Abstract
We have identified a novel family of synaptic adhesion-like molecules (SALMs). The family members, SALM1–SALM4, have a single transmembrane (TM) domain and contain extracellular leucine-rich repeats, an Ig C2 type domain, a fibronectin type III domain, and an intracellular postsynaptic density-95 (PSD-95)/Discs large/zona occludens-1 (PDZ) binding domain, which is present on all members except SALM4. SALM1 interacts with PSD-95, synapse-associated protein 102 (SAP102), and SAP97 based on coimmunoprecipitation of detergent-solubilized brain. Distribution studies show that SALM1 is present in synaptic membrane and postsynaptic density fractions but is also distributed in axons and dendrites. Transfection of hippocampal neurons for 4 d in vitro (DIV) with SALM1 more than doubles the dendritic lengths of neurons after 48 h, whereas transfection of neurons 14 DIV has no significant effect on neurite outgrowth. Overexpression of SALM1 in 14 DIV neurons recruits NMDA receptors (NR) and PSD-95 to dendritic puncta. This effect is dependent on the PDZ-binding domain of SALM1. SALM1 also enhances surface expression of transfected NR2A subunit. Immunoprecipitation of detergent-solubilized brain membranes with anti-SALM1 antibodies shows coimmunoprecipitation of NR1 and NR2 subunits. After transfection of heterologous cells with NR1 and NR2 cDNAs, through coimmunoprecipitation analyses, we find that SALM1 also interacts with the NMDA receptor NR1 subunit through its extracellular or TM1 domains.
Keywords: adhesion, PDZ domains, NMDA receptors, synaptic plasticity, glutamate receptor, synapse
Introduction
The formation of functional synapses is a multistep process that includes the recruitment of adhesion molecules, the formation of presynaptic release sites, the organization of postsynaptic scaffolding proteins, and the incorporation of ion channels into postsynaptic membranes. Several adhesion molecules are early residents of the synapse and play roles in both the formation and maintenance of the synapse (Scheiffele, 2003; Yamagata et al., 2003). Although most are also present at nonsynaptic sites and may be expressed by non-neuronal cells as well as by neurons, some adhesion molecules, such as synaptic cell adhesion molecule (SynCAM), are highly concentrated at the synapse (Biederer et al., 2002). Many adhesion molecules that function in synapse development are also expressed in the adult and are likely to continue to play a role in the plasticity and organization of the mature synapse.
A relatively late step in synapse formation is the recruitment of receptors and ion channels to the postsynaptic membrane. Many of these proteins interact either directly or indirectly with postsynaptic density-95 (PSD-95)/Discs large/zona occludens-1 (PDZ) proteins, thus linking them to the postsynaptic scaffold and providing a mechanism for both retaining the protein at the synapse and keeping its proximity to signaling molecules known to associate with PDZ proteins (Nourry et al., 2003; Kim and Sheng, 2004; Montgomery et al., 2004). In the development of excitatory synapses, NMDA receptors (NR) are relatively early residents and generally precede the recruitment of AMPA receptors (Malenka and Nicoll, 1997; Malinow and Malenka, 2002). It is thought that NMDA receptors are retained at the synapse through a direct interaction between the C termini of their NR2 subunits and PDZ proteins of the PSD-95 family, although additional interactions may also secure receptors at the synapse. In addition to their roles in excitatory neurotransmission, activation of NMDA receptors has been shown to influence neurite outgrowth and synapse development; calcium influx through NMDA receptors indirectly regulates cytoskeletal dynamics in dendrites (Luo, 2002). Adhesion molecules, such as SynCAM and neuroligin, are also anchored to PDZ proteins at the synapse, and, because of the multiple domains found on many PDZ proteins, these PDZ interactions may indirectly link adhesion molecules to ion channels at the synapse (Yamagata et al., 2003). Neuroligin, for example, binds to the third PDZ domain of PSD-95, whereas NR2 subunits bind to PDZ domains one and two and forms a link with the presynaptic terminal through β-neurexin (Nourry et al., 2003; Graf et al., 2004; Kim and Sheng, 2004; Prange et al., 2004; Chih et al., 2005). Thus, PDZ proteins play a central role in organizing functionally diverse membrane proteins at the synapse. We focused on identifying adhesion molecules that may be indirectly linked to NMDA and AMPA receptors through PDZ proteins of the PSD-95 family. We identified a novel class of synaptic adhesion-like molecules (SALMs) that interacts with PSD-95 family members and is present at the synapse. SALM1 overexpression in cultured hippocampal neurons clusters NMDA receptors and PSD-95 in dendrites and promotes neurite outgrowth. SALMs also interact with the NMDA receptor subunit NR1 through their extracellular domains.
Materials and Methods
Yeast two-hybrid screening.
The yeast two-hybrid screening was performed using Matchmaker Two-Hybrid System 3 (BD Biosciences Clontech, Palo Alto, CA). The cDNA corresponding to amino acids 1–543 of synapse-associated protein 97 (SAP97) was subcloned into pGBKT7 bait vector (BD Biosciences Clontech) and transformed into yeast strain AH109. The yeast AH109 containing the SAP97 PDZ domain was mated to the pretransformed yeast strain Y187 containing adult mouse brain cDNA library (BD Biosciences Clontech). The screening procedures followed the instructions of the manufacturer with modifications. The mating yeast were grown on agar plates containing complete minimal dropout base with glucose minus adenine, leucine, histidine, tryptophan, and α-5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (α-X-gal) (Teknova, Hollister, CA). The α-X-gal-positive colonies were regrown on quadruple dropout agar plates. The positive clones were extracted from yeast and transformed into DH5α, and plasmids were isolated, sequenced, and analyzed. The sequences of the selected clones were analyzed using National Center for Biotechnology Information bioinformatics software, BLAST, SMART, CLUSTAL, and PAUP.
Cloning of SALM1–SALM4 and cDNA constructs.
SALM1, SALM2, and SALM4 were obtained by PCR cloning from a mouse brain cDNA library (BD Biosciences Clontech). SALM3 was obtained from the Integrated Molecular Analysis of Genomes and their Expression (IMAGE) Consortium (clone MGC:36545; IMAGE:4951386). The primers for cloning SALM1 are 5′-TTG AAT TCA TGG AGA CTC TGC TTG GTG GGC-3′ for the forward primer and 5′-CGC TCG AGC TAC ACA GTA CTT TCC ATT ACC-3′ for the reverse primer. The primers for SALM2 are 5′-TTA CCC GGA AGC TTC CCT GCA CCA TGG CTC CA-3′ for the forward primer and 5′-TTA CCC GGG AAT TCC GCC CGT CGC TCA CAC GG-3′ for the reverse primer. The primers for SALM4 are 5-CCC GCG ATG GCC GTC CTT CCA CTA-3′ for the forward primer and 5′-CTA TGG TTC CTC TGG TCC CAG GG-3′ for the reverse primer. The PCR products were subcloned into the Topo vectors (Invitrogen, Carlsbad, CA) and then into the mammalian expression vector pcDNA3.1 (+) (Invitrogen). The sequences for SALM1, SALM2, and SALM4 were submitted to GenBank (accession numbers DQ070869, DQ070870, and DQ078787). The accession number for SALM3 (clone MGC:36545; IMAGE:4951386) is BC023156. SALM1 was epitope tagged by inserting myc immediately after the predicted signal peptide using Quick-Change (Stratagene, La Jolla, CA) mutagenesis method. The deletion of the PDZ-binding motif (SALM1-Δ4myc) was generated by inserting a stop codon immediately before the PDZ-binding motif, resulting in termination at M784 for SALM1. The C-terminal deletion of SALM1-myc (SALM1-ΔCT) was constructed by inserting a stop codon immediately after the transmembrane (TM) domain, resulting in termination at K558. NR1-1TM-yellow fluorescent protein (YFP) was generated by inserting a stop codon immediately after the first transmembrane domain, resulting in termination at K590 (NR1-1a) using Quick-Change mutagenesis (Stratagene). cDNA constructs were generously provided by the following: PSD-95-green fluorescent protein (GFP), David Bredt (Eli Lilly, Greenfield, IN); SAP102 and SAP97, Craig Garner (Stanford, Palo Alto, CA); NR1-1b, Michael Hollmann (Ruhr University Bochum, Bochum, Germany); and NR1-1b-YFP, NR2A-GFP, NR2B-GFP, and NR1-4b-YFP, Stefano Vicini (Georgetown University, Washington, DC).
Anti-SALM antibody characterization and purification.
The peptide sequence used for generating a SALM1 N-terminal antibody is [NH2]-NSTSRMAPPKSRLS-[COOH] (amino acids 384–397) and that for the C-terminal antibody is [NH2]-GAGTSSRGHHSDREPL-[COOH] (amino acids 684–699). Peptides were synthesized and conjugated to KLH through a cysteine added to the N terminus of the peptide. Antisera were produced in rabbits (Princeton BioMolecules, Langhorne, PA). Antibodies were purified by antigen affinity columns using peptides coupled to SulfoLink gel (Pierce, Rockford, IL) following the instructions of the manufacturer.
Preparation of crude synaptosomal proteins and brain subcellular fractions.
The crude synaptosomal proteins and brain subcellular fractions were prepared as described previously (Carlin et al., 1980; Blackstone et al., 1992). All of the PSD fractions were resuspended in 40 mm Tris-HCl and assayed for protein content (Bio-Rad, Hercules, CA).
Immunoprecipitation and Western blotting.
Heterologous cell lysates and rat brain tissues were solubilized with 1% sodium deoxycholate (DOC) in 50 mm Tris-HCl, pH 7.5 or pH 9.0. The solubilized fraction was dialyzed against 50 mm Tris-HCl with 0.1% Triton X-100 overnight at 4°C or was directly immunoprecipitated. Before immunoprecipitation, NaCl was added to the protein extracts to a final concentration of 150 mm. For immunoprecipitation, antibodies were added to the solubilized fractions and incubated for 2 h at 4°C and then incubated with protein A/G agarose beads at 4°C overnight. The beads were washed with 0.1% Triton X-100/TBS four times and resuspended in 2× SDS loading buffer. Samples were boiled and loaded onto 4–20% Tris-glycine gels (Invitrogen). Proteins were transferred to polyvinylidene difluoride membranes, incubated with primary and secondary antibodies, and detected with ECL using BioMax MR x-ray films (Eastman Kodak, Rochester, NY).
Hippocampal neuronal cultures.
The hippocampal neuronal cultures were prepared as described previously (Sans et al., 2003) with modification. Embryonic day 18 (E18) hippocampi from Sprague Dawley rats were dissected and dissociated with trypsin EDTA (Invitrogen). A total of 0.25 million cells per well were plated onto poly-ornithine/fibronectin-coated coverslips in 2% fetal bovine serum/DMEM medium (Invitrogen) in six-well culture plates (Corning, Corning, NY). The culture media was changed to Neurobasal media (Invitrogen) plus B27 (Invitrogen) the next day. For transfection, 2 μg of plasmid DNA was diluted into 100 μl of DMEM and mixed with 100 μl of DMEM containing 4 μl of Lipofectamine 2000 (Invitrogen). The mixture was incubated at room temperature for 20–30 min and added to the cells. Cells were incubated at 37°C for 2–3 h and then washed. Cells were cultured for 2 additional days in Neurobasal medium plus B27 before processing. For the neurite outgrowth studies, neurons were transfected after 4 d in vitro (DIV) with GFP plus SALM1 or SALM1-ΔCT and fixed 48 h later with 4% paraformaldehyde. For other studies, 14 DIV neurons were transfected and fixed 48 h later with 4% paraformaldehyde. For heterologous cell transfection, 1 μg of plasmid DNA was mixed with 1 μl of Lipofectamine 2000 per well for 12-well cultures. Cells were incubated with DNA-Lipofectamine 2000 mixture overnight, and the medium was changed the next day. Forty-eight hours later, cells were either fixed and processed for immunostaining or harvested for protein extraction.
Immunocytochemistry, image acquisition, and analysis.
For immunocytochemical studies, cells were fixed with 4% paraformaldehyde, washed with PBS, permeabilized with 0.2% Triton X-100/PBS, blocked with 10% normal goat serum in PBS for 1 h, incubated with primary antibody for 1 h, stained with Alexa Fluor 488, 568, or 647 secondary antibody for 0.5 h, and mounted onto slides. For surface staining, cells were live stained with primary and secondary antibodies in culture medium at room temperature and then washed with medium and fixed with 4% paraformaldehyde. For immunocytochemistry, anti-PSD-95 antibodies (Affinity BioReagents, Golden, CO) were used at 1:500 dilution, anti-NR2A (Upstate Biotechnology, Lake Placid, NY) at 1:500, and anti-NR1 (clone 54.2) at 1:1000. For image acquisition, images from neurons and HeLa cells were captured using a Zeiss (Oberkochen, Germany) LSM510 confocal microscope. The image data were analyzed and quantified using MetaMorph software (Universal Imaging Corporation, West Chester, PA). For neurite outgrowth analysis, whole dendritic images were traced using MetaMorph to obtain the total length of neurites; the number of neurons for each count is specified in the figure legends. For determining surface expression levels versus total expression levels, the ratio was taken from the intensity of fluorescence derived from each parameter. For the surface fluorescence quantification, the average total intensity and the pixel area of fluorescent puncta per 10 μm were measured. For these analyses, neurites in a micrograph (taken using 63× oil objective, 1.4 numerical aperture, at 2× zoom) were outlined, measured, and combined to make a single count. In most cases, each micrograph was taken from a different neuron; n in the figures represents the number of micrographs counted. Data are expressed as the mean ± SD. All animal procedures were done in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (National Institutes of Health publication 85-23) under National Institute on Deafness and Other Communication Disorders protocol 1022-01.
Results
The molecular structure of the SALM family
We used the first 543 amino acids of SAP97, which contains an LN27 (a domain in the receptor targeting protein Lin2 and Lin7) domain and three PDZ domains, to screen a mouse brain cDNA library using the yeast two-hybrid system. We obtained 265 positive clones, which included molecules involved in protein trafficking and signal transduction, channels/receptors, cytoskeletal proteins, and those with characteristics of cell adhesion molecules. In this study, we focused on two cell adhesion-like molecules, previously identified as KIAA1246 and KIAA1484 (Nagase et al., 1999). Based on motif analysis, the molecular structure of KIAA1246 includes one signal peptide, one leucine-rich-N-terminal domain (LRRNT), six leucine-rich repeat domains (LRRs), one leucine-rich-C-terminal domain (LRRCT), one Ig C2-like domain (IgC2), one fibronectin type III domain (FNIII), one TM, and a PDZ-binding motif, ESTV. A preliminary analysis of KIAA1246 showed that this molecule is most highly expressed in brain, spleen, and testis (Nagase et al., 1999). In brain, KIAA1246 is widely distributed. The chromosome location of this gene is 6p21.32. By sequence analysis and GenBank searching, we identified two additional family members. We named this protein family SALM1–SALM4. The GenBank accession numbers for mouse orthologs of SALM family members are AK017594 for SALM1, BC023156 for SALM3, and NM_175478 for SALM4. No full-length mouse cDNA corresponding to SALM2 was present in the current database. This sequence was submitted with GenBank accession number DQ070870. By sequence alignment, each member contains a signal peptide, one LRRNT, six LRRs, one LRRCT, one IgC2, one FNIII, one TM, and a PDZ-binding motif, ESTV. SALM4 is the only family member that does not contain an ESTV motif (Fig. 1A).
Figure 1.
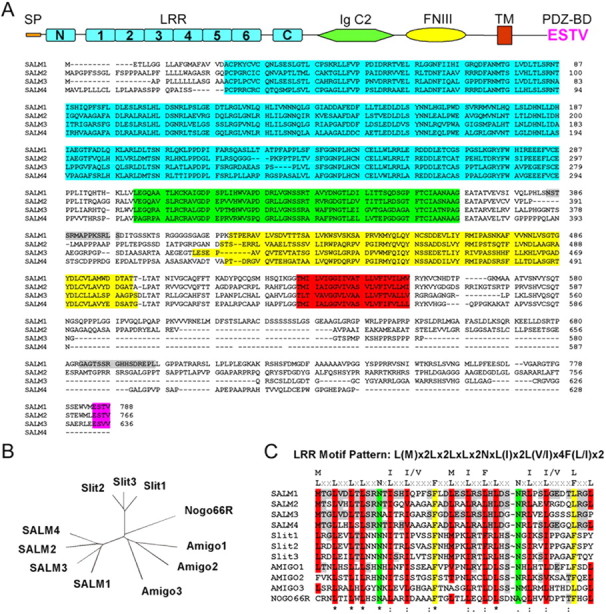
The molecular structure of the SALM family. A, The alignment of four SALM proteins. Each member contains a signal peptide, six LRR domains, N- and C termini of the LRR, one IgC2, one FNIII domain, one TM domain, and one C-terminal PDZ-binding motif, ESTV. SALM4 is the only family member that does not contain an ESTV motif. The amino acid sequence identities among SALM family members range from 51% (SALM2 and SALM3) to 59% (SALM2 and SALM4). The molecular structure domains are highlighted as follows: LRR domain, light blue; IgC2 domain, green; FNIII domain, yellow; TM domain, red; PDZ-binding domain, magenta; the peptide sequences used for generating SALM-specific antibodies are highlighted in gray. cDNA sequences are from mouse. B, An unrooted N-J phylogeny tree analyzed by the CLUSTAL multiple sequence alignment program indicates that the sequence of the SALM family is highly related to the Slit family, the Amigo family, and the Nogo 66 receptor. Percentage of amino acid identities of SALM1 to Slit1–Slit3, Amigo1–Amigo3, and Nogo receptor are 24–25, 24–28, and 28%, respectively. C, The leucine-rich repeat domains of the SALM family are highly conserved, and the repeat consensus is L(M)x2Lx2LxLx2NxL(I)x2L(V/I)x4F(L/I)x2. This consensus is similar in the Slit family, Amigo family, and Nogo 66 receptor.
An unrooted N-J phylogeny tree analyzed by the CLUSTAL multiple sequence alignment program indicates that the N terminus of the LRR sequence of the SALM family is highly related to that of the Slit family, the Amigo family, and the Nogo 66 receptor (Fig. 1B). The leucine-rich repeat domains of the SALM family are highly conserved, and the repeat consensus is L(M)x2Lx2LxLx2NxL(I)x2L(V/I)x4F(L/I)x2 (Fig. 1C). Leucine-rich repeats, IgC2, and FNIII domains are involved in protein–protein interactions and are found in many adhesion molecules. Leucine-rich repeat proteins, such as connectin, Slit, chaoptin, and Kekkon, have been shown to play important roles in neuronal development in Drosophila (Reinke et al., 1988; Rothberg et al., 1990; Nose et al. 1994; Ghiglione et al., 1999). Slit is a midline repellent molecule, is involved in the formation of the commissural axon pathway, and is also involved in cortical dendrite development (Rothberg et al., 1990; Whitford et al., 2002). Chaoptin is involved in photoreceptor cell morphogenesis (Reinke et al., 1988). Connectin is an adhesion molecule important in neuronal pathfinding and recognition (Nose et al., 1992, 1994). Kekkon, which contains six LRRs and a single Ig loop, is also expressed in neurons as they differentiate in the embryonic CNS (Ghiglione et al., 1999). In addition to the above motifs, the SALM family also contains a PDZ interacting motif, a characteristic of many proteins associated with the postsynaptic density.
SALM1 interacts with the PSD-95 family of membrane-associated guanylate kinase proteins
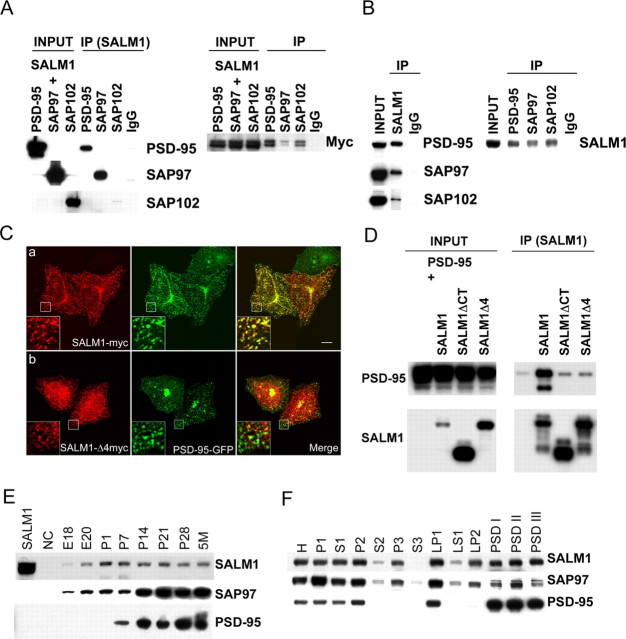
SALM1 contains a type I PDZ-binding motif, ESTV, at its C terminus. To verify its interaction with PDZ proteins, we cotransfected SALM1-myc with PSD-95, SAP97, or SAP102 in HEK293 cells, immunoprecipitated detergent-solubilized extracts with anti-myc antibodies, and probed with antibodies to PSD-95, SAP97, or SAP102. Immunoprecipitation was also done with anti-PSD-95, SAP97, or SAP102 antibodies and probed with anti-myc antibodies (Fig. 2A). These results showed that SALM1 is able to bind to PSD-95 family members in heterologous cells. To verify that the interaction occurs in brain, we performed immunoprecipitation using detergent extracts of crude synaptosomal fractions from 2-week-old rat brains. Coimmunoprecipitation was seen after immunoprecipitation with SALM1 antibodies or with antibodies to PSD-95, SAP97, and SAP102 (Fig. 2B). These results show that SALM1 binds to PSD-95, SAP97, and SAP102 in the brain.
Figure 2.
SALM1 interacts with the PSD-95 family of proteins. The interaction of SALM1 with the PSD-95 family of proteins was verified in heterologous cells (A, C, D) and in brain (B). A, HEK293 cells were cotransfected with cDNAs of SALM1-myc and PSD-95, SAP97, or SAP102. The cell lysate proteins were solubilized with 1% DOC, immunoprecipitated (IP) with anti-myc, anti-SALM1, or normal IgG as a control, and probed with anti-PSD-95, SAP97, or SAP102 antibodies (left). The DOC-solubilized proteins were immunoprecipitated with antibodies to PSD-95, SAP97, SAP102, or IgG and probed with anti-myc antibodies (right). B, Crude synaptosomal proteins were isolated from 2-week-old rat brains, solubilized with 1% DOC, immunoprecipitated with anti-SALM1 antibodies, and probed with anti-PSD-95, SAP97, or SAP102 antibodies individually (left). The proteins were immunoprecipitated with anti-PSD-95, SAP97, or SAP102 antibodies and probed with anti-SALM1 antibodies (right). C, The interaction of SALM1 with the PSD-95 family of proteins was further verified by their colocalization in HeLa cells. HeLa cells were cotransfected with SALM1-myc plus PSD-95-GFP (a) or SALM1-Δ4myc (the construct with the deletion of last four amino acids, ESTV) plus PSD-95-GFP (b). Cells were immunostained with anti-myc (red) for SALM1 and anti-GFP (green) for PSD-95. SALM1 is colocalized with PSD-95 (a). The deletion of PDZ-binding motif of SALM1 disrupts this colocalization with PSD-95 (b). Scale bar, 10 μm. D, PSD-95 interacts with SALM1 through its PDZ-binding domain. HEK293 cells were transfected with PSD-95-GFP alone or with SALM1, SALM1-ΔCT, or SALM1Δ4. After detergent solubilization, immunoprecipitation was done using anti-SALM1-N antibodies, and blots were probed with anti-PSD-95. E, Developmental profile of SALM1 in brain. SALM1 is detectable from E18 through adult (5-month-old). F, Subcellular distribution of SALM1. SALM1 can be detected in most subcellular fractions, including the plasma membrane (P2), intracellular organelle membrane (P3), synaptosome (LP1), and postsynaptic density (PSDI/II/III) fractions. Fractions were obtained from 2-week-old rat brains. H, Homogenate; P1, nucleic fraction; S1, soluble fraction; S3, organelle soluble fraction; LS1, soluble synaptosomal fraction; LP2, synaptic vesicle fraction.
Immunocytochemical studies on HeLa cells also support an interaction between SALM1 and PSD-95. Cells were cotransfected with PSD-95-GFP and SALM1-myc or SALM1-Δ4myc (the construct with the deletion of last four amino acids, ESTV) and immunostained with anti-myc in red for SALM1 and anti-GFP in green for PSD-95. SALM1 colocalized with PSD-95 (Fig. 2Ca), whereas deletion of the PDZ-binding motif of SALM1 disrupted this colocalization (Fig. 2Cb,D).
Developmental profile and subcellular distribution of SALM1 in the brain
Two antibodies were made to SALM1: SALM1-N and SALM1-C. On Western blots, SALM1 antibodies detected a band in both transfected HEK293 cells and in brain extracts migrating at 98 kDa. The predicted molecular weight of SALM1 is 85 kDa, indicating significant posttranslational modification. We studied the developmental expression pattern of SALM1 and compared it with those of PSD-95 and SAP97 (Fig. 2E). Subfractionation of 2-week-old rat brains showed that SALM1 is present in all membrane fractions as well as postsynaptic density fractions (Fig. 2F).
Overexpression of SALM1 promotes neurite outgrowth in hippocampal neurons
Several proteins, including Pal, ZfNLRR, and XNLRR-1 that contain domains similar to those found in SALM, have been shown to be involved in neural development (Hayata et al., 1998; Bormann et al., 1999; Gomi et al., 2000). This suggests that SALMs may also play a role in neuronal development. To test this, we overexpressed SALM1 along with GFP in young neurons (4 DIV) and analyzed its effects compared with neurons that were transfected with GFP alone. Forty-eight hours after transfection, neurons were fixed and immunostained with anti-GFP and anti-SALM1 antibodies. Transfected neurons were clearly identified compared with nontransfected neurons; very light staining of endogenous SALM1 was seen in nontransfected neurons (Fig. 3a–c). The total length of neurites in an individual neuron transfected with GFP plus SALM1, GFP plus SALM1-ΔCT (in which the entire C terminus was deleted), or GFP control were measured and analyzed with MetaMorph software. The total length of neurites in neurons transfected with SALM1 plus GFP was two times greater than that of neurons transfected with GFP alone (Fig. 3a–c,g). Overexpression of SALM1-ΔCT had no effect on neurite outgrowth. Overexpression of SALM1 in 14 DIV neurons had no significant effect on neurite outgrowth (n = 5; data not shown), but, interestingly, SALM1-ΔCT had an effect on dendrite morphology by increasing the number of protrusions found on dendrites compared with neurons transfected with GFP alone or SALM1 plus GFP (Fig. 3d–f,h).
Figure 3.
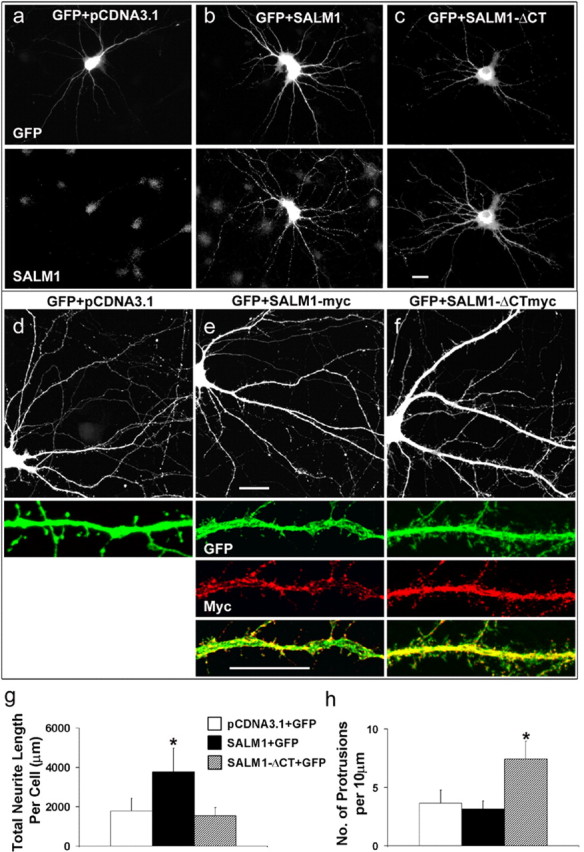
SALM1 promotes neurite outgrowth in cultured hippocampal neurons. Cultured hippocampal neurons at 4 DIV were cotransfected with GFP plus pcDNA3.1 vector (a), GFP plus SALM1 (b), or GFP plus SALM1-ΔCT (c) and stained with anti-SALM1 and anti-GFP antibodies. Cells were processed 48 h after transfection. The total neurite length of each neuron was traced based on GFP distribution and measured using MetaMorph software. The total neurite length per cell is 1795 ± 649 μm for GFP plus pcDNA3.1 vector, 3807 ± 1219 μm for GFP plus SALM1, and 1558 ± 403 μm for GFP plus SALM1-ΔCT (g), obtained from 42, 54, and 10 neurons, respectively. Hippocampal neurons at 14 DIV were also transfected with GFP plus SALM1-myc and GFP plus SALM1-ΔCTmyc (d–f, h). Overexpression of SALM1-ΔCTmyc had a dramatic effect on dendritic morphology of these neurons (f) with the appearance of more filopodia-like structures. The number of protrusions per 10 μm for GFP plus pcDNA3.1 is 3.7 ± 1.1, for GFP plus SALM1-myc is 3.2 ± 0.7, and for GFP plus SALM1-ΔCTmyc is 7.4 ± 1.5 (h). Data were obtained from 10 neurons for each condition. Scale bars: a–c, d–f, top panels, 20 μm; e, f, bottom panels, 10 μm. *p < 0.001 compared with GFP alone. Values are mean ± SD.
The PDZ-binding motif of SALM1 plays an important role in its surface targeting and the recruitment of PSD-95
Many membrane proteins that contain a PDZ-binding domain require an interaction with a PDZ protein for correct targeting to the cell surface. For example, the NMDA receptor requires an interaction with a PDZ protein through its NR2 subunits for proper synaptic expression of the receptor (Wenthold et al., 2003). Interaction with the PDZ protein CAL/PIST can block surface expression of the β1-adrenergic receptor (He et al., 2004), and deletion of the PDZ-binding motif on TGF receptor has been shown to affect its surface targeting (Blobe et al., 2001). To determine whether the PDZ-binding domain of SALM1 is important for surface expression, we measured surface expression of SALM1-myc or SALM1-Δ4myc relative to total expression in HeLa cells. We found that the deletion of the PDZ-binding motif of SALM1 dramatically decreased the level of surface expression of SALM1 in HeLa cells (Fig. 4Aa–Ac) and in neurons (Fig. 4Ad–Af). We also observed that the immunostaining of SALM1 showed larger puncta in neurons transfected with SALM1-myc than those with SALM1-Δ4myc, with the area of SALM1 staining being reduced by >50% (Fig. 4Ag). These data suggest that the PDZ interaction plays an important role in surface expression and molecular organization of SALM1. The puncta observed in neurons overexpressing SALM1 were also immunostained with anti-PSD-95 antibodies to detect endogenous PSD-95 (Fig. 4B). The percentage of the area of colocalization of SALM1 and PSD-95 was measured as the percentage of the total immunostaining of SALM1 that contains PSD-95 staining and was compared with that of SALM1-Δ4myc. For SALM1-Δ4myc, there was a large decrease in the area of colocalization with PSD-95, indicating that SALM1 can recruit PSD-95 through a PDZ interaction in neurons. Furthermore, 35% of SALM1 puncta observed in neurons transfected with SALM1-myc colocalized with the synaptic marker synapsin (data not shown).
Figure 4.
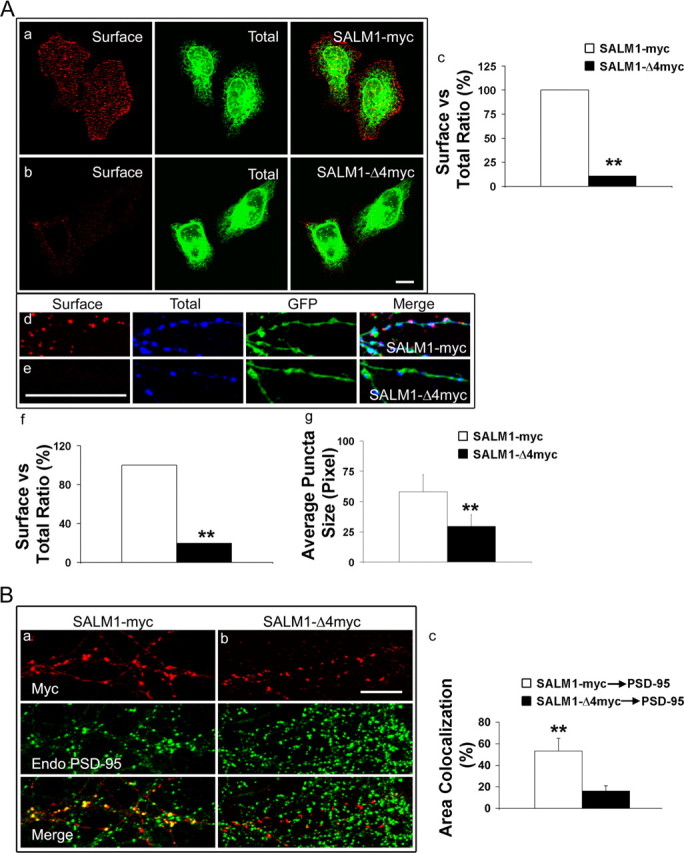
The PDZ-binding motif of SALM1 plays an important role in SALM1 surface targeting and the recruitment of PSD-95. A, The deletion of the PDZ-binding motif of SALM1 changes the level of the surface expression of SALM1 in HeLa cells and in neurons. HeLa cells (a, b) and neurons (d, e) were transfected with SALM1-myc (a, d) or SALM1-Δ4myc (b, e). Neurons were also transfected with GFP (d, e; green fluorescence). Surface staining with anti-SALM1 N-terminus antibody is shown in red (left panels), and permeabilized staining with anti-myc is in green (a, b) or blue (d, e) for total SALM1 (middle panels). Surface expression and total expression were analyzed using MetaMorph software and graphed by Excel (Microsoft, Seattle, WA) (c, f). The ratio of surface to total staining for full-length SALM1 was arbitrarily assigned 100%. The surface staining of cells transfected with SALM1-Δ4myc is dramatically reduced when compared with that of cells transfected with SALM1-myc (11.0% in HeLa cells and 19.8% in neurons). Neurons transfected with SALM1-myc show larger puncta than those transfected with SALM1-Δ4myc. The size of puncta is 58.1 ± 14.0 pixels with transfection of SALM1-myc and 29.5 ± 9.6 pixels with transfection of SALM1-Δ4myc. **p < 0.001. Scale bar, 10 μm. Data were obtained from 14 DIV cultures 48 h after transfection. For SALM1-myc and SALM1-Δ4myc, results presented in f are based on n = 22 and 5, and in g on n = 17 and 12, respectively; values are mean ± SD. B, SALM1 is colocalized with PSD-95. Neurons at 14 DIV were transfected with SALM1-myc or SALM1-Δ4myc, and, 48 h after transfection, cells were fixed and stained for endogenous PSD-95. The percentage of area colocalization for SALM1-myc and PSD-95 is 57.8 ± 3.7% and for SALM1-Δ4myc and PSD-95 is 16.1 ± 4.9%. The area of colocalization is defined as the percentage of SALM that has PSD-95 staining. **p < 0.001. Scale bar, 10 μm. For SALM1-myc and SALM1-Δ4myc, results presented in c are based on n = 14 and 24, respectively; values are mean ± SD.
Colocalization of SALM1 and the NMDA receptor
Because the NMDA receptor and PSD-95 are colocalized in neurons, it is reasonable to expect that NMDA receptor complexes are also recruited to SALM1 clusters with overexpression of SALM1. Neurons transfected with SALM1-myc or SALM1-Δ4myc were permeabilized and stained with anti-NR2A antibodies to detect endogenous NMDA receptors (Fig. 5A,B). The area of colocalization of SALM1 and NR2A and the average punctum size were analyzed. There was a large reduction in the area of colocalization of NR2A with SALM1-Δ4myc with 83% colocalization with SALM1-myc and 13% for SALM1-Δ4myc (Fig. 5C). The average punctum size of NR2A was larger for SALM1-myc than for SALM1-Δ4myc (30.3 and 12.2 pixels, respectively) (Fig. 5D). These data show that overexpressed SALM1 can recruit endogenous NMDA receptors and that this is dependent on a PDZ interaction of SALM1.
Figure 5.
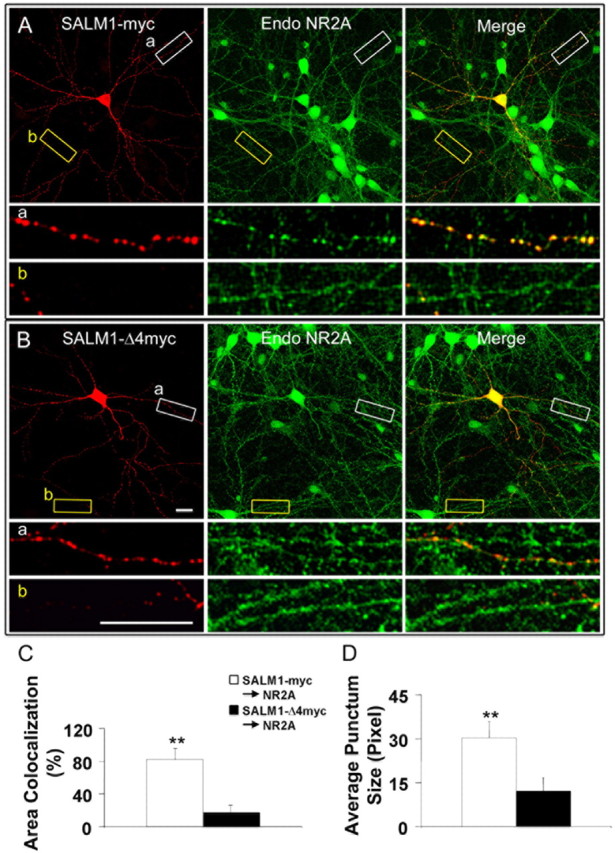
SALM1 colocalizes with the NMDA receptor. Neurons at 14 DIV were transfected with SALM1-myc (A) or SALM1-Δ4myc (B) and were permeabilized and stained with anti-NR2A antibody to detect endogenous NMDA receptors. The area of colocalization of SALM1 and NR2A (C) and the average punctum size of NR2A (D) were analyzed. There is a dramatic reduction in colocalization of SALM1-Δ4myc and NR2A (13.4 ± 2.9%) compared with SALM1-myc and NR2A (83.3 ± 28.3%) (C). The area of colocalization between SALM1 and NR2A is defined as the percentage of SALM1-myc or SALM1-Δ4myc that has NR2A staining. The average punctum size of NR2A in SALM1-myc-transfected neurons (30.3 ± 5.5 pixels) is larger than that in SALM1-Δ4myc-transfected neurons (12.2 ± 4.5 pixels) (D). *p < 0.001. Scale bar, 10 μm. Data were obtained 48 h after transfection. For SALM1-myc and SALM1-Δ4myc, results presented in C are based on an n = 20 and 21, and in D on n = 14 and 11, respectively; values are mean ± SD.
We investigated in more detail the nature of the SALM1/NMDA receptor interaction and, in particular, whether the NR2A that colocalized with SALM1 is expressed on the cell surface. Because reliable antibodies for surface labeling of endogenous NMDA receptors are not available, we cotransfected neurons with NR2A-GFP plus vector, SALM1-myc, or SALM1-Δ4myc and determined the immunocytochemical distribution of surface and total NR2A-GFP. Cells were surface stained with anti-GFP in red, permeabilized, and stained with anti-myc for SALM1-myc or SALM1-Δ4myc in blue (Fig. 6a–c). The expression of NR2A-GFP is shown in green. The level of surface fluorescence was analyzed with MetaMorph software. We were interested in whether the NR2A that is recruited to SALM1 puncta is on the surface, whether SALM1 enhances surface expression of NR2A, and whether SALM1 enhances NR2A clustering on the cell surface. To address these questions, the number of puncta for NR2A-GFP surface staining, the average total intensity of NR2A-GFP surface staining, the percentage of area colocalization of SALM1 and NR2A-GFP surface and total, and the average punctum size of NR2A-GFP surface staining were analyzed from segments of dendrites. Each 30 μm length of dendritic segment was analyzed, and the data were obtained from dendrites of 10 neurons. Both the number of NR2A surface puncta and the intensity of NR2A surface staining were significantly higher for neurons that were transfected with SALM1-myc compared with those transfected with SALM1-Δ4myc or vector alone (Fig. 6d,e), suggesting that SALM1 is able to enhance surface NR2A expression through a PDZ-dependent interaction. The area of colocalization of SALM1 with NR2A-GFP surface was 48.4 ± 9.6% and with NR2A-GFP total was 56.5 ± 10.4%, and that for SALM1-Δ4myc with NR2A-GFP surface was 18.0 ± 6.0% and with NR2A-GFP total was 29.9 ± 10.4% (Fig. 6f). These data indicate that SALM1 is able to recruit surface NR2A into SALM1 clusters. Finally, SALM1 overexpression increased the size of NR2A surface puncta (Fig. 6g). To confirm our observation that SALM1 enhances NR2A surface expression and clustering, the number of puncta and the average total intensity of NR2A-GFP surface staining with a pixel area >15 were analyzed. The number of puncta for NR2A-GFP, NR2A-GFP plus SALM1-myc, and NR2A-GFP plus SALM1-Δ4myc was 2.5 ± 0.7, 6.5 ± 1.4, and 1.8 ± 0.7, respectively (Fig. 6h); the average total intensity for NR2A-GFP, NR2A-GFP plus SALM1-myc, and NR2A-GFP plus SALM1-Δ4myc was 1077.7 ± 190.3, 1516.0 ± 244.1, and 1155.5 ± 295.0 individually (Fig. 6i). Therefore, these results show that overexpression of SALM1 clusters both endogenous and transfected NMDA receptors and increases their surface expression through PDZ-dependent interactions.
Figure 6.
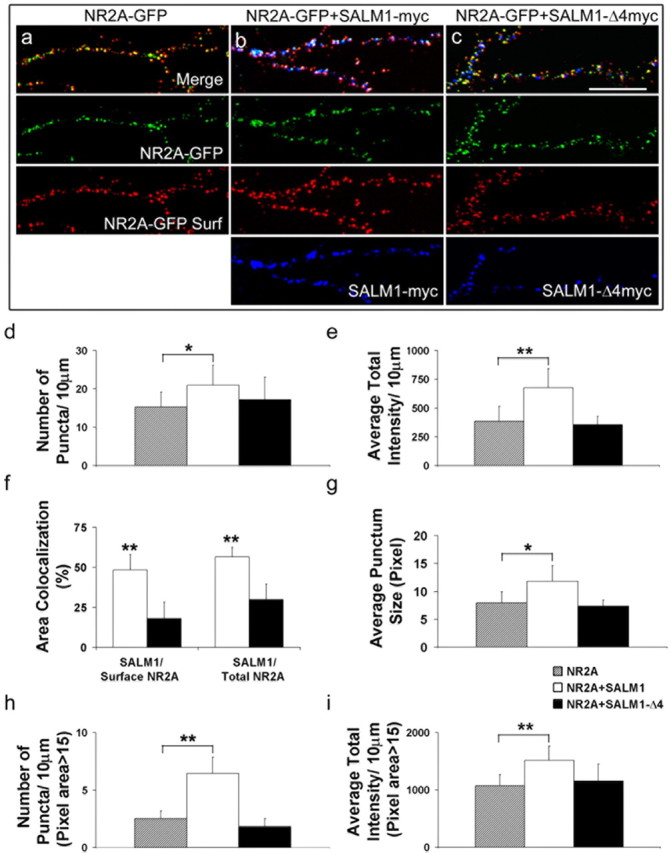
SALM1 recruits surface NMDA receptors through its interaction with PDZ proteins. Cultured hippocampal neurons at 14 DIV were cotransfected with NR2A-GFP and vector (a), NR2A-GFP and SALM1-myc (b), or NR2A-GFP and SALM1-Δ4myc (c). Cells were surface stained with anti-GFP (red) and permeabilized and stained with anti-myc (blue) for SALM1-myc or SALM1-Δ4myc. The expression of NR2A-GFP is shown in green. The level of surface fluorescence was analyzed with MetaMorph software. The number of puncta for NR2A-GFP surface staining (d), the average total intensity of NR2A-GFP surface staining (e), the percentage of area of colocalization for SALM1 and NR2A-GFP surface and NR2A-GFP total (f), and the average punctum size of NR2A-GFP surface staining (g) were analyzed. In addition, the number of puncta (h) and the average total intensity (i) of NR2A-GFP surface staining with the pixel area >15 were analyzed. The number of puncta/10 μm is 15.3 ± 3.9, 20.9 ± 5.3, and 17.2 ± 5.9 for cells transfected with NR2A alone, NR2A/SALM1, and NR2A/SALM1Δ4, respectively (d). The average total intensity is 386.3 ± 129.1, 675.8 ± 165.9, and 355.8 ± 71.7 units for cells transfected with NR2A alone, NR2A/SALM1, and NR2A/SALM1Δ4, respectively (e). The area of colocalization for SALM1 and NR2A surface is 48.4 ± 9.6% and of NR2A total is 56.5 ± 10.4%. The area of colocalization for SALM1-Δ4myc and NR2A surface is 18.0 ± 6.0% and for NR2A total is 29.9 ± 10.4% (f). The average punctum size is 7.9 ± 2.0, 11.8 ± 2.8, and 7.3 ± 1.1 pixels for cells transfected with NR2A alone, NR2A/SALM1, and NR2A/SALM1Δ4, respectively (g). For pixel areas larger than 15, the number of puncta for NR2A-GFP, NR2A-GFP/SALM1-myc, and NR2A-GFP/SALM1-Δ4myc is 2.5 ± 0.7, 6.5 ± 1.4, and 1.8 ± 0.7, respectively (h); the average total intensity for NR2A-GFP, NR2A-GFP/SALM1-myc, and NR2A-GFP/SALM1-Δ4myc is 1077.7 ± 190.3, 1516.0 ± 244.1, and 1155.5 ± 295.0, respectively (i). The area of colocalization is defined as the percentage of SALM1-myc or SALM1-Δ4myc that has NR2A-GFP surface staining or NR2A-GFP total expression. *p < 0.01; **p < 0.001. Scale bar, 10 μm. Results presented in d, e, and g–i are based on n = 23, 20, and 12 for NR2A, NR2A plus SALM1, and NR2A plus SALM1-Δ4, respectively, and in f on n = 18 and 12 for surface and total NR2A, respectively; values are mean ± SD.
SALM1 also interacts with NR1
The ability of SALM1 to recruit the NMDA receptor in neurons in a PDZ-dependent manner suggests that these two molecules are linked through PDZ proteins such as members of the PSD-95 family. To investigate this further, we performed immunoprecipitation studies using detergent extracts of crude synaptosomal fractions isolated from 2-week-old rat brains. Two different anti-SALM1 antibodies, made to the N- and C-terminal domains, were used for immunoprecipitation, and the bound fraction was probed with anti-NMDA receptor antibodies, anti-NR1, NR2A, and NR2B (Fig. 7A, left). Immunoprecipitation was also done with anti-NR1 and anti-NR2 antibodies and probed with anti-SALM1 antibodies (Fig. 7A, right). These results show that a SALM1/NMDA receptor complex exists in brain and support our observation that SALM1 is able to recruit NMDA receptor/PSD-95 complexes. No significant immunoprecipitation of AMPA receptors was seen (data not shown).
Figure 7.
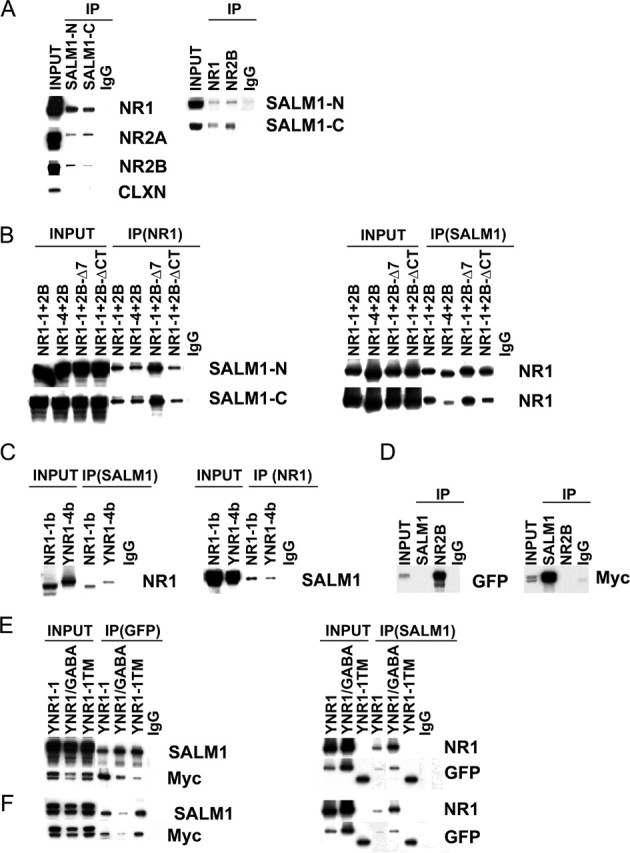
SALM1 interacts directly with the NR1 subunit. A, The interaction of SALM1 with the NMDA receptor complex in the brain was shown by immunoprecipitation (IP) using 1% DOC solubilized crude synaptosomal proteins from 2-week-old rat brains. Proteins were immunoprecipitated with anti-SALM1-N (N-terminal antibody), anti-SALM1-C (C-terminal antibody), or IgG and probed with anti-NMDA receptor antibodies, anti-NR1, anti-NR2A, and anti-NR2B (left). The DOC-soluble proteins also were immunoprecipitated with anti-NR1, anti-NR2B, or IgG and probed with anti-SALM1-N/C antibodies (right). As a control for nonspecific interaction, calnexin (CLXN) does not coimmunoprecipitate with SALM1. An IgG control was not included with calnexin. The Western blot data show that the SALM1/NMDA receptor complexes are present in vivo in the brain. B, The interaction of SALM1 with the NMDA receptor was studied in heterologous cells. NR1 and NR2 subunits must assemble to form functional receptors on the cell surface. HEK293 cells were cotransfected with SALM1myc plus NR1-1 and NR2B, NR1-4 and NR2B, NR1-1 and NR2B-Δ7 (NR2B with the deletion of the last 7 amino acids), or NR1-1 and NR2B-ΔCT (NR2B with the deletion of the entire C-terminal domain). The cell lysates were solubilized with 1% DOC, immunoprecipitated with anti-NR1 antibodies or IgG, and probed with anti-SALM1-N antibodies (left). The soluble proteins were also immunoprecipitated with anti-SALM1 antibodies or IgG and probed with anti-NR1 (right). C, The interaction of SALM1 with individual NMDA receptor subunits was also studied. HEK293 cells were cotransfected with SALM1-myc plus NR1-1b or NR1-4b-YFP (YNR1). The proteins were solubilized with 1% DOC, immunoprecipitated with anti-SALM1 or IgG, and probed with anti-NR1 antibodies. Proteins were also immunoprecipitated with anti-NR1 antibodies or IgG and probed with anti-SALM1-N. A direct interaction with SALM1 and NR1 is seen. D, SALM1-myc was cotransfected with the NR2B-GFP, followed by immunoprecipitation with anti-SALM1-N, anti-NR2B, or IgG and probed with anti-myc to detect SALM1 (right) or anti-GFP to detect NR2B (left). No coimmunoprecipitation is seen. E, F, The domain of NR1 that interacts with SALM1 was studied. HEK293 cells were transfected with SALM1-myc (E) or SALM1-ΔCTmyc (F) plus NR1-1-YFP, NR1-1/GABA-YFP (the intracellular C-terminal domain of YFP-NR1-1 was replaced by the C-terminal domain of the GABAB receptor), or NR1-1TM-YFP (NR1-1-YFP with the deletion after first transmembrane domain). The proteins were solubilized with 1% DOC, immunoprecipitated with anti-GFP antibodies or IgG, and probed with anti-SALM1-N or anti-Myc antibodies (left). The proteins were also immunoprecipitated with anti-SALM1-N antibodies or IgG and probed with anti-NR1 or anti-GFP antibodies (right). The lack of staining with the anti-NR1 antibody for YNR1-1TM is attributable to the loss of the epitope recognized by the NR1 antibody (54.2), which is made to the extracellular loop. These results show that SALM1 interacts directly with the N-terminal extracellular domain of NR1.
The relatively robust coimmunoprecipitation of SALM1 and NMDA receptor subunits raised the interesting possibility that SALM1 and the NMDA receptor were selectively associated with the same population of PSD-95 molecules or that SALM1 interacts directly with the NMDA receptor. This led us to investigate the SALM1/NMDA receptor interaction further. Heterologous cells were cotransfected with SALM1 and NR1 and NR2B subunits, followed by immunoprecipitation of detergent extracts. The interaction of SALM1 with the NMDA receptor was verified in heterologous cells (Fig. 7B). NR2B cotransfected with NR1-1 or NR1-4 forms functional NMDA receptors and both coimmunoprecipitated with SALM1, using either C- or N-terminal antibodies. Coimmunoprecipitation was also seen after transfection with NR2A (data not shown). Using NR2B constructs without the PDZ-binding domain, 2B-Δ7, which lacks the last seven amino acids, and 2B-ΔCT, which lacks the entire C terminus, also coimmunoprecipitated, indicating that the interaction with SALM1 is not occurring through the PDZ domain or exclusively through the PDZ domain. To study this further, SALM1 was cotransfected with NR1 and NR2 subunits alone. SALM1 coimmunoprecipitated with NR1 (Fig. 7C) but not NR2B (Fig. 7D). Two different splice variants of NR1 were used, NR1-1, which is retained in the endoplasmic reticulum (ER) when not assembled with NR2, and NR1-4, which is not retained in the ER and also has a PDZ-binding domain. Both forms coimmunoprecipitated with SALM1, indicating that the NR1/SALM1 interaction can occur in the ER. This interaction is similar to that seen between NR1 and the EphB receptor (Dalva et al., 2000), which also can occur with NR1-1 alone, indicating an association in the ER. We next determined the area of the NR1 molecule that interacts with SALM1 by using constructs that contain parts of the NR1 subunit. These included a chimera of NR1 through TM4 and the C terminus of the GABAB receptor, YNR1/GABA, and a construct in which NR1 is terminated immediately after TM1, Y-NR1-1TM. All of these constructs are tagged with YFP at their N termini for immunoprecipitation and immunoblotting. Immunoprecipitation in both directions showed that both of these constructs, as well as full-length NR1-1, interact with SALM1 (Fig. 7E,F), suggesting that the SALM1/NR1 interaction occurs at the level of the N-terminal extracellular domain or TM1.
Discussion
In this study, we report the characterization of a family of proteins with many properties of adhesion molecules that are involved in neurite outgrowth and synapse formation, including the following. (1) Like other proteins that have been identified to play a role in synapse formation, including SynCAM, fasciclin II, and Aplysia cell adhesion molecule, SALMs contain extracellular Ig domains and intracellular PDZ-binding domains. (2) SALMs have significant identity to Amigo and Slit family members, which are involved in neurite outgrowth, and contain multiple LRR domains and an Ig domain in their extracellular domains like SALMs. Thus, SALMs are members of both the LRR and Ig superfamilies. (3) SALMs interact with the NR1 subunit of the NMDA receptor, like the EphB receptor, and this interaction may play a role in the localization or stability of the receptor on the membrane or may modulate its function. (4) Like many adhesion proteins, SALMs are expressed early in the developing brain and continue to be expressed in the adult. These properties place SALMs into a large group of proteins involved in neuronal development and synapse function.
Both extracellular and intracellular signals play a role in neuron development. The extracellular domains, including LRR, Ig, and FNIII domains, and the cytoplasmic PDZ-binding domain present in SALMs provide multiple mechanisms for interactions with other proteins. Slit, a secreted protein that contains several LRR domains and an Ig domain, like the SALMs, interacts with ROBO through its LRR domains (Howitt et al., 2004). Nogo 66 receptor, a glycosylphosphatidylinositol-linked protein, has multiple LRR domains and interacts with the 66 amino acid extracellular domain of Nogo (Fournier et al., 2001). The L1 family of IgCAMs contains extracellular FNIII domains that are involved in homomultimerization and integrin recruitment (Silletti et al., 2000; Falk et al., 2004). The synaptic adhesion molecule SynCAM forms homophilic interactions through its extracellular Ig domains and can link presynaptic and postsynaptic membranes (Biederer et al., 2002). Amigo and other members of the Ig superfamily also can form homophilic interactions. Preliminary results suggest that SALMs can form homomeric or heteromeric complexes (G. K. Seabold, Y.-X. Wang, K. Chang, C.Y. Wang, and R. J. Wenthold, unpublished observation), raising the possibility that SALMs could also link presynaptic and postsynaptic membranes through homophilic interactions or interactions with other proteins. It is interesting that the amino acid sequences are most divergent in the C-terminal intracellular domain, which is most apparent in SALM4, which lacks a PDZ-binding domain. This suggests that the four SALMs may have different interacting partners and distinct signaling mechanisms associated with their cytoplasmic domains, although their extracellular interactions may be similar. Although the enhancement of neurite outgrowth with overexpression of SALM1 appears to apply only to young neurons, overexpression of SALM1 lacking the PDZ-binding domain led to an increase in filopodia-like structures in older (14 DIV) neurons (Fig. 3). Thus, SALM4 may have a role quite different from that of the remaining SALMs.
Our results linking the NMDA receptor to a family of adhesion molecules in neurons is consistent with the extensive literature supporting a role for the NMDA receptor in neuronal development and plasticity. Numerous studies have implicated the NMDA receptor in neurite outgrowth, spine development, and synapse formation, and effects on these processes are often attributed to calcium entering through the NMDA receptor channel and affecting downstream signaling. However, direct interactions of signaling molecules with the NMDA receptor may also be important. A recent report links the Rac1-guanine nucleotide exchange factor Tiam1 with NMDA receptor-dependent actin remodeling in dendrites (Tolias et al., 2005), with a direct interaction between the NMDA receptor and Tiam1. The EphB receptor interacts directly with the extracellular domain of the NR1 subunit and coclusters with the NMDA receptor in neurons. Activation of EphB receptors by ephrinB1 increases the number of postsynaptic specializations, as well as the number of presynaptic release sites (Dalva et al., 2000).
SALM coimmunoprecipitates with the NMDA receptor in detergent extracts from brain. A molecular complex of SALM and the NMDA receptor could be formed by an indirect link through PDZ proteins, because both the NR2 subunit and SALM can bind to the PSD-95 family of proteins, or by an interaction between NR1 and SALM. In heterologous cells, we showed an interaction between SALM1 and the N-terminal extracellular domain of NR1, whereas no interaction was seen with SALM and the NR2 subunit. The NR1/SALM1 interaction may be direct between the two proteins or through an unknown third protein. This interaction can occur in the ER as shown by its interaction with NR1-1, which is retained in the ER in the absence of assembly with an NR2 subunit. Thus, one possible role for this interaction is that SALM may associate with the NMDA receptor early in the biosynthetic pathway and the two proteins may be trafficked together. Overexpression of SALM1 increased the number of NMDA receptor puncta and their size in dendrites, whereas overexpression of SALM1Δ4, in which the PDZ interacting domain is deleted, had no effect on number of puncta or their size, showing that an interaction with a PDZ protein is required to form clusters of SALM and NMDA receptor in dendrites. The PDZ-binding domain of SALM1 also is important for its colocalization with both endogenous and transfected NR2. These results suggest that the indirect PDZ interaction is necessary for colocalization of SALM and the NMDA receptor in neurons, although the role of the NR1 interaction is unclear.
The direct interaction of ion channels and adhesion molecules is not unprecedented. The extracellular domain of the NR1 subunit interacts directly with the EphB receptor (Dalva et al., 2000), and this interaction is similar to its interaction with SALM1. This interaction is important in the Fyn catalyzed phosphorylation of the NR2B subunit in response to ephrinB2 to ensure that the two molecules are in close proximity (Takasu et al., 2002). Common to both EphB receptors and SALMs are fibronectin type III motifs in their extracellular domains, which may be candidates for interaction with the NR1 subunit. Voltage-gated sodium channels interact directly with members of the Ig superfamily of cell adhesion molecules, including neurofascin, neuron–glia-related cell adhesion molecule, and contactin (Ratcliffe et al., 2001; McEwen and Isom, 2004; Shah et al., 2004), through an Ig loop in the extracellular domain of the β subunit. This interaction is important for regulating the cell surface expression of the channel. No known adhesion domain is present on the NR1 subunit to account for its interaction with SALM or the EphB receptor.
Our results bring together two classes of proteins that are essential for neuron development and synapse function, the NMDA receptor and adhesion molecules. Because adhesion molecules are among the earliest residents of the synapse and are often maintained at the synapse in adults, interactions between adhesion molecules and ion channels at the synapse may be an effective way of both recruiting the ion channel to the synapse and maintaining it there. Although overexpression of SALM1 leads to enhanced neurite outgrowth and clustering of NMDA receptors, it is unlikely that the clustering of NMDA receptors alone would explain the neurite outgrowth properties of SALM. A more likely explanation is that the interaction between SALM and the NMDA receptor, which may occur after they have been delivered to the synapse, is either functionally important and modulates the ligand binding or channel properties of the receptor, like the EphB receptor interaction (Takasu et al., 2002), or it stabilizes one or both molecules on the cell surface.
Footnotes
This work was supported by the National Institute on Deafness and Other Communication Disorders Intramural Program.
References
- Biederer T, Sara Y, Mozhayeva M, Atasoy D, Liu X, Kavalali ET, Sudhof TC (2002). SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science 297:1525–1531. [DOI] [PubMed] [Google Scholar]
- Blackstone CD, Moss SJ, Martin LJ, Levey AI, Price DL, Huganir RL (1992). Biochemical characterization and localization of a non-N-methyl-d-aspartate glutamate receptor in rat brain. J Neurochem 58:1118–1126. [DOI] [PubMed] [Google Scholar]
- Blobe GC, Schiemann WP, Pepin MC, Beauchemin M, Moustakas A, Lodish HF, O’Connor-McCourt MD (2001). Functional roles for the cytoplasmic domain of the type III transforming growth factor beta receptor in regulating transforming growth factor beta signaling. J Biol Chem 276:24627–24637. [DOI] [PubMed] [Google Scholar]
- Bormann P, Roth LW, Andel D, Ackermann M, Reinhard E (1999). zfNLRR, a novel leucine-rich repeat protein is preferentially expressed during regeneration in zebrafish. Mol Cell Neurosci 13:167–179. [DOI] [PubMed] [Google Scholar]
- Carlin RK, Grab DJ, Cohen RS, Siekevitz P (1980). Isolation and characterization of postsynaptic densities from various brain regions: enrichment of different types of postsynaptic densities. J Cell Biol 86:831–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih B, Engelman H, Scheiffele P (2005). Control of excitatory and inhibitory synapse formation by neuroligins. Science 307:1324–1328. [DOI] [PubMed] [Google Scholar]
- Dalva MB, Takasu MA, Lin MZ, Shamah SM, Hu L, Gale NW, Greenberg ME (2000). EphB receptors interact with NMDA receptors and regulate excitatory synapse formation. Cell 103:945–956. [DOI] [PubMed] [Google Scholar]
- Falk J, Thoumine O, Dequidt C, Choquet D, Faivre-Sarrailh C (2004). NrCAM coupling to the cytoskeleton depends on multiple protein domains and partitioning into lipid rafts. Mol Biol Cell 15:4695–4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier AE, GrandPre T, Strittmatter SM (2001). Identification of a receptor mediating Nogo-66 inhibition of axonal regeneration. Nature 409:341–346. [DOI] [PubMed] [Google Scholar]
- Ghiglione C, Carraway KL III, Amundadottir LT, Boswell RE, Perrimon N, Duffy JB (1999). The transmembrane molecule kekkon 1 acts in a feedback loop to negatively regulate the activity of the Drosophila EGF receptor during oogenesis. Cell 96:847–856. [DOI] [PubMed] [Google Scholar]
- Gomi F, Imaizumi K, Yoneda T, Taniguchi M, Mori Y, Miyoshi K, Hitomi J, Fujikado T, Tano Y, Tohyama M (2000). Molecular cloning of a novel membrane glycoprotein, pal, specifically expressed in photoreceptor cells of the retina and containing leucine-rich repeat. J Neurosci 20:3206–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM (2004). Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell 119:1013–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayata T, Uochi T, Asashima M (1998). Molecular cloning of XNLRR-1, a Xenopus homolog of mouse neuronal leucine-rich repeat protein expressed in the developing Xenopus nervous system. Gene 221:159–166. [DOI] [PubMed] [Google Scholar]
- He J, Bellini M, Xu J, Castleberry AM, Hall RA (2004). Interaction with cystic fibrosis transmembrane conductance regulator-associated ligand (CAL) inhibits beta1-adrenergic receptor surface expression. J Biol Chem 279:50190–50196. [DOI] [PubMed] [Google Scholar]
- Howitt JA, Clout NJ, Hohenester E (2004). Binding site for Robo receptors revealed by dissection of the leucine-rich repeat region of Slit. EMBO J 23:4406–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Sheng M (2004). PDZ domain proteins of synapses. Nat Rev Neurosci 5:771–781. [DOI] [PubMed] [Google Scholar]
- Luo L (2002). Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annu Rev Cell Dev Biol 18:601–635. [DOI] [PubMed] [Google Scholar]
- Malenka RC, Nicoll RA (1997). Silent synapses speak up. Neuron 19:473–476. [DOI] [PubMed] [Google Scholar]
- Malinow R, Malenka RC (2002). AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci 25:103–126. [DOI] [PubMed] [Google Scholar]
- McEwen DP, Isom LL (2004). Heterophilic interactions of sodium channel beta1 subunits with axonal and glial cell adhesion molecules. J Biol Chem 279:52744–52752. [DOI] [PubMed] [Google Scholar]
- Montgomery JM, Zamorano PL, Garner CC (2004). MAGUKs in synapse assembly and function: an emerging view. Cell Mol Life Sci 61:911–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase T, Ishikawa K, Kikuno R, Hirosawa M, Nomura N, Ohara O (1999). Prediction of the coding sequences of unidentified human genes. XV. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res 6:337–345. [DOI] [PubMed] [Google Scholar]
- Nose A, Mahajan VB, Goodman CS (1992). Connectin: a homophilic cell adhesion molecule expressed on a subset of muscles and the motoneurons that innervate them in Drosophila. Cell 70:553–567. [DOI] [PubMed] [Google Scholar]
- Nose A, Takeichi M, Goodman CS (1994). Ectopic expression of connectin reveals a repulsive function during growth cone guidance and synapse formation. Neuron 13:525–539. [DOI] [PubMed] [Google Scholar]
- Nourry C, Grant SG, Borg JP (2003). PDZ domain proteins: plug and play!. Sci STKE 2003:RE7. [DOI] [PubMed] [Google Scholar]
- Prange O, Wong TP, Gerrow K, Wang YT, El-Husseini A (2004). A balance between excitatory and inhibitory synapses is controlled by PSD-95 and neuroligin. Proc Natl Acad Sci USA 101:13915–13920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe CF, Westenbroek RE, Curtis R, Catterall WA (2001). Sodium channel beta1 and beta3 subunits associate with neurofascin through their extracellular immunoglobulin-like domain. J Cell Biol 154:427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke R, Krantz DE, Yen D, Zipursky SL (1988). Chaoptin, a cell surface glycoprotein required for Drosophila photoreceptor cell morphogenesis, contains a repeat motif found in yeast and human. Cell 52:291–301. [DOI] [PubMed] [Google Scholar]
- Rothberg JM, Jacobs JR, Goodman CS, Artavanis-Tsakonas S (1990). Slit: an extracellular protein necessary for development of midline glia and commissural axon pathways contains both EGF and LRR domains. Genes Dev 4:2169–2187. [DOI] [PubMed] [Google Scholar]
- Sans N, Prybylowski K, Petralia RS, Chang K, Wang YX, Racca C, Vicini S, Wenthold RJ (2003). NMDA receptor trafficking through an interaction between PDZ proteins and the exocyst complex. Nat Cell Biol 5:520–530. [DOI] [PubMed] [Google Scholar]
- Scheiffele P (2003). Cell-cell signaling during synapse formation in the CNS. Annu Rev Neurosci 26:485–508. [DOI] [PubMed] [Google Scholar]
- Shah BS, Rush AM, Liu S, Tyrrell L, Black JA, Dib-Hajj SD, Waxman SG (2004). Contactin associates with sodium channel Nav1.3 in native tissues and increases channel density at the cell surface. J Neurosci 24:7387–7399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silletti S, Mei F, Sheppard D, Montgomery AM (2000). Plasmin-sensitive dibasic sequences in the third fibronectin-like domain of L1-cell adhesion molecule (CAM) facilitate homomultimerization and concomitant integrin recruitment. J Cell Biol 149:1485–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasu MA, Dalva MB, Zigmond RE, Greenberg ME (2002). Modulation of NMDA receptor-dependent calcium influx and gene expression through EphB receptors. Science 295:491–495. [DOI] [PubMed] [Google Scholar]
- Tolias KF, Bikoff JB, Burette A, Paradis S, Harrar D, Tavazoie S, Weinberg RJ, Greenberg ME (2005). The Rac1-GEF Tiam1 couples the NMDA receptor to the activity-dependent development of dendritic arbors and spines. Neuron 45:525–538. [DOI] [PubMed] [Google Scholar]
- Wenthold RJ, Prybylowski K, Standley S, Sans N, Petralia RS (2003). Trafficking of NMDA receptors. Annu Rev Pharmacol Toxicol 43:335–358. [DOI] [PubMed] [Google Scholar]
- Whitford KL, Marillat V, Stein E, Goodman CS, Tessier-Lavigne M, Chedotal A, Ghosh A (2002). Regulation of cortical dendrite development by Slit-Robo interactions. Neuron 33:47–61. [DOI] [PubMed] [Google Scholar]
- Yamagata M, Sanes JR, Weiner JA (2003). Synaptic adhesion molecules. Curr Opin Cell Biol 15:621–632. [DOI] [PubMed] [Google Scholar]