Abstract
Objective:
Gut microorganisms contribute to the metabolism of environmental toxicants, including methylmercury (MeHg). Our main objective was to investigate whether associations between biomarkers for prenatal MeHg exposure and maternal gut microbiota differed between early and late gestation.
Methods:
Maternal blood and stool samples were collected during early (8.3–17 weeks, n=28) and late (27–36 weeks, n=24) gestation. Total mercury and MeHg concentrations were quantified in biomarkers, and inorganic mercury was estimated by subtraction. The diversity and structure of the gut microbiota were investigated using 16S rRNA gene profiling (n=52). Biomarkers were dichotomized, and diversity patterns were compared between high/low mercury concentrations. Spearman's correlation was used to assess bivariate associations between MeHg biomarkers (stool, blood, and meconium), and 23 gut microbial taxa (genus or family level, >1% average relative abundance).
Results:
Within-person and between-person diversity patterns in gut microbiota differed between early/late gestation. The overall composition of the microbiome differed between high/low MeHg concentrations (in blood and stool) during early gestation, but not late gestation. Ten (of 23) taxa were significantly correlated with MeHg biomarkers (increasing or decreasing); however, associations differed, depending on whether the sample was collected during early or late gestation. A total of 43% of associations (69/161) reversed the direction of correlation between early/late gestation.
Conclusions:
The time point at which a maternal fecal sample is collected may yield different associations between gut microorganisms and MeHg biomarkers, which may be due in part to remodeling of maternal microbiota during pregnancy. Our results suggest the effectiveness of dietary interventions to reduce prenatal MeHg exposure may differ between early and late gestation.
Keywords: methylmercury, gut microbiota, meconium, prenatal
INTRODUCTION1
The human microbiota refers to the tens of trillions of microorganisms that colonize the body and their genes, referred to as the microbiome. Of all the body sites, the majority of microbiota live in the gastrointestinal tract, particularly in the colon. Methylmercury (MeHg) is a potent neurotoxin, and the fetal period is the most vulnerable period for exposure (Clarkson and Magos, 2006). There is significant variability in MeHg metabolism, which has been attributed to differences in the structure and function of the gut microbiome (Rowland et al., 1984, 1986).
Much of the research on MeHg metabolism is focused on methylation/demethylation by gut microbiota. In the large intestine, gut microbes are hypothesized to demethylate MeHg and produce less toxic inorganic mercury (IHg), which is excreted in feces (Clarkson and Magos, 2006). Stool MeHg is not entirely demethylated before excretion (Caito et al., 2018; Ishihara, 2000; Rand et al., 2016; Rothenberg et al., 2016), suggesting other mechanisms may limit MeHg absorption from the large intestine. Methylation of IHg by gut microbes is also possible; to date, one commensal methanogen (Methanomassiliicoccus luminyensis) isolated from human feces contained the gene cluster (hgcA and hgcB) required for inorganic Hg methylation (Parks et al., 2013). However, this gene cluster was not detected in fecal samples from 297 healthy individuals in the Human Microbiome Project (Podar et al., 2015), 145 elderly Swiss women (Podar et al., 2015), or six pregnant mothers in South Carolina (Rothenberg et al., 2016), suggesting stool MeHg most likely originated from dietary sources of MeHg. In animal and human studies, exposure to lead, cadmium, and arsenic impacted the structure and/or function of gut microbiota (Breton et al., 2013; Dong et al., 2017; Lu et al., 2014), and thus MeHg may also impact the gut microbial community.
A better understanding of the interplay between gut microorganisms and MeHg metabolism is needed, especially during pregnancy. Pregnancy is marked by changes in maternal physiology, anatomy, and immune function, and gut microbiota may contribute to these changes (Koren et al., 2012). Previously, we reported associations between gut microbiota taxa and MeHg biomarkers during late gestation (Rothenberg et al., 2016). In the present study, we investigated whether associations between biomarkers for prenatal MeHg exposure and gut microbiota differed between early and late gestation. We also hypothesized associations would differ between MeHg and IHg. Maternal blood total Hg (THg), MeHg, and IHg concentrations were previously published for 24 (of 28) mothers (Donohue et al., 2018); all other data have not been previously published.
MATERIALS AND METHODS
Enrollment and biomarker collection
From October 2014 through March 2016, mothers were recruited at the Medical University of South Carolina (MUSC) in Charleston, South Carolina, USA. Eligible mothers were in good general health, between 18–45 years old, expecting a singleton birth, and within 14 weeks of the last menstrual period. Mothers receiving antibiotic treatment within the previous two weeks were ineligible. All mothers provided written informed consent prior to participation. Protocols were approved by the Institutional Review Board at the Medical University of South Carolina and approved by cooperative review at the University of South Carolina. A total of 28 mothers were enrolled, including 20 mothers who were initially enrolled in a double-blind vitamin D supplementation study (ClinicalTrials.gov, NCT01932788). After collection of the first blood sample, participants in the vitamin D supplementation study were randomized into placebo (400 IU) and supplemented (4400 IU) groups.
Biomarker collection.
Upon enrollment and during late gestation, a non-fasting blood sample was collected by venipuncture for analysis of whole blood THg and MeHg (Becton Dickinson, K2EDTA, Royal Blue). A second vial of blood was collected for analysis of total circulating plasma 25(OH)D (Becton Dickinson, K2EDTA, Lavender) from all mothers enrolled in the vitamin D supplementation study (n=20) and from some of the other mothers (early gestation: n=2; late gestation: n=4). Vials of whole blood for THg and MeHg analyses were frozen at −20°C, and transported to the University of South Carolina in a Credo Cube (−20°C) (20M, Pelican, MN, USA), where they were stored frozen (−25°C). For 25(OH)D analyses, plasma was separated by centrifugation, and aliquots were archived at MUSC (−80°C).
Mothers were given a stool collection kit including sterile collection containers (Thermo Fisher 02-544-208) and detailed instructions adapted from the Human Microbiome Project protocols (Human Microbiome Project, 2010). Participants shipped stool samples overnight to the University of South Carolina. Upon receipt, in an anaerobic AtmosBag (Sigma Aldrich, Z530212) filled with N2, stool samples were aliquoted using sterile microspatulas (Corning 3012, New York, USA) into 2 mL sterile cryovials and 50 mL polypropylene vials, and then frozen (−80°C). A maternal hair sample was collected from the occipital region using stainless steel scissors, which was tied with a string and stored in a plastic bag at room temperature. While in the hospital, the first diaper with meconium was put into a plastic bag and frozen (−20°C). Frozen diapers were transferred to the University of South Carolina in a Credo Cube, and archived (−80°C). Before analysis, meconium was gently loosened from the frozen diaper, and transferred into 50 mL polypropylene vials. Of 22 diapers, there was sufficient meconium to analyze THg and MeHg concentrations in 14 and 17 diapers, respectively. Table A1 includes the timeframe for collection of blood, stool, and hair samples.
Fish consumption
During their second trimester, mothers completed at home a food frequency questionnaire (FFQ) including 30 varieties of fish/shellfish commonly consumed in Charleston, S.C. The questionnaire included nine options, ranging from "never" to "almost every day," which were converted to servings/day: 0 = never, 1/365 = once per year, 2.5/365 = two or three times per year, 6/365 = about every other month, 12/365 = once per month, 2.5/30.5 = two or three times per month, 1/7 = once per week, 2.5/7 = two or three times per week, and 1 = almost every day. Daily servings were summed, providing total fish/shellfish intake.
Other covariates
Mothers completed a sociodemographic questionnaire, including race/ethnicity, maternal age, education level, parity, and previous alcohol and tobacco use. Height and weight were recorded at the first visit, and weight was recorded at subsequent visits. First trimester body mass index (BMI) was calculated, including underweight (<18.5 kg/m2), normal weight (18.5–24.9 kg/m2), overweight (25.0–29.9 kg/m2), and obese (≥30.0 kg/m2). Two mothers were classified as underweight, and were combined with normal weight mothers. Gestational weight gain was classified as below, within, or above recommended weight gain based on guidelines from the Institute of Medicine of the National Academies (Institute of Medicine, 2009). Antibiotic treatment between the first and second stool sample was obtained from the medical record, and categorized (0 times, 1–4 times). Mothers reported whether they received the flu vaccine and the date of the vaccine, and whether they had amalgam fillings. In animal studies and human studies, antibiotic treatment increased the MeHg elimination rate (Caito et al., 2018; Rowland et al., 1984; Seko et al., 1981). Amalgam fillings contain elemental Hg, and the flu vaccine is usually preserved with thimerosal, which contains ethyl Hg (Clarkson and Magos, 2006).
Laboratory analyses
Hair preparation.
THg and MeHg concentrations were analyzed at the Rothenberg Mercury lab. Hair samples were collected at the beginning of the third trimester and were sectioned corresponding to the previous three, six, and nine months. On average, hair samples corresponded to 21–32 weeks gestation, 7.6–21 weeks gestation, and 5.2 before pregnancy to 7.6 weeks gestation, respectively. Hair samples corresponding to 7.6–21 and 21–32 weeks gestation represented early and late gestation, respectively. Hair samples were cut using the specific monthly growth rate of hair for African American women (0.87 cm/month) and Caucasian women (1.12 cm/month) (Loussouarn et al., 2005); we assumed the hair growth rate for Hispanic women was the same as Caucasian women. To remove exogenous Hg, sectioned hair samples were washed using 0.1% (v/v) 2-mercaptoethanol, gently shaken for one hour, then triple-rinsed with Milli-Q H20 (>18.0 MΩ cm−1), and air-dried in a biosafety cabinet, as previously described (Hong et al., 2016).
THg analyses.
THg concentrations in hair, stool, blood, and meconium were analyzed following U.S. Environmental Protection Agency (USEPA) Method 1631, although digestion methods differed slightly (USEPA, 2001a, 2002). Briefly, each sample was aliquoted into a 40 mL borosilicate vial with a Teflon-lined cap. For blood samples, cold digestion was used (Donohue et al., 2018). Blood (~0.5 g) was digested in 4 mL of hydrochloric acid (HCl) and 1 mL of nitric acid (HNO3) overnight. For stool, meconium, and hair samples, hot digestion was used (Rothenberg et al., 2016, 2017). Stool (~0.75 g) and meconium (~1.2 g) were digested in 5 mL of freshly prepared sulfuric acid (H2SO4):HNO3 [3:7 (v/v)], while hair (~0.02 g) was digested in HNO3:HCL [(3:1 (v/v)]. Vials were heated in a water bath at 70–80°C (stool and meconium: 2–3 hours, hair: 6 hours).
After digestion, bromine monochloride (BrCl) was added (1% v/v) to oxidize all Hg to Hg(II), and samples sat overnight. Just before analysis, 0.1 mL of hydroxylamine hydrochloride (30% w/v) was added, followed by 0.1 mL of tin (II) chloride to reduce Hg(II) to volatile Hg(0). De-foaming agent (Antifoam AF, Spectrum, USA) (2% v/v, 1 mL) was added to analysis vials to reduce foaming. THg concentrations were analyzed using cold vapor atomic fluorescence spectrometry (CVAFS) (Merx-T with Model III Detector, Brooks Rand Instruments).
MeHg analyses.
Blood MeHg was extracted using methods from Liang et al. (2000) and Donohue et al. (2018). Approximately 0.5 g of thawed blood was weighed into a 50 mL polypropylene tube, and dried in an oven overnight at 70°C. Samples were digested in 2 mL of 25% potassium hydroxide:methanol (w/v) for 3 hours at 75°C. Then 10 mL of dichloromethane (CH2Cl2) and 2 mL of hydrochloric acid were added to each sample. Samples were shaken for 30 minutes, then left overnight to complete phase separation. Approximately 8 mL of the CH2Cl2 layer was transferred to a pre-weighed 50-mL polypropylene tube, and Milli-Q H2O was added to 30 mL. Vials were heated in a water bath (60–70°C) for 1.5 hours to evaporate the CH2Cl2 layer, and then the volume was raised to 40 ml using Milli-Q H2O.
For stool and meconium samples, MeHg was extracted using methods from Bloom et al. (1997). Stool samples (~3 g) and meconium samples (~0.9 g) were leached for one hour in 5 mL of 18% (w/v) potassium bromide + 5% (v/v) H2SO4, and 1 mL of 1 M copper sulfate solution. Then 10 mL of CH2Cl2 was added, samples were shaken for one hour, centrifuged (4000 RPM, 30 min), and phases separated (Whatman 1-PS). The CH2Cl2 layer was evaporated by heating vials in a water bath, as described above, and the final volume was raised to 40 mL using Milli-Q H2O.
Following solvent extraction and back extraction into water, MeHg was quantified by gas chromatography-CVAFS, according to U.S. EPA Method 1630 (USEPA, 2001b) (Model-III Detector, Brooks Rand Instruments).
There was insufficient volume of hair to analyze MeHg. However, hair THg is considered an accurate measure of dietary MeHg intake because MeHg is the Hg species that is accumulated in the hair shaft (Zareba et al., 2008). Stool samples were dried overnight at 105°C, and Hg concentrations were reported in dry weight. There was insufficient meconium mass to measure the wet:dry ratio; however, all meconium samples had the same tar-like consistency, and thus meconium Hg concentrations were reported in wet weight.
THg and MeHg quality assurance/quality control (QA/QC).
QA/QC is summarized in Table A2. At least two standard reference materials were analyzed alongside samples, as well as matrix spikes and sample replicates. Average recoveries for THg and MeHg in standard reference materials and matrix spikes ranged from 81–118%, and the relative standard deviation between replicate digests for THg and MeHg ranged from 3.3–22%. Six values for blood %MeHg (of THg) exceeded 100% (Table 1), which is mainly attributed to variability in the analytical methods (Donohue et al., 2018). The method detection levels for THg and MeHg were based on the ratio between the lowest point on the calibration curve and the average mass analyzed (USEPA, 2011) (Table A2). The detection levels for stool THg and MeHg were converted to dry weight using the average wet:dry ratio [=4.18 (unitless)]. MeHg concentrations in two meconium samples of 17 (12%) were below the detection level, and half the detection level was imputed. All other values exceeded the method detection levels.
Table 1.
Summary statistics for biomarkers, collected during early and late gestation.
| a. Blood, stool, meconium and hair | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Maternal blood (μg/L) |
Maternal stool (ng/g dry weight) |
Meconium (ng/g wet weight) |
Hair (μg/g) |
|||||||||||
| Early Median (range) |
Early (n) |
Late Median (range) |
Late (n) |
Early Median (range) |
Early (n) |
Late Median (range) |
Late (n) |
Median (range) |
(n) | −5.2–7.6 wks Median (range) |
7.6–21 wks Median (range) |
21–32 wks Median (range) |
(n) | |
| THg | 0.63 (0.017, 2.5) | 28 | 0.48 (0.075, 1.5) | 24 | 28 (1.1, 1400) | 28 | 19 (1.0,400) | 24 | 3.9 (0.64, 11) | 14 | 0.078 (0.0054, 0.57) | 0.071 (0.0050, 0.66) | 0.080 (0.0030, 0.57) | 24 |
| MeHg | 0.37 (0.0099, 1.6) | 28 | 0.29 (0.014, 1.4) | 24 | 0.035 (0.0029,2.1) | 28 | 0.0093 (0.0013, 1.7) | 24 | 0.0078 (<MDL, 0.078) | 17 | NA | NA | NA | NA |
| IHg | 0.25 (0, 0.90) | 28 | 0.069 (0, 0.69) | 24 | 28 (1.1, 1400) | 28 | 19 (1.0,400) | 24 | 3.9 (0.64, 11) | 14 | NA | NA | NA | NA |
| %MeHg (of THg) | 59 (16, 114) | 28 | 72 (16, 117) | 24 | 0.12 (0.00031, 8.9) | 28 | 0.050 (0.0025, 2.0) | 24 | 0.34 (0.055, 0.84) | 14 | NA | NA | NA | NA |
| %IHg (of THg) | 41 (0, 84) | 28 | 28 (0, 84) | 24 | 100 (91, 100) | 28 | 100 (98, 100) | 24 | 100 (99, 100) | 14 | NA | NA | NA | NA |
| IHg (inorganic mercury, THg - MeHg), MDL (method detection level), MeHg (methylmercury), THg (total mercury) | ||||||||||||||
| b. Plasma 25(OH)D | ||||||||||||||
| 25(OH)D (ng/mL) | ||||||||||||||
| Early Median (range) |
Early (n) |
Late Median (range) |
Late (n) |
|||||||||||
| All | 28.7 (10.5, 40.9) | 22 | 40.9 (12.9, 87.1) | 21 | ||||||||||
| Unsupplemented | 28.1 (10.5, 40.9) | 13 | 35.7 (12.9, 56.2) | 13 | ||||||||||
| Supplemented | 29.7 (17.1,37.6) | 9 | 50.9 (31.2, 87.1) | 8 | ||||||||||
| 25(OH)D (25-hydroxy-vitamin D) | ||||||||||||||
Hematocrit.
The hematocrit (%) is part of the Complete Blood Count, and is defined as the ratio between the volume of red blood cells over the total volume of blood. About 80% of blood MeHg binds to the red blood cells (Clarkson and Magos, 2006); plasma volume increases from early to late gestation (Hytten, 1985), and may contribute to a dilution effect for blood MeHg. To control for hemodilution, blood Hg was normalized by hematocrit (Donohue et al., 2018).
Vitamin D.
For 20 mothers enrolled in the vitamin D supplementation study, total circulating 25-hydroxy-vitamin D [25(OH)D] concentrations in plasma were analyzed using a rapid, direct radioimmunoassay, which was developed by the Hollis lab at MUSC (Hollis et al., 1993). For eight mothers not enrolled in the vitamin D supplementation study, plasma 25(OH)D2 and 25(OH)D3 concentrations were analyzed using liquid chromatography/mass spectrometry at the MUSC Clinical Chemistry lab, and then summed to obtain total plasma 25(OH)D concentrations (van den Ouweland et al., 2010). In a comparison of the two methods, 25(OH)D concentrations were highly correlated (r-squared=0.90) (van den Ouweland et al., 2010). Both assays have a coefficient of variation of ≤10%. The radioimmunoassay detection level is 2.8 ng/mL (Hollis et al., 1993), and the detection levels using liquid chromatography/mass spectrometry are <1 ng/mL (van den Ouweland et al., 2010). All 25(OH)D concentrations exceeded the detection levels.
Data analysis for biomarkers
Pairwise associations between Hg parameters, including stool, blood, and meconium THg and MeHg, fish consumption, and maternal characteristics were examined using Spearman’s correlation, Pearson's correlation, one way analysis of variance (ANOVA), Wilcoxon rank-sum test, Student's two-tailed t-test, Kruskal-Wallis test, or Fisher's exact test, depending on the distribution and properties of each variable. For ANOVA, multiple comparisons were estimated using the Sidak test, and p-values for pairwise comparisons are reported in the text.
Associations between stool and blood Hg were potentially biased by the differences in biomarker collection dates (Table A1). To assess potential bias, we used weighted linear regression relating the Hg parameter (independent variable) and gestational period (dependent variable). The inverse of the absolute value of the number of days between stool and blood collection was used as the analytical weight. Then results were compared between the unweighted and weighted regression models.
Longitudinal trends were assessed using a repeated measures mixed model adjusting for within-person correlation, including gestational period (early versus late gestation) and vitamin D supplementation status (coded 0, 1) as covariates. P-values reported in the text are for the effect estimates for gestational period. For Pearson's correlation, regression analysis, and repeated measures, right-skewed variables were log10 transformed. Inorganic Hg was estimated by subtraction (THg-MeHg); six negative values of blood IHg were assigned 0. Before log10-transformation, a value of 0.001 was added to all blood IHg concentrations, so these observations were not dropped. Diagnostics for model fit included examination of residual plots to ensure assumptions for residuals were met (no evidence of non-linearity, constant variance, and normal distribution).
One mother (3.6%) did not give blood, and two mothers (7.1%) did not complete the FFQ. Blood THg and MeHg concentrations and total fish consumption (servings/weekly) were imputed using multiple imputation based on the multivariate normal distribution (Schafer, 1997), conditional on maternal characteristics, and maternal stool, blood, and hair THg and MeHg concentrations. As a sensitivity analysis, data were checked with and without imputed observations, and results did not differ.
An alpha-level of 0.05 was used as a guide for statistical significance. Statistical analyses were performed using Stata (Version 9.2, College Station, TX, USA), and the R-platform (version 3.2.1) (R Core Team, 2017).
DNA extraction, sequencing, and bioinformatics
DNA isolation.
Maternal stool samples were shipped overnight in a Credo Cube to the University of North Carolina at Chapel Hill Microbiome Core Facility. Genomic DNA was isolated on a King Fisher Flex automated instrument (Thermo Fisher Scientific, Grand Island, NY) using the MagMAX™ DNA protocol, following manufacturer's instructions (see Appendix). The quality of the isolated DNA was assessed by agarose gel electrophoresis and purity verified using 260/280 and 260/230 ratios measured by NanoDrop 1000 instrument (Thermo Fisher Scientific, Waltham, MA). DNA concentration was quantified using Quant-iTTM PicoGreen dsDNA Reagent (Molecular Probes, Thermo Fisher Scientific division, Eugene, OR). DNA was stored in elution buffer at −20°C prior until further processing.
16S rRNA amplicon sequencing.
DNA was amplified using primers targeting the V1/V2 variable regions of the bacterial 16S rRNA gene on the Illumina MiSeq platform, as previously described (Allali et al., 2017) (Table A3). The two Illumina sequencing runs (MiSeq1 and MiSeq2) were converted to multiplexed Illumina version 1.9 FASTQ format using CASAVA 1.8.2. Paired-end reads were joined and all downstream analyses were performed using R statistical programming environment (version 3.4.1) (R Core Team, 2017).
Bioinformatics.
Bioinformatic analysis of bacterial 16S rRNA amplicon data was conducted using R package DADA2 (version 1.5.2) (Callahan et al., 2016, 2017) and R package phyloseq (version 1.22.3) (McMurdie and Holmes, 2013). The reads were assessed for chimeras to identify side effects of incomplete PCR amplification. Approximately 200 taxa were identified to be chimeras and subsequently removed. A water sample was included as negative control to account for the effects of possible sample handling contaminants. Any amplicon sequence variants (ASVs) observed within negative control were subsequently removed from all samples.
Alpha diversity (within person) was assessed using three indices, including the observed number of species, the Shannon index, and Fisher's index. Fisher's index accounts for species richness, i.e., number of species, while the Shannon index accounts for both richness and evenness. Differences in alpha diversity between categorical variables were assessed using Student's two-tailed t-test or ANOVA. Robust beta diversity analyses were performed using an R implementation of multivariate Welch Tw2 test on Jansen-Shannon divergence distances, and p-values were computed using permutation testing (Alekseyenko, 2016).
Pairwise associations between gut microbiota taxa and Hg parameters, as well as 25(OH)D levels, were determined using Spearman's correlation. Multiple comparisons were checked using the Benjamini-Hochberg False Discovery Rate (FDR) procedure (Yekutieli and Benjamini, 1999); however there were no significant values after correction, assuming q=0.20. Longitudinal changes in alpha diversity and gut microbial taxa were assessed using a repeated measures mixed model, including gestational period and vitamin D supplementation status as covariates, as described above. For gut microbial taxa, a value of 0.001 was added to the relative abundance before log10-transformation, so that observations with 0 relative abundance were not excluded.
Accession numbers
This study has been deposited to the U.S. National Institutes of Health Database for Genotypes and Phenotypes (dbGaP) and the data are available through accession number phs001768.v1.p1.
RESULTS
Longitudinal trends in Hg.
Summary statistics for all Hg biomarkers (stool, blood, hair, and meconium) are in Table 1. For comparison, during early gestation median blood THg (0.63 μg/L, n=28) was lower compared to the median value for reproductive age women enrolled in the U.S. National Health and Nutrition Examination Survey 2009–2010 (median: 0.84 μg/L, n=1786) (USEPA, 2013).
Median concentrations of blood THg, MeHg, and IHg decreased between early and late gestation by 24%, 22%, and 72%, respectively (n=24–28). In stool samples, the same parameters decreased by 32%, 16%, and 32%, respectively (n=24–28). Using a mixed model adjusted for gestational period and vitamin D supplementation status, most biomarkers (blood THg and IHg, and stool THg, MeHg, and IHg) were significantly lower during late gestation compared to early gestation (when log10-transformed) (p<0.05, n=52) (Table 2). The exception was blood MeHg, which did not differ (when log10-transformed) (p=0.50, n=52). The results for blood THg, MeHg, and IHg did not differ when using the hematocrit-adjusted values (Table 2), suggesting the declines in blood THg and IHg between early/late gestation were not entirely due to hemodilution. Log10 hair THg concentrations did not differ between the earliest gestational period, when compared to the other two gestational periods (p=0.73–0.76, n=72). Stool IHg (% of THg) was previously correlated with the MeHg excretion rate (Caito et al., 2018; Rand et al., 2016). In the present study, stool IHg (% of THg) did not differ between early/late gestation (p=0.99, n=52). Vitamin D supplementation status was non-significant in all models (p=0.42–0.98, n=50–72).
Table 2.
Results for repeated measures mixed model, relating biomarkers (dependent variables) with gestational period and vitamin D supplementation status (independent variables) (n=52 for stool and blood, n=50 for blood normalized, n=72 for hair).
| Dependent variable | Gestational period |
Beta (95% CI) |
|---|---|---|
| Log10 Stool THg | Early | Referent |
| Late | −0.19 (−0.35, −0.03)* | |
| Log10 Stool MeHg | Early | Referent |
| Late | −0.29 (−0.56, −0.03)* | |
| Log10 Stool IHg | Early | Referent |
| Late | −0.19 (−0.35, −0.03)* | |
| Stool Percent IHg | Early | Referent |
| Late | 0.002 (−0.19, 0.19) | |
| Log10 Blood THg | Early | Referent |
| Late | −0.14 (−0.25, −0.02)* | |
| Log10Blood MeHg | Early | Referent |
| Late | −0.04 (−0.16, 0.08) | |
| Log10Blood IHg | Early | Referent |
| Late | −0.62 (−1.1, −0.18)** | |
| Log10 Blood THg, normalized1 | Early | Referent |
| Late | −0.13 (−0.25, −0.02)* | |
| Log10 Blood MeHg, normalized1 | Early | Referent |
| Late | −0.04 (−0.16, 0.09) | |
| Log10 Blood IHg, normalized1 | Early | Referent |
| Late | −0.68 (−1.2, −0.16)* | |
| Log10Hair THg | Pre-pregnancy | Referent |
| Early | −0.02 (−0.14, 0.10) | |
| Late | 0.02 (−0.12, 0.16) |
p<0.05,
p<0.01, confidence interval (CI), IHg (inorganic mercury, THg-MeHg), methylmercury (MeHg), THg (total mercury)
Normalized = 100*(blood mercury/hematocrit)
Associations between Hg parameters.
Mothers ingested on average 1.1 ± 1.2 fish meals/weekly (n=28) (Table 3). Fish consumption (servings/week) was strongly positively correlated with stool THg and MeHg, blood THg and MeHg, and stool IHg during early gestation (Spearman's rho: 0.51–0.56, p<0.01 for all, n=28), and was positively correlated with stool MeHg during late gestation (Spearman's rho: 0.50, p<0.05, n=24) (Table A4). Fish consumption was also positively correlated with meconium MeHg (Spearman's rho: 0.54, p<0.03, n=17) (Table A4). The correlation between fish consumption and maternal hair THg increased between pre-pregnancy, early gestation, and late gestation (Spearman's rho: 0.50, 0.61, and 0.63, respectively, p<0.05, p<0.01, p<0.001, respectively, n=24 for each period). Hair THg reflects average exposure over three months, while blood and stool Hg represents a single time point, which potentially contributed to differences in their associations with fish consumption.
Table 3.
Maternal characteristics, including comparison between vitamin D supplemented and unsupplemented mothers (n=28 mothers total).1
| All (n=28) |
Supplemented (n=9) |
Unsupplemented (n=19) |
||
|---|---|---|---|---|
| Mean ± 1 SD (range) or N (%) |
Mean ± 1 SD (range) or N (%) |
Mean ± 1 SD (range) or N (%) |
p- value2 |
|
| Age (years) | 31 ± 6.5 (19, 42) | 30 ± 6.0 (21, 41) | 31 ± 6.9 (19, 42) | 0.46 |
| First trimester BMI (kg/m2) | 29 ± 7.1 (18, 51) | 28 ± 8.0 (18, 45) | 29 ± 6.8 (20, 51) | 0.17 |
| Weight change (kg) | 8.6 ± 4.4 (−2, 18) | 8.0 ± 4.7 (−2, 18) | 9.7 ± 4.0 (4.5, 17) | 0.29 |
| Fish consumption (servings/weekly) | 1.1 ± 1.1 (0, 4.1) | 1.1 ± 1.1 (0, 4.1) | 1.0 ± 1.1 (0, 4.1) | 0.98 |
| Fish consumption (servings/weekly) | ||||
| 0–0.9 | 17 (61) | 6 (67) | 11 (58) | 0.53 |
| 1–1.9 | 7 (25) | 1 (11) | 6 (32) | |
| ≥2 | 4 (14) | 2 (22) | 2 (11) | |
| Ethnicity/Race | ||||
| African-American | 5 (18) | 1 (11) | 4 (21) | 0.54 |
| Caucasian | 8 (29) | 4 (44) | 4 (21) | |
| Hispanic | 15 (54) | 4 (44) | 11 (58) | |
| Education | ||||
| ≥ High school | 18 (64) | 5 (56) | 13 (68) | 0.73 |
| College (some or completed) | 5 (18) | 2 (22) | 3 (16) | |
| Graduate school | 5 (18) | 2 (22) | 3 (16) | |
| Parity | ||||
| 0 | 4 (14) | 1 (11) | 3 (16) | 1.0 |
| 1 | 11 (39) | 4 (44) | 7 (37) | |
| 2–5 | 13 (46) | 4 (44) | 9 (47) | |
| BMI class | ||||
| Underweight or normal weight | 8 (29) | 4 (44) | 4 (21) | 0.53 |
| Overweight | 11 (39) | 3 (33) | 8 (42) | |
| Obese | 9 (32) | 2 (22) | 7 (37) | |
| IOM weight class | ||||
| Below | 10 (42) | 5 (63) | 5 (31) | 0.30 |
| Within | 11 (46) | 2 (25) | 9 (56) | |
| Above | 3 (13) | 1 (13) | 2 (13) | |
| Antibiotic treatment between first and second stool collection | ||||
| 0 times | 22 (85) | 5 (63) | 17 (94) | 0.07 |
| 1–4 times | 4 (15) | 3 (38) | 1 (5.6) | |
| Flu vaccine between first and second stool collection | ||||
| No | 21 (88) | 6 (86) | 15 (88) | 0.66 |
| Yes | 3 (13) | 1 (14) | 2 (12) | |
| Did you drink alcohol prior to this pregnancy | ||||
| No | 22 (79) | 6 (67) | 16 (84) | 0.28 |
| Yes | 6 (21) | 3 (33) | 3 (16) | |
| Were you ever a smoker? | ||||
| No | 23 (82) | 7 (78) | 16 (84) | 0.53 |
| Yes | 5 (18) | 2 (22) | 3 (16) | |
| Do you have amalgam fillings? | ||||
| No | 13 (54) | 3 (38) | 10 (63) | 0.39 |
| Yes | 11 (46) | 5 (63) | 6 (38) |
BMI (Body mass index), IOM (Institute of Medicine)
Sample size for weight change, IOM weight class, and amalgam fillings: all (n=24), supplemented (n=8), and unsupplemented (n=16). Sample size for antibiotic treatment: all (n=26), supplemented (n=8), and unsupplemented (n=18). Sample size for flu vaccine: all (n=24), supplemented (n=7), and unsupplemented (n=17).
p-value is for Wilcoxon rank sum test or Fisher's exact test.
Stool and blood THg and MeHg were strongly correlated (Table A4); however, collection dates differed on average by 3.7 weeks and 0.3 weeks for the first and second stool collections, respectively (Table A1). Weighted linear regression was used to investigate this further (Table A5). During early gestation, when the timeframe between stool and blood collection was wider, the effect estimates for gestational period increased by 1.1–7.5%, and the r-squared increased by 3–7%. During late gestation, the effect estimates decreased by 6.1% (for THg) and increased by 12% (for MeHg), while the r-squared increased by 1-10%. Overall, the results did not differ between weighted and unweighted linear regression, i.e., THg and MeHg concentrations in blood and stool were strongly positively correlated during both gestational periods. This may be attributed to the slow elimination rate of MeHg. The mean biological half-life of MeHg is approximately 70 days, although reported values vary from 35 to >150 days (Caito et al., 2018; Rand et al., 2016). Nearly all stool and blood samples were collected within 70 days of each other, while just one stool sample was collected 78 days after the blood sample (Table A1).
Meconium Hg.
In comparison with other studies reporting meconium THg concentrations, the median meconium THg concentration was similar to values reported in Austria (Gundacker et al., 2010), and Manila, the Philippines (Ostrea et al., 2002), and 7.3–23 times lower compared to values reported in Taiwan (Hsi et al., 2014; Jiang et al., 2010, 2014), Croatia (Knezovic et al., 2016), China (Peng et al., 2015), and Tagum, the Philippines (Ramirez et al., 2000) (Table A6). In five of the six latter studies, meconium was lyophilized, which would have increased THg concentrations. To the best of our knowledge, there were no other studies reporting meconium MeHg concentrations. Our results suggest the developing fetus metabolizes MeHg, some of which is excreted.
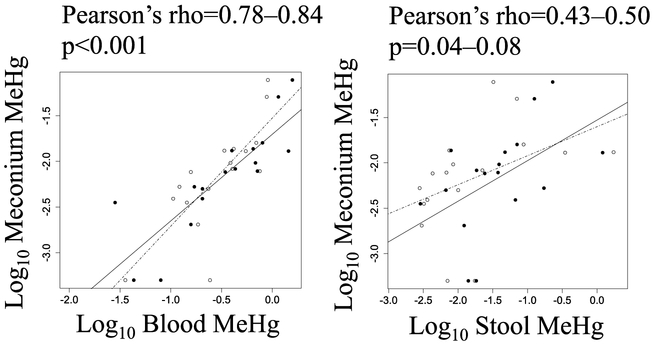
Meconium MeHg was more strongly correlated with maternal blood MeHg during both early and late gestation, compared to maternal stool MeHg (Table A4, Fig. 1). It was surprising that associations between meconium and maternal stool and blood were similar during early/late gestation. However, meconium is formed by the fetus as early as the 12th week of gestation (Ostrea et al., 2006), which would have preceded the collection dates for all stool samples and 18 (of 27) blood samples (Table A1).
Figure 1.
Bivariate associations between log10 meconium MeHg and a) log10 maternal blood MeHg and b) log10 maternal stool MeHg. Maternal stool and blood samples were collected during early (black circles, solid line) and late (open circles, broken line) gestation (n=17 paired samples at each time point). P-values are for Pearson's correlation.
Other covariates and Hg.
Among the covariates in Table 3, stool THg and stool IHg (early and late gestation) and meconium THg were positively correlated with maternal amalgam fillings (Wilcoxon rank-sum, p≤0.05 for all, n=23–24 for stool, n=13 for meconium). Stool MeHg (during late gestation) was associated with higher parity (Kruskal-Wallis, p=0.05, n=24). Blood THg and MeHg concentrations (during late gestation) were higher among mothers with one previous birth compared to mothers with no previous births and mothers with at least two previous births (Kruskal-Wallis, p<0.05, n=24). First trimester BMI (continuous) was inversely associated with meconium THg (Spearman's rho: −0.63, p<0.05, n=14). Blood THg and MeHg were significantly higher among mothers who drank alcohol before becoming pregnant compared to mothers who did not (Wilcoxon rank-sum, p<0.01, n=24 in total). There were no other significant associations between parameters in Table 3 and Hg biomarkers.
Vitamin D.
During early gestation (baseline), plasma 25(OH)D concentrations did not differ between supplemented and control mothers (Wilcoxon rank-sum test, p=0.35, n=22). Plasma 25(OH)D concentrations for all mothers increased between early and late gestation, due to prenatal supplements (Table 1). As expected, 25(OH)D concentrations were significantly higher during late gestation for supplemented mothers (4400 IU) compared to unsupplemented mothers (400 IU) (Wilcoxon rank-sum test, p=0.05, n=21).
Concentrations of plasma 25(OH)D were positively correlated with fish consumption during early gestation (Spearman's rho: 0.43, p<0.05, n=22) and borderline significant during late gestation (Spearman's rho: 0.42, p=0.06, n=21) (Table A4). Fish tissue is a source of vitamin D (Wu et al., 2013), which explains why positive associations between 25(OH)D and fish consumption were observed. Plasma 25(OH)D was positively correlated with stool MeHg, blood THg and blood MeHg during early gestation (Spearman's rho: 0.45–0.52, p<0.05, n=22–23), and was positively correlated with meconium MeHg during early and late gestation (Spearman's rho: 0.59–0.76, p<0.05, n=12–14) (Table A4). During early gestation, plasma 25(OH)D was significantly higher for Caucasian mothers compared to African-American and Hispanic mothers (ANOVA, p<0.05 for both, n=22); however, differences were attenuated during late gestation (ANOVA, p=0.81–1.0 for all, n=21). During late gestation, mothers gaining excessive weight had significantly higher 25(OH)D concentrations compared to mothers gaining normal weight (ANOVA, p<0.05, n=21). All mothers gaining excessive weight were Caucasian or Hispanic, which explains this association. Four mothers that received antibiotic treatment between early and late gestation had significantly higher 25(OH)D concentrations during late gestation (two-tailed t-test, p<0.05, n=21).
Gut microbiota diversity.
After pre-processing, an average of 72,012 reads (range 16,719–750,420) were identified across 2400 ASVs in all samples. During early and late gestation, five phyla comprised on average 92% of gut microbiota: Firmicutes (early: 48%, late: 51%), Bacteroidetes (early: 33%, late: 26%), Actinobacteria (early: 4.1%, late: 6.7%), Proteobacteria (early: 3.5%, late: 4.5%), and Verrucomicrobia (early: 4.0%, late: 3.8%). Using a mixed model, Actinobacteria significantly increased between early and late gestation (when log10-transformed) (p<0.05, n=52).
Alpha diversity was significantly lower during late gestation compared to early gestation for the observed number of taxa and for Fisher's index, when using a mixed model (p<0.05 for both, n=52) (Fig. A1). During early gestation, alpha diversity was higher for mothers with at least two previous births compared to nulliparous mothers (for the observed number of taxa, Shannon Index, and Fisher's index) (ANOVA, p<0.05 for all, n=28). However, differences in parity were attenuated during late gestation using all three indices (ANOVA, p>0.85 for all, n=24). Alpha diversity was significantly higher for mothers who attended college compared to mothers who attended graduate school (late gestation), when using the observed number of species and Fisher's index (ANOVA, p<0.05, n=24). Alpha diversity was higher for mothers with amalgam fillings, using the observed number of species (late gestation) and Fisher's index (early and late gestation) (two-tailed t-test, p<0.05, n=24–28).
Beta diversity patterns differed significantly between early and late gestation, when stratified by mothers (Multivariate Welch Tw2 test on Jansen-Shannon divergence distances, p=0.03, n=52) (Fig. 2). Using pairwise associations, beta diversity patterns differed by race/ethnicity, including Caucasian and Hispanic mothers (early and late gestation), African-American and Hispanic mothers (early gestation), and Caucasian and African-American mothers (late gestation) (Tw2 test, p≤0.05, n=24–28). Similar to alpha diversity, beta diversity patterns differed between mothers with at least two previous births compared to nulliparous mothers during early gestation only (Tw2 test, p<0.05, n=28), while pairwise differences were no longer significant during late gestation (Tw2 test, p=0.61, n=24). Beta diversity also differed between mothers with amalgam fillings compared to no amalgams (early gestation only) (Tw2 test, p<0.05, n=28).
Figure 2.
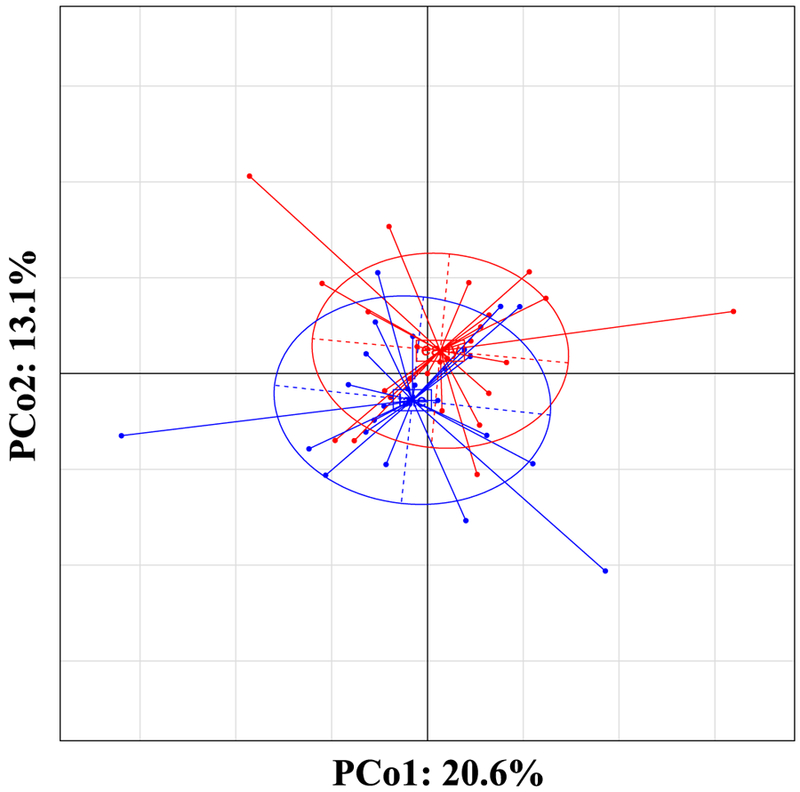
Within class Principal Coordinate Analysis (PCoA) showing community structure (β-diversity) assessed by Jensen-Shannon diversity distances controlling for subject effect. Composition of bacterial communities during late gestation (n=24) were significantly different compared to early gestation (n=28) when accounting for the repeated measures of same subjects (Tw2 test, p=0.03).
Hg biomarkers (stool, blood, and hair) were dichotomized at the median, and alpha and beta diversity values were compared between high/low Hg concentrations, during early and late gestation (Table 4). The overall composition of the microbiome differed significantly between high/low stool MeHg concentrations and high/low blood MeHg concentrations during early gestation (Tw2 test, p=0.01 and p=0.05, respectively); however, these differences were attenuated during late gestation (p=0.86 and p=0.25, respectively). There were no other differences between the composition of the microbiota in correlation to THg, MeHg, and IHg concentrations in stool, blood, and hair during early or late gestation. For the same dichotomized Hg parameters, alpha diversity patterns did not differ using all three indices (Wilcoxon rank-sum test, p=0.10–0.95). These results support potential differences in metabolism between Hg species (THg, MeHg, and IHg), and between early/late gestation.
Table 4.
Comparison of beta diversity patterns between early/late gestation using dichotomized mercury biomarkers, and multivariate Welch Tw2 test on Jansen-Shannon divergence distances.
| Early | Late | |||
|---|---|---|---|---|
| Tw2 statistic | P-value | Tw2 statistic | P-value | |
| Blood (μg/L) | N=28 | N=24 | ||
| THg | 1.00 | 0.45 | 0.86 | 0.67 |
| MeHg | 1.49 | 0.05* | 1.15 | 0.25 |
| IHg | 1.00 | 0.45 | 1.13 | 0.29 |
| Stool (ng/g) | N=28 | N=28 | ||
| THg | 1.30 | 0.13 | 0.73 | 0.85 |
| MeHg | 1.77 | 0.01** | 0.73 | 0.86 |
| IHg | 1.30 | 0.13 | 0.73 | 0.85 |
| Hair (μg/g) | N=24 | N=24 | ||
| THg | 1.21 | 0.21 | 1.03 | 0.41 |
IHg (THg - MeHg), MeHg (methylmercury), THg (total mercury),
p≤0.05
p≤0.01
As noted in the Methods, 20 (of 28) mothers were initially enrolled in a vitamin D supplementation study, including nine mothers in the treatment group (n=8 mothers during late gestation). When alpha and beta diversity patterns were compared by supplementation status (late gestation only), we did not observe differences between treatment and control groups (alpha diversity, Wilcoxon rank sum test, p=0.71–0.98 for all three indices, n=24; beta diversity, Tw2 test, p=0.34, n=24). Using a mixed model, alpha diversity decreased over time (using the observed number of species and Fisher's index) (p<0.05 for both, n=52), while the effect estimate for vitamin D supplementation status was non-significant (p=0.20–0.30, n=52). In addition, associations between 25(OH)D concentrations and alpha diversity indices were non-significant during early gestation [Spearman's rho range: (−0.19, 0.13), p=0.39–0.56, n=22] and late gestation [Spearman's rho range: (−0.12, 0.07), p=0.60–0.88, n=21]. Therefore, observed changes in diversity patterns between early and late gestation were not likely attributed to vitamin D supplementation. It is possible a larger sample size may yield a different result.
Associations between gut microbiota and Hg parameters.
During early and late gestation, pairwise associations were assessed between seven Hg parameters and the five most abundant phyla (Fig. 3). Hg parameters included 1) fish meals (servings/week), 2) stool MeHg, 3) stool IHg, 4) stool IHg (% of THg), 5) meconium MeHg, 6) blood MeHg, and 7) blood IHg. Stool IHg was negatively correlated with the phylum Bacteroidetes (early gestation) (Spearman's rho: −0.47, p<0.05, n=28). However, this association was no longer significant during late gestation, and the direction of correlation changed from negative to positive (Spearman's rho: 0.20, p=0.36, n=24). In total, 54% (18/35) of the correlations between phyla and Hg parameters changed direction of association between early and late gestation.
Figure 3.
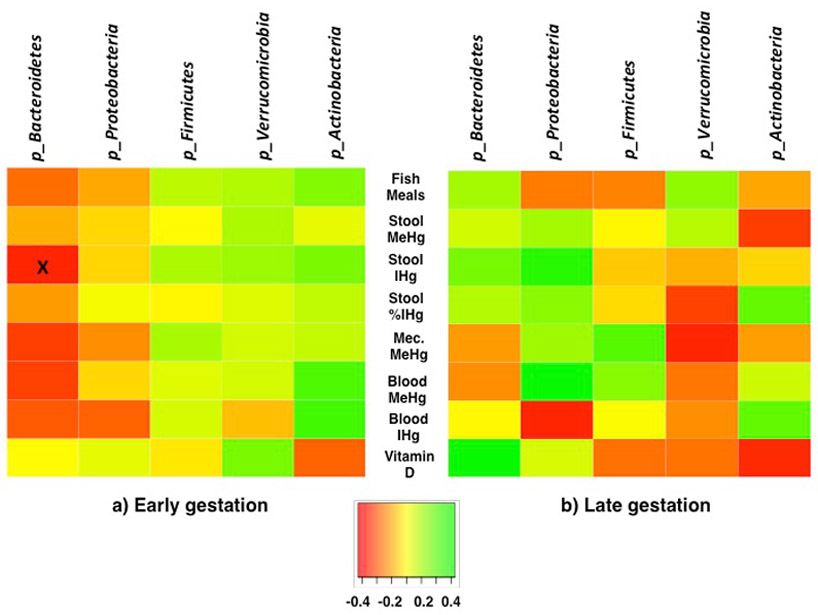
Spearman's correlation between gut microbiota phyla, and fish meals (meals/weekly), stool methylmercury (MeHg) (ng/g), stool inorganic mercury (IHg) (ng/g), stool IHg (% of THg), meconium (Mec) MeHg, blood MeHg (μg/L), blood IHg (μg/L), and plasma vitamin D (25-hydroxy-vitamin D) (ng/mL), during a) early gestation and b) late gestation. During early gestation n=28, except vitamin D (n=22), and meconium MeHg (n=17). During late gestation n=24, except vitamin D (n=21), and meconium (n=17). Note that fish meals and meconium did not differ between early and late gestation, whereas biomarkers for stool and blood MeHg and IHg, and plasma vitamin D were analyzed at each time point. "x" p<0.05.
The same Hg parameters were compared with 23 taxa (at the genus or family level, average relative abundance >1%) (Fig. 4). During early gestation, significant associations (positive and negative) were observed between Hg parameters and the following six taxa: Lachnoclostridium, Collinsella, Ruminococcaceae_ucg002, Ruminococcaceae_ucg013, Prevotella_9, and Streptococcus (Spearman's rho range: ∣0.39–0.60∣, p<0.05). During late gestation, significant positive associations were observed between Hg parameters and the following five taxa: Faecalibacterium, Lachnospiraceae_nk4a136, Megasphaera, Ruminococcaceae_ucg013, and Parabacteroides (Spearman's rho range: 0.44–0.59, p<0.05). The direction of correlation changed for 43% (69/161) of the associations between gut microbiota and Hg parameters, although a majority of these trends were non-significant. Overall, the percentage of correlations that changed direction of association was higher for stool (IHg and MeHg), compared to blood (IHg and MeHg) (52% versus 33%).
Figure 4.
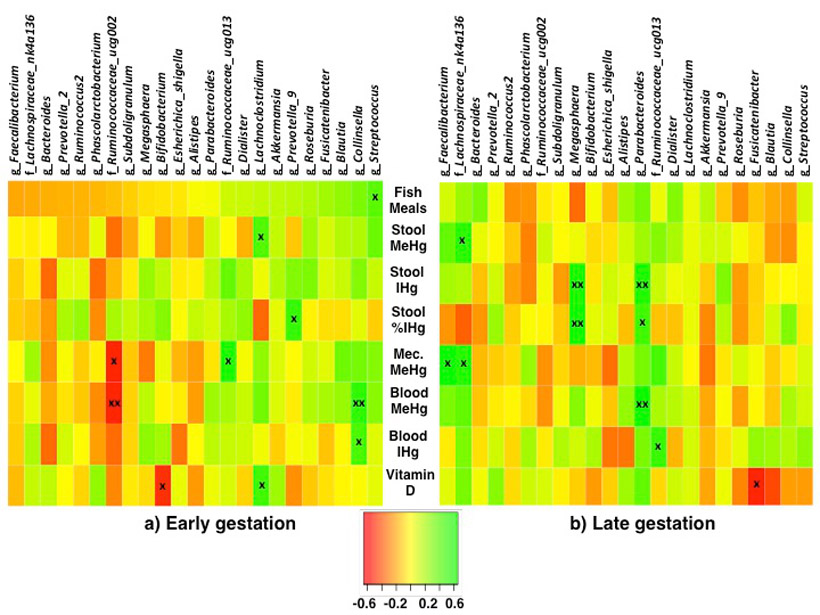
Spearman's correlation between gut microbiota taxa (family or genus level, >1% average abundance), and fish meals (meals/weekly), stool methylmercury (MeHg) (ng/g), stool inorganic mercury (IHg) (ng/g), stool IHg (% of THg), meconium (Mec) MeHg, blood MeHg (μg/L), blood IHg (μg/L), and plasma vitamin D (25-hydroxy-vitamin D) (ng/mL), during a) early gestation and b) late gestation. During early gestation n=28, except vitamin D (n=22), and meconium MeHg (n=17). During late gestation n=24, except vitamin D (n=21), and meconium (n=17). Note that fish meals and meconium did not differ between early and late gestation, whereas biomarkers for stool and blood MeHg and IHg, and plasma vitamin D were analyzed at each time point. "x" p<0.05, "xx" p<0.01.
One question is whether the changes in associations between early/late gestation reflected changes in taxa relative abundance, Hg parameters, or both. Using a mixed model, the relative abundance for three (of 23) taxa increased or decreased between early and late gestation, including two taxa significantly correlated with Hg biomarkers (when taxa were log10-transformed) (Ruminococcaceae_ucg002 and Megasphaera) (p≥0.05, n=52). As noted in Table 2, stool MeHg, stool IHg, and blood IHg decreased significantly between early/late gestation, while stool IHg (% of THg) and blood MeHg did not differ between early/late gestation. In addition, meconium was collected once, and mothers were queried just one time concerning fish consumption, and therefore, there were no differences between early/late gestation.
Changes in nine (of 17) associations between early/late gestation possibly reflected shifts in the relative abundance of the taxa, or a decrease in the concentration of the Hg parameter, or both. The exceptions included the associations between Ruminococcaceae_ucg013 and meconium MeHg (early gestation), Prevotella_9 and stool IHg (% of THg) (early gestation), Collinsella and blood MeHg (early gestation), Streptococcus and fish meals (early gestation), Faecalibacterium and meconium MeHg (late gestation), Lachnospiraceae_nk4a136 and meconium MeHg (late gestation), Parabacteroides and blood MeHg (late gestation), and Parabacteroides and stool IHg (% of THg) (late gestation). Of these seven taxa, Ruminococcaceae_ucg013, Collinsella, Faecalibacterium, Prevotella_9, and Parabacteroides did not differ between early/late gestation (when log10-transformed, using a mixed model) (p=0.46–0.82, n=52), while Streptococcus and Lachnospiraceae_nk4a136 differed during late gestation, but not significantly (when log10-transformed, using a mixed model) (p=0.06 for both, n=52). Results suggested in six cases (or eight, including Streptococcus and Lachnospiraceae_nk4a136), changes in Spearman's correlation between early/late gestation potentially reflected a random shift in the gut microbiota, which altered the associations with these Hg parameters.
Gut microbiota and vitamin D.
Pairwise associations were also assessed between plasma 25(OH)D concentrations and the relative abundances of five phyla (Fig. 3). There were no significant associations between phyla and 25(OH)D concentrations, and just one (20%) association changed the direction of correlation between early and late gestation.
When compared with 23 taxa (Fig. 4), during early gestation, 25(OH)D was significantly inversely associated with Bifidobacterium and positively correlated with Lachnoclostridium (Spearman's rho = −0.49 and 0.45, respectively, p<0.05 for both, n=22). During late gestation, 25(OH)D was inversely associated with Fusicatenibacter (Spearman's rho: −0.51, p<0.05, n=21). Of the three taxa, only Bifidobacterium significantly changed between early and late gestation (increasing in relative abundance) (when log10-transformed) (p<0.01, n=52). Similar to Hg parameters, 10 (of 23) (43%) pairwise associations reversed direction of association between early and late gestation.
DISCUSSION
Longitudinal trends in Hg biomarkers.
Most Hg biomarkers (stool and blood) significantly decreased between early and late gestation. On average, blood MeHg also decreased; however, the trend was non-significant. Differences in blood Hg between early/late gestation were not entirely due to hemodilution. Mothers completed the FFQ just one time (after the first stool collection), and therefore it is uncertain whether fish consumption was reduced. However, maternal diets do not change much from early to late pregnancy, including fish consumption (Crozier et al., 2009). In addition, blood MeHg would have likely significantly decreased if mothers reduced their fish consumption.
Longitudinal trends in gut microbiota.
Gut microbiota is considered stable over time (David et al., 2014; Faith et al., 2013). However, during pregnancy the structure and function of maternal gut microbial communities may change, potentially reflecting the energy and nutritional requirements of the developing fetus (Koren et al., 2012). In a study of 91 Finnish pregnant mothers, changes in gut microbiota were reported between the first and third trimesters, including greater diversity, reduced species richness, and increases in the phyla, Proteobacteria and Actinobacteria (Koren et al., 2012). Conversely, among 40 pregnant mothers in California, USA, no changes were observed in gut microbiota (collected weekly) from early to late pregnancy, including alpha diversity, beta diversity, and taxonomic composition of communities (DiGiulio et al., 2015). Likewise, in stool samples collected from 86 Norwegian mothers during early and late gestation, maternal microbial composition was stable (Avershina et al., 2014). Differences between studies investigating longitudinal trends during pregnancy have been attributed to diet, or BMI, or other environmental factors, which influence the gut microbiome (Walker et al., 2017).
In the present study, alpha diversity (using two indices) and beta diversity patterns (stratified by mothers) differed between early and late gestation, including greater abundance of the phylum, Actinobacteria, similar to Koren et al. (2012). These trends persisted after controlling for maternal vitamin D supplementation status. More importantly, associations between intestinal taxa and Hg biomarkers also changed between early/late gestation. Most Hg biomarkers decreased between early/late gestation, and these declines would presumably not change the direction of association with intestinal taxa. However, 45% of the associations between gut taxa (genus or family level) and Hg biomarkers reversed direction. These results are suggestive that remodeling of the host microbiota during pregnancy potentially contributed to changes in associations with Hg biomarkers.
Gut microbiota and Hg biomarkers.
It is possible the associations between Hg biomarkers and gut taxa may reflect contributions of gut microbiota to MeHg metabolism. As noted above, stool IHg (% of THg) is positively associated with the MeHg elimination rate (Caito et al., 2018; Rand et al., 2016). During early gestation, stool IHg (% of THg) was significantly positively correlated with the relative abundance of Prevotella_9. Prevotella spp. are associated with a high-fiber, plant-based diet (Jeffrey and O'Toole, 2013). For example, Prevotella_9 includes the species, Prevotella copri; using an in vitro model, the addition of insoluble wheat bran particles was associated with an enrichment of P. copri (De Paepe et al., 2018). Among 39 healthy adults, ingestion of barley bread improved glucose metabolism and was associated with enrichment of P. copri (Kovatcheva-Datchary et al., 2015). Our results suggest that mothers ingesting a high-fiber plant-based diet may also have significantly higher MeHg elimination rates. This is consistent with earlier Hg studies, which reported high-fiber diets (in animals) and higher fruit consumption (in humans) were associated with lower concentrations of MeHg in biomarkers and tissues (Passos et al., 2007; Rowland et al, 1986). However, the association between stool IHg (% of THg) and Prevotella_9 was attenuated during late gestation, suggesting this potential protective effect was limited to early gestation.
During early gestation, blood MeHg and meconium MeHg were negatively correlated with members of the Ruminococcaceae_ucg002 family. In the human gut, several strains of Ruminococcacea produce butyrate, which serves as an energy source for coloncytes, and contributes to maintaining gut homeostasis (Vital et al., 2014). Ruminococcaceae is also associated with fiber-degradation (Gamage et al., 2017). These results support the hypothesis that a fiber-rich diet may reduce prenatal MeHg exposure (Rowland et al., 1986); however, associations between Ruminococcaceae_ucg002 and MeHg biomarkers were attenuated during late gestation.
During late gestation, stool IHg (continuous), stool IHg (% of THg), and blood MeHg were significantly positively correlated with the relative abundance of Parabacteroides. Other researchers reported Parabacteroides spp. were enriched following an increase in monounsaturated fats, although the function of Parabacteroides was unknown (Pu et al., 2016). It is unclear why all three Hg parameters had strong positive associations with Parabacteroides, given that stool IHg (% of THg) is associated with the MeHg excretion rate, while blood MeHg suggests higher MeHg exposure.
Gut microbiota and vitamin D.
It is notable that the genus Bifidobacterium was not strongly correlated with Hg parameters during early or late gestation. In the present study, Bifidobacterium was significantly inversely correlated with 25(OH)D during early gestation. Similarly, an inverse association between serum 25(OH)D and the relative abundance of Bifidobacterium in gut microbiota was observed among 150 non-pregnant adults, in both unadjusted models and models adjusted for age, sex, BMI, and season (Luthold et al., 2017). Among 913 one-month old infants, there was a statistically significant inverse association between infant gut Bifidobacterium spp. and maternal plasma 25(OH)D quintiles (assessed at 36 weeks gestation) (Talsness et al., 2017).
During the first few months of life, Bifidobacterium is one of the dominant taxa in the infant gut, which is attributed to its ability to digest breast milk oligosaccharides (LaTuga et al., 2014). In a comparison of maternal feces, breast milk, and neonatal feces (feces collected 10 days postpartum), Bifidobacterium was one of the few taxa observed in all three matrices, suggesting vertical transfer of maternal gut microbiota to the neonate gut (Jost et al., 2014). In the present study, during late gestation, the inverse association between Bifidobacterium and maternal plasma 25(OH)D levels was attenuated, which would be considered beneficial to neonatal health.
Limitations.
This study has several strengths; however, there are some limitations to note. First, primers targeted the V1/V2 variable regions of the 16S rRNA gene, which did not amplify Archaea (Wear et al., 2018). Therefore it was not possible to detect one commensal methanogen (M. luminyensis), which has the potential to methylate IHg (Parks et al., 2013). However, the contribution of M. luminyensis to stool MeHg was non-detectable in several studies (Podar et al., 2015; Rothenberg et al., 2016), suggesting minimal impact on our results. Also, we do not anticipate the choice of the primer to impact the main results of the study, i.e., longitudinal trends in microbe-biomarker associations. Second, associations between gut taxa and Hg biomarkers were non-significant at a FDR of 20%; however modest associations (in Figs. 3 & 4) were suggestive that correlations between Hg biomarkers and gut taxa differed between early/late gestation, and should be further investigated. Correlations were observed between Hg biomarkers and gut taxa, which are frequently associated with a high fiber, plant-based diet. However, we did not have detailed dietary data (fiber intake, etc.) to test this hypothesis, which should be confirmed in future studies. Strengths of the study included collection of maternal biomarkers (stool and blood) at two time points, and analysis of THg and MeHg concentrations in maternal blood, maternal stool, and meconium. Many studies rely on concentrations of THg as an estimate of MeHg; however, our results demonstrate that associations between Hg parameters and gut microbiota will be missed if MeHg is not analyzed.
Conclusions.
During pregnancy, associations between MeHg biomarkers and gut taxa differed between early and late gestation, which may have implications for reducing fetal MeHg exposure. In animal and human studies, co-consumption of certain foods (e.g., fruit, fiber, or yogurt) was associated with lower MeHg exposure (Bisanz et al., 2014; Passos et al., 2007; Rowland et al., 1986). However, our results suggest dietary interventions to reduce prenatal MeHg exposure may be limited to certain windows, thus hindering their effectiveness. Early gestation may be a more effective window; however, fetal brain growth is more rapid during the third trimester. More studies are needed to optimize the timing of dietary interventions to reduce prenatal MeHg exposure.
Supplementary Material
Highlights.
Pregnant mothers donated blood and stool samples during early/late gestation.
Total mercury and methylmercury (MeHg) were determined in both biomarkers.
Gut microbiota community structure was determined using 16S rRNA gene profiling.
Within- and between-person diversity patterns differed between early/late gestation.
Associations between gut microbiota and MeHg biomarkers differed longitudinally.
ACKNOWLEDGEMENTS
The authors with to thank G. Carly McCalla and Allison M. Emmons for their assistance in the lab.
FUNDING
This study was supported in part by grants to S. Rothenberg from the U.S. National Institute of Environmental Health Sciences of the U.S. National Institute of Health (R21 ES026412), the U.S. National Institute of Health Loan Replacement Program (L30 ES023165), and the University of South Carolina Office of Research. This study was also supported in part by a grant to C. Wagner from the W.K. Kellogg Foundation. B. Hamidi and A.V. Alekseyenko were supported in part by a grant to A.V. Alekseyenko from the U.S. National Library of Medicine of the U.S. National Institute of Health (R01 LM012517). M.A. Azcarate-Peril was supported in part by the U.S. National Institute of Diabetes and Digestive and Kidney Diseases of Health of the U.S. National Institute of Health (P30 DK34987). The content is solely the responsibility of the authors and does not necessarily represent the official views of the study sponsors. The study sponsors did not play a role in the study design, in the collection, analysis, and interpretation of data, in the writing of the report, or in the decision to submit the paper for publication. There are no conflicts to declare.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
25(OH)D (25-hydroxy-vitamin D), (ASV) amplicon sequence variants, ANOVA (one way analysis of variance), BMI (body mass index), BrCl (bromine monochloride), CVAFS (cold vapor atomic fluorescence spectrometry), CH2Cl2 (dichloromethane), dbGaP (U.S. National Institutes of Health Database for Genotypes and Phenotypes), FDR (False Discovery Rate), FFQ (food frequency questionnaire), Hg (mercury), HCl (hydrochloric acid), HNO3 (nitric acid), H2SO4 (sulfuric acid), MeHg (methylmercury), MUSC (Medical University of South Carolina), QA/QC (quality assurance/quality control), SRA (short-read archives), THg (total mercury), USEPA (U.S. Environmental Protection Agency)
REFERENCES
- Alekseyenko AV. 2016. Multivariate Welch t-test on distances. Bioinformatics 32:3552–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allali I, Arnold JW, Roach J, Cadenas MB, Butz N, Hassan HM, Koci M. Ballou A, Mendoza M, Ali R, Azcarate-Peril MA. 2017. A comparison of sequencing platforms and bioinformatics pipelines for compositional analysis of the gut microbiome. BMC Microbiol 17:194 10.1186/s12866-017-1101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avershina E, Storro O, Oien T, Johnsen R, Rope P, Rudi K. 2014. Major faecal microbiota shifts in composition and diversity with age in a geographically restricted cohort of mothers and their children. FEMS Microbiol 87:280–290. [DOI] [PubMed] [Google Scholar]
- Bisanz JE, Enos MK, Mwanga JR, Changalucha J, Burton JP, Gloor GB, Reid G. 2014. Randomized open-label pilot study of the influence of probiotics and the gut microbiome on toxic metal levels in Tanzanian pregnant women and school children. mBio 5, doi: 10.1128/mBio.01580-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom NS, Colman JA, Barber L. 1997. Artifact formation of methyl mercury during aqueous distillation and alternative techniques for the extraction of methyl mercury from environmental samples. Fresenius J. Anal. Chem. 358:371–377. [Google Scholar]
- Breton J, Massart S, Vandamme P, De Brandt E, Pot B, Foligne B. 2013. Ecotoxicology inside the gut: impact of heavy metals on the mouse microbiome. BMC Pharmacol Toxicol 14: doi: 10.1186/2050-6511-14-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caito SW, Jackson BP, Punshon T, Scrimale T, Grier A, Gill SR, Love TM, Watson GE, van Wijngaarden E, Rand MD. 2018. Variation in methylmercury metabolism and elimination status in humans following fish consumption. Toxicol Sci 161:443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: High resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Holmes SP. 2017. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J 11:2639–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson TW, Magos L. 2006. The toxicology of mercury and its chemical compounds. Crit Rev Toxicol 36:609–662. [DOI] [PubMed] [Google Scholar]
- Crozier SR, Robinson SM, Godfrey KM, Cooper C, Inskip HM. 2009. Women's dietary patterns change little from before to during pregnancy. J Nutr 139:1956–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, Erdman SE, Alm EJ. 2014. Host lifestyle affects human microbiota on daily timescales. Genome Biol 15:R89, 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paepe K, Verspreet J, Verbeke K, Raes J, Courtin CM, Van de Wiele T. 2018. Introducing insoluble wheat bran as a gut microbiota niche in an in vitro dynamic gut model stimulates propionate and butyrate production and induces colon region specific shifts in the luminal and mucosal microbial community. Environ Microbiol 20:3406–3426. [DOI] [PubMed] [Google Scholar]
- DiGiulio DB, Callahan BJ, McMurdie PJ, Costello EK, Lyell DJ, Robaczewska A, Sun CL, Goltsman DSA, Wong RJ, Shaw G, Stevenson DK, Holmes SP, Relman DA. 2015. Temporal and spatial variation of the human microbiota during pregnancy. Proc Natl Acad Sci USA 112:11060–11065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Shulzhenko N, Lemaitre J, Greer RL, Peremyslova K, Quamruzzaman Q, Rahman M, Sharif O, Hasan I, Joya SA, Golam M, Christiani DC, Morgun A, Kile ML. 2017. Arsenic exposure and intestinal microbiota in children from Sirajdikhan, Bangladesh. Plos One 12: 10.1371/journal.pone.0188487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue A, Wagner CL, Burch JB, Rothenberg SE. 2018. Blood total mercury and methylmercury among pregnant mothers in Charleston, South Carolina, USA. J Expo Sci Environ Epidemiol 28:494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI. 2013. The long-term stability of the human gut microbiota. Science 341:1237439 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamage HKAH, Tetu SG, Chong RWW, Ashton J, Packer NH, Paulsen IT. 2017. Cereal products derived from wheat, sorghum, rice and oats alter the infant gut microbiota in vitro. Sci Rep 7: doi: 10.1038/s41598-017-14707-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundacker C, Frolich S, Graf-Rohrmeister K, Eibenberger B, Jessenig V, Gicic D, Prinz S, Wittmann KJ, Zeisler H, Vallant B, Pollak A, Husslein P. 2010. Perinatal lead and mercury exposure in Austria. Sci Total Environ 408:5744–5749. [DOI] [PubMed] [Google Scholar]
- Hollis BW, Kamerud JQ, Selvaag SR, Lorenz JD, Napoli JL. 1993. Determination of vitamin D status by radioimmunoassay with and 125I-labeled trcer. Clin Chem 39:529–533. [PubMed] [Google Scholar]
- Hong C, Yu X, Liu J, Cheng Y, Rothenberg SE. 2016. Low-level methylmercury exposure through rice ingestion in a cohort of pregnant mothers in rural China. Environ Res 150:519–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsi HC, Jiang CB, Yang TH, Chien LC. 2014. The neurological effects of prenatal and postnatal mercury/methylmercury exposure on three-year-old children in Taiwan. Chemosphere 100:71–76. [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project. 2010. Manual of Procedures, Core Microbiome Sampling, Protocol A, Version 12.0, July 29, 2010. https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/document.cgi?study_id=phs000228.v3.p1&phd=3190. Last accessed November 24, 2018.
- Hytten F 1985. Blood volume changes in normal pregnancy. Clin Haematol 14:601–612. [PubMed] [Google Scholar]
- Institute of Medicine. 2009. Weight gain during pregnancy: reexamining the guidelines. http://www.nationalacademies.org/hmd/~/media/Files/Report%20Files/2009/Weight-Gain-During-Pregnancy-Reexamining-the-Guidelines/Report%20Brief%20-%20Weight%20Gain%20During%20Pregnancy.pdf. Last accessed November 24, 2018.
- Ishihara N 2000. Excretion of methyl mercury in human feces. Arch Environ Health 55:44–47. [DOI] [PubMed] [Google Scholar]
- Jeffrey IB, O'Toole PW. 2013. Diet-microbiota interactions and their implications for healthy living. Nutrients 5:234–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang CB, Yeh CY, Lee HC, Chen MJ, Hung FY, Fang SS, Chien LC. 2010. Mercury concentration in meconium and risk assessment of fish consumption among pregnant women in Taiwan. Sci Total Environ 408:518–523. [DOI] [PubMed] [Google Scholar]
- Jiang CB, Hsi HC, Fan CH, Chien LC. 2014. Fetal exposure to environmental neurotoxins in Taiwan. PLoS ONE 9(10):e109984 10.1371/journal.pone.0109984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost T, Lacroix C, Braegger CP, Rochat F, Chassard C. 2014. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ Microbiol 16:2891–2904. [DOI] [PubMed] [Google Scholar]
- Knezovic Z, Trgo M, Sutlovic D. 2016. Monitoring mercury pollution through bioaccumulation in meconium. Process Saf Environ Prot 101:2–8. [Google Scholar]
- Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Backhed HK, Gonzalez A, Werner JJ, Angenent LT, Knight R, Backhed F, Isolauri E, Salminen S, Ley RE. 2012. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 150:470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, Hallen A, Martens E, Bjorck I, Backhed F. 2015. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab 22:971–982. [DOI] [PubMed] [Google Scholar]
- LaTuga MS, Stuebe A, Seed PC. 2014. A review of the source and function of microbiota in breast milk. Semin Reprod Med 32:68–73. [DOI] [PubMed] [Google Scholar]
- Liang L, Evens C, Lazoff S, Woods JS, Cernichiari E, Horvat M, Martin MD, DeRouen T. 2000. Determination of methyl mercury in whole blood by ethylation-GC-CVAFS after alkaline digestion-solvent extraction. J Anal Toxicol 24:328–332. [DOI] [PubMed] [Google Scholar]
- Loussouarn G, El Rawadi C, Genain G. 2005. Diversity of hair growth profiles. Intl J Dermatol 44(Suppl):6–9. [DOI] [PubMed] [Google Scholar]
- Lu K, Abo RP, Schlieper KA, Graffam ME, Levine S, Wishnok JS, Swenberg JA, Tannenbaum SR, Fox JG. 2014. Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: an integrated metagenomics and metabolomics analysis. Environ Health Perspect 122:284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthold RV, Fernandes GR, Franco-de-Moraea AC, Folchetti LGD, Ferreira SRG. 2017. Gut microbiota interactions and the immunomodulatory role of vitamin D in normal individuals. Metab Clin Exp 69:76–86. [DOI] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. 2013. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrea EM Jr., Morales V, Ngoumgna E, Prescilla R, Tan E, Hernandez E, Ramirez GB, Cifra HL, Manlapaz ML. 2002. Prevalence of fetal exposure to environmental toxins as determined by meconium analysis. Neurotoxicology 23:329–339. [DOI] [PubMed] [Google Scholar]
- Ostrea EM Jr., Bielawski DM, Posecion NC Jr.. 2006. Meconium analysis to detect fetal exposure to neurotoxicants. Arch Dis Child 91:628–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks JM, Johs A, Podar M, Bridou R, Hurt RA, Smith SD, Tomanicek SJ, Qian Y, Brown SD, Brandt CC, Palumbo AV, Smith JC, Wall JD, Elias DA, Liang LY. 2013. The genetic basis for bacterial mercury methylation. Science 339:1332–1335. [DOI] [PubMed] [Google Scholar]
- Passos CJ, Mergler D, Fillion M, Lemire M, Mertens F, Guimaraes JRD, Philibert A. 2007. Epidemiologic confirmation that fruit consumption influences mercury exposure in riparian communities in the Brazilian Amazon Environ Res 105:183–193. [DOI] [PubMed] [Google Scholar]
- Peng S, Liu L, Zhang X, Heinrich J, Zhang J, Schramm KW, Huang Q, Tian M, Eqani SAMAS, Shen H. 2015. A nested case-control study indicating heavy metal residues in meconium associate with maternal gestational diabetes mellitus risk. Environ Health 14:19 10.1186/s12940-015-0004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar M, Gilmour CC, Brandt CC, Soren A, Brown SD, Crable BR, Palumbo AV, Somenahally AC, Elias DA. 2015. Global prevalence and distribution of genes and microorganisms involved in mercury methylation. Sci Adv 9:e1500675 10.1126/sciadv.1500675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu S, Khazanehei H, Jones PJ, Khafipour E. 2016. Interactions between obesity status and dietary intake of monosaturated and polyunsaturated oils on human gut microbiome profiles in the canola oil multicenter intervention trial (COMIT). Front Microbiol 7:1612 10.3389/fmicb.2016.01612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2017. R: A language and environment for statistical computing. R Foundation for Statistical Computing:Vienna, Austria: https://www.R-project.org/. Last accessed November 24, 2018. [Google Scholar]
- Ramirez GB, Cruz MCVC, Pagulayan O, Ostrea E, Dalisay C. 2000. The Tagum Study I: Analysis and clinical correlates of mercury in maternal and cord blood, breast milk, meconium, and infants' hair. Pediatrics 106:774–781. [DOI] [PubMed] [Google Scholar]
- Rand MD, Vorojeikina D, van Wijngaarden E, Jackson BP, Scrimale T, Zareba G, Love TM, Myers GJ, Watson GE. 2016. Methods for individualized determination of methylmercury elimination rate and de-methylation status in humans following fish consumption. Toxicol Sci 149:385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg SE, Keiser S, Ajami NJ, Wong MC, Gesell J, Petrosino JF, Johs A. 2016. The role of gut microbiota in fetal methylmercury exposure: Insights from a pilot study. Toxicol Lett 242:60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg SE, Yin R, Hurley JP, Krabbenhoft DP, Ismawati Y, Hong C, Donohue A. 2017. Stable mercury isotopes in polished rice (Oryza sativa L.) and hair from rice consumers. Environ Sci Technol. 51:6480–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland IR, Robinson RD, Doherty RA. 1984. Effects of diet on mercury metabolism and excretion in mice given methylmercury: Role of gut flora. Arch Environ Health 39:401–408. [DOI] [PubMed] [Google Scholar]
- Rowland IR, Mallett AK, Flynn J, Hargreaves RJ. 1986. The effect of various dietary fibres on tissue concentration and chemical form of mercury after methylmercury exposure in mice. Arch Toxicol 59:94–98. [DOI] [PubMed] [Google Scholar]
- Schafer JL. 1997. Analysis of incomplete multivariate data. Chapman & Hall: London. [Google Scholar]
- Seko Y, Miura T, Takahashi M, Koyama T. 1981. Methyl mercury decomposition in mice treated with antibiotics. Acta Pharmacol Toxicol 49:259–265. [DOI] [PubMed] [Google Scholar]
- Talsness CE, Penders J, Jansen EHJM, Damoiseaux J, Thijs C, Mommers M. 2017. Influence of vitamin D on key bacterial taxa in infant microbiota in the KOALA Birth Cohort study. PLoS ONE 12:e0188011 10.1371/journal.pone.0188011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Environmental Protection Agency (USEPA). 2001a. Appendix to Method 1631 Total mercury in tissue, sludge, sediment, and soil by acid digestion and BrCl Oxidation. EPA-821-R-01-013. Office of Water, Washington D.C. [Google Scholar]
- U.S. Environmental Protection Agency (USEPA). 2001b. Method 1630, Methyl mercury in water by distillation, aqueous ethylation, purge and trap, and cold vapor atomic spectrometry. Washington, D.C. [Google Scholar]
- U.S. Environmental Protection Agency (USEPA). 2002. Method 1631, Revision E: Mercury in water by oxidation, purge and trap, and cold vapor atomic fluorescence spectrometry. Washington, DC. [Google Scholar]
- U.S. Environmental Protection Agency (USEPA). 2011. 40 Code of Federal Regulation, Appendix B to Part 136: Definition and procedure for the determination of the method detection limit-revision 1.11. https://www.gpo.gov/fdsys/granule/CFR-2011-title40-vol23/CFR-2011-title40-vol23-part136-appB/content-detail.html. Last accessed November 24, 2018.
- U.S. Environmental Protection Agency (USEPA). 2013. Trends in blood mercury concentrations and fish consumption among U.S. women of childbearing age NHANES, 1999–2010. EPA-823-R-13-002. Washington, DC. [Google Scholar]
- van den Ouweland JMW, Beijers AM, Demacker PNM, van Daal H. 2010. Measurement of 25-OH-vitamin D in human serum using liquid chromatography tandem-mass spectrometry with comparison to radioimmunoassay and automated immunoassay. J Chromatogr B 878:1163–1168. [DOI] [PubMed] [Google Scholar]
- Vital M, Howe AC, Tiedje JM. 2014. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio 5: doi: 10.1128/mBio.00889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RW, Clemente JC, Peter I, Loos RJF, 2017. The prenatal gut micriome: are we colonized with bacteria in utero. Pediatric Obesity 12, 3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wear EK, Wilbanks EG, Nelson CE, Carlson CA. 2018. Primer selection impacts specific population abundances but not community dynamics in a monthly time-series 16S rRNA gene amplicon analysis of coastal marine bacterioplankton. Environ Microbiol 20:2709–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu BT, Dyer RA, King DJ, Innis SM. 2013. Low fish intake is associated with low blood concentrations of vitamin D, choline and n-3 DHA in pregnant women. Br J Nutr 109:936–943. [DOI] [PubMed] [Google Scholar]
- Yekutieli D, Benjamini Y. 1999. Resampling-based false discovery rate controlling multiple test procedures for correlated test statistics. J. Stat. Plann. Inference 82:171–196. [Google Scholar]
- Zareba G, Cernichiari E, Goldsmith LA, Clarkson TW. 2008. Validity of methyl mercury hair analysis: mercury monitoring in human scalp/nude mouse model. J Appl Toxicol 28:535–542. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.