Abstract
Expansion of a polyglutamine tract in the huntingtin protein causes neuronal degeneration and death in Huntington's disease patients, but the molecular mechanisms underlying polyglutamine-mediated cell death remain unclear. Previous studies suggest that expanded polyglutamine tracts alter transcription by sequestering glutamine rich transcriptional regulatory proteins, thereby perturbing their function. We tested this hypothesis in Caenorhabditis elegans neurons expressing a human huntingtin fragment with an expanded polyglutamine tract (Htn-Q150). Loss of function alleles and RNA interference (RNAi) were used to examine contributions of C. elegans cAMP response element-binding protein (CREB), CREB binding protein (CBP), and histone deacetylases (HDACs) to polyglutamine-induced neurodegeneration. Deletion of CREB (crh-1) or loss of one copy of CBP (cbp-1) enhanced polyglutamine toxicity in C. elegans neurons. Loss of function alleles and RNAi were then used to systematically reduce function of each C. elegans HDAC. Generally, knockdown of individual C. elegans HDACs enhanced Htn-Q150 toxicity, but knockdown of C. elegans hda-3 suppressed toxicity. Neuronal expression of hda-3 restored Htn-Q150 toxicity and suggested that C. elegans HDAC3 (HDA-3) acts within neurons to promote degeneration in response to Htn-Q150. Genetic epistasis experiments suggested that HDA-3 and CRH-1 (C. elegans CREB homolog) directly oppose each other in regulating transcription of genes involved in polyglutamine toxicity. hda-3 loss of function failed to suppress increased neurodegeneration in hda-1/+;Htn-Q150 animals, indicating that HDA-1 and HDA-3 have different targets with opposing effects on polyglutamine toxicity. Our results suggest that polyglutamine expansions perturb transcription of CREB/CBP targets and that specific targeting of HDACs will be useful in reducing associated neurodegeneration.
Keywords: Huntington, polyglutamine, HDAC, polyQ, neurodegeneration, CBP
Introduction
Huntington's disease (HD) is one of several neurodegenerative diseases caused by expansion of a polyglutamine (polyQ) tract (Zoghbi and Orr, 2000; Ross, 2002). In Huntington's disease, the expansion occurs in the huntingtin protein. Understanding the primary changes that occur in neurons expressing expanded huntingtin could lead to therapeutic interventions. Several hypotheses have been proposed to explain how expansions of polyglutamine domains lead to neurodegeneration. Although, multiple pathways likely contribute to neuronal death, many studies suggest that expanded polyglutamine domains interfere with transcriptional regulation. In patient tissue, as well as in cell culture, vertebrate, and invertebrate models, the expanded polyglutamine form of huntingtin protein binds the acetyltransferase cAMP response element-binding protein (CREB) binding protein (CBP); both huntingtin and CBP are sequestered into aggregates (McCampbell et al., 2000; Nucifora et al., 2001; Steffan et al., 2001; Jiang et al., 2003). Moreover, acetyltransferase activity is reduced in cell culture models of Huntington's disease (Nucifora et al., 2001). Increased CBP expression restores histone acetylation, altered transcription, and retinal degeneration in a Drosophila model of polyglutamine disease (Taylor et al., 2003), supporting the hypothesis that CBP is a critical target of expanded polyQ tracts.
Histone acetyltransferase activity is directly opposed by histone deacetylases (HDACs), which remove acetyl groups from core histones. HDACs are divided into three classes based on structure and function. Class I and class II HDACs are similar in sequence and are part of numerous repressor complexes (Grozinger et al., 1999; Knoepfler and Eisenman, 1999). Class III HDACs share little homology with class I and class II HDACs and are homologous to the NAD+-dependent yeast HDAC, SIR2 (silent information regulator 2) (Brachmann et al., 1995), and are implicated in chromatin silencing, cellular metabolism, and aging (Guarente, 2000; Tanner et al., 2000). HDAC proteins have diverse functions and targets, suggesting that inhibition of individual proteins could have different effects on neuronal survival and resistance to polyglutamine toxicity.
Chemical inhibitors of class I and class II HDACs reduce degeneration in cell culture, yeast, Caenorhabditis elegans, and Drosophila models of polyglutamine toxicity (McCampbell et al., 2001; Steffan et al., 2001; Hockly et al., 2003; Parker et al., 2005). One HDAC inhibitor, suberoylanilide hydroxamic acid, partially rescues motor defects in a mouse model of Huntington's disease (Hockly et al., 2003). However, chemical inhibitors of enzymes are not necessarily specific. Most HDAC inhibitors indiscriminately target class I and class II HDACs, complicating the identification of critical targets.
Our C. elegans model expresses exon 1 of human Htn with an expanded polyglutamine (Htn-Q150) reminiscent of the R6/2 mouse model of HD (Mangiarini et al., 1996; Faber et al., 1999). Expression of Htn-Q150 in the ASH sensory neurons causes progressive neurodegeneration. As in patients, aggregation of mutant protein and neurodegeneration are directly related to the length of the polyglutamine tract and age of the animal (Faber et al., 1999). We found that loss of CBP and CREB function increases polyglutamine toxicity. We surveyed C. elegans HDACs to address possible differential contributions to Htn-Q150 toxicity in neurons and found that C. elegans HDACs have divergent effects on polyglutamine toxicity.
Materials and Methods
RNA interference.
To target individual C. elegans HDACs and decrease off-target effects, unique fragments of HDAC exonic sequences were amplified by PCR from genomic DNA using primers listed in the supplemental material (supplemental Table 1, available at www.jneurosci.org). Restriction sites were designed into primers when necessary for cloning into the L4440 plasmid (Timmons et al., 2001). Constructs were amplified in DH5 α cells, and HT115 bacteria were transformed to yield strains for feeding RNA interference (RNAi). RNAi feeding strains for hda-1 (C53A5.3), hda-2 (C08B11.2), and hda-3 (R06C1.1) were provided by Iva Greenwald (Columbia University, New York, NY) (Jarriault and Greenwald, 2002). RNAi feeding strain for hda-10 (Y51H1A.5) was provided by Mark Vidal (Harvard Medical School, Boston, MA). All bacteria feeding strains were cultured as described previously to induce expression of double-stranded RNA (dsRNA) (Timmons et al., 2001). Gravid adult hermaphrodites were lysed using a 20% bleach and 2.5% 10N NaOH solution to release eggs of similar ages. Larva (stage L1) from these egg preparations were placed onto RNAi bacterial strains and passaged to new RNAi plates daily starting at young adult stage. On the eighth day, animals were washed off plates with M9 buffer, dye filled with dioctadecyl tetramethylindodicarbocyanine (DiD) (Perkins et al., 1986), and allowed to recover for at least 20 min before scoring.
To confirm ASH neuron sensitivity to RNAi, green fluorescent protein (GFP) fluorescence was visually assessed in rtIs11[osm-10::Htn-Q150, osm-10::GFP] young adult animals that were fed bacteria expressing GFP dsRNA from L1 stage. In addition, degeneration of ASH neurons was assessed in 8-d-old aged rtIs11[osm-10::Htn-Q150, osm-10::GFP] fed bacteria expressing pqe-1 (Faber et al., 2002) dsRNA starting at L1 stage.
Strains and aging.
Animals were cultured at 25°C using standard methods unless otherwise indicated (Brenner, 1974). rtIs18[Htn-Q150], rtIs14[Htn-Q150], and rtIs11[Htn-Q150] transgenic animals express osm-10::Htn-Q150 as described previously (Faber et al., 1999). sir-2.1(OEx) are animals that carry the integrated array geIn3[sir-2.1] (Tissenbaum and Guarente, 2001). deg-1(rt70) (Faber et al., 2002) animals were aged to 4 d at 20°C when 9 ± 4% of the ASH neurons degenerate. For each genetic strain generated, control lines [lacking loss of function (lf) mutation or Htn-Q150 transgene] were scored in parallel. To look for suppression of necrotic neuronal death, deg-1(rt70) animals were aged to 7 d at 15°C when 78 ± 4% of the ASH neurons degenerate. After reaching reproductive maturity (day 3), all animals were transferred to new plates daily until the day of scoring. The day of scoring was selected to maximize detection of suppression or enhancement. Each experiment consists of at least three separate days of scoring that total 150 or more neurons when combined.
Overexpression and rescue lines. srb-6::hda-1
(pHA#442) was constructed by inserting hda-1 cDNA between the KpnI and SacI sites behind the srb-6 promoter of plasmid pHA#355 (Baran et al., 1999). hda-1(e1795)/unc-76(e911);rtIs18[Htn-Q150] animals were injected with 50 ng/μl srb-6::hda-1and 25 ng/μl myo-2::GFP (pPD48.33) (Thatcher et al., 1999) to rescue hda-1(e1795) polyglutamine toxicity enhancement. hda-1(e1795);Htn-Q150 homozygous animals carrying the extrasomal srb-6::hda-1 transgene were selected and aged to 6 d before scoring for degeneration. As a control, hda-1(e1795)/unc-76(e911);rtIs18[Htn-Q150] animals were injected with 50 ng/μl srb-6::empty pHA#352 and 25 ng/μl myo-2::GFP (pPD48.33) and scored in parallel. rtIs18[Htn-Q150] animals were injected with 50 ng/μl srb-6::hda-1 and 25 ng/μl myo-2::GFP to create hda-1(OEx);Htn-Q150 transgenic lines overexpressing hda-1. srb-6::hda-3 (pHA#401) rescue constructs were generated by insertion of hda-3 cDNA into pHA#355 plasmid between the KpnI and SacI sites and injected at 30 ng/μl srb-6::hda-3 pHA#401 with 120 ng/μl pha-1 pBX#1 into hda-3;pha-1;rtIs11[Htn-Q150] animals. pha-1 rescue serves as a transformation marker. hda-3 overexpression lines were generated by injection of 100 ng/μl srb-6::hda-3 and 120 ng/μl pha-1 pBX#1 rescue construct in hda-3(+);pha-1;Htn-Q150 animals. At least three transgenic lines were scored and pooled for all genotypes.
Chemical inhibition of HDAC function. pha-1;rtIs11 [Htn-Q150]
L1 animals were raised in a humidified chamber at 25°C in S-basal medium with (2%) DMSO with or without 150 μg/ml trichostatin A (TSA) (Sigma, St. Louis, MO) for 8 d before scoring.
Quantitative PCR.
Gravid adults were lysed to collect staged eggs. Embryos were hatched and synchronized to stage L1 larva by starvation overnight in M9 buffer (Johnson et al., 1984). Animals were plated on RNAi nematode growth media plates seeded with HT115 bacteria expressing double-stranded RNA of the desired target gene. Animals developed to young adults at 25°C. A subset of the adults was aged to confirm degeneration phenotypes. The remaining young adult animals were washed off plates with M9 buffer. After centrifugation, supernatant was replaced with 4× volume RNAzol (Iso-Tex Diagnostics, Friendswood, TX). Solution was vortexed and flash frozen in liquid N2, allowed to thaw at 37°C, vortexed, and flash frozen again. This process was repeated twice before tubes were stored at −80°C. RNA was isolated using standard procedure by freeze/thaw in RNAzol and phenol/chloroform/isoamylalcohol extraction. cDNA was generated using polyA primers and Super II single-strand reverse transcription (RT)-PCR (Invitrogen, San Diego, CA). RNA was quantified using TaqMan quantitative PCR (PE Biosystems, Foster City, CA). Primers were designed to span sequence from two exons to prevent genomic DNA amplification. Htn-Q150 transcripts were amplified using primers: forward, CAAGGTTACAGCTCGACGAGATC; reverse, CCTCCAAGGGTCCTCCTTTG. All primers anneal at 58 or 59°C. Transcript levels were detected using TaqMan Htn Probe: CCGGGATTGGCC. Transcripts of two neuronally expressed genes, osm-6 and cat-1, were measured as comparisons for loading controls (Perkins et al., 1986; Zhang et al., 2004). osm-6 primers are as follows: forward primer, GCCGGAAATTAATGATTACACAAA, and reverse primer, CAATTCTCCTTCGTACATACAAACCTT, which anneals with a melting temperature of 59°C. osm-6 transcript levels were detected with TaqMan Probe: TCATATGTCCCAGCAGAT. cat-1: forward primer, TCCGGACCACTTGTCAAG AGT; reverse primer, CTGATGACGGCAATGATGTACA. cat-1 transcript levels were measured with Taqman probe: TAGGATTCCCAACGATG. The reaction contained 5 nm template DNA and 50 μm primer DNA in 50 mm salt and 1 mm Mg2+. A three-step PCR was performed for 35 cycles. Denaturation temperature was 94°C for 20 s. Annealing was performed at 55°C for 20 s. Extension was performed at 72°C for 30 s.
Statistical analysis.
Error bars represent the SD for multiple determinations of each genotype; each genotype was examined in independent experiments undertaken on more than 1 d as indicated in the text. n > 150 neurons for each genotype. Pairwise analysis using the two-tailed t test or ANOVA were used as indicated to determine statistical significance. Microsoft Excel (Microsoft, Seattle, WA) was used for all statistical calculations.
Results
Htn-Q150 expression perturbs CBP-1/CRH-1 function
Previous studies have suggested that expanded polyglutamine tracts perturb CBP and/or CREB function, thereby contributing to transcriptional dysregulation and eventual neuronal death. Based on these findings we hypothesized that decreasing CBP or CREB function in neurons expressing an expanded huntingtin protein should increase neurodegeneration. This hypothesis was tested in a C. elegans model of neuronal polyglutamine toxicity (Faber et al., 1999). The first exon of human huntingtin was expressed in a small subset of C. elegans neurons, including the two ASH sensory neurons. Each ASH neuron has a sensory process that directly contacts the environment allowing neurons to take up the lipophilic, fluorescent dye DiD. Perturbations in the sensory process prevent dye uptake.
Expression of expanded polyQ tracts leads to neuronal dysfunction, perturbations in ASH sensory processes, and, eventually, cell death. Therefore, dye filling is a rapid, sensitive assay that can detect either degeneration or neuronal death caused by polyQ toxicity; this study does not distinguish between them.
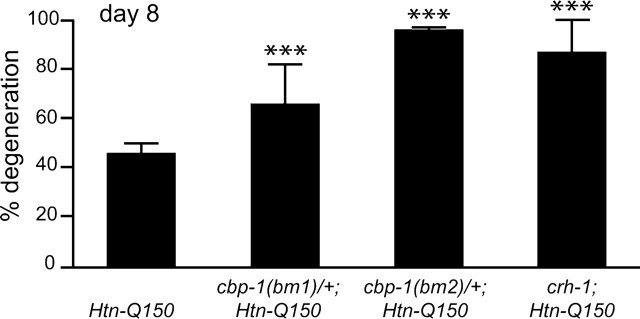
To test the hypothesis that transcriptional dysregulation contributes to polyglutamine toxicity, the requirement for cbp-1 function was examined. Each of the two deletion alleles of the C. elegans ortholog of CBP, cbp-1, were introduced to rtIs18[Htn-Q150] transgenic animals. cbp-1(bm1) generates a stable, truncated protein that lacks the conserved histone acetyltransferase (HAT) domain and bromodomain. The cbp-1(bm2) allele deletes the promoter and first four exons of cbp-1. cbp-1(bm2) is a complete loss of function deletion allele; no CBP-1 protein is detected by immunostaining in cbp-1(bm2) homozygous embryos. Both deletion alleles cause embryonic lethality when homozygous, whereas animals that are heterozygous for cbp-1 deletion alleles are indistinguishable from wild-type animals in morphology and viability (Victor et al., 2002). Therefore, Htn-Q150 animals carrying one wild-type cbp-1 allele and one cbp-1 deletion allele are compared with Htn-Q150 animals with normal cbp-1 function (two wild-type alleles). By 8 d, expression of Htn-Q150 causes 46 ± 4% of the ASH neurons to degenerate (Fig. 1) (all error bars presented are SDs). Neurodegeneration increased in Htn-Q150;cbp-1(bm1)/+ animals (67 ± 16%) and in Htn-Q150; cbp-1(bm2)/+ animals (97 ± 1%). Both CBP mutant alleles increased Htn-Q150 toxicity, suggesting that CBP-1 HAT activity is critical for protection against polyglutamine toxicity. This is consistent with the hypothesis that expanded polyglutamine tracts interfere with CBP-1-mediated transcriptional regulation.
Figure 1.
cbp-1(ys2)/+, cbp-1(ys4)/+, and crh-1(3315) enhance Htn-Q150-induced degeneration. Sibling controls for cbp-1(ys2)/+; Htn-Q150, cbp-1(ys4)/+;Htn-Q150, and crh-1(e3315); Htn-Q150 were combined, because degeneration levels were not significantly different. No degeneration was detected in cbp-1(ys2)/+, cbp-1(ys4)/+, or crh-1 animals that did not express Htn-Q150. Error bars represent SD. ***p < 0.005 versus column 1. At least three trials were conducted for each experiment to total n > 150.
CREB is one of the DNA binding proteins that interacts with CBP to activate transcription. Reducing CBP-1 function increased polyglutamine toxicity in C. elegans, suggesting that CREB function may also be important. To test this hypothesis, we examined the effect of CREB loss of function on Htn-Q150 toxicity. A loss of function deletion allele of a C. elegans CREB homolog, crh-1(n3315), resulted in homozygous viable animals with no overt developmental defects (Alkema and Horvitz, manuscript in preparation). Loss of crh-1 function enhanced Htn-Q150-mediated degeneration to 86 ± 13% in Htn-Q150 animals versus 45 ± 8% in control Htn-Q150 lines (Fig. 1). These results are consistent with the hypothesis that polyglutamine expansions perturb CBP and CREB function.
If polyglutamine expansions cause neuronal degeneration solely by decreasing CBP and CREB function, then reducing function of either CBP or CREB individually could cause degeneration independent of Htn-Q150 expression. To test this, ASH degeneration was assayed in cbp-1 and crh-1 homozygous mutant animals lacking the Htn-Q150 transgene. No degeneration was observed in heterozygous cbp-1(bm1), heterozygous cbp-1(bm2) animals nor in homozygous crh-1(n3315) animals in the absence of Htn-Q150 transgene (n = 120). Decreasing CBP or eliminating CREB function in C. elegans increases polyglutamine-induced degeneration, but loss of their function is not sufficient to cause degeneration independent of Htn-Q150 expression. This suggests that other cellular pathways are perturbed by polyglutamine expansion. Combined, these data suggest that transcriptional dysregulation is a critical factor in Htn-Q150 toxicity, consistent with previous results suggesting that expanded polyglutamine tracts interfere with CBP-mediated transcription.
Chemical inhibition of class I and class II HDACs decreases degeneration
Pharmacological manipulation of HDACs alters polyQ toxicity. Sirtuins increase class III HDAC activity and suppress polyQ toxicity in many organisms. Broadly acting class I and class II chemical HDAC inhibitors decrease polyQ-mediated neurodegeneration in vertebrate and Drosophila neurons (McCampbell et al., 2001; Steffan et al., 2001; Hockly et al., 2003). We found that TSA, a chemical inhibitor of class I and class II HDACs, reduces degeneration in C. elegans neurons. pha-1;rtIs11 [Htn-Q150] L1 animals were cultured for 8 d with or without TSA. Degeneration was reduced from 61 ± 9% in control animals to 26 ± 3% in animals incubated with 150 μg/ml TSA (n > 150 neurons; two trials) (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). TSA suppression was dose dependent; degeneration was moderately reduced by 100 μg/ml TSA (41 ± 6%; n = 100). These results confirm HDACs modulate polyQ toxicity in C. elegans, but broadly acting HDAC inhibitors cannot reveal the contributions of specific HDACs to polyQ toxicity.
Decreasing HDAC gene function has differing effects on Htn-Q150 toxicity
The activity of histone acetyltransferases, including CBP, is opposed by histone deacetylases. Each of the three HDAC classes is represented in C. elegans (supplemental Fig. 2 and supplemental Table 2, available at www.jneurosci.org as supplemental material). Class I HDACs include hda-1 (C53A5.3), hda-2 (C08B11.2), and hda-3 (R06C1.1) (Shi and Mello, 1998). Mammalian orthologs of C. elegans class I HDACs are ubiquitously expressed and predominantly localized to the nucleus. Previous analysis revealed a role for hda-1 in development. Homozygous hda-1(e1794) animals are defective in gonadogenesis and are, consequently, sterile (Dufourcq et al., 2002). C. elegans class II HDACs include hda-4 (C10E2.3), hda-5 (F43G6.11), hda-6 (F41H10.6), hda-10 (Y51H1A.5), and hda-11 (C35A5.9) (Dufourcq et al., 2002). C. elegans class III HDACs include sir-2.1 (R11A8.4), sir-2.2 (F46G10.7), and sir-2.3 (F46G10.3). Decreased function of sir-2.1 reduces lifespan, whereas overexpression of sir-2.1 increases lifespan (Tissenbaum and Guarente, 2001). The relative contribution of these HDACs to polyglutamine toxicity is unclear. Perturbation or modulation of individual HDAC genes could have different effects on polyglutamine toxicity.
In C. elegans, gene function can be systematically reduced using either lf alleles or RNAi. Only a few loss of function alleles exist for HDAC genes; RNAi knockdown was used to examine the other C. elegans HDAC genes. Although neurons are relatively insensitive to RNAi, we found that feeding animals bacteria expressing dsRNA reduced expression of genes in the ASH neurons. When animals were fed bacteria expressing GFP dsRNA, GFP expression was lost in 88% of the ASH neurons (n = 100), whereas all control animals expressed GFP in both ASH neurons. In addition, when function of a previously described polyQ enhancer-1 (pqe-1) (Faber et al., 2002) was reduced by feeding pqe-1 dsRNA, Htn-Q150-induced ASH neuron degeneration was increased from 26 ± 13% in control animals to 81 ± 9% (n = 100 for each). Thus, ASH neurons are susceptible to RNAi. Loss of function alleles and/or RNAi was used to examine the ability of each C. elegans HDAC gene to modulate polyglutamine toxicity.
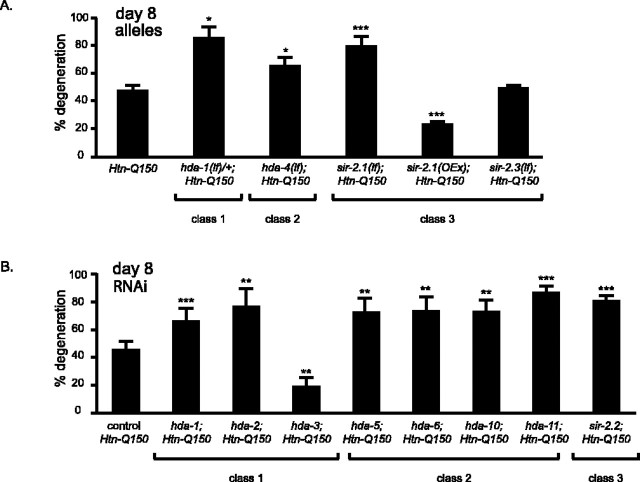
Four HDAC loss of function alleles were tested for their ability to modify Htn-Q150 toxicity; three of the four enhanced toxicity. Heterozygous loss of hda-1 increased degeneration of ASH neurons in Htn-Q150 animals (86 ± 6 from 46 ± 4% in control animals at day 8) (Fig. 2A). Similarly, reducing function of a class II HDAC, hda-4, increased degeneration (65 ± 7%). Deletion alleles exist for two of the class III HDAC genes (Tissenbaum and Guarente, 2001; Parker et al., 2005). Although complete loss of sir-2.3 function had no effect on Htn-Q150-induced degeneration (48 ± 2%), complete loss of sir-2.1 function enhanced degeneration (79 ± 6%). Consistent with this result, sir-2.1 overexpression in sir-2.1(OEx);Htn-Q150) animals suppressed degeneration (23 ± 2%) (Fig. 2A).
Figure 2.
A, hda-1(e1795)/+, hda-4, sir-2.1(ok434) enhance Htn-Q150 toxicity. sir-2.1 overexpression suppresses Htn-Q150 toxicity. sir-2.3 has no effect on Htn-Q150 toxicity. Controls were combined, because they are not significantly different. No degeneration was detected in hda-1(e1795)/+, hda-4, sir-2.1(ok434), or sir-2.3 animals without expression of Htn-Q150. At least three trials were conducted for each experiment to total n > 150. B, RNAi of class I HDACs, hda-1 and hda-2, enhances Htn-Q150 toxicity. RNAi of class I HDAC, hda-3, suppresses Htn-Q150 toxicity. RNAi of class II HDACs enhanced degeneration, as did RNAi of sir-2.2, a class III HDAC. Controls were combined, because they are not significantly different. At least three trials were conducted for each experiment to total n > 150. Error bars represent SD. *p < 0.05, **p < 0.005, ***p < 0.0005 in pairwise comparison with column 1 using a two-tailed t test.
RNAi was used to examine the contributions of remaining HDACs, because mutant alleles were not available. Consistent with preceding analysis, degeneration is increased to 66 ± 8% in hda-1(RNAi);Htn-Q150 animals (from 44 ± 7% in Htn-Q150 animals on control RNAi) (Fig. 2B). Similarly, RNAi of the class I HDAC, hda-2, and all class II HDACs increased degeneration (Fig. 2B) (quantified in supplemental Table 2, available at www.jneurosci.org as supplemental material). In contrast, hda-3(RNAi) reduced degeneration (20 ± 5%). RNAi of the class III HDAC, sir-2.2, increased degeneration to 81 ± 4%. Given the similarity between class I HDACs at the molecular level, the dramatically different effect of hda-3 knockdown is striking. To address the cellular specificity of class I HDACs on polyglutamine toxicity, additional analysis of hda-1 and hda-3 was undertaken.
HDA-1 activity protects Htn-Q150-expressing neurons
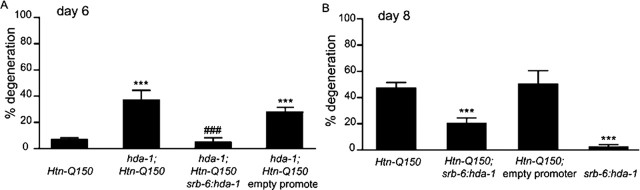
hda-1(e1795) homozygous animals are sterile and die prematurely (Dufourcq et al., 2002). The overall poor health of homozygous hda-1(e1795) animals could contribute to the enhanced polyglutamine degeneration observed in hda-1(e1795);Htn-Q150 animals. To address this issue and to determine whether hda-1(lf) enhancement of polyglutamine toxicity was cell autonomous, an hda-1 cDNA was expressed in ASH neurons using the srb-6 promoter, which drives expression in a limited number of C. elegans neurons (Troemel et al., 1995). Degeneration in homozygous hda-1(e1795) animals was evaluated at day 6, because hda-1(e1795) homozygous animals rarely survive longer. In Htn-150 animals, 6 ± 2% of the ASH neurons degenerated by day 6 (Fig. 3A). Homozygous loss of hda-1 increases toxicity (36 ± 2%). Expression of the hda-1 cDNA in 6-d-old homozygous hda-1(e1795);Htn-Q150 animals restored neuronal survival to original levels (4 ± 3%) (Fig. 3A). Injection of equal concentrations of an empty srb-6 promoter plasmid, srb-6::empty, and the transgenic marker fail to rescue the homozygous hda-1(e1795) enhancement of polyglutamine toxicity (29 ± 2%). No ASH neurons degenerated in hda-1(e1795) homozygous animals lacking the Htn-Q150 transgene. We conclude that loss of hda-1 function is deleterious in neurons that express expanded polyglutamine proteins. Restoring hda-1 levels in a small subset of neurons, including the ASH neurons, is sufficient to ameliorate hda-1 loss of function enhancement of Htn-Q150 toxicity. This suggests that, normally, HDA-1 acts within the affected neuron and can protect against Htn-Q150 toxicity; reducing HDA-1 function sensitizes neurons to polyglutamine assault.
Figure 3.
A, Expression of HDA-1 in ASH neurons restores Htn-Q150 toxicity to normal levels. Injection of equal concentrations of an empty srb-6 promoter plasmid and the transgenic marker fail to rescue the hda-1(e1795) enhancement of polyQ toxicity (29 ± 2%). B, In Htn-Q150 animals without the srb-6::hda-1 transgene, 47 ± 4% of the ASH neurons degenerate by day 8. Degeneration is decreased to 20 ± 4% by increased expression of HDA-1 (50 ng/μl) in Htn-Q150 ASH neurons. Expression of HDA-1 does not cause detectible degeneration of ASH neurons that do not express Htn-Q150 (2 ± 2%). No significant change in neurodegeneration is detectable in animals that carry an empty srb-6 promoter and the transgenic marker myo-2::GFP (50 ± 10%). ***p < 0.0005 compared with corresponding Htn-Q150 animals in column 1; ###p < 0.0005 when comparing hda-1;Htn-Q150; srb-65::hda-1 with hda-1;Htn-Q150 in column 2 (to assess rescue) in A. Empty promoter transgenes did not significantly change polyglutamine toxicity. At least three trials were conducted for each experiment to total n > 150. Error bars represent SD.
Decreasing hda-1 function could negatively affect neuronal survival by a nonspecific mechanism. However, if overexpression of hda-1 protects against Htn-Q150-induced neurodegeneration, then HDA-1 targets are likely directly involved in protecting polyglutamine-expressing neurons. To test this hypothesis, hda-1 was overexpressed in ASH neurons. Htn-Q150 animals were injected with srb-6::hda-1 to create hda-1 overexpression transgenic animals [hda-1(OEx);Htn-Q150]. Suppression is most readily detected by comparing degeneration in Htn-Q150 animals, Htn-Q150; hda-1(OEx) animals, and Htn-Q150; srb-6::empty transgenic control animals at day 8. In Htn-Q150 animals without the srb-6::hda-1 transgene, 47 ± 4% of the ASH neurons degenerate by day 8. Degeneration is decreased to 20 ± 4% by increased expression of HDA-1 in Htn-Q150 ASH neurons (Fig. 3B). Expression of HDA-1 does not cause significant degeneration in the absence of the Htn-Q150 transgene (1 ± 2%). Increased HDA-1 activity in the neurons protects them against Htn-Q150 toxicity, suggesting that HDA-1 targets may be critical factors in decreasing polyglutamine toxicity.
HDA-3 promotes degeneration in response to Htn-Q150 expression
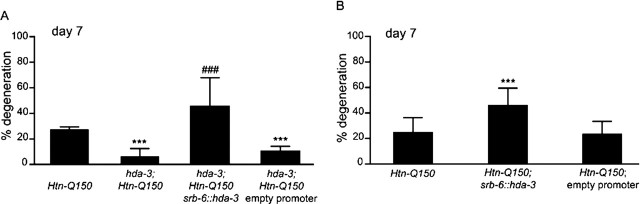
The hda-3(RNAi) analysis suggested that reducing HDA-3 function could decrease Htn-Q150-induced degeneration. To confirm the role of hda-3 in Htn-Q150-induced neurodegeneration, a deletion allele, tm1374, was obtained from the C. elegans Gene Knockout Center (Tokyo, Japan). The tm1374 deletion begins in the second exon of hda-3, omits DNA sequence encoding the FPGTGDL residues that are critical for deacetylase activity (Finnin et al., 1999), and results in a frame shift that is likely to eliminate hda-3 function. The integrated rtIs18[Htn-Q150] array maps are very close to the hda-3 gene on chromosome I. Therefore, it was necessary to use a different Htn-Q150 transgene for hda-3(tm1374) analysis. Degeneration was assessed in rtIs11[Htn-Q150] and rtIs14[Htn-Q150] animals on day 7 when either enhancement or suppression of Htn-Q150 toxicity could be detected. hda-3(tm1374) recapitulated the suppression of Htn-Q150 toxicity observed in hda-3(RNAi) animals. Degeneration was reduced from 27 ± 2% in rtIs11[Htn-Q150] animals to 6 ± 5% in homozygous hda-3(tm1374);rtIs11[Htn-Q150] animals (Fig. 4A).
Figure 4.
HDA-3 specifically rescues hda-3(tm1374) suppression of Htn-Q150 in ASH neurons. Degeneration is reduced from 27 ± 2% in rtIs11Htn-Q150 animals to 6 ± 6% in hda-3(tm1374); rtIs11(Htn-Q150) animals at day 7. Degeneration ranges from 14 to 63%, depending on the transgenic line (average, 45 ± 22%) in hda-3(tm1374); rtIs11(Htn-Q150); srb-6:hda-3 transgenic animals. As a control, a plasmid carrying the empty srb-6 promoter was injected along with transgenic markers. The empty promoter has no significant effect on polyQ toxicity (10 ± 3%). Overexpression of hda-3 in ASH using the srb-6 promoter increases degeneration from 24 ± 11 to 45 ± 13% (p = 0.03). ***p < 0.0005 compared with column 1 for each graph; ###p < 0.0005 when comparing hda-3;Htn-Q150; srb-6::hda3 with hda-3;Htn-Q150. At least three trials were conducted for each experiment to total n > 150. Error bars represent SD.
To confirm that HDA-3 acts in the polyglutamine expressing neurons to modulate toxicity, we examined the HDA-3 cellular expression pattern. Immunohistochemistry revealed that HDA-3 is ubiquitously expressed. Costaining with an OSM-10 antisera that recognizes an endogenous protein expressed in the ASH (Hart et al., 1999) confirmed that HDA-3 protein is found in the ASH neurons. Endogenous HDA-3 predominantly localizes to the nucleus in C. elegans, where it is likely to function in transcriptional regulation (supplemental Fig. 3, available at www.jneurosci.org as supplemental material) (Whetstine et al., 2005).
To determine whether hda-3 modulation of Htn-Q150 toxicity occurs in the neurons and to confirm that reduced hda-3 function is responsible for suppression, an srb-6::hda-3 transgene was introduced into hda-3;rtIs11[Htn-Q150] animals. Polyglutamine-induced degeneration of ASH neurons was restored when hda-3 was expressed in a small subset of neurons, including ASH. Degeneration averaged 31 ± 8% in hda-3(tm1374);rtIs11[Htn-Q150];srb-6::hda-3(+) transgenic animals compared with 27 ± 2% in rtIs11[Htn-Q150] control animals and 10 ± 3% in hda-3;rtIs11[Htn-Q150] animals (Fig. 4A). Thus, decreased HDA-3 activity decreases the toxicity of Htn-Q150 specifically in neurons in which polyglutamine is expressed.
To determine whether HDA-3 contributes directly to Htn-Q150-induced degeneration in ASH neurons, srb-6::hda-3 was introduced into Htn-Q150 animals that retain normal hda-3 activity. Overexpression of hda-3 in ASH using the srb-6 promoter increased degeneration from 24 ± 11 to 45 ± 13% (Fig. 4B). We concluded that suppression of polyglutamine toxicity in hda-3(tm1374);rtIs11[Htn-Q150] animals is attributable to loss of hda-3 function and that hda-3 acts cell autonomously to modulate polyglutamine toxicity in these neurons.
Loss of hda-3 or hda-1 specifically modulates polyglutamine-induced neurotoxicity
To determine whether hda-1 or hda-3 modulation of neurotoxicity is specific to polyglutamine-induced degeneration, we examined possible roles of hda-1 and hda-3 in necrotic neurodegeneration independent of polyglutamine toxicity. Hyperactive ion channels disrupt cellular osmotic balance and cause necrotic neuronal degeneration. A recessive, cold-sensitive mutation in a sodium channel, deg-1(rt70), causes temperature-dependent ASH degeneration (Faber et al., 2002). hda-1 and hda-3 functions were individually reduced by RNAi in deg-1(rt70) mutant animals. deg-1(rt70) animals were aged to 4 d at 20°C when 9 ± 4% of the ASH neurons degenerate or were aged to 7 d at 15°C when 78 ± 6% of the ASH neurons degenerate to assess enhancement or suppression, respectively. Neither hda-1(RNAi) nor hda-3(RNAi) altered necrotic degeneration (supplemental Fig. 4, available at www.jneurosci.org as supplemental material). These data suggest that transcriptional dysregulation selectively contributes to polyglutamine-induced degeneration.
Reducing function of hda-3 or hda-1 does not significantly alter Htn-Q150 expression
Reducing function of specific proteins found in HDAC complexes can alter transgene expression levels in many C. elegans tissues, but neurons are generally protected from these alterations (Kim et al., 2005). To address the possibility that Htn-Q150 transgene levels were changed by loss of HDAC function in the ASH neurons, we took three experimental approaches. First, to demonstrate that osm-10::Htn-Q150 transgene levels are not altered by hda-3 loss of function, we quantified expression of osm-10::GFP in homozygous hda-3(tm1374);osm-10::GFP animals. GFP expression in ASH neurons was indistinguishable from control osm-10::GFP animals (n = 380). To directly determine whether genes implicated in transgene silencing decrease osm-10 promoter activity, we examined GFP expression in 3-d-old rtIs11[osm-10::GFP, osm-10::Htn-Q150] animals lacking an RNA-directed RNA polymerase, rrf-3, or lacking a ribosomal RNA repressor, ncl-1 (Frank and Roth, 1998; Simmer et al., 2002). Both of these genes are expressed in ASH neurons and have been implicated previously in transgene silencing in C. elegans (Simmer et al., 2002; Kim et al., 2005). Neither rrf-3(pk1426) nor ncl-1(e1865) silence expression of GFP in ASH neurons (n = 200 and n = 150, respectively). Additionally, to determine specifically whether rrf-3(pk1426) or ncl-1(e1865) could suppress or enhance polyglutamine-induced degeneration, ASH neuron degeneration was assessed in corresponding animals. Degeneration was not altered in rrf-3 or ncl-1 homozygous mutant backgrounds (supplemental Fig. 5A, available at www.jneurosci.org as supplemental material). Therefore, we consider it unlikely that transgene silencing contributes to decreased neurodegeneration in hda-3 mutant animals.
To directly address the possibility that reducing HDAC function alters Htn-Q150 expression levels, consequently altering polyglutamine toxicity, Htn-Q150 mRNA levels were measured using quantitative RT-PCR. Transcript levels were measured in hda-3(RNAi), hda-1(RNAi), and control RNAi rtIs18[Htn-Q150] animals. Levels were assessed in young adult animals before the onset of neuronal degeneration. There was no significant difference in Htn-Q150 mRNA levels (supplemental Fig. 5B, available at www.jneurosci.org as supplemental material). We conclude that changes in Htn-Q150 toxicity are not attributable to changes in Htn-Q150 transcript levels.
HDA-3 loss suppresses CREB modulation Htn-Q150-induced degeneration
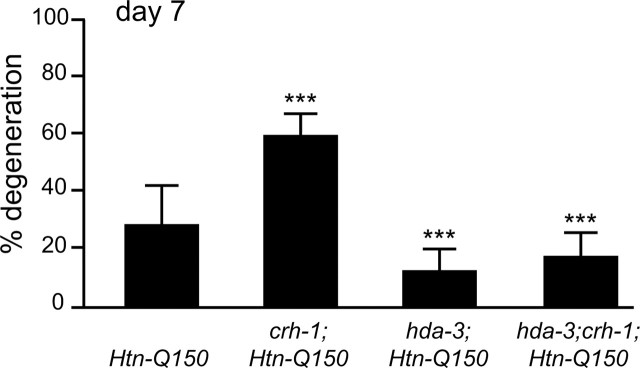
Histone deacetylases are critical for transcriptional regulation, but HDACs are not limited to histones as targets (Hubbert et al., 2002). C. elegans HDACs could modulate polyglutamine toxicity via transcriptional regulation or by deacetylating proteins that are directly involved with polyglutamine toxicity. Genetic epistasis analysis was undertaken to determine whether hda-3(tm1374) suppression of Htn-Q150 toxicity is via regulation of CBP/CREB transcription. Double homozygous hda-3;crh-1;Htn-Q150 animals were generated and compared with homozygous hda-3;Htn-Q150, homozygous crh-1;Htn-Q150, and Htn-Q150 sibling lines. If HDA-3 and CRH-1 participate in independent pathways, their combined effects could be additive, resulting in polyglutamine degeneration levels between crh-1 enhanced degeneration and hda-3 suppressed degeneration. If HDA-3 and CRH-1 (C. elegans CREB homolog) act in the same pathway, either the phenotype of hda-3 (suppression) or the phenotype of crh-1 (enhancement) should predominate. 28 ± 13% of the ASH neurons degenerate in control Htn-Q150 animals. As expected, degeneration increased to 59 ± 7% in crh-1;Htn-Q150 animals and decreased by loss of hda-3 to 12 ± 7% in Htn-Q150 animas (Fig. 5). Interestingly, hda-3 loss of function suppressed the crh-1 loss of function phenotype in hda-3(tm1374);crh-1;Htn-Q150 animals to 17 ± 8%. Thus, hda-3 is epistatic to crh-1, suggesting that HDA-3, directly or indirectly, opposes crh-1 activity in regulating transcription of genes that protect against Htn-Q150-induced toxicity.
Figure 5.
HDA-3 modulates Htn-Q150 toxicity via transcriptional regulation. Loss of hda-3 function suppresses the deleterious effects of loss of crh-1 function. ASH neuron degeneration is increased in crh-1;rtIs11[Htn-Q150] animals and decreased in hda-3(tm1374);rtIs11[Htn-Q150] animals compared with rtIs11[Htn-Q150]. ASH degeneration is suppressed in hda-3(tm1374);crh-1; rtIs11[Htn-Q150] animals and is not significantly different from degeneration levels in hda-3(tm1374);rtIs11[Htn-Q150] animals at day 7. Error bars represent SD. ***p < 0.0005 compared with column 1 of each graph. At least three trials were conducted for each determination to total n > 150.
Suppression of Htn-Q150 toxicity in hda-3 mutant animals requires HDA-1 function
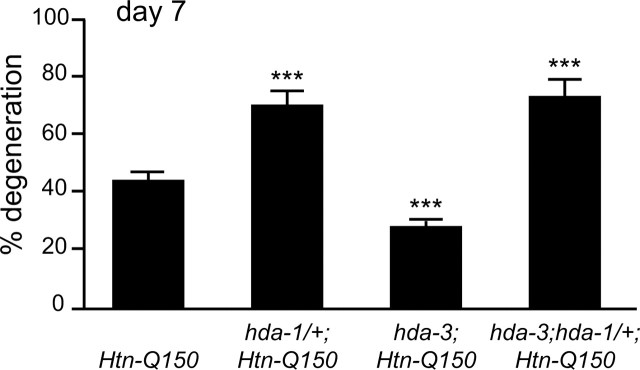
Reducing hda-1 function enhances Htn-Q150 toxicity. In contrast, reducing hda-3 function suppresses Htn-Q150 toxicity. To determine whether hda-3 loss of function could suppress hda-1 enhanced polyglutamine toxicity, ASH neuron degeneration was assessed in hda-3;hda-1/+;Htn-Q150 animals. As expected, degeneration was reduced from 45 ± 4% in control rtIs14[Htn-Q150] animals to 29 ± 2% in hda-3;rtIs14[Htn-Q150] animals at day 7. Consistent with results described above, degeneration was increased in hda-1/+;rtIs14[Htn-Q150] animals to 76 ± 6% (Fig. 6). Interestingly, degeneration was also increased in hda-3(tm1374);hda-1(e1795)/+;rtIs14[Htn-Q150] animals to 74 ± 7% versus controls. hda-3 loss of function can suppress degeneration when hda-1 function is intact but cannot suppress Htn-Q150-induced degeneration when hda-1 levels are compromised. One possible interpretation of these results is that HDA-1 activity is required, directly or indirectly, for suppression of Htn-Q150 toxicity by hda-3 loss of function.
Figure 6.
Normal hda-1 levels are critical for hda-3 modulation of polyglutamine toxicity. Loss of one copy of hda-1 eliminates the salubrious effects of hda-3 loss of function. ASH neuron degeneration is increased in hda-1(e1795)/+; rtIs14[Htn-Q150] animals and decreased in hda-3(tm1374);rtIs14[Htn-Q150i] animals compared with rtIs14[Htn-Q150] animals. Degeneration levels are increased in hda-3(tm1374);hda-1(e1795)/+; rtIs14[Htn-Q150] and are not significantly different from degeneration levels in hda-1(e1795)/+; rtIs14[Htn-Q150] animals. Error bars represent SD. ***p < 0.0005 compared with column 1. At least three trials were conducted for each determination to total n > 150.
Discussion
This analysis suggests that expression of expanded Htn fragments interferes with CBP and/or CREB function, thereby contributing to neuronal degeneration. Reducing function of most C. elegans histone deacetylases enhances Htn-Q150-induced degeneration. Additional analysis confirms that HDA-1 protects Htn-Q150 expressing neurons from degeneration. HDA-3 is unique; loss of HDA-3 suppresses polyglutamine toxicity.
Reducing cbp-1 and crh-1 activity enhances Htn-Q150-induced degeneration. This suggests that expanded polyglutamine perturbs transcription leading to neuronal degeneration. Histone acetyltransferase and Q-rich domains of human CBP interact with expanded Htn (Steffan et al., 2001). C. elegans cbp-1(bm1), the allele that deletes the histone acetyltransferase and bromodomains, significantly enhances polyglutamine toxicity. cbp-1(bm1) encodes a stable, truncated CBP-1 protein that lacks acetyltransferase activity (Victor et al., 2002). This suggests that acetyltransferase activity is important for CBP-1 protection against Htn-Q150 toxicity.
Reducing crh-1 (CREB) function is sufficient to enhance polyglutamine toxicity, implicating crh-1 regulated genes in neuronal survival. Deletion of CREB combined with deletion of its cofactor, CREM (cAMP-responsive element modulator), causes a phenotype similar to Huntington's disease in mice (Mantamadiotis et al., 2002). However, reducing C. elegans cbp-1 or eliminating crh-1 function does not cause degeneration independent of the Htn-Q150 transgene. We conclude that Htn-Q150 perturbs additional transcriptional pathways and/or cellular processes that contribute to degeneration. In vertebrates, TATA binding protein, specificity protein 1, and p53 have all been found in polyglutamine aggregates (Cha, 2000; Sugars and Rubinsztein, 2003). In addition, proteasomes, ubiquitin, and chaperones have been found in aggregates, suggesting disruption of the protein folding and degradation pathways (Sakahira et al., 2002; Ciechanover and Brundin, 2003). The conclusion that multiple pathways contribute to polyglutamine toxicity is strengthened by the recent finding that protective effects of increased CREB and heat-shock protein 70 are additive in Drosophila (Iijima-Ando et al., 2005). We conclude that histone acetylation activity of CBP-1 and transcriptional activation by CRH-1 are likely perturbed by Htn-Q150 expression contributing to the process of neurodegeneration.
Chemical and genetic manipulation of HDAC function yield distinctly different results for polyQ toxicity. This likely reflects their different specificity and efficacy. polyQ toxicity is reduced by broadly acting chemical inhibitors of class I and class II HDACs. In contrast, eliminating specific HDACs can increase or reduce degeneration depending on the gene targeted. At first glance, this discrepancy may be puzzling. However, at the concentrations tested, broadly acting chemical inhibitors like TSA likely cause a partial decrement in all class I and class II HDACs. These HDACs likely regulate many genes with various and divergent impacts on polyQ toxicity. It is reasonable to assume that eliminating specific HDACs using genetic techniques might then yield dramatically different results; loss of a specific HDAC likely yields specific and unique changes in gene expression (Dangond and Gullans, 1998). Additionally, chemical or genetic manipulation may differ in their impact on the compensatory mechanisms that operate when HDAC function is decreased (Ajamian et al., 2004). And, chemical inhibitors may have off-target effects, perturbing proteins other than HDACs. Previous analyses using broadly acting chemical inhibitors may have obscured the relative importance of specific HDACs in polyQ toxicity.
Decreasing the function of most class I and class II HDACs increased polyglutamine toxicity in C. elegans. Mammalian HDAC1 and HDAC2 are found together in numerous complexes [Sin3, νcleosome remodeling and deacetylation, retinoblastoma, and corepressor to RE1 silencing transcription factor/neural restrictive silencing factor corepressor complexes (Knoepfler and Eisenman, 1999; Humphrey et al., 2001)]. In mammals, Sin3 is important for the regulation of fatty acid synthesis, which is critical for mitochondrial function and vesicle formation, both of which have been implicated in polyglutamine expansion disease (Schweizer and Hofmann, 2004). Sin3 and nuclear repressor protein (NCoR) interact with Htn when the polyglutamine tract is expanded and have been found in polyglutamine protein aggregates (Boutell et al., 1999). Although mammalian HDAC3 is found in complexes with NCoR and SMRT (silencing mediator of retinoic acid and thyroid hormone receptor), it is not yet clear in which complexes C. elegans hda-3 takes part (Guenther et al., 2000, 2001; Li et al., 2000; Wen et al., 2000). If ectopic polyglutamine interactions with Sin3 and NCoR titrate away functions of these repressor complexes, then reducing the function of HDACs in the Sin3 and NCoR complexes could further decrease repressor complex function and thereby impinge on neuronal survival.
In this study, reducing function of most class II HDACs increases polyglutamine toxicity. Many class II HDACs have myocyte enhancer factor-2 (MEF-2) interaction domains (Choi et al., 2002). Mammalian MEF-2 proteins play important roles in neuronal survival (Mao and Wiedmann, 1999; Youn et al., 1999). Thus, decreasing function of class II HDACs could compromise survival of neurons. HDAC4 and HDAC5 repression of MEF-2 is dependent on calcium, suggesting a possible link between calcium signaling and polyglutamine toxicity (Youn et al., 2000). Thus, C. elegans HDA-1, HDA-2, and class II HDACs could be important for maintaining survival of neurons under polyglutamine stress.
In contrast, loss of Ce-hda-3 suppresses polyglutamine toxicity. Mammalian HDAC3 is found in different complexes from HDAC1 and HDAC2 (Guenther et al., 2000, 2001; Wen et al., 2000). Thus, specific HDAC complexes could regulate genes critical for protection against polyglutamine toxicity or neuronal survival, whereas other complexes regulate genes that contribute to polyglutamine toxicity.
Class III HDACs have been implicated previously in polyglutamine toxicity and aging. In C. elegans and Drosophila, increased Sir2 function extends the lifespan (Tissenbaum and Guarente, 2001; Wood et al., 2004). In this study, reducing function of two of the three C. elegans class III HDACs enhanced polyglutamine toxicity. Interestingly, Ce-sir-2.1 deletion increases Htn-Q150-induced degeneration, whereas overexpression suppresses degeneration. Our results are consistent with recent findings that increasing activity of Ce-sir-2.1 protects against polyglutamine-induced neuronal dysfunction (Parker et al., 2005). Based on the results presented herein, Ce-sir-2.1 is not unique. Decreasing function of most Ce-HDACs increases polyglutamine toxicity, and increasing both Ce-hda-1 and Ce-sir-2.1 suppresses toxicity. The results presented here suggest that SIR-2.1 HDAC activity is critical for polyglutamine toxicity but not necessarily increased longevity or aging associated with activity of sir-2.1.
Is the activity of Ce-hda-3 as a transcriptional regulator critical for its role in polyglutamine toxicity? Some HDACs target proteins that are not involved in transcription. For instance, mammalian HDAC6 and Sir2 deacetylate tubulin (Hubbert et al., 2002; Matsuyama et al., 2002; North et al., 2003; Zhang et al., 2003). Therefore, at least two functional targets exist for HDACs in polyglutamine toxicity. First, specific HDACs could regulate the transcription of genes that encode protective proteins or proteins that assist in degeneration. Second, specific HDACs could deacetylate proteins that play nontranscriptional roles underlying polyglutamine toxicity. Based on the ability of Ce-hda-3 deletion to suppress increased polyglutamine toxicity of crh-1 mutants, genetic epistasis suggests that HDA-3 modulates polyglutamine toxicity through transcriptional regulation. crh-1 loss of function can no longer enhance polyglutamine toxicity in hda-3 mutant animals, suggesting that other transcriptional activators are able to compensate for crh-1 loss of function when hda-3 function is reduced. HDA-3 might normally repress transcription of C. elegans genes that protect against expanded polyglutamine or promote neuronal survival. Therefore, release from negative regulation via hda-3 loss of function protects against polyglutamine toxicity.
Here, we demonstrate that normal Ce-hda-1 function is required for Ce-hda-3(tm1374) suppression of Htn-Q150-induced neurodegeneration. There are at least two molecular mechanisms that could underlie alterations in polyglutamine toxicity. Ce-hda-1 protein could negatively regulate cell death genes, and/or HDA-3 could repress transcription of protective genes. One possible interpretation of the epistasis analysis is that Ce-hda-1 target genes are downstream of Ce-HDA-3 targets. A second possible explanation is that Ce-hda-1 transcription may normally be directly repressed by Ce-HDA-3 protein in ASH neurons. Therefore, when Ce-hda-3 function is reduced, Ce-hda-1 transcription is increased. Our results demonstrate that increased Ce-hda-1 dosage suppresses Htn-Q150-induced neurodegeneration. Therefore, Ce-hda-3 loss of function could suppress polyglutamine toxicity by increasing HDA-1 activity. Thus, in Ce-hda-3(tm1374);Ce-hda-1(e1795);rtIs14[Htn-Q150] animals, neurodegeneration may not be suppressed, because Ce-hda-1 activity cannot be sufficiently increased. However, we cannot rule out the possibility that hda-3(lf) cannot compensate for the severity of hda-1(lf) degeneration.
In C. elegans, genes positively regulated by CREB/CBP and negatively regulated by hda-3 likely play important roles protecting against expanded polyglutamine. We speculate that in mammals, specific HDACs could negatively regulate transcription of bcl-2, BDNF, chaperones, or other proteins necessary for neuronal survival, whereas other HDACs regulate transcription of degeneration promoting genes such as caspases.
In summary, using a C. elegans neuronal model of polyglutamine disease, we found that CBP and CREB functions are critical for protecting against polyglutamine-induced neurodegeneration. Results presented here suggest that HDACs play opposing roles in polyglutamine toxicity in C. elegans neurons. Therefore, it is likely that in mammals, specific HDACs will also have opposing effects on polyglutamine-induced degeneration. These C. elegans results suggest that finding downstream target genes of specific HDAC complexes will help to determine the mechanism of polyglutamine-induced degeneration. The development of inhibitors targeted to specific HDACs or HDAC complexes could be both more effective and less toxic therapeutic agents in polyglutamine expansion diseases.
Footnotes
This work was supported by National Institutes of Health–National Institute of Neurological Disorders and Stroke Grant 40048 (A.C.H.). We thank the C. elegans gene knockout consortium (Japan and Oklahoma) and the C. elegans Genetic Center for strains; Mark Alkema and Robert Horvitz for the crh-1(e3315) animals; Beth Muir and Denis Sgroi for assistance with quantitative RT-PCR results; and Johnathan Whetstine, Anders Näär, and members of the Hart and van den Heuvel laboratories for reagents and discussion.
References
- Ajamian F, Salminen A, Reeben M (2004). Selective regulation of class I and class II histone deacetylases expression by inhibitors of histone deacetylases in cultured mouse neural cells. Neurosci Lett 365:64–68. [DOI] [PubMed] [Google Scholar]
- Baran R, Aronoff R, Garriga G (1999). The C. elegans homeodomain gene unc-42 regulates chemosensory and glutamate receptor expression. Development 126:2241–2251. [DOI] [PubMed] [Google Scholar]
- Boutell JM, Thomas P, Neal JW, Weston VJ, Duce J, Harper PS, Jones AL (1999). Aberrant interactions of transcriptional repressor proteins with the Huntington's disease gene product, huntingtin. Hum Mol Genet 8:1647–1655. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD (1995). The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev 9:2888–2902. [DOI] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans Genetics 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha JH (2000). Transcriptional dysregulation in Huntington's disease. Trends Neurosci 23:387–392. [DOI] [PubMed] [Google Scholar]
- Choi KY, Ji YJ, Jee C, Kim do H, Ahnn J (2002). Characterization of CeHDA-7, a class II histone deacetylase interacting with MEF-2 in Caenorhabditis elegans Biochem Biophys Res Commun 293:1295–1300. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Brundin P (2003). The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron 40:427–446. [DOI] [PubMed] [Google Scholar]
- Dangond F, Gullans SR (1998). Differential expression of human histone deacetylase mRNAs in response to immune cell apoptosis induction by trichostatin A and butyrate. Biochem Biophys Res Commun 247:833–837. [DOI] [PubMed] [Google Scholar]
- Dufourcq P, Victor M, Gay F, Calvo D, Hodgkin J, Shi Y (2002). Functional requirement for histone deacetylase 1 in Caenorhabditis elegans gonadogenesis. Mol Cell Biol 22:3024–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber PW, Alter JR, MacDonald ME, Hart AC (1999). Polyglutamine-mediated dysfunction and apoptotic death of a Caenorhabditis elegans sensory neuron. Proc Natl Acad Sci USA 96:179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber PW, Voisine C, King DC, Bates EA, Hart AC (2002). Glutamine/proline-rich PQE-1 proteins protect Caenorhabditis elegans neurons from huntingtin polyglutamine neurotoxicity. Proc Natl Acad Sci USA 99:17131–17136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP (1999). Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401:188–193. [DOI] [PubMed] [Google Scholar]
- Frank DJ, Roth MB (1998). ncl-1 is required for the regulation of cell size and ribosomal RNA synthesis in Caenorhabditis elegans J Cell Biol 140:1321–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozinger CM, Hassig CA, Schreiber SL (1999). Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci USA 96:4868–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L (2000). Sir2 links chromatin silencing, metabolism, and aging. Genes Dev 14:1021–1026. [PubMed] [Google Scholar]
- Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R (2000). A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev 14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Barak O, Lazar MA (2001). The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol 21:6091–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AC, Kass J, Shapiro JE, Kaplan JM (1999). Distinct signaling pathways mediate touch and osmosensory responses in a polymodal sensory neuron. J Neurosci 19:1952–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockly E, Richon VM, Woodman B, Smith DL, Zhou X, Rosa E, Sathasivam K, Ghazi-Noori S, Mahal A, Lowden PA, Steffan JS, Marsh JL, Thompson LM, Lewis CM, Marks PA, Bates GP (2003). Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington's disease. Proc Natl Acad Sci USA 100:2041–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP (2002). HDAC6 is a microtubule-associated deacetylase. Nature 417:455–458. [DOI] [PubMed] [Google Scholar]
- Humphrey GW, Wang Y, Russanova VR, Hirai T, Qin J, Nakatani Y, Howard BH (2001). Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J Biol Chem 276:6817–6824. [DOI] [PubMed] [Google Scholar]
- Iijima-Ando K, Wu P, Drier EA, Iijima K, Yin JC (2005). cAMP-response element-binding protein and heat-shock protein 70 additively suppress polyglutamine-mediated toxicity in Drosophila Proc Natl Acad Sci USA 102:10261–10266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarriault S, Greenwald I (2002). Suppressors of the egg-laying defective phenotype of sel-12 presenilin mutants implicate the CoREST corepressor complex in LIN-12/Notch signaling in C. elegans Genes Dev 16:2713–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Nucifora FC Jr, Ross CA, DeFranco DB (2003). Cell death triggered by polyglutamine-expanded huntingtin in a neuronal cell line is associated with degradation of CREB-binding protein. Hum Mol Genet 12:1–12. [DOI] [PubMed] [Google Scholar]
- Johnson TE, Mitchell DH, Kline S, Kemal R, Foy J (1984). Arresting development arrests aging in the nematode Caenorhabditis elegans Mech Aging Dev 28:23–40. [DOI] [PubMed] [Google Scholar]
- Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, Rual JF, Kennedy S, Dybbs M, Bertin N, Kaplan JM, Vidal M, Ruvkun G (2005). Functional genomic analysis of RNA interference in C. elegans Science 308:1164–1167. [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Eisenman RN (1999). Sin meets NuRD and other tails of repression. Cell 99:447–450. [DOI] [PubMed] [Google Scholar]
- Li J, Wang J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J (2000). Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J 19:4342–4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, Bates GP (1996). Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87:493–506. [DOI] [PubMed] [Google Scholar]
- Mantamadiotis T, Lemberger T, Bleckmann SC, Kern H, Kretz O, Martin Villalba A, Tronche F, Kellendonk C, Gau D, Kapfhammer J, Otto C, Schmid W, Schutz G (2002). Disruption of CREB function in brain leads to neurodegeneration. Nat Genet 31:47–54. [DOI] [PubMed] [Google Scholar]
- Mao Z, Wiedmann M (1999). Calcineurin enhances MEF2 DNA binding activity in calcium-dependent survival of cerebellar granule neurons. J Biol Chem 274:31102–31107. [DOI] [PubMed] [Google Scholar]
- Matsuyama A, Shimazu T, Sumida Y, Saito A, Yoshimatsu Y, Seigneurin-Berny D, Osada H, Komatsu Y, Nishino N, Khochbin S, Horinouchi S, Yoshida M (2002). In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J 21:6820–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCampbell A, Taylor JP, Taye AA, Robitschek J, Li M, Walcott J, Merry D, Chai Y, Paulson H, Sobue G, Fischbeck KH (2000). CREB-binding protein sequestration by expanded polyglutamine. Hum Mol Genet 9:2197–2202. [DOI] [PubMed] [Google Scholar]
- McCampbell A, Taye AA, Whitty L, Penney E, Steffan JS, Fischbeck KH (2001). Histone deacetylase inhibitors reduce polyglutamine toxicity. Proc Natl Acad Sci USA 98:15179–15184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E (2003). The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell 11:437–444. [DOI] [PubMed] [Google Scholar]
- Nucifora FC Jr, Sasaki M, Peters MF, Huang H, Cooper JK, Yamada M, Takahashi H, Tsuji S, Troncoso J, Dawson VL, Dawson TM, Ross CA (2001). Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science 291:2423–2428. [DOI] [PubMed] [Google Scholar]
- Parker JA, Arango M, Abderrahmane S, Lambert E, Tourette C, Catoire H, Neri C (2005). Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat Genet 37:349–350. [DOI] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG (1986). Mutant sensory cilia in the nematode Caenorhabditis elegans Dev Biol 117:456–487. [DOI] [PubMed] [Google Scholar]
- Ross CA (2002). Polyglutamine pathogenesis: emergence of unifying mechanisms for Huntington's disease and related disorders. Neuron 35:819–822. [DOI] [PubMed] [Google Scholar]
- Sakahira H, Breuer P, Hayer-Hartl MK, Hartl FU (2002). Molecular chaperones as modulators of polyglutamine protein aggregation and toxicity. Proc Natl Acad Sci USA 99:>[Suppl 4], 16412–16418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer E, Hofmann J (2004). Microbial type I fatty acid synthases (FAS): major players in a network of cellular FAS systems. Microbiol Mol Biol Rev 68:501–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Mello C (1998). A CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans Genes Dev 12:943–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RH (2002). Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol 12:1317–1319. [DOI] [PubMed] [Google Scholar]
- Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol BL, Kazantsev A, Schmidt E, Zhu YZ, Greenwald M, Kurokawa R, Housman DE, Jackson GR, Marsh JL, Thompson LM (2001). Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila Nature 413:739–743. [DOI] [PubMed] [Google Scholar]
- Sugars KL, Rubinsztein DC (2003). Transcriptional abnormalities in Huntington disease. Trends Genet 19:233–238. [DOI] [PubMed] [Google Scholar]
- Tanner KG, Landry J, Sternglanz R, Denu JM (2000). Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci USA 97:14178–14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JP, Taye AA, Campbell C, Kazemi-Esfarjani P, Fischbeck KH, Min KT (2003). Aberrant histone acetylation, altered transcription, and retinal degeneration in a Drosophila model of polyglutamine disease are rescued by CREB-binding protein. Genes Dev 17:1463–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher JD, Haun C, Okkema PG (1999). The DAF-3 Smad binds DNA and represses gene expression in the Caenorhabditis elegans pharynx. Development 126:97–107. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A (2001). Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans Gene 263:103–112. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L (2001). Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans Nature 410:227–230. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Chou JH, Dwyer ND, Colbert HA, Bargmann CI (1995). Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans Cell 83:207–218. [DOI] [PubMed] [Google Scholar]
- Victor M, Bei Y, Gay F, Calvo D, Mello C, Shi Y (2002). HAT activity is essential for CBP-1-dependent transcription and differentiation in Caenorhabditis elegans EMBO Rep 3:50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen YD, Perissi V, Staszewski LM, Yang WM, Krones A, Glass CK, Rosenfeld MG, Seto E (2000). The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc Natl Acad Sci USA 97:7202–7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetstine JR, Ceron J, Ladd B, Dufourcq P, Reinke V, Shi Y (2005). Regulation of tissue-specific and extracellular matrix-related genes by a class I histone deacetylase. Mol Cell 18:483–490. [DOI] [PubMed] [Google Scholar]
- Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D (2004). Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature 430:686–689. [DOI] [PubMed] [Google Scholar]
- Youn HD, Sun L, Prywes R, Liu JO (1999). Apoptosis of T cells mediated by Ca2+-induced release of the transcription factor MEF2. Science 286:790–793. [DOI] [PubMed] [Google Scholar]
- Youn HD, Grozinger CM, Liu JO (2000). Calcium regulates transcriptional repression of myocyte enhancer factor 2 by histone deacetylase 4. J Biol Chem 275:22563–22567. [DOI] [PubMed] [Google Scholar]
- Zhang S, Sokolchik I, Blanco G, Sze JY (2004). Caenorhabditis elegans TRPV ion channel regulates 5HT biosynthesis in chemosensory neurons. Development 131:1629–1638. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li N, Caron C, Matthias G, Hess D, Khochbin S, Matthias P (2003). HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J 22:1168–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi HY, Orr HT (2000). Glutamine repeats and neurodegeneration. Annu Rev Neurosci 23:217–247. [DOI] [PubMed] [Google Scholar]