Abstract
Objective:
Alcoholism arises from combined effects of multiple biological factors including genetic and non-genetic causes with gene/environmental interaction. Intensive research and advanced genetic technology has generated a long list of genes and biomarkers involved in alcoholism neuropathology. These markers reflect complex overlapping and competing effects of possibly hundreds of genes which impact brain structure, function, biochemical alcohol processing, sensitivity and risk for dependence.
Method:
We compiled a tabular list of clinically relevant genetic biomarkers for alcoholism targeting expression disturbances in the human brain through an extensive search of keywords related to alcoholism, alcohol abuse, and genetics from peer reviewed medical research articles and related nationally sponsored websites. Gene symbols were then placed on high resolution human chromosome ideograms with gene descriptions in tabular form.
Results:
We identified 337 clinically relevant genetic biomarkers and candidate genes for alcoholism and alcohol-responsiveness from human brain research. Genetic biomarkers included neurotransmitter pathways associated with brain reward processes for dopaminergic (e.g., DRD2, MAOA, and COMT), serotoninergic (e.g., HTR3A, HTR1B, HTR3B, and SLC6A4), GABAergic (e.g., GABRA1, GABRA2, and GABRG1), glutaminergic (GAD1, GRIK3, and GRIN2C) and opioid (e.g., OPRM1, OPRD1, and OPRK1) pathways which presumably impact reinforcing properties of alcohol. Gene level disturbances in cellular and molecular networks impacted by alcohol and alcoholism pathology include transketolase (TKT), transferrin (TF), and myelin (e.g., MBP, MOBP, and MOG).
Conclusions:
High resolution chromosome ideograms provide investigators, physicians, geneticists and counselors a convenient visual image of the distribution of alcoholism genetic biomarkers from brain research with alphabetical listing of genes in tabular form allowing comparison between alcoholism-related phenotypes, and clinically-relevant alcoholism gene(s) at the chromosome band level to guide research, diagnosis, and treatment. Chromosome ideograms may facilitate gene-based personalized counseling of alcohol dependent individuals and their families.
Keywords: High resolution chromosome ideograms, Alcoholism, Genetic biomarkers, Brain, Chromosome band location, Gene
1. Introduction
Alcoholism arises from the combined effects of multiple biological factors including a wide range of possible genetic variations and/or abnormalities, as well as non-genetic causes including interpersonal and psychosocial relationships, neuroadaptive responses, and gene/environmental interaction (epigenetics)( Enoch, 2013; Koob, 2013; Mayfield et al., 2008; Morozova et al., 2014; Rietschel and Treutlein, 2013). The risk to develop alcoholism is strongly related to the family history, childhood environment particularly the number of life stressors and presence or absence of co-occurring mental disorders (Bierut et al., 1998; Brady and Back, 2012; Brady and Sinha, 2005; Enoch, 2013; Keyes et al., 2012; Schepis et al., 2011). The interface of these relationships are complex and involve overlapping and competing effects of many genes impacting brain development, structure, and function, as well as, alcohol processing and sensitivity (Enoch, 2013; Koob, 2013).
1.1. Candidate gene strategies
Alcohol-responsive brain networks have been reported and identified by case–control, population and family-based studies incorporating candidate gene approach with whole-genome methodology in individuals with and without alcoholism (Edenberg, 2012; Edenberg and Foroud, 2006, 2013; Enoch, 2013; Morozova et al., 2014; Yan et al., 2014). Candidate gene strategies have considered neurobiological and neurodevelopmental pathways which are key to brain function by emphasizing emotion and reward pathways as well as biochemical processes associated with the physiologic effects and molecular targets of alcohol itself particularly those impacting alcohol metabolism (Blum et al., 1990; Bolos et al., 1990; Chai et al., 2005; Edenberg et al., 2006, 2008; Enoch, 2013; Feinn et al., 2005; Kapoor et al., 2014; Luo et al., 2005; Macgregor et al., 2009; Radel et al., 2005; Tolstrup et al., 2008; Wetherill et al., 2014; Zhang et al., 2008). Further, genetic biomarkers including chromosome regions have been identified by overlapping genetic linkage and functional data with alcoholism-related phenotypes of interest and refined to develop a molecular signature of alcoholism phenotype (Ehlers et al., 2004; Enoch, 2013; Kapoor et al., 2014; Wetherill et al., 2014). A representative example of a candidate gene/biomarker analysis is the alcohol dehydrogenase enzyme (ADH) which is responsible for alcohol metabolism and linked to the ADH gene family located in the 4q23 chromosome band (Chai et al., 2005; Edenberg et al., 2006; Macgregor et al., 2009; Tolstrup et al., 2008). The 4q23 chromosome band has been strongly associated with alcoholism vulnerability in several genome-wide linkage studies (Long et al., 1998; Reich et al., 1998). The activity of alcohol metabolizing enzymes (e.g., ADH) influences sensitivity to alcohol, its effects on the body and the accumulation of metabolites (e.g., acetaldehyde) which may be toxic. Genetic variants that increase production or slow the processing of alcohol intermediates (e.g., ADH1B, ADH1C, ALDH2) influence the likelihood of developing a drinking problem (Chai et al., 2005; Edenberg et al., 2006; Macgregor et al., 2009; Tolstrup et al., 2008). Human genetic studies have similarly identified alcoholism candidate genes involving neurotransmitter pathways associated with brain reward processes including the dopaminergic (e.g., DRD2, MAOA, COMT), serotoninergic (e.g., HTR3A, HTR1B, HTR3B), GABAergic (e.g., GABRA1, GABRA2), glutaminergic (GAD1, GRIK3, GRIN2C) and opioid (e.g., OPRM1, OPRD1; Blum et al., 1990; Bolos et al., 1990; Edenberg, 2012; Edenberg and Foroud, 2013; Edenberg et al., 2008; Feinn et al., 2005; Luo et al., 2005; Radel et al., 2005; Zhang et al., 2008). These genes presumably directly impact the reinforcing properties of alcohol driving the motivation to seek and use alcohol to excess. Whole-genome studies have shown abnormalities of chromosomes and identified specific regions (e.g., 4p12, 7q31.32, 13q14.2) where known or candidate genes for alcoholism are located and for alcoholism-related phenotypes such as age of drinking onset (3q26.1, 5q11.2, and 12q32.2; Edenberg, 2012; Edenberg and Foroud, 2013; Enoch, 2013; Kapoor et al., 2014; Morozova et al., 2014; Radel et al., 2005; Yan et al., 2014).
1.2. Whole genome investigation
Advances in genetic technology beyond linkage or cytogenetic analysis of affected families with alcoholism or other complex disorders using COGA, SAGE, and other resources have led to genome-wide association studies (GWAS) involving hundreds of affected and control individuals analyzing the distribution and clustering of hundreds and thousands of SNPs to search for candidate genes (Edenberg, 2012; Edenberg and Foroud, 2013; Enoch, 2013; Morozova et al., 2014; Rietschel and Treutlein, 2013; Yan et al., 2014). GWAS studies have identified genetic linkage to primary disease risk, alcoholism-related phenotypes and responsiveness such as consumption, level of response to alcohol’s effects and event-related potential. Examples of candidate genes identified by GWAS studies include functional roles in cell adhesion important for brain development and implicated in Autism Spectrum Disorder (e.g., AUTS2, CDH13, EFNA5), molecular transporters (e.g., SLC1A3, SLC5A11, SLC6A4), and growth factors (e.g., BDNF) (Edenberg, 2012; Edenberg and Foroud, 2006, 2013; Enoch, 2013; Morozova et al., 2014; Rietschel and Treutlein, 2013; Yan et al., 2014).
1.3. Functional genomic biomarkers from brain
Additional functional genomic biomarker studies using high resolution microarrays have identified regionally distinct gene and exon level expression disturbances in post-mortem brain impacting neuronal growth and function which may influence alcoholism onset and progression (Flatscher-Bader et al., 2005, 2006, 2010; Lewohl et al., 2000; Liu et al., 2004, 2006; Manzardo et al., 2014; Mayfield et al., 2002; Sokolov et al., 2003). Functional gene expression profiling has identified disturbances in selected brain regions associated with reward processing and prefrontal inhibitory control mechanisms relevant to the development and propagation of abuse behaviors (Flatscher-Bader et al., 2005, 2006, 2010; Lewohl et al., 2000; Liu et al., 2004, 2006; Manzardo et al., 2014; Mayfield et al., 2002; Sokolov et al., 2003). An overrepresentation of down-regulated vs up-regulated genes at the mRNA level have been reported in addiction-related brain regions and functional disturbances impacting myelination, cellular signaling and energy production influencing brain structure, function, growth and development (Lewohl et al., 2000; Liu et al., 2004, 2006; Manzardo et al., 2014; Mayfield et al., 2002; Sokolov et al., 2003). These effects have been correlated with medical and psychiatric co-morbidities such as cirrhotic liver disease, smoking status and/or nutritional deficiency (Liu et al., 2007) and become potential genetic biomarkers for considerations in assessment and treatment development.
Advanced genetic platforms incorporating sophisticated bioinformatics techniques have enhanced our ability to identify SNPs and bio-markers involved in the genetics of alcoholism including risk factors for the development and progression of illness and have provided invaluable insight into the molecular and cellular mechanisms underlying the pathophysiology of alcoholism and the addictions. This insight has contributed to the development of the first targeted, clinically-validated and FDA approved drug treatments for alcoholism. The field will continue to advance through the use of next generation sequencing (whole genome or exome) which will yield additional valuable information on the location and description of genes contributing to alcoholism, enabling the identification of specific and recurring mutations of single genes impacting upon alcoholism-related phenotypes such as tolerance, withdrawal sensitivity, and craving which may provide novel therapeutic targets for future interventions. A current list of clinically relevant genetic biomarkers from brain in alcoholism are summarized and incorporated into high resolution chromosome ideograms (850 band level) to facilitate research, diagnostic testing and genetic counseling options for families in the clinical setting. The location of the 337 genes now recognized by searching literature and website information as playing a role in alcoholism are also presented in tabular form listing the individual gene symbol, name and chromosome location.
2. Materials and methods
We searched key words such as genetics, genes, alcoholism, alcohol abuse and alcohol dependence, mutations or gene variants and gene expression related to molecular disturbances in humans and alcoholism using computer-based internet sources including peer-reviewed medical literature (e.g., PubMed), federally sponsored (e.g., National Center for Biotechnology Information) and other informative websites (e.g., Online Mendelian Inheritance in Man; Ethanol-Related Gene Resource) (Guo et al., 2009) to compile a list of genetic biomarkers. The research articles ascertained were examined for evidence of gene or genetic biomarker involvement in alcoholism causation or pathology. These searches included whole-genome microarray and sequencing data and results from genome-wide association studies (GWAS) of alcoholism and families with and without a history of alcoholism as well as functional gene expression profiles using human brain. This list included a total of 337 genes identified with at least one mechanism related to alcoholism, or that could contribute to behavioral or neurological disturbances seen in alcoholism although not necessarily implicated in disease risk or causality (Table 1). The genetic biomarkers recognized, to date, as playing a role in alcoholism susceptibility or causation generally appear to impact alcohol metabolism and major neuro-transmitter pathways related to brain reward such as dopamine, serotonin, GABA, glutamate and opiate receptors and their metabolic pathways. Additional influences are noted for neurodevelopmental, molecular transport and cellular signaling pathways associated with alcohol exposure and pathophysiology and functionally disturbed in brain expression studies with possible impact on disease course. We include gene symbols representing each biomarker, their expanded name and chromosome band location in Table 1 for the 337 separate genes with a possible role in alcoholism. The position for each gene is plotted on high resolution chromosome ideograms (850 band level) as shown in Figure 1.
Table 1.
Genetic biomarkers for alcoholism and alcohol-responsiveness in human brain.
| Gene symbol | Gene name | Location | References |
|---|---|---|---|
| AADAC | Arylacetamide deacetylase | 3q25.1 | Flatscher-Bader et al. (2005) |
| ABCA1 | ATP-binding cassette, subfamily A, member 1 | 9q31.1 | Liu et al. (2007) |
| ABCA8 | ATP-binding cassette, subfamily A, member 8 | 17q24.2 | Manzardo et al. (2014) |
| ACACB | Acetyl-CoA carboxylase-beta | 12q24.11 | Manzardo et al. (2014) |
| ACN9 | ACN9, S. cerevisiae, homolog of | 7q21.3 | Dick et al. (2008) |
| ACO1 | Aconitase 1, soluble | 9p21.1 | Flatscher-Bader et al. (2005) |
| ACOT12 | Acyl-CoA thioesterase 12 | 5q14.1 | Flatscher-Bader et al. (2005) |
| ACTG1 | Actin, gamma-1 | 17q25.3 | Hill et al. (2004) |
| ACTN4 | Actinin, alpha-4 | 19q13.2 | Joslyn et al. (2010); Yu et al. (2008) |
| ADH1A | Alcohol dehydrogenase 1A, class I, alpha polypeptide | 4q23 | Morozova et al. (2014) |
| ADH1B | Alcohol dehydrogenase 1B, class I, beta polypeptide | 4q23 | Edenberg (2007) |
| ADH1C | Alcohol dehydrogenase 1C, class I, gamma polypeptide | 4q23 | Rietschel and Treutlein (2013) |
| ADH4 | Alcohol dehydrogenase 4 | 4q23 | Zuo et al. (2011) |
| ADH5 | Alcohol dehydrogenase 5, chi polypeptide | 4q23 | Zuo et al. (2011) |
| ADH6 | Alcohol dehydrogenase 6 | 4q23 | Flatscher-Bader et al. (2005) |
| ADO | 2-aminoethanethiol dioxygenase | 10q21.3 | Guo et al. (2009) |
| AGT | Angiotensinogen | 1q42.2 | Liu et al. (2006) |
| ALDH1A1 | Aldehyde dehydrogenase 1 family, member A1 | 9q21.13 | Morozova et al. (2014) |
| ALDH1B1 | Aldehyde dehydrogenase 1 family, member B1 | 9p13.2 | Linneberg et al. (2010) |
| ALDH2 | Aldehyde dehydrogenase 2 family | 12q24.12 | Cui et al. (2009) |
| ALDOC | Aldolase C, fructose-bisphosphate | 17q11.2 | Guo et al. (2009) |
| ALK | Anaplastic lymphoma kinase | 2p23.2-p23.1 | Wang et al. (2011b) |
| AMPH | Amphiphysin | 7p14.1 | Flatscher-Bader et al. (2005) |
| ANKK1 | Ankyrin repeat and kinase domain containing 1 | 11q23.2 | Yang et al. (2008) |
| ANKRD7 | Ankyrin repeat domain 7 | 7q31.31 | Chen et al. (2012) |
| ANKS1A | Ankyrin repeat and sterile alpha motif domain containing 1A | 6p21.31 | Heath et al. (2011) |
| ANLN | Anillin, actin binding protein | 7p14.2 | Manzardo et al. (2014) |
| ANP32E | Acidic leucine-rich nuclear phosphoprotein 32 family, member E | 1q21.2 | Liu et al. (2006) |
| APOA1 | Apolipoprotein A-I | 11q23.3 | Mayfield et al. (2002) |
| APOD | Apolipoprotein D | 3q29 | Mayfield et al. (2002) |
| APP | Amyloid beta A4 precursor protein | 21q21.3 | Flatscher-Bader et al. (2005) |
| AQP4 | Aquaporin 4 | 18q11.2 | Liu et al. (2007) |
| ARF1 | ADP-ribosylation factor 1 | 1q42.13 | Liu et al. (2004) |
| ARF4 | ADP-ribosylation factor 4 | 3p14.3 | Liu et al. (2004) |
| ARID5A | AT rich interaction domain-containing protein 5A (MRF1-like) | 2q11.2 | Zlojutro et al. (2010) |
| ARIH1 | Ariadne, Drosophila, homolog of, 1 | 15q24.1 | Morozova et al. (2014) |
| ARHGAP11A | RHO (rhodopsin) GTPase-activating protein 11A | 15q13.3 | Morozova et al. (2012) |
| ARHGAP28 | RHO (rhodopsin) GTPase-activating protein 28 | 18p11.31 | Morozova et al. (2012) |
| ARL2 | ADP-ribosylation factor-like 2 | 11q13.1 | Liu et al. (2004) |
| ARL4D | ADP-ribosylation factor-like 4D | 17q21.31 | Liu et al. (2004) |
| ARL15 | ADP-ribosylation factor-like 15 | 5q11.2 | Kapoor et al. (2014) |
| ART3 | ADP-ribosyltransferase 3 | 4q21.1 | Mayfield et al. (2002) |
| ASPA | Aspartoacylase | 17p13.2 | Manzardo et al. (2014) |
| ATP5A1 | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 | 18q21.1 | Guo et al. (2009) |
| ATP6V1B2 | ATPase, H+ transporting, lysosomal 56/58 kDa, V1 subunit B2 | 8p21.3 | Guo et al. (2009) |
| ATXN1 | Ataxin 1 | 6p22.3 | Mayfield et al. (2002) |
| AUTS2 | Autism susceptibility 2 | 7q11.22 | Schumann et al. (2011) |
| BBX | Bobby sox homolog (Drosophila) | 3q13.12 | Edenberg et al. (2010) |
| BDNF | Brain-derived neurotrophic factor | 11p14.1 | Zhao et al. (2012) |
| BNIP3 | BCL2/adenovirus E1B 19 kDa interacting protein 3 | 10q26.3 | Flatscher-Bader et al. (2005) |
| C15ORF32 | Chromosome 15 open reading frame 32 | 15q26.1 | Heath et al. (2011) |
| C4BPB | Complement component 4-binding protein, beta chain | 1q32.2 | Liu et al. (2006) |
| CA2 | Carbonic Anhydrase II | 8q21.2 | Flatscher-Bader et al. (2005) |
| CACNG1 | Calcium channel, voltage-dependent, gamma-1 subunit | 17q24.2 | Mayfield et al. (2002) |
| CALM2 | Calmodulin 2 | 2p21 | Mayfield et al. (2002) |
| CANX | Calnexin | 5q35.3 | Liu et al. (2006) |
| CAPN3 | Calpain 3 | 15q15.1 | Liu et al. (2006) |
| CAST | Calpastatin | 5q15 | Morozova et al. (2014) |
| CAT | Catalase | 11p13 | Lind et al. (2012) |
| CCR1 | Chemokine (C–C motif) receptor 1 | 3p21.31 | Mayfield et al. (2002) |
| CCT3 | Chaperonin containing TCP1 (T-complex 1), subunit 3 | 1q22 | Guo et al. (2009) |
| CD33 | CD33 Antigen | 19q13.41 | Flatscher-Bader et al. (2005) |
| CDC14A | Cell division cycle 14, S. Cerevisiae, homolog A | 1p21.2 | Flatscher-Bader et al. (2005) |
| CDKL5 | Cyclin-dependent kinase-like 5 | Xp22.13 | Liu et al. (2006)() |
| CDH13 | Cadherin 13 | 16q23.3 | Rietschel and Treutlein (2013) |
| CDHR2 | Cadherin-related family member 2 | 5q35.2 | Wetherill et al. (2014) |
| CEBPG | CCAAT/enhancer binding protein (C/EBP), gamma | 19q13.11 | Flatscher-Bader et al. (2005) |
| CHRM2 | Cholinergic receptor, muscarinic, 2 | 7q33 | Luo et al. (2005) |
| CHRNA3 | Cholinergic receptor, neuronal nicotinic, alpha polypeptide 3 | 15q25.1 | Zuo et al. (2011) |
| CHRNA5 | Cholinergic receptor, neuronal nicotinic, alpha polypeptide 5 | 15q25.1 | Wang et al. (2009) |
| CHRNB2 | Cholinergic receptor, neuronal nicotinic, beta polypeptide 2 | 1q21.3 | Ehringer et al. (2007) |
| CKB | Creatine kinase, brain type | 14q32.32 | Hill et al. (2004) |
| CLCN4 | Chloride channel 4 | Xp22.2 | Flatscher-Bader et al. (2005) |
| CLDN11 | Claudin 11 | 3q26.2 | Liu et al. (2004); Manzardo et al. (2014) |
| CNP | Cyclic nucleotide phosphodiesterase | 17q21.2 | Liu et al. (2006) |
| COMT | Catechol-O-methyltransferase | 22q11.21 | Yang et al. (2008) |
| COPB2 | Coatomer protein complex, subunit beta-2 | 3q23 | Morozova et al. (2014) |
| CPE | Carboxypeptidase E | 4q32.3 | Mayfield et al. (2002) |
| CREM | cAMP responsive element modulator | 10p11.21 | Mayfield et al. (2002) |
| CRMP1 | Collapsin response mediator protein 1 | 4p16.2 | Guo et al. (2009) |
| CRYAA | Crystallin, alpha-A | 21q22.3 | Mayfield et al. (2002) |
| CRYAB | Crystallin, alpha-B | 11q23.1 | Mayfield et al. (2002) |
| CSMD1 | CUB (complement C1r/C1s, Uegf, Bmp1) and sushi multiple domains 1 | 8p23.2 | Morozova et al. (2012) |
| CSMD2 | CUB and sushi multiple domains 2 | 1p34.3 | Morozova et al. (2012) |
| CTNNA3 | Catenin, alpha-3 | 10q21.3 | Manzardo et al. (2014) |
| CTNND2 | Catenin, delta-2 | 5p15.2 | Morozova et al. (2012) |
| CXCL12 | Chemokine (C-X-C motif) ligand 12 | 10q11.21 | Flatscher-Bader et al. (2005) |
| CYP2E1 | Cytochrome p450, subfamily IIE | 10q26.3 | Lind et al. (2012) |
| DDC | Dopa decarboxylase | 7p12.1 | Pan et al. (2013) |
| DKK2 | Dickkopf WNT signaling pathway inhibitor 2 | 4q25 | Kalsi et al. (2010) |
| DLG2 | Discs large, Drosophila, homolog of, 2 | 11q14.1 | Morozova et al. (2014) |
| DPYSL2 | Dihydropyrimidinase-like 2 | 8p21.2 | Taylor and Wang (2014) |
| DRD2 | Dopamine receptor D2 | 11q23.2 | Yang et al. (2008) |
| DSCAML1 | Down syndrome cell adhesion molecule-like 1 | 11q23.3 | Wang et al. (2011a) |
| DTWD2 | DTW domain containing 2 | 5q23.1 | Pan et al. (2013) |
| EDIL3 | EGF (epidermal growth factor)-like repeats and discoidin I-like domains 3 | 5q14.3 | Liu et al. (2006) |
| EDNRB | Endothelin receptor, type B | 13q22.3 | Wang et al. (2011a) |
| EEF1G | Eukaryotic translation elongation factor 1, gamma | 11q12.3 | Sokolov et al. (2003) |
| EFNA5 | Ephrin-A5 | 5q21.3 | Wang et al. (2012) |
| ENO1 | Enolase 1 | 1p36.23 | Guo et al. (2009) |
| ENO3 | Enolase 3 | 17p13.2 | Lind et al. (2012) |
| ENPP2 | Ectonucleotide pyrophosphatase/phosphodiesterase 2 | 8q24.12 | Liu et al. (2004); Manzardo et al. (2014) |
| ETFA | Electron-transfer-flavoprotein, alpha polypeptide | 15q24.3 | Flatscher-Bader et al. (2005) |
| ETNPPL | Ethanolamine-phosphate phospho-lyase | 4q25 | Manzardo et al. (2014) |
| EVI2A | Ectropic viral integration site 2A | 17q11.2 | Manzardo et al. (2014) |
| FGFR2 | Fibroblast growth factor receptor 2 | 10q26.13 | Flatscher-Bader et al. (2010) |
| FH | Fumarate hydratase | 1q43 | Guo et al. (2009) |
| FN1 | Fibronectin 1 | 2q35 | Liu et al. (2004) |
| FSIP2 | Fibrous sheath interacting protein 2 | 2q32.1 | Wang et al. (2012) |
| FZD9 | Frizzled class receptor 9 | 7q11.23 | Mayfield et al. (2002) |
| GABBR1 | GABA (gamma-aminobutyric acid) B receptor 1 | 6p21.31 | Flatscher-Bader et al. (2005) |
| GABRA1 | GABA (gamma-aminobutyric acid) receptor, alpha-1 | 5q34 | Enoch (2008) |
| GABRA2 | GABA (gamma-aminobutyric acid) receptor, alpha-2 | 4p12 | Agrawal et al. (2006) |
| GABRB1 | GABA (gamma-aminobutyric acid) receptor, beta-1 | 4p12 | Liu et al. (2006) |
| GABRG1 | GABA (gamma-aminobutyric acid) receptor, gamma-1 | 4p12 | Enoch (2008) |
| GABRG2 | GABA (gamma-aminobutyric acid) receptor, gamma-2 | 5q34 | Zuo et al. (2011) |
| GABRG3 | GABA (gamma-aminobutyric acid) receptor, gamma-3 | 15q12 | Edenberg (2012) |
| GAD1 | Glutamate decarboxylase 1 | 2q31.1 | Zuo et al. (2011) |
| GATA4 | GATA binding protein | 8p23.1 | Rietschel and Treutlein (2013) |
| GFAP | Glial fibrillary acidic protein | 17q21.31 | Liu et al. (2004) |
| GJB6 | Gap junction protein, beta-6 | 13q12.11 | Manzardo et al. (2014) |
| GKN1 | Gastrokine 1 | 2p13.3 | Flatscher-Bader et al. (2005) |
| GLDN | Gliomedin | 15q21.2 | Manzardo et al. (2014) |
| GLUD1 | Glutamate dehydrogenase 1 | 10q23.2 | Joslyn et al. (2010) |
| GOSR1 | Golgi SNAP (NSF attachment protein) receptor complex member 1 | 17q11.2 | Mayfield et al. (2002) |
| GOT1 | Glutamate oxaloacetate transaminase, soluble | 10q24.2 | Lind et al. (2012) |
| GPM6B | Glycoprotein M6B | Xp22.2 | Mayfield et al. (2002) |
| GRB2 | Growth factor receptor-bound protein 2 | 17q25.1 | Guo et al. (2009) |
| GRIA1 | Glutamate receptor, ionotropic, AMPA (alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate) 1 | 5q33.2 | Mayfield et al. (2013) |
| GRIA4 | Glutamate receptor, ionotropic, AMPA (alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate) 4 | 11q22.3 | Mayfield et al. (2013) |
| GRIK3 | Glutamate receptor, ionotropic, kainate 3 | 1p34.3 | Zuo et al. (2011) |
| GRIN1 | Glutamate receptor, ionotropic, N-methyl-D-aspartate, subunit 1 | 9q34.3 | Mayfield et al. (2013) |
| GRIN2B | Glutamate receptor, ionotropic, N-methyl-D-aspartate, subunit 2B | 12p13.1 | Mayfield et al. (2013) |
| GRIN2C | Glutamate receptor, ionotropic, N-methyl-D-aspartate, subunit 2C | 17q25.1 | Zuo et al. (2011) |
| GRM8 | Glutamate receptor, metabotropic, 8 | 7q31.33 | Chen et al. (2009) |
| GSN | Gelsolin | 9q33.2 | Kuo et al. (2006) |
| HIP1 | Huntingtin-interacting protein 1 | 7q11.23 | Heath et al. (2011) |
| HLA-DRA | Major histocompatibility complex, class II, DR alpha | 6p21.32 | Flatscher-Bader et al. (2005) |
| HLA-DRB1 | Major histocompatibility complex, class II, DR beta 1 | 6p21.32 | Flatscher-Bader et al. (2005) |
| HLA-DRB4 | Major histocompatibility complex, class II, DR beta 4 | 6p21.3 | Flatscher-Bader et al. (2005) |
| HMGCR | 3-hydroxy-3-methylglutaryl-CoA reductase | 5q13.3 | Mayfield et al. (2002) |
| HMOX1 | Heme oxygenase (decycling) 1 | 22q13.1 | Mayfield et al. (2002) |
| HSP90B1 | Heat-shock protein, 90-kDa, beta, 1 | 12q23.3 | Guo et al. (2009) |
| HSPA9 | Heat-shock 70-kDa protein 9 | 5q31.2 | Guo et al. (2009) |
| HTR1A | 5-hydroxytryptamine receptor 1A | 5q12.3 | Zuo et al. (2013b) |
| HTR1B | 5-hydroxytryptamine receptor 1B | 6q14.1 | Cao et al. (2013) |
| HTR3A | 5-hydroxytryptamine receptor 3A | 11q23.2 | Cao et al. (2013) |
| HTR3B | 5-hydroxytryptamine receptor 3B | 11q23.2 | Cao et al. (2013) |
| HTR7 | 5-hydroxytryptamine receptor 7, adenylate cyclase-coupled | 10q23.31 | Zlojutro et al. (2010) |
| IFRD1 | Interferon-related developmental regulator 1 | 7q31.1 | Liu et al. (2006) |
| IGF1 | Insulin-like growth factor I | 12q23.2 | Hill et al. (2004) |
| IGF1R | Insulin-like growth factor 1 receptor | 15q26.3 | Liu et al. (2004) |
| IGF2R | Insulin-like growth factor II receptor | 6q25.3 | Morozova et al. (2014) |
| IKBKB | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta | 8q11.21 | Flatscher-Bader et al. (2005) |
| IKZF1 | Ikaros family zinc finger 1 | 7p12.2 | Flatscher-Bader et al. (2005) |
| IL10RA | Interleukin 10 receptor, alpha | 11q23.3 | Mayfield et al. (2002) |
| IMPA2 | Myo-Inositol monophosphatase 2 | 18p11.21 | Morozova et al. (2014) |
| INADL | InaD-like | 1p31.3 | Heath et al. (2011) |
| IPO11 | Importin 11 | 5q12.1 | Zuo et al. (2013b) |
| ITGA2 | Integrin, alpha-2 | 5q11.2 | Manzardo et al. (2014) |
| ITGAV | Integrin, alpha-V | 2q32.1 | Flatscher-Bader et al. (2010) |
| ITPKA | Inositol 1,4,5-trisphosphate 3-kinase A | 15q15.1 | Flatscher-Bader et al. (2005) |
| KCNB2 | Potassium channel, voltage-gated, Shab-related subfamily, member 2 | 8q13.3 | Pan et al. (2013) |
| KCNE3 | Potassium channel, voltage-gated, Isk-related subfamily, member 2 | 11q13.4 | Flatscher-Bader et al. (2005) |
| KCNIP1 | Potassium channel-interacting protein 1 | 5q31.1 | Morozova et al. (2012) |
| KCNJ6 | Potassium channel, inwardly rectifying, subfamily j, member 6 | 21q22.13 | Kang et al. (2012) |
| KCNJ10 | Potassium channel, inwardly rectifying, subfamily j, member 6 | 11q24.3 | Lewohl et al. (2000) |
| KDM4C | Lysine (K)-specific demethylase 4C | 9p24.1 | Wang et al. (2012) |
| KIAA0040 | Uncharacterized protein | 1q25.1 | Zuo et al. (2012) |
| KLK6 | Kallikrein-related peptidase 6 | 19q13.41 | Manzardo et al. (2014) |
| LAMP1 | Lysosomal-associated membrane protein 1 | 13q34 | Flatscher-Bader et al. (2005) |
| LAMP2 | Lysosomal-associated membrane protein 2 | Xq24 | Liu et al. (2006) |
| LDHB | Lactate dehydrogenase B | 12p12.1 | Guo et al. (2009) |
| LGR5 | Leucine-rich repeat-containing G protein-coupled receptor 5 | 12q21.1 | Manzardo et al. (2014) |
| LIMS1 | LIM and senescent cell antigen-like domains 1 | 2q12.3 | Mayfield et al. (2002) |
| LMO1 | LIM domain only 1 | 11p15.4 | Kapoor et al. (2013) |
| LPIN1 | Lipin 1 | 2p25.1 | Flatscher-Bader et al. (2005) |
| LPXN | Leupaxin | 11q12.1 | Flatscher-Bader et al. (2005) |
| LRP2 | Low density lipoprotein receptor-related protein 2 | 2q31.1 | Manzardo et al. (2014) |
| LRRN1 | Leucine rich repeats neuronal 1 | 3p26.2 | Wetherill et al. (2014) |
| LTF | Lactotransferrin | 3p21.31 | Mayfield et al. (2002) |
| MAG | Myelin-associated glycoprotein | 19q13.12 | Mayfield et al. (2002); Manzardo et al.(2014) |
| MAOA | Monoamine oxidase A | Xp11.3 | Tikkanen et al. (2009) |
| MAPT | Microtubule-associated protein tau | 17q21.31 | Flatscher-Bader et al. (2005) |
| MAX | MAX protein | 14q23.3 | Morozova et al. (2014) |
| MBP | Myelin basic protein | 18q23 | Liu et al. (2004), Manzardo et al. (2014) |
| MCM5 | Minichromosome maintenance complex component 5 | 22q13.3 | Liu et al. (2006) |
| MDK | Midkine | 11p11.2 | Flatscher-Bader et al. (2005) |
| MGST1 | Glutathione S-transferase, microsomal, 1 | 12p12.3 | Liu et al. (2007) |
| MOBP | Myelin-associated oligodendrocyte basic protein | 3p21.33 | Mayfield et al. (2002) |
| MOG | Myelin-oligodendrocyte glycoprotein | 6p22.1 | Liu et al. (2004); Manzardo et al. (2014) |
| MPDZ | Multiple PDZ (PSD95, Dlg1, & ZO-1) domain protein | 9p23 | Zhao et al. (2012) |
| MT1L | metallothionein 1 L | 16q12.2 | Mayfield et al. (2002) |
| MUS81 | MUS81 structure-specific endonuclease subunit | 11q13.1 | Flatscher-Bader et al. (2005) |
| MYT1L | Myelin transcription factor 1-like | 2p25.3 | Liu et al. (2004) |
| NALCN | Sodium leak channel, non-selective | 13q33.1 | Wetherill et al. (2014) |
| NCAM1 | Cell adhesion molecule, neural, 1 | 11q23.2 | Yang et al. (2008) |
| NDUFA1 | NADH (nicotinamide adenine dinucleotide) dehydrogenase (ubiquinone) 1 | Xq24 | Flatscher-Bader et al. (2005) |
| NDUFA11 | NADH (nicotinamide adenine dinucleotide) dehydrogenase 1 alpha subcomplex, 11 | 19p13.3 | Liu et al. (2006) |
| NDST4 | N-deacetylase/N-sulfotransferase 4 | 4q26 | Pan et al. (2013) |
| NFKB1 | Nuclear factor kappa-B, subunit 1 | 4q24 | Morozova et al. (2014) |
| NKAIN1 | Na+/K+ transporting ATPase interacting 1 | 1p35.2 | Zuo et al. (2013a) |
| NKAIN2 | Na+/K+ transporting ATPase-interacting 2 | 6q22.31 | Wang et al. (2011a) |
| NPAS3 | Neuronal PAS (Per-Arnt-Sim) domain protein 3 | 14q13.1 | Morozova et al. (2012) |
| NPY | Neuropeptide Y | 7p15.3 | Kauhanen et al. (2000); Mayfield et al. (2002) |
| NPY1R | Neuropeptide Y receptor Y1 | 4q32.2 | Zuo et al. (2011) |
| NPY2R | Neuropeptide Y receptor Y2 | 4q32.1 | Zhao et al. (2012) |
| NPY5R | Neuropeptide Y receptor Y5 | 4q32.2 | Wetherill et al. (2008) |
| NR4A2 | Nuclear receptor subfamily 4, group A, member 2 | 2q24.1 | Pan et al. (2013) |
| NRD1 | Nardilysin | 1p32.3 | Wang et al. (2011b) |
| NRXN3 | Neurexin 3 | 14q24.3-q31 | Hishimoto et al. (2007) |
| NSMAF | Neutral sphingomyelinase activation-associated factor | 8q12.1 | Liu et al. (2004) |
| NTRK2 | Neurotrophic tyrosine kinase, receptor, type 2 | 8q21.33 | Mayfield et al. (2002) |
| OMG | Oligodendrocyte-myelin glycoprotein | 17q11.2 | Hill et al. (2004) |
| OPRD1 | Opioid receptor, delta-1 | 1p35.3 | Ashenhurst et al. (2012); Zhang et al. (2008) |
| OPRM1 | Opioid receptor, mu-1 | 6q25.2 | Ray and Hutchison (2004) |
| OR51L1 | Olfactory receptor, family 51, subfamily L, member 1 | 11p15.4 | Wetherill et al. (2014) |
| PARK7 | Parkinson protein 7 | 1p36.23 | Guo et al. (2009) |
| PARP1 | Poly(ADP-ribose) polymerase 1 | 1q42.12 | Mayfield et al. (2002) |
| PARP4 | Poly(ADP-ribose) polymerase 4 | 13q12.12 | Mayfield et al. (2002) |
| PDHB | pyruvate dehydrogenase, beta polypeptide | 3p14.3 | Guo et al. (2009) |
| PDIA3 | Protein disulfide isomerase, family A, member 3 | 15q15.3 | Morozova et al. (2014) |
| PDYN | Prodynorphin | 20p13 | Williams et al. (2007) |
| PEA15 | Phosphoprotein enriched in astrocytes, 15-kDa | 1q23.2 | Liu et al. (2007) |
| PEBP1 | Phosphatidylethanolamine-binding protein 1 | 12q24.23 | Manzardo et al. (2014) |
| PGM2L1 | Phosphoglucomutase 2-like 1 | 11q13.4 | Flatscher-Bader et al. (2005) |
| PHF3 | PHD (plant homeodomain) finger protein 3 | 6q12 | Zuo et al. (2011) |
| PIBF1 | Progesterone-induced blocking factor 1 | 13q22.1 | Flatscher-Bader et al. (2005) |
| PIEZO2 | Piezo-type mechanosensitive ion channel component 2 | 18p11.22 | Manzardo et al. (2014) |
| PIPOX | Pipecolic acid oxidase | 17q11.2 | Flatscher-Bader et al. (2005) |
| PKN1 | Protein kinase N1 | 19p13.12 | Guo et al. (2009) |
| PKNOX2 | PBX(pre-B-cell leukemia homeobox)/knotted 1 homeobox 2 | 11q24.2 | Bierut et al. (2010) |
| PLA2G1B | Phospholipase A2, group IB (pancreas) | 12q24.31 | Mayfield et al. (2002) |
| PLCL1 | Phospholipase C-like 1 | 2q33.1 | Kapoor et al. (2013) |
| PLP1 | Proteolipid protein 1 | Xq22.2 | Mayfield et al. (2002) |
| PML | Promyelocytic leukemia | 15q24.1 | Flatscher-Bader et al. (2005) |
| PMP22 | Peripheral myelin protein 22 | 17p12 | Liu et al. (2004); Manzardo et al. (2014) |
| POLD2 | Polymerase (DNA-directed), delta 2, regulatory subunit | 7p13 | Flatscher-Bader et al. (2005) |
| POMC | Proopiomelanocortin | 2p23.3 | Zuo et al. (2011) |
| PPAN | Peter pan, Drosophila, homolog of | 19p13.2 | Flatscher-Bader et al. (2005) |
| PRDX6 | Peroxiredoxin 6 | 1q25.1 | Guo et al. (2009) |
| PRKCA | Protein kinase C, alpha | 17q24.2 | Morozova et al. (2014) |
| PSMB2 | Proteasome subunit, beta type, 2 | 1p34.3 | Liu et al. (2006) |
| PTP4A1 | Protein-tyrosine phosphatase, type 4A, 1 | 6q12 | Zuo et al. (2009) |
| PTPRD | Protein-tyrosine phosphatase, receptor-type, delta | 9p23 | Morozova et al. (2012) |
| PURA | Purine-rich element-binding protein A | 5q31.2 | Liu et al. (2004) |
| QDPR | Quinoid dihydropteridine reductase | 4p15.32 | Joslyn et al. (2010); Yu et al. (2008) |
| RAB1A | RAS-associated protein RAB1 | 2p14 | Guo et al. (2009) |
| RAB11B | RAB11B, member RAS oncogene family | 19p13.2 | Mayfield et al. (2002) |
| RAB3A | RAS-associated protein RAB3A | 19p13.11 | Guo et al. (2009) |
| RANBP3L | RAN (Ras-related nuclear protein) Binding Protein 3-like | 5p13.2 | Manzardo et al. (2014) |
| RASGRP3 | RAS guanyl nucleotide-releasing protein 3 | 2p22.3 | Manzardo et al. (2014) |
| RGS8 | Regulator of G protein signaling 8 | 1q25.3 | Flatscher-Bader et al. (2005) |
| RHOA | RAS homolog gene family, member A | 3p21.31 | Flatscher-Bader et al. (2010) |
| RHOB | RAS homolog gene family, member B | 2p24.1 | Flatscher-Bader et al. (2010) |
| RPE | Ribulose 5-phosphate 3-epimerase | 2q34 | Flatscher-Bader et al. (2005) |
| RPL22 | Ribosomal protein L22 | 1p36.31 | Flatscher-Bader et al. (2005) |
| RPS29 | Ribosomal protein S29 | 14q21.3 | Flatscher-Bader et al. (2005) |
| RPS6KA5 | Ribosomal protein S6 kinase, 90 kDa, polypeptide 5 | 14q32.11 | Flatscher-Bader et al. (2005) |
| RTN1 | Reticulon 1 | 14q23.1 | Mayfield et al. (2002) |
| RTN4 | Reticulon 4 | 2p16.1 | Liu et al. (2007) |
| SAMHD1 | SAM (sterile alpha-motif) domain and HD (Huntington disease) domain 1 | 20q11.23 | Flatscher-Bader et al. (2005) |
| SCD | Stearoyl-CoA desaturase | 10q24.31 | Liu et al. (2006) |
| SDC2 | Syndecan 2 | 8q22.1 | Mayfield et al. (2002) |
| SEMA3E | Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E | 7q21.11 | Kerner et al. (2011) |
| SEMA5A | Semaphorin 5A | 5p15.31 | Wang et al. (2011b) |
| SERINC2 | Serine incorporator 2 | 1p35.2 | Zuo et al. (2013a) |
| SERTAD3 | SERTA (SEI-1, RBT1, TARA) domain containing 3 | 19q13.2 | Flatscher-Bader et al. (2005) |
| SESN2 | Sestrin 2 | 1p35.3 | Flatscher-Bader et al. (2005) |
| SGOL1 | Shugoshin-like 1 | 3p24.3 | Pan et al. (2013) |
| SHC3 | SHC (Src homology 2 domain containing) transforming protein 3 | 9q22.1 | Morozova et al. (2014) |
| SI | Sucrase-isomaltase (alpha-glucosidase) | 3q26.1 | Guo et al. (2009) |
| SLC1A3 | Solute carrier family 1 (glial high affinity glutamate transporter), member 3 | 5p13.2 | Flatscher-Bader et al. (2005) |
| SLC12A2 | Solute carrier family 12 (sodium/potassium/chloride transporter), member 2 | 5q23.3 | Liu et al. (2006) |
| SLC30A5 | Solute carrier family 30 (zinc transporter), member 5 | 5q13.1-q13.2 | Hill et al. (2004); Flatscher-Bader et al. (2005) |
| SLC5A11 | Solute carrier family 5 (sodium/glucose cotransporter), member 11 | 16p12.1 | Manzardo et al. (2014) |
| SLC6A4 | Solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 | 17q11.2 | Feinn et al. (2005) |
| SMARCA5 | SWI/SNF (switch/sucrose nonfermentable) related, matrix associated, actin dependent regulator of chromatin, subfamily A, member 5 | 4q31.21 | Liu et al. (2006) |
| SMPD2 | Sphingomyelin phosphodiesterase 2, neutral membrane | 6q21 | Liu et al. (2004) |
| SMPDL3A | Sphingomyelin phosphodiesterase, acid-like, 3A | 6q22.31 | Liu et al. (2004) |
| SMPDL3B | Sphingomyelin phosphodiesterase, acid-like, 3B | 1p35.3 | Liu et al. (2004) |
| SNCA | Synuclein, alpha | 4q22.1 | Bönsch et al. (2005) |
| SOCS1 | Suppressor of cytokine signaling 1 | 16p13.13 | Guo et al. (2009) |
| SOX9 | SRY (sex determining region Y)-box 9 | 17q24.3 | Mayfield et al. (2002) |
| SPARC | Secreted protein, acidic, cysteine-rich | 5q33.1 | Flatscher-Bader et al. (2010) |
| SPOCK1 | Sparc/osteonectin, CWCV (cys–trp–cys–val) and kazal-like domains proteoglycan (testican) 1 | 5q31.2 | Liu et al. (2006) |
| SPP1 | Secreted phosphoprotein 1 | 4q22.1 | Manzardo et al. (2014) |
| SRSF10 | Splicing factor, serine/arginine-rich, 10 | 1p36.11 | Flatscher-Bader et al. (2005) |
| ST18 | Suppression of tumorigenicity 18, zinc finger | 8q11.23 | Manzardo et al. (2014) |
| STXBP2 | Syntaxin binding protein 2 | 19p13.2 | Mayfield et al. (2002) |
| SYN2 | Synapsin II | 3p25.2 | Mayfield et al. (2002) |
| SYT1 | Synaptotagmin 1 | 12q21.2 | Flatscher-Bader et al. (2005) |
| TACR3 | Tachykinin receptor 3 | 4q24 | Foroud et al. (2008) |
| TAS2R16 | Taste receptor, type 2, Member 16 | 7q31.32 | Hinrichs et al. (2006) |
| TAS2R38 | Taste receptor, type 2, Member 38 | 7q34 | Wang et al. (2007) |
| TF | Transferrin | 3q22.1 | Liu et al. (2004) |
| TGFB1 | Transforming growth factor, beta-1 | 19q13.2 | Mayfield et al. (2002) |
| THRB | Thyroid hormone receptor, beta | 3p24.2 | Mayfield et al. (2002) |
| THSD7B | Thrombospondin, type I, domain containing 7B | 2q22.1 | Wang et al. (2011b) |
| TIMM10B | Translocase of inner mitochondrial membrane 10 homolog B (yeast) | 11p15.4 | Flatscher-Bader et al. (2005) |
| TIMP1 | TIMP (human tissue inhibitor of metalloproteinase) Metallopeptidase Inhibitor 1 | Xp11.23 | Mayfield et al. (2002) |
| TIMP2 | Tissue inhibitor of metallopeptidase 2 | 17q25.3 | Liu et al. (2006) |
| TIMP3 | Tissue inhibitor of metallopeptidase 3 | 22q12.3 | Flatscher-Bader et al. (2005) |
| TIPARP | TCDD (tetrachlorodibenzo-p-dioxin)-inducible poly(ADP-ribose) polymerase | 3q25.31 | Wang et al. (2011a) |
| TKT | Transketolase | 3p21.1 | Liu et al. (2006) |
| TMED10 | Transmembrane emp24-like trafficking protein 10 (yeast) | 14q24.3 | Mayfield et al. (2002) |
| TMEM63A | Transmembrane protein 63A | 1q42.12 | Manzardo et al. (2014) |
| TMEM108 | Transmembrane protein 108 | 3q22.1 | Heath et al. (2011) |
| TMX1 | Thioredoxin-related transmembrane protein 1 | 14q22.1 | Flatscher-Bader et al. (2005) |
| TPI1 | Triosephosphate isomerase 1 | 12p13.31 | Liu et al. (2006) |
| TSPAN7 | Tetraspanin 7 | Xp11.4 | Liu et al. (2007) |
| TTC12 | Tetratricopeptide repeat domain-containing protein 12 | 11q23.2 | Yang et al. (2008) |
| TXN | Thioredoxin | 9q31.3 | Flatscher-Bader et al. (2005) |
| TUBGCP2 | Tubulin, gamma complex associated protein 2 | 10q26.3 | Mayfield et al. (2002) |
| TULP1 | Tubby like protein 1 | 6p21.31 | Mayfield et al. (2002) |
| UBP1 | Upstream binding protein 1 | 3p22.3 | Flatscher-Bader et al. (2005) |
| UGT8 | Uridine diphosphate glycosyltransferase 8 | 4q26 | Manzardo et al. (2014) |
| ULK2 | Unc51-like kinase 2 | 17p11.2 | Liu et al. (2006) |
| UPP1 | Uridine phosphorylase 1 | 7p12.3 | Flatscher-Bader et al. (2005) |
| UQCRC2 | Ubiquinol-cytochrome c reductase core protein II | 16p12.2 | Liu et al. (2006) |
| UQCRFS1 | Ubiquinol-cytochrome C reductase, Rieske iron–sulfur polypeptide 1 | 19q12 | Liu et al. (2006) |
| UROD | Uroporphyrinogen decarboxylase | 1p34.1 | Flatscher-Bader et al. (2005) |
| USH2A | Usher syndrome, type IIA | 1q41 | Morozova et al. (2012) |
| UTP20 | Small subunit (SSU) processome component, homolog (yeast) | 12q23.2 | Kapoor et al. (2014) |
| VAMP3 | Vesicle-associated membrane protein 3 | 1p36.23 | Mayfield et al. (2002) |
| VCP | Valosin-containing protein | 9p13.3 | Guo et al. (2009) |
| WASF1 | Wiskott–Aldrich syndrome protein family, member 1 | 6q21 | Flatscher-Bader et al. (2005) |
| WIF1 | WNT (wingless-type MMTV integration site family) inhibitory factor 1 | 12q14.3 | Manzardo et al. (2014) |
| XRCC5 | X-ray repair, complementing defective, in Chinese hamster cells 5 (double-strand-break rejoining) | 2q35 | Joslyn et al. (2010); Yu et al. (2008) |
| ZBTB11 | Zinc finger and BTB domain containing 11 | 3q12.3 | Flatscher-Bader et al. (2005) |
| ZBTB49 | Zinc finger and BTB domain containing 49 | 4p16.3 | Flatscher-Bader et al. (2005) |
| ZNF24 | Zinc finger protein 24 | 18q12.2 | Flatscher-Bader et al. (2005) |
| ZSCAN32 | Zinc finger and SCAN domain containing 32 | 16p13.3 | Flatscher-Bader et al. (2005) |
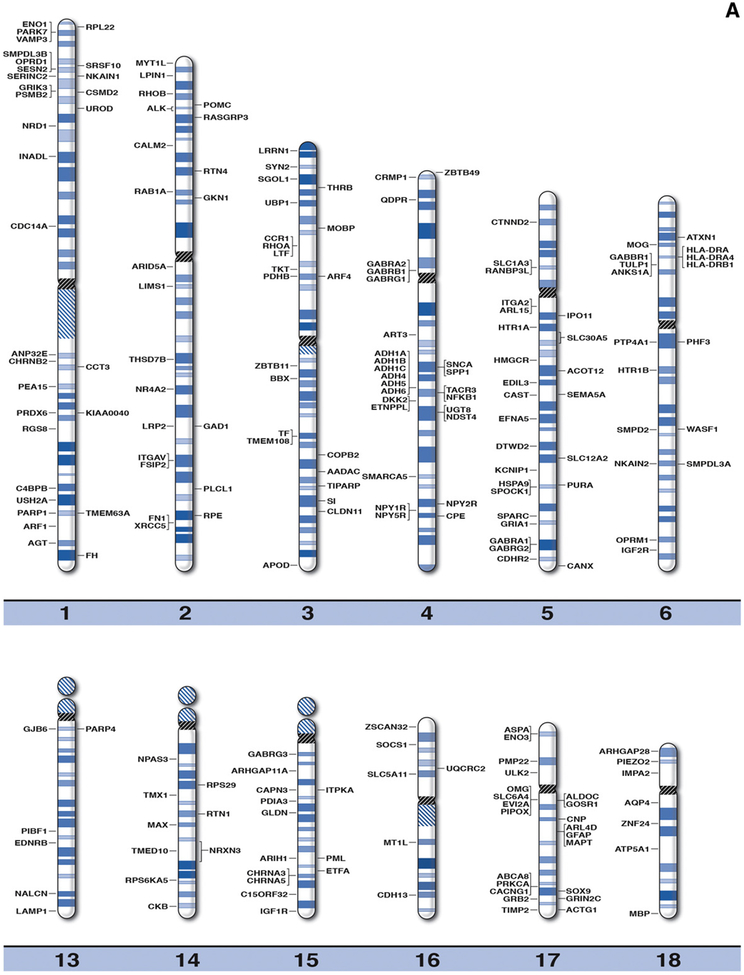
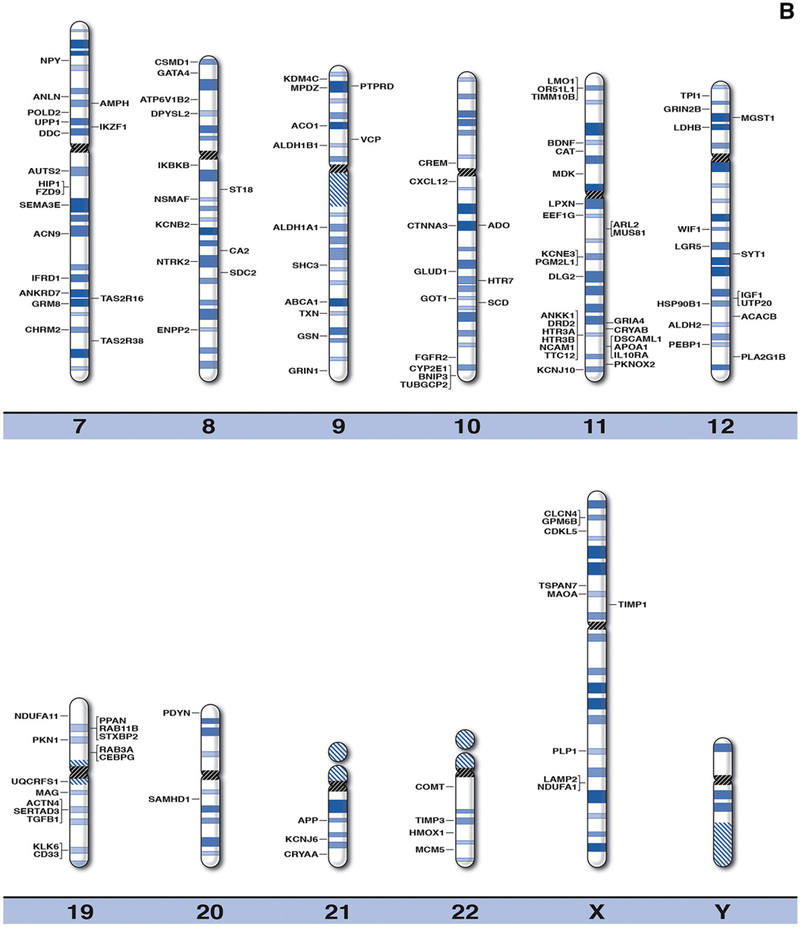
Fig. 1.
High resolution human chromosome ideograms (850 band level) with alcoholism gene symbols positioned at the chromosome band location. The upper ‘p’ and lower ‘q’ arms for each chromosome are separated by the centromere area highlighted in black. The gene symbol in alphabetical order, expanded name of the gene and precise chromosome band position are found in Table 1.
3. Discussions and conclusion
Decades of intensive research involving international collaborative networks utilizing large databases and tissue banks and advanced genetic technology and bioinformatics have led to significant discoveries in recognition of genetic variants and biomarkers implicated in the causation and course of alcoholism. Improved commercial platforms available for alcoholism research with potential application in the clinical setting have permitted the identification and characterization of the molecular signatures for novel or disturbed gene or exon expression and disease-specific profiles and patterns of interconnected disturbed gene pathways for alcoholism and other psychiatric or aberrant behavioral disorders in a growing body of genetic data and evidence. Significant discoveries have been made using microarray technology and now next generation sequencing with readily available tissue such as peripheral blood, autopsy specimens, established lymphoblastoid cell lines and/or saliva hold promise for more advances in alcoholism biomarker research by enabling the identification of new, clinically relevant genes impacting causation and disease mechanisms stimulating discovery of treatment modalities.
We identified 337 clinically relevant or known gene biomarkers for alcoholism based on computer searches of key words from peer reviewed medical literature reports or nationally sponsored web sites. These genetic biomarkers may be causative in nature or transient reflecting physiological responses to alcohol use or environmental influences such as poor nutrition, sanitation, and exposure to other drugs, illnesses, and viruses, common in chronic alcoholism. Transient, potentially modifiable genomic imbalances are important considerations in discovery, treatment and assessment of phenotypic outcomes in alcoholism. Future studies should also consider the impact of copy number variation, segmental deletions and duplications and regions of homozygosity in the genome for determination of identical by descent for calculation of inbreeding coefficients or consanguinity status along with uniparental disomy of individual chromosomes and areas of genomic imprinting on alcoholism risk and course of illness.
Our summary of the current status and number of clinically relevant human genetic biomarkers associated with alcoholism — targeting disturbed brain networks along with their position on high resolution chromosome ideograms will enhance the development of genetic testing options including DNA or gene testing panels for alcoholism and encourage genetic counseling of family members with a high density of alcoholism in biological relatives. The number of genes identified may vary in future studies based upon size and selected subject characteristics, tissue type and genetic technique used. The authors encourage the use of this current collection of genetic biomarkers from brain in alcoholism in their evaluation of patients and families to improve diagnosis and genetic counseling of selected patients, at-risk individuals and their families.
Acknowledgments
We thank Lorie Gavulic for excellent artistic design and preparation of chromosome ideograms. Support for this study was made available by grants from the Hanlon Charitable Trust, NICHD 02528 and the Headley Family Scholarship.
Abbreviations:
- COGA
collaborativestudyonthegeneticsofalcoholism
- DNA
deoxyribonucleic acid
- FDA
Food and Drug Administration
- GWAS
genome-wide association studies
- mRNA
messenger ribonucleic acid
- SAGE
study of addiction: genetics and environment
- SNPs
single nucleotide polymorphisms
References
- Agrawal A, Edenberg HJ, Foroud T, Bierut LJ, Dunne G, Hinrichs AL, Nurnberger JI, Crowe R, Kuperman S, Schuckit MA, Begleiter H, Porjesz B, Dick DM, 2006. Association of GABRA2 with drug dependence in the collaborative study of the genetics of alcoholism sample. Behav. Genet 36, 640–665. [DOI] [PubMed] [Google Scholar]
- Ashenhurst JR, Bujarski S, Ray LA, 2012. Delta and kappa opioid receptor polymorphisms influence the effects of naltrexone on subjective responses to alcohol. Pharmacol. Biochem. Behav 103 (2), 253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Dinwiddie SH, Begleiter H, Crowe RR, Hesselbrock V, Nurnberger JI Jr., Porjesz B, Schuckit MA, Reich T, 1998. Familial transmission of substance dependence: alcohol, marijuana, cocaine, and habitual smoking: a report from the Collaborative Study on the Genetics of Alcoholism. Arch. Gen. Psychiatry 55, 982–988 (Erratum in: Arch Gen Psychiatry 2002 59, 153). [DOI] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, Hinrichs AL, Almasy L, Breslau N, Culverhouse RC, Dick DM, Edenberg HJ, Foroud T, Grucza RA, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Krueger RF, Kuperman S, Lynskey M, Mann K, Neuman RJ, Nöthen MM, Nurnberger JI Jr., Porjesz B, Ridinger M, Saccone NL, Saccone SF, Schuckit MA, Tischfield JA, Wang JC, Rietschel M, Goate AM, Rice JP, 2010. A genome-wide association study of alcohol dependence. Proc. Natl. Acad. Sci. U. S. A 107 (11), 5082–5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum K, Noble EP, Sheridan PJ, Montgomery A, Ritchie T, Jagadeeswaran P, Nogami H, Briggs AH, Cohn JB, 1990. Allelic association of human dopamine D(2) receptor gene in alcoholism. J. Am. Med. Assoc 263, 2055–2060. [PubMed] [Google Scholar]
- Bolos AM, Dean M, Lucas-Derse S, Ramsburg M, Brown GL, Goldman D, 1990. Population and pedigree studies reveal a lack of association between the dopamine D(2) receptor gene and alcoholism. J. Am. Med. Assoc 264, 3156–3160. [PubMed] [Google Scholar]
- Bönsch D, Lederer T, Reulbach U, Hothorn T, Kornhuber J, Bleich S, 2005. Joint analysis of the NACP-REP1 marker within the alpha synuclein gene concludes association with alcohol dependence. Hum. Mol. Genet 14, 967–971. [DOI] [PubMed] [Google Scholar]
- Brady KT, Back SE, 2012. Childhood trauma, posttraumatic stress disorder, and alcohol dependence. Alcohol Res. 34, 408–413. [PMC free article] [PubMed] [Google Scholar]
- Brady KT, Sinha R, 2005. Co-occurring mental and substance use disorders: the neuro-biological effects of chronic stress. Am. J. Psychiatry 162, 1483–1493. [DOI] [PubMed] [Google Scholar]
- Cao J, LaRocque E, Li D, 2013. Associations of the 5-hydroxytryptamine (serotonin) receptor 1B gene (HTR1B) with alcohol, cocaine, and heroin abuse. Am. J. Med. Genet. B Neuropsychiatr. Genet 162B (2), 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y-G, Oh D-Y, Chung EK, Kim GS, Kim L, Lee Y-S, Choi I-G, 2005. Alcohol and aldehyde dehydrogenase polymorphisms in men with type I and type II alcoholism. Am. J. Psychiatry 162, 1003–1005. [DOI] [PubMed] [Google Scholar]
- Chen AC, Tang Y, Rangaswamy M, Wang JC, Almasy L, Foroud T, Edenberg HJ, Hesselbrock V, Nurnberger J Jr., Kuperman S, O’Connor SJ, Schuckit MA, Bauer LO, Tischfield J, Rice JP, Bierut L, Goate A, Porjesz B, 2009. Association of single nucleotide polymorphisms in a glutamate receptor gene (GRM8) with theta power of event-related oscillations and alcohol dependence. Am. J. Med. Genet. B Neuropsychiatr. Genet 150B (3), 359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XD, Xiong DH, Yang TL, Pei YF, Guo YF, Li J, Yang F, Pan F, Tan LJ, Yan H, Liu XG, Lei SF, Li X, Ning LL, Zhu XZ, Levy S, Kranzler HR, Farrer LA, Gelernter J, Recker RR, Deng HW, 2012. ANKRD7 and CYTL1 are novel risk genes for alcohol drinking behavior. Chin. Med. J. (Engl.) 125, 1127–1134. [PMC free article] [PubMed] [Google Scholar]
- Cui R, Kamatani Y, Takahashi A, Usami M, Hosono N, Kawaguchi T, Tsunoda T, Kamatani N, Kubo M, Nakamura Y, Matsuda K, 2009. Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology 137, 1768–1775. [DOI] [PubMed] [Google Scholar]
- Dick DM, Aliev F, Wang JC, Saccone S, Hinrichs A, Bertelsen S, Budde J, Saccone N, Foroud T, Nurnberger J Jr., Xuei X, Conneally PM, Schuckit M, Almasy L, Crowe R, Kuperman S, Kramer J, Tischfield JA, Hesselbrock V, Edenberg HJ, Porjesz B, Rice JP, Bierut L, Goate A, 2008. A systematic single nucleotide polymorphism screen to fine-map alcohol dependence genes on chromosome 7 identifies association with a novel susceptibility gene ACN9. Biol. Psychiatry 63 (11), 1047–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, 2007. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol. Res. Health 30 (1), 5–13. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, 2012. Genes contributing to the development of alcoholism: an overview. Alcohol Res. 34, 336–338. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T, 2006. The genetics of alcoholism: identifying specific genes through family studies. Addict. Biol 11, 386–396. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T, 2013. Genetics and alcoholism. Nat. Rev. Gastroenterol. Hepatol 10, 487–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Xuei X, Chen H-J, Tian H, Wetherill LF, Dick DM, Almasy L, Bierut L, Bucholz KK, Goate A, Hesselbrock V, Kuperman S, Nurnberger J, Porjesz B, Rice J, Schuckit M, Tischfield J, Begleiter H, Foroud T, 2006. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Hum. Mol. Genet 15, 1539–1549. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Wang J, Tian H, Pochareddy S, Xuei X, Wetherill L, Goate A, Hinrichs T, Kuperman S, Nurnberger JI, Schuckit M, Tischfield JA, Fououd T, 2008. A regulatory variation in OPRK1, the gene encoding the kappa-opioid receptor, is associated with alcohol dependence. Hum. Mol. Genet 17, 1783–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, Dick D, Hesselbrock V, Hinrichs A, Kramer J, Kuperman S, Nurnberger JI Jr., Rice JP, Schuckit MA, Taylor R, Todd Webb B, Tischfield JA, Porjesz B, Foroud T, 2010. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol. Clin. Exp. Res 34, 840–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Gilder DA, Wall TL, Phillips E, Feiler H, Wilhelmsen KC, 2004. Genomic screen for loci associated with alcohol dependence in Mission Indians. Am. J. Med. Genet. B Neuropsychiatr. Genet 129B, 110–115. [DOI] [PubMed] [Google Scholar]
- Ehringer MA, Clegg HV, Collins AC, Corley RP, Crowley T, Hewitt JK, Hopfer CJ, Krauter K, Lessem J, Rhee SH, Schlaepfer I, Smolen A, Stallings MC, Young SE, Zeiger JS, 2007. Association of the neuronal nicotinic receptor beta2 subunit gene (CHRNB2) with subjective responses to alcohol and nicotine. Am. J. Med. Genet. B Neuropsychiatr. Genet 144B (5), 596–604. [DOI] [PubMed] [Google Scholar]
- Enoch MA, 2008. The role of GABA(A) receptors in the development of alcoholism.Pharmacol. Biochem. Behav 90 (1), 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, 2013. Genetic influences on the development of alcoholism. Curr. Psychiatry Rep 15, 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinn R, Nellissery M, Kranzler HR, 2005. Meta-analysis of the association of a functional serotonin transporter promoter polymorphism with alcohol dependence. Am. J. Med. Genet. B Neuropsychiatr. Genet 133B, 79–84. [DOI] [PubMed] [Google Scholar]
- Flatscher-Bader T, van der Brug M, Hwang JW, Gochee PA, Matsumoto I, Niwa S, Wilce PA, 2005. Alcohol-responsive genes in the frontal cortex and nucleus accumbens of human alcoholics. J. Neurochem 93, 359–370. [DOI] [PubMed] [Google Scholar]
- Flatscher-Bader T, van der Brug MP, Landis N, Hwang JW, Harrison E, Wilce PA, 2006. Comparative gene expression in brain regions of human alcoholics. Genes Brain Behav 5 (Suppl. 1), 78–84. [DOI] [PubMed] [Google Scholar]
- Flatscher-Bader T, Harrison E, Matsumoto I, Wilce PA, 2010. Genes associated with alcohol abuse and tobacco smoking in the human nucleus accumbens and ventral tegmental area. Alcohol. Clin. Exp. Res 34, 1291–1302. [DOI] [PubMed] [Google Scholar]
- Foroud T, Wetherill LF, Kramer J, Tischfield JA, Nurnberger J, Schuckit MA, Xuei X, Edenberg HJ, 2008. The tachykinin receptor 3 is associated with alcohol and cocaine dependence. Alcohol. Clin. Exp. Res 32, 1023–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo AY, Webb BT, Miles MF, Zimmerman MP, Kendler KS, Zhao Z, 2009. ERGR: an ethanol-related gene resource. Nucleic Acids Res. 37, D840–D845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA, McEvoy BP, Schrage AJ, Grant JD, Chou YL, Zhu R, Henders AK, Medland SE, Gordon SD, Nelson EC, Agrawal A, Nyholt DR, Bucholz KK, Madden PA, Montgomery GW, 2011. A quantitative-trait genome-wide association study of alcoholism risk in the community: findings and implications. Biol. Psychiatry 70 (6), 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SY, Shen S, Zezza N, Hoffman EK, Perlin M, Allan W, 2004. A genome wide search for alcoholism susceptibility genes. Am. J. Med. Genet. B Neuropsychiatr. Genet 128B (1), 102–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs A, Wang JC, Bufe B, Kwon JM, Budde J, Allen R, Bertelsen S, Evans W, Dick D, Rice J, Foroud T, Nurnberger J, Tischfield JA, Kuperman S, Crowe R, Hesselbrock V, Schuckit M, Almasy L, Porjesz B, Edenberg HJ, Begleiter H, Meyerhof W, Bierut LJ, Goate AM, 2006. Functional variant in a bitter-taste receptor (hTAS2R16) influences risk of alcohol dependence. Am. J. Hum. Genet 78, 103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishimoto A, Liu Q-R, Drgon T, Pletnikova O, Walther D, Zhu XG, Troncoso JC, Uhl GR, 2007. Neurexin 3 polymorphisms are associated with alcohol dependence and altered expression of specific isoforms. Hum. Mol. Genet 16, 2880–2891. [DOI] [PubMed] [Google Scholar]
- Joslyn G, Ravindranathan A, Brush G, Schuckit M, White RL, 2010. Human variation in alcohol response is influenced by variation in neuronal signaling genes. Alcohol. Clin. Exp. Res 34, 800–812. [DOI] [PubMed] [Google Scholar]
- Kalsi G, Kuo P-H, Aliev F, Alexander J, McMichael O, Patterson DG, Walsh D, Zhao Z, Schuckit M, Nurnberger J Jr., Edenberg H, Kramer J, Hesselbrock V, Tischfield JA, Vladimirov V, Prescott CA, Dick DM, Kendler KS, Riley BP, 2010. A systematic gene-based screen of chr4q22-q32 identifies association of a novel susceptibility gene, DKK2, with the quantitative trait of alcohol dependence symptom counts. Hum. Mol. Genet 19, 2497–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SJ, Rangaswamy M, Manz N, Wang JC, Wetherill L, Hinrichs T, Almasy L, Brooks A, Chorlian DB, Dick D, Hesselbrock V, Kramer J, Kuperman S, Nurnberger J Jr., Rice J, Schuckit M, Tischfield J, Bierut LJ, Edenberg HJ, Goate A, Foroud T, Porjesz B, 2012. Family-based genome-wide association study of frontal θ oscillations identifies potassium channel gene KCNJ6. Genes Brain Behav 11 (6), 712–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M, Wang JC, Wetherill L, Le N, Bertelsen S, Hinrichs AL, Budde J, Agrawal A, Bucholz K, Dick D, Harari O, Hesselbrock V, Kramer J, Nurnberger JI Jr., Rice J, Saccone N, Schuckit M, Tischfield J, Porjesz B, Edenberg HJ, Bierut L, Foroud T, Goate A, 2013. A meta-analysis of two genome-wide association studies to identify novel loci for maximum number of alcoholic drinks. Hum. Genet 132 (10), 1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M, Wang JC, Wetherill L, Le N, Bertelsen S, Hinrichs AL, Budde J, Agrawal A, Almasy L, Bucholz K, Dick DM, Harari O, Xiaoling X, Hesselbrock V, Kramer J, Nurnberger JI Jr., Rice J, Schuckit M, Tischfield J, Porjesz B, Edenberg HJ, Bierut L, Foroud T, Goate A, 2014. Genome-wide survival analysis of age at onset of alcohol dependence in extended high-risk COGA families. Drug Alcohol Depend. 142C, 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauhanen J, Karvonen MK, Pesonen U, Koulu M, Tuomainen T-P, Uusitupa MIJ, Salonen JT, 2000. Neuropeptide Y polymorphism and alcohol consumption in middle-aged men. Am. J. Med. Genet 93, 117–121. [DOI] [PubMed] [Google Scholar]
- Kerner B, Lambert CG, Muthen BO, 2011. Genome-wide association study in bipolar patients stratified by co-morbidity. PLoS One 6, e28477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes KM, Hatzenbuehler ML, Grant BF, Hasin DS, 2012. Stress and alcohol: epidimiologic evidence. Alcohol Res. 34, 391–400. [PMC free article] [PubMed] [Google Scholar]
- Koob GF, 2013. Theoretical frameworks and mechanistic aspects of alcohol addiction: alcohol addiction as a reward deficit disorder. Curr. Top. Behav. Neurosci 13, 3–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo PH, Neale MC, Riley BP, Webb BT, Sullivan PF, Vittum J, Patterson DG, Thiselton DL, van den Oord EJ, Walsh D, Kendler KS, Prescott CA, 2006. Identification of susceptibility loci for alcohol-related traits in the Irish Affected Sib Pair Study of Alcohol Dependence. Alcohol. Clin. Exp. Res 30 (11), 1807–1816. [DOI] [PubMed] [Google Scholar]
- Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA, 2000. Gene expression in human alcoholism: microarray analysis of frontal cortex. Alcohol. Clin. Exp. Res 24, 1873–1882. [PubMed] [Google Scholar]
- Lind PA, Macgregor S, Heath AC, Madden PA, Montgomery GW, Martin NG, Whitfield JB, 2012. Association between in vivo alcohol metabolism and genetic variation in pathways that metabolize the carbon skeleton of ethanol and NADH reoxidation in the Alcohol Challenge Twin Study. Alcohol. Clin. Exp. Res 36 (12), 2074–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linneberg A, Gonzalez-Quintela A, Vidal C, Jorgensen T, Fenger M, Hansen T, Pedersen O, Husemoen LL, 2010. Genetic determinants of both ethanol and acetaldehyde metabolism influence alcohol hypersensitivity and drinking behaviour among Scandinavians. Clin. Exp. Allergy 40 (1), 123–130. [DOI] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Dodd PR, Randall PK, Harris RA, Mayfield RD, 2004. Gene expression profiling of individual cases reveals consistent transcriptional changes in alcoholic human brain. J. Neurochem 90, 1050–1058. [DOI] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Lyer VR, Dodd PR, Randall PK, Mayfield RD, 2006. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology 31, 1574–1582. [DOI] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Dodd PR, Mayfield RD, 2007. Altered gene expression profiles in the frontal cortex of cirrhotic alcoholics. Alcohol. Clin. Exp. Res 31, 1460–1466. [DOI] [PubMed] [Google Scholar]
- Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, Bennett PH, Goldman D, 1998. Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am. J. Med. Genet 81, 216–221. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Wang S, Blumberg HP, Gelernter J, 2005. CHRM2 gene predisposes to alcohol dependence, drug dependence and affective disorders: results from an extended case–control structured association study. Hum. Mol. Genet 14, 2421–2434. [DOI] [PubMed] [Google Scholar]
- Macgregor S, Lind PA, Bucholz KK, Hansell NK, Madden PAF, Richter MM, Montgomery GW, Martin NG, Heath AC, Whitfield JB, 2009. Associations of ADH and ALDH2 gene variation with self report alcohol reactions, consumption and dependence: an integrated analysis. Hum. Mol. Genet 18, 580–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzardo AM, Gunewardena S, Wang K, Butler MG, 2014. Exon microarray analysis of human dorsolateral prefrontal cortex in alcoholism. Alcohol. Clin. Exp. Res 38, 1594–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield RD, Lewohl JM, Dodd PR, Herlihy A, Liu J, Harris RA, 2002. Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics.J. Neurochem 81, 802–813. [DOI] [PubMed] [Google Scholar]
- Mayfield RD, Harris RA, Schuckit MA, 2008. Genetic factors influencing alcohol dependence. Br. J. Pharmacol 154, 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield J, Ferguson L, Harris RA, 2013. Neuroimmune signaling: a key component of alcohol abuse. Curr. Opin. Neurobiol 23, 513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova TV, Goldman D, Mackay TFC, Anholt RRH, 2012. The genetic basis of alcoholism: multiple phenotypes, many genes, complex networks. Genome Biol. 13(2), 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova TV, Mackay TF, Anholt RR, 2014. Genetics and genomics of alcohol sensitivity. Mol. Genet. Genomics 289, 253–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Luo X, Liu X, Wu LY, Zhang Q, Wang L, Wang W, Zuo L, Wang KS, 2013. Genome-wide association studies of maximum number of drinks. J. Psychiatr. Res 47 (11), 1717–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radel M, Vallejo RL, Iwata N, Aragon R, Long JC, Virkkunen M, Goldman D, 2005. Haplotype-based localization of an alcohol dependence gene to the 5q34 gammaaminobutyric acid type A gene cluster. Arch. Gen. Psychiatry 62, 47–55. [DOI] [PubMed] [Google Scholar]
- Ray LA, Hutchison KE, 2004. A polymorphism of the mu-opioid receptor gene (OPRM1) and sensitivity to the effects of alcohol in humans. Alcohol. Clin. Exp. Res 28 (12), 1789–1795. [DOI] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li TK, et al. , 1998. Genomewide search for genes affecting the risk for alcohol dependence. Am. J. Med. Genet 81, 207–215. [PubMed] [Google Scholar]
- Rietschel M, Treutlein J, 2013. The genetics of alcohol dependence. Ann. N. Y. Acad. Sci 1282, 39–70. [DOI] [PubMed] [Google Scholar]
- Schepis TS, Rao U, Yadav H, Adinoff B, 2011. The limbic-hypothalamic-pituitary-adrenal axis and the development of alcohol use disorders in youth. Alcohol. Clin. Exp. Res 35, 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, Desrivières S, Aliev FA, Khan AA, Amin N, Aulchenko YS, Bakalkin G, Bakker SJ, Balkau B, Beulens JW, Bilbao A, de Boer RA, Beury D, Bots ML, Breetvelt EJ, Cauchi S, Cavalcanti-Proença C, Chambers JC, Clarke TK, Dahmen N, de Geus EJ, Dick D, Ducci F, Easton A, Edenberg HJ, Esko T, Fernández-Medarde A, Foroud T, Freimer NB, Girault JA, Grobbee DE, Guarrera S, Gudbjartsson DF, Hartikainen AL, Heath AC, Hesselbrock V, Hofman A, Hottenga JJ, Isohanni MK, Kaprio J, Khaw KT, Kuehnel B, Laitinen J, Lobbens S, Luan J, Mangino M, Maroteaux M, Matullo G, McCarthy MI, Mueller C, Navis G, Numans ME, Núñez A, Nyholt DR, Onland-Moret CN, Oostra BA, O’Reilly PF, Palkovits M, Penninx BW, Polidoro S, Pouta A, Prokopenko I, Ricceri F, Santos E, Smit JH, Soranzo N, Song K, Sovio U, Stumvoll M, Surakk I, Thorgeirsson TE, Thorsteinsdottir U, Troakes C, Tyrfingsson T, Tönjes A, Uiterwaal CS, Uitterlinden AG, van der Harst P, van der Schouw YT, Staehlin O, Vogelzangs N, Vollenweider P, Waeber G, Wareham NJ, Waterworth DM, Whitfield JB, Wichmann EH, Willemsen G, Witteman JC, Yuan X, Zhai G, Zhao JH, Zhang W, Martin NG, Metspalu A, Doering A, Scott J, Spector TD, Loos RJ, Boomsma DI, Mooser V, Peltonen L, Stefansson K, van Duijn CM, Vineis P, Sommer WH, Kooner JS, Spanagel R, Heberlein UA, Jarvelin MR, Elliott P, 2011. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc. Natl. Acad. Sci. U. S. A 108, 7119–7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolov BP, Jiang L, Trivedi NS, Aston C, 2003. Transcription profiling reveals mitochondrial, ubiquitin and signaling systems abnormalities in postmortem brains from subjects with a history of alcohol abuse or dependence. J. Neurosci. Res 72, 756–767. [DOI] [PubMed] [Google Scholar]
- Taylor A, Wang KS, 2014. Association between DPYSL2 gene polymorphisms and alcohol dependence in Caucasian samples. J. Neural Transm 121 (1), 105–111. [DOI] [PubMed] [Google Scholar]
- Tikkanen R, Sjoberg RL, Ducci F, Goldman D, Holi M, Tiihonen J, Virkkunen M, 2009. Effects of MAOA-genotype, alcohol consumption, and aging on violent behavior. Alcohol. Clin. Exp. Res 33 (3), 428–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolstrup JS, Nordestgaard BG, Rasmussen S, Tybjaerg-Hansen A, Gronbaek M, 2008. Alcoholism and alcohol drinking habits predicted from alcohol dehydrogenase genes. Pharmacogenomics J. 8, 220–227. [DOI] [PubMed] [Google Scholar]
- Wang JC, Hinrichs AL, Bertelsen S, Stock H, Budde JP, Dick DM, Bucholz KK, Rice J, Saccone N, Edenberg HJ, Hesselbrock V, Kuperman S, Schuckit MA, Bierut LJ, Goate AM, 2007. Functional variants in TAS2R38 and TAS2R16 influence alcohol consumption in high-risk families of African–American origin. Alcohol. Clin. Exp. Res 31, 209–215. [DOI] [PubMed] [Google Scholar]
- Wang JC, Grucza R, Cruchaga C, Hinrichs AL, Bertelsen S, Budde JP, Fox L, Goldstein E, Reyes O, Saccone N, Saccone S, Xuei X, Bucholz K, Kuperman S, Nurnberger J Jr., Rice JP, Schuckit M, Tischfield J, Hesselbrock V, Porjesz B, Edenberg HJ, Bierut LJ, Goate AM, 2009. Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. Mol. Psychiatry 14 (5), 501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KS, Liu X, Aragam N, Jian X, Mullersman JE, Liu Y, Pan Y, 2011a. Family-based association analysis of alcohol dependence in the COGA sample and replication in the Australian twin-family study. J. Neural Transm 118 (9), 1293–1299. [DOI] [PubMed] [Google Scholar]
- Wang KS, Liu X, Zhang Q, Pan Y, Aragam N, Zeng M, 2011b. A meta-analysis of two genome-wide association studies identifies 3 new loci for alcohol dependence.J. Psychiatr. Res 45 (11), 1419–1425. [DOI] [PubMed] [Google Scholar]
- Wang KS, Liu X, Zhang Q, Wu LY, Zeng M, 2012. Genome-wide association study identifies 5q21 and 9p24.1 (KDM4C) loci associated with alcohol withdrawal symptoms. J. Neural Transm 119 (4), 425–433. [DOI] [PubMed] [Google Scholar]
- Wetherill L, Schuckit MA, Hesselbrock V, Xuei X, Liang T, Dick DM, Kramer J, Nurnberger JI Jr., Tischfield JA, Porjesz B, Edenberg HJ, Foroud T, 2008. Neuropep-tide Y receptor genes are associated with alcohol dependence, alcohol withdrawal phenotypes, and cocaine dependence. Alcohol. Clin. Exp. Res 32 (12), 2031–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetherill L, Kapoor M, Agrawal A, Bucholz K, Koller D, Bertelsen SE, Le N, Wang JC, Almasy L, Hesselbrock V, Kramer J, Nurnberger JI Jr., Schuckit M, Tischfield JA, Xuei X, Porjesz B, Edenberg HJ, Goate AM, Foroud T, 2014. Family-based association analysis of alcohol dependence criteria and severity. Alcohol. Clin. Exp. Res 38, 354–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TJ, LaForge KS, Gordon D, Bart G, Kellogg S, et al. , 2007. Prodynorphin gene promoter repeat associated with cocaine/alcohol codependence. Addict. Biol 12, 496–502. [DOI] [PubMed] [Google Scholar]
- Yan J, Aliev F, Webb BT, Kendler KS, Williamson VS, Edenberg HJ, Agrawal A, Kos MZ, Almasy L, Nurnberger JI Jr., Schuckit MA, Kramer JR, Rice JP, Kuperman S, Goate AM, Tischfield JA, Porjesz B, Dick DM, 2014. Using genetic information from candidate gene and genome-wide association studies in risk prediction for alcohol dependence. Addict. Biol 19, 708–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang BZ, Kranzler HR, Zhao H, Gruen JR, Luo X, Gelernter J, 2008. Haplotypic variants in DRD2, ANKK1, TTC12, and NCAM1 are associated with comorbid alcohol and drug dependence. Alcohol. Clin. Exp. Res 32 (12), 2117–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Gwinn M, Clyne M, Yesupriya A, Khoury MJ, 2008. A navigator for human genome epidemiology. Nat. Genet 40 (2), 124–125. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kranzler HR, Yang B-Z, Luo X, Gelernter J, 2008. The OPRD1 and OPRK1 loci in alcohol or drug dependence: OPRD1 variation modulates substance dependence risk. Mol. Psychiatry 13, 531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Guo AY, van den Oord EJ, Aliev F, Jia P, Edenberg HJ, Riley BP, Dick DM, Bettinger JC, Davies AG, Grotewiel MS, Schuckit MA, Agrawal A, Kramer J, Nurnberger JI Jr., Kendler KS, Webb BT, Miles MF, 2012. Multi-species data integration and gene ranking enrich significant results in an alcoholism genome-wide association study. BMC Genomics 13 (Suppl. 8), S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlojutro M, Manz N, Rangaswamy M, Xuei X, Flury-Wetherill L, Koller D, Bierut LJ, Goate A, Hesselbrock V, Kuperman S, Nurnberger J Jr., Rice JP, Schuckit MA, Foroud T, Edenberg HJ, Porjesz B, Almasy L, 2010. Genome-wide association study of theta band event-related oscillations identifies serotonin receptor gene HTR7 influencing risk of alcohol dependence. Am. J. Med. Genet. B Neuropsychiatr. Genet 156B, 44–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Luo X, Listman JB, Kranzler HR, Wang S, Anton RF, Blumberg HP, Stein MB, Pearlson GD, Covault J, Charney DS, van Kammen DP, Price LH, Lappalainen J, Cramer J, Krystal JH, Gelernter J, 2009. Population admixture modulates risk for alcohol dependence. Hum. Genet 125, 605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Zhang CK, Wang F, Li CS, Zhao H, Lu L, Zhang XY, Zhang H, Zhang F, Krystal JH, Luo X, 2011. A novel, functional and replicable risk gene region for alcohol dependence identified by genome-wide association study. PLoS One 6 (11), e26726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Gelernter J, Zhang CK, Zhao H, Lu L, Kranzler HR, Malison RT, Li CS, Wang F, Zhang XY, Deng HW, Krystal JH, Zhang F, Luo X, 2012. Genome-wide association study of alcohol dependence implicates KIAA0040 on chromosome 1q. Neuropsychopharmacology 37 (2), 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Wang K, Zhang XY, Krystal JH, Li CS, Zhang F, Zhang H, Luo X, 2013a. NKAIN1-SERINC2 is a functional, replicable and genome-wide significant risk gene region specific for alcohol dependence in subjects of European descent. Drug Alcohol Depend. 129 (3), 254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Zhang XY, Wang F, Li CS, Lu L, Ye L, Zhang H, Krystal JH, Deng HW, Luo X, 2013b. Genome-wide significant association signals in IPO11-HTR1A region specific for alcohol and nicotine codependence. Alcohol. Clin. Exp. Res 37 (5), 730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]