Abstract
Dyslipidemia is a well-established risk factor for cardiovascular diseases. Although, advances in genome-wide technologies have enabled the discovery of hundreds of genes associated with blood lipid phenotypes, most of the heritability remains unexplained. Here we performed targeted resequencing of 13 bona fide candidate genes of dyslipidemia to identify the underlying biological functions. We sequenced 940 Sikh subjects with extreme serum levels of hypertriglyceridemia (HTG) and 2,355 subjects were used for replication studies; all 3,295 participants were part of the Asian Indians Diabetic Heart Study. Gene-centric analysis revealed burden of variants for increasing HTG risk in GCKR (p = 2.1x10-5), LPL (p = 1.6x10-3) and MLXIPL (p = 1.6x10-2) genes. Of these, three missense and damaging variants within GCKR were further examined for functional consequences in vivo using a transgenic zebrafish model. All three mutations were South Asian population-specific and were largely absent in other multiethnic populations of Exome Aggregation Consortium. We built different transgenic models of human GCKR with and without mutations and analyzed the effects of dietary changes in vivo. Despite the short-term of feeding, profound phenotypic changes were apparent in hepatocyte histology and fat deposition associated with increased expression of GCKR in response to a high fat diet (HFD). Liver histology of the GCKRmut showed severe fatty metamorphosis which correlated with ~7 fold increase in the mRNA expression in the GCKRmut fish even in the absence of a high fat diet. These findings suggest that functionally disruptive GCKR variants not only increase the risk of HTG but may enhance ectopic lipid/fat storage defects in absence of obesity and HFD. To our knowledge, this is the first transgenic zebrafish model of a putative human disease gene built to accurately assess the influence of genetic changes and their phenotypic consequences in vivo.
Introduction
Dyslipidemia is a well-established risk factor for cardiovascular disease and a principal cause of mortality in individuals with type 2 diabetes (T2D). Circulating blood lipid phenotypes are heritable risk factors for the development of atherosclerosis and their measurements are used clinically to predict future coronary artery disease (CAD) risk and therapy for primary prevention [1,2]. Epidemiological studies suggest that elevated serum triglyceride (TG) concentration is a strong independent risk factor for CAD [1,3]. There is an inverse correlation between serum TG and serum high-density cholesterol (HDL-C) that is associated with increased risk of cardiovascular dysfunction, despite the level of low density cholesterol (LDL-C) being normal. This combination of lipid alterations is defined as atherogenic dyslipidemia, which is a significant risk factor for the development of CAD [4]. Lowering of LDL-C has been the major focus in CAD prevention following treatment with HMG-CoA reductase inhibitors (statins). However, the mortality rate of CAD remains elevated particularly in the patients with T2D and insulin resistance, and reasons for their discordant effects in diabetics remain unknown [5].
Family and twin studies have shown that TG and lipoprotein levels aggregate in families [6]. Relatives of individuals with hyperlipidemia/dyslipidemia will have a 2.5- to 7-fold increase in risk of death due to premature CAD compared to relatives of control individuals [7,8]. The principal lipid alterations observed in these patients include high TG and low HDL-C. Cincinnati Lipid Research Clinic Family Study showed that low HDL-C and high TG occur conjointly and are transmitted across generations as a "combined phenotype" or "conjoint trait” [9]. Genome-wide association studies (GWAS) and meta-analyses studies on multiethnic populations including Punjabi Sikhs have uncovered more than 200 genetic loci associated with circulating blood lipid phenotypes [10–13]. However, despite the high clinical heritability (50–80%) of many of the lipid traits [14], these and several other studies have only explained up to10% of heritability in these genes. To identify putative functional with larger effects, in this study, we have performed targeted sequencing of 13 bona fide candidate gene regions (~2.9 Mb) (S1 Table/S1 Fig) on 940 Sikh individuals [572 cases with high serum triglycerides (TG) (~95th percentile for their age and gender) and 368 controls with low TG (below the 20th percentile for their age and gender), using subjects from the Asian Indians Diabetic Heart Study (AIDHS)/Sikh Diabetes Study (SDS) [15–17].
Materials and methods
Study subjects of discovery (sequencing) cohort
Genomic DNA samples of individuals including HTG cases (TG>150 mg/dl) and healthy controls with TG (<100 mg/dl) were sequenced with custom Nimblegen probes designed for targeted resequencing of 13 confirmed candidate genes for diabetic dyslipidemia in Sikhs. Diagnosis of T2D was confirmed by scrutinizing medical records for symptoms, use of medication, and measuring fasting glucose levels following the guidelines of American Diabetes Association [18]. The diagnosis for normo-glycemic controls was based on a fasting glycemia <110 mg/dL or a 2-h glucose <140 mg/dL. CAD was assigned when there was a documented prior diagnosis of heart disease, electrocardiographic evidence of angina pain, coronary angiographic evidence of severe (>50%) stenosis, or echocardiographic evidence of myocardial infarction. HTG is broadly defined as fasting serum TG concentrations above the ninety-fifth percentile [19], and was classified as mild HTG (150–399 mg/dL), high HTG (400–875 mg/dL), and severe HTG (>875 mg/dL).
The non-HTG control participants were recruited from the same Punjabi Sikh community and from the same geographic location as the HTG participants. They were selected on the basis of a fasting glycemia <100.8 mg/dL or a 2h glucose <141.0 mg/dL. BMI was calculated as weight (kg)/[height (m)2]. Education, socio-economic status, dietary, and physical activity data were recorded. Smoking information was collected regarding past smoking, current smoking status, and length of time, number of cigarettes smoked /day. The vast majority of Sikhs were non-smokers, details are described elsewhere [17]. The individuals on lipid lowering medication are excluded from this cohort. All participants provided a written informed consent for investigations.
The study was reviewed and approved by the University of Oklahoma Health Sciences Center’s Institutional Review Board, as well as the Human Subject Protection Committees of Hero Dayanand Medical College and Heart Institute, Ludhiana and Guru Nanak Dev University, Amritsar in India. Metabolic estimations of fasting serum lipids [total cholesterol, LDL-C, HDL-C, and TG] were quantified by using standard enzymatic methods (Roche, Basel, Switzerland) as previously described [17].
In this study, we only included those individuals who self-reported having no South Indian admixture and exclusively belonged to the North Indian Punjabi Sikh community, who reported that all of their four grandparents were of North Indian origin, and spoke the Punjabi language. Excluded were individuals of South, East, or Central Indian origin; those of non-Sikh/non-Punjabi origin; those with rare forms of lipid disorders including very low serum TG (abetalipoprotenemia, homozygous hypobetalipoprotenemia, familial combined hypobetalipoproteinemia), or severe HTG (extremely high serum TG >1,000 mg/dL); those with familial chylomicronemia, hemochromatosis, or pancreatitis; those on lipid lowering medication; and those with excessive alcohol intake (>400 mL/day). About 50% of HTG patients had T2D and ~9% had CAD. Clinical characteristics of discovery- (resequencing) and replication cohort are summarized in Table 1.
Table 1. Phenotypic attributes of discovery (sequencing) and replication cohorts as part of Asian Indian Diabetic Heart Study (AIDHS).
| Traits | Discovery cohort (n = 820) |
Replication cohort (n = 1,769) |
||
|---|---|---|---|---|
| HTG cases | Controls | T2D cases | Controls | |
| N | 572 | 248 | 1,074 | 695 |
| Females (%) | 39 | 49 | 44 | 46 |
| T2D (%) | 61 | 53 | N/A | N/A |
| Age (years) | 52.0 ± 12.4 | 55.0 ± 10.6 | 55.0 ± 11.7 | 45.7 ± 14.5 |
| BMI (kg/m2) | 27.6 ± 4.3 | 25.7 ± 4.4 | 27.1 ± 4.8 | 26.0 ± 4.6 |
| TG (mg/dl) | 314.9 ± 134.6 | 78.0 ± 16.9 | 158.1 ± 79.1 | 132.2 ± 67.6 |
| TC (mg/dl) | 203.3 ± 52.3 | 167.2 ± 42.7 | 178.3 ± 46.3 | 184.6 ± 90.2 |
| HDL-C (mg/dl) | 41.1 ± 17.5 | 41.9 ± 14.0 | 40.7 ± 14.1 | 42.6 ± 12.8 |
| LDL-C (mg/dl) | 110.6 ± 43.2 | 106.6 ± 36.2 | 107.9 ± 38.3 | 114.9 ± 35.7 |
HTG- hypertriglyceridemia, T2D-type 2 diabetes, BMI-body mass index, TG-triglyceride, TC-total cholesterol, HDL-C high density lipoprotein cholesterol, LDL-C low density lipoprotein cholesterol.
Targeted sequencing
Targeted sequencing was performed at the Northwest Genomics Center in the department of Genome Sciences at the University of Washington through the RS&G Service sponsored by the National Heart Lung Blood Institute of the National Institutes of Health.
Library production, targeted capture, sequencing
Genomic DNA was extracted from whole blood or buffy coats using Qiagen kits (Qiagen, Chatsworth, CA, USA) or salting out procedures described previously [20,21]. 1 ug of genomic DNA was sent to the Core lab at Northwest Genomics Center at the University of Washington for sequencing. The quality and integrity of DNA was checked at the Core lab using Agilent’s Analyzer and Tape Station reagents before target capture and library preparation. Library construction and custom capture have been automated (Perkin-Elmer Janus II) in a 96-well plate format. The purified DNA was subjected to a series of shotgun library construction steps, including fragmentation through acoustic sonication (Covaris), end-polishing and A-tailing, ligation of sequencing adaptors, and PCR amplification with 8 bp barcodes for multiplexing. Libraries undergo capture using the Roche/Nimblegen SeqCap EZ custom designed probe. Prior to sequencing, the library concentration was determined by triplicate qPCR and molecular weight distributions verified on the Agilent Bioanalyzer. Barcoded libraries were pooled using liquid handling robotics prior to clustering (Illumina cBot) and loading. Massively parallel sequencing-by-synthesis with fluorescently labeled, reversibly terminating nucleotides was carried out on the HiSeq sequencer.
Read processing
Our sequencing pipeline is a combined suite of Illumina software and other “industry standard” software packages (i.e., Genome Analysis ToolKit [GATK], Picard, BWA, SAMTools, and in-house custom scripts) and consists of base calling, alignment, local realignment, duplicate removal, quality recalibration, data merging, variant detection, genotyping and annotation. The overall processing pipeline consists of the following elements: (1) base calls generated in real-time on the HiSeq2500 instrument (RTA 1.13.48.0) (2) demultiplexed, unaligned BAM files produced by Picard ExtractIlluminaBarcodes and IlluminaBasecallsToSam and (3) BAM files aligned to a human reference using BWA (Burrows-Wheeler Aligner; v0.6.2). Read data from a flow cell lane is treated independently for alignment and QC purposes in instances where the merging of data from multiple lanes is required (e.g., for sample multiplexing). The samples were sequenced using paired-end ~140 to 150bp reads and the insert sizes were at least 100 bp in length. Therefore, we expected to see ~240 to 250bp on the Bioanalyzer. Read-pairs not mapping within ± 2 standard deviations of the average library size (~150 ± 15 bp for the targeted region) were removed. All aligned read data are subject to the following steps: (1) “duplicate removal” was performed, (i.e., the removal of reads with duplicate start positions; Picard MarkDuplicates; v1.70) (2) indel realignment was performed (GATK IndelRealigner; v1.6-11-g3b2fab9) resulting in improved base placement and lower false variant calls and (3) base qualities were recalibrated (GATK TableRecalibration; v1.6-11-g3b2fab9).
Sequence data analysis QC
All sequence data underwent a QC protocol before they were released to the annotation group for further processing. This included an assessment of: (1) total reads; (2) library complexity—the ratio of unique reads to total reads mapped to target. DNA libraries exhibiting low complexity are not cost-effective to finish; (3) capture efficiency—the ratio of reads mapped to human versus reads mapped to target; (4) coverage distribution—80% at 20X required for completion; (5) capture uniformity; (6) raw error rates; (7) Transition/Transversion ratio (Ti/Tv)—typically ~3 for known sites and ~2.5 for novel sites; (8) distribution of known and novel variants relative to dbSNP—typically < 7% using dbSNP build 129 in samples of European ancestry [22]; (9) fingerprint concordance > 99%; (10) sample homozygosity and heterozygosity and (11) sample contamination validation. All QC metrics for both single-lane and merged data were reviewed by a sequence data analyst to identify data deviations from known or historical norms. Lanes/samples that failed QC were flagged in the system and could be re-queued for library prep (< 5% failure) or further sequencing (< 2% failure), depending upon the QC issue. Completion was defined as having > 80% of the target at >20X coverage.
Variant detection
Variant detection and genotyping were performed using the UnifiedGenotyper (UG) tool from GATK (v1.6-11-g3b2fab9). Variant data for each sample were formatted (variant call format [VCF]) as “raw” calls that contain individual genotype data for one or multiple samples and flagged using the filtration walker (GATK) to mark sites that were of lower quality/false positives [e.g., low quality scores (Q50), allelic imbalance (ABHet 0.75), long homopolymer runs (HRun> 3) and/or low quality by depth (QD < 5)].
Variant annotation
We used an automated pipeline for annotation of variants derived from targeted sequencing data, the SeattleSeq Annotation Server (http://gvs.gs.washington.edu/ SeattleSeqAnnotation/). These publically accessible server returns annotations including dbSNP rsID (or whether the coding variant is novel), gene names and accession numbers, predicted functional effect (e.g., splice-site, nonsynonymous, missense, etc.), protein positions and amino-acid changes, PolyPhen predictions, conservation scores (e.g., PhastCons, GERP), ancestral allele, dbSNP allele frequencies, and known clinical associations. The annotation process has also been automated into our analysis pipeline to produce a standardized, formatted output (VCF-variant call format, described above).
Replication studies, population characteristics, and SNP genotyping
We replicated the association of three functional variants in additional 2355 individuals of Punjabi Sikh ancestry. These included 1000 individuals from Sikh families and the remaining 1355 were unrelated; all were part of the AIDHS/SDS described previously [17,21,23,24]. Recruitment and diagnostic details of the Sikh replication cohort are similar as described above for the discovery cohort. Clinical and demographical details of these cohorts are provided in Table 1. Genotyping for selected GCKR SNPs (rs774930016 (S105N), rs760427565 (R297Q), and rs755537970 (R553W)) (Table 2) was performed using TaqMan pre-designed or TaqMan made-to-order SNP genotyping assays from Applied Biosystems Inc. (ABI, Foster City, USA) as described previously [25]. Genotyping reactions were performed on Quant Studio6 genetic analyzer using 2 uL of genomic DNA (10 ng/uL), following manufacturers’ instructions. For quality control, 8–10% replicative controls and negative controls were used in each 384 well plate to match the concordance. Genotyping call rate was 96% or more in all the SNPs studied.
Table 2. Carrier counts for three population-specific variants in the GCKR gene in AIDHS and multiethnic populations.
| Population | rs774930016 (S105N) | rs760427565 (R297Q) | rs755537970 (R553W) |
|---|---|---|---|
| European | 0/33356 | 1/33359 | 1/33358 |
| Latino | 0/5788 | 1/5787 | 0/5788 |
| African | 0/5197 | 0/5200 | 0/5197 |
| East Asian | 0/4324 | 0/4326 | 0/4327 |
| South Asian | 3/8250 | 11/8255 | 1/8255 |
| AIDHS (Sikhs) | 9/3132 | 18/3016 | 8/2950 |
Data of all non-Sikh populations are from Exome Aggregation Consortium (ExAC).
Functional studies using zebrafish (ZF) model
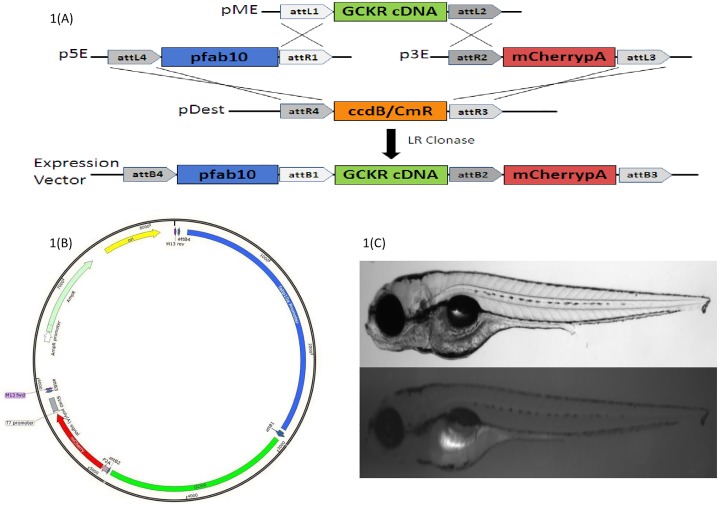
To test the phenotypic effects of this and other novel variants in vivo, we created transgenic ZF (Danio rerio) models of the glucokinase regulatory protein (GCKR)mut and GCKRwt using TAB-5 strain, a commonly used strain derived from two commonly used fish lines (Tubingen and AB). Heterozygous human carriers of this mutation exhibit HTG and high rates of T2D, so we examined whether GCKRmut induces features of this phenotype in ZF. To build our transgenic models, we employed the Tol2 system, which mediates highly-efficient transgenesis [26]. The Tol2kit system uses site-specific recombination-based cloning with 5’, middle and 3’ entry clones first described (Hartley et al., 2000) to allow quick, modular assembly of [promoter]–[coding sequence]–[3_ tag] constructs in a Tol2 transposon backbone using multisite Gateway technology (Invitrogen, Grand Island, NY, USA). The expression construct were generated using “LR reaction” (in which attL and attR sites recombine); and transformed into bacteria following the protocol described (Kwan et al, 2007) [26]. The GCKR full CDNA was purchased from AddGene (Watertown, MA, USA). Mutations were created using site-directed mutagenesis kit (New England Biolabs, Ipswich, MA, USA) as described [26]. We used a promoter construct that drives human GCKRmut expression only in hepatocytes, while simultaneously labeling those cells fluorescently. To achieve hepatocyte-specific expression in D. rerio, we used the D. rerio liver fatty acid binding protein (L-FABP) promoter (courteously provided by Dr. Schlegel, University of Utah) [27]. To label GCKRmut expressing D. rerio hepatocytes, we joined the cDNA for human GCKRmut to the enhanced red fluorescent protein (mCherryFP), separated by a 2A peptide linker [26,28]. The 2A linker is an auto-cleaving peptide, resulting in the GCKRmut and mCherryFP proteins being expressed in a 1:1 stoichiometric ratio. Expression of mCherryFP by the ZF liver confirms expression of GCKRmut hepatocytes (Fig 1A–1C). Transformation of TOP10 cells with an LR recombination reaction yielded two classes of colonies: clear and opaque. Clear colonies yield the correct recombination product and were selected following the protocol described previously [26].
Fig 1.
A-C. Fig 1A Multisite Gateway-based plasmid construction using Tol2 system; Fig 1B Bacterial plasmid construct (p3E-2A-mCherrypA); Fig 1C larvae (4 day post fertilization) of wild type and transgenic fish showing hepatic expression (fluorescence) after successful transgenesis of human GCKR with three disruptive mutations.
After building GCKRmut transgenic ZF, we evaluated the in vivo metabolic consequences of these human GCKR mutations by feeding a high fat diet to 5 day old larvae of wildtype (WT) and transgenic ZF with and without GCKR mutations. Our protocol for building transgenic lines of zebrafish for studying post-GWAS quantitative trait loci (QTL) for diabetes and cardiovascular traits has been approved by the Institutional Animal Care and Use Committee (IACUC) and Institutional Bio-safety Committee (IBC) of the University of Oklahoma Health Sciences Center (Protocol # 01550-16-067-SSHITF).
Diet experiments
For all feeding studies, 5-day post-fertilization (dpf) homozygous humanized GCKR-mutant or −WT transgenic zebrafish larvae were studied. For comparison to WT, we also studied 5 dpf larvae of Tab5 fish (the parental line used to construct transgenic constructs). All WT and mutant fish were distributed in 3 liter tanks (20 fish per tank) and fed defined diets for 14 days. Animals were housed in the main aquarium of the ZF Animal Resource core facility of the University of Oklahoma and maintained on a 14-hour light, 10-hour dark cycle. Animals were anesthetized and killed by immersion in ice water [29]. For control studies, 5 dpf homozygous GCKR mutant (GCKRmut) or homozygous WT (GCKRwt) fish were reared on a conventional diet (commercial powder and newly-hatched Artemia salina nauplii), twice-daily for 14 days. The high fat diet (HFD) groups were fed with a special fish diet (with 24% fat, 43% protein, and 4% fiber from Purina Aqua Mix) thrice-daily for 14 days. Three larvae from each feeding group were euthanized and their livers were dissected using a dissection microscope and sent for transmission electron microscopy at the Oklahoma Medical Research Foundations imaging core facility.
Larvae tissue embedding and hematoxylin and eosin (H&E) staining
A Leica TP1020 tissue processor was used to process the tissue, following the manufacturer protocol. Briefly, tissue in 10% neutral formalin buffer (NBF) are moved into labelled tissue blocks. The tissue in the blocks are progressively dehydrated with increasing concentrations of ethanol, then in xylene and imbibed with paraffin liquid. Due to fragility of Zebrafish larvae, they were placed in biospecimen bags and 5 minutes in each step of processing was adapted. The paraffin imbibed tissue is taken out and embedded according to orientation as needed using a 10X dissection microscope. The formalin-fixed paraffin-embedded tissues were sectioned at desired thickness (4 μm) and mounted on positively charged slides. The slides were dried overnight at room temperature and incubated at 60°C for 45 minutes. The Hematoxylin and Eosin were purchased from Leica biosystems and staining was performed utilizing Leica ST5020 Automated Multistainer following the Hematoxylin-Eosin (HE) staining protocol at the SCC Tissue Pathology Shared Resource.
Transmission electron microscopy
Zebrafish larvae were extracted using the dissection microscope and were fixed with 4% Paraformaldehyde (EM grade), 2% Gluteraldehyde (EM grade), in 0.1M Sodium Cacodylate buffer for 48 hours at 4°C. Samples were then post fixed for 90 minutes in 1% Osmium tetroxide (OsO4) in 0.1M Sodium Cacodylate buffer, and rinsed three times for five minutes each in 0.1M Sodium Cacodylate buffer following dehydration in a graded acetone series-(50%, 60%, 75%, 85%, 95%, 100%) and kept in each concentration for 15 minutes on a rocker. Then the samples had two 15 minute treatments in 100% Propylene Oxide. Following dehydration, the samples were infiltrated in a graded Epon/Araldite (EMS) resin /Propylene Oxide series (1:3, 1:1, 3:1) for 60 minutes,120 minutes, and overnight respectfully. The following day samples were further infiltrated with pure resin for 45 minutes, 90 minutes, and then overnight. The livers were then embedded in resin plus BDMA (accelerator) and polymerized at 60°C for 48 hours. Semithin sections were stained with toluidine blue and were imaged on a Zeiss Axiovert 200M microscope. Ultrathin sections were stained with Lead Citrate and Uranyl Acetate before viewing on a Hitachi H7600 Transmission Electron Microscope at 80 kV equipped with a 2k X 2k AMT digital camera.
Quantitative gene expression studies
Gene expression studies for quantifying GCKR mRNA were performed on ZF larvae fed with normal and HFD. Total RNA was isolated using Absolutely RNA Mini Prep Kit (Agilent Technologies Inc., Santa Clara, CA), and was reverse transcribed using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories), according to the manufacturers’ protocols. For the quantification of GCKR mRNA quantitative PCR (qPCR) was performed using SsoAdvanced SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA). Real Time qPCR was then performed using Quant Studio6 in conjunction with GCKR forward and reverse primers (Integrated DNA Technologies, Skokie, Illinois, USA) and Bio-Rad’s SYBR Green Supermix with ROX) (Supplementary Table XX). Beta-actin was used as a normalizing control. Results were analyzed using ABI’s RQ Manager (v.1.2.1) software. Statistically significant difference in fold change was determined using the two-tailed t-test.
Bioinformatics and statistical analysis
Missense variants were designated as damaging using the in-silico predictions generated by tools like PolyPhen [30], SIFT [31], BONGO [32], LRT [33], Mutation Taster [34], and PolyPhen-2 [35]. The variants with score of four of six defined by these algorithms were considered potentially damaging. Data quality for SNP genotyping was checked by establishing reproducibility of control DNA samples. Departure from HWE of common variants in controls was tested using the Pearson chi-square test.
Gene-centric association analysis
For gene-centric analysis, we performed gene-centric burden tests to jointly analyze multiple non-synonymous or other likely functional variants including singleton variants by Combined Multivariate and Collapsing (CMC) method [36], to collapse rare variants in different MAF categories and evaluate the joint effect of common and rare variants using SVS, v 2.0 (Golden Helix, Bozeman, MT, USA). We also used the variance-component test within a random-effects model including the sequence kernel association test (SKAT) [37], which tests for association by evaluating the distribution of genetic effects for a group of variants instead of aggregating variants.
Single SNP association analysis
The genotype and allele frequencies in T2D cases were compared to those in control subjects using the chi-square test. Statistical evaluation of genetic effects on T2D risk used multivariate logistic regression analysis with adjustments for age, gender, and other covariates. Continuous traits with skewed sampling distributions (e.g., triglycerides or fasting glucose) were log-transformed before statistical analysis. However, for illustrative purposes, values were re-transformed into the original measurement scale. General mixed linear models were used to test the impact of genetic variants on transformed continuous traits using the variance-component test adjusted for the random-effects of relatedness and fixed effects of age, gender, BMI and disease implemented in SVS, v 2.0 (Golden Helix, Bozeman, MT, USA). Other significant covariates for each dependent trait were identified by Spearman’s correlation and step-wise multiple linear regression with an overall 5% level of significance using SPSS for Windows statistical package (version 18.0) (SPSS Inc., Chicago, USA). Mean values between cases and controls were compared by using an unpaired t test. To adjust for multiple testing, we used Bonferroni’s correction (0.05/number of tests performed).
Results
Of a total of 2,709 individuals studied, targeted sequencing was performed on 940 subjects and 1,769 subjects were used for the replication studies. All these participants were part of the AIDHS/SDS [15–17]. Of the 940 sequenced samples, 820 passed the stringent QC based on multiple parameters and were used for further analysis. S1 Table describes details of the lipid candidate gene regions selected for targeted resequencing. A summary of high-quality variants analyzed for their distribution and association with lipid-related traits, diabetes and other cardiometabolic risk factors is provided in S2 Table.
Our results revealed accumulation of several unknown rare (<1%) and less common variants (<10%) that were not found in any of the existing variant databases. For instance, our results of GCKR sequencing in Sikhs revealed clustering of 13 rare mutations and many of these were predicted to be damaging/ deleterious based on the in-silico prediction methods (Fig 2A). Gene-centric analysis for studying the aggregate effects of clustered variants within each gene, revealed significant burden of in the GCKR (p = 2.1x10-5) along with LPL (p = 1.6x10-3) and MLXIPL (p = 1.6x10-2) for increasing the risk for HTG (S1 Table).
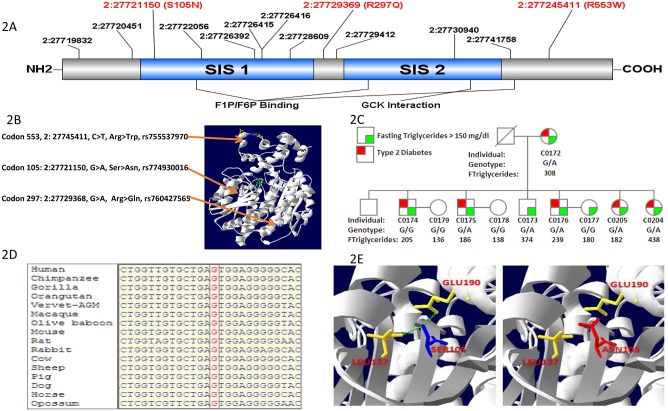
Fig 2.
A-E. Fig 2A. Targeted sequencing in the GCKR gene reveals three novel damaging mutations in SIS domains in Sikhs; Fig 2B. Crystal structure of human GCKR showing mutant residues (S105N) mapping to SIS-1 FIP/F6P binding domain while R297Q is located between SIS -1 and SIS-2 domains, while R553W maps near the GCK interaction domain; Fig 2C. Pedigree figure of a Sikh family showing overrepresentation of a rare damaging variant rs774930016 (S105N) segregating with T2D and hypertriglyceridemia; Fig 2D. Sequence alignment of SIS-1 domain reveals absolute conservation of rs774930016 at position S105N of the GCKR gene across species; Fig 2E. The wild-type residue (blue) forms hydrogen bonds with Glutamic Acid at position 190 and Leucine at position 137. However, the mutant residue (red) destabilize the folding of Fructose Binding domain by the loss of hydrogen bond with Glutamic Acid 190 and Leucine at position 137.
The present study is further mainly focused on 3 population-specific rare variants identified in GCKR gene (Fig 2B). The first functional variant (S105N), located on the Sugar Isomerase domain -1 (SIS-1), was functionally disruptive, and absent in Caucasians (n = 33,356), Africans (n = 5,197), Hispanic/ Latinos (5,788), East Asians (n = 4,324) in a large Exome Aggregation Consortium (ExAC) of multiethnic populations (Table 2). Two additional rare functional variants (R297Q and R553W) were also confined to this Sikh population only and were with high HTG in most carriers (Fig 2A and 2B, Table 2). Two of these three disruptive missense variants (S105N near fructose binding site and R553W near GCK interaction domain) were highly conserved across species (Figs 2D and 3B-1), while the mutant allele of R297Q variant was also found in cow and sheep in addition to its predominant presence in South Asians (Fig 3A-1).
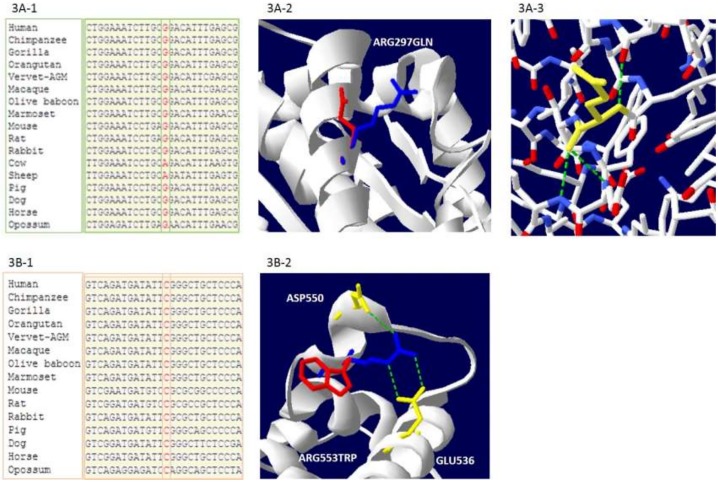
Fig 3.
A-B. Fig 3A-1. Sequence alignment of 297Gln reveals partial conservation of ancestral ‘G’ allele at positionArg297 to Gln297 of the GCKR gene across species. Fig 3A-2. Protein modeling of Arg297Gln showing the wild-type blue (Arg297) and mutant red (Gln297) residues. Fig 3A-3 The wild-type Arg297 forms a salt bridge with glutamic acid at position 63 glutamic acid at position 300, however, the new mutant residue is not in the correct position to make the same hydrogen bond as the original wild-type residue did. Fig 3B-1. Sequence alignment reveals a complete conservation of wild type ‘C’ allele at position of Arg553Trp of the GCKR gene across species. Fig 3B-2. Protein modeling of and Arg 553Trp mutations in the GCKR gene. The wild-type residue (blue) forms a hydrogen bond with glutamic acid at position 536, and aspartic acid at position 550. The size difference between wild-type and mutant residue makes that the new residue is not in the correct position to make the same hydrogen bond as the original wild-type residue. The wild-type residue was positively charged, the mutant residue is neutral. The mutant residue is more hydrophobic than the wild-type residue.
The three functionally tested damaging rare mutations in GCKR were at the fructose binding site and GCK binding site at or near the sugar isomerase (SIS-1-2) domains (Fig 2A). The disruptive allele at codon 105 is predicted to destabilize the folding of the fructose binding domain that results in the loss of hydrogen bond between Serine (105) and Glutamine (190) (Fig 2E). Interestingly, this variant was monomorphic in Europeans, East Asians, Africans and Latinos of the ExAC consortium and only 3 of 8250 South Asians from Pakistan were carriers (genotype frequency 0.00036) whereas 9 out of 3132 Sikhs were carriers of this variant (0.0029). This variant co-segregated between heterozygous carriers, HTG- and T2D phenotypes in one Sikh family. Of these over 83% of carriers in this family had HTG (ranging from 148mg/dl to 530 mg/dl) and 75% of carriers were diabetic. Similarly, two more rare functional variants (R297Q and R553W) were confined to this population only and were with high TG in most individuals (S3B and S3C Table).
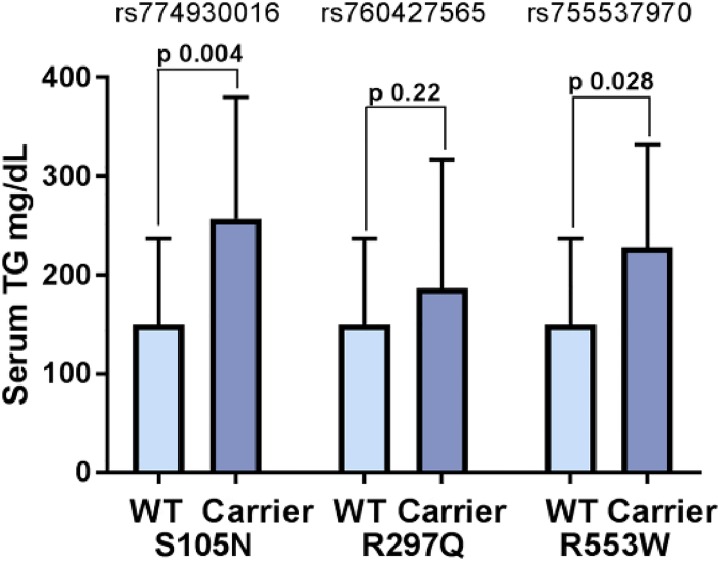
We investigated single variant association of each rare variant with diabetes and quantitative risk phenotypes (e.g. fasting glucose, body mass index (BMI), total cholesterol, LDL-C, HDL-C and TG) in discovery and replication cohorts. None of these variants showed any significant association with diabetes, fasting glucose or lipid traits except TG. As shown in Table 3 carriers of S105N (rs774930016) variant had a significant increased levels serum TG (β 0.59 ± 0.17; p = 0.001) after adjusting for age, gender and BMI. This association remained significant even after including T2D and family relatedness in the model (β 0.59 ± 0.17; p = 4.97 x 10−4) in replication cohort and in combined (discovery and replication) samples (β 0.55 ± 0.19; p = 0.004). Similar but marginally significant association of R553W (rs755537970) variant was observed in combined samples with triglycerides (β 0.51± 0.23; p = 0.028). However, no significant association was observed in R297Q (rs760427565) with TG (Table 3, Fig 4).
Table 3. Multivariate linear regression analysis showing association of three population-specific rare GCKR variants with serum triglycerides in discovery and replication cohortsGCKR S105N (rs774930016).
| Cohort | N | Carriers | Beta (SE) | P value (adj. age, gender, BMI) | Beta (SE) | P value (adj. age, gender, BMI, relatedness, T2D) |
| Discovery | 820 | 2 | 0.36 (0.49) | 0.46 | 0.31 (0.49) | 0.53 |
| Replication | 1769 | 7 | 0.59 (0.17) | 7 x 10−4 | 0.60 (0.17) | 4.32 x 10−4 |
| Combined | 2589 | 9 | 0.55 (0.19) | 0.004 | 0.55 (0.19) | 0.004 |
| GCKR R297Q (rs760427565) | ||||||
| Cohort | N | Carriers | Beta (SE) | P value (adj. age, gender, BMI) | Beta (SE) | P value (adj. age, gender, BMI, relatedness, T2D) |
| Discovery | 820 | 6 | 0.21 (0.31) | 0.49 | 0.21 (0.31) | 0.49 |
| Replication | 1769 | 12 | 0.14 (0.17) | 0.42 | 0.13 (0.17) | 0.45 |
| Combined | 2589 | 18 | 0.21 (0.17) | 0.20 | 0.20 (0.17) | 0.22 |
| GCKR R553W (rs755537970) | ||||||
| Cohort | N | Carriers | Beta (SE) | P value (adj. age, gender, BMI) | Beta (SE) | P value (adj. age, gender, BMI, relatedness, T2D) |
| Discovery | 820 | 4 | 0.45 (0.34) | 0.20 | 0.52 (0.34) | 0.13 |
| Replication | 1769 | 4 | 0.06 (0.32) | 0.86 | 0.16 (0.32) | 0.62 |
| Combined | 2589 | 8 | 0.41 (0.24) | 0.08 | 0.51 (0.23) | 0.028 |
Fig 4. Serum Triglyceride mean distribution among carriers and non-carriers of missense variants.
Bar graph shows the differences in the distribution of mean serum triglycerides among carriers and non-carriers of missense mutations (S105N, R297Q, and R553W) represented by rs774930016, rs760427565, rs755537970 variants, respectively. Data are shown in mean and standard deviation of means. P values are derived from the general mixed linear models used to test the impact of genetic variants on transformed continuous trait (TG) using the variance-component test adjusted for the random-effects of relatedness and fixed effects of age, gender, BMI and type 2 diabetes. Only association of S105N would remain significant after applying Bonferroni correction.
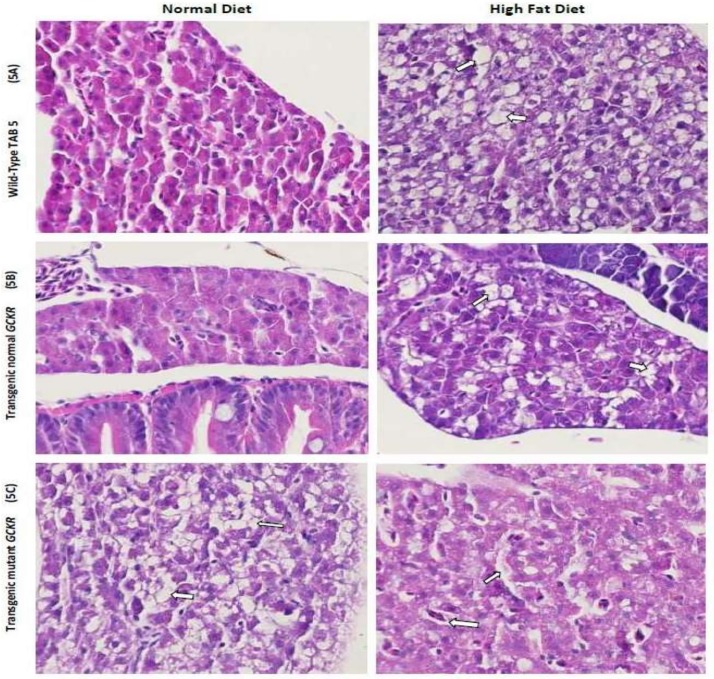
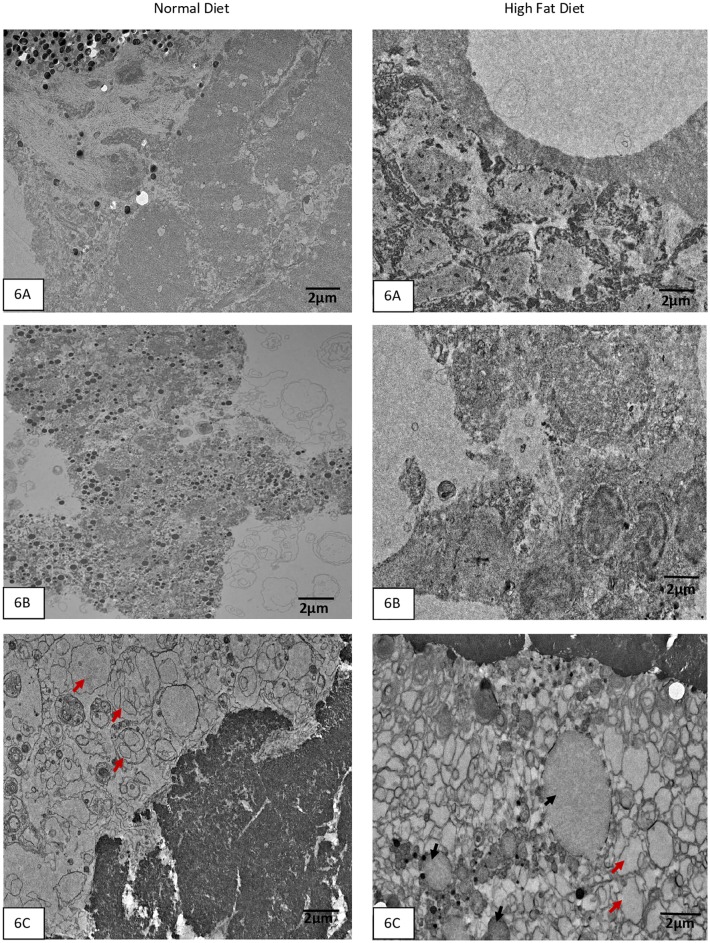
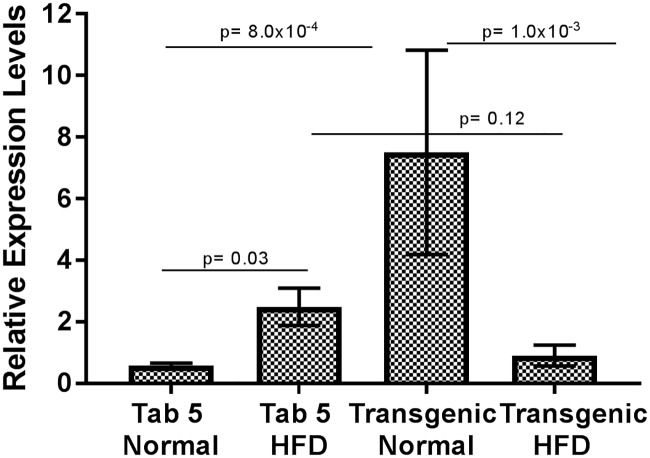
Based on significant association of these variants with HTG, we next evaluated the functional consequences of three South Asian population-specific variants by designing a humanized GCKR ZF model. The H&E images of liver of TAB-5, transgenic GCKRwt, and GCKRmut groups with normal diet and HFD are shown in Fig 5A–5C and S3A–S3C Fig. The fat disposition in liver hepatocytes of TAB-5 larvae was increased 3-4-fold in response to HFD. A similar increase in response to HFD was noticed in transgenic fish with wild type GCKR. However, in mutant transgenic fish exhibited a 3-fold increase in ectopic fat in hepatocytes with normal diet, with 80% hepatocytes having fat deposition, while transgenic mutants on HFD had hepatocytes loaded with fat showing a marked degeneration of hepatocyte nuclei with possible steatosis (Figs 5C and 6C). In response to HFD, mRNA expression of GCKR but increased about two folds in normal TAB-5 compared to normal diet. On the other hand, there was 7-fold increase in GCKR mut larvae even in the absence of HFD; whereas, the GCKR mut mRNA levels were restored to normal when fed on HFD (Fig 7).
Fig 5.
A-C. Fig 5A. Zebrafish liver histology of Hematoxylin and Eosin stained sections showing cellular differences in normal and high fat feeding for 2 weeks. A. A wild-type Tab-5 liver with normal hepatic cells and HFD showing normal liver with scattered fat cells (white arrow) showing 3+ fat without apoptosis. Fig 5B. Transgenic normal GCKR with normal hepatocytes in larvae fed on normal diet and normal liver with scattered fat cells (white arrow) in transgenic larvae fed on HFD with no apoptosis. Fig 5C. Transgenic mutant GCKR with normal diet shows liver with fatty metamorphosis and scattered hepatic cell apoptosis (white arrow) with 4+ fat. Transgenic mutant GCKR with HFD shows 4+ fat with severe cellular damage and higher fatty infiltration than the normal and transgenic normal fish. Most hepatic cells also contain vacuoles of fat and there is severe disorganization of the hepatic structure.
Fig 6.
A-C. Transmission Electron Microscopy of zebrafish liver sections at medium magnification show the presence of neutral fat with HFD in normal Tab-5 Fig 6A, transgenic with normal GCKR Fig 6B, and transgenic with mutant GCKR Fig 6C. Neutral fat appears as small round vesicles with no structure. There is abnormal fat accumulation in the transgenic mutant fish with HFD (Fig 6C). Black arrows point to round vesicle like structures with empty content which are fat droplets. The red arrows point to two huge abnormal structures which appear like fusion of large neutral fat vesicles in the transgenic mutant with HFD. However, the transgenic mutant with normal diet shows abnormal structures with possible accumulation of phospholipids (red arrows) in hepatic cells suggesting abnormal metabolic function.
Fig 7. GCKR Relative Expression Levels.
Graph summarizes the relative mRNA expression levels of GCKR normalized by beta actin in zebrafish larvae of wildtype (WT) Tab-5 and transgenic mutant fish fed on a normal and high fat diet (HFD). Data are presented as mean and standard error of mean using Tab-5 WT as reference. Significant differences in the relative expression of GCKR mRNA were detected between Tab-5 (WT) and the transgenic mutant on a normal diet; and between transgenic mutants on a normal diet vs. HFD.
Discussion
In this investigation, we have attempted to identify functional variants by resequencing 13 known candidate genes of dyslipidemia using an endogamous population of Punjabi Sikhs known to have high risk for cardiovascular diseases [11,12,38–41]. Despite considerable success of GWAS, whole-genome, and exome sequencing, including studies from our group [15,42–44], the genetic mechanisms that predispose people to metabolic and cardiovascular disease risk factors remain poorly understood. Of these 13 selected loci with prior evidence of association with three major lipids (HDL cholesterol, LDL cholesterol, and TG) in European populations [10,12,45,46], variants in ANGPTL3, GCKR, MLXIPL, LPL, TRIB1 and APOE genes have been shown to be associated with lipid phenotypes in South Asians [12]. Fine mapping of ~195 kb region encompassing Chr11q23.3 [APO-A1-C3-A4-A5, ZNF259, and BUD13] by targeted genotyping revealed a strong association of this region with HTG (rs964184; p = 1.6x10-39) in Punjabi Sikhs and South Asians) [11]. Here we have intended to capture putatively functional rare and less common variants from coding, non-coding, and intergenic regions including variants influencing gene regulation and expression within and around these known candidate genes. The degree of clinical heterogeneity existing in the CAD or cardiometabolic phenotypes imposes serious limitations in our ability to effectively measure genetic risk, environmental exposure, and their interactions. Additionally, most post-GWA studies on candidate gene sequencing have predominantly been focused on European populations which provide limited information on the usefulness of variants in populations of non-European ancestry. Moreover, the post-GWAS exome arrays could capture the majority of low-frequency variants in European populations only when the sample size exceeded >300,000 [47]. However, such studies in other disparate populations are sparse. The current investigation in family and population based sample from the AIDHS/SDS is an effort to identify missing heritability associated with GWAS-driven loci of dyslipidemia, specifically the HTG by using candidate gene resequencing.
As expected, the AIDHS/SDS, being an endogamous and relatively homogenous population, was enriched with rare and less common variants. Enrichment of functional variants in cases with HTG along with our focus on individuals with extreme trait values (TG) increased our power to discover pathogenic variants and aided the discovery of multiple rare and common known and novel variants in splice regions, 5’UTRs, 3’UTRs, intronic, and missense (loss-of-function) variants. Moreover, this ethnic subgroup of Sikhs of North India were enrolled from one single geographic location with shared environmental and cultural traits, which further has reduced the environmental and cultural heterogeneity.
Gene-centric analysis of the identified variants revealed a significant burden of variants for increasing HTG risk in GCKR (p = 2.1x10-5), LPL (p = 1.6x10-3) and MLXIPL (p = 1.6x10-2). The GCKR is glucokinase regulatory protein that inhibits glucokinase (GCK) by forming a complex with the enzyme in the liver, which plays a role in glucose homeostasis [48]. Fructose 6- phosphate (F6P) enhance while fructose 1-phosphate (F1P) reduce the GCKR-mediated inhibition of GCK [49]. Lipoprotein lipase (LPL) has long been recognized as an enzyme that hydrolysis of triglyceride- rich lipoproteins to release free fatty acids for energy metabolism [50]. The MLX1PL encodes a basic helix-loop-helix leucine zipper transcription factor of the Myc/Max/Mad superfamily. This protein forms a heterodimeric complex and binds and activates carbohydrate response element (ChoRE) motifs in the promoters of triglyceride synthesis genes [51]. Common variants within and around these genes are associated with increased levels of TG and CAD in multiethnic GWAS and metanalysis studies including Sikhs [10,39]. To test their phenotypic effects and to evaluate metabolic consequences in vivo, we focused on three putative variants identified in the human GCKR gene by building four transgenic humanized ZF models. These variants were located near the fructose binding site or GCK binding sites at the sugar isomerase domains of the human GCKR gene. Evidently, the human GCKR is about 3 times larger than the ZF GCKR and it only shows 41% similarity with humans (S2-B Fig). Due to the absence of 386 amino acids in the ZF GCKR gene, our three functional variants fall outside the ZF GCKR protein.
Despite this dissimilarity, ZF are a well-suited model for studies involving human energy metabolism because the pathways of lipid storage and transport are conserved across species [52]. Further, the dietary studies performed in a ZF model for developing atherosclerosis and hepatic steatosis in response to a high-cholesterol diet revealed the potential strength of this model for analyzing diet-induced phenotypes [53]. In this study, the ZF larvae exposed to HFD and normal diet revealed a 2 to 3 fold increase in the fat accumulation in hepatocytes in response to HFD both in TAB-5, and control transgenic fish (with normal human GCKR) with no apoptosis. However, the observed 4+fold increase in liver fat accumulation with at least 1 apoptotic cell every hundred hepatic cells, even in the absence of a HFD in transgenic GCKRmut, suggests the impaired function of GCKR due to mutations, which may impair GCKR to act promptly in response to the increased concentration of fructose 6-phosphate. This consequently would lead to uninterrupted release of GCK in the liver, resulting in increased uptake of glucose and eventually leading to de novo lipogenesis [54]. Alternatively, studies suggest that GCKR stabilizes and protects GCK from degradation. Thus, the increase expression with impaired function of GCKR may result in reduced GCK activity or function, which would give rise to impaired glucose tolerance and hepatic fat accumulation [55]. Evidently, from these studies it appears that the GCKR could be a thrifty gene and the functionally disrupted variants in the GCKR in Punjabi Sikhs may enhance ectopic fat storage defects even in the absence of HFD, as revealed in the transgenic ZF. Not only did most hepatic cells contain vacuoles of fat, but the structure of hepatocytes was disorganized due to fatty metamorphosis and severe disorganization of the hepatic structure and hepatocyte nuclei with possible steatosis in the absence of HFD in GCKRmut compared to GCKRwt or WT (TAB-5) Fig 5C. Also, there was abnormal accumulation of lipids (phospholipids) other than the neutral fat in mutant transgenic in the absence of HFD (Fig 6C). Phospholipid accumulation in hepatic cells are often seen in people with metabolic disorders.
A previously known functional variant (Proline to Leucine) at position 446 of the GCKR (rs1260326) was identified as a novel locus for TG metabolism in Caucasian GWAS, and has since been robustly replicated in multiple genome-wide studies of plasma TG [39]. The same variant has also been shown to influence fatty liver disease in children and adults [56]. Of note, the minor risk allele frequency differed significantly between Sikhs (0.27) and other South Asians [e.g. Gujarati Indians (GIH 0.19) and South Asians from Pakistan (ExAC) 0.20], also European Caucasians (0.36). Although our study confirmed the association of this SNP rs1260326 with TG in Sikhs (β 0.09, ± 0.02, p = 3.42x10-5), the genetic variance explained by this variant was <2% in Sikhs. Whereas, the rs774930016 (representing codon 105) explained 38% of HTG. Both rs760427565 for codon 297 and rs75537970 for codon 553 explained ~25% of HTG genetic variance among carriers. The association of codon 105 with TG remained statistically significant even after controlling for BMI, age, gender and T2D (Table 3), suggesting its functional role in increasing HTG risk independent of T2D. Notably, these variants were only restricted to South Asian populations; indeed the rs755537970 (of codon 553) appears to be confined to Punjabi Sikhs (Table 2).
We and others have shown that Asian Indian populations may possess a different physiology of obesity [17,57–59]. South Asians generally have a non-obese BMI with lower muscle mass and increased visceral fat, which is also associated with their high rates of T2D in the absence of obesity [58,60–65]. Even results of computed tomography (CT) scans show that Asian Indians have 30% more body fat than age- and BMI-matched African American men, and 21% more body fat than Swedish men [66–68]. Thus, Asian Indians are metabolically obese despite a non-obese BMI. The uneven distribution of fat in insulin sensitive organs like the liver or pancreas increases the risk for development of insulin resistance, T2D, and non-alcoholic fatty liver disease (NAFLD) [69], which are common in Indians. Based on the results of this study, carriers of these evolutionarily conserved variants (specifically S105N and R553W) will have a high risk of ectopic fat deposition and increased risk for NAFLD in the absence of overt obesity.
Overall, successful humanized transgenic GCKR mut-expressing D. rerio has provided a platform for our ongoing studies to define the precise mechanisms of metabolic derangement perhaps by modulating the GCKR-GCK complex leading to HTG and T2D in humans. Limitations of our study include the lack of data on GCK mRNA, GCKR/GCK protein quantification and GCK activity, which may provide more insight on the putative effects of functional mutations on regulation of GCKR and clarify the effects of exposure of HFD on the humanized ZF. Our results agree with the earlier reports of targeted improvement of GCK activity by liver–specific GCKR inhibition which may lower the risk of HTG [49]
In summary, our study for the first time reports a causal role of rare disruptive variants in GCKR for increasing serum TG levels independent of T2D in Punjabi Sikhs. These results may also partly support the “non-obese-metabolic obese phenotype” of Asian Indians linked to increased risk for developing cardiovascular diseases.
Supporting information
Flow Chart summarizes research design and targeted sequencing and replication and functional studies workflow.
(PDF)
S2 Fig A. Upper gray horizontal bar is the protein sequence is human GCKR (NP_001477.2) with 625 amino acids and lower red horizontal block is the zebrafish GCKR (XP_002665191.3) protein sequence. S2 Fig B. Protein sequence alignment of human and zebrafish GCKR. Upper rows represent human residues and lower rows represent zebrafish residues starting at codon 387 of human GCKR. The black spaces and + symbols indicate low degree of homolog between human and zebrafish, only 94 out of 233 (41%) residues showed complete alignment.
(DOCX)
General observation of zebrafish larvae from three groups fed on a normal and high fat diet at 4X magnification.
(DOCX)
(DOCX)
(DOCX)
A-C. (S3 Table-A). Demographic and clinical traits of carriers of GCKR S105N (rs774930016) variant in subjects from the AIDHS (S3 Table-B). Demographic and clinical traits of carriers of GCKR R297Q (rs760427565) variant in subjects from the AIDHS. (S3 Table-C). Demographic and clinical traits of carriers of GCKR R553W (rs755537970) variant in AIDHS.
(DOCX)
(DOCX)
Acknowledgments
Authors thank all the participants of AIDHS/SDS who made this study possible. We thank the Stephenson Cancer Center at the University of Oklahoma, Oklahoma City for the use of Histology and Immunohistochemistry Core, which provided Processing and Embedding and Tissue staining service and Nikon Microscopic imaging service. Authors thank Dr. Amnon Dr. Schlegel, University of Utah for providing us D. rerio liver fatty acid binding protein (L-FABP) promoter. Technical help provided by Dr. Bishwa Sapkota, Jayaraman Muralidharan, Dr. Anil Singh, Louisa Williams, and Sheeja Aravindan is duly acknowledged.
Data Availability
Data set containing fragment coordinates for TargetSeq Custom Enrichment Kit which was designed to target sequencing the region containing the complete genomic sequence of the GCKR gene in locus 2p23.3 (chr2: 20996301-21494945; GRCh37/ hg19 reference human genome) are within the Supporting Information files. Public sharing of other variant data presented in the article will be made available on dbGap https://www.ncbi.nlm.nih.gov/gap/docs/submissionguide/. These data are available from the Principal investigator through collaborations by contacting dharambir-sanghera@ouhsc.edu and/or the head of Institutional Data Access / Ethics Committee (contact Donna Hogan via email IRB@ouhsc.edu) for researchers who meet the criteria for access to confidential data.
Funding Statement
D.K.S: The Sikh Diabetes Study/ Asian Indian Diabetic Heart Study was supported by National Institutes of Health grants-R01DK082766 (NIDDK) (https://www.niddk.nih.gov/), NOT-HG-11-009 (NHGRI) (https://www.genome.gov/), and grants from the Presbyterian Health Foundation (http://phfokc.com/). Sequencing services were provided through the RS&G Service by the Northwest Genomics Center at the University of Washington, Department of Genome Sciences, under U.S. Federal Government contract number HHSN268201100037C from the National Heart, Lung, and Blood Institute of the National Institutes of Health (https://www.nhlbi.nih.gov/). Cancer Core Lab: Funding for the use of Histology and Immunohistochemistry Core was provided by an Institutional Development Award (IDeA) grant number P20 GM103639 from the National Institute of General Medical Sciences of the National Institutes of Health (https://www.nigms.nih.gov/Research/DRCB/IDeA/pages/INBRE.aspx), and Tissue Pathology Shared Resources by National Cancer Institute Grant P30CA225520 of the National Institutes of Health (https://www.cancer.gov/). The ZebraFish laboratory of J.K.F. was funded by grants from Hyundai Hope On Wheels (https://hyundaihopeonwheels.org/), the Oklahoma Center for the Advancement of Science and Technology (HRP-067) (https://www.ok.gov/ocast/), a National Institute of General Medical Sciences (P20 GM103447) INBRE pilot project award (https://www.nigms.nih.gov/Research/DRCB/IDeA/pages/INBRE.aspx), and the E.L. & Thelma Gaylord Endowed Chair of the Children’s Hospital Foundation (https://chfkids.com/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Snieder H, van Doornen LJ, Boomsma DI (1999) Dissecting the genetic architecture of lipids, lipoproteins, and apolipoproteins: lessons from twin studies. Arterioscler Thromb Vasc Biol 19: 2826–2834. [DOI] [PubMed] [Google Scholar]
- 2.Brunham LR, Kruit JK, Hayden MR, Verchere CB (2010) Cholesterol in beta-cell dysfunction: the emerging connection between HDL cholesterol and type 2 diabetes. Curr Diab Rep 10: 55–60. 10.1007/s11892-009-0090-x [DOI] [PubMed] [Google Scholar]
- 3.Hokanson JE, Austin MA (1996) Plasma triglyceride level is a risk factor for cardiovascular disease independent of high-density lipoprotein cholesterol level: a meta-analysis of population-based prospective studies. J Cardiovasc Risk 3: 213–219. [PubMed] [Google Scholar]
- 4.Drenos F, Talmud PJ, Casas JP, Smeeth L, Palmen J, et al. (2009) Integrated associations of genotypes with multiple blood biomarkers linked to coronary heart disease risk. Hum Mol Genet 18: 2305–2316. 10.1093/hmg/ddp159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Libby P (2005) The forgotten majority: unfinished business in cardiovascular risk reduction. J Am Coll Cardiol 46: 1225–1228. 10.1016/j.jacc.2005.07.006 [DOI] [PubMed] [Google Scholar]
- 6.Sing CF, Orr JD (1978) Analysis of genetic and environmental sources of variation in serum cholesterol in Tecumseh, Michigan. IV. Separation of polygene from common environment effects. Am J Hum Genet 30: 491–504. [PMC free article] [PubMed] [Google Scholar]
- 7.Robertson FW (1981) The genetic component in coronary heart disease—a review. Genet Res 37: 1–16. [DOI] [PubMed] [Google Scholar]
- 8.Rissanen AM, Nikkila EA (1977) Coronary artery disease and its risk factors in families of young men with angina pectoris and in controls. Br Heart J 39: 875–883. 10.1136/hrt.39.8.875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sprecher DL, Hein MJ, Laskarzewski PM (1994) Conjoint high triglycerides and low HDL cholesterol across generations. Analysis of proband hypertriglyceridemia and lipid/lipoprotein disorders in first-degree family members. Circulation 90: 1177–1184. 10.1161/01.cir.90.3.1177 [DOI] [PubMed] [Google Scholar]
- 10.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, et al. (2013) Discovery and refinement of loci associated with lipid levels. Nat Genet 45: 1274–+. 10.1038/ng.2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braun TR, Been LF, Singhal A, Worsham J, Ralhan S, et al. (2012) A Replication Study of GWAS-Derived Lipid Genes in Asian Indians: The Chromosomal Region 11q23.3 Harbors Loci Contributing to Triglycerides. PLoS One 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, et al. (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466: 707–713. 10.1038/nature09270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schierer A, Been LF, Ralhan S, Wander GS, Aston CE, et al. (2012) Genetic variation in cholesterol ester transfer protein, serum CETP activity, and coronary artery disease risk in Asian Indian diabetic cohort. Pharmacogenetics and Genomics 22: 95–104. 10.1097/FPC.0b013e32834dc9ef [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weissglas-Volkov D, Pajukanta P (2010) Genetic causes of high and low serum HDL-cholesterol. J Lipid Res 51: 2032–2057. 10.1194/jlr.R004739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saxena R, Saleheen D, Been LF, Garavito ML, Braun T, et al. (2013) Genome-Wide Association Study Identifies a Novel Locus Contributing to Type 2 Diabetes Susceptibility in Sikhs of Punjabi Origin From India. Diabetes 62: 1746–1755. 10.2337/db12-1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saxena R, Bjonnes A, Prescott J, Dib P, Natt P, et al. (2014) Genome-wide association study identifies variants in casein kinase II (CSNK2A2) to be associated with leukocyte telomere length in a Punjabi Sikh diabetic cohort. Circ Cardiovasc Genet 7: 287–295. 10.1161/CIRCGENETICS.113.000412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanghera DK, Bhatti JS, Bhatti GK, Ralhan SK, Wander GS, et al. (2006) The Khatri Sikh Diabetes Study (SDS): Study design, methodology, sample collection, and initial results. Hum Biol 78: 43–63. 10.1353/hub.2006.0027 [DOI] [PubMed] [Google Scholar]
- 18.American Diabetes Association (2004) Diagnosis and classification of diabetes mellitus. Diabetes Care 27 Suppl 1: S5–S10. [DOI] [PubMed] [Google Scholar]
- 19.Yuan G, Al-Shali KZ, Hegele RA (2007) Hypertriglyceridemia: its etiology, effects and treatment. CMAJ 176: 1113–1120. 10.1503/cmaj.060963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16: 1215 10.1093/nar/16.3.1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanghera DK, Ortega L, Han S, Singh J, Ralhan SK, et al. (2008) Impact of nine common type 2 diabetes risk polymorphisms in Asian Indian Sikhs: PPARG2 (Pro12Ala), IGF2BP2, TCF7L2 and FTO variants confer a significant risk. BMC Med Genet 9: 59 10.1186/1471-2350-9-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, et al. (2009) Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461: 272–276. 10.1038/nature08250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanghera DK, Been LF, Ralhan S, Wander GS, Mehra NK, et al. (2011) Genome-wide linkage scan to identify loci associated with type 2 diabetes and blood lipid phenotypes in the Sikh Diabetes Study. PLoS One 6: e21188 10.1371/journal.pone.0021188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sapkota BR, Hopkins R, Bjonnes A, Ralhan S, Wander GS, et al. (2016) Genome-wide association study of 25(OH) Vitamin D concentrations in Punjabi Sikhs: Results of the Asian Indian diabetic heart study. J Steroid Biochem Mol Biol 158: 149–156. 10.1016/j.jsbmb.2015.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Been LF, Ralhan S, Wander GS, Mehra NK, Singh J, et al. (2011) Variants in KCNQ1 increase type II diabetes susceptibility in South Asians: A study of 3,310 subjects from India and the US. Bmc Medical Genetics 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, et al. (2007) The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn 236: 3088–3099. 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- 27.Her GM, Chiang CC, Chen WY, Wu JL (2003) In vivo studies of liver-type fatty acid binding protein (L-FABP) gene expression in liver of transgenic zebrafish (Danio rerio). FEBS Lett 538: 125–133. 10.1016/s0014-5793(03)00157-1 [DOI] [PubMed] [Google Scholar]
- 28.Langenau DM, Ferrando AA, Traver D, Kutok JL, Hezel JP, et al. (2004) In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proc Natl Acad Sci U S A 101: 7369–7374. 10.1073/pnas.0402248101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson JM, Bunte RM, Carty AJ (2009) Evaluation of rapid cooling and tricaine methanesulfonate (MS222) as methods of euthanasia in zebrafish (Danio rerio). J Am Assoc Lab Anim Sci 48: 785–789. [PMC free article] [PubMed] [Google Scholar]
- 30.Ramensky V, Bork P, Sunyaev S (2002) Human non-synonymous SNPs: server and survey. Nucleic Acids Res 30: 3894–3900. 10.1093/nar/gkf493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee W, Zhang Y, Mukhyala K, Lazarus RA, Zhang Z (2009) Bi-directional SIFT predicts a subset of activating mutations. PLoS One 4: e8311 10.1371/journal.pone.0008311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng TM, Lu YE, Vendruscolo M, Lio P, Blundell TL (2008) Prediction by graph theoretic measures of structural effects in proteins arising from non-synonymous single nucleotide polymorphisms. PLoS Comput Biol 4: e1000135 10.1371/journal.pcbi.1000135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chun S, Fay JC (2009) Identification of deleterious mutations within three human genomes. Genome Res 19: 1553–1561. 10.1101/gr.092619.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwarz JM, Cooper DN, Schuelke M, Seelow D (2014) MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 11: 361–362. 10.1038/nmeth.2890 [DOI] [PubMed] [Google Scholar]
- 35.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, et al. (2010) A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li B, Leal SM (2008) Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet 83: 311–321. 10.1016/j.ajhg.2008.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ionita-Laza I, Lee S, Makarov V, Buxbaum JD, Lin X (2013) Sequence kernel association tests for the combined effect of rare and common variants. Am J Hum Genet 92: 841–853. 10.1016/j.ajhg.2013.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang WS, Nevin DN, Peng R, Brunzell JD, Deeb SS (1995) A mutation in the promoter of the lipoprotein lipase (LPL) gene in a patient with familial combined hyperlipidemia and low LPL activity. Proc Natl Acad Sci U S A 92: 4462–4466. 10.1073/pnas.92.10.4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, et al. (2007) Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316: 1331–1336. 10.1126/science.1142358 [DOI] [PubMed] [Google Scholar]
- 40.Baroukh N, Bauge E, Akiyama J, Chang J, Afzal V, et al. (2004) Analysis of apolipoprotein A5, c3, and plasma triglyceride concentrations in genetically engineered mice. Arterioscler Thromb Vasc Biol 24: 1297–1302. 10.1161/01.ATV.0000130463.68272.1d [DOI] [PubMed] [Google Scholar]
- 41.Johansen CT, Kathiresan S, Hegele RA (2011) Genetic determinants of plasma triglycerides. J Lipid Res 52: 189–206. 10.1194/jlr.R009720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kooner JS, Saleheen D, Sim X, Sehmi J, Zhang WH, et al. (2011) Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet 43: 984–U994. 10.1038/ng.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Replication DIG, Meta-analysis C, Asian Genetic Epidemiology Network Type 2 Diabetes C, South Asian Type 2 Diabetes C, Mexican American Type 2 Diabetes C, et al. (2014) Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet 46: 234–244. 10.1038/ng.2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reddivari Lavanya Sapkota Bishwa R R A, Liang Yundi, Aston Christopher, Sidorov Evgeny, Vanamala Jairam KP, Sanghera Dharambir K (2017) Metabolite signatures of diabetes with cardiovascular disease: a pilot investigation. Metabolomics 13: 154. [Google Scholar]
- 45.Talmud PJ, Drenos F, Shah S, Shah T, Palmen J, et al. (2009) Gene-centric association signals for lipids and apolipoproteins identified via the HumanCVD BeadChip. Am J Hum Genet 85: 628–642. 10.1016/j.ajhg.2009.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, et al. (2008) Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet 40: 189–197. 10.1038/ng.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahajan A, Wessel J, Willems SM, Zhao W, Robertson NR, et al. (2018) Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes. Nat Genet 50: 559–571. 10.1038/s41588-018-0084-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hayward BE, Dunlop N, Intody S, Leek JP, Markham AF, et al. (1998) Organization of the human glucokinase regulator gene GCKR. Genomics 49: 137–142. 10.1006/geno.1997.5195 [DOI] [PubMed] [Google Scholar]
- 49.Raimondo A, Rees MG, Gloyn AL (2015) Glucokinase regulatory protein: complexity at the crossroads of triglyceride and glucose metabolism. Curr Opin Lipidol 26: 88–95. 10.1097/MOL.0000000000000155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Havel RJ (2010) Triglyceride-rich lipoproteins and plasma lipid transport. Arterioscler Thromb Vasc Biol 30: 9–19. 10.1161/ATVBAHA.108.178756 [DOI] [PubMed] [Google Scholar]
- 51.Yamashita H, Takenoshita M, Sakurai M, Bruick RK, Henzel WJ, et al. (2001) A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc Natl Acad Sci U S A 98: 9116–9121. 10.1073/pnas.161284298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlegel A, Stainier DY (2007) Lessons from "lower" organisms: what worms, flies, and zebrafish can teach us about human energy metabolism. PLoS Genet 3: e199 10.1371/journal.pgen.0030199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schlegel A (2012) Studying non-alcoholic fatty liver disease with zebrafish: a confluence of optics, genetics, and physiology. Cell Mol Life Sci 69: 3953–3961. 10.1007/s00018-012-1037-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brouwers M, Jacobs C, Bast A, Stehouwer CDA, Schaper NC (2015) Modulation of Glucokinase Regulatory Protein: A Double-Edged Sword? Trends Mol Med 21: 583–594. 10.1016/j.molmed.2015.08.004 [DOI] [PubMed] [Google Scholar]
- 55.Lloyd DJ, St Jean DJ Jr., Kurzeja RJ, Wahl RC, Michelsen K, et al. (2013) Antidiabetic effects of glucokinase regulatory protein small-molecule disruptors. Nature 504: 437–440. 10.1038/nature12724 [DOI] [PubMed] [Google Scholar]
- 56.Santoro N, Zhang CK, Zhao H, Pakstis AJ, Kim G, et al. (2012) Variant in the glucokinase regulatory protein (GCKR) gene is associated with fatty liver in obese children and adolescents. Hepatology 55: 781–789. 10.1002/hep.24806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanghera DK, Blackett PR (2012) Type 2 Diabetes Genetics: Beyond GWAS. J Diabetes Metab 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McKeigue PM, Pierpoint T, Ferrie JE, Marmot MG (1992) Relationship of glucose intolerance and hyperinsulinaemia to body fat pattern in south Asians and Europeans. Diabetologia 35: 785–791. [DOI] [PubMed] [Google Scholar]
- 59.Sanghera DK, Dodani S. (2016) Cardiovascular disease in South Asians; Risk factors, genetics and environment Medicine Update 2016-1 New Delhi, London, Philadelphia, Panama: The Health Sciences Publishers. [Google Scholar]
- 60.Abate N, Chandalia M (2001) Ethnicity and type 2 diabetes: focus on Asian Indians. J Diabetes Complications 15: 320–327. [DOI] [PubMed] [Google Scholar]
- 61.Zimmet PZ (1992) Kelly West Lecture 1991. Challenges in diabetes epidemiology—from West to the rest. Diabetes Care 15: 232–252. 10.2337/diacare.15.2.232 [DOI] [PubMed] [Google Scholar]
- 62.Nakagami T, Qiao Q, Carstensen B, Nhr-Hansen C, Hu G, et al. (2003) Age, body mass index and Type 2 diabetes-associations modified by ethnicity. Diabetologia 46: 1063–1070. 10.1007/s00125-003-1158-9 [DOI] [PubMed] [Google Scholar]
- 63.Karter AJ, Mayer-Davis EJ, Selby JV, D’Agostino RB Jr., Haffner SM, et al. (1996) Insulin sensitivity and abdominal obesity in African-American, Hispanic, and non-Hispanic white men and women. The Insulin Resistance and Atherosclerosis Study. Diabetes 45: 1547–1555. 10.2337/diab.45.11.1547 [DOI] [PubMed] [Google Scholar]
- 64.Wang J, Thornton JC, Russell M, Burastero S, Heymsfield S, et al. (1994) Asians have lower body mass index (BMI) but higher percent body fat than do whites: comparisons of anthropometric measurements. Am J Clin Nutr 60: 23–28. 10.1093/ajcn/60.1.23 [DOI] [PubMed] [Google Scholar]
- 65.McKeigue PM, Shah B, Marmot MG (1991) Relation of central obesity and insulin resistance with high diabetes prevalence and cardiovascular risk in South Asians. Lancet 337: 382–386. 10.1016/0140-6736(91)91164-p [DOI] [PubMed] [Google Scholar]
- 66.Banerji MA, Faridi N, Atluri R, Chaiken RL, Lebovitz HE (1999) Body composition, visceral fat, leptin, and insulin resistance in Asian Indian men. J Clin Endocrinol Metab 84: 137–144. 10.1210/jcem.84.1.5371 [DOI] [PubMed] [Google Scholar]
- 67.Banerji MA, Chaiken RL, Gordon D, Kral JG, Lebovitz HE (1995) Does intra-abdominal adipose tissue in black men determine whether NIDDM is insulin-resistant or insulin-sensitive? Diabetes 44: 141–146. 10.2337/diab.44.2.141 [DOI] [PubMed] [Google Scholar]
- 68.Chowdhury B, Lantz H, Sjostrom L (1996) Computed tomography-determined body composition in relation to cardiovascular risk factors in Indian and matched Swedish males. Metabolism 45: 634–644. [DOI] [PubMed] [Google Scholar]
- 69.Kahn HS (1993) Choosing an index for abdominal obesity: an opportunity for epidemiologic clarification. J Clin Epidemiol 46: 491–494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow Chart summarizes research design and targeted sequencing and replication and functional studies workflow.
(PDF)
S2 Fig A. Upper gray horizontal bar is the protein sequence is human GCKR (NP_001477.2) with 625 amino acids and lower red horizontal block is the zebrafish GCKR (XP_002665191.3) protein sequence. S2 Fig B. Protein sequence alignment of human and zebrafish GCKR. Upper rows represent human residues and lower rows represent zebrafish residues starting at codon 387 of human GCKR. The black spaces and + symbols indicate low degree of homolog between human and zebrafish, only 94 out of 233 (41%) residues showed complete alignment.
(DOCX)
General observation of zebrafish larvae from three groups fed on a normal and high fat diet at 4X magnification.
(DOCX)
(DOCX)
(DOCX)
A-C. (S3 Table-A). Demographic and clinical traits of carriers of GCKR S105N (rs774930016) variant in subjects from the AIDHS (S3 Table-B). Demographic and clinical traits of carriers of GCKR R297Q (rs760427565) variant in subjects from the AIDHS. (S3 Table-C). Demographic and clinical traits of carriers of GCKR R553W (rs755537970) variant in AIDHS.
(DOCX)
(DOCX)
Data Availability Statement
Data set containing fragment coordinates for TargetSeq Custom Enrichment Kit which was designed to target sequencing the region containing the complete genomic sequence of the GCKR gene in locus 2p23.3 (chr2: 20996301-21494945; GRCh37/ hg19 reference human genome) are within the Supporting Information files. Public sharing of other variant data presented in the article will be made available on dbGap https://www.ncbi.nlm.nih.gov/gap/docs/submissionguide/. These data are available from the Principal investigator through collaborations by contacting dharambir-sanghera@ouhsc.edu and/or the head of Institutional Data Access / Ethics Committee (contact Donna Hogan via email IRB@ouhsc.edu) for researchers who meet the criteria for access to confidential data.