peptide I, a PCR strategy was used on cDNA of a single neuron, i.e., RPD2, which also contains peptide I (Jimenez et al., 1998). Using triplicate reactions of 5′ RACE and 3′ RACE, a compiled peptide I encoding cDNA of 633 nt was obtained (Fig. 5B,D). The longest open reading frame of 222 nt is flanked by a 113 nt 5′ leader and a 298 nt 3′ untranslated region. The proposed initiation codon Met-1 is preceded by an upstream in-frame stop codon. Taking into account that Northern blot analysis detected a transcript of ∼1.8 kb (Fig. 5C), we conclude that the peptide I encoding cDNA of 633 nt represents a cDNA that is not full length. Nevertheless, only 5′ and 3′ untranslated sequences are missing because the cDNA clone comprises the complete open-reading frame. The deduced amino acid sequence of the peptide I cDNA contains a hydrophobic leader sequence that is predicted to be cleaved after Ala-20 (von Heijne, 1983) and a single peptide domain of 53 amino acids. The predicted protonated mass of the peptide I is 6027 Da, whereas the measured protonated mass is 6019.1/6020.2 Da (mass accuracy of ∼0.05%). Therefore, most likely the six Cys residues are connected via three disulphide bridges, yielding a protonated molecular weight of 6021 Da (Table 1).
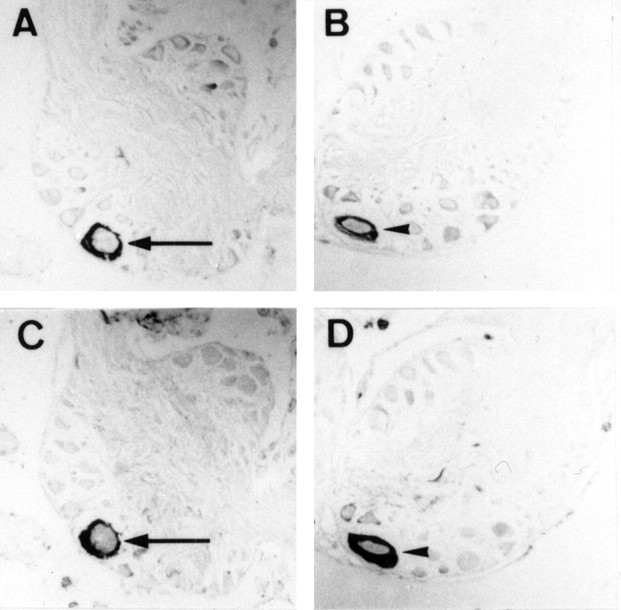
The cellular localization of mRNA of peptide I was studied in sections of the visceral and right parietal ganglia of L. stagnalis by in situ hybridization (Fig. 6A,B, respectively). VD1 and RPD2 neurons were identified on alternating sections by immunocytochemistry using an anti-α1 peptide antibody (Fig. 6C,D, respectively). The transcripts of peptide I are present in both neurons, confirming the peptide profiles obtained by MALDI-TOF-MS of single VD1 and RPD2 (Jimenez et al., 1998).
Figure 6.
Localization of the peptide I transcript. A–D, Cellular localization of the peptide I transcript by in situ hybridization in VD1 (A) and RPD2 (B) and identification of VD1 and RPD2 by immunostaining (C, D, respectively) with an anti-α peptide antibody on parallel sections of the brain. Arrows indicate VD1, and arrowheads indicate RPD2.
Functional connectivity of VD1 to the heart
Previous morphological (Kerkhoven et al., 1991), physiological, pharmacological (Bogerd et al., 1994), and peptide chemical (Jimenez et al., 1998) studies indicated that VD1 and RPD2 may modulate heart beat via the use of its cardioactive peptide transmitters. These in vivo studies do not, however, provide unequivocal evidence that these effects were indeed mediated directly.
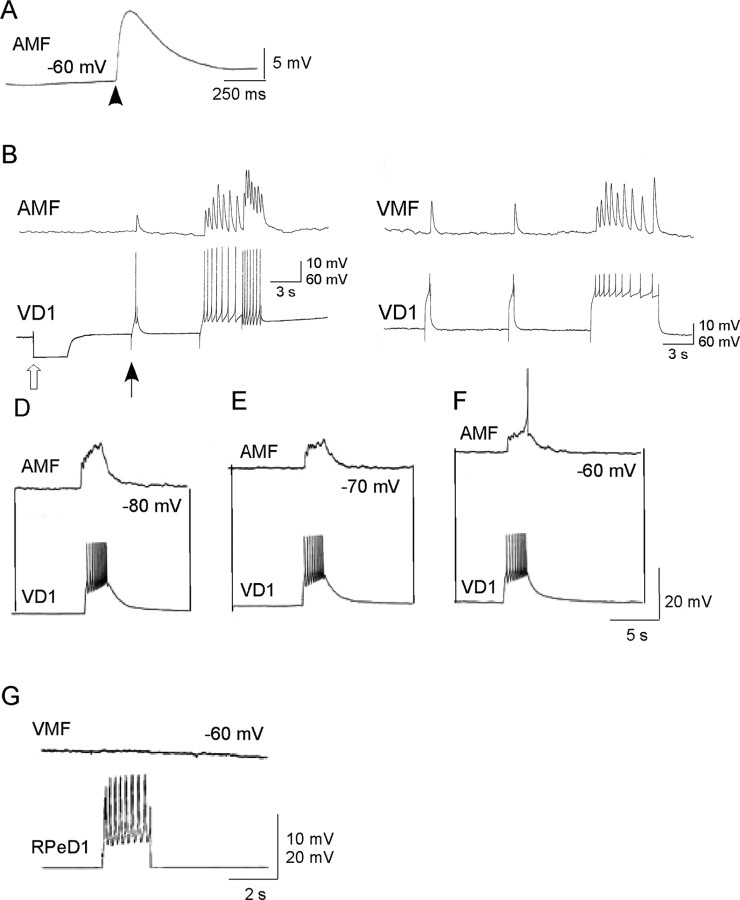
To further investigate the role of VD1 in modulating heart cell activity, we cocultured VD1, isolated from the brain, with enzymatically dissociated auricle and ventricle cells. First, we confirmed that the dissociated auricle and ventricle muscle fibers were functionally viable. Specifically, muscle contractions were induced by applying α2 peptide exogenously. An example of the effect of α2 peptide on the ventricle membrane potential is shown in Figure 7A (n = 12). Next, the neuron VD1 was isolated and cocultured with the dissociated muscle fibers. After 24 h of coculture, simultaneous intracellular recordings clearly show the presence of synaptic connections between VD1 and the heart muscle fibers (Fig. 7B–F) (n = 17). Spiking activity in VD1 induced by depolarizing current resulted in 1:1 excitatory junction potential (EJP) in the muscle cells (Fig. 7), indicating the presence of a chemical synapse. These synapses were clearly chemical because hyperpolarizing current pulses in VD1 (Fig. 7B) were not registered in the muscle fiber and the synaptic transmission was completely blocked by zero Ca2+ saline (data not shown). Moreover, in the in vitro coculture situation, the effects of VD1 on isolated heart muscle cells were voltage dependent. As the resting membrane potential of the postsynaptic muscle was brought closer to its resting membrane, the VD1-induced EJPs in the target became smaller, albeit generating spikes in the muscle fiber (Fig. 7D–F) (n = 7). To test for the specificity of synapse formation, the heart muscle was paired with RPeD1 (does not innervate heart), and synapses were tested electrophysiologically. We found that, despite extensive physical contacts, RPeD1 failed to establish synapse with the ventricular muscle fiber (Fig. 7G) (n = 5).
Figure 7.
Synaptic transmission between VD1 and heart muscle cells in vitro. A, Effect of synthetic α2 peptide (10–8 m) applied by pressure application on a ventricle muscle fiber (VMF). B–F, Chemical synapses between VD1 and auricle muscle fiber (AMF) and ventricle muscle fiber reform in cell culture. Simultaneous intracellular recordings from VD1 and auricle muscle fiber did not reveal electrical coupling between the cells. However, depolarizing current injected in VD1 (at filled arrow) triggered spikes in this cell, which induced 1:1 EJPs in the auricle muscle fiber. Open arrow represents hyperpolarizing current injection to determine the incidence of electrical coupling between the cells. C, Similarly, induced action potentials in VD1 induced 1:1 EJPs in the ventricle muscle fiber. D–F, Voltage dependence of the muscle cell response to VD1 stimulation. To show that the synapse between VD1 and the auricle muscle fiber is indeed excitatory, the postsynaptic potentials were tested at various different membrane potentials (–80 to –60 mV). Raising the membrane potential closer to its threshold (–60 mV; F) generated action potential in the muscle fiber. G, A heart muscle fiber cocultured with a nonsynaptic partner, RPeD1, did not develop synapses because the induced action potentials in RPeD1 failed to elicit any response in the ventricle muscle fiber despite physical contacts between the cells.
Convergence of VD1 peptides onto the same calcium channel in dissociated heart muscle cells
To unequivocally establish the presence of receptors for the different colocalized VD1 and RPD2 peptides on muscle cells of the heart and to study their functional role (including the glycosylated forms of the α2 peptide), we measured the electrophysiological effects of the differentially modified α2 peptides, SCPb, peptide I, and the phosphorylated β peptide on dissociated heart muscle cells. Under whole-cell voltage-clamp control conditions, muscle cells from the auricle responded to step depolarizations with variable time- and voltage-dependent outward currents. Ventricle muscle cells displayed reproducible responses to step depolarizations. Therefore, the effects of the VD1/RPD2 peptides were compared on dissociated ventricle cells. All VD1/RPD2 peptides tested caused most prominent effects on the inward currents (Figs. 8, 9). Using saline containing barium ions, we observed both a low-voltage-activated (LVA) and a high-voltage-activated (HVA) current in the ventricle cells (Fig. 8A,B). The LVA current was rapidly inactivating and insensitive to dihydropyridines and appeared not to be modulated by the VD1/RPD2 peptides (Fig. 8B). The HVA current was suppressed by the dihydropyridine nicardipine (81 ± 12% block by 1 μm; means ± SEM; n = 4) (data not shown). Therefore, the HVA current can be classified as a typical L-current, in agreement with a previous study in which this current was analyzed in more detail (Yeoman et al., 1999).
Figure 8.
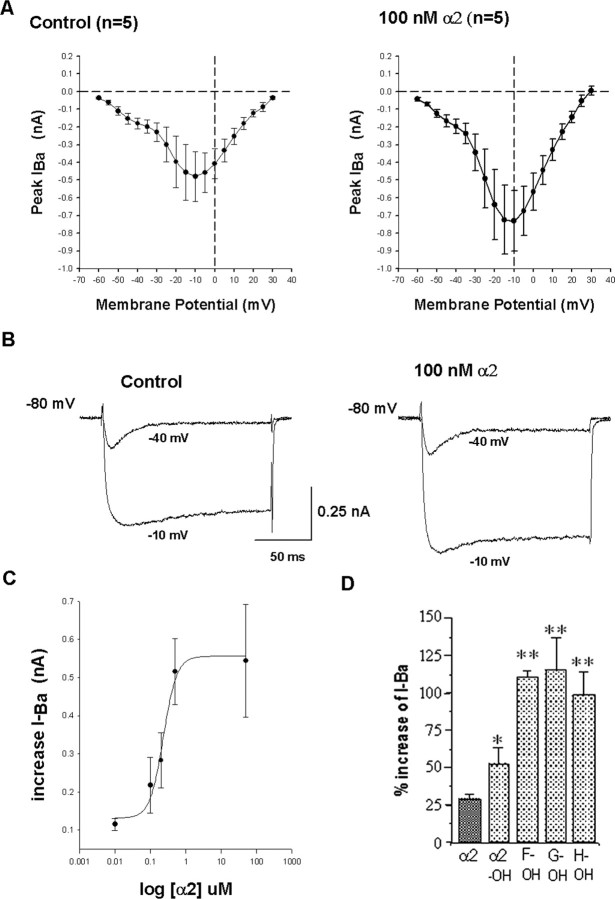
Voltage-dependent barium currents in Lymnaea ventricular cells and modulation by the α2 peptide family. A, I–V relationships of currents recorded inventricle cell. Left, Control; right, in the presence of 100 nm α2 peptide. B, Voltage step protocols were applied from a –80 mV holding potential to –40 and –10 mV. The current traces of a representative ventricle cell show the LVA and HVA current in the control condition and in the presence of α2 peptide. C, Dose–response curve of the α2 peptide. Effects of α2 peptide were tested in the range of 10–9 to 10–6 m. Each concentration was tested on different cells: n = 4 for 10–9 m; n = 5 for 10–8 m; n = 8 for 10–7 m; n = 6 for 2 × 10–7 m; n = 5 for 5 × 10–7 m; and n = 5 for 10–6 m. The data are described by a four-parameter Hill function (r = 0.9908). D, Quantitative comparison of the effects of the modified α2 peptides, HyP-α2 (E2), monoglycosylated HyP-α2 peptide (F2), diglycosylated HyP-α2 peptide (G2), and triglycosylated HyP-α2 peptide (H2), on the ventricle cell HVA-barium current, each applied at 2.5 × 10–8 m; n = 5 for each peptide. Error bars indicate SEM values. Peptides were tested on different cells. The enhancement of the barium current by the glycosylated-hydroxy-proline α2 peptide variants was significantly larger than the effect of the hydroxy-proline variant of α2 peptide that in turn was significantly larger than the effect of unmodified α2 peptide. OH, Hydroxy-proline. **p < 0.05, t test.
Figure 9.
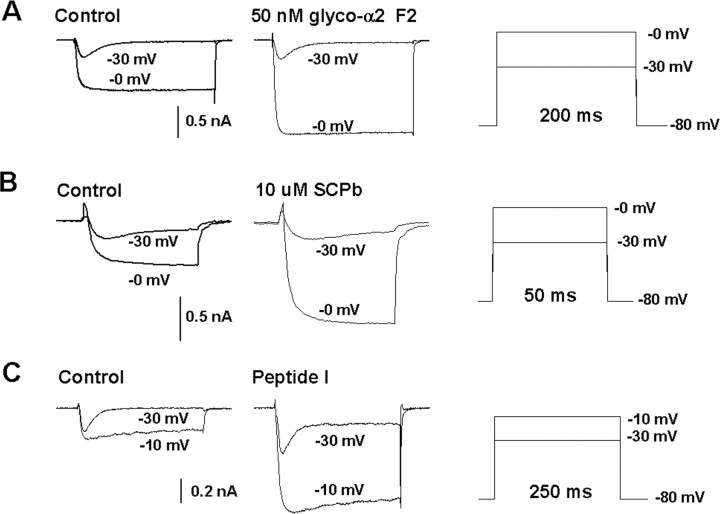
Selective increase of the HVA-barium current in ventricle cells by monoglycosylated HyP-α2 peptide F2, SCPb, and peptide I/LyCCAP. A–C, Family of current traces at –40 to +20 mV (10 mV steps) in the absence (left) and presence (middle) of monoglycosylated HyP-α2 peptide F2 (A), SCPb (B), and peptide I/LyCCAP (C). Voltage step protocols were applied as indicated in the right panel. Currents of representative ventricle cells are shown. Application of each peptide caused a strong increase in the HVA current.
The α2 peptide caused a consistent and dose-dependent increase in the amplitude of the L-type current (Fig. 8). We did not observe any changes in kinetic properties or voltage dependence. The dose dependence was determined for the range of 10–9 to 10–6 m (Fig. 8C). Between 10–7 and 10–6 m, the dose–effect curve was very steep, with a maximal effect of the barium current observed at 5 × 10–6 m (Fig. 8C). The dose–response curve can be described by a four-parametric Hill function (r = 0.9908), and the effect of concentration is significant (one-way ANOVA, p = 0.0037). Because saturation of the response did not yet occur within this concentration range, the EC50 cannot be determined with certainty but is estimated to be ∼2 × 10–7 m. Responses to higher concentration of α2 peptide could not be recorded because of the contractions of the muscle fibers. Next, we investigated whether the posttranslationally modified α2 peptides mimicked the effect of the unmodified α2 peptide. Like unmodified α2 peptide, the different modified α2 peptides (HyP-α2 peptide E2, monoglycosylated HyP-α2 peptide F2, diglycosylated HyP-α2 peptide G2, and triglycosylated HyP-α2 peptide H2) all increased the amplitude of the HVA-barium current without altering the kinetics or voltage dependence. An example of the response of the monoglycosylated HyP-α2 peptide F2 is shown in Figure 9A. Interestingly, the modified α2 peptides yielded larger responses than the α2 peptide itself (Student's t test, p < 0.05). Comparing the effect of the same concentration (2.5 × 10–8 m) demonstrates that the differentially glycosylated HyP-α2 peptides F2, G2, and H2 are equipotent and approximately twofold more potent than HyP-α2 peptide E2 (Student's t test, p < 0.05), which in turn is approximately twofold more potent than unmodified α2 peptide (Student's t test, p < 0.05) (Fig. 8D) (n = 5 for each peptide).
Testing the effect of SCPb on the isolated barium currents showed a striking effect of SCPb on the HVA-barium current (Fig. 9B). The threshold concentration for this effect was 1 μm (n = 4). At 10 μm SCPb, the increase of this current amounted to 300% (n = 3) (Fig. 9B). Finally, purified peptide I and purified phosphorylated β peptide (peptide K) were applied to dissociated ventricle cells at a concentration of ∼5 × 10–7 m. Peptide I clearly enhanced the HVA-barium currents of ventricle cells by ∼100% (n = 5). An example of the current traces and I–V relationships is shown in Figure 9C. Therefore, we renamed peptide I to Lymnaea calcium current activating peptide (LyCCAP). The phosphorylated β peptide did not have any effect on the HVA-barium currents of ventricle cells (n = 5; data not shown). The unphosphorylated β peptide (peptide J) and the C-terminal peptide of the SCP precursor (peptide L) were not tested because they could not be purified in large enough quantities for physiological experiments.
Discussion
Peptide diversity in VD1 and RPD2
In this study, we structurally and functionally characterized hitherto unidentified peptides in VD1 and RPD2 that were initially discovered by direct mass spectrometric peptide profiling of single neurons (Jimenez et al., 1994, 1998; and this study, Fig. 1). A summary of the precursors and proposed processing pathways of the peptides is shown in supplemental Figure 1 (available at www.jneurosci.org as supplemental material).
Diversity generated by different genes, alternative splicing, and processing of precursors
VD1 and RPD2 express in total three genes that encode four peptide precursors with distinct structural organization. The novel LyCCAP precursor expressed in both neurons contains only a single functional peptide domain, with no homology to other sequences in the NCBI database. The processing of LyC-CAP and SCP precursors to their respective peptides conform to the consensus sequences for proteolytic processing encoded in their precursors. The α peptide pre-mRNA is alternatively spliced to generate two mRNAs, giving rise to the α1 and α2 peptide precursors (Bogerd et al., 1993). Not all predicted dibasic cleavage sites (Lys-Arg or Arg-Arg) (Steiner et al., 1992) in the α peptide precursors are cleaved by the prohormone convertases. Because we have detected a peptide at m/z of 1160.0 with the primary sequence DRRLNGDTVN, the double Arg residues at positions 38 and 39 are not used as a cleavage site.
Peptide diversity generated by posttranslational modification
We characterized multiple posttranslational modifications of the α and β peptides derived from the α peptide precursors. The α1 and α2 peptides are highly similar and differ only in the N terminal, in which the Tyr-Glu residues at positions 3 and 4 inα1 are replaced by Val-Thr-Thr-Thr-Arg-Ile-Gly-Thr-Gly in α2. Using tryptic peptide mapping and tandem MS analysis, we demonstrated the presence of three differentially glycosylatedα2 peptides, most likely differentially modified on the three Thr residues in the N terminal. Absence of these residues in the α1 peptide explains the lack of glycosylation of α1. Tandem MS of modifiedα2 peptides demonstrated the presence of HexNac, which is commonly found in O-linked sugars attached to Ser and Thr residues. The carbohydrate of 379 Da may consist of HexNac–hexuronic acid, because this is the only structure predicted by the GlycoMod tool. Glycosylation of protein hormones has been implicated in protein folding, stability (Baudys et al., 1995; Kihlberg et al., 1995; Haneda et al., 2001), intracellular transport, enhanced receptor binding, and biological potency (Flack et al., 1994; Nissen, 1994; Furuhashi et al., 1995). However, few reports exist on the presence and functions of naturally occurring glycosylated peptides (Ivell et al., 1981; Nakakura et al., 1992). Therefore, in addition to their increased signaling properties, the differentially glycosylatedα2 peptides may have several other features than the unmodified α2 peptide.
Besides differential glycosylation, the Pro residue 25 of each α2 peptide is differentially modified into a HyP variant. Hydroxylation is a common posttranslational modification of Pro residues that occur in collagens (McCormack et al., 1994; Gaill et al., 1995). Because the HyP residues are especially important for noncovalent interactions between proteins, it may be speculated that the higher potency of the HyP-containing α2 peptide is caused by increased receptor binding.
Edman degradation in conjunction with phosphatase treatment and tryptic peptide mass mapping demonstrated that the majority of β peptide is phosphorylated. In the C-terminal part of the β peptide, the casein kinase consensus site (i.e., -Ser/Thr-X-X-Glu/Asp-) at the Ser residue at position 53 is most likely phosphorylated (NetPhos score of 0.992). Casein kinases are present at the luminal side of the Golgi apparatus (Vegh and Varro, 1994), and, therefore, they may phosphorylate the β peptide during passage through the Golgi apparatus.
VD1 modulates heartbeat via a direct neuromuscular connection
In vitro experiments in this study support the idea that VD1 modulates heartbeat via the use of its peptides. Coculture experiments of VD1 and RPeD1 and dissociated heart auricle and ventricle cells show that excitatory synapses are selectively formed in vitro between VD1 and heart cells. The successful synapse formation between VD1 and heart muscle cells is in line with previous immunocytochemical findings (Kerkhoven et al., 1991) that showed the presence of numerous α1 peptide-positive varicosities terminating on auricle trabecular muscle fibers, which are reminiscent of synapses. Moreover, VD1 and RPD2 peptides derived from all prohormones can be detected in biopsies of heart tissue using mass spectrometry (Jimenez et al., 1998). However, VD1 is not the only neuron that sends projections to the heart. Several peptidergic and nonpeptidergic heart motor neurons have been described in the past (Buckett et al., 1990a,b), indicating a rich innervation of the myogenic heart of Lymnaea.
Differentially modified α2 peptides, SCPb, and LyCCAP converge onto calcium channels in dissociated heart ventricle cells
We compared the effects of selected peptides of VD1 and RPD2 on membrane currents of heart ventricle muscle fibers, because they displayed more reproducible voltage-activated currents than the auricle fibers. The voltage-clamp recordings showed that a dihydropyridine-sensitive HVA current belonging to the L-type family is modulated by all tested variant α2 peptides, but significant quantitative differences were observed between them, with the posttranslationally modified forms being more potent. The SCPb peptide modulated the same HVA L-type calcium current as the α2 peptides, but the threshold of the response was higher than that for α2 and the modulatory effect reversed very slowly. Interestingly, the novel peptide LyCCAP also activated this current, with kinetics similar to the α2 peptides. Together, our voltage-clamp experiments show that seven peptides, representing three peptide families, exhibit convergent activation of a HVA L-type calcium current. Interestingly, this current is also modulated by FMRFamide (Brezden et al., 1999), from heart motor neurons (Buckett et al., 1990). Recently, the possible role of this current was investigated in detail in heart ventricle cells of Lymnaea by Yeoman and Benjamin (1999). They showed that the L-type current is important for pacemaking in generating action potentials and in providing calcium for contraction-excitation coupling. Therefore, modulation of the L-type current by VD1 peptides may alter the myogenic beating rate of the heart.
Convergent and divergent neuromodulation
The frequent occurrence of colocalized neurotransmitters suggests that the combination of transmitters released by a neuron attributes specificity to its actions (Kupfermann, 1991; Brezina and Weiss, 1997). The results reported here are interesting and perhaps surprising for several reasons: first, because VD1 and RPD2 cosynthesize such a large array of chemically diverse peptides; and second, because most of these peptides converge onto a single type of calcium channel to modulate a single type of Ca current. To the best of our knowledge, this study is the first to demonstrate that single neurons exhibit such a complex pattern of peptide gene expression, precursor processing, and modifications. The present results indicate that neuropeptides derived from three different genes (α peptides, SCPs, and LyCCAP) can exhibit activation of the same effector protein, the L-type HVA calcium channel. This remarkable degree of convergence in neuromodulation was also observed in the crustacean somatogastric ganglion (Swensen and Marder, 2000; Nusbaum et al., 2001). Convergence might prevent overmodulation by multiple modulatory inputs.
In addition to their convergent effects, it is feasible that α peptides and SCPs (and possibly LyCCAP) may also exert divergent effects. Based on the complexity of the whole auricle response and the differential effects of α2 peptide and SCPb application (Jimenez et al., 1998), it is likely that these peptides influence more than one channel type in Lymnaea heart muscle cells. This notion is consistent with the observation that the synaptic effects of VD1 in the coculture were evident even at varying hyperpolarizing potentials, suggesting the involvement of other ion channels, in addition to the HVA calcium channel. The potential discrepancy between sharp electrode and patch-clamp recordings may also be attributable to differential sensitivities of synaptic (coculture) and extrasynaptic (single cells) peptidergic receptors. Regarding the molecule mediating VD1-induced EJPs in heart muscle fibers, we believe this be a (combination of) peptide(s) because there is no evidence for the presence of small molecule transmitters in either VD1 somata or its axonal branches (Boer et al., 1979; Winlow et al., 1998; Feng et al., 2000). Together, it might be speculated that, through a process of combinatorial convergence and divergence (Brezina and Weiss, 1997), flexibility in neuronal output can be generated.
The full functional implications of the expression of such a variety of neuropeptides in VD1 must await detailed morphological and physiological studies. For example, it will be interesting to investigate whether the three different classes of cardioactive peptides in VD1 are colocalized in the same granules and thus obligatory coreleased or whether they are packaged in distinct granule types. In the latter case, differential release depending on the firing pattern may dynamically vary the ratio of the released peptides. Finally, the complexity of the peptide profiles in VD1 may be a general feature of widely acting modulatory neurons that, through extensive projection patterns, modulate a large number of targets. Possibly, in these modulatory neurons, a large array of different types of peptides is needed for (fine tuning of) the coordination of diverse target cells.
Footnotes
We thank Dr. H. D. Mansvelder for assisting in statistical analysis and Drs. K. F. Medzihradszky and M. Wuhrer for interpretation of the MS/MS data of the α2 peptide. N.I.S. is supported by Canadian Institute of Health Research (CIHR) and is the recipient of a CIHR Investigator Award.
Correspondence should be addressed to Dr. Connie R. Jiménez, Department of Molecular and Cellular Neurobiology, Center for Neurogenomics and Cognitive Research, Vrije Universiteit Amsterdam, DE Boelelaan 1085, 1081 HV Amsterdam, The Netherlands. E-mail: connie.jimenez@falw.vu.nl.
DOI:10.1523/JNEUROSCI.2566-05.2006
Copyright © 2006 Society for Neuroscience 0270-6474/06/260518-12$15.00/0
References
- Baudys M, Uchio T, Mix D, Wilson D, Kim SW (1995) Physical stabilization of insulin by glycosylation. J Pharmacol Sci 84: 28–33. [DOI] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S (1999) Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol 294: 1351–1362. [DOI] [PubMed] [Google Scholar]
- Boer HH, Schot LP, Roubos EW, ter Maat A, Lodder JC, Reichelt D, Swaab DF (1979) ACTH-like immunoreactivity in two electronically coupled giant neurons in the pond snail Lymnaea stagnalis Cell Tissue Res 202: 231–240. [DOI] [PubMed] [Google Scholar]
- Bogerd J, Van Kesteren RE, Van Heerikhuizen H, Geraerts WP, Veenstra J, Smit AB, Joosse J (1993) Alternative splicing generates diversity of VD1/RPD2 alpha peptides in the central nervous system of Lymnaea stagnalis Cell Mol Neurobiol 13: 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd J, Li KW, Jimenez CR, van der Schors RC, Ebberink RH, Geraerts WP (1994) Processing, axonal transport and cardioregulatory functions of peptides derived from two related prohormones generated by alternative splicing of a single gene in identified neurons VD1 and RPD2 of Lymnaea Brain Res Mol Brain Res 23: 66–72. [DOI] [PubMed] [Google Scholar]
- Brezden BL, Yeoman MS, Gardner DR, Benjamin PR (1999) FMRFamide-activated Ca2+ channels in Lymnaea heart cells are modulated by “SEEPLY,” a neuropeptide encoded on the same gene. J Neurophysiol 81: 1818–1826. [DOI] [PubMed] [Google Scholar]
- Brezina V, Weiss KR (1997) Analyzing the functional consequences of transmitter complexity. Trends Neurosci 20: 538–543. [DOI] [PubMed] [Google Scholar]
- Buckett KJ, Peters M, Dockray GJ, Minnen J van, Benjamin PR (1990a) Regulation of heart beat in Lymnaea by motoneurones containing FMRFamidergic-like peptides. J Neurophysiol 63: 1426–1435. [DOI] [PubMed] [Google Scholar]
- Buckett KJ, Peters M, Benjamin PR (1990b) Excitation and inhibition of the heart of the snail Lymnaea by non-FMRFamidergic motoneurones. J Neurophysiol 63: 1436–1447. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159. [DOI] [PubMed] [Google Scholar]
- Church PJ, Lloyd PE (1991) Expression of diverse neuropeptide cotransmitters by identified motor neurons in Aplysia J Neurosci 11: 618–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cropper EC, Lloyd PE, Reed W, Tenenbaum R, Kupfermann I, Weiss KR (1987) Multiple neuropeptides in cholinergic motor neurons of Aplysia: evidence for modulation intrinsic to the motor circuit. Proc Natl Acad Sci USA 84: 3486–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dockray GJ, Varro A, Dimaline R (1996) Gastric endocrine cells: gene expression, processing, and targeting of active products. Physiol Rev 76: 767–798. [DOI] [PubMed] [Google Scholar]
- Eipper BA, Mains RE (1980) Structure and biosynthesis of pro-adrenocorticotropin/endorphin and related peptides. Endocr Rev 1: 1–27. [DOI] [PubMed] [Google Scholar]
- Feng ZP, Hasan SU, Lukowiak K, Syed NI (2000) Target cell contact suppresses neurite outgrowth from soma-soma paired Lymnaea neurons. J Neurobiol 42: 357–369. [DOI] [PubMed] [Google Scholar]
- Flack MR, Bennet AP, Froehlich J, Anasti JN, Nisula BC (1994) Increased biological activity due to basic isoforms in recombinant human follicle-stimulating hormone produced in a human cell line. J Clin Endocrinol Metab 79: 756–760. [DOI] [PubMed] [Google Scholar]
- Furuhashi M, Shikone T, Fares FA, Sugahara T, Hsueh AJ, Boime I (1995) Fusing the carboxy-terminal peptide of the chorionic gonadotropin (CG) beta-subunit to the common alpha-subunit: retention of O-linked glycosylation and enhanced in vivo bioactivity of chimeric human CG. Mol Endocrinol 9: 54–63. [DOI] [PubMed] [Google Scholar]
- Gaill F, Mann K, Wiedemann H, Engel J, Timpl R (1995) Structural comparison of cuticle and interstitial collagens from annelids living in shallow sea-water and at deep-sea hydrothermal vents. J Mol Biol 246: 284–294. [DOI] [PubMed] [Google Scholar]
- Haneda K, Inazu T, Mizuno M, Iguchi R, Tanabe H, Fujimori K, Yamamoto K, Kumagai H, Tsumori K, Munekata E (2001) Chemo-enzymatic synthesis of a bioactive peptide containing a glutamine-linked oligosaccharide and its characterization. Biochim Biophys Acta 1526: 242–248. [DOI] [PubMed] [Google Scholar]
- Hekimi S, Fischer-Lougheed J, O'Shea M (1991) Regulation of neuropeptide stoichiometry in neurosecretory cells. J Neurosci 11: 3246–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokfelt T, Broberger C, Xu ZQ, Sergeyev V, Ubink R, Diez M (2000) Neuropeptides—an overview. Neuropharmacology 39: 1337–1356. [DOI] [PubMed] [Google Scholar]
- Ivell R, Schmale H, Richter D (1981) Glycosylation of the arginine vasopressin/neurophysin II common precursor. Biochem Biophys Res Commun 102: 1230–1236. [DOI] [PubMed] [Google Scholar]
- Janse C, Van der Wilt GJ, Van der Plas J, Van der Roest M (1985) Central and peripheral neurones involved in oxygen perception in the pulmonate snail Lymnaea stagnalis Comp Biochem Physiol 82A: 459–469. [Google Scholar]
- Jimenez CR, Burlingame AL (1998) Ultramicroanalysis of peptide profiles in biological samples using MALDI mass spectrometry. Exp Nephrol 6: 421–428. [DOI] [PubMed] [Google Scholar]
- Jimenez CR, van Veelen PA, Li KW, Wildering WC, Geraerts WP, Tjaden UR, van der Greef J (1994) Neuropeptide expression and processing as revealed by direct matrix-assisted laser desorption ionization mass spectrometry of single neurons. J Neurochem 62: 404–407. [DOI] [PubMed] [Google Scholar]
- Jimenez CR, Li KW, Dreisewerd K, Spijker S, Kingston R, Bateman RH, Burlingame AL, Smit AB, van Minnen J, Geraerts WP (1998) Direct mass spectrometric peptide profiling and sequencing of single neurons reveals differential peptide patterns in a small neuronal network. Biochemistry 37: 2070–2076. [DOI] [PubMed] [Google Scholar]
- Kerkhoven RM, Croll RP, Van Minnen J, Bogerd J, Ramkema MD, Lodder H, Boer HH (1991) Axonal mapping of the giant peptidergic neurons VD1 and RPD2 located in the CNS of the pond snail Lymnaea stagnalis, with particular reference to the innervation of the auricle of the heart. Brain Res 565: 8–16. [DOI] [PubMed] [Google Scholar]
- Kihlberg J, Ahman J, Walse B, Drakenberg T, Nilsson A, Soderberg-ahlm C, Bengtsson B, Olsson H (1995) Glycosylated peptide hormones: pharmacological properties and conformational studies of analogues of [1-desamino,8-d-arginine]vasopressin. J Med Chem 38: 161–169. [DOI] [PubMed] [Google Scholar]
- Kupfermann I (1991) Functional studies of cotransmission. Physiol Rev 71: 683–732. [DOI] [PubMed] [Google Scholar]
- Kupfermann I, Weiss KR (2001) Motor program selection in simple model systems. Curr Opin Neurobiol 11: 673–677. [DOI] [PubMed] [Google Scholar]
- Lee TKM, Leung AAC, Brezden BL, Lukowiak K, Syed N (2002) Specificity of synapse formation between Lymnaea heart motor neuron and muscle fiber is maintained in vitro in a soma-muscle configuration. Synapse 46: 66–71. [DOI] [PubMed] [Google Scholar]
- Li L, Garden RW, Sweedler JV (2000) Single-cell MALDI: a new tool for direct peptide profiling. Trends Biotechnol 18: 151–160. [DOI] [PubMed] [Google Scholar]
- Loh YP (1992) Molecular mechanisms of beta-endorphin biosynthesis. Biochem Pharmacol 44: 843–849. [DOI] [PubMed] [Google Scholar]
- Matsui NM, Smith DM, Clauser KR, Fichmann J, Andrews LE, Sullivan CM, Burlingame AL, Epstein LB (1997) Immobilized pH gradient two-dimensional gel electrophoresis and mass spectrometric identification of cytokine-regulated proteins in ME-180 cervical carcinoma cells. Electrophoresis 18: 409–417. [DOI] [PubMed] [Google Scholar]
- McCormack FX, Calvert HM, Watson PA, Smith DL, Mason RJ, Voelker DR (1994) The structure and function of surfactant protein A. Hydroxyproline- and carbohydrate-deficient mutant proteins. J Biol Chem 269: 5833–5841. [PubMed] [Google Scholar]
- Merighi A (2002) Costorage and coexistence of neuropeptides in the mammalian CNS. Prog Neurobiol 66: 161–190. [DOI] [PubMed] [Google Scholar]
- Nakakura N, Hietter H, Van Dorsselaer A, Luu B (1992) Isolation and structural determination of three peptides from the insect Locusta migratoria Identification of a deoxyhexose-linked peptide. Eur J Biochem 204: 147–153. [DOI] [PubMed] [Google Scholar]
- Nissen C (1994) Glycosylation of recombinant human granulocyte colony stimulating factor: implications for stability and potency. Eur J Cancer 30A: S12–S14. [PubMed] [Google Scholar]
- Nusbaum MP, Blitz DM, Swensen AM, Wood D, Marder E (2001) The roles of co-transmission in neural network modulation. Trends Neurosci 24: 146–154. [DOI] [PubMed] [Google Scholar]
- Peaker M (1992) Chemical signalling systems: the rules of the game. J Endocrinol 135: 1–4. [DOI] [PubMed] [Google Scholar]
- Ridgway RL, Syed NI, Lukowiak K, Bulloch AG (1991) Nerve growth factor (NGF) induces sprouting of specific neurons of the snail, Lymnaea stagnalis J Neurobiol 22: 377–390. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74: 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders SE, Kellett E, Bright K, Benjamin PR, Burke JF (1992) Cell-specific alternative RNA splicing of an FMRFamide gene transcript in the brain. J Neurosci 12: 1033–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader M, Schulz-Knappe P (2001) Peptidomics technologies for human body fluids. Trends Biotechnol 19: S55–S60. [DOI] [PubMed] [Google Scholar]
- Smit AB, Spijker S, Van Minnen J, Burke JF, De Winter F, Van Elk R, Geraerts WP (1996) Expression and characterization of molluscan insulin-related peptide VII from the mollusc Lymnaea stagnalis Neuroscience 70: 589–596. [DOI] [PubMed] [Google Scholar]
- Spijker S, Smit AB, Eipper BA, Malik A, Mains RE, Geraerts WP (1999) A molluscan peptide alpha-amidating enzyme precursor that generates five distinct enzymes. FASEB J 13: 735–748. [DOI] [PubMed] [Google Scholar]
- Steiner DF, Smeekens SP, Ohagi S, Chan SJ (1992) The new enzymology of precursor processing endoproteases. J Biol Chem 267: 23435–23438. [PubMed] [Google Scholar]
- Swensen AM, Marder E (2000) Multiple peptides converge to activate the same voltage-dependent current in a central pattern-generating circuit. J Neurosci 20: 6752–6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed NI, Bulloch AG, Lukowiak K (1990) In vitro reconstruction of the respiratory central pattern generator of the mollusk Lymnaea Science 250: 282–285. [DOI] [PubMed] [Google Scholar]
- Tensen CP, Van Kesteren ER, Planta RJ, Cox KJ, Burke JF, van Heerikhuizen H, Vreugdenhil E (1994) A G protein-coupled receptor with low density lipoprotein-binding motifs suggests a role for lipoproteins in G-linked signal transduction. Proc Natl Acad Sci USA 91: 4816–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegh M, Varro A (1994) Phosphorylation of human preprogastrin 93–101 by a Golgi membrane kinase from rat mammary gland. Biochim Biophys Acta 1205: 49–53. [DOI] [PubMed] [Google Scholar]
- Vilim FS, Price DA, Lesser W, Kupfermann I, Weiss KR (1996) Costorage and corelease of modulatory peptide cotransmitters with partially antagonistic actions on the accessory radula closer muscle of Aplysia californica J Neurosci 16: 8092–8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G (1983) Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem 133: 17–21. [DOI] [PubMed] [Google Scholar]
- Winlow W, Spencer GE, Syed NI, Qazzaz MM (1998) Modulation of reconstructed peptidergic synapses and electrical synapses by general anaesthetics. Toxicol Lett 100–101: 77–84. [DOI] [PubMed] [Google Scholar]
- Wood DE, Stein W, Nusbaum MP (2000) Projection neurons with shared cotransmitters elicit different motor patterns from the same neural circuit. J Neurosci 20: 8943–8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoman MS, Brezden BL, Benjamin PR (1999) LVA and HVA Ca2+ currents in ventricular muscle cells of the Lymnaea heart. J Neurophysiol 82: 2428–2440. [DOI] [PubMed] [Google Scholar]
- Zupanc GK (1996) Peptidergic transmission: from morphological correlates to functional implications. Micron 27: 35–91. [DOI] [PubMed] [Google Scholar]