Abstract
Bipolar disorder (BPD) is genetically heterogeneous with a growing list of BPD associated genes reported in recent years resulting from increased genetic testing using advanced genetic technology, expanded genomic databases, and better awareness of the disorder. We compiled a master list of recognized susceptibility and genes associated with BPD identified from peer-reviewed medical literature sources using PubMed and by searching online databases, such as OMIM. Searched keywords were related to bipolar disorder and genetics. Our compiled list consisted of 290 genes with gene names arranged in alphabetical order in tabular form with source documents and their chromosome location and gene symbols plotted on high-resolution human chromosome ideograms. The identified genes impacted a broad range of biological pathways and processes including cellular signaling pathways particularly cAMP and calcium (e.g., CACNA1C, CAMK2A, CAMK2D, ADCY1, ADCY2); glutamatergic (e.g., GRIK1, GRM3, GRM7), dopaminergic (e.g., DRD2, DRD4, COMT, MAOA) and serotonergic (e.g., HTR1A, HTR2A, HTR3B) neurotransmission; molecular transporters (e.g., SLC39A3, SLC6A3, SLC8A1); and neuronal growth (e.g., BDNF, IGFBP1, NRG1, NRG3). The increasing prevalence of BPD calls for better understanding of the genetic etiology of this disorder and associations between the observed BPD phenotype and genes. Visual representation of genes for bipolar disorder becomes a tool enabling clinical and laboratory geneticists, genetic counselors, and other health care providers and researchers easy access to the location and distribution of currently recognized BPD associated genes. Our study may also help inform diagnosis and advance treatment developments for those affected with this disorder and improve genetic counseling for families.
Keywords: Bipolar disorder, Chromosome ideograms, Major affective disorder, Manic-depressive illness, Susceptibility genes
1. Introduction
Bipolar disorder (BPD) or major affective disorder is largely undiagnosed but known to cause unusual shifts in behavior, including mood, activity levels, and ability to perform everyday tasks. According to the Diagnostic and Statistical Manual for Mental Disorders, fifth edition (DSM-5), BPD can be separated into four basic types: Bipolar I Disorder (BP-I), Bipolar II Disorder (BP-II), Bipolar Disorder Not Otherwise Specified (BP-NOS), and Cyclothymic Disorder. The diagnosis is dependent on severity and length of the manic and depressive phases [Hirschfeld et al., 2000, 2003a, 2003b; Juvenile Bipolar Research Foundation (http://www.jbrf.org/diagnosis-by-the-dsm/); National Institute of Mental Health (http://www.nimh.nih.gov/health/topics/bipolar-disorder/index.shtml#part_145403)]. The twelve month prevalence of BPD is 1.5% for the adult population in the U.S., 0.4% for BP-I, 0.3% for BP-II, and 0.8% for BP-NOS. The lifetime prevalence of BPD in the U.S. population is 2.4% for the total population; 0.6% for BP-I, 0.4% for BP-II, and 1.4% for BP-NOS (Merikangas et al., 2011). Men and women are equally affected by BPD, but women are three times more likely than men to experience the rapid cycling process and tend to experience the depressive and mixed episodes more often than men (Merikangas et al., 2011). Most individuals are diagnosed with bipolar disorder around the age of 25 years, but some individuals are diagnosed as young as six years of age or as late as 40 and 50 years [Birmaher et al., 2006; Geller et al., 2004; Depression and Bipolar Support AllianceDepression and Bipolar Support Alliance (http://www.dbsalliance.org/site/PageServer?pagename=education_statistics_bipolar_disorder)]. Given the nature of the disorder, it is common for individuals with BPD to be misdiagnosed as having another mental illness, which can hinder recognition and treatment [Depression and Bipolar Support Alliance (http://www.dbsalliance.org/site/PageServer?pagename=education_statistics_bipolar_disorder)]. BPD often coexists with other Axis I and Axis II disorders [e.g. substance abuse, anxiety disorders (such as agoraphobia, post-traumatic stress disorder, and social phobia), and eating disorders] with reported rates of lifetime psychiatric comorbidity in bipolar I samples ranging from 50% to 70% (McElroy et al., 2001). Approximately 20% of the children and adolescents experiencing major depression will develop bipolar disorder within five years of being diagnosed with depression (Birmaher et al., 2006; McClellan et al., 2007). Almost one-third of adolescents with depression are actually experiencing early onset of bipolar disorder.
Numerous twin and family studies have supported a genetic contribution to the risk of developing BPD. Monozygotic twins show greater concordance with psychopathology than dizygotic twins and individuals with affected relatives show an increased chance of developing bipolar and/or unipolar depression (Smoller and Finn, 2003). One affected parent is associated with a 15 to 25% elevation in risk of BPD while two affected parents shows a 50 to 75% increase [Smoller and Finn, 2003; Depression and Bipolar Support Alliance (http://www.dbsalliance.org/site/PageServer?pagename=education_statistics_bipolar_disorder)]. Never-the-less, bipolar disorder is a complex genetic disorder that does not follow Mendelian inheritance patterns with no identifiable “gene of major effect” found for the majority of BPD cases. There are also several chromosomal regions, susceptibility loci and implicated genes for BPD that have been repeatedly reported as associated with the disorder through linkage, candidate and genome-wide association studies (Craddock and Jones, 1999; Craddock et al., 2005). Interactions between numerous genes in multiple overlapping pathways and functions reduce the likelihood of one-to-one correspondence for any single “causal” bipolar disorder gene mutation (Potash and DePaulo, 2000).
Over the last few decades, researchers have identified numerous candidate genes associated with BPD using various methodologies, including genotyping single-nucleotide polymorphisms (SNPs), identifying cytogenetic abnormalities (i.e., chromosomal breakpoints which lead to the loss or alteration of specific genes), or convergent functional genomics (Cichon et al., 2009). We used existing literature and genomic databases to obtain evidence to compile a master list of currently recognized genes with their locations plotted on high-resolution chromosome ideograms (850 band level) associated with BPD. In tabular form, we listed the individual gene symbol, expanded name, and chromosome location alongside the reference source providing the information.
2. Material and methods
We used computer-based internet websites and PubMed (https://www.ncbi.nlm.nih.gov/pubmed) to search for key words based on the genetics of bipolar disorder. Articles were obtained by searching PubMed and Online Mendelian Inheritance in Man (OMIM: http://www.ncbi.nlm.nih.gov/omim) databases with the following search words: bipolar disorder (BPD), bipolar, major affective disorder, bipolar affective disorder (BPAD), bipolar syndrome, and manic depressive psychosis. We examined literature found in medical journals after our search for genetic involvement for BPD. Some articles we found had their own compilation of susceptibility genes for BPD with references to their own sources which we then further studied (Thomson et al., 2005; Le-Niculescu et al., 2009; Cichon et al., 2009; Shinozaki and Potash, 2014). The articles were then prioritized based on the following considerations: sample size, use of standardized diagnostic criteria, types of genetic testing such as genome wide association studies and validated methods (e.g., convergent functional genomics, genotyping SNPs or identifying cytogenetic abnormalities), and quality with reliability of the genetic data and presentation. GeneCards (http://www.genecards.org/) was the primary source for determining the location of the gene or gene locus. The cytogenetic location of the gene was provided by Ensembl, Entrez Gene, or the Human Genome Organization Gene Nomenclature Committee (HGNC).
BPD is a heterogeneous disorder involving many genes acting individually or in combination and responsive to environmental stimuli. We compiled a list of genes from the major sources and their references for a total of 290 genes. Our paper focused on genes associated with BPD by at least one mechanism of proven association with or susceptibility to BPD. Research articles were not limited to causal relationships to BPD. For example, Le-Niculescu and others used convergence of microsatellite markers for which at least one published study showed evidence for linkage for BPD, or a positive association study for the gene itself reported in previous literature (Le-Niculescu et al., 2009). Many of the genes on our list were found in multiple research studies and were reported more than once for being associated with BPD. Some studies considered target neurochemical pathways rather than susceptibility loci. These genes were included on our list if they were also recognized by a peer-reviewed publication (e.g., PubMed) with supporting evidence (e.g., GWAS, informative SNPs, genetic linkage, or identified gene mutations) as a BPD associated gene (Le-Niculescu et al., 2009; Cichon et al., 2011). Other supporting genetic evidence can be found in the National Center for Biotechnology Resources at https://www.ncbi.nlm.nih.gov/gene.
3. Results
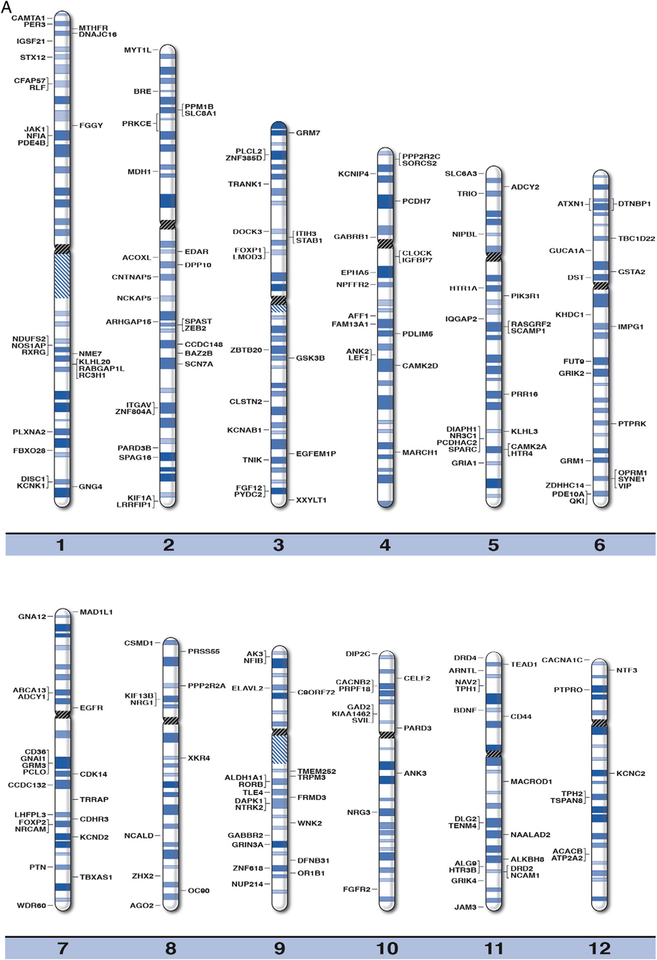
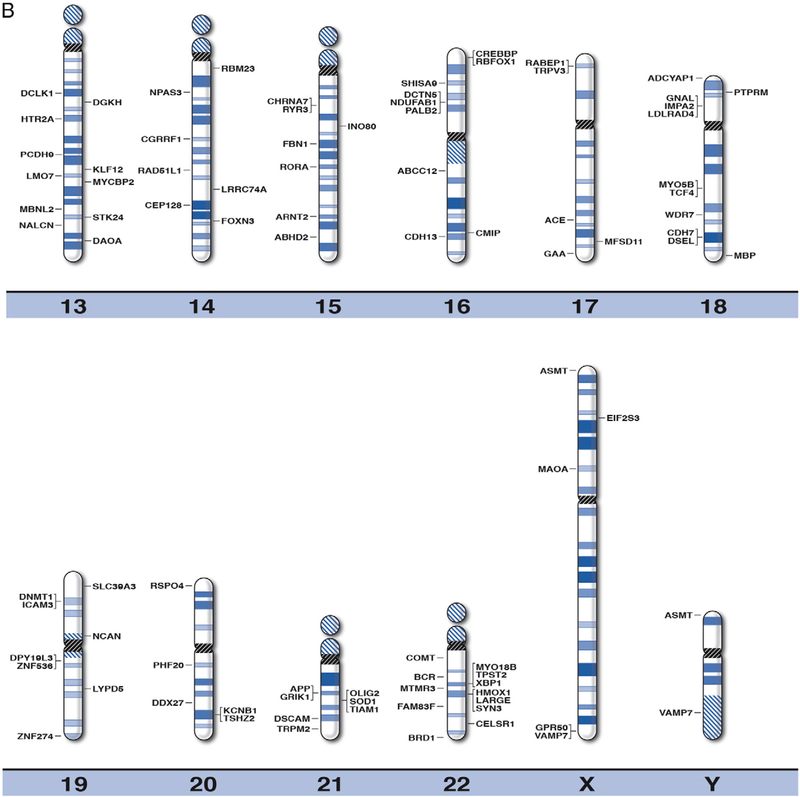
By using online databases and existing literature in peer-reviewed journals, we were able to compile a master list of 290 recognized genes associated with BPD. These genes were clinically relevant, or related to susceptibility of BPD. The position for each of the recognized or susceptibility genes for BPD were plotted on the high-resolution chromosome ideograms (850 band level), as shown in Fig. 1. We have also included gene symbols, expanded names, chromosome band location and reference sources in Table 1 for the 290 genes recognized as playing a role in BPD. The distribution of BPD genes is shown in Table 2 among individual chromosomes and chromosome arms arranged by the size for each chromosome (largest chromosome represented by the smallest number) in relationship to the proportion of total genes for BPD.
Fig. 1.
High-resolution chromosome ideograms (850 band level) with the BPD gene symbol placed at the chromosomal band location. The centromere area, highlighted in black, separates the upper short “p” arm and the lower long “q” arm for each chromosome. The gene symbols are arranged in alphabetical order with the expanded name and chromosome band position listed in Table 1.
Table 1.
Known and candidate genes for bipolar disorder (BPD) and overlap with schizophrenia and autism.
| Gene symbol | Gene name | Location | Source | ||
|---|---|---|---|---|---|
| ABCA13* | ATP-binding cassette, subfamily A, member 13 | 7p12.3F | Knightet al. (2009) | ||
| ABCCI2 | ATP-binding cassette, subfamily C, member 12 | 16q12.1 | Xu et al. (2014) | ||
| ABHD2 | Abhydrolase domain containing 2 | 15q26.1 | Xu et al. (2014) | ||
| ACACB | Acetyl-coenzyme A carboxylase beta | 12q24.11 | Le-Niculescu et al. (2009) | ||
| ACE* | Angiotensin I converting enzyme | 17q23.3 | Zou et al. (2011) | ||
| ACOXL | Acyl-CoA oxidase-like | 2q13 | Xu et al. (2014) | ||
| ADCY1* | Adenylate cyclase 1 (brain) | 7p12.3 | Le-Niculescu et al. (2009) | ||
| ADCY2 | Adenylate cyclase 2 | 5p15.31 | Mühleisen et al. (2014) | ||
| ADCYAP1* | Adenylate cyclase activating polypeptide 1 (pituitary) | 18p11.32 | Le-Niculescu et al. (2009) | ||
| AFF1 | AF4/FMR2 family, member 1 | 4q21.3 | Xu et al. (2014) | ||
| AGO2 | Argonate RISC catalytic component 2 | 8q24.3 | Le-Niculescu et al. (2009) | ||
| AK3 | Adenylate kinase 3 | 9p24.1 | Le-Niculescu et al. (2009) | ||
| ALDHIAI | Aldehyde dehydrogenase 1 family, member A1 | 9q21.13 | Le-Niculescu et al. (2009) | ||
| ALG9 | ALG9, alpha-1,2-mannosyltransferase | 11q23.1 | Baysal et al. (2002) | ||
| ALKBH8 | AlkB, alkylation repair homolog 8 (Escherichia coli) | 11q22.3 | Xu et al. (2014) | ||
| ANK2# | Ankyrin 2, neuronal | 4q25 | Le-Niculescu et al. (2009) | ||
| ANK3*# | Ankyrin 3 | 10q21.2 | Takata et al. (2011) | ||
| APP# | Amyloid beta (A4) precursor protein | 21q21.3 | Le-Niculescu et al. (2009) | ||
| ARHGAP15# | Rho GTPase activating protein 15 | 2q22.2 | Xu et al. (2014) | ||
| ARNT2# | Aryl-hydrocarbon receptor nuclear translocator 2 | 15q25.1 | Xu et al. (2014) | ||
| ARNTL* | Aryl hydrocarbon receptor nuclear translocator-like | 11p15.3 | Le-Niculescu et al. (2009) | ||
| ASMT# | Acetylserotonin O-methyltransferase | Xp22.33 or Yp11.32 | Etain et al. (2012) | ||
| ATP2A2 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | 12q24.11 | Xu et al. (2014) | ||
| ATXN1* | Ataxin 1 | 6p22.3 | Le-Niculescu et al. (2009) | ||
| BAZ2B | Bromodomain adjacent to zinc finger domain, 2B | 2q24.2 | Xu et al. (2014) | ||
| BCR | Breakpoint cluster region | 22q11.23 | Hashimoto et al. (2005) | ||
| BDNF*# | Brain-derived neurotrophic factor | 11p14.1 | Fan and Sklar (2008) | ||
| BRD1* | Bromodomain containing 1/zinc finger, BED-type containing 4 | 22q13.33 | Nyegaard et al. (2010) | ||
| BRE | Brain and reproductive organ-expressed (TNFRSF1Amodulator) | 2p23.2 | Cichon et al. (2011) | ||
| C9ORF72 | Chromosome 9 open reading frame 72 | 9p212 | Meisler et al. (2013) | ||
| CACNA1C*# | Calcium channel, voltage-dependent, L type, alpha 1C subunit | 12p13.33 | Ou et al. (2015) | ||
| CACNB2*# | Calcium channel, voltage dependent, beta 2 subunit | 10p12.33 | Le-Niculescu et al. (2009) | ||
| CAMK2A | Calcium/calmodulin-dependent protein kinase II, alpha | 5q32 | Le-Niculescu et al. (2009) | ||
| CAMK2D | Calcium/calmodulin-dependent protein kinase II, delta | 4q26 | Le-Niculescu et al. (2009) | ||
| CAMTA1# | Calmodulin binding transcription activator 1 | 1p36.31 | Cichon et al. (2011) | ||
| CCDC132 | Coiled-coil domain containing 132 | 7q21.3 | Cichon et al. (2011) | ||
| CCDC148 | Coiled-coil domain containing 148 | 2q24.1 | Xu et al. (2014) | ||
| CD36 | CD36 molecule (Thrombospondin receptor) | 7q21.11 | Blair et al. (2006) | ||
| CD44# | CD44 molecule (Indian blood group) | 11p13 | Le-Niculescu et al. (2009) | ||
| CDH7 | Cadherin 7, type 2 | 18q22.1 | Soronen et al. (2010) | ||
| CDH13 | Cadherin 13 | 16q23.3 | Le-Niculescu et al. (2009) | ||
| CDHR3 | Cadherin-related family member 3 | 7q22.3 | Xu et al. (2014) | ||
| CDK14 | Cyclin-dependent kinase 14 | 7q21.13 | Le-Niculescu et al. (2009) | ||
| CELF2 | CUGBP, elav-like family member 2 | 10p13 | Le-Niculescu et al. (2009) | ||
| CELSR1 | Cadherin, EGF LAG seven-pass G-type receptor 1 | 22q13.31 | Le-Niculescu et al. (2009) | ||
| CEP128 | Centrosomal protein 128 kDa | 14q31.1 | Le-Niculescu et al. (2009) | ||
| CFAP57 | Cilia and flagella associated protein 57 | 1p342 | Xu et al. (2014) | ||
| CGRRF1 | Cell growth regulator with ring finger domain 1 | 14q22.2 | Xu et al. (2014) | ||
| CHRNA7*# | Cholinergic receptor, nicotinic, alpha 7 (neuronal) | 15q13.3 | Le-Niculescu et al. (2009) | ||
| CLOCK* | Clock circadian regulator | 4q12 | Benedetti et al. (2003) | ||
| CLSTN2 | Calsyntenin 2 | 3q23 | Le-Niculescu et al. (2009) | ||
| CMIP# | C-Maf inducing protein | 16q23.2 | Xu et al. (2014) | ||
| CNTNAP5*# | Contactin associated protein-like 5 | 2q14.3 | Baum et al. (2008a, 2008b) | ||
| COMT* | Catechol-O-methyltransferase | 22q11.21 | Zhang et al. (2009) | ||
| CREBBP# | CREB binding protein | 16p13.3 | Le-Niculescu et al. (2009) | ||
| CSMD1*# | CUB and sushi multiple domains 1 | 8p23.2 | Sklar et al. (2008) | ||
| DAOA* | D-Amino acid oxidase activator | 13q33.2 | Detera-Wadleigh and McMahon (2006) | ||
| DAPK1 # | Death-associated protein kinase 1 | 9q21.33 | Le-Niculescu et al. (2009) | ||
| DCLK1 | Doublecortin-like kinase 1 | 13q13.3 | Le-Niculescu et al. (2009) | ||
| DCTN5# | Dynactin 5 (P25) | 16p12.2 | Burton et al. (2007) | ||
| DDX27 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 | 20q13.13 | Xu et al. (2014) | ||
| DFNB31 | Deafness, autosomal recessive 31 | 9q32 | Ollila et al. (2009) | ||
| DGKH | Diacylglycerol kinase, eta | 13q14.11 | Baum et al. (2008a, 2008b) | ||
| DIAPH1 | Diaphanous-related formin 1 | 5q31.3 | Le-Niculescu et al. (2009) | ||
| DIP2C | Disco-interacting protein 2 homolog C (Drosophila) | 10p15.3 | Djurovic et al. (2010) | ||
| DISC1 *# | Disruption in schizophrenia 1 | 1q42.2 | Hodgkinson et al. (2004) | ||
| DLG2* | Discs, large homolog 2 (Drosophila) | 11q14.1 | Xu et al. (2014) | ||
| DNAJC16 | DnaJ (Hsp40) homolog, subfamily C, member 16 | 1p36.21 | Xu et al. (2014) | ||
| DNMT1 * | DNA methyltransferase 1 | 19p13.2 | Veldic et al.(2005) | ||
| DOCK3 | Dedicator of cytokinesis 3 | 3p212 | Baum et al. (2008a, 2008b) | ||
| DPP10*# | Dipeptidyl-peptidase 10 (non-functional) | 2q14.1 | Le-Niculescu et al. (2009) | ||
| DPY19L3 | Dpy-19-like 3 (Caenorhabditis elegans) | 19q13.11 | Smith et al. (2009) | ||
| DRD2*# | Dopamine receptor D2 | 11q23.2 | Le-Niculescu et al. (2009) | ||
| DRD4* | Dopamine receptor D4 | 11p15.5 | López León et al. (2005) | ||
| DSCAM# | Down syndrome cell adhesion molecule | 21q22.2 | Xu et al. (2014) | ||
| DSEL | Dermatan sulfate epimerase-like | 18q22.1 | Goossens et al. (2003) | ||
| DST# | Dystonin | 6p12.1 | Le-Niculescu et al. (2009) | ||
| DTNBP1* | Dystrobrevin binding protein 1 | 6p22.3 | Gaysina et al. (2009) | ||
| EDAR | Ectodysplasin A receptor | 2q12.3 | Xu et al. (2014) | ||
| EGFEM1P | EGF-Like and EMI domain containing 1, pseudogene | 3q26.2 | Xu et al. (2014) | ||
| EGFR | Epidermal growth factor receptor | 7p11.2 | Sklaret al.(2008) | ||
| EIF2S3# | Eukaryotic translation initiation factor 2, subunit 3 gamma, 52 kDa | Xp22.11 | Cichon et al., (2011) | ||
| ELAVL2 | ELAV like neuron-specific RNA binding protein 2 | 9p21.3 | Le-Niculescu et al. (2009) | ||
| EPHA5 | EPH receptor 5 | 4q13.1 | Le-Niculescu et al. (2009) | ||
| FAM13A1 | Family with sequence similarity 13, member A | 4q22.1 | Le-Niculescu et al. (2009) | ||
| FAM83F | Family with sequence similarity 83, member F | 22q13.1 | Xu et al. (2014) | ||
| FBN1 | Fibrillin 1 | 15q21.1 | Djurovic et al. (2010) | ||
| FBXO28 | F-Box protein 28 | 1q42.11 | Xu et al. (2014) | ||
| FGF12 | Fibroblast growth factor 12 | 3q28 | Cichon et al. (2011) | ||
| FGFR2 | Fibroblast growth factor receptor 2 | 10q26.13 | Xu et al. (2014) | ||
| FGGY | FGGY carbohydrate kinase domain containing | 1p32.1 | Kerner et al. (2013) | ||
| FRMD3 | FERM domain containing 3 | 9q21.32 | Djurovic et al. (2010) | ||
| FOXN3 | Forkhead box N3 | 14q32.11 | Baum et al. (2008a, 2008b) | ||
| FOXP1# | Forkhead box P1 | 3p14.1 | Le-Niculescu et al. (2009) | ||
| FOXP2*# | Forkhead box P2 | 7q31.1 | Xu et al. (2014) | ||
| FUT9 | Fucosyltransferase 9 (alpha (1,3) fucosyltransferase) | 6q16.1 | Le-Niculescu et al. (2009) | ||
| GAA | Glucosidase, alpha; acid | 17q25.3 | Le-Niculescu et al. (2009) | ||
| GABBR2 | Gamma-aminobutyric acid (GABA) B receptor 2 | 9q22.33 | Djurovic et al. (2010) | ||
| GABRB1# | Gamma-aminobutyric acid (GABA) A receptor, beta 1 | 4p12 | Burton et al. (2007) | ||
| GAD2* | Glutamate decarboxylase 2 (pancreatic islets and brain, 65 kDa) | 10p11.23 | Heckers et al. (2002) | ||
| GNAI1 | Guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 | 7q21.11 | Le-Niculescu et al. (2009) | ||
| GNA12 | Guanine nucleotide binding protein (G protein) alpha 12 | 7p22.2 | Le-Niculescu et al. (2009) | ||
| GNAL | Guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type | 18p11.21 | Corradi et al. (2005) | ||
| GNG4 | Guanine nucleotide binding protein (G protein), gamma 4 | 1q42.3 | Cichon et al. (2011) | ||
| GPR50* | G protein-coupled receptor 50 | Xq28 | Thomson et al. (2005) | ||
| GRIA1* | Glutamate receptor, ionotropic, AMPA1 (alpha 1) | 5q33.2 | Le-Niculescu et al. (2009) | ||
| GRIK1 | Glutamate receptor, ionotropic, kainate 1 | 21q21.3 | Le-Niculescu et al. (2009) | ||
| GRIK2# | Glutamate receptor, ionotropic, kainite 2 | 6q16.3 | Shaltiel et al. (2008) | ||
| GRIK4* | Glutamate receptor, ionotropic, kainite 4 | 11q23.3 | Blackwood et al. (2007) | ||
| GRIN3A* | Glutamate receptor, ionotropic, N-methyl-D-aspartate 3A | 9q31.1 | Cichon et al. (2011) | ||
| GRM1# | Glutamate receptor, metabotropic 1 | 6q24.3 | Le-Niculescu et al. (2009) | ||
| GRM3* | Glutamate receptor, metabotropic 3 | 7q21.11 | Le-Niculescu et al. (2009) | ||
| GRM7* | Glutamate receptor, metabotropic 7 | 3p26.1 | Burton et al. (2007) | ||
| GSK3B*# | Glycogen synthase kinase 3 beta | 3q13.33 | Le-Niculescu et al. (2009) | ||
| GSTA2 | Glutathione S-transferase alpha 2 | 6p12.2 | Le-Niculescu et al. (2009) | ||
| GUCA1A | Guanylate cyclase activator 1 A | 6p21.1 | Xu et al. (2014) | ||
| HMOX1 | Heme oxygenase 1 | 22q12.3 | Le-Niculescu et al. (2009) | ||
| HTR1A* | 5-Hydroxytryptamine (serotonin) receptor 1A, G protein-coupled | 5q12.3 | Kishi et al. (2011) | ||
| HTR2A*# | 5-Hydroxytryptamine (serotonin) receptor 2A, G-protein-coupled | 13q14.2 | Le-Niculescu et al. (2009) | ||
| HTR3B* | 5-Hydroxytryptamine (serotonin) receptor 3B, ionotropic | 11q23.1 | Hammer et al. (2012) | ||
| HTR4* | 5-Hydroxytryptamine (serotonin) receptor 4, G-protein coupled | 5q32 | Ohtsuki et al. (2002) | ||
| ICAM3 | Intercellular adhesion molecule 3 | 19p13.2 | Cichon et al. (2011) | ||
| IGFBP7 | Insulin-like growth factor binding protein 7 | 4q12 | Xu et al. (2014) | ||
| IGSF21 | Immunoglobin superfamily, member 21 | 1p36.13 | Cichon et al. (2011) | ||
| IMPA2* | Myo-inositol monophosphatase 2 | 18p11.21 | Sjøholt et al. (2004) | ||
| IMPG1 | Interphotoreceptor matrix proteoglycan 1 | 6q14.1 | Xu et al. (2014) | ||
| INO80 | INO80 complex subunit | 15q15.1 | Xu et al. (2014) | ||
| IQGAP2 | IQmotif containing GTPase activating protein 2 | 5q13.3 | Le-Niculescu et al. (2009) | ||
| ITGAV | Integrin, alpha V | 2q32.1 | Le-Niculescu et al. (2009) | ||
| ITIH3* | Inter-alpha-trypsin inhibitor, heavy chain 3 | 3p21.1 | Hamshere et al. (2013) | ||
| JAK1 | Janus kinase 1 | 1p31.3 | Xu et al. (2014) | ||
| JAM3 | Junctional adhesion molecule 3 | 11q25 | Baum et al. (2008a, 2008b) | ||
| KCNAB1 | Potassium channel, voltage gated subfamily A regulatory beta subunit 1 | 3q25.31 | Le-Niculescu et al. (2009) | ||
| KCNB1 | Potassium channel, voltage gated Shab-related subfamily B, member 1 | 20q13.2 | Le-Niculescu et al. (2009) | ||
| KCNC2 | Potassium channel, voltage gated Shaw related subfamily C, member 2 | 12q14.1 | Burton et al. (2007) | ||
| KCND2# | Potassium voltage-gated channel, Shal-related family, member 2 | 7q31.31 | Le-Niculescu et al. (2009) | ||
| KCNIP4 | Kv channel interacting protein 4 | 4p15.32 | Xu et al. (2014) | ||
| KCNK1 | Potassium channel, subfamily K, member 1 | 1q422 | Le-Niculescu et al. (2009) | ||
| KHDC1 | KH homology domain containing 1 | 6q13 | Xu et al. (2014) | ||
| KIAA1462 | KIAA1462 gene | 10p11.23 | Xu et al. (2014) | ||
| KIF13B | Kinesin family member 13B | 8p12 | Xu et al. (2014) | ||
| KLF12 | Kruppel-like factor 12 | 13q22.1 | Le-Niculescu et al. (2009) | ||
| KIF1A | Kinesin family member 1 A | 2q37.3 | Le-Niculescu et al. (2009) | ||
| KLHL3 | Kelch-like family member 3 | 5q312 | Cichon et al. (2011) | ||
| KLHL20 | Kelch-like family member 20 | 1q25.1 | Xu et al. (2014) | ||
| LARGE | Like-glycosyltransferase | 22q12.3 | Le-Niculescu et al. (2009) | ||
| LDLRAD4 | Low density lipoprotein receptor class A domain containing 4 | 18p11.21 | Le-Niculescu et al. (2009) | ||
| LEF1 | Lymphoid enhancer-binding factor 1 | 4q25 | Le-Niculescu et al. (2009) | ||
| LHFPL3 | Lipoma HMGIC fusion partner-like 3 | 7q22.2 | Cichon et al. (2011) | ||
| LMO7 | LIM domain 7 | 13q22.2 | Le-Niculescu et al. (2009) | ||
| LMOD3 | Leiomodin 3 | 3p14.1 | Xu et al. (2014) | ||
| LRRC74A | Leucine rich repeat containing 74A | 14q24.3 | Xu et al. (2014) | ||
| LRRFIP1 | Leucine rich repeat (In FLII) interacting protein 1 | 2q37.3 | Xu et al. (2014) | ||
| LYPD5 | LY6/PLAUR domain containing 5 | 19q13.31 | Cichon et al. (2011) | ||
| MACROD1 | MACRO domain containing 1 | 11q13.1 | Cichon et al. (2011) | ||
| MAD1L1* | MAD1 mitotic arrest deficient-like 1 (yeast) | 7p22.3 | Cichon et al. (2011) | ||
| MAOA*# | Monoamine oxidase A | Xp11.3 | Fan et al. (2010) | ||
| MARCH1 | Membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase | 4q32.2 | Xu et al. (2014) | ||
| MBNL2 | Musclebind-like splicing regulator 2 | 13q32.1 | Le-Niculescu et al. (2009) | ||
| MBP | Myelin basic protein | 18q23 | Le-Niculescu et al. (2009) | ||
| MDH1 | Malate dehydrogenase 1, NAD (soluble) | 2p13.3 | Le-Niculescu et al. (2009) | ||
| MFSD11 | Major facilitator superfamily domain containing 11 | 17q25.1 | Xu et al. (2014) | ||
| MTHFR*# | Methylenetetrahydrofolate reductase (NAD(P)H) | 1p36.22 | El-Hadidy et al. (2014) | ||
| MTMR3 | Myotubularin related protein 3 | 22q12.2 | Xu et al. (2014) | ||
| MYCBP2 | MYC binding protein 2, E3 ubiquitin protein ligase | 13q22.3 | Le-Niculescu et al. (2009) | ||
| MYO5B | Myosin VB | 18q21.1 | Sklar et al. (2008) | ||
| MYO18B | Myosin XVIIIB | 22q12.1 | Xu et al. (2014) | ||
| MYT1L# | Myelin transcription factor 1-like | 2p25.3 | Le-Niculescu et al. (2009) | ||
| NAALAD2 | N-acetylated alpha-linked acidic dipeptidase 2 | 11q14.3 | Xu et al. (2014) | ||
| NALCN* | Sodium leak channel, non-selective | 13q32.3 | Sklar et al. (2008) | ||
| NAV2 | Neuron navigator 2 | 11p15.1 | Le-Niculescu et al. (2009) | ||
| NCALD | Neurocalcin delta | 8q22.3 | Xu et al. (2014) | ||
| NCAM1* | Neural cell adhesion molecule 1 | 11q23.2 | Le-Niculescu et al. (2009) | ||
| NCAN | Neurocan | 19p12 | Cichon et al. (2011) | ||
| NCKAP5# | NCK-associated protein 5 | 2q21.2 | Smith et al. (2009) | ||
| NDUFAB1 | NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1, 8 kDa | 16p12.2 | Burton et al. (2007) | ||
| NDUFS2 | NADH dehydrogenase (ubiquinone) Fe-S protein 2,49 kDa (NADH-coenzyme Qreductase) | 1q23.3 | Le-Niculescu et al. (2009) | ||
| NFIA# | Nuclear factor I/A | 1p31.3 | Le-Niculescu et al. (2009) | ||
| NFIB | Nuclear factor I/B | 9p24.1 | Le-Niculescu et al. (2009) | ||
| NIPBL | Nipped-B homolog (Drosophila) | 5p132 | Xu et al. (2014) | ||
| NME7 | NME/NM23 family member 7 | 1q24.2 | Xu et al. (2014) | ||
| NOS1AP*# | Nitric oxide synthase 1 (neuronal) adaptor protein | 1q23.3 | Xu et al. (2014) | ||
| NPAS3* | Neuronal PAS domain protein 3 | 14q13.1 | Pickard et al. (2009) | ||
| NPFFR2 | Neuropeptide FF receptor 2 isoform 1 | 4q13.3 | Xu et al. (2014) | ||
| NR3C1 | Nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | 5q31.3 | Le-Niculescu et al. (2009) | ||
| NRCAM# | Neuronal cell adhesion molecule | 7q31.1 | (Le-Niculescu et al., (2009) | ||
| NRG1 *# | Neuregulin 1 | 8p12 | Prata et al. (2009) | ||
| NRG3* | Neuregulin 3 | 10q23.1 | Xu et al. (2014) | ||
| NTF3* | Neurotrophin 3 | 12p13.31 | Cichon et al. (2011) | ||
| NTRK2* | Neurotrophic tyrosine kinase, receptor, type 2 | 9q21.33 | Smith et al. (2009) | ||
| NUP214 | Nucleoporin 214 kDa | 9q34.13 | Xu et al. (2014) | ||
| OC90 | Otoconin 90 | 8q24.22 | Xu et al. (2014) | ||
| OLIG2* | Oligodendrocyte lineage transcription factor 2 | 21q22.11 | Le-Niculescu et al. (2009) | ||
| OPRM1 # | Opioid receptor, mu 1 | 6q25.2 | Le-Niculescu et al. (2009) | ||
| OR1B1 | Olfactory receptor, family 1, subfamily B, member 1 | 9q33.2 | Xu et al. (2014) | ||
| PALB2 | Partner and localizer of BRCA2 | 16p12.2 | Tesli et al. (2010) | ||
| PARD3 | Par-3 family cell polarity regulator | 10p11.21 | Le-Niculescu et al. (2009) | ||
| PARD3B# | Par-3 family cell polarity regulator beta | 2q33.3 | Cichon et al. (2011) | ||
| PCDHAC2# | Protocadherin-alpha, subfamily C, 2 | 5q31.3 | Pedrosa et al. (2008) | ||
| PCDH7 | Protocadherin 7 | 4p15.1 | Le-Niculescu et al. (2009) | ||
| PCDH9# | Protocadherin 9 | 13q21.32 | Le-Niculescu et al. (2009) | ||
| PCLO | Piccolo presynaptic cytomatrix protein | 7q21.11 | Choi et al. (2011) | ||
| PDE10A | Phosphodiesterase 10 A | 6q26 | Le-Niculescu et al. (2009) | ||
| PDE4B*# | Phosphodiesterase 4B, CAMP-specific | 1p31.3 | Xu et al. (2014) | ||
| PER3* | Period circadian clock 3 | 1p3623 | Benedetti et al. (2008) | ||
| PDLIM5* | PDZ and LIM domain 5 | 4q22.3 | Shi et al. (2008) | ||
| PHF20 | PHD finger protein 20 | 20q11.22 | Xu et al. (2014) | ||
| PIK3R1 | Phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | 5q13.1 | Le-Niculescu et al. (2009) | ||
| PLCL2 | Phospholipase C-like 2 | 3p24.3 | Xu et al. (2014) | ||
| PLXNA2* | Plexin A2 | 1q32.2 | Le-Niculescu et al. (2009) | ||
| PPM1B | Protein phosphatase, Mg2+/Mn2+ dependent, 1B | 2p22.1 | Le-Niculescu et al. (2009) | ||
| PPP2R2A | Protein phosphatase 2, regulatory subunit B, alpha | 8p212 | Xu et al. (2014) | ||
| PPP2R2C | Protein phosphatase 2, regulatory subunit B, gamma | 4p16.1 | Baum et al. (2008a, 2008b) | ||
| PRKCE | Protein kinase C, epsilon | 2p21 | Le-Niculescu et al. (2009) | ||
| PRPF18 | Pre-MRNA processing factor 18 | 10p12.33 | Xu et al. (2014) | ||
| PRR16 | Proline rich 16 | 5q23.1 | Xu et al. (2014) | ||
| PRSS55 | Protease, serine, 55 | 8p23.1 | Xu et al. (2014) | ||
| PTN | Pleiotrophin | 7q33 | Le-Niculescu et al. (2009) | ||
| PTPRK | Protein tyrosine phosphatase, receptor type, K | 6q22.33 | Le-Niculescu et al. (2009) | ||
| PTPRM# | Protein tyrosine phosphatase, receptor type, M | 18p11.23 | Le-Niculescu et al. (2009) | ||
| PTPRO | Protein tyrosine phosphatase, receptor type, O | 12p12.3 | Xu et al. (2014) | ||
| PYDC2 | Pyrin domain containing 2 | 3q28 | Cichon et al. (2011) | ||
| QKI* | Quaking homolog, KH domain RNA binding | 6q26 | Le-Niculescu et al. (2009) | ||
| RABEP1 | Rabaptin, RAB GTPase binding effector protein 1 | 17p13.2 | Djurovic et al. (2010) | ||
| RABGAP1L | RAB GTPase activating protein 1-like | 1q25.1 | Xu et al. (2014) | ||
| RAD51L1 | RAD51, paralog B | 14q24.1 | Xu et al. (2014) | ||
| RASGRF2 | Ras protein-specific guanine nucleotide-releasing factor 2 | 5q14.1 | Le-Niculescu et al. (2009) | ||
| RBFOX1 # | RNAbindingprotein, fox-1 homolog (C. elegans) 1 | 16p13.3 | Le-Niculescu et al. (2009) | ||
| RBM23 | RNA binding motif protein 23 | 14q11.2 | Xu et al. (2014) | ||
| RC3H1 | Ring finger and CCCH-type domains 1 | 1q25.1 | Xu et al. (2014) | ||
| RLF | Rearranged L-Myc fusion | 1p34.2 | Xu et al. (2014) | ||
| RORA# | RAR-related orphan receptor alpha | 15q22.2 | Le-Niculescu et al. (2009) | ||
| RORB | RAR-related orphan receptor beta | 9q21.13 | Le-Niculescu et al. (2009) | ||
| RSPO4 | R-spondin 4 | 20p13 | Xu et al. (2014) | ||
| RXRG | Retinoid × receptor, gamma | 1q23.3 | Le-Niculescu et al. (2009) | ||
| RYR3 | Ryanodine receptor 3 | 15q13.3 | Le-Niculescu et al. (2009) | ||
| SCAMP1 | Secretory carrier membrane protein 1 | 5q14.1 | Le-Niculescu et al. (2009) | ||
| SCN7A# | Sodium channel, voltage gated, type VII alpha subunit | 2q24.3 | Xu et al. (2014) | ||
| SHISA9 | Shisa family member 9 | 16p13.12 | Xu et al. (2014) | ||
| SLC39A3 | Solute carrier family 39 (Zinc transporter), member 3 | 19p13.3 | Baum et al. (2008a, 2008b) | ||
| SLC6A3*# | Solute carrier family 6 (neurotransmitter transporter, dopamine), member 3 | 5p15.33 | Greenwood et al. (2006) | ||
| SLC8A1 | Solute carrier family 8 (sodium/calcium exchanger), member 1 | 2p22.1 | Le-Niculescu et al. (2009) | ||
| SOD1 # | Superoxide dismutase 1, soluble | 21q22.11 | Le-Niculescu et al. (2009) | ||
| SORCS2 | Sortilin-related VPS10 domain containing receptor 2 | 4p16.1 | Soronen et al. (2010) | ||
| SPAG16 | Sperm associated antigen 16 | 2q34 | Xu et al. (2014) | ||
| SPARC | Secreted protein, acidic, cysteine-rich (osteonectin) | 5q31.3 | Le-Niculescu et al. (2009) | ||
| SPAST# | Spastin | 2q22.3 | Le-Niculescu et al. (2009) | ||
| STAB1 | Stabilin 1 | 3p21.1 | Baum et al. (2008a, 2008b) | ||
| STK24 | Serine/threonin kinase 24 | 13q32.2 | Le-Niculescu et al. (2009) | ||
| STX12 | Syntaxin 12 | 1p35.3 | Xu et al. (2014) | ||
| SVIL | Supervillin | 10p11.23 | Purcell et al. (2009) | ||
| SYN3*# | Synapsin III | 22q12.3 | Le-Niculescu et al. (2009) | ||
| SYNE1 # | Spectrin repeat containing, nuclear envelope 1 | 6q252 | Ferreira et al. (2008) | ||
| TBC1D22B | TBC1 domain family, member 22B | 6p21.2 | Xu et al. (2014) | ||
| TBXAS1 | Thromboxane A synthase 1 | 7q34 | Xu et al. (2014) | ||
| TCF4*# | Transcription factor 4 | 18q21.1 | Del-Favero et al. (2002) | ||
| TEAD1 | TEA domain family member 1 | 11p15.4 | Xu et al. (2014) | ||
| TENM4 | Teneurin transmembrane protein 4 | 11q14.1 | Sklar et al., 2011 | ||
| TIAM1 | T-cell lymphoma invasion and metastasis 1 | 21q22.11 | Le-Niculescu et al. (2009) | ||
| TLE4 | Transducin-like enhancer ofsplit 4 | 9q21.31 | Cichon et al. (2011) | ||
| TMEM252 | Transmembrane protein 252 | 9q21.11 | Xu et al. (2014) | ||
| TNIK | TRAF2 and NCK interacting kinase | 3q26.31 | Le-Niculescu et al. (2009) | ||
| TPH1 | Tryptophan hydroxylase 1 | 11p15.1 | Chen et al. (2012) | ||
| TPH2*# | Tryptophan hydroxylase 2 | 12q21.1 | Cichon et al. (2008) | ||
| TPST2 | Tyrosylprotein sulfotransferase 2 | 22q12.1 | Le-Niculescu et al. (2009) | ||
| TRANK1 | Tetratricopeptide repeat and ankyrin repeat containing 1 | 3p22.2 | Mühleisen et al. (2014) | ||
| TRIO# | Trio rho guanine nucleotide exchange factor | 5p15.2 | Xu et al. (2014) | ||
| TRPM2 | Transient receptor potential cation channel, subfamily M, member 2 | 21q22.3 | McQuillin et al. (2006) | ||
| TRPM3 | Transient receptor potential cation channel, subfamily M, member 3 | 9q21.12 | Le-Niculescu et al. (2009) | ||
| TRPV3 | Transient receptor potential cation channel, subfamily V member 3 | 17p13.2 | Cichon et al. (2011) | ||
| TRRAP | Transformation/transcription domain-associated protein | 7q22.1 | Xu et al. (2014) | ||
| TSHZ2 | Teashirt zinc finger homeobox 2 | 20q13.2 | Le-Niculescu et al. (2009) | ||
| TSPAN8 | Tetraspanin 8 | 12q21.1 | Scholz et al. (2010) | ||
| VAMP7 | Vesicle-associated membrane protein 7 | Xq28 or Yq12 | Saito et al. (2000) | ||
| VIP# | Vasoactive Intestinal Peptide | 6q252 | Soria et al. (2010) | ||
| WDR60 | WD repeat domain 60 | 7q36.3 | Xu et al. (2014) | ||
| WDR7 | WD repeat domain 7 | 18q21.31 | Xu et al. (2014) | ||
| WNK2 | WNK lysine deficient protein kinase 2 | 9q22.31 | Cichon et al. (2011) | ||
| XBP1* | X box-binding protein 1 | 22q12.1 | Kakiuchi et al. (2003) | ||
| XKR4* | XK, Kell blood group complex subunit-related family, member 4 | 8q12.1 | Djurovic et al. (2010) | ||
| XXYLT1 | Xyloside xylosyltransferase 1 | 3q29 | Xu et al. (2014) | ||
| ZEB2* | Zinc finger E-box binding homeobox 2 | 2q22.3 | Cichon et al. (2011) | ||
| ZBTB20# | Zinc finger and BTB domain containing 20 | 3q13.31 | Xu et al. (2014) | ||
| ZDHHC14 | Zinc finger, DHHC-type containing 14 | 6q25.3 | Le-Niculescu et al. (2009) | ||
| ZHX2 | Zinc fingers and homeoboxes 2 | 8q24.13 | Le-Niculescu et al. (2009) | ||
| ZNF274 | Zinc finger protein 274 | 19q13.43 | Xu et al. (2014) | ||
| ZNF385D* | Zinc finger protein 385D | 3p24.3 | Xu et al. (2014) | ||
| ZNF536 | Zinc finger protein 536 | 19q13.11 | Djurovic et al. (2010) | ||
| ZNF618 | Zinc finger protein 618 | 9q33.1 | Cichon et al. (2011) | ||
| ZNF804A*# | Zinc finger protein 804 A | 2q32.1 | Williams et al.(2011) | ||
denotes BPD genes that overlap with clinically relevant genes for schizophrenia.
denotes BPD genes that overlap with clinically relevant genes for autism spectrum disorder.
Table 2.
Distribution of bipolar disorder (BPD) genes among chromosomes.
| Chromosome | Total | Proportion of total BPD genes | P arm | Q arm |
|---|---|---|---|---|
| 1 | 24 | 8.3% | 12 | 12 |
| 2 | 23 | 7.9% | 6 | 17 |
| 3 | 18 | 6.2% | 9 | 9 |
| 4 | 16 | 5.5% | 5 | 11 |
| 5 | 18 | 6.2% | 4 | 14 |
| 6 | 18 | 6.2% | 6 | 12 |
| 7 | 20 | 6.9% | 5 | 15 |
| 8 | 10 | 3.4% | 5 | 5 |
| 9 | 19 | 6.6% | 4 | 15 |
| 10 | 11 | 3.8% | 8 | 3 |
| 11 | 18 | 6.2% | 7 | 11 |
| 12 | 8 | 2.8% | 3 | 5 |
| 13 | 11 | 3.8% | NA | 11 |
| 14 | 7 | 2.4% | NA | 7 |
| 15 | 7 | 2.4% | NA | 7 |
| 16 | 9 | 3.1% | 6 | 3 |
| 17 | 5 | 1.7% | 2 | 3 |
| 18 | 11 | 3.8% | 5 | 6 |
| 19 | 8 | 2.8% | 4 | 4 |
| 20 | 5 | 1.7% | 1 | 4 |
| 21 | 7 | 2.4% | NA | 7 |
| 22 | 12 | 4.1% | NA | 12 |
| X | 3 | 1.0% | 2 | 1 |
| Y | 0 | 0.0% | 0 | 0 |
| Both X and Y | 2 | 0.7% | NA | NA |
| Total | 290 | 100% | 94 | 194 |
Total number of BPD genes were counted for each chromosome and chromosome arm.
p = short arm; q = long arm.
NA = not applicable due to chromosome structure or location.
The majority of the genes listed in Table 1 are located on chromosomes 1, 2, 3, 4, 5 6, 7, 8, 9, 10, 11, 13, 16, 18, and 22 in accordance with previous reports examining polymorphisms and chromosomal breakpoints of known and candidate genes for BPD (Potash and DePaulo, 2000; Etain et al., 2012). One of the smallest chromosomes (i.e., 22) contained more BPD genes in relationship to its size (i.e., 4.1%) than other comparable chromosomes. The X chromosome which accounts for about 5.6% of the genome contained only 1.0% of the BPD genes. The genes identified in our investigation impacted a broad range of biological pathways and processes including cellular signaling pathways for cAMP and calcium (e.g., CACNA1C, CAMK2A, CAMK2D, ADCY1, ADCY2); glutamatergic (e.g., GRIK1, GRM3, GRM7), dopaminergic (e.g., DRD2, DRD4, COMT, MAOA) and serotonergic (e.g., HTR1A, HTR2A, HTR3B) neurotransmission; molecular transporters (e.g., SLC39A3, SLC6A3, SLC8A1); and neuronal growth (e.g., BDNF, IGFBP1, NRG1, NRG3). These gene classes influence neurotransmission and psychological functioning through direct and indirect effects on neuronal activity, growth, development, maintenance and remodeling. These genes overlap with biological markers and genetic factors associated with other psychiatric disorders such as schizophrenia, (e.g., BDNF, CACNA1, DISC1), major depressive and anxiety disorders (e.g., TPH2, HTR2A) involving developmental processes (e.g., NRG1, NRG3).
4. Discussion
The advent of genetic testing to identify the predisposition for BPD has increased understanding of the genetic etiology of BPD and the application of genetic testing should facilitate research and treatment development. We illustrated a master list of susceptibility genes and genes that are associated with BPD in our results by plotting the individual genes on high-resolution chromosome ideograms and generated a tabular form with references in order to inform more individuals about the necessity for genetic testing and treatment for BPD.
The transmission of BPD does not appear to manifest through a simple Mendelian inheritance with a single allele (dominant) or two alleles (recessive) pattern instead showing incomplete penetrance, etiological heterogeneity and high prevalence (Kerner, 2015). Similarly, multivariate threshold models conceptualized as an accumulation of traits normally distributed throughout the genome are not strongly supported to achieve some threshold for expression. Rather, BPD appears to be a product of a combination of genetic, neurochemical, and environmental influences. The expression or development of bipolar disorder may be due to epistasis (interaction of multiple genes) or other complex mechanisms such as genomic imprinting or dynamic mutations with environmental contributions (Craddock and Jones, 1999). An intricate oligogenic quasi-Mendelian pattern has been proposed whereby a small number of mutations accumulate in a select biological pathway that is only tied to expression of the phenotype when released by environmental influences (Kerner, 2015). Symptoms of bipolar disorder parallel other genetically influenced psychiatric disorders including schizophrenia, depression and anxiety (Krishnan, 2005) and is, not surprisingly, impacted by overlapping genetic constructs (e.g., BDNF, DISC1, HTR2A, TPH2).
Our list of genes for BPD reflects the current status of recognized genes with clinical relevance but susceptibility and new genes are continually being identified. Not all genes in the list are equally significant or certainly causative for all individuals with BPD. Similarly, our results reflect gene-level associations and do not provide evidence of individual SNP- or CNV-level contributions to pathology. The list is particularly suited to the evaluation of structural genomic data (e.g., DNA microarrays) for copy number variations impacting genomic regions and genes of interest which may involve large regions and multiple candidate genes or encompass known genetic syndromes. Phenotype and severity can be predicted to be proportional to the number of candidate genes impacted by the CNV – highlighting the clinical value of spatial representation of the gene list and whether the region is duplicated or deleted. The effect of any individual SNP depends upon both the physiological relevance of the gene to neurodevelopmental processes and the impact of the specific sequence variation or mutation on the expression and function of the gene product (i.e., synonymous vs. non-synonymous). Non-synonymous variations leading to a change of the codon or reading frame (e.g., frameshift mutation, stop codon generation) have greater predicted impact on gene expression and thus stronger ties to pathology. Advances in genomic technology will facilitate the identification and characterization of novel SNPs among the gene candidates and improve understanding of the relative contributions of selected SNP or CNVs to the general disease prevalence.
Combinations of single nucleotide polymorphisms (SNPs) which alter the genetic code, creating a higher likelihood of developing bipolar disorder, are reportedly more common than chromosomal breakpoints at the susceptibility regions for BPD. For example, SNPs within the brain derived neurotrophic factor (BDNF) gene, located in the 11p14, region are reported to result in a modification of the processing and trafficking of the BDNF gene and increased susceptibility to BPD (Craddock et al., 2005). The ABCA13 gene was also identified as a candidate gene for both bipolar and schizophrenia disorders once it was initially discovered by a chromosome abnormality in a schizophrenic patient (Knight et al., 2009). The ABCA13 gene was then resequenced and multiple rare coding variants identified. These variants were genotyped in bipolar cohorts, which led to the conclusion that 4.0% of the population contained a variant which could contribute to bipolar disorder (Knight et al., 2009). Multiple genome-wide association studies (GWAS) testing for SNPs in independent bipolar cases and controls often generate strong signals for gene associations with bipolar disorder (Sklar et al., 2011). For example, results from the first GWAS for BPD reported in 2007 determined that the strongest signals for an association were found in five genes: BDNF at 11p14, DAOA at 13q33, DTNBP1 at 6p22, DISC1 at 1q42, and NRG1 at 8p12, and the SNP rs420259 at 16p12 (Burton et al., 2007). Since this initial study, additional GWAS reports (e.g., Sklar et al., 2008; Ferreira et al., 2008) and a collection of genetic studies examining bipolar disorder did implicate genes that were consolidated in 2014 by Shinozaki and Potash (Shinozaki and Potash, 2014) with a list of candidate genes summarized from 15 studies (e.g., Baum et al., 2008a, 2008b; Sklar et al., 2011; Ou et al., 2015). These genes overlapped with biological markers and genetic factors associated with other psychiatric disorders such as schizophrenia, (e.g., BDNF, CACNA1, DISC1), major depressive and anxiety disorders (e.g., TPH2, HTR2A) and involved in developmental processes.
Our list of candidate genes and visual representation of their locations on high-resolution chromosome ideograms for bipolar disorder provide an informative perspective of the pathophysiology of BPD to facilitate research, accurate diagnosis and genetic counseling, treatment development and classification of BPD (Craddock and Jones, 1999). The authors encourage the use of this collection of currently associated recognized susceptibility genes for BPD in the evaluation of patients presenting for genetic services and for a more accurate understanding of the role of genetics in BPD, a genetically heterogeneous disorder.
Acknowledgments
We thank Lorie Gavulic for excellent artistic design and preparation of chromosome ideograms and Maaz Hassan for assistance in manuscript preparation.
Financial support
This work was supported by the Consortium for Translational Research on Aggression and Drug Abuse and Dependence (ConTRADA) (grant QB864900) and the National Institute of Child Health and Human Development (NICHD) (grant HD02528).
Abbreviations:
- BPD
bipolar disorder
- BP-I
Bipolar I Disorder
- BP-II
Bipolar II Disorder
- BP-NOS
Bipolar Disorder Not Otherwise Specified
- cAMP
cyclic adenosine monophosphate
- DNA
deoxyribonucleic acid
- DSM-5
Diagnostic and Statistical Manual for Mental Disorders, fifth edition
- FDA
Food and Drug Administration
- GWAS
genome-wide association studies
- OMIM
online Mendelian inheritance in man
- SNPs
single nucleotide polymorphisms
Footnotes
Conflicts of interest
The authors declare no conflict of interest.
References
- Baum AE, Akula N, Cabanero M, Cardona I, Corona W, Klemens B, Schulze TG, Cichon S, Rietschel M, Nöthen MM, Georgi A, Schumacher J, Schwarz M, Abou Jamra R, Höfels S, Propping P, Satagopan J, Detera-Wadleigh SD, Hardy J, McMahon FJ, 2008a. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol. Psychiatry 13, 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A, Hamshere M, Green E, Cichon S, Rietschel M, Noethen M, McMahon F, 2008b. Meta-analysis of two genome-wide association studies of bipolar disorder reveals important points of agreement. Mol. Psychiatry 13 (5), 466–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysal BE, Willett-Brozick JE, Badner JA, Corona W, Ferrell RE, Nimgaonkar VL, Detera-Wadleigh SD, 2002. A mannosyltransferase gene at 11q23 is disrupted by a translocation breakpoint that co-segregates with bipolar affective disorder in a small family. Neurogenetics 4, 43–53. [DOI] [PubMed] [Google Scholar]
- Benedetti F, Serretti A, Colombo C, Barbini B, Lorenzi C, Campori E, Smeraldi E, 2003. Influence of CLOCK gene polymorphism on circadian mood fluctuation and illness recurrence in bipolar depression. Am. J. Med. Genet. B Neuropsychiatr. Genet 123B, 23–26. [DOI] [PubMed] [Google Scholar]
- Benedetti F, Dallaspeziaa S, Colomboa C, Pirovanoa A, Marinoa E, Smeraldia E, 2008. A length polymorphism in the circadian clock gene Per3 influences age at onset of bipolar disorder. Neurosci. Lett 445 (2), 184–187. [DOI] [PubMed] [Google Scholar]
- Birmaher B, Axelson D, Strober M, Gill MK, Valeri S, Chiappetta L, Ryan N, Leonard H, Hunt J, Iyengar S, Keller M, 2006. Clinical course of children and adolescents with bipolar spectrum disorders. Arch. Gen. Psychiatry 63 (2), 175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood DH, Pickard BJ, Thomson PA, Evans KL, Porteous DJ, Muir WJ, 2007. Are some genetic risk factors common to schizophrenia, bipolar disorder and depression? Evidence from DISC1, GRIK4 and NRG1. Neurotox. Res 11 (1), 73–83. [DOI] [PubMed] [Google Scholar]
- Blair IP, Chetcuti AF, Badenhop RF, Scimone A, Moses MJ, Adams LJ, Craddock N, Green E, Kirov G, Owen MJ, Kwok JB, Donald JA, Mitchell PB, Schofield PR, 2006. Positional cloning, association analysis and expression studies provide convergent evidence that the cadherin gene FAT contains a bipolar disorder susceptibility allele. Mol. Psychiatry 11 (4), 372–383. [DOI] [PubMed] [Google Scholar]
- Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A, Kwiatkowski DP, McCarthy MI, Ouwehand WH, Samani NJ, Todd JA, Donnelly P, Barrett JC, Burton PR, Davison D, Donnelly P, Easton D, Evans D, Leung HT, Marchini JL, Morris AP, Spencer CC, Tobin MD, Cardon LR, Clayton DG, Attwood AP, Boorman JP, Cant B, Everson U, Hussey JM, Jolley JD, Knight AS, Koch K, Meech E, Nutland S, Prowse CV, Stevens HE, Taylor NC, Walters GR, Walker NM, Watkins NA, Winzer T, Todd JA, Ouwehand WH, Jones RW, McArdle WL, Ring SM, Strachan DP, Pembrey M, Breen G, St Clair D, Caesar S, Gordon-Smith K, Jones L, Fraser C, Green EK, Grozeva D, Hamshere ML, Holmans PA, Jones IR, Kirov G, Moskvina V, Nikolov I, O’Donovan MC, Owen MJ, Craddock N, Collier DA, Elkin A, Farmer A, Williamson R, McGuffin P, Young AH, Ferrier IN, Ball SG, Balmforth AJ, Barrett JH, Bishop DT, Iles MM, Maqbool A, Yuldasheva N, Hall AS, Braund PS, Burton PR, Dixon RJ, Mangino M, Suzanne S, Tobin MD, Thompson JR, Samani NJ, Bredin F, Tremelling M, Parkes M, Drummond H, Lees CW, Nimmo ER, Satsangi J, Fisher SA, Forbes A, Lewis CM, Onnie CM, Prescott NJ, Sanderson J, Mathew CG, Barbour J, Mohiuddin MK, Todhunter CE, Mansfield JC, Ahmad T, Cummings FR, Jewell DP, Webster J, Brown MJ, Clayton DG, Lathrop GM, Connell J, Dominczak A, Samani NJ, Marcano CA, Burke B, Dobson R, Gungadoo J, Lee KL, Munroe PB, Newhouse SJ, Onipinla A, Wallace C, Xue M, Caulfield M, Farrall M, Barton A, Bruce IN, Donovan H, Eyre S, Gilbert PD, Hider SL, Hinks AM, John SL, Potter C, Silman AJ, Symmmons DP, Thomson W, Worthington J, Clayton DG, Dunger DB, Nutland S, Stevens HE, Walker NM, Widmer B, Todd JA, Frayling TA, Freathy RM, Lango H, Perry JR, Shields BM, Weedon MN, Hattersley AT, Hitman GA, Walker M, Elliott KS, Groves CJ, Lindgren CM, Rayner NW, Timpson NJ, Zeggini E, McCarthy MI, Newport M, Sirugo G, Lyons E, Vannberg F, Hill AV, Bradbury LA, Farrar C, Pointon JJ, Wordsworth P, Brown MA, Franklyn JA, Heward JM, Simmonds MJ, Gough SC, Seal S, Stratton MR, Rahman N, Ban M, Goris A, Sawcer SJ, Compston A, Conway D, Jallow M, Newport M, Sirugo G, Rockett KA, Kwiatowski DP, Bumpstead SJ, Chaney A, Downes K, Ghori MJ, Gwilliam R, Hunt SE, Inouye M, Keniry A, King E, McGinnis R, Potter S, Ravindrarajah R, Whittaker P, Widden C, Withers D, Deloukas P, Leung HT, Nutland S, Stevens HE, Walker NM, Todd JA, Easton D, Clayton DG, Burton PR, Tobin MD, Barrett JC, Evans D, Morris AP, Cardon LR, Cardin NJ, Davison D, Ferreira T, Pereira-Gale J, Hallgrimsdottir IB, Howie BN, Marchini JL, Spencer CC, Su Z, Teo YY, Vukcevic D, Donnelly P, Bentley D, Brown MA, Gordon LR, Caulfield M, Clayton DG, Compston A, Craddock N, Deloukas P, Donnelly P, Farrall M, Gough SC, Hall AS, Hattersley AT, Hill AV, Kwiatkowski DP, Mathew C, McCarthy MI, Ouwehand WH, Parkes M, Pembrey M, Rahman N, Samani NJ, Stratton MR, Todd JA, Worthington J, Consortium, Wellcome Trust Case Control, 2007. Genomewide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447 (7145), 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Liu F, Yang C, Liang X, Shang Q, He W, Wang Z, 2012. Association between the TPH1 A218C polymorphism and risk of mood disorders and alcohol dependence: evidence from the current studies. J. Affect. Disord 138 (1–2), 27–33. [DOI] [PubMed] [Google Scholar]
- Choi KH, Higgs BW, Wendland JR, Song J, McMahon FJ, Webster MJ, 2011. Gene expression and genetic variation data implicate PCLO in bipolar disorder. Biol. Psychiatry 69 (4), 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichon S, Winge I, Mattheisen M, Georgi A, Karpushova A, Freudenberg J, Freudenberg-Hua Y, Babadjanova G, Van Den Bogaert A, Abramova LI, Kapiletti S, Knappskog PM, McKinney J, Maier W, Jamra RA, Schulze TG, Schumacher J, Propping P, Rietschel M, Haavik J, Nöthen MM, 2008. Brain-specific tryptophan hydroxylase 2 (TPH2): a functional Pro206Ser substitution and variation in the 5-prime region are associated with bipolar affective disorder. Hum. Mol. Genet 17, 87–97. [DOI] [PubMed] [Google Scholar]
- Cichon S, Craddock N, Daly M, Faraone SV, Gejman PV, Kelsoe J, Lehner T, Levinson DF, Moran A, Sklar P, Sullivan PF, Faraone S, Anney R, Buitelaar J, Elia J, Franke B, Gill M, Hakonarson H, Kent L, McGough J, Mick E, Nisenbaum L, Smalley S, Thapar A, Todd R, Todorov A, Devlin B, Daly M, Anney R, Arking D, Buxbaum JD, Chakravarti A, Cook E, Gill M, Peltonen L, Piven J, Rouleau G, Santangelo S, Schellenberg G, Scherer S, Sutcliffe J, Szatmari P, Vieland V, Kelsoe J, Sklar P, Andreassen OA, Blackwood D, Boehnke M, Breuer R, Burmeister M, Cichon S, Corvin A, Craddock N, Ferreira M, Flickinger M, Greenwood T, Guan W, Gurling H, Li J, Mick E, Moskvina V, Muglia P, Muir W, Noethen M, Nurnberger J, Purcell S, Rietschel M, Ruderfer D, Schork N, Schulze T, Scott L, Steffens M, Upmanyu R, Wienker T, Smoller J, Craddock N, Kendler K, Nurnberger J, Perlis R, Purcell S, Rietschel M, Santangelo S, Thapar A, Sullivan P, Blackwood D, Boomsma D, Breuer R, Cichon S, Coryell W, de Geus E, Hamilton S, Hoogendijk W, Kloiber S, Lawson WB, Levinson D, Lewis C, Lucae S, Martin N, McGrath P, McGuffin P, Muglia P, Muir W, Noethen M, Offord J, Penninx B, Potash JB, Rietschel M, Scheftner WA, Schulze T, Slager S, Tozzi F, Weissman MM, Willemsen AH, Wray N, Gejman P, Andreassen OA, Blackwood D, Cichon S, Corvin A, Daly M, Fanous A, Gill M, Gurling H, Holmans P, Hultman C, Kendler K, Kivikko S, Laurent C, Lencz T, Levinson D, Malhotra A, Mowry B, Noethen M, O’Donovan M, Ophoff R, Owen M, Peltonen L, Pulver A, Rietschel M, Riley B, Sanders A, Schulze T, Schwab S, Sklar P, St Clair D, Sullivan P, Suvisaari J, van den Oord E, Wray N, Wildenaver D, Daly M, Awadalla P, Devlin B, Dudbridge F, Frigessi A, Holliday E, Holmans P, Lencz T, Levinson D, Lewis C, Lin D, Moskvina V, Mowry B, Neale B, Pickering E, Posthuma D, Purcell S, Rice J, Ripke S, Schork N, Sebat J, Steffens M, Stone J, Tzeng JY, van den Oord E, Vieland V, Psychiatric GWAS Consortium Coordinating Committee, 2009. Genomewide association studies: history, rationale, and prospects for psychiatric disorders. Am. J. Psychiatry 166 (5), 540–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichon S, Mühleisen TW, Degenhardt FA, Mattheisen M, Miró X, Strohmaier J, Steffens M, Meesters C, Herms S, Weingarten M, Priebe L, Haenisch B, Alexander M, Vollmer J, Breuer R, Schmäl C, Tessmann P, Moebus S, Wichmann HE, Schreiber S, Müller-Myhsok B, Lucae S, Jamain S, Leboyer M, Bellivier F, Etain B, Henry C, Kahn JP, Heath S, Bipolar Disorder Genome Study (BiGS) Consortium, Hamshere M, O’Donovan MC, Owen MJ, Craddock N, Schwarz M, Vedder H, Kammerer-Ciernioch J, Reif A, Sasse J, Bauer M, Hautzinger M, Wright A, Mitchell PB, Schofield PR, Montgomery GW, Medland SE, Gordon SD, Martin NG, Gustafsson O, Andreassen O, Djurovic S, Sigurdsson E, Steinberg S, Stefansson H, Stefansson K, Kapur-Pojskic L, Oruc L, Rivas F, Mayoral F, Chuchalin A, Babadjanova G, Tiganov AS, Pantelejeva G, Abramova LI, Grigoroiu-Serbanescu M, Diaconu CC, Czerski PM, Hauser J, Zimmer A, Lathrop M, Schulze TG, Wienker TF, Schumacher J, Maier W, Propping P, Rietschel M, Nöthen MM, 2011. Genome-wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am. J. Hum. Genet 88 (3), 372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi JP, Ravyn V, Robbins AK, Hagan KW, Peters MF, Bostwick R, Buono RJ, Berrettini WH, Furlong ST, 2005. Alternative transcripts and evidence of imprinting of GNAL on 18p11.2. Mol. Psychiatry 10, 1017–1025. [DOI] [PubMed] [Google Scholar]
- Craddock N, Jones I, 1999. Genetics of bipolar disorder. J. Med. Genet 36, 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock N, O’Donovan MC, Owen MJ, 2005. The genetics of schizophrenia and bipolar disorder: dissecting psychosis. J. Med. Genet 42, 193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del-Favero J, Van Gestel S, Borglum AD, Muir W, Ewald H, Mors O, Ivezic S, Oruc L, Adolfsson R, Blackwood D, Kruse T, Mendlewicz J, Schalling M, Van Broeckhoven C, 2002. European combined analysis of the CTG18.1 and the ERDA1 CAG/CTG repeats in bipolar disorder. Eur. J. Hum. Genet 10, 276–280. [DOI] [PubMed] [Google Scholar]
- Detera-Wadleigh SD, McMahon FJ, 2006. G72/G30 in schizophrenia and bipolar disorder: review and meta-analysis. Biol. Psychiatry 60, 106–114. [DOI] [PubMed] [Google Scholar]
- Djurovic S, Gustafsson O, Mattingsdal M, Athanasiu L, Bjella T, Tesli M, Agartz I, Lorentzen S, Melle I, Morken G, Andreassen OA, 2010. A genome-wide association study of bipolar disorder in Norwegian individuals, followed by replication in Icelandic sample. J. Affect. Disord 126, 312–316. [DOI] [PubMed] [Google Scholar]
- El-Hadidy MA, Abdeen HM, Abd El-Aziz SM, Al-Harrass M, 2014. MTHFR gene polymorphism and age of onset of schizophrenia and bipolar disorder. Biomed. Res. Int 2014, 318483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etain B, Dumaine A, Bellivier F, Pagan C, Francelle L, Goubran-Botros H, Moreno S, Deshommes J, Moustafa K, Le Dudal K, Mathieu F, Henry C, Kahn JP, Launay JM, Mühleisen TW, Cichon S, Bourgeron T, Leboyer M, Jamain S, 2012. Genetic and functional abnormalities of the melatonin biosynthesis pathway in patients with bipolar disorder. Hum. Mol. Genet 21 (18), 4030–4037. [DOI] [PubMed] [Google Scholar]
- Fan J, Sklar P, 2008. Genetics of bipolar disorder: focus on BDNF Val66Met polymorphism. Novartis Found. Symp 289, 60–72. [DOI] [PubMed] [Google Scholar]
- Fan M, Liu B, Jiang T, Jiang X, Zhao H, Zhang J, 2010. Meta-analysis of the association between the monoamine oxidase-A gene and mood disorders. Psychiatr. Genet 20 (1), 1–7. [DOI] [PubMed] [Google Scholar]
- Ferreira MA, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, Fan J, Kirov G, Perlis RH, Green EK, Smoller JW, Grozeva D, Stone J, Nikolov I, Chambert K, Hamshere ML, Nimgaonkar VL, Moskvina V, Thase ME, Caesar S, Sachs GS, Franklin J, Gordon-Smith K, Ardlie KG, Gabriel SB, Fraser C, Blumenstiel B, Defelice M, Breen G, Gill M, Morris DW, Elkin A, Muir WJ, McGhee KA, Williamson R, MacIntyre DJ, MacLean AW, St CD, Robinson M, Van Beck M, Pereira AC, Kandaswamy R, McQuillin A, Collier DA, Bass NJ, Young AH, Lawrence J, Ferrier IN, Anjorin A, Farmer A, Curtis D, Scolnick EM, McGuffin P, Daly MJ, Corvin AP, Holmans PA, Blackwood DH, Gurling HM, Owen MJ, Purcell SM, Sklar P, Craddock N, Consortium, Wellcome Trust Case Control, 2008. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat. Genet 40 (9), 1056–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaysina D, Cohen-Woods S, Chow PC, Martucci L, Schosser A, Ball HA, Tozzi F, Perry J, Muglia P, Craig IW, McGuffin P, Farmer A, 2009. Association of the dystrobrevin binding protein 1 gene (DTNBP1) in a bipolar case–control study (BACCS). Am. J. Med. Genet. B Neuropsychiatr. Genet 150B (6), 836–844. [DOI] [PubMed] [Google Scholar]
- Geller B, Tillman R, Craney JL, Bolhofner K, 2004. Four-year prospective outcome and natural history of mania in children with a prepubertal and early adolescent bipolar disorder phenotype. Arch. Gen. Psychiatry 61 (5), 459–467. [DOI] [PubMed] [Google Scholar]
- Goossens D, Van Gestel S, Claes S, De Rijk P, Souery D, Massat I, Van den Bossche D, Backhovens H, Mendlewicz J, Van Broeckhoven C, Del-Favero J, 2003. A novel CpG-associated brain-expressed candidate gene for chromosome 18q-linked bipolar disorder. Mol. Psychiatry 8 (1), 83–89. [DOI] [PubMed] [Google Scholar]
- Greenwood TA, Schork NJ, Eskin E, Kelsoe JR, 2006. Identification of additional variants within the human dopamine transporter gene provides further evidence for an association with bipolar disorder in two independent samples. Mol. Psychiatry 11 (2), 125–133. [DOI] [PubMed] [Google Scholar]
- Hammer C, Cichon S, Mühleisen TW, Haenisch B, Degenhardt F, Mattheisen M, Breuer R, Witt SH, Strohmaier J, Oruc L, Rivas F, Babadjanova G, Grigoroiu-Serbanescu M, Hauser J, Röth R, Rappold G, Rietschel M, Nöthen MM, Niesler B, 2012. Replication of functional serotonin receptor type 3A and B variants in bipolar affective disorder: a European multicenter study. Transl. Psychiatry 2, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamshere ML, Walters JT, Smith R, Richards AL, Green E, Grozeva D, Jones I, Forty L, Jones L, Gordon-Smith K, Riley B, O’Neill FA, Kendler KS, Sklar P, Purcell S, Kranz J, Schizophrenia Psychiatric Genome-wide Association Study Consortium, Wellcome Trust Case Control Consortium, Wellcome Trust Case Control Consortium 2, Morris D, Gill M, Holmans P, Craddock N, Corvin A, Owen MJ, O’Donovan MC, 2013. Genome-wide significant associations in schizophrenia to ITIH3/4, CACNA1C and SDCCAG8, and extensive replication of associations reported by the schizophrenia PGC. Mol. Psychiatry 18, 708–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto R, Okada T, Kato T, Kosuga A, Tatsumi M, Kamijima K, Kunugi H, 2005. The breakpoint cluster region gene on chromosome 22q11 is associated with bipolar disorder. Biol. Psychiatry 57 (10), 1097–1102. [DOI] [PubMed] [Google Scholar]
- Heckers S, Stone D, Walsh J, Shick J, Koul P, Benes FM, 2002. Differential hippocampal expression of glutamic acid decarboxylase 65 and 67 messenger RNA in bipolar disorder and schizophrenia. Arch. Gen. Psychiatry 59 (6), 521–529. [DOI] [PubMed] [Google Scholar]
- Hirschfeld RM, Williams JB, Spitzer RL, Calabrese JR, Flynn L, Keck PE Jr., Lewis L, McElroy SL, Post RM, Rapport DJ, Russell JM, Sachs GS, Zajecka J, 2000. Development and validation of a screening instrument for bipolar spectrum disorder: the Mood Disorder Questionnaire. Am. J. Psychiatry 157 (11) (1873–1502). [DOI] [PubMed] [Google Scholar]
- Hirschfeld RM, Calabrese JR, Weissman MM, Reed M, Davies MA, Frye MA, Keck PE Jr., Lewis L, McElroy SL, McNulty JP, Wagner KD, 2003a. Screening for bipolar disorder in the community. J. Clin. Psychiatry 64 (1), 53–59. [DOI] [PubMed] [Google Scholar]
- Hirschfeld RM, Lewis L, Vornik LA, 2003b. Perceptions and impact of bipolar disorder: how far have we really come? Results of the National Depressive and Manic-Depressive Association 2000 survey of individuals with bipolar disorder. J. Clin. Psychiatry 64 (2), 161–174. [PubMed] [Google Scholar]
- Hodgkinson CA, Goldman D, Jaeger J, Persaud S, Kane JM, Lipsky RH, Malhotra AK, 2004. Disrupted in schizophrenia 1 (DISC1): association with schizophrenia, schizoaffective disorder, and bipolar disorder. Am. J. Hum. Genet 75, 862–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakiuchi C, Iwamoto K, Ishiwata M, Bundo M, Kasahara T, Kusumi I, Tsujita T, Okazaki Y, Nanko S, Kunugi H, Sasaki T, Kato T, 2003. Impaired feedback regulation of XBP1 as a genetic risk factor for bipolar disorder. Nat. Genet 35, 171–175. [DOI] [PubMed] [Google Scholar]
- Kerner B, 2015. Toward a deeper understanding of the genetics of bipolar disorder. Front. Psychiatry 6, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner B, Rao AR, Christensen B, Dandekar S, Yourshaw M, Nelson SF, 2013. Rare genomic variants link bipolar disorder with anxiety disorders to CREB-regulated intracellular signaling pathways. Front. Psychiatry 4, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi T, Okochi T, Tsunoka T, Okumura T, Kitajima T, Kawashima K, Yamanouchi Y, Kinoshita Y, Naitoh H, Inada T, Kunugi H, Kato T, Yoshikawa T, Ujike H, Ozaki N, Iwata N, 2011. Serotonin 1A receptor gene, schizophrenia and bipolar disorder: an association study and meta-analysis. Psychiatry Res. 185 (1–2), 20–26. [DOI] [PubMed] [Google Scholar]
- Knight HM, Pickard BS, Maclean A, Malloy MP, Soares DC, McRae AF, Condie A, White A, Hawkins W, McGhee K, van Beck M, MacIntyre DJ, Starr JM, Deary IJ, Visscher PM, Porteous DJ, Cannon RE, St Clair D, Muir WJ, Blackwood DH, 2009. A cytogenetic abnormality and rare coding variants identify ABCA13 as a candidate gene in schizophrenia, bipolar disorder, and depression. Am. J. Hum. Genet 85 (6), 833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan KR, 2005. Psychiatric and medical comorbidities of bipolar disorder. Psychosom. Med 67 (1), 1–8. [DOI] [PubMed] [Google Scholar]
- Le-Niculescu H, Patel SD, Bhat M, Kuczenski R, Faraone SV, Tsuang MT, McMahon FJ, Schork NJ, Nurnberger JI Jr., Niculescu AB 3rd, 2009. Convergent functional genomics of genome-wide association data for bipolar disorder: comprehensive identification of candidate genes, pathways and mechanisms. Am. J. Med. Genet. B Neuropsychiatr. Genet 150B (2), 155–181. [DOI] [PubMed] [Google Scholar]
- López León S, Croes EA, Sayed-Tabatabaei FA, Claes S, Van Broeckhoven C, van Duijn CM, 2005. The dopamine D4 receptor gene 48-base-pair-repeat polymorphism and mood disorders: a meta-analysis. Biol. Psychiatry 57, 999–1003. [DOI] [PubMed] [Google Scholar]
- McClellan J, Kowatch R, Findling RL, Work Group on Quality Issues, 2007. Practice parameter for the assessment and treatment of children and adolescents with bipolar disorder. J. Am. Acad. Child Adolesc. Psychiatry 46 (1), 107–125. [DOI] [PubMed] [Google Scholar]
- McElroy SL, Altshuler LL, Suppes T, Keck PE, Frye MA, Denicoff KD, Nolen WA, Kupka RW, Leverich GS, Rochussen JR, Rush AJ, Post RM, 2001. Axis I psychiatric comorbidity and its relationship to historical illness variables in 288 patients with bipolar disorder. Am. J. Psychiatry 158, 420–426. [DOI] [PubMed] [Google Scholar]
- McQuillin A, Bass NJ, Kalsi G, Lawrence J, Puri V, Choudhury K, Detera-Wadleigh SD, Curtis D, Gurling HM, 2006. Fine mapping of a susceptibility locus for bipolar and genetically related unipolar affective disorders, to a region containing the C21ORF29 and TRPM2 genes on chromosome 21q22.3. Mol. Psychiatry 11, 134–142. [DOI] [PubMed] [Google Scholar]
- Meisler MH, Grant AE, Jones JM, Lenk GM, He F, Todd PK, Kamali M, Albin RL, Lieberman AP, Langenecker SA, McInnis MG, 2013. C9ORF72 expansion in a family with bipolar disorder. Bipolar Disord. 15, 326–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merikangas KR, Jin R, He JP, Kessler RC, Lee S, Sampson NA, Viana MC, Andrade LH, Hu C, Karam EG, Ladea M, Medina-Mora ME, Ono Y, Posada-Villa J, Sagar R, Wells JE, Zarkov Z, 2011. Prevalence and correlates of bipolar spectrum disorder in the world mental health survey initiative. Arch. Gen. Psychiatry 68 (3), 241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühleisen TW, Leber M, Schulze TG, Strohmaier J, Degenhardt F, Treutlein J, Mattheisen M, Forstner AJ, Schumacher J, Breuer R, Meier S, Herms S, Hoffmann P, Lacour A, Witt SH, Reif A, Müller-Myhsok B, Lucae S, Maier W, Schwarz M, Vedder H, Kammerer-Ciernioch J, Pfennig A, Bauer M, Hautzinger M, Moebus S, Priebe L, Czerski PM, Hauser J, Lissowska J, Szeszenia-Dabrowska N, Brennan P, McKay JD, Wright A, Mitchell PB, Fullerton JM, Schofield PR, Montgomery GW, Medland SE, Gordon SD, Martin NG, Krasnow V, Chuchalin A, Babadjanova G, Pantelejeva G, Abramova LI, Tiganov AS, Polonikov A, Khusnutdinova E, Alda M, Grof P, Rouleau GA, Turecki G, Laprise C, Rivas F, Mayoral F, Kogevinas M, Grigoroiu-Serbanescu M, Propping P, Becker T, Rietschel M, Nöthen MM, Cichon S, 2014. Genome-wide association study reveals two new risk loci for bipolar disorder. Nat. Commun 5, 3339. [DOI] [PubMed] [Google Scholar]
- Nyegaard M, Severinsen JE, Als TD, Hedemand A, Straarup S, Nordentoft M, McQuillin A, Bass N, Lawrence J, Thirumalai S, Pereira AC, Kandaswamy R, Lydall GJ, Sklar P, Scolnick E, Purcell S, Curtis D, Gurling HM, Mortensen PB, Mors O, Børglum AD, 2010. Support of association between BRD1 and both schizophrenia and bipolar affective disorder. Am. J. Med. Genet. B Neuropsychiatr. Genet 153B (2), 582–591. [DOI] [PubMed] [Google Scholar]
- Ohtsuki T, Ishiguro H, Detera-Wadleigh SD, Toyota T, Shimizu H, Yamada K, Yoshitsugu K, Hattori E, Yoshikawa T, Arinami T, 2002. Association between serotonin 4 receptor gene polymorphisms and bipolar disorder in Japanese case–control samples and the NIMH Genetics Initiative bipolar pedigrees. Mol. Psychiatry 7, 954–961. [DOI] [PubMed] [Google Scholar]
- Ollila HM, Soronen P, Silander K, Palo OM, Kieseppä T, Kaunisto MA, Lönnqvist J, Peltonen L, Partonen T, Paunio T, 2009. Findings from bipolar disorder genome-wide association studies replicate in a Finnish bipolar family-cohort. Mol. Psychiatry 14, 351–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X, Crane DE, MacIntosh BJ, Young LT, Arnold P, Ameis S, Goldstein B, 2015. CACNA1C Rs1006737 genotype and bipolar disorder: focus on intermediate phenotypes and cardiovascular comorbidity. Neurosci. Biobehav. Rev 55, 198–210. [DOI] [PubMed] [Google Scholar]
- Pedrosa E, Stefanescu R, Margolis B, Petruolo O, Lo Y, Nolan K, Novak T, Stopkova P, Lachman HM, 2008. Analysis of protocadherin alpha gene enhancer polymorphism in bipolar disorder and schizophrenia. Schizophr. Res 102, 210–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard BS, Christoforou A, Thomson PA, Fawkes A, Evans KL, Morris SW, Porteous DJ, Blackwood DH, Muir WJ, 2009. Interacting haplotypes at the NPAS3 locus alter risk of schizophrenia and bipolar disorder. Mol. Psychiatry 14 (9), 874–884. [DOI] [PubMed] [Google Scholar]
- Potash JB, DePaulo JR Jr., 2000. Searching high and low: a review of the genetics of bipolar disorder. Bipolar Disord. 2, 8–26. [DOI] [PubMed] [Google Scholar]
- Prata DP, Breen G, Osborne S, Munro J, St Clair D, Collier DA, 2009. An association study of the neuregulin 1 gene, bipolar affective disorder and psychosis. Psychiatr. Genet 19 (3), 113–116. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P, 2009. International Schizophrenia Consortium: common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Parsia S, Papolos DF, Lachman HM, 2000. Analysis of the pseudoautosomal X-linked gene SYBL1 in bipolar affective disorder: description of a new candidate allele for psychiatric disorders. Am. J. Med. Genet 96B, 317–323. [DOI] [PubMed] [Google Scholar]
- Scholz CJ, Jacob CP, Buttenschon HN, Kittel-Schneider S, Boreatti-Hümmer A, Zimmer M, Walter U, Lesch KP, Mors O, Kneitz S, Deckert J, Reif A, 2010. Functional variants of TSPAN8 are associated with bipolar disorder and schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet 153B (4), 967–972. [DOI] [PubMed] [Google Scholar]
- Shaltiel G, Maeng S, Malkesman O, Pearson B, Schloesser RJ, Tragon T, Rogawski M, Gasior M, Luckenbaugh D, Chen G, Manji HK, 2008. Evidence for the involvement of the kainate receptor subunit GluR6 (GRIK2) in mediating behavioral displays related to behavioral symptoms of mania. Mol. Psychiatry 19 (9), 858–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Badner JA, Liu C, 2008. PDLIM5 and susceptibility to bipolar disorder: a family-based association study and meta-analysis. Psychiatr. Genet 18 (3), 116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki G, Potash JB, 2014. New developments in the genetics of bipolar disorder. Curr. Psychiatry Rep 16 (11), 493. [DOI] [PubMed] [Google Scholar]
- Sjøholt G, Ebstein RP, Lie RT, Berle JØ, Mallet J, Deleuze JF, Levinson DF, Laurent C, Mujahed M, Bannoura I, Murad I, Molven A, Steen VM, 2004. Examination of IMPA1 and IMPA2 genes in manic-depressive patients: association between IMPA2 promoter polymorphisms and bipolar disorder. Mol. Psychiatry 9 (6), 621–629. [DOI] [PubMed] [Google Scholar]
- Sklar P, Smoller JW, Fan J, Ferreira MA, Perlis RH, Chambert K, Nimgaonkar VL, McQueen MB, Faraone SV, Kirby A, de Bakker PI, Ogdie MN, Thase ME, Sachs GS, Todd-Brown K, Gabriel SB, Sougnez C, Gates C, Blumenstiel B, Defelice M, Ardlie KG, Franklin J, Muir WJ, McGhee KA, MacIntyre DJ, McLean A, VanBeck M, McQuillin A, Bass NJ, Robinson M, Lawrence J, Anjorin A, Curtis D, Scolnick EM, Daly MJ, Blackwood DH, Gurling HM, Purcell SM, 2008. Whole-genome association study of bipolar disorder. Mol. Psychiatry 13 (6), 558–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, Edenberg HJ Jr., Nurnberger JI, Rietschel M, Blackwood D, Corvin A, Flickinger M, Guan W, Mattingsdal M, McQuillen A, Kwan P, Wienker TF, Daly M, Dudbridge F, Holmans PA, Lin D, Burmeister M, Greenwood TA, Hamshere ML, Muglia P, Smith EN, Zandi PP, Nievergelt CM, McKinney R, Shilling PD, Schork NJ, Bloss CS, Foroud T, Koller DL, Gershon ES, Liu C, Badner JA, Scheftner WA, Lawson WB, Nwulia EA, Hipolito M, Coryell W, Rice J, Byerley W, McMahon FJ, Schulze TG, Berrettini W, Lohoff FW, Potash JB, Mahon PB, McInnis MG, Zöllner S, Zhang P, Craig DW, Szelinger S, Barrett TB, Breuer R, Meier S, Strohmaier J, Witt SH, Tozzi F, Farmer A, McGuffin P, Strauss J, Xu W, Kennedy JL, Vincent JB, Matthews K, Day R, Ferreira MA, O’Dushlaine C, Perlis R, Raychaudhuri S, Ruderfer D, Lee PH, Smoller JW, Li J, Absher D, Bunny WE, Barchas JD, Schatzberg AF, Jones EG, Meng F, Thompson RC, Watson SJ, Myers RM, Akil H, Boehnke M, Chambert K, Moran J, Scolnick E, Djurovic S, Melle I, Morken G, Gill M, Morris D, Quinn E, Mühleisen TW, Degenhardt FA, Mattheisen M, Schumacher J, Maier W, Steffans M, Propping P, Nöthen MM, Anjorin A, Bass N, Gurling H, Kandaswamy R, Lawrence J, McGhee K, McIntosh A, McLean AW, Muir WJ, Pickard BS, Breen G, St Clair D, Caesar S, Gordon-Smith K, Jones L, Fraser C, Green EK, Frozeva D, Jones IR, Kirov G, Moskvina V, Nikolov I, O’Donovan MC, Owen MJ, Collier DA, Elkin A, Williamson R, Young AH, Ferrier IN, Stefansson K, Stefansson H, Porgeirsson P, Steinberg S, Gustafsson Ó, Bergen SE, Nimgaonkar V, Hultman C, Landén M, Lichtenstein P, Sullivan P, Schalling M, Osby U, Backlund L, Frisén L, Langstrom N, Jamain S, Leboyer M, Etain B, Bellivier F, Petursson H, Sigurđsson E, Müller-Mysok B, Lucae S, Schwarz M, Fullerton JM, Schofield PR, Martin N, Montgomery GW, Lathrop M, Óskarsson H, Bauer M, Wright A, Mitchell PB, Hautzinger M, Reif A, Kelsoe JR, Purcell SM, Psychiatric GWAS Consortium Bipolar Disorder Working Group, 2011. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet 43, 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EN, Bloss CS, Badner JA, Barrett T, Belmonte PL, Berrettini W, Byerley W, Coryell W, Craig D, Edenberg HJ, Eskin E, Foroud T, Gershon E, Greenwood TA, Hipolito M, Koller DL, Lawson WB, Liu C, Lohoff F, McInnis MG, McMahon FJ, Mirel DB, Murray SS, Nievergelt C, Nurnberger J, Nwulia EA, Paschall J, Potash JB, Rice J, Schulze TG, Scheftner W, Panganiban C, Zaitlen N, Zandi PP, Zöllner S, Schork NJ, Kelsoe JR, 2009. Genome-wide association study of bipolar disorder in European American and African American individuals. Mol. Psychiatry 14 (8), 755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoller JW, Finn CT, 2003. Family, twin, and adoption studies of bipolar disorder. Am. J. Med. Genet. C: Semin. Med. Genet 123C (1), 48–58. [DOI] [PubMed] [Google Scholar]
- Soria V, Martínez-Amorós E, Escaramís G, Valero J, Pérez-Egea R, García C, Gutiérrez-Zotes A, Puigdemont D, Bayés M, Crespo JM, Martorell L, Vilella E, Labad A, Vallejo J, Pérez V, Menchón JM, Estivill X, Gratacòs M, Urretavizcaya M, 2010. Differential association of circadian genes with mood disorders: CRY1 and NPAS2 are associated with unipolar major depression and CLOCK and VIP with bipolar disorder. Neuropsychopharmacology 35, 1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soronen P, Ollila HM, Antila M, Silander K, Palo OM, Kieseppä T, Lönnqvist J, Peltonen L, Tuulio-Henriksson A, Partonen T, Paunio T, 2010. Replication of GWAS of bipolar disorder: association of SNPs near CDH7 with bipolar disorder and visual processing. Mol. Psychiatry 15, 4–6. [DOI] [PubMed] [Google Scholar]
- Takata A, Kim SH, Ozaki N, Iwata N, Kunugi H, Inada T, Ujike H, Nakamura K, Mori N, Ahn YM, Joo EJ, Song JY, Kanba S, Yoshikawa T, Kim YS, Kato T, 2011. Association of ANK3 with bipolar disorder confirmed in East Asia. Am. J. Med. Genet. B Neuropsychiatr. Genet 156B (3), 312–315. [DOI] [PubMed] [Google Scholar]
- Tesli M, Athanasiu L, Mattingsdal M, Kähler AK, Gustafsson O, Andreassen BK, Werge T, Hansen T, Mors O, Mellerup E, Koefoed P, Jönsson EG, Agartz I, Melle I, Morken G, Djurovic S, Andreassen OA, 2010. Association analysis of PALB2 and BRCA2 in bipolar disorder and schizophrenia in a scandinavian case–control sample. Am. J. Med. Genet. B Neuropsychiatr. Genet 153B (7), 1276–1282. [DOI] [PubMed] [Google Scholar]
- Thomson PA, Wray NR, Thomson AM, Dunbar DR, Grassie MA, Condie A, Walker MT, Smith DJ, Pulford DJ, Muir W, Blackwood DH, Porteous DJ, 2005. Sex-specific association between bipolar affective disorder in women and GPR50, and X-linked orphan G protein-coupled receptor. Mol. Psychiatry 10 (5), 470–478. [DOI] [PubMed] [Google Scholar]
- Veldic M, Guidotti A, Maloku E, Davis JM, Costa E, 2005. In psychosis, cortical inter-neurons overexpress DNA-methyltransferase 1. Proc. Natl. Acad. Sci. U. S. A 102, 2152–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams HJ, Norton N, Dwyer S, Moskvina V, Nikolov I, Carroll L, Georgieva L, Williams NM, Morris DW, Quinn EM, Giegling I, Ikeda M, Wood J, Lencz T, Hultman C, Lichtenstein P, Thiselton D, Maher BS, Molecular Genetics of Schizophrenia Collaboration (MGS) International Schizophrenia Consortium (ISC), SGENE-plus, GROUP, Malhotra AK, Riley B, Kendler KS, Gill M, Sullivan P, Sklar P, Purcell S, Nimgaonkar VL, Kirov G, Holmans P, Corvin A, Rujescu D, Craddock N, Owen MJ, O’Donovan MC, 2011. Fine mapping of ZNF804A and genome-wide significant evidence for its involvement in schizophrenia and bipolar disorder. Mol. Psychiatry 16 (4), 429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Cohen-Woods S, Chen Q, Noor A, Knight J, Hosang G, Parikh SV, De Luca V, Tozzi F, Muglia P, Forte J, McQuillin A, Hu P, Gurling HM, Kennedy JL, McGuffin P, Farmer A, Strauss J, Vincent JB, 2014. Genome-wide association study of bipolar disorder in Canadian and UK populations corroborates disease loci including SYNE1 and CSMD1. BMC Med. Genet 15, 2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lindpaintner K, Che R, He Z, Wang P, Yang P, Feng G, He L, Shi Y, 2009. The Val/Met functional polymorphism in COMT confers susceptibility to bipolar disorder: evidence from an association study and a meta-analysis. J. Neural Transm 116 (10), 1193–1200. [DOI] [PubMed] [Google Scholar]
- Zou YF, Wang F, Feng XL, Li WF, Pan FM, Huang F, 2011. Meta-analysis of ACE gene I/D polymorphism and bipolar disorder susceptibility. Nord. J. Psychiatry 65 (4), 276–282. [DOI] [PubMed] [Google Scholar]
Web Resources
- Depression and Bipolar Support Alliance, 2015. Bipolar disorder statistics. http://www.dbsalliance.org/site/PageServer?pagename=education_statistics_bipolar_disorder (Accessed 12 October).
- GeneCards, 2015. http://www.genecards.org (Accessed 12 October).
- Juvenile Bipolar Research Foundation, 2015. http://www.jbrf.org/diagnosis-by-the-dsm/ (Accessed 12 October).
- National Center for Biotechnology Information, 2015. https://www.ncbi.nlm.nih.gov/gene (Accessed 12 October). [DOI] [PMC free article] [PubMed]
- National Institute of Mental Health, 2015. Bipolar disorder. http://www.nimh.nih.gov/health/topics/bipolar-disorder/index.shtml#part_145403 (Accessed 12 October).
- Online Mendelian Inheritance in Man (OMIM), 2015. http://www.ncbi.nlm.nih.gov/omim (Accessed 12 October).
- PubMed, 2015. https://www.ncbi.nlm.nih.gov/pubmed Accessed 12 October.