Abstract
The nervous system plays a critical role in adaptation to a new environment. In Caenorhabditis elegans, reduced access to food requires both changes in behavior as well as metabolic adaptation for survival, which is postulated to involve the bioamine octopamine. The transcription factor cAMP response element-binding protein (CREB) is generally activated by G-protein-coupled receptors (GPCRs) that activate Gαs and is known to play an important role in long-term changes, including synaptic plasticity. We show that, in C. elegans, the CREB ortholog CRH-1 (CREB homolog family member 1) activates in vivo a cAMP response element–green fluorescent protein fusion reporter in a subset of neurons during starvation. This starvation response is mediated by octopamine via the GPCR SER-3 (serotonin/octopamine receptor family member 3) and is fully dependent on the subsequent activation of the Gαq ortholog EGL-30 (egg-laying defective family member 30). The signaling cascade is only partially dependent on the phospholipase Cβ (EGL-8) and is negatively regulated by Gαo [GOA-1 (G-protein, O, α subunit family member 1)] and calcium/calmodulin-dependent kinase [UNC-43 (uncoordinated family member 43)]. Nonstarved animals in a liquid environment mediate a similar response that is octopamine independent. The results show that the endogenous octopamine system in C. elegans is activated by starvation and that different environmental stimuli can activate CREB through Gαq.
Keywords: aminergic, C. elegans, CREB, GPCR, Gq, octopamine, starvation
Introduction
cAMP response element-binding protein (CREB) is a signal-activated transcription factor that, after phosphorylation, induces expression of genes from promoters containing the cAMP-response element (CRE) enhancer (Mayr and Montminy, 2001). CREB is activated by various physiological stimuli and plays important roles in many biological processes (Lonze and Ginty, 2002; Johannessen et al., 2004). The bioamine neurotransmitters dopamine, serotonin, and norepinephrine can activate CREB (Hyman, 1996; Mayford and Kandel, 1999; Simonneaux and Ribelayga, 2003). Most studies show that bioamine-induced CREB activation is dependent on Gαs-coupled bioamine receptors through an adenylyl cyclase–protein kinase A (PKA) signaling pathway. However, little is known about in vivo mechanisms and the possible involvement of alternative cascades leading to CREB activation. Some in vitro studies suggest that Gαq can activate CREB (Lin et al., 1998; Chalecka-Franaszek et al., 1999; Thonberg et al., 2002), although it is unknown whether the Gαq–CREB pathway is of physiological relevance.
Caenorhabditis elegans is a model organism that is particularly suitable for genetic studies. The bioamines dopamine (Sulston et al., 1975), serotonin (Horvitz et al., 1982), tyramine (Alkema et al., 2005), and octopamine (Horvitz et al., 1982) have been found in C. elegans and are believed to function as both neurotransmitters as well as neurohumoral factors. Octopamine is considered to be the invertebrate biological equivalent of mammalian norepinephrine (Roeder, 1999). In C. elegans, exogenous application of octopamine induces behavioral changes that are similar to those observed during starvation (Horvitz et al., 1982). It is also shown that exogenous octopamine phenocopies starvation in a temperature–starvation association learning paradigm (Mohri et al., 2005). However, the functional role of endogenous octopamine and its targets, including receptors and neurons that mediate the octopamine responses, are unknown.
In C. elegans, CRE-mediated gene expression can be visualized in vivo using a CRE promoter fused to the green fluorescent protein (GFP) sequence (cre::gfp) (Kimura et al., 2002). In animals carrying the cre::gfp reporter, CRE-mediated gene expression is detected by GFP fluorescence. Using this reporter system, it has been shown that the introduction of a constitutively active form of CMK-1, a calcium/calmodulin-dependent protein kinase (CaMK) I in C. elegans, induces CRE-mediated gene expression in neurons and that this gene expression is dependent on the CREB ortholog CRH-1 (CREB homolog family member 1) (Kimura et al., 2002).
The cre::gfp reporter provides a unique opportunity to understand how environmental stimuli or bioamine neurotransmitters can induce or enhance activation of CREB in C. elegans. In this study, we used the strain carrying the cre::gfp reporter to address this question and found that starvation of C. elegans induces CRE-mediated gene expression in a distinct set of neurons known as the SIA neurons. By reverse genetic approaches, we reveal that this starvation-induced gene expression is mediated by octopamine through the octopamine receptor SER-3 (serotonin/octopamine receptor family member 3) and that a Gαq signaling pathway is responsible for the CREB activation.
Materials and Methods
Strains.
Culturing and genetic manipulation of C. elegans was performed as described previously (Brenner, 1974). The alleles used in this study are as follows: egl-30(ad806, n686, js126gf) I (Trent et al., 1983; Lackner et al., 1999; Hawasli et al., 2004), goa-1(sa734) I (Robatzek and Thomas, 2000), ser-3(ad1774) I (a gift from Drs. Niacaris and Avery, University of Texas Southwestern Medical Center, Dallas, TX), unc-13(e51) I (Brenner, 1974), unc-64(e246) III (Brenner, 1974), cmk-1(ok287) IV (Kimura et al., 2002), crh-1(tz2) IV (Kimura et al., 2002), unc-43(e408, n498gf) IV (Park and Horvitz, 1986; Reiner et al., 1999), egl-8(md1971, n488) V (Trent et al., 1983; Miller et al., 1999), tbh-1(ok1196) X (isolated by C. elegans Gene Knockout Project, Oklahoma Medical Research Foundation, Oklahoma City, OK), and tzIs3[cre::gfp; lin-15(+)] (Kimura et al., 2002). The strains carrying cre::gfp reporter were constructed by mating tzIs3 males with other mutants. goa-1(sa734);unc-43(n498gf);tzIs3 was constructed as described previously (Robatzek and Thomas, 2000) using goa-1(sa734);tzIs3 and unc-43(n498gf);tzIs3. The strain was confirmed by crossing with wild-type males and re-isolating both mutations. Males carrying ser-3::gfp transgene were used to construct egl-30 (egg-laying defective family member 30), crh-1, and unc-43(n498gf) mutants carrying ser-3::gfp.
Analyses of expression patterns.
For the analyses of expression patterns of tbh-1 (tyramine-β-hydroxylase family member 1) and ser-3, transcriptional reporter fusion genes were constructed by the fusion PCR method (Hobert, 2002). Nucleotides 25375–22542 of the cosmid H13N06 and nucleotides 26575–17996 of the cosmid K02F2 were fused to 2–1876 of pPD95.75 to obtain tbh-1::gfp and ser-3::gfp, respectively. The reporter fusions were injected to wild-type N2 animals together with pRF4 as described previously (Mello et al., 1991). Resulting Rol animals were stained with DiI (Herman and Hedgecock, 1990) and examined under a confocal laser microscope (LSM510; Zeiss, Oberkochen, Germany). ceh-17 (C. elegans homeobox family member 17) gene is shown to be expressed in one ALA neuron and four SIA neurons (Pujol et al., 2000). ceh-17::dsred fusion gene was constructed by inserting the 2.2 kb promoter region of ceh-17 gene to HindIII and KpnI digested pPR1-1 (a gift from Dr. Roy, University of Toronto, Toronto, Ontario, Canada), and we confirmed that ceh-17::dsred induce Discosoma red (DsRed) expression in five neurons. ser-3::gfp and ceh-17::dsred were coinjected together with the marker gene and determined that DsRed expressing sublateral neurons (SIA neurons) also express GFP in animals carrying both plasmids.
Analysis of CRE-mediated gene expression.
Strains carrying cre::gfp were synchronized and grown for 3 d after hatching at 20°C on nematode growth media (NGM) plates seeded with the bacteria strain OP50 (Brenner, 1974). Animals were collected and washed twice with water. Animals in a drop of water were transferred onto NGM plates with or without bacteria. The water remaining on plates was immediately removed with absorbent paper. For the soaking condition, 5 ml of water was poured onto 60 mm Petri dishes to cover the surface of NGM plates seeded with bacteria. After 6 h, animals were collected in M9 buffer (Brenner, 1974) containing 50 mm NaN3 to immobilize the animals. The animals were examined under a fluorescent microscope (DMIRB; Leica, Nussloch, Germany) equipped with a CCD camera (MicroMAX system; Princeton Instruments, Trenton, NJ) for counting or the confocal laser microscope for the presented images. The frequency of animals that showed any detectable expression of GFP in the ventral ganglia was determined. For octopamine treatment, the assay plates contained 1.7% agar with or without 3 mg/ml dl-octopamine hydrochloride (Fluka, Buchs, Switzerland). An overnight culture of OP50 in lysogeny broth was spun down and resuspended in 1:20 volume of water. Fifty microliters of the concentrated bacteria was spread on the assay plates. The plates were dried for at least 2 h without lids. Animals were incubated on the assay plates overnight (∼16 h) before they were examined by fluorescence microscopy. Because the overnight incubation of egl-30(js126gf) and goa-1(sa734) mutants on both octopamine-containing plates and control plates resulted in frequent death of the animals, the treatment was limited to 2 h for these mutants. Two hour treatments of tzIs3 animals resulted in a comparable induction of GFP with that of the overnight treatment. The neurons expressing GFP after starvation, soaking, and octopamine treatment were determined to be SIA neurons by colocalization of DsRed expression in the animals carrying both cre::gfp and ceh-17::dsred.
For rescue experiments, the region corresponding to nucleotides 18423–25422 of the cosmid H13N06 was amplified with primers tbh-1 forward (ccggaatcttccttatgtatctc) and tbh-1 reverse (gacgttttcgacgacaagatc) from genomic DNA and the region corresponding to nucleotides 14691–26568 of the cosmid K02F2 was obtained as a BamHI–NcoI fragment. These DNA fragments were gel purified and injected into tbh-1 and ser-3 mutant animals carrying cre::gfp together with pRF4.
The coding region of ser-3 was amplified from the cosmid K02F2 using primer ser-3 forward (gaaaaacaaaaccatggaatggatgagaaatacg) and ser-3 reverse (taagttcatagcggccgcttagttggttggagtg). The PCR product was digested with KpnI and NotI and cloned into KpnI and NotI digested ceh-17::dsred to obtain ceh-17::ser-3. The coding region of egl-30 was amplified from pGADT7–EGL-30 (a gift from Dr. Plasterk, Hubrecht Laboratory, Utrecht, The Netherlands) using egl-30 forward (ggccatggaaccggtcgccaccatggcctgctgtttatcc) and egl-30 reverse (cacccggggcggccgcttacaccaagttgtactcc). The PCR product was cloned into the AgeI and NotI site of ceh-17::dsred to obtain ceh-17::egl-30. ceh-17::ser-3 and ceh-17::egl-30 were injected into ser-3 and egl-30 mutants carrying cre::gfp together with pRF4. Rol animals were analyzed in the rescue experiments.
Results
Starvation induces CREB activation in SIA neurons
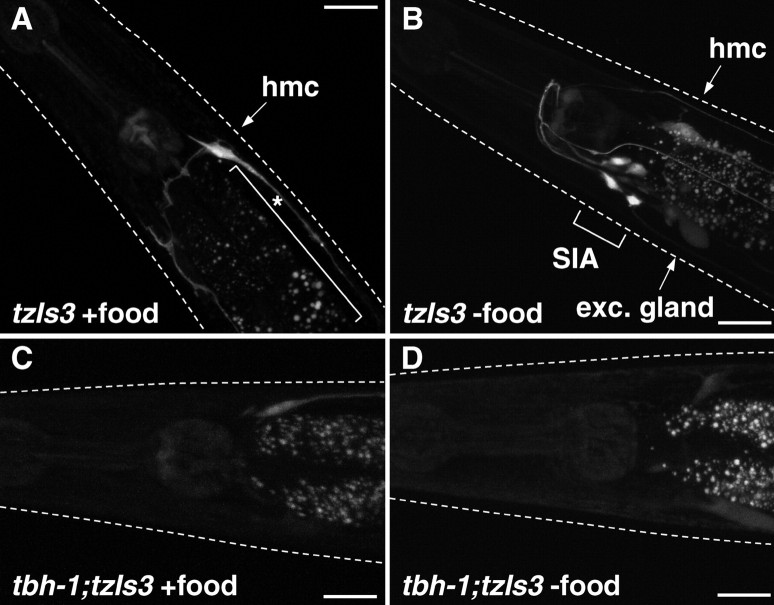
To assess CREB activity in C. elegans under various physiological conditions, we used the transgene tzIs3[cre::gfp; lin-15(+)] that contains an integrated array for a cre::gfp reporter. For the strain carrying tzIs3, it has been shown that GFP expression is dependent on the CREB CRH-1 (Kimura et al., 2002). When cultured on standard NGM seeded with bacteria (OP50) as the food source, sporadic GFP expression was seen in the head mesodermal cell, some pharyngeal cells, and the excretory glands (Fig. 1A). When animals are transferred to NGM plates without bacteria, GFP expression is induced in four neurons in the ventral ganglia as early as 2 h after transfer (Fig. 1B). These neurons were identified as the SIA neurons, because the GFP expression colocalized with DsRed when animals were injected with ceh-17::dsred fusion gene, which induces DsRed expression in the SIA neurons and the ALA neuron (data not shown). After prolonged starvation (6 h in the absence of food), 84% of the animals displayed GFP-positive SIA neurons, whereas no GFP expression was seen in the SIA neurons of nonstarved animals (Table 1). Furthermore, the consistently high frequency of GFP induction was unique for the SIA neurons and was not seen in other neurons.
Figure 1.
GFP expression of control animals and tbh-1 mutant animals carrying cre::gfp in the well fed and starved conditions. Fluorescent images were taken from control (A, B) and tbh-1 mutant (C, D) animals carrying tzIs3[cre::gfp; lin-15(+)] placed on NGM plates with (A, C) or without (B, D) food for 6 h. A, Neuronal GFP expression was rarely observed in the head region in the well fed condition. B, GFP expression was observed in SIA neurons in starved animals. In tbh-1 mutants, GFP expression in SIA neurons was rarely observed in either well fed (C) or starved (D) condition. White dotted lines indicate the outline of the heads of the animals. The bracket with asterisk indicates autofluorescence of the intestine. hmc, Head mesodermal cell; exc. gland, excretory gland. Scale bars, 20 μm.
Table 1.
The percentages of animals expressing GFP in SIA neurons
| Background | % Animals expressing GFP in SIA |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Well fed | Starvation | Soaking | Control | Octopamine | ||||||
| Control | 0 | (50) | 84 | (50) | 92 | (50) | 2 | (50) | 94 | (50) |
| 0a | (50) | 98a | (50) | |||||||
| tbh-1(ok1196) | 0 | (50) | 2 | (50) | 72 | (50) | 0 | (52) | 94 | (50) |
| tbh-1(ok1196) + tbh-1 gene | 11 | (55) | 65 | (55) | 96 | (55) | 9 | (55) | 75 | (55) |
| ser-3(ad1774) | 0 | (50) | 14 | (50) | 86 | (50) | 0 | (50) | 0 | (50) |
| ser-3(ad1774) + ser-3 gene | 1 | (70) | 37 | (70) | 79 | (42) | 0 | (55) | 42 | (62) |
| ser-3(ad1774) + ceh-17::ser-3 | 6 | (50) | 74 | (50) | 98 | (50) | 4 | (50) | 84 | (50) |
| unc-13(e51) | 0 | (50) | 2 | (50) | 0 | (50) | 2 | (50) | 96 | (50) |
| unc-64(e246) | 0 | (50) | 0 | (50) | 0 | (50) | 0 | (50) | 96 | (50) |
| egl-30(ad806) | 0 | (53) | 0 | (60) | 0 | (60) | 0 | (62) | 6 | (62) |
| egl-30(n686) | 0 | (52) | 0 | (52) | 0 | (52) | 0 | (55) | 5 | (55) |
| egl-30(n686) + ceh-17::egl-30 | 0 | (75) | 1 | (75) | 4 | (75) | 0 | (84) | 44 | (87) |
| egl-30(js126gf) | 52 | (50) | 66 | (50) | 98 | (50) | 56a | (50) | 92a | (50) |
| egl-8(n488) | 0 | (50) | 28 | (50) | 12 | (50) | 0 | (76) | 46 | (87) |
| egl-8(md1971) | 0 | (50) | 14 | (50) | 6 | (50) | 0 | (60) | 53 | (60) |
| crh-1(tz2) | 0 | (50) | 0 | (50) | 22 | (50) | 0 | (50) | 4 | (50) |
| egl-30(js126gf);crh-1(tz2) | 0 | (50) | 0 | (50) | 10 | (50) | 0a | (50) | 2a | (50) |
| cmk-1(ok287) | 2 | (50) | 56 | (50) | 94 | (50) | 0 | (50) | 71 | (51) |
| goa-1(sa734) | 30 | (70) | 33 | (70) | 60 | (70) | 34a | (50) | 88a | (50) |
| unc-43(n498gf) | 0 | (50) | 0 | (50) | 0 | (50) | 0 | (50) | 4 | (50) |
| unc-43(e408) | 0 | (50) | 94 | (50) | 100 | (50) | 0 | (50) | 100 | (50) |
| unc-43(n498gf);goa-1(sa734) | 2 | (50) | 4 | (50) | 4 | (50) | 4a | (50) | 60a | (50) |
Animals carrying cre::gfp in various genetic backgrounds were subjected to well fed, starvation, and soaking conditions for 6 h or control and octopamine-containing plates for ∼16 h. Percentages of animals showing detectable GFP expression in SIA neurons were counted. The numbers in the parentheses indicate the number of animals tested.
aIncubation was limited to 2 h.
TBH-1 is required for starvation-induced CREB activation
Behavioral changes attributable to starvation in C. elegans are phenocopied by exogenously added octopamine (Horvitz et al., 1982; Mohri et al., 2005). We therefore postulated that endogenous octopamine may be involved in the starvation-induced CREB activation. Octopamine is synthesized from tyramine by tyramine-β-hydroxylase. In C. elegans, the tbh-1 gene encodes this enzyme and, as expected, tbh-1 mutant animals are octopamine deficient (Alkema et al., 2005). In transgenic animals carrying an extrachromosomal array of tbh-1::gfp in which 2.8 kb immediately upstream from the tbh-1 coding sequence is fused to the gfp reporter gene, GFP expression was observed in RIC neurons and gonadal sheath cells (supplemental Fig. S1, available at www.jneurosci.org as supplemental material), as also reported by Alkema et al. (2005), using immunostaining of TBH-1 protein and a 4.5 kb promoter reporter fusion transgene. To establish whether octopamine mediates the starvation response, the cre::gfp reporter in tzIs3 was transferred into the deletion mutant tbh-1(ok1196). After starvation, induction of GFP expression in the SIA neurons of the crossed animals was comparable with baseline (Fig. 1C,D; Table 1). Reintroduction of the tbh-1 gene as an extrachromosomal array rescued the starvation-induced GFP expression, further demonstrating that the tbh-1 gene is required for this response (Table 1).
Exogenous octopamine activates CREB in SIA neurons
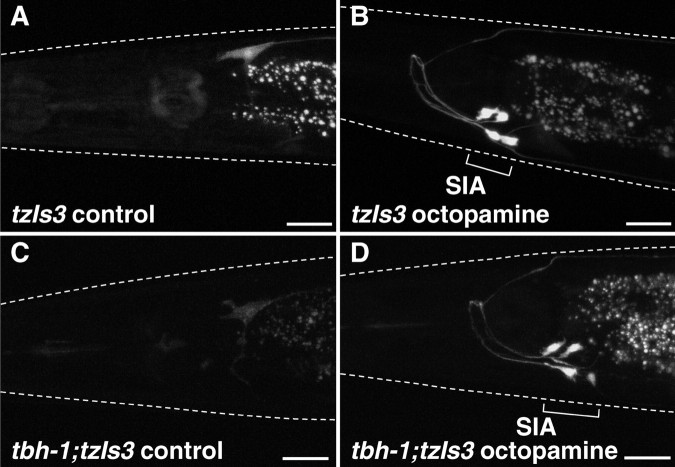
The necessity of the tbh-1 gene for starvation-induced CREB activation in the SIA neurons predicts that exogenously applied octopamine could activate CREB in these neurons independently of both TBH-1 expression and starvation state. Indeed, exposure of well fed cre::gfp reporter animals to exogenously applied octopamine results in a high frequency of GFP expression in SIA neurons (98 and 94% after 2 h and overnight incubation, respectively), whereas animals not exposed to octopamine rarely displayed GFP expression in these neurons (Fig. 2A,B; Table 1). The neurons expressing GFP was confirmed to be SIA neurons by colocalization of GFP and DsRed in the animal carrying cre::gfp and ceh-17::dsred (Fig. 3). As seen for starvation-induced CREB activation, exogenous octopamine did not induce detectable GFP expression in other neurons. The tbh-1 mutants, which do not synthesize octopamine and fail to show starvation-induced CREB activation, display a strong induction of GFP expression after octopamine treatment when carrying the cre::gfp transgene (94%; untreated 0%) (Fig. 2C,D; Table 1).
Figure 2.
Octopamine-induced GFP expression in control animals and tbh-1 mutants carrying cre::gfp. Control animals (A, B) and tbh-1 mutants (C, D) carrying tzIs3[cre::gfp; lin-15(+)] were placed on agar plates containing no (A, C) or 3 mg/ml (B, D) octopamine in the presence of food and examined for GFP-mediated fluorescence. GFP expression in SIA neurons was observed in both control animals and tbh-1 mutants in the octopamine-containing plates but not in control plates. White dotted lines indicate the outline of the heads of the animals. Scale bars, 20 μm.
Figure 3.
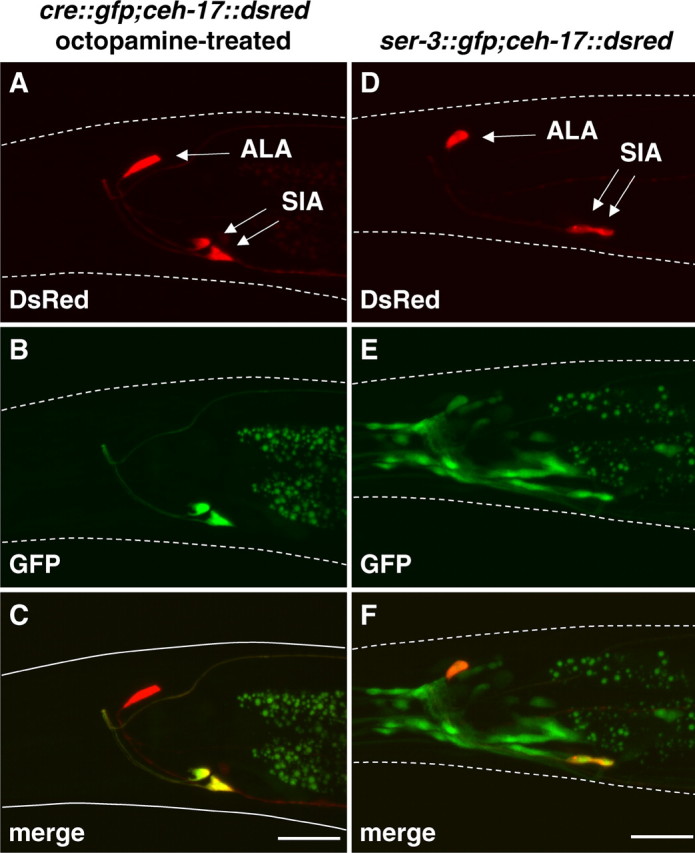
Colocalization of GFP and DsRed in an octopamine-treated animal carrying cre::gfp and ceh-17::dsred and an animal carrying ser-3::gfp and ceh-17::gfp. A, B, D, E, The fluorescent images for DsRed expression (A, D) and GFP expression (B, E) were taken from the octopamine-treated animal carrying cre::gfp and ceh-17::dsred (A, B) and the animal carrying ser-3::gfp and ceh-17::dsred (D, E). C, F, Merged images show the colocalization of GFP and DsRed. White dotted lines indicate the outline of the head of the animal. Scale bars, 20 μm.
SER-3 mediates starvation- and octopamine-induced CREB activation
In silico analysis has identified several putative bioamine receptors in C. elegans (Wintle and Van Tol, 2001). Additional analyses revealed that the putative bioamine receptor SER-3 (K02F2.6) is highly homologous to the Drosophila melanogaster and Apis mellifera octopamine receptor OAMB (octopamine receptor in mushroom bodies) (Han et al., 1998; Grohmann et al., 2003) (Fig. 4) and less so to other invertebrate receptors. With respect to sequence homology to mammalian receptors, SER-3 is most closely related to the Gαq-coupled α1 adrenergic receptors (Wu et al., 1992). The insect octopamine receptors are likely to be Gαq-coupled receptors and have been shown to increase intracellular Ca2+ and cAMP during activation (Han et al., 1998; Grohmann et al., 2003)
Figure 4.
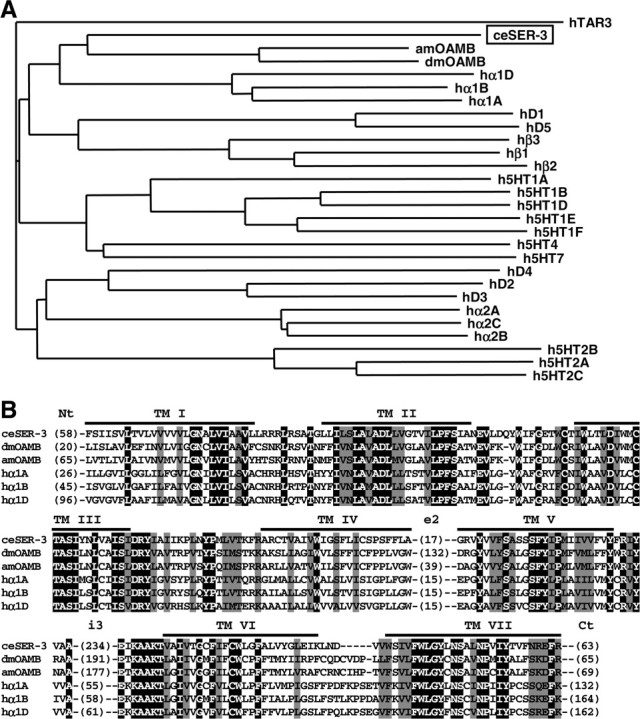
A phylogenetic tree and an alignment of invertebrate and human amine receptors. A, The deduced amino acid sequence of SER-3 was compared with invertebrate and human amine receptors using ClustalW, and a phylogenetic tree was calculated by the NJ method at DNA Data Bank of Japan web site. Relatively well conserved blocks (the region excluding the N terminus, second extracellular loops, third intracellular loops, and the C terminus) of sequences were used for the analysis. Receptor sequences used and the GenBank accession numbers are as follows: C. elegans SER-3 (ceSER-3, NP491954), Drosophila octopamine receptor (dmOAMB, AAC17442), Apis mellifera octopamine receptor (amOAMB, CAD67999), human dopamine receptors (hD1, P21728; hD2, P14416; hD3, P35462; hD4, P21917; and hD5, P21918), human serotonin receptors (h5HT1a, I38209; h5HT1b, JN0268; h5HT1d, A53279; h5HT1e, A45260; h5HT1f, A47321; h5HT2a, A43956; h5HT2b, S43687; h5HT2c, JS0616; h5HT4, Q13639; and h5HT7, A48881), and human adrenergic receptors (hα1A, NP000671; hα1B, NP000670; hα1D, NP000669; hα2A, A34169; hα2B, A37223; hα2C, A31237; hβ1, QRHUB1; hβ2, QRHUB2; and hβ3, QRHUB3). A human trace amine receptor 3 (hTAR3, AAO24660) was used as an out group. B, Alignment of SER-3 with insect OAMBs and human α1 adrenergic receptors. Amino acid sequences of C. elegans (ce) SER-3, Drosophila melanogaster (dm), and Apis mellifera (am) octopamine receptor OAMB and human α1 adrenergic receptors (hα1A, hα1B, and hα1D) were aligned with ClustalW. Residues that were identical and similar in all proteins are highlighted with black and gray, respectively. The positions of the putative transmembrane domains (TM) are indicated. The numbers in parentheses indicate the numbers of residues in the N terminus (Nt), second extracellular loops (e2), third intracellular loops (i3), and the C terminus (Ct), which were not aligned. The identities with ceSER-3 in transmembrane domains and flanking regions were as follows: dmOAMB, 51%; amOAMB, 53%; hα1A, 48%; hα1B, 45%; and hα1D, 41%.
We created a ser-3::gfp reporter fusion transgene in which 8.6 kb of upstream sequence plus exon 1, intron 1, and part of exon 2 are fused to the gfp reporter gene to analyze the expression pattern of the ser-3 gene (Fig. 5). In animals carrying this transgene as an extrachromosomal array, GFP expression was observed in various tissues, including many neurons in the head and tail, head muscles, intestine, phasmid socket cells, spermatheca, and eggs. A weak expression was also observed in the cells of the gonad and vulva. The neurons expressing ser-3 promoter-driven GFP in the tail were identified as PHA, PHB, and PVQ neurons. SIA neurons are among the many neurons that were GFP positive in the head, which was confirmed by colocalization of GFP and DsRed in the animal carrying ser-3::gfp and ceh-17::dsred (Fig. 3). This suggests that SER-3 may mediate the starvation- and octopamine-induced activation of CREB in these neurons. To test this, we transferred the cre::gfp reporter in tzIs3 into the deletion mutant ser-3(ad1774) in which the region corresponding to the putative transmembrane domains III to VII of SER-3 had been removed from the ser-3 gene and therefore likely represents a null allele. These animals had a much reduced activation of GFP expression in the SIA neurons after starvation (14%) and exposure to exogenously applied octopamine (0%) (Table 1). The reduced induction of CREB activation after starvation and octopamine exposure was in part rescued by the reintroduction of a transgene containing the wild-type ser-3 gene as an extrachromosomal array. To examine whether SER-3 protein is working within the SIA neurons, SER-3 was expressed in the ser-3 mutant under control of the ceh-17 promoter (ceh-17::ser-3) that induces expression only in SIA neurons and the ALA neuron. Introduction of ceh-17::ser-3 rescued the starvation- and octopamine-induced GFP expression in the ser-3 mutant (84%). To further show that octopamine is working directly on the SIA neurons and that release of other transmitters from other cells is not required, unc-13 (uncoordinated family member 13) and unc-64 mutants were tested for the GFP induction. unc-13 and unc-64 genes encode UNC-13 protein [or Munc13 (mammalian unc)] and syntaxin, respectively, which are required for vesicle release, including neurotransmitter release. As expected, these mutants failed to respond to starvation. However, both unc-13 and unc-64 mutants responded normally to exogenous octopamine, indicating that endogenous neurotransmitter release is not required for the response to exogenous octopamine. Collectively, the data show that octopamine is working directly on the SIA neurons and that SER-3 is the octopamine receptor in SIA neurons that mediates activation of CREB in response to a starvation-mediated octopamine signal.
Figure 5.
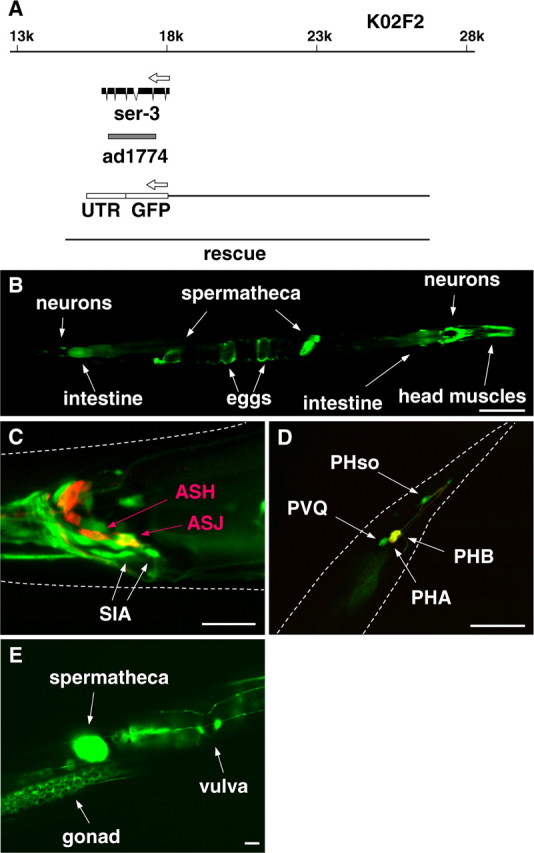
Gene structure and expression pattern of ser-3. A, ser-3 gene is located on the cosmid K02F2. The gray bar indicates the region deleted in the ad1774 allele. The regions used for the reporter fusion (ser-3::gfp) and in the rescue experiments are also indicated. B–E, The fluorescent images were taken from animals carrying ser-3::gfp. The expression of GFP and DiI staining of sensory neurons are shown in green and red, respectively. ASH, ASJ, PHA, and PHB neurons are stained by DiI. UTR, Untranslated region. B, GFP expression was observed in various tissues, including neurons, head muscles, intestine, spermatheca, and eggs. C, In the head region, GFP expression was observed in several neurons, including SIA neurons. D, In the tail region, GFP expression was observed in PHA, PHB, and PVQ neurons and phasmid socket cells. PHA and PHB appear yellow because they are both stained with DiI and are expressing GFP. E, Weak GFP expression was observed in the gonad and the vulva. White dotted lines indicate the outlines of the head (C) and the tail (D) of the animals. Scale bars: B, 100 μm; C–E, 20 μm.
Octopamine activates an EGL-30–CRH-1 signaling cascade
SER-3 protein is most homologous to the insect OAMB and mammalian α1 adrenergic receptor family that signals through Gαq. To investigate whether CREB activation through SER-3 is dependent on a Gαq signaling cascade, we analyzed expression from the cre::gfp reporter in two near loss-of-function mutants for Gαq, namely egl-30(ad806) and egl-30(n686) (Trent et al., 1983; Lackner et al., 1999). None of the animals showed any induction of GFP after starvation, and only ∼5% of the animals showed GFP expression in SIA neurons after octopamine exposure (Table 1). Expression of EGL-30 protein in SIA neurons by ceh-17 promoter using ceh-17::egl-30 restored in part the response to exogenous octopamine in egl-30(n686) mutants (Table 1), showing that EGL-30 is working in SIA neurons downstream of octopamine. However, introduction of ceh-17::egl-30 did not rescue the response to starvation. A simple explanation for this would be that the rescue was partial and therefore the CREB activation was not induced by the endogenous level of octopamine, which might be lower than in the exogenous application. Another possible explanation for this is that EGL-30 also plays a role in other cells upstream from SIA neurons. In this case, EGL-30 might be working upstream of UNC-13 similarly as reported for the regulation of neurotransmitter release (Lackner et al., 1999).
We also tested the gain-of-function mutant egl-30(js126gf) (Hawasli et al., 2004) and found that ∼50% of the mutant animals show the GFP expression in the well fed condition (Table 1). The frequency of the GFP expression was further increased during octopamine treatment but only slightly increased by starvation. This high basal frequency of the GFP expression in the absence of stimuli further supports the involvement of Gαq in the GFP expression in SIA neurons.
Phospholipase Cβ (PLCβ) is a common effector for Gαq, and therefore we analyzed the activation of CREB in two near loss-of-function PLCβ mutants, egl-8(n488) and egl-8(md1971) (Trent et al., 1983; Miller et al., 1996). After starvation, the egl-8(n488) mutant showed a reduction in the frequency of GFP induction to 28%, whereas in the egl-8(md1971) mutant, the GFP induction was reduced to 14% (Table 1). The induction of GFP in response to exogenously applied octopamine in egl-8 mutants was also reduced but still two to three times more frequent than observed after starvation (Table 1). These results suggest that EGL-8 plays a role as a downstream effector of EGL-30 as well as a role upstream of octopamine release. It is also possible that the exogenous application of octopamine resulted in stronger activation of the SER-3 signal than that induced by the starvation, and therefore the egl-8 mutants showed less reduction of the response to the octopamine treatment.
To confirm that the cre::gfp-mediated induction of GFP by starvation and octopamine is indeed dependent on the CREB homolog CRH-1, we analyzed the cre::gfp reporter in the crh-1(tz2) mutant background. As expected, these animals did not show induction of GFP expression by starvation or exogenously applied octopamine (Table 1). To establish that CRH-1 is working downstream of EGL-30, we tested an egl-30(js126gf);crh-1 double mutant and found that the mutation in the crh-1 gene completely suppressed the high basal GFP expression observed in egl-30(js126gf) mutants. Considering that a constitutively active CMK-1 (CaMKI) can induce CREB-dependent gene expression (Kimura et al., 2002), we also tested the ability of the deletion mutant cmk-1(ok287) to induce GFP expression from the cre::gfp reporter. Both starvation as well as octopamine exposure showed only a mild suppression of the frequency of GFP expression, indicating that CMK-1 plays a minor role in the cascade. Together, these data indicate that octopamine initiates an EGL-30 signaling cascade that activates CRH-1 in the SIA neurons.
GOA-1 suppresses EGL-30-mediated CREB activation
It has been shown previously in C. elegans that Gαo [GOA-1 (G-protein, O, α subunit family member 1)] activation suppresses signaling by Gαq (EGL-30) (Hajdu-Cronin et al., 1999; Miller et al., 1999; Nurrish et al., 1999; Matsuki et al., 2006). To test whether GOA-1 also acts in the starvation-induced CREB activation, we examined the induction of GFP expression of the cre::gfp transgene in the loss-of-function mutant goa-1(sa734). These animals showed an increased basal frequency of GFP expression of ∼30% in the SIA neurons, which was not increased by starvation (Table 1). Octopamine treatment of goa-1(sa734) mutants resulted in an induction of CREB activation in the SIA neurons in 88% of the animals. The increased basal frequency of GFP expression in the SIA neurons of the goa-1 mutant suggests that GOA-1 is normally suppressing CREB activation in the SIA neurons.
A gain-of-function mutant of UNC-43 CaMKII has been found to activate GOA-1 (Robatzek and Thomas, 2000). If GOA-1 can suppress the activity of EGL-30 signaling in the SIA neurons, we would predict that unc-43(n498gf) gain-of-function mutant animals would suppress both starvation as well as exogenous octopamine-mediated CREB activation. Indeed, we found that the unc-43(n498gf) mutant carrying the cre::gfp reporter showed an almost complete suppression of GFP expression by starvation and octopamine exposure (Table 1). The loss-of-function unc-43(e408) mutant showed normal, or even enhanced, frequency of GFP expression. If UNC-43 is suppressing CREB activation through activation of GOA-1, an unc-43(n498gf);goa-1 double mutant is predicted not to show suppression of CREB activation seen in the unc-43(n498gf) mutant. Sixty percent of the double mutants responded to the exogenous octopamine with expression of GFP in the SIA neurons, showing that the suppression of CREB activation by UNC-43 is dependent on GOA-1. This indicates that GOA-1 and UNC-43 negatively regulate the EGL-30–CRH-1 signaling cascade downstream of octopamine. However, the unc-43(n498gf);goa-1 double mutant did not show the high basal frequency of GFP expression, which was seen in the goa-1 mutant. This suggests that UNC-43 may also function independently of GOA-1.
“Soaking” activates an EGL-30–CRH-1 cascade independently of octopamine
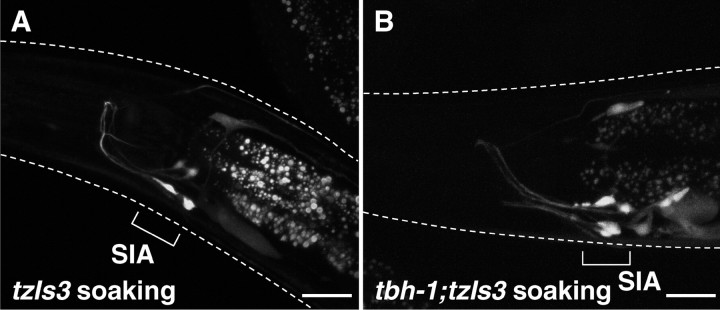
It was noticed that the basal frequency of GFP expression from the cre::gfp reporter was enhanced in the SIA neurons when animals were placed on NGM agar media, of which the surface was not completely dried. This was further tested by incubating the animals on NGM agar plates seeded with bacteria on which the entire surface was covered with 5 mm of water. This “soaking” condition induced GFP expression in the SIA neurons in >90% of the animals carrying the cre::gfp reporter after 6 h, similar to what was seen by starvation and octopamine exposure (Fig. 6, Table 1). The soaking-induced GFP expression in the SIA neurons is unlikely to be caused by starvation, because the animals appear to eat bacteria from the surface of the agar plate and did not display the pale appearance resulting from loss of fat stores (McKay et al., 2003). In testing the different mutants that delineate the starvation-mediated octopamine–SER-3–EGL-30–CRH-1 cascade, we find that the soaking-induced response is independent of octopamine and SER-3 signaling because both tbh-1 and ser-3 mutants responded to this stimulus (Table 1). However, the expression of GFP attributable to this stimulus was strongly suppressed in the egl-30, egl-8, and crh-1 mutants, indicating that the soaking stimulus also uses the same EGL-30 signaling cascade as seen for starvation (Table 1). The soaking-induced signaling cascade is likely also under negative control by GOA-1 because the gain-of-function mutant unc-43(n498gf) suppresses GFP expression, whereas the goa-1 mutant still responds to this stimulus, albeit with a reduced frequency compared with control animals. In addition, unc-13 and unc-64 mutants failed to respond to soaking, suggesting that vesicle release is also involved in the soaking signaling cascade.
Figure 6.
Soaking-induced GFP expression in control animals and tbh-1 mutants carrying cre::gfp. Fluorescent images were taken from control animals (A) and tbh-1 mutants (B) carrying cre::gfp that were soaked in water in the presence of food. GFP expression in SIA neurons was observed in both control and tbh-1 mutants after soaking. White dotted lines indicate the outline of the heads of the animals. Scale bars, 20 μm.
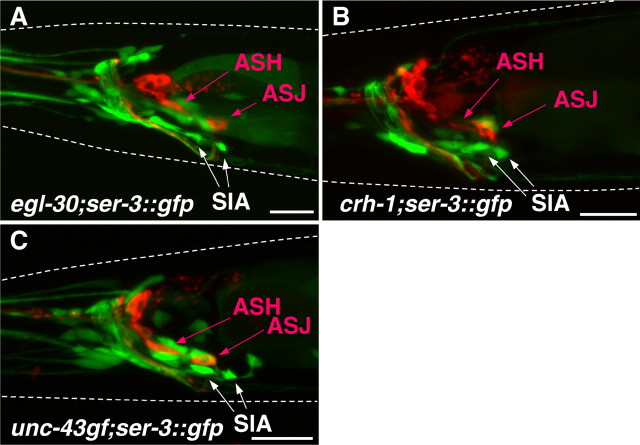
An important question is whether the loss of stimulus-induced GFP expression in the different cre::gfp transgene containing mutants is caused by a loss of the SIA neurons attributable to developmental defects rather than signaling deficits. To address this, we introduced the ser-3::gfp transgene, which induces GFP expression in the SIA neurons, into the egl-30(ad806), egl-30(n686), crh-1(tz2), and unc-43(n498gf) single mutants. All of these mutants showed the presence of GFP expression in the SIA neurons (Fig. 7). Furthermore, because the tbh-1 and ser-3 mutants still responded by expressing GFP from the cre::gfp transgene in the SIA neurons to the soaking stimulus, these animals also have no developmental defect, resulting in the loss of the SIA neurons.
Figure 7.
SIA neurons in egl-30, crh-1, and unc-43 mutants. The fluorescent images were taken from egl-30(n686), crh-1(tz2), and unc-43(n498gf) mutants carrying ser-3::gfp. The expression of GFP and DiI staining of sensory neurons are shown in green and red, respectively. GFP expression was observed in the position of SIA neurons in egl-30 (A), crh-1 (B), and unc-43(n498gf) (C) mutants. White dotted lines indicate the outlines of the heads of the animals. Scale bars, 20 μm.
Discussion
In this study, a novel in vivo mechanism for CREB activation was elucidated by using a CRE reporter and reverse genetic approaches in C. elegans. We have shown that octopamine signals starvation to activate CREB through EGL-30 (Gαq) selectively in SIA neurons. This clearly demonstrates that an amine neurotransmitter can activate CREB through Gαq in vivo. Furthermore, our results provide evidence that two unrelated environmental stimuli, starvation and soaking, converge to induce CRE-mediated gene expression in the same neurons, suggesting a role for CREB in the integration of environmental stimuli.
Octopamine signals starvation
The feeding state of C. elegans is a strong environmental stimulus that is important for behavioral adaptive changes to maximize foraging (Sawin et al., 2000; Hills et al., 2004) and initiates the developmental program to enter the dauer stage for long-term survival under environmentally unfavorable conditions (Golden and Riddle, 1982). Exogenous application of the neurotransmitter octopamine will cause C. elegans to mimic behaviors seen during starvation (Horvitz et al., 1982; Mohri et al., 2005). This has suggested that the endogenous octopamine system plays an important role in these behaviors. Here we show that starvation indeed activates the octopamine system, which results in activation of the C. elegans CREB ortholog CRH-1 in SIA neurons. The role of octopamine in the starvation response was demonstrated by the fact that loss of the tyramine-β-hydroxylase TBH-1, which is essential for octopamine synthesis (Alkema et al., 2005), prevents starvation-induced GFP expression in SIA neurons, which can be rescued by exogenously applied octopamine. The receptor mediating the starvation–octopamine response on the SIA neurons was found to be SER-3, which, based on sequence homology, is most closely related to previously identified invertebrate octopamine receptors and the mammalian α1 adrenergic receptors. ser-3 deletion mutants fail to show GFP expression in the SIA neurons after both starvation and exogenous application of octopamine. This indicates that SER-3 acts downstream from octopamine. Because ser-3 gene is expressed in SIA neurons and because expression of SER-3 protein in SIA neurons restores the response to both starvation and octopamine treatment in ser-3 mutants, SER-3 works within SIA neurons to induce CREB activation. Moreover, mutants deficient in neurotransmission respond normally to exogenously applied octopamine. The results indicate that octopamine is working directly on the SIA neurons. Because there is no known synaptic output from the octopaminergic RIC neurons to SIA neurons (White et al., 1986), octopamine is likely working as a neurohumoral factor in the regulation of CREB activation.
CREB is known to be activated by various stimuli, and multiple extracellular signals may converge into CREB activation in the same cells. CREB activation in the mammalian pineal gland is shown to be regulated by different transmitters, including norepinephrine and neuropeptides (Simonneaux and Ribelayga, 2003). We found that soaking in C. elegans results in the activation of CREB in SIA neurons, as seen after starvation. However, unlike the starvation response, the soaking response is octopamine independent. Because the soaking pathway that leads to CREB activation also requires Gαq, EGL-30, and the genes essential for vesicle release, it suggests that soaking results in the activation of another non-octopaminergic G-protein-coupled receptor. Our results demonstrate that CREB in the same cells can be activated by distinct extracellular signals in C. elegans.
C. elegans is a soil-dwelling nematode, and it encounters a soaking environment during periods of precipitation. After entering liquid, animals rapidly switch their movement from sinusoidal movement to “thrashing” movement, which is characterized by exaggerated body bends (Miller et al., 1996). Liquid environment may also affect the metabolism of the animals. Because oxygen levels quickly become limiting in water, it is suggested that the animals use a low oxygen consuming metabolic state (Rea and Johnson, 2003). It is noteworthy that the same metabolic state is likely to be induced in starved animals (Rea and Johnson, 2003).
EGL-30 (Gαq) activation of CRH-1 (CREB)
We show that the Gαq EGL-30 works downstream of octopamine, most likely through SER-3, to activate the CREB/CRH-1 in the SIA neurons. We found that EGL-30 in SIA neurons is required for octopamine-induced CREB activation, and the gain-of-function mutant of egl-30 results in increased CREB activity in the absence of stimuli. Moreover, this increased basal gene expression in egl-30 gain-of-function mutants was suppressed in crh-1 mutants. We also found that EGL-8, a PLCβ, plays a role in octopamine-induced CREB activation. However, EGL-8 does not seem to be the only downstream effector of EGL-30 because the response to octopamine in egl-8 mutants was not as strongly reduced, as seen for egl-30 mutants. This is similar to previous findings in which the action of EGL-30 is shown to be not entirely dependent on EGL-8 in the regulation of C. elegans behaviors (Lackner et al., 1999; Bastiani et al., 2003) and predicts the existence of other effectors of EGL-30 that have yet to be identified.
Gαq signaling may potentially activate CREB through alternative pathways. It was shown previously that the C. elegans CaMKI homolog CMK-1 can activate the cre::gfp reporter through the CREB CRH-1 (Kimura et al., 2002). However, we found that CMK-1 plays only a minor role in starvation/octopamine-mediated activation of CRH-1 in SIA neurons, suggesting that another kinase may be involved. The only other known CaMK homolog in C. elegans is UNC-43. However, CRH-1 can still be activated in an unc-43 loss-of-function mutant, excluding a major role for CaMKs in starvation/octopamine-mediated CRH-1 activation in the SIA neurons. Gαq signaling can activate mitogen-activated protein kinases, which in turn can stimulate the kinases ribosomal protein S6 kinase (RSK), mitogen- and stress-activated kinase (MSK), and mitogen-activated protein kinase-activated protein kinase 2 (MAPKAPK2) to activate CREB (Shaywitz and Greenberg, 1999). The C. elegans genome contains the genes rskn-1 (RSK–p90 kinase homolog family member 1), rskn-2, and mak-2 (MAP kinase activated protein kinase family member 2) which encode putative homologs of RSK, MSK, and MAPKAPK2, respectively, but their role in CRH-1 activation in C. elegans is thus far unknown. In addition, our results do not exclude the possibility that PKA is involved in Gαq-mediated CREB activation, which was implicated in the α1 adrenergic receptor, a Gq-coupled receptor-mediated activation of CRE-dependent gene expression in Rat1 cells (Lin et al., 1998).
In the motor neurons of C. elegans, EGL-30 signals through EGL-8 to control neurotransmitter release at the neuromuscular junction, and this pathway is negatively regulated by GOA-1, the α subunit of Go (Hajdu-Cronin et al., 1999; Nurrish et al., 1999; Hawasli et al., 2004). unc-43 gene encodes a CaMKII that acts upstream of GOA-1 and is postulated to directly activate GOA-1 by phosphorylation, which in turn suppresses EGL-30 (Reiner et al., 1999; Robatzek et al., 2000). We provide evidence that CRH-1 activation in the SIA neurons is subject to a comparable dynamic control between Gαq and Gαo in which EGL-30 and EGL-8 promote CREB activation, whereas it is suppressed by GOA-1 and UNC-43. Inhibition of CREB activation by an unc-43 gain-of-function mutation was partially suppressed by a goa-1 mutation, which suggest that UNC-43 is working upstream of GOA-1 as seen in the regulation of neurotransmitter release at the neuromuscular junction. However, the unc-43(n498gf);goa-1 double mutant or the unc-43 loss-of-function mutant did not show high basal CRE-mediated gene expression as seen for the goa-1 mutant. It has been shown in mammals that CaMKII could be a negative regulator of CRE-mediated gene expression by direct phosphorylation of an inhibitory site of CREB (Shaywitz and Greenberg, 1999; Mayr and Montminy, 2001). It remains possible that UNC-43 also works independently of GOA-1 to act directly on CRH-1 to control the CRE-mediated gene expression.
Amine neurotransmitter-mediated CREB activation is shown to play important roles in learning and memory (Mayford and Kandel, 1999) and the regulation of the biological clock (Simonneaux and Ribelayga, 2003) and is also implicated in mental disorders such as addiction (Berke and Hyman, 2000; Nestler, 2001) and depression (Nestler et al., 2002). A plethora of studies in various systems have shown that Gαs signaling plays a predominant role in amine neurotransmitter-mediated CREB activation, and it was unknown whether other G-proteins activate CREB in vivo. Nevertheless, some studies have shown that Gαq signaling can also result in CREB activation (Lin et al., 1998; Chalecka-Franaszek et al., 1999; Thonberg et al., 2002). However, these studies depend on pharmacological approaches, and the experiments were conducted using cultured cells. Our results demonstrate that endogenous octopamine is released during starvation and activates CREB through Gαq-mediated signaling, which is under negative control of Gαo, in the neurons of C. elegans, providing genetic evidence that an amine neurotransmitter can work through Gαq to activate CREB in vivo.
Footnotes
This work was supported by the Canadian Psychiatric Research Foundation, Astra-Zeneca, and the Canadian Institutes of Health Research. H.H.M.V.T. was a holder of a Canadian Research Chair in Neurobiology. S.S. is a recipient of Centre for Addiction and Mental Health fellowship. We thank Drs. Niacaris and Avery for the ser-3 mutant, Dr. Plasterk for the egl-30 vector, Dr. Fire for the GFP vector, Dr. Coulson for the cosmids, Dr. Roy for the DsRed vector, and Dr. Clark for comments on this manuscript. We gratefully acknowledge Dr. Culotti for helpful discussions and comments on this manuscript. tbh-1(ok1196) and cmk-1(ok287) were isolated by the C. elegans Gene Knockout Project at Oklahoma Medical Research Foundation (Oklahoma City, OK). C. elegans strains were obtained through Caenorhabditis Genetic Center (University of Minnesota, Minneapolis, MN). We also thank S. H. Ordog for excellent technical assistance. This paper is dedicated to the memory of Dr. Hubert H. M. Van Tol.
References
- Alkema MJ, Hunter-Ensor M, Ringstad N, Horvitz HR. Tyramine functions independently of octopamine in the Caenorhabditis elegans nervous system. Neuron. 2005;46:247–260. doi: 10.1016/j.neuron.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Bastiani CA, Gharib S, Simon MI, Sternberg PW. Caenorhabditis elegans Gαq regulates egg-laying behavior via a PLCβ-independent and serotonin-dependent signaling pathway and likely functions both in the nervous system and in muscle. Genetics. 2003;165:1805–1822. doi: 10.1093/genetics/165.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke JD, Hyman SE. Addiction, dopamine, and the molecular mechanisms of memory. Neuron. 2000;25:515–532. doi: 10.1016/s0896-6273(00)81056-9. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalecka-Franaszek E, Chen H, Chuang DM. 5-Hydroxytryptamine2A receptor stimulation induces activator protein-1 and cAMP-responsive element binding with cyclic AMP-responsive element-binding protein and Jun D as common components in cerebellar neurons. Neuroscience. 1999;88:885–898. doi: 10.1016/s0306-4522(98)00269-3. [DOI] [PubMed] [Google Scholar]
- Golden JW, Riddle DL. A pheromone influences larval development in the nematode Caenorhabditis elegans. Science. 1982;218:578–580. doi: 10.1126/science.6896933. [DOI] [PubMed] [Google Scholar]
- Grohmann L, Blenau W, Erber J, Ebert PR, Strunker T, Baumann A. Molecular and functional characterization of an octopamine receptor from honeybee (Apis mellifera) brain. J Neurochem. 2003;86:725–735. doi: 10.1046/j.1471-4159.2003.01876.x. [DOI] [PubMed] [Google Scholar]
- Hajdu-Cronin YM, Chen WJ, Patikoglou G, Koelle MR, Sternberg PW. Antagonism between Goα and Gqα in Caenorhabditis elegans: the RGS protein EAT-16 is necessary for Goα signaling and regulates Gqα activity. Genes Dev. 1999;13:1780–1793. doi: 10.1101/gad.13.14.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han KA, Millar NS, Davis RL. A novel octopamine receptor with preferential expression in Drosophila mushroom bodies. J Neurosci. 1998;18:3650–3658. doi: 10.1523/JNEUROSCI.18-10-03650.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawasli AH, Saifee O, Liu C, Nonet ML, Crowder CM. Resistance to volatile anesthetics by mutations enhancing excitatory neurotransmitter release in Caenorhabditis elegans. Genetics. 2004;168:831–843. doi: 10.1534/genetics.104.030502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman RK, Hedgecock EM. Limitation of the size of the vulval primordium of Caenorhabditis elegans by lin-15 expression in surrounding hypodermis. Nature. 1990;348:169–171. doi: 10.1038/348169a0. [DOI] [PubMed] [Google Scholar]
- Hills T, Brockie PJ, Maricq AV. Dopamine and glutamate control area-restricted search behavior in Caenorhabditis elegans. J Neurosci. 2004;24:1217–1225. doi: 10.1523/JNEUROSCI.1569-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Horvitz HR, Chalfie M, Trent C, Sulston JE, Evans PD. Serotonin and octopamine in the nematode Caenorhabditis elegans. Science. 1982;216:1012–1014. doi: 10.1126/science.6805073. [DOI] [PubMed] [Google Scholar]
- Hyman SE. Addiction to cocaine and amphetamine. Neuron. 1996;16:901–904. doi: 10.1016/s0896-6273(00)80111-7. [DOI] [PubMed] [Google Scholar]
- Johannessen M, Delghandi MP, Moens U. What turns CREB on? Cell Signal. 2004;16:1211–1227. doi: 10.1016/j.cellsig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Corcoran EE, Eto K, Gengyo-Ando K, Muramatsu MA, Kobayashi R, Freedman JH, Mitani S, Hagiwara M, Means AR, Tokumitsu H. A CaMK cascade activates CRE-mediated transcription in neurons of Caenorhabditis elegans. EMBO Rep. 2002;3:962–966. doi: 10.1093/embo-reports/kvf191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner MR, Nurrish SJ, Kaplan JM. Facilitation of synaptic transmission by EGL-30 Gqα and EGL-8 PLCβ: DAG binding to UNC-13 is required to stimulate acetylcholine release. Neuron. 1999;24:335–346. doi: 10.1016/s0896-6273(00)80848-x. [DOI] [PubMed] [Google Scholar]
- Lin RZ, Chen J, Hu ZW, Hoffman BB. Phosphorylation of the cAMP response element-binding protein and activation of transcription by α1 adrenergic receptors. J Biol Chem. 1998;273:30033–30038. doi: 10.1074/jbc.273.45.30033. [DOI] [PubMed] [Google Scholar]
- Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Matsuki M, Kunitomo H, Iino Y. Goα regulates olfactory adaptation by antagonizing Gqα-DAG signaling in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2006;103:1112–1117. doi: 10.1073/pnas.0506954103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Kandel ER. Genetic approaches to memory storage. Trends Genet. 1999;15:463–470. doi: 10.1016/s0168-9525(99)01846-6. [DOI] [PubMed] [Google Scholar]
- Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- McKay RM, McKay JP, Avery L, Graff JM. C. elegans: a model for exploring the genetics of fat storage. Dev Cell. 2003;4:131–142. doi: 10.1016/s1534-5807(02)00411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KG, Alfonso A, Nguyen M, Crowell JA, Johnson CD, Rand JB. A genetic selection for Caenorhabditis elegans synaptic transmission mutants. Proc Natl Acad Sci USA. 1996;93:12593–12598. doi: 10.1073/pnas.93.22.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KG, Emerson MD, Rand JB. Goα and diacylglycerol kinase negatively regulate the Gqα pathway in C. elegans. Neuron. 1999;24:323–333. doi: 10.1016/s0896-6273(00)80847-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohri A, Kodama E, Kimura KD, Koike M, Mizuno T, Mori I. Genetic control of temperature preference in the nematode Caenorhabditis elegans. Genetics. 2005;169:1437–1450. doi: 10.1534/genetics.104.036111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ. Molecular basis of long-term plasticity underlying addiction. Nat Rev Neurosci. 2001;2:119–128. doi: 10.1038/35053570. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Barrot M, DiLeone RJ, Eisch AJ, Gold SJ, Monteggia LM. Neurobiology of depression. Neuron. 2002;34:13–25. doi: 10.1016/s0896-6273(02)00653-0. [DOI] [PubMed] [Google Scholar]
- Nurrish S, Segalat L, Kaplan JM. Serotonin inhibition of synaptic transmission: Gα(o) decreases the abundance of UNC-13 at release sites. Neuron. 1999;24:231–242. doi: 10.1016/s0896-6273(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Park EC, Horvitz HR. Mutations with dominant effects on the behavior and morphology of the nematode Caenorhabditis elegans. Genetics. 1986;113:821–852. doi: 10.1093/genetics/113.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol N, Torregrossa P, Ewbank JJ, Brunet JF. The homeodomain protein CePHOX2/CEH-17 controls antero-posterior axonal growth in C. elegans. Development. 2000;127:3361–3371. doi: 10.1242/dev.127.15.3361. [DOI] [PubMed] [Google Scholar]
- Rea S, Johnson TE. A metabolic model for life span determination in Caenorhabditis elegans. Dev Cell. 2003;5:197–203. doi: 10.1016/s1534-5807(03)00242-9. [DOI] [PubMed] [Google Scholar]
- Reiner DJ, Newton EM, Tian H, Thomas JH. Diverse behavioural defects caused by mutations in Caenorhabditis elegans unc-43 CaM kinase II. Nature. 1999;402:199–203. doi: 10.1038/46072. [DOI] [PubMed] [Google Scholar]
- Robatzek M, Thomas JH. Calcium/calmodulin-dependent protein kinase II regulates Caenorhabditis elegans locomotion in concert with a G(o)/G(q) signaling network. Genetics. 2000;156:1069–1082. doi: 10.1093/genetics/156.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder T. Octopamine in invertebrates. Prog Neurobiol. 1999;59:533–561. doi: 10.1016/s0301-0082(99)00016-7. [DOI] [PubMed] [Google Scholar]
- Sawin ER, Ranganathan R, Horvitz HR. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron. 2000;26:619–631. doi: 10.1016/s0896-6273(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- Simonneaux V, Ribelayga C. Generation of the melatonin endocrine message in mammals: a review of the complex regulation of melatonin synthesis by norepinephrine, peptides, and other pineal transmitters. Pharmacol Rev. 2003;55:325–395. doi: 10.1124/pr.55.2.2. [DOI] [PubMed] [Google Scholar]
- Sulston J, Dew M, Brenner S. Dopaminergic neurons in the nematode Caenorhabditis elegans. J Comp Neurol. 1975;163:215–226. doi: 10.1002/cne.901630207. [DOI] [PubMed] [Google Scholar]
- Thonberg H, Fredriksson JM, Nedergaard J, Cannon B. A novel pathway for adrenergic stimulation of cAMP-response-element-binding protein (CREB) phosphorylation: mediation via α1-adrenoceptors and protein kinase C activation. Biochem J. 2002;364:73–79. doi: 10.1042/bj3640073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent C, Tsuing N, Horvitz HR. Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics. 1983;104:619–647. doi: 10.1093/genetics/104.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the Caenorhabditis elegans nervous system. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Wintle RF, Van Tol HH. Dopamine signaling in Caenorhabditis elegans-potential for parkinsonism research. Parkinsonism Relat Disord. 2001;7:177–183. doi: 10.1016/s1353-8020(00)00055-9. [DOI] [PubMed] [Google Scholar]
- Wu D, Katz A, Lee CH, Simon MI. Activation of phospholipase C by α1-adrenergic receptors is mediated by the α subunits of Gq family. J Biol Chem. 1992;267:25798–25802. [PubMed] [Google Scholar]