Abstract
Progression of fibrosis and the development of cirrhosis are responsible for the liver-related morbidity and mortality associated with chronic liver diseases. There is currently a great unmet need for effective antifibrotic strategies. Stem cells play a central role in wound-healing responses to restore liver homeostasis following injury. Here we tested the hypothesis that extracellular vesicles (EVs) isolated from induced pluripotent stem cells (iPSCs) modulate hepatic stellate cell (HSC) activation and may have antifibrotic effects. Human iPSCs were generated by reprogramming primary skin fibroblasts. EVs were isolated by differential centrifugation, quantified by flow cytometry (FACS), and characterized by dynamic light scattering and transmission electron microscopy (TEM). Primary human HSCs were activated with TGF-β (10 ng/mL) and exposed to iPSC-EVs. Efficacy of iPSC-EVs was tested on HSCs in vitro and in 2 murine models of liver injury (CCl4 and bile duct ligation). Characterization of iPSC-derived EVs by flow cytometry identified a large population of EVs released by iPSCs, primarily with a diameter of 300 nm, that could be visualized by TEM as round, cup-shaped objects. Fluorescent tracing assays detected iPSC-EVs in HSC cytosol after a short incubation, and EV uptake by HSCs resulted in both decrease of profibrogenic markers α–smooth muscle actin, CollagenIα1, fibronectin, and tissue inhibitor of metalloproteinases–1 and HSC profibrogenic responses, such as chemotaxis and proliferation. Genomics analyses of iPSC-EV miRNA cargo revealed 22 highly expressed miRNAs, among which miR-92a-3p was the most abundant. Transcriptome analysis identified 60 genes downmodulated and 235 upregulated in TGF-β–primed HSCs in the presence or absence of iPSC-EVs. Intravenous injection of iPSC-EVs in CCl4-induced and bile duct ligation–induced liver fibrosis resulted in antifibrotic effects at protein and gene levels. Results of this study identify iPSC-EVs as a potentially novel antifibrotic approach that may reduce or reverse liver fibrosis in patients with chronic liver disease.
Keywords: Cell Biology, Hepatology
Keywords: Fibrosis, Human stem cells
Stem cell–derived extracellular vesicles shuttle miRNAs and reduce hepatic stellate cell activation and liver fibrosis in vivo.
Introduction
Chronic liver disease represents a major cause of morbidity and mortality worldwide (1–3). Fibrosis is an intrinsic response to chronic persistent liver injury that results in a wound-healing process to mitigate the damage but that also can lead to scar formation. Progression of fibrosis and the development of cirrhosis represent the central events responsible for the liver-related morbidity and mortality associated with chronic liver disease. Thus, there is currently a great unmet need for effective antifibrotic strategies. Stem cells, and in particular induced pluripotent stem cells (iPSCs), represent a promising therapeutic approach for various diseases, such as fibrotic diseases (4). Although the mechanisms are only partially understood, studies report that iPSCs play a role in downregulating the expression of proinflammatory and profibrogenic genes (5).
Despite the enormous potential of stem cells as therapeutics, iPSCs remain constrained by safety issues, including immunogenic risks, tumorigenesis potential, and incomplete differentiation, as well as by difficulties in scaling up production (4). These issues may be avoided by generating and harvesting ex vivo stem cell–derived extracellular vesicles (EVs) that can be administered either as an autologous or allogeneic (off-the-shelf) product (6).
EVs are membrane-bound structures efficiently produced and released by parental cells that carry a variety of bioactive molecules, including noncoding RNA (short and long noncoding RNAs), DNA, proteins, and lipids (7, 8). EVs contain a footprint of the cell of origin, they facilitate cell-to-cell communication, and after being internalized by the target cell, they can modulate several cell responses as well as gene expression (9, 10). Extensive studies have highlighted the role of embryonic, hematopoietic, or progenitor stem cell–derived EVs in dedifferentiation and pluripotency (11–16), in healing of infarcted hearts (17), in revascularization of injured tissue (18, 19), and as potential antifibrotic agents. EVs’ relatively small size, prolonged activity after release, composition, and efficient cargo capacity make them less likely to get entrapped in filter organs, more likely to pass through biological barriers, and highly biologically active in cell phenotype regulation. In this study, we explore whether (a) iPSCs produce and release EVs and to what extent, (b) iPSC-EVs can reduce or reverse hepatic stellate cell (HSC) activation in vitro and through which mechanism, (c) bioactive molecules are encapsulated in iPSC-EVs (e.g., miRNAs) and whether a specific miRNA signature can be determined, and (d) iPSC-EVs have therapeutic antifibrotic effects in 2 experimental murine models of liver fibrosis.
Our findings show that iPSCs physiologically produce and release EVs that are internalized by HSCs and modulate the HSC profibrogenic phenotype by reducing the expression of profibrogenic markers. Genomics and efficacy studies reported here show that iPSC-EVs carry a significant number of miRNAs, accumulate in the mouse liver, and reduce liver fibrosis and HSC activation in 2 murine models of liver injury and fibrosis. Altogether these findings suggest that EVs can be isolated from iPSCs and can have potential therapeutic effects for fibrotic diseases, such as chronic liver diseases.
Results
iPSCs produce and release EVs in vitro.
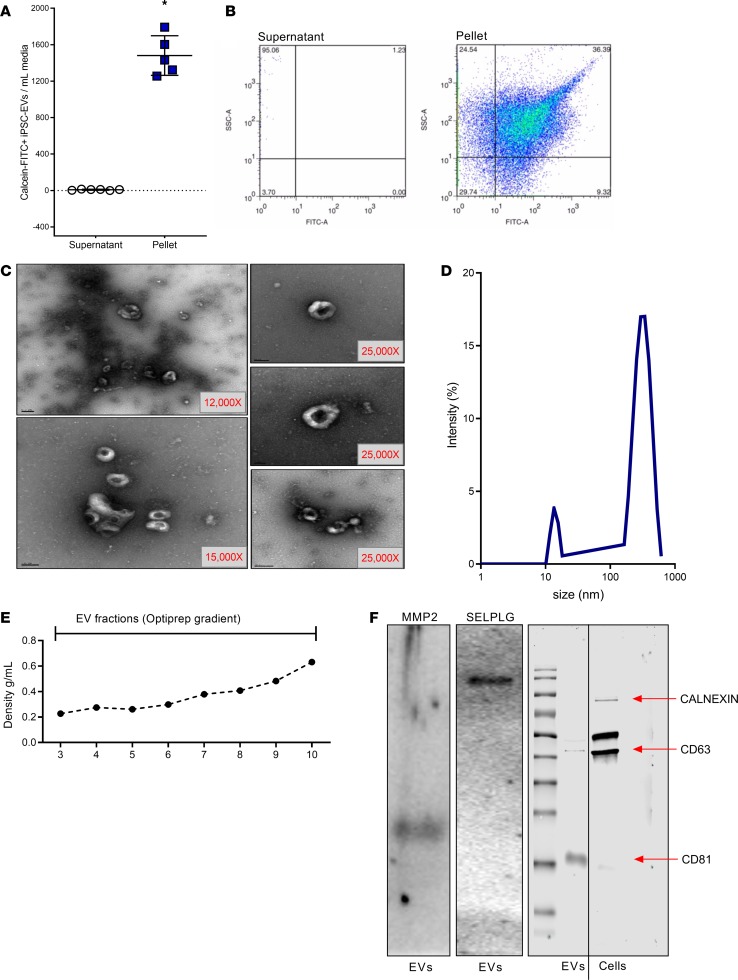
To determine whether iPSCs produce and release EVs in the extracellular space, we collected iPSC-conditioned media and isolated EVs by differential centrifugation. After removal of cell debris and apoptotic bodies, iPSC-EVs were separated from the EV-free supernatant by ultracentrifugation and quantified. Flow cytometry identified a large number of calcein-FITC+ EVs released by iPSCs in culture that were efficiently isolated from the rest of the cell-conditioned supernatant (Figure 1A). Scatter plots generated after sorting the isolated EV pellet confirm the presence of a large number of FITC+ events in a size range between 200 and 1000 nm, as determined by using a cocktail of calibration beads (Figure 1B). The presence and ultrastructure of iPSC-EVs in the pelleted fraction was further assessed by transmission electron microscopy (TEM). Representative microphotographs show a heterogeneous population of round, cup-shaped EVs with diameter of about 300 nm (Figure 1C). The heterogeneous nature of EVs was also confirmed by dynamic light scattering (DLS), which identified an EV size range between 200 and 900 nm in diameter, with a main peak at around 400 nm (Figure 1D). Several elegant studies have identified the presence of a group of surface proteins that can be considered markers of EVs (20, 21). To confirm the presence of some of these surface markers, we applied purified EV samples on an Optiprep density gradient to separate the different populations of vesicles (Figure 1E). Fractions containing purified iPSC-EV fractions were pooled and protein isolation was performed. Protein samples were resolved on an SDS-PAGE, and common markers of EVs were identified, including MMP-2, P-selectin glycoprotein ligand-1 (PSGL1), tetraspanins CD63 (only mildly expressed), and CD81 (Figure 1F). To further confirm the purity of our EV samples and the absence of any cell contamination, we detected cell chaperone Calnexin in iPSC-EVs and in cell lysates, as reported previously (22). Western blot analysis identified Calnexin only in cell lysates but not in the EV sample, indicating that iPSC-EV samples were fairly pure (Figure 1F). Based on the EV isolation and purification techniques available and used for this report, we decided to study the antifibrotic role of a population of EVs with a size ranging between 200 and 1000 nm in diameter. Taken together, these data support our hypothesis and indicate that iPSCs physiologically produce and release a large number of heterogeneous EVs in conditioned media.
Figure 1. Characterization of human iPSC-derived EVs.
(A) Bar graph and (B) scatter plot indicate the level of calcein-FITC+ iPSC-derived EVs isolated by ultracentrifugation (pellet) from pooled iPSC culture media (supernatant). (C) Representative TEM microphotographs of EVs isolated by differential centrifugation from culture media of human iPSCs. Original magnification, ×12–25,000. (D) Curve graph of DLS performed to determine the size of EVs isolated from iPSC-conditioned media. The majority of EVs range between 150 and 1000 nm in diameter. (E) Density (g/mL) of fractions collected after isolation of iPSC-EVs by density gradient centrifugation. (F) Representative immunoblots for purified EV markers MMP-2, PSGL1, CD63, and CD81 and cell marker Calnexin. Values represent mean ± SD from at least 3 independent experiments. *P < 0.05; Kruskal-Wallis test with post hoc Mann-Whitney test and Bonferroni’s correction.
iPSC-derived EVs are internalized by HSCs and reduce the expression of profibrogenic markers.
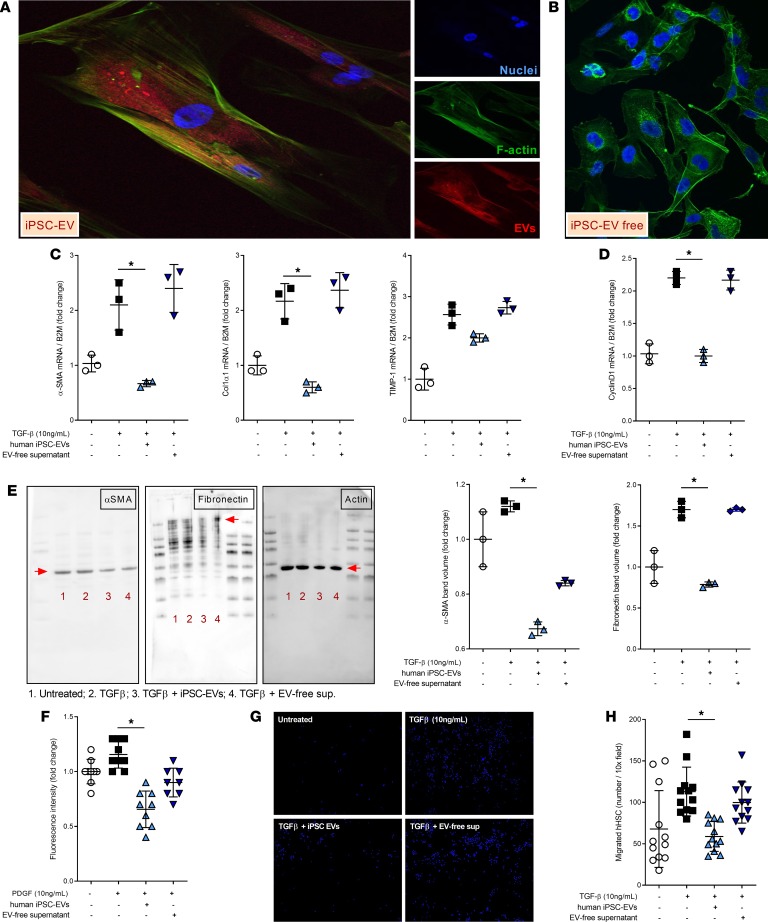
In this study we hypothesize that iPSC-EVs have an antifibrotic or reprogramming capacity that may reverse activation of HSCs. Hence, we investigated the potential effect of EVs released by iPSCs on the profibrogenic mechanisms of primary HSCs, primed with 10 ng/mL TGF-β to enhance their activation. Before studying the iPSC-EV–dependent gene regulation of HSCs, we investigated the mechanism of action (MOA) of iPSC-EVs. Our group and others have demonstrated that cell-derived EVs are effectively engulfed by target cells and modulate a variety of cell responses (23–26). Based on this evidence, we assessed whether this is true for iPSC-EVs as well. As expected, iPSC-EVs fluorescently labeled with PKH26 were internalized by HSCs after 6 hours of exposure, supporting the previously described MOA. Confocal microscopy imaging detected PKH26+ EVs in the cytosol of HSCs, particularly localized in the perinuclear area (Figure 2A), while no PKH26+ EV events were identified in HSCs exposed to EV-free supernatant (Figure 2B). This suggests that HSCs take up iPSC-EVs in vitro and that HSC biology might be regulated by EVs following the internalization. In a previously published study, Kisseleva and colleagues elegantly demonstrated that myofibroblasts revert to inactivated cells where the main profibrogenic genes are significantly downregulated (27). To investigate whether iPSC-EVs could potentially have any effect on the regulation of profibrogenic genes, we performed a focused gene expression analysis of some well-described markers of HSC activation in the presence or absence of iPSC-EVs. The expression of selected genes, such as α–smooth muscle actin (αSMA), CollagenIα1, and tissue inhibitor of metalloproteinases–1 (TIMP-1), was significantly reduced when TGF-β–activated HSCs were exposed to iPSC-EVs for 24 hours compared with HSCs treated with EV-free supernatant (Figure 2C). This suggests that iPSC-EVs may play an important role in posttranscriptional regulation of genes associated with profibrogenic TGF-β–dependent activation of HSCs. In addition to regulating profibrogenic genes, iPSC-EV–treated HSCs also reduced gene expression of cyclin D, a common marker of cell proliferation normally upregulated in activated HSCs during liver fibrosis (Figure 2D). To test the ability of iPSC-EVs to modulate HSC biology posttranslationally, we detected the protein levels of profibrogenic markers αSMA and fibronectin. Immunoblot analyses for these 2 markers showed a mild reduction of protein expression in TGF-β–activated HSCs exposed to iPSC-EVs compared with TGF-β alone or with HSCs exposed to EV-free supernatant (Figure 2E). Based on these findings, we believe that modulation of protein expression by iPSC-EVs may require an incubation longer than 24 hours or that iPSC-EVs may play their antifibrotic role only on a pretranscriptional or immediately posttranscriptional level. Follow-up studies need to be performed to address this hypothesis. As indicated in Kisseleva and colleagues’ report (27), activated HSCs can either revert to an inactivated phenotype or undergo cell death during liver fibrosis resolution. We tested whether treatment of HSCs with iPSC-EVs induced cell death, and we did not observe any apoptosis or necrosis. These findings suggest that iPSC-EVs may play an important role in the phenotypic switch of activated HSCs toward a partially or fully inactivated status that may reverse or resolve liver fibrosis.
Figure 2. iPSC-derived EVs are internalized by HSCs and reduce HSC profibrogenic phenotype.
Representative confocal microphotographs of human HSCs exposed to (A) PKH26-labeled EVs (red) or (B) EV-free supernatant for 6 hours. Original magnification, ×40 (A), ×10 (B) (by confocal microscopy). (C) Quantitative PCR expression graphs of profibrogenic genes α–smooth muscle actin (αSMA), CollagenIα1, and tissue inhibitor of metalloproteinases–1 (TIMP-1) in HSCs primed with the profibrogenic agent transforming growth factor–β (TGF-β) and exposed to iPSC-EVs or EV-free supernatant for 24 hours. β2m, β2-microglobulin. (D) Gene expression levels of proliferation marker cyclin D determined by quantitative PCR. β2m was used as a housekeeping gene for quantitative PCR. (E) Representative Western blot gels and corresponding densitometry analyses of fibrosis markers αSMA and fibronectin in HSCs primed with TGF-β and exposed to iPSC-EVs or EV-free supernatant for 24 hours. Actin was used as a loading control. (F) Quantification graph of proliferation assay assessed in human HSCs primed with TGF-β and exposed to iPSC-EVs or EV-free supernatant for 48 hours. (G) Representative microphotographs and (H) quantitation histogram of chemotaxis assay performed on human HSCs primed with TGF-β and exposed to iPSC-EVs or EV-free supernatant for 6 hours. Original magnification, ×10. Values represent mean ± SD from at least 3 independent experiments. *P < 0.05; Kruskal-Wallis test with post hoc Mann-Whitney test and Bonferroni’s correction were used for statistical analysis.
iPSC-EVs reduce functional HSC proliferation and chemotaxis in vitro.
Initiation and perpetuation of liver fibrosis are characterized by an initial genetic switch of quiescent adipocyte-like HSCs to myofibroblast-like activated and αSMA+ HSCs. HSC transdifferentiation during liver injury results in activation of various profibrotic cell responses that sustain and perpetuate liver fibrosis. Among several profibrogenic cell responses, HSC proliferation and chemotaxis are particularly crucial for the severity of fibrosis. Based on the importance of HSC proliferation and chemotaxis for liver fibrosis, we investigated whether iPSC-EVs could block or reduce functional HSC responses, such as proliferation and chemotaxis. Established proliferation BrdU assay performed on HSCs indicated that proliferation of activated HSCs was significantly reduced after exposure to iPSC-EVs for 48 to 72 hours (Figure 2F). Equally important during perpetuation of liver fibrosis is HSC-oriented migration toward the area of injury. Similarly as assessed for proliferation, we investigated the potential inhibition of migration by iPSC-EVs. HSCs primed with TGF-β and exposed to iPSC-EVs showed a markedly decreased chemotaxis compared with TGF-β–primed HSCs exposed to EV-free supernatant. A qualitative staining of the nuclei of HSCs trapped in the Transwell filter showed how many cells migrated in the presence of EV-free supernatant while only few cells migrated in the presence of iPSC-EVs, despite the exposure to TGF-β in both conditions (Figure 2, G and H). These results indicate that iPSC-EVs not only regulate HSC markers’ expression but may also regulate secondary cell responses, such as proliferation and migration, which ultimately perpetuate and sustain the progression of liver fibrosis.
Administration of murine iPSC-EVs shows efficacious therapeutic effects in experimental models of liver injury and fibrosis.
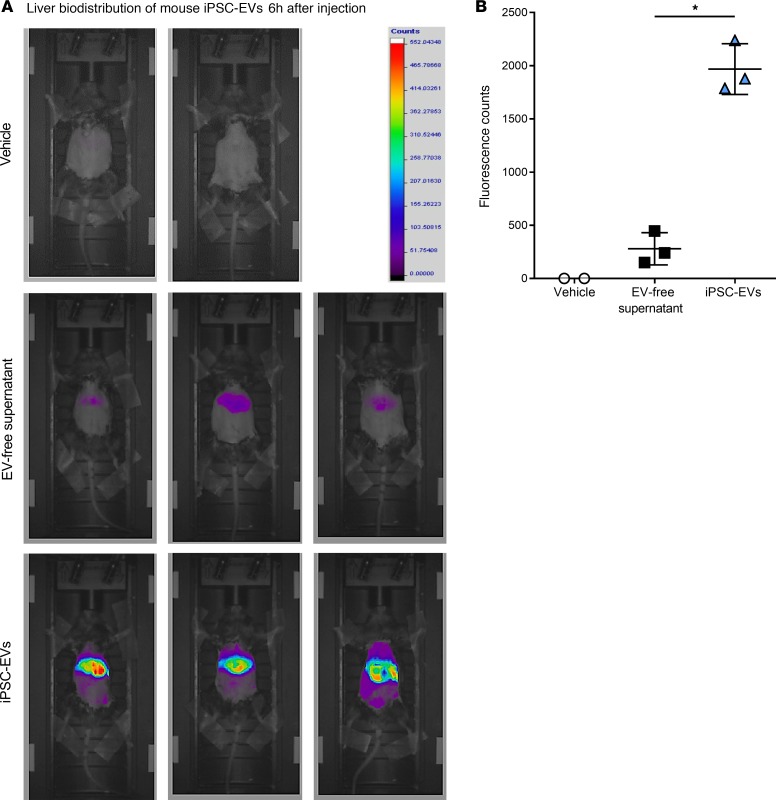
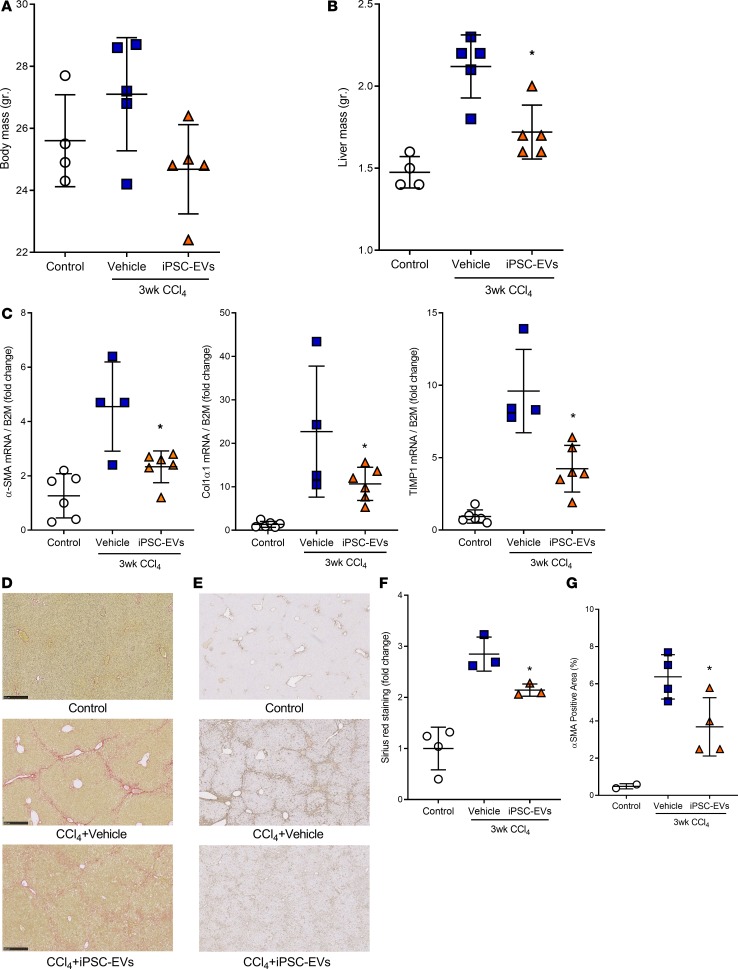
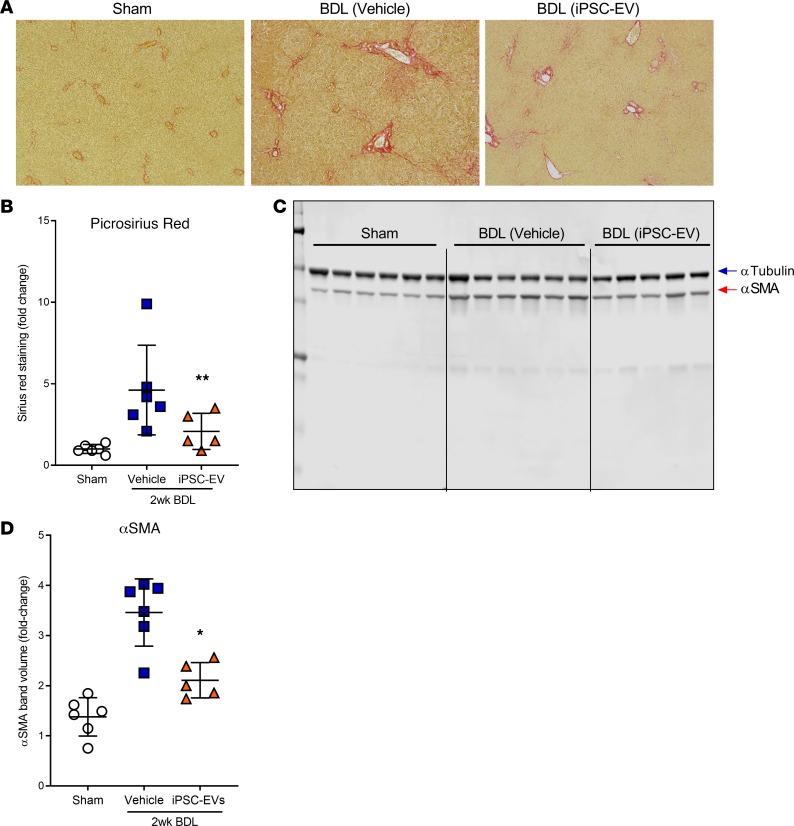
Based on the positive in vitro data, we investigated the potential therapeutic role of iPSC-EVs in 2 experimental in vivo models of liver fibrosis. Before performing our efficacy studies, we measured the liver accumulation of iPSC-EVs after intravenous (i.v.) injection. We intravenously injected C57BL/6 mice with a single volume of DiR-labeled murine iPSC-EVs (m–iPSC-EVs) by tail vein. Six hours after a single i.v. injection of 1.5 × 106 DiR+ m–iPSC-EVs, most of the fluorescent signal was localized in the liver and only slightly in the spleen. EVs were not detected in any other organ or tissue. Mice injected with control vehicle and DiR+ EV-free supernatant showed only minimal or did not show any fluorescent signal compared with EV-treated mice (Figure 3, A and B). Based on these encouraging results, we tested the efficacy of m–iPSC-EVs in a CCl4-induced murine model of liver injury and fibrosis. Mice were gavaged with CCl4 for 3 weeks and were administered m–iPSC-EVs or vehicle 3 times a week for the last 2 weeks of the study. Macroscopic postmortem assessments of body and liver mass showed reduced body and liver mass in CCl4-EV mice, compared with CCl4-saline mice (Figure 4, A and B). Molecular analysis of whole-liver gene expression showed a significant downregulation of key profibrogenic genes αSMA, CollagenIα1, and TIMP-1 in mice injected with m–iPSC-EVs compared with vehicle-injected mice (Figure 4C). To have a closer look at liver fibrosis, we stained liver sections and measured collagen types I and III fiber deposition by Picrosirius red. Quantification of Picrosirius red staining showed a significant decrease of collagen deposition in livers of mice injected with m–iPSC-EVs compared with saline-injected mice (Figure 4, D and F). Administration of EVs did not fully prevent or reverse liver fibrosis but reduced it significantly by about 20% to 30%. Additional assessment of HSC activation by immunohistochemistry for αSMA confirmed a 40% decrease in activated αSMA+ HSCs in the livers of mice injected with m–iPSC-EVs compared with vehicle-administered mice (Figure 4, E and G). To test the efficacy of m–iPSC-EVs in a second model of cholestatic liver fibrosis, we performed a double ligation of common bile duct (BDL) in mice for 2 weeks. The last 6 days of the study, mice were administered daily, by tail vein, 1.5 × 106 murine iPSC-EVs or vehicle control. Assessment of collagen types I and III fiber deposition by Picrosirius red staining showed approximately 2-fold reduction of collagen in livers of mice treated with iPSC-EVs compared with vehicle-treated mice (Figure 5, A and B). Immunostaining for HSC activation marker αSMA confirmed iPSC-EV–dependent decrease of HSC activation in livers, even though HSC activation was not completely abrogated (Figure 5, C and D). A longer exposure to a higher number of iPSC-EVs may result in better efficacy, but we believe that the antifibrotic signals we observed are promising and open the field to follow-up studies testing iPSC-derived or other stem cell–derived EVs as antifibrotic agents for various chronic liver diseases, such as nonalcoholic and alcoholic steatohepatitis.
Figure 3. Liver accumulation of fluorescently labeled m–iPSC-EVs in vivo.
(A) In vivo fluorescence microphotographs and (B) quantitation of liver accumulation of fluorescently labeled m–iPSC-EVs (n = 3 mice) 6 hours after their administration by tail vein in C57BL/6 wild-type mice. EV-free supernatant (n = 3 mice) and vehicle (n = 2 mice) were used as controls. Values represent mean ± SD from at least 3 independent experiments. *P < 0.05; Kruskal-Wallis test with post hoc Mann-Whitney test and Bonferroni’s correction were used for statistical analysis.
Figure 4. Intravenous administration of m–iPSC-EVs mitigates development of fibrosis and HSC activation in a CCl4 experimental model of hepatic fibrosis.
(A) Body mass and (B) liver mass of C57BL/6 mice exposed twice a week to CCl4 for 3 weeks that received 1.5 × 106 EVs/100 μL or vehicle control (n = 5 mice/group) 3 times a week after 1 week of CCl4 and for the last 2 weeks. (C) Quantitative PCR expression graphs of profibrogenic genes αSMA, CollagenIα1, and TIMP-1 in livers harvested from control mice or CCl4-treated mice that received tail vein injections of iPSC-EVs or vehicle. β2m was used as a housekeeping gene. Representative microphotographs (original magnification, ×10) and corresponding quantification graph of (D and F) Picrosirius red staining for determination of collagen deposition and (E and G) immunohistochemistry for HSC activation marker αSMA in liver specimens harvested from mice administered CCl4 for 3 weeks and treated with mouse iPSC-EVs or vehicle for the last 2 weeks of the study. Values represent mean ± SD. *P < 0.05; Kruskal-Wallis test with post hoc Mann-Whitney test and Bonferroni’s correction were used for statistical analysis.
Figure 5. Intravenous administration of m–iPSC-EVs reduces development of fibrosis and HSC activation in an experimental model of cholestatic liver fibrosis.
(A) Representative microphotographs and (B) corresponding quantification graph of Picrosirius red staining of liver sections harvested from mice that received common bile duct ligation (BDL) for 2 weeks (n = 5–6 mice/group) and were administered, by tail vein, mouse iPSC-EVs (BDL iPSC-EVs) or vehicle control (BDL vehicle) for the last 6 days of the study. Mice that underwent abdomen laparotomy but did not receive BDL were used as no-disease controls (sham). Original magnification, ×10. (C) Immunoblots and (D) corresponding quantification graph of αSMA protein levels over 2 weeks in BDL mice treated with mouse iPSC-EVs or with vehicle for the last 6 days of the study. Tubulin was used as a loading control for Western blots. Values represent mean ± SD. *P < 0.05, and **P < 0.005; Kruskal-Wallis test with post hoc Mann-Whitney test and Bonferroni’s correction were used for statistical analysis.
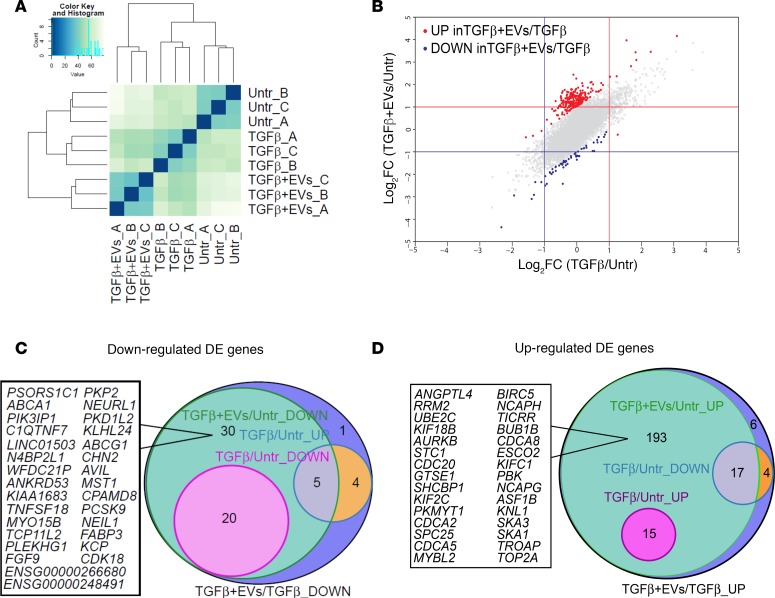
Transcriptomic profile of activated HSCs and miRNA profile of iPSC-EVs show a potential modulation of the profibrogenic machinery in activated HSCs.
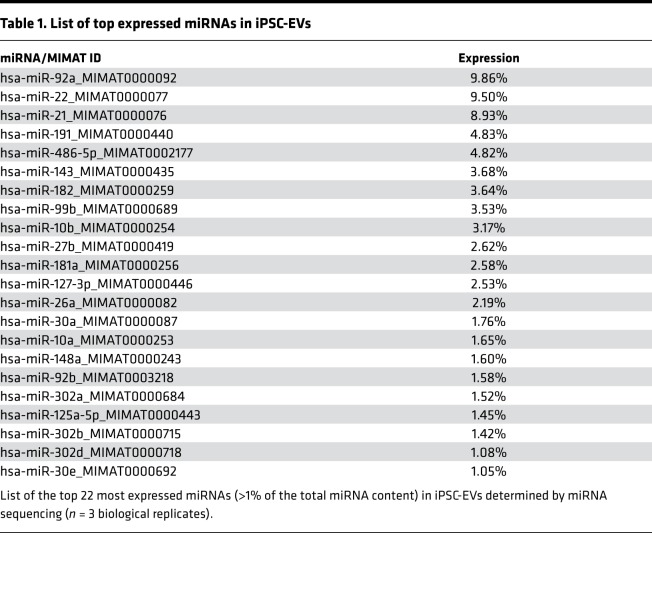
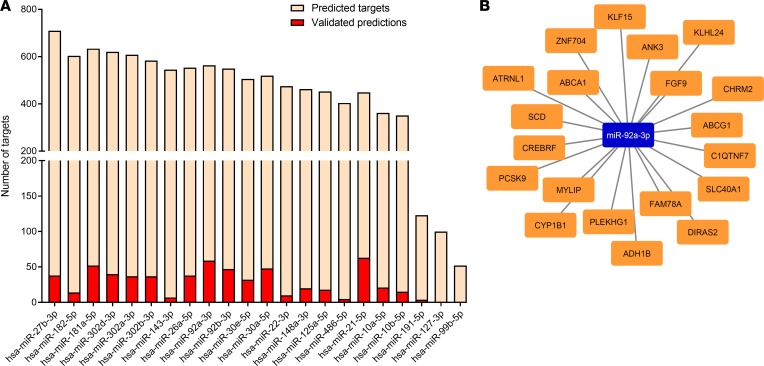
Based on our in vitro and in vivo results, we investigated the potential regulatory role of iPSC-EVs in the HSC gene profile. A complete transcriptomic analysis was conducted on activated HSCs exposed to iPSC-EVs. RNA-Seq was performed in triplicate on untreated or TGF-β–activated HSCs in the presence (TGF-β plus iPSC-EVs) or absence (TGF-β) of iPSC-derived EVs. The Euclidean clustering of gene expression levels showed a good reproducibility within the experimental triplicates and a clear separation between the 3 experimental groups (Figure 6A). As the first step, we evaluated the log2 fold change (log2FC) generated when comparing TGF-β–treated cells with untreated HSCs (x axis) and TGF-β plus iPSC-EV–treated cells with untreated HSCs (y axis). Both treatment conditions led to changes of gene expression ranging from –4 to 4.5 log2FC, when compared with untreated cells. Importantly, the observed changes in gene expression were also highly correlated, as indicated by the diagonal shape on the dot plot (Figure 6B). Then we compared TGF-β–activated HSCs exposed to iPSC-EVs with cells exposed to TGF-β only. Our analysis identified 60 downregulated genes (marked in blue) and 235 upregulated genes (marked in red; Figure 6B). We then subclassified the 60 genes downregulated in TGF-β–activated HSCs with iPSC-EVs versus TGF-β–activated HSCs without iPSC-EVs and compared the gene expression pattern in each of the 2 groups with that of untreated cells. This analysis showed 20 genes already downmodulated at baseline by the TGF-β exposure itself, while 30 genes were specifically downmodulated by the exposure to iPSC-EVs (Figure 6C). Among the remaining 10 genes, 9 were upmodulated by TGF-β treatment itself while 1 gene was unchanged between untreated cells and those exposed to TGF-β with or without iPSC-EVs. The same subclassification was performed for the 235 upregulated genes. This analysis showed that 15 genes were already upregulated by TGF-β exposure alone while 193 genes were specifically upregulated in human HSCs preactivated with TGF-β and then incubated with iPSC-EVs. Among the remaining 27 genes, 21 were downmodulated by the pretreatment with TGF-β alone, and 6 genes were completely unchanged between treated and untreated cells (Figure 6D). Data comparison of the transcriptomic profiles of activated human HSCs exposed or not to iPSC-EVs suggests that iPSC-EVs may play a crucial role in the regulation of profibrogenic genes (e.g., collagens, growth factors, or TIMPs), whose expression may be downmodulated or switched off by bioactive material shuttled by EVs and released in the target cells. Extensive data from our group and other groups showed that EVs carry various bioactive molecules, such as proteins, coding and noncoding RNAs, and lipids (28, 29). Based on this, we performed miRNA sequencing of iPSC-EVs, and we identified the most abundant EV miRNAs. About 700 miRNAs were identified in iPSC-EVs. However, considering that highly abundant miRNAs shuttled into the target cells are more likely to regulate the cell gene expression machinery, because they compete with endogenous miRNAs, we selected only the miRNAs representing at least 1% of the total miRNA content for further characterization. This selection resulted in 22 miRNAs (Table 1), and numerous validated and predicted targets were identified among the genes downmodulated (adjusted P value < 0.01) following TGF-β plus EV exposure (Figure 7A). Among many, miR-92a-3p was the top expressed miRNA, with almost 10% of total miRNAs and an abundant series of predicted targets among the downregulated DEGs (Figure 7B). Other miRNAs abundantly expressed and of interest for our hypothesis include miR-10b-5p, miR-302-3p, and miR-92b-3p, which all have potential antifibrotic properties. These findings suggest that iPSC-EVs may influence the pro- or antifibrotic gene machinery of HSCs by transferring bioactive molecules, such as miRNAs, that may modulate HSC biology.
Figure 6. Differentially expressed genes in TGF-β–activated HSCs in the presence or absence of iPSC-EVs.
(A) Euclidean clustering of the samples based on variance stabilizing transformation. (B) Effect of iPSC-EV treatment on HSCs. Gray circles represent log2FC values of TGF-β–activated versus untreated HSCs in the presence (x axis) or absence (y axis) of iPSC-EVs for each analyzed gene. Differentially expressed genes (DEGs) induced (red dots) or repressed (blue dots) in the comparison between TGF-β–activated HSCs stimulated with iPSC-EVs and TGF-β–activated HSCs without iPSC-EVs (log2FC > 1; adjusted P value < 0.01). Gray dots correspond to nondifferential genes. Venn diagrams reporting the number of DEGs (C) upregulated and (D) downregulated in HSCs activated with 10 ng/mL of TGF-β and stimulated with iPSC-EVs, compared with unstimulated cells. Genes were further assigned into subgroups according to their expression trend, based on their log2FC with P value less than 0.01 and with respect to the untreated cells.
Table 1. List of top expressed miRNAs in iPSC-EVs.
Figure 7. Analysis of top expressed miRNAs in iPSC-EVs.
(A) Histogram integrating mRNA and miRNA sequencing results. For each of the 22 most expressed miRNAs, we generated the list of the target genes predicted by at least 2 databases among Targetscan, Miranda, DIANA-microT, PITA, Pictar, and RNA22, as well as the list of the validated target genes identified by Tarbase. Bars represent the number of downmodulated (P value < 0.01) genes in TGF-β–activated HSCs, stimulated versus unstimulated, with iPSC-EVs subdivided into predicted genes (pink) and validated genes (red). (B) miRNA target network of miR-92a-3p–predicted targets showing log2FC less than –1 in the comparison between TGF-β plus EVs versus TGF-β only.
Discussion
The main findings of this study provide insights on the molecular signature and on the modulatory mechanisms of stem cell–derived EVs on HSC biology and liver fibrosis, suggesting a potential role for stem cell–derived EVs as novel therapeutic biologics for treatment of liver fibrosis. Our complete characterization analyses show that iPSCs physiologically produce and release EVs that can be isolated from cell supernatant, identified and quantified by flow cytometry, and imaged by electron microscopy. An additional purification and characterization of iPSC-EVs identified some key EV protein markers commonly described on cell-derived vesicles in various studies (21), which confirms the ability of iPSCs to produce and release EVs. The regenerative potential of stem cell–derived EVs has been previously described mainly in mesenchymal stem cells (6) and in various diseases, but very little is currently known about iPSC-derived EVs and their potential role in liver fibrosis. Using iPSCs would reduce or perhaps eliminate risks related to undesired immune response after a cell transplant, risk of tumorigenesis, and issues related to cell sourcing and production. HSCs are vitamin A–storing cells in normal liver but transdifferentiate in myofibroblast-like cells, producing and releasing extracellular matrix, during liver injury. This transdifferentiation initiates and sustains liver fibrosis, which is associated with high liver-related morbidity and mortality if not reversed or resolved. A plethora of very elegant studies have demonstrated that HSC activation and perpetuation is multifactorial and a variety of signaling pathways, receptors, cytokines, and MAPK families are involved synergistically, antagonistically, or additively. We hypothesize that using biologically produced particles that contain different bioactive molecules (miRNAs, proteins, and lipids), and are biologically active and stable upon release, may modulate HSC biology. As a consequence, this EV-dependent mechanism may lead to greater efficacy than small molecules or synthetic biologics. Additionally, small membrane structures, like EVs that have a diameter between 100 and 1000 nm, elicit effective penetration properties, can efficiently shuttle bioactive molecules, can penetrate cells by endocytosis or through receptors, and are very stable compared with large stem cells that have been used experimentally for regenerative medicine. Through our initial characterization of iPSC-EVs, we were able to detect EVs by flow cytometry at an average concentration of 600 EVs/μL of iPSC media. Our initial analyses were further supported by morphological and molecular data, which not only identified EVs by electron microscopy, providing important information on the physical aspect of EVs, but also detected key EV protein markers, confirming the presence of EVs in our samples and excluding any cell contamination. In our previous reports, we demonstrated that EVs are actively and efficiently incorporated in target cells and modulate their phenotype. Here, we could demonstrate a similar mechanism, where primary human HSCs take up iPSC-EVs shortly after being exposed to EVs. This uptake resulted in a significant reduction of the key profibrogenic genes and proteins, such as αSMA, CollagenIα1, TIMP-1, and fibronectin, in HSCs activated by the most potent profibrogenic agent, TGF-β. iPSC-EVs not only modulated gene expression but also regulated and, interestingly, reduced crucial HSC responses occurring during liver fibrosis, such as proliferation and chemotaxis. After initial liver damage, HSCs undergo a phase of activation, which includes cell expansion and migration toward the area of injury to release matrix that will replace the damaged tissue. As a consequence, blocking or reducing these 2 responses of activated HSCs is extremely important for antifibrotic therapeutic interventions. The promising and interesting in vitro data needed to be confirmed in vivo in an experimental murine model of liver fibrosis. A biweekly injection of mouse iPSC-EVs in a CCl4-induced liver fibrosis model for 3 weeks resulted in significant improvement of liver functionality, reduction of hepatic expression of key profibrogenic genes and proteins, reduction of hepatic collagen deposition, and HSC activation by about 30% to 40%, which is particularly marked when considering the severity of the CCl4 model and the short administration of iPSC-EVs. Similar antifibrotic results were obtained in a cholestatic liver fibrosis model (BDL) for 2 weeks, where iPSC-EVs were injected once daily for the last 6 days of the study. Future studies on chronic liver diseases, such as nonalcoholic and alcoholic steatohepatitis, are warranted.
In this proof-of-concept report, we provide further insights into iPSC-EVs’ cargo and potential gene regulation in HSCs. By using a nonbiased approach, we performed miRNA profiling of iPSC-EVs and identified a signature of the most abundant miRNAs. Bioinformatics analysis showed that miR-92a-3p was the top expressed in iPSC-EVs (Table 1). A previous report showed that miR-92a-3p and miR-302-3p, another miRNA particularly abundant in our iPSC-EV profile, are highly and specifically expressed in iPSCs, have differentiation capacity, and may trigger HSC differentiation into an inactive or quiescent-like phenotype (30). In previous studies, miR-92a-3p elicited antifibrotic effects by suppressing profibrotic mediators in both human fibroblasts (31) and a bleomycin model of lung fibrosis with miR-92b-3p, also abundant in our iPSC-EVs (32). Other miRNAs with antifibrotic effects include miR-26a-5p, which reduces both phosphorylation of SMAD3 and connective tissue growth factor (CTGF), attenuating fibroblast transdifferentiation (33) and cyclin D2 and TGF-β receptors and suppressing cell proliferation (34); miR-27b-3p, which reduces Gremlin 1, attenuating epithelial-mesenchymal transition (35); miR-22-3p, which negatively regulates angiotensin II–induced cardiac fibrosis by suppressing the expression of TGF-β receptor type I in the heart (36); and miR-486-5p, which targets and suppresses SMAD2, decreasing lung lesions in lung fibrosis (37). We also performed an RNA-Seq analysis of HSCs exposed to TGF-β with or without iPSC-EVs. We found that many genes commonly associated with fibrosis were downregulated in TGF-β–activated HSCs exposed to iPSC-EVs compared with TGF-β–treated cells alone. Our bioinformatics analyses identified several DEGs, up- or downregulated in TGF-β–treated HSCs or TGF-β plus iPSC-EV–treated cells. These findings suggest that iPSC-EVs can modulate activated HSCs in vitro and that, at least in part, the antifibrotic effects observed in vivo with iPSC-EV treatment might be mediated via shuttling of specific miRNAs into HSCs. A particularly important aspect of this study is that fluorescently labeled iPSC-EVs could be detected mainly in the liver only 6 hours after i.v. injection in wild-type mice. This is a very important proof of concept demonstrating that EVs accumulate in the liver in vivo, supporting a potential use of iPSC-EVs as therapeutic biologics for liver diseases. On the other hand, further studies are needed to elucidate EV cellular localization after they accumulate in the liver. In summary, our study identifies iPSC-derived EVs as potential novel antifibrotic biologics and has important translational implications for the treatment of various liver diseases associated with fibrosis.
Methods
Animal studies.
Male C57BL/6 wild-type mice, 7 to 8 weeks old, were purchased from The Jackson Laboratory and were maintained under specific pathogen–free conditions at UCSD with ad libitum access to water and normal chow. For EV in vivo tracing studies, murine iPSC–conditioned media was centrifuged at 3000 g for 15 minutes at 10°C to remove cell debris (pellet). Supernatant was transferred in new tubes and incubated with 1 μM of the lipophilic carbocyanine DiOC18 (7) or DiR dye (Thermo Fisher Scientific) at room temperature for 15 minutes to label EVs present in the supernatant. DiR+ EVs were then isolated by ultracentrifugation at 100,000 g for 1.5 hours at 4°C. Pelleted EVs were resuspended in 100 μL of sterile saline solution, while supernatant was concentrated in Amicon Ultra 0.5-mL centrifugal filters with 3 kDa MWCO (MilliporeSigma) to 100–200 μL and used as EV-free control. Mice (n = 2–3/group) received a single injection of 1.5 × 106 DiR+ iPSC-EVs per mouse by tail vein or 100 uL of DiR+ cell-free supernatant or 100 μL of saline. Mice were imaged under anesthesia 1 hour or 6 hours after injection with the eXplore Optix in vivo preclinical fluorescence imager (GE Healthcare). Liver injury was induced in mice by oral gavage of 10 μL/g of CCl4 in mineral oil (MilliporeSigma) 3 times a week for 3 weeks. After 1 week of CCl4 administration, mice (n = 5/group) received 1.5 × 106 murine iPSC-EVs/100 μL of saline or vehicle control by tail vein twice a week for the last 2 weeks of the study. In addition, cholestatic liver injury and fibrosis was induced in mice by BDL for 2 weeks, as described previously (38, 39). A group of control mice (sham control) underwent abdominal laparotomy but no BDL was performed. Nine days after surgery, BDL mice (n = 5–6/group) were injected by tail vein with 1.5 × 106 murine iPSC-EVs/100 μL saline or vehicle control once daily for the last 6 days of the study. At the end of each study, mice were euthanized by intraperitoneal injection of a mixture of 100 mg/kg of ketamine and 10 mg/kg of xylazine dissolved in a 0.9% saline solution (23). Body weight and liver weight were determined and tissues harvested for molecular and histopathological assessments.
Cell culture.
Human primary HSCs were provided in-house at the UCSD and were cultured in DMEM supplemented with 10% FBS and antibiotic/antimycotic (Gibco). Human and mouse iPSCs were provided by Lawrence Goldstein at the UCSD and cultured as previously reported (40).
Isolation and characterization of iPSC-derived EVs.
Cell culture medium was harvested every 24 to 48 hours from unstimulated, normally cultured human or mouse iPSCs and stored at –80°C until isolation of EVs. EVs were isolated as described previously (9). Briefly, iPSC-conditioned medium was centrifuged for 15 minutes at 3000 g to remove cell debris and aggregates. Supernatant was transferred into new tubes and ultracentrifuged at 100,000 g for 90 minutes at 4°C by Beckman L8-70M ultracentrifuge using the SW41 Ti swinging rotor (Beckman Coulter). EV-free supernatant was collected and used as negative control for in vitro assays. Pelleted EVs were resuspended in PBS for characterization, in FBS-free media for functional in vitro studies or in saline solution for in vivo studies. The number of calcein-positive iPSC-EVs per microliter of media was determined by flow cytometry using a BD LSRII FACS machine, as described previously (23). EV size was determined by measuring DLS with the Zetasizer system (Malvern Panalytical), and EV morphology was determined by TEM provided by the TEM facility at UCSD, as previously described (23, 41). For additional characterization, isolated iPSC-EVs were further purified by using Optiprep density gradient medium (MilliporeSigma) for detection of EV protein markers. Briefly, pelleted EVs were resuspended in 500 μL of PBS and slowly seeded on a gradient of 40%, 20%, 10%, and 5% of Optiprep medium dissolved in 0.25 mM sucrose, 1 mM EDTA, and 10 mM Tris-HCl. EV samples were ultracentrifuged at 100,000 g for 18 hours at 4°C. A volume of 1 mL per fraction was collected, transferred into a new tube, and diluted in 12 mL of PBS. Small vesicles eluted in fractions 6–7 (density: 1.10 g/mL), while large vesicles eluted in fractions 9–10 (density: 1.14–1.20 g/mL). Diluted Optiprep fractions in PBS were further ultracentrifuged at 100,000 g for 3 hours at 4°C to remove any Optiprep residues. Pelleted and Optiprep-free EVs were resuspended in RIPA lysis buffer for protein isolation and Western blot analyses of EV protein markers.
Internalization of iPSC-EVs.
An aliquot of purified iPSC-EVs was used for EV internalization into HSCs. EVs were resuspended in PBS containing the fluorescent tracing dye PKH26 (MilliporeSigma) and were labeled according to the manufacturer’s instructions. PKH26-labeled iPSC-EVs were incubated with 1.5 × 105 serum-starved HSCs for up to 6 hours in a 4-well tissue culture slide. Cells were washed with cold PBS and fixed/permeabilized with 4% paraformaldehyde. To visualize F-actin fibers in cytosol and cell nuclei, cells were stained with FITC-labeled phalloidin (MilliporeSigma) and 4′,6-diamidino-2-phenylindole (DAPI) (Vector Labs), respectively. Cells were imaged at ×10 or ×40 by Olympus FV1000 Spectral Confocal microscope.
RNA isolation and quantitative real-time PCR.
Unlabeled human iPSC-EVs or EV-free supernatant was incubated with human HSCs in combination with 10 ng/mL of profibrogenic agent TGF-β (R&D Systems, 240-B) for 24 hours at 4°C. Cells or liver samples were homogenized in TRIzol, and total RNA was isolated using RNeasy kit (Qiagen) and reverse-transcribed by iScript cDNA synthesis kit (Bio-Rad) according to the manufacturers’ instructions and as previously reported (9). Quantitative real-time PCR was performed on a Bio-Rad Cycler by using SYBR Green real-time PCR master mix (Kapa Biosystems, Roche) according to the manufacturers’ instructions. The housekeeping gene β2m was used as an internal control. The PCR primers used to amplify each gene are listed in Supplemental Table 1 (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.125652DS1).
Immunoblotting.
Unlabeled human iPSC-EVs or EV-free supernatant was incubated with human HSCs in combination with 10 ng/mL of profibrogenic agent TGF-β (R&D Systems) for 24 hours at 4°C. Total protein content of purified iPSC-EVs and HSCs was isolated and quantified, respectively, by Micro BCA and BCA Protein assay kit (both from Thermo Fisher Scientific), according to the manufacturer’s instructions. For immunoblotting, equivalent amounts of protein were separated by SDS-PAGE and blotted onto nitrocellulose membranes. Primary antibodies anti-MMP2 (ABclonal Biotech, catalog A-6247, lot P00009, rabbit polyclonal, 1:100), anti-PSGL1 (ABclonal Biotech, catalog A-1660, lot P00009, rabbit polyclonal, 1:100), anti-Calnexin (Santa Cruz Biotechnology, catalog sc-23954, mouse monoclonal, 1:200), anti-CD63 (GeneTex, catalog GTX37555, rabbit polyclonal, 1:100–1000), anti-CD81 (GeneTex, catalog GTX31381, rabbit polyclonal, 2.5 μg/mL), anti-αSMA (MilliporeSigma, catalog A2547, monoclonal mouse, 1:500), antifibronectin (Abcam, catalog Ab2413, monoclonal rabbit, 1:1000), and anti–type I collagen (SouthernBiotech, catalog 1310-01, goat polyclonal, 1:200) were used for immune detection with HRP-conjugated secondary antibodies (1:5000, Cell Signaling Technology). Proteins were visualized by Supersignal West Pico chemiluminescence reagents (Pierce Biotechnology). Western blot data were obtained from 3 independent experiments, and representative gels are reported in the figures.
Migration assay.
Chemotaxis was assessed in human primary HSCs by using the Transwell chamber assay, as described previously (9). Briefly, each well (lower chamber) of a 24-well plate was filled with 500 μL of FBS-free DMEM with or without 10 ng/mL TGF-β (R&D Systems, catalog 240-B). Transwell polycarbonate membrane cell culture inserts (Corning) were placed on top of each well (upper chamber) of the 24-well plate, and a mixture of 5 × 104 HSCs with or without iPSC-EVs or EV-free supernatant was added on the insert (upper chamber). Plates were incubated for 6 hours at 37°C, and then the filters were removed and stained with Vectashield mounting medium with DAPI (Vector Labs). Migrated cells were detected with a fluorescence microscope and DAPI+ nuclei counted.
Proliferation assay.
HSC proliferation was assessed by detecting DNA synthesis as incorporation of BrdU using CyQuant Direct Cell Proliferation Assay (Thermo Fisher Scientific) according to the manufacturer’s instructions. Briefly, HSCs were plated onto a 96-well plate (0.15 × 105 cells/well), treated with or without 10 ng/mL of PDGF (R&D Systems, 220-B), and exposed to iPSC-EVs or EV-free media for 48 hours at 37°C. Fluorescence was detected by Eppendorf M2 plate reader, and relative fluorescence units were reported in the quantification graphs as FC from control.
Apoptosis/necrosis assay.
Apoptosis-associated cytotoxicity events and cell death due to necrosis were detected simultaneously with the Necrosis vs. Apoptosis Assay kit (Immunochemistry Technologies). Primary human HSCs were plated in a 24-well plate and treated with or without 10 ng/mL TGF-β, followed by exposure to iPSC-EVs or EV-free supernatant for 16 hours in FBS-free media. Necrosis and apoptosis were assessed according to the manufacturer’s instructions. Incubation with 1 μM staurosporine (MilliporeSigma) for 5 hours was used as a positive control for apoptosis while 90% ethanol for 5 minutes was used as a positive control for necrosis. Stained cells were imaged by epifluorescence on an Olympus microscope and representative images reported.
miRNA sequencing and data analysis.
Isolated iPSC-EVs (n = 3 biological replicates) were resuspended in 700 μL of QIAzol lysis reagent (Qiagen), and total RNA, including small RNA, was isolated by miRNeasy Mini kit (Qiagen). Quantity and quality of small RNA were assessed by a NanoDrop ND-1000 and Agilent Tapestation, respectively. Libraries were generated from 1 to 5 μg of total RNA, using TruSeq SmallRNA Sample Prep Kit (Illumina), following the manufacturer’s instructions. Library quality was determined by High Sensitivity DNA Kit (Agilent). Libraries were multiplexed and sequenced at 13 pM to an average depth of 7.5 M reads with 50–base pair (50-bp) single-end (SE) reads on an Illumina HiSeq 4000. The raw sequencing reads in FASTQ format were preprocessed, aligned, quantified, and normalized as previously described (42). The top 22 most expressed miRNAs (>1% of the total miRNA content) were determined and listed in Table 1. Complete FASTQ files of miRNA sequencing were deposited in the National Center for Biotechnology Information’s (NCBI’s) SRA (Bioproject ID PRJNA497133) and can be viewed at this link: http://www.ncbi.nlm.nih.gov/bioproject/497133 A complete list of miRNAs identified in iPSC-EVs has been deposited in figshare and can be viewed at this link: https://doi.org/10.6084/m9.figshare.7215356.v1
RNA-Seq and data analysis.
HSCs were incubated with or without (untreated) 10 ng/mL of TGF-β, in the presence or absence of iPSC-EVs for 24 hours (n = 3/group). Total RNA was isolated as described in RNA isolation and quantitative real-time PCR. Quantity and quality of RNA were determined by a NanoDrop ND-1000 and Agilent Tapestation, respectively. Libraries were generated from 2 to 3 μg of total RNA using TruSeq SmallRNA Sample Prep Kit (Illumina), following the manufacturer’s instructions. Library quality was determined by High Sensitivity DNA Kit (Agilent). RNA samples were sequenced by using a HiSeq 4000 instrument (Illumina). For each sample and replicate, a minimum of 25 M of 50-bp-long SE reads were generated, aligned to the human genome using STAR (43), and assigned to genes using HtSeq (44) and human genome annotation (GENCODE release 19). Differential expression analysis was performed in R environment using DeSEQ2 package (45). DEGs were defined using log2FC more than 1 and adjusted P value less than 0.01 as cutoff for statistical significance. Complete FASTQ files of mRNA-Seq were deposited in the NCBI’s SRA (Bioproject ID PRJNA497290) and can be viewed at this link: http://www.ncbi.nlm.nih.gov/bioproject/497290 The complete list of DEGs has been deposited in figshare and can be viewed at this link: https://figshare.com/s/65828daf9e27f7caf634
Histopathology and immunohistochemistry.
Liver tissue was fixed in 10% formalin, embedded in paraffin, and sectioned at 10 μm. Immunohistochemistry for detection of profibrogenic and HSC activation marker αSMA were incubated overnight with anti-αSMA antibody (Abcam, ab7817, mouse monoclonal, 1:25), and immune complexes were detected by using HRP-conjugated goat anti-mouse antibody, as described previously (23). For determination of collagen types I and III fiber deposition, liver sections were stained with Picrosirius red solution (saturated picric acid containing 0.1% Direct Red 80 and 0.1% Fast Green FCF [Sigma-Aldrich]) for 2 hours. Tissue was washed and mounted for imaging. Stained sections were imaged by using NanoZoomer S360 (Hamamatsu) and quantified with the latest version of ImageJ software (NIH). Representative ×10 or ×20 images are reported in figures.
Statistics.
All data were expressed as the mean ± SD unless otherwise indicated. Differences between 3 or more groups were compared by a nonparametric Kruskal-Wallis ANOVA test. If a significant effect was detected, post hoc pairwise comparisons were performed using Mann-Whitney tests with Bonferroni’s correction. Differences between 2 groups were compared by a 2-tailed Student’s t test if data had a normal distribution or a Mann-Whitney test if the data deviated from the normal distribution. Differences were considered statistically significant at P < 0.05. All statistical analyses were performed using GraphPad Prism 4.0c or R v3.0.2 (www.r-project.org).
Study approval.
The studies were approved by the UCSD Institutional Animal Care and Use Committee and followed the NIH guidelines outlined in Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011).
Author contributions
AEF and DP conceived the idea and designed the experiments. DP, AL, TN, DK, and LDAH performed the experiments. DP, EMP, AL, and JK analyzed the data. DP, EMP, AL, AE, NPG, PMO, and TK contributed reagents, materials, and analysis tools. DP, EMP, and AEF wrote and revised the paper.
Supplementary Material
Acknowledgments
Financial support was received from NIH grants R01 DK113592 and U01 AA024206 to AEF, American Liver Foundation Postdoctoral Research Fellowship to DP, and Gilead Sciences Research Scholars Program in Liver Disease to AE.
We would like to acknowledge Jennifer Santini of the UCSD Microscopy Core with grant NS047101 for her support and troubleshooting. We would also like to acknowledge Timo Meerlo and Vanessa Taupin of the UCSD Electron Microscopy Facility for their precious help with imaging EVs. We would like to thank Kristen Jepsen and Eugenia Ricciardelli of the IGM Genomics Center, UCSD, for conducting the miRNA and mRNA sequencing and for their grant P30CA023100. We thank Lawrence Goldstein of UCSD for providing human and mouse iPSCs.
Version 1. 06/11/2019
In-Press Preview
Version 2. 07/25/2019
Electronic publication
Version 3. 08/01/2019
Updated metadata.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(14):e125652. https://doi.org/10.1172/jci.insight.125652.
Contributor Information
Davide Povero, Email: dapovero@gmail.com.
Eva M. Pinatel, Email: evapinatel@gmail.com.
Aleksandra Leszczynska, Email: aleszczynska@ucsd.edu.
Nidhi P. Goyal, Email: npandhoh@ucsd.edu.
Takahiro Nishio, Email: tnishio@ucsd.edu.
Jihoon Kim, Email: j5kim@ucsd.edu.
David Kneiber, Email: kneiber@wustl.edu.
Lucas de Araujo Horcel, Email: lucashorcel@uol.com.br.
Akiko Eguchi, Email: pordonez@ucsd.edu.
Paulina M. Ordonez, Email: akieguchi@clin.medic.mie-u.ac.jp.
Tatiana Kisseleva, Email: tkisseleva@ucsd.edu.
Ariel E. Feldstein, Email: afeldstein@ucsd.edu.
References
- 1.Bellentani S, Tiribelli C. Is it time to change NAFLD and NASH nomenclature? Lancet Gastroenterol Hepatol. 2017;2(8):547–548. doi: 10.1016/S2468-1253(17)30146-2. [DOI] [PubMed] [Google Scholar]
- 2.Mahady SE, George J. Predicting the future burden of NAFLD and NASH. J Hepatol. 2018;69(4):774–775. doi: 10.1016/j.jhep.2018.06.025. [DOI] [PubMed] [Google Scholar]
- 3.Adams LA, Feldstein AE. Nonalcoholic steatohepatitis: risk factors and diagnosis. Expert Rev Gastroenterol Hepatol. 2010;4(5):623–635. doi: 10.1586/egh.10.56. [DOI] [PubMed] [Google Scholar]
- 4.Fox IJ, Daley GQ, Goldman SA, Huard J, Kamp TJ, Trucco M. Stem cell therapy. Use of differentiated pluripotent stem cells as replacement therapy for treating disease. Science. 2014;345(6199):1247391. doi: 10.1126/science.1247391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Srijaya TC, Ramasamy TS, Kasim NH. Advancing stem cell therapy from bench to bedside: lessons from drug therapies. J Transl Med. 2014;12:243. doi: 10.1186/s12967-014-0243-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riazifar M, Pone EJ, Lötvall J, Zhao W. Stem cell extracellular vesicles: extended messages of regeneration. Annu Rev Pharmacol Toxicol. 2017;57:125–154. doi: 10.1146/annurev-pharmtox-061616-030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 9.Povero D, et al. Lipid-induced hepatocyte-derived extracellular vesicles regulate hepatic stellate cell via microRNAs targeting PPAR-γ. Cell Mol Gastroenterol Hepatol. 2015;1(6):646–663.e4. doi: 10.1016/j.jcmgh.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol. 2014;14(3):195–208. doi: 10.1038/nri3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katsman D, Stackpole EJ, Domin DR, Farber DB. Embryonic stem cell-derived microvesicles induce gene expression changes in Müller cells of the retina. PLoS ONE. 2012;7(11):e50417. doi: 10.1371/journal.pone.0050417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kourembanas S. Exosomes: vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu Rev Physiol. 2015;77:13–27. doi: 10.1146/annurev-physiol-021014-071641. [DOI] [PubMed] [Google Scholar]
- 13.Quesenberry PJ, Aliotta J, Deregibus MC, Camussi G. Role of extracellular RNA-carrying vesicles in cell differentiation and reprogramming. Stem Cell Res Ther. 2015;6:153. doi: 10.1186/s13287-015-0150-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katsuda T, Kosaka N, Takeshita F, Ochiya T. The therapeutic potential of mesenchymal stem cell-derived extracellular vesicles. Proteomics. 2013;13(10-11):1637–1653. doi: 10.1002/pmic.201200373. [DOI] [PubMed] [Google Scholar]
- 15.Lamichhane TN, Sokic S, Schardt JS, Raiker RS, Lin JW, Jay SM. Emerging roles for extracellular vesicles in tissue engineering and regenerative medicine. Tissue Eng Part B Rev. 2015;21(1):45–54. doi: 10.1089/ten.teb.2014.0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rani S, Ryan AE, Griffin MD, Ritter T. Mesenchymal stem cell-derived extracellular vesicles: toward cell-free therapeutic applications. Mol Ther. 2015;23(5):812–823. doi: 10.1038/mt.2015.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan M, et al. Embryonic stem cell-derived exosomes promote endogenous repair mechanisms and enhance cardiac function following myocardial infarction. Circ Res. 2015;117(1):52–64. doi: 10.1161/CIRCRESAHA.117.305990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deregibus MC, et al. Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood. 2007;110(7):2440–2448. doi: 10.1182/blood-2007-03-078709. [DOI] [PubMed] [Google Scholar]
- 19.Teixeira JH, Silva AM, Almeida MI, Barbosa MA, Santos SG. Circulating extracellular vesicles: their role in tissue repair and regeneration. Transfus Apher Sci. 2016;55(1):53–61. doi: 10.1016/j.transci.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 20.Choi DS, Kim DK, Kim YK, Gho YS. Proteomics of extracellular vesicles: exosomes and ectosomes. Mass Spectrom Rev. 2015;34(4):474–490. doi: 10.1002/mas.21420. [DOI] [PubMed] [Google Scholar]
- 21.D’Souza-Schorey C, Di Vizio D. Biology and proteomics of extracellular vesicles: harnessing their clinical potential. Expert Rev Proteomics. 2014;11(3):251–253. doi: 10.1586/14789450.2014.874290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haga H, Yan IK, Takahashi K, Matsuda A, Patel T. Extracellular vesicles from bone marrow-derived mesenchymal stem cells improve survival from lethal hepatic failure in mice. Stem Cells Transl Med. 2017;6(4):1262–1272. doi: 10.1002/sctm.16-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Povero D, et al. Lipid-induced toxicity stimulates hepatocytes to release angiogenic microparticles that require Vanin-1 for uptake by endothelial cells. Sci Signal. 2013;6(296):ra88. doi: 10.1126/scisignal.2004512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abels ER, Breakefield XO. Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake. Cell Mol Neurobiol. 2016;36(3):301–312. doi: 10.1007/s10571-016-0366-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Czernek L, Chworos A, Duechler M. The uptake of extracellular vesicles is affected by the differentiation status of myeloid cells. Scand J Immunol. 2015;82(6):506–514. doi: 10.1111/sji.12371. [DOI] [PubMed] [Google Scholar]
- 26.Lasič E, et al. Time-dependent uptake and trafficking of vesicles capturing extracellular S100B in cultured rat astrocytes. J Neurochem. 2016;139(2):309–323. doi: 10.1111/jnc.13754. [DOI] [PubMed] [Google Scholar]
- 27.Kisseleva T, et al. Myofibroblasts revert to an inactive phenotype during regression of liver fibrosis. Proc Natl Acad Sci USA. 2012;109(24):9448–9453. doi: 10.1073/pnas.1201840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crescitelli R, et al. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J Extracell Vesicles. 2013;2(10):3402/jev.v2i0.20677. doi: 10.3402/jev.v2i0.20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yáñez-Mó M, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson KD, Venkatasubrahmanyam S, Jia F, Sun N, Butte AJ, Wu JC. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev. 2009;18(5):749–758. doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berschneider B, et al. miR-92a regulates TGF-β1-induced WISP1 expression in pulmonary fibrosis. Int J Biochem Cell Biol. 2014;53:432–441. doi: 10.1016/j.biocel.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 32.Dakhlallah D, et al. Epigenetic regulation of miR-17~92 contributes to the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med. 2013;187(4):397–405. doi: 10.1164/rccm.201205-0888OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang H, et al. The antifibrotic effects and mechanisms of microRNA-26a action in idiopathic pulmonary fibrosis. Mol Ther. 2014;22(6):1122–1133. doi: 10.1038/mt.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Zhao X, Shan H, Liang H. MicroRNAs in idiopathic pulmonary fibrosis: involvement in pathogenesis and potential use in diagnosis and therapeutics. Acta Pharm Sin B. 2016;6(6):531–539. doi: 10.1016/j.apsb.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Graham JR, Williams CM, Yang Z. MicroRNA-27b targets gremlin 1 to modulate fibrotic responses in pulmonary cells. J Cell Biochem. 2014;115(9):1539–1548. doi: 10.1002/jcb.24809. [DOI] [PubMed] [Google Scholar]
- 36.Hong Y, et al. MiR-22 may suppress fibrogenesis by targeting TGFβR I in cardiac fibroblasts. Cell Physiol Biochem. 2016;40(6):1345–1353. doi: 10.1159/000453187. [DOI] [PubMed] [Google Scholar]
- 37.Ji X, et al. The antifibrotic effects and mechanisms of microRNA-486-5p in pulmonary fibrosis. Sci Rep. 2015;5:14131. doi: 10.1038/srep14131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tag CG, et al. Bile duct ligation in mice: induction of inflammatory liver injury and fibrosis by obstructive cholestasis. J Vis Exp. 2015;96(96):52438. doi: 10.3791/52438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koyama Y, et al. Mesothelin/mucin 16 signaling in activated portal fibroblasts regulates cholestatic liver fibrosis. J Clin Invest. 2017;127(4):1254–1270. doi: 10.1172/JCI88845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Israel MA, et al. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature. 2012;482(7384):216–220. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Povero D, et al. Circulating extracellular vesicles with specific proteome and liver microRNAs are potential biomarkers for liver injury in experimental fatty liver disease. PLoS ONE. 2014;9(12):e113651. doi: 10.1371/journal.pone.0113651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eguchi A, et al. Extracellular vesicles released by hepatocytes from gastric infusion model of alcoholic liver disease contain a microRNA barcode that can be detected in blood. Hepatology. 2017;65(2):475–490. doi: 10.1002/hep.28838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.