Abstract
INTRODUCTION:
DNA methylation is a key epigenetic mechanism in brain aging and Alzheimer’s disease (AD). The newly discovered 5-hydroxymethylcytosine (5hmC) mediates DNA demethylation, is highly abundant in the brain, and is dynamically regulated by life experiences. However, little is known about its genome-wide patterns and potential role in AD.
METHODS:
Using a genome-wide capture followed by high-throughput sequencing, we studied the genome-wide distribution of 5hmC at specific genomic loci in human AD brain and identified differentially hydroxymethylated regions (DhMRs) associated with AD pathology.
RESULTS:
We identified 517 DhMRs significantly associated with neuritic plaques and 60 DhMRs associated with neurofibrillary tangles. DNA hydroxymethylation in gene bodies was predominantly positively correlated with cis-acting gene expression. Moreover, genes showing differential hydroxymethylation were significantly enriched in neurobiological processes and clustered in functional gene ontology categories.
DISCUSSION:
Our results reveal a critical role of DNA hydroxymethylation in AD pathology and provide mechanistic insight into the molecular mechanisms underlying AD.
Keywords: Alzheimer’s disease, postmortem brain, DNA hydroxymethylation, genome-wide association
1. Background
Alzheimer’s disease (AD) is characterized clinically by progressive cognitive decline, and histologically by neuritic plaques, neurofibrillary tangles, and loss of neurons in the brain. Epigenetic factors mediate the effects of age and environmental factors independently of DNA sequence, and epigenetic dysregulation has been implicated in brain aging and neurodegenerative disorders including AD [1]. Elucidating epigenetic pathways is likely to provide mechanistic insight into disease etiology and holds promise for discovering novel strategies for early detection and therapeutic intervention at preclinical stages of the disease.
DNA methylation on the fifth carbon of the cytosine base (5-methylcytosine, 5mC) is the most extensively studied epigenetic modification that is essential for neurogenesis [2], learning and memory [3], and synaptic plasticity [4]. Altered levels of 5-methylcytosine (5mC) have been associated with AD diagnosis and numerous neuropathological phenotypes, [5–8] though results are somewhat conflicting.[5, 9–11] Notably, in a large collection of postmortem human brains, our group has recently identified an association between altered levels of 5mC with the burden of AD pathology [12–15].
The newly discovered 5-hydroxymethylcytosine (5hmC) is an oxidative product of 5mC catalyzed by the ten-eleven translocation (TET) family of proteins. This conversion is an important mechanism underlying the active demethylation of DNA [16]. 5hmC acts as an intermediate in DNA demethylation and also serves as a stable epigenetic mark during development of disease [17]. Studies of mouse brain indicated that 5hmC is particularly enriched in the brain [18, 19] [9, 20], dynamically regulated during neurodevelopment [21–23], and accumulates with age across the lifespan [21, 23–26]. Unlike 5mC which is often located in CpG-rich regions (e.g., promoters), 5hmC occurs primarily in gene bodies and exon-intron boundaries. As with 5mC, 5hmC levels vary substantially between different cell types and tissues, and its enrichment in gene bodies positively influences gene expression in the human brain [27]. These findings suggest that 5hmC represents a new dimension of epigenetic regulation that may play an important role in brain aging and neurodegenerative disorders. However, there is little research examining the genome-wide patterns of 5hmC in human AD brain and its association with AD pathology in human populations. Here we report findings from a genome-wide profiling of 5hmC in human postmortem brain tissue from a community-based cohort of aging and dementia with brain donation at the time of death with the goal of identifying differentially hydroxymethylated regions (DhMRs) associated with quantitative measures of neuropathological burden on AD.
2. Methods
2.1. Subject population and postmortem brain sample
Postmortem human dorsolateral prefrontal cortex (DLPFC) tissue (N=30) were obtained from deceased participants in two community-based, ongoing prospective cohort studies of older individuals: the Religious Orders Study (ROS) and the Rush Memory and Aging Project (MAP). Detailed study design and assessment procedures have been described previously [28, 29]. In brief, ROS recruited more than 1,300 older Catholic priests, nuns and brothers from across the USA. MAP recruited more than 1,800 older men and women from across the Chicagoland area. All participants are free of dementia at enrollment, and agree to annual clinical evaluations and brain donation upon death. The clinical evaluation includes detailed neurologic examination and clinical classification of dementia and Alzheimer’s disease (AD). At the time of death, a summary diagnosis of AD is made based on review of all clinical data without access to the post-mortem data [30]. Both studies were approved by the Institutional Review Board of Rush University Medical Center. Written informed consent was obtained from all subjects, followed by an Anatomic Gift Act for organ donation.
2.2. Assessment of neuropathological phenotypes
Detailed procedures for postmortem brain examination and neuropathological phenotyping have been described previously [31, 32]. Briefly, Bielschowsky silver stain was used to visualize neuritic plaques (NP) and neurofibrillary tangles (NFTs) in the frontal, temporal, parietal, and entorhinal cortices, and the hippocampus. The quantitative NP and NFT burden was based on counts of the lesions in the maximum density in each region, as described [33, 34].
2.3. Genome-wide profiling of brain DNA hydroxymethylome
5hmC-capture sequencing.
Genomic DNA was isolated from 10 mg frozen dorsolateral prefrontal cortex with proteinase K digestion. 5hmC enrichment was performed using previously described selective chemical labeling technique [25]. This method used the T4 bacteriophage β-glucosyltransferase (T4-BGT) to specifically modify 5hmC residues by adding glucose moiety to 5hmC. 5hmC-captured libraries were generated following the Illumina protocol for “Preparing Samples for ChIP Sequencing of DNA”. We used 25 ng of input genomic DNA or 5hmC-captured DNA to initiate the protocol. DNA fragments were gel-purified after adapter ligation. PCR-amplified DNA libraries were quantified on Agilent 2100 Bioanalyzer and diluted to 6-8 pM for cluster generation and sequencing. We performed 38-cycle single-end sequencing generation on Illumina HiSeq 2000 to obtain 5hmC-enriched DNA fragment sequence.
Sequence alignment, 5hmC quantification and distribution.
FASTQ files were aligned to the human genome (hg19) with Bowtie, retaining only non-duplicate genomic matches with no more than 2 mismatches in the first 25 bp [35]. Model-based Analysis of ChIP-Seq (MACS) software was used to estimate the level of 5hmC enrichment in each sample by directly comparing to the input DNA (effective genome size =1.87 × 109, tag size = 38, bandwidth = 200). Unique, non-duplicate reads were counted in 10kb bins and normalized to the total number of non-duplicate reads. Genome-wide patterns of 5hmC were evaluated by counting mapped reads per 10kb bin, which were then normalized to sequencing coverage.
2.4. Statistical and bioinformatics analysis
DhMRs analysis.
To identify differentially hydroxymethylated regions (DhMRs) associated with AD pathology, we separately constructed zero-inflated negative binomial regression models for quantitative measures of NP or NFTs, adjusting for potential confounding factors including age at death, sex, education level, neuronal proportions, and the first 3 principal components (derived from GWAS [36]) that capture the effect of population substructure in European ancestry among our study participants. This analysis was performed using the R package pscl (http://CRAN.R-project.org/package=pscl) by scanning the genome in 10 kb sliding windows. The zero-inflated negative binomial model was used here to account for the overdispersed count outcome variables for the neuropathological phenotypes. To facilitate data interpretation, we further categorized each trait into tertiles and compared 5hmC patterns between subjects with high (top tertile) versus low (bottom tertile) burden of AD pathology. Brain neuronal proportions were inferred using the computer package CETS 3.03 in R [37]. Multiple testing was corrected for the total number of 10-kb bins by false-discovery rate (FDR) and a cutoff of FDR-adjusted P-value (i.e., q-value) <0.05 was used to determine statistical significance. Homer [38] was used to annotate DhMRs to genomic features in hg19.
Relationship between 5hmC and gene expression.
Using brain gene expression data quantified by RNA-Seq in the same brain neocortex, we assessed the functional importance of putative DhMRs by calculating partial correlations (corrected for age at death, sex, and education level) between levels of 5hmC enrichment and cis or trans-acting gene expression. Methods for RNA-seq data collection, data pre-processing, QC and normalization were described in online Supplementary material. Here cis was defined as correlation between 5hmC enrichment of a DhMR gene with its own expression and trans otherwise. Multiple testing was corrected by FDR and FDR<0.05 was used to determine statistical significant. To further examine the role of altered 5hmC in gene regulation, we also conducted gene overlapping analysis with the gene lists of differential hydroxymethylation with that of gene expression using GeneOverlap software in R.
Co-hydroxymethylation network analysis.
To identify co-regulated genes associated with AD, we conducted network analyses using the Weighted Gene Correlation Network Analysis (WGCNA) [39]. This method allows for identifying co-hydroxymethylation or co-expression modules associated with AD pathology and hub gene(s) that may play key roles in disease pathogenesis. For this analysis, we compiled lists of genes showing nominally significant association for DNA hydroxymethylation (P< 0.01, a total of 1,261 genes for NP, 702 genes for NFTs) or gene expression (P< 0.01, total genes = 1,074) obtained from the above described regression models. Co-hydroxymethylation or co-expression modules (subnetworks) were constructed separately among samples with high (upper tertile) or low (bottom tertile) burden of AD pathology. Differential networks were then identified by comparing the two groups. Network visualization was performed using CytoScape [40].
Gene ontology (GO) enrichment analysis.
To functionally annotate putative DhMR genes, we conducted functional enrichment analysis for the identified DhMR genes using the functional annotation tool DAVID.[41] Gene symbols corresponding to putative DhMRs (P<0.05) were used as input of the gene ontology (GO) enrichment analysis. Here we used this method to characterize genes corresponding to modules and the list of genes that are hyper-hydroxymethylated (or hypo-hydroxymethylated) in relation to AD pathologic burden.
3. Results
Our 5hmC-enriched sequencing in 30 brain samples resulted in a total of 349 million non-duplicated reads, covering 5,792 unique genes. The mean age of brain donors was 91.0 (SD 4.6), and about 70% of the donors were females. The mean post-mortem interval (PMI) was 6.5 hours. Table 1 shows the clinical characteristics of brain donors at the age of death.
Table 1.
Characteristics of study participants at the time of death (N=30)
| Characteristic | % or Mean ± SD |
|---|---|
| Age (years) | 91.1±4.6 |
| Male sex (%) | 30% |
| Education level (years) | 18.2± 2.0 |
| NCI (%) | 20.0% |
| MCI (%) | 13.3% |
| AD (%) | 66.7% |
| MMSE score | 16.1±10.9 |
| Neuritic plaques (NPs) | 11.6±12.7 |
| Neurofibrillary tangles (NFTs) | 1.6±4.1 |
| Post-mortem interval (PMI, hours) | 6.5±3.4 |
NCI, no cognitive impairment; MCI, mild cognitive impairment; MMSE, mini-mental status examination; PMI, post-mortem interval (in hours) from time of death to autopsy.
3.1. DhMRs associated with AD neuropathology
After adjusting for covariates (age at death, sex, education, brain cellular heterogeneity and the first 3 PCs derived from GWAS) and multiple testing (q-value < 0.05), we identified 517 DhMRs (311 hyper-hydroxymethylated, 206 hypo-hydroxymethylated) that were significantly associated with NP. Homer annotated these DhMRs to 321 distinct genes (203 hyper-hydroxymethylated, 118 hypo-hydroxymethylated). At q-value < 0.05, 60 DhMRs (35 hyper-hydroxymethylated, 25 hypo-hydroxymethylated) were significantly associated with NFTs. These NFTs-related DhMRs were annotated to 49 distinct genes. Tables 2 and 3 list the top 50 DhMRs regions associated with NP and NFTs, respectively. A full lists of the identified DhMRs along with the nearest annotated genes associated with NP and NFTs are shown in Tables S1 and S2, respectively. Figure S1 displays DhMRs that are positively or negatively associated with NP (Supplementary Fig 1A) and NFTs (Supplementary Fig 1B), respectively.
Table 2.
Top 50 DhMRs associated with quantitative neuritic plaque in human postmortem brain (N=30)
| Chromosome | Start (bp) | End (bp) | Distance to TSS (bp) of the nearest gene |
Nearest gene | Effect size* | P-value* | q-value † |
|---|---|---|---|---|---|---|---|
| 16 | 12220000 | 12230000 | 19258 | CACNA1H | 0.0152 | 5.36×10−20 | 1.01×10−5 |
| 10 | 105550000 | 105560000 | −51187 | LOC101928322 | 0.0139 | 2.09×10−15 | 1.36×10−11 |
| 9 | 6600000 | 6610000 | −46307 | KANK1 | 0.0132 | 2.17×10−15 | 1.36×10−11 |
| 9 | 127960000 | 127970000 | 17889 | LURAP1L-AS1 | 0.0148 | 2.07×10−10 | 9.71×10−7 |
| 5 | 124180000 | 124190000 | −156470 | LINC01194 | 0.0133 | 3.15×10−10 | 1.18×10−6 |
| 17 | 4310000 | 4320000 | −135768 | FAM101B | 0.0127 | 5.83×10−10 | 1.82×10−6 |
| 15 | 75730000 | 75740000 | −12914498 | CHEK2P2 | 0.0083 | 1.09×10−9 | 2.75×10−6 |
| 11 | 470000 | 480000 | 84421 | LINC01001 | −0.0297 | 1.32×10−9 | 2.75×10−6 |
| 1 | 234550000 | 234560000 | 39852 | LUZP1 | 0.0127 | 1.32×10−9 | 2.75×10−6 |
| 11 | 64740000 | 64750000 | −12245 | HPX | 0.0111 | 1.60×10−9 | 3.00×10−6 |
| 12 | 117740000 | 117750000 | −28289 | ETV6 | 0.0119 | 1.91×10−9 | 3.26×10−6 |
| 7 | 98990000 | 99000000 | 225599 | PER4 | 0.0136 | 2.31×10−9 | 3.34×10−6 |
| 17 | 47020000 | 47030000 | 3060 | PSMB6 | 0.0090 | 2.67×10−9 | 3.34×10−6 |
| 20 | 10430000 | 10440000 | −50407 | PSMF1 | 0.0147 | 2.45×10−9 | 3.34×10−6 |
| 1 | 227050000 | 227060000 | −72845 | ZBTB40 | 0.0140 | 2.67×10−9 | 3.34×10−6 |
| 22 | 31960000 | 31970000 | −12965567 | BMS1P17 | 0.0115 | 3.41×10−9 | 3.55×10−6 |
| 10 | 61550000 | 61560000 | 24190 | RBM17 | 0.0098 | 3.19×10−9 | 3.55×10−6 |
| 12 | 30790000 | 30800000 | 10751 | TEAD4 | 0.0114 | 3.31×10−9 | 3.55×10−6 |
| 19 | 20150000 | 20160000 | 203 | BTBD2 | 0.0083 | 4.37×10−9 | 4.29×10−6 |
| 17 | 3480000 | 3490000 | −52768 | FAM101B | 0.0123 | 4.57×10−9 | 4.29×10−6 |
| 19 | 4150000 | 4160000 | −6329 | C2CD4C | 0.0118 | 5.20×10−9 | 4.65×10−6 |
| 2 | 116030000 | 116040000 | 2804 | E2F6 | 0.0141 | 6.20×10−9 | 4.90×10−6 |
| 7 | 73290000 | 73300000 | 34714 | LOC101927354 | 0.0120 | 6.27×10−9 | 4.90×10−6 |
| 6 | 49530000 | 49540000 | 50798 | RPP40 | 0.0104 | 5.87×10−9 | 4.90×10−6 |
| 13 | 20240000 | 20250000 | 17289740 | LINC00417 | 0.0124 | 7.13×10−9 | 5.35×10−6 |
| 22 | 31060000 | 31070000 | −13055567 | BMS1P17 | 0.0080 | 7.85×10−9 | 5.67×10−6 |
| 2 | 233150000 | 233160000 | −292799 | KLHL29 | 0.0192 | 8.22×10−9 | 5.71×10−6 |
| 3 | 162930000 | 162940000 | 12997 | DPH3 | 0.0136 | 9.03×10−9 | 6.05×10−6 |
| 17 | 1700000 | 1710000 | −10497 | LOC100506388 | 0.0118 | 9.45×10−9 | 6.11×10−6 |
| 14 | 34450000 | 34460000 | −15932095 | OR11H12 | 0.0122 | 9.91×10−9 | 6.20×10−6 |
| 9 | 35990000 | 36000000 | −68587 | RFX3-AS1 | 0.0131 | 1.16×10−8 | 6.98×10−6 |
| 7 | 23480000 | 23490000 | 5600 | SNX8 | 0.0129 | 1.19×10−8 | 6.98×10−6 |
| 3 | 48810000 | 48820000 | −58411 | BHLHE40-AS1 | 0.0132 | 1.70×10−8 | 9.11×10−6 |
| 7 | 75670000 | 75680000 | 7961 | COL28A1 | 0.0078 | 1.66×10−8 | 9.11×10−6 |
| 17 | 1100000 | 1110000 | −70497 | LOC100506388 | 0.0080 | 1.70×10−8 | 9.11×10−6 |
| 20 | 25410000 | 25420000 | 24246 | TMC2 | 0.0143 | 1.94×10−8 | 1.01×10−5 |
| 12 | 64620000 | 64630000 | −11216 | TNFRSF1A | −0.0259 | 2.42×10−8 | 1.23×10−5 |
| 14 | 22020000 | 22030000 | −17175095 | OR11H12 | 0.0114 | 2.55×10−8 | 1.26×10−5 |
| 7 | 106580000 | 106590000 | 321314 | NDUFA4 | −0.0222 | 2.82×10−8 | 1.36×10−5 |
| 7 | 150760000 | 150770000 | −195424 | DGKB | −0.0363 | 3.07×10−8 | 1.44×10−5 |
| 6 | 17820000 | 17830000 | 171818 | FOXC1 | 0.0057 | 3.86×10−8 | 1.77×10−5 |
| 3 | 150350000 | 150360000 | −45551 | FGD5-AS1 | 0.0164 | 4.39×10−8 | 1.96×10−5 |
| 6 | 107460000 | 107470000 | −1493 | TMEM14B | 0.0105 | 6.87×10−8 | 3.00×10−5 |
| 1 | 1430000 | 1440000 | −2933 | LOC729737 | −0.0206 | 7.11×10−8 | 3.03×10−5 |
| 8 | 22100000 | 22110000 | 185830 | MIR7160 | 0.0306 | 1.13×10−7 | 4.71×10−5 |
| 22 | 31960000 | 31970000 | −12965567 | BMS1P17 | 0.0094 | 1.62×10−7 | 6.61×10−5 |
| 8 | 144620000 | 144630000 | 248520 | MIR383 | 0.0304 | 2.92×10−7 | 1.17×10−4 |
| 1 | 39230000 | 39240000 | −77173 | LINC01346 | 0.0165 | 3.07×10−7 | 1.20×10−4 |
| 5 | 56520000 | 56530000 | 229713 | ICE1 | 0.0097 | 3.51×10−7 | 1.34×10−4 |
| 15 | 75730000 | 75740000 | −12914498 | CHEK2P2 | 0.0088 | 4.51×10−7 | 1.69×10−4 |
Obtained from zero-inflated negative binomial regression, adjusting for age at death, sex, education level, neuronal proportions and the first 3 PC derived from previous GWAS;
Adjusting for a total number of 18,767 10-kb bins.
Table 3.
Top 50 DhMRs associated with quantitative NFTs in human postmortem brain (N=30)
| Chromosome | Start (bp) | End (bp) | Distance to TSS (bp) of nearest gene |
Nearest gene | Effect size* | P-value* | q-value† |
|---|---|---|---|---|---|---|---|
| 1 | 16490000 | 16500000 | 6292 | CDK11A | 0.2579 | 6.87× 10−11 | 8.03×10−7 |
| 9 | 131920000 | 131930000 | 87064 | MPDZ | 0.2350 | 1.47× 10−10 | 8.57×10−7 |
| 16 | 3360000 | 3370000 | 3381 | PDIA2 | −0.1322 | 3.62× 10−10 | 1.41×10−6 |
| 9 | 79400000 | 79410000 | −140700 | TMEM261 | 0.1143 | 6.30× 10−10 | 1.84×10−6 |
| 7 | 105090000 | 105100000 | 470314 | NDUFA4 | −0.4217 | 1.42× 10−9 | 3.33×10−6 |
| 22 | 39410000 | 39420000 | −12220567 | BMS1P17 | −0.4868 | 1.93× 10−9 | 3.76×10−6 |
| 3 | 39330000 | 39340000 | 92378 | LRRN1 | 0.1198 | 3.29× 10−9 | 5.49×10−6 |
| 14 | 55520000 | 55530000 | −13825095 | OR11H12 | −0.3974 | 4.14× 10−9 | 6.05×10−6 |
| 16 | 68960000 | 68970000 | 72689 | RBFOX1 | 0.1142 | 1.60× 10−8 | 2.08×10−5 |
| 11 | 62200000 | 62210000 | 45 | OR52W1 | 0.1312 | 2.05× 10−8 | 2.39×10−5 |
| 2 | 101040000 | 101050000 | 12707 | GRHL1 | 0.0958 | 2.34× 10−8 | 2.49×10−5 |
| 10 | 105400000 | 105410000 | −36187 | LOC101928322 | 0.0677 | 2.75× 10−8 | 2.68×10−5 |
| 12 | 12620000 | 12630000 | 125585 | ERC1 | 0.1227 | 2.99× 10−8 | 2.69×10−5 |
| 10 | 99380000 | 99390000 | −162186 | LOC101928298 | 0.2970 | 5.93× 10−8 | 4.95×10−5 |
| 22 | 37930000 | 37940000 | −12368567 | BMS1P17 | 0.1362 | 7.53× 10−8 | 5.87×10−5 |
| 16 | 72910000 | 72920000 | −91252 | RBFOX1 | 0.0806 | 8.49× 10−8 | 6.20×10−5 |
| 16 | 70610000 | 70620000 | 237689 | RBFOX1 | 0.1096 | 9.82× 10−8 | 6.75×10−5 |
| 3 | 171010000 | 171020000 | 126811 | MIR3714 | −0.5098 | 1.20× 10−7 | 7.76×10−5 |
| 17 | 57950000 | 57960000 | 119945 | LOC339166 | −0.2200 | 1.49× 10−7 | 9.17×10−5 |
| 13 | 52980000 | 52990000 | 14015740 | LINC00417 | 0.0757 | 2.07× 10−7 | 1.21×10−4 |
| 10 | 123250000 | 123260000 | −66084 | CAMK1D | 0.2151 | 2.26× 10−7 | 1.26×10−4 |
| 12 | 121210000 | 121220000 | −42392 | RNU6-19P | 0.1551 | 3.43× 10−7 | 1.82×10−4 |
| 1 | 117610000 | 117620000 | 9718 | DRAXIN | 0.2147 | 3.92× 10−7 | 1.99×10−4 |
| 2 | 198390000 | 198400000 | −229116 | LINC00954 | 0.0826 | 4.25× 10−7 | 2.07×10−4 |
| 3 | 72980000 | 72990000 | 278472 | GRM7-AS1 | −0.0571 | 4.78× 10−7 | 2.23×10−4 |
| 14 | 105970000 | 105980000 | −8780095 | OR11H12 | 0.1601 | 5.75× 10−7 | 2.59×10−4 |
| 14 | 58890000 | 58900000 | −13488095 | OR11H12 | 0.0832 | 1.18× 10−6 | 5.11×10−4 |
| 8 | 2250000 | 2260000 | 42949 | ZNF596 | 0.0922 | 1.84× 10−6 | 7.7×10−4 |
| 4 | 7380000 | 7390000 | 37137 | LOC100129917 | 0.1447 | 2.21× 10−6 | 8.91×10−4 |
| 6 | 3770000 | 3780000 | −14240 | IRF4 | −0.0850 | 2.38× 10−6 | 9.28×10−4 |
| 13 | 49890000 | 49900000 | 14324740 | LINC00417 | 0.0853 | 2.78× 10−6 | 1.05×10−3 |
| 1 | 1460000 | 1470000 | −5933 | LOC729737 | −0.0401 | 4.59× 10−6 | 1.67×10−3 |
| 16 | 87810000 | 87820000 | 13055 | ABAT | 0.0724 | 6.32× 10−6 | 2.24×10−3 |
| 9 | 131730000 | 131740000 | 106064 | MPDZ | 0.0576 | 6.92× 10−6 | 2.38×10−3 |
| 4 | 83310000 | 83320000 | 60007 | HTRA3 | −0.3446 | 7.61× 10−6 | 2.54×10−3 |
| 17 | 57500000 | 57510000 | 74945 | LOC339166 | 0.0973 | 8.58× 10−6 | 2.78×10−3 |
| 8 | 6450000 | 6460000 | 35727 | ERICH1 | 0.1302 | 1.02× 10−5 | 3.22×10−3 |
| 17 | 41910000 | 41920000 | −16310 | LOC103021295 | 0.1064 | 1.26× 10−5 | 3.86×10−3 |
| 3 | 179080000 | 179090000 | −95565 | LOC339862 | −0.1694 | 1.29× 10−5 | 3.86×10−3 |
| 22 | 24060000 | 24070000 | −13755567 | BMS1P17 | 0.1000 | 1.32× 10−5 | 3.87×10−3 |
| 8 | 1400000 | 1410000 | −23475 | OR4F21 | −0.0197 | 1.61× 10−5 | 4.50×10−3 |
| 14 | 102410000 | 102420000 | −9136095 | OR11H12 | 0.0694 | 1.82× 10−5 | 5.06×10−3 |
| 19 | 56660000 | 56670000 | −14277 | HSD11B1L | −0.0424 | 1.91× 10−5 | 5.18×10−3 |
| 20 | 11520000 | 11530000 | 12618 | TMEM74B | −0.2219 | 2.13× 10−5 | 5.67×10−3 |
| 4 | 187210000 | 187220000 | −698016 | LCORL | −0.0566 | 2.32× 10−5 | 6.02×10−3 |
| 1 | 93200000 | 93210000 | 20596 | H6PD | −0.0479 | 2.84× 10−5 | 7.21×10−3 |
| 17 | 64440000 | 64450000 | 15378 | PITPNM3 | −0.1952 | 2.91× 10−5 | 7.23×10−3 |
| 1 | 2220000 | 2230000 | −81933 | LOC729737 | 0.0403 | 3.22× 10−5 | 7.84×10−3 |
| 16 | 910000 | 920000 | 12133 | POLR3K | −0.0652 | 3.55× 10−5 | 8.46×10−3 |
| 8 | 19490000 | 19500000 | 27455 | KBTBD11 | −0.0337 | 3.71×10−5 | 8.67×10−3 |
Obtained from zero-inflated negative binomial regression, adjusting for age at death, sex, education, neuronal proportions and the first 3 PCs derived from GWAS;
Adjusting for a total number of 18,767 10-kb bins.
Of the 321 annotated DhMR genes associated with neuritic plaques, 47 genes also showed differential expression at the cutoff of q-value ≤ 0.05 (Table 4). Of these, many genes such as ATXN1, RAF-1, DGKB, TP73, TNFRSF1A, MME, DUSP22, CSNK2A1, SYN2, APC2, CSMD1, etc. are known to be associated with AD-related pathological processes including Aβ degradation, tau phosphorylation, synaptic plasticity, cognition, glucose transporter, and oxidative stress, etc. Four of these 47 genes (ABAT, CAMK1D, HTRA3 and LRRN1) were also significantly associated with NFTs. Moreover, these genes are enriched for GO biological processes such as learning (long term memory) and neurotrophin signaling pathway, indicating functional relevance of these genes to AD pathology. Further, gene overlapping analysis revealed that these DhMR genes are 2.2 times more likely to also show changes in expression (P<1.6×10−9). Together, these results suggest that altered DNA hydroxymethylation may contribute to AD neuropathology.
Table 4.
Genes showing both differential hydroxymethylation and expression in relation to burden of neuritic plaque
| Gene name | q-value for DhMR analysis | q-value for differential expression analysis |
|---|---|---|
| CAMK1D* | 2.93×10−4 | 1.83×10−5 |
| ENO1 | 2.57×10−5 | 2.21×10−5 |
| ADCYAP1 | 8.26×10−4 | 2.37×10−5 |
| NBAS | 6.21×10−4 | 4.51×10−5 |
| KCNA6 | 3.18×10−4 | 5.24×10−5 |
| DKK3 | 1.29×10−3 | 7.51×10−5 |
| SYN2 | 1.58×10−3 | 9.88×10−5 |
| ADCY2 | 1.86×10−4 | 1.01×10−4 |
| MTRR | 1.81×10−3 | 1.36×10−4 |
| DTNBP1 | 1.54×10−3 | 1.38×10−4 |
| CSNK2A1 | 9.65×10−5 | 1.72×10−4 |
| ABAT* | 3.18×10−4 | 1.86×10−4 |
| BASP1 | 4.72×10−6 | 2.33×10−4 |
| ATP1B2 | 1.04×10−3 | 2.34×10−4 |
| CTNND2 | 1.09×10−3 | 2.36×10−4 |
| TP73 | 5.99×10−5 | 2.84×10−4 |
| CSMD1 | 2.06×10−5 | 3.35×10−4 |
| EMP1 | 1.88×10−5 | 3.76×10−4 |
| MPG | 1.29×10−3 | 3.83×10−4 |
| ASPA | 8.85×10−7 | 4.01×10−4 |
| PER1 | 7.61×10−4 | 4.12×10−4 |
| VAMP2 | 2.53×10−4 | 4.60×10−4 |
| KCNA5 | 5.49×10−5 | 4.78×10−4 |
| MAP2K2 | 8.97×10−4 | 5.03×10−4 |
| FUT6 | 1.14×10−3 | 6.44×10−4 |
| HPX | 1.60×10−9 | 7.62×10−4 |
| COLEC12 | 1.53×10−3 | 7.74×10−4 |
| DUSP22 | 1.62×10−3 | 8.75×10−4 |
| ETV1 | 1.73×10−4 | 9.42×10−4 |
| LRRN1* | 1.20×10−3 | 1.11×10−3 |
| SMARCA2 | 8.95×10−4 | 1.19×10−3 |
| MSRA | 1.30×10−4 | 1.42×10−3 |
| EPHB2 | 3.84×10−5 | 1.47×10−3 |
| APC2 | 5.76×10−5 | 1.51×10−3 |
| NFIB | 8.31×10−5 | 1.69×10−3 |
| ATXN1 | 5.52×10−4 | 1.76×10−3 |
| HTRA3* | 7.77×10−4 | 3.55×10−3 |
| SLC2A14 | 3.54×10−4 | 3.67×10−3 |
| TCF3 | 1.21×10−3 | 3.98×10−3 |
| GRM7-AS3 | 3.48×10−4 | 6.91×10−3 |
| TNFRSF1A | 2.42×10−8 | 8.15×10−3 |
| RPL32 | 4.18×10−5 | 1.10×10−2 |
| PTBP1 | 2.43×10−4 | 1.54×10−2 |
| DGKB | 3.07×10−8 | 1.62×10−2 |
| RAF1 | 9.21×10−4 | 1.69×10−2 |
| SKI | 6.97×10−4 | 2.14×10−2 |
| MMEL1 | 2.36×10−4 | 2.15×10−2 |
Also associated with NFTs
3.2. Genomic distribution of 5hmC in human AD brain
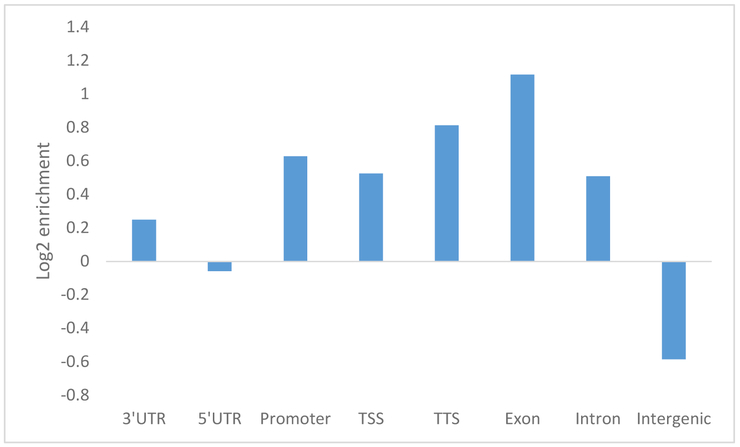
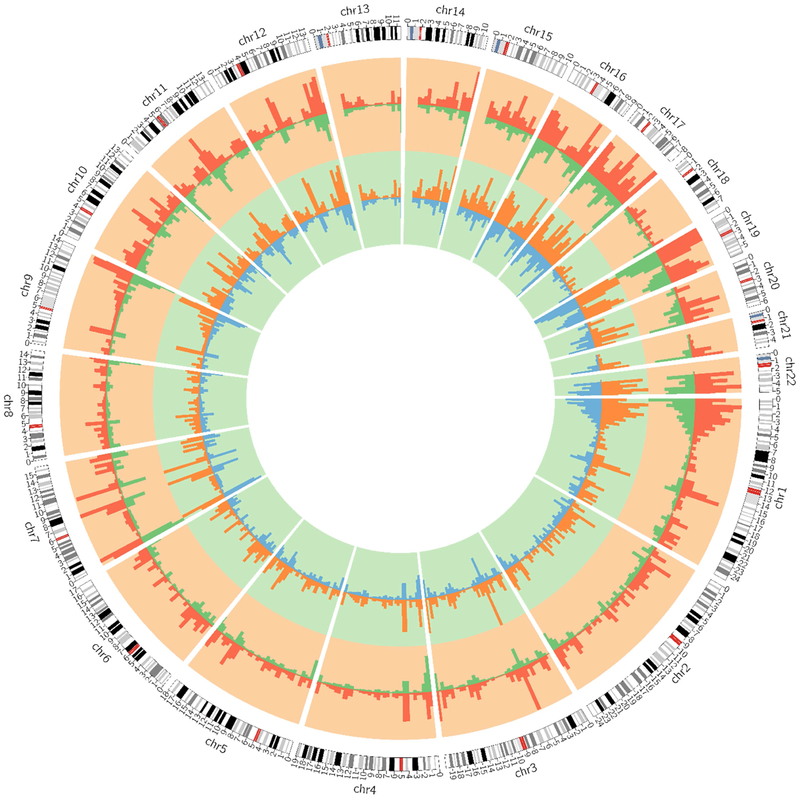
To examine the distribution of 5hmC enrichment in human brain, we annotated each putative DhMR to the human genome (hg19) using Homer,[38] which assigns each specified region to genomic features. Consistent with previous studies in human brain [27], we found that AD-related DhMRs were largely enriched in intragenic regions (e.g., exons, introns, promoters, transcription termination sites) and depleted in intergenic regions (Figure 1). The genome-wide patterns of brain 5hmC enrichment in relation to AD pathology are shown in Figure 2.
Figure 1.
Log2 enrichment of normalized 5hmC densities overlapping with various genomic features in hg19
Figure 2.
Genome-wide distribution of brain 5hmC enrichment in relation to burden of AD pathology. The outermost circle displays the human chromosomes. The middle cycle represents 5hmC enrichment in subjects with high (red) and low (green) burden of NP. The inner most cycle represents 5hmC enrichment in subjects with high (orange) and low (blue) burden of NFTs. The height of the histogram bins indicates the level of 5hmC enrichment.
3.3. Relationship between DNA hydroxymethylation and gene expression in human AD brain
To determine whether altered DNA hydroxymethylation influences gene expression, we calculated partial correlation coefficients (corrected for age at death, sex and education level) between 5hmC enrichment and overall gene expression level quantified by RNA-Seq in the same brain cortex. Of the 1,261 genes nominally associated with NP (P<0.01), we identified 228 positive and 74 negative cis correlation pairs between DNA hydroxymethylation and gene expression after correction for multiple testing (q-value<0.05). These correlations involved 234 different hydroxymethylated loci (from 199 unique genes) and 228 corresponding expression probes (representing 199 unique genes). For trans regulation, we found 164 positive and 389 negative trans correlation pairs between DNA hydroxymethylation and gene expression, including 243 different hydroxymethylation probes (from 220 distinct genes) and 316 different expression probes (representing 298 unique genes) at the level of q-value < 0.05. Significant correction pairs for DNA hydroxymethylation with cis- and trans-acting gene expression are given in Tables S3 and S4, respectively. There appeared to be a trend that cis correlations between DNA hydroxymethylation and gene expression are largely positive (76% overall) while negative correlations are more frequently observed with trans effects (70% overall). The genome-wide correlations between 5hmC enrichment and gene expression for the identified DhMR genes are shown in Figure S2.
Of the 321 DhMR genes showing significant associations with neuritic plaques (q-value<0.05), 5hmC enrichment in majority of the genes (>75%) was positively correlated with cis-acting gene expression. These results are consistent with previous studies showing that enrichment of DNA hydroxymethylation in gene bodies was correlated with higher gene expression in mouse [25] and human brains [27], and indicated that the role of DNA hydroxymethylation in gene regulation depends on genomic locations.
3.4. Co-hydroxymethylation modules in human AD brain
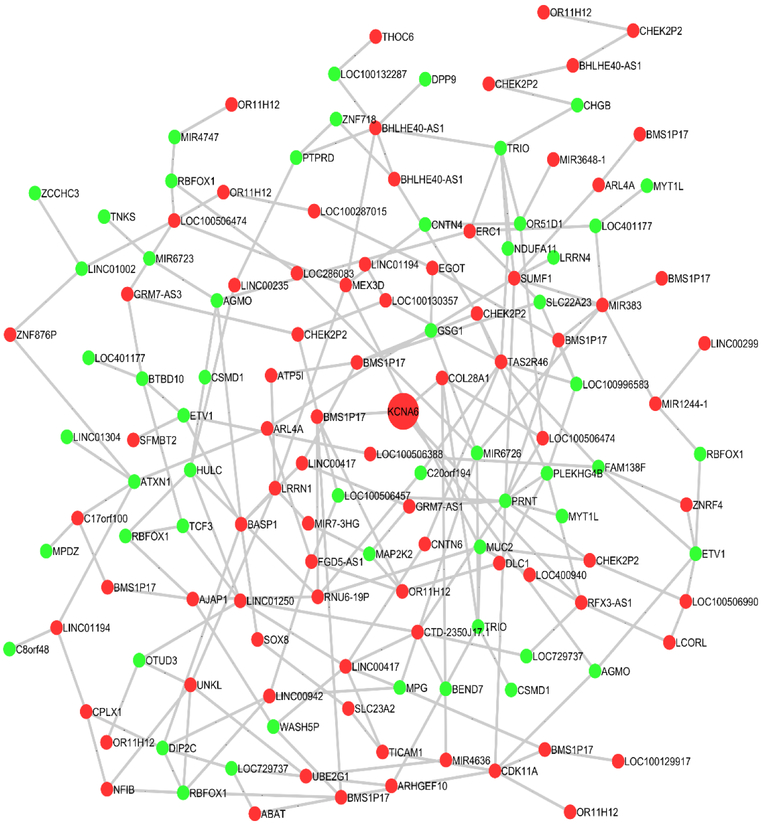
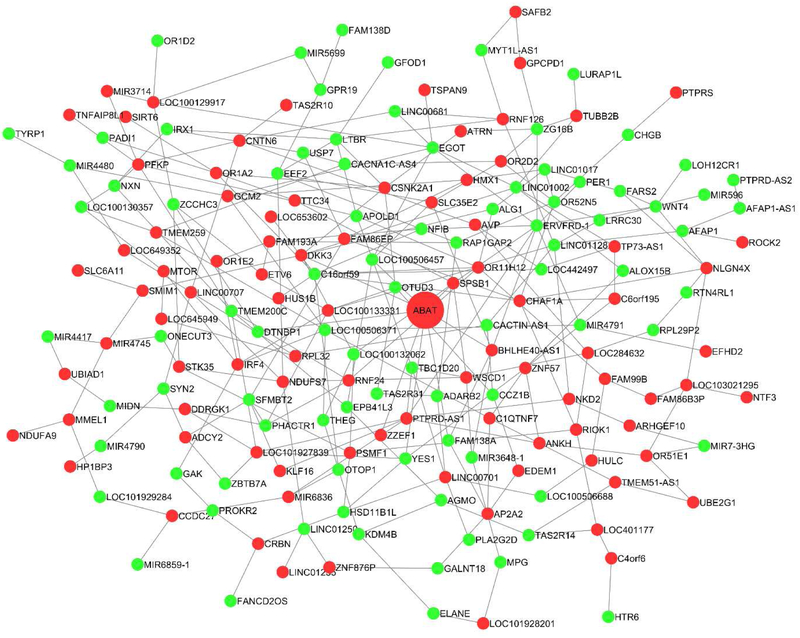
Identifying co-regulated gene modules is crucial for understanding the complex gene regulatory pathways involved in disease etiology. As such, we constructed co-hydroxymethylation networks for NP and NFTs, respectively. We identified 10 differential co-hydroxymethylation modules (labeled 1 to 10) of sizes ranging from 7 to 144 genes for NP, and 4 co-hydroxymethylation modules (labeled 1 to 5) of sizes ranging from 81 to 196 genes for NFTs. For NP, a total of 832 (66% of total) genes were assigned to a module, while 437 were not assigned. In contrast, a total of 590 (84% of total) genes were assigned to a module, while 112 were not assigned for NFTs. Tables 5 provide the top 5 co-hydroxymethylation modules along with hub genes (genes with highest module membership) in each module for AD pathology. Table S5 and S6 list the module membership of all genes in DNA hydroxymethylation modules for NP and NFTs, respectively. Figures 3 and 4 schematically illustrate two of the identified co-hydroxymethylation modules associated with NP and NFTs, respectively.
Table 5.
Differential co-hydroxymethylation modules and top enriched GO terms in each module for AD pathology.
| Module | Size | q-value* | Top enriched GO terms in each module | Hub gene |
|---|---|---|---|---|
| Neuritic plaques (NP) | ||||
| 1 | 144 | 2.69×10−5 | Ion channel activity; Voltage-gated potassium channel activity; Neuron projection development | KCNA6 |
| 2 | 128 | 5.25×10−5 | Neuron development | TP73 |
| 3 | 116 | 6.92×10−7 | Glutamatergic neuron; Neuron differentiation | MSRA |
| 4 | 99 | 4.28×10−3 | Neurotransmitter transport; Neuron cell-cell adhesion | PRKCZ |
| 5 | 88 | 5.14×10−4 | Calcium ion-dependent exocytosis of neurotransmitter; Neuron differentiation; Glutamatergic synapse | NDUFA4 |
| Neurofibrillary tangles (NFTs) | ||||
| 1 | 196 | 7.48×10−4 | GABA transaminase; Neuron development | ABAT |
| 2 | 167 | 2.39×10−3 | Protein binding | LRRN1 |
| 3 | 146 | 5.22×10−3 | Nucleotide binding; | RBFOX1 |
| 4 | 81 | 1.14×10−2 | Ion channel activity; Low voltage-gated calcium channel activity | CACNA1H |
q-value for the association between AD pathology and the first three eigenvalues of the module (adjusting for a total number of modules).
Figure 3.
An example of the identified differential co-hydroxymethylation network for NP. Genes in red indicate hyper-hydroxymethylation and those in green represent hypo-hydroxymethylation relative to burden of neuritic plaques.
Figure 4.
An example of the identified differential co-hydroxymethylation network for NFTs. Genes in red indicate hyper-hydroxymethylation and those in green represent hypo-hydroxymethylation relative to burden of NFTs.
Using RNA-Seq data from the same brain cortex, we also constructed co-expression networks in subjects with high or low burden of neuritic plaques, separately. We identified 9 co-expression modules with sizes ranging from 18 to 192 genes. Table S7 provides the top ranked differential co-expression modules in relation to AD pathology along with hub genes (genes with highest module membership) and corresponding GO terms in each module for NP and NFTs. A total of 612 (57% of total) genes were assigned to a module while 461 background genes were not assigned to a module. Of the 832 genes in the 10 identified co-hydroxymethylation modules in relation to neuritic plaques, 158 genes overlapped with the co-expression modules. However, only 5 co-hydroxymethylation modules and 2 co-expression modules are shared by the two groups.
3.5. Enrichment of sets of genes involved in neurological functions by gene ontology (GO) analysis
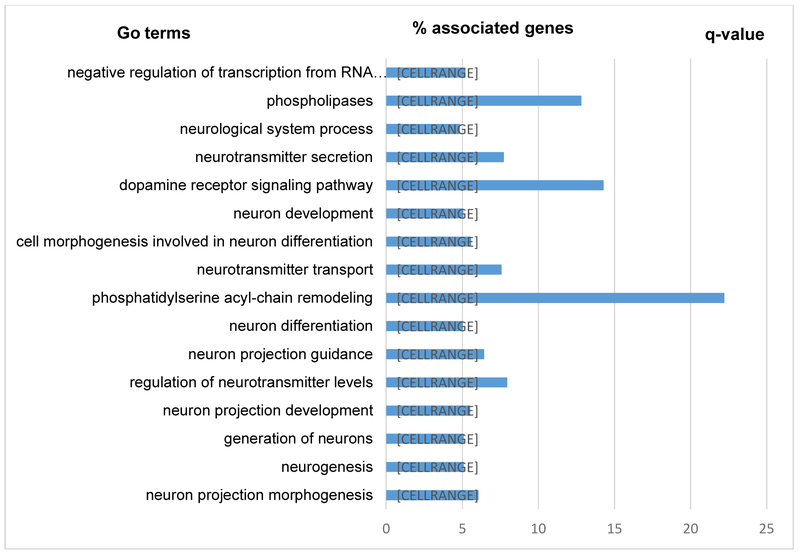
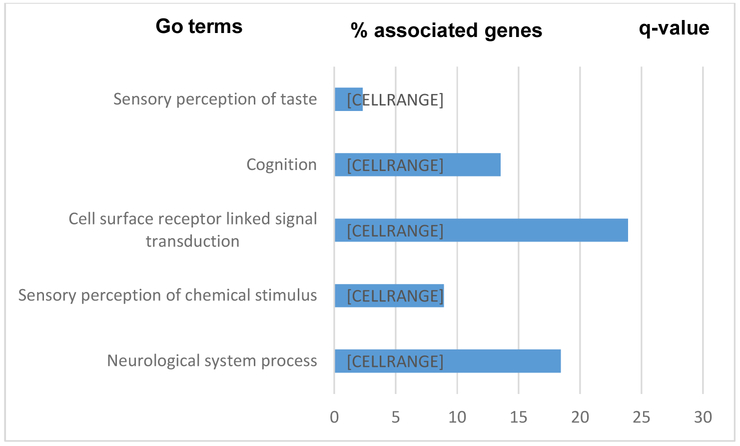
To determine whether candidate genes showing differential hydroxymethylation in relation to AD pathology are involved in neurological pathways, we performed GO analysis using DAVID [41]. As expected, the most significantly enriched GO terms for the identified DhMR genes include biological processes related to neurogenesis, neuron differentiation, neuron projection development, neurotransmitter transport, etc. Other enriched groups include neurotransmitter secretion, dopamine receptor signaling, phospholipases, phosphatidylserine acyl-chain remodeling, etc. GO analysis of genes showing co-hydroxymethylation revealed multiple significantly enriched categories, indicating that these modules are biologically relevance to brain functions or AD. These results provide further support for a potential functional role of altered 5hmC in brain functions and AD pathology. Significant GO categories (FDR<0.05) for NP and NFTs can be seen in Figures 5 and 6, respectively.
Figure 5.
Significant GO categories identified by the functional annotation tool in DAVID for NP. Only GO terms with a gene count >10 and q-value <0.05 are shown. Blue bar represents the percent of each gene set occurred in the identified DhMR genes submitted to DAVID. q-value of the corresponding term are shown at the right end.
Figure 6.
Significant GO categories identified by the functional annotation tool in DAVID for NFTs. Only GO terms with a gene count >10 and q-value <0.05 are shown. Blue bar represents the percent of each gene set occurred in the identified DhMR genes submitted to DAVID. The q-value of the corresponding term are shown at the right end.
4. Discussion
Using a genome-wide 5hmC profiling, we found that alterations in brain DNA hydroxymethylation (both hyper- and hypo-hydroxymethylation, predominantly hyper-hydroxymethylation) is associated with AD pathology. Specifically, after correction for potential confounders and multiple testing, we identified 517 DhMRs (annotated to 321 distinct genes) and 60 DhMRs (annotated to 49 distinct genes) significantly associated with NP and NFTs, respectively. Four genes (ABAT, CAMK1D, HTRA3 and LRRN1) were associated with both traits. These DhMRs are enriched in intragenic regions and depleted in intergenic regions. Functional validation using brain RNA-Seq data from the same individuals demonstrated that DNA hydroxymethylation in gene bodies is predominantly positively correlated with cis-acting gene expression. However, 5hmC enrichment in some cis-regions and majority of the trans-regions can also negatively affect gene expression. Moreover, genes showing differential hydroxymethylation are more likely to be differentially expressed and are enriched in neurobiological processes, indicating functional relevance to AD pathogenesis. We also identified co-hydroxymethylation modules that are enriched in functional GO terms, revealing that the identified DhMR genes are co-regulated and clustered in neurobiological pathways. Together, in terms of proof of concept, our results identified AD-related DhMRs containing both known and novel candidate genes, highlight the critical role of dysregulated DNA hydroxymethylation in AD pathology, and provide mechanistic insights into the molecular mechanisms underlying AD pathogenesis.
4.1. Associations between DNA hydroxymethylation and AD neuropathology
Several studies using immunohistochemistry have reported altered level of global DNA hydroxymethylation in postmortem AD brain, but results were mixed with some studies reporting decreased [9, 20] whereas others showing increased [42, 43] or no change [44] in global 5hmC level in AD patients compared to controls, probably due to the use of samples from different brain regions and/or different stages of AD. Moreover, the use of immunohistochemical staining did not provide locus-specific information for 5hmC distribution, and thus was unable to identify genomic regions and candidate genes associated with AD pathogenesis. In this study, we employed a chemical labeling technique followed by high-throughput sequencing to determine the genome-wide patterns of brain 5hmC at specific locus and identified a list of DhMRs associated with AD pathologic burden. These DhMRs are enriched in genes involved in neurogenesis, synaptic plasticity, neuronal signal transduction and other neurobiological processes essential for brain functions. Of particular interest was the evidence for altered 5hmC in several genes related to ion channel activities (e.g., CACNA1H, CAMK1D, KCNA6, KCNA5, DGKB, BASP1), a mechanism known to be involved in cognition, memory and learning as well as AD pathology [45]. Among these, the CACNA1H gene showed the most significant differential hydroxymethylation relative to burden of neuritic plaque (q-value =1.01×10−15). The 5hmC enrichment level in this gene was two times higher among samples in the top tertile of neuritic plaque distribution compared to those in the bottom tertile (Figure S1). This gene regulates the T-type calcium channels that are essential for the regulation of brain functions [46]. It is likely that altered 5hmC in calcium channel genes influences neuronal excitability, resulting in changes in neurotransmission and/or neurotransmitter secretion via mediating intracellular calcium influx in response to depolarization. Mutations in CACNA1H have been associated with idiopathic epilepsy [47]. Another two potassium channel genes (KCNA6, KCNA5) may influence AD susceptibility via regulating neurotransmitter release, neuronal excitability, or brain insulin secretion [45]. The KCNA6 gene was also found to be the hub gene of a co-hydroxymethylation network associated with neuritic plaque. Another two ion channel related genes, DGKB and BASP1, are overexpressed in the brain. Together, these findings provide further support for an important role of ion channels in AD pathology, and reveal novel molecular mechanisms through which altered ion channel activities contribute to AD.
Of the 321 DhMR genes associated with neuritic plaque, 47 genes also showed significant differential expression. Of these, many genes are known to be involved in Aβ degradation or clearance (e.g., MMEL1, COLEC12, ATXN1), tau phosphorylation (e.g., DUSP22, CSNK2A1), synaptic plasticity (e.g., SYN2, VAMP2), cognition (e.g., APC2, CSMD1), glucose transporter (e.g., ENO1, SLC2A14), and oxidative stress (e.g., MSRA, RAF1). Several genes, such as EPHB2, ENO1, and ATXN1, were also associated with AD or neuropsychiatric disorders in previous GWAS. Interestingly, we identified altered 5hmC enrichment in two genes (ENO1, SLC2A14) involved in glucose metabolism, a mechanism previously implicated in tau phosphorylation and neurofibrillary tangle pathogenesis [48]. The ENO1 gene was reported to be upregulated in AD brain [49], and SLC2A14 was found to be causative in a fly model of AD.[50] Moreover, four differentially hydroxymethylated and expressed genes (ABAT, CAMK1D, HTRA3, LRRN1) relative to Aβ plaque were also significantly associated with NFTs after adjusting for covariates and multiple comparisons (q-value <0.05). Of these, ABAT catalyzes the neurotransmitter gamma-aminobutyric acid (GABA), dysfunction of which has been associated with several neurological disorders [51] including early onset AD [52]. The CAMK1D gene encodes the calcium/calmodulin-dependent protein kinase, which is essential for the calcium-triggered CaMKK-CaMK1 signaling pathway that is known to be involved in memory and AD pathogenesis [45]. The HTRA3 gene belongs to heat shock-induced serine proteases involved in protein quality control and cell fate. Although its role in AD pathogenesis has not been reported previously, two of its homologs, HTRA1 and HTRA2, were previously associated with Aβ plaque formation [41] and Parkinson’s disease [39]. The LRRN1 gene belongs to the leucine rich repeat (LRR) neuronal family, which is involved in multiple biological functions including neural development and regeneration. Mutations in the LRR family genes have been associated with several neurological disorders such as epilepsy [53], Parkinson disease [54], and autism.[55] Together, we confirmed the role of many previously reported genes and also identified novel genomic regions/genes in AD pathology. These findings reveal a critical role of DNA hydroxymethylation in AD and provide novel mechanistic insights into the understanding of the molecular mechanism underlying AD pathogenesis. Those genes showing both differential hydroxymethylation and expression represent key regulators that may play causal roles in AD pathology.
4.2. Relationship between 5hmC and gene expression in human AD brain
Although it is generally believed that 5mC causes gene silencing whereas 5hmC activates gene transcription, the relationship between DNA (hydroxy)methylation and gene expression may be more complicated than appreciated. To date, a global analysis of the extent and pattern of DNA hydroxymethylation with gene expression in human AD brain is lacking. Using brain RNA-Seq data from the same individuals, we found that brain DNA hydroxymethylation can be both positively and negatively correlated with gene expression, although there appears to be a trend that cis correlations are predominantly positive whereas trans effects are largely negative. These results are in agreement with previous studies reporting both positive and negative correlations between DNA methylation and gene expression in blood and brain [56-58]. Moreover, our finding that 5hmC enrichment was largely correlated with higher level of cis-acting gene expression is consistent with previous research demonstrating that 5hmC enrichment in gene bodies was correlated with higher gene expression in mouse [25] and human brains [27]. Possible mechanisms for the negative correlations between DNA (hydroxy)methylation and gene expression may include interference with the binding of transcription factors or through the recruitment of repressors such as histone deacetylases [56]. Together, our integrative analysis of DNA hydroxymethylation and gene expression confirms the importance of DNA hydroxymethylation in gene regulation and allows for identifying key candidate genes whose role in AD are modulated by epigenetic changes.
4.3. Co-hydroxymethylation modules in human AD brain
AD is a complex disorder involving the functions of many genes that jointly or interactively contribute to disease etiology. Traditional methods that model the effect of a single gene cannot capture the complicated biological pathways implicated in AD. We hypothesize that alterations in DNA hydroxymethylation across different genomic regions are interdependent and that the brain hydroxymethylome comprises a complex network of interacting processes. Using a network-based approach, we detected co-hydroxymethylated modules containing coordinated genes across different genomic loci, and identified distinct modularities in ion channel activity, neuron differentiation and development, neurotransmitter transporter, glutamatergic synapse, calcium ion-dependent exocytosis of neurotransmitter, etc. Specifically, we identified several co-hydroxymethylated modules, each of which represents a group of co-regulated genes involved in a biological process perturbed in AD. AD-related genes, such as CSMD1, KCNA6, BASP1, MAP2K2 and PTPRD, are congregated in a single module. Some of the AD-related genes were hub genes. Of the identified co-hydroxymethylated subnetworks for NP, hub genes in two largest modules are of particular interest. KCNA6, a potassium channel gene associated with neuronal excitability and neurotransmitter secretion, was identified as a central regulatory hub gene up-regulated in brain of subjects with higher NP burden. Another gene TP73, a paralogue of the p53 tumor suppressor gene that regulates neurodegeneration and phosphor-tau accumulation in AD [59], was also up-regulated in samples with higher than those with lower burden of NP. ABAT is a hub gene for one of the identified co-hydroxymethylation modules for NFTs. This gene encodes the key enzyme responsible for catabolism of a major inhibitory neurotransmitter γ-aminobutyric acid (GABA), altered function of which has been reported in AD patients as well as transgenic AD mouse models [60]. We also conducted co-expression analysis using brain RNA-Seq data of same individuals. Consistent with a previous study on DNA methylation and gene expression in whole blood of healthy human subjects [58], only a few co-expressed modules correspond to modules of co-hydroxymethylation, suggesting that genes could be co-regulated at different levels or through different molecular mechanisms.
4.4. Functional relevance of the identified DhMR genes by GO analysis
Results of GO analysis highlight several functionally related biological pathways, including those involved in neurogenesis, neuron projection development or morphogenesis, neurotransmitter regulation, neuron differentiation, dopamine receptor signaling, phospholipases, etc. In addition, given the postulated link between perturbed lipids metabolism, oxidative stress, and neurodegeneration [61], it is plausible that a number of lipids related GO terms (e.g., phosphatidylserine acyl-chain remodeling, phospholipases) were identified as functional categories related to AD pathology.
To our knowledge, this is the first genome-wide profiling of 5hmC at specific locus in human postmortem brain from a community-based cohort of aging and dementia. Moreover, our analyses corrected for potential confounders and stringently controlled for multiple testing. However, several limitations of the study are worth mentioning. First, as with all postmortem human brain studies, our sample size is small, and thus our results should be considered as proof of concept rather than conclusive. In addition, we cannot exclude the possibilities for false positives and/or false negatives due to the small sample size. Second, the human prefrontal cortex includes many different types of cells such as neurons, astrocytes and other glia cells, each may have different epigenetic profiles. Thus, our results could be confounded by cellular heterogeneity inherent in the brain tissue. However, we corrected for neural compositions in the statistical analyses. Moreover, our previous epigenome-wide analysis showed that the proportion of neurons in each brain sample was not related to AD [12], indicating that cellular heterogeneity is unlikely to confound the association between epigenetic changes and AD pathology. Third, the current study only investigated genome-wide patterns of DNA hydroxymethylome in the prefrontal cortex, but brain hydroxymethylation is likely to be region-specific [62]. Thus, our results derived from one region may not reflect the 5hmC profiles in other regions. Brain region-specific DNA hydroxymethylome should be investigated in future research. Fourth, our study population comprises highly educated older individuals of European ancestry, so we should be cautious in generalizing our findings to other settings or populations. Fifth, our analysis cannot differentiate 5hmC and 5mC due to the low resolution of our sequencing analysis. Single base resolution technique, such as Tet-assisted bisulfite sequencing (TAB-Seq), is required to elucidate the relationship between 5hmC and 5mC. Finally, it remains unclear whether the differences in DNA hydroxymethylation seen in AD brain are the cause or an early consequence of neuropathologic changes. It is possible that the observed alteration in 5hmC enrichment actually results from Aβ plaque or NFTs induced changes in the brain. The correlation between 5hmC enrichment and gene expression observed in our study does not establish causality either. Longitudinal studies are required to establish the causality between epigenetic changes and AD pathology.
Supplementary Material
Systematic review: We used PubMed and Google Scholar to search for articles related to DNA hydroxymethylation for Alzheimer’s disease (AD). In addition, we searched for literature relating our identified differentially hydroxymethylated regions (DhMRs) to AD pathology and other neurological disorders through searching for genes showing differential hydroxymethylation using the terms “DNA methylation”, “DNA hydroxymethylation”, “Alzheimer’s disease”, and “neurodegeneration”. Relevant work relating our significant genomic regions/genes to AD and neurodegeneration was cited.
Interpretation: Using a genome-wide capture followed by high-throughput sequencing, we studied the genome-wide patterns of brain DNA hydroxymethylome at specific genomic loci in human AD brain and identified significant genomic regions harboring putative genes that are differentially hydroxymethylated in relation to AD pathology.
Future directions: Identification of AD-related DhMRs provides mechanistic insight into AD pathology and holds promise for discovering novel strategies for early detection and therapeutic intervention at preclinical stages of this debilitating disorder.
Acknowledgements
We would like to thank the participants of the ROS and MAP studies for their participation in these studies, and the staff of the Rush Alzheimer’s Disease Center.
Funding sources
This work was supported by the National Institutes of Health grants: R01DK091369, R01MH097018, P30AG10161, RF1AG15819, R01AG17917, R01AG16042, U01AG46152, and R01AG36836.
Footnotes
Conflicts of interest: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Bennett DA, Yu L, Yang J, Srivastava GP, Aubin C, De Jager PL. Epigenomics of Alzheimer’s disease. Translational research. 2015;165:200–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Covic M, Karaca E, Lie DC. Epigenetic regulation of neurogenesis in the adult hippocampus. Heredity. 2010;105:122–34. [DOI] [PubMed] [Google Scholar]
- [3].Yu NK, Baek SH, Kaang BK. DNA methylation-mediated control of learning and memory. Molecular brain. 2011;4:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Feng J, Fan G. The role of DNA methylation in the central nervous system and neuropsychiatric disorders. International review of neurobiology. 2009;89:67–84. [DOI] [PubMed] [Google Scholar]
- [5].Bakulski KM, Dolinoy DC, Sartor MA, Paulson HL, Konen JR, Lieberman AP, et al. Genome-wide DNA methylation differences between late-onset Alzheimer’s disease and cognitively normal controls in human frontal cortex. Journal of Alzheimer’s disease. 2012;29:571–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic changes in Alzheimer’s disease: decrements in DNA methylation. Neurobiology of aging. 2010;31:2025–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Rao JS, Keleshian VL, Klein S, Rapoport SI. Epigenetic modifications in frontal cortex from Alzheimer’s disease and bipolar disorder patients. Translational psychiatry. 2012;2:e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mastroeni D, McKee A, Grover A, Rogers J, Coleman PD. Epigenetic differences in cortical neurons from a pair of monozygotic twins discordant for Alzheimer’s disease. PloS one. 2009;4:e6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chouliaras L, Mastroeni D, Delvaux E, Grover A, Kenis G, Hof PR, et al. Consistent decrease in global DNA methylation and hydroxymethylation in the hippocampus of Alzheimer’s disease patients. Neurobiology of aging. 2013;34:2091–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Numata S, Ye T, Hyde TM, Guitart-Navarro X, Tao R, Wininger M, et al. DNA methylation signatures in development and aging of the human prefrontal cortex. American journal of human genetics. 2012;90:260–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Akbarian S, Beeri MS, Haroutunian V. Epigenetic determinants of healthy and diseased brain aging and cognition. JAMA neurology. 2013;70:711–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].De Jager PL, Srivastava G, Lunnon K, Burgess J, Schalkwyk LC, Yu L, et al. Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nature neuroscience. 2014;17:1156–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yu L, Chibnik LB, Srivastava GP, Pochet N, Yang J, Xu J, et al. Association of brain DNA methylation in SORL1, ABCA7, HLA-DRB5, SLC24A4, and BIN1 with pathological diagnosis of Alzheimer disease. JAMA neurology. 2015;72:15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yu L, De Jager PL, Yang J, Trojanowski JQ, Bennett DA, Schneider JA. The TMEM106B locus and TDP-43 pathology in older persons without FTLD. Neurology. 2015;84:927–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yang J, Yu L, Gaiteri C, Srivastava GP, Chibnik LB, Leurgans SE, et al. Association of DNA methylation in the brain with age in older persons is confounded by common neuropathologies. The international journal of biochemistry & cell biology. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hahn MA, Szabo PE, Pfeifer GP. 5-Hydroxymethylcytosine: a stable or transient DNA modification? Genomics. 2014;104:314–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139:1895–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Khare T, Pai S, Koncevicius K, Pal M, Kriukiene E, Liutkeviciute Z, et al. 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nature structural & molecular biology. 2012;19:1037–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Condliffe D, Wong A, Troakes C, Proitsi P, Patel Y, Chouliaras L, et al. Cross-region reduction in 5-hydroxymethylcytosine in Alzheimer’s disease brain. Neurobiology of aging. 2014;35:1850–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell reports. 2013;3:291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nature neuroscience. 2011;14:1607–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chouliaras L, van den Hove DL, Kenis G, Keitel S, Hof PR, van Os J, et al. Age-related increase in levels of 5-hydroxymethylcytosine in mouse hippocampus is prevented by caloric restriction. Current Alzheimer research. 2012;9:536–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nature biotechnology. 2011;29:68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Munzel M, Globisch D, Bruckl T, Wagner M, Welzmiller V, Michalakis S, et al. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angewandte Chemie. 2010;49:5375–7. [DOI] [PubMed] [Google Scholar]
- [27].Jin SG, Wu X, Li AX, Pfeifer GP. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic acids research. 2011;39:5015–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bennett DA, Schneider JA, Arvanitakis Z, Wilson RS. Overview and findings from the religious orders study. Current Alzheimer research. 2012;9:628–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bennett DA, Schneider JA, Buchman AS, Barnes LL, Boyle PA, Wilson RS. Overview and findings from the rush Memory and Aging Project. Current Alzheimer research. 2012;9:646–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bennett DA, Schneider JA, Aggarwal NT, Arvanitakis Z, Shah RC, Kelly JF, et al. Decision rules guiding the clinical diagnosis of Alzheimer’s disease in two community-based cohort studies compared to standard practice in a clinic-based cohort study. Neuroepidemiology. 2006;27:169–76. [DOI] [PubMed] [Google Scholar]
- [31].Bennett DA, Schneider JA, Arvanitakis Z, Kelly JF, Aggarwal NT, Shah RC, et al. Neuropathology of older persons without cognitive impairment from two community-based studies. Neurology. 2006;66:1837–44. [DOI] [PubMed] [Google Scholar]
- [32].Bennett DA, De Jager PL, Leurgans SE, Schneider JA. Neuropathologic intermediate phenotypes enhance association to Alzheimer susceptibility alleles. Neurology. 2009;72:1495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bennett DA, Schneider JA, Tang Y, Arnold SE, Wilson RS. The effect of social networks on the relation between Alzheimer’s disease pathology and level of cognitive function in old people: a longitudinal cohort study. The Lancet Neurology. 2006;5:406–12. [DOI] [PubMed] [Google Scholar]
- [34].Bennett DA, Wilson RS, Schneider JA, Evans DA, Aggarwal NT, Arnold SE, et al. Apolipoprotein E epsilon4 allele, AD pathology, and the clinical expression of Alzheimer’s disease. Neurology. 2003;60:246–52. [DOI] [PubMed] [Google Scholar]
- [35].Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].De Jager PL, Shulman JM, Chibnik LB, Keenan BT, Raj T, Wilson RS, et al. A genome-wide scan for common variants affecting the rate of age-related cognitive decline. Neurobiology of aging. 2012;33:1017 e1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Guintivano J, Aryee MJ, Kaminsky ZA. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics : official journal of the DNA Methylation Society. 2013;8:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Patterson VL, Zullo AJ, Koenig C, Stoessel S, Jo H, Liu X, et al. Neural-specific deletion of Htra2 causes cerebellar neurodegeneration and defective processing of mitochondrial OPA1. PloS one. 2014;9:e115789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Grau S, Baldi A, Bussani R, Tian X, Stefanescu R, Przybylski M, et al. Implications of the serine protease HtrA1 in amyloid precursor protein processing. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:6021–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Coppieters N, Dieriks BV, Lill C, Faull RL, Curtis MA, Dragunow M. Global changes in DNA methylation and hydroxymethylation in Alzheimer’s disease human brain. Neurobiology of aging. 2014;35:1334–44. [DOI] [PubMed] [Google Scholar]
- [43].Bradley-Whitman MA, Lovell MA. Epigenetic changes in the progression of Alzheimer’s disease. Mechanisms of ageing and development. 2013;134:486–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lashley T, Gami P, Valizadeh N, Li A, Revesz T, Balazs R. Alterations in global DNA methylation and hydroxymethylation are not detected in Alzheimer’s disease. Neuropathology and applied neurobiology. 2015;41:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Etcheberrigaray E, Gibson GE, Alkon DL. Molecular mechanisms of memory and the pathophysiology of Alzheimer’s disease. Annals of the New York Academy of Sciences. 1994;747:245–55. [DOI] [PubMed] [Google Scholar]
- [46].Cheong E, Shin HS. T-type Ca2+ channels in normal and abnormal brain functions. Physiological reviews. 2013;93:961–92. [DOI] [PubMed] [Google Scholar]
- [47].Eckle VS, Shcheglovitov A, Vitko I, Dey D, Yap CC, Winckler B, et al. Mechanisms by which a CACNA1H mutation in epilepsy patients increases seizure susceptibility. The Journal of physiology. 2014;592:795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Liu Y, Liu F, Iqbal K, Grundke-Iqbal I, Gong CX. Decreased glucose transporters correlate to abnormal hyperphosphorylation of tau in Alzheimer disease. FEBS letters. 2008;582:359–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Butterfield DA, Lange ML. Multifunctional roles of enolase in Alzheimer’s disease brain: beyond altered glucose metabolism. Journal of neurochemistry. 2009;111:915–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Shulman JM, Chipendo P, Chibnik LB, Aubin C, Tran D, Keenan BT, et al. Functional screening of Alzheimer pathology genome-wide association signals in Drosophila. American journal of human genetics. 2011;88:232–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Barnby G, Abbott A, Sykes N, Morris A, Weeks DE, Mott R, et al. Candidate-gene screening and association analysis at the autism-susceptibility locus on chromosome 16p: evidence of association at GRIN2A and ABAT. American journal of human genetics. 2005;76:950–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hooli BV, Kovacs-Vajna ZM, Mullin K, Blumenthal MA, Mattheisen M, Zhang C, et al. Rare autosomal copy number variations in early-onset familial Alzheimer’s disease. Molecular psychiatry. 2014;19:676–81. [DOI] [PubMed] [Google Scholar]
- [53].Morante-Redolat JM, Gorostidi-Pagola A, Piquer-Sirerol S, Saenz A, Poza JJ, Galan J, et al. Mutations in the LGI1/Epitempin gene on 10q24 cause autosomal dominant lateral temporal epilepsy. Human molecular genetics. 2002;11:1119–28. [DOI] [PubMed] [Google Scholar]
- [54].Singleton AB. Altered alpha-synuclein homeostasis causing Parkinson’s disease: the potential roles of dardarin. Trends in neurosciences. 2005;28:416–21. [DOI] [PubMed] [Google Scholar]
- [55].Wang K, Zhang H, Ma D, Bucan M, Glessner JT, Abrahams BS, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome biology. 2011;12:R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS genetics. 2010;6:e1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].van Eijk KR, de Jong S, Boks MP, Langeveld T, Colas F, Veldink JH, et al. Genetic analysis of DNA methylation and gene expression levels in whole blood of healthy human subjects. BMC genomics. 2012;13:636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Wetzel MK, Naska S, Laliberte CL, Rymar VV, Fujitani M, Biernaskie JA, et al. p73 regulates neurodegeneration and phospho-tau accumulation during aging and Alzheimer’s disease. Neuron. 2008;59:708–21. [DOI] [PubMed] [Google Scholar]
- [60].Wu Z, Guo Z, Gearing M, Chen G. Tonic inhibition in dentate gyrus impairs long-term potentiation and memory in an Alzheimer’s [corrected] disease model. Nature communications. 2014;5:4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Cutler RG, Kelly J, Storie K, Pedersen WA, Tammara A, Hatanpaa K, et al. Involvement of oxidative stress-induced abnormalities in ceramide and cholesterol metabolism in brain aging and Alzheimer’s disease. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2070–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Lunnon K, Smith R, Hannon E, De Jager PL, Srivastava G, Volta M, et al. Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer’s disease. Nature neuroscience. 2014;17:1164–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.