Abstract
Cellular senescence is a tumor-suppressive mechanism that can paradoxically contribute to aging pathologies. Despite evidence of immune clearance in mouse models, it is not known how senescent cells (SnCs) persist and accumulate with age or in tumors in individuals. Here, we identify cooperative mechanisms that orchestrate the immunoevasion and persistence of normal and cancer human SnCs through extracellular targeting of natural killer receptor signaling. Damaged SnCs avoided immune recognition through MMP-dependent shedding of NKG2D ligands reinforced via paracrine suppression of NKG2D receptor–mediated immunosurveillance. These coordinated immunoediting processes were evident in residual, drug-resistant tumors from cohorts of more than 700 prostate and breast cancer patients treated with senescence-inducing genotoxic chemotherapies. Unlike in mice, these reversible senescence subversion mechanisms were independent of p53/p16 and exacerbated in oncogenic RAS-induced senescence. Critically, the p16INK4A tumor suppressor could disengage the senescence growth arrest from the damage-associated immune senescence program, which was manifest in benign nevus lesions, where indolent SnCs accumulated over time and preserved a non-proinflammatory tissue microenvironment maintaining NKG2D-mediated immunosurveillance. Our study shows how subpopulations of SnCs elude immunosurveillance and reveals potential secretome-targeted therapeutic strategies to selectively eliminate — and restore the clearance of — the detrimental SnCs that actively persist after chemotherapy and accumulate at sites of aging pathologies.
Keywords: Aging, Oncology
Keywords: Cellular senescence, Proteases, Tumor suppressors
The immunoediting mechanisms that cause senescent cells to persist in chemotherapy-resistant tumors are reversible and offer potential targeted senolytic strategies to restore senescent cell clearance.
Introduction
Cellular senescence is best known as a cell-intrinsic mechanism that prevents cancer by limiting the proliferation of damaged or malignant cells (1–3). Tumor suppressor pathways involving p53/p21CIP1/CDKN1A, p16INK4A/CDKN2A/pRb, and DNA damage repair factors enforce cell growth arrest in response to genotoxic stress and oncogenic insults.
Recent reports in mice show that cellular senescence can also regulate immune processes leading to the elimination of senescent cells (SnCs). In mouse models of hepatocarcinoma and liver fibrosis, restoring p53 function enables senescent tumor cells and stellate cells to be eliminated by NK cells in part via NKG2D detection, while oncogenic RAS-induced senescence of hepatocytes promotes immune responses involving CD4+ T cells, neutrophils, and macrophages that lead to SnC removal (4–9). In mouse models of cutaneous wound repair, p53/p21- and p16-proficient SnCs are cleared after healing is complete (10, 11). Notably, however, p53 and p16 are not required to trigger cellular senescence in human tissues/cells, and many senescence features are p53 or p16 independent (1, 3, 12), suggesting that additional mechanisms may regulate the interplay between SnCs and the immune system.
Despite evidence of immune surveillance and clearance of SnCs in mice, SnCs accumulate with age in patients and are found in inflamed and damaged tissues, premalignant lesions, and arrested tumors and after chemo- or radiotherapy (12–21). Persistent SnCs can contribute to age-associated pathologies and tissue dysfunction, including cancer (18, 22–26). These effects have been attributed to the senescence-associated secretory phenotype (SASP; refs. 12, 27, 28), which includes inflammatory factors secreted by tissue-resident SnCs (1–3).
Importantly, elimination of persistent SnCs by genetic or pharmacological strategies has been shown to have beneficial effects in mouse models (22, 23, 29–32). For example, these approaches reduced chemotherapy side effects and tumor recurrence, and reversed the decline in hematopoiesis after irradiation or aging (18, 20, 23, 31, 32). However, results from transgenic models such as INK-ATTAC and p16-3MR may not be directly translatable in humans, and purging SnCs from tissues by blocking antiapoptotic pathways activated in SnCs may have side effects limiting their therapeutic use in patients (33, 34). Alternatively, strategies designed to potentiate the immune recognition of persistent SnCs could help augment their physiological clearance. For instance, the specific cell surface overexpression of DPP4 by human senescent fibroblasts supported antibody-dependent cell-mediated cytotoxicity (ADCC) against SnCs (35).
Understanding the signaling pathways involved in the persistence of SnCs may open new intervention avenues to boost their elimination. Here, we identify immunoediting mechanisms by which persistent SnCs evade immune clearance, with the potential for developing novel SnC-killing therapies (senolytics) to restore immune detection and elimination of SnCs, and to improve the outcome of cancer treatments and mitigate the detrimental effects of aging.
Results
Expression of NKG2D ligands by tissue-resident SnCs in patients.
NKG2D ligands (NKG2D-Ls) are cell surface semaphores that mediate the immune recognition and clearance of cells that are transformed, damaged, stressed, or infected (36–39). NKG2D-Ls are mostly absent in healthy tissues. Others and we previously observed an increase in NKG2D-L expression upon senescence induction in vitro in normal human fibroblasts (8, 40). However it is not known whether the senescence-associated upregulation of NKG2D-Ls happens in human tissues where SnCs reside.
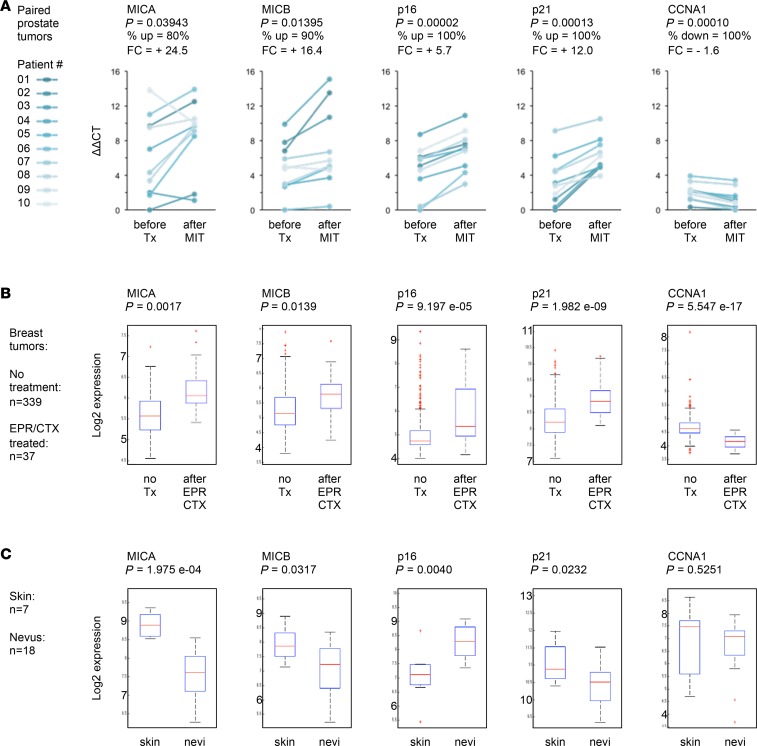
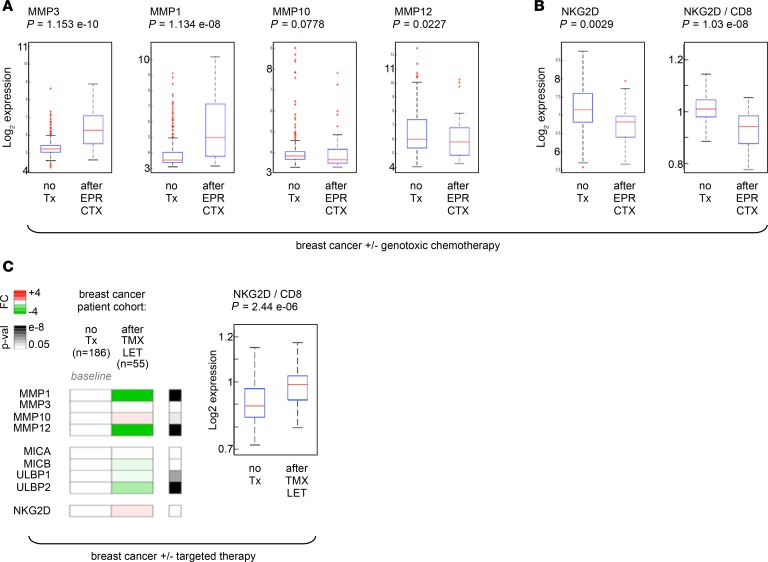
To address this, we first measured the expression of NKG2D-L MICA and MICB in laser-captured, paired tumor samples from 10 patients with prostate cancer before and after mitoxantrone (MIT) treatment, which we previously showed induces cellular senescence based on cell cycle arrest and SASP markers (12, 41). We found that after senescence-inducing genotoxic chemotherapy, residual tumors expressed significantly higher levels of MICA/B (Figure 1A). To test whether or not this was limited to prostate cancer, we performed a meta-analysis of data from breast cancer clinical trials to compare gene expression profiles of patients who underwent epirubicin/cyclophosphamide (EPR/CTX) therapy versus untreated patients (376 patients; refs. 42–44). Tumor tissues remaining after genotoxic chemotherapy showed significantly elevated expression of MICA/B and hallmarks of senescence-associated growth arrest (Figure 1B). These results show that DNA-damaging chemotherapies induce tumors to develop a senescence phenotype associated with elevated levels of NKG2D-Ls. Although this may agree with the notion that SnCs upregulate NKG2D-Ls, it is surprising because NKG2D-Ls should promote the immune detection and clearance of those cells. Thus, other characteristics likely allow these SnCs to elude immune recognition and persist while expressing elevated levels of NKG2D-Ls.
Figure 1. Expression of NKG2D ligands in human senescent tissues.
(A) Pairwise comparison of gene expression before and after genotoxic chemotherapy in prostate cancer patients. For each patient, laser-captured tumor areas were isolated from prostate biopsies obtained before chemotherapy and from prostate tissue after four 4-week cycles of neoadjuvant mitoxantrone (MIT) treatment followed by radical prostatectomy. mRNA levels were measured by qRT-PCR for 20 paired tumor specimens collected from 10 individuals (linked dots before vs. after treatment [Tx]). Left panels show expression levels of NKG2D ligands MICA and MICB. Right panels show expression levels of senescence-associated growth arrest genes p16, p21, and cyclin A1 (CCNA1). P value, Student’s t test; paired; 2 tails. FC, fold change (averaged across patients. Percentage of tumors following the main trend in changes associated with MIT-treatment is indicated. up, upregulated; down, downregulated (B) Gene expression in tumors from breast cancer patients treated or not with genotoxic therapy (37 vs. 339 patients). Each box plot displays the median (horizontal red lines), first to third quartile range (Q1–Q3 or interquartile range [IQR]; blue boxes), minimum to maximum (dashed lines), outliers (red marks). FDR-corrected P values are shown. EPR/CTX, epirubicin/cyclophosphamide treatment. (C) Gene expression in nevi compared with normal skin (18 vs. 7 individuals).
Intrigued by these observations, we asked whether a similar phenomenon occurs in cutaneous nevi, in which cells arrest and senesce largely due to p16 expression and persist for long periods in vivo (45, 46). Using transcriptome data comparing normal skin with nevus samples (25 patients; ref. 47), we found that MICA and -B were not upregulated in nevi (Figure 1C). Not only are these results opposite to what we found in tumors after genotoxic chemotherapy, but nevi also did not show increased levels of p21 (Figure 1C), which is a known downstream effector of activated p53 and DNA damage response (DDR) pathways (3, 48). This suggests that in individuals, some SnCs may not express NKG2D-Ls or may not signal their presence to the immune system.
These findings show that different kinds of tissue-resident SnCs exist and show distinct immunogenic phenotypes, hence persisting through different mechanisms. Understanding how SnCs persist could define new therapeutic interventions to eliminate them where and when needed, for instance, to help restore therapeutic sensitivity, prevent cancer relapse, or mitigate aging pathologies (2, 34, 49–51). So we undertook to test a wide panel of senescence-inducing conditions and senescence regulators (including p53, p16, and p21), and then developed coculture systems to explore and resolve mechanisms driving the persistence of SnCs.
Severe genotoxic stress induces NKG2D-L upregulation independently of p53/p16.
As a first model, we induced cellular senescence by DNA damage (10 Gy X-ray [XRA]; or replicative senescence [REP]) in normal human WI-38, IMR-90, and HCA2 fibroblasts expressing WT p53/p16, or exogenously inactivated p53 (p53–), or knocked-down p16 (p16–). Controls are provided in Supplemental Figure 1, A–D, and Supplemental Table 1 (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.124716DS1).
We found that mRNA levels of NKG2D-L MICA/B and ULBP-1/2/3 were increased in p53/p16-proficient XRA and REP SnCs (Figure 2A). Cell-surface abundance of NKG2D-Ls was elevated in SEN (XRA) compared with presenescent (PRE) cells (Figure 2B). NKG2D-L expression developed over time (5–7 days after 10 Gy exposure), coinciding with the expression of SASP components (12), such as IL-7 (Supplemental Figure 2A).
Figure 2. p53/p16-independent upregulation of NKG2D ligands in damaged SnCs, but not in CDKI-induced SnCs.
(A, C, E, and G) NKG2D ligand mRNA levels measured by quantitative real-time PCR in fibroblasts. For each gene transcript (MICA/B, ULBP-1, -2, -3), fold changes were first normalized to the average expression levels across PRE cells, and then values averaged across cell types for each condition. The number of individual samples (n) tested at least in quadruplicate by TaqMan assay is indicated. Profiles are subdivided into 4 blocks: (A) p53/p16 WT XRA or REP senescence. CON, control empty lentiviral vector. (C) p53/p16-deficient XRA or REP senescence, where the sequence of interventions is annotated as follows: p53/p16-induced deficiency followed by XRA/REP-induced senescence — SEN (p53–/p16– XRA/REP) (i.e., loss of p53 or p16 before senescence); XRA/REP-induced senescence followed by p53-induced deficiency — SEN (XRA/REP → p53–) (i.e., loss of p53 after senescence). (E) Transient DDR and ATM-deficient senescence, annotated as 0.5 Gy and ATM–. (G) p16/p21-induced senescence growth arrest, annotated as +p16 or +p21. (B, D, F, and H) Cell surface presentation of NKG2D ligands. (B) Immunofluorescence of cell surface NKG2D ligands (using recombinant NKG2D receptor coupled to an Fc fragment (rNKG2D/Fc) to detect all NKG2D ligands [NKG2D-Ls]; top; green) or MICA (antibody against MICA; bottom; red), in PRE versus SEN (XRA) WI-38 cells. Original magnification, ×20. Box plots overlaid on bar graphs of averages show the signal intensity/cell quantification for PRE (n = 580) and XRA (n = 190) cells (box plot length: 25% and 75% of data; centerline: median; whiskers: 25% – (or 75% +) 1.5 × IQR; dots: outliers; color bars: average (Ave) ± SD; P value, 2-tailed Student’s t test. Immunofluorescence panels in D show cell surface NKG2D ligands in p53-deficient or p16-deficient XRA SnCs; (F) transiently damaged cells (10 days after low-dose [0.5 Gy] radiation); (H) p16-induced SnCs. Original magnification, ×20.
Although the p53/p21 and p16/pRb pathways are important effectors of cellular senescence, the upregulation of NKG2D-Ls in fibroblasts occurred regardless of p53 loss before or after senescence-inducing damage, and irrespective of their p16 status (Figure 2, C and D; fold changes detailed in Supplemental Table 2, A and B). We observed the same phenomenon in epithelial cells. NKG2D-Ls were upregulated in p53-deficient prostate cells (PC-3, BPH1, DU145) and breast cells (MCF10A) upon DNA damage–induced senescence (10 Gy [XRA]; MIT; etoposide [ETO]; Supplemental Figure 2B). Therefore, p53 and p16 were not required to induce senescence-associated NKG2D-L expression in response to radiotherapy, chemotherapy, or telomere shortening (i.e., replicative life span) in normal fibroblasts, and in immortalized or cancer epithelial cells.
Next, we asked whether transient DDR was sufficient to induce NKG2D-Ls in our model. Fibroblasts exposed to low-dose irradiation (0.5 Gy), which transiently activates a DDR and ATM (52, 53), did repair their DNA damage to levels similar to unirradiated controls after ≥2 days based on numbers of 53BP1 foci (Supplemental Figure 1E) and did not express elevated levels of NKG2D-L expression (Figure 2, E and F). This contrasted with NKG2D-L expression in irreversibly damaged fibroblasts exposed to 10 Gy (Figure 2A). We validated these results by stably depleting ATM using shRNA (ref. 52 and Supplemental Figure 1F); this was followed by 10 Gy irradiation, which resulted in largely reduced NKG2D-L expression in SnCs (Figure 2E). So, NKG2D-L expression by SnCs required a sustained DDR without necessitating p53 or p16 tumor suppressors.
p16/p21-induced senescence growth arrest is disconnected from NKG2D-L expression.
In contrast to these results, our observations in nevi (Figure 1C), where cells arrested and senesced with high levels of p16 but low levels of NKG2D-Ls, led us to postulate that the induction of NKG2D-L expression by SnCs may depend on the DDR but not on cell growth arrest per se. SnC cycle arrest is carried out by cyclin-dependent kinase inhibitor (CDKI) p16 or p21 (1, 28). To mimic the senescence arrest elicited by these CDKIs, we exogenously overexpressed p16 or p21 in WI-38 and IMR-90 fibroblasts (controls in Supplemental Figure 1, A, D, G, and H). We found that these cells showed limited changes in levels of NKG2D-L mRNA and protein (Figure 2, G and H).
This demonstrates that the expression of NKG2D-Ls is not a consequence of CDKIs’ activation or senescence per se, but rather a response to damage that is separable from the growth arrest. These findings also indicate that, in vivo, p16+ cells/SnCs may not necessarily express NKG2D-Ls, as we observed in patients’ nevi (Figure 1C). Hence, p16 neither establishes nor triggers NKG2D-L expression, and the immunogenic program of cellular senescence can be dissociated from other senescence characteristics, including cell cycle arrest and p16 expression.
NKG2D-L expression regulates the balance between clearance and persistence of SnCs.
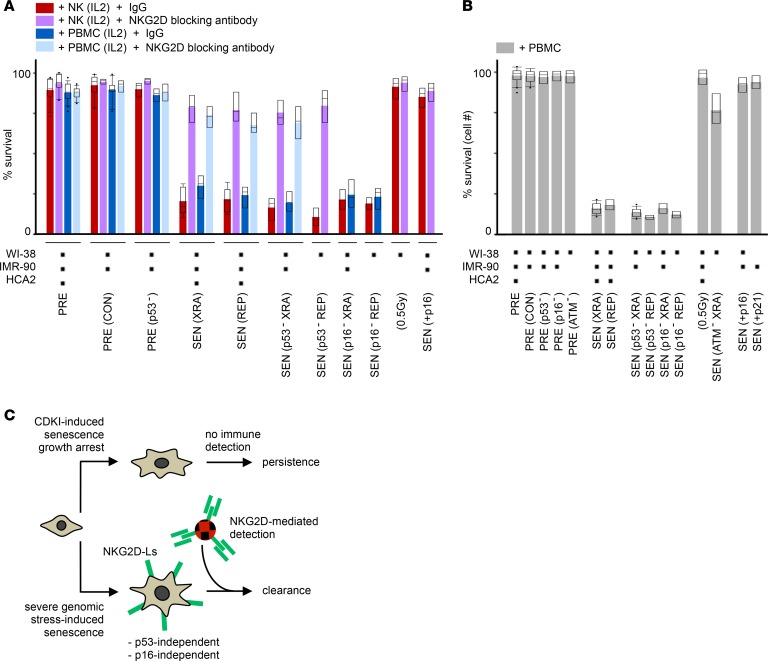
To explore how the fate of these different types of SnCs may depend on NKG2D-Ls, we cocultured leukocytes with SnCs or their presenescent counterparts, and measured lactate dehydrogenase (LDH) release as a reporter of cytotoxicity. We found that IL-2–preactivated primary NK cells were the main effectors of XRA or REP SnC cytolytic killing (Figure 3A, red bars, and Supplemental Figure 3A). Blocking the NKG2D receptor significantly prevented the killing of SnCs (Figure 3A, purple bars; increased SnC persistence). Sensitivity to NKG2D-mediated killing was independent of SnCs’ p53 and p16 status (Figure 3A). Moreover, SnCs pretreated with recombinant NKG2D receptor/Fc fusion protein were more susceptible to killing (Supplemental Figure 3B; decreased SnC persistence). These results show that NKG2D-Ls are key limiting factors that mediate the immune detection of damaged SnCs and orchestrate the balance between elimination/clearance and survival/persistence of SnCs.
Figure 3. In damaged SnCs, but not in CDKI-induced SnCs, the p53/p16-independent upregulation of NKG2D ligands regulates the balance between NKG2D-mediated immune detection and clearance of SnCs versus SnC survival and persistence.
(A) Survival of cells senesced under different conditions, and cocultured with IL-2–preactivated PBMCs or NK cells treated with a NKG2D receptor-blocking antibody, or IgG control, prior to coculture. Survival was evaluated by LDH release after 16 hours (effector/target ratio, 1:5). Black squares below the bar plot indicate which cells were tested in ≥2 independent experiments per cell condition and coculture setting. Box plot length: 25% and 75% of data; centerline: median; whiskers: 25% – (or 75% +) 1.5 × IQR, dots: outliers; color bars, averages. (B) Survival of cells senesced under different conditions and cocultured with PBMCs. Survival in triplicate was scored as the number of fibroblasts after 10 days in coculture relative to fibroblast monoculture. Effector/target ratio, 10:1. (C) Schematic summary.
These results were recapitulated using freshly collected, IL-2–preactivated PBMCs (Figure 3A, blue and turquoise bars). Similar results were obtained with prostate and mammary epithelial cells induced to senesce with genotoxic agents (MIT, ETO, XRA; Supplemental Figure 3, C and D). Cells that recovered from low-dose irradiation (0.5 Gy) remained impervious to killing, further supporting that severe genotoxic stress is a driver of NKG2D-L–dependent immunosurveillance.
Additionally, other cell surface molecules such as DNAM-1 ligands (DNAM-1-Ls) (CD112, CD155), which can also trigger targeted cytolytic responses of NK and T cells (54, 55), were increased in XRA or REP SnCs (Supplemental Table 2A). However, blocking the DNAM-1 receptor had limited impact on SnC clearance/persistence in coculture (Supplemental Figure 3E), thus indicating that NKG2D-Ls — but not DNAM-1-Ls — were primary mediators of SnC recognition and cytolysis in our model. Since the fate of SnCs is likely tightly regulated by a fine-tuned balance between activating and inhibitory ligands (37), we also tested whether the senescence program may induce a loss of inhibitory ligands HLA-E and -C. The limited changes we observed (Supplemental Table 2A) further suggest that the levels of NKG2D-Ls may act as the main molecular threshold driving SnC clearance in our model system.
To further assess these SnC clearance results, we maintained SnCs or presenescent cells in long-term coculture conditions with primary PBMCs (Figure 3B). After 10 days in direct coculture, nearly 90% of XRA or REP SnCs had been cleared, regardless of their p53 or p16 status, while ATM-deficient XRA SnCs and 0.5 Gy–irradiated cells remained mostly resilient to clearance (Figure 3B and Supplemental Figure 3F). The killing of damaged SnCs was associated with high levels of granzyme B (GRZB), IFN-γ, and TNF-α in the media of direct cocultures, regardless of SnCs’ p53 status (Supplemental Figure 3, G and H), indicating activation of innate immune cells (54). Treatment with NKG2D receptor/Fc fusion protein increased GRZB levels and SnC elimination (Supplemental Figure 3I), while inhibiting GRZB/perforin-mediated (GRZB/PRF-mediated) cytolysis using concanamycin A treatment blocked the killing of SnCs in cocultures (Supplemental Figure 3J).
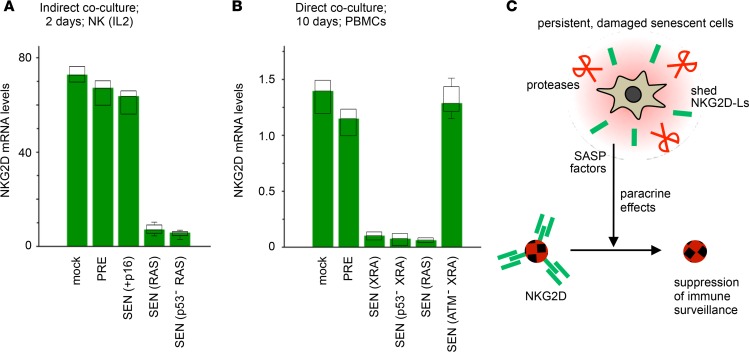
Importantly, cells that were induced to senesce by exogenous overexpression of p16 or p21 remained largely impervious to immune clearance in cocultures (Figure 3, A and B, far right bars). Survival levels were similar to those of NKG2D receptor–blocked XRA/REP SnCs or 0.5 Gy–irradiated cells. These p16 results mirrored observations in patients’ nevi, where indolent SnCs persist, and indicated that the senescence growth arrest could be uncoupled from mechanisms of genotoxic stress response, including the immune clearance program. By not upregulating cell surface antigens that would promote their elimination, p16-senesced cells remain undetected. Conversely, severe genotoxic stress promotes NKG2D-L expression, which allows SnCs to be mostly cleared (summarized in Figure 3C).
Yet what causes the persistence of damaged SnCs? Based on patient data (Figure 1, A and B; high levels of NKG2D-Ls after chemotherapy) and coculture experiments (Figure 3A; antibody-based targeting of NKG2D-Ls/NKG2D, modulating levels of survival/persistence), we hypothesized that a subset of damaged SnCs may actively evade immune recognition, possibly via mechanisms preventing NKG2D-L/NKG2D functions.
A subset of damaged SnCs actively evades leukocyte recognition and killing.
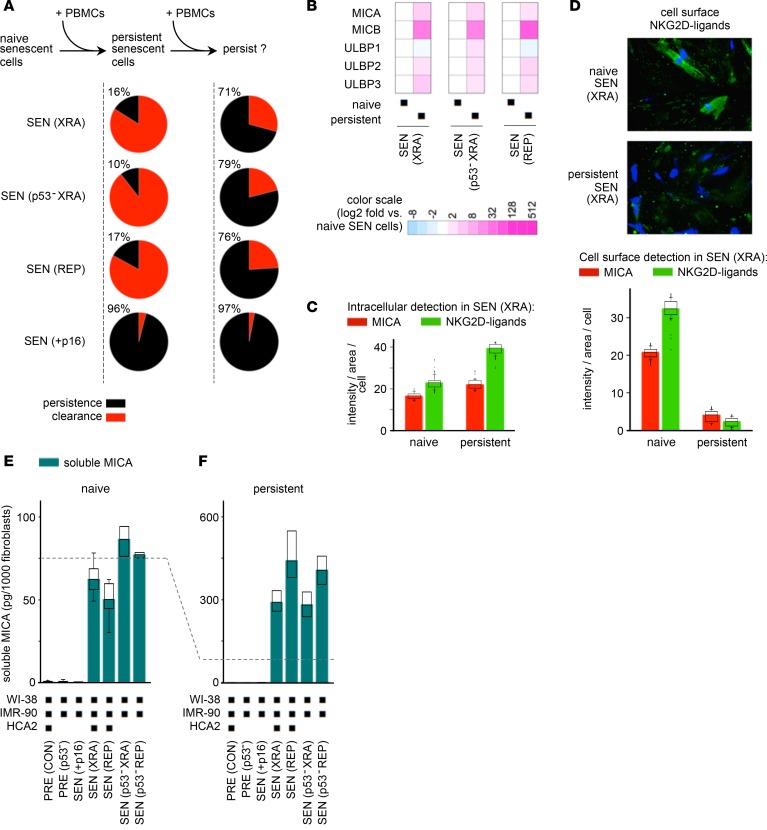
We had initially noticed that the elimination of damaged SnCs in PBMC cocultures was never complete (Figure 3B). So, we treated these persistent SnCs with fresh batches of PBMCs, and scored survival. We found that 70%–80% of the original persistent SnCs remained impervious to killing (Figure 4A; up to 8-fold increase in resistance to immune clearance). Thus, persistent SnCs possessed inherent properties that allowed them to actively evade recognition and cytolysis.
Figure 4. A subset of SnCs evade immune clearance by shedding NKG2D ligands.
(A) Quantification of naive and persistent SnCs. Naive SnCs are mock-treated monocultures. Persistent SnCs survive exposure to PBMCs. SnCs that persisted after 10 days of coculture with PBMCs were treated again with fresh PBMCs, and survival was assessed after an additional 10 days. Shown are the fractions of persistent SnCs after 1 (left) or 2 (right) rounds of PBMC exposure. (B) NKG2D ligand mRNA levels measured by quantitative real-time PCR in naive and persistent SnCs. Values were normalized to the fibroblast marker CD90 and then compared between persistent and naive SnCs (fold change relative to naive SnC expression levels). (C) Immunofluorescence signal intensity per cell of intracellular MICA (red bars) or NKG2D ligands (green bars), detected as in Figure 2B, in naive and persistent XRA SEN WI-38 cells. (D) Immunofluorescence detection of cell surface NKG2D ligands in naive and persistent SnCs. Original magnification, ×20. The average intensity per cell (n > 90) for both MICA and NKG2D ligands measured in nonpermeabilized senescent WI-38 cells is shown in the graph. Persistent SnCs display 10- to 35-fold fewer NKG2D ligands at their surface than naive SnCs. Two-tailed Student’s t test P values for differences between naive and persistent cells were 1.3 × 10–24 and 1.2 × 10–20 for MICA and NKG2D ligands, respectively. (E and F) Soluble MICA in media from naive PRE and SEN cell monocultures (E) or from persistent cells after direct coculture with PBMCs (F), as measured by ELISA. In C, D, E, and F, box plot length: 25% and 75% of data; centerline: median; whiskers: 25% – (or 75% +) 1.5 × IQR, dots: outliers; color bars: median.
Persistent SnCs cleave NKG2D-Ls from their cell surfaces.
To characterize persistent SnCs, we compared NKG2D-L expression in SnCs that had not been exposed (naive) or had been exposed (persistent) to PBMCs. Surprisingly, persistent SnCs expressed equal or greater levels of NKG2D-L mRNA and intracellular protein compared with naive cells (Figure 4, B and C, and Supplemental Figure 4A). However, immunofluorescence showed strikingly diminished levels of NKG2D-Ls on the surface of persistent SnCs relative to naive ones (Figure 4D).
Since cancer cells can promote their immunoevasion by shedding NKG2D-Ls (56–60), SnCs may also shed NKG2D-Ls to elude immune detection and persist. We found that the cell culture media of senescent fibroblasts and epithelial cells contained soluble NKG2D-L MICA (sMICA; Figure 4E and Supplemental Figure 4B) and that the media from persistent SnCs contained markedly higher levels of soluble NKG2D-Ls compared with naive counterparts (Figure 4F, dashed line). Thus, SnCs shed NKG2D-Ls regardless of cell type and p53 status (Figure 4, E and F, and Supplemental Figure 4B), and this was amplified in persistent SnCs that avoided killing.
MMP inhibition restores the clearance of persistent SnCs.
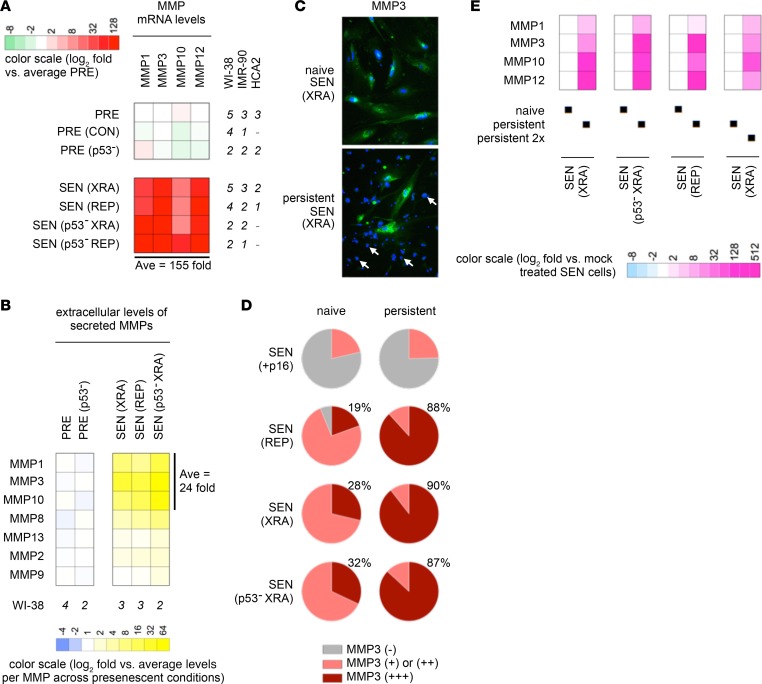
MMPs coordinate extracellular proteolytic cascades, signaling pathways, and shedding of cell-surface receptors and ligands, including NKG2D-Ls (36). We found that p53 WT and p53-deficient XRA or REP SnCs expressed and secreted multiple MMPs at higher levels than PRE cells (Figure 5, A and B, and Supplemental Table 2C). Because MMP3 was among the most upregulated MMPs across damaged SnCs, we used it as a marker of senescence detectable by immunofluorescence. In contrast to the high but variable MMP3 levels observed among naive SnCs (Figure 5C, top), persistent SnCs consistently displayed intense MMP3 staining (Figure 5C, bottom). Quantitation showed that persistent SnCs systematically expressed the highest levels of MMP3 (Figure 5D and Supplemental Figure 4C). Focusing on transcriptional activity of the MMP-1, -3, -10, -12 locus, which we previously showed increases upon senescence (27), we found that mRNA levels from this chromosomal locus were collectively and substantially far higher in persistent SnCs compared with naive cells (Figure 5E and Supplemental Figure 4C; >100-fold).
Figure 5. Persistent SnCs express high levels of MMPs regardless of their p53/p16 status.
(A) MMP mRNA levels measured by quantitative real-time PCR. (B) Secreted MMP levels measured by antibody arrays in conditioned media from fibroblast monocultures. (C) Immunofluorescence of MMP3 staining in naive (top) and persistent (bottom) XRA SEN WI-38 cells. Of note, the vast majority of DAPI-stained nuclei evident in the bottom panel correspond to the nuclei of PBMCs, i.e. not the persistent SnCs (exemplified by arrows indicating PBMC nuclei). Original magnification, ×20. (D) MMP3 expression levels in naive (left) versus persistent (right) cells senesced owing to p16 overexpression versus REP versus XRA versus p53-deficient XRA. MMP3 was detected by immunofluorescence and binned into one of 3 categories: undetectable or very low [MMP3 (–), gray], low to high [MMP3 (+) or (++), pink], and very high [MMP3 (+++), red]. WI-38 and IMR-90 cells were tested. (E) MMP mRNA levels measured by quantitative real-time PCR in SnCs that persisted after 1 or 2 (2x) rounds of PBMCs separated by 10 days. Values were normalized to the fibroblast marker CD90 and then compared between persistent and naive SnCs (fold change relative to naive SnCs). MMPs were not detected in PBMCs (Supplemental Table 2D).
To test the possibility that MMPs inhibition might preserve the cell surface presentation of NKG2D-Ls and thus enhance the killing of persistent SnCs, we used the broad-spectrum MMP inhibitor GM6001. GM6001 effectively blocked MICA shedding in a dose-dependent manner (Figure 6A) and increased cell surface NKG2D-Ls (Figure 6B). Critically, GM6001 treatment of fibroblast and epithelial cancer SnCs prior to coculture with IL-2–activated NKs or PBMCs markedly decreased SnC survival (Figure 6C; 4- to 9-fold reduction in persistence, Supplemental Figure 4D). Moreover, GM6001 treatment of already-persistent SnCs prior to a second round of PBMCs led to their near complete clearance (Figure 6D).
Figure 6. High levels of MMPs in persistent SnCs are responsible for NKG2D ligand shedding and cause SnCs to evade immune detection, but can be targeted to rescue the clearance of residual SnCs, regardless of their p53/p16 status.
(A) Soluble MICA quantified by ELISA in media from senescent WI-38 cells treated with increasing concentrations of GM6001 or mock treated (DMSO). (B) Immunofluorescence detection of NKG2D ligands after mock (top) or GM6001 (bottom) treatment of XRA WI-38 cells. Original magnification, ×40. (C) Survival assessed by LDH release in WI-38 and IMR-90 SnCs treated with GM6001 prior to 12 hour coculture with IL-2–preactivated PBMCs or NK cells (effector/target ratio, 1:3). (D) Percent survival assessed by number of SnCs remaining after coculture with PBMCs for 20 days, with 2 rounds of PBMCs separated by 10 days, with either mock or GM6001 treatment before the second round of PBMC addition. In A, C, and D, box plot length: 25% and 75% of data; centerline: median; whiskers: 25% – (or 75% +) 1.5 × IQR; dots: outliers; color bars: average. (E) Schematic summary: p53/p16-independent clearance rescue of persistent SnCs via coordinated targeting of MMP/NKG2D ligand signaling. Stabilizers of cell surface presentation of NKG2D ligands, such as extracellular protease inhibitors, are effective senolytics that restore clearance.
Importantly, persistent SnCs harboring a p53 or p16 deficiency were also highly susceptible to clearance rescue by protease inhibitor treatment in both coculture models (Figure 6, C and D). Other extracellular proteases such as plasmin and cathepsin B were not consistently identified as prosurvival factors (data not shown), and PDIA6/ERp5 and ADAM17 only increased slightly at senescence compared with MMPs (Supplemental Table 2C). Thus, MMPs were key factors that enabled damaged SnCs to evade immune killing by reducing cell-surface presentation of NKG2D-Ls (summarized in Figure 6E), and the use of protease inhibitors may be an effective therapeutic strategy to restore the immune detection and clearance of persistent, deleterious SnCs.
Oncogene-induced SnCs are hyperresistant to immune killing.
Oncogenic mutations causing DNA replication stress are another senescence modality (3, 46, 61). Surprisingly, we found that fibroblasts senesced due to oncogenic HRAS(G12V) expression (RAS; Supplemental Figure 5, A–C) were largely refractory (>90% survival) to killing by PBMCs (Figure 7A), which contrasted with XRA or REP SnCs (~10% survival). RAS cocultures were also devoid of GRZB (Supplemental Figure 5D). RAS SnCs displayed only limited changes in expression of the inhibitory ligands HLA–E and -C (Supplemental Table 2A), which are thus unlikely to mediate the bulk of the resistance to killing in our in vitro model of RAS SnC persistence. This was also not due to a downregulation or loss of NKG2D-Ls, because p53-proficient and p53-deficient RAS SnCs expressed NKG2D-Ls at even greater levels than XRA or REP SnCs (Figure 7B; >2-fold; Supplemental Table 2A).
Figure 7. RAS SnCs evade immune clearance with high MMP expression and robust NKG2D ligand shedding.
(A) Survival of RAS-induced senescent WI-38, IMR-90, and HCA-2 cells with active or inactive p53 (p53–) assessed by cell number after 10 days of direct coculture with PBMCs. (B) mRNA levels of NKG2D ligands and MMPs in RAS SnCs were measured by quantitative real-time PCR, and normalized to the average expression in control and p53-deficient presenescent cells. The range of the color scale is 64-fold greater than in Figure 2, A, C, E, and G, and Figure 5A. The ratio of the average expression levels of MMPs relative to NKG2D ligands is ~130 in RAS SnCs compared with ~7 in XRA or REP SnCs. (C) MMP-1, -3, -10, -12 immunofluorescence comparing PRE, XRA, and RAS-induced senescent WI-38 cells. Original magnification, ×20. (D) Detection of soluble MICA by ELISA in conditioned media of RAS-induced senescent WI-38, IMR-90, and HCA2 cells (p53-deficient or WT). Left: MICA levels in monocultures (naive), with SEN (XRA) and PRE cells for comparison. Right, MICA levels in supernatants collected after 10 days of direct coculture with PBMCs (persistent SnCs). In A and D, box plot length: 25% and 75% of data; centerline: median; whiskers: 25% – (or 75% +) 1.5 × IQR; dots: outliers; color bars: average in A, median in D. (E) Proportion of cells with different MMP3 levels quantified from immunofluorescence of naive and persistent RAS SnCs (top: p53 WT; bottom: p53-deficient; quantified as in Figure 5D).
However, RAS SnCs expressed a significantly wider set of MMPs and at levels that far exceeded those of XRA or REP SnCs both at the mRNA and protein levels (Figure 7, B and C; >20 fold; Supplemental Table 2C and Supplemental Figure 5, E and F). Media from naive RAS-senescent fibroblasts and epithelial cancer cells contained substantially more soluble MICA/B (>10 fold) than naive REP or XRA cells (Figure 7D and Supplemental Figure 5, G and H). These high levels were similar to levels in media from persistent XRA or REP SnCs and were maintained by persistent RAS SnCs (Figure 7D). All RAS SnCs that evaded immune killing expressed high levels of the senescence marker MMP3 (Figure 7E). Although the extremely high levels of proteases produced by RAS SnCs may prevent protease inhibitors from fully restoring their immune detection and clearance, and even though the expanded secretome of RAS SnCs (12) may impact immune processes in other detrimental ways, we still found that GM6001 treatment increased the immune clearance of RAS SnCs by up to 20% (Supplemental Figure 5I). These results support the notion that immune evasion and persistence of RAS SnCs was mediated, at least in part, via high protease secretion and high levels of NKG2D-L shedding.
Persistent SnCs suppress NKG2D receptor–mediated immunosurveillance in a paracrine manner.
Reduced NKG2D receptor expression by NK/T cells impairs tumor immunosurveillance in patients (36, 37, 56). To determine whether SnCs could also influence NKG2D receptor expression as an additional mechanism of immunoevasion, we treated IL-2–activated NK cells with media from PRE, p16 SEN, or RAS SEN cells, and measured NKG2D levels. Media from PRE and p16 SEN cells had no effect, but media from RAS SnCs reduced NKG2D mRNA levels by more than 10-fold (Figure 8A). Likewise, NKG2D expression declined sharply when PBMCs were in direct coculture with persistent senescent XRA or RAS cells, regardless of their p53 status (Figure 8B). Consistent with ATM-dependent upregulation of SASP factors (52) and NKG2D-Ls (Figure 2E), ATM-deficient XRA SnCs failed to repress NKG2D expression (Figure 8B). Thus, NKG2D-Ls shed in an autocrine manner by SnCs facilitate SnC immunoevasion by preventing NKG2D recognition, and SnCs further suppress immunosurveillance by decreasing NKG2D expression in a paracrine manner in cytolytic effector cells (summarized in Figure 8C).
Figure 8. Damaged SnCs that persist suppress NKG2D-dependent immunosurveillance.
(A) NKG2D mRNA levels were measured by quantitative real-time PCR (qRT-PCR) in IL-2–activated NK cells maintained for 48 hours in media supplemented with conditioned media from the indicated cells (4 rounds of treatment). mRNA levels were normalized to GUS; mock indicates regular medium. (B) Expression of NKG2D measured by qRT-PCR in PBMCs after 10 days of direct coculture with PRE or SEN fibroblasts. Expression of NKG2D was undetectable in fibroblasts (data not shown). Box plot length: 25% and 75% of data, centerline: median, whiskers: 25% – (or 75% +) 1.5 × IQR; color bars: median. (C) Schematic summary.
Concerted expression of MMPs and NKG2D in residual tumors after chemotherapy.
Results in Figures 2–8 show that SnCs can elude immune recognition via extracellular targeting of NKG2D-L/NKG2D signaling, so we asked whether chemotherapy-treated tumors recapitulated these cooperative mechanisms of SnC persistence.
In prostate cancer tissues from patients treated with MIT that showed elevated levels NKG2D-Ls (Figure 1A), we found that residual tumors expressed significantly higher levels of MMPs after genotoxic chemotherapy (Figure 9A, blue lines; controls for senescence markers provided in Supplemental Figure 6A). We validated these results in an additional cohort of 10 patients (Figure 9A, purple lines; 20 laser-captured paired tumors). We confirmed high MMP3 levels in tumor cells after MIT plus docetaxel (DTX) chemotherapy by IHC in a separate cohort of 76 patients (Figure 9B). We then measured NKG2D receptor expression across all prostate cancer specimens (20 patients, 40 tumors total) and found a striking significant reduction in NKG2D after chemotherapy (Figure 9C). This reduction was not due to fewer NK/T cells after chemotherapy, as shown by unchanged expression of the NK/T cell markers CD94, CD8, and CD57 (Supplemental Figure 6, B and C). Notably, individuals who did not overexpress MMPs (Supplemental Figure 6D; patient 8) or NKG2D-Ls (Supplemental Figure 6D; patients 9 and 10) did not show downregulated NKG2D expression.
Figure 9. Coordinated upregulation of MMPs and downregulation of NKG2D occurs in prostate tumors where NKG2D-L–expressing SnCs persist after genotoxic chemotherapy, but not in adjacent noncancerous normal tissues.
(A) Pairwise comparison of MMP expression before and after genotoxic chemotherapy in patients with prostate cancer. Analyses and displays are explained in the Figure 1A legend. An additional set of 20 laser-captured tumor areas isolated from 10 patients before versus after neoadjuvant mitoxantrone therapy is included for validation (purple markers/lines). (B) MMP3 detection by IHC in prostate cancer tissues from patients treated or not with mitoxantrone/docetaxel (MIT/DTX; original magnification, ×4). Tumors from 26 patients not subjected to therapy were compared with tumors from 50 patients receiving MIT/DTX. Histopathology and staining assessment are color coded as 0 (no staining), +1 (detectable), +2 (intermediate), +3 (high). Distribution of each staining intensity level is shown as pie charts below a representative image of intense staining in either condition. (C) Pairwise comparison of NKG2D expression before and after chemotherapy, as described in A. (D) Comparison of MMP, NKG2D ligand, and NKG2D expression profiles in tumor versus normal prostate tissue (top vs. bottom panels) before and after chemotherapy. The 2 sets of 20 paired tissues collected from 10 patients were pooled and plotted by gene type: MMPs (MMP-1, -3, -10, -12; left), NKG2D ligands (MICA, -B, ULBP-1, -2, -3; middle), NKG2D receptor (right). Results are displayed as values before (x axis) compared with after (y axis) chemotherapy. Overall fold change (FC; averaged across patients) and significance (P value, Student’s t test, paired, 2 tails) across MMPs, NKG2D ligands, and NKG2D are shown.
Remarkably, adjacent normal prostate tissues, which are much less susceptible to MIT damage owing to low cycling rates, did not show significant changes in MMPs, NKG2D-Ls, or NKG2D after chemotherapy (Figure 9D, bottom panels).
We extended our studies to breast cancer patients treated with EPR/CTX who showed high levels of NKG2D-Ls (Figure 1B). We found that tumor tissues remaining after genotoxic chemotherapy expressed elevated levels of MMPs (Figure 10A), which was accompanied by significantly reduced expression of NKG2D (Figure 10B and Supplemental Figure 7A).
Figure 10. The coordinated upregulation of MMPs and downregulation of NKG2D occurs in breast tumors where NKG2D ligand–expressing SnCs persist after genotoxic chemotherapy, but not after targeted therapy.
(A and B) MMP and NKG2D expression in breast tumors from patients treated or not with genotoxic therapy (dose-intense epirubicin and cyclophosphamide regimen [EPR CTX]). Analyses and displays are explained in the Figure 1B legend. In the right panel in B, NKG2D receptor expression was further normalized to CD8 expression (significant decrease). (C) MMP, NKG2D ligand, and NKG2D expression in breast tumors from patients treated or not with non-DNA-damaging, targeted therapy. Patient cohorts were identified and analyzed using the same methods used for Figure 1B (see Methods). 186 untreated patients were compared with 55 patients who underwent tamoxifen (TMX) or letrozole (LET) estrogen receptor–targeted therapy. Results are displayed as color heatmaps of median-level expression of each gene in untreated (baseline) versus TMX/LET-treated (green-to-white-to-red scale indicates mRNA levels lower-to-equal-to-higher compared with untreated; details in Supplemental Figure 7, C and D). FDR P values (p-val) are displayed as a gray scale on the right. In the box plot, NKG2D receptor expression was further normalized to CD8 expression (significant increase). Box plot length: 25% and 75% of data; centerline: median; whiskers: 25% – (or 75% +) 1.5 × IQR, dots: outliers.
To address whether these findings were specific to DNA-damaging therapy, we performed an additional transcriptional meta-analysis of tissues from breast cancer patients who underwent tamoxifen/letrozole (TMX/LET) targeted therapy, i.e., nongenotoxic hormone therapy. We found no increase in MMP or NKG2D-L expression and no decrease in NKG2D receptor expression after treatment (Figure 10C, 241 patients; Supplemental Figure 7, B and C).
Collectively, results from these multiple tissue types (prostate, breast, tumor, normal) and more than 700 patients treated with several therapeutic regimens (MIT, MIT/DTX, EPR/CTX, TMX/LET), demonstrated that after DNA-damaging chemotherapy, residual tumors harboring persistent SnCs recapitulate the cooperative mechanisms that drive SnC immunoevasion. Our work elucidates how damaged SnCs can persist in patients while paradoxically expressing high levels of NKG2D-Ls.
Unchanged MMP levels and maintained NKG2D immunosurveillance in persistent nevi.
Finally, in nevi, where persistent senescent melanocytes did not upregulate NKG2D-Ls (Figure 1C), we found no upregulation of MMPs and no downregulation of NKG2D (Figure 11, A and B). We also found that DDR-associated SASP inflammatory factors were not upregulated in patients’ nevi (Supplemental Figure 8, A and B).
Figure 11. There is no coordinated regulation of MMPs and NKG2D in long-lasting benign nevi.
(A and B) MMPs and NKG2D expression in nevi versus normal skin. In the right panel in B, NKG2D levels were normalized to CD8 expression (unchanged). Box plot length: 25% and 75% of data; centerline: median, whiskers: 25% – (or 75% +) 1.5 × IQR; dots: outliers.
These results mirrored our findings in cell culture models and indicated that some p16-induced SnCs can persist for long periods of time owing to minimal changes in expression of NKG2D-Ls, MMPs, and inflammatory factors, and without impairing NKG2D-dependent immunosurveillance. These results suggest that not all SnCs promote their immune detection and self-elimination, and different mechanisms leading to SnC persistence allow different kinds of tissue-resident SnCs to exist in individuals.
Discussion
Although cellular senescence is a safeguard against malignant transformation and can promote tissue repair, persistent SnCs can also drive aging and contribute to tumor relapse and adverse effects of chemotherapy (1–3, 18, 20, 22, 23). While recent studies have focused on the clearance of SnCs in transgenic mouse models, our work concentrates on mechanisms causing SnCs to persist in humans and reveals new senolytic strategies to augment the killing of detrimental, immune-evading, residual SnCs. Our data show how oncogenic and tumor-suppressive drivers of cellular senescence regulate surveillance processes that can be circumvented to enable SnCs to elude immune recognition but can be reversed by cell surface–targeted interventions to purge the SnCs that persist in vitro and in patients.
We found that the senescence program couples the sensing of genotoxic damage with extracellular mechanisms by which chemotherapy- and oncogene-induced SnCs subvert immune clearance through concerted MMP-dependent shedding of NKG2D-Ls and suppression of NKG2D-mediated immunosurveillance, leading to SnC persistence. This process was conserved in vitro in normal fibroblasts and in immortalized or cancer epithelial cells, and in vivo in residual tumors from patients with breast and prostate cancer who underwent senescence-inducing genotoxic chemotherapy.
Critically, neither the p53 nor the p16 tumor suppressor was required to establish or maintain the senescence-associated expression of NKG2D-Ls and MMPs in response to senescence-inducing genotoxic stresses caused by telomere shortening (replicative senescence), radiotherapy, chemotherapy, or oncogenic stress (e.g., RAS[G12mut]). In addition, p53 and p16 were dispensable to successfully clear residual SnCs. This is crucial, because in patients, the majority of tumors are p53 or p16 deficient. Thus, the targetable vulnerabilities we uncovered offer the possibility to inhibit proteases or block ligand shedding or stabilize ligand presentation to restore the immune detection and clearance of chemotherapy-resistant senescent cancer cells, regardless of tumors’ p53 or p16 status (Figure 12, right side). Furthermore, we found that these immunogenic and proteolytic mechanisms did not occur in noncancerous tissues. So these evasion processes were localized to genotoxic-resistant tumors, which suggest that senolytics directed at these actionable dependencies could avoid undesirable side effects while effectively and specifically removing deleterious SnCs. Finally, our finding that, in human cells and tissues, the occurrence, clearance, and persistence of SnCs do not depend on p16 or p53 expands the scope of targetable mechanisms, as well as the kind of persistent SnCs to study beyond those currently testable in transgenic mouse models that depend on p16 induction or p53 restoration (4–7, 11, 20, 22, 23, 29, 30, 62).
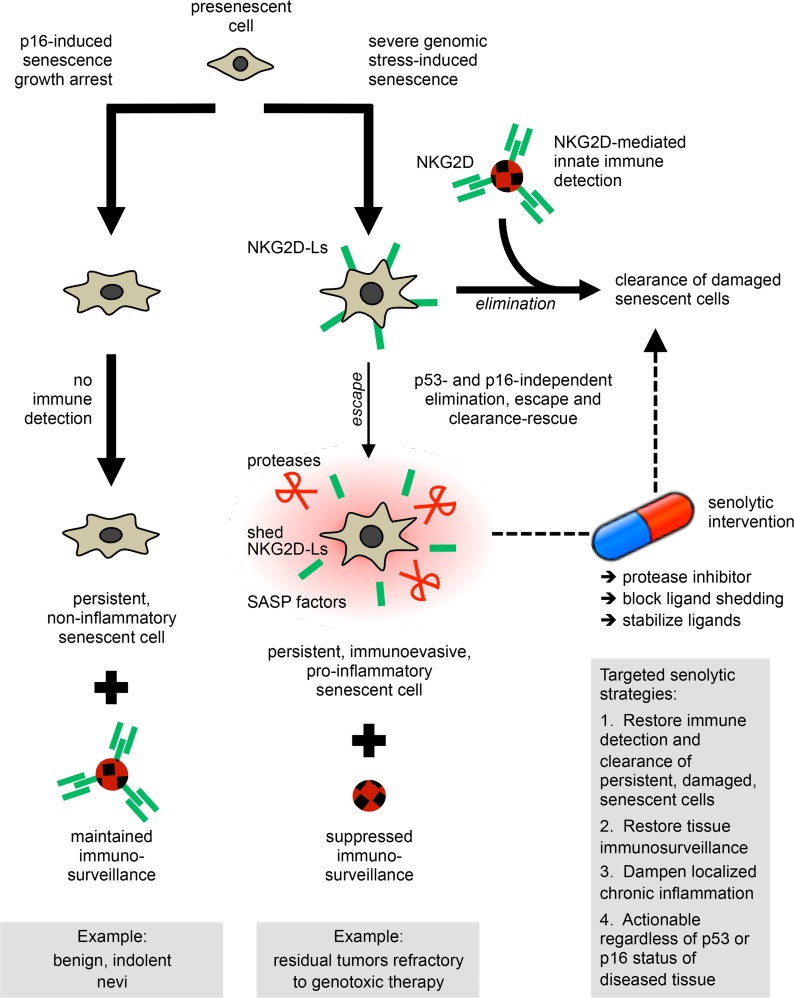
Figure 12. p53- and p16-independent immunoediting mechanisms engaged by damaged senescent cells define targeted senolytic strategies to restore immunosurveillance and eliminate deleterious senescent cells that evade clearance and persist.
The mechanisms that cause detrimental senescent cells (SnCs) to escape and persist can be turned into actionable targets. Such a rescue strategy allows augmentation of the physiological removal of residual SnCs (see cascade on the right and the senolytic rescue loop on the far right). Since mechanisms directing both the elimination and escape of SnCs are independent of p53 or p16, the immunoevasion of SnCs is reversible and targetable in tissues where p53 or p16 are defective, such as tumors. This offers new opportunities to treat cancer by combining standard genotoxic drugs with targeted senolytic agents (e.g., a protease blocker). The nondamaged and non-proinflammatory persistent SnCs, including cells that senesced owing high p16 expression, would remain insensitive to — and be spared by — such senolytic intervention (see cascade on the left). Since SnCs are in a state of final growth arrest, acquired senolytic resistance, or adverse selection of genetically adapting SnCs is not anticipated. Since the secretome of residual damaged SnCs is often inflammatory, such senolytic strategy would also help restore immunosurveillance in tissues where these SnCs would otherwise accumulate, such as chemotherapy-resilient tumors and possibly at sites of aging pathologies, including atherosclerotic lesions, osteoarthritic joints, or pulmonary fibrotic tissue.
Importantly, senescence-associated cell cycle arrest per se was not sufficient to trigger NKG2D-L expression, and cells that exogenously overexpressed p16 or p21 remained impervious to immune clearance. This mirrored the lack of inflammatory secretory phenotype by these cells (28). Likewise, we found that human nevi, which arrest largely due to the expression of the aging biomarker p16 and persist for long periods in vivo (45, 46), did not upregulate NKG2D-Ls and did not display inflammatory SASP features (Figure 12, left side). Although the p16 promoter has been used to eliminate SnCs from diseased tissues in mice (18, 22, 23, 29, 62), our previous (28) and present cell culture, nevus, and tumor data suggest that two kinds of p16-expressing, persistent SnCs may exist. On one hand, some p16+ cells might not activate mechanisms of immune clearance and inflammation, thus remaining relatively neutral with respect to tissue microenvironment. This scenario may be relevant in adult organisms, where retaining arrested yet functional cells may help maintain tissue homeostasis (1, 3, 63, 64). This also raises the possibility that p16 can prevent tumor formation not only by triggering senescence arrest as an early sensor of cellular stress (65), but also by precluding the induction of immune signals that may become detrimental over time if unresolved or unwarranted. This p16-driven protective effect may relate to observations in benign cutaneous nevi. In the alternative scenario, SnCs residing at sites of damage or experiencing severe genotoxic stress induce both p16 and sustained DDR signaling that cause cell growth arrest and elicit a complex inflammatory program, which includes the coordinated expression of SASP factors and immune stress ligands. Our results in tumors from patients treated with chemotherapies (MIT, MIT/DTX, EPR/CTX) and in cell culture models (REP, XRA, ETO, MIT, RAS) support this latter situation. While both scenarios may reflect the context-specific functions of the p16 tumor suppressor and underline the multifaceted, dynamic nature of senescence-regulated signaling networks in tissues, it remains clear that patients could benefit from therapeutic interventions that would eliminate the p16+ SnCs that accumulate with age in inflamed or damaged tissues or persist in malignant lesions (2), as demonstrated in mice (18, 22, 23, 29, 62).
In vivo studies have shown that SnCs are present in damaged tissues (1, 14), suggesting that SnCs circumvent immune clearance and persist. We consistently found that a subset of damaged SnCs is not eliminated by leukocytes. We identified 2 cooperative mechanisms of immunoevasion that drive SnCs persistence through extracellular targeting of NK cell receptor signaling.
First, secreted MMPs, which are prominent SASP factors, determined the level of cell surface NKG2D-Ls, and through autocrine ligand shedding rendered SnCs unrecognizable by leukocytes. Persistent SnCs systematically expressed high levels of MMPs. Oncogenic RAS-induced SnCs, which develop an exacerbated SASP (12, 28), secreted very high levels of MMPs and shed NKG2D-Ls, and largely evaded immune recognition. This protease-dependent immune subversion mechanism was conserved in nontransformed cells and cancer cells regardless of their p53 or p16 status, and in residual tumors refractory to genotoxic chemotherapy in patients with breast and prostate cancer. NKG2D-L stabilization by MMPs inhibition led to almost complete clearance (Figure 12, right side). The fraction of NKG2D-Ls cleaved from the cell surface by secreted proteases may ultimately control whether damaged SnCs persist.
Since eliminating SnCs can prevent tumor progression, delay the onset of degenerative diseases, and restore fitness (18, 20, 22, 23, 29, 32); since NKG2D-Ls are not widely expressed in healthy human tissues and NKG2D-L shedding is an evasion mechanism also employed by tumor cells (36, 56–58, 66–68); and since increasing numbers of B cells express NKG2D ligands in NKG2D receptor–deficient mice as they age (69), we propose that therapeutic interventions designed to increase cell surface presentation of NKG2D-Ls could be effective senolytic strategies to resensitize persistent SnCs to immune detection and rescue their clearance, whether in cancer or aging settings. Importantly, such a senolytic strategy would spare the nondamaged p16+ SnCs that do not develop the MMPs/NKG2D-L–dependent immunoevasion phenotype and that may be needed to carry on normal tissue functions, as reported in p16+ aging pancreatic β cells (63) or as we found in nevi.
This actionable immunoevasion mechanism implies that differences in expression of NKG2D-Ls and MMPs may define which types of SnCs can persist in patient tissues, including damaged and nondamaged cells, but it also points at possible threshold effects likely driven by the fine-tuned balance between levels and types of ligands and proteases, and their combinatorial effects. Since different senescence-inducing stresses likely trigger distinct senescence programs, it will be critical to explore whether SnCs present in age-related diseases such as osteoarthritis, pulmonary fibrosis, and atherosclerosis, in tissues from frail or obese patients, or in other drug-resistant cancers engage subversion mechanisms similar to those we found, or instead regulate alternative proteolytic networks (e.g., uPA, ADAMs) or express higher or lower levels of other ligands that promote — or inhibit — their immune recognition and killing (e.g., DNAM-1-Ls, HLA-E, ICAMs). How such threshold dependencies may regulate SnC immune selection and persistence suggests that cellular senescence could be a pivotal component of cancer immunoediting (70). Considering that the loss of p53 and p16 is a cancer event that can also enable cells to overcome or revert senescence arrest while amplifying their inflammatory secretory activities (1, 3, 12, 71), it is possible that some of the paradoxical cell-intrinsic and -extrinsic effects of cellular senescence may be involved in the elimination, equilibrium, and escape phases (67, 70) that underlie tumorigenesis, as well as acquired drug resistance or cancer recurrence.
In line with this concept, and as a second mechanism of senescence immunoevasion, we found that the SASP of persistent SnCs can in a paracrine manner suppress NKG2D-dependent immunosurveillance. Besides the possibility that chronically released soluble NKG2D-Ls may directly block NKG2D or act as decoys for effector leukocytes, we found that persistent SnCs decreased NK cell functionality by reducing NKG2D expression in vitro and in chemotherapy-resistant tumors (Figure 12, right side). This corroborates previously observed effects of shed NKG2D-Ls (56, 57, 66, 67, 72) and likely involves the non-cell-autonomous activities of other SASP factors, including TGF-β (60, 73–76), especially in the context of p53-deficient and/or oncogene-activated SnCs that develop an amplified secretome (12). Elevated levels of LDH released by RAS-SnCs (data not shown) may also disrupt tissue surveillance, as observed in glioblastoma, where LDH-induced NKG2D-Ls on patrolling myeloid cells cause NKG2D downregulation and subversion of immune responses (77). The SASP can create a proinflammatory microenvironment, disrupting tissue homeostasis (2) and generating locally immunosuppressed tissue niches that may promote cancer relapse and impact response to therapy (16, 20, 24). So, therapeutic manipulations dampening the SASP, such as inhibitors of BET motifs (9), NF-κB (78), mTOR (79), or cGAS/STING signaling (80), may mitigate the detrimental effects of residual SnCs and restore tumor immunosurveillance.
Because cellular senescence is not necessarily detrimental and SASP components can facilitate tissue repair or support immune functions (4–7, 10) or may communicate the compromised state of cells via MULT1 release in mice (81), it will be important to design therapeutic strategies that specifically target the deleterious SnCs of diseased tissues (2). Our work suggests that therapies blocking mechanisms of NKG2D-L shedding could effectively eradicate persistent senescent tumor cells, and concurrently restore immune surveillance. Harnessing the immunosuppressive effects of residual SnCs may help restore therapeutic sensitivity and mitigate aging pathologies. Specifically, inhibitors of SnCs’ secretome could be used in combination with neoadjuvant chemotherapies to reduce tumor burden, avert drug resistance, lower the risk of tumor recurrence, and ultimately improve patient outcomes.
Methods
Quantitative real-time PCR
Gene expression analyses were performed with Applied Biosystems TaqMan assays. Catalog IDs for each gene are listed in Supplemental Table 3A.
Meta-analysis of microarray data
We queried microarray data from clinical studies deposited in the NIH/Gene Expression Omnibus or ArrayExpress website. Briefly, we used terms/key words to filter studies of interest (e.g., “breast cancer and not treated/untreated and chemotherapy treated,” or “nevi and normal skin”). Next, we identified which studies could be merged by applying a bias analysis. Finally, the selected datasets were analyzed. Supplemental Table 3B lists genes, NCBI Gene IDs, microarray probe set IDs, and availability on Affymetrix platforms U133A or U133 Plus 2.0. Supplemental Table 4 lists study IDs and references.
Breast cancer treated or not with DNA-damaging chemotherapy.
Two studies included gene expression data from 339 biopsies before treatment (NCBI Gene Expression Omnibus [GEO] GSE23988, GSE20194; refs. 42, 43). A third study included 37 locally advanced breast carcinoma patients treated with EPR and CTX, followed by mastectomy (EMBL-EBI ArrayExpress, E-TABM-43; ref. 44). Pre- and posttreated breast tumors were heterogeneous for ER and HER2 status and grade. Data were from the Affymetrix GeneChip Human Genome U133A array. Data (376 patients) were preprocessed with the robust multi-array average (RMA) algorithm in R. Probe sets were first filtered based on the following criteria: (i) minimum 20% of samples with >100 raw value; (ii) 1 < COV < 10 (coefficient of variation). The maximum varying probe set per gene was then selected, reducing 22,283 probe sets to 12,789 unique genes. Unsupervised hierarchical clustering with Euclidean distance and average linkage was used to investigate potential bias between data sets. Genes of interest (proteases, innate immune ligands, and receptors) were each represented by a single probe set on the U133A platform of the selected studies (Supplemental Figure 7A and Supplemental Table 3B). Expression levels were compared between the 339 pretreated and 37 posttreated samples, using the nonparametric Wilcoxon’s rank-sum test. For NKG2D, levels were normalized to CD8. To correct for multiple testing, the FDR was applied to the combined set of 9 P values (82). FDR P values less than 0.05 were deemed significant. Analyses were performed in MATLAB (MathWorks) and R.
Breast cancer treated or not with endocrine therapy.
A cohort of 186 untreated breast cancer patients was compared with 55 breast cancer patients treated with TMX or LET (merged study accession nos. GSE16391, GSE13787, GSE22035, GSE16446; results in Supplemental Figure 7, B and C, and refs. 83–86). We applied the same process as described above, with data from the Affymetrix GeneChip Human Genome U133 Plus 2.0 array.
Normal skin versus nevi.
We used data from the study GSE3189 (47), which had the largest number of specimens (from 25 patients), comprising 7 normal skin and 18 nevus samples. Data were from the Affymetrix GeneChip Human Genome U133A array, and processed and analyzed as described above. To validate our results, we analyzed another, smaller clinical study, where normal skin and benign nevi were included (GSE4587; profiles could not be merged with GSE3189 based on bias analysis of microarray profiles), and we found that gene expression profiles of nevi versus normal skin followed the same trends as in GSE3189 (data not shown).
Cells
WI-38, IMR-90, and HCA2 human diploid fibroblasts; PC-3, RWPE1, BPH1, and DU145 human prostate carcinoma cells; and MCF10A human immortalized mammary epithelial cells were purchased from ATCC or produced by members of our research team, and were cultured in 3% O2 conditions (or as otherwise indicated when maintained in ambient O2 conditions) and media as previously described (12, 28).
Cells were induced to senesce by ionizing radiation (10 Gy X-ray [XRA]), DNA-damaging agent MIT treatment (25 nM; Sigma-Aldrich, M6545), ETO treatment (2.5 μM; Sigma-Aldrich, E1383), or expression of oncogenic HRAS(G12V) or CDKI tumor suppressors p16INK4a or p21CIP1/CDKN1A, and used 10 days later. Alternatively, cells were subcultured until proliferation ceased (replicative senescence owing to telomere shortening; REP). Senescence was assessed using senescence-associated β-gal (SA-βgal) staining and BrdU incorporation (Supplemental Figures 1 and 5, and refs. 12, 13, 53). To express p16INK4a, p21CDKN1A, HRAS(G12V), shp16, shATM, or GSE22 (genetic suppressor element that blocks p53 functions), cells were transduced with lentiviruses carrying no insert or a GFP insert (controls), or the indicated cDNAs, or shRNAs including control shRNA (12, 28, 52).
Freshly isolated human PBMCs were maintained in supplemented RPMI media and cultured in 3% O2 conditions. Monocytes were obtained from ATCC (U934 and Thp1) or AllCells (CD36 selection; >99%pure). Purified NK, T, and DC cells were from AllCells (>99%pure). PBMCs, NK cells, and T cells were activated by IL-2 treatment (1000 U/mL for 3 days). Monocyte differentiation into macrophages was achieved using PMA or LPS (3–4 days).
Survival assays.
LDH release assays were performed in phenol red–free media, and calculation of killing was done according to the manufacturer’s protocol (Promega, CytoTox). Effector/target cell ratios ranged from 1:3 to 1:15. Direct coculture was performed by adding effector leukocytes directly to adherent target cells. Indirect coculture was performed by adding effector leukocytes in upper chambers of Transwell culture plates (Millipore), where adherent target cells were maintained in the bottom well. All survival assays were performed in 3% O2 culture conditions. All survival assay data are the average of ≥3 independent experiments run in quadruplicate.
ELISA, immunofluorescence, antibodies, inhibitors, and recombinant proteins
ELISA kits were used to measure MICA (R&D Systems, DY1300), MICB (R&D Systems, DY1599), MICA/B (R&D Systems, DY990), MMP3 (R&D Systems, DMP300), TNF-α (R&D Systems, DTA00C), IFN-γ (R&D Systems, DIF50), IL-2 (R&D Systems, D2050), IL-6 (R&D Systems, D6050), IL-7 (R&D Systems, M7000), IL-8 (R&D Systems, D8000C), IL-12 (R&D Systems, D1200), IL-17 (R&D Systems, D1700), and GRZB (R&D Systems, DY2906-05; Thermo Fisher Scientific, BMS2027). Alternatively, secretory profiles were assessed by antibody arrays (RayBiotech, Cytokine and MMP Arrays; refs. 12, 27, 28). We used the following primary antibodies: p53 (Oncogene Research Products, DO-1), 53BP1 (Bethyl, BL182), RAS (BD Biosciences, 610001), p16INK4a (Neomarkers, MS-889-P1 and MS-218-P1), ATM (Abcam Y-170), phospho-ATM (Upstate, 05-740), IL-6 (R&D Systems, MAB2061 and AF-206-NA), IL-8 (R&D Systems, MAB208), MICA/B (R&D Systems, MAB13001; Santa Cruz Biotechnology Inc., sc-5460), MICA (R&D Systems, AF1300 and MAB1300), MICB (R&D Systems, AF1599 and MAB1599), NKG2D (R&D Systems, MAB139; Santa Cruz Biotechnology Inc., sc-9621), NKG2D blocking peptide (Santa Cruz Biotechnology Inc., sc-9621p), CD155 (R&D Systems, MAB2530), CD112 (Abcam, 502-57), DNAM-1 (R&D Systems, MAB666), IL-7 (Abcam, ab175380), MMP1 (R&D Systems, AF901; Oncogene Research Products, IM35L), MMP3 (R&D Systems, AF513; Oncogene Research Products, IM70T; BioVision, 3523-100), MMP10 (R&D Systems, AF910; Oncogene Research Products, IM75T), MMP12 (R&D Systems, MAB917), IgG control (R&D Systems, G-101-C-ABS; MAB002). Donkey secondary antibodies conjugated to Alexa Fluor were purchased from Molecular Probes (Alexa Fluor 350, 488, and 594). DAPI was used to stain nuclei. Immunofluorescence using anti-MICA/B or rNKG2D/Fc was performed without permeabilization (to monitor the presence of NKG2D-Ls at the cell surface) or after permeabilization with 0.5% Triton X-100 (to monitor intracellular levels of NKG2D-Ls). All ELISA and antibody array data are the average of ≥3 independent experiments run in quadruplicate. We used the MMP inhibitor GM6001 (Chemicon, CC1000) and recombinant chimeric NKG2D protein rNKG2D/Fc (R&D Systems, 1299-NK). NKG2D-blocking antibody is listed above along with other antibodies. Leukocytes were preincubated with concanamycin A (Sigma-Aldrich) for 3 hours before use in direct coculture. Before and after use, leukocytes were checked for viability and ability to mediate cell killing.
Human tissue microarrays and IHC
Tissue microarrays from prostate cancer patient biopsies were made at the University of Washington. Antibodies used in IHC are listed in the ELISA, immunofluorescence, antibodies, inhibitors, and recombinant proteins. All slides were reviewed and scored by a pathologist.
Statistics
Two-tailed Student’s t test was used to determine significance of differences, as indicated in figure legends. P values less than 0.05 were considered significant.
Study approval
With regard to patient consent and human tissues, prostate cancer patients with high-risk localized prostate cancer were enrolled and treated in a phase I–II neoadjuvant chemotherapy trial at Oregon Health & Science University, Portland VA Medical Center, Kaiser Permanente Northwest Region, Legacy Health System, and University of Washington. Written informed consent was obtained from all patients. The protocol was approved by the Institutional Review Boards of Oregon Health and Science University, Portland Veterans Affairs Medical Center, the University of Washington, and the Portland Legacy Health Systems. Prostate biopsies were obtained prior to chemotherapy. At the time of radical prostatectomy following chemotherapy, cancer-containing tissue samples were obtained and frozen as described (12). Cancerous epithelium from pretreated biopsies and posttreated prostatectomy specimens were captured separately, and histology was verified by review of H&E-stained sections and laser confocal microscopy images.
Author contributions
SMY, DPM, MK, AMF, FR, and JPC developed experimental methods, generated samples, and acquired data. For prostate cancer data, YS and PSN collected patient samples and ran gene expression profiles; FVL and JDW generated tissue microarrays and analyzed IHC profiles. For breast cancer and skin nevi, AD analyzed gene expression array studies from clinical trials. SMY, DPM, AD, DHR, PSN, LJvtV, JC, and JPC contributed result analysis and discussion. JPC and SMY conceptualized hypotheses and designed the study. DPM, SMY, JC, and JPC wrote the manuscript, and PYD helped with editing references. All authors reviewed the manuscript.
Supplementary Material
Acknowledgments
We thank patients and their families for participating in these studies, Tomasz Beer, Celestia Higano and other team members for assisting in clinical trials and biospecimen collections. We thank the Genome Analysis Core at the Helen Diller Family Comprehensive Cancer Center for the TaqMan assays. We thank Christian Beauséjour (Hopital St Justine, Université de Montreal) and Tim Nice (University of California, Berkeley) for scientific discussions; Christina Yau (UCSF) for complementary computational analyses; Christopher K. Patil (Lawrence Berkeley National Laboratory) for technical help scanning antibody arrays; Lawrence D. True (University of Washington, Seattle) for assistance with histopathology. This study was in part supported by grants from the NIH (AG09909 and AG017242 to JC; CA097186 and CA165573 to PSN; R01CA093678 and R01AI113041 to DHR; R01CA204021 to JW), SENS Foundation (to JC), US Department of Defense Prostate Cancer Research Program (PC111703 to YS), the Breast Cancer Research Foundation (to LJvtV), the Angela and Shu Kai Chan Endowed chair (to LJvtV), Swim Across America Funds (to DPM), the Tri-Valley SOCKs program (to JPC), UCSF Breast Oncology Research and Development Program (to AD and JPC), the Give Breast Cancer The Boot Program (to JPC).
Version 1. 06/11/2019
In-Press Preview
Version 2. 07/25/2019
Electronic publication
Footnotes
AD’s present address is: Bioinformatics and Computational Biology, Genentech Inc., South San Francisco, California, USA.
YS’s present address is: Key Lab of Stem Cell Biology, Institute of Health Sciences, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China.
AMF’s present address is: Calico Life Sciences, LLC, South San Francisco, California, USA.
FR’s present address is: Université de Montréal, Department of Radiology, Radio-Oncology and Nuclear Medicine, and CRCHUM et Institut du cancer de Montréal, Montreal, Quebec, Canada.
JDW’s present address is: Feinberg School of Medicine and Robert Lurie Comprehensive Cancer Center, Northwestern University, Chicago, Illinois, USA.
Conflict of interest: JC is cofounder and shareholder of Unity Biotechnology.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(14):e124716. https://doi.org/10.1172/jci.insight.124716.
Contributor Information
Denise P. Muñoz, Email: dmunoz@chori.org.
Anneleen Daemen, Email: daemena@gene.com.
Yu Sun, Email: sunyu@sibs.ac.cn.
Funda Vakar-Lopez, Email: fvakar@uw.edu.
Misako Kawahara, Email: Misako.Stillion@ucsf.edu.
Adam M. Freund, Email: afreund@gmail.com.
Francis Rodier, Email: rodierf@mac.com.
Jennifer D. Wu, Email: Jennifer.wu@northwestern.edu.
Pierre-Yves Desprez, Email: pydesprez@cpmcri.org.
David H. Raulet, Email: raulet@berkeley.edu.
Peter S. Nelson, Email: pnelson@fhcrc.org.
Judith Campisi, Email: jcampisi@buckinstitute.org.
Jean-Philippe Coppé, Email: jean-philippe.coppe@ucsf.edu.
References
- 1.Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Childs BG, et al. Senescent cells: an emerging target for diseases of ageing. Nat Rev Drug Discov. 2017;16(10):718–735. doi: 10.1038/nrd.2017.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He S, Sharpless NE. Senescence in health and disease. Cell. 2017;169(6):1000–1011. doi: 10.1016/j.cell.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xue W, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445(7128):656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krizhanovsky V, et al. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134(4):657–667. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang TW, et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011;479(7374):547–551. doi: 10.1038/nature10599. [DOI] [PubMed] [Google Scholar]
- 7.Iannello A, Thompson TW, Ardolino M, Lowe SW, Raulet DH. p53-dependent chemokine production by senescent tumor cells supports NKG2D-dependent tumor elimination by natural killer cells. J Exp Med. 2013;210(10):2057–2069. doi: 10.1084/jem.20130783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sagiv A, et al. NKG2D ligands mediate immunosurveillance of senescent cells. Aging (Albany NY) 2016;8(2):328–344. doi: 10.18632/aging.100897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tasdemir N, et al. BRD4 connects enhancer remodeling to senescence immune surveillance. Cancer Discov. 2016;6(6):612–629. doi: 10.1158/2159-8290.CD-16-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jun JI, Lau LF. The matricellular protein CCN1 induces fibroblast senescence and restricts fibrosis in cutaneous wound healing. Nat Cell Biol. 2010;12(7):676–685. doi: 10.1038/ncb2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demaria M, et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev Cell. 2014;31(6):722–733. doi: 10.1016/j.devcel.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coppé JP, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6(12):2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92(20):9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erusalimsky JD, Kurz DJ. Cellular senescence in vivo: its relevance in ageing and cardiovascular disease. Exp Gerontol. 2005;40(8–9):634–642. doi: 10.1016/j.exger.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Herbig U, Ferreira M, Condel L, Carey D, Sedivy JM. Cellular senescence in aging primates. Science. 2006;311(5765):1257. doi: 10.1126/science.1122446. [DOI] [PubMed] [Google Scholar]
- 16.Gilbert LA, Hemann MT. DNA damage-mediated induction of a chemoresistant niche. Cell. 2010;143(3):355–366. doi: 10.1016/j.cell.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanoff HK, et al. Effect of cytotoxic chemotherapy on markers of molecular age in patients with breast cancer. J Natl Cancer Inst. 2014;106(4):dju057. doi: 10.1093/jnci/dju057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang J, et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat Med. 2016;22(1):78–83. doi: 10.1038/nm.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeon HY, et al. Irradiation induces glioblastoma cell senescence and senescence-associated secretory phenotype. Tumour Biol. 2016;37(5):5857–5867. doi: 10.1007/s13277-015-4439-2. [DOI] [PubMed] [Google Scholar]
- 20.Demaria M, et al. Cellular senescence promotes adverse effects of chemotherapy and cancer relapse. Cancer Discov. 2017;7(2):165–176. doi: 10.1158/2159-8290.CD-16-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le ON, et al. Ionizing radiation-induced long-term expression of senescence markers in mice is independent of p53 and immune status. Aging Cell. 2010;9(3):398–409. doi: 10.1111/j.1474-9726.2010.00567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker DJ, et al. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530(7589):184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker DJ, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479(7372):232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruhland MK, et al. Stromal senescence establishes an immunosuppressive microenvironment that drives tumorigenesis. Nat Commun. 2016;7:11762. doi: 10.1038/ncomms11762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci USA. 2001;98(21):12072–12077. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eggert T, et al. Distinct functions of senescence-associated immune responses in liver tumor surveillance and tumor progression. Cancer Cell. 2016;30(4):533–547. doi: 10.1016/j.ccell.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coppé JP, et al. A human-like senescence-associated secretory phenotype is conserved in mouse cells dependent on physiological oxygen. PLoS ONE. 2010;5(2):e9188. doi: 10.1371/journal.pone.0009188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coppé JP, Rodier F, Patil CK, Freund A, Desprez PY, Campisi J. Tumor suppressor and aging biomarker p16(INK4a) induces cellular senescence without the associated inflammatory secretory phenotype. J Biol Chem. 2011;286(42):36396–36403. doi: 10.1074/jbc.M111.257071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeon OH, et al. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat Med. 2017;23(6):775–781. doi: 10.1038/nm.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roos CM, et al. Chronic senolytic treatment alleviates established vasomotor dysfunction in aged or atherosclerotic mice. Aging Cell. 2016;15(5):973–977. doi: 10.1111/acel.12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yosef R, et al. Directed elimination of senescent cells by inhibition of BCL-W and BCL-XL. Nat Commun. 2016;7:11190. doi: 10.1038/ncomms11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baar MP, et al. Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell. 2017;169(1):132–147.e16. doi: 10.1016/j.cell.2017.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Keizer PL. The fountain of youth by targeting senescent cells? Trends Mol Med. 2017;23(1):6–17. doi: 10.1016/j.molmed.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 34.Ovadya Y, Krizhanovsky V. Strategies targeting cellular senescence. J Clin Invest. 2018;128(4):1247–1254. doi: 10.1172/JCI95149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim KM, et al. Identification of senescent cell surface targetable protein DPP4. Genes Dev. 2017;31(15):1529–1534. doi: 10.1101/gad.302570.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raulet DH, Gasser S, Gowen BG, Deng W, Jung H. Regulation of ligands for the NKG2D activating receptor. Annu Rev Immunol. 2013;31:413–441. doi: 10.1146/annurev-immunol-032712-095951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morvan MG, Lanier LL. NK cells and cancer: you can teach innate cells new tricks. Nat Rev Cancer. 2016;16(1):7–19. doi: 10.1038/nrc.2015.5. [DOI] [PubMed] [Google Scholar]
- 38.Cerwenka A, Baron JL, Lanier LL. Ectopic expression of retinoic acid early inducible-1 gene (RAE-1) permits natural killer cell-mediated rejection of a MHC class I-bearing tumor in vivo. Proc Natl Acad Sci USA. 2001;98(20):11521–11526. doi: 10.1073/pnas.201238598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436(7054):1186–1190. doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Coppé J-P. Aging and Cancer: Cell Non-Autonomous Effects of Senescent Fibroblasts on Tissue Microenvironment [dissertation]. Berkeley, California, USA: University of California, Berkeley; 2007. [Google Scholar]
- 41.Sun Y, et al. Treatment-induced damage to the tumor microenvironment promotes prostate cancer therapy resistance through WNT16B. Nat Med. 2012;18(9):1359–1368. doi: 10.1038/nm.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iwamoto T, et al. Gene pathways associated with prognosis and chemotherapy sensitivity in molecular subtypes of breast cancer. J Natl Cancer Inst. 2011;103(3):264–272. doi: 10.1093/jnci/djq524. [DOI] [PubMed] [Google Scholar]
- 43.Popovici V, et al. Effect of training-sample size and classification difficulty on the accuracy of genomic predictors. Breast Cancer Res. 2010;12(1):R5. doi: 10.1186/bcr2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bertheau P, et al. Exquisite sensitivity of TP53 mutant and basal breast cancers to a dose-dense epirubicin-cyclophosphamide regimen. PLoS Med. 2007;4(3):e90. doi: 10.1371/journal.pmed.0040090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michaloglou C, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436(7051):720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 46.Chandeck C, Mooi WJ. Oncogene-induced cellular senescence. Adv Anat Pathol. 2010;17(1):42–48. doi: 10.1097/PAP.0b013e3181c66f4e. [DOI] [PubMed] [Google Scholar]
- 47.Talantov D, et al. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res. 2005;11(20):7234–7242. doi: 10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
- 48.Cazzalini O, Scovassi AI, Savio M, Stivala LA, Prosperi E. Multiple roles of the cell cycle inhibitor p21(CDKN1A) in the DNA damage response. Mutat Res. 2010;704(1–3):12–20. doi: 10.1016/j.mrrev.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 49.Jeon OH, David N, Campisi J, Elisseeff JH. Senescent cells and osteoarthritis: a painful connection. J Clin Invest. 2018;128(4):1229–1237. doi: 10.1172/JCI95147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Childs BG, Li H, van Deursen JM. Senescent cells: a therapeutic target for cardiovascular disease. J Clin Invest. 2018;128(4):1217–1228. doi: 10.1172/JCI95146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baker DJ, Petersen RC. Cellular senescence in brain aging and neurodegenerative diseases: evidence and perspectives. J Clin Invest. 2018;128(4):1208–1216. doi: 10.1172/JCI95145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodier F, et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11(8):973–979. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodier F, et al. DNA-SCARS: distinct nuclear structures that sustain damage-induced senescence growth arrest and inflammatory cytokine secretion. J Cell Sci. 2011;124(pt 1):68–81. doi: 10.1242/jcs.071340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu Rev Immunol. 2013;31:227–258. doi: 10.1146/annurev-immunol-020711-075005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soriani A, et al. ATM-ATR-dependent up-regulation of DNAM-1 and NKG2D ligands on multiple myeloma cells by therapeutic agents results in enhanced NK-cell susceptibility and is associated with a senescent phenotype. Blood. 2009;113(15):3503–3511. doi: 10.1182/blood-2008-08-173914. [DOI] [PubMed] [Google Scholar]
- 56.Groh V, Wu J, Yee C, Spies T. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature. 2002;419(6908):734–738. doi: 10.1038/nature01112. [DOI] [PubMed] [Google Scholar]
- 57.Wu JD, Higgins LM, Steinle A, Cosman D, Haugk K, Plymate SR. Prevalent expression of the immunostimulatory MHC class I chain-related molecule is counteracted by shedding in prostate cancer. J Clin Invest. 2004;114(4):560–568. doi: 10.1172/JCI22206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chitadze G, et al. Shedding of endogenous MHC class I-related chain molecules A and B from different human tumor entities: heterogeneous involvement of the “a disintegrin and metalloproteases” 10 and 17. Int J Cancer. 2013;133(7):1557–1566. doi: 10.1002/ijc.28174. [DOI] [PubMed] [Google Scholar]
- 59.Liu G, et al. Perturbation of NK cell peripheral homeostasis accelerates prostate carcinoma metastasis. J Clin Invest. 2013;123(10):4410–4422. doi: 10.1172/JCI69369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klöß S, et al. Increased sMICA and TGFβ. Oncoimmunology. 2015;4(11):e1055993. doi: 10.1080/2162402X.2015.1055993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bartkova J, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444(7119):633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 62.Childs BG, Baker DJ, Wijshake T, Conover CA, Campisi J, van Deursen JM. Senescent intimal foam cells are deleterious at all stages of atherosclerosis. Science. 2016;354(6311):472–477. doi: 10.1126/science.aaf6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Helman A, et al. p16(Ink4a)-induced senescence of pancreatic beta cells enhances insulin secretion. Nat Med. 2016;22(4):412–420. doi: 10.1038/nm.4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Diekman BO, et al Expression of p16(INK)(4a) is a biomarker of chondrocyte aging but does not cause osteoarthritis [published online ahead of print May 9, 2018]. Aging Cell. doi: 10.1111/acel.12771. [Google Scholar]
- 65.Collado M, Serrano M. Senescence in tumours: evidence from mice and humans. Nat Rev Cancer. 2010;10(1):51–57. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oppenheim DE, et al. Sustained localized expression of ligand for the activating NKG2D receptor impairs natural cytotoxicity in vivo and reduces tumor immunosurveillance. Nat Immunol. 2005;6(9):928–937. doi: 10.1038/ni1239. [DOI] [PubMed] [Google Scholar]
- 67.Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat Rev Immunol. 2006;6(11):836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 68.Ferrari de Andrade L, et al. Antibody-mediated inhibition of MICA and MICB shedding promotes NK cell-driven tumor immunity. Science. 2018;359(6383):1537–1542. doi: 10.1126/science.aao0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Raju S, et al. NKG2D-NKG2D Ligand interaction inhibits the outgrowth of naturally arising low-grade B cell lymphoma in vivo. J Immunol. 2016;196(11):4805–4813. doi: 10.4049/jimmunol.1501982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3(11):991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 71.Beauséjour CM, et al. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J. 2003;22(16):4212–4222. doi: 10.1093/emboj/cdg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Song H, Kim J, Cosman D, Choi I. Soluble ULBP suppresses natural killer cell activity via down-regulating NKG2D expression. Cell Immunol. 2006;239(1):22–30. doi: 10.1016/j.cellimm.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 73.Lee JC, Lee KM, Kim DW, Heo DS. Elevated TGF-beta1 secretion and down-modulation of NKG2D underlies impaired NK cytotoxicity in cancer patients. J Immunol. 2004;172(12):7335–7340. doi: 10.4049/jimmunol.172.12.7335. [DOI] [PubMed] [Google Scholar]
- 74.Mamessier E, et al. Human breast cancer cells enhance self tolerance by promoting evasion from NK cell antitumor immunity. J Clin Invest. 2011;121(9):3609–3622. doi: 10.1172/JCI45816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wiemann K, et al. Systemic NKG2D down-regulation impairs NK and CD8 T cell responses in vivo. J Immunol. 2005;175(2):720–729. doi: 10.4049/jimmunol.175.2.720. [DOI] [PubMed] [Google Scholar]
- 76.Barker HE, Paget JT, Khan AA, Harrington KJ. The tumour microenvironment after radiotherapy: mechanisms of resistance and recurrence. Nat Rev Cancer. 2015;15(7):409–425. doi: 10.1038/nrc3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Crane CA, et al. Immune evasion mediated by tumor-derived lactate dehydrogenase induction of NKG2D ligands on myeloid cells in glioblastoma patients. Proc Natl Acad Sci USA. 2014;111(35):12823–12828. doi: 10.1073/pnas.1413933111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chien Y, et al. Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev. 2011;25(20):2125–2136. doi: 10.1101/gad.17276711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Laberge RM, et al. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat Cell Biol. 2015;17(8):1049–1061. doi: 10.1038/ncb3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dou Z, et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature. 2017;550(7676):402–406. doi: 10.1038/nature24050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Deng W, et al. Antitumor immunity. A shed NKG2D ligand that promotes natural killer cell activation and tumor rejection. Science. 2015;348(6230):136–139. doi: 10.1126/science.1258867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19(3):368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 83.Cizkova M, et al. Gene expression profiling reveals new aspects of PIK3CA mutation in ERalpha-positive breast cancer: major implication of the Wnt signaling pathway. PLoS ONE. 2010;5(12):e15647. doi: 10.1371/journal.pone.0015647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Desmedt C, et al. Multifactorial approach to predicting resistance to anthracyclines. J Clin Oncol. 2011;29(12):1578–1586. doi: 10.1200/JCO.2010.31.2231. [DOI] [PubMed] [Google Scholar]
- 85.Desmedt C, et al. The Gene Expression Grade Index: a potential predictor of relapse for endocrine-treated breast cancer patients in the BIG 1-98 trial. BMC Med Genomics. 2009;2:40. doi: 10.1186/1755-8794-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marty B, et al. Frequent PTEN genomic alterations and activated phosphatidylinositol 3-kinase pathway in basal-like breast cancer cells. Breast Cancer Res. 2008;10(6):R101. doi: 10.1186/bcr2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.