Abstract
Variation in gray matter (GM) and white matter (WM) volume of the adult human brain is primarily genetically determined. Moreover, total brain volume is positively correlated with general intelligence, and both share a common genetic origin. However, although genetic effects on morphology of specific GM areas in the brain have been studied, the heritability of focal WM is unknown. Similarly, it is unresolved whether there is a common genetic origin of focal GM and WM structures with intelligence. We explored the genetic influence on focal GM and WM densities in magnetic resonance brain images of 54 monozygotic and 58 dizygotic twin pairs and 34 of their siblings. For genetic analyses, we used structural equation modeling and voxel-based morphometry. To explore the common genetic origin of focal GM and WM areas with intelligence, we obtained cross-trait/cross-twin correlations in which the focal GM and WM densities of each twin are correlated with the psychometric intelligence quotient of his/her cotwin. Genes influenced individual differences in left and right superior occipitofrontal fascicle (heritability up to 0.79 and 0.77), corpus callosum (0.82, 0.80), optic radiation (0.69, 0.79), corticospinal tract (0.78, 0.79), medial frontal cortex (0.78, 0.83), superior frontal cortex (0.76, 0.80), superior temporal cortex (0.80, 0.77), left occipital cortex (0.85), left postcentral cortex (0.83), left posterior cingulate cortex (0.83), right parahippocampal cortex (0.69), and amygdala (0.80, 0.55). Intelligence shared a common genetic origin with superior occipitofrontal, callosal, and left optical radiation WM and frontal, occipital, and parahippocampal GM (phenotypic correlations up to 0.35). These findings point to a neural network that shares a common genetic origin with human intelligence.
Keywords: genetics, twins, neuroanatomy, voxel-based morphometry, intelligence, cognition
Introduction
Variation in total gray and white matter volume of the adult human brain is primarily (70–80%) genetically determined (Pennington et al., 2000; Pfefferbaum et al., 2000; Baaré et al., 2001). Moreover, total brain volume is positively correlated with general intelligence, and both share a common genetic origin (Posthuma et al., 2002). However, this finding does not necessarily mean that genes influence focal brain structures in the same manner throughout the brain. Moreover, it does not necessarily mean that a common genetic origin with general intelligence is shared by all structures throughout the brain.
Determining the extent to which focal brain morphology and its association with cognitive functioning are influenced by genes (or environment) is important for both our understanding of healthy functioning and elucidating the causes of brain disease. More specifically, it enhances our knowledge of individual variation in brain functioning and aids in the interpretation of the morphological changes as found in psychiatric disorders such as schizophrenia. Also, it allows future efforts to find particular genes responsible for brain structures and functions to be concentrated on brain areas that are under considerable genetic influence.
To date, most studies focused on the genetic influence on variation in total brain volume. A few studies have examined genetic effects on more specific brain areas throughout the brain. Morphology of focal frontal and temporal gray matter (GM) areas is particularly influenced by genetic factors (Thompson et al., 2001; Wright et al., 2002). However, the genetic influence on focal white matter (WM) has not been examined. It has been suggested that frontal GM volume and intelligence share genetic factors, but it also remains unresolved whether this association shares a common genetic origin (Thompson et al., 2001; Toga and Thompson, 2005).
To determine the relative contributions of genetic, common, and unique environmental influences on variation in focal brain structures and their common origin with intelligence, the (extended) twin model is a powerful approach (Posthuma and Boomsma, 2000), particularly because morphological (Hulshoff Pol et al., 2002) and cognitive (Posthuma et al., 2000) findings in twins can be extended toward the general singleton population. For genetic influences, the extent to which monozygotic (MZ) twin pairs resemble each other more than is the case for dizygotic (DZ) twin pairs is the determining factor. However, in addition to genetic influences, common (or shared) environmental influences may play a role in explaining resemblances. The presence of shared environmental factors is suggested when correlations in DZ twins are larger than half the MZ correlation (Boomsma et al., 2002). A first impression of the importance of unique environmental factors is obtained from the extent to which MZ twins do not resemble each other. In a similar manner, the extent to which genetic and environmental factors influence the association of two traits such as brain structure and intelligence can be determined by comparing the cross-trait/cross-twin correlation between brain structure and intelligence in MZ and DZ twins. If cross-trait/cross-twin correlations are larger in MZ twins, this suggests genetic mediation of the association.
Materials and Methods
Magnetic resonance brain images were analyzed using voxel-based morphometry (VBM) (Collins et al., 1995; Hulshoff Pol et al., 2001, 2004). The extent to which GM and WM densities of MZ twin pairs correlate with each other compared with the correlations of DZ twin pairs and siblings enables the calculation of brain maps of the genetic contribution to focal GM and WM areas throughout the brain. For the genetic analyses, structural equation modeling (SEM) was used implemented in Mx software (Neale, 1997) in each voxel separately. The significance level was corrected for multiple comparisons according to random field theory (Worsley et al., 1996; Ashburner and Friston, 2000). To define which anatomical WM areas were involved, the genetic WM maps were superimposed onto histologically defined human anatomical WM brain maps (Bürgel et al., 1999; Rademacher et al., 2001). The cross-trait/cross-twin correlations were obtained between focal GM and WM density and intelligence, verbal (VIQ) and nonverbal (performance) (PIQ) intelligence quotient measures of the Dutch version of the Wechsler Adult Intelligence Scale III-Revised (WAIS-IIIR). We chose to include VIQ and PIQ separately because they correlate ∼0.6 and both are highly associated to general intelligence. Also, by including both VIQ and PIQ, we could assess possible differential cross-trait/cross-twin correlations with focal GM and WM density in particular brain areas. The SEM analyses included IQ data of 688 subjects from 271 extended twin families (Posthuma et al., 2001) that overlapped for 135 subjects with the sample of 258 subjects who had magnetic resonance image (MRI) brain data. SEM allows for estimates to be based on all available data. Thus, the cross-trait/cross-twin correlations were based on the 135 subjects who had both MRI as well as IQ data. The estimates of means and variances for cognitive and MRI traits were based on all data, regardless of the pattern of bivariate missingness. The approach adds to the statistical power of the analysis.
We controlled for sex, handedness, and age throughout our analyses. Males and females were reported to have differences in brain density (Good et al., 2001b), and, compared with males, women were shown to have more white matter and fewer gray matter areas related to intelligence (Haier et al., 2005). However, because no differences in genetic or environmental influences on brain volumes (Baaré et al., 2001) or intelligence (Posthuma et al., 2001) were found in the males and females included in our sample, we estimated genetic and environmental influences on brain density for both the sexes combined. Although handedness was not of significant influence on brain density in a previous study using VBM (Good et al., 2001b), it has been associated with subtle changes in the hippocampus (Maguire et al., 2000) and in sulcal topography (Amunts et al., 2000). Therefore, we added it as a covariate to exclude for possible differences in the relationship between brain density and intelligence. Age significantly contributes to changes in brain density as we (Hulshoff Pol et al., 2001) and others have repeatedly found.
When interpreting the findings, limitations of our chosen approach have to be considered. The VBM procedure involves several steps. VBM requires registration of the separate brain images onto a model brain to allow for statistical inferences in focal brain areas. The choice of the model brain may have influenced the results. However, when comparing several model brains with a similar dataset in a previous study, we found that the results remained the same. Thus, the influence of the model brain on our results seems to have been of minimal influence. Moreover, registration error will occur during this procedure and is unavoidable. Therefore, some caution in interpreting these results is warranted. However, because we registered each individual brain separately to the model brain, it seems unlikely that a bias occurred that behaved differently in MZ and DZ twins other than the similarities that we expected to find attributable to differences in the overlap of their genetic makeup. Also, note that, because we were especially interested in the influence of genes on focal brain densities, thus after correction for the known overall effect of genes onto GM and WM volumes, we did not include a volume-preserving step (Good et al., 2001a) in the procedure. Thus, the GM and WM that we now find to be highly heritable include those areas in which the individual variation is influenced by genes on top of the genetic influences on global brain volume. One also has to keep in mind that the focal brain areas that we do not find to be genetically determined may still be determined by genes that influence overall brain volume. Alternatively, these focal brain areas may be influenced by environmental factors. Finally, our overall age correction may be only relevant for the age range as included in the sample. Aged and early development samples can and probably will give different patterns. Recently, a longitudinal study in children and adolescent singletons revealed a nonlinear trajectory of change in cortical thickness, which was associated with intelligence (Shaw et al., 2006). Within the age range of 19–69 years, changes in the genetic and environmental influences on brain morphology and in the relationship between brain morphology and intelligence may also occur. Analyses of possible changes in genetic and environmental influences with age await longitudinal follow-up measurements in these subjects.
Demographics.
For magnetic resonance brain imaging, a total number of 258 family members from 112 families participated in the study after written consent was obtained (Baaré et al., 2001) (Table 1). They consisted of 33 MZ male, 17 DZ male, 21 MZ female, 20 DZ female, and 21 DZ opposite-sex twin pairs, and 19 male and 15 female full siblings. Subjects were between 19 and 69 years of age at the time of the scan (mean ± SD, 30.7 ± 9.6 years). Handedness was determined by the Cognitive Assessment for Handedness and Laterality Data from the Comprehensive Assessment of Symptoms and History (Andreasen et al., 1992): 213 subjects were right-handed, 34 were left-handed, and 11 were mixed-handed. Twins were recruited from the (healthy) twin sample of the Department of Psychiatry of the University Medical Centre Utrecht and the Netherlands Twin Registry (http://www.tweelingenregister.org/index_uk.html). DNA testing using the polymorphic markers D06S474, D07S1804, D07S1870, D12S811, D13S119, D13S126, D13S788, D20S119, D22S683, DXS1001, and ELN, or D13S317, VWA, D74520, D35158, TH01, TP0X, CSF1P0, and D55818 determined zygosity. Except for one twin pair, all twins and their siblings were reared together. Two twin pairs were born by cesarean section delivery. Mental and physical health was assessed by means of the Family Interview for Genetic Studies (Nurnberger et al., 1994) and a medical history inventory, respectively. All subjects' consent was obtained according to the declaration of Helsinki. The Scientific and Ethical Committee of the University Medical Centre Utrecht, in which the study was performed, approved the study.
Table 1.
Demographics
| MZ | DZ | SIB | |
|---|---|---|---|
| n individuals (MRI/IQ) | 49/213 | 65/285 | 34/190 |
| Sex, M/F (MRI) | 34/15 | 33/32 | 19/15 |
| Sex, M/F (IQ) | 98/115 | 117/168 | 91/99 |
| Hand, R/L, A (MRI) | 41/8 | 54/11 | 29/5 |
| Hand, R/L, A (IQ) | 183/30 | 251/34 | 175/15 |
| Age, years (MRI) | 28.3 (3.6) | 29.2 (8.7) | 29.6 (4.8) |
| Age, years (VIQ, PIQ) | 37.7 (36.6) | 36.6 (13.3) | 36.7 (13.2) |
| VIQ (% correct) | 63.3 (13.3) | 64.9 (12.2) | 65.4 (13.3) |
| PIQ (% correct) | 76.0 (12.8) | 75.0 (14.0) | 75.6 (13.2) |
Values are shown as mean (SD). M, Male; F, female; R, right; L, left; A, ambidexter; SIB, singleton siblings of twin pairs.
Image acquisition.
Magnetic resonance images were acquired on a Philips (Amsterdam, The Netherlands) NT scanner operating at 1.5 T in all subjects. T1-weighted three-dimensional (3D) fast-field echo scans with 160–180 1.2 mm contiguous coronal slices [echo time (TE), 4.6 ms; repetition time (TR), 30 ms; flip angle, 30°; field of view (FOV), 256 mm/80%; in-plane voxel sizes, 1 × 1 mm2] and T2-weighted dual-echo turbo spin echo (DE-TSE) scans with 120 1.6 mm contiguous coronal slices (TE1, 14 ms; TE2, 80 ms; TR, 6350 ms; flip angle, 90°; FOV, 256 mm/80%; in-plane voxel sizes, 1 × 1 mm2) of the whole head were used for quantitative measurements. In addition, T2-weighted DE-TSE scans with 17 axial 5 mm slices and 1.2 mm gap (TE1, 9 ms; TE2, 100 ms; flip angle, 90°; FOV, 250 mm/80%; in-plane voxel sizes, 0.98 × 0.98 mm2) were used for clinical neurodiagnostic evaluation.
Image processing.
Processing was done on the neuroimaging computer network of the Department of Psychiatry, which includes Hewlett Packard (Palo Alto, CA) UNIX 9000 workstations, a compute server, and Pentium-3 personal computers. All images were coded to ensure blindness for subject identification. As part of our image processing pipeline, the T1-weighted images were transformed (no scaling) to the Talairach frame with software developed in house. The transformation used information gathered from the placement of a midline in coronal and axial views and the marking of the superior edge of the anterior commissure and the inferior edge of the posterior commissure in the sagittal view. The images were corrected for inhomogeneities in the magnetic field (Sled et al., 1998). Binary masks of GM and WM were made based on histogram analyses and series of mathematical morphology operators to connect all voxels of interest in the T1-weighted images. The GM and WM separation was done for voxels within the cranium. The intra-cranium was extracted from the T2-weighted images and superimposed onto the T1-weighted images to remove nonbrain tissue voxels, as developed and validated previously (Schnack et al., 2001). The binary GM and WM masks were then analyzed separately, using a voxel-based morphometry procedure that was based on the ANIMAL algorithm (Collins et al., 1995) in a similar manner as was done previously for the GM (Hulshoff Pol et al., 2001) and WM (Hulshoff Pol et al., 2004) masks (for detailed information, see http://www.smri.nl/techniques_description.html).
The binary GM masks with voxels of 1 × 1 × 1.2 mm3 were blurred using an isotropic Gaussian kernel (full-width at half-maximum of 8 mm) and resampled to a voxel size of 2 × 2 × 2.4 mm3 to generate GM “density maps.” After blurring the binary GM and WM maps, it is not useful to analyze the data in the original high resolution of the scans. We chose to double the voxel size in each direction. The downsampling ensured that the features were independent of the original position, scale and orientation of one dataset with respect to the other (Collins et al., 1995), and to decrease the number of statistical tests by a factor of 8. The GM density maps represent the local concentration of GM (between 0 and 1) per voxel. Each of the MRI images was transformed into a standardized coordinate system in a two-stage process using the ANIMAL algorithm (Collins et al., 1995). In the first step, a linear transformation was found by minimizing a mutual information joint entropy objective function (Maes et al., 1997) computed for each of the gray level images with the standardized brain, using nine parameters (three for rotation, three for translation, and three for scaling).
The standardized brain to which these images were transformed was selected earlier among 200 brain images of healthy subjects between 16 and 70 years of age. To select the standardized brain, all 200 brain images were registered to the Montreal Neurological Institute (MNI) standard brain (T1-weighted individual reference brain of the MNI, http://www.bic.mni.mcgill.ca/software/distribution) and averaged yielding one average brain image. The standardized brain was the brain image, with the smallest mean square error. In the second step, a nonlinear transformation was computed by maximizing the correlation of the subject's image with that of a standardized brain reference. The nonlinear transformation is run up to a scale (full-width at half-maximum of 4 mm) that aligns global anatomical regions but minimally affects local volume changes. The mean square error on the normalized intensity values was computed between each of the brain images and the average brain image. The linear and nonlinear transformations were then applied to the GM density maps to remove global differences in the size and shape of individual brains. Thus, all GM density maps were transformed to the standardized brain. The procedure for GM density was repeated for the WM binary maps. Thus, after the procedure, all of the WM density maps were transformed to the standardized brain.
To obtain a good estimate of areas of significance, the WM statistical maps were overlaid onto the maps of histologically identified long fiber tracts in human cerebrum (Bürgel et al., 1999; Rademacher et al., 2001). These maps are based on the delineation of fiber tracts in serial myeloarchitectonic sections (Bürgel et al., 1997) of 10 human postmortem brains, their 3D reconstruction, and registration to the T1-weighted individual MRI reference brain of the MNI (Zilles et al., 2002; Amunts et al., 2004). As a result, probabilistic maps of fiber tracts have been calculated that quantify intersubject variability in extent and location of the fiber tracts in each voxel of the reference space (http://www.fz-juelich.de/ime/Structure/). To be part of a fiber tract, a voxel had to have a probability of at least 50%. When a voxel was found to be part of more than one fiber tract, both the probability of inclusion and the inclusion of neighboring voxels were taking into consideration for labeling.
Cognitive assessment.
Measures of VIQ and PIQ were obtained by the WAIS-IIIR in 688 subjects from 271 extended twin families (Posthuma et al., 2001) (Table 1). A heritability of VIQ of 0.84 and PIQ of 0.67 was found.
Structural equation modeling in each voxel separately.
To estimate the contribution of additive genetic (A), common environmental (C), and unique environmental (E) factors to the variance in GM and WM density for each GM and WM voxel separately, we used structural equation modeling implemented in Mx software (Neale, 1997). In more than 95% of the ∼122,000 voxels, it was possible to obtain an estimate for A, C, and E. Voxels with an average GM density below 0.1 were excluded from the GM density voxel-based morphometry analysis. Likewise, voxels with an average WM density below 0.1 were excluded from the WM density analysis. Analyses were controlled for age, sex, and handedness (right, left, or ambidextrous). The extent to which A, C, and E explained the variance in GM and WM density was expressed as percentage of the total variance in each voxel. The relative fit of nested models (e.g., no influence of C compared with a model including C) was determined by likelihood ratio test. Twice the difference between the log-likelihoods of two models is asymptotically distributed as χ2. The degrees of freedom for these tests are equal to the difference in numbers of parameters being estimated. Using the principle of parsimony, the most restrictive model was accepted as the best fitting one in case the difference between a nested and a more comprehensive model was not significant (Neale, 1997).
Additionally, we explicitly tested several common assumptions of the twin method: equal environment assumption (homogeneity of means and variances across MZ and DZ twins) and generalizability of twin data to the singleton population (homogeneity of means and variances across twins and siblings).
Given the number of subjects, data resolution, voxel size, and volume of the search region, the critical threshold χ2 value for p < 0.05 after correcting for multiple comparisons is χ2 > 25.3 for difference df = 1 and χ2 > 29.3 for difference df = 2, according to random field theory (Worsley et al., 1996; Ashburner and Friston, 2000) (the uncorrected significance levels would have been χ2 > 3.8 for difference df = 1 and χ2 > 6.0 for difference df = 2). Voxels that reached significant results for both GM and WM maps were forced into GM and WM based on the highest value of the average density.
Statistical analysis of common origin between GM and WM density and intelligence.
To test whether verbal (VIQ) and nonverbal (PIQ) intelligence and GM and WM density share a common origin attributable to genetic, common, or unique environmental factors, the “observed correlation” (rp) between GM and WM density and VIQ/PIQ was decomposed into genetic and environmental components, but only in those voxels in which density was significantly determined by genetic factors.
Decomposition of the association was based on the comparison of cross-trait/cross-twin correlations for MZ and DZ twins (or sibling pairs) (Posthuma et al., 2002). If the correlation between GM and WM density of twin 1 and VIQ or PIQ of twin 2 is larger in MZ than in DZ twins, this indicates that the genes influencing GM and WM density partly overlap with the genes that influence VIQ/PIQ. The extent of the overlap is reflected by the magnitude of the genetic correlation (rg). rg reflects the correlation between the set of genes that influences focal GM and WM density and the set of genes that influences VIQ/PIQ. When the cross-trait/cross-twin correlations are similar for MZ and DZ twins, this suggests that environmental factors contribute to the observed phenotypic correlation between GM and WM density and VIQ/PIQ. Given a heritability of 0.80 for brain density and a heritability of 0.80 for VIQ/PIQ and a correlation between brain volume and VIQ/PIQ of 0.25, at least 16 MZ and 16 DZ twin pairs are needed.
In 24 MZ pairs, 31 DZ pairs, and 25 additional siblings (in these 135 subjects, both volumetric and IQ data were available), the correlations between GM and WM density and VIQ/PIQ were decomposed into genetic (A) and unique environmental components (E) using structural equation modeling for a trivariate genetic design for GM and WM density separately. Observed correlations with absolute values below 0.10 were not considered for additional analysis.
Results
Sources of variation in focal GM and WM areas
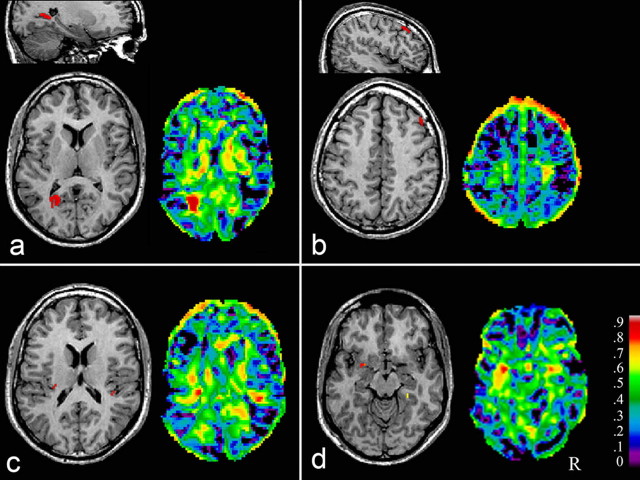
A heritability with a peak value of 0.83 was found for GM density in the left and right medial frontal cortex and right superior frontal cortex, of 0.80 and 0.77 in the left and right Heschl's gyrus, of 0.83 in the left occipital cortex, and of 0.83 in the left posterior cingulate (Table 2, Fig. 1) (see Fig. 4). In addition, heritabilities of 0.80 and 0.55 were found in the left and right amygdala, respectively, and of 0.69 in the right parahippocampal gyrus. Heritabilities with a peak value of 0.79 were found for WM density in the superior occipitofrontal fascicle bilaterally, of up to 0.82 throughout the corpus callosum in the left and right hemisphere (thus including off-midline callosal tract areas in the left and right hemisphere) but hardly involving the midline structure, up to 0.79 in the optic radiation, predominantly in the left hemisphere, and up to 0.79 in the corticospinal tract bilaterally (Table 2, Figs. 2, 4) (for a 3D representation, see Fig. 3 and supplemental movie 1, available at www.jneurosci.org as supplemental material).
Table 2.
Heritability estimates for focal gray and white matter areas with significant genetic contribution
| Left hemisphere |
Right hemisphere |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Talairacha |
Heritability | Talairach |
Heritability | ||||||
| x | y | z | x | y | z | ||||
| Gray matter | |||||||||
| Superior frontal | −5 | 58 | 35 | 0.76 | Superior frontalb | 7 | 43 | 51 | 0.80 |
| Medial frontal | −37 | 34 | 39 | 0.78 | Superior frontal | 21 | 53 | 33 | 0.76 |
| Postcentral gyrus | −55 | −29 | 57 | 0.83 | Medial frontalb | 29 | 43 | 37 | 0.82 |
| Posterior cingulate | −5 | −33 | 23 | 0.83 | Medial frontal | 43 | 22 | 43 | 0.83 |
| Heschl's gyrus | −27 | −31 | 21 | 0.80 | Heschl's gyrus | 37 | −36 | 19 | 0.77 |
| Amygdala | −29 | −5 | −7 | 0.80 | Amygdala | 21 | −5 | −15 | 0.55 |
| Occipital cortex | −25 | −60 | 11 | 0.85 | Parahippocampal | 25 | −31 | −11 | 0.69 |
| White matter | |||||||||
| Superior occipitofrontal fascicle | −17 | 10 | 25 | 0.79 | Superior occipitofrontal fascicle | 23 | 22 | 15 | 0.77 |
| Corpus callosum | −21 | −9 | 35 | 0.82 | Corpus callosum | 3 | 19 | 17 | 0.80 |
| Optic radiation | −35 | −48 | 17 | 0.69 | Optic radiation | 37 | −36 | 17 | 0.79 |
| Corticospinal tract | −17 | −12 | 21 | 0.78 | Corticospinal tract | 21 | −16 | 37 | 0.79 |
aThe x values indicate the distance in millimeters from the left (negative values) to the right (positive values) side of the brain passing through the anterior commissure. The y values indicate the distance from posterior (negative values) to anterior (positive values). The z values indicate the distance from inferior (negative values) to superior (positive values).
bTwo separate genetically determined areas were identified within the superior and medial frontal cortices.
Figure 1.
Genetically influenced focal GM density brain areas. Heritability estimates of GM density in focal brain areas in healthy adult humans are shown for the significance level thresholded A-map superimposed on axial and sagittal sections through the magnetic resonance image of the standardized reference brain (left) and for the complete A-map (right). The significance level thresholded A-map shows the voxels that had a significant fit of the genetic model compared with the other models based on the likelihood ratio test and using structural equation modeling. Moreover the level of significance was set at χ2 > 25.3 for Δdf = 1 and χ2 > 29.3 for Δdf = 2 after correction for multiple comparisons according to random field theory (the uncorrected significance levels would have been χ2 > 3.8 and χ2 > 6.0). Analyses were controlled for age, sex, and handedness. a, Left occipital cortex. b, Right medial frontal gyrus. c, Left and right Heschl's gyrus. d, Left amygdala and right parahippocampal gyrus. R, Right.
Figure 4.
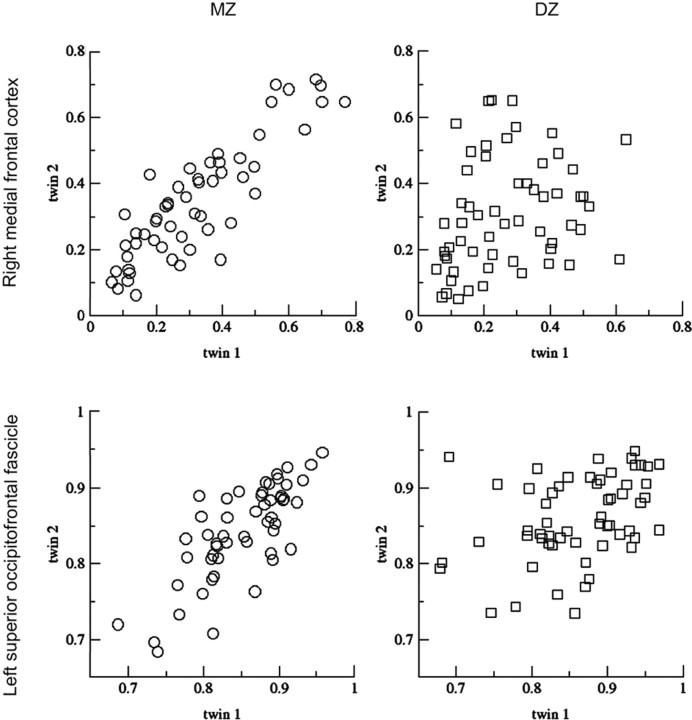
GM density in the right medial frontal cortex (top) and WM density in the left superior occipitofrontal fascicle (bottom). Dots represent values of GM and WM density (varying between 0 and 1) for individual MZ (left) and DZ (right) twin pairs at (x, y, z) (39, 43, 37) (top) and at (−17, 10, 25) (bottom). Twin 1 and twin 2 represent the two individuals of a twin pair. The correlation within MZ twin pairs is much higher than that in DZ twin pairs, indicating that the individual variation in GM and WM density in these areas is primarily determined by genes.
Figure 2.
Genetically influenced focal WM density brain areas superimposed on the histologically defined map of the superior occipitofrontal fascicle. Heritability of WM density in focal brain areas in healthy adult humans is shown for the significance level thresholded A-map superimposed on axial and sagittal sections in the left (L) and right (R) hemisphere through the magnetic resonance image of the standardized reference brain (left) and superimposed on the histologically defined map of the occipitofrontal superior fascicle (middle) (reproduced with the permission of K.Z, K.A., and U.B.). The complete A-maps are shown on the right. Note that several of the genetically influenced focal WM density voxels shown on the left also overlapped with the histologically defined maps of the corpus callosum, optic radiation, and corticospinal tract, which are not shown in the middle figures because relatively few voxels overlapped with these white matter tracts. a, Axial section. b, c, Sagittal sections.
Figure 3.
3D representation of genetically influenced focal WM density brain areas superimposed on the histologically defined map of the superior occipitofrontal fascicle. Three-dimensional glass brain representations (top left, left side view; top right, right side view; bottom left, superior view; bottom right, posterior view) of the variance in WM density found to be significantly influenced by genetic factors in healthy adult humans (orange) with the histologically defined map of the occipitofrontal superior fascicle (green) and lateral and third ventricles (blue). See also supplemental movie 1, available at www.jneurosci.org as supplemental material.
Common and unique environmental factors also had their impact on focal GM and WM areas. Common environmental factors were only of limited influence on small areas in the left amygdala (up to 0.34), the left anterior corpus callosum (up to 0.43), as well as in the right corticospinal tract and optic radiation. Unique environmental factors influenced vast GM and WM areas surrounding the lateral ventricles (up to 0.50) and included the thalamus, hypothalamus, inferior frontal cortex, medial and interior temporal cortices, lingual, cingulate gyrus, corpus callosum, internal capsule, optical tract, and corticospinal tract.
Genetically determined association between focal GM and WM density and VIQ/PIQ
Indications were found for a shared genetic origin between GM density and VIQ/PIQ in the right medial frontal cortex (positively associated with PIQ; the phenotypic and genetic correlations are rp = 0.25, rg = 0.37), occipital cortex (positively associated with PIQ, rp = 0.17, rg = 0.23), and right parahippocampal gyrus (positively associated with PIQ, rp = 0.23, rg = 0.40; positively associated with VIQ, rp = 0.11, rg = 0.19) (Table 3, Fig. 5). Also, a shared genetic origin was suggested between WM density and VIQ/PIQ in parts of the superior occipitofrontal fascicle with genetically determined associations that were positive for PIQ on the left (rp = 0.17, rg = 0.24) and on the right (rp = 0.35, rg = 0.32). In addition, positive genetically determined associations were found in the corpus callosum on the left with PIQ (rp = 0.18, rg = 0.22) and on the right with VIQ (rp = 0.14, rg = 0.15), as well in the optic radiation on the left with PIQ (rp = 0.26, rg = 0.39). Note that there were positive genetic associations with VIQ in the superior occipitofrontal fascicle bilaterally. However, the strict criterion set in the study prevented these genetically determined associations from reaching statistical significance.
Table 3.
Observed (phenotypic) correlations, cross-trait/cross-twin correlations, and genetic correlations
| Left hemisphere, IQ (x, y, z) | Correlation |
Right hemisphere, IQ (x, y, z) | Correlation |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Observed (rp) | Cross-trait/cross-twin |
Genetic (rg) | Observed (rp) | Cross-trait/cross-twin |
Genetic (rg) | ||||
| MZ | DZ | MZ | DZ | ||||||
| Gray matter | Gray matter | ||||||||
| Occipital cortex, PIQ (−21, −62, 13) | 0.17 | 0.08 | −0.06 | 0.23 | Medial frontal, PIQ (41, 22, 43) | 0.25 | 0.32 | 0.02 | 0.37 |
| White matter | Parahippocampal, PIQ (23, −36, −9) | 0.23 | 0.24 | −0.04 | 0.40 | ||||
| Superior occipitofrontal fascicle, PIQ (−21, −24, 35) | 0.17 | 0.18 | 0.04 | 0.24 | Parahippocampal, VIQ (23, −38, −7) | 0.11 | 0.15 | −0.02 | 0.19 |
| Corpus callosum, PIQ (−15, 3, 33) | 0.18 | 0.17 | 0.17 | 0.22 | White matter | ||||
| Optic radiation, PIQ (−35, −48, 17) | 0.26 | 0.28 | 0.01 | 0.39 | Superior occipitofrontal fascicle, PIQ (21, −29, 31) | 0.35 | 0.25 | 0.08 | 0.32 |
| Corpus callosum, VIQ (15, 3, 37) | 0.14 | 0.13 | 0.02 | 0.15 | |||||
The observed correlation (rp) reflects the correlation that is found between GM and WM density and VIQ/PIQ within individuals. The cross-trait/cross-twin correlations show the correlations of focal GM and WM densities of a twin with the intelligence quotient of his/her cotwin and vice versa for MZ and DZ twins (or sibling pairs) separately. If the correlation between GM and WM density of a twin and VIQ or PIQ in the cotwin is larger in MZ than in DZ twins, this indicates that the genes influencing GM and WM density partly overlap with the genes that influence VIQ/PIQ. The extent of the overlap is reflected by the magnitude of the genetic correlation (rg). rg reflects the correlation between the set of genes that influences focal GM and WM density and the set of genes that influences VIQ/PIQ. rp = rgh1h2 + ree1e2 (i.e., the phenotypic correlation is rg weighted by the square roots of the heritabilities h1 for GM and WM density and h2 for VIQ/PIQ plus the unique environmental component ree1e2).
Figure 5.

Cross-trait/cross-twin correlations for GM and WM density and VIQ/PIQ in MZ and DZ twin pairs ranging from 0 to 0.5. The cross-trait/cross-twin correlations were significant for GM density with VIQ in the right parahippocampal gyrus and for WM density with PIQ in the right superior occipitofrontal fascicle. A significant cross-trait/cross-twin correlation indicates that the genes influencing GM and WM density partly overlap with the genes that influence VIQ/PIQ. Note that, for illustration purposes, positive cross-correlations as shown here were not thresholded for significance. By definition, the cross-correlations in voxels that were not significantly determined by genetic factors could not become significant (because both factors, i.e., GM and WM density and VIQ and PIQ measures, have to be determined by genes to allow for inferences that possible mutual genes determine that association). Negative cross-correlations (data not shown) were present, but none of these reached significance. For details on these and other significant cross-trait/cross-twin correlations in the sample, see Table 3. Rt, Right.
Discussion
Our data indicate that, even after correcting for overall brain size (through scaling brains during the transformation procedure), and thus for the considerable genetic influences on total brain volume, and after stringent correction for multiple comparisons of the voxel-based genetic analyses, we found genes to significantly influence WM density of the superior occipitofrontal fascicle, corpus callosum, optic radiation, and corticospinal tract, as well as GM density of the medial frontal, superior frontal, superior temporal, occipital, postcentral, posterior cingulate, and parahippocampal cortices. Moreover, our results show that VIQ/PIQ share a common genetic origin with an anatomical neural network involving the frontal, occipital, and parahippocampal GM and connecting GM of the superior occipitofrontal fascicle, and corpus callosum.
Variation in the superior occipitofrontal fascicle was found to be highly heritable. Voxel-based morphometry analysis on anatomical magnetic resonance images does not allow for white matter tract tracing, such as is possible with diffusion tensor images. However, the majority of the WM voxels that we found to be highly heritable did essentially overlap with and follow the anatomical pathway of the superior occipitofrontal fascicle as shown by its superposition onto the PM histological probability maps. In contrast, relatively few voxels overlapped with the probability maps of other white matter tracts. Moreover, there was no indication that these voxels followed, for example, the right-to-left hemisphere direction of the corpus callosum, the superior-to-inferior direction of the corticospinal tract, or the distinguished pattern of the optic tract.
The anatomical and functional knowledge of the superior occipitofrontal fiber tract, as with many other brain fiber tracts, is still sparse. However, the GM areas that we (and others) found to be highly heritable may communicate through this pathway, forming a neural network. The superior occipitofrontal fascicle is a corticocortical association pathway. Fibers arise in the lateral prefrontal cortex of the inferior and middle frontal gyri and join to form a tight bundle at the level of the anterior horn of the lateral ventricle. The tract then runs posterior, superolateral to the caudate and can be easily seen in coronal sections at the apex of the lateral ventricle (Dejerine, 1895). It is considered to carry fibers that connect occipital, temporal (Crosby et al., 1962), and parietal (Cantani et al., 2002) regions with the frontal cortex. In monkeys, this tract is known to project to areas 46 and 8 (Petrides and Pandya, 1988). In addition, it is through the superior occipitofrontal fascicle that the prefrontal cortex maintains strong connections with limbic and paralimbic association areas, particularly the cingulate gyrus, retrosplenial cortex, parahippocampal area, and the presubiculum (Nieuwenhuys et al., 1988) (but see Ture et al., 1997).
The finding of a highly heritable medial frontal cortex bilaterally, Heschl's gyrus bilaterally, and left postcentral gyrus is in agreement with previous reports (Thompson et al., 2001; Wright et al., 2002), underscoring the relevance of these brain areas when searching for genes influencing brain structure and function. Additionally, we found the superior frontal cortex bilaterally, the occipital (striate and extrastriate) cortex in the left hemisphere, the left posterior cingulate, the amygdala bilaterally, and the right parahippocampal gyrus to be highly heritable. Thus, it seems that the individual variation in morphology of areas involved in attention, language, visual, and emotional processing, as well as in sensorimotor processing are strongly genetically influenced. Unique environmental factors influenced vast GM and WM areas surrounding the lateral ventricles (up to 0.50). This finding coincides with the significant environmental influences on lateral ventricle volume [common (0.58) and unique (0.42) with no significant contributions of genes] that we reported previously in this sample (Baaré et al., 2001).
However, we also found differences in our study compared with previous findings in healthy adult twin samples. We did not find significant heritability for Broca's language area (Thompson et al., 2001), ventrolateral prefrontal cortex, and anterior cingulate gyrus (Wright et al., 2002). There may be several reasons for these differences. First, the transformation procedures used in our study (linear followed by nonlinear warping) differed from the transformation procedures used in the other two studies, which applied cortical brain mapping (Thompson et al., 2001) and linear warping (Wright et al., 2002) procedures. Second, our structural equation modeling procedure with correction for multiple comparisons based on random field theory differed from the approach used in the other two studies. Thompson et al. (2001) applied twice the difference between MZ and DZ intraclass correlation coefficients and a permutation analysis to correct for multiple comparisons (Thompson et al., 2001). Wright et al. (2002) adopted a modeling strategy similar to the one used in this study but did not correct for multiple comparisons. Thus, although our method of structural equation modeling, allowing for separate estimates of additive genetic, common environmental, and unique environmental influences, can be considered an optimal model for genetic studies in twin pairs, the conservative approach of using the random field theory for correction for multiple comparisons in these dependent samples may have prevented some areas from reaching significant heritability but allowed the areas with significant heritability to be considered reliable.
A common genetic origin of brain structure shared with PIQ was found to be maximal in the superior occipitofrontal fascicle bilaterally, left optic radiation, right medial frontal cortex, and right parahippocampal gyrus and was still considerable in the left occipital cortex and left corpus callosum. The observed (phenotypic) correlations of up to 0.35 and the genetic correlations of up to 0.39 were higher than those found for the observed and genetically determined associations between total GM and WM volume and intelligence (Posthuma et al., 2002) and comparable with the observed correlations found for total frontal GM with intelligence (Thompson et al., 2001). A common genetic origin of VIQ shared with brain structure was small but significant in the right parahippocampal gyrus and anterior corpus callosum.
The finding of a common genetic origin of frontal and occipital GM with intelligence is corroborated by findings from two magnetic resonance imaging studies in singletons, in which several brain areas were found to be correlated with intelligence (Frangou et al., 2004; Haier et al., 2004). Interestingly, frontal and occipital GM has been associated with general fluid intelligence (Spearman's g) in a functional positron emission tomography study in singletons (Duncan et al., 2000). Moreover, in a functional magnetic resonance imaging study, greater fluid intelligence was associated with larger event-related neural activity in the prefrontal cortex, quite comparable with that found to be genetically determined in our study (Gray et al., 2003). A role for the superior occipitofrontal fascicle in (nonverbal) intelligence is corroborated by a study suggesting this pathway to be implicated in spatial attention and visuospatial functioning (Filley, 2001). We found evidence that (nonverbal) intelligence shares common genes with frontal and occipital GM areas, in concert with the connecting superior occipitofrontal fascicle and interhemispheric corpus callosum.
Most focal brain areas that were primarily influenced by genes were represented on both sides of the brain. Thus, both the left- and right-hemisphere representations of anatomical brain regions shared the extent to which their individual differences in GM and WM density were genetically determined. Indeed, GM was found to be a good predictor of the density of the homotopic region in the contralateral hemisphere, with the striking exception of the primary visual cortex (Mechelli et al., 2005). Our findings suggest that the compatibility of GM density of several homotopic areas in the contralateral hemisphere is genetically determined. Moreover, our findings suggest that individual variation in GM density of the primary visual cortex is genetically mediated.
In this study, we measured the extent of genetic contributions and not the influence of individual genes on brain morphology. However, our findings are supported by a study in which a functional variation in the val66met locus in the 5′ prodomain of brain-derived neurotrophic factor (BDNF) was found to influence brain morphology in normal humans. Individuals who were met-BDNF carriers compared with individuals who were val/val-BDNF carriers had reduced volumes of the medial frontal GM bilaterally and the right hippocampus, which has close anatomical connections with the parahippocampal gyrus (Pezawas et al., 2004). Considering that the BDNF gene has a role in human learning and memory, this gene may be one of the candidates implicated in a positive association between the medial frontal and medial temporal cortices and intelligence that is attributable to common genes.
In conclusion, we found that specific focal GM and WM areas in the human brain are highly heritable. Moreover, some of these GM and WM areas, including the superior occipitofrontal fascicle, corpus callosum, and medial frontal and occipital cortices, share common genes with intelligence, forming a heritable neural network in the human brain that is involved in intelligence.
Footnotes
This work was supported in part by Netherlands Organization for Scientific Research/Netherlands Organization for Health Research and Development VIDI Program Grant 917.46.370 (H.E.H.), the Federation for European Neuroscience Societies (H.E.H), the Human Frontiers Science Program Grant rg0154/1998-B (D.I.B.), and the Universitair Stimulerings Fonds (D.I.B.). This Human Brain Project/Neuroinformatics research is funded by the National Institute of Biomedical Imaging and Bioengineering, the National Institute of Neurological Disorders and Stroke, and the National Institute of Mental Health (K.Z., K.A.). Additional support by the Brain Imaging Center West (Bundesministerium für Bildung und Forschung Grant 01GO0204) is gratefully acknowledged (K.Z.).
Parts of this work have been presented previously at the 34th Annual Meeting of the Society for Neuroscience, San Diego, CA, October 23–27, 2004.
References
- Amunts K, Jancke L, Mohlberg H, Steinmetz H, Zilles K. Interhemispheric asymmetry of the human motor cortex related to handedness and gender. Neuropsychologia. 2000;38:304–312. doi: 10.1016/s0028-3932(99)00075-5. [DOI] [PubMed] [Google Scholar]
- Amunts K, Weiss PH, Mohlberg H, Pierperhoff P, Eickhoff S, Gurd JM, Marshall JC, Shah NJ, Fink GR, Zilles K. Analysis of the neural mechanisms underlying verbal fluency in cytoarchitectonically defined stereotactic space: the role of Brodmann's areas 44 and 45. NeuroImage. 2004;22:42–56. doi: 10.1016/j.neuroimage.2003.12.031. [DOI] [PubMed] [Google Scholar]
- Andreasen NC, Flaum M, Arndt S. The comprehensive assessment of symptoms and history (CASH). An instrument for assessing diagnosis and psychopathology. Arch Gen Psychiatry. 1992;49:615–623. doi: 10.1001/archpsyc.1992.01820080023004. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Voxel-based morphometry: the methods. NeuroImage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Baaré WFC, Hulshoff Pol HE, Boomsma DI, Posthuma D, de Geus EJC, Schnack HG, van Haren NEM, van Oel CJ, Kahn RS. Quantitative genetic modeling of variation in human brain morphology. Cereb Cortex. 2001;11:816–824. doi: 10.1093/cercor/11.9.816. [DOI] [PubMed] [Google Scholar]
- Boomsma D, Busjahn A, Peltonen L. Classical twin studies and beyond. Nat Rev Genet. 2002;3:872–882. doi: 10.1038/nrg932. [DOI] [PubMed] [Google Scholar]
- Bürgel U, Mecklenburg I, Blohm U, Zilles K. Histological visualization of long fiber tracts in the white matter of adult human brains. J Brain Res. 1997;3:393–404. [PubMed] [Google Scholar]
- Bürgel U, Schormann T, Schleicher A, Zilles K. Mapping of histologically identified long fiber tracts in human cerebral hemispheres to the MRI volume of a reference brain: position and spatial variability of the optic radiation. NeuroImage. 1999;10:489–499. doi: 10.1006/nimg.1999.0497. [DOI] [PubMed] [Google Scholar]
- Cantani M, Howard RJ, Pajevic S, Jones DK. Virtual in vivo interactive dissection of white matter fasciculi in the human brain. NeuroImage. 2002;17:77–94. doi: 10.1006/nimg.2002.1136. [DOI] [PubMed] [Google Scholar]
- Collins DL, Holmes CJ, Peters TM, Evans AC. Automatic 3-D model-based neuroanatomical segmentation. Hum Brain Mapp. 1995;4:190–208. [Google Scholar]
- Crosby EC, Humphrey T, Lauer EW. New York: Macmillan; 1962. Correlative anatomy of the nervous system. [Google Scholar]
- Dejerine J. Vol 1. Paris: Rueff et Cie; 1895. Anatomie des Centres Nerveux. [Google Scholar]
- Duncan J, Seitz RJ, Kolodny J, Bor D, Herzog H, Ahmed A, Newell FN, Emslie H. A neural basis for general intelligence. Science. 2000;289:457–460. doi: 10.1126/science.289.5478.457. [DOI] [PubMed] [Google Scholar]
- Filley CM. New York: Oxford UP; 2001. The behavioral neurology of white matter. [Google Scholar]
- Frangou S, Chitnis X, Williams SCR. Mapping IQ and grey matter density in healthy young people. NeuroImage. 2004;23:800–805. doi: 10.1016/j.neuroimage.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Good CD, Johnsrude IS, Ashburner J, Henson RN, Friston KJ, Frackowiak RS. A voxel-based morphometric study of ageing in 465 normal adult human brains. NeuroImage. 2001a;14:21–36. doi: 10.1006/nimg.2001.0786. [DOI] [PubMed] [Google Scholar]
- Good CD, Johnsrude I, Ashburner J, Henson RN, Friston KJ, Frackowiak RS. Cerebral asymmetry and the effects of sex and handedness on brain structure: a voxel-based morphometric analysis of 465 normal adult human brains. NeuroImage. 2001b;14:685–700. doi: 10.1006/nimg.2001.0857. [DOI] [PubMed] [Google Scholar]
- Gray JR, Chabris CF, Braver TS. Neural mechanisms of general fluid intelligence. Nat Neurosci. 2003;6:316–322. doi: 10.1038/nn1014. [DOI] [PubMed] [Google Scholar]
- Haier RJ, Jung RE, Yeo RA, Head K, Alkire MT. Structural brain variation and general intelligence. NeuroImage. 2004;23:425–433. doi: 10.1016/j.neuroimage.2004.04.025. [DOI] [PubMed] [Google Scholar]
- Haier RJ, Jung RE, Yeo RA, Head K, Alkire MT. The neuroanatomy of general intelligence: sex matters. NeuroImage. 2005;25:320–327. doi: 10.1016/j.neuroimage.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Hulshoff Pol HE, Schnack HG, Mandl RCW, van Haren NEM, Koning H, Collins DL, Evans AC, Kahn RS. Focal gray matter density changes in schizophrenia. Arch Gen Psychiatry. 2001;58:1118–1125. doi: 10.1001/archpsyc.58.12.1118. [DOI] [PubMed] [Google Scholar]
- Hulshoff Pol HE, Baaré WFC, Posthuma D, de Geus EJC, Schnack HG, van Haren NEM, Oel CJ, Kahn RS, Boomsma DI. Twin-singleton differences in brain structure using structural equation modelling. Brain. 2002;125:384–390. doi: 10.1093/brain/awf035. [DOI] [PubMed] [Google Scholar]
- Hulshoff Pol HE, Schnack HG, Mandl RCW, Cahn W, Collins DL, Evans AC, Kahn RS. Focal white matter density changes in schizophrenia: reduced inter-hemispheric connectivity. NeuroImage. 2004;21:27–35. doi: 10.1016/j.neuroimage.2003.09.026. [DOI] [PubMed] [Google Scholar]
- Maes F, Collignon A, Vandermeulen D, Marchal G, Suetens P. Multimodality image registration by maximization of mutual information. IEEE Trans Med Imag. 1997;16:187–198. doi: 10.1109/42.563664. [DOI] [PubMed] [Google Scholar]
- Maguire EA, Gadian DG, Johnsrude IS, Good CD, Ashburner J, Frackowiak RS, Frith CD. Navigation-related structural change in the hippocampi of taxi drivers. Proc Natl Acad Sci USA. 2000;97:4398–4403. doi: 10.1073/pnas.070039597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechelli A, Friston KJ, Frackowiak RS, Price CJ. Structural covariance in the human cortex. J Neurosci. 2005;25:8303–8310. doi: 10.1523/JNEUROSCI.0357-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MC. Ed 3. Richmond, VA: Medical College of Virginia; 1997. Mx: statistical modeling. [Google Scholar]
- Nieuwenhuys R, Voogd J, van Huijzen C. Ed 3. Berlin: Springer; 1988. The human central nervous system. [Google Scholar]
- Nurnberger JI, Jr, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, Severe JB, Malaspina D, Reich T. Diagnostic interview for genetic studies. Rationale, unique features, and training, NIMH genetics initiative. Arch Gen Psychiatry. 1994;51:849–859. doi: 10.1001/archpsyc.1994.03950110009002. [DOI] [PubMed] [Google Scholar]
- Pennington BF, Filipek PA, Lefly D, Chhabildas N, Kennedy DN, Simon JH, Filley CM, Galaburda A, DeFries JC. A twin MRI study of size variations in the human brain. J Cogn Neurosci. 2000;12:223–232. doi: 10.1162/089892900561850. [DOI] [PubMed] [Google Scholar]
- Petrides M, Pandya DN. Association fiber pathways to the frontal cortex from the superior temporal region in the rhesus monkey. J Comp Neurol. 1988;27:52–66. doi: 10.1002/cne.902730106. [DOI] [PubMed] [Google Scholar]
- Pezawas L, Verchinski BA, Mattay VS, Callicott JH, Kolachana BS, Straub RE, Egan MF, Meyer-Lindenberg A, Weinberger DR. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci. 2004;24:10099–10102. doi: 10.1523/JNEUROSCI.2680-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferbaum A, Sullivan EV, Swan GE, Carmeli D. Brain structure in men remains highly heritable in the seventh and eighth decades of life. Neurobiol Aging. 2000;21:63–74. doi: 10.1016/s0197-4580(00)00086-5. [DOI] [PubMed] [Google Scholar]
- Posthuma D, Boomsma DI. A note on the statistical power in extended twin designs. Behav Genet. 2000;30:147–158. doi: 10.1023/a:1001959306025. [DOI] [PubMed] [Google Scholar]
- Posthuma D, de Geus EJC, Bleichrodt N, Boomsma DI. Twin-singleton differences in intelligence? Twin Res. 2000;3:83–87. doi: 10.1375/136905200320565535. [DOI] [PubMed] [Google Scholar]
- Posthuma D, de Geus EJC, Boomsma DI. Perceptual speed and IQ are associated through common genetic factors. Behav Genet. 2001;31:593–602. doi: 10.1023/a:1013349512683. [DOI] [PubMed] [Google Scholar]
- Posthuma D, De Geus EJC, Baaré WFC, Hulshoff Pol HE, Kahn RS, Boomsma DI. The association between brain volume and intelligence is of genetic origin. Nat Neurosci. 2002;5:83–84. doi: 10.1038/nn0202-83. [DOI] [PubMed] [Google Scholar]
- Rademacher J, Burgel U, Geyer S, Schormann T, Schleicher A, Freund HJ, Zilles K. Variability and asymmetry in the human precentral motor system. A cytoarchitectonic and myeloarchitectonic brain mapping study. Brain. 2001;124:2232–2258. doi: 10.1093/brain/124.11.2232. [DOI] [PubMed] [Google Scholar]
- Schnack HG, Hulshoff Pol HE, Baaré WFC, Staal WG, Viergever MA, Kahn RS. Automated separation of gray and white matter from MR images of the human brain. NeuroImage. 2001;13:230–237. doi: 10.1006/nimg.2000.0669. [DOI] [PubMed] [Google Scholar]
- Shaw P, Greenstein D, Lerch J, Clasen L, Lenroot R, Gogtay N, Evans A, Rapoport J, Giedd J. Intellectual ability and cortical development in children and adolescents. 2006;440:676–679. doi: 10.1038/nature04513. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imag. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Cannon TD, Narr KL, van Erp T, Poutanen VP, Huttunen M, Lonnqvist J, Standertskjold-Nordenstam CG, Kaprio J, Khaledy M, Dail R, Zoumalan CI, Toga AW. Genetic influences on brain structure. Nat Neurosci. 2001;4:1253–1258. doi: 10.1038/nn758. [DOI] [PubMed] [Google Scholar]
- Toga AW, Thompson PM. Genetics of brain structure and intelligence. Annu Rev Neurosci. 2005;28:1–23. doi: 10.1146/annurev.neuro.28.061604.135655. [DOI] [PubMed] [Google Scholar]
- Ture U, Yasargil MG, Pait TG. Is there a superior occipitofrontal fasciculus? A microsurgical anatomic study. Neurosurgery. 1997;40:1226–1232. doi: 10.1097/00006123-199706000-00022. [DOI] [PubMed] [Google Scholar]
- Worsley KJ, Marrett S, Neelin P, Vandal AC, Friston KJ, Evans AC. A unified statistical approach for determining significant signals in images of cerebral activation. Hum Brain Mapp. 1996;4:58–73. doi: 10.1002/(SICI)1097-0193(1996)4:1<58::AID-HBM4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Wright IC, Sham P, Murray RM, Weinberger DR, Bullmore ET. Genetic contributions to regional variability in human brain structure: methods and preliminary results. NeuroImage. 2002;17:256–271. doi: 10.1006/nimg.2002.1163. [DOI] [PubMed] [Google Scholar]
- Zilles K, Schleicher A, Palomero-Gallagher N, Amunts K. Quantitative analysis of cyto- and receptor architecture of the human brain. In: Mazziotta JC, Toga A, editors. Brain mapping: the methods. New York: Elsevier; 2002. pp. 573–602. [Google Scholar]