Abstract
Immunoglobulin E (IgE) is the least abundant antibody isotype in mammalians, yet it plays a critical role in allergy and asthma. IgE-producing (IgE+) B cells are rare and difficult to detect, which have hindered progress to understand their generation and differentiation. Recently developed new fluorescent IgE reporter mice have enabled better understanding of the biology of IgE+ B cells. We here describe the usage of the Verigem IgE reporter mouse to study IgE+ B cells and plasma cells by flow cytometry and microscopy.
Keywords: IgE, B cells, IgE reporter, Verigem, Flow cytometry, Microscopy, Imaging, Germinal center, Plasma cells
1. Introduction
Immunoglobulin, or antibody, is an important component of humoral immunity. Mammals produce five major isotypes of antibodies - IgM, IgD, IgG, IgE, and IgA. Among these isotypes, IgE is the least abundant isotype, ranging from undetectable to micrograms per milliliter in the serum [1]. Despite its low abundance, IgE plays a critical role in allergy and asthma [2, 3]. Understanding how IgE-producing (IgE+) B cells are produced and differentiated under normal and pathophysiological conditions is crucial for developing new therapies for IgE-mediated allergic diseases.
Upon activation by binding of their surface B cell receptors (BCR) to cognate antigens, naïve B cells may undergo class-switch recombination from IgM/IgD to other isotypes of BCR. Some of these activated B cells may differentiate into short-lived plasma cells (PCs) extrafollicularly [4]. Alternatively, activated B cells can form or enter germinal centers (GCs), where class-switch recombination, affinity maturation, and differentiation into memory B cells or long-lived PCs take place [5]. Due to the technical difficulties to directly study IgE+ B cells, further explained below, it has been unclear whether IgE+ B cells follow the same route of activation and differentiation as B cells expressing other BCR isotypes or display unique characteristics in these processes.
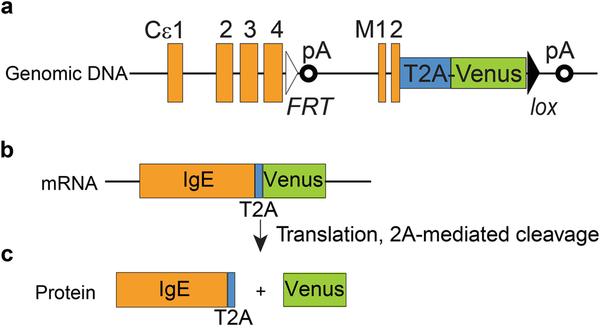
As IgE is the least abundant antibody isotype, the IgE+ PCs that produce these antibodies, and their precursor IgE+ B cells, are accordingly very rare. An even more confounding fact is that in addition to IgE+ B cells, several types of immune cells are also coated with IgE. For example, some B cells serve as reservoir for IgE through their surface low-affinity IgE receptor CD23 [6]. Basophils, mast cells, and dendritic cells in humans also absorb IgE through high-affinity IgE receptor, FcεRI, on their surface [7]. Thus it is almost impossible to identify genuine IgE+ B cells by conventional direct surface staining with anti-IgE antibodies. To overcome these challenges, special staining methods for flow cytometry analysis have been applied or developed (reviewed in [8]). For example, an acid wash method was employed to strip IgE noncovalently bound to CD23 [9, 10]. Two techniques have been developed recently to stain intracellular IgE, which is exclusively present in IgE-producing B cells [11, 12]. Around the same time, at least three reporter mouse strains were generated for tracking IgE+ B cells with fluorescent protein reporters (reviewed in [8]). Two of the reporter strains, M1′/GFP and CeGFP, placed the IRES-GFP cassette 3′ of the last exon of membrane IgE, M2 [13, 14]. The third IgE reporter mouse strain, Verigem, was generated in our laboratory [12]. In the Verigem reporter, a T2A-Venus cassette is placed immediately before the stop codon of the IgE M2 exon (see Fig. 1). T2A is a type of 2A peptide that allows separation of two or more proteins encoded in a single mRNA into individual proteins during the process of translation [15], and Venus is a brighter variant of yellow fluorescent protein [16]. Therefore, all B cells expressing the membrane IgE BCR in homozygous Verigem reporter mice will be labeled by Venus.
Fig. 1.
Diagram of the genomic allele, mRNA, and protein of the Verigem reporter. The relative location of exons and polyadenylation sites (pA) of the IgE genomic locus are shown (a). The coding sequence of T2A-Venus is fused in frame to the coding sequence of the membrane IgE M2 exon. Also shown are one FRT site placed immediately 3′ of the coding sequence of Cε4s exon (the last exon for secreted IgE) and one loxP site placed immediately 3′ of the Venus coding sequence. The coding sequences for membrane IgE, T2A, and Venus will be transcribed and spliced into single mRNA molecules (b), but the IgE BCR and Venus will be separated into individual proteins during translation at the T2A site (c)
The abovementioned IgE reporters can be used for flow cytometric characterization of IgE+ B cells by directly tracking the fluorescent reporters [12–14]. Furthermore, the fluorescent reporters can also be used as a readout of IgE+ B cells in immunohistochemistry and immunofluorescence imaging, bypassing the interference of other immune cells decorated with surface IgE [12, 13]. A unique advantage of these IgE reporters is that they can be used for live cell imaging in intact tissues, making the monitoring of interaction of IgE+ B cells and other cells possible [12, 13]. In this chapter, we describe detailed protocols for using of the Verigem reporter to track IgE+ B cells by flow cytometry and microscopy. These protocols may also be suitable for the analysis of the other reporter mice.
2. Materials
2.1. Materials for Cell Culture, Immunization, and Flow Cytometry Analyses
5–3/4″ Pasteur pipettes.
100 mm diameter, 60 mm diameter Petri dishes.
Complete DMEM medium (cDMEM): DMEM high glucose medium with 10% fetal bovine serum (FBS), 10 mM HEPES, 1× penicillin/streptomycin/l-glutamine.
DNase I (10 mg/mL).
Complete RPMI growth medium (cRPMI): RPMI 1640 medium with 10% FBS, 10 mM HEPES, 1× penicillin/streptomycin/l-glutamine, 50 μM β-mercaptoethanol.
Recombinant murine interleukin-4 (IL-4).
Anti-CD40 (FGK-45, Miltenyi Biotec, 2 mg/mL).
U-bottom 96-well tissue culture plate.
U-bottom 96-well assay plate.
FACS buffer: 1× PBS, 2% FBS, 1 mM EDTA, 0.1% sodium azide (omit sodium azide for CD138 staining [17]).
TruStain fcX (anti-mouse CD16/32, clone 93) antibody (BioLegend).
Peanut agglutinin (PNA), biotinylated.
A fluorescent streptavidin conjugate such as Qdot 605 streptavidin conjugate.
Fixable Viability Dye eFluor 780 (Thermo Fisher).
Propidium iodide (PI) in H2O (1 mg/mL).
DAPI (4′,6-diamidino-2-phenylindole) in H2O (1 mM).
Alum adjuvant (Alhydrogel, Accurate Chemical and Scientific).
4-hydroxy-3-nitrophenylacetyl conjugated to chicken gamma globulin (NP-CGG, ratio 30–39, Biosearch Technologies).
Tuberculin syringe with permanently attached needle, 1/2 cc, 27G 1/2.
Swinging bucket centrifuge that can accommodate 15/50 mL tubes and plates.
Tissue culture incubator.
Flow cytometer.
2.2. Materials for Preparation and Microscopic Imaging of Tissue Sections
1× phosphate buffered saline (1× PBS).
4% w/v paraformaldehyde (PFA) in 1× PBS. The PFA must be methanol-free; typically this is prepared from pure powder or from sealed ampules (such as from Electron Microscopy Sciences). To avoid degradation, 4% PFA/PBS should be frozen in aliquots at ≤−20 °C and thawed freshly for each use.
30% w/v sucrose in 1× PBS.
1000 mL pipette tip box lid.
Dry ice pellets.
Ethanol, 200 proof.
Optimal cutting temperature (O.C.T.) compound.
Cryomold 10 × 10 × 5 mm (biopsy size).
Cryostat.
Good quality microtome blades (Tissue-Tek Accu-Edge High Profile Microtome Blades).
Superfrost Plus Microscope Slides or other treated slides optimized for tissue adherence.
Acetone.
Humidity chamber for slide staining.
Glass container with rack to hold slides (we typically use containers that hold 300–500 mL volume).
Section staining buffer: 1× PBS, 1% normal mouse serum, and 0.1% bovine serum albumin. However, if rat anti-mouse IgG1 antibody will be used, normal mouse serum should be substituted with normal rat serum, since the former contains free mouse IgG1.
Kimwipes tissue paper or equivalent.
Fluoromount-G mounting medium (Thermo Fisher Scientific).
Slip-Rite coverslips, 24 × 55.
Rabbit anti-GFP tag polyclonal antibody (Thermo Fisher Scientific).
Clear nail polish.
3. Methods
Flow cytometry can be used for characterization of IgE+ B cells generated in vitro or in vivo. Some general notes are listed in Notes section (see Note 1).
3.1. Flow Cytometric Analysis of IgE+ B Cells Generated by In Vitro Cell Culture
Euthanize a Verigem reporter mouse, and harvest the spleen into a 15 mL conical tube with 5 mL cDMEM medium on ice. Mash the spleen in medium on a 40 μm cell strainer in a 100 mm diameter Petri dish using the bulb of a 3 cc syringe. Pipette the cell suspension three times through a 5–3/4″ Pasteur pipette to break up tissue particles. Then pass the cell suspension through the cell strainer again to remove unbroken cell aggregates. Centrifuge the cell suspension at 400 × g-force at 4 °C for 5 min.
Remove the supernatant and add 10 μL DNase I to the cell pellet, and then resuspend the cell pellet in 1 mL cDMEM by pipetting slowly and carefully using a P1000 pipette.
Count the cells. The splenocyte suspension can be used directly for cell culture or can be used for B cell purification before setting up the culture (see Note 2). Adjust the final cell concentration to 1 million cells per mL in cRPMI.
Add 100 μL cRPMI with 2× final concentration of IL-4 and anti-CD40 to each well of a U-bottom 96-well plate (see Note 3). Then add 100 μL cRPMI containing 100,000 splenocytes or purified B cells.
Place the culture in a humidified tissue culture incubator at 37 °C with 5% CO2.
After culture for desired amount of time (see Note 4), centrifuge the 96-well plate with cultured cells at 700 × g-force at 4 °C for 2 min.
Dump the culture media into a sink by thrusting the plate downward quickly while holding the plate firmly. Hold the plate horizontally, and tap against your palm gently until the cell pellet is fully resuspended in the residual liquid. To block non-specific binding of antibodies to Fc receptors, add 25 μL FACS buffer with 1:50 diluted TruStain fcX™ to each well. Incubate on ice for 10 min.
Add 200 μL FACS buffer to each well. Centrifuge the 96-well plate at 700 × g-force at 4 °C for 2 min. Dump the supernatant and resuspend the cell pellet as in step 7. Add 25 μL FACS buffer with diluted antibodies, such as anti-IgM, anti-IgD, anti-B220, anti-CD138, anti-IgG1, and anti-IgE (see Note 4). Incubate on ice for 20–60 min.
Add 200 μL FACS buffer to each well. Centrifuge the 96-well plate at 700 × g-force at 4 °C for 2 min, and then dump the supernatant. Wash the cell pellet again with 200 μL FACS buffer/well.
Resuspend the cell pellets in 50 μL FACS buffer with 1:3500 DAPI or 1:2000 PI.
Load the samples for flow cytometer analysis (Fig. 2a, b; see Note 5).
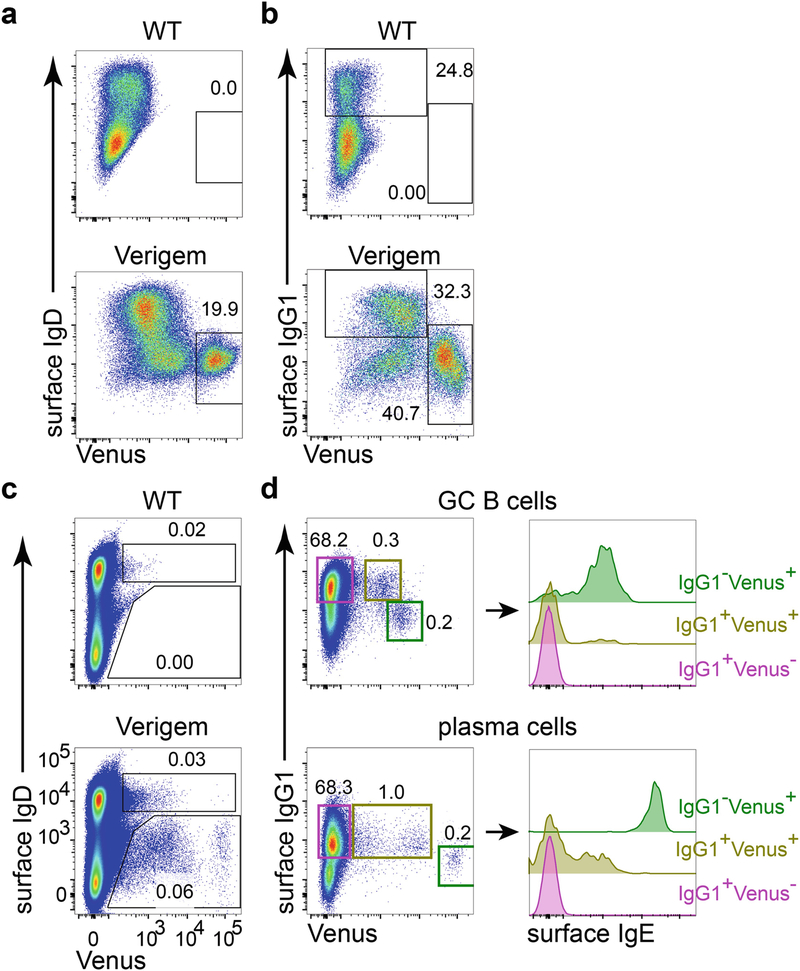
Fig. 2.
Flow cytometric characterization of IgE+ B cells generated in vitro or in vivo from Verigem reporter and control mice. (a, b) B cells from WT control or homozygous Verigem mice were cultured in vitro with anti-CD40 (250 ng/mL) and murine IL-4 (25 ng/mL) for 4 days. (c, d) Cell suspensions from draining LNs of WT control or homozygous Verigem mice 7 days after subcutaneous immunization with NP-CGG in alum adjuvant were stained and analyzed by flow cytometry. Cells in panels a and c were gated on live singlets; cells in panel b were gated on live singlets that are IgD−, IgM−, and CD19+. Cells in panel d were pooled from the draining LNs of five homozygous Verigem mice and were pre-gated as GC B cells (live singlets that are CD138−B220highCD 19+CD38lowGL7highIgDlow), and PCs (live singlets that are CD138+B220−CD38lowIgDlow) were further gated according to their expression of Venus and surface IgG1 (panel d, left), and histograms of surface IgE staining on these gated populations are further plotted (panel d, right)
3.2. Flow Cytometric Analysis of IgE+ B Cells Generated In Vivo (See Note 6)
Normally IgE+ B cells are nearly undetectable in naïve mice maintained in a clean-specific pathogen-free facility. But these cells can be induced either by immunization with an antigen or by infection with a parasite. Here we describe using NP-CGG immunization as a model to induce IgE+ B cells in vivo.
Dissolve lyophilized NP-CGG in sterile water to the concentration of 2.5 mg/mL. The reagent may be sonicated for 5 min to facilitate dissolving it. The NP-CGG solution can be aliquoted and frozen for future use.
Prepare NP-CGG and alum mixture for immunization. For injection at each site, mix 5 μL NP-CGG, 1.5 μL 10× PBS, and 8.5 μL water. Then add 15 μL alum. Mix by tapping or inverting the container.
Inject 30 μL NP-CGG:alum mixture subcutaneously using a tuberculin syringe to each site.
After immunization for desired amount of time (see Note 7), sacrifice immunized mice according to approved institutional protocol. Collect draining lymph nodes (LNs) of each mouse into a 1.5 mL Eppendorf tube with 1.2 mL cDMEM. Store samples on ice until all the samples have been collected.
Mash the draining LNs from each animal on a 70 μm cell strainer in a 60 mm Petri dish using the bulb of a 1 cc syringe. Pipette the cell suspension two times through a 5–3/4″ Pasteur pipette to break tissue particles. Then pass the cell suspension through the cell strainer twice to remove unbroken cell aggregates. Measure cell concentration of each sample.
Add 3 × 106 cells to each well of a round bottom 96-well plate. If necessary, add FACS buffer to bring the volume of each sample to about 200 μL. Centrifuge the plate at 700 × g-force at 4 °C for 2 min. Dump the supernatant, and then resuspend cell pellet of each sample in 50 μL FACS buffer with 1:50 diluted TruStain fcX. If desired, to identify GC B cells, PNA biotin (diluted 1:2000) can be added at the same time as the TruStain fcX, since the PNA reagent is a lectin and not an antibody. Alternatively, a biotin-conjugated antibody can be added directly to the samples after incubation with TruStain fcX for 10 min, allowing time for the Fc receptors to become blocked. Incubate on ice for 20–60 min.
Add 200 μL FACS buffer to each well. Centrifuge the 96-well plate at 700 × g-force at 4 °C for 2 min, and then dump the supernatant. Wash the cell pellet again with 200 μL FACS buffer/well.
Resuspend cell pellet of each sample in 50 μL FACS buffer, with a diluted fluorescent conjugate of streptavidin (such as 1:400 Qdot 605 streptavidin conjugate) and other appropriately diluted antibodies for cell surface staining. If desired, 1:500 diluted eFluor 780 viability dye may be included in the antibody mixture to label dead cells (we now routinely use this reagent to exclude nonviable cells even for surface staining and have found it can be added in our normal FACS buffer at the same time as antibodies). Incubate on ice for 20–60 min.
Add 200 μL FACS buffer to each well. Centrifuge the 96-well plate at 700 ×g-force at 4 °C for 2 min, and then dump the supernatant. Wash the cell pellet again with 200 μL FACS buffer/well.
Resuspend the cell pellet in 25 μL FACS buffer. If no fixable viability dye was included in step 8, then DAPI or PI should be included at this step (see Note 1). Combine staining replicates in one tube.
3.3. Staining of IgE+ B Cells in LN Cryostat Sections
3.3.1. Preparation of Whole LNs (See Note 8)
Remove LN(s) and place in a 1.5–5.0 mL tube containing 4% PFA/PBS, pre-chilled to 4 °C.
Rotate the tubes on a rotator at 4 °C for 2 h.
Remove LNs from PFA and wash three times with 1× PBS. Allow LNs to soak in PBS for at least 5 min for each wash.
Transfer washed LNs to new tubes containing 30% sucrose/PBS solution pre-chilled to 4 °C.
Rotate the tubes on a rotator at 4 °C overnight.
Transfer the LNs to a cryomold and fill with O.C.T. compound, avoiding bubbles.
Freeze the tissue rapidly. We prepare a dry ice pellet and ethanol bath in the lid of a 1000 mL pipette tip box. The cryomold should be held or positioned such that it is floating in the bath during the freezing process (avoid letting the ethanol reach the unfrozen tissue/OCT). Once frozen, you may leave the block temporarily on dry ice while preparing other samples and then absorb excess ethanol with a Kimwipes and transfer to a −80 °C freezer for long-term storage.
3.3.2. Cryostat Sectioning of Frozen LNs
Section frozen specimen in a cryostat with the chamber temperature set at −22 to −20 °C. If the system has the ability to separately adjust the object temperature, set this to about 3 °C warmer than the chamber temperature, which will assist with capturing the sections (we typically use an object temperature of −18 °C and a chamber temperature of −21 °C). The sections should be 5–10 μm thick (we typically cut 7 μm sections).
Mount LN sections on microscope glass slides (can typically mount ten or more sections on a single slide); allow to air-dry for at least 1 h.
Place slides with mounted tissue in a glass chamber with prechilled 4 °C acetone for 10 min.
After removing from acetone, allow slides to air-dry for at least 1 h before staining. Slides may alternatively be stored at −80 °C with desiccant (avoid getting the tissue wet). Best results are obtained by directly proceeding to the staining steps.
3.3.3. Staining Sectioned Tissue
Use a scriber to etch the glass around a region containing all the tissue on each slide (see Note 9).
Hydrate the tissue by placing glass slides with tissue in a glass chamber with 1× PBS for at least 15 min.
In the meantime, prepare primary antibody master mixture (see Note 10) containing rabbit anti-GFP antibody at a dilution of 1:400 (see Note 8) in section staining buffer. Each slide will need 350 μL of section staining buffer.
Remove each glass slide one at a time from the glass chamber containing PBS, run the edge of the slide along the glass chamber to remove excess fluid and dry the back of the slide on a paper towel, quickly dry a region around the tissue with a piece of folded Kimwipes (see Note 11), place the slide in humidity chamber, and quickly add the primary stain solution (it is important for the tissue not to dry out during this process).
5. Stain for 2–3 h at room temperature.
Prepare the secondary antibody mixtures (see Notes 10 and 12) in section staining buffer.
Flick off the staining solution onto a paper towel, and wash each slide by placing it on a rack in a glass chamber with an excess of PBS. Allow at least 3 min to wash each slide (see Note 13).
Remove one slide at a time from the PBS wash container, run the edge of the slide along the glass chamber to remove excess fluid and dry the back of the slide on a paper towel, quickly dry a region around the tissue, place slide in humidity chamber, and quickly add the secondary stain solution (again, it is imperative that the tissue does not dry out during this process). Repeat for additional slides.
Stain for 2–3 h at room temperature in the dark (covering the humidity chamber with foil is adequate for light protection).
Flick off the staining solution and wash slides as described in step 7.
Remove one slide at a time from the PBS wash container, run the edge of the slide along the glass chamber to remove excess fluid and dry the back of the slide on a paper towel, quickly dry a region around the tissue, and then add mounting media and coverslip carefully avoiding bubbles. (You may store the slides in a box at 4 °C in the dark overnight if needed). Prior to imaging, let the mounting media begin to solidify by leaving the slides exposed to air at room temperature, protected from light, for 1–2 h. You may seal the coverslips using clear nail polish. Proceed to step 12 or store the slides in a box at 4 °C in the dark until ready to image. Best results are obtained when imaging within 24–48 h after staining.
Collect images on a suitable microscope. Care must be taken in the adjustment of exposure times and display levels (see Fig. 3).
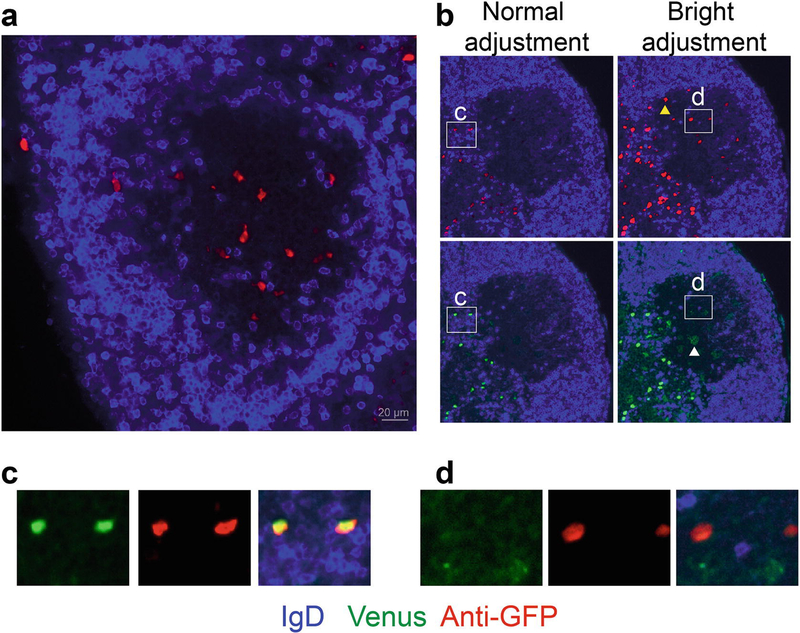
Fig. 3.
Representative microscopy of the Verigem reporter in LN cryostat sections. Heterozygous Verigem reporter mice were immunized subcutaneously with NP-KLH in alum adjuvant, and LNs were collected 7 d later. (a) Example of a GC (IgDlow region) with IgE+ B cells detected by anti-GFP staining (red). (b) Demonstration of the difference in sensitivity between anti-GFP staining and Venus fluorescence and the importance of the display adjustments to detect IgE+ PCs (Venushigh) versus IgE+ GC B cells (Venuslow). All four panels show the same image with either anti-GFP staining (red, upper panels) or with Venus fluorescence (green, lower panels). The image display levels are adjusted in a standard fashion to show the full range of signal detected (left panels) versus an exaggerated narrow range making the red/green signal appear bright (right panels). Note the GC B cells (e.g., see the yellow arrowhead) can only be detected by anti-GFP staining on the right panels with the bright adjustment of display levels. While PCs can be directly detected by either Venus fluorescence or anti-GFP staining (inset c) with normal display levels, the GC B cells cannot be visualized by Venus fluorescence but can be visualized by the anti-GFP staining with the bright adjustment of display levels (inset d). Within the GC, the bright display adjustment of the Venus channel instead reveals autofluorescent cells, such as tingible body macrophages (e.g., see the white arrowhead in (b, lower right panel)). The bright adjustment of display levels also results in oversaturation of the PCs; thus it may be difficult to properly visualize PCs and GC B cells with the same display settings. Anti-GFP was detected in the far red channel (see Note 10)
3.4. Live Microscopy of the Verigem Reporter
The Verigem reporter mice may also be used for live imaging. Two-photon laser scanning microscopy is the preferred method because this enables visualization deep within living tissues with minimal phototoxicity. In live imaging, since the tissue is not fixed, Venus fluorescence is detectable in both IgE+ GC B cells (Venuslo) and in IgE+ PCs (Venushi). Since detailed protocols have been provided elsewhere for the preparation of explanted tissues [18] and intravital two-photon microscopy [19], we simply provide the following advice for visualization of the Verigem reporter by two- photon microscopy.
A sensitive detector, such as a gallium arsenide phosphide (GaAsP) detector, will facilitate the detection of IgE+ GC B cells with weak Venus fluorescence. A rather high detector voltage may be needed to visualize weak Venus fluorescence, and this voltage may result in oversaturation of IgE+ PCs with bright Venus fluorescence. Given the pronounced green/yellow autofluorescence of some cell types, it is recommended to include a Verigem-negative control mouse tissue in the experiment to validate that the observed signal originates from Venus fluorescence.
The use of a 525/50 filter with high efficiency transmission characteristics will maximize signal to noise. This filter would also detect green (e.g., GFP) signal. To distinguish GFP from Venus, narrow band filters can be used, such as 510/20 for GFP and 535/30 for Venus. However, note that the use of narrow band filters will greatly reduce the Venus signal.
We have determined Venus has two distinct two-photon excitation peaks near 940 nm and 1020 nm (unpublished observations). In our experience, the 1020 nm peak gives the strongest excitation on systems with suitable optical properties. However, we have observed that some pre-compensation (“pre-chirp”) devices, found on many two-photon laser setups, may interfere with laser excitation in the 1020 nm range. Some microscope optics, such as the objective, may also lose transmission efficiency above 1000 nm. We routinely use a Coherent Ultra II Ti:Sapphire laser without a pre-compensation device and an objective optimized for transmission up to 1300 nm, and we consistently observe the best excitation at 1020 nm. Overall, the two-photon excitation of Venus can occur over a broad spectrum between 890 nm and 1040 nm, and thus other wavelengths may be preferred for optimal co-excitation of other fluorophores. Another important consideration is that second harmonic generation, such as from collagen in the lymph node capsule, will appear at half the excitation wavelength. This means that excitation at 1020 nm will result in second harmonic generation at 510 nm, which will appear in the typical green channel (typically the same channel as the Venus signal), which may be undesirable in some cases.
4. Notes
General notes for flow cytometry analysis: (1) Tissues should be collected in media pre-chilled to 4 °C and stored on ice. Samples should be kept on ice throughout the staining procedure and before flow cytometric analysis, and centrifuged at 4 °C. (2) Dead cells in the samples should be excluded for further analysis. Dead cells can be identified by washable viability dyes such as Fixable Viability Dye eFluor 780 or by DNA-binding dyes DAPI or PI. The eFluor 780 dye can be included during the last cell surface staining step (this reagent can be added together with other antibodies to surface markers), whereas DAPI or PI should be added after the completion of staining procedures. (3) The information of reagents used for flow cytometry staining can be found in our previous publication [20]. (4) The surface membrane IgE expression on IgE+ B cells from Verigem reporter mice is about two- to threefold higher than normal. This could be due to the insertion of an FRT site and/or T2A-Venus coding sequence into the IgE genomic locus. Although our previous study found little difference between wild-type (WT) IgE+ B cells and Verigem IgE+ B cells in terms of their differentiation and kinetics [12], readers are still advised to keep in mind that Verigem IgE+ cells have a higher level of membrane IgE than WT IgE+ cells.
We have found that comparable numbers of IgE+ B cells can be induced from purified B cells or from total splenocytes [12], and IgE+ B cells generated either way demonstrated the same propensity for antigen-independent PC differentiation [20]. Thus, efforts in purifying B cells can be saved if the in vitro culture is only going to be analyzed by flow cytometry. However, if the effect of other immune cells could potentially interfere with your assay, purified B cells should be used for culture.
IgE+ B cells can be generated from naive B cells activated with LPS and/or anti-CD40 together with IL-4 in in vitro culture. We have found that anti-CD40 plus IL-4 is more potent than LPS plus IL-4 in inducing IgE+ B cells. Additionally, LPS from different batches can vary dramatically in inducing B cell class switching and differentiation (unpublished observations). The generation and differentiation of IgE+ B cells is relatively insensitive to the amount of IL-4 (ranging from 10 to 100 ng/mL) but is sensitive to the amount of anti-CD40 (≥62.5 ng/mL) [20]. Therefore the concentration of anti-CD40 should be tested empirically under different experimental settings.
IgE+ and IgG1+ cells will start to appear when naive B cells have been cultured for 3 days, and peak numbers are observed when cells have been cultured for 4 days. We routinely stain in vitro cultured cells with anti-IgM, anti-IgD, anti-B220, anti-CD138, anti-IgG1, and anti-IgE. Anti-B220 staining can help gate out B220− cells of non-B cell lineages. Some of the cultured B cells will differentiate into PCs, and CD 138 is a marker for these cells (PCs will be stained more weakly for B220 but will still appear positive compared to non-B cells). Note that a recent study reported that sodium azide negatively affects CD138 staining and thus sodium azide should be omitted from buffers for optimal staining of this marker [17]. Staining with anti-IgE antibody for cells expressing Venus is optional but can help to corroborate the specificity of the Verigem reporter. Note that in step 3.1.8, the initial washing step after Fc block, prior to antibody staining, is optional, but is included in the protocol to minimize the staining volume and thus reduce the amount of antibodies needed.
Contrary to mouse B cells expressing other isotypes of BCR, IgE+ PCs express higher surface IgE BCR than IgE+ B cells. Accordingly, the IgE+ PCs carrying the Verigem reporter express much higher Venus than IgE+ B cells. The resultant fluorescence of Venus in IgE+ PCs B cells is so strong that it can exceed the detection range of flow cytometry with standard detector settings. This is true for IgE+ PCs generated in vitro and in vivo. Thus it may be necessary to lower the voltage of the “green channel” by 5–10% to make sure that the Venus fluorescence falls within the range of detection of the cytometer.
Examples of flow cytometry of B cells from in vitro culture and cells from draining LN are presented in Fig. 2 to make the following points. (1) Control animals should be included for in vivo experiments. Although expression of Venus in the Verigem reporter correlates tightly with IgE BCR expression, there are several types of cells, such as eosinophils, that have autofluorescence in the green channel. While there are almost no cells that appear to be Venus+ from the WT B cells cultured in vitro (Fig. 2a), autofluorescent cells are readily detected from the analysis of tissue samples. For example, immunized WT control and Verigem reporter mice have a similar percentage of IgDhigh cells that appear to be Venus+ (Fig. 2c) in lymph nodes. However, only the sample from the Verigem reporter mouse has Venus+ cells in IgDlow population. Therefore Venus fluorescence alone is not sufficient for specifically identifying IgE+ B cells. Inclusion of non-Verigem control animals in the experiment and staining of additional surface markers [12] will render more confidence in identifying genuine IgE+ B cells. (2) It is advisable to test the FACS settings for detecting the Verigem reporter with cultured Verigem cells before using the mice for in vivo studies. Under optimal in vitro culture conditions, the abundance of IgE+ cells can be comparable to that of IgG1+ cells (Fig. 2b). However, even at the peak of in vivo response, the number of IgE+ B cells is usually about 100-fold less than that of IgG1+ B cells (Fig. 2d). Therefore the technical conditions for assessing the Verigem reporter can be most easily tested through in vitro cell cultures to generate a large frequency of IgE+ B cells. (3) Flow cytometry analysis of in vivo IgE response should start with sufficient amount of cells. Since IgE+ cells are still rare even at the peak of responses to immunization or parasite infection, we usually start with four wells of cells from each sample for one staining scheme, with three million cells/well. These wells are then pooled for flow cytometric analysis. About 30–50% of cells will be retained at the completion of staining. As an example, in Fig. 2d, out of about 22 million events pooled from 5 Verigem reporter mice 7 days after immunization, there are only 895 Venus+ GC B cells and 237 Venus+ PCs. Therefore, it is very important to start the staining with a sufficient amount of cells to avoid stochastic effects. (4) Some B cells expressing other isotypes of BCRs may also be Venus+. For example, all B cells generated during in vitro cell culture from the Verigem reporter have a background shift in the Venus channel comparing to the WT control (Fig. 2b), which may be due to low-level translation of the germline transcript (unpublished observations). There are also small fractions of IgG1+ GC B cells and PCs generated in vivo from Verigem reporter mice that are weakly Venus positive (Fig. 2d). These Verigemlow cells that also express other isotypes of BCR could be transitional cells that recently switched to IgE, as suggested by surface IgE staining. Alternatively, they could have switched to IgE in the nonproductive IgH allele, as was described with the CεGFP reporter mouse [14], although translation of this allele is not expected [8]. The frequency of Verigemlow cells expressing other isotypes seems to vary depending on immunization conditions and time points. We are actively investigating the identity of these cells. Nevertheless, this again underscores the importance of including non-Verigem controls and additional cell surface markers (such as IgG1) to establish whether bona fide IgE+ B cells are being detected.
The IgE response in the immunization model described here will peak at around 7 days after immunization and quickly decline over time. In other models, such as Nippostrongylus brasiliensis infection, the peak IgE response may be closer to 10–14 days after immunization [12].
Fixation with PFA is essential to preserve Venus (YFP) inside the cells during sectioning. However, the fluorescence of Venus from IgE+ B cells with Verigem reporter will be greatly diminished after fixation by PFA, which precludes the visualization of IgE+ GC B cells (see Fig. 3). Thus we include rabbit anti-GFP antibody in the primary antibody staining solution. As a YFP derivative, Venus is recognized by this anti-GFP antibody. It is important to include LNs from an immunized WT mouse as a negative control. In some cases we have observed that non-specific staining can occur. This non-specific staining is particularly prominent at the peak of the extrafollicular PC response, as some PCs will encode antibodies that cross-react with the reagents used for staining (this is particularly notable in our experience in mice immunized with antigens containing NP and with the use of Cy/Alexa dye fluorophores).
Prior to the hydration step, we use an etching pen (diamond scriber) to etch a region around the tissue(s). This makes it easier to dry a region around the tissue prior to adding the staining mix since the tissue becomes difficult to see after rehydration. The dried region around the tissue keeps the staining solution contained and enclosed for the duration of the staining step.
Suggested additional complementary staining reagents are PNA biotin to identify GC B cells, rat anti-mouse CD35 (BD Biosciences, clone 8C12) to identify follicular dendritic cells, rat anti-mouse CD138 (BD Biosciences, clone 281–2) to identify PCs, rat anti-mouse IgD (BioLegend, clone 11–26c.2a) or goat anti-mouse IgD (Fc) (Nordic-MUbio) to detect follicular B cells, rat anti-mouse IgE biotin (BD Biosciences, clone R35–118), rat anti-mouse IgE PE (BioLegend, clone RME-1), and rat anti-mouse IgG1 (BD Biosciences, clone A85–1). Some of the aforementioned reagents can be used when directly conjugated to fluorophores if the signal has sufficient intensity; alternatively, purified antibodies can be detected with fluorescently conjugated secondary antibodies. We typically obtain polyclonal F(ab′)2 secondary reagents, made in donkeys, that are conjugated to AMCA, Cy3, or Alexa Fluor 647 from Jackson ImmunoResearch and are absorbed against many species (must be absorbed against mouse), such as Cy3 F(ab′)2 donkey anti-rat IgG. Good strep-tavidin conjugates that can be paired with biotin staining are streptavidin-Cy3 (Jackson ImmunoResearch) and streptavidin- Alexa Fluor 647 (Thermo Fisher Scientific).
A folded Kimwipes or paper towel is sufficient for the drying step. For hydration and washing steps, we dip our slides in glass containers of 400–500 mL PBS.
For detection of anti-rabbit GFP antibody staining, we have found good success using donkey anti-rabbit Alexa Fluor 488 (Jackson ImmunoResearch) and donkey anti-rabbit Alexa Fluor 647 (Jackson ImmunoResearch). Detection in the far red channel (such as with Alexa Fluor 647) gives the least background due to low tissue autofluorescence in the far red region of the visible light spectrum. Detection in the far red channel also allows simultaneous detection of Venus fluorescence (see Fig. 3). A dilution of 1:300 for both reagents is sufficient.
The duration of washes may affect staining results. We aim to achieve a 3–5 min wash time for each slide. This is accomplished by staggering the start time for putting the slides in the wash container. For example, add one slide to the wash container, wait 1 min, add the next slide to the wash container, and repeat until four slides have been added (at this time, the first slide will have been washed for about 3 min). Now remove the first slide, and proceed to the next staining step as described in Subheading 3.3.3, step 8. When you have added the antibody stain to this slide, put another slide in the wash container, and then remove the slide that has been washing for the longest time, and then follow Subheading 3.3.3, step 8 again. Continue repeating by adding one slide and removing one slide from the wash container about every 1 min until all slides have been washed for about 3–5 min each.
Acknowledgments
These protocols were developed for research projects supported by the UCSF Sandler Asthma Basic Research Center, the UCSF Cardiovascular Research Institute, the Weston Havens Foundation, and grants DP2HL117752 and R01AI103146 from the National Institutes of Health. C.D.C.A. is a Pew Scholar in the Biomedical Sciences, supported by The Pew Charitable Trusts. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or of the Pew Charitable Trusts.
References
- 1.Dullaers M, De Bruyne R, Ramadani F, Gould HJ, Gevaert P, Lambrecht BN (2012) The who, where, and when of IgE in allergic airway disease. J Allergy Clin Immunol 129(3):635–645. 10.1016/j.jaci.2011.10.029 [DOI] [PubMed] [Google Scholar]
- 2.Graham MT, Nadeau KC (2014) Lessons learned from mice and man: mimicking human allergy through mouse models. Clin Immunol 155(1):1–16. 10.1016/j.clim.2014.08.002 [DOI] [PubMed] [Google Scholar]
- 3.Gould HJ, Sutton BJ (2008) IgE in allergy and asthma today. Nat Rev Immunol 8(3):205–217. 10.1038/nri2273 [DOI] [PubMed] [Google Scholar]
- 4.MacLennan IC, Toellner KM, Cunningham AF, Serre K, Sze DM, Zuniga E, Cook MC, Vinuesa CG (2003) Extrafollicular antibody responses. Immunol Rev 194:8–18. 10.1034/j.1600-065X.2003.00058.x [DOI] [PubMed] [Google Scholar]
- 5.Allen CD, Okada T, Cyster JG (2007) Germinal-center organization and cellular dynamics. Immunity 27(2):190–202. 10.1016/j.immuni.2007.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng LE, Wang ZE, Locksley RM (2010) Murine B cells regulate serum IgE levels in a CD2 3-dependent manner. J Immunol 185(9):5040–5047. 10.4049/jimmunol.1001900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shin JS, Greer AM (2015) The role ofFc epsilon RI expressed in dendritic cells and monocytes. Cell Mol Life Sci 72(12):2349–2360. 10.1007/s00018-015-1870-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z, Robinson MJ, Allen CD (2014) Regulatory constraints in the generation and differentiation of IgE-expressing B cells. Curr Opin Immunol 28:64–70. 10.1016/j.coi.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katona IM, Urban JF Jr, Scher I, Kanellopoulos-Langevin C, Finkelman FD (1983) Induction of an IgE response in mice by Nippostrongylus brasiliensis: characterization of lymphoid cells with intracytoplasmic or surface IgE. J Immunol 130(1):350–356 [PubMed] [Google Scholar]
- 10.Erazo A, Kutchukhidze N, Leung M, Christ AP, Urban JF Jr, Curotto de Lafaille MA, Lafaille JJ (2007) Unique maturation program of the IgE response in vivo. Immunity 26(2):191–203. 10.1016/j.immuni.2006.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wesemann DR, Magee JM, Boboila C, Calado DP, Gallagher MP, Portuguese AJ, Manis JP, Zhou X, Recher M, Rajewsky K, Notarangelo LD, Alt FW (2011) Immature B cells preferentially switch to IgE with increased direct Smu to Sepsilon recombination. J Exp Med 208(13):2733–2746. 10.1084/jem.20111155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z, Sullivan BM, Allen CD (2012) Fluorescent in vivo detection reveals that IgE(+) B cells are restrained by an intrinsic cell fate predisposition. Immunity 36(5):857–872. 10.1016/j.immuni.2012.02.009 [DOI] [PubMed] [Google Scholar]
- 13.Talay O, Yan D, Brightbill HD, Straney EE, Zhou M, Ladi E, Lee WP, Egen JG, Austin CD, Xu M, Wu LC (2012) IgE(+) memory B cells and plasma cells generated through a germinal-center pathway. Nat Immunol 13(4):396–404. 10.1038/ni.2256 [DOI] [PubMed] [Google Scholar]
- 14.He JS, Meyer-Hermann M, Xiangying D, Zuan LY, Jones LA, Ramakrishna L, de Vries VC, Dolpady J, Aina H, Joseph S, Narayanan S, Subramaniam S, Puthia M, Wong G, Xiong H, Poidinger M, Urban JF, Lafaille JJ, Curotto de Lafaille MA (2013) The distinctive germinal center phase of IgE+ B lymphocytes limits their contribution to the classical memory response. J Exp Med 210(12):2755–2771. 10.1084/jem.20131539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Felipe P, Luke GA, Hughes LE, Gani D, Halpin C, Ryan MD (2006) E unum pluribus: multiple proteins from a self-processing polyprotein. Trends Biotechnol 24(2):68–75. 10.1016/j.tibtech.2005.12.006 [DOI] [PubMed] [Google Scholar]
- 16.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A (2002) A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20(1):87–90. 10.1038/nbt0102-87 [DOI] [PubMed] [Google Scholar]
- 17.Wilmore JR, Jones DD, Allman D (2017) Protocol for improved resolution of plasma cell subpopulations by flow cytometry. Eur J Immunol 47(8):1386–1388. 10.1002/eji.201746944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matheu MP, Parker I, Cahalan MD (2007) Dissection and 2-photon imaging of peripheral lymph nodes in mice. J Vis Exp 7:265 10.3791/265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murooka TT, Mempel TR (2012) Multiphoton intravital microscopy to study lymphocyte motility in lymph nodes. Methods Mol Biol 757:247–257. 10.1007/978-1-61779-166-6_16 [DOI] [PubMed] [Google Scholar]
- 20.Yang Z, Robinson MJ, Chen X, Smith GA, Taunton J, Liu W, Allen CD (2016) Regulation of B cell fate by chronic activity of the IgE B cell receptor. eLife 5:e21238 10.7554/eLife.21238 [DOI] [PMC free article] [PubMed] [Google Scholar]