Abstract
PURPOSE
To determine the predictive and prognostic value of the consensus molecular subtypes (CMSs) of colorectal cancer (CRC) that represent a merging of gene expression–based features largely in primary tumors from six independent classification systems and provide a framework for capturing the intrinsic heterogeneity of CRC in patients enrolled in CALGB/SWOG 80405.
PATIENTS AND METHODS
CALGB/SWOG 80405 is a phase III trial that compared the addition of bevacizumab or cetuximab to infusional fluorouracil, leucovorin, and oxaliplatin or fluorouracil, leucovorin, and irinotecan as first-line treatment of advanced CRC. We characterized the CMS classification using a novel NanoString gene expression panel on primary CRCs from 581 patients enrolled in this study to assess the prognostic and predictive value of CMSs in these patients.
RESULTS
The CMSs are highly prognostic for overall survival (OS; P < .001) and progression-free survival (PFS; P < .001). Furthermore, CMSs were predictive for both OS (P for interaction < .001) and PFS (P for interaction = .0032). In the CMS1 cohort, patients treated with bevacizumab had a significantly longer OS than those treated with cetuximab (P < .001). In the CMS2 cohort, patients treated with cetuximab had a significantly longer OS than patients treated with bevacizumab (P = .0046).
CONCLUSION
These findings highlight the possible clinical utility of CMSs and suggests that refinement of the CMS classification may provide a path toward identifying patients with metastatic CRC who are most likely to benefit from specific targeted therapy as part of the initial treatment.
INTRODUCTION
Colorectal cancer (CRC) is the second leading cause of cancer death in the United States.1-3 The goal of CALGB/SWOG 80405 was to determine whether the addition of cetuximab or bevacizumab to chemotherapy leads to superior outcomes, with one or the other as first-line therapy in metastatic CRC (mCRC).4 In the primary analysis, 1,137 patients with KRAS wild-type (codons 12 and 13) mutations were randomly assigned to either bevacizumab or cetuximab plus patient/physician choice of infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX) or fluorouracil, leucovorin, and irinotecan (FOLFIRI). The primary end point was overall survival (OS), which was not different between the arms (cetuximab v bevacizumab, −30.1 v −29.0 months, respectively; stratified hazard ratio [HR], 0.88; 95% CI, 0.77 to 1.01; P = .08).4
An OS that exceeds 30 months in a large population of patients with mCRC is encouraging but is inflated by the exclusion of patients with RAS mutations. Even when restricted to those with tumors that have wild-type RAS, a significant proportion of patients do not respond to cetuximab. CRCs that originate from the right side of the colon portend a significantly worse prognosis than those that develop in the left-side colon.5-7 We observed a survival benefit in the cohort of patients with tumors that originated in the left-side colon enrolled in this frontline study of targeted therapy who were treated with the chemotherapy regimens coupled with cetuximab over those treated with bevacizumab.8 This finding highlights the necessity of identifying biomarkers to better understand the therapeutic vulnerabilities and exploit the heterogeneity of mCRCs. These insights have the potential to help treating physicians to use an individual’s tumor molecular makeup to predict likely sensitivity and resistance to targeted therapies.9,10
Consensus molecular subtypes (CMSs) provide an integrated framework to capture the intrinsic heterogeneity of CRC at the gene expression level.11 Developed using transcriptome-wide analyses of primarily early-stage tumors, this framework merges insights garnered from clinically relevant anatomic, genetic, and carcinogenic classification systems. On the basis of the biology of the disease rather than on the outcomes, this convention divides CRCs into one of four CMS groups: CMS1, microsatellite instability (MSI) immune; CMS2, canonical; CMS3, metabolic; and CMS4, mesenchymal.11 The utility of comprehensive gene expression analyses is that such analyses reflect the functional implications of mutations rather than just their presence or absence. Thus, they evaluate the status of redundant pathways and upregulations of signaling pathways that are independent of DNA mutations. This has the potential to provide a better characterization of the molecular status of the cancer with therapeutic implications.
The theoretical power of the CMS classification lies in the possibility that it not only is a prognostic tool but also has become a tool for drug development. However, its translation into clinical practice is subject to several obstacles. To be useful for clinical decision making, the testing must be done in Clinical Laboratory Improvement Amendments (CLIA)–approved laboratories that ensure quality laboratory testing and are overseen by the Centers for Medicare & Medicaid Services. In addition, the classification is based on the biology of stage II to III cancers, and its applicability in patients with metastatic cancer has not been elucidated. Trinh et al12 showed CMSs to be predictive in CAIRO2, but in that study, CMS status was ascertained by immunohistochemistry rather than by gene expression. They used five immunohistochemistry markers, assessed for MSI, and showed an 87% concordance with the transcriptome analyses. One of the limitations of this approach is a lack of a clear distinction between CMS2 and CMS3. The current report represents, to our knowledge, the first assessment of the prognostic and predictive value of CMSs using a novel NanoString (NanoString Technologies, Seattle, WA) gene expression platform on tumor samples from patients with mCRC enrolled in the CALGB/SWOG 80405 trial (Alliance substudy A151425) and treated with one of the two targeted agents along with standard-of-care chemotherapy.
PATIENTS AND METHODS
CALGB/SWOG 80405
This trial was conducted to determine the relative efficacy of cetuximab versus bevacizumab when added to standard modified FOLFOX or FOLFIRI as first-line therapy in advanced or KRAS wild-type mCRC.4 The trial was initially designed to compare three strategies: chemotherapy plus cetuximab, chemotherapy plus bevacizumab, and chemotherapy plus cetuximab and bevacizumab. Three years after the start of the trial, data on the lack of efficacy of epidermal growth factor receptor (EGFR) antibodies in KRAS mutant tumors emerged, and KRAS wild-type (codons 12 and 13) status became an eligibility criterion. The combined treatment group (chemotherapy plus cetuximab and bevacizumab) was discontinued because of lack of efficacy. In 2015, a revised two-arm trial (cetuximab v bevacizumab with chemotherapy regimens) had a mature primary end point. The full protocol is provided in the Data Supplement.
Gene Expression Analysis by NanoString
Gene expression analyses were included in the original protocol as a potential predictive and prognostic marker. Custom-designed CRC NanoString code sets were used to measure gene expression using 250 ng of total RNA from formalin-fixed paraffin-embedded samples in a non–CLIA-approved laboratory. These panels consisted of genes that were known to regulate key aspects of CRC biology (Data Supplement). Positive and negative control probes also were included for hybridization efficiency and background calculations. Gene expression was quantified using the nCounter Analysis System, and raw counts were generated by nSolver software (NanoString Technologies).
CMS Classification
Because of a lack of overlap in gene contents between the custom NanoString panel for the CALBG/SWOG 80405 cohort and the official CMS classifier software, we redeveloped a CMS classifier using some of the large data sets with published gold standard CMS labels,11 The Cancer Genome Atlas, PETACC-3, and Marisa et al.13 Only genes that are common to these three data sets and those assessed in the CALGB/SWOG 80405 panel are used. A multinomial logistic regression model using GLMNET was used to derive the classifiers.14 The NanoString data were log-transformed, and normalization was achieved by parameterizing the features to use all possible pairwise differences in log2 counts to achieve a self-normalizing linear predictor. The BRAF/KRAS signatures were newly derived from the same data sets as in the CMS classifiers. Patients were assigned into three groups: KRAS wild type, BRAF wild type, or double wild type. Multinomial logistic regression using GLMNET was used to classify CALBG/SWOG 80405 samples into one of these classes.
Statistical Analysis
Patient baseline clinical characteristics were compared between patients who had NanoString data and the CALBG/SWOG 80405 primary analysis population as well as across CMSs. Descriptive statistics are presented and compared using Wilcoxon rank sum test for continuous variables and Pearson χ2 tests for categorical variables.15,16
Kaplan-Meier method17 and log-rank test18 were used to estimate and compare time-to-event end points. The association between CMS and time-to-event end points19 was assessed using multivariable Cox proportional hazards regression models. The models are adjusted by the following variables: age, sex, race, Eastern Cooperative Oncology Group performance status, treatment arm, prior adjuvant chemotherapy, protocol chemotherapy, tumor location, number of metastatic sites, and disease diagnosis (synchronous or metachronous). Interaction between CMSs and treatment arm was tested to determine the potential predictive effect of CMSs.
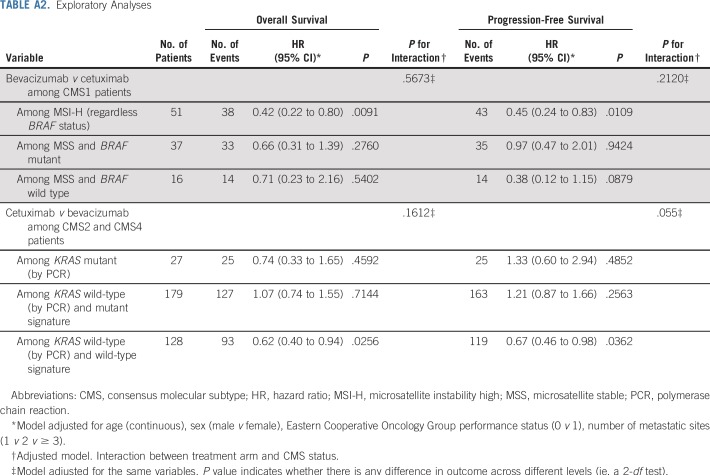
Exploratory analyses were done to test the hypothesis that bevacizumab may have greater efficacy in MSI-high (MSI-H) tumors (eg, patients with CMS1) on the basis of reported differential effects of the two agents on tumor-associated macrophages.20 Patients with CMS1 tumors were separated into three groups: MSI-H (regardless of BRAF status), BRAF mutant (with microsatellite stable [MSS] tumors), and BRAF wild-type (with MSS tumors). Finally, we conducted exploratory analyses to test whether CMS2 and CMS4 patients responded to cetuximab differently on the basis of KRAS signature. Patients were separated into three groups: KRAS mutant (by polymerase chain reaction [PCR]), KRAS wild type (by PCR) with KRAS signature (determined by NanoString), and KRAS wild type (by PCR) without KRAS signature. For exploratory analyses, only age, sex, Eastern Cooperative Oncology Group performance status, and number of metastases were adjusted.
A two-sided P < .05 was considered statistically significant for all tests (P < .1 was used for interaction tests). No adjustments were made for multiple comparisons given the exploratory nature of the analyses. All analyses were performed using SAS 9.4 software (SAS Institute, Cary, NC).
Data collection and statistical analyses were conducted by the Alliance Statistics and Data Center. Data quality was ensured by review of data by the Alliance Statistics and Data Center and by the study chairperson while following Alliance policies. All analyses were based on the study database frozen on December 15, 2015.
RESULTS
CMS Is Prognostic in CALGB/SWOG 80405
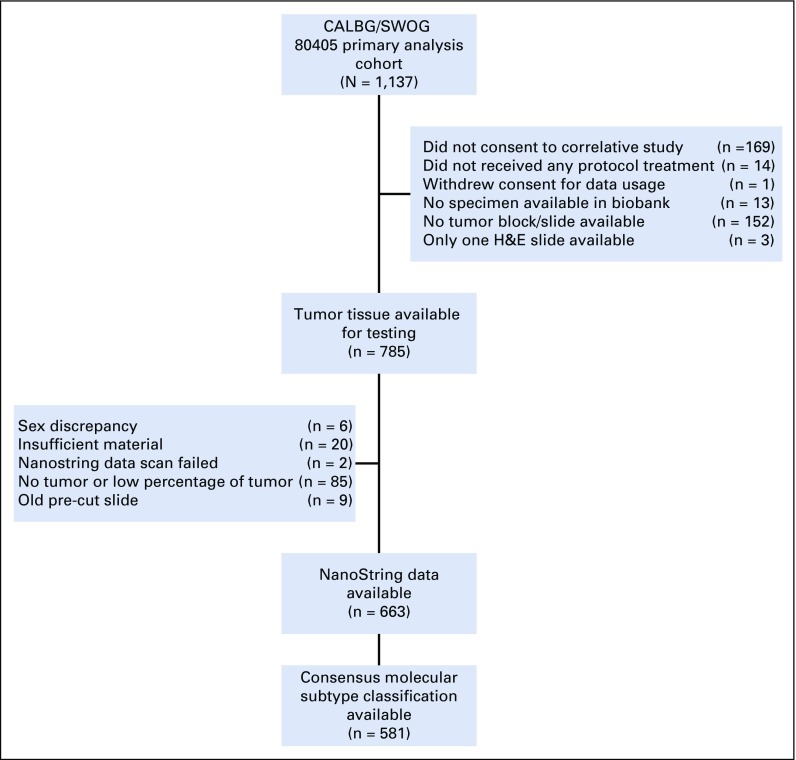
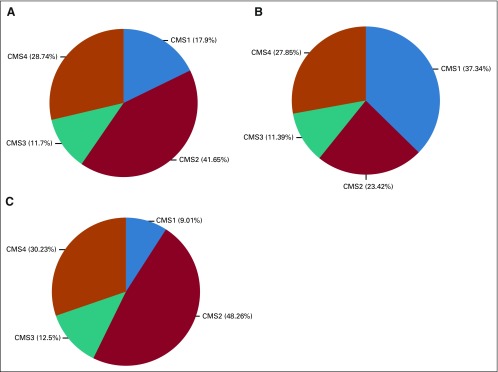
Of 1,137 patients enrolled in the primary analysis cohort of CALGB/SWOG 80405, CMS classification was possible on 5816 (Fig 1). Figure 2 shows the frequency of CMS groups in CALGB/SWOG 80405 overall and by tumor location. The median follow-up for the 581 patients was 61.1 months.
FIG 1.
Study flow diagram.
FIG 2.
Consensus molecular subtype (CMS) proportion. (A) Overall, (B) Right-sided tumors, and (C) left-sided tumors.
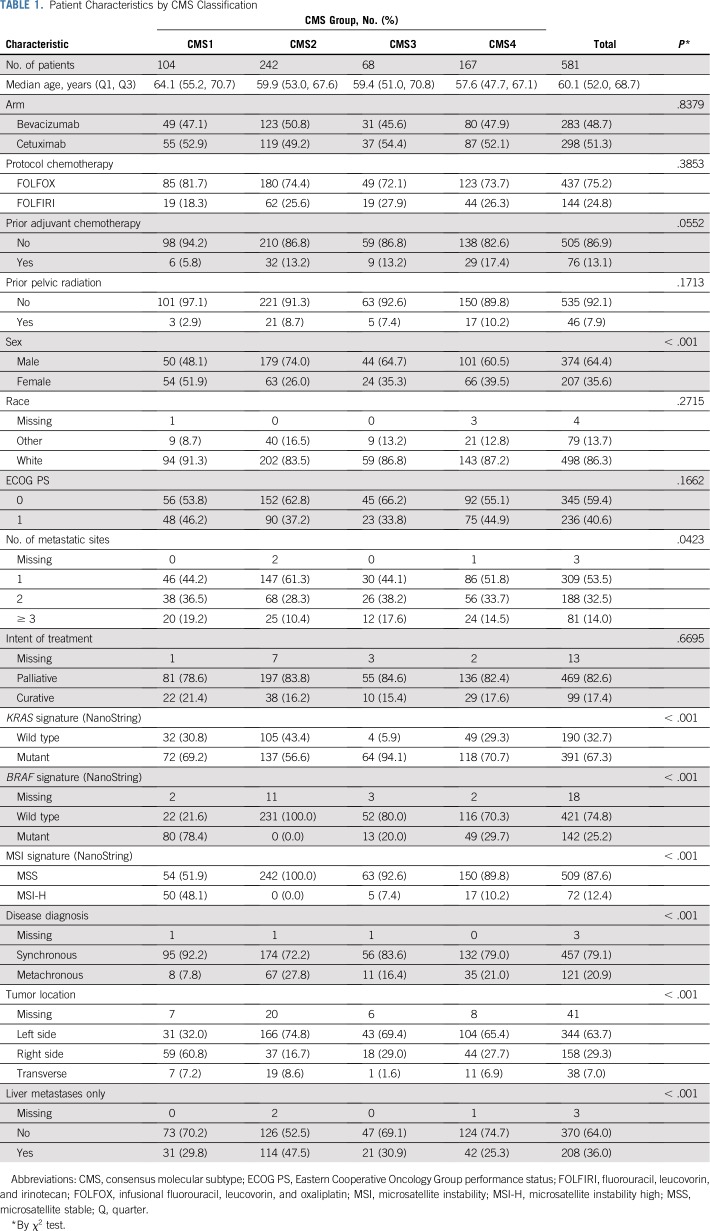
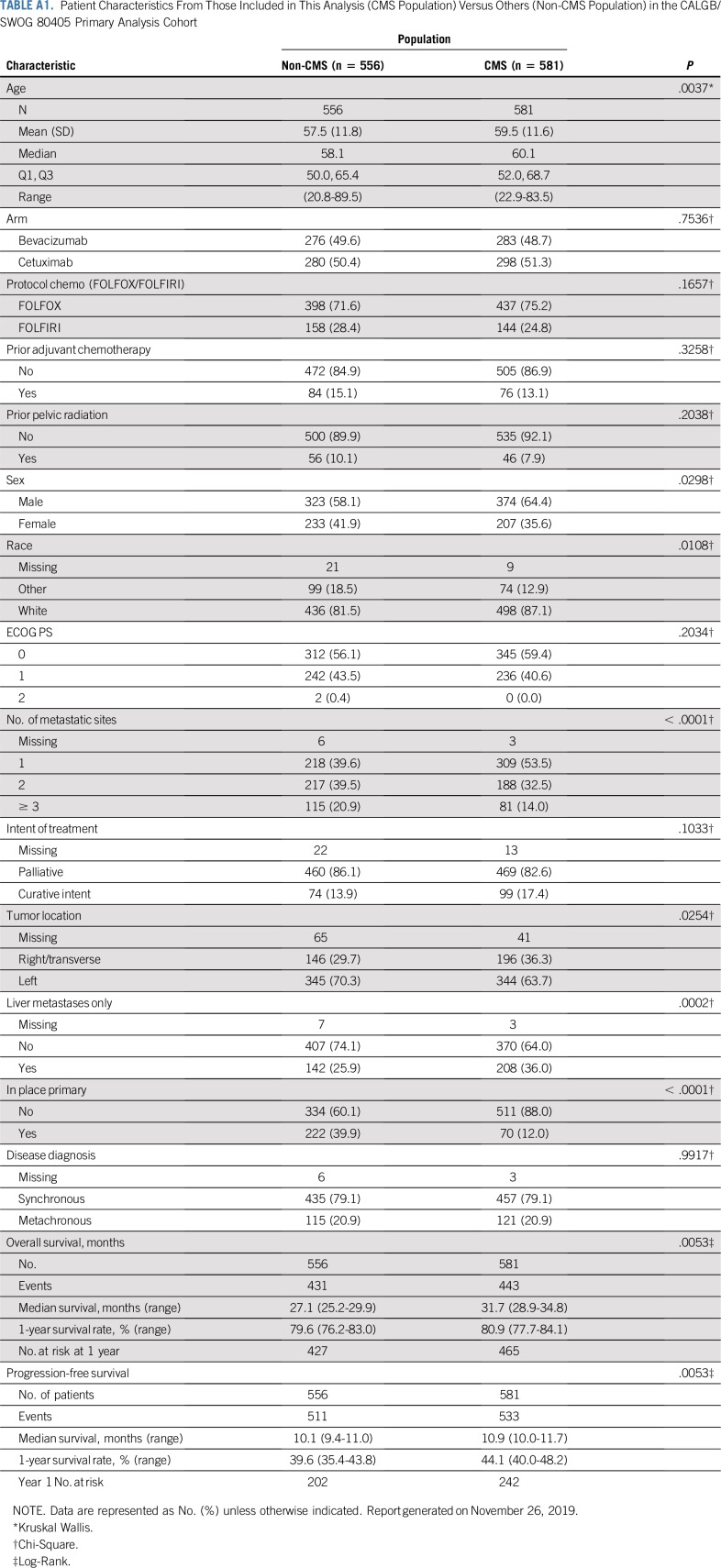
Appendix Table A1 (online only) lists the baseline features of patients for whom CMS classification was available and those in CALGB/SWOG 80405. Across CMS groups, both the molecular signatures and the tumor characteristics were significantly different as expected, as were age, sex, and number of metastatic sites (Table 1).
TABLE 1.
Patient Characteristics by CMS Classification
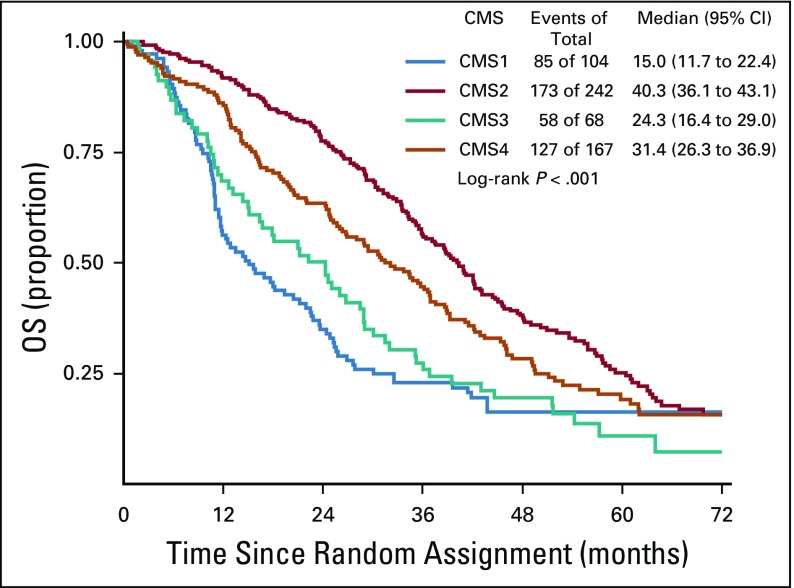
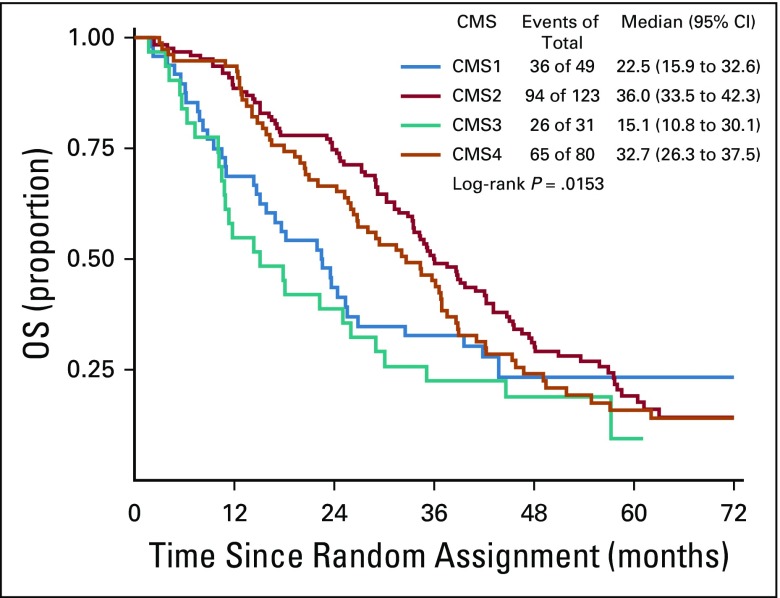
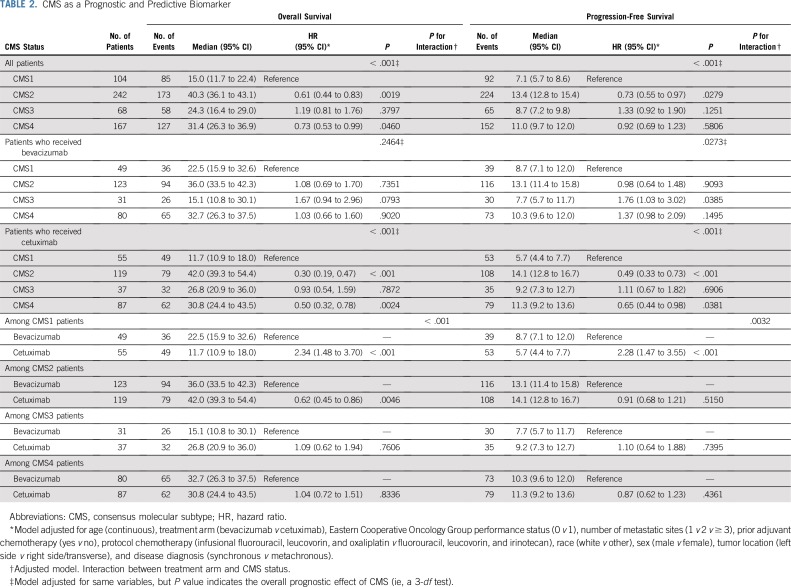
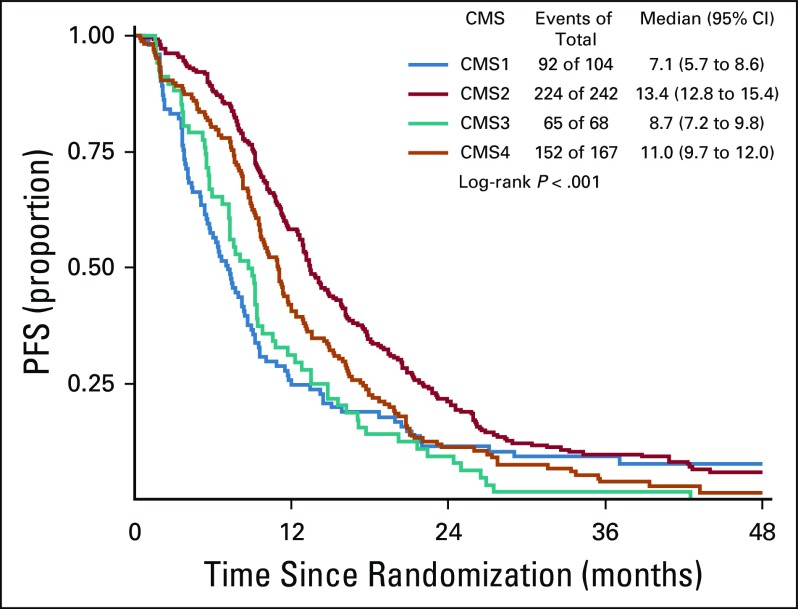
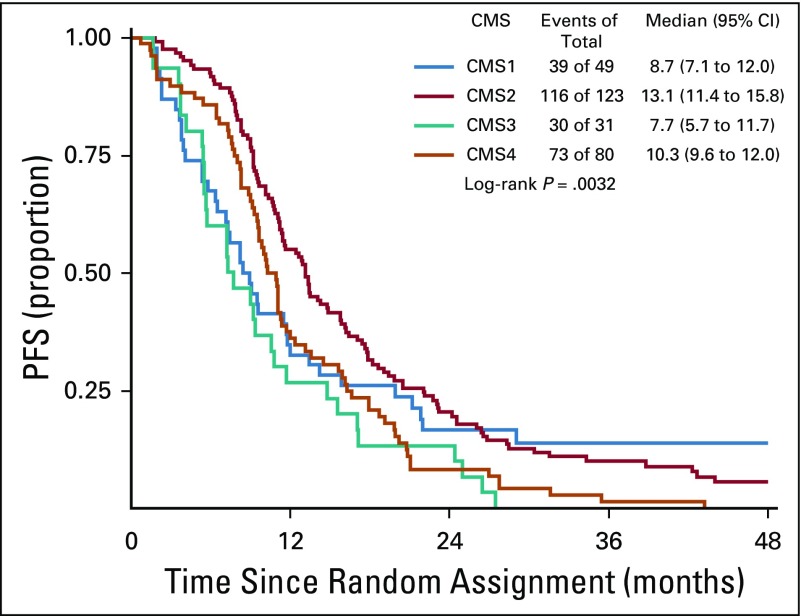
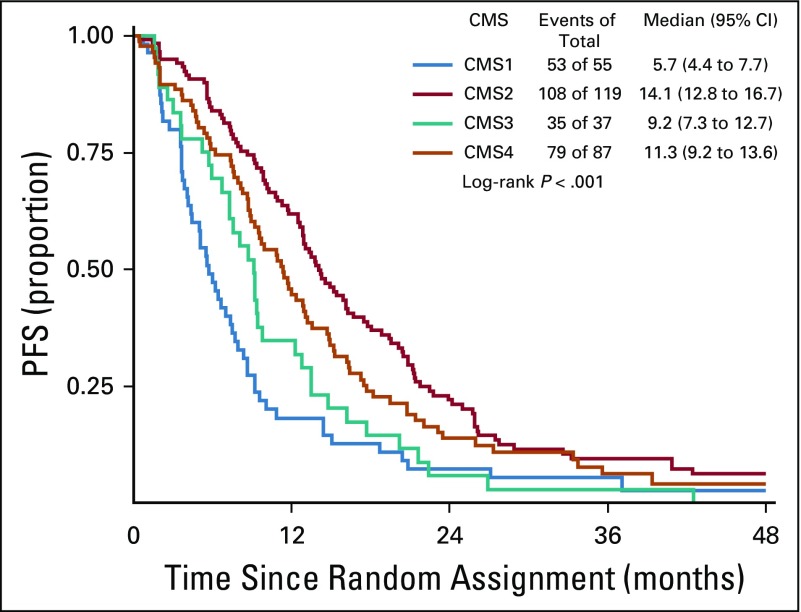
The CMS classification was a significant prognostic marker for OS (P < .001), with a median survival of 15, 40.3, 24.3, and 31.4 months for CMS1, CMS2, CMS3, and CMS4, respectively (Fig 3). In patients who received bevacizumab, CMS was a significant prognostic marker for OS before multivariable adjustment (log-rank P = .015; Fig 4); however, the association between CMSs and OS in patients who received bevacizumab was attenuated after multivariable adjustment (P = .2464). In patients who received cetuximab, CMS was a significant prognostic marker for OS (P < .001), with a median survival of 11.7, 42, 26.8, and 30.8 months for CMS1, CMS2, CMS3, and CMS4, respectively (Fig 5). CMS was also a significant prognostic marker for PFS in the overall cohort (P < .001) and in the bevacizumab (P = .027) and cetuximab (P < .001) treatment arms separately (Table 2; Appendix Figs A1, A2, and A3, online only). Altogether, these results support a strong prognostic link between CMS and OS/PFS in the CALGB/SWOG 80405 cohort overall as well as in the individual treatment arms.
FIG 3.
Overall survival (OS) among patients included in this analysis. CMS, consensus molecular subtype.
FIG 4.
Overall survival (OS) among patients who received bevacizumab. CMS, consensus molecular subtype.
FIG 5.
Overall survival (OS) among patients who received cetuximab. CMS, consensus molecular subtype.
TABLE 2.
CMS as a Prognostic and Predictive Biomarker
KRAS Gene Expression Signature Predicts Response to Cetuximab in CALGB/SWOG 80405
With consideration that RAS mutation is a predictive and clinically actionable biomarker in CRC,5 we examined the impact of RAS mutation and RAS mutant–like gene expression signature on the predictive value of CMSs. We tested whether exclusion of a gene expression signature of mutant KRAS in KRAS wild-type would increase the benefit of EGFR inhibitors.21,22 Patients with KRAS wild type and a KRAS wild-type gene expression signature who received cetuximab had a significantly longer OS (adjusted HR, 0.62; P = .0256) and PFS (adjusted HR, 0.67; P = .0362) than those who received bevacizumab (Appendix Table A2, online only). Cetuximab did not show a significant benefit for patients with CMS2 or CMS4 tumors who were either KRAS mutant (by PCR) or exhibited a KRAS mutant signature. For PFS, the treatment/KRAS signature interaction is statistically significant (P for interaction = .055), indicating that RAS signaling is critical for cetuximab activity and that assessment for mutations of RAS do not predict all the patients who will not benefit from cetuximab.
CMS Classification Predicts Response to Cetuximab and Bevacizumab in CALGB/SWOG 80405
CMS classification is a predictive marker for cetuximab and bevacizumab in both OS (P for interaction < .001) and PFS (P for interaction = .0032). More specifically, in the CMS1 cohort, patients who received bevacizumab had a significantly longer OS (P < .001), with a median survival of 22.5 months compared with 11.7 months for patients who received cetuximab (Table 2). Furthermore, a significantly longer PFS (P < .001) was observed for patients with CMS1 tumors who received bevacizumab compared with those who received cetuximab, with a median PFS of 8.7 months compared with 5.7 months, respectively (Table 2).
In the CMS2 cohort, patients who received cetuximab had a significantly longer OS (P = .0046), with a median OS of 42 months compared with 36 months for patients who received bevacizumab (Table 2). Patients with CMS2 tumors tended to exhibit a slightly improved PFS profile compared with those who receive bevacizumab, although this did not reach statistical significance (P = .52; Table 2).
Patients with CMS1, MSI-H cancers who received bevacizumab had a longer OS than those who received cetuximab (adjusted HR, 0.42; P = .0091; Appendix Table A2). Similarly, patients with CMS1, MSI-H CRC who received bevacizumab had a longer PFS than those who received cetuximab (HR, 0.45; P = .0109). In patients with CMS1, MSS cancers, there was no significant difference in OS and PFS among those who received bevacizumab versus cetuximab for both BRAF mutant and BRAF wild-type tumors (Appendix Table A2). These data suggest that the predictive effect of CMS1 seems to depend on MSI-H.
DISCUSSION
Our findings suggest that CMS is an independent prognostic marker in patients with mCRC who undergo first-line chemotherapy in combination with bevacizumab or cetuximab. Patients with CMS2 tumors had the lowest risk of death and progression compared with patients with other CRC subtypes, whereas those with CMS1 had the shortest PFS and OS. After adjustment for known clinical prognostic factors, the prognostic effect of CMSs remained significant and clinically relevant.
This analysis used a custom CRC-focused gene expression panel that was based on the NanoString platform. The same CMS classifier used in CALBG/SWOG 80405 was used in the FIRE-3 trial which used an Affymetrix-based technology (developed by the independent Swiss Institute for Bioinformatics) that allowed for cross-trial comparison. Our ability to classify CMSs with a custom CRC NanoString panel on paraffin-embedded tumor specimens supports the possibility that a CLIA-approved assay may be developed in the future.
Our data on the predictive value of CMSs raise the possibility that may help to guide treatment in the future. For example, patients with CMS1 tumors had significantly longer PFS and OS when chemotherapy was combined with bevacizumab as opposed to cetuximab. This result was mostly driven by the subset of CMS1 patients with MSI-H tumors. These data may be explained in part through the impact of bevacizumab on inflammatory tumor-associated macrophages on their M1/2 polarization.23 These data support a possible immunomodulatory effect of vascular endothelial growth factor inhibition, which may involve vessel normalization triggered by T-lymphocyte infiltration or activity.23,24 Becht et al25 reported that microenvironmental signatures are highly correlated with CMS1. These data support that CMS classification will help to identify subgroups with immune stimulatory pathways that may lead to novel treatment strategies.
Our data also demonstrate that patients with CMS2 cancers receive a significant benefit from treatment with cetuximab-based chemotherapy. The CMS2 group is enriched for left-sided tumors, which have been shown to preferentially benefit from cetuximab therapy in a recent pooled analysis.26 These tumors are characterized by activated EGFR pathways that render them sensitive to cetuximab.27,28 Even in KRAS wild-type tumor, not all patient respond to cetuximab. In our exploratory analyses, we showed that in using a KRAS mutant gene expression signature in KRAS wild-type tumors, a significant interaction with benefit from cetuximab therapy in CMS2 and CMS4 cancers for PFS was seen. These findings suggest that further refinement of these signatures may lead to the ability to use these agents strategically rather than empirically.
The findings presented here must be considered as preliminary and in need of validation in an independent cohort. The prognostic value of CMS classification was consistent between FIRE-3 and CALGB/SWOG 80405 using the same classification. Recently, the Mitomycin C, Avastin and Xeloda in Patients With Untreated Colorectal Cancer study was published and confirmed the prognostic value of CMSs; however, this study is limited because both treatment arms included bevacizumab.29 However, CMS was not predictive in the same direction across these two studies, which strongly suggests that the OS superiority of the cetuximab-treated compared with the bevacizumab-treated patients in FIRE-3 (OS difference of 8 months) was not the result of a major CMS imbalance between the two trials. This discrepancy may be the result of heterogeneity among the CMSs; differences in the proportion of subtypes within a CMS classification; or other factors such as patterns of care, the differences in the chemotherapy backbone, or the simple play of chance. Indeed, there may be a CMS-chemotherapy interaction that could explain the different outcomes. Although only 25% of the patients in CALGB/SWOG 80405 received first-line FOLFIRI, all patients treated on FIRE-3 started with FOLFIRI.
The choice of the use of irinotecan versus oxaliplatin could lead to distinct interactions between antibodies and chemotherapy. Bevacizumab not only inhibits angiogenesis to allow for T-cell homing but also inhibits dendritic cell maturation, and the extent of bevacizumab-induced dendritic cell suppression has been associated with survival in patients with CRC.30,31 Similarly, oxaliplatin induces immunogenic cancer cell death partly through calreticulin exposure on the dendritic cell surface.32,33 These data suggest that oxaliplatin in combination with bevacizumab may be synergistic and, therefore, clinically beneficial within CMS1. Conceivably, the differences in findings between FIRE-3 and CALGB/SWOG 80405 may be reconciled by data that suggest increased sensitivity of mesenchymal and stem-like tumors to irinotecan and relative resistance to oxaliplatin.34,35
Certain limitations of our study should be acknowledged. The study was limited to patients with KRAS wild-type tumors, which resulted in a relative minority of CMS3 cancers. Moreover, given the design of CALGB/SWOG 80405, a robust stratification of CMS effect by chemotherapy backbone was not possible. This will be integral to discerning interactions between cytotoxic and targeted agents, which may affect the predictive effect of CMS classification. Finally, only one half of the primary analysis cohorts were included in this analysis. Appendix Table A1 shows the comparison of patient characteristics where the difference in primary tumor in place (yes v no) is pronounced but reasonable because patients with a resected primary tumor are more likely to have sufficient tumor tissue for NanoString analysis. The clinical end points are similar across cohorts.
In conclusion, our study demonstrates that CMS classification is an independent prognostic factor in patients with mCRC who undergo first-line therapy. CMS may also have predictive value that may have the potential to guide selection of anti–vascular endothelial growth factor and anti-EGFR therapy. These findings warrant validation in prospective trials using CMS grouping as a stratification factor. In this regard, the CMS platform represents a fundamental step toward the translation of CRC biology into pathway-driven drug and trial development. However, CMSs represent a static assessment of tumor behavior, and heterogeneity exists within each subtype. Ideally, a panomics approach that integrates CMS with surrogates of the tumor immune and stromal environment and that captures dynamic treatment-induced changes in tumor biology will become feasible and will provide tools that enable us to advance precision medicine in mCRC.
Appendix
FIG A1.
Progression-free survival (PFS) among patients included in this analysis. CMS, consensus molecular subtype.
FIG A2.
Progression-free survival (PFS) among patients who received bevacizumab. CMS, consensus molecular subtype.
FIG A3.
Progression-free survival (PFS) among patients who received cetuximab. CMS, consensus molecular subtype.
TABLE A1.
Patient Characteristics From Those Included in This Analysis (CMS Population) Versus Others (Non-CMS Population) in the CALGB/SWOG 80405 Primary Analysis Cohort
TABLE A2.
Exploratory Analyses
Footnotes
Clinical trial information: NCT00265850.
Supported by National Cancer Institute grants U10CA180821 and U10CA180882 (Alliance for Clinical Trials in Oncology), U10CA180790 (Mayo Clinic), U10CA180791 (Memorial Sloan Kettering), U10CA180795 (Indiana University), U10CA180799 (University of Wisconsin), U10CA180830 (University of Southern California Norris Cancer Hospital), U10CA180836 (The University of Chicago), U10CA180838 (The University of North Carolina), U10CA180850 (The Ohio State University), U10CA180867 (Dana-Farber Cancer Institute), and U10CA180888 (SWOG). The trial was also supported in part by Bristol-Myers Squibb, Eli Lilly, Genentech, Pfizer, and Sanofi.
Presented at the American Society of Clinical Oncology 2017 Annual Meeting, Chicago, IL, June 2-6, 2017.
See accompanying Editorial on page 1847
AUTHOR CONTRIBUTIONS
Conception and design: Heinz-Josef Lenz, Fang-Shu Ou, Alan P. Venook, Richard M. Goldberg, Monica M. Bertagnolli, Charles D. Blanke, Sabine Tejpar, Federico Innocenti, Omar Kabbarah
Financial support: Monica M. Bertagnolli
Administrative support: Heinz-Josef Lenz, Monica M. Bertagnolli
Provision of study materials or patients: Heinz-Josef Lenz, Federico Innocenti
Collection and assembly of data: Heinz-Josef Lenz, Fang-Shu Ou, Alan P. Venook, Howard S. Hochster, Donna Niedzwiecki, Monica M. Bertagnolli, Charles D. Blanke, Xueping Qu, Sabine Tejpar, Federico Innocenti, Omar Kabbarah
Data analysis and interpretation: Heinz-Josef Lenz, Fang-Shu Ou, Alan P. Venook, Howard S. Hochster, Richard M. Goldberg, Robert J. Mayer, Monica M. Bertagnolli, Charles D. Blanke, Tyler Zemla, Pratyaksha Wirapati, Sabine Tejpar, Federico Innocenti, Omar Kabbarah
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Impact of Consensus Molecular Subtype on Survival in Patients With Metastatic Colorectal Cancer: Results From CALGB/SWOG 80405 (Alliance)
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Heinz-Josef Lenz
Honoraria: Merck Serono, Roche, Bayer AG, Boehringer Ingelheim
Consulting or Advisory Role: Merck Serono, Roche, Bayer AG, Pfizer
Travel, Accommodations, Expenses: Merck Serono, Bayer AG, Roche
Alan P. Venook
Consulting or Advisory Role: Taiho Pharmaceutical, Bayer AG, Halozyme, Eisai
Research Funding: Genentech (Inst), Roche (Inst), Bristol-Myers Squibb (Inst)
Patents, Royalties, Other Intellectual Property: Royalties from Now-UpToDate for authoring and maintaining two chapters
Travel, Accommodations, Expenses: Genentech, Roche, Halozyme, Bayer AG
Howard S. Hochster
Consulting or Advisory Role: Bayer AG, Genentech, Amgen, Exelixis
Richard M. Goldberg
Consulting or Advisory Role: Merck, Taiho Pharmaceutical, Merck KGaA, Novartis
Research Funding: Bristol-Myers Squibb
Travel, Accommodations, Expenses: Merck KGaA, Merck
Robert J. Mayer
Honoraria: Taiho Pharmaceutical
Consulting or Advisory Role: CASI Pharmaceuticals
Monica M. Bertagnolli
Leadership: Leap Therapeutics
Consulting or Advisory Role: Syntimmune, Syntalogic
Research Funding: AbbVie (Inst), Agenus (Inst), Astellas Pharma (Inst), AstraZeneca (Inst), Breast Cancer Research Foundation (Inst), Bristol-Myers Squibb (Inst), Celgene (Inst), Complion (Inst), Exelixis (Inst), Genentech (Inst), GHI Pharma (Inst), Gilead Sciences (Inst), GlaxoSmithKline (Inst), Incyte (Inst), Jazz Pharmaceuticals (Inst), Leidos (Inst), Eli Lilly (Inst), Matrex (Inst), Mayo Clinic (Inst), MGH (Inst), Millennium Pharmaceuticals (Inst), Novartis (Inst), Patient-Centered Outcomes Research Institute (Inst), Pfizer (Inst), Robert Wood Johnson Foundation (Inst), Sagerock Advisors (Inst), Taiho Pharmaceutical (Inst), Bayer AG (Inst), Eisai (Inst), Lexicon (Inst), Merck (Inst), Pharmacyclics (Inst), Takeda Pharmaceuticals (Inst), Tesaro (Inst), Baxalta (Inst), Sanofi (Inst), TEVA Pharmaceuticals Industries (Inst), Janssen Pharmaceuticals (Inst)
Xueping Qu
Employment: Genentech, BeiGene (I)
Stock and Other Ownership Interests: Genentech, Roche, BeiGene (I)
Honoraria: Genentech, Roche, BeiGene (I)
Pratyaksha Wirapati
Consulting or Advisory Role: Roche (Inst), Genentech (Inst), Sanofi (Inst), Bayer AG (Inst)
Sabine Tejpar
Honoraria: Merck Serono, Regeneron Pharmaceuticals
Consulting or Advisory Role: Merck Serono, Regeneron Pharmaceuticals, Bayer AG, Roche, Genentech, Boehringer Ingelheim
Speakers’ Bureau: Merck Serono, Sanofi, Roche, Bristol-Myers Squibb, MSD
Research Funding: Regeneron Pharmaceuticals (Inst), Merck Serono (Inst), Bayer AG (Inst)
Travel, Accommodations, Expenses: Sanofi, Amgen, Roche, Merck Serono, Bayer AG
Federico Innocenti
Patents, Royalties, Other Intellectual Property: Royalties from the Mayo Foundation on UGT1A1 testing
Omar Kabbarah
Employment: Genentech
Stock and Other Ownership Interests: Genentech
No other potential conflicts of interest were reported.
REFERENCES
- 1.Siegel R, DeSantis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–241. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis CE, Lin CC, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64:252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 4.Venook AP, Niedzwiecki D, Lenz HJ, et al. Effect of first-line chemotherapy combined with cetuximab or bevacizumab on overall survival in patients with KRAS wild-type advanced or metastatic colorectal cancer: A randomized clinical trial. JAMA. 2017;317:2392–2401. doi: 10.1001/jama.2017.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Modest DP, Stintzing S, von Weikersthal LF, et al. Exploring the effect of primary tumor sidedness on therapeutic efficacy across treatment lines in patients with metastatic colorectal cancer: Analysis of FIRE-3 (AIOKRK0306) Oncotarget. 2017;8:105749–105760. doi: 10.18632/oncotarget.22396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stintzing S, Tejpar S, Gibbs P, et al. Understanding the role of primary tumour localisation in colorectal cancer treatment and outcomes. Eur J Cancer. 2017;84:69–80. doi: 10.1016/j.ejca.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loupakis F, Yang D, Yau L, et al. Primary tumor location as a prognostic factor in metastatic colorectal cancer. J Natl Cancer Inst. 2015;107:dju427. doi: 10.1093/jnci/dju427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinemann V, von Weikersthal LF, Decker T, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): A randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15:1065–1075. doi: 10.1016/S1470-2045(14)70330-4. [DOI] [PubMed] [Google Scholar]
- 9.Punt CJ, Koopman M, Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat Rev Clin Oncol. 2017;14:235–246. doi: 10.1038/nrclinonc.2016.171. [DOI] [PubMed] [Google Scholar]
- 10.Cremolini C, Loupakis F, Antoniotti C, et al. FOLFOXIRI plus bevacizumab versus FOLFIRI plus bevacizumab as first-line treatment of patients with metastatic colorectal cancer: Updated overall survival and molecular subgroup analyses of the open-label, phase 3 TRIBE study. Lancet Oncol. 2015;16:1306–1315. doi: 10.1016/S1470-2045(15)00122-9. [DOI] [PubMed] [Google Scholar]
- 11.Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trinh A, Trumpi K, De Sousa E Melo F, et al. Practical and robust identification of molecular subtypes in colorectal cancer by immunohistochemistry. Clin Cancer Res. 2017;23:387–398. doi: 10.1158/1078-0432.CCR-16-0680. [DOI] [PubMed] [Google Scholar]
- 13.Marisa L, de Reyniès A, Duval A, et al. Gene expression classification of colon cancer into molecular subtypes: Characterization, validation, and prognostic value. PLoS Med. 2013;10:e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman J, Hastie T, Tibshirani R. Regularization Paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 15.Mann HB, Whitney DR. On a Test of whether one of two random variables is stochastically larger than the other. Ann Math Stat. 1947;18:50–60. [Google Scholar]
- 16.Cochran WG. The χ2 test of goodness of fit. Ann Math Stat. 1952;23:315–345. [Google Scholar]
- 17.Kaplan EL, Meier P. Nonparametric estimation form incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 18.Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep. 1966;50:163–170. [PubMed] [Google Scholar]
- 19.Cox D. Regression models and life-tables. J R Stat Soc B. 1972;34:187–220. [Google Scholar]
- 20.Kather JN, Halama N, Jaeger D. Genomics and emerging biomarkers for immunotherapy of colorectal cancer. Semin Cancer Biol. 2018;52:189–197. doi: 10.1016/j.semcancer.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 21. doi: 10.1093/annonc/mdz387. Stintzing S, Wirapati P, Lenz HJ, et al: Consensus molecular subgroups (CMS) of colorectal cancer (CRC) and first-line efficacy of FOLFIRI plus cetuximab or bevacizumab in the FIRE3 (AIO KRK-0306) trial. J Clin Oncol 35, 2017 (suppl; abstr 3510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sartore-Bianchi A, Trusolino L, Martino C, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): A proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17:738–746. doi: 10.1016/S1470-2045(16)00150-9. [DOI] [PubMed] [Google Scholar]
- 23.Peterson TE, Kirkpatrick ND, Huang Y, et al. Dual inhibition of Ang-2 and VEGF receptors normalizes tumor vasculature and prolongs survival in glioblastoma by altering macrophages. Proc Natl Acad Sci U S A. 2016;113:4470–4475. doi: 10.1073/pnas.1525349113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian L, Goldstein A, Wang H, et al. Mutual regulation of tumour vessel normalization and immunostimulatory reprogramming. Nature. 2017;544:250–254. doi: 10.1038/nature21724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becht E, de Reyniès A, Giraldo NA, et al. Immune and stromal classification of colorectal cancer is associated with molecular subtypes and relevant for precision immunotherapy. Clin Cancer Res. 2016;22:4057–4066. doi: 10.1158/1078-0432.CCR-15-2879. [DOI] [PubMed] [Google Scholar]
- 26.Arnold D, Lueza B, Douillard JY, et al. Prognostic and predictive value of primary tumour side in patients with RAS wild-type metastatic colorectal cancer treated with chemotherapy and EGFR directed antibodies in six randomized trials. Ann Oncol. 2017;28:1713–1729. doi: 10.1093/annonc/mdx175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Missiaglia E, Jacobs B, D’Ario G, et al. Distal and proximal colon cancers differ in terms of molecular, pathological, and clinical features. Ann Oncol. 2014;25:1995–2001. doi: 10.1093/annonc/mdu275. [DOI] [PubMed] [Google Scholar]
- 28.Bertotti A, Papp E, Jones S, et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature. 2015;526:263–267. doi: 10.1038/nature14969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mooi JK, Wirapati P, Asher R, et al. The prognostic impact of consensus molecular subtypes (CMS) and its predictive effects for bevacizumab benefit in metastatic colorectal cancer: Molecular analysis of the AGITG MAX clinical trial. Ann Oncol. 2018;29:2240–2246. doi: 10.1093/annonc/mdy410. [DOI] [PubMed] [Google Scholar]
- 30.Motz GT, Santoro SP, Wang LP, et al. Tumor endothelium FasL establishes a selective immune barrier promoting tolerance in tumors. Nat Med. 2014;20:607–615. doi: 10.1038/nm.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michielsen AJ, Noonan S, Martin P, et al. Inhibition of dendritic cell maturation by the tumor microenvironment correlates with the survival of colorectal cancer patients following bevacizumab treatment. Mol Cancer Ther. 2012;11:1829–1837. doi: 10.1158/1535-7163.MCT-12-0162. [DOI] [PubMed] [Google Scholar]
- 32.Tesniere A, Schlemmer F, Boige V, et al. Immunogenic death of colon cancer cells treated with oxaliplatin. Oncogene. 2010;29:482–491. doi: 10.1038/onc.2009.356. [DOI] [PubMed] [Google Scholar]
- 33.Tel J, Hato SV, Torensma R, et al. The chemotherapeutic drug oxaliplatin differentially affects blood DC function dependent on environmental cues. Cancer Immunol Immunother. 2012;61:1101–1111. doi: 10.1007/s00262-011-1189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan CW, Chen T, Shang YN, et al. Cancer-initiating cells derived from human rectal adenocarcinoma tissues carry mesenchymal phenotypes and resist drug therapies. Cell Death Dis. 2013;4:e828. doi: 10.1038/cddis.2013.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sadanandam A, Lyssiotis CA, Homicsko K, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19:619–625. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]