Abstract
In skeletal muscle, brain-derived neurotrophic factor (BDNF) has long been thought to serve as a retrograde trophic factor for innervating motor neurons throughout their lifespan. However, its localization in mature muscle fibers has remained elusive. Given the postulated roles of BDNF in skeletal muscle, we performed a series of complementary experiments aimed at defining the localization of BDNF and its transcripts in adult muscle. By reverse transcription-PCR, in situ hybridization, and immunofluorescence, we show that BDNF, along with the receptor p75NTR, is not expressed at significant levels within mature myofibers and that it does not accumulate preferentially within subsynaptic regions of neuromuscular junctions. Interestingly, expression of BDNF correlated with that of Pax3, a marker of muscle progenitor cells, in several different adult skeletal muscles. Additionally, BDNF was expressed in Pax7+ satellite cells where it colocalized with p75NTR. In complementary cell culture experiments, we detected high levels of BDNF and p75NTR in myoblasts. During myogenic differentiation, expression of BDNF became drastically reduced. Using small interfering RNA (siRNA) technology to knock down BDNF expression, we demonstrate enhanced myogenic differentiation of myoblasts. This accelerated rate of myogenic differentiation seen in myoblasts expressing BDNF siRNA was normalized by administration of recombinant BDNF. Collectively, these findings show that BDNF plays an important regulatory function during myogenic differentiation. In addition, the expression of BDNF in satellite cells is coherent with the notion that BDNF serves a key role in maintaining the population of muscle progenitors in adult muscle.
Keywords: diaphragm, Pax3, Pax7, p75NTR, TrkB, myoblasts
Introduction
Neurotrophins are known to modulate various neuronal processes throughout development. The mammalian family of neurotrophins consists of nerve growth factor, neurotrophin-3, neurotrophin-4/5 (NT-4/5), and brain-derived neurotrophic factor (BDNF). Depending on the developmental context and availability of their putative receptors, these neurotrophins promote neuronal survival, differentiation, synaptic potentiation, and depression as well as apoptosis. Among these neurotrophins, BDNF has been the focus of intense studies, because it has been associated with various neurological disorders. Accordingly, via binding to its receptors tyrosine receptor kinase B (TrkB) and/or p75NTR, BDNF initiates intracellular events central to neuronal development and maturation (Huang and Reichardt, 2003; Nykjaer et al., 2005).
In the neuromuscular system, it has been hypothesized that skeletal muscle-derived BDNF enhances the survival of innervating motor neurons throughout their lifespan and potentiates neuromuscular transmission (Oppenheim et al., 1992; Sendtner et al., 1992; Lohof et al., 1993; Yan et al., 1993; Zhang and Poo, 2002). This hypothesis is supported by several lines of evidence including, for example, the expression of BDNF in skeletal muscle and retrograde transport of exogenously applied BDNF to distinct motor neuron cell bodies (DiStefano et al., 1992; Funakoshi et al., 1993; Yan et al., 1993; Griesbeck et al., 1995; Mousavi et al., 2004). Moreover, BDNF enhances the survival of motor neurons in culture and rescues motor neurons after severing their connection with target skeletal muscles in neonates (Sendtner et al., 1992; Yan et al., 1992; Koliatsos et al., 1993; Mousavi et al., 2004). Collectively, these results suggest that skeletal muscle-derived BDNF acts as a retrograde survival factor for motor neurons in the neuromuscular system.
During embryonic development, primordial skeletal muscle cells express relatively high levels of BDNF with subsequent downregulation with maturation (Griesbeck et al., 1995). Moreover, the expression of BDNF is associated with developing myofibers, which later express myosin heavy chain (MyHC) IIB (Griesbeck et al., 1995; Mousavi et al., 2004). In contrast to developing muscle, the localization and role of BDNF in mature skeletal muscle has remained controversial (Copray et al., 2000; Liem et al., 2001). Here, we examined the expression and localization of BDNF in synaptic and extrasynaptic regions of mature diaphragm skeletal muscle. We demonstrate that expression of BDNF occurs in skeletal muscle satellite cells as well as in myoblasts in culture. Furthermore, we show that reduction in BDNF levels results in early myogenic differentiation of myoblasts in culture, which can be rescued with exogenous BDNF treatment. We conclude by proposing that the primary role of BDNF in mature skeletal muscle is to inhibit myogenic differentiation of satellite cells or muscle progenitor cells.
Materials and Methods
Animals and tissues.
Adult (1–2 months of age) female Sprague Dawley rats (Charles River, Quebec, Canada) were housed in the animal care facility at the University of Ottawa and were provided with unlimited access to food and water. All surgical procedures and skeletal muscle biopsies were approved by the University of Ottawa Animal Care and Use Committee.
Cell culture.
All cell cultures were incubated at 37°C with 5% CO2 and maximum humidity. Satellite cells were isolated in culture as described previously (Rosenblatt et al., 1995). In brief, diaphragm skeletal muscle fibers were dissociated using collagenase (10 mg/ml) in DMEM for 30 min at 37°C. Dissociated fibers were placed onto Matrigel-treated (Invitrogen, Burlington, Ontario, Canada) plates in growth media (GM) containing 10% fetal calf serum (FCS), 100 U/ml penicillin/streptomycin, and 292 ng/ml l-glutamine in DMEM for 1–2 weeks or until satellite cells began to detach from fibers. Satellite cells were subsequently fixed in cold (−20°C) methanol and used for immunofluorescence (IF) thereafter (see below).
L6 myoblasts (American Type Culture Collection, Manassas, VA) were plated at a density of 10,000 cells/ml in GM until harvest. GM was changed every 48 h. For RNA and protein extraction, L6 myoblasts were harvested at 60–70% confluency. For myogenicity and fusion index, cells were processed for immunofluorescence as described below. For differentiation, 80–90% confluent cultures were placed in differentiation media containing 10 nm insulin, 2% FCS, 100 U/ml penicillin/streptomycin, and 292 ng/ml l-glutamine in DMEM for 4 d.
RNA extraction and quantitative reverse transcription-PCR.
Total RNA extraction and quantitative reverse transcription (RT)-PCR was performed as described previously (Mousavi et al., 2004). Briefly, total RNA was extracted using Trizol extraction according to the protocol of the manufacturer (Invitrogen). Total RNA was quantified and diluted to 50 and 500 ng/μl. Relative expression of genes was compared at two different starting RNA concentrations. Similar results were obtained using both starting RNA concentrations. Reverse transcription protocol was followed as described previously (Mousavi et al., 2004).
Sequences of primers have been published previously: skeletal muscle α-actin and BDNF (Mousavi et al., 2004), ε subunit of acetylcholine receptor (ε-AChR) (Boudreau-Lariviere et al., 1996), rat p75NTR (Yabe et al., 2004), rat TrkB (Yabe et al., 2004), exon-specific primers of BDNF (Tabuchi et al., 2002), and Pax3 (Nakagawa et al., 1996). The content of PCR mix has been described previously (Mousavi et al., 2004); except for PCRs starting with 50 ng/μl total RNA, 8.0 pmol/μl primers were used to account for template:primer ratio. All PCRs were preheated to 94°C for 2 min followed by repetitive cycles of denaturation (94°C for 30 s), annealing (60°C for 30 s), and extension (72°C for 30 s). The annealing temperature for BDNF exons, Pax3, and ε-AChR was 55, 56, and 62°C, respectively. PCR cycle numbers are shown in parentheses for the following genes: α-actin (18), BDNF (30), ε-AChR (30), p75NTR (30), TrkB (50), BDNF exon-1 (40), BDNF exon-2 (40), BDNF exon-3 (36), BDNF exon-4 (32), and Pax3 (32). Finally, PCRs were extended for 10 min at 72°C followed by storage at 4°C until use. For quantitative purposes, we made certain that all PCRs were within linear range of amplification as described previously (Mousavi et al., 2004).
In situ hybridization.
The BDNF cDNA construct was kindly provided by Regeneron Pharmaceuticals (Tarrytown, NY). Acetylcholinestrase (AChE) cDNA encoding exon 6 and 3′-UTR (nucleotides 1476–1987 of the AChE mRNA sequence; GenBank accession number S50879) was inserted into pBS II-SK and used as in situ hybridization (ISH) probes. ISH was performed as described previously (Young et al., 1998). Briefly, BDNF cDNA in pBSII-SK- was linearized with EcoRI or KpnI for in vitro transcription of antisense and sense probes, respectively. In the case of AChE, plasmid construct was linearized with EcoRI or XbaI for transcription of antisense and sense probes, respectively. Radiolabeled probes were generated using the following in vitro transcription mix: 1× transcription buffer, 10 mm DTT, 0.6 mm ATP and GTP, 0.5 μg/μl linearized DNA template, 10 U of RNase inhibitor; and 1.5 μCi/μl 35S-UTP and 35S-CTP (Amersham Biosciences, Piscataway, NJ). The reactions were incubated at 37°C for 2 h, after which cDNA template was digested with RNase-free DNase I (10 U). Unincorporated nucleotides were removed from the mixture using a Sephadex column (NAP-5; Amersham Biosciences). After ethanol precipitation, riboprobes were suspended in DTT and hybridization buffer as described previously (Young et al., 1998). Activity of the radioprobes was measured by scintillation counting, and the probes were stored at −80°C until use.
Skeletal muscles were cryosectioned at 10 μm thick and subjected to prehybridization protocol described previously (Young et al., 1998). Briefly, sections were fixed in 2% paraformaldehyde (PFA), placed in 100 mm citrate buffer, pH 6.0, containing 5′-bromoindoxyl acetate and hexazotized pararosaniline at 4°C for 1 h for staining the neuromuscular junctions (NMJs), and then placed in 4% PFA. Subsequently, sections were treated with proteinase K (20 μg/ml) in 50 mm Tris-HCl, 5 mm EDTA for 7.5 min, and acetylated in a solution containing 2.5% (w/v) acetic anhydride in 100 mm triethanolamine, pH 7.5. After dehydration in graded alcohol solutions, sections were left to dry for 1 h. BDNF or AChE antisense and sense probes (40 μl of 20,000–30,000 cpm/μl) were placed on sections and mounted with a sterile coverslip. After an overnight incubation at 50°C in a moist chamber, samples were processed for posthybridization and autoradiography as described previously (Young et al., 1998). For all ISH, sections were exposed to autoradiographic emulsion for 2–3 weeks in the dark at 4°C.
In ISH experiment using postnatal day 10 (P10) diaphragm, alternative serial sections were processed for Karnovsky and Roots (1964) method of staining the NMJs. In short, sections were placed in a solution containing 65 mm sodium acetate, 5 mm sodium citrate, 0.5 mm potassium ferricyanide, 3 mm copper sulfate, and 0.1 mg of acetylthiocholine iodide for 10–15 min for color development.
ELISA.
BDNF ELISA was performed as described previously (Mousavi et al., 2004). In short, junctional and extrajunctional regions of diaphragm skeletal muscle samples were weighed and homogenized in lysis buffer (1:2 w/v) (Mousavi et al., 2004). L6 cultures were lysed with 100 μl of lysis buffer (per 75 mm dish). In all cases, samples were vortexed, diluted with four times Dulbecco’s PBS (v/v), sonicated, and spun at 14,000 × g for 20 min. ELISA was performed following the protocol of the manufacturer (Promega, Madison, WI) and is described previously (Mousavi et al., 2004).
Immunofluorescence.
Antibodies were polyclonal anti-BDNF (Promega), anti-myosin heavy chain (MyHC) (MF20), anti-MyHC slow (A4–840), anti-Desmin (D3), anti-Pax7 (Developmental Studies Hybridoma Bank, Iowa City, IA), and anti-p75NTR (192-IgG). Anti-Pax7 was kindly provided by Dr. Michael A. Rudnicki (Ottawa Health Research Institute, Ottawa, Ontario, Canada). 192-IgG was a gift from Dr. Philip A. Barker (McGill University, Montreal, Quebec, Canada). Secondary antibody to anti-BDNF was FITC-conjugated donkey anti-chicken (Jackson ImmunoResearch, West Grove, PA). Secondary antibody for MF20, A4–840, D3, anti-Pax7, and 192-IgG was goat anti-mouse Texas Red (Invitrogen).
Skeletal muscle sections were washes in PBS between every step. First, sections were fixed with 1% PFA for 10 min at room temperature and blocked (1% BSA, 0.1% Triton X-100 in PBS) for an additional hour. Samples were incubated overnight (4°C) with primary antibodies. For blocked experiments, anti-BDNF was incubated with human recombinant BDNF (Promega) in a 1:1 ratio for 30 min at 37°C before placing on sections. The next day, sections were incubated with appropriate secondary antibodies for an extra hour. Sections were incubated with α-bungratoxin 594 (Invitrogen) for 1 h to stain acetylcholine receptor densities at NMJs. Finally, slides were mounted with Vectashield (Vector Laboratories, Burlington, Ontario, Canada) and coverslips.
Western blotting.
Total protein was extracted as in ELISA protocol (Mousavi et al., 2004). Protein concentration was determined using Bradford Assay (Bio-Rad, Hercules, CA). Samples (25 μg) were run on 6% SDS-PAGE and electroblotted onto polyvinylidene difluoride membranes. After blocking with 5% nonfat milk in Tris-buffered saline Triton X-100, blots were incubated with appropriate antibodies directed against MyHC or tubulin (Sigma, St. Louis, MO). After incubation with goat anti-mouse conjugated to horse-radish peroxidase (Chemicon, Temecula, CA), protein bands were visualized using SuperSignal Substrate (Pierce Biotechnology, Rockford, IL) on a Kodak (Rockford, NY) imaging station.
BDNF small interfering RNA and stable cell lines.
The small interfering RNA (siRNA) fragments were designed using BDNF cDNA sequence (GenBank accession number NM_012513) and Ambion (Austin, TX) design guidelines/Target Finder. Fragments were designed to have 19–22 bases, 30–50% guanine cytosine content while avoiding sequence ending with four or higher consecutive thymidines. Two different siRNA fragments were designed and named after their first starting nucleotide within NM_012513 sequence. In addition, siRNA Wizard (Invivogen, San Diego, CA) was used to design hairpin fragments: BDNF599 5′-TCCC-ATTACCTGGATGCCGCAAA-TCAAGAG-TTTGCGGCATCCAGGTAAT-TT-3′ and BDNF948 5′-TCCC-AAGGATAGACACTTCCTGTGT-TCAAGAG-ACACAGGAAGTGTCTATCCTTTT-3′. LacZ siRNA (5′TCCC-AAATCGTCTGACCGATGATCCGT-TCAAGAGACGGATCATCGGTCAGACGATT-TT-3′) was used as scrambled control sequence in these experiments (Shafey et al., 2005). LacZ siRNA construct was a gift from Dr. Robin J. Parks (Ottawa Health Research Institute, Ottawa, Ontario, Canada). The siRNA fragments were inserted into psiRNA-hH1neo construct following the protocol of the manufacturer (Invivogen). Accordingly, siRNA fragments were annealed with their complementary strands, ligated to BbsI-digested psiRNA-hH1neo construct, and used for plasmid amplification. After plasmid purification, constructs were sequenced to establish correct insertion.
BDNF599, BDNF948, and LacZ siRNA constructs were transfected into L6 myoblasts following Lipofectamine 2000 transfection protocol (Invitrogen). A ratio of 1:1 [DNA (μg):lipofectamine (μl)] in Opti-MEM (Invitrogen) was used to transfect cells overnight. Then, cells were placed in GM for 48 h and subsequently in selective media (SM) containing 600 μg/ml G418 (Invitrogen) in GM. In parallel, nontransfected cells were also placed in SM to ensure the potency and selectivity of G418. Clonal populations were isolated from G418-resistant cells for additional analysis. For PCR, total DNA was extracted using the Trizol method according to the protocol of the manufacturer (see above). PCR mix consisted of reagents previously mentioned (see above, RNA extraction and quantitative reverse transcription-PCR), except nucleotides were added directly to PCRs. PCR primers were OL178 and OL381 (Invivogen). For rescue experiments, BDNF599 and BDNF948 cells were treated daily with 100 ng/ml human recombinant BDNF (Regeneron Pharmaceuticals) for 6 d in GM.
Statistical analysis.
Student’s t test was used to determine the significant difference between two groups. For comparison between three or more groups, one-way ANOVA and least square difference tests were used to determine statistical significance. The level of significance was set at p < 0.05.
Results
Expression and localization of BDNF in mature skeletal muscle
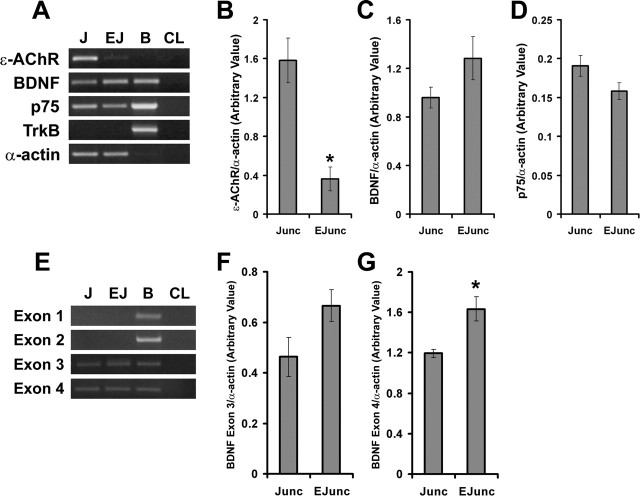
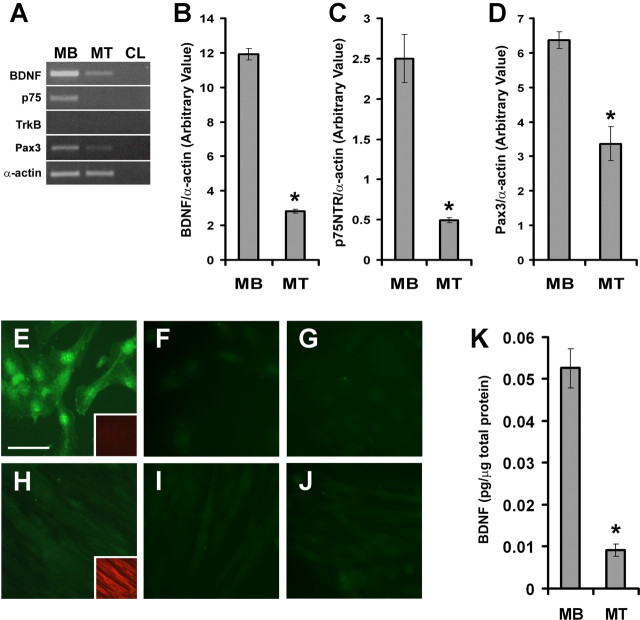
Initially, we began examining the distribution of BDNF transcripts and protein in synaptic and extrasynaptic regions of mature skeletal muscle. For this, we surgically dissected adult diaphragm into its junctional and extrajunctional regions. To ascertain proper surgical separation, the levels of ε-AChR mRNA was quantified to mark for junctional regions (Boudreau-Lariviere et al., 1996) (Fig. 1A). As expected, the levels of ε-AChR mRNA were approximately fourfold higher in junctional compared with extrajunctional regions (Fig. 1B). Using primers to amplify all splice variants, we detected BDNF transcripts in both compartments of diaphragm skeletal muscle (Fig. 1A,C). In fact, there was a tendency toward higher levels of BDNF transcripts in extrajunctional regions of mature diaphragm skeletal muscle (Fig. 1A,C). As well, p75NTR transcripts were equally distributed in the two compartments of mature diaphragm (Fig. 1A,D), whereas full-length TrkB transcript was not detected, even after 50 cycles of PCR (Fig. 1A).
Figure 1.
Expression of BDNF and p75NTR is observed in junctional and extrajunctional regions of adult diaphragm skeletal muscle. A, Representative ethidium bromide gels showing RT-PCR products of ε-AChR, BDNF, p75NTR, TrkB, and α-actin in junctional (J) and extrajunctional (EJ) regions. Also, total RNA from brain (B) and no RNA (CL) were used as positive and negative controls, respectively. B–D, Arbitrary levels of ε-AChR, BDNF, and p75NTR as normalized to α-actin in junctional (Junc) versus extrajunctional (EJunc) regions (n = 4; *p < 0.05). E, Representative ethidium bromide gels showing RT-PCR products of BDNF exon variants in junctional and extrajunctional regions of adult diaphragm skeletal muscle. F, Arbitrary levels of BDNF transcript containing exon-3 (normalized to α-actin levels). G, BDNF transcript containing exon-4 was significantly higher in extrajunctional regions than in the junctional compartment (n = 4; *p < 0.05). Error bars represent SE.
The genomic structure of BDNF consists of four different 5′ exons, which are alternatively spliced into coding exon-5 (Timmusk et al., 1993, 1995). Each of these 5′ exons contains different promoters, which drive expression of BDNF in the CNS (Timmusk et al., 1993, 1995). In peripheral tissues including skeletal muscle, promoters 3 and 4 are used to drive the expression of BDNF (Timmusk et al., 1993, 1995). Using exon-specific primers, we found BDNF transcripts containing exon-3 and exon-4 in adult diaphragm skeletal muscle, suggesting that expression of BDNF in this tissue is driven by promoters upstream of these exons as it has been suggested previously (Timmusk et al., 1995) (Fig. 1E). Moreover, similar to the trend observed when using primers to amplify all BDNF transcripts (Fig. 1C), there was a tendency toward higher levels of BDNF transcripts containing exon-3 or exon-4 in extrajunctional regions of mature diaphragm skeletal muscle (Fig. 1E–G).
To determine the precise localization of BDNF transcripts, we performed radioactive ISH with long exposure time (see Materials and Methods) on transverse sections of adult skeletal muscles. Accumulations of silver grain densities representative of BDNF mRNA were barely above sense background levels across sections (Fig. 2). Furthermore, we did not observe concentrations of silver grains overlaying mature NMJs in diaphragm (Fig. 2A, arrows). This trend was also observed in adult soleus and extensor digitorum longus (EDL) skeletal muscles (Fig. 2C,E, arrows). To ascertain that our hybridization approach and probe were appropriate, we performed two sets of control experiments. First, we used an antisense probe against AChE mRNA to demonstrate our ability to localize transcripts in muscle fibers and at the NMJs (Fig. 2G, arrows) as demonstrated previously (Michel et al., 1994). Second, coronal sections of neonatal brain were processed for BDNF ISH, which, as expected, displayed high levels of BDNF mRNA in the CA1–3 regions of the hippocampus (Fig. 2I, arrows). Additionally, serial coronal brain slices processed for BDNF sense control probe showed minimal homogenous silver grains across sections that were considered as background signal (data not shown).
Figure 2.
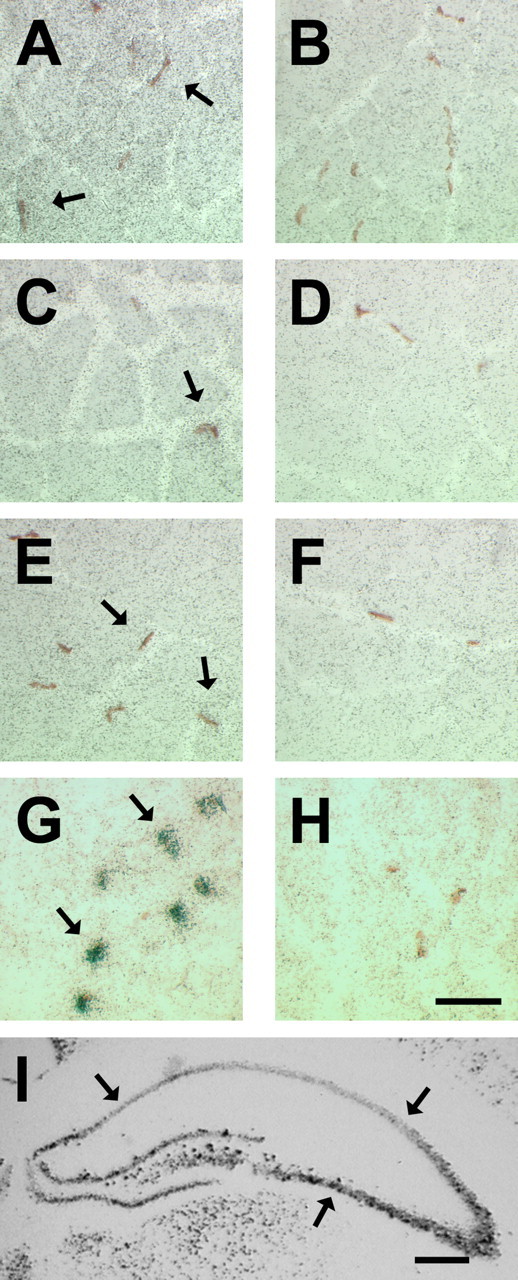
BDNF is low or absent in mature myofibers and not restricted to NMJs. ISH of BDNF probes on transverse sections of adult skeletal muscles. A, C, E, Representative micrographs showing hybridization of BDNF antisense probe on diaphragm, soleus, and EDL, respectively. B, D, F, BDNF sense control on diaphragm, soleus, and EDL, respectively. Arrows point to NMJs (brown staining). G, Representative micrograph showing hybridization of AChE antisense probe on adult diaphragm muscle sections. Arrows point to silver grain densities overlaying mature NMJs. H, Representative micrograph showing hybridization of AChE sense probe. Scale bar, 50 μm. I, A micrograph of coronal sections of neonatal (P10) brain showing hybridization of BDNF antisense probe. Scale bar, 500 μm. Arrows point to CA1–3 hippocampal regions.
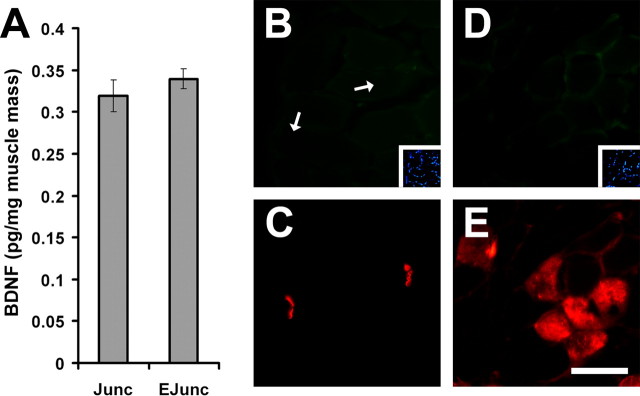
To establish whether the levels of BDNF transcript(s) were correlated with that of its protein within diaphragm, we measured the levels of BDNF protein using ELISA. For this, total protein was extracted from junctional and extrajunctional regions of adult diaphragm and processed for BDNF ELISA. There were equal levels of BDNF protein in junctional and extrajunctional regions of the diaphragm, albeit with a trend toward higher levels in extrajunctional compartment (Fig. 3A). It is important to note that in these assays, the concentrations of BDNF detected in junctional and extrajunctional regions were ∼75 pg/ml, which corresponds to values that are approximately fivefold above the sensitivity threshold of this particular ELISA assay. Furthermore, BDNF was not immunolocalized to myofibers or to subsynaptic regions of NMJs (Fig. 3B, arrows, D). In parallel to BDNF localization experiments, muscle sections were also processed for detection of MyHC slow or β/I (Fig. 2E) to ascertain our ability to detect muscle antigen in these experiments. Specificity of BDNF antibody was tested on myogenic cell cultures (see below). Thus, the above results suggest that expression and localization of BDNF is low or absent in myofibers and not restricted to mature NMJs.
Figure 3.
BDNF protein is found in junctional and extrajunctional compartments of adult diaphragm skeletal muscle but not in mature myofibers or within the subsynaptic regions of NMJs. A, Levels of BDNF protein, per milligram of wet muscle mass, in junctional (Junc) and extrajunctional (EJunc) regions of diaphragm (n = 4). B, D, Immunofluorescence of BDNF on transverse sections of the diaphragm. Arrows point to subsynaptic areas of NMJs. Insets are 4′,6-diamidino-2-phenylindole staining of myonuclei. C, Same section as in B showing the localization of postsynaptic acetylcholine receptors stained with α-bungratoxin depicting the presence of NMJs. E, Same section as in D but stained for MyHC slow or β/I. Scale bar, 50 μm. Error bars represent SE.
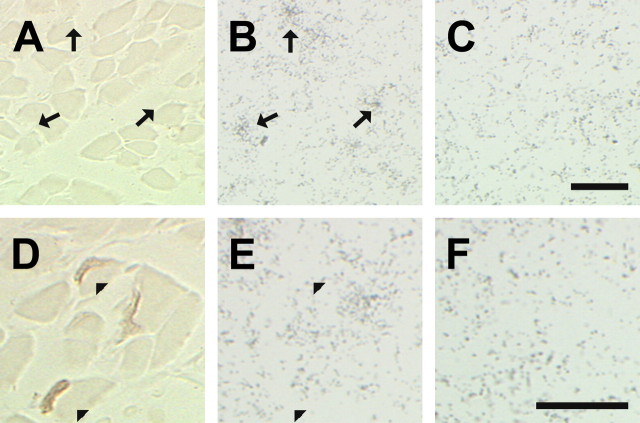
Because developing skeletal muscle contains comparatively higher levels of BDNF (Funakoshi et al., 1995; Griesbeck et al., 1995), we decided to examine its mRNA localization in neonatal (P10) diaphragm. For this, alternative serial sections of neonatal diaphragm were processed for BDNF ISH and NMJ staining (Fig. 4). Silver grain densities representative of BDNF mRNAs were observed in regions of the diaphragm, which were above background sense control levels (Fig. 4, compare B and C). Moreover, BDNF silver grain densities (Fig. 4B, arrows, E, arrowheads) were detected in restricted areas not associated with developing NMJs (Fig. 4A, arrows; or E, arrowheads).
Figure 4.
Concentrations of BDNF transcript(s) do not colocalize with NMJs in neonatal diaphragm skeletal muscle. A, Micrograph of a transverse section of a neonatal diaphragm stained for NMJs. This micrograph is devoid of any NMJ. Arrows point to the same regions as in B. B, Serial section to A showing hybridization of BDNF antisense probe. Arrows point to concentrations of silver grains representing BDNF transcripts. C, Serial section to A processed for BDNF sense control. Scale bar, 25 μm. D, A micrograph showing NMJs in the developing diaphragm. E, Serial section to D showing hybridization of BDNF antisense probe. Arrowheads point toward NMJs. F, Serial section to D processed for BDNF sense probe. Scale bar, 25 μm.
BDNF localization in muscle satellite cells
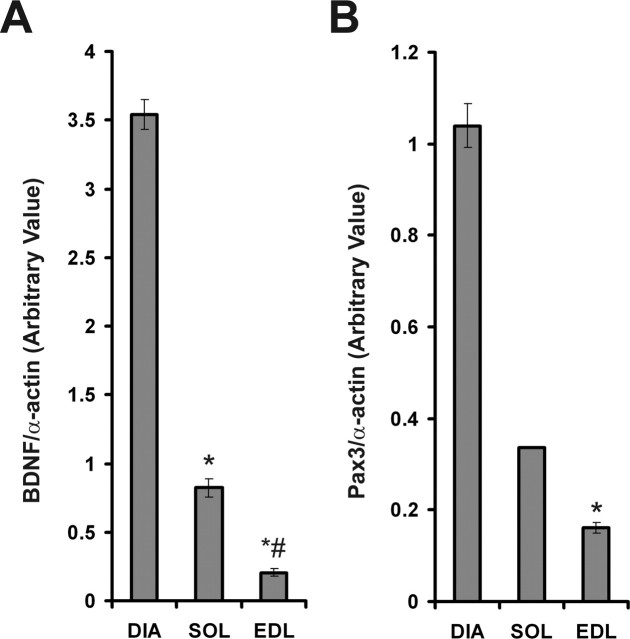
Because BDNF transcript(s) and protein were detected in skeletal muscle but not associating with mature myofibers or NMJs, we questioned whether its synthesis occurs in other components of skeletal muscles and, in particular, in satellite cells. To this end, we quantified the levels of BDNF mRNA in adult diaphragm, soleus, and EDL skeletal muscles, which are known to contain a different number of satellite cells (Schmalbruch and Hellhammer, 1977). Diaphragm contained approximately fourfold and ∼17-fold the levels of BDNF mRNA compared with soleus and EDL, respectively (Fig. 5A). Furthermore, to establish whether levels of BDNF in these skeletal muscles correlate with their respective populations of satellite cells, we measured the levels of a skeletal muscle satellite cell marker, Pax3 (Relaix et al., 2005). Levels of Pax3 mRNA in the diaphragm were approximately threefold and ∼6.5-fold higher than in soleus and EDL, respectively, thus corresponding to levels of BDNF mRNA in these muscles (Fig. 5B). Overall, levels of BDNF were highly correlated (R2 = 0.98) to that of Pax3 in adult skeletal muscles.
Figure 5.
Levels of BDNF are highly correlated to those of a muscle progenitor marker, Pax3. A, Arbitrary levels of BDNF mRNA (normalized to α-actin levels) in adult diaphragm (DIA), soleus (SOL), and extensor digitorum longus (EDL) skeletal muscles (n = 3). B, Arbitrary levels of Pax3 (relative to α-actin) in DIA, SOL, and EDL (*p < 0.05 vs DIA; #p < 0.05 vs soleus). Error bars represent SE.
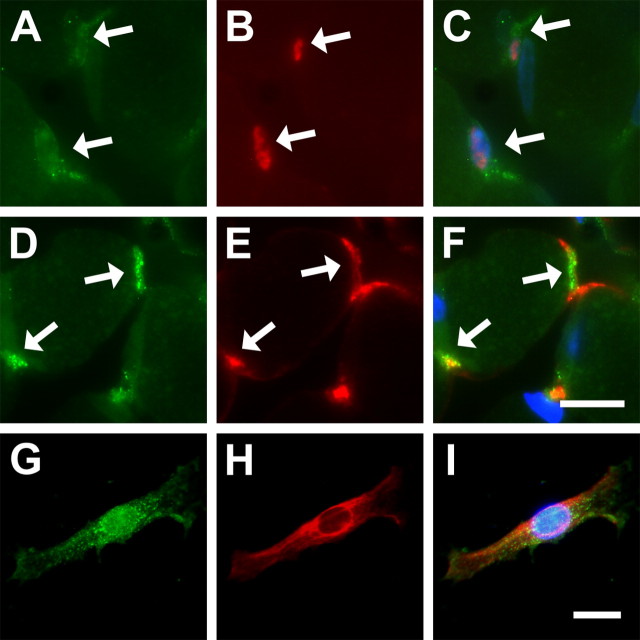
To determine whether expression of BDNF does indeed occur in skeletal muscle satellite cells, we performed immunolocalization studies. To identify skeletal muscle satellite cells in these experiments, we used an antibody directed against Pax7, a paralog of Pax3, which has recently been shown to be expressed in all skeletal muscle-derived Pax3+ cells (Seale et al., 2000; Montarras et al., 2005). BDNF was immunolocalized to the cytoplasmic portion of Pax7+ skeletal muscle satellite cells (Fig. 6A–C, arrows). Approximately 64% (of 111) of Pax7+ cells were immunoreactive for BDNF. Moreover, isolated desmin+ satellite cells, which were harvested from dissociated mature diaphragm myofibers in culture (see Materials and Methods) were also immunoreactive for BDNF (Fig. 6G–I) (Rosenblatt et al., 1995).
Figure 6.
BDNF is localized to skeletal muscle satellite cells. A–C, BDNF is colocalized to Pax7+ cells. A, BDNF IF in adult diaphragm skeletal muscle. B, Same section stained for Pax7. C, Overlay of A, B, and 4′,6-diamidino-2-phenylindole (DAPI) staining of nuclei (blue), showing BDNF in cytoplasmic regions of Pax7+ cells (arrows). D, E, Colocalization of BDNF with p75NTR in adult diaphragm skeletal muscle. D, BDNF IF. E, p75NTR IF. F, Overlay of D, E, and DAPI (blue), showing the colocalization of BDNF with p75NTR (arrows). Scale bar, 12.5 μm. G–I, Isolated diaphragm skeletal muscle satellite cells express BDNF in culture. G, BDNF IF. H, Same cell as in G stained for the myogenic marker desmin. I, Overlay of G, H, and DAPI (blue). Scale bar, 12.5 μm.
BDNF and p75NTR transcripts were expressed in diaphragm skeletal muscle, suggesting that BDNF signaling is mediated by p75NTR in this tissue (Fig. 1). To further examine whether BDNF was spatially restricted to p75NTR-positive regions of skeletal muscle, we performed immunofluorescence experiments, which illustrated that BDNF was indeed colocalized with p75NTR (Fig. 6D–F).
Expression of BDNF during myogenic differentiation
To elucidate the role of BDNF in satellite cells, we decided to perform experiments using cell cultures. To this end, we used rat myogenic cells L6, which have been demonstrated to retain their ability to undergo myogenic differentiation after several months in culture (Yaffe, 1968). Thus, extracts from L6 myoblasts and 4 d differentiated myotubes were used to investigate the expression of BDNF, p75NTR, TrkB, and Pax3 during myogenic differentiation.
BDNF and p75NTR transcripts were detected in myoblast cultures (Fig. 7A). Furthermore, their expression was decreased by approximately fivefold after myogenic differentiation (Fig. 7B,C). Levels of BDNF transcripts containing exon-3 and exon-4 were also downregulated to an extent similar to levels of all BDNF transcripts after myogenic differentiation (data not shown) (Fig. 7B). Pax3 levels were also considerably reduced after differentiation (Fig. 7A,D). As in diaphragm muscle, full-length TrkB transcripts were not present in either myoblasts or 4 d differentiated myotubes (Fig. 7A).
Figure 7.
Repression of BDNF and p75NTR expression during myogenic differentiation. A, Representative ethidium bromide gels of BDNF, p75NTR, TrkB, Pax3, and α-actin RT-PCR products from L6 myoblasts (MB) and myotubes (MT). No RNA (CL) was used as negative control. TrkB was not detected in MB or MT samples. B, There was an approximate fivefold reduction in the levels of BDNF mRNA, as normalized to α-actin after myogenic differentiation (*p < 0.05). C, The expression of p75NTR followed the same trend as BDNF during differentiation (*p < 0.05). D, Pax3 levels are reduced by ∼50% after myogenic differentiation (n = 6; *p < 0.05). E–J, BDNF protein is detected in myoblasts and not in myotubes. E, H, BDNF IF of myoblasts and myotubes, respectively. Scale bar, 50 μm. Insets are staining for all isoforms of MyHC (red). F, I, Same as in E and H, except BDNF antibody was blocked with BDNF before incubation (see Materials and Methods). G, J, No primary control. K, BDNF protein levels per microgram of total protein in MB and MT (n = 3; *p < 0.05). Error bars represent SE.
To determine whether BDNF protein followed the same trend as its transcript during myogenic differentiation, we performed immunofluorescence experiments on myoblasts and 4 d differentiated myotubes. BDNF was immunolocalized to the cytoplasmic region of myoblasts (Fig. 7E). As expected, differentiated myotubes did not stain (or stained weakly) for BDNF (Fig. 7H). To establish the specificity of our primary antibody, anti-BDNF was blocked with recombinant BDNF before incubation with samples (Fig. 7F,I). In a parallel experiment, we also omitted incubation with the primary BDNF antibody to show specificity of our secondary antibody (Fig. 7G,J). Total BDNF protein levels were also decreased after myogenic differentiation, replicating the extent of decrease in mRNA levels (Fig. 7, compare B, K). Collectively, these results demonstrate that expression of BDNF, including its exon variants, is similar in skeletal muscle satellite cells and L6 myoblasts in culture. Furthermore, expression of BDNF decreases in myotubes and is hardly detectable in adult skeletal myofibers.
BDNF inhibits myogenic differentiation
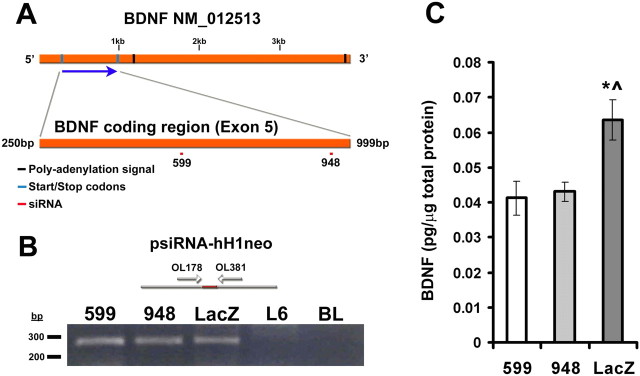
To decipher the role of BDNF in myoblasts, we used siRNA technology to decrease the endogenous levels of BDNF in myoblasts. Two siRNA fragments were designed to target the coding region of BDNF (Fig. 8A). After stably transfecting L6 myoblasts, clonal populations containing the siRNA constructs were isolated as verified by PCR (Fig. 8B). BDNF siRNA clonal populations, BDNF599 and BDNF948, contained significantly reduced levels of BDNF compared with parallel control cells containing siRNA for a bacterial gene (LacZ) (Fig. 8C). Levels of BDNF did not significantly differ between LacZ and L6 parental cells (compare Figs. 8C and 7K).
Figure 8.
siRNA design for BDNF. A, Representative map of BDNF NM_012513 sequence. BDNF599 and BDNF948 siRNA fragments are shown to be targeted to the coding regions (exon 5) of BDNF. B, Amplification of siRNA fragments from stably transfected siRNA cell lines. Using psiRNA-hH1neo specific primers (arrows), siRNA fragments (red) were amplified using DNA preparations from stable cell lines. A representative ethidium bromide gel showing amplified PCR products of psiRNAhH1neo in transfected cells. PCR product length is ∼290 bp. siRNA constructs are present in stably transfected clonal cells. L6 extracts and no DNA (BL) were used controls. C, Reduction of BDNF, per microgram of total protein, in BDNF599 (n = 9) and BDNF948 (n = 6) clonal cells compared with LacZ (n = 9) control cells (*p < 0.05 vs 599; ⁁p < 0.05 vs 948). Error bars represent SE.
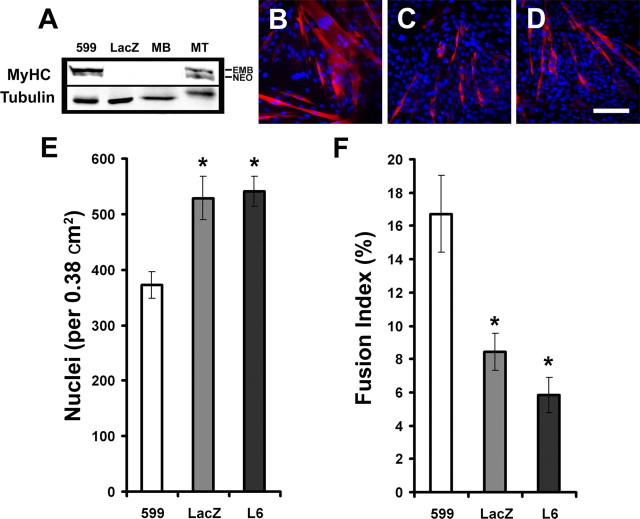
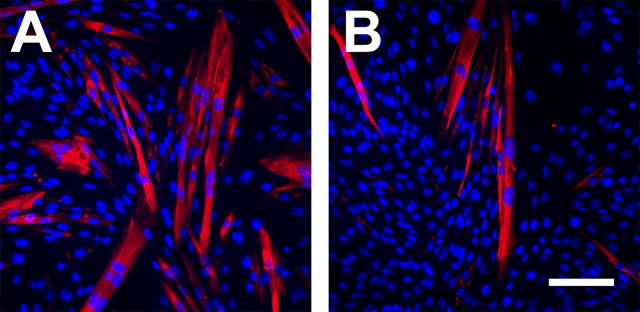
Remarkably, 2–3 d after equal plating of cells in growth media, we noticed significant morphological differences between BDNF599 clonal cells compared with LacZ or parental (L6) control cells (see below). BDNF599 clonal cells consistently exhibited an elongated mononucleated form and even formed multinucleated myotubes after 2–3 d in growth media. In contrast, parallel LacZ and L6 cultures consisted of relatively rounder mononucleated cells with no apparent multinucleated myotubes (see below). The above observations suggested that BDNF599 clonal cells were exhibiting a relatively faster rate of myogenic differentiation and fusion in growth media. To quantify the extent of myogenic differentiation between BDNF599 cells and parallel controls, cultures were analyzed for the expression of a terminal differentiation marker, MyHC. Using an antibody that recognizes all isoforms of MyHC (MF20), we detected MyHC in BDNF599 myoblast cultures but not in parallel LacZ or parental L6 myoblast cultures 6 d after equal plating in growth media (Fig. 9A). In agreement with previous findings (Conway et al., 2004), we detected two isoforms of MyHC in differentiated cultures of control myotubes. BDNF599 cultures were expressing high levels of embryonic MyHC isoform, whereas 4 d differentiated myotubes expressed similar levels of embryonic and neonatal isoforms of MyHC (Fig. 9A). These observations were confirmed using specific antibodies directed against embryonic and neonatal MyHC isoforms (data not shown).
Figure 9.
Suppressing BDNF synthesis enhances myogenic differentiation. A, A Western blot showing expression of MyHC in BDNF599, LacZ, and parental (MB) after 6 d of incubation in growth media. Four day differentiated myotubes (MT) were used as MyHC+ control. Tubulin is shown as a loading control. Right markers show the position of embryonic (EMB) and neonatal (NEO) MyHC bands on the blot. B–D, Representative micrographs of BDNF599, LacZ, and parental cultures, respectively, stained for MyHC (red) after 6 d of incubation in growth media. Nuclei are stained with 4′,6-diamidino-2-phenylindole (blue). Scale bar, 100 μm. E, Number of nuclei in BDNF599, LacZ, and parental cultures after 6 d of incubation in growth media (n = 5; *p < 0.05). F, BDNF599 clonal cells exhibit a higher rate of fusion compared with LacZ and parental controls (n = 5; *p < 0.05). Error bars represent SE.
After 6 d incubation in growth media, BDNF599 cultures consistently exhibited an elevated number of MyHC+ myotubes compared with LacZ or parental L6 cultures (Fig. 9B–D, red staining). Moreover, the number of nuclei in BDNF599 cultures was ∼70% of LacZ or parental L6 cultures after the 6 d incubation period (Fig. 9E). To assess the extent of myogenic fusion in growth media, the number of myonuclei within myotubes (more than two nuclei) was expressed as a percentage of total nuclei per field of view (i.e., fusion index). BDNF599 cultures contained approximately twofold the number of nuclei within myotubes compared with LacZ or parental L6 cultures after 6 d incubation in growth media (Fig. 9F). Finally, we measured the levels of BDNF mRNA at the end of 6 d incubation in growth media. BDNF599 clonal cultures contained ∼55% less BDNF mRNA (normalized to α-actin) compared with parallel LacZ control cultures (supplemental figure, available at www.jneurosci.org as supplemental material). Together, the above observations suggest that the reduction in levels of BDNF promotes myoblasts to exit the cell cycle and initiate the myogenic differentiation program despite still being in growth media.
To determine whether the enhanced differentiation of BDNF599 clonal cells was exclusively caused by a reduction in BDNF, we sought to rescue their relatively rapid rate of myogenic differentiation with daily doses of exogenous BDNF during the 6 d incubation period in growth media. Treated BDNF599 cells exhibited a reduced number of MyHC+ mononucleated cells and myotubes compared with parallel, vehicle-treated BDNF599 cultures (Fig. 10, compare red staining in A and B). Furthermore, treatment of these cells with recombinant BDNF resulted in a 50% reduction in the fusion compared with parallel, vehicle-treated cultures. Vehicle-treated cultures contained 18.2 ± 1.6% fused cells, whereas those treated with recombinant BDNF displayed 9.7 ± 1.1% of total nuclei within myotubes. The number of nuclei (per 0.38 cm2) in BDNF (325 ± 22) and vehicle (306 ± 15)-treated cultures did not differ significantly.
Figure 10.
Recombinant BDNF rescues the accelerated myogenic differentiation of BDNF599 siRNA clonal cells. A, A representative micrograph of vehicle-treated BDNF599 cells stained for MyHC (red) after 6 d of incubation in growth media. Nuclei are stained with 4′,6-diamidino-2-phenylindole (blue). B, A micrograph of parallel BDNF599 cultures, which were treated with daily dose of BDNF, stained for MyHC (red). Scale bar, 100 μm.
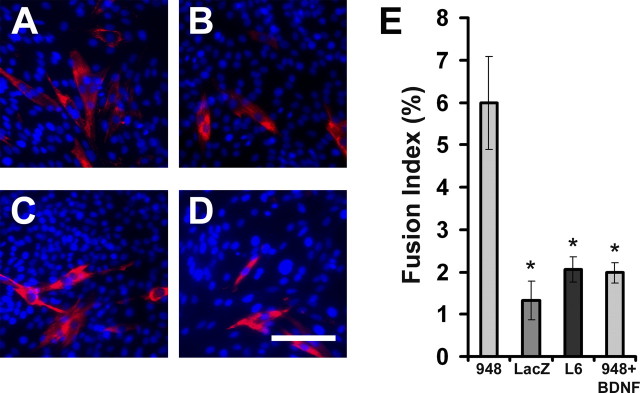
Similar to BDNF599 cultures, we observed enhanced myogenic differentiation and fusion of BDNF948 clonal cells (Fig. 11). After 6 d incubation in growth media, there was a higher percentage of MyHC+ myotubes in BDNF948 (Fig. 11A) compared with LacZ (Fig. 11B) or parental L6 (Fig. 11C). The number of nuclei in BDNF948 cultures (380 ± 22 per 0.38 cm2) was significantly lower than those of LacZ (544 ± 23) or parental L6 cells (842 ± 45) after 6 d incubation in growth media. Moreover, there were higher incidents of myogenic fusion in BDNF948 cultures compared with LacZ or parental L6 cells (Fig. 11E). BDNF948 cultures contained ∼66% less BDNF mRNA (normalized to α-actin) at the end of the 6 d incubation in growth media (supplemental figure, available at www.jneurosci.org as supplemental material). Administration of daily doses of BDNF during the 6 d incubation period delayed the relatively rapid rate of differentiation as evident by reduced number of MyHC+ cells (Fig. 11D) and decreased fusion of BDNF948 cells (Fig. 11E). Altogether, the above results suggest that BDNF impedes the differentiation and fusion of myoblasts.
Figure 11.
BDNF impedes the myogenic differentiation program. A–D, Micrographs of BDNF948, LacZ, parental (L6), and BDNF-treated 948 cultures, respectively, stained for MyHC (red) after 6 d in growth media. Scale bar, 100 μm. Nuclei are stained with 4′,6-diamidino-2-phenylindole (blue). E, BDNF948 cells fused at a higher rate than LacZ and parental (L6) cells (*p < 0.05 vs BDNF948). BDNF treatment of 948 cells reduced the fusion of these cells (n = 12). Error bars represent SE.
Discussion
Skeletal muscle-derived BDNF has been hypothesized previously to act as a retrograde signaling factor for innervating motor neurons throughout their lifespan. However, its localization in mature myofibers has remained enigmatic. Here, we report that expression of BDNF was not detectable in mature myofibers or within the subsynaptic regions of NMJs. Most notably, BDNF was found in a Pax7+ muscle satellite cell, and the overall levels of BDNF mRNA were highly correlated with that of a progenitor marker, Pax3, in mature muscles. In complementary cell culture experiments, BDNF was found in myoblasts, and its expression was repressed in differentiated myotubes. By reducing the endogenous levels of BDNF using siRNA, myoblasts engaged in early myogenic differentiation while still being in growth media. This effect could be reversed with the addition of recombinant BDNF. Our data provide evidence indicating that the primary role of BDNF in mature skeletal muscle is to maintain the population of satellite cells by preventing their myogenic differentiation. Because TrkB was not detected in skeletal muscle, satellite cell-derived BDNF signaling is proposed to occur via p75NTR.
BDNF mRNA and protein were detected in junctional and extrajunctional regions of mature diaphragm muscle, albeit with a tendency toward higher levels in extrajunctional compartment. Moreover, closer examination indicated that BDNF transcript(s) and protein were absent within subsynaptic regions of NMJs, thus suggesting that muscle-derived BDNF may not serve as a retrograde survival factor for innervating motor neurons throughout their lifespan. In fact, higher levels of BDNF in extrajunctional regions clearly argue against the retrograde signaling role of BDNF in mature neuromuscular system. In agreement with our observations, Liem et al. (2001) reported that subsynaptic regions of NMJs in soleus muscle were devoid of BDNF transcript(s). Here, we also demonstrated the absence of BDNF protein at the subsynaptic regions of mature NMJs. This is in contrast to what is observed in adult CNS, where BDNF and TrkB associate with synapses (Bramham and Messaoudi, 2005). Therefore, we postulate that unlike CNS synapses, mature NMJs are stable arrangements that do not require BDNF signaling for remodeling and modulation of neuromuscular transmission.
Expression of BDNF in skeletal muscle satellite cells has been controversial (Copray et al., 2000; Liem et al., 2001). However, considering that levels of BDNF were highly correlated with that of Pax3 in adult skeletal muscles, and that BDNF was present in 64% of Pax7+ satellite cells, our results strongly suggest that BDNF is indeed expressed in satellite cells. To our knowledge, this is the first time BDNF has been specifically localized to immunologically characterized satellite cells in mature skeletal muscle. Our findings also support the notion of BDNF expression in muscle progenitor cells. In fact, paraxial somitic muscle progenitors express BDNF during embryogenesis (Jungbluth et al., 1997).
Repression of BDNF synthesis was found to be differentiation dependent in skeletal muscle. BDNF was highly expressed in muscle satellite cells and myoblasts in culture, whereas its expression was absent in myofibers (or very low) and repressed after myogenic differentiation. Our results are consistent with the findings of Seidl et al. (1998), who reported expression of BDNF in C2C12 myoblasts. BDNF transcripts containing exon-3 and exon-4 were detected in skeletal muscle satellite cells and myoblasts in culture, suggesting that expression of BDNF is driven by upstream promoters of these exons. Although the activity of these promoters is regulated by calcium-mediated signaling in neurons, this type of regulation may not be relevant for repression of BDNF during myogenic differentiation (Tao et al., 1998). That is, the repression of BDNF coincides with increased levels of intracellular calcium needed for myogenic differentiation (Shainberg et al., 1969). Thus, one plausible mechanism by which BDNF transcription is repressed during myogenic differentiation involves the induction of methyl CpG binding proteins (MeCP2 and/or MBD2) (Brero et al., 2005). MeCP2, by recruiting histone modifying enzymes, binds to methylated promoter regions upstream of exon-3, thereby silencing BDNF expression in neurons (Chen et al., 2003; Martinowich et al., 2003). The induction of MeCP2 and MBD2 in differentiated myotubes enhances the clustering of heterochromatin (i.e., silenced genes), which may contribute to repression of BDNF (Brero et al., 2005).
An important question raised by our findings is whether indeed BDNF also inhibits myogenic differentiation of satellite cells in skeletal muscle in vivo. Although we do not currently have data to address this issue directly, there are converging lines of evidence in the literature that suggest that BDNF acts as a potent inhibitor of myogenic differentiation in vivo. For example, whereas the levels of BDNF are significantly reduced in gastrocnemius skeletal muscle during postnatal development, those of NT-4/5 are increased (Funakoshi et al., 1995). Similarly, levels of BDNF are decreased in mature gastrocnemius muscle hours after electrical stimulation of sciatic nerve, whereas those of NT-4/5 are increased, suggesting that BDNF and NT-4/5 may have different functions in skeletal muscle (Funakoshi et al., 1995). Given our findings that BDNF, conceivably by acting on p75NTR, inhibits myogenic differentiation, its decline in skeletal muscle during postnatal development is required for the progression of myogenic differentiation. Moreover, the reduction in BDNF levels after electrical stimulation suggests an enhanced myogenic differentiation of skeletal muscle satellite cells. In fact, electrical stimulation or exercise promotes activation and incorporation of satellite cells into skeletal myofibers (Schultz, 1989).
Similar to electrical stimulation, denervation results in incorporation of satellite cells into myofibers concomitant with the reduction in BDNF levels in adult skeletal muscle, further supporting the role of this neurotrophin as a potent inhibitor of myogenic differentiation (Funakoshi et al., 1993; Jejurikar and Kuzon, 2003). In contrast, the rise in the levels of NT-4/5 in skeletal muscle during development and after electrical stimulation suggests it may, by binding to p75NTR, act as a mitogen for satellite cells. Consistent with this notion, our recent unpublished data show higher expression of NT-4/5 in myoblasts than in myotubes. Altogether, the above observations suggest the functional diversity of neurotrophins in mature muscle with BDNF as a potent inhibitor of myogenic differentiation and NT-4/5 as a mitogen of satellite cells.
Although full-length TrkB has been implicated in the clustering of postsynaptic acetylcholine receptors, we could not detect its transcript, even at high PCR cycles, in mature skeletal muscle (Gonzalez et al., 1999; Wells et al., 1999). Gonzalez et al. (1999) reported the disruption of acetylcholine receptors by overexpression of dominant-negative truncated TrkB in adult sternomastoid muscle, thereby concluding that TrkB signaling is involved in the maintenance and/or aggregation of postsynaptic NMJ components (Gonzalez et al., 1999). However, considering that BDNF has been observed in satellite cells, the overexpression of truncated TrkB may result in depletion of BDNF, thus exhausting the normal regenerative capacity and the maintenance of postsynaptic acetylcholine receptor densities. In a separate study, Wells et al. (1999) illustrated the disruption of agrin-induced acetylcholine receptor clusters with BDNF stimulation. Given our findings that BDNF inhibits myogenic differentiation, it is plausible that the effects of BDNF on acetylcholine receptor clustering is observed as a consequence of delayed myogenic differentiation.
Our current observations, in conjunction with previous findings, allow for formulation of a model regarding the role of BDNF in the development of neuromuscular system. During embryonic development, motor axons enter the primordial skeletal muscle comprised of mainly mononucleated muscle progenitors (Allan and Greer, 1997; Babiuk et al., 2003). Furthermore, these progenitors express BDNF and p75NTR as their innervating motor neurons in the spinal cord express p75NTR and TrkB (Ernfors et al., 1988; Griesbeck et al., 1995; Allan and Greer, 1997; Wheeler et al., 1998). As axons begin to traverse through the primordial skeletal muscle, they form synapses with muscle progenitors and promote their myogenic differentiation (Ross et al., 1987a; Allan and Greer, 1997). Given that BDNF potentiates this nerve-muscle synapse formation in culture (Lohof et al., 1993; Zhang and Poo, 2002), progenitor-derived BDNF may in fact facilitate this initial synaptogenesis between motor axons and primordial skeletal muscle. Motor axons, in turn, promote the commencement of myogenic differentiation program and myotube formation in skeletal muscles (Ross et al., 1987b; Allan and Greer, 1997; Greer et al., 1999). Given our findings that downregulation of BDNF promote myogenic differentiation and fusion, we envisage that motor axons, by attracting progenitor-derived BDNF to their neuropodia, promote the differentiation and fusion of muscle progenitors during this critical period of neuromuscular contact formation. This mechanism allows for synchronized neuromuscular differentiation thereby potentiating presynaptic acetylcholine release and, at the same time, upregulating postsynaptic acetylcholine receptors as a result of myogenic differentiation (Paterson and Prives, 1973; Lohof et al., 1993; Kues et al., 1995; Zhang and Poo, 2002). Subsequently, after establishing contact and the differentiation of muscle progenitors (i.e., in the absence of BDNF signaling), anterograde signals (i.e., acetylcholine, agrin, and neuregulin) from motor axons initiate the specialization of postsynaptic densities (Sanes and Lichtman, 2001).
Footnotes
This work was supported by research grants from the Canadian Institutes of Health Research to B.J.J. We thank Dr. David J. Parry for comments and suggestions. We also thank Amanda Bradford and John Lunde for their assistance in performing the fusion index. We are grateful to Dr. Mark Stocksley for his initial guidance in the ISH experiments and to Drs. Philip A. Barker, Robin J. Parks, and Michael A. Rudnicki and Regeneron Pharmaceuticals for providing reagents used in this study.
References
- Allan DW, Greer JJ (1997). Embryogenesis of the phrenic nerve and diaphragm in the fetal rat. J Comp Neurol 382:459–468. [DOI] [PubMed] [Google Scholar]
- Babiuk RP, Zhang W, Clugston R, Allan DW, Greer JJ (2003). Embryological origins and development of the rat diaphragm. J Comp Neurol 455:477–487. [DOI] [PubMed] [Google Scholar]
- Boudreau-Lariviere C, Sveistrup H, Parry DJ, Jasmin BJ (1996). Ciliary neurotrophic factor: regulation of acetylcholinesterase in skeletal muscle and distribution of messenger RNA encoding its receptor in synaptic versus extrasynaptic compartments. Neuroscience 73:613–622. [DOI] [PubMed] [Google Scholar]
- Bramham CR, Messaoudi E (2005). BDNF function in adult synaptic plasticity: the synaptic consolidation hypothesis. Prog Neurobiol 76:99–125. [DOI] [PubMed] [Google Scholar]
- Brero A, Easwaran HP, Nowak D, Grunewald I, Cremer T, Leonhardt H, Cardoso MC (2005). Methyl CpG-binding proteins induce large-scale chromatin reorganization during terminal differentiation. J Cell Biol 169:733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME (2003). Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science 302:885–889. [DOI] [PubMed] [Google Scholar]
- Conway K, Pin C, Kiernan JA, Merrifield P (2004). The E protein HEB is preferentially expressed in developing muscle. Differentiation 72:327–340. [DOI] [PubMed] [Google Scholar]
- Copray S, Liem R, Brouwer N, Greenhaff P, Habens F, Fernyhough P (2000). Contraction-induced muscle fiber damage is increased in soleus muscle of streptozotocin-diabetic rats and is associated with elevated expression of brain-derived neurotrophic factor mRNA in muscle fibers and activated satellite cells. Exp Neurol 161:597–608. [DOI] [PubMed] [Google Scholar]
- DiStefano PS, Friedman B, Radziejewski C, Alexander C, Boland P, Schick CM, Lindsay RM, Wiegand SJ (1992). The neurotrophins BDNF, NT-3, and NGF display distinct patterns of retrograde axonal transport in peripheral and central neurons. Neuron 8:983–993. [DOI] [PubMed] [Google Scholar]
- Ernfors P, Hallbook F, Ebendal T, Shooter EM, Radeke MJ, Misko TP, Persson H (1988). Developmental and regional expression of beta-nerve growth factor receptor mRNA in the chick and rat. Neuron 1:983–996. [DOI] [PubMed] [Google Scholar]
- Funakoshi H, Frisen J, Barbany G, Timmusk T, Zachrisson O, Verge VM, Persson H (1993). Differential expression of mRNAs for neurotrophins and their receptors after axotomy of the sciatic nerve. J Cell Biol 123:455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi H, Belluardo N, Arenas E, Yamamoto Y, Casabona A, Persson H, Ibanez CF (1995). Muscle-derived neurotrophin-4 as an activity-dependent trophic signal for adult motor neurons. Science 268:1495–1499. [DOI] [PubMed] [Google Scholar]
- Gonzalez M, Ruggiero FP, Chang Q, Shi YJ, Rich MM, Kraner S, Balice-Gordon RJ (1999). Disruption of Trkb-mediated signaling induces disassembly of postsynaptic receptor clusters at neuromuscular junctions. Neuron 24:567–583. [DOI] [PubMed] [Google Scholar]
- Greer JJ, Allan DW, Martin-Caraballo M, Lemke RP (1999). An overview of phrenic nerve and diaphragm muscle development in the perinatal rat. J Appl Physiol 86:779–786. [DOI] [PubMed] [Google Scholar]
- Griesbeck O, Parsadanian AS, Sendtner M, Thoenen H (1995). Expression of neurotrophins in skeletal muscle: quantitative comparison and significance for motoneuron survival and maintenance of function. J Neurosci Res 42:21–33. [DOI] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF (2003). Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem 72:609–642. [DOI] [PubMed] [Google Scholar]
- Jejurikar SS, Kuzon WM Jr (2003). Satellite cell depletion in degenerative skeletal muscle. Apoptosis 8:573–578. [DOI] [PubMed] [Google Scholar]
- Jungbluth S, Koentges G, Lumsden A (1997). Coordination of early neural tube development by BDNF/trkB. Development 124:1877–1885. [DOI] [PubMed] [Google Scholar]
- Karnovsky MJ, Roots L (1964). A “direct-coloring” thiocholine method for cholinestrases. J Histochem Cytochem 12:219–221. [DOI] [PubMed] [Google Scholar]
- Koliatsos VE, Clatterbuck RE, Winslow JW, Cayouette MH, Price DL (1993). Evidence that brain-derived neurotrophic factor is a trophic factor for motor neurons in vivo. Neuron 10:359–367. [DOI] [PubMed] [Google Scholar]
- Kues WA, Sakmann B, Witzemann V (1995). Differential expression patterns of five acetylcholine receptor subunit genes in rat muscle during development. Eur J Neurosci 7:1376–1385. [DOI] [PubMed] [Google Scholar]
- Liem RS, Brouwer N, Copray JC (2001). Ultrastructural localisation of intramuscular expression of BDNF mRNA by silver-gold intensified non-radioactive in situ hybridisation. Histochem Cell Biol 116:545–551. [DOI] [PubMed] [Google Scholar]
- Lohof AM, Ip NY, Poo MM (1993). Potentiation of developing neuromuscular synapses by the neurotrophins NT-3 and BDNF. Nature 363:350–353. [DOI] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE (2003). DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 302:890–893. [DOI] [PubMed] [Google Scholar]
- Michel RN, Vu CQ, Tetzlaff W, Jasmin BJ (1994). Neural regulation of acetylcholinesterase mRNAs at mammalian neuromuscular synapses. J Cell Biol 127:1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montarras D, Morgan J, Collins C, Relaix F, Zaffran S, Cumano A, Partridge T, Buckingham M (2005). Direct isolation of satellite cells for skeletal muscle regeneration. Science 309:2064–2067. [DOI] [PubMed] [Google Scholar]
- Mousavi K, Parry DJ, Jasmin BJ (2004). BDNF rescues myosin heavy chain IIB muscle fibers after neonatal nerve injury. Am J Physiol Cell Physiol 287:C22–C29. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Kaneko T, Ogura T, Suzuki T, Torii M, Kaibuchi K, Arai K, Nakamura S, Nakafuku M (1996). Roles of cell-autonomous mechanisms for differential expression of region-specific transcription factors in neuroepithelial cells. Development 122:2449–2464. [DOI] [PubMed] [Google Scholar]
- Nykjaer A, Willnow TE, Petersen CM (2005). p75NTR–live or let die. Curr Opin Neurobiol 15:49–57. [DOI] [PubMed] [Google Scholar]
- Oppenheim RW, Yin QW, Prevette D, Yan Q (1992). Brain-derived neurotrophic factor rescues developing avian motoneurons from cell death. Nature 360:755–757. [DOI] [PubMed] [Google Scholar]
- Paterson B, Prives J (1973). Appearance of acetylcholine receptor in differentiating cultures of embryonic chick breast muscle. J Cell Biol 59:241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F, Rocancourt D, Mansouri A, Buckingham M (2005). A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 435:948–953. [DOI] [PubMed] [Google Scholar]
- Rosenblatt JD, Lunt AI, Parry DJ, Partridge TA (1995). Culturing satellite cells from living single muscle fiber explants. In Vitro Cell Dev Biol Anim 31:773–779. [DOI] [PubMed] [Google Scholar]
- Ross JJ, Duxson MJ, Harris AJ (1987a). Formation of primary and secondary myotubes in rat lumbrical muscles. Development 100:383–394. [DOI] [PubMed] [Google Scholar]
- Ross JJ, Duxson MJ, Harris AJ (1987b). Neural determination of muscle fibre numbers in embryonic rat lumbrical muscles. Development 100:395–409. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Lichtman JW (2001). Induction, assembly, maturation and maintenance of a postsynaptic apparatus. Nat Rev Neurosci 2:791–805. [DOI] [PubMed] [Google Scholar]
- Schmalbruch H, Hellhammer U (1977). The number of nuclei in adult rat muscles with special reference to satellite cells. Anat Rec 189:169–175. [DOI] [PubMed] [Google Scholar]
- Schultz E (1989). Satellite cell behavior during skeletal muscle growth and regeneration. Med Sci Sports Exerc 21:S181–S186. [PubMed] [Google Scholar]
- Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA (2000). Pax7 is required for the specification of myogenic satellite cells. Cell 102:777–786. [DOI] [PubMed] [Google Scholar]
- Seidl K, Erck C, Buchberger A (1998). Evidence for the participation of nerve growth factor and its low-affinity receptor (p75NTR) in the regulation of the myogenic program. J Cell Physiol 176:10–21. [DOI] [PubMed] [Google Scholar]
- Sendtner M, Holtmann B, Kolbeck R, Thoenen H, Barde YA (1992). Brain-derived neurotrophic factor prevents the death of motoneurons in newborn rats after nerve section. Nature 360:757–759. [DOI] [PubMed] [Google Scholar]
- Shafey D, Cote PD, Kothary R (2005). Hypomorphic Smn knockdown C2C12 myoblasts reveal intrinsic defects in myoblast fusion and myotube morphology. Exp Cell Res 311:49–61. [DOI] [PubMed] [Google Scholar]
- Shainberg A, Yagil G, Yaffe D (1969). Control of myogenesis in vitro by Ca2+ concentration in nutritional medium. Exp Cell Res 58:163–167. [DOI] [PubMed] [Google Scholar]
- Tabuchi A, Sakaya H, Kisukeda T, Fushiki H, Tsuda M (2002). Involvement of an upstream stimulatory factor as well as cAMP-responsive element-binding protein in the activation of brain-derived neurotrophic factor gene promoter I. J Biol Chem 277:35920–35931. [DOI] [PubMed] [Google Scholar]
- Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME (1998). Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron 20:709–726. [DOI] [PubMed] [Google Scholar]
- Timmusk T, Palm K, Metsis M, Reintam T, Paalme V, Saarma M, Persson H (1993). Multiple promoters direct tissue-specific expression of the rat BDNF gene. Neuron 10:475–489. [DOI] [PubMed] [Google Scholar]
- Timmusk T, Lendahl U, Funakoshi H, Arenas E, Persson H, Metsis M (1995). Identification of brain-derived neurotrophic factor promoter regions mediating tissue-specific, axotomy-, and neuronal activity-induced expression in transgenic mice. J Cell Biol 128:185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells DG, McKechnie BA, Kelkar S, Fallon JR (1999). Neurotrophins regulate agrin-induced postsynaptic differentiation. Proc Natl Acad Sci USA 96:1112–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler EF, Gong H, Grimes R, Benoit D, Vazquez L (1998). p75NTR and Trk receptors are expressed in reciprocal patterns in a wide variety of non-neural tissues during rat embryonic development, indicating independent receptor functions. J Comp Neurol 391:407–428. [PubMed] [Google Scholar]
- Yabe T, Herbert JT, Takanohashi A, Schwartz JP (2004). Treatment of cerebellar granule cell neurons with the neurotrophic factor pigment epithelium-derived factor in vitro enhances expression of other neurotrophic factors as well as cytokines and chemokines. J Neurosci Res 77:642–652. [DOI] [PubMed] [Google Scholar]
- Yaffe D (1968). Retention of differentiation potentialities during prolonged cultivation of myogenic cells. Proc Natl Acad Sci USA 61:477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Elliott J, Snider WD (1992). Brain-derived neurotrophic factor rescues spinal motor neurons from axotomy-induced cell death. Nature 360:753–755. [DOI] [PubMed] [Google Scholar]
- Yan Q, Elliott JL, Matheson C, Sun J, Zhang L, Mu X, Rex KL, Snider WD (1993). Influences of neurotrophins on mammalian motoneurons in vivo. J Neurobiol 24:1555–1577. [DOI] [PubMed] [Google Scholar]
- Young C, Lindsay S, Vater R, Slater CR (1998). An improved method for the simultaneous demonstration of mRNA and esterase activity at the human neuromuscular junction. Histochem J 30:7–11. [DOI] [PubMed] [Google Scholar]
- Zhang X, Poo MM (2002). Localized synaptic potentiation by BDNF requires local protein synthesis in the developing axon. Neuron 36:675–688. [DOI] [PubMed] [Google Scholar]