Abstract
The mechanisms regulating the generation of cell diversity in the mammalian cerebral cortex are beginning to be elucidated. In that regard, Hairy/Enhancer of split (Hes) 1 and 5 are basic helix-loop-helix (bHLH) factors that inhibit the differentiation of pluripotent cortical progenitors into neurons. In contrast, a related Hes family member termed Hes6 promotes neurogenesis. It is shown here that knockdown of endogenous Hes6 causes supernumerary cortical progenitors to differentiate into cells that exhibit an astrocytic morphology and express the astrocyte marker protein GFAP. Conversely, exogenous Hes6 expression in cortical progenitors inhibits astrocyte differentiation. The negative effect of Hes6 on astrocyte differentiation is independent of its ability to promote neuronal differentiation. We also show that neither its proneuronal nor its anti-gliogenic functions appear to depend on Hes6 ability to bind to DNA via the basic arm of its bHLH domain. Both of these activities require Hes6 to be localized to nuclei, but only its anti-gliogenic function depends on two short peptides, LNHLL and WRPW, that are conserved in all Hes6 proteins. These findings suggest that Hes6 is an important regulator of the neurogenic phase of cortical development by promoting the neuronal fate while suppressing astrocyte differentiation. They suggest further that separate molecular mechanisms underlie the proneuronal and anti-gliogenic activities of Hes6 in cortical progenitor cells.
Keywords: astrocyte, cortical development, Groucho, Hes6, LXXLL motif, neurogenesis
Introduction
During mammalian cerebral cortex (cortical) development, pluripotent neural progenitor cells located in the ventricular zone lining the lateral ventricles give rise to neurons, astrocytes, and oligodendrocytes (Job and Tan, 2003). Neurogenesis occurs first, followed by astrocyte and then oligodendrocyte cell differentiation. A number of extrinsic and intrinsic cues regulate the mechanisms that promote or inhibit the differentiation of different neural cell types (Bertrand et al., 2002; Ross et al., 2003). In particular, the Notch signaling pathway plays an important role in inhibiting/delaying cortical neuron differentiation and promoting the undifferentiated progenitor state (Gaiano and Fishell, 2002, Kageyama et al., 2005). In the embryonic cerebral cortex, multipotent progenitors correspond to radial glia, a specific subtype of astrocyte-like cells that possess neurogenic potential (Fishell and Kriegstein, 2003; Malatesta et al., 2003). Notch signaling promotes the cortical radial glia fate (Gaiano and Fishell, 2002; Kageyama et al., 2005). It also induces the differentiation of Mueller glial cells in the retina (Furukawa et al., 2000) and astrocytes in the spinal cord (Wu et al., 2003).
Most of the neural functions of the Notch pathway involve the activity of the basic helix-loop-helix (bHLH) proteins Hairy/Enhancer of split (Hes) 1 and 5 (Gaiano and Fishell, 2002; Kageyama et al., 2005). Hes1 and Hes5 are transcriptional repressors, the expression of which is activated in response to Notch stimulation. They act by recruiting Groucho(Gro)/transducin-like Enhancer of split (TLE) transcriptional corepressors to specific DNA sites to repress the expression of target genes (Ju et al., 2004; Nuthall et al., 2004). In agreement with the ability of Hes1 repressor complexes to suppress the expression of proneuronal genes (Chen et al., 1997; Ross et al., 2003; Ju et al., 2004), Hes1 inhibits neuronal differentiation in vivo (Ishibashi et al., 1994, 1995). Moreover, Hes1 drives undifferentiated progenitors to an astrocyte cell fate in the spinal cord (Wu et al., 2003) and promotes the differentiation of Mueller glial cells in the retina (Furukawa et al., 2000). Recent studies have shown further that Hes1 facilitates complex formation between Janus kinase 2 and signal transducer and activator of transcription 3 (STAT3) thus promoting STAT3 activation (Kamakura et al., 2004). Activation of STAT3 promotes astrocyte differentiation from pluripotent cortical progenitor cells (Sun et al., 2001; Kamakura et al., 2004). Together, these observations suggest an involvement of Hes1 and Hes5 in the acquisition of the astrocyte cell fate in the CNS.
In contrast to Hes1/Hes5, a related Hes family member termed Hes6 promotes neuronal differentiation when exogenously expressed in murine retinal explants (Bae et al., 2000) and Xenopus neural plate (Koyano-Nakagawa et al., 2000). Hes6 is expressed in the developing cerebral cortex (Bae et al., 2000; Pissarra et al., 2000; Vasiliauskas and Stern, 2000), and exogenous Hes6 expression in undifferentiated cortical progenitor cells promotes the differentiation of supernumerary neurons (Gratton et al., 2003). Here, we describe studies aimed at testing whether Hes6 is involved in the regulation of cortical astrocyte differentiation, in addition to promoting neurogenesis. Our results show that Hes6 inhibits astrocyte differentiation and that the molecular mechanisms underlying this function are different from those involved in its neurogenic activity.
Materials and Methods
DNA plasmids.
Plasmids pCMV2- hemagglutinin (HA)-Hes6WT, pCMV2-HA-Hes6ΔWRPW (Gratton et al., 2003), and pcTOPO-Hes6-VP16 (Gao et al., 2001) were described. A PCR-based strategy was used to generate the mutated constructs pCMV2-HA-Hes6BAD [primers basic arm defective (BAD)-F, 5′-GTGGAGGAGAAGGAACGCGCACGGATCAA-3′, and BAD-R, 5′-GATCCGTGCGCGTTCCTTCTCCTCCAC-3′] and pCMV2-HA-Hes6LMD [primers LNHLL-motif-defective (LMD)-F, 5′-CTGAACTCGCGAACCACGCGCTAGAATCC-3′ and LMD-R, 5′-GGATTCTAGCGCGTGGTTCGCGAGTTCAG-3′]. Plasmid pCMV2-HA-Hes6AQ was obtained using the QuickChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) with primers AQ-F, 5′-GTGGAGAAGAAGCGAGACGGACGCGCACGGATCA-3′ and AQ-R, 5′-TGATCCGTGCGCGTCCGTCTCGCTTCTTCTCCAC-3′. Wild-type and mutated Hes6 sequences were subcloned from pCMV2-HA into pCMV2-FLAG vector by restriction digestion with HindIII and BamHI. All mutations were verified by DNA sequencing. Plasmids pGEX1-Hes6WT and pGEX1-Hes6AQ were obtained by digesting pCMV2-FLAG-Hes6WT or pCMV2-FLAG-Hes6AQ with BglII and BamHI, and subcloning into the BamHI site of pGEX1. Constructs pCMV2-FLAG-Hes1, pGEX1-Hes1, pEGFP, pCol-Luc, p2xESE-Col-Luc, pNgn3-promoter-Luc, pNgn3-ΔNbox-promoter-Luc, and pRSV-β-galactosidase were described previously (McLarren et al., 2000; Cossins et al., 2002; Nuthall et al., 2002; Gratton et al., 2003).
Cortical neural progenitor cell cultures.
Primary neural progenitor cell cultures were established from dorsal telencephalic cortices obtained from mouse embryos collected at stage 11.5–12.5 [embryonic day 11.5 (E11.5) to E12.5] and cultured in four-well chamber slides as described previously (Marcal et al., 2005; Theriault et al., 2005). For RNA interference (RNAi) studies, ∼2.0 × 105 cells/ml were seeded and cultured for 2 d in vitro (DIV) in Neurobasal medium supplemented with 1% N2, 2% B27, 0.5 mm glutamine, 1% penicillin-streptomycin, and 40 ng/ml FGF2. After this time, cells were transfected as described previously (Marcal et al., 2005) with an enhanced green fluorescent protein (EGFP) expression plasmid (pEGFP; 300 ng/transfection) in the absence or presence (30 nm) of validated small interfering RNA (siRNA) duplexes (Ambion, Austin, TX); these included either negative control siRNA (identification number 4615), Hes6 siRNA (identification numbers 185198, 185199, and 73594), or Grg6 siRNA (Marcal et al., 2005). Five days after transfection, cells were fixed and subjected to double-label immunocytochemistry and quantitation of the percentage of GFP-positive cells that were also positive for the expression of proliferation or differentiation markers using the following antibodies: mouse anti-Ki67 (cell proliferation marker; 1:100; BD PharMingen, San Diego, CA), mouse anti-neuron specific nuclear protein (NeuN) (neuron marker; 1:100; Chemicon, Temecula, CA), and mouse anti-glial fibrillary acidic protein (GFAP) (astrocyte marker; 1:300; Sigma, St. Louis, MO) (identical results were obtained using a rabbit anti-GFAP antibody from Chemicon). All images were captured with a black-and-white DVC camera mounted on an Axioskop 2 fluorescence microscope (Zeiss, Toronto, Ontario, Canada). Grayscale images were digitally assigned to the appropriate red or green channel using Northern Eclipse software (Empix, Missisauga, Ontario, Canada). For gain of function studies, cortical progenitor cells were cultured in the presence of FGF2 for 24 h and then transfected with pEGFP alone (200 ng/transfection) or in combination with Hes6 expression plasmids (800 ng/transfection) as described previously (Theriault et al., 2005). After transfection, cells were first cultured in the presence of FGF2 for 12–24 h and then either incubated with FGF2 for an additional 48 h (assays to measure effect of Hes6 on neuronal differentiation) or switched to a medium lacking FGF2 but containing 50 ng/ml ciliary neurotrophic factor (CNTF) and leukemia inhibitory factor (LIF) (R&D Systems, Minneapolis, MN) to induce astrocyte differentiation (Sun et al., 2001). Cells were treated with CNTF/LIF for various lengths of time (see figures for details) to examine the effect of Hes6 on astrocyte differentiation at different time points. Quantitation of the percentage of transfected cells that were expressing markers of proliferation or differentiation was as described above, except that a mouse anti-nestin antibody (neural progenitor cell marker; 1:400; Chemicon) was used in addition to the anti-Ki67 antibody.
Transient transfection/transcription assays.
Cortical progenitor cells were seeded in six-well dishes (1.0 × 106 cells/ml) and cultured throughout the experiment in the presence of FGF2. Cells were transfected with the appropriate plasmids 24 h after seeding as described previously (Theriault et al., 2005). In each case, a β-galactosidase expression plasmid was cotransfected to provide a means of normalizing the assays. Cells were harvested 24 h after transfection, followed by measurement of luciferase and β-galactosidase activities (Gratton et al., 2003; Theriault et al., 2005). Human embryonic kidney 293 (HEK293) cells were grown in DMEM supplemented with 10% fetal bovine serum, transfected with the appropriate plasmids, and subjected to analysis of reporter gene expression as described previously (McLarren et al., 2000, 2001). Results were expressed as mean values ± SD.
Coimmunoprecipitation assays and Western blot analysis.
Coimmunoprecipitation studies were performed by transfecting HEK293 cells for 24 h with plasmids encoding FLAG- or HA-tagged forms of wild-type or mutated Hes6 (500 ng/transfection), followed by cell lysis, immunoprecipitation with anti-FLAG antibodies (Sigma), and Western blotting (Gratton et al., 2003; Nuthall et al., 2004) with either anti-FLAG, anti-HA (Roche, Indianapolis, IN), or anti-Hes1 (Yao et al., 2001) antibodies.
Immunofluorescence.
HEK293 cells were transfected with HA- or FLAG-tagged forms of Hes6 (500 ng/transfection), cultured on four-well chamber slides, fixed in HEPES-buffered saline containing 4% paraformaldehyde, and permeabilized in 0.1% Igepal as described previously (Nuthall et al., 2004). Cells were then incubated with anti-HA or anti-FLAG antibodies followed by FITC-conjugated goat anti-mouse secondary antibody (Nuthall et al., 2004; Marcal et al., 2005). Hoechst 33258 reagent (Sigma) was used to counterstain nuclei.
Electrophoretic mobility shift assays.
Fusion proteins of glutathione S-transferase (GST) and either Hes6WT, Hes6AQ, or Hes1 were expressed and purified from bacteria as described previously (Gratton et al., 2003). Each fusion protein was incubated with a double-stranded oligonucleotide containing two Enhancer of split E (ESE) box motifs (underlined), top strand 5′-GAAGGGTGGCACGTGCCATTTGGCACGTGCCATG-3′ (Cossins et al., 2002). Probes were labeled at both ends by filling in with Klenow DNA polymerase in the presence of [α-32P]dCTP, and electrophoretic mobility shift assay (EMSA) experiments were performed as described previously (Yao et al., 2001; Cossins et al., 2002).
Results
Hes6 inhibits astrocyte differentiation in addition to promoting neurogenesis
To examine the role of Hes6 during cortical neuron and glial cell differentiation, primary cultures of pluripotent neural progenitor cells were established from the dorsal telencephalon of E11.5–E12.5 mouse embryos. This defined primary culture system (“cortical progenitor cells”) has been used extensively to study the functions of extrinsic and intrinsic factors during the progenitor-to-neuron transition (Ghosh and Greenberg, 1995; Morrow et al., 2001; Barnabe-Heider et al., 2005; Marcal et al., 2005). Cultures of pluripotent cortical progenitors generate both neurons and glia, and these cells appear at days in vitro approximately equivalent to their time of appearance in vivo (Morrow et al., 2001; Shen et al., 2006).
An RNAi approach was used to knock down the expression of endogenous Hes6 in cortical progenitor cells. siRNA oligonucleotides specific for Hes6, but not control siRNA, were first shown to cause a significant decrease in Hes6 protein levels in transfected HEK293 cells without affecting the levels of other proteins (Fig. 1A–C). Hes6 siRNA was transfected into cortical progenitors (together with enhanced GFP to mark the transfected cells) after 2 DIV, and individual cells were examined immunocytochemically 5 d later (7 DIV) for the expression of proliferation and differentiation markers (Fig. 1D) (data not shown). Compared with cells transfected with GFP alone or in combination with control siRNA, treatment with Hes6 siRNA was correlated with a decrease in the number of postmitotic neurons, detected by cell morphology (data not shown) (see Fig. 4A) and the expression of the neuron-specific marker protein NeuN (Fig. 1E, bars 1–3). This result is in agreement with the previously demonstrated proneuronal activity of Hes6 in mouse cortical progenitor cells (Gratton et al., 2003). The decrease in neuron cell number associated with transfection of Hes6 siRNA was not correlated with an increased number of Ki67-positive mitotic progenitors, but rather with a significant increase in the number of cells expressing the astrocyte marker protein GFAP and displaying astrocyte-like morphology (Fig. 1D,E, bars 5–7, 9–11). The increased astrocyte differentiation was not simply a consequence of decreased neurogenesis, because knock down of the Groucho-related protein Grg6 also caused a decrease in NeuN-positive neurons, as shown previously (Marcal et al., 2005), but had no significant effect on astrocyte differentiation (Fig. 1E, bars 4, 8), and instead resulted in an expansion of the mitotic cell pool (Fig. 1E, bar 12). These results strongly suggest that Hes6 is involved in mechanisms that inhibit cortical astrocyte differentiation, in addition to promoting neurogenesis.
Figure 1.
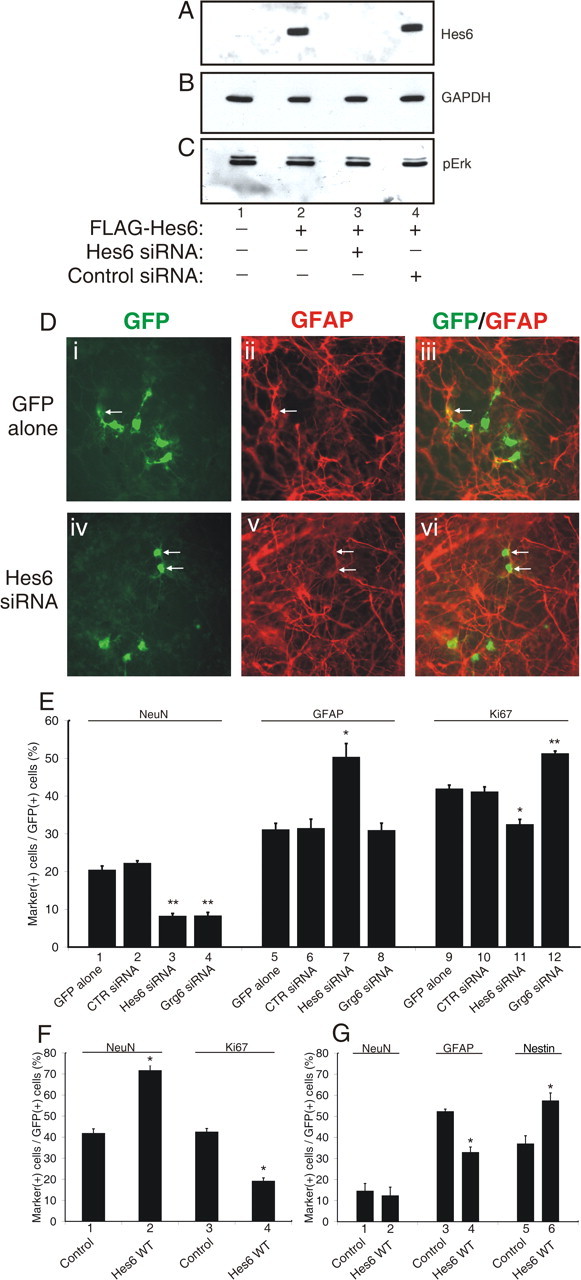
Inhibition of astrocyte differentiation and promotion of neurogenesis by Hes6. A–C, HEK293 cells were transfected with FLAG-Hes6WT in the absence (lane 2) or presence of either Hes6 siRNA (lane 3) or control siRNA (lane 4). Forty-eight hours later, cell lysates were subjected to sequential Western blotting with anti-FLAG (A), anti-glyceraldehyde-3-phosphate dehydrogenase (B), or anti-phosphoErk (C) antibodies; lane 1 was loaded with lysate from untransfected cells. D, Mouse cortical progenitor cells were transfected with either GFP alone (i–iii) or GFP in combination with Hes6 siRNA (iv–vi), followed by double-labeling analysis of the expression of GFP (i, iv) and the astrocyte marker GFAP (ii, v) after 7 DIV. The combined GFP and GFAP staining is shown in iii and vi. Arrows point to double-labeled cells. E, Cortical progenitor cells were transfected with GFP alone as control (bars 1, 5, 9) or in combination with the indicated siRNA reagents, followed by double-labeling analysis to calculate the percentage of GFP-positive cells that were also positive for the expression of either the neuronal-specific protein NeuN (bars 1–4), GFAP (bars 5–8), or the mitotic cell marker protein Ki67 (bars 9–12). The results are shown as the mean ± the SD (*p < 0.005; **p < 0.0005). F, G, Cortical progenitor cells were transfected with GFP alone (Control) or in combination with Hes6WT, as indicated, and cultured in the presence of either FGF2 for 2 d (F) or CNTF/LIF for 4 d (G). Cells were then subjected to double-labeling analysis with the indicated antibodies and quantitation as described above (*p < 0.001).
Figure 4.
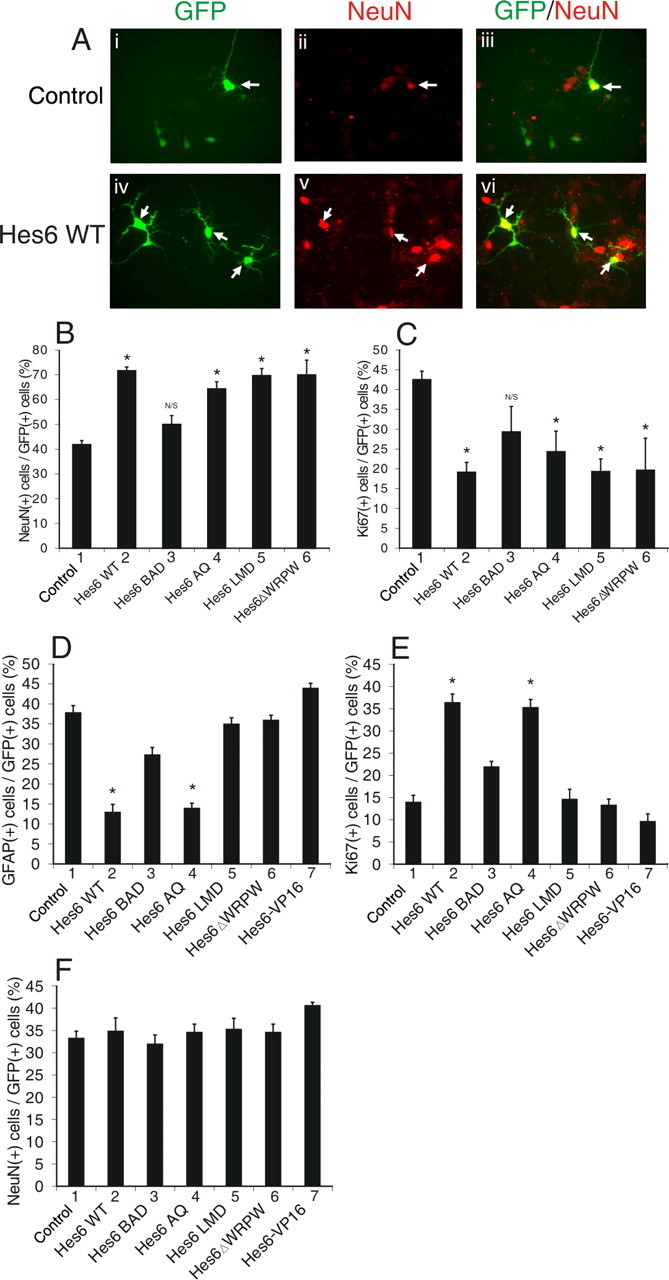
Analysis of the proneuronal and anti-gliogenic activities of mutated Hes6 proteins. A, Cortical progenitor cells were transfected with GFP alone (i–iii) or in combination with Hes6WT (iv–vi), followed by double-labeling analysis of the expression of GFP (i, iv) and the neuronal marker NeuN (ii, v) after 4 DIV. The combined GFP and NeuN staining is shown in iii and vi. B–F, Cortical progenitor cells were transfected with GFP alone (Control) or in combination with the indicated Hes6 proteins and cultured in the presence of either FGF2 (B, C) or CNTF/LIF (D–F) for 2 d. Cells were then subjected to double-labeling analysis and quantitation of the percentage of GFP-positive cells that also expressed the indicated marker proteins (*p < 0.005; NS, not significant).
To examine this possibility further, Hes6 was exogenously expressed in cortical progenitor cells cultured in the absence or presence of CNTF and LIF, two potent inducers of the astrocyte fate (Sun et al., 2001). In the absence of CNTF/LIF (but in the presence of FGF2), exogenous expression of wild-type Hes6 (Hes6WT) significantly induced neuronal differentiation compared with GFP alone (Fig. 1F, bars 1, 2). This effect was accompanied by a decrease in proliferating cells (Fig. 1F, bars 3,4), suggesting that Hes6 promotes the recruitment of supernumerary precursors into the neuronal lineage. Only small numbers of GFAP-positive cells were observed under these experimental conditions (data not shown). In contrast, exogenous expression of Hes6WT in cultures treated with CNTF/LIF did not have an effect on the number of differentiated neurons (Fig. 1G, bars 1, 2) but instead caused a significant decrease in the number of GFAP-positive cells (Fig. 1G, bars 3, 4). This effect was correlated with a parallel increase in the number of transfected cells that expressed the neural progenitor marker protein nestin (Fig. 1G, bars 5, 6). Similar results were obtained when Ki67 was used to label proliferating progenitors (data not shown). Together, these findings show that Hes6 is sufficient to promote the precursor-to-neuron transition in the absence of gliogenic stimuli. In the presence of CNTF/LIF, Hes6 can prevent the precursor-to-astrocyte transition but does not increase neuronal differentiation, resulting in an expansion of the neural progenitor cell pool. These observations strongly suggest that Hes6 both promotes neurogenesis and inhibits astrocyte differentiation during cortical neurogenesis.
Nuclear localization mediated by the basic arm is important for both neurogenic and anti-gliogenic activities of Hes6
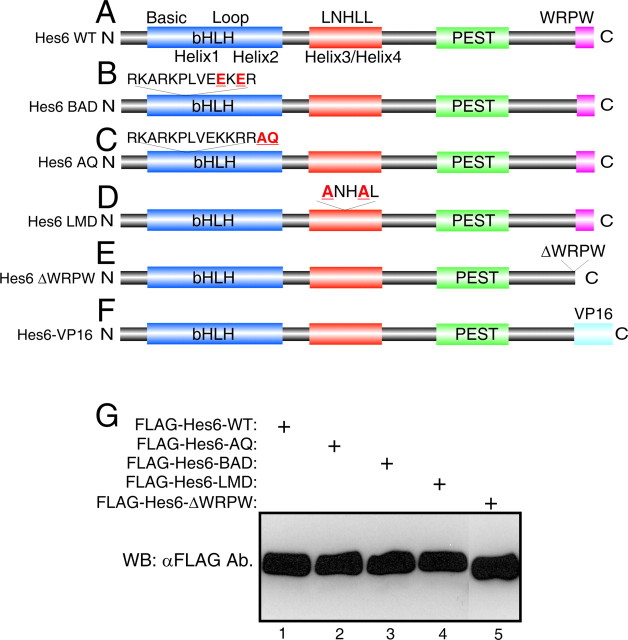
Hes6 is a unique Hes family member characterized by an N-terminal bHLH domain containing a shortened loop region, followed by a helix3/helix4 domain, a PEST region containing consensus phosphorylation sites, and a C-terminal WRPW tetrapeptide that mediates binding to Gro/TLE transcriptional corepressors (Fig. 2A) (Bae et al., 2000; Gratton et al., 2003). To determine whether Hes6 regulates neuron and astrocyte differentiation through similar or different mechanisms, a number of mutated forms of Hes6 were generated to test the contributions of individual domains to its proneuronal and anti-gliogenic activities.
Figure 2.
Depiction of wild-type and mutated Hes6 proteins. A, Schematic representation of Hes6: shown are the N-terminal bHLH domain, followed by the helix3/helix4 domain containing the LNHLL motif, the putative PEST region, and the WRPW motif at the C-terminal end (Bae et al., 2000; Gratton et al., 2003). B, Hes6BAD contains a 35KKRR-to-35EKER mutation within the basic arm of the bHLH domain. C, Hes6AQ contains a two amino acid insertion (AQ) at position 39 at the end of the basic arm. D, Hes6LMD contains a 126LNHLL-to-126ANHAL mutation near the end of the helix3/helix4 domain. E, Hes6ΔWRPW lacks the last four C-terminal residues, WRPW. F, In the Hes6-VP16 protein, the WRPW motif was deleted and replaced with the VP16 transcription activation domain. G, The indicated FLAG-tagged Hes6 proteins were transfected into HEK293 cells, followed by Western blotting analysis (WB) with anti-FLAG antibody (Ab.).
Previous studies have shown that Hes6 interacts with neither N box sequences (CACNAG), which are bound with high affinity by Notch-activated Hes proteins like Hes1, nor canonical E boxes (CANNTG), to which Hes1 binds with lower affinity (Bae et al., 2000; Koyano-Nakagawa et al., 2000; Cossins et al., 2002). However, Hes6 was shown to interact in vitro with a specific type of E box sequence, the ESE box (TGGCACGTGCCA) (Jennings et al., 1999; Cossins et al. 2002). The functional significance of this interaction was unclear, because studies in Xenopus embryos suggested that the proneuronal function of Hes6 is not dependent on DNA binding mediated by its bHLH domain (Koyano-Nakagawa et al., 2000; Cossins et al., 2002). To examine further the contribution of direct DNA binding to the neural functions of Hes6, we generated a mutation in the basic arm of mouse Hes6 by changing two basic residues to acidic amino acids (Fig. 2B). This mutated protein, Hes6BAD, was properly expressed (Fig. 2G, lane 3) but displayed an impaired nuclear translocation compared with Hes6WT when transfected into COS7 cells (Fig. 3, compare A–C and G–I). This defect was correlated with the finding that, when compared with Hes6WT, Hes6BAD had a reduced ability to promote neurogenesis (Fig. 4B,C, bars 1–3). Hes6BAD also displayed a decreased capacity to inhibit astrocyte differentiation (Fig. 4D,E, bars 1–3). Hes6BAD was competent to dimerize with both Hes6WT (Fig. 3M–P, lane 2) and itself and interacted normally with Gro/TLE (data not shown). These results indicate that the basic arm mutation does not perturb Hes6 homodimerization ability. Together, these findings show that an intact basic arm is important for the nuclear localization of Hes6 and provide a correlation between the neural functions of Hes6 and its ability to become localized to nuclei.
Figure 3.
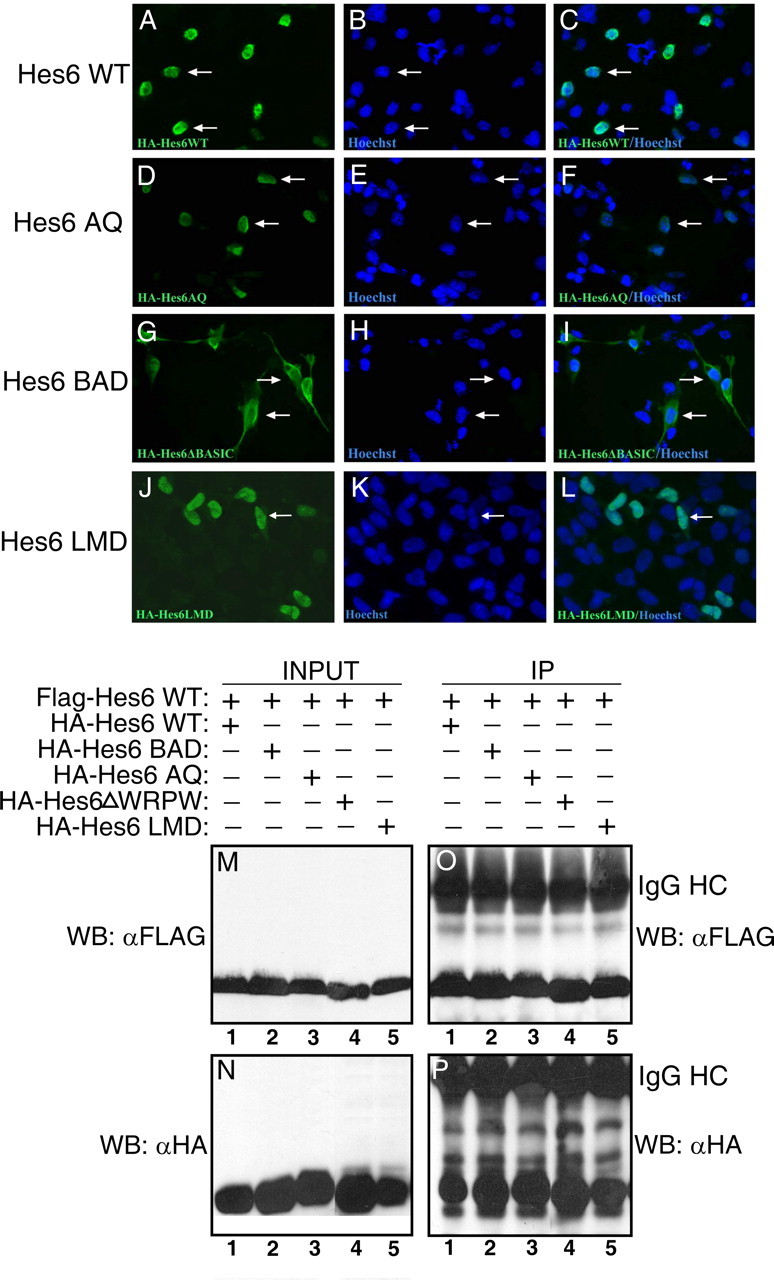
Analysis of mutated Hes6 proteins. A–L, Subcellular localization. COS7 cells were transfected with the indicated HA-tagged Hes6 proteins, followed by immunocytochemistry with anti-HA antibody (left panels) and counterstaining with Hoechst (middle panels). Results of combined anti-HA antibody/Hoechst staining are shown in the right panels. Arrows point to representative immunoreactive cells. M–P, Hes6 homodimerization ability. HEK293 cells were transfected with FLAG-Hes6WT (M) and the indicated HA-tagged Hes6 proteins (N), followed by immunoprecipitation (IP) with anti-FLAG antibody. Immunoprecipitates (O, P) were analyzed together with one-tenth of each input lysate (M, N) by Western blotting with either anti-FLAG (M, O) or anti-HA (N, P) antibodies. IgG HC indicates the immunoglobulin G heavy chain.
Direct DNA binding through the bHLH domain is not important for the neurogenic and anti-gliogenic activities of Hes6
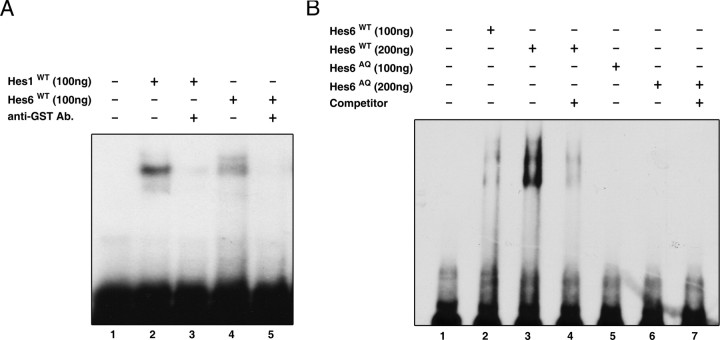
A decreased nuclear transport may reduce/prevent functions of Hes6 that depend on DNA binding. Alternatively (or in addition), it may preclude protein–protein interactions that occur in the nucleus. To examine these possibilities, we generated a mutation aimed at interfering with DNA binding mediated by the basic arm of the bHLH domain (if this indeed occurred in vivo) without perturbing nuclear localization. This mutation consisted of the insertion of two amino acids, alanine and glutamine, between the basic arm and helix 1 and was termed Hes6AQ (Fig. 2C). It was based on the previous demonstration that similar AQ insertions block the DNA binding of a number of other bHLH proteins (Sun et al., 2001; Lee and Pfaff, 2003). The Hes6AQ mutant was properly expressed and localized to the nucleus (Figs. 2G, lane 2, 3D–F) and displayed a normal homodimerization ability (Fig. 3M–P, lane 3). To determine whether the AQ insertion affected the ability of Hes6 to interact with the ESE box, EMSA experiments were performed with bacterially purified fusion proteins of GST and either Hes6WT or Hes6AQ. These proteins were expressed at equal levels and exhibited the same electrophoretic mobility on SDS-PAGE (data not shown). Hes6WT bound in a dose-dependent manner to a double-stranded oligonucleotide containing two ESE box motifs (Fig. 5A, compare lanes 1 and 4; B, lanes 1–3). This binding was decreased when an anti-GST antibody was included in the reaction mixture (Fig. 5A, compare lanes 4 and 5). Purified Hes1 also bound to the ESE box under the same conditions (Fig. 5A, lanes 1–3), in agreement with the previous demonstration that Hes1 binds to E boxes, albeit with low affinity (Sasai et al., 1992). The binding of both Hes6WT (Fig. 5B, compare lanes 3 and 4) and Hes1 (data not shown) to the ESE box was significantly reduced in the presence of an excess of unlabeled oligonucleotide. In contrast to Hes6WT, an equal amount of Hes6AQ failed to bind to the oligonucleotide probe, indicating that the AQ insertion disrupts the ability of Hes6 to interact with the ESE box sequence (Fig. 5B, lanes 5–7).
Figure 5.
EMSA experiments. A, A [32P]-labeled double-stranded oligonucleotide containing two tandem ESE boxes was incubated in the absence (lane 1) or presence of either GST-Hes1 (lanes 2 and 3) or GST-Hes6WT (lanes 4 and 5). An anti-GST antibody was included in the incubation mixtures in lanes 3 and 5. B, EMSA experiments were performed with Hes6WT (100 ng in lane 2 and 200 ng in lanes 3 and 4) or Hes6AQ (100 ng in lane 5 and 200 ng in lanes 6 and 7) purified from bacteria as fusion proteins with GST. For competition assays, reactions included labeled oligonucleotide in the presence of a 200-fold excess of unlabeled oligonucleotide (lanes 4 and 7).
We next determined whether there was a correlation between the negative effect of the AQ insertion on the ability of Hes6 to bind to the ESE box and its ability to promote neurogenesis and/or inhibit astrocyte differentiation. Both proneuronal (Fig. 4B,C, bars 1, 2, 4) and anti-gliogenic (Fig. 4D–F, bars 1, 2, 4) activities of Hes6AQ were essentially equivalent to those of Hes6WT. These results strongly suggest that bHLH motif-mediated interactions with ESE box sequences are not important for the neural functions of Hes6, either because Hes6 acts in a DNA binding-independent manner or because it does not bind with high affinity to ESE box sequences in vivo.
To examine these possibilities further, we tested whether Hes6 would repress transcription from ESE box-containing promoters in cortical progenitor cells. A previously described (Cossins et al., 2002) reporter construct harboring the luciferase gene under the control of a fragment of the collagenase promoter linked to two ESE boxes (“2xESE-collagenase promoter”) was used. This promoter exhibited a robust basal activity in cortical progenitor cells, which was designated as 100% (Fig. 6A, bar 1). Coexpression of Hes6 had no detectable effect on the activity of either the 2xESE-collagenase promoter (Fig. 6A, bar 2) or a control collagenase promoter lacking the tandem ESE boxes (Fig. 6A, bars 4, 5). This lack of repressive effect was observed over a range of increasing Hes6 concentrations (data not shown). Similar to Hes6, Hes1 also failed to repress transcription from the 2xESE-collagenase promoter (Fig. 6A, bar 3). In contrast, Hes1 repressed expression of a reporter gene under the control of the Ngn3 promoter, which contains tandem N boxes that act as high-affinity binding sites for Hes1 (Lee et al., 2001) (Fig. 6A, bars 6, 7). This result shows that Hes1 was competent to repress transcription under those experimental conditions. We found that Hes6 also failed to suppress (or activate) the 2xESE-collagenase promoter in HEK293 cells (Fig. 6B). Together, these results suggest that Hes6 does not mediate transcriptional repression from promoters containing ESE boxes in cortical progenitor cells, although it can bind to this sequence in vitro.
Figure 6.
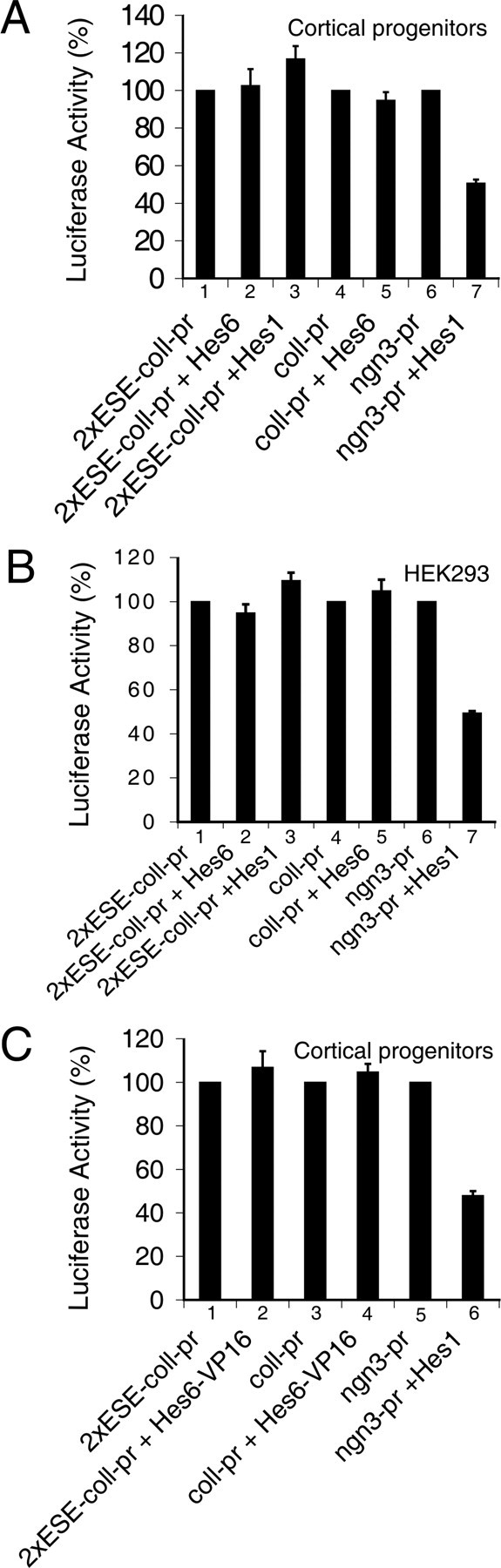
Analysis of the effect of Hes6 on transcription driven by an ESE box-containing promoter. Either cortical progenitor (A, C) or HEK293 (B) cells were transfected with the reporter constructs p2xESE-Col-Luc (bars 1–3 in A and B and bars 1 and 2 in C), pCol-Luc (bars 4 and 5 in A and B and bars 3 and 4 in C), or pNgn3-Luc (bars 6 and 7 in A and B and bars 5 and 6 in C). Reporter constructs (1.0 μg/transfection) were transfected alone or in combination with plasmids encoding the indicated proteins. In all panels, luciferase activity in the absence of effector plasmids was considered 100%; activities in the presence of effector plasmids are expressed as the mean ± the SD (n = 4).
A lack of repressive effect suggests either that Hes6 does not bind to ESE boxes in cortical progenitor cells or that transcriptional cofactors required by Hes6 may not be present, or may not form complexes with Hes6, in these cells. To address this question, a mutated protein was examined in which the C-terminal WRPW motif of Hes6 was replaced with the potent transcription activation domain of the herpes simplex virus protein 16. This chimeric protein was predicted to mediate transcriptional activation if bound to DNA. In agreement with this possibility, the same Hes6-VP16 fusion protein was shown to mediate transcriptional activation in muscle cells (Gao et al., 2001). We found that Hes6-VP16 failed to activate the 2xESE-collagenase promoter (Fig. 6C, bars 1, 2) although it was properly expressed in the transfected cells (data not shown). Together, these results suggest that the DNA-binding ability of Hes6 is cell type-dependent and that Hes6 does not bind to, and does not mediate transcriptional repression or activation from, ESE (or N) boxes in cortical progenitor cells. These observations suggest further that Hes6 regulates neuron and astrocyte differentiation through mechanisms that are not dependent on DNA binding through its own bHLH motif but require nuclear localization mediated by the basic arm.
The C-terminal WRPW motif is required for the anti-gliogenic, but not neurogenic, activity of Hes6
The lack of a correlation between the neural activities of Hes6 and DNA binding mediated by its own bHLH motif does not exclude the possibility that Hes6 might be recruited to DNA by either alternative DNA-binding sequences or interactions with other DNA-binding proteins. In either case, DNA-associated Hes6 is hypothesized to mediate transcriptional repression given its demonstrated ability to form complexes with Gro/TLE corepressors through its C-terminal WRPW tetrapeptide (Gratton et al., 2003). Previous studies have suggested that the WRPW motif is dispensable for the neurogenic function of Hes6 (Koyano-Nakagawa et al., 2000). In agreement with this notion, we found that a mutated form lacking the WRPW sequence (Hes6ΔWRPW) (Fig. 2E) displayed a proneuronal activity similar to Hes6WT (Fig. 4B,C, bars 1, 2, 6). These results strongly suggest that transcriptional repression involving Gro/TLE proteins is not important for the proneuronal activity of Hes6.
In contrast to neuronal differentiation, Hes6ΔWRPW failed to inhibit astrocyte differentiation (Fig. 4D–F, bars 1, 2, 6), suggesting a role for transcriptional repression in that event. This possibility was further supported by the finding that, like Hes6ΔWRPW, Hes6-VP16 did not suppress astrocyte differentiation (Fig. 4D–F, bars 1, 6, 7). These results suggest that distinct mechanisms underlie different neural functions of Hes6. More specifically, they suggest that WRPW motif-dependent mechanisms are important for Hes6-mediated inhibition of astrocyte differentiation but not promotion of neurogenesis. At least some of those mechanisms may involve transcriptional repression; if so, it is likely that Hes6 does not bind to DNA through its own bHLH domain and is instead recruited to DNA via alternative mechanisms, possibly through interactions with other DNA-binding proteins.
An LNHLL sequence within the helix3/helix4 domain of Hes6 is important for inhibition of astrocyte differentiation but not promotion of neurogenesis
In all Hes6 proteins, the helix3/helix4 domain contains a short sequence (LNHLL) that resembles LXXLL motifs found in a number of transcriptional regulators. In several cases, LXXLL motifs were shown to act as short protein–protein interaction sites (Savkur and Burris, 2004; Mahajan and Samuels, 2005). To examine the role of the LNHLL motif in Hes6 functions, we generated an LNHLL-motif-defective protein, Hes6LMD, in which this sequence was mutated to ANHAL (Fig. 2D). A similar type of mutation abolishes protein–protein interactions mediated by LXXLL motifs of other proteins (Litterst and Pfitzner, 2002). Hes6LMD was localized to the nucleus, formed homodimers, and interacted with Gro/TLE corepressors similar to Hes6WT, suggesting that these functions are not perturbed by this mutation (Figs. 2G, 3J–L, 3M–P) (data not shown). When expressed in cortical progenitor cells, Hes6LMD displayed a proneuronal activity essentially equivalent to Hes6WT (Fig. 4B,C, bars 1, 2, 5). In contrast, Hes6LMD failed to inhibit astrocyte differentiation (Fig. 4D–F, bars 1, 2, 5). These results suggest that the LNHLL motif contributes specifically to the anti-gliogenic, but not proneuronal, activity of Hes6, possibly by mediating specific protein–protein interactions.
Correlation between ability to suppress Hes1-mediated transcriptional repression and Hes6 anti-gliogenic, but not proneuronal, activity
Hes6 can suppress the ability of Hes1 to mediate transcriptional repression (Bae et al., 2000; Gratton et al., 2003). This finding suggests that Hes6 may promote neurogenesis and/or inhibit astrocyte differentiation, at least in part, by antagonizing the transcription repression function of Hes1 and/or other related Hes family members. Transient transfection assays in HEK293 cells showed that Hes6WT completely suppressed Hes1-mediated repression of the Ngn3 promoter (Fig. 7A, bars 1–4). Hes6WT caused increased promoter activity even in the absence of transfected Hes1 (Fig. 7A, bars 1, 7, 8). This effect was not observed when a mutated form of the promoter lacking Hes1 binding sites was used (Lee et al., 2001) (Fig. 7B, bars 1–3, 6–8). Hes1 had no repressive effect on the activity of this mutated promoter (Fig. 7B, bars 1, 6). Because Hes1 is endogenously expressed in HEK293 cells (Fig. 7D, lane 2), it is likely that exogenous Hes6WT caused promoter activation in the absence of transfected Hes1 by inhibiting the activity of endogenous Hes1. In contrast to Hes6WT, Hes6LMD had no significant effect on Hes1-mediated repression when expressed at levels equivalent to the former (Fig. 7A, bars 1–6, 9, 10; C). Similarly, both Hes6BAD and Hes6-VP16 failed to de-repress the Ngn3 promoter in the presence of Hes1 (Fig. 7E, bars 1, 5–8). These results show that the ability of Hes6 to translocate to nuclei is important for its inhibitory effect on Hes1. It is likely that Hes6 must be localized to the nucleus to participate in protein–protein interactions that affect Hes1 transcription repression functions (see Fig. 8 for possible mechanisms of Hes6 activity). In addition, Hes6 requires intact LNHLL and WRPW motifs to antagonize Hes1 activity. Because both Hes6LMD and Hes6-VP16 fail to inhibit astrocyte differentiation, our results suggest a correlation between the ability of Hes6 to antagonize transcriptional repression by Hes1 (and/or other functionally related family members) and its anti-gliogenic activity. In contrast, because Hes6LMD promotes neurogenesis like Hes6WT (Fig. 4), inhibition of Hes1-mediated transcriptional repression does not appear to be important for the neurogenic function of Hes6.
Figure 7.
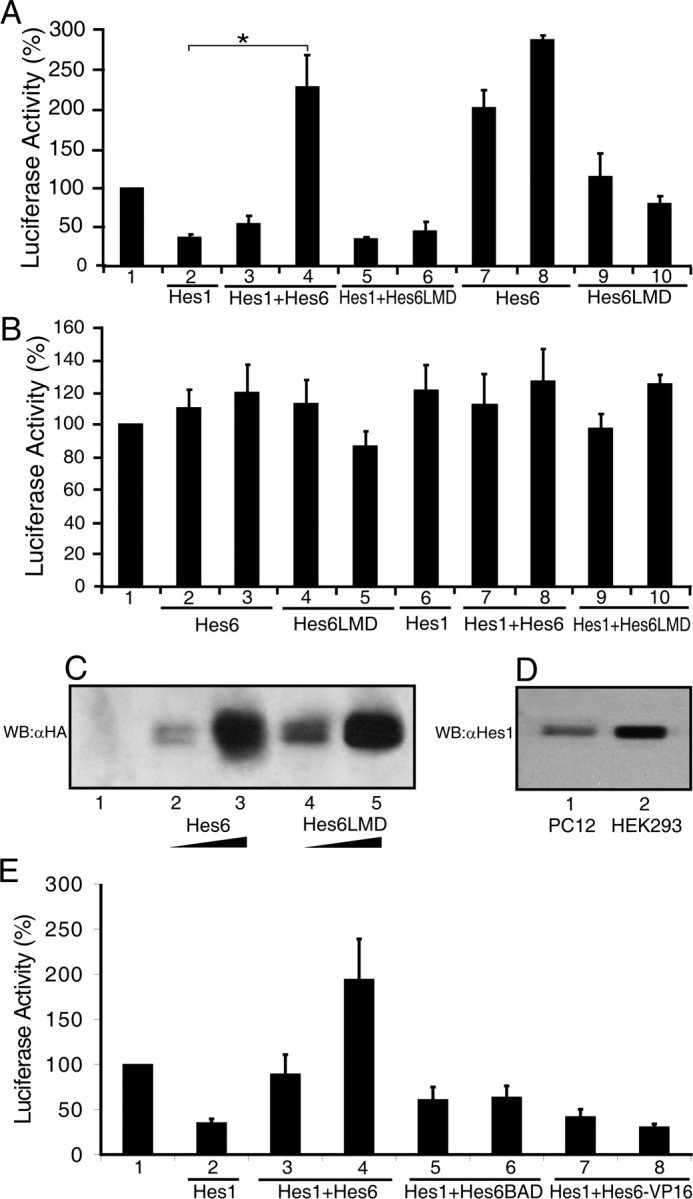
Effect of wild-type and mutated forms of Hes6 on Hes1-mediated transcriptional repression. A, B, E, Transcription assays. HEK293 cells were transfected with luciferase reporter constructs (1.0 μg/transfection) harboring either a 3.0 kb fragment of the Ngn3 promoter containing Hes1 binding sites (A, E) or a truncated version of the Ngn3 promoter lacking Hes1 binding sites (B). Reporter constructs were transfected alone (bar 1) (in which case luciferase activity was considered 100%) or in combination with the following expression plasmids: Hes1, 20 ng/transfection in each case; Hes6WT, 200 ng/transfection (bars 3 and 7 in A; bars 2 and 7 in B; bar 3 in E) and 600 ng/transfection (bars 4 and 8 in A; bars 3 and 8 in B; bar 4 in E); HES6LMD 200 ng/transfection (bars 5 and 9 in A; bars 4 and 9 in B) and 600 ng/transfection (bars 6 and 10 in A; bars 5 and 10 in B); Hes6BAD, 200 ng/transfection (bar 5 in E) and 600 ng/transfection (bar 6 in E); or Hes6-VP16, 300 ng/transfection (bar 7 in E) and 900 ng/transfection (bar 8 in E). Luciferase activities are expressed as the mean ± the SD (n ≥ 4; *p < 0.01). C, Western blot analysis of the expression of either Hes6WT or Hes6LMD transfected into HEK293 cells at the same amounts used in A. D, Expression of endogenous Hes1 in PC12 (lane 1) or HEK293 (lane 2) cells.
Figure 8.
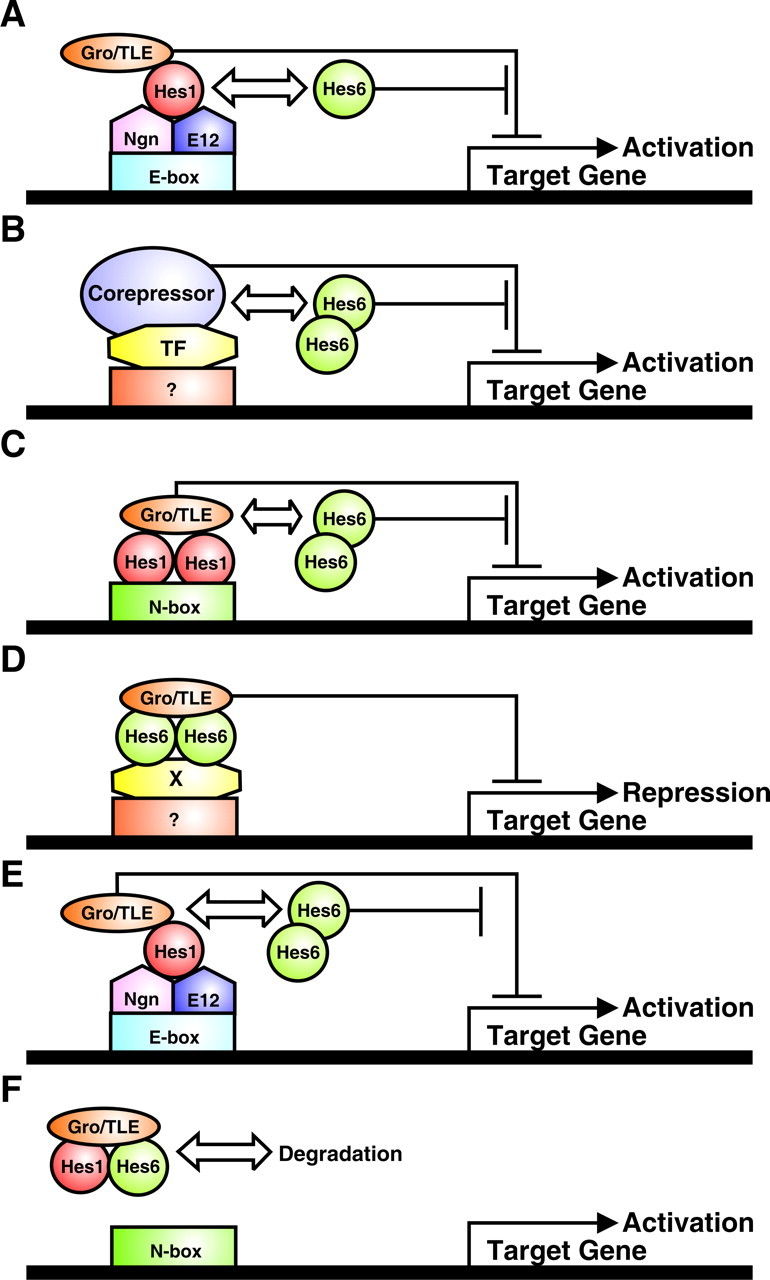
Possible mechanisms of Hes6 activity. A, Hes6 forms heterodimers with Hes1 in committed neuronal precursors and prevents Hes1 from recruiting transcriptional corepressors like Gro/TLE to DNA-bound Ngn:E12 heterodimers, thereby relieving repression and promoting the transactivating functions of the latter. Target genes encode factors that induce neuronal differentiation. This effect is WRPW motif independent. B, Hes6 homodimers titrate away transcriptional corepressors other than Gro/TLE from a yet to be characterized DNA-bound transcription factor(s) (TF) that inhibits neurogenesis, causing increased neuronal differentiation. This de-repression is WRPW motif independent. Target genes may encode factors that induce neuronal differentiation and/or factors that inhibit astrocyte differentiation. C, Hes6 homodimers titrate away Gro/TLE corepressors from DNA-bound Hes1 homodimers. This effect depends on the WRPW motif of Hes6 and prevents transcriptional repression of target genes that inhibit astrocyte differentiation, possibly including Ngn1. D, Hes6 homodimers form complexes with Gro/TLE, and these complexes are recruited to DNA by a yet to be identified transcription factor(s) (X). Targeting of Hes6:Gro/TLE complexes to DNA results in the transcriptional repression of genes that promote astrocyte differentiation. This mechanism requires the WRPW motif; the LNHLL sequence of Hes6 may be involved in binding to transcription factor X. E, Hes6 homodimers titrate away Gro/TLE corepressors from Hes1:Ngn:E12 complexes, promoting activation of Ngn1 target genes (possibly including autoactivation of Ngn1 itself). This mechanism depends on Hes6 WRPW motif. F, Hes6 forms heterodimers with Hes1 and these dimers bind to Gro/TLE in a WRPW motif-dependent manner, resulting in the formation of Hes1:Hes6:Gro/TLE complexes that are targeted for degradation. This effect causes the de-repression of Hes1 target genes and the suppression of other functions of Hes1 that rely on protein–protein interactions.
Discussion
In both mouse and Xenopus, Hes6 is preferentially expressed between the proliferative and postmitotic zones. The domain of Hes6 expression resembles that of the neuronal precursor gene, NeuroD (Bae et al., 2000; Koyano-Nakagawa et al., 2000). Hes6 expression is not activated by the Notch signaling pathway, as is the case for repressor-type Hes genes (i.e., Hes1 and Hes5), but instead is positively regulated by Ngn1 (Koyano-Nakagawa et al., 2000). These observations suggest that Hes6 is activated in committed neuronal precursors in which proneural genes have escaped repression by Notch-induced Hes proteins and are competent to drive neuronal differentiation. In this study, we provided evidence suggesting that the expression of Hes6 in committed neuroblasts has two important consequences: Hes6 promotes neuronal differentiation and suppresses astrocytogenesis. We also show that Hes6 nuclear localization ability is important for both of these functions. In contrast, an internal LNHLL motif and a C-terminal WRPW tetrapeptide are required for inhibition of astrocyte differentiation but not for induction of neurogenesis. Mutation of the LNHLL or WRPW sequences also impairs the ability of Hes6 to suppress Hes1-mediated transcriptional repression. These findings suggest that distinct mechanisms underlie Hes6 ability to promote or inhibit the differentiation of neurons or astrocytes, respectively.
Promotion of neurogenesis by Hes6 is correlated with neither Gro/TLE recruitment nor inhibition of Hes1-mediated transcriptional repression
We have shown that the proneuronal activity of Hes6 requires nuclear localization. Because Hes6 interacts with transcriptional corepressors of the Gro/TLE family through its WRPW tetrapeptide (Gratton et al., 2003), a requirement for nuclear localization may suggest a role for DNA binding and Gro/TLE-mediated transcriptional repression. In possible agreement with this hypothesis, Hes6 represses transcription in a WRPW motif-dependent manner when targeted to DNA as a fusion protein with a heterologous DNA-binding domain (Gao et al., 2001). Moreover, Hes6 was reported to repress transcription in transfected muscle cells, although it was not determined whether that ability was dependent on an intact WRPW motif (Cossins et al., 2002). However, several previous and present observations argue against the possibility that mechanisms involving direct DNA-binding and transcriptional repression are important for the proneuronal function of Hes6. Mutation of the basic arm of the bHLH domain of Xenopus Hes6 (presumably in a way that did not decrease its nuclear localization) did not impair Hes6 neurogenic ability (Koyano-Nakagawa et al., 2000). In agreement with that observation, we found that the insertion of alanine and glutamine residues between the basic arm and helix 1 of the bHLH domain, a mutation that disrupts binding of Hes6 to the ESE box sequence in vitro, does not perturb the neurogenic effect of Hes6. Moreover, although Hes6 binds to the ESE box in vitro, it does not mediate transcriptional repression from an ESE box-containing promoter in cortical progenitor cells. This situation may reflect the fact that either Hes6 does not bind to ESE sequences in vivo or high-affinity binding is modulated by cell type-specific cofactors that are not present in cortical progenitors.
The notion that Hes6 promotes neurogenesis in a DNA binding-independent manner is also consistent with investigations showing that removal of the WRPW motif has no effect on Hes6 proneuronal activity in Xenopus neural plate (Koyano-Nakagawa et al., 2000) and mouse cortical progenitors (our study). These results argue against a model in which Hes6 promotes neurogenesis by binding directly to ESE box sequences (or other related sequences yet to be characterized) and recruiting Gro/TLE proteins to repress transcription. Because a fusion protein of Hes6 and the DNA binding domain of GAL4 does not mediate transcriptional activation (Gao et al., 2001), it also appears unlikely that Hes6 induces neuronal differentiation by transactivating gene expression. Thus, these combined observations suggest that Hes6 regulates neuronal differentiation via mechanisms that rely on protein–protein interactions rather than Hes6 intrinsic DNA-binding ability via its bHLH domain. It is worth noting that other regulators of neuronal development, most notably the members of the Id protein family, were shown to require nuclear localization and protein–protein interactions, but not DNA binding, for their biological activity (Ross et al., 2003).
It is possible that Hes6 acts by binding to, and inhibiting, other bHLH factors that exert a negative influence on the transactivating functions of the proneural proteins. A likely target is Hes1, which is known to both interact with Hes6 (Bae et al., 2000) and reduce the transcriptional activity of proneural proteins via protein–protein interactions (Sasai et al., 1992; Bae et al., 2000). Hes6 may act by dimerizing with Hes1 and titrating the latter away from Ngn:E protein complexes (Fig. 8A). This effect would antagonize the transcription repression activity of Hes1:Gro/TLE complexes bound to Ngn:E protein dimers (Giagtzoglou et al., 2003), thereby increasing the transactivating functions of Ngns (and/or other proneural proteins) and resulting in a promotion of neuronal differentiation. Another possibility is that Hes6 may induce neurogenesis by forming homodimers that have no intrinsic DNA binding ability in cortical progenitors but are able to associate with specific cofactors that are required by other anti-neurogenic DNA binding proteins. In doing so, Hes6 may sequester those cofactors away from DNA, thus limiting their availability and influencing the transcriptional functions of other proteins (Fig. 8B).
The present results also show that a number of mutations that block the inhibitory effect of Hes6 on Hes1-mediated transcriptional repression have no consequence on Hes6 neurogenic function, although they impair Hes6 anti-gliogenic ability. This situation is consistent with the temporal profile of Hes1 and Hes6 expression during neurogenesis. Hes1 is expressed in uncommitted progenitors and generally precedes the expression of Hes6 (Koyano-Nakagawa et al., 2000). Together, these observations strongly suggest that Hes6 promotes neuronal differentiation by increasing the biochemical activity of proneural proteins and not by suppressing the repression of proneural gene expression by Notch-activated Hes proteins.
Hes6 inhibits astrocyte differentiation, and this function is correlated with the ability to interact with Gro/TLE and to suppress Hes1-mediated transcriptional repression
Using a combination of RNAi and gain-of-function studies, we obtained the first evidence suggesting that Hes6 suppresses the differentiation of cortical astrocytes, in addition to promoting neurogenesis. In contrast to the latter function, the anti-gliogenic effect of Hes6 involves the WRPW and LNHLL motifs. Mutations of these sequences also impair the ability of Hes6 to antagonize Hes1-mediated transcriptional repression.
A requirement for the WRPW tetrapeptide was observed previously during the study of Hes6 activity in the developing Xenopus myotome (Cossins et al., 2002). Exogenous expression of both wild-type Hes6 and Hes6 containing a mutation in the basic arm resulted in a significant expansion of the myotome. In contrast, Hes6ΔWRPW did not lead to an expansion of the myotome, although it was competent to promote neuronal differentiation. These results suggest that transcription repression mechanisms involving the recruitment of Gro/TLE corepressors may be involved in only certain functions of Hes6 in a context-dependent manner. During the neurogenic phase of cortical development, Hes6 may be recruited to specific regulatory sequences from which it mediates the repression of genes that promote astrocyte differentiation. In this model, DNA targeting would likely not be mediated by the Hes6 bHLH domain, because the AQ insertion that blocks binding to the ESE box does not impair the anti-gliogenic activity of Hes6. Instead, recruitment of Hes6 to DNA might involve interaction with another DNA-binding protein(s) (Fig. 8D). This interaction could be mediated by the LNHLL sequence of Hes6 that resembles the LXXLL motif that mediates protein–protein interactions in several other proteins. The LNHLL motif is required for inhibition of astrocyte differentiation but not for promotion of neurogenesis, suggesting a specific involvement in some but not all of Hes6 activities.
The importance of the WRPW motif for the anti-gliogenic function of Hes6 may underscore additional mechanisms. It is possible that Hes6 competes with repressor-type Hes proteins for limiting amounts of Gro/TLE. By antagonizing the formation of complexes of Gro/TLE and Hes1 (or Hes5), Hes6 may reduce the ability of the latter to repress the expression (Fig. 8C) and/or transcriptional activity (Fig. 8E) of proneural proteins like Ngn1. Although the latter effect may not be directly relevant to the Ngn1 anti-gliogenic activity, which was suggested to be DNA-binding independent (Sun et al., 2001), it may influence other transcriptional functions of Ngn1 that may have a role in regulating the neuronal versus glial cell fate choice.
A third possibility is suggested by the demonstration that deletion of the WRPW tetrapetide renders Hes6 more resistant to proteasome-mediated degradation (Kang et al., 2005). In agreement with this observation, Hes6:Hes1 heterodimers become gradually more stable if either one or both of the WRPW motifs in the heterodimers are deleted (Gratton et al., 2003). The requirement for the WRPW motif of Hes6 during astrocyte differentiation may thus underscore a role for Hes6 in decreasing the expression levels of Hes1 (Fig. 8F), resulting in a general inhibition of all Hes1 functions, including those mediated by DNA binding or protein–protein interactions.
Hes6 may negatively regulate the onset of astrocyte differentiation by preventing the premature inhibition of proneural gene expression by repressor-type Hes proteins
Previous work has demonstrated that Ngn1 suppresses astrocyte differentiation, in addition to inducing neurogenesis (Sun et al., 2001). Inactivation of both Mash1 and Ngn2 in vivo is correlated with increased generation of astrocytes (Nieto et al., 2001). Conversely, Notch-activated Hes proteins promote astrocyte differentiation in hippocampal, retinal, and spinal cord progenitor cells (Ross et al., 2003). Bone morphogenetic protein treatment of cultured cortical progenitor cells induces astrocyte differentiation and causes an induction of Hes5 expression and downregulation of Ngn genes (Nakashima et al., 2001). Moreover, Hes1 facilitates STAT3 activation (Kamakura et al., 2004), which in turn promotes astrocyte differentiation from pluripotent cortical progenitor cells (Sun et al., 2001; Kamakura et al., 2004). These results suggest that repressor-type Hes proteins (which are downregulated during the progenitor to neuron transition) are reactivated during astrocyte differentiation. They suggest further that Hes1/Hes5 upregulation may signal the end of the neurogenic period and the start of glial cell differentiation by antagonizing the anti-gliogenic activity of Ngn proteins through a repression of Ngn gene transcription. We propose that Hes6 delays astrocyte differentiation, at least in part, by preventing a premature suppression of Ngn gene expression by the Notch-activated Hes proteins. Because neurogenesis dominates over glial differentiation, even in the presence of gliogenic stimuli, as long as Ngn expression is maintained (Park et al., 1999; Sun et al., 2001), Hes6 may represent an important regulator of the correct timing of the transition from neurogenesis to astrocyte differentiation.
Footnotes
This work was supported by Canadian Institutes of Health Research Grants MOP-13957 and MGC-14971 to S.S., who is a Chercheur National of the Fonds de la Recherche en Sante du Quebec. We thank Dr. Phil Jones (Medical Research Council Cancer Cell Unit, Cambridge, UK) for providing the pCol-Luc and p2xESE-Col-Luc plasmids and Manuel Buscarlet for invaluable assistance during the preparation of this manuscript.
References
- Bae SK, Bessho Y, Hojo M, Kageyama R. The bHLH gene Hes6, an inhibitor of Hes1, promotes neuronal differentiation. Development. 2000;127:2933–2943. doi: 10.1242/dev.127.13.2933. [DOI] [PubMed] [Google Scholar]
- Barnabe-Heider F, Wasylnka JA, Fernandes KJ, Porsche C, Sendtner M, Kaplan DR, Miller FD. Evidence that embryonic neurons regulate the onset of cortical gliogenesis via cardiotrophin-1. Neuron. 2005;48:253–265. doi: 10.1016/j.neuron.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Chen HA, Thiagalingam A, Chopra H, Borges MW, Feder JN, Nelkin BD, Baylin SB, Ball DW. Conservation of the Drosophila lateral inhibition pathway in human lung cancer: a hairy-related protein (HES-1) directly represses achaete-scute homolog-1 expression. Proc Natl Acad Sci USA. 1997;94:5355–5360. doi: 10.1073/pnas.94.10.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossins J, Vernon AE, Zhang Y, Philpott A, Jones PH. Hes6 regulates myogenic differentiation. Development. 2002;129:2195–2207. doi: 10.1242/dev.129.9.2195. [DOI] [PubMed] [Google Scholar]
- Fishell G, Kriegstein AR. Neurons from radial glia: the consequences of asymmetric inheritance. Curr Opin Neurobiol. 2003;13:34–41. doi: 10.1016/s0959-4388(03)00013-8. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Mukherjee S, Bao ZZ, Morrow EM, Cepko CL. rax, Hes1, and Notch1 promote the formation of Mueller glia by postnatal retinal progenitor cells. Neuron. 2000;26:383–394. doi: 10.1016/s0896-6273(00)81171-x. [DOI] [PubMed] [Google Scholar]
- Gaiano N, Fishell G. The role of Notch in promoting glial and neural stem cell fates. Annu Rev Neurosci. 2002;25:471–490. doi: 10.1146/annurev.neuro.25.030702.130823. [DOI] [PubMed] [Google Scholar]
- Gao X, Chandra T, Gratton MO, Quelo I, Prud'homme J, Stifani S, St-Arnaud R. Hes6 acts as a transcriptional repressor in myoblasts and can induce the myogenic differentiation program. J Cell Biol. 2001;154:1161–1171. doi: 10.1083/jcb.200104058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Greenberg ME. Distinct roles for bFGF and NT-3 in the regulation of cortical neurogenesis. Neuron. 1995;15:89–103. doi: 10.1016/0896-6273(95)90067-5. [DOI] [PubMed] [Google Scholar]
- Giagtzoglou N, Alifragis P, Koumbanakis KA, Delidakis C. Two modes of recruitment of E(spl) repressors onto target genes. Development. 2003;130:259–270. doi: 10.1242/dev.00206. [DOI] [PubMed] [Google Scholar]
- Gratton ME, Torban E, Belanger-Jasmin S, Theriault FM, German MS, Stifani S. Hes6 promotes cortical neurogenesis and inhibits Hes1 transcription repression activity by multiple mechanisms. Mol Cell Biol. 2003;23:6922–6935. doi: 10.1128/MCB.23.19.6922-6935.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi M, Moriyoshi K, Sasai Y, Shiota K, Nakanishi S, Kageyama R. Persistent expression of helix-loop-helix factor HES-1 prevents mammalian neural differentiation in the central nervous system. EMBO J. 1994;13:1799–1805. doi: 10.1002/j.1460-2075.1994.tb06448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi M, Ang SL, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Targeted disruption of mammalian hairy and Enhancer of split homolog 1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- Jennings BH, Tyler DM, Bray SJ. Target specificities of Drosophila enhancer of split basic helix-loop-helix proteins. Mol Cell Biol. 1999;19:4600–4610. doi: 10.1128/mcb.19.7.4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job C, Tan SS. Constructing the mammalian cortex: the role of intrinsic factors. Dev Biol. 2003;257:221–232. doi: 10.1016/s0012-1606(03)00070-8. [DOI] [PubMed] [Google Scholar]
- Ju BG, Solum D, Song EJ, Lee KJ, Rose DW, Glass CK, Rosenfeld MG. Activating the PARP-1 sensor component of the Groucho/TLE1 corepressor complex mediates a CaMKinase IIδ-dependent neurogenic gene activation pathway. Cell. 2004;119:815–829. doi: 10.1016/j.cell.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Hatakeyama J, Ohsawa R. Roles of bHLH genes in neural stem cell differentiation. Exp Cell Res. 2005;306:343–348. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Kamakura S, Oishi K, Yoshimatsu T, Nakafuku M, Masuyama N, Gotoh Y. Hes binding to STAT3 mediates crosstalk between Notch and JAK-STAT signalling. Nat Cell Biol. 2004;6:547–554. doi: 10.1038/ncb1138. [DOI] [PubMed] [Google Scholar]
- Kang SA, Seol JH, Kim J. The conserved WRPW motif of Hes6 mediates proteasomal degradation. Biochem Biophys Res Comm. 2005;332:33–36. doi: 10.1016/j.bbrc.2005.04.089. [DOI] [PubMed] [Google Scholar]
- Koyano-Nakagawa N, Kim J, Anderson D, Kintner C. Hes6 acts in a positive feedback loop with the neurogenins to promote neuronal differentiation. Development. 2000;127:4203–4216. doi: 10.1242/dev.127.19.4203. [DOI] [PubMed] [Google Scholar]
- Lee JC, Smith SB, Watada H, Lin J, Scheel D, Wang J, Mirmira RG, German MS. Regulation of the pancreatic pro-endocrine gene Neurogenin3. Diabetes. 2001;50:928–936. doi: 10.2337/diabetes.50.5.928. [DOI] [PubMed] [Google Scholar]
- Lee SK, Pfaff SL. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron. 2003;38:731–745. doi: 10.1016/s0896-6273(03)00296-4. [DOI] [PubMed] [Google Scholar]
- Litterst CM, Pfitzner E. An LXXLL motif in the transactivation domain of STAT6 mediates recruitment of NcoA-1/SRC-1. J Biol Chem. 2002;277:36052–36060. doi: 10.1074/jbc.M203556200. [DOI] [PubMed] [Google Scholar]
- Mahajan MA, Samuels HH. Nuclear hormone receptor coregulator: role in hormone action, metabolism, growth, and development. Endocr Rev. 2005;26:583–597. doi: 10.1210/er.2004-0012. [DOI] [PubMed] [Google Scholar]
- Malatesta P, Hack MA, Hartfuss E, Kettenmann H, Klinkert W, Kirchhoff F, Gotz M. Neuronal or glial progeny:regional differences in radial glia fate. Neuron. 2003;37:751–764. doi: 10.1016/s0896-6273(03)00116-8. [DOI] [PubMed] [Google Scholar]
- Marcal N, Patel H, Dong Z, Belanger-Jasmin S, Hoffman B, Helgason CD, Dang J, Stifani S. Antagonistic effects of Grg6 and Groucho/TLE on the transcription repression activity of brain factor 1/FoxG1 and cortical neuron differentiation. Mol Cell Biol. 2005;25:10916–10929. doi: 10.1128/MCB.25.24.10916-10929.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLarren KW, Lo R, Grbavec D, Thirunavukkarasu K, Karsenty G, Stifani S. The mammalian basic helix loop helix protein HES-1 binds to and modulates the transactivating function of the Runt-related factor Cbfa1. J Biol Chem. 2000;275:530–538. doi: 10.1074/jbc.275.1.530. [DOI] [PubMed] [Google Scholar]
- McLarren KW, Theriault FM, Stifani S. Association with the nuclear matrix and interaction with Groucho and RUNX proteins regulate the transcription repression activity of the basic helix loop helix factor Hes1. J Biol Chem. 2001;276:1578–1584. doi: 10.1074/jbc.M007629200. [DOI] [PubMed] [Google Scholar]
- Morrow T, Song MR, Ghosh A. Sequential specification of neurons and glia by developmentally regulated extracellular factors. Development. 2001;128:3585–3594. doi: 10.1242/dev.128.18.3585. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Takizawa T, Occhiai W, Yanagisawa M, Hisatune T, Nakafuku M, Miyazono K, Kishimoto T, Kageyama R, Taga T. BMP2-mediated alteration in the developmental pathway of fetal mouse brain cells from neurogenesis to astrocytogenesis. Proc Natl Acad Sci USA. 2001;98:5868–5873. doi: 10.1073/pnas.101109698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M, Schuurmans C, Britz O, Guillemot F. Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron. 2001;29:401–413. doi: 10.1016/s0896-6273(01)00214-8. [DOI] [PubMed] [Google Scholar]
- Nuthall HN, Husain J, McLarren KW, Stifani S. Role for Hes1-induced phosphorylation in Groucho-mediated transcriptional repression. Mol Cell Biol. 2002;22:389–399. doi: 10.1128/MCB.22.2.389-399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuthall HN, Joachim K, Stifani S. Phosphorylation of serine 239 of Groucho/TLE1 by protein kinase CK2 is important for inhibition of neuronal differentiation. Mol Cell Biol. 2004;24:8395–8407. doi: 10.1128/MCB.24.19.8395-8407.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JK, Williams BP, Alberta JA, Stiles CD. Bipotent cortical progenitor cells process conflicting cues for neurons and glia in a hierarchical manner. J Neurosci. 1999;19:10383–10389. doi: 10.1523/JNEUROSCI.19-23-10383.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pissarra L, Henrique D, Duarte A. Expression of hes6, a new member of the Hairy/Enhancer of split family, in mouse development. Mech Dev. 2000;95:275–278. doi: 10.1016/s0925-4773(00)00348-8. [DOI] [PubMed] [Google Scholar]
- Ross SE, Greenberg ME, Stiles CD. Basic helix-loop-helix factors in cortical development. Neuron. 2003;39:13–25. doi: 10.1016/s0896-6273(03)00365-9. [DOI] [PubMed] [Google Scholar]
- Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- Savkur RS, Burris TP. The coactivator LXXLL nuclear receptor recognition motif. J Pept Res. 2004;63:207–212. doi: 10.1111/j.1399-3011.2004.00126.x. [DOI] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, Ivanova NB, Stifani S, Morrisey EE, Temple S. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- Sun Y, Nadal-Vicens M, Misono S, Lin MZ, Zubiaga A, Hua X, Fan G, Greenberg ME. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104:365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- Theriault FM, Nuthall HN, Dong Z, Lo R, Barnabe-Heider F, Miller FD, Stifani S. Role for Runx1 in the proliferation and neuronal differentiation of selected progenitor cells in the mammalian nervous system. J Neurosci. 2005;25:2050–2061. doi: 10.1523/JNEUROSCI.5108-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiliauskas D, Stern C. Expression of mouse HES-6, a new member of the Hairy/Enhancer of split family of bHLH transcription factors. Mech Dev. 2000;98:133–137. doi: 10.1016/s0925-4773(00)00443-3. [DOI] [PubMed] [Google Scholar]
- Wu Y, Liu Y, Levine EM, Rao MS. Hes1 but not Hes5 regulates an astrocyte versus oligodendrocyte fate choice in glial restricted precursors. Dev Dyn. 2003;226:675–689. doi: 10.1002/dvdy.10278. [DOI] [PubMed] [Google Scholar]
- Yao J, Lai E, Stifani S. The winged helix protein brain factor 1 interacts with Groucho and Hes proteins to repress transcription. Mol Cell Biol. 2001;21:1962–1972. doi: 10.1128/MCB.21.6.1962-1972.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]