Abstract
Drosophila phototransduction serves as a model for phosphoinositide (PI) signaling and for characterizing the mechanisms regulating transient receptor potential (TRP) channels in vivo. Activation of TRP and TRP-like (TRPL) requires hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2), resulting in the generation of inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG). Although a role for IP3 has been excluded, TRP channels have been proposed to be activated by either a reduction of inhibitory PIP2 or production of DAG/polyunsaturated fatty acids. Here, we characterize a protein, phosphatidylinositol synthase (dPIS), required for a key step during PIP2 regeneration, the production of phosphatidylinositol. Overexpression of dPIS suppressed the retinal degeneration resulting from two other mutations affecting PIP2 cycling, rdgB (retinal degeneration B) and cds (CDP-diacylglycerol synthase). To characterize the role of dPIS, we generated a mutation in dpis, which represented the first mutation in a gene encoding a PI synthase in an animal. In contrast to other mutations that reduce PIP2 regeneration, the dpis1 mutation eliminated all PI synthase activity in flies and resulted in lethality. In mosaic animals, we found that dPIS was essential for maintaining the photoresponse. Because the dpis1 mutation eliminates production of an enzyme essential for PIP2 regeneration, our data argue against activation of TRP and TRPL through a reduction of inhibitory PIP2.
Keywords: TRP channels, phototransduction, Drosophila, calcium, retinal degeneration, PIP2
Introduction
Phosphoinositides (PIs) regulate a diversity of events ranging from sensory signaling to cell proliferation, actin organization, membrane trafficking, cell differentiation, and cell migration (for review, see Downes et al., 2005; Niggli, 2005). A pivotal PI is phosphatidylinositol 4,5-bisphosphate (PIP2), because it directly modulates a host of effectors and is the substrate for phospholipase C (PLC), which leads to production of the second messengers, inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG). A group of cation channels that depend on PLC stimulation are classical transient receptor potential (TRPC) proteins (for review, see Montell, 2005). In vitro studies indicate that TRPC channels are activated by DAG, IP3-induced Ca2+ release, or reductions in levels of inhibitory PIP2. However, the mechanisms that regulate these channels in vivo are poorly understood.
A model for characterizing a PI signaling cascade coupled to TRP channel activation is Drosophila phototransduction. The cascade is initiated by light activation of rhodopsin (Rh), engagement of a Gq, and stimulation of the PLC encoded by norpA (no receptor potential A) (for review, see Montell, 1999). Fly phototransduction culminates with the opening of the TRP and TRPL channels. Despite the established requirement for norpA (Bloomquist et al., 1988), the mechanism through which PLC activates these channels is controversial. Although roles for IP3 or IP3-mediated Ca2+ release have been excluded (Acharya et al., 1997; Hardie and Raghu, 1998; Raghu et al., 2000b), either DAG or its metabolites, such as polyunsaturated fatty acids (PUFAs), are proposed to activate the channels (Chyb et al., 1999; Raghu et al., 2000a). Alternatively, PLC activity could promote TRP activation by relieving PIP2-mediated inhibition of the channels. A variation of these models is that a decline in PIP2 activates the channels in combination with a rise in DAG or PUFAs (Estacion et al., 2001; del Pilar Gomez and Nasi, 2005).
Sustained exposure to agonists coupled to PLC activity potentially can lead to depletion of PIP2 and loss of responsiveness. Therefore, mechanisms that promote the regeneration of PIP2 are critical for maintaining signaling capacity. Mutations in three Drosophila proteins that impinge on PIP2 regeneration have been described (see Fig. 1A). These include an enzyme, retinal degeneration A (RDGA), that catalyzes the phosphorylation of DAG and production of phosphatidic acid (PA) (Masai et al., 1993). However, rdgA does not eliminate PA because PA is also generated through phospholipase D-dependent hydrolysis of phosphatidlylcholine (LaLonde et al., 2005). A mutation disrupting the CDP-diacylglycerol synthase (CDS), required for the subsequent conversion of PA to CDP–DAG, has been reported to affect phototransduction but not development or morphogenesis (Wu et al., 1995). Because CDS is the only CDP-diacylglycerol synthase predicted in Drosophila, the questions arise as to whether the cds1 mutation eliminates CDP–DAG synthase activity and the effects on phototransduction resulting from a profound reduction in PIP2 regeneration. Mutations in the PI transfer protein RDGB cause severe retinal degeneration (Harris and Stark, 1977; Vihtelic et al., 1993). Although the precise role of RDGB is not known, it is localized to the base of the rhabdomeres and the underlying submicrovillar cisternae, which is a specialized region of the endoplasmic reticulum in which the early events in PIP2 regeneration are thought to occur (Vihtelic et al., 1993; Suzuki and Hirosawa, 1994). As such, it may be involved in the transfer of PIs from the submicrovillar cisternae to the rhabdomeral membranes.
Figure 1.
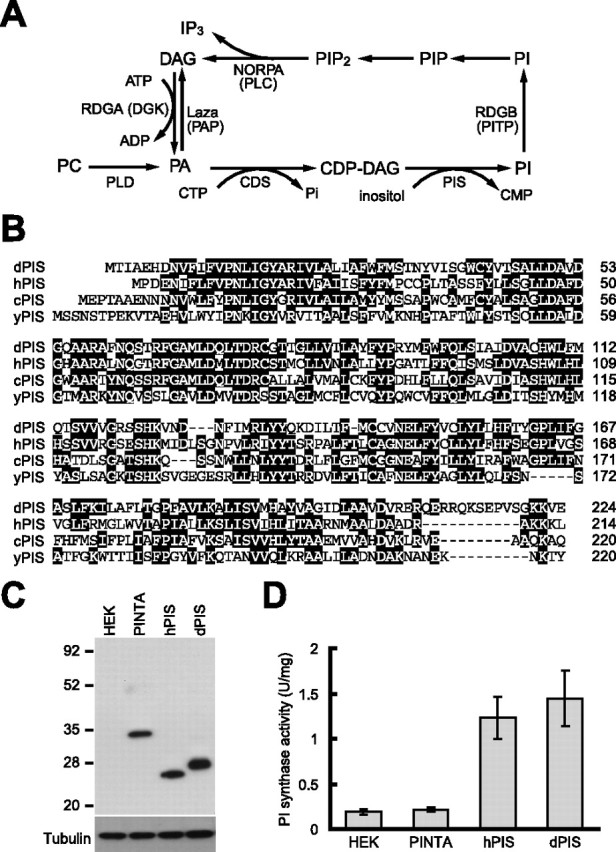
dPIS is a Drosophila phosphatidylinositol synthase. A, The PIP2 regeneration cycle that operates in fly phototransduction. During light stimulation, the NORPA PLC hydrolyzes PIP2 to yield DAG and IP3. DAG is phosphorylated by the RDGA DAG kinase (DGK) to produce PA. CDS adds CMP to PA to generate CDP–DAG. PIS catalyzes the incorporation of inositol and CDP–DAG to produce PI. The RDGB PI-transfer protein (PITP) is proposed to promote the transfer of PIs from the submicrovillar cisternae to the base of the rhabdomeres (Vihtelic et al., 1993; Suzuki and Hirosawa, 1994), in which the PI is phosphorylated by a PI kinase to generate PIP, which in turn is phosphorylated by a PIP kinase to yield PIP2. A second two-step pathway for generation of DAG is shown. This pathway involves the phospholipase D (PLD)-stimulated conversion of phosphatylcholine (PC) to PA, followed by the Lazaro PA phosphatase (PAP)-promoted dephosphorylation of PA to generate DAG. B, Alignment of dPIS, hPIS, C. elegans PIS (cPIS), and yeast PIS (yPIS; Saccharomyces cerevisiae) protein sequences. Identical residues, found in at least two proteins, are highlighted by the black boxes, and the running tallies of amino acids are indicated to the right. C, Expression of dPIS and hPIS in HEK293 cells. The cells were transfected with cDNAs encoding PINTA (Wang and Montell, 2005), hPIS (Lykidis et al., 1997), or dPIS fused to C-terminal Myc tags. Extracts were prepared from untransfected cells (HEK) and from the transfected cells, and a Western blot was probed with anti-Myc antibodies and reprobed with anti-tubulin antibodies. D, PI synthase activities in untransfected HEK293 cells or cells expressing Myc-tagged versions of PINTA, dPIS, or hPIS. The error bars indicate SDs (n = 3).
Here, we report the characterization of the Drosophila PI synthase (dPIS), which functions directly downstream of CDS in the conversion of CDP–DAG to PI. Overexpression of dPIS increased PI synthase activity and suppressed the retinal degeneration in cds and rdgB mutant flies. A dpis1 loss-of-function mutation eliminated PI synthase activity and caused lethality. We rescued dpis activity during development and found that dPIS had an activity-dependent role in photoreceptor cells. dPIS was required to prevent retinal degeneration but only in the presence of light. During short light exposures, dPIS was dispensable but was essential for maintaining the response during sustained light exposures. Because dpis1 disrupts an essential step in PIP2 regeneration and causes loss of the light response, these results support the model that depletion of PIP2 alone does not lead to activation of the TRP and TRPL channels.
Materials and Methods
Fly stocks.
The following stocks were obtained from the Bloomington Stock Center: (1) y w; P[70FLP]11 P[70I-SceI]2B nocSco/CyO, (2) y w1118 P[neoFRT]19A; P[ey-FLP.N]5, and (3) w1118 sn3 P[neoFRT]19A. The rdgBK222 flies (Harris and Stark, 1977) were obtained from Dr. W. Stark (Saint Louis University, St. Louis, MO), and the cds1 flies (Wu et al., 1995) were obtained from Dr. C. Zuker (University of California at San Diego, La Jolla, CA).
Generation of the dpis1 mutation.
We produced the dpis1 mutation using ends-out homologous recombination (Gong and Golic, 2003). Two genomic fragments (3.0 and 3.3 kb) were subcloned into the pw35 plasmid such that they were separated by the w marker gene. The 3.0 kb fragment (extending from −3140 to −112 bp) was subcloned between the SphI and NotI sites and the 3.3 kb fragment, which extended from nucleotide +406 (downstream of the start codon of dpis), was inserted into the BamHI site (see Fig. 4A). The targeting construct (pw35–dpis) was injected into w1118 embryos, and transformants were identified on the basis of eye pigmentation. Flies containing the targeting construct (P[pw35–dpis]) on the second chromosome were crossed to y w; P[70FLP]11 P[70I-SceI]2B nocSco/CyO flies carrying the transgenes encoding the FLP and I-SceI enzymes. The progeny were heat shocked at 37°C for 1 h during the first-instar larval stage, and, ∼10 d later, the adult females were crossed to w1118 males. The subsequent w+ progeny were screened by PCR to identify flies with precise gene targeting into the dpis gene. Confirmation that the phenotypes observed in the dpis1 mutant flies were attributable to the gene targeting was obtained by rescuing the phenotypes with a wild-type dpis transgene.
Figure 4.

dPIS is required for photoreceptor cell survival. A, Scheme for generation of the dpis knock-out by gene targeting. The organization of the dpis locus (top), the targeting construct (middle), and the expected structure of the targeted gene (bottom) are shown. A scale bar (kilobases) and the positions of the DNA primers used for PCR (short horizontal lines with arrowheads) are indicated. B, PCR product obtained on successful gene targeting. Shown are the results of PCR reactions using the primers indicated in A and genomic DNAs prepared from wild-type (wt), P[pw35–dpis], and dpis1 flies. The positive PCR product is 3.8 kb and a 1 kb DNA size ladder (SM0313; Fermentas Life Sciences, Burlington, Ontario, Canada) is shown to the left. The bottom shows the 4.2 kb product generated using primers to the trp gene and the same genomic DNAs used as in the top. C–F, Scanning electron micrographs of compound eyes. C, The dpis+ flies were of the following genotype: GMR-hid P[neoFRT]19A/P[neoFRT]19A;EGUF. D, The dpis1 flies were of the following genotype: GMR-hid P[neoFRT]19A/dpis1 P[neoFRT]19A;EGUF. E, dpis1;P[dpis]. F, dpis1;P[hpis]. G, H, Transmission EMs of cross sections from the distal region of the compound eyes. G, The dpis+ flies were of the following genotype: GMR-hid P[neoFRT]19A/P[neoFRT]19A;EGUF. Scale bar, 10 μm. H, The dpis1 flies were of the following genotype: GMR-hid P[neoFRT]19A/dpis1 P[neoFRT]19A;EGUF.
Generation of transgenic flies.
The dpis and human pis (hpis) cDNAs (Expressed Sequence Tag clones RH33540 and 5753633, respectively) were expressed under control of the dpis promoter by fusing the cDNAs at the 5′ end to a 2.0 kb dpis genomic region (−2100 to −10 bp 5′ to the initiator codon). These dpis promoter–cDNA constructs were subcloned into the pCaspeR4 vector (Thummel and Pirrotta, 1992) between the XhoI and XbaI sites.
To express dPIS under the control of the ninaE (neither inactivation nor afterpotential E) promoter, the dpis cDNA was subcloned between NotI and XbaI sites of the pCNX vector. The pCNX vector was constructed as follows. First, the pNX vector (provided by Dr. J. O'Tousa, University of Notre Dame, Notre Dame, IN) was digested with KpnI and NotI, which liberated a 3 kb ninaE promoter sequence, and this fragment was subsequently introduced between the same sites of pCaspeR4. Second, to provide the signals for polyadenylation and 3′ end cleavage of RNAs, a simian virus 40 3′ region (0.5 kb) was excised from pCaspeR–heat shock protein 70 promoter (hs) and introduced between the XbaI and XhoI sites of the vector.
To express dPIS under the control of the heat shock protein 70 promoter, the sequence encoding a Myc tag was fused to the 3′ end of the dpis cDNA, which was subcloned between the NotI and XbaI sites of pCaspeR–hs (Thummel and Pirrotta, 1992). The constructs were injected into of w1118 embryos, and transformants were identified on the basis of eye color.
Heat shock regimens.
The dpis1;P[hs–dpis] flies were subjected to daily heat-shock treatments for 30–45 min at 37°C beginning during the embryonic stage and ending during the late pupal period. To maintain dPIS protein in adults, the dpis1;P[hs–dpis] flies were heat shocked daily for 30 min at 37°C.
Generation of recombination proteins and anti-dPIS antibodies.
To generate anti-dPIS antibodies, a cDNA fragment encoding the C-terminal end of dPIS (residues 191–224) was subcloned into the pGEX5X-1 vector (GE Healthcare, Little Chalfont, UK). The glutathione S-transferase fusion protein was expressed in Escherichia coli BL21 codon-plus (Stratagene, La Jolla, CA), purified using glutathione agarose beads (GE Healthcare), and introduced into rabbits (Covance, Princeton, NJ).
PI synthase and CDP–DAG synthase assays.
To express dPIS in vitro, the full-length dpis and hpis cDNAs were subcloned into the pcDNA3–Myc vector (constructed by inserting the sequences encoding the 9E11 Myc tag between the BamHI and XbaI sites). The constructs were transfected into HEK293 cells using Fugene 6 reagents (Roche, Indianapolis, IN). At 48 h after transfection, the cells were isolated and ruptured in PBS buffer plus 1% Triton X-100. To prepare extracts from fly heads and bodies, the tissues were homogenized with Pellet Pestles (Kimble/Kontes, Vineland, NJ) in PBS buffer plus 1% Triton X-100.
The PI synthase assay was performed essentially as described previously (Carman and Fischl, 1992). Briefly, the assay was conduced in 100 μl total volume containing 0.2 mm CDP–DAG, 0.5 mm myo-[3H]inositol (5000 cpm/nmol), 2 mm MnCl2, 50 mm Tris-HCl, pH 8.0, 0.15% Triton X-100, and 50 μg of total protein isolated either from tissue culture cells or fly heads or bodies. After 1 h incubation at 37°C, the reaction was terminated by adding 0.35 ml of methanol (in 0.1N HCl). A phase separation was obtained by adding 0.5 ml of chloroform and 0.5 ml of 1 m MgCl2, and the radioactivity in the organic phase was measured using a scintillation counter. One unit of PI synthase activity is defined as the amount of enzyme that catalyzes the formation of 1 nmol of PI/min.
The CDP–DAG synthase activity was measured by generation of CDP–DAG in 50 mm Tris-HCl, pH 8.0, 20 mm MgCl2, 1% Triton X-100, 1.0 mm [3H]CTP (2000 cpm/nmol), 0.75 mm phosphatidic acid, and 50 μg of total protein in a volume of 100 μl at 37°C for 1 h (Nigou and Besra, 2002). The reactions were terminated by addition of 0.7 ml of chloroform/methanol/water (10:10:3). Immediately thereafter, we added 0.3 ml of chloroform and 0.3 ml of water, and the organic phase was removed and washed twice in chloroform/methanol/water (3:47:48). The radioactivity in the organic phase was measured by scintillation counting. One unit of CDS activity is defined as the amount of enzyme that catalyzes the formation of 1 nmol of CDP–DAG per minute.
Immunolocalizations and Western blots.
To perform immunolocalization experiments, the adult fly heads were hemisected, fixed in paraformaldehyde, and embedded in LR White resin as described previously (Porter et al., 1992). Cross sections (0.5 μm) of compound eyes were obtained from the distal region of the retina, which included the R7 cells, and were stained with primary antibodies (1:200) and one of the following secondary antibodies (1:200): goat anti-rabbit IgG (Alexa Fluor 568) or goat anti-mouse IgG (Alexa Fluor 488).
To perform Western blots, fly heads and bodies were homogenized in SDS sample buffer with a Pellet Pestle (Kimble/Kontes). The proteins were fractionated by SDS-PAGE and transferred overnight at 25 V to Immobilon-P transfer membranes (Millipore, Bedford, MA) in Tris-glycine buffer. The blots were probed with rabbit anti-dPIS primary antibodies, rabbit anti-Myc antibodies (Santa Cruz Biotechnology, Santa Cruz, CA), mouse anti-Rh1 antibodies (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), or mouse anti-α-tubulin antibodies (Developmental Studies Hybridoma Bank) and then subsequently with anti-rabbit IgG peroxidase conjugate (Sigma, St. Louis, MO) or anti-mouse IgG peroxidase conjugate (Sigma). The signals were detected using ECL reagents (GE Healthcare).
PIS antibody depletion assays.
The dPIS polyclonal antibodies (described above) were cross linked to cyanogen bromide-activated Sepharose 4 Fast Flow (GE Healthcare) to generate anti-dPIS Sepharose. Fly tissue extracts (prepared as described above) were flowed over the anti-dPIS Sepharose at least three times, and the effectiveness of the procedure was monitored by probing Western blots with anti-dPIS antibodies.
Electroretinogram recordings.
Electroretinogram (ERG) recordings were performed as described previously (Wes et al., 1999). Briefly, two glass microelectrodes filled with Ringer's solution were inserted into small drops of electrode cream placed on the surfaces of the compound eye and the thorax. A Newport light projector (model 765) was used for stimulation. The ERGs were amplified with a Warner Instruments (Hamden, CT) electrometer IE-210 and recorded with a MacLab/4 s analog-to-digital converter and the Chart version 3.4/s program (ADInstruments, Colorado Springs, CO). All recordings were performed at room temperature.
Transmission electron microscopy.
Heads were dissected from flies reared under a 12 h light/dark cycle or in constant darkness fixed in glutaraldehyde and embedded in LR White resin as described previously (Porter et al., 1992). Thin sections (85 nm) prepared at a depth of 30 μm were examined by transmission electron microscopy (EM) using a Zeiss (Oberkochen, Germany) FEI Tecnai 12 electron microscope. The images were acquired using a Gatan (Pleasanton, CA) camera (model 794) and Gatan Digital Micrograph software and converted into tiff files.
Results
CG9245 is a dPIS
To identify a candidate Drosophila PI synthase that functions in the regeneration of PIP2 during phototransduction (Fig. 1A), we queried the Drosophila sequence database for predicted PIS proteins. One Drosophila gene, CG9245, encoded a putative protein, which was 53% identical to the human PIS (Lykidis et al., 1997), 47% identical to a possible Caenorhabditis elegans PIS (National Center for Biotechnology Information accession number CAB60350), and 36% identical to the yeast PIS (Nikawa et al., 1987b) (Fig. 1B). We refer to the Drosophila protein as dPIS.
Based on our previous microarray analysis, which compared the RNA expression of genes in wild-type heads and the heads of a mutant missing eyes (sine oculis) (Xu et al., 2004), it appeared that expression of dpis was slightly eye enriched (wild-type head/sine oculis head ratio was 2.4 and wild-type head/wild-type body ratio was 3.8). Modest levels of eye enrichment are also characteristic of three other Drosophila genes that function in the regeneration of PIP2 (Xu et al., 2004). These include the genes encoding a DAG kinase (RDGA; wild-type head/sine oculis head ratio is 2.8) (Hotta and Benzer, 1970; Harris and Stark, 1977), CDP–DAG synthase (CDS; wild-type head/sine oculis head ratio is 3.0) (Wu et al., 1995), and a PI transfer protein (RDGB; wild-type head/sine oculis head ratio is 1.2) (Harris and Stark, 1977; Vihtelic et al., 1993).
To test whether dPIS had PI synthase activity, we expressed the protein in HEK293 cells and performed PIS assays. In addition to Myc-tagged dPIS, we expressed a Myc-tagged retinal binding protein PINTA (PDA is not apparent)(Wang and Montell, 2005) as a negative control, and a Myc-tagged hPIS as a positive control (Fig. 1C). In whole-cell extracts, dPIS increased total cellular PI synthase activity to a similar extent as hPIS (Fig. 1D) (7.6- and 6.5-fold respectively; mock-transfected HEK293 cells, 0.19 ± 0.3 U/mg; dPIS, 1.45 ± 0.2 U/mg; hPIS, 1.23 ± 0.21 U/mg; n = 3), whereas expression of PINTA did not affect PI synthase activity (Fig. 1D) (PINTA, 0.23 ± 0.02 U/mg). Therefore, we propose that dPIS is a bona fide PI synthase required for a key step (conversion of CDP–DAG to PI) necessary for the regeneration of PIP2 from DAG.
Enrichment of dPIS in photoreceptor cell bodies
To address whether dPIS is a candidate for functioning in the PI cycle during phototransduction, we examined whether the dPIS protein is expressed in photoreceptor cells. Drosophila compound eyes consist of ∼800 repetitive units, referred to as ommatidia, each of which contains 20 cells, including seven photoreceptor cells in any plane of section. Each photoreceptor cell contains a microvillar structure, the rhabdomere, which is the site for light reception and phototransduction. Although proteins that function directly in phototransduction are found primarily in the rhabdomeres (Montell, 1999), those that participate in PIP2 cycling appear to be localized to the extra-rhabdomeral cell bodies (Vihtelic et al., 1993; Suzuki and Hirosawa, 1994; Masai et al., 1997).
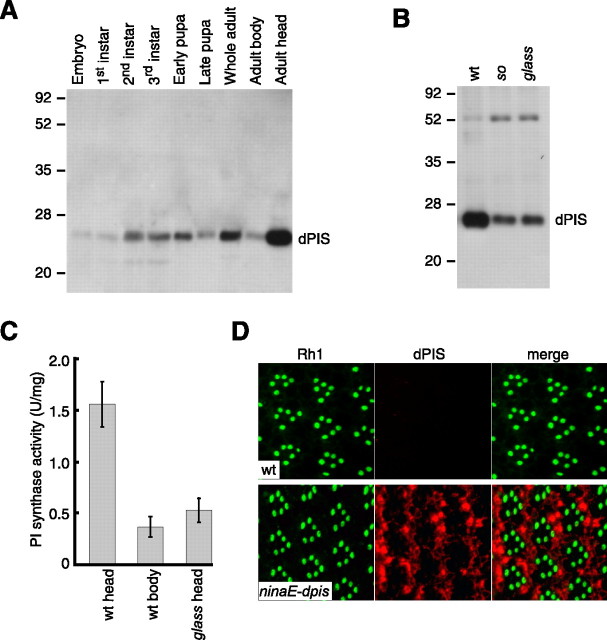
To examine the expression pattern of the dPIS protein, we used a combined biochemical and genetic approach. We generated anti-dPIS polyclonal antibodies (see Materials and Methods), which recognized a polypeptide in wild-type adult heads (Fig. 2A) that matched the predicted molecular weight of 26 kDa. The dPIS protein was expressed throughout the fly's lifecycle and at higher levels in wild-type heads than in wild-type bodies (Fig. 2A) (supplemental Fig. 1A, available at www.jneurosci.org as supplemental material). Expression of dPIS was decreased in sine oculis heads (Fig. 2B) (supplemental Fig. 1B, available at www.jneurosci.org as supplemental material), indicating that dPIS was enriched in the compound eyes. These data were consistent with microarray analyses demonstrating that dpis mRNA was present at higher levels in heads than bodies (wild-type head/wild-type body ratio of 3.8) (Xu et al., 2004); within the head, dpis mRNA was moderately enriched in the eyes (wild-type head/sine oculis head ratio of 2.4).
Figure 2.
Expression and localization of dPIS. A, Developmental Western blot probed with anti-dPIS antibodies. Samples were prepared from the indicated developmental stages. Molecular weight markers in kilodaltons are indicated to the left. The membrane was stained with Ponceau S to compare the relative concentrations of proteins loaded in each lane (supplemental Fig. 1A, available at www.jneurosci.org as supplemental material). B, Expression of dPIS is enriched in photoreceptor cells. The Western blot, which contained extracts prepared from the heads of wild-type, sine oculis (so; flies missing eyes), and glass (missing photoreceptor cells), was probed with anti-dPIS antibodies. The membrane was subsequently stained with Ponceau S (supplemental Fig. 1B, available at www.jneurosci.org as supplemental material). C, PI synthase activity is enriched in photoreceptor cells. PI synthase activity was assayed using extracts prepared from wild-type heads, wild-type bodies, and glass heads. The error bars indicate SDs (n = 4). D, Localization of dPIS in the cell bodies of the photoreceptor cells. Cross sections from the distal regions of compound eyes prepared from wild-type (wt) and from transgenic flies expressing increased levels of dpis under the control of the ninaE promoter (ninaE–dpis) were stained with mouse anti-Rh1 (green) and rabbit anti-dPIS (red). Merged images are included to the right.
Two lines of evidence indicate that dPIS was expressed in photoreceptor cells. First, expression of dPIS was reduced in glass mutant heads, which are missing photoreceptor cells (Fig. 2B) (supplemental Fig. 1B, available at www.jneurosci.org as supplemental material) (Moses et al., 1989). Within the eye, dPIS appeared to be primarily expressed in photoreceptor cells because the concentration of the protein was similar in sine oculis and glass mutant heads (Fig. 2B) (supplemental Fig. 1B, available at www.jneurosci.org as supplemental material). Second, we compared PI synthase levels in wild-type heads/bodies and glass mutant heads and found that the PI synthase activity was 4.3-fold higher in wild-type heads than in wild-type bodies (Fig. 2C) (wild-type heads, 1.55 ± 0.22 U/mg; wild-type bodies, 0.36 ± 0.10 U/mg; n = 4) and 3.0-fold higher in wild-type heads than in glass heads (Fig. 2C) (glass, 0.52 ± 0.12 U/mg; n = 4).
We were not able to examine the subcellular distribution of dPIS in wild-type photoreceptor cells, because the antibodies did not detect a signal on sections of compound eyes. Because this may have been attributable to relatively low protein levels, we introduced a transgene into flies (P[ninaE–dpis]) in which the dpis coding region was fused to the ninaE (rh1) promoter, which directs strong expression in the six outer photoreceptor cells (Mismer and Rubin, 1987). In contrast to the Rh1 protein that localized exclusively to the rhabdomeres, we found that dPIS was present only in the photoreceptor cell bodies (Fig. 2D). Evidence that the dPIS protein expressed in P[ninaE–dpis] flies is functional is presented below.
Overexpression of dPIS suppressed retinal degeneration in cds and rdgB
If dPIS functions in PIP2 regeneration, then overexpression of dPIS potentially could suppress the phenotypes resulting from mutations disrupting other steps in the PIP2 cycle. To conduct these experiments, we used the P[ninaE–dpis] flies, which overexpressed dPIS ∼12-fold (Fig. 3A). The overexpressed dPIS appeared to be functional because the increase in PI synthase activity in ninaE–dpis fly heads paralleled the elevation in protein levels (Fig. 3B) (11.5-fold; wild-type, 1.67 ± 0.22 U/mg; ninaE–dpis, 19.2 ± 3.6 U/mg). To test for genetic interactions between dpis and other genes required for PIP2 cycling, we introduced the P[ninaE–dpis] transgene into cds1 flies, which contain a mutation in the enzyme functioning in the step directly upstream of PI synthase (Fig. 1A) (Wu et al., 1995). We also introduced the transgene into the rdgBKS222 mutant, which has a defect in a PI transfer protein (Harris and Stark, 1977; Vihtelic et al., 1993) and therefore might reduce the movement of PI into the rhabdomeres (Fig. 1A). Thus, the RDGB protein would be predicted to function subsequent to dPIS in PIP2 cycling (Fig. 1A).
Figure 3.

Overexpression of dPIS rescued the retinal degeneration caused by either the rdgBKS222 or cds1 mutations. A, Western blot showing overexpression of dPIS in P[ninaE–dpis] flies. Extracts from the heads of wild-type and transgenic flies were fractionated by SDS-PAGE, and a Western blot was probed with anti-dPIS antibodies and reprobed with anti-tubulin antibodies. B, PI synthase activity was elevated in ninaE–dpis heads compared with wild-type heads. The error bars represent the SDs (n = 4). C, D, Suppression of retinal degeneration in rdgBKS222 and cds1 by overexpression of dPIS (as a result of introduction of the ninaE–dpis transgene). The time course of retinal degeneration was assayed using the optical neutralization technique. Flies were reared under a 12 h light/dark cycle. Each point is based on ≥60 ommatidia from three separate flies. The error bars indicate SDs. E, EM images of 25-d-old wild-type (wt) and ninaE–dpis flies, 6-d-old rdgBKS222 and rdgBKS222;ninaE–dpis flies, and 5-d-old cds1 and ninaE-dpis;cds1 flies. All of the flies were raised under a 12 h light/dark cycle. Scale bar, 2 μm.
To address whether PIS may function in the same pathway as CDS and RDGB, we tested whether overexpression of dPIS suppressed the retinal degeneration resulting from mutations in rdgB and cds (Harris and Stark, 1977; Wu et al., 1995). Flies were maintained under a 12 h light/dark cycle, and retinal degeneration was assayed in intact fly heads using the optical neutralization technique (Franceschini and Kirschfeld, 1971). In wild-type ommatidia, seven rhabdomeres were detected regardless of the age of the flies (Fig. 3C), whereas rdgBKS222 flies underwent age-dependent loss of the rhabdomeres (Fig. 3C). However, rdgBKS222;ninaE–dpis showed a dramatic reduction in the rate of degeneration compared with rdgBKS222 (Fig. 3C). A reduction in the rate of retinal degeneration attributable to the cds1 mutation was also observed in ninaE–dpis;cds1 flies (Fig. 3D).
The suppression of degeneration in the rdgBKS222 and cds1 retinas by ninaE–dpis was examined further by transmission EM. The eyes from wild-type and ninaE–dpis flies contained intact ommatidia with seven rhabdomeres even after 25 d under a light/dark cycle (Fig. 3E). Few rhabdomeres were left after 6 d in rdgBKS222 flies (Fig. 3E). In contrast, the retinal degeneration was greatly suppressed in rdgBKS222;ninaE–dpis flies (Fig. 3E). Almost no rhabdomeres were observed by transmission EM of cds1 ommatidia after 5 d under a light/dark cycle, whereas ommatidia from ninaE–dpis;cds1 flies contained several rhabdomeres (Fig. 3E). The partial suppression of the retinal degeneration phenotypes in the cds1 and rdgBKS222 mutants, by overexpression of dPIS, is consistent with the predicted function of dPIS as a PI synthase and the biochemical evidence that dPIS has such activity.
Mutation of dpis caused embryonic lethality and absence of photoreceptor cells in mosaic adults
To characterize the requirement for dpis for phototransduction, we generated a mutation in the dpis gene by homologous recombination (Gong and Golic, 2003) (see Materials and Methods) (Fig. 4A). We screened for fly lines containing a disruption of the dpis locus by PCR using primers corresponding to the w marker gene and the genomic region outside of the targeting construct (Fig. 4A). A PCR product was detected only in the dpis1 mutant line but not in wild-type flies or in the original transgenic line, P[pw35–dpis], which was used to mobilize the knock-out construct (Fig. 4B). The dpis1 mutation represents the first disruption of a pis gene in animals, although a mutation in a yeast pis has been described previously (Nikawa et al., 1987a).
The dpis1 mutation caused embryonic lethality, and introduction of either the dpis or the hpis gene expressed under the control of the dpis promoter (P[dpis] and P[hpis], respectively) rescued the lethality of dpis1 (data not shown). Therefore, to assess the role of dpis for the visual response, we generated mosaic animals using a mitotic recombination approach that leads to the generation of fully homozygous mutant eyes in otherwise heterozygous animals (Stowers and Schwarz, 1999). The eyes in the dpis1 mosaic flies displayed a rough eye morphology and were reduced in size (Fig. 4D) compared with dpis+ control flies (Fig. 4C). Furthermore, no photoreceptor cells were detected in the mutant ommatidia (Fig. 4G,H). The abnormal morphology of the dpis1 eyes was rescued by either the Drosophila or human pis transgenes (Fig. 4E,F). The rescue by the human pis transgene was complete, because the ERG response in dpis1;P[hpis] flies was also indistinguishable from wild type (supplemental Fig. 2, available at www.jneurosci.org as supplemental material). These results indicated that PI synthase activity was required during development of the compound eye.
dPIS required for maintaining the light response and rapid recovery
To assess the requirement for dpis for the light response, we generated dpis1 mutant flies that formed anatomically normal photoreceptor cells by supplying dpis function during development. To obtain dpis1 mutant flies with photoreceptor cells, we expressed a Myc-tagged version of dPIS in dpis1 flies under control of the heat shock protein 70 promoter (hs–dpis). Expression of the dpis gene was induced between the embryonic and the late pupal stages by daily heat-shock treatments. To assess the level of dPIS–Myc expression in adult dpis1 flies, we performed Western blots using anti-PIS antibodies. We found that dPIS–Myc was detected in dpis1;P[hs–dpis] flies immediately after eclosion (D0), although at a lower level than dPIS in wild-type flies (Fig. 5A). We then incubated the dpis1;P[hs–dpis] flies in the dark without heat-shock treatments. By 7 d after eclosion (D7), no dPIS–Myc protein was detected on Western blots (Fig. 5A), and a full complement of photoreceptor cells was still present (Fig. 6A,B).
Figure 5.

dPIS is required for phototransduction. A, The dPIS protein is not detected in 7-d-old dpis1;P[hs–dpis] flies, which were heat shocked during development. The dpis1;P[hs–dpis] flies underwent daily heat shock before eclosion, resulting in expression of dPIS–Myc. After eclosion, the flies were maintained in the dark at 25°C and were not exposed further to heat shock treatments. The extracts used for the Western blots were prepared from wild-type (wt) flies and dpis;P[hs–dpis] flies after the following times after eclosion: (1) 12 h (D0), (2) 3 d (D3), and (3) 7 d (D7). The Western blot was probed with anti-dPIS antibodies and reprobed with anti-Rh1 antibodies. Molecular weight markers (kilodaltons) are indicated to the left. B–E, ERGs of wild-type as well as D0, D3, and D7 dpis1;P[hs–dpis] flies. The flies were dark adapted for 2 min and then exposed to three pulses of orange light (∼1800 lux): P1 (5 s), P2 (30 s), and P3 (5 s). The maintained component (mc) and on- and off-transients are indicated. F, Comparison of the P1 and P3 ERG amplitudes from B–E (n ≥7; error bars represent SDs). G–J, ERGs of wild-type as well as D0, D3, and D7 dpis1;P[hs–dpis] flies after 2 min exposure to orange light and a 3 s dark adaptation. K, Quantification of the ERG amplitudes. The flies were light adapted for 2 min (∼1800 lux) and then dark adapted for the times indicated. The amplitudes are indicated in millivolts ± SDs (n ≥8).
Figure 6.

Light-dependent retinal degeneration in dpis1 flies. A, The time course of retinal degeneration in dpis1;P[hs–dpis] flies assayed using the optical neutralization technique. The dpis1;P[hs--dpis] flies were heat shocked until eclosion and reared in the dark at 25°C without heat shock for 7 d after eclosion. The 7-d-old dpis1;P[hs–dpis] flies were maintained under a 12 h light/dark cycle either without heat-shock treatment or with daily 30 min heat-shock treatment at 37°C. The error bars indicate SDs (n ≥50 ommatidia from ≥2 flies each). B–E, Transmission EM images of cross sections from dpis;P[hs–dpis] flies. All of the flies were heat shocked during development and then maintained for 7 d in the dark after eclosion without heat-shock treatments. The flies were then either immediately examined by transmission EM (B) or subsequently treated as follows before the EM analyses: C, days 7–14 in the dark without heat shock; D, days 7–14 under a 12 h light/dark cycle without heat-shock treatments; E, days 7–14 under a 12 h light/dark cycle with daily heat-shock treatments. Scale bar, 2 μm.
We examined the consequences on the photoresponse resulting from reducing the levels of dPIS in dpis1;P[hs–dpis] flies, by performing ERG recordings. This assay, which measures the summed responses of all retinal cells to light, results in two discernible light-induced components. These include a maintained corneal negative component (Fig. 5B) that arises primarily from phototransduction, and on- and off-transients that are attributable to light-induced activity in the second-order neurons (Fig. 5B) (on-transients are not consistently observed as a result of limitations in the sampling rate).
We found that dPIS was required for a sustained light response and for rapid recovery. In wild-type or newly eclosed dpis1;P[hs–dpis] flies (D0), the amplitudes of the ERG were similar during exposure to successive pulses (P) of light, including a long 30 s pulse (Fig. 5B,C, compare P1, P2, and P3 amplitudes). In D3 dpis1;P[hs–dpis] flies, the maintained component declined slightly during the 30 s light stimulation (Fig. 5D, P2). In addition, the ERG amplitude resulting from exposure to the subsequent 5 s light pulse was decreased compared with the first 5 s light pulse (Fig. 5D,F, compare P1 and P3 amplitudes). The reduction in the light response was most obvious in D7 dpis1;P[hs–dpis] flies, which declined to baseline during the 30 s pulse of light (Fig. 5E, P2). A very small photoresponse was observed in these flies during exposure to the third light pulse (Fig. 5E,F, P3). These data indicated that there was a defect in sustaining and recovery of the photoresponse in dpis1 flies.
To assay the rate of recovery, we exposed the flies to light for 2 min and then performed ERG recordings after dark adapting the flies for different periods of time. The ERG amplitudes in wild-type and D0 dpis1;P[hs–dpis] flies were the same, regardless of whether or not they have been dark adapted (Fig. 5G,H,K). However, in D3 dpis1;P[hs–dpis] flies, which underwent a dramatic decrease in the concentration of dPIS protein, the ERG amplitude was much smaller than wild-type after the 2 min light adaptation (Fig. 5I,K). After incubating the flies in the dark, the ERG response was gradually restored. A full recovery of the ERG amplitude in D3 dpis1;P[hs–dpis] flies required between 1.5 and 2 min of dark adaptation (Fig. 5K) (ERG amplitude: 3 s dark, 2.4 ± 0.4 mV; 0.5 min dark, 5.1 ± 1.1 mV; 1 min dark, 7.5 ± 0.6 mV; 1.5 min dark, 8.3 ± 0.9 mV; 2 min dark, 9.2 ± 1.6 mV). The restoration of the light response was even slower in D7 dpis1;P[hs–dpis] flies, which did not express detectable dPIS protein. The photoresponse was nearly undetectable in light-adapted D7 dpis1;P[hs–dpis], and these flies required a minimum of 2 min of dark adaptation to recover the light response (Fig. 5J,K) (ERG amplitude: 3 s dark, 0.7 ± 0.5 mV; 0.5 min dark, 2.9 ± 0.5 mV; 1 min dark, 4.2 ± 0.4 mV; 1.5 min dark, 6.7 ± 1.0 mV; 2 min dark, 8.0 ± 1.3 mV). These results demonstrated that dPIS was essential for maintaining and recovering the photoresponse. The very slow dark recovery (rather than a complete absence of recovery) could be attributable to very low levels of residual dPIS, which is not detected on Western blots, or to a very minor alternative mechanism for generation of PI or PIP2.
dpis1 flies undergo activity-dependent retinal degeneration
To determine whether dpis is required to prevent retinal degeneration, we examined the morphology of dpis1 photoreceptor cells. The dpis1;P[hs–dpis] flies were heat shocked until the late pupal period, and the newly eclosed flies were reared in the dark without additional heat shocks for 7 d, resulting in the elimination of detectable dPIS protein (Fig. 5A). The 7-d-old dpis1;P[hs–dpis] flies were subsequently exposed to a 12 h light/dark cycle or maintained in the dark for an additional 7 d. The number of photoreceptor cell rhabdomeres was then assayed in 7- to 14-d-old flies using the optical neutralization technique. We found that dpis1;P[hs–dpis] flies exposed to the light/dark cycle underwent gradual loss of rhabdomeres, whereas those animals maintained in the dark did not (Fig. 6A). Induction of daily heat-shock treatments after day 7 after eclosion prevented the rhabdomere loss (Fig. 6A).
The light- and age-dependent photoreceptor cell death in dpis1;P[hs–dpis] flies were examined in more detail by transmission EM. The ommatidia from dpis1;P[hs–dpis] flies maintained for either 7 or 14 d in the dark contained the full complement of seven intact rhabdomeres (Fig. 6B,C). In contrast to these results, all of the rhabdomeres were either missing or dramatically smaller and distorted in appearance in 14-d-old mutant flies exposed to the 7 d light/dark cycle (Fig. 6D). Normal morphology was maintained in 14-d-old dpis1;P[hs–dpis] flies, which were given daily heat-shock treatments beginning on day 7 and were exposed to a light/dark cycle (Fig. 6E). These results demonstrate that disruption of dPIS leads to light-dependent photoreceptor cell death.
dPIS is the only PI synthase in Drosophila
Based on homologies to known enzymes, only one PI synthase and one CDP–DAG synthase appear to be encoded in the Drosophila genome. Given that PI is an essential lipid for eukaryotic cells, both the cds1 and dpis1 mutations would be expected to be lethal (Nikawa et al., 1987a; Shen et al., 1996). However, the cds1 mutant flies are viable, whereas the dpis1 mutation causes embryonic lethality. Because CDS functions immediately upstream of the PI synthase, there may be another CDP–DAG synthase in Drosophila, which is not revealed by examination of the Drosophila genome databases. Alternatively, the cds1 mutation may not completely eliminate CDP–DAG synthase activity in adult flies. Before examining CDP–DAG synthase activity in cds1 heads, we first tested whether dPIS is responsible for all of the PI synthase activity. Because dpis1 flies are lethal, we used an antibody depletion method to abolish the dPIS protein in extracts from wild-type flies. No dPIS protein was detected in either head or body extracts after the antibody depletion (Fig. 7A), and other proteins were not affected (supplemental Fig. 1C, available at www.jneurosci.org as supplemental material). Of significance here, there were nearly no PI synthase activities in the head or body extracts, in which we removed dPIS (Fig. 7B). The antibody depletion specifically eliminated PI synthase activity because it did not reduce CDP–DAG synthase activity (Fig. 7C). These data indicate that dPIS is the only PI synthase in Drosophila.
Figure 7.

dPIS is the only PI synthase in Drosophila. A, Depletion of dPIS with anti-dPIS antibodies. A Western blot, consisting of wild-type head and body extracts before and after the dPIS depletion, was probed with anti-dPIS antibodies. The total proteins in each lane were detected by staining with Ponceau S staining (supplemental Fig. 1C, available at www.jneurosci.org as supplemental material). B, PI synthase activities in fly head and body extracts before and after depletion of dPIS. C, CDP–DAG synthase activities in head and body extracts of cds1 mutant flies compared with wild type. Also shown are the CDP–DAG synthase activities in wild-type extracts before and after depletion of dPIS.
To determine whether there was CDP–DAG synthase activity remaining in cds1 (Wu et al., 1995), we compared enzyme activities in wild-type and mutant extracts. CDP–DAG synthase activities were found in the head and body extracts of cds1 flies, although the activities were decreased slightly more than threefold relative to wild type (Fig. 7C) (CDS activity: wild-type heads, 0.85 ± 0.11 U/mg; cds1 heads, 0.26 ± 0.07 U/mg; wild-type bodies, 0.23 ± 0.04 U/mg; cds body 0.07 ± 0.02 U/mg). These results demonstrate that the cds1 mutation does not eliminate CDP–DAG synthase activity.
Discussion
To characterize PIP2 regeneration pathways, we and others have focused on Drosophila phototransduction. However, existing mutations affecting this pathway reduce rather than eliminate the generation of intermediates produced during PIP2 recycling. Mutation of rdgA, which encodes one of eight DAG kinases encoded in the fly genome (Masai et al., 1993), does not eliminate PA production because it can also be produced through the activity of phospholipase D (LaLonde et al., 2005) (Fig. 1A). Moreover, mutation of rdgA can result in accumulation of DAG and have impacts on protein kinase C or TRP activity. Thus, the effects of the rdgA mutation are difficult to interpret. The cds1 mutation has been reported to have no phenotype other than effects on the visual response, leading to the proposal that distinct pools of PIP2 are involved in phototransduction and other functions in photoreceptor cells (Wu et al., 1995). The cds1 mutation is considered a null allele, because no CDS protein expression is detected on Western blots (Wu et al., 1995). However, we found that the cds1 mutation results in a threefold reduction rather than an elimination of CDP–DAG synthase activity. Given that CDS is the only Drosophila protein with homology to known CDP–DAG synthases, these data raise the possibility that there is at least one additional protein with a similar enzymatic activity that does not share obvious sequence identity with characterized CDP–DAG synthases.
In the current work, we created a loss-of-function mutation in dpis, which encodes the enzyme that functions in the step after CDS and promotes the conversion of CDP–DAG to PI. Several observations support the conclusions that dPIS is a bona fide PI synthase, that the dpis1 mutation eliminates dpis function, and that dPIS is the only PI synthase in Drosophila. First, the deletion in dpis eliminated the initiating ATG and the N-terminal region of the protein. Second, the dPIS protein is not detected in the dpis1 mutant. Third, overexpression or depletion of dPIS from wild-type flies caused an increase or elimination of PI synthase activity, respectively. The elimination of PI synthase activity by depletion with anti-dPIS antibodies is consistent with basic local alignment search tool searches indicating that dpis is the only gene encoding a PI synthase in Drosophila. Fourth, a known PI synthase (human PIS) can functionally substitute for dPIS in vivo. Fifth, dPIS is not detected in the rhabdomeres but is found in the extra-rhabdomeral cell bodies. RDGA and RDGB are also detected in the photoreceptor cell bodies, in a portion of the endoplasmic reticulum in which DAG is proposed to be converted into PI (Vihtelic et al., 1993; Suzuki and Hirosawa, 1994; Masai et al., 1997). Sixth, overexpression of dPIS suppresses the retinal degeneration in cds1 and in rdgBKS222, further supporting the conclusion that dPIS operates in vivo in the PIP2 regeneration pathway. Seventh, evidence that dPIS is the only PI synthase in Drosophila is supported not only by the biochemical assays after antibody depletion but also by the phenotype. Unlike the rdgA, rdgB, and cds1 phenotypes, the dpis1mutation results in lethality.
The observation that dPIS is the only PI synthase in Drosophila provides an opportunity to assess the effects on phototransduction resulting from a severe interruption in the PIP2 regeneration pathway. To characterize the phenotype of dpis1, we generated mosaic flies with mutant eyes because dpis is required for viability. However, dpis was also required for eye development and production of photoreceptor cells. To analyze a role for dpis for visual transduction, we produced dpis1 eyes with a normal complement of photoreceptor cells by expressing dPIS during development in mosaic animals.
We found that dpis has an activity-dependent role during phototransduction. After dark adapting the flies, the photoresponses in wild-type and dpis1 were indistinguishable. However, the ability to maintain a light response was rapidly eliminated in dpis1 flies. The depletion of the light response was even more severe than in cds1 (our unpublished observation), consistent with our findings that significant CDP–DAG synthase activity is retained in these flies. Additional evidence that dPIS had an activity-dependent function was that the dpis1 flies underwent retinal degeneration during a light/dark cycle but not if they were maintained in the dark.
An unresolved question concerns the mechanism underlying the activation of the TRP and TRPL channels, which function in phototransduction. PLC catalyzes production of IP3 and DAG from PIP2. IP3 does not appear to function in the activation of the channels because IP3 cannot excite phototransduction alone and a normal photoresponse takes place in an IP3 receptor mutant background (Hardie and Minke, 1995; Acharya et al., 1997; Hardie and Raghu, 1998; Raghu et al., 2000b). DAG or PUFAs have been suggested to contribute to excitation in the Drosophila photoresponse. In the rdgA mutant, which disrupts a DAG kinase, TRP and TRPL are constitutively active (Raghu et al., 2000a). Moreover, PUFAs, which are derived from DAG, can activate TRP and TRPL channels in photoreceptors cells (Chyb et al., 1999). A PA phosphatase, Lazaro, is expressed in the photoreceptor cells (Garcia-Murillas et al., 2006; Kwon and Montell, 2006), and a putative reduction in DAG levels resulting from mutation of lazaro results in a smaller light response (Kwon and Montell, 2006). However, it remains to be determined whether DAG or PUFAs are reduced in the lazaro mutant. Nevertheless, in the rdgA mutant, PIP2 regeneration is also affected. Thus, it cannot be excluded that excitation of TRP and TRPL results from reduction in PIP2 levels.
The model that PIP2 is inhibitory to the TRP and TRPL channels is based on both in vitro and genetic analyses. Recombinant TRPL channels have been reported to be inhibited by application of PIP2 (Estacion et al., 2001). In the rdgB mutant, which decreases transfer of PI to the rhabdomeres, the channels fail to close quickly after cessation of the light stimulus (Milligan et al., 1997; Hardie et al., 2001). A similar failure to terminate the phototransduction cascade is also be caused by removal of extracellular Ca2+, which also leads to PIP2 depletion (Hardie et al., 2001).
Our analyses of dpis1 argue against a role for a reduction of PIP2 on activation of the light response. In dpis mutant flies, the light response is not sustained, despite the absence of the PI synthase, which is an activity essential for PIP2 regeneration. Although the cds and rdgB mutations reduce PIP2 cycling, their effects on PIP2 recycling are only partial. In fact, the dpis1 mutation, but not cds1 or rdgB, prevents formation of the eye. Thus, the dpis1 offers the best genetic approach to address the effect of interfering with PIP2 cycling on TRP channel activity in vivo. Although our data indicate that PIP2 depletion alone is insufficient to activate TRP and TRPL, they do not exclude that a reduction of PIP2 may contribute in concert with an increase in DAG and/or PUFAs or that accumulation of CDP–DAG contributes to channel activation. Nevertheless, accumulation of CDP–DAG is unlikely to lead to channel inactivation because the threefold lower CDP–DAG level in the cds1 mutant causes a light-dependent loss of the photoresponse, whereas overexpression of CDS causes an increase in the response amplitude (Wu et al., 1995).
Disruption of PIP2 regeneration by mutations in the genes coding RDGB, CDS (Harris and Stark, 1977; Wu et al., 1995), and dPIS causes light-dependent retinal degeneration. Thus, the question arises as to roles in photoreceptor cells of the mammalian homologs of these proteins. It is unlikely that proteins, which function in PIP2 cycling, participate in phototransduction in rods and cones because the cascades in these cells operate through cGMP- rather than PI-mediated signaling. Moreover, mutation of mouse rdgB2 has no effect on photoreceptor cell function or on the survival of photoreceptor cells in the retina (Lu et al., 2001). Rather, we suggest that mutations that affect PIP2 regeneration in mammals may disrupt phototransduction in the intrinsically photosensitive retinal ganglion cells (ipRGCs) that function in circadian rhythm (for review, see Peirson and Foster, 2006). Consistent with this proposal are recent pharmacological and electrophysiological studies suggesting that that light receptor in the ipRGCs, referred to as melanopsin, couples to a pathway involving a Gq/PLC and a TRPC channel (Isoldi et al., 2005; Panda et al., 2005; Qiu et al., 2005).
Footnotes
This work was supported by National Eye Institutes Grants EY08117 and EY10852 (C.M.). We thank Mike Sepanski for preparing sections of compound eyes, the Bloomington Stock Center and Drs. W. Stark and C. Zuker for fly stocks, Drs. B. Tu-Sekine and D. Raben for advice, and members of the C. Montell laboratory for helpful discussions.
References
- Acharya et al., 1997.Acharya JK, Jalink K, Hardy RW, Hartenstein V, Zuker CS. InsP3 receptor essential for growth and differentiation but not for vision in Drosophila. Neuron. 1997;18:881–887. doi: 10.1016/s0896-6273(00)80328-1. [DOI] [PubMed] [Google Scholar]
- Bloomquist et al., 1988.Bloomquist BT, Shortridge RD, Schneuwly S, Perdew M, Montell C, Steller H, Rubin G, Pak WL. Isolation of a putative phospholipase C gene of Drosophila, norpA, and its role in phototransduction. Cell. 1988;54:723–733. doi: 10.1016/s0092-8674(88)80017-5. [DOI] [PubMed] [Google Scholar]
- Carman and Fischl, 1992.Carman GM, Fischl AS. Phosphatidylinositol synthase from yeast. Methods Enzymol. 1992;209:305–312. doi: 10.1016/0076-6879(92)09038-5. [DOI] [PubMed] [Google Scholar]
- Chyb et al., 1999.Chyb S, Raghu P, Hardie RC. Polyunsaturated fatty acids activate the Drosophila light-sensitive channels TRP and TRPL. Nature. 1999;397:255–259. doi: 10.1038/16703. [DOI] [PubMed] [Google Scholar]
- del Pilar Gomez and Nasi, 2005.del Pilar Gomez M, Nasi E. A direct signaling role for phosphatidylinositol 4,5-bisphosphate (PIP2) in the visual excitation process of microvillar receptors. J Biol Chem. 2005;280:16784–16789. doi: 10.1074/jbc.M414538200. [DOI] [PubMed] [Google Scholar]
- Downes et al., 2005.Downes CP, Gray A, Lucocq JM. Probing phosphoinositide functions in signaling and membrane trafficking. Trends Cell Biol. 2005;15:259–268. doi: 10.1016/j.tcb.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Estacion et al., 2001.Estacion M, Sinkins WG, Schilling WP. Regulation of Drosophila transient receptor potential-like (TrpL) channels by phospholipase C-dependent mechanisms. J Physiol (Lond) 2001;530:1–19. doi: 10.1111/j.1469-7793.2001.0001m.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini and Kirschfeld, 1971.Franceschini N, Kirschfeld K. Etude optique in vivo des éléments photorécepteurs dans l'oeil composé de Drosophila. Kybernetik. 1971;8:1–13. doi: 10.1007/BF00270828. [DOI] [PubMed] [Google Scholar]
- Garcia-Murillas et al., 2006.Garcia-Murillas I, Pettitt T, Macdonald E, Okkenhaug H, Georgiev P, Trivedi D, Hassan B, Wakelam M, Raghu P. lazaro encodes a lipid phosphate phosphohydrolase that regulates phosphatidylinositol turnover during Drosophila phototransduction. Neuron. 2006;49:533–546. doi: 10.1016/j.neuron.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Gong and Golic, 2003.Gong WJ, Golic KG. Ends-out, or replacement, gene targeting in Drosophila. Proc Natl Acad Sci USA. 2003;100:2556–2561. doi: 10.1073/pnas.0535280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie and Minke, 1995.Hardie RC, Minke B. Phosphoinositide-mediated phototransduction in Drosophila photoreceptors: the role of Ca2+ and trp. Cell Calcium. 1995;18:256–274. doi: 10.1016/0143-4160(95)90023-3. [DOI] [PubMed] [Google Scholar]
- Hardie and Raghu, 1998.Hardie RC, Raghu P. Activation of heterologously expressed Drosophila TRPL channels: Ca2+ is not required and InsP3 is not sufficient. Cell Calcium. 1998;24:153–163. doi: 10.1016/s0143-4160(98)90125-7. [DOI] [PubMed] [Google Scholar]
- Hardie et al., 2001.Hardie RC, Raghu P, Moore S, Juusola M, Baines A, Sweeney ST. Calcium influx via TRP channels is required to maintain PIP2 levels in Drosophila photoreceptors. Neuron. 2001;30:149–159. doi: 10.1016/s0896-6273(01)00269-0. [DOI] [PubMed] [Google Scholar]
- Harris and Stark, 1977.Harris WA, Stark WS. Heriditary retinal degeneration in Drosophila melanogaster: a mutant defect associated with the phototransduction process. J Gen Physiol. 1977;69:261–291. doi: 10.1085/jgp.69.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta and Benzer, 1970.Hotta Y, Benzer S. Genetic dissection of the Drosophila nervous system by means of mosaics. Proc Natl Acad Sci USA. 1970;67:1156–1163. doi: 10.1073/pnas.67.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isoldi et al., 2005.Isoldi MC, Rollag MD, de Lauro Castrucci AM, Provencio I. Rhabdomeric phototransduction initiated by the vertebrate photopigment melanopsin. Proc Natl Acad Sci USA. 2005;102:1217–1221. doi: 10.1073/pnas.0409252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon and Montell, 2006.Kwon Y, Montell C. Dependence on the Lazaro phosphatidic acid phosphatase for the maximum light response. Curr Biol. 2006;16:723–729. doi: 10.1016/j.cub.2006.02.057. [DOI] [PubMed] [Google Scholar]
- LaLonde et al., 2005.LaLonde MM, Janssens H, Rosenbaum E, Choi SY, Gergen JP, Colley NJ, Stark WS, Frohman MA. Regulation of phototransduction responsiveness and retinal degeneration by a phospholipase D-generated signaling lipid. J Cell Biol. 2005;169:471–479. doi: 10.1083/jcb.200502122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu et al., 2001.Lu C, Peng YW, Shang J, Pawlyk BS, Yu F, Li T. The mammalian retinal degeneration B2 gene is not required for photoreceptor function and survival. Neuroscience. 2001;107:35–41. doi: 10.1016/s0306-4522(01)00337-2. [DOI] [PubMed] [Google Scholar]
- Lykidis et al., 1997.Lykidis A, Jackson PD, Rock CO, Jackowski S. The role of CDP-diacylglycerol synthetase and phosphatidylinositol synthase activity levels in the regulation of cellular phosphatidylinositol content. J Biol Chem. 1997;272:33402–33409. doi: 10.1074/jbc.272.52.33402. [DOI] [PubMed] [Google Scholar]
- Masai et al., 1993.Masai I, Okazaki A, Hosoya T, Hotta Y. Drosophila retinal degeneration A gene encodes an eye-specific diacylglycerol kinase with cysteine-rich zinc-finger motifs and ankyrin repeats. Proc Natl Acad Sci USA. 1993;90:11157–11161. doi: 10.1073/pnas.90.23.11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai et al., 1997.Masai I, Suzuki E, Yoon CS, Kohyama A, Hotta Y. Immunolocalization of Drosophila eye-specific diacylgylcerol kinase, rdgA, which is essential for the maintenance of the photoreceptor. J Neurobiol. 1997;32:695–706. doi: 10.1002/(sici)1097-4695(19970620)32:7<695::aid-neu5>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Milligan et al., 1997.Milligan SC, Alb JG, Jr, Elagina RB, Bankaitis VA, Hyde DR. The phosphatidylinositol transfer protein domain of Drosophila retinal degeneration B protein is essential for photoreceptor cell survival and recovery from light stimulation. J Cell Biol. 1997;139:351–363. doi: 10.1083/jcb.139.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mismer and Rubin, 1987.Mismer D, Rubin GM. Analysis of the promoter of the ninaE opsin gene in Drosophila melanogaster. Genetics. 1987;116:565–578. doi: 10.1093/genetics/116.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell, 1999.Montell C. Visual transduction in Drosophila. Annu Rev Cell Dev Biol. 1999;15:231–268. doi: 10.1146/annurev.cellbio.15.1.231. [DOI] [PubMed] [Google Scholar]
- Montell, 2005.Montell C. The TRP superfamily of cation channels. Sci STKE. 2005;2005:re3. doi: 10.1126/stke.2722005re3. [DOI] [PubMed] [Google Scholar]
- Moses et al., 1989.Moses K, Ellis MC, Rubin GM. The glass gene encodes a zinc-finger protein required by Drosophila photoreceptor cells. Nature. 1989;340:531–536. doi: 10.1038/340531a0. [DOI] [PubMed] [Google Scholar]
- Niggli, 2005.Niggli V. Regulation of protein activities by phosphoinositide phosphates. Annu Rev Cell Dev Biol. 2005;21:57–79. doi: 10.1146/annurev.cellbio.21.021704.102317. [DOI] [PubMed] [Google Scholar]
- Nigou and Besra, 2002.Nigou J, Besra GS. Cytidine diphosphate-diacylglycerol synthesis in Mycobacterium smegmatis. Biochem J. 2002;367:157–162. doi: 10.1042/BJ20020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa et al., 1987a.Nikawa J, Kodaki T, Yamashita S. Primary structure and disruption of the phosphatidylinositol synthase gene of Saccharomyces cerevisiae. J Biol Chem. 1987a;262:4876–4881. [PubMed] [Google Scholar]
- Nikawa et al., 1987b.Nikawa J, Tsukagoshi Y, Kodaki T, Yamashita S. Nucleotide sequence and characterization of the yeast PSS gene encoding phosphatidylserine synthase. Eur J Biochem. 1987b;167:7–12. doi: 10.1111/j.1432-1033.1987.tb13297.x. [DOI] [PubMed] [Google Scholar]
- Panda et al., 2005.Panda S, Nayak SK, Campo B, Walker JR, Hogenesch JB, Jegla T. Illumination of the melanopsin signaling pathway. Science. 2005;307:600–604. doi: 10.1126/science.1105121. [DOI] [PubMed] [Google Scholar]
- Peirson and Foster, 2006.Peirson S, Foster RG. Melanopsin: another way of signaling light. Neuron. 2006;49:331–339. doi: 10.1016/j.neuron.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Porter et al., 1992.Porter JA, Hicks JL, Williams DS, Montell C. Differential localizations of and requirements for the two Drosophila ninaC kinase/myosins in photoreceptor cells. J Cell Biol. 1992;116:683–693. doi: 10.1083/jcb.116.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu et al., 2005.Qiu X, Kumbalasiri T, Carlson SM, Wong KY, Krishna V, Provencio I, Berson DM. Induction of photosensitivity by heterologous expression of melanopsin. Nature. 2005;433:745–749. doi: 10.1038/nature03345. [DOI] [PubMed] [Google Scholar]
- Raghu et al., 2000a.Raghu P, Usher K, Jonas S, Chyb S, Polyanovsky A, Hardie RC. Constitutive activity of the light-sensitive channels TRP and TRPL in the Drosophila diacylglycerol kinase mutant, rdgA. Neuron. 2000a;26:169–179. doi: 10.1016/s0896-6273(00)81147-2. [DOI] [PubMed] [Google Scholar]
- Raghu et al., 2000b.Raghu P, Colley NJ, Webel R, James T, Hasan G, Danin M, Selinger Z, Hardie RC. Normal phototransduction in Drosophila photoreceptors lacking an InsP3 receptor gene. Mol Cell Neurosci. 2000b;15:429–445. doi: 10.1006/mcne.2000.0846. [DOI] [PubMed] [Google Scholar]
- Shen et al., 1996.Shen H, Heacock PN, Clancey CJ, Dowhan W. The CDS1 gene encoding CDP-diacylglycerol synthase in Saccharomyces cerevisiae is essential for cell growth. J Biol Chem. 1996;271:789–795. doi: 10.1074/jbc.271.2.789. [DOI] [PubMed] [Google Scholar]
- Stowers and Schwarz, 1999.Stowers RS, Schwarz TL. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics. 1999;152:1631–1639. doi: 10.1093/genetics/152.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki and Hirosawa, 1994.Suzuki E, Hirosawa K. Immunolocalization of a Drosophila phosphatidylinositol transfer protein (rdgB) in normal and rdgA mutant photoreceptor cells with special reference to the subrhabdomeric cisternae. J Electron Microsc. 1994;43:183–189. [PubMed] [Google Scholar]
- Thummel and Pirrotta, 1992.Thummel CS, Pirrotta V. New pCaSpeR P element vectors. Dros Info Serv. 1992;71:150. [Google Scholar]
- Vihtelic et al., 1993.Vihtelic TS, Goebl M, Milligan S, O'Tousa JE, Hyde DR. Localization of Drosophila retinal degeneration B, a membrane-associated phosphatidylinositol transfer protein. J Cell Biol. 1993;122:1013–1022. doi: 10.1083/jcb.122.5.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang and Montell, 2005.Wang T, Montell C. Rhodopsin formation in Drosophila is dependent on the PINTA retinoid-binding protein. J Neurosci. 2005;25:5187–5194. doi: 10.1523/JNEUROSCI.0995-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wes et al., 1999.Wes PD, Xu X-ZS, Li H-S, Chien F, Doberstein SK, Montell C. Termination of phototransduction requires binding of the NINAC myosin III and the PDZ protein INAD. Nat Neurosci. 1999;2:447–453. doi: 10.1038/8116. [DOI] [PubMed] [Google Scholar]
- Wu et al., 1995.Wu L, Niemeyer B, Colley N, Socolich M, Zuker CS. Regulation of PLC-mediated signalling in vivo by CDP-diacylglycerol synthase. Nature. 1995;373:216–222. doi: 10.1038/373216a0. [DOI] [PubMed] [Google Scholar]
- Xu et al., 2004.Xu H, Lee SJ, Suzuki E, Dugan KD, Stoddard A, Li HS, Chodosh LA, Montell C. A lysosomal tetraspanin associated with retinal degeneration identified via a genome-wide screen. EMBO J. 2004;23:811–822. doi: 10.1038/sj.emboj.7600112. [DOI] [PMC free article] [PubMed] [Google Scholar]