Abstract
The late larval brain consists of embryonically produced primary neurons forming a deep core cortex, surrounded at the surface by ∼100 secondary lineages. Each secondary lineage forms a tract (secondary lineage tract) with an invariant and characteristic trajectory. Within the neuropile, tracts of neighboring lineages bundle together to form secondary tract systems. In this paper, we visualized secondary lineages by the global marker BP106 (neurotactin), as well as green fluorescent protein-labeled clones and thereby establish a comprehensive digital atlas of secondary lineages. The information contained in this atlas is the location of the lineage within the cortex, the neuropile compartment contacted by the lineage tract, and the projection pattern of the lineage tract within the neuropile. We have digitally mapped the expression pattern of three genes, sine oculis, period, and engrailed into the lineage atlas. The atlas will enable us and others to analyze the phenotype of mutant clones in the larval brain. Mutant clones can only be interpreted if the corresponding wild-type clone is well characterized, and our lineage atlas, which visualizes all wild-type lineages, will provide this information. Secondly, secondary lineage tracts form a scaffold of connections in the neuropile that foreshadows adult nerve connections. Thus, starting from the larval atlas and proceeding forward through pupal development, one will be able to reconstruct adult brain connectivity at a high level of resolution. Third, the atlas can serve as a repository for genes expressed in lineage-specific patterns.
Keywords: Drosophila, brain, neuroblast, lineage, axon, connectivity
Introduction
The Drosophila brain is formed by a stereotyped set of neuroblasts that appear in the early embryo. The brain contains ∼100 neuroblasts (Younossi-Hartenstein et al., 1996; Urbach and Technau, 2003). Neuroblasts divide in a stem cell mode; each neuroblast produces a lineage of 10–20 primary neurons during the embryonic period. Primary neurons and glial cells form the functional circuitry controlling larval behavior. Starting during the second instar, neuroblasts become reactivated and produce secondary lineages (Ito and Hotta, 1992). Rough estimates suggest that each neuroblast undergoes anywhere between 40 and 60 additional rounds of mitosis that produce lineages of 100–150 secondary neurons. Secondary lineage tracts do not terminally differentiate in the larva. Thus, unlike primary neurons of the late embryo, dendritic and axonal branches are not formed (Truman, 1990; Dumstrei et al., 2003). Differentiation takes place during the pupal period, when remodeled primary neurons become integrated with evolving secondary lineages into adult circuits (Truman, 1990, 1996).
Neurons that belong to one lineage form a coherent cluster (Dumstrei et al., 2003; Truman net al., 2004; Younossi-Hartenstein et al., 2006; this study), and axons emitted by neurons of one lineage form one fascicle, the primary and secondary lineage axon tract (PAT and SAT), respectively. This implies that neurons of one lineage share their principal trajectory, forming a “unit of projection.” The initial trajectories of secondary lineages of the ventral cord have been carefully mapped in a recent study by Truman and colleagues (2004). Here, we present a complete atlas of the secondary lineages of the late larval brain, based on global labelings of these lineages with the antibody against neurotactin (BP106) (de la Escalera et al., 1990), as well as individually labeled clones induced in the early larva. The data were prepared as digital three-dimensional (3D) models, which allow one to demonstrate the spatial relationships of lineages and their tracts to each other and to landmark structures, in particular the mushroom body and the commissures, which occupy a central position in the brain neuropile (Dumstrei et al., 2003; Pereanu and Hartenstein, 2004).
An important application of the atlas is the interpretation of clonal data. The clonal strategy is widely used in the Drosophila imaginal disc-derived tissues, in particular eyes and wings, and has yielded important insight into gene function during pattern formation. Mutant clones can only be interpreted if the corresponding wild-type clone is well characterized, which is the case for structures such as the eye disc, but not at all the central brain, with the notable exception of the four virtually identical lineages that make up the mushroom body of the larval brain (Lee and Luo, 2001). The lineage atlas presented here visualizes all wild-type lineages, which will enable us and others to interpret mutant clones. Furthermore, reconstructing the pattern of SATs provides inroads into the study of connectivity of the adult brain. Previous works (Stocker, 1994; Nassif et al., 1998, 2003) and preliminary data (W. Pereanu and V. Hartenstein, unpublished observations) indicate that SATs of secondary lineages remain visible through metamorphosis; the atlas of larval tracts therefore forms an essential step toward reconstructing adult brain connectivity. Finally, an important application of a lineage atlas rests in its use as a repository for functional and genetic data. This application is very actively pursued by many groups working with the human brain (Mazziotta et al., 2001; Toga et al., 2001; Thompson et al., 2002), as well as numerous animal model organisms (Carson et al., 2002; Lein et al., 2004; Visel et al., 2004), including Drosophila (Rein et al., 2002; Pereanu and Hartenstein, 2004). Used in this way, digital models may serve as three-dimensional archives of function, pathology, and gene/protein expression patterns.
Materials and Methods
Markers and stocks.
The neuropile of the larval brain was labeled with a monoclonal antibody against the Syntaxin protein (8C3; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA). Neuroblasts and a subset of secondary neurons were labeled with a monoclonal antibody against Shotgun/DE-cadherin (Dumstrei et al., 2003). To clearly visualize lineages of secondary neurons and their axon tract, a monoclonal antibody against the Neurotactin protein (BP106; Developmental Studies Hybridoma Bank) was used. Primary cholinergic neurons and their axons forming the neuropile of the larval brain were labeled by the transgene ChAT-Gal4;UAS-GFP (Salvaterra and Kitamoto, 2001). Gene expression mapping was performed with the following fly lines: so-gal4 (Chang et al., 2003), per-gal4 (Kaneko and Hall, 2000), and ryxho25lacZ [an engrailed (en) reporter line] (Hama et al., 1990). Flippase recombinase/flippase recombinase target (FLP/FRT) clones were induced with the following fly lines: elav-Gal4 (Bloomington Stock Center, Indiana University, Bloomington, IN) and hs-FLP; UAS-taulacz (Ito et al., 1997). Mosaic analysis with a repressible cell marker (MARCM) clones were generated using hs-flp,elav-Gal4,UAS-GFP/FM7 (kindly provided by Dr. U. Tepass, University of Toronto, Toronto, Ontario, Canada) and FRT42Dgal80 (Bloomington Stock Center) fly lines.
Clonal analysis.
We used the FLP/FRT technique to induce labeled clones in early larval brains (Ito et al., 1997). Briefly, in one series of experiments, a UAS-srcGFP or UAS-cd8GFP [both targeting green fluorescent protein (GFP) to membrane] containing a FRT flanked flip-out cassette was driven by elav-Gal4, which is expressed in neurons as well as secondary neuroblasts (in the larval brain). Beside these two chromosomes, the stock contains a hs-FLP construct that allows one to induce the flip-out event, leading to a GFP-expressing cell, by applying short, 30–40 min heat pulses. We also used the MARCM technique to generate labeled clones (Lee and Luo, 2001).
Immunohistochemistry and histology.
The anti-Syntaxin, anti-Neurotactin antibodies were diluted 1:10-fold. Anti-DE-cadherin was diluted 1:100-fold. Secondary antibodies were Alexa 546-conjugated anti-rabbit Ig (A11030; Invitrogen, San Diego, CA) used at a 1:500 dilution and Alexa 568-conjugated anti-rat Ig (A11077; Invitrogen) used at a 1:50 dilution. For antibody labeling, standard procedures were followed (Ashburner, 1989). Staged Drosophila dissected brains were viewed as whole mounts in the confocal microscope (40× oil lens; MRC 1024ES microscope with Radiance 2000 using Lasersharp 2000, version 5.2 build 824 software; Bio-Rad, Hercules, CA). Complete series of optical sections were taken at 2 μm intervals for the clonal preparations and 1 μm intervals for the digital model construction.
Generation of three-dimensional models.
Staged Drosophila larval brains labeled with anti-FasII and other suitable markers were viewed as whole mounts by confocal microscopy [Bio-Rad MRC 1024ES microscope using Bio-Rad Lasersharp, version 3.2 software; 40× oil lens (numerical aperture, 1.0; working distance, 0.17 mm)]. Complete series of optical sections were taken at 2 μm intervals. Digitized images of confocal sections were imported into the Amira program. Because sections were taken from focal planes of one and the same embryo preparation, there was no need for alignment of different sections. All models were generated using the Amira software package. Surface-rendered digital atlas models were created by manually labeling each lineage and neuropile compartment within a series of confocal images taken from a ChAT-Gal4+UAS-GFP third-larval instar brain labeled with anti-Neurotactin. For clonal analysis, the surface rendered neuropile compartments were brought into registration with the dataset of the clone by warping (Pereanu and Hartenstein, 2004). The Amira program also allows one to adjust virtual lighting, camera angle, transparency, reflection, and other parameters in a straightforward manner.
Results
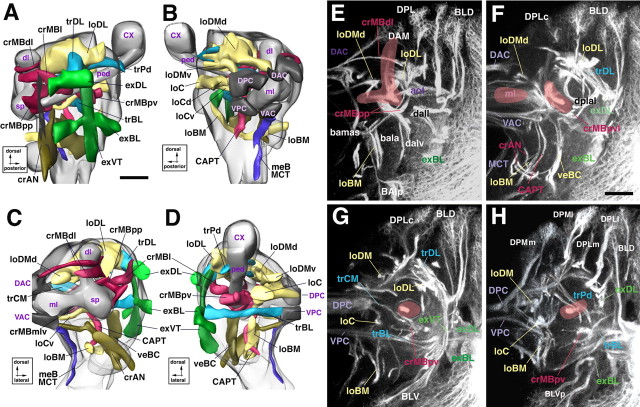
Global markers of neuroblasts and neuronal precursors such as anti-DE-cadherin or monoclonal antibody BP106 reveal ∼100 secondary lineages distributed over the surface of the central brain (Dumstrei et al., 2003). Axons formed by a given lineage fasciculate with each other, thereby forming a discrete bundle within the brain cortex (SAT). Because secondary axon tracts travel radially (centripetally), they reach the neuropile surface at a position that is usually closest to the lineage of origin. Tracts penetrate the neuropile glial sheath or travel along the neuropile surface for variable distance (Fig. 1). Typically, tracts of several neighboring lineages bundle together in the neuropile to form what we will call “secondary tract systems” in the following. The pattern of secondary tract systems of the larval brain is shown as a series of Z-projections of confocal sections in Figure 1; a digital 3D model of this pattern is presented in Figure 2. It is apparent that some secondary tract systems extend along the glial sheaths in between neuropile compartments; others penetrate the center of compartments, typically following pioneer tracts previously laid down by primary axons at an earlier stage (Nassif et al., 2003). It is important to note that each secondary lineage forms a tract with an invariant and characteristic trajectory within the neuropile. In other words, a given lineage tract reaches the neuropile at a characteristic position and then joins one or more (in case the tract branches) secondary tract systems. The low degree of variability can be gleaned from Figure 4, in which the position of lineages in the cortex (Fig. 3A–C) and lineage tracts in the neuropile (Fig. 3D–F) is shown for three different brains.
Figure 1.
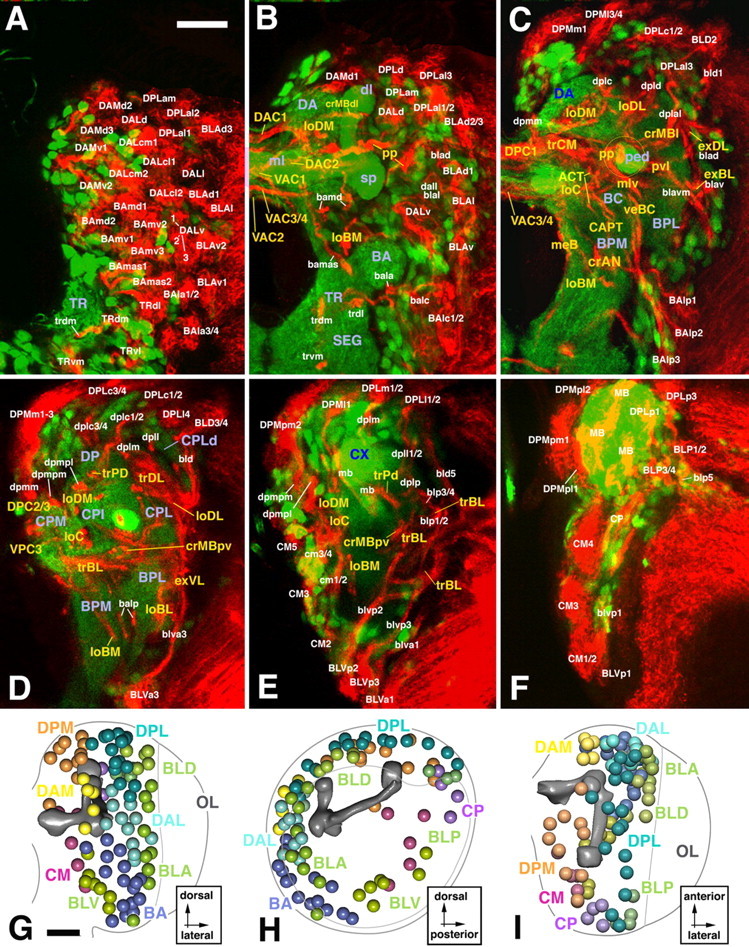
Atlas of secondary neural lineages and their projections in the late third instar brain. A–F, Z-projections prepared from transverse sections of a brain labeled with the BP106 antibody (secondary lineages and their axon tracts; red) and a ChAT-Gal4; UAS-GFP construct (primary neurons and neuropile; green). In this and the following figures depicting cross sections, panels show one brain hemisphere, with lateral pointing to the right and dorsal up. Z-projections were prepared from ∼10 adjacent 1 μm confocal sections. Intervals in between neighboring Z-projections are 10–15 μm. The first projection (A) represents a slice through the anterior cortex; B, anterior neuropile (lobes of mushroom body, antennal lobe); C, D, center of the neuropile with supraesophageal commissures; E, posterior neuropile (calyx); F, posterior cortex. Lineages and their tracts are identified by white lettering; fiber tracts in the neuropile are annotated in yellow, and neuropile compartments in violet. G–I, 3D models of brain hemisphere (G, anterior view; H, lateral view; I, dorsal view) showing location of lineages located by colored spheres. The outline of brain hemisphere and optic lobe (OL) is indicated by the gray line; the mushroom body (mb; shaded dark gray), located in the center of the neuropile, is provided as landmark. SEG, Subesophageal ganglion; CX, calyx of mushroom body; sp, spur of mushroom body; dl, dorsal lobe of mushroom body; ml, medial lobe of mushroom body. Scale bars, 25 μm.
Figure 2.
3D digital models of larval brain neuropile showing pattern of secondary axon tract systems. A, Lateral view; B, medial view; C, anterior view; D, posterior view. Surface of neuropile and mushroom body are shaded light and intermediate gray, respectively. Commissural tracts forming the supraesophageal commissure are dark gray. Longitudinal tract systems are colored yellow; transverse systems are colored blue; circumferential systems around mushroom body (lobes, peduncle) are red; circumferential systems around antennal lobe (BA compartment) are olive green; external systems (at neuropile surface) are bright green; medial cervical tract is violet. E–H, Z-projections prepared from confocal cross sections of BP106-labeled brain. Fiber tracts within the neuropile, formed by the confluence of secondary axon tracts labeled by anti-BP106, were identified on the basis of location and axonal trajectory. As explained for Figure 1, Z-projections were prepared from 10 successive sections at different anteroposterior levels. E, Anterior neuropile (lobes of mushroom body, antennal lobe); F, anterocentral neuropile right behind lobes of mushroom body; G, center of the neuropile with posterior supraesophageal commissures; H, posterior neuropile. CX, Calyx of mushroom body; sp, spur of mushroom body; dl, dorsal lobe of mushroom body; ml, medial lobe of mushroom body; ped, peduncle of the mushroom body; aot, anterior optic tract. Scale bars, 20 μm.
Figure 4.
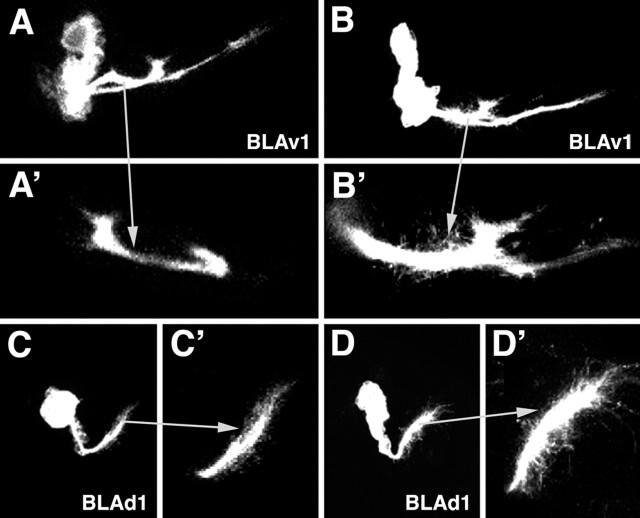
Comparison of clones generated by flip-out (A, C) and MARCM (B, D). The panels show Z-projections of two clones identified as corresponding to the BLAv1 lineage (A, B) and the BLAd1 lineage (C, D). Note that the tip of the SA, shown at higher magnification in adjacent panels (A′–D′), appear relatively thin and smooth in flip-out clones (A′, C′) and are thick and tufted in the corresponding MARCM clones (B′, D′).
Figure 3.
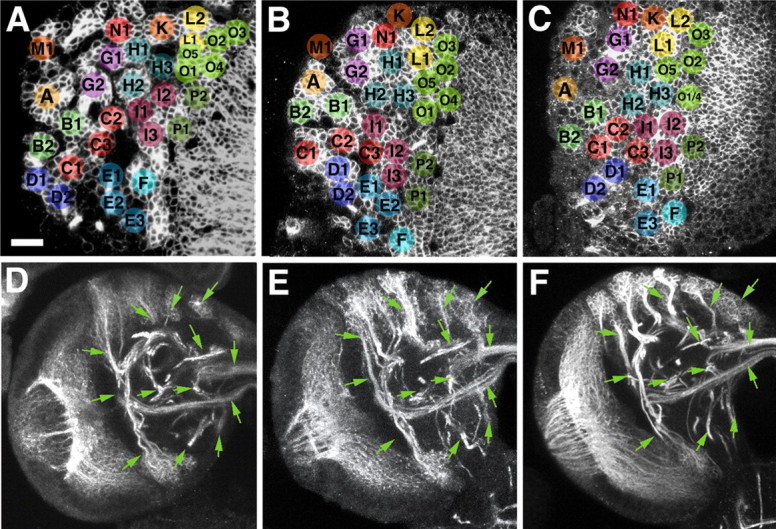
Variability of location and axonal trajectory of secondary brain lineages. A–C, Representative confocal cross sections of three different larval brains at level of anterior cortex. Secondary lineages, labeled with BP106 antibody, were identified on the basis of location and axonal trajectory (not visible on figure). Corresponding lineages are annotated with the same letter and color. With one exception (lack of “E2” in C), each one of the 32 lineage clusters visible at this level of the brain could be identified in all three specimens. Neighborhood relationships between lineages are also strongly conserved, although a degree of variability is evident. For example, lineage “K” can always found in between “N1,” “H1,” and “L2.” However, in A, “K” directly borders “H1,” whereas in B and C, “N1” or “L1,” respectively, are interposed between “K” and “H1.” D–F, Z-projections prepared from 10 successive confocal cross sections at level of central neuropile, prepared from three different specimens. Several axon tract systems (labeled with BP106 antibody) are shown. As shown in the previous three panels for the cell bodies of secondary lineages located in the cortex, the degree of similarity between axon trajectories is also considerable. Green arrows, placed at identical positions and angles, point at details of the tract systems that are evidently alike. Scale bar, 20 μm.
Secondary neurons differentiate during metamorphosis, producing terminal axonal and dendritic branches. During the larval stage, terminal branching appears to be absent from brain lineages (with the exception of the mushroom body lineages) (Lee et al., 1999). Larval SATs, visualized in clones generated by the flipping out of a stop-cassette on a UAS-srcGFP construct (Ito et al., 1997), typically terminate in a slightly enlarged, club-shaped tip (Fig. 4A–D). When inducing labeled wild-type clones using the MARCM technique (removing Gal-80 by somatic recombination) (Lee and Luo, 2001), we often observed that SAT endings appeared thicker and “fuzzier” (Fig. 4A′–D′). This fuzzy appearance is probably caused by the labeling of filopodia formed at the SAT growing tips. Tufts of filopodia could sometimes be also observed at points where the SAT made a turn. The GFP construct used in the MARCM experiments is UAS-cd8GFP, which, like srcGFP, targets GFP to the membrane. It appears that cd8GFP results in stronger membrane-associated GFP signal, thereby visualizing fine filopodial processes.
In the following, we will first describe the pattern of secondary tract systems in the late larval brain neuropile. We deem it worthwhile to reconstruct these tracts systems in great anatomical detail because they appear to form the primordial scaffold of fiber tracts of the adult brain. Thus, many of the SAT systems in the neuropile of late larvae are topologically very close to fiber systems of the adult brain, and for select cases, such as the antenno-cerebral tract (ACT), the continuity between larval and adult structure has been proven (Stocker, 1994) (see Discussion for more detail). Table 1, which lists all secondary lineages and the tract systems, indicates the adult fiber tracts (Strausfeld, 1976), which we tentatively assign to some of the larval secondary tract systems.
Table 1.
Secondary lineages of the larval brains and their axonal trajectories
| Lineage | Number of Branches | Tract vortex | Adult term | Tract transverse | Adult term | Tract longitudinal | Adult term | Tract vertical | Adult term |
|---|---|---|---|---|---|---|---|---|---|
| DAMv1 | 1 | DAC1 | anterodorsal commissure | ||||||
| DAMv2 | 1 | DAC1 | anterodorsal commissure | ||||||
| DAMd1 | 1 | DAC1 | anterodorsal commissure | ||||||
| DAMd2 | 1 | loDM | dorsal-horizontal tract | ||||||
| DAMd3 | 1 | loDM | dorsal-horizontal tract | ||||||
| DALv1 | 1 | crMBpvl | |||||||
| DALv2 | 2 | crMBmlv | VAC3/4 | inferior ventral body commissure | |||||
| crMBmlv | median fascicle | trCM/DPC1 | central complex primordium | ||||||
| DALv3 | 2 | crMBmlv | VAC3/4 | inferior ventral body commissure | |||||
| crMBmlv | median fascicle | trCM/DPC1 | central complex primordium | ||||||
| DALcl1 | 2 | crMBpp | VAC3/4 | inferior ventral body commissure | CAPT | subesophageal ventrolateral tract | |||
| DALcl2 | 2 | crMBpp | VAC3/4 | inferior ventral body commissure | CAPT | subesophageal ventrolateral tract | |||
| DALl1 | 1 | crMBpvl | |||||||
| DALcm1 | 2 | crMBpp | DAC2 | frontal medial commissure | CAPT | subesophageal ventrolateral tract | |||
| DALcm2 | 2 | crMBpp | DAC2 | frontal medial commissure | CAPT | subesophageal ventrolateral tract | |||
| DALd1 | 1 | crMBpp | CAPT | subesophageal ventrolateral tract | |||||
| BAmd1 | 2 | VAC2 | antennal commissure | ||||||
| DAC2 | frontal medial commissure | ||||||||
| BAmd2 | 1 | VAC2 | antennal commissure | ||||||
| BAmv1 | 1 | loBM | |||||||
| BAmv2 | 1 | loBM | |||||||
| BAmv3 | 1 | ACT | antenno-cerebral tract | ||||||
| BAmas1 | 1 | DAC1 | anterodorsal commissure | MeBd | median bundle | ||||
| BAmas2 | 1 | DAC1 | anterodorsal commissure | MeBd | median bundle | ||||
| BAla1 | 1 | crAN | root 1 ACT | ACT | antenno-cerebral tract | ||||
| BAla2 | 1 | crAN | root 1 ACT | ACT | antenno cerebral tract | ||||
| BAla3 | 1 | crAN | root 1 ACT | ACT | antenno-cerebral tract | ||||
| BAla4 | 1 | crAN | root 1 ACT | ACT | antenno-cerebral tract | ||||
| BAlc1 | 2 | crAN | root 2 ACT | ACT | antenno-cerebral tract | ||||
| BAlc2 | 2 | crAN | root 2 ACT | ACT | antenno-cerebral tract | ||||
| BAlp1 | 1 | balpm | |||||||
| BAlp2 | 2 | balpp | veBC | root 3 ACT | |||||
| BAlp3 | 2 | balpp | veBC | root 3 ACT | |||||
| BAlv | 1 | crAN | |||||||
| CM1 | 2 | loBM | ? | ||||||
| loC | posterior tract ventral body | ||||||||
| CM2 | 3 | VPC3 | ? | loBM | ? | ||||
| VAC3/4 | subellipsoid body commissure | loC | posterior tract ventral body | ||||||
| CM3 | 2 | . | VAC3/4 | subellipsoid body commissure | loC | posterior tract ventral body | |||
| trCM/DPC1 | central complex primordium | ||||||||
| CM4 | 1 | VAC3/4 | subellipsoid body commissure | loC | posterior tract ventral body | ||||
| CM5 | 1 | VAC3/4 | subellipsoid body commissure | loC | posterior tract ventral body | ||||
| TRdm | 1 | trdm | |||||||
| TRdl | 1 | trdl | |||||||
| TRvm | 1 | trvm | |||||||
| TRvl | 1 | trvl | |||||||
| Trco | 1 | trco | |||||||
| DPMpl1 | 1 | loDM | dorsal-horizontal tract | ||||||
| DPMpl2 | 1 | loDM | dorsal-horizontal tract | ||||||
| DPMpl3 | 1 | loDM | dorsal-horizontal tract | ||||||
| DPMpl4 | 1 | loDM | dorsal-horizontal tract | ||||||
| DPMpm1 | 2 | DPC2 | lateral horn commissure | loDM | dorsal-horizontal tract | ||||
| DPMpm2 | 2 | DPC2 | lateral horn commissure | loDM | dorsal-horizontal tract | ||||
| DPMm1 | 3 | DPC2 | lateral horn commissure | MCT | |||||
| DPC1/VPC3 | central complex primordium | ||||||||
| DPMm2 | 3 | DPC2 | lateral horn commissure | MCT | |||||
| DPC1/VPC3 | central complex primordium | ||||||||
| DPMm3 | 2 | DPC2 | lateral horn commissure | ||||||
| DPC1/VPC3 | central complex primordium | ||||||||
| DPMl1 | 1 | DPPT/veBL | |||||||
| DPMl2 | 1 | ||||||||
| DPMl3 | 1 | DPC2 | lateral horn commissure | ||||||
| DPMl4 | 1 | DPC2 | lateral horn commissure | ||||||
| DPLam | 2 | trCM | central complex primordium | ||||||
| >DPC1d | superior arch commissure | ||||||||
| DPLal1 | 3 | crMBl | DAC1 | anterior dorsal commissure | lateral dorsal horizontal tract | ||||
| trCM | central complex primordium | ||||||||
| DPLal2 | 3 | crMBl | DAC1 | anterior dorsal commissure | loDL | lateral dorsal horizontal tract | |||
| trCM | central complex primordium | ||||||||
| DPLal3 | 3 | crMBl | DAC1 | anterior dorsal commissure | loDL | lateral dorsal horizontal tract | |||
| trCM | central complex primordium | ||||||||
| DPLd | 1 | crMBdl | anterior collar | DAC1 | anterior dorsal commissure | ||||
| DPLc1 | 1 | crMBdlt | anterior collar | loDM | dorsal-horizontal tract | ||||
| DPLc2 | 1 | dplc | medial intracerebral cascade | loDM | dorsal-horizontal tract | ||||
| DPLc3 | 1 | dplc | medial intracerebral cascade | loDM | dorsal-horizontal tract | ||||
| DPLc4 | 1 | dplc | medial intracerebral cascade | loDM | dorsal-horizontal tract | ||||
| DPLc5 | 1 | dplc | medial intracerebral cascade | loDM | dorsal-horizontal tract | ||||
| DPLl1 | 1 | dpll | lateral intracerebral cascade | loDLv | lateral dorsal horizontal tract | ||||
| DPLl2 | 2 | dpll | lateral intracerebral cascade | loDLd/v | lateral dorsal horizontal tract | ||||
| DPLl3 | 2 | dpll | lateral intracerebral cascade | loDLd/v | lateral dorsal horizontal tract | ||||
| DPLp1 | 2 | trDL | loDL | lateral dorsal horizontal tract | |||||
| trDp > DPC2 | lateral horn commissure | ||||||||
| DPLp2 | 2 | trDL | loDL | lateral dorsal horizontal tract | |||||
| trDp > DPC2 | lateral horn commissure | ||||||||
| DPLm1 | 1 | trDL to lateral | |||||||
| DPLm2 | 1 | trDL to lateral | |||||||
| DPLcv | 2 | crMBpv | |||||||
| CP1 | 2 | cp1v | trDp | lateral horn commissure | loDM | dorsal-horizontal tract | |||
| CP2 | 2 | crMBpv | trDp | lateral horn commissure | loDM | dorsal-horizontal tract | |||
| CP3 | 2 | crMBpv | trDp | lateral horn commissure | loDM | dorsal-horizontal tract | |||
| CP4 | 1 | crMBpv | |||||||
| BLAv1 | 1 | trBL > VPC2 | optic foci commissure anterior | exBL | fascicle optic foci? | ||||
| BLAv2 | 1 | trBL > VPC2 | optic foci commissure anterior | exBL | fascicle optic foci? | ||||
| BLAd1 | 1 | trDL | exDL | ||||||
| BLAd2 | 1 | trDL | exDL | ||||||
| BLAd3 | 1 | trDL | exDL | ||||||
| BLAd4 | 1 | trDL | exDL | ||||||
| BLAl | 2 | crMBpvl | trDL | exBL > exDL | |||||
| BLAvm | 1 | crMBpvl | exBL | ||||||
| BLD1 | 2 | trDL | exDL | ||||||
| BLD2 | 1 | trDL | exDL | ||||||
| BLD3 | 1 | trDL | exDL | ||||||
| BLD4 | 2 | trDL | exDL | ||||||
| trBL | |||||||||
| BLD5 | 2 | trBL | optic foci commissure anterior | Lobula B2 | |||||
| BLP1 | 1 | exBL | fascicle optic foci? | ||||||
| BLP2 | 1 | exBL | fascicle optic foci? | ||||||
| BLP3 | 1 | trDL | exDL | ||||||
| BLP4 | 1 | trDL | exDL | ||||||
| BLP5 | 1 | exBL | fascicle optic foci? | ||||||
| BLVa1 | 1 | exVT | |||||||
| BLVa2 | 1 | exVT | |||||||
| BLVa3 | 1 | exVT | |||||||
| BLVp1 | 2 | trBL | optic foci commissure anterior | exBL | fascicle optic foci? | ||||
| LVp2 | 2 | trBL | optic foci commissure anterior | exBL | fascicle optic foci? | ||||
| BLVp5 | exBL | fascicle optic foci? |
The names of lineages are given in column 1; column 2 indicates the number of branches formed by SAT at point of entry into neuropile. All other columns specify the neuropile tract systems joined by the SATs. SAT trajectories are ordered by cardinal direction. If the SAT of a given lineage (see, for example, DALv2) enters one of the circumferential systems of the mushroom body or antennal lobe, this is listed in column 3/4. If the same tract has also an element (e.g., a branch or simply the continuation of the circumferential tract) that runs in a transverse direction, this is listed in column 5/6. Likewise, longitudinal and vertical elements of SAT trajectories are given in columns 7/8 and 9/10, respectively. Columns 3, 5, 7, and 9 give names introduced in this paper for larval tracts; the adjacent columns (4, 6, 8, 10) provide the names of the adult counterpart, where known or suspected. Lineages and lineage tracts: BAla1-4, Basoanterior lineages, anterior-lateral subgroup, 1-4; BAlc1-2, basoanterior lineages, caudo-lateral subgroup, 1-2; BAlp1-3, basoanterior lineages, posterolateral subgroup, 1-3; BAlv, basoanterior lineages, ventrolateral subgroup; BAmas1-2, basoanterior lineages, medial ascending subgroup, 1-2; BAmd1-2, basoanterior lineages, dorsomedial subgroup, 1-2; BAmv1-3, basoanterior lineages, ventromedial subgroup, 1-3; BLAd1-4, anterior basolateral lineages, dorsal subgroup, 1-4; BLAl1-2, anterior basolateral lineages, lateral subgroup, 1-2; BLAv1-2, ventral basolateral lineages, ventral subgroup, 1-2; BLAvm, anterior basolateral lineages, ventromedial; BLD1-5, dorsal basolateral lineages 1-5; BLP1-5, posterior basolateral lineages 1-5; BLVa1-3, ventral basolateral lineages, anterior subgroup, 1-3; BLVp1-3, ventral basolateral lineages, posterior subgroup, 1-3; CM1-5, centro-medial lineages 1-5; CP1-4, centro-posterior lineages 1-4; DALcl1-2, lateral dorsoanterior lineages, centro-lateral subgroup, 1-2; DALcm1-2, lateral dorsoanterior lineages, centro-medial subgroup, 1-2; DALd, lateral dorsoanterior lineages, dorsal subgroup; DALl, lateral dorsoanterior lineages, lateral subgroup; DALv1-3, lateral dorsoanterior lineages, ventral subgroup, 1-3; DAMd1-3, medial dorsoanterior lineages, dorsal subgroup, 1-3; DAMv1-2, medial dorsoanterior lineages, ventral subgroup, 1-2; DPLal1-3, lateral dorsoposterior lineages, antero-lateral subgroup, 1-3; DPLam, lateral dorsoposterior lineages, antero-medial subgroup; DPLc1-5, lateral dorsoposterior lineages, central subgroup, 1-5; DPLd, lateral dorsoposterior lineages, dorsal subgroup; DPLl1-3, lateral dorsoposterior lineages, lateral subgroup, 1-3; DPLm1-2, lateral dorsoposterior lineages, medial subgroup, 1-2; DPLp1-2, lateral dorsoposterior lineages, posterior subgroup, 1-3; DPMl1-4, medial dorsoposterior lineages, lateral subgroup, 1-4; DPMm1-3, medial dorsoposterior lineages, medial subgroup, 1-3; DPMpl1-4, medial dorsoposterior lineages, posterolateral subgroup, 1-4; DPMpm 1-2, medial dorsoposterior lineages, posteromedial subgroup, 1-2; MB 1-4, mushroom body lineages 1-4; TRco, commissural tritocerebral lineage; TRdl, dorsolateral tritocerebral lineage; TRdm, dorsomedial tritocerebral lineage; TRvl, ventrolateral tritocerebral lineage; TRvm, ventromedial tritocerebral lineage. Axon tract systems: ABT, Antero-basal tracts (crAN, veBC, et al.); ACO, antennal commissure (VAC2); ACT, antenno-cerebral tract; bm, basomedial component of antenno-cerebral tract; ATT, anterior transverse tracts (crMBdl, trDL, et al.); CAPT, centro-anterior protocerebral tract; crAN, circumferential tracts of antennal lobe; crMB, circumferential tracts of the mushroom body; dl, around dorsal lobe; l, lateral; mlv, ventral of medial lobe; pp, around proximal peduncle; pv, ventral of peduncle; pvl, ventrolateral of peduncle; DAC, 1-4, dorsoanterior commissures 1-4; DACd, DAC3/4; DACr, DAC1; DPC 1-3, dorsoposterior commissures 1-3; DPPT, veBC; exBL, external basolateral tract system; exDL, external dorsolateral tract system; exVT, external vertical tract system; LCOTd, lateral commissural optic tract (trPd); LCT, lateral cervical tract (continuation of CAPT); LCTco, commissural component of LCT (trBL); loBM, basomedial longitudinal tract system; loC, central longitudinal tract system (ACT, et al.); loDL, longitudinal dorsolateral tract system; loDM, dorsomedial longitudinal tract system; d, dorsal component of dorsomedial longitudinal tract system; v, ventral component of dorsomedial longitudinal tract system; meB, median bundle; MCT, medial cervical tract (meB); MOT, medial optic tract (exBL, crMBpp, et al.); trBL, transverse basolateral tract system; trCM, transverse centro-medial tract system; trDL, transverse dorsolateral tract system; trPd, transverso posterodorsal tract system; VAC 1-4, ventroanterior commissures 1-4; veBC, vertical basocentral tract system; VPC 1-3, ventroposterior commissures 1-3. Neuropile compartments: BA, Basal anterior (antennal) compartment; BC, basal central compartment; BCv, basal cervical compartment; BPL, basal posterolateral compartment; BPM, basal posteromedial compartment; CA, central anterior compartment; CPI, central postero-intermediate compartment; CPL, central posterolateral compartment; d, dorsal component of CPL; CPM, central posteromedial compartment; CX, calyx of mushroom body; DA, dorsal anterior compartment; dl, dorsal lobe of mushroom body; DP, dorsal posterior compartment; LON, larval optic neuropile; ml, medial lobe of mushroom body; OL, optic lobe; ped, peduncle of mushroom body; SEG, subesophageal ganglion; sp, spur of mushroom body; TR, tritocerebrum.
Secondary tract systems of the late larval brain
Tract systems at the cortex–neuropile interface
Tract systems that follow the neuropile surface are the external dorsolateral (exDL), external basolateral (exBL), and external vertical-lateral (exVT) (Figs. 1C,D, 2A,C,D,F–H). The external lateral systems are formed by longitudinally oriented tracts of the BL lineages. Basolateral anterior (BLA) tracts travel posteriorly, basolateral posterior (BLP), and basolateral dorsal (BLD) tracts anteriorly. Where they converge, axon bundles turn medially to join the massive axon bundles that originate in the lobula primordium and form the basolateral transverse system (trBL) (Fig. 2A,D,G,H). The exVT system is formed by tracts of the ventrally located basolateral ventral (BLV) group of lineages. Reaching upward toward the level of the central posterolateral compartment, dorsal component (CPLd), the exVT makes a sharp medial turn and becomes the transverse dorsolateral (trDL) system (Fig. 2A,C,G).
Tract systems of the dorsomedial neuropile
The longitudinal dorsomedial system (loDM) (Figs. 1B–E, 2B–H) extends through the center of the dorsal compartments, dorsoanterior compartment (DA), and dorsoposterior compartment (DP). From anteriorly, axons of several dorsoanterior medial group (DAM) lineages enter the loDM: medial dorsoposterior (DPM), centromedial (CM), and centroposterior (CP) tracts originating at the posterior cortex travel in the opposite direction. Posteriorly, two main components of the loDM are recognized, a thick dorsal and a less voluminous ventral component. The ventral component (loDMv) bends medially and forms the dorsal posterior commissural system (DPC) (Fig. 2B,D). The dorsal component (loDMd) continues anteriorly and enters the DA, where it is joined by DAM lineage tracts originating in the anterior brain.
The longitudinal central system (loC) is located medially and ventrally of the loDM (Figs. 1C–E, 2B,D,G,H). From anteriorly, tracts formed by a subset of basoanterior (BA) lineages, as well as tracts ascending from the ventral nerve cord, join to form the anterior section of the loC. These axon bundles follow the course of the ACT (Fig. 1C), which projects posteriorly and dorsally toward the calyx of the mushroom body. Axons entering the loC from posteriorly are the tracts of a subset of the CM and DPM lineages. This posterior part of the loC splits into dorsal and ventral component. The dorsal component curves medially and projects into the DPC1 commissural system (Figs. 1C, 2B,D,G). The ventral part of the loC continues forward before it also turns medially to form a commissural bundle underneath the medial lobes of the mushroom body [ventroanterior commissural system 3/4(VAC3/4)] (Figs. 1B,C, 2B,C).
A transverse system (trCM) is formed by axon tracts of lateral dorsoanterior (DAL), lateral dorsoposterior (DPL), DPM, and CM lineages, which all converge medially behind the medial lobe of the mushroom body. The trCM continues medially as the DPC1 commissure (Figs. 1C, 2C,G). TrCM and DPC1 constitute the larval primordium of the central complex.
Tract systems focused on the mushroom body
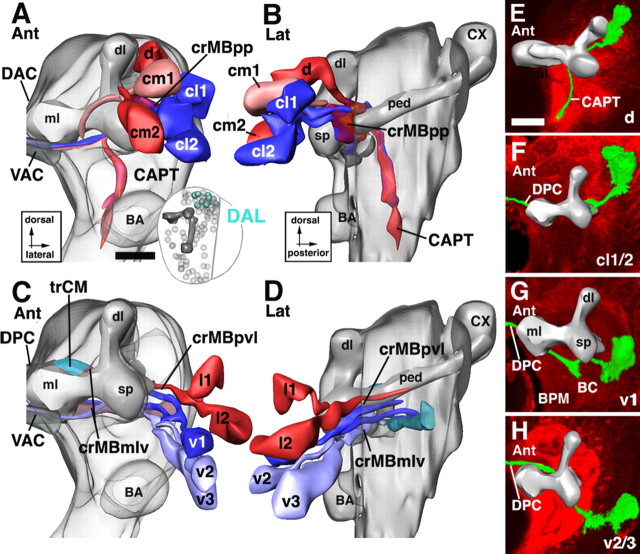
The circumferential system of the mushroom body (crMB) represents a complex system of tracts that converge on the mushroom body from various directions and form “swirls” of fibers around the peduncle, spur, and lobes. The crMB is formed mainly by axon bundles of DAL lineages that reach the neuropile from anteriorly and dorsally. Most of these fibers pass through the crMB and continue medially toward the midline and contribute to the primordium of the central complex (trCM; see above). Certain DAL and the DPLd lineages form a cuff of fibers around the dorsal lobe (crMBdl) (Figs. 1B, 2A,C,E). DALcl and DALd lineage tracts swirl around the proximal peduncle (crMBpp) (Figs. 1B,C, 2A,C,E). DALv tracts form a thick bundle (crMBmlv) (Figs. 1C, 2C) that approaches the crMBpp from anteriorly, weaves through it, and continues posteromedially toward the primordium of the central complex (trCM and DPC1). Tracts of DALl and BLAvm form a bundle coming from lateral and curving underneath the proximal peduncle (crMBpvl) (Figs. 1C, 2F) to merge with the crMBpp.
Aside from the above-mentioned anterior axons tracts, other lineages located more posteriorly add to the crMB. A subset of CP tracts traveling forward underneath the peduncle form a posteroventral branch of the crMB system (crMBpv) (Figs. 1D,E, 2A,D,G,H). Several bundles of forward directed DPL axon tracts (DPLal) converge on the peduncle from dorsolaterally (crMBl) (Figs. 1C, 2A,C,D).
Tract systems of the dorsolateral neuropile
The loDL extends within the neuropile along the compartment boundary of CPLd and DP (Figs. 1C,D, 2A,C–G). Anteriorly, the loDL communicates with the crMB system; posteriorly, it merges with the trDL (see below). Most tracts making up the loDL originate from DPL lineages that enter the neuropile from dorsally and posteriorly and travel forward. From anteriorly, some DAL bundles pass through the crMB system and reach into the loDL system where they continue backward.
The anterior trDL occupies the center of the CPLd compartment (Figs. 1D, 2A,C,F,G). This fiber system effectively separates a narrow external layer of CPLd neuropile from a thicker internal layer. The core of the trDL system is formed by the exVT (Fig. 1C) (see above), a highly conspicuous axon tract emanating from several contiguous ventrally located BLV lineages. This tract follows a straight vertical course at the outer surface of the basal posterolateral (BPL) and CPL compartments. The BLV tract then turns medially and penetrates the dorsal CPL as the trDL. Additional axons joining the trDL branch off the BLD, BLA, and BLP tracts (forming the dorsolateral external system; see above), as well as the DPL tracts that form the loDL.
A posterior transverse system (trPd) is formed by tracts of the CP lineages, located at the posterior aspect of the brain. Additional components are branches of posterior DPL lineages. The trPd passes dorsal of the peduncle, right in front of the calyx (Figs. 1D,E, 2A,D,H). It continues medially and merges with the loDM system.
Tract systems of the basal neuropile
The basal neuropile contains one medial longitudinal system (loBM), which passes from all the way anterior to posterior, and a lateral system [circumferential antennal system (crAN)], which is restricted to the anterior part of the neuropile. In addition, vertical systems organized in three planes (medial, intermediate, lateral) penetrate the basal neuropile.
The crAN is formed predominantly by tracts of the BA (antennal lobe) group. These tracts approach the BA neuropile from anterolateral and posterolateral directions. Posterior of the BA these axon tracts converge and form a crescent-shaped bundle that extends along the boundary between BA and basocentral (BC) compartment (Figs. 1C, 2A,C,F). Axons spinning out of the crAN continue dorsomedially and dorsoposteriorly to join the antennal commissure (VAC2; see below) and antenno-cerebral tract. In addition to these major fiber systems, two thin bundles exit the crAN and form their own tracts: the basomedial antenno-cerebral tract (bmACT; not shown) and the lateral accessory antenno-cerebral tract (laACT; not shown).
The trBL represents the forerunner of the commissural output tract of the optic lobe and the optic foci (Figs. 1D,E, 2A,D,G,H) of the adult brain. Axons emanating from the lobula primordium of the optic lobe form the most abundant source of the trBL. In addition, axon tracts of the BL lineages converge on the trBL. The trBL grows toward and crosses the midline in the ventroposterior commissure (VPC2; named the commissure of the optic foci in the adult) (Strausfeld, 1976).
The loBM extends medially of the crAN system, forming the boundary between basal posteromedial (BPM) and BCv compartments (Figs. 1B,C,E, 2B–H). The loBM system includes SATs from medial members of the BA group of lineages (BAm) that enter the neuropile from anteriorly; from posteriorly, a subset of CM (CMv) tracts grow into the loBM system.
Three vertical basal systems can be distinguished. The central basal vertical system (veBC) (Figs. 1C, 2C,D,F) forms a massive plate-like assembly of fibers in between the BPL and BPM compartments. It is formed by tracts of the BAlp lineages. Continuing dorsally, the system splits into an anterior and posterior branch. The anterior branch extends toward the crMB system; the posterior branch follows a more medial course and joins the ACT. The second vertical system is the central anterior protocerebral tract (CAPT), which follows the trajectory laid down by primary axons already in the late embryo (Nassif et al., 2003) (Figs. 1C, 2B–D,F). Axon tracts of several DAL lineages (DALcm, DALcl, DALd) projecting from anterolaterally over the peduncle and then turning ventral form the CAPT. Finally, a system of vertical fibers formed by several DPM lineages, as well as ascending tracts of unidentified lineages located in the subesophageal ganglion, form a medial vertical system that follows the trajectory of the medial cervical tract (MCT)/median bundle described for the embryo (Nassif et al., 2003) (Figs. 1C, 2B,C,F).
Commissural systems
Four major subdivisions of the supraesophageal commissure, which interconnects the two brain hemispheres, can be recognized from embryonic stages onward. These subdivisions are defined with respect to the medial lobe of the mushroom body, which forms the central core of the commissural systems, although its fibers stop short before reaching the midline (Fig. 2B–F). These are the VAC, dorsal anterior (DAC), VPC, and DPC commissural tracts. Secondary lineage tracts identify smaller tracts within these subdivisions, and in many instances (indicated in Table 1), these tracts prefigure commissures defined for the adult fly brain.
The VAC commissural tracts are located ventrally in front of the medial lobe. These tracts stem from lineages located in the deuterocerebrum and anterior protocerebrum. VAC1 is the crossed median bundle (Fig. 1B), strictly speaking not a commissure but a chiasm (the crossing axons connecting noncorresponding points of the two sides of the brain). VAC1 has numerous ascending BP106-positive axons from unidentified lineages in the subesophageal ganglion (the anterior part of the ventral nerve cord), as well as descending branches from one or two DPM lineages. VAC2 corresponds to the antennal commissure, located ventrally behind VAC1 (Fig. 1B). The antennal commissure carries crossing axons of BAmd lineages. VAC3/4 are closely adjacent bundles formed by crossing branches of several DALv and DALcl lineages, as well as posteriorly located CM lineages (Fig. 1C). Commissural tracts with these trajectories and location were identified in the adult fly brain as inter-ventral body connective and subellipsoid commissure, respectively (Strausfeld, 1976).
The DAC subdivision of the commissure lies anterior and dorsal of the medial lobe and contains crossing fibers from all over the protocerebrum and deuterocerebrum. DAC1 (tentatively assigned to the anterodorsal commissure of the adult brain) carries axons from the DAMv, BAmas, and DPLd lineages (Fig. 1B). DAC2 (frontal medial commissure of adult), closely behind the VAC1 chiasm, is formed by DALcm tracts (Fig. 1B). DAC3 (forerunner of the arched commissure of the ventral body) carries DALv tracts; DAC4 (anterior cross) is formed by medial protocerebral lineages of the DPLc group.
BP106-positive secondary axon tracts located posterior of the medial lobe of the mushroom body prefigure “true” posterior commissures of the adult brain, but in part also constitute the primordium of the central complex. The central complex is a conspicuous median compartment of the adult insect brain that differentiates during metamorphosis (Hanesch et al., 1989; Renn et al., 1999). The major components, ellipsoid body, fan shaped body, noduli, and protocerebral bridge can be clearly recognized from the first day of pupariation onwards (Schneider et al., 1993; Renn et al., 1999) (Pereanu and Hartenstein, unpublished observations) in BP106-labeled preparations. In the late larval brain, the distinction of individual components of the central complex is difficult. However, it is clear that a large BP106-positive domain (DPC1 and trCM) located right behind the medial lobe will further grow and become the central complex of the pupal brain (Figs. 1C, 2B,C,G). DPC1 is formed by tracts of DALv and DPMpl lineages. A distinct commissure right on top of DPC1 is DPC1d, the forerunner of the superior arch commissure. It is formed by fibers that spin out of the crMB system and can be traced back to branches of DPLal lineages (see below). DPC2 is a conspicuous H-shaped commissure posterior to DPC1 (Fig. 1D, 2B,D). It carries tracts of the dorsoposterior protocerebrum (DPMm, DPMpm, possibly CP and DPL lineages). It is likely that part of DPC2 will also become incorporated into the central complex; in addition, it will give rise to the commissure of the lateral horns of the adult brain. DPC3, at the posterior tip of the supraesophageal commissure, is formed by branches of DPM and CM lineages.
The most conspicuous element of the ventral posterior commissural system is VPC2, a thick, straight commissure formed by tracts from the lateral protocerebrum. This commissure represents the continuation of the trBL tract that carries axons from many BL lineages, as well as the developing lobula (Fig. 2D,F). This commissure will become the “great commissure” or “commissure of the optic foci” of the adult brain (Strausfeld, 1976). Two discrete, thinner BP106-positive bundles flank the VPC2 anteriorly (VPC1) and posteriorly (VPC3). The latter is formed by a subset of CM lineages (Fig. 1D); the origin of the former has not been determined.
The groups of secondary lineages and their projection
In the following, we will describe eleven topologically defined groups of lineages. Each group is represented by its own figure (see Figs. 5–15), which includes digital models of the lineages of the group in relationship to landmarks, as well as confocal images of representative lineages. In this work, we do not describe in detail the four lineages forming the mushroom body, because they have been covered in several previous publications (Lee et al., 1999; Zhu et al., 2003; Watts et al., 2004).
Figure 5.
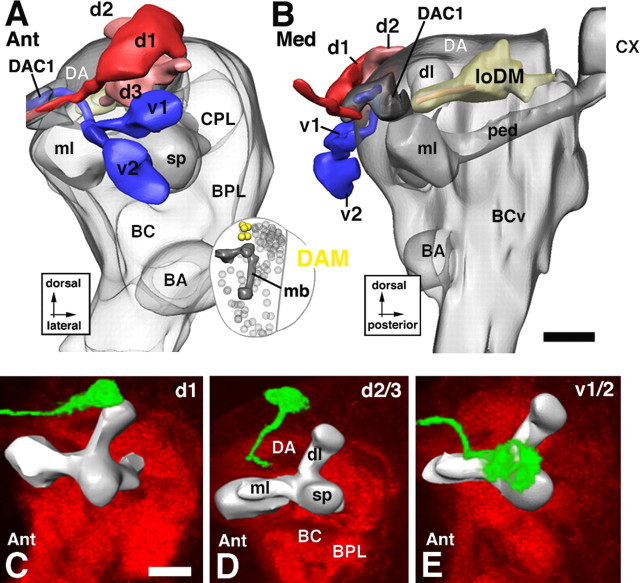
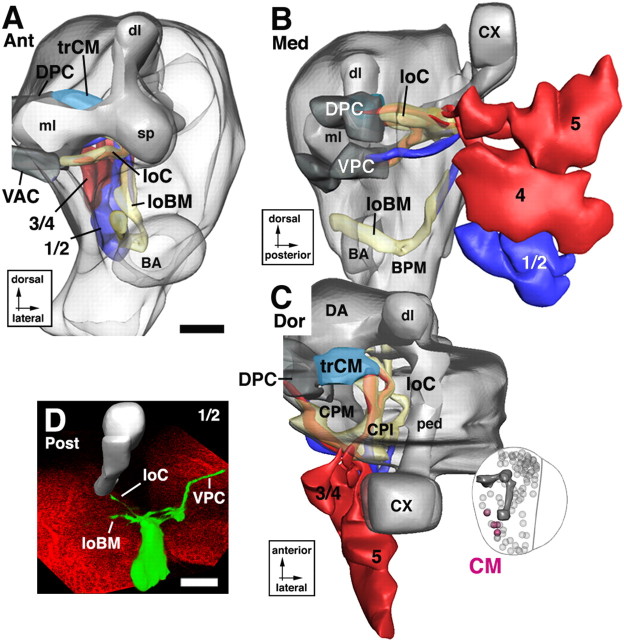
Lineages of the DAM group. In this and subsequent figures (Figs. 6–15), a group of lineages is illustrated in the form of a digital 3D model, supplemented by volume renderings of representative GFP-labeled lineages. The top two panels represent an anterior view (A) and medial view (B) of the larval brain neuropile (gray shading) on which the lineages of the DAM group, shown in different colors, are superimposed. The orientation at which the model is shown is indicated in this and the following figures by identifiers underneath the panel letter (Ant, anterior; Post, posterior; Lat, lateral; Med, medial; Dor, dorsal). On the model, lineages are identified by their suffix (see Table 1); “d1-d3” and “v1-v2” stand for “DAMd1-DAMd3” and “DAMv1-DAMv2,” respectively. Additional details indicated in the models are the lobes of the mushroom body (dl, dorsal lobe; ml, medial lobe; sp, spur; ped, peduncle; CX, calyx), as well as the location of neuropile compartments. Furthermore, axon tract systems that are targeted by the DAM lineages (loDM longitudinal dorsomedial system; B) are shown. The inset at bottom right of A shows “thumbnail” view of a brain hemisphere (dorsal view), which helps the viewer to locate the DAM lineages (yellow) in relationship to all other lineages (gray spheres) and the mushroom body (mb). The bottom panels, C–E, are volume renderings of confocal stacks of larval brains in which individual DAM lineages (identified by letters at top right of panels; indicating 2 lineages, such as d2/3, indicates that both have the same trajectory and cannot be told apart) are labeled by GFP through hs-Flp-activated somatic recombination. Lineages appear in green; brains were double labeled with anti-Syntaxin (red) to visualize neuropile. For better orientation, a digital model of the mushroom body (gray shading) was warped into the confocal stack containing the labeled lineage. Scale bars, 20 μm.
Figure 6.
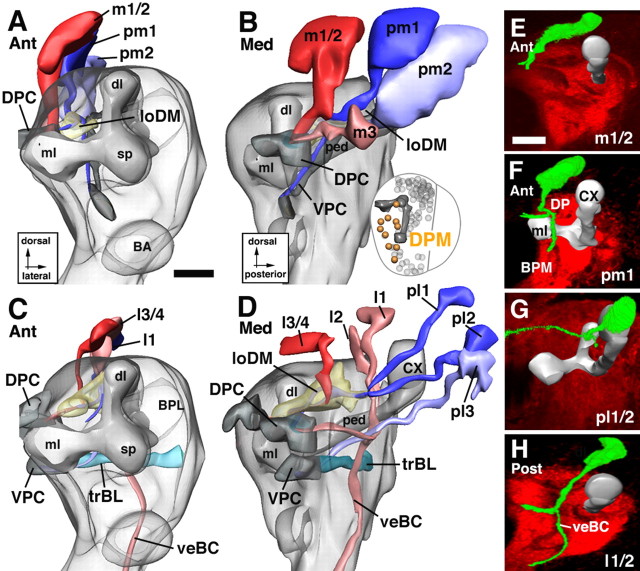
Lineages of the DAL group. The top panels (A, B) illustrate the dorsal subgroup of DAL lineages (DALd, DALcm, DALcl); the bottom panels (C, D) show ventral subgroup (DALl, DALv). The right panels (E–H) show representative members of the DAL family of lineages. For additional details explaining the figure, see the legend for Figure 5. Scale bars, 20 μm.
Figure 7.
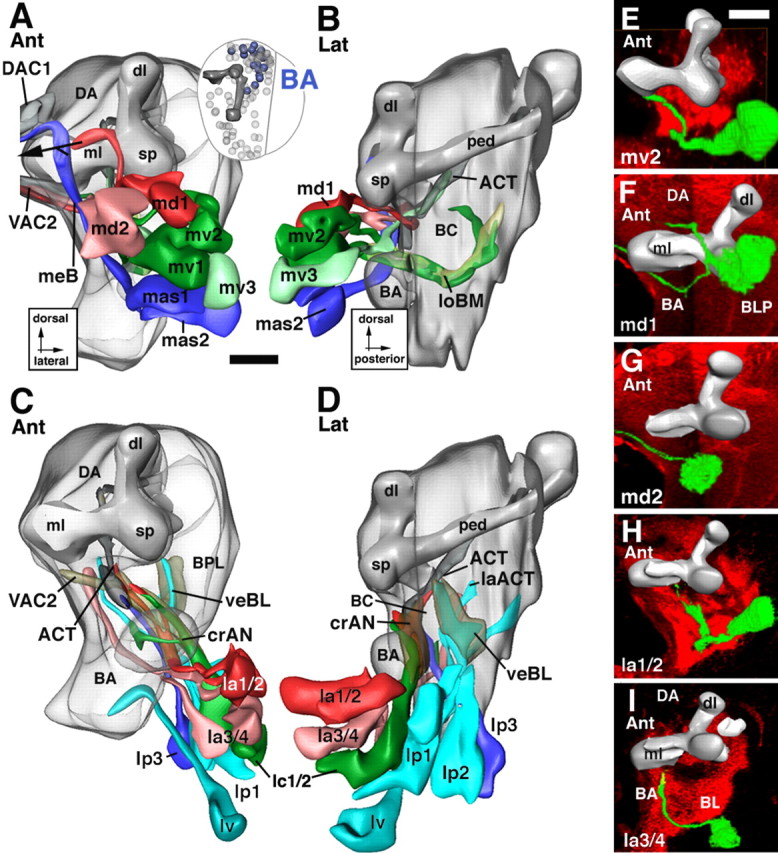
Lineages of the BA group. The top panels (A, B) illustrate the dorsal subgroup of BA lineages (BAmd, BAmv, BAmas); the bottom panels (C, D) show ventral subgroup (BAla, BAlc, BAlp, BAlv). The right panels (E–I) show representative members of the BA group. For additional details explaining this figure, see the legend for Figure 5. Scale bars, 20 μm.
Figure 8.
Lineages of the CM group. A–C illustrate three views of a digital 3D neuropile model highlighting the position and trajectory of CM lineages; D shows a representative member of this group. For additional details explaining this figure, see the legend for Figure 5. Scale bars, 20 μm.
Figure 9.
Lineages of the DPM group. The top panels (A, B) illustrate the medial subgroup of DPM lineages (DPMm, DPMpm); the bottom panels (C, D) show lateral members (DPMl, DPMpl). The right panels (E–H) show representative members of the DPM group. For additional details explaining this figure, see the legend for Figure 5. Scale bars, 20 μm.
Figure 10.
Lineages of the DPL group. The top panels (A, B) illustrate the anterior set of DPL lineages (DPLam, DPLal, DPLd, DPLc); the bottom panels (C, D) show posterior members (DPLl, DPLm, DPLp). The right panels (E–H) show representative members of the DPL group. For additional details explaining this figure, see the legend for Figure 5. Scale bars, 20 μm.
Figure 11.
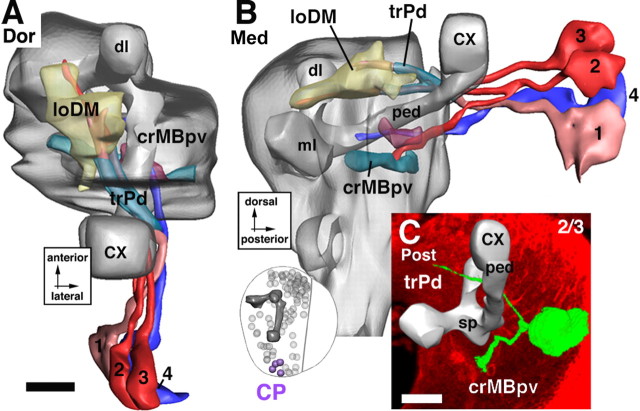
Lineages of the CP group. A and B illustrate a dorsal and medial view of a digital 3D neuropile model highlighting the position and trajectory of CP lineages; C shows a representative member of this group. For additional details explaining this figure, see the legend for Figure 5. Scale bars, 20 μm.
Figure 12.
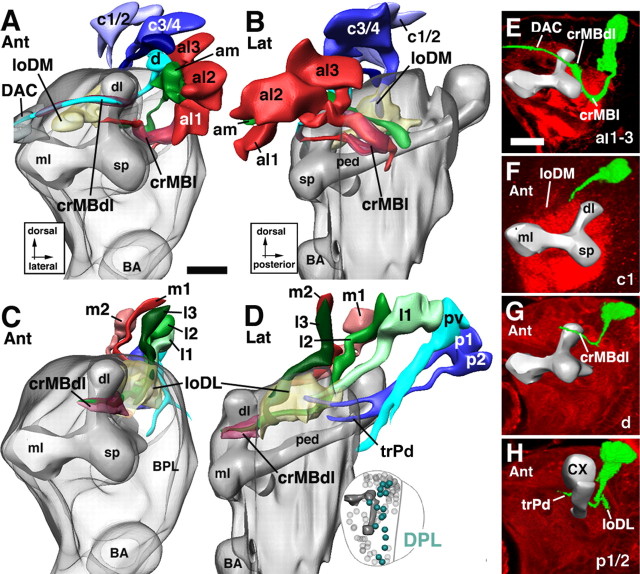
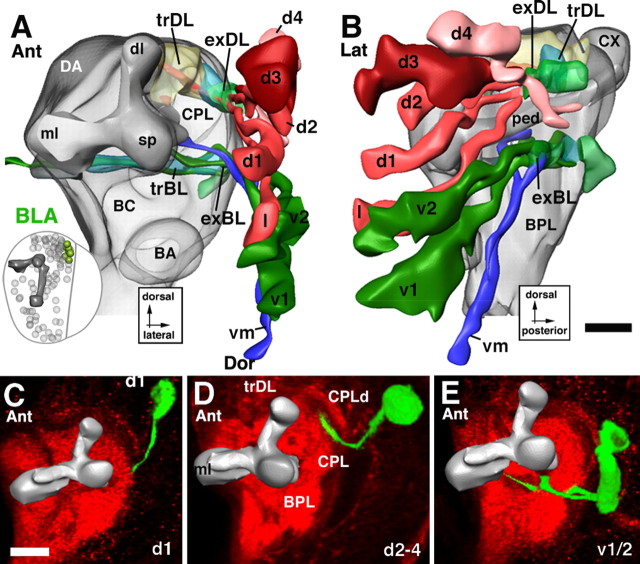
Lineages of the BLA group. A and B illustrate an anterior and lateral view of a digital 3D neuropile model highlighting the position and trajectory of BLA lineages; C–E show representative members of this group. For additional details explaining this figure, see the legend for Figure 5. Scale bars, 20 μm.
Figure 13.
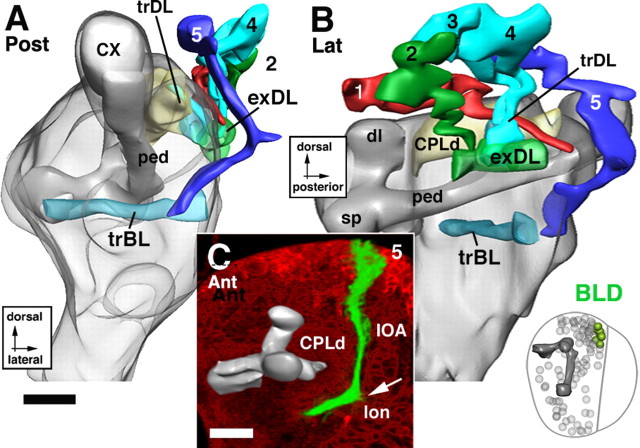
Lineages of the BLD group. A and B illustrate a posterior and lateral view of a digital 3D neuropile model highlighting the position and trajectory of BLD lineages; C shows a representative member of this group. For additional details explaining this figure, see the legend for Figure 5. Scale bars, 20 μm.
Figure 14.
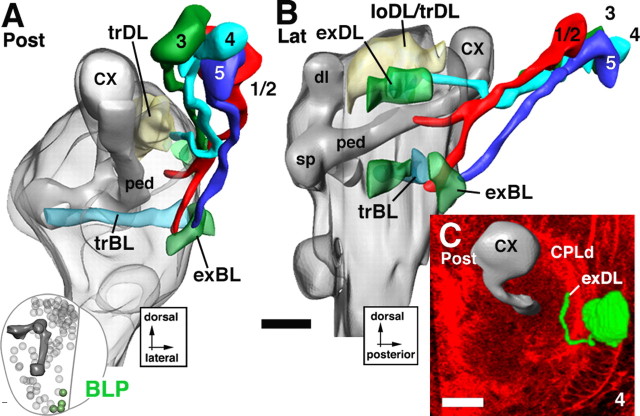
Lineages of the BLP group. A and B illustrate a posterior and lateral view of a digital 3D neuropile model highlighting the position and trajectory of BLP lineages; C–E show representative members of this group. For additional details explaining this figure, see the legend for Figure 5. Scale bars, 20 μm.
Figure 15.
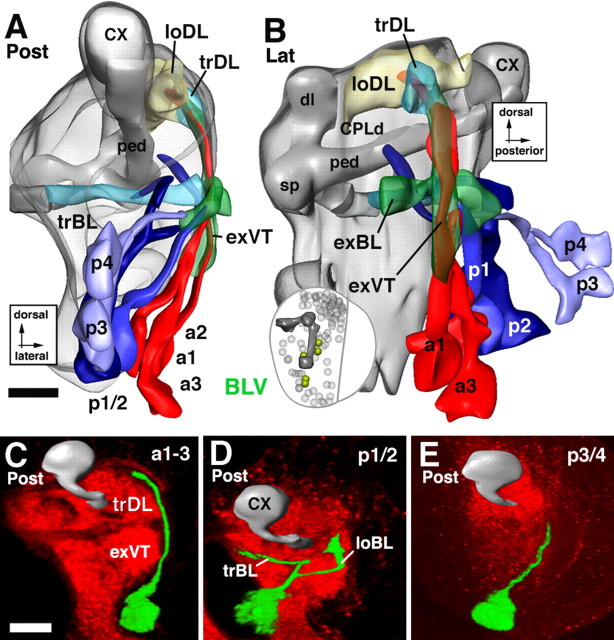
Lineages of the BLV group. A and B illustrate a posterior and lateral view of a digital 3D neuropile model highlighting the position and trajectory of BLV lineages; C–E show representative members of this group. For additional details explaining this figure, see the legend for Figure 5. Scale bars, 20 μm.
Dorsoanterior medial group
DAM lineages are located anterior and medial of the dorsal lobe of the mushroom body and send their axon tracts toward the DA neuropile compartment (Fig. 1G–I). Two subgroups, a ventral DAMv and a dorsal DAMd, with two to three lineages each, are distinguished (Fig. 5). DAMv1–2 flank the spur anteriorly (Figs. 1A, 5A). Axon tracts extend posteromedially and cross in the posterior part of the dorsal anterior commissure DAC (Figs. 1B, 5A,E). DAMd1–3 are located dorsal of the mushroom body spur (Figs. 1A, 5A). DAMd1 represents a conspicuous anterodorsal landmark. Its tract grows straight medially and crosses in the DAC (Fig. 5A,C). Some labeled clones of DAMd1 reveal a conspicuous “hairpin” loop: reaching the contralateral hemisphere, the DAMd1 tract makes a 180° turn and crosses back toward ipsilaterally at a more ventral level (data not shown). DAMd2/3, located ventrally of DAMd1, project their tracts straight posteriorly into the loDM (Fig. 5A,B,D).
Dorsoanterior lateral group
DAL lineages flank the spur and dorsal lobe of the MB anteriorly and laterally (Figs. 1G–I, 6). Their axon tracts are directed posteromedially, toward the spur of the mushroom body, the BC compartment (ventral of the spur), and the CPL compartment (dorsoposterior of the spur). Passing through the crMB system, many DAL tracts continue toward the midline where they contribute to the primordium of the central complex. Five DAL subgroups are distinguished: DALv, DALcm, DALl, DALcl, and DALd.
The DALv lineages (DALv1–3) are the most ventral components of the DAL group (Figs. 1A,B, 6C). Projecting straight posteriorly, tracts of DALv1–3 reach the crMB system ventral of the spur (Fig. 6D). The DALv1 tract continues posteriorly as part of the crMBpvl system and ends in the CPL compartment (Figs. 1C, 6D,G). DALv2/3 are located ventral of DALv1. Their tracts pass underneath the medial lobe in the crMBmlv system (Figs. 1C, 6C,D,H) and then turn medially. The tracts split into a dorsal and a ventral branch. The dorsal branch (dalvd) continues medially through the centromedial transverse system (trCM) toward the DAC3 and DPC1 commissures (Figs. 1C, 6C). The ventral branch crosses the midline at a more ventral level in the VAC3/4, located in between the medial lobe of the MB and the antennal commissure (Fig. 6C) (see below). DALv3 is the dorsal-most one of the deuterocerebral lineages that expresses engrailed, thereby demarcating a central location within the deuterocerebral realm of neurons and their axons.
The tracts of DALcl1/2 tracts project onto the spur where they bifurcate into a dorsal and a ventral branch. This bifurcation forms part of the ring-shaped crMBpp system that surrounds the proximal peduncle (Figs. 1B,C, 2A, 6B). The dorsal branch passes over the peduncle and then turns ventrally as part of the CAPT that projects to the ventral nerve cord (Fig. 6A,B). The ventral branch (Fig. 6A,B,F) passes underneath the peduncle and projects medially, joining the DALv2/3 tracts in forming the inferior commissure of the ventral body (VAC3/4).
DALl1 is located adjacent to DALcl and grows straight posteriorly, approaching the peduncle from laterally as part of the crMBpvl system (Fig. 6D).
The two DALcm lineages are medially adjacent to DALcl (Figs. 1A, 6A). Their tracts bifurcate in the horizontal plane, one branch extending medially, the other laterally (Fig. 1B). The medial branch grows toward the midline, closely attached to the anterior surface of the MB medial lobe (Fig. 6A). It forms part of the VAC. The lateral branch joins tracts of DALcl (see above) into the crMBpp ring, which encircles the base of the peduncle of the mushroom body. Leaving the ring, axons continue downward through the CAPT tract (Fig. 6A,B)
The DALd subgroup is located dorsal of DALcl. It comprises one or two lineages that project toward the crMBpp system (Fig. 6A,B,E). Joining branches of DALcm tracts they project into the CAPT toward the anterior ventral nerve cord that will become the subesophageal ganglion.
Basoanterior group
These lineages surround the BA (antennal) compartment (Figs. 1A,B, 7). Their tracts being directed toward the BA and BPM compartments, the BA lineages form the ventral deuterocerebrum. Five subgroups are distinguished: BAmd, BAmv, and BAmas, all located medially; BAla, BAlc, BAlp, and BAlv, located more laterally and posteriorly.
The BAmd subgroup comprises two lineages located ventrally of DAMv, in front of the central anterior (CA) compartment and medial lobe (Figs. 1A, 7A). The BAmd1 tract grows straight posteriorly and meets the CA compartment. The tract bifurcates in the vertical plane. The dorsal branch passes over the medial lobe and, after meeting the medial branch formed by the DALcm lineages (see above), curves medially to approach the midline in close association with the DAC2 commissure (Figs. 1B, 7A,F). The ventral branch of BAmd1 passes underneath the medial lobe before turning medially. It crosses the midline, forming the primordium of the antennal commissure (VAC2) (Fig. 7A,F). BAmd2 lies at the medial edge of the ventral deuterocerebrum (Figs. 1A, 7A). The BAmd2 tract travels straight posteriorly and then medially, joins the ventral BAmd1 branch, and crosses the midline in the antennal commissure (VAC2) (Fig. 7A,G).
Three BAmv lineages are located ventrally of BAmd (Figs. 1A, 8A). The tracts of BAmv1/2 curve posteriorly and ventrally, forming the loBM system (Fig. 7B,E), reaching as far backward as the commissural optic fibers of the trBL system. BAmv3 is located laterally of BAmv1/2. Its tract extends posteriorly laterally adjacent to the loBM system. After passing underneath the medial lobe, the BAmv3 tract turns dorsally and joins the ACT (Fig. 7B). From its position and axonal projection, we conclude that BAmv3 represents one of the three lineages that produce the antennal projection neurons, with dendrites in the antennal glomeruli and axons directed toward the calyx and lateral horn (Homberg et al., 1989; Malun et al., 1993; Marin et al., 2002).
Two BAmas lineages, localized ventrally of BAmv, form a highly conspicuous tract that projects upward along the medial edge of the brain along the median bundle (Figs. 1A,B, 7A,B). The tract crosses the midline as the most anterior and dorsal component of the DAC1 commissure.
The BAla subgroup is formed by two pairs of closely associated lineages located laterally and ventrally of BAmas, right in front of the BA (antennal) compartment (Figs. 1A, 7C). BAla1/2 form a tract that grows along the ventrolateral surface of the BA compartment before turning dorsally and medially to project into the crAN system (Figs. 1B,C, 7C,D,H). A smaller branch of the BAla1/2 tract appears to join the ACT. Based on this trajectory, BAla1/2 corresponds to the lineage that produces the lateral cluster of antennal projection neurons (Homberg et al., 1989; Malun et al., 1993; Marin et al., 2002). The BAla3/4 lineage contains engrailed-positive (primary) neurons and represents the ventral one of the two deuterocerebral engrailed clusters. The BAla3/4 tract extends posteriorly closely adjacent to the BAla1/2 tract. Passing through the crAN, BAla3/4 axons continue straight posteriorly and terminate in the posterior reaches of the BPM compartment (Fig. 7C,D,I).
The BAlc subgroup possesses two lineages, located lateral and posterior of BAla (Figs. 1B, 7C,D). The BAlc tract passes along the lateral and posterior surface of the BA compartment, forming a major component of the crAN system. It branches into a dorsal and ventral component (Fig. 7C,D). The dorsal component terminates in the posterior realm of the BA compartment. The ventral branch leaves the crAN in posterior direction and then curves upward, joining the antenno-cerebral tract (Fig. 7D). In view of this pattern, one or both of the BAlc lineages could correspond to the lineages described to produce the third group of antennal projection neurons that project toward the dorsal protocerebrum lateral of the calyx (Homberg et al., 1989; Malun et al., 1993; Marin et al., 2002).
BAlp has three lineages located along the lateral boundary of the ventral deuterocerebrum (Figs. 1C, 7C,D). The tracts of BAlp1–3 form a thick bundle projecting posteromedially. The bundle reaches the neuropile at the posterior boundary of the BC compartment (Fig. 7D). The BAlp1 tract extends posteromedially into the BPM compartment. The BAlp2 and BAlp3 tracts branch into a dorsal and ventral component. The major dorsal branch turns upward and forms the lateral vertical fiber system (veBL; see above), which passes along the prominent cleft between BC and BPL (Figs. 1C, 7D) and radiates from ventrally into the crMB system. The ventral component of BAlp2/3 (balpp) grows posteriorly into the BLP compartment.
Ventral to BAlp are one or two additional lineages, BAlv. Extending straight posteriorly, the BAlv tract grows along the boundary between deuterocerebrum and tritocerebrum and seems to send a branch into the tritocerebral compartment (Fig. 7C). In some specimens, a second BAv lineage exists that sends a tract along BAla dorsally toward the crMBpp (data not shown).
Centromedial group
These large lineages occupy the post ero-medial part of the basal brain, forming the posterior deuterocerebrum (Figs. 1F, 8A–C). CM1–2 are the ventral-most members of the CM group. Their axon tracts bifurcate and send one branch anteriorly through the loBM; the second branch runs parallel to the first one at a more dorsal level, forming part of the loC (Fig. 8A,B). CM2 has a third branch directed medially toward the VPC3 commissure (Fig. 8B,D).
The CM3–5 lineages are located dorsally of CM1/2. Their axon tracts form a dense tangle when entering the central posteromedial (CPM) compartment (Figs. 1E, 8B). All three tracts have a main anteriorly directed branch that projects with the loC (Fig. 8B). CM3 (and possibly CM4/5 as well) has a second, dorsally directed branch that projects onto the DPC1/DPC1d commissural system (Fig. 8C).
Tritocerebral group
The tritocerebrum forms a neuropile compartment that borders the BA compartment (anterior deuterocerebrum) medioventrally. Toward posteriorly, no clear boundary between tritocerebrum and subesophageal ganglion (anterior ventral nerve cord) has been defined. One can recognize five tritocerebral (TR) lineages located ventrally of the BA lineages. All TR tracts grow posteriorly into the TR and subesophageal neuropile compartment (Fig. 1A,B). The TR lineages have unique trajectories: TRdm forms straight posterior tract that ends with a T-shaped bifurcation; TRdl is located laterally of TRdm, close to the BAmas lineages; its tract makes a sharp medial turn to meet the tract of TRdm. Three additional TR lineages, TRvm, TRvl, and TRco, are farther ventrally. TRvm/vl project straight posteriorly into the subesophageal ganglion; TRco crosses in the subesophageal (tritocerebral) commissure.
Dorsoposterior medial group
The DPM group of lineages is spread out over a long and narrow domain covering the dorsoposterior-medial aspect of the protocerebrum (Fig. 9). DPM axon tracts are directed anteromedially toward the DP and CPM compartments. Many tracts cross the midline in the dorsal posterior commissures. Four subgroups are distinguished: DPMpm, DPMpl, DPMl, and DPMm.
Two subgroups, DPMpl (three to four lineages) and DPMpm (two lineages), form the posterior realm of the DPM clusters (Figs. 1F, 9B,D). The tracts of these lineages approach the DP compartment from posteriorly and travel along the loDM system toward the DA compartment (Fig. 9B,D,F,G). DPMpm tracts form additional commissural branches that cross in the dorsoposterior commissure DPC2 (Fig. 9A,F).
The DPMm subgroup lies anterior and medial to the DPMpm lineages (Figs. 1E, 9A,B). They project anteromedially along the medial surface of the neuropile and then branch into multiple commissural branches (it is not clear whether all three DPMm lineages contribute to all of these branches). The main branch crosses in the DPC2 commissure (Fig. 9A,B,E); other branches contribute to the DPC1, VPC3, or VPC1 commissures. At least one of the DPMm lineages has a descending branch that follows the MCT toward the subesophageal ganglion (data not shown).
DPMl is the group located most anteriorly, wedged in between the DAMd lineages anteriorly and the DPLc lineages posterolaterally (Figs. 1E, 9C,D). DPMl axon tracts travel anteroventrally, flanking the calyx medially (Fig. 9D). Axons from the DPMl1 lineage grow ventrolaterally along the dorsoposterior protocerebral tract (DPPT; parallel and close to the veBC system), one of the tracts established in the embryo that connects the protocerebrum with the ventral nerve cord (Fig. 9C,D,H). DPMl3/4 project axon tracts posteromedially toward the loDM and the DPC2 commissure (Fig. 9C,D).
Dorsoposterior lateral group
DPL lineages cover the dorsolateral aspect of the protocerebrum, and their axon tracts extend ventromedially toward the CPLd compartment (Fig. 10). Eight DPL subgroups are distinguished: DPLam and DPLal anteriorly, DPLd and DPLc dorsally, and DPLp, DPLpv, DPLm, and DPLl posteriorly.
The DPLam subgroup comprises one lineage located laterally adjacent to the dorsal lobe of the mushroom body (Figs. 1A, 10A). Its tract projects straight posteriorly, dorsally and laterally of the peduncle. At a mid-peduncular level, the DPLam tract bifurcates. Both branches turn medially; the dorsal branch passes over the peduncle and approaches the primordium of the central complex, and the ventral branch terminates in the CPL compartment. DPLam expresses engrailed. It emerges from the en-positive “head spot” that in the late embryonic brain demarcates the boundary between protocerebrum and deuterocerebrum (see below).
Laterally adjacent to DPLam are three DPLal lineages (Figs. 1A, 10A). Their axon tracts bundle together and grow posteroventrally toward the lateral face to the peduncle. These DPLal tracts form the crMBl component of the circumferential mushroom body system (Figs. 1C, 2C, 10B). The crMBl turns medially and splits into three branches (Fig. 10A,B,E); it is unclear whether all three lineages contribute to each of these branches. The dorsal branch makes a turn dorsoanteriorly (turning back toward where the lineage is located) toward the loDL. It follows this system anteriorly. The ventral branch (dplalv) passes underneath the peduncle and then dorsomedially toward the primordium of the central complex. The intermediate branch passes the peduncle dorsally and also enters the primordium of the central complex (trCM/DPC1) (data not shown).
The DPLd and DPLc subgroups are located on the crown of the brain, posteriorly adjacent to the tip of the dorsal lobe (Figs. 1B,C, 10A,B). DPLd, represented by one lineage, forms a conspicuous tract that initially grows posteriorly toward the loDL before turning back on itself and looping around the tip of the dorsal lobe as the crMBdl (Fig. 10A,G). DPLc has four to five lineages with tracts extending ventromedially toward the loDM. The tracts turn posteriorly and join the loDM; some branches appear to reach the midline in the DAC4 commissure (Fig. 10A,B,F).
Three of the four posterior DPL subgroups have rather uniform patterns of projection. Their tracts are targeted from posteriorly onto the trDL. Many tracts can be followed through trDL into the anterior part of loDL. The DPLl subgroup has three to four lineages. DPLl tracts invade the CPLd neuropile posterior to the trDL (Figs. 1D,E, 10C,D). In most specimens, DPLl2 and 3 bifurcate on impact with the neuropile; both branches extend anteriorly along the dorsal and ventral surface of the loDL, respectively. DPLl1 projects along the ventral surface of loDL (Fig. 10D). Located farther posteriorly than DPLl, laterally and posterior of the Kenyon cells of the mushroom body, the DPLp subgroup has two lineages (Figs. 1F, 10D,H). Their tracts bifurcate after meeting the neuropile; the medial branch forms part of the trPd; the anterior branch follows the loDL forward (Fig. 10D). Two lineages form the DPLm subgroup (Figs. 1E, 10C,D). Their axon tracts grow anteriorly along the loDL and then turn laterally with the trDL.
The fourth posterior DPL subgroup is typically represented by one lineage, DPLpv, the tract of which projects farther ventrally and, along with several of the CP lineage tracts (see below), joins the crMBpv system, located ventral of the peduncle (Fig. 10D).
Centroposterior group
CP lineages are located over the posterior apex of the brain hemisphere (Fig. 11A,B). They are in between the mushroom body and the BLP lineages. CP axon tracts run close to the peduncle of the mushroom body. Three CP lineages, CP1–3, project an axon tract anteromedially into the trPd (Fig. 11A,C). After crossing over the peduncle, the axons join the loDM, continue forward, and terminate in the DA compartment. One or more of these three CP lineages produces an additional thin bundle directed forward, parallel to and ventral to the peduncle (Fig. 11B, CP2). This fiber system, called crMBpv, is joined by the unbranched tract formed by CP4, the most lateral member of the CP cluster (Fig. 11B).
Basolateral lineages
The four basolateral groups of lineages described in the following are arranged around the medial edge of the outer optic anlage. Basolateral lineages are all characterized by axon tracts that travel superficially over the lateral surface of the neuropile, forming the exBL, the exDL, and the exVT systems.
The basolateral anterior group.
BLA lineages are located anterior to the outer optic anlage, lateral of the spur of the mushroom body (Figs. 1A,B, 12A). Four subgroups, BLAd, BLAl, BLAv, and BLAvm, are distinguished. Tracts of the two BLAv lineages project straight posteriorly, forming the exBL system (Fig. 12B,E). After reaching the trBL system, BLAv tracts turn medially and join this system (Fig. 12A,E). The BLAd subgroup has four to five lineages, the tracts of which project straight posteriorly, forming the exDL system (Fig. 12B–D). This system reaches the trDL from anteriorly. Turning medially, it follows the trDL into the center of the CPLd compartment (Fig. 12A,D). BLAl is located in between the BLAv and BLAd cluster (Figs. 1A,B, 12A); its tract initially travels along exBL, but then interchanges dorsally toward the exBL and exDL (Fig. 12B). BLAvm is located ventroposterior of BLAv. BLAvm axons initially travel with the exBL, but then turn medially toward the peduncle, joining the crMBpvl system (Fig. 12A,B). BLAl also seems to have a medially directed branch toward crMBpvl in some specimens.
The basolateral dorsal group.
The BLD lineages are located dorsally of the optic lobe and form tracts that converge on the exDL (Figs. 1C,D, 13A,B). BLD1 represents the most anterior BLD lineage. Its tract runs along the exDL into the trDL (Fig. 13B). The tracts of BLD2–3 reach the neuropile at a position where the exDL system meets trDL. They project mediodorsally into the trDL (Fig. 13A,B). Among BLD1–3, an additional branch continuing anteriorly in the exDL is frequently seen. Slightly more posterior, a tract of one to three individual fascicles (BLD4, 4a, 4b) reaches the lateral edge of trDL and, like the axons of BLD2/3, project dorsomedially into this system (Fig. 13B). One or more of the BLD1–4 lineages often send a second branch ventrally and medially toward the trBL system (data not shown). BLD5 represents the most posterior lineages of the BLD group. The massive BLD5 tract is directed ventrolaterally toward the emerging lobula neuropile (Figs. 1E, 13A–C). The tract bifurcates, extending a lateral branch into the lobula neuropile and a medial branch into the trBL.
The basolateral posterior lineages.
The BLP lineages flank the posterior slope of the optic lobe (Figs. 1F, 14A,B). Their tracts approach the exBL and exDL from posteriorly. BLP1/2 are located most dorsally. Their tracts, however, extend ventrally and travel along the exBL to a fairly anterior level (Fig. 14B). BLP3/4 are located ventromedially of BLP1/2. Their tracts project straight dorsoanteriorly along the exDL; they turn dorsomedially after reaching the trDL (Fig. 14A–C). BLP5 is the most ventral BLP lineage and forms a tract that extends along the exBL toward the level of the trBL (Fig. 14A,B).
The basolateral ventral lineages.
The BLV lineages are spatially isolated from all other secondary lineages. They are situated far ventrally, medially adjacent to the outer optic anlage (Figs. 1D–F, 15A,B). BLV axon tracts project anteriorly and dorsally along the neuropile surface. Two subgroups, a posterior BLVp and an anterior BLVa, can be distinguished. The BLVa subgroup contains three lineages, the tracts of which fasciculate to form the external vertical system (Figs. 1D, 15A–C). BLVp has three to four lineages, the tracts of which remain at a more ventral level than those of the BLVa group (Fig. 15B). The tracts of BLVp1/2 bifurcate and send one branch medially toward the trBL (Fig. 15A,B,D). The other branch continues anteriorly along the exBL or exDL (Fig. 15B). BLVp3/4 have unbranched tracts that project anteriorly along the exBL (Fig. 15B,E).
Mapping gene expression data onto the lineage atlas model
From published reports, it is clear that many genes with expression in the larval nervous system are expressed in “packets” that include most (or all) of the secondary neurons from one or several lineages. Because secondary lineages represent units of gene expression, the digital 3D model of secondary lineages can function as a suitable framework to compare and record gene expression patterns in a consistent and systematic way. We have reconstructed the pattern of three genes, engrailed (en) (Fig. 16A–C), period (per) (Fig. 16D–F), and sine oculis (so) (Fig. 16G–I), using the digital atlas model of secondary lineages in the third-instar larval brain. In all cases, stacks of late third larval instar brains were recorded in standard orientation (posterior to anterior) and imported into Amira. Volume renderings of the clusters of labeled cells were computed and warped into the atlas model (Fig. 16B,E,H). We then established which lineages of the atlas model had maximum overlap with the volume renderings (Fig. 16C,F,I). Subsequently, the lineages identified as candidates expressing the gene were checked in double-label experiments in which brains were double labeled with BP106 antibody. In all cases, the lineages that overlapped most in the warping approach turned out to be the lineages really expressing the gene.
Figure 16.
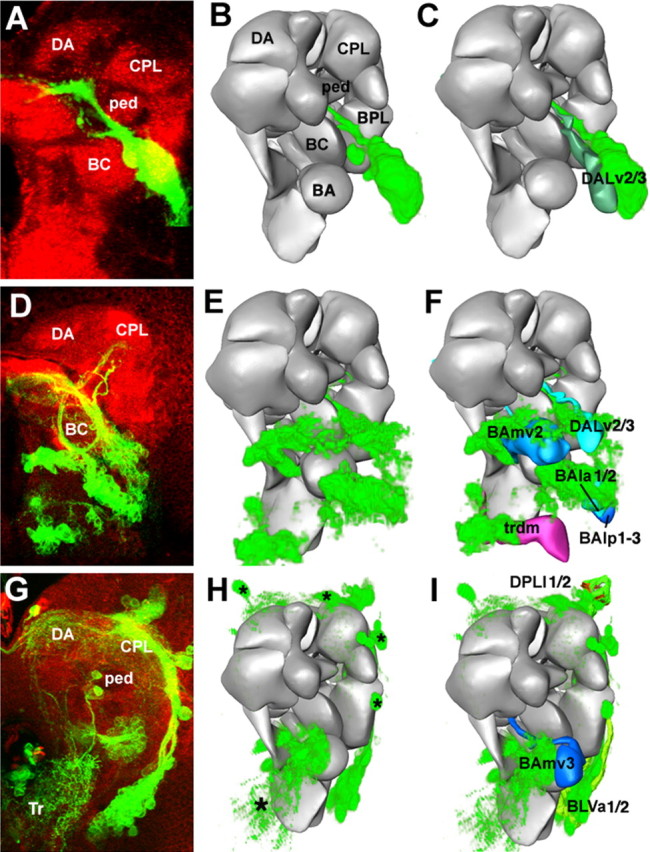
Mapping gene expression patterns within the central brain. The first row (A–C) shows the expression mapping for the engrailed reporter line ryxho25 (only the dorsal deuterocerebral cluster is shown); the second row (D–F) shows the period expression pattern, and the third row (G–I) shows the sine oculis expression pattern. The left column (A, D, G) shows Z-projections of confocal stacks taken from a frontal orientation; dorsal is up and lateral is to the right. For engrailed, the gene expression is visualized using antibody against LacZ; period and sine oculis are visualized through the use of a reporter construct driving GFP. The asterisks in G indicate primary neurons expressing sine oculis. The middle column (B, E, H) shows a volume rendering of the gene expression from the first column that has been warped into the atlas model. This step was performed by applying rigid transformations (translation, rotations, and scaling) to match landmark structures (such as the position of mushroom body substructures and the neuropile surface) within the two datasets. The right column (C, F, I) shows the volume-rendered gene expression pattern superimposed on the models of the secondary lineages that they overlap the most. The engrailed-expressing secondary lineage most closely matches the DALv2/3 lineages from the model. Period is expressed by five lineages: BAlp3, BAla2, BAmv2, DALv3, and trdm. Sine oculis is expressed by four lineages: BLVa1/2, BLVmv3, and DPLl1/2. ped, Peduncle of mushroom body.
A reporter line of engrailed, ryxho25lacZ, is expressed in small domains in the protocerebrum, deuterocerebrum, and tritocerebrum in Drosophila and other insects (Hama et al., 1990; Schmidt-Ott and Technau, 1992; Boyan et al., 1995; Rogers and Kaufman, 1996). In the late larval brain, we distinguish a cluster of primary neurons projecting to the tritocerebrum, three lineages in the deuterocerebrum (BAla1, DALv2/3), and one in the basal protocerebrum (DPLam). In Figure 17A, only the large cluster formed by en-positive DALv2/3 is shown and morphed into the model. A period-Gal4 driving the GFP reporter was developed and described by Kaneko and Hall (2000). The neurons described in detail by these authors are several clusters of primary neurons associated with the optic lobe and dorsolateral protocerebrum (the LN and DN neurons involved in the circadian rhythm). In addition, a number of secondary lineages located in the basal brain also express the reporter under period-Gal4 control (Fig. 17D). These lineages were matched to and confirmed as BAmv2, DALv2/3, BAla1/2, BAlp1–3, and trdm (Fig. 16F). Sine oculis is expressed in a number of lineages, most of which are derived from the optic lobe placode of the embryo (Chang et al., 2003) (Fig. 16G). Warping the secondary lineages expressing so into the model showed that they correspond to BLVa1/2, BAmv3, and DPLl1/2 (Fig. 16I).
Figure 17.
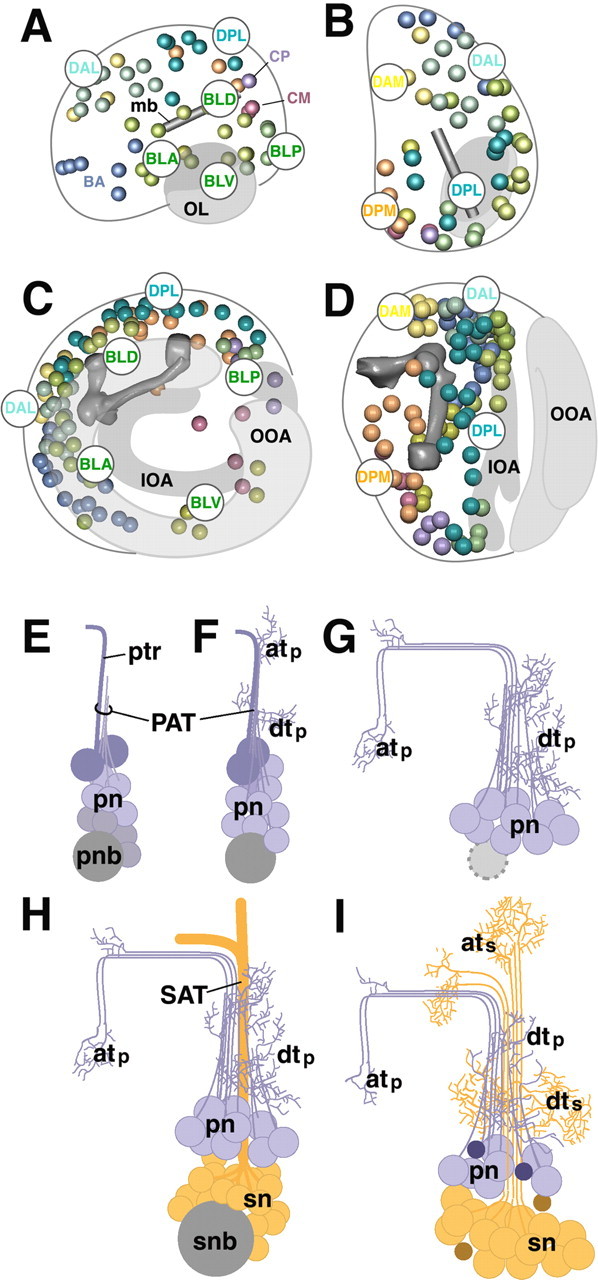
Topography of Drosophila brain lineages. A–D, Schematic representation of late embryonic brain (A, B) and larval brain (C, D). A and C show brain in lateral view, B and D in dorsal view. Location of primary lineages (in A, B) [adapted from Younossi-Hartenstein, et al. (2006)] and secondary lineages (C, D) is indicated by colored spheres. The mushroom body (mb) and optic lobe (OL) is depicted in gray. IOA, Inner optic anlage; OOA, outer optic anlage. Note the posterior-to-anterior shift of lineages (compare location of DPL and DPM in B and D), as well as centripetal movement of lateral (BL) lineages caused by the expansion of optic lobe (compare BLA, BLD, BLP, and BLV in A and C). E–I, Morphogenesis of brain lineages. The panels schematically depict generic lineage at the mid-embryonic stage (E), late embryo (F), early larva (G), late larva (H), and pupa/adult (I). The neuroblast (pnb, primary neuroblast; snb, secondary neuroblast) is shaded gray; primary neurons (pn) are purple, and secondary neurons (sn) are orange. For details, see Discussion. atp, Axonal terminal arborization of primary neurons; ats, axonal terminal arborization of secondary neurons; dtp, dendritic terminal arborization of primary neurons; dts, dendritic terminal arborization of secondary neurons; ptr, pioneer tract.
Discussion
We presented an atlas of the secondary lineages of the Drosophila larval brain. A fundamental property of the developing fly nervous system that made the approach of lineage reconstruction used in this paper possible (and useful) is its modular composition of lineages. Each lineage is formed by the progeny of one neuroblast. All neurons belonging to the same lineage stay together as a coherent cell cluster and, more importantly, form axons that fasciculate with each other in one tract, the SAT. The trajectory of the SAT within the neuropile, even more than the location of the lineage within the brain cortex, makes it possible to faithfully identify each lineage.
Neural lineages as modular units of neuronal localization and projection have been described at different levels of detail for other insects (Malzacher, 1968; Witten and Truman, 1991; Condron and Zinn, 1994; Boyan et al., 1995; Ludwig et al., 1999), as well as other parts and stages of the Drosophila nervous system (Bossing et al., 1996; Schmidt et al., 1997; Truman et al., 2004; Younossi-Hartenstein et al., 2006). Truman et al. (2004) mapped the secondary lineages of the ventral nerve cord in a similar manner as done here for the brain lineages. It is apparent that secondary lineages of the cord and the brain behave very similarly, in respect to approximate size (reflecting cell cycle of the neuroblast) and to the cohesiveness of the SAT produced by the neurons of each lineage. Similar to what is described here, lineages of the cord typically produce one unbranched SAT, which radially projects into the neuropile where it associates with one of the major longitudinal or commissural fiber systems that were laid down in the embryo (Landgraf et al., 2003; Nassif et al., 2003).
Primary and secondary lineages of the Drosophila brain
A preliminary map of the primary lineages recognizable in the late Drosophila brain has been presented recently (Younossi-Hartenstein et al., 2006). In the embryonic brain, lineages are not yet separated from each other by glial sheaths, which makes it truly difficult to separate lineage clusters in the cortex. On the basis of the number of PATs, which can be recognized more easily, ∼80 primary lineages were recognized in the late embryonic brain (Fig. 17A,B). It is possible that some PATs are formed by more than one lineage; alternatively, there could exist some neuroblasts in the embryo that divide symmetrically into two daughter neuroblasts, which then produce separate secondary lineages in the larval brain. This and other open questions can only be answered once we have followed, with the help of specific markers, individual lineages from their beginnings in the early embryo to the late larva. What is important to state already at this point is that the comparison of late embryonic (primary) lineages and larval secondary lineages described here reveals a solid similarity in regard to position and lineage trajectories (compare Fig. 17A,B with C,D). Without any difficulty, one can recognize groups of primary and secondary lineages located at similar position in the cortex and extending axon tracts in a similar pattern.
How do primary and secondary lineages relate to each other topologically and in terms of their axonal projection? To answer this question, the analysis of clones induced in the early embryo and analyzed at late larval stages would represent an appropriate approach. This approach is currently being taken (Pereanu and Hartenstein, unpublished observations). Preliminary data suggest the following picture (Fig. 17E–I): primary lineages are born during early and mid-embryonic stages (stage 10–14; 4.5–11 h) and start to differentiate toward the end of this interval. Initially, a highly stereotyped subset of early differentiating neurons forms a scaffold of pioneer tracts (Nassif et al., 1998). Soon thereafter, later-born neurons produce axon bundles (PATs) that orient themselves along the preexisting pioneer scaffold (Fig. 17E) (Younossi-Hartenstein et al., 2006). Primary axons then elaborate interlaced axonal and dendritic arbors, which, together with sheath-like processes formed by glial cells, establish the neuropile compartments of the larval brain (Fig. 17F). In the early larva, branching of axons increases. Furthermore, the close packing of cell bodies and their axons has loosened up, although somata and neurites of one lineage are still close to each other (Fig. 17G). Most of the compact PATs visible in late embryos are not any longer in existence in the early larva, although at least some PATs remain, such as the ones formed by the mushroom body (these four PATs make up the peduncle).
During mid-larval stages, the same neuroblasts that had produced primary lineages are reactivated and form the secondary lineages. In general, these secondary neuroblasts and their secondary lineages are still close to the elements of the primary lineages they had made earlier (Fig. 17H). This has been established for the mushroom body lineages (which proliferate uninterruptedly), but also for other central brain lineages, such as some of BLV and CP lineages that express the transcription factor sine oculis (Chang et al., 2003). Correspondingly, the SATs that appear in the larva have a trajectory similar to the corresponding PATs. One major difference is, of course, that the neuropile has grown substantially as a result of the arborization of primary neurons. As a result, most SATs travel for considerable distances within the neuropile, generating systems of fiber tracts that did not exist in the minute late embryonic brain.
During the pupal period, secondary lineages differentiate, and many primary neurons that had already differentiated and functioned as larval neurons are remodeled (Truman, 1990; Marin et al., 2005). It appears as if a substantial number of SATs remain tight fiber bundles in the neuropile that make the transition into the long axon tracts of the adult brain. However, similar to what happened to primary lineages in the transition from late embryo to early larva, the lineage related clustering of somata disappears during late pupal stages, and SATs disappear from the cortex (Fig. 17I).
Digital models of the developing Drosophila brain
Today’s digital modeling programs provide the opportunity to generate sophisticated three-dimensional models of complex biological structures (for recent review, see Mazziotta et al., 1995, 2001; Toga, 2002). The versatile surface rendering capabilities, along with the ability of generating virtual sections at any desired plane, allow for the use of computer models as digital atlases. Probably even more important is the use of digital models as repositories for functional and genetic data. A question of great importance for any model is what elements of a biological structure should be included in the model. More generally speaking, what should be the resolution or “granularity” of the model? The answer obviously depends on the size and complexity of the biological structure, as well as the purpose for which the model is constructed. In vertebrates, the resolution is typically that of morphologically defined compartments and tissues. Thus, models of the rodent brain (Toga et al., 1995; Leergaard et al., 2003; MacKenzie-Graham et al., 2004) or human brain (Mazziotta et al., 1995, 2001) show the outlines of brainstem nuclei or cortical domains in relationship to internal and external surfaces. Our current technical abilities make it impracticable to generate models at the level of resolution of individual neurons and their connections, although this goal is obviously envisioned in the future.
Brain models of invertebrates, notably the fly brain (Rein et al., 2002; Younossi-Hartenstein et al., 2003), are currently at a qualitatively similar level of resolution. Structurally defined brain compartments, visualized in a pool of samples, are “normalized” and can be assembled into a “standard brain” (Rein et al., 1999, 2002). By adding neural lineages and relating them to compartments and other landmarks, we have increased the granularity of the model substantially. The pattern and size of secondary lineages proved to be sufficient for the semiautomated mapping of gene expression patterns, as shown in this paper for per, so, and en. We consider it to be realistic to expect that other expression patterns that coincide with discrete lineages (or groups of lineages) can be deposited in the same manner into the larval brain model.
Future directions include generating models of earlier and later stages and “linking” the objects within these models. The usefulness of the larval model will be greatly enhanced if lineage identity is provided. To that end, models of the early to mid embryonic neuroblast population for which genetic addresses are provided (Urbach and Technau, 2003) have to be generated. This will allow one to transfer neuroblast identity to the corresponding lineage in the late embryo, and from there to the larva. Future studies can then refer to a lineage and approach details of its development with the full knowledge of its genetic history. To this genetic record of known genes, the large number of genes activated in the larva and pupa can be added.
Footnotes
This work was supported by National Institutes of Health Grant NS29367 to V.H. and the United States Public Health Service National Research Service Award GM07185 to W.P. We thank Dr. Siaumin Fung and Shana Fawcett for helpful hints with this manuscript. We are grateful to Drs. U. Tepass and J. Hall for providing us with fly stocks.
References
- Ashburner M (1989). In: Drosophila. A laboratory manual pp. 214–217. New York: Cold Spring Harbor Laboratory.
- Bossing T, Udolph G, Doe CQ, Technau GM (1996). The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev Biol 179:41–64. [DOI] [PubMed] [Google Scholar]
- Boyan GS, Williams JL, Reichert H (1995). Morphogenetic reorganization of the brain during embryogenesis in the grasshopper. J Comp Neurol 361:429–440. [DOI] [PubMed] [Google Scholar]
- Carson JP, Thaller C, Eichele G (2002). A transcriptome atlas of the mouse brain at cellular resolution. Curr Opin Neurobiol 12:562–565. [DOI] [PubMed] [Google Scholar]
- Chang T, Younossi-Hartenstein A, Hartenstein V (2003). Development of neural lineages derived from the sine oculis positive eye field of Drosophila. Arthropod Struct Dev 32:303–317. [DOI] [PubMed] [Google Scholar]
- Condron BG, Zinn K (1994). The grasshopper median neuroblast is a multipotent progenitor cell that generates glia and neurons in distinct temporal phases. J Neurosci 14:5766–5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Escalera S, Bockamp EO, Moya F, Piovant M, Jimenez F (1990). Characterization and gene cloning of neurotactin, a Drosophila transmembrane protein related to cholinesterases. EMBO J 9:3593–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumstrei K, Wang F, Nassif C, Hartenstein V (2003). Early development of the Drosophila brain. V. Pattern of postembryonic neuronal lineages expressing DE-cadherin. J Comp Neurol 455:451–462. [DOI] [PubMed] [Google Scholar]
- Hama C, Ali Z, Kornberg TB (1990). Region-specific recombination and expression are directed by portions of the Drosophila engrailed promoter. Genes Dev 4:1079–1093. [DOI] [PubMed] [Google Scholar]
- Hanesch U, Fischbach KF, Heisenberg M (1989). Neuronal architecture of the central complex in Drosophila melanogaster. Cell Tissue Res 257:343–366. [Google Scholar]
- Homberg U, Mantague RA, Hildebrand JG (1989). Anatomy of antenno-cerebral pathways in the brain of the sphinx moth Manduca sexta. Cell Tissue Res 254:255–281. [DOI] [PubMed] [Google Scholar]
- Ito K, Hotta Y (1992). Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Dev Biol 149:134–148. [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D (1997). The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development 124:761–771. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Hall JC (2000). Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol 422:66–94. [DOI] [PubMed] [Google Scholar]
- Landgraf M, Sanchez-Soriano N, Technau GM, Urban J, Prokop J (2003). A charting the Drosophila neuropile: a strategy for the standardised characterisation of genetically amenable neurites. Dev Biol 260:207–225. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L (2001). Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci 24:251–254. [DOI] [PubMed] [Google Scholar]
- Lee T, Lee A, Luo L (1999). Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development 126:4065–4076. [DOI] [PubMed] [Google Scholar]
- Leergaard TB, Bjaalie JG, Devor A, Wald LL, Dale AM (2003). In vivo tracing of major rat brain pathways using manganese-enhanced magnetic resonance imaging and three-dimensional digital atlasing. NeuroImage 20:1591–1600. [DOI] [PubMed] [Google Scholar]
- Lein ES, Zhao X, Gage FH (2004). Defining a molecular atlas of the hippocampus using DNA microarrays and high-throughput in situ hybridization. J Neurosci 24:3879–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig P, Williams JL, Lodde E, Reichert H, Boyan GS (1999). Neurogenesis in the median domain of the embryonic brain of the grasshopper Schistocerca gregaria. J Comp Neurol 414:379–390. [PubMed] [Google Scholar]
- MacKenzie-Graham A, Lee EF, Dinov ID, Bota M, Shattuck DW, Ruffins S, Yuan H, Konstantinidis F, Pitiot A, Ding Y, Hu G, Jacobs RE, Toga AW (2004). A multimodal, multidimensional atlas of the C57BL/6J mouse brain. J Anat 204:93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malun D, Waldow U, Kraus D, Boeckh J (1993). Connections between the deutocerebrum and the protocerebrum, and neuroanatomy of several classes of deutocerebral projection neurons in the brain of male Periplaneta Americana. J Comp Neurol 329:143–162. [DOI] [PubMed] [Google Scholar]
- Malzacher P (1968). Die Embryogenese des Gehirns paurometaboler Insekten. Untersuchungen an Carausius morosus und Periplaneta americana. Z Morph Tiere 62:103–161. [Google Scholar]
- Marin EC, Jefferis GS, Komiyama T, Zhu H (2002). Representation of the glomerular olfactory map in the Drosophila brain. Cell 109:243–255. [DOI] [PubMed] [Google Scholar]
- Marin EC, Watts RJ, Tanaka NK, Ito K, Luo L (2005). Developmentally programmed remodeling of the Drosophila olfactory circuit. Development 132:725–737. [DOI] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, et al (2001). A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM). Philos Trans R Soc Lond B Biol Sci 356:1293–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotta JC, Toga AW, Evans A, Fox P, Lancaster J (1995). A probabilistic atlas of the human brain: theory and rationale for its development. The International Consortium for Brain Mapping (ICBM). NeuroImage 2:89–101. [DOI] [PubMed] [Google Scholar]
- Nassif C, Noveen A, Hartenstein V (1998). Embryonic development of the Drosophila brain I. The pattern of pioneer tracts. J Comp Neurol 402:10–31. [PubMed] [Google Scholar]
- Nassif C, Noveen A, Hartenstein V (2003). Early development of the Drosophila brain III. The pattern of neuropile founder tracts during the larval period. J Comp Neurol 455:417–434. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Hartenstein V (2004). Digital three-dimensional models of Drosophila development. Curr Opin Genet Dev 14:382–391. [DOI] [PubMed] [Google Scholar]
- Rein K, Zockler M, Heisenberg M (1999). A quantitative three-dimensional model of the Drosophila optic lobes. Curr Biol 9:93–96. [DOI] [PubMed] [Google Scholar]
- Rein K, Zockler M, Mader MT, Grubel C, Heisenberg M (2002). The Drosophila standard brain. Curr Biol 12:227–231. [DOI] [PubMed] [Google Scholar]
- Renn SC, Armstrong JD, Yang M, Wang Z, An X, Kaiser K, Taghert PH (1999). Genetic analysis of the Drosophila ellipsoid body neuropil: organization and development of the central complex. J Neurobiol 41:189–207. [PubMed] [Google Scholar]
- Rogers BT, Kaufman TC (1996). Structure of the insect head as revealed by the EN protein pattern in developing embryos. Development 122:3419–3432. [DOI] [PubMed] [Google Scholar]
- Salvaterra PM, Kitamoto T (2001). Drosophila cholinergic neurons and processes visualized with Gal4/UAS-GFP. Brain Res Gene Expr Patterns 1:73–82. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Rickert C, Bossing T, Vef O, Urban J, Technau GM (1997). The embryonic central nervous system lineages of Drosophila melanogaster. II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Dev Biol 189:186–204. [DOI] [PubMed] [Google Scholar]
- Schmidt-Ott U, Technau GM (1992). Expression of en and wg in the embryonic head and brain of Drosophila indicates a refolded band of seven segment remnants. Development 116:111–125. [DOI] [PubMed] [Google Scholar]
- Schneider LE, Sun ET, Garland DJ, Taghert PH (1993). An immunocytochemical study of the FMRFamide neuropeptide gene products in Drosophila. J Comp Neurol 337:446–460. [DOI] [PubMed] [Google Scholar]
- Stocker RF (1994). The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res 275:3–26. [DOI] [PubMed] [Google Scholar]
- Strausfeld N (1976). In: Atlas of an insect brain Berlin: Springer.
- Thompson P, Cannon TD, Toga AW (2002). Mapping genetic influences on human brain structure. Ann Med 34:523–536. [DOI] [PubMed] [Google Scholar]
- Toga AW (2002). Imaging databases and neuroscience. The Neuroscientist 8:423–436. [DOI] [PubMed] [Google Scholar]
- Toga AW, Santori EM, Hazani R, Ambach K (1995). A 3D digital map of rat brain. Brain Res Bull 38:77–85. [DOI] [PubMed] [Google Scholar]
- Toga AW, Thompson PM, Mega MS, Narr KL, Blanton RE (2001). Probabilistic approaches for atlasing normal and disease-specific brain variability. Anat Embryol (Berl) 204:267–282. [DOI] [PubMed] [Google Scholar]
- Truman JW (1990). Metamorphosis of the central nervous system of Drosophila. J Neurobiol 21:1072–1084. [DOI] [PubMed] [Google Scholar]
- Truman JW (1996). Steroid receptors and nervous system metamorphosis in insects. Dev Neurosci 18:87–101. [DOI] [PubMed] [Google Scholar]
- Truman JW, Schuppe H, Shepherd D, Williams DW (2004). Developmental architecture of adult-specific lineages in the ventral CNS of Drosophila. Development 131:5167–5184. [DOI] [PubMed] [Google Scholar]
- Urbach R, Technau GM (2003). Molecular markers for identified neuroblasts in the developing brain of Drosophila. Development 130:3621–3637. [DOI] [PubMed] [Google Scholar]
- Visel A, Thaller C, Eichele G (2004). GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res 32:D552–D556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts RJ, Schuldiner O, Perrino J, Larsen C, Luo L (2004). Glia engulf degenerating axons during developmental axon pruning. Curr Biol 14:678–684. [DOI] [PubMed] [Google Scholar]
- Witten JL, Truman JW (1991). The regulation of transmitter expression in postembryonic lineages in the moth Manduca sexta. I. Transmitter identification and developmental acquisition of expression. J Neurosci 11:1980–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Nassif C, Green P, Hartenstein V (1996). Early neurogenesis of the Drosophila brain. J Comp Neurol 370:313–329. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Salvaterra PM, Hartenstein V (2003). Early development of the Drosophila brain: IV. Larval neuropile compartments defined by glial septa. J Comp Neurol 455:435–450. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Nguyen B, Shy D, Hartenstein V (2006). The embryonic origin of the Drosophila brain neuropile. J Comp Neurol in press. [DOI] [PubMed]
- Zhu S, Chiang AS, Lee T (2003). Development of the Drosophila mushroom bodies: elaboration, remodeling and spatial organization of dendrites in the calyx. Development 130:2603–2610. [DOI] [PubMed] [Google Scholar]