Abstract
Native mass spectrometry (MS) has become an important tool for the analysis of membrane proteins. Although detergent micelles are the most commonly used method for solubilizing membrane proteins for native MS, nanoscale lipoprotein complexes such as nanodiscs are emerging as a promising complementary approach because they solubilize membrane proteins in a lipid bilayer environment. However, prior native MS studies of intact nanodiscs have employed only a limited set of phospholipids that are similar in mass. Here, we extend the range of lipids that are amenable to native MS of nanodiscs by combining lipids with masses that are simple integer multiples of each other. Although these lipid combinations create complex distributions, overlap between resonant peak series allows interpretation of nanodisc spectra containing glycolipids, sterols, and cardiolipin. We also investigate the gas-phase stability of nanodiscs with these new lipids towards collisional activation. We observe that negative ionization mode or charge reduction stabilize nanodiscs and are essential to preserving labile lipids such as sterols. These new approaches to native MS of nanodiscs will enable future studies of membrane proteins embedded in model membranes that more accurately mimic natural bilayers.
Introduction
Native or noncovalent mass spectrometry (MS) has become a powerful technique to characterize membrane protein interactions [1–3]. Conventional native MS relies on detergent micelles to solubilize membrane proteins for electrospray ionization (ESI). Collisional activation inside the mass spectrometer releases the membrane protein from the detergent micelle for mass analysis. To address limitations with detergent micelles, alternative membrane mimetics have been explored [4], including amphipols [5–7], bicelles [8], styrene maleic acid lipid particles (SMALPs) [9], and even vesicles of natural membranes [10].
Due to their relatively low polydispersity and high homogeneity, lipoprotein nanodiscs have emerged as a promising membrane mimetic for native MS. Nanodiscs have been used to deliver integral membrane proteins [8, 11, 12], peripheral membrane proteins [13, 14], and transmembrane peptides [15] for native MS and to characterize large numbers of lipids bound to membrane protein complexes [16]. Recent work found that charge manipulation reagents enable a wide range of dissociation pathways, allowing the membrane protein to be either ejected from the nanodisc or characterized within a nearly intact nanodisc complex [17].
One advantage of nanodiscs is that they can be assembled with a precisely defined composition of lipids. However, most prior native MS used homogeneous nanodiscs formed with a single phosphatidyl-choline (PC) lipid. Hoi et al. explored native MS of nanodiscs containing mixtures of palmitoyl-oleoyl-phosphatidyl-choline (POPC), palmitoyl-oleoyl-phosphatidylglycerol (POPG), and palmitoyl-oleoyl-phosphatidyl-serine (POPS) lipids. Because these lipids are similar in mass, with a maximum difference of 11 Da, nanodiscs with mixtures of POPC/POPG and POPC/POPS showed a well-resolved series of peaks separated by the average mass of the two lipids [18]. Peaks from nanodiscs with the same total number of lipids but different populations of each specific type of lipid overlapped into a single peak. However, natural membranes are far more complex in composition and contain lipids with very different masses. As the complexity of lipids in the nanodisc increases, the intrinsic distribution in the number of lipids per nanodisc leads to hundreds of mass states that could blur into a broad unresolvable hump.
To expand the types of lipids that are amenable for native MS of nanodiscs, we hypothesized that mixed lipid nanodiscs would be resolvable if the lipid masses are resonant, meaning their masses are simple rational multiples of each other. Although hundreds of mass states are present, constructive overlap of resonant states keeps the peaks resolvable. POPC, POPG, and POPS are intrinsically resonant because their masses are very close, a multiplier of one. Here, we extend the resonance approach to a wider range of lipid combinations and multipliers. For example, glycolipid GM1 is about twice the mass of POPC, and cholesterol is about half the mass of POPC. Cardiolipin has a more complex resonance because twice the mass of tetra-myristoyl-cardiolipin (TMCL) is about three times the mass of POPG. We discovered that each of these combinations shows well-resolved native mass spectra.
Exploring the stability of these novel lipid combinations towards collision-induced dissociation (CID), we found that nanodiscs containing GM1 or TMCL are relatively stable against CID, but sterols need to be stabilized by chemical modifications, charge-reducing reagents, or negative ionization mode. In addition to conventional lipoprotein nanodiscs formed with ApoAI-based membrane scaffold proteins (MSP), we demonstrated lipid resonance with Saposin A (SapA) lipoprotein nanoparticles, referred to as Salipro nanoparticles, SapNPs, or picodiscs [19–21]. Klassen and coworkers have shown the potential of picodiscs as an alternative membrane mimetic for native MS applications [22–25]. Ultimately, using resonant lipids lays the foundation for creating lipoprotein complexes that are suitable for native MS but are more accurate models of natural membranes.
Methods
Materials
Imidazole, ammonium acetate, monosialoganglioside-GM1 from bovine brain, and Amberlite XAD-2 were purchased from Sigma Aldrich. 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1’-rac-glycerol) (POPG), 1’,3’-bis[1,2-dimyristoyl-sn-glycero-3-phospho]-glycerol (TMCL), and cholesterol were purchased from Avanti Polar Lipids. Cholesteryl hemisuccinate (CHS) was purchased from Anatrace. Ergosterol was purchased from Arcos Organics. Lipid structures are shown in Figure S1, and key data is provided in Table S1. Membrane scaffold proteins MSPD1 and MSP1D1T1 [12] were expressed in E. coli and purified by immobilized metal affinity chromatography (IMAC) as previously described [12, 26]. MSP1D1T1 is identical to MSP1D1 except for an extra threonine residue near the N-terminus. The additional mass of the MSP1D1T1 belts was used to confirm that the nanodiscs retain both MSP belts during mass analysis in initial control POPC nanodiscs [12]. TEV protease was utilized to cleave the polyhistidine tag from the MSPs to form MSP1D1(−) and MSP1D1T1(−) respectively. SapA was expressed and purified as previously described [20, 21, 27]. Briefly, SapA was expressed in E. coli and purified via IMAC. TEV protease was added to cleave the polyhistidine tag.
Nanodisc Assembly
Nanodiscs were prepared as previously described [12, 16, 28]. Briefly, nanodiscs were self-assembled by removal of detergents by Amberlite XAD-2 hydrophobic beads from a mixture of lipids and MSP. Lipids were mixed as chloroform stocks and dried under nitrogen together before resuspension into 0.1 M sodium cholate. POPC/GM1 mixtures were prepared at a molar ratio of 98/2 and used the MSP1D1T1(−) variant. All others used MSP1D1(−). Lipid-sterol mixtures of POPC/cholesterol, POPC/ergosterol, and POPC/CHS were each prepared at a molar ratio of 90/10. Lipid mixtures of POPG/TMCL were prepared at a molar ratio of 95/5. Nanodiscs were assembled at a molar ratio of 65/1 or 70/1 total lipid/MSP. Following addition of MSP and removal of detergent by hydrophobic beads, assembled nanodiscs were purified with size exclusion chromatography (SEC) using a Superose 6 Increase 10/300 column (GE Healthcare) into 0.2 M ammonium acetate at pH 6.8. Final concentrations of nanodiscs were around 1–5 μM.
Saposin A Nanoparticle Assembly
Saposin A nanoparticles (SapNPs) were prepared as previously described [20] using the same reconstitution approach as MSP nanodiscs. Lipids were mixed as chloroform stocks and dried before resuspension into 0.1 M sodium cholate. Lipid mixtures of POPG/TMCL were prepared at a molar ratio of 95/5. SapNPs were assembled at a molar ratio of 12/1 total lipid/SapA. Detergent was removed by Amberlite XAD-2 hydrophobic beads from a mixture of lipids and SapA to drive self-assembly. Following self-assembly, assembled SapNPs were purified via SEC using a Superose 6 Increase 10/300 column into 0.2 M ammonium acetate at pH 6.8. The final concentration of SapA in purified SapNPs was typically around 15–25 μM.
Mass Spectrometry
For charge reduction experiments, samples were mixed 9:1 v/v with 400 mM (pH 7) imidazole (IM) for a final concentration of 40 mM IM [17]. Nano-electrospray ionization was performed using clipped borosilicate needles pulled with a P-1000 micropipette puller (Sutter Instrument, Novato, CA). Mass spectrometry was performed using a Q-Exactive HF quadrupole-Orbitrap mass spectrometer equipped with the Ultra-High Mass Range (UHMR) research modifications (Thermo Fisher Scientific) [29]. Instrumental parameters were used as previously described [12, 28]. Important parameters included 0.9–1.6 kV for capillary voltage and 200 °C capillary temperature. Scans were collected from 2,000 and 25,000 m/z at a target resolution of 15,000 with 10 microscans summed into one scan. The collision voltage was applied in the high collisional dissociation (HCD) cell and ramped from 0–100 V or 0–200 V in 5 V or 10 V increments respectively with 1-minute acquisitions for each step. Source fragmentation was set to 20 V for positive ionization mode with no additives. For negative ionization mode and imidazole experiments, the source fragmentation was set to 50 V. Three separate nanodisc or SapNP assembly reactions were measured for each sample to validate the reproducibility of the results, and data is shown for one representative replicate.
Mass Spectrometry Data Analysis
Native mass spectra were deconvolved using MetaUniDec as previously described with modifications [28]. The mass range for nanodiscs was set from 20–200 kDa with a charge range from 1–30 and peak width of 10 m/z. The mass range for SapNPs was set from 1–130 kDa with a charge range from 1–15 and a peak width of 5 m/z. The mass of a proton was used as the ESI charge carrier. The mass of the charge carrier multiplied by the charge was subtracted from the measured mass in positive ionization mode (standard in UniDec) and added to the mass in negative mode using the new negative mode option in UniDec.
A new point smooth feature of the UniDec deconvolution algorithm was used to improve the deconvolution. The point smooth assumes that neighboring data points should have similar charge states. This is implemented as a mean filter of width ±n applied to each extracted charge state at the start of each iteration, before the charge state smooth and Bayesian deconvolution are applied [30]. Here, we used a point smooth width of 1, so the mean filter averages the probabilities of each data point with one neighboring point on each side for each charge state. We combined this new point smooth with a log mean mass smooth width of 2 and a Gaussian charge state smooth of width 0.5. UniDec conventionally uses a log mean filter for both mass and charge state smoothing, but the Gaussian filter is available by adding a negative flag (an input value of −0.5). It is not clear why this combination of filters is optimal, but these provided the slightly fewer deconvolution artifacts with nanodiscs and SapNPs in this study.
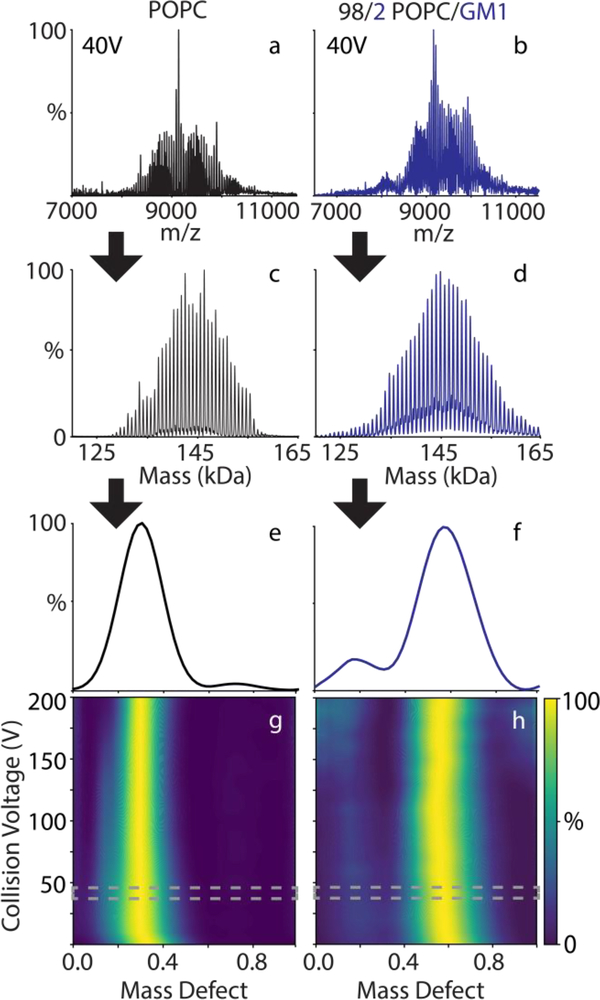
Each lipoprotein complex sample yields a distribution of peaks with different numbers of lipids per complex, which are separated by the mass of a lipid. To help visualize and analyze the spectra, we utilized macromolecular mass defect analysis [16]. We define the mass defect as the remainder of the measured mass divided by a reference lipid mass. For this study, we have defined the reference lipid as either POPC (with sterols and GM1) or POPG (with TMCL) because these are the most abundant lipids in each complex. Except where noted, we summed the mass defects across all measured masses and collision voltages. The mass defect will always be between 0 and 1, and mass defect values combine using modular arithmetic. Because the mass defect of the reference lipid is 0, shifts in mass defect of the complex must be caused by molecules that are not the reference lipid, in this case either the lipoprotein belts or the other lipid component. An example workflow for deconvolution and mass defect analysis is shown in Figure 1.
Figure 1:
The positive ionization mode mass spectra (a, b) at 40 V CID of POPC-only (a, c, e, g) and POPC/GM1 (b, d, f, h) nanodiscs are deconvolved into zero-charge mass distributions (c, d). The mass defect summed over all the measured spectra is shown for the 40 V step (e, f) and compared as a function of collision voltage (g, h), where the 40 V step is highlighted in gray.
Results and Discussion
Mass Resonance
Because nanodiscs are formed by a random self-assembly process, they have an intrinsic distribution in the number of lipids per complex (Figure 1C). Mixed lipid nanodiscs have an additional distribution in the number of each lipid type that will be binomial assuming ideal mixing [18]. For example, a nanodisc with 140 total lipids could have 70 of one lipid and 70 of the other. The nanodisc could also have lipid populations of 69/71, 71/69, and so on. The overlap between the distribution in number of total lipids and the distribution in types of lipids can become very complex and can prevent nanodisc spectra from being resolvable by native MS. The ability to resolve these complex distributions depends on three factors: 1) width of the peaks, 2) the difference in mass between the two lipids, and 3) the relative molar ratio of each lipid.
To illustrate the effects of these parameters on an idealized system, we simulated the mass distributions for nanodiscs with 140 total lipids/nanodisc. POPC was chosen as the reference lipid. A second theoretical lipid was simulated at different mass defects ranging from 0.5 to 0.99 times the mass of POPC and at different levels of incorporation ranging from 1 to 25%. We simulated peaks with a full width at half max of 100 Da (Figure S2 and S3) or 200 Da (Figure S4).
We observed several trends in the simulated mass distributions. First, narrower peaks obviously reduced the overlap between masses. Second, lower molar ratios of the added lipid had less overlap due to their reduced complexity. At only 1% loading, all mass defect combinations were resolvable, and peaks overlap most at 50% loading. Finally, particular mass defect values were favorable at any loading level. These “resonant” masses occurred when the mass of one lipid is a simple integer multiple of the other. In the simplest case, the mass defect of 0.99 was very close to 1/1. Similarly, a mass defect of 0.5 corresponded to a resonance of 1/2. A mass defect of 0.66 had a resonance of 2/3. A similar approach can be useful in polymer analysis [31].
If we define a peak series as a repeating pattern of peaks separated by the mass of the reference lipid, the denominator of the resonance specifies the number of peak series present, as shown in Figure S3. For example, a denominator of 1 shows a single peak series spaced by the mass of the reference lipid. A resonance of 1/2 gives an additional series with the same spacing but shifted by half the mass of the reference lipid. As the denominator gets larger, the pattern eventually leads to complete overlap and unresolvable peaks (Figure S2 and S4). We hypothesized that a careful choice of lipids with resonant masses would allow more complex combinations of lipids to be assembled into lipoprotein complexes that are still experimentally resolvable by native MS.
GM1 Nanodiscs
To test the hypothesis that lipid combinations with resonant masses would be resolvable by native MS, we created nanodiscs with a 98/2 mixture of POPC/GM1. GM1 is an important glycolipid receptor for pathogens [13] and is thought to partition into lipid rafts [32, 33]. Comparing control POPC-only nanodiscs to POPC/GM1 nanodiscs, the spectra are well resolved and show similar deconvolved mass distributions (Figure 1a–d). Mass defects for control nanodiscs agreed well with predicted values (Table S1), indicating adduction was minimal.
The mass of POPC is 760.08 Da, and GM1 is primarily a mixture of two masses at 1,545 and 1,573 Da [13] (Table S1). Because the mass of two POPC molecules (1,520.16 Da) is close to the mass of one GM1, nanodiscs that differ in the number of GM1 per complex overlap in mass with nanodiscs with different numbers of POPC per nanodisc. For example, POPC/GM1 nanodiscs have a peak at 150,139 Da. After subtracting the mass of the two MSP belts, this peak could be assigned to 133 POPC and 3 GM1 (a mass of 150,099 Da, assuming the most abundant form of GM1). It could also be assigned to 131 POPC and 4 GM1 (150,152 Da). Because the peak full width half max is around 150–200 Da, it is likely a combination of these states and others that overlap to form the peak at an average mass of 150,139 Da.
Extracting the intensity of the deconvolved mass distribution at different combinations of POPC and GM1, we find that the nanodiscs contain around 4 GM1 and 120–140 observed POPC molecules, slightly higher than the expected 2% GM1 (Figure S5). The added mass from the GM1 molecules shifts the observed mass defect values from 0.29 to 0.57 (Figure 1e and 1f). The mass defect of the most abundant GM1 is 0.07 (1,573 Da / 760.08 Da), as shown in Table S2. Thus, 4 GM1 molecules should shift the mass defect by 0.28, in excellent agreement with the observed shift. Thus, both mass defects or more traditional peak assignment can be used to determine the lipid stoichiometries.
Because the mass defect does not shift as POPC/GM1 nanodiscs are collisionally activated (Figure 1h), GM1 is stable towards dissociation, and POPC is the only lipid lost during CID. The preference for ejection of POPC in positive mode is likely because POPC is more readily able to become positively charged and hence labile. GM1, in contrast, has a sialic acid (Figure S1), so acquiring a net positive charge is less favorable. This agrees with data from Klassen and coworkers showing that GM1 is readily ejected as an anion in negative mode [13]. The preferential dissociation of POPC may contribute to the higher than expected observed levels of GM1. Together, these data demonstrate that heterogeneous nanodiscs containing lipids with very different masses are amenable to native MS provided that the masses create an overlapping pattern of peaks.
Sterol Nanodiscs
Cholesterol is a critical membrane lipid for the animal kingdom and is thought to have important interactions with G-protein coupled receptors [34, 35]. To explore native MS of nanodiscs with cholesterol, we assembled nanodiscs with a 90/10 mixture of POPC/cholesterol. The mass of cholesterol is 386.66 Da, which is nearly half that of POPC (760.08 Da / 2 = 380.04 Da). As with POPC/GM1 nanodiscs, we expected that nanodiscs with different numbers of POPC and cholesterol per complex would constructively overlap. Because two cholesterol molecules have nearly the same mass as one POPC molecule, nanodiscs with an even number of cholesterol molecules should be similar to control POPC-only nanodiscs. Nanodiscs with an odd number of cholesterol molecules should have a similar peak series but shifted by the mass of cholesterol, as shown in Table S2 and Figure S3.
However, using positive ionization mode, we found that nanodiscs with cholesterol had the same mass distribution as controls with only POPC (Figure 2a and 2b). POPC has strong electrostatic intermolecular interactions that likely help stabilize nanodiscs in the gas phase. In contrast, cholesterol has only one polar hydroxy moiety (Figure S1), so intermolecular interactions in the bilayer are primarily driven by nonpolar interactions that will likely become less relevant in the absence of solvent. Thus, we suspected that cholesterol was dissociating from the nanodisc in the gas phase. Imidazole has been shown to stabilize nanodiscs for native MS by charge reduction, so we investigated whether addition of imidazole would stabilize nanodiscs with cholesterol [17]. After adding 40 mM imidazole to the same samples, a second series was observed with a mass defect shift of 0.5, which indicated that cholesterol was retained in the nanodiscs. However, the series of peaks around mass defect 0.1 (an even number of cholesterol molecules) was greater than the series around mass defect 0.6 (an odd number of cholesterol molecules). Because the evens and odds should be the same height in a random mixture with this level of incorporation (see Figure S3), the higher series of evens indicates low levels of cholesterol were retained in the nanodiscs, which leads to more POPC-only nanodiscs with no cholesterol.
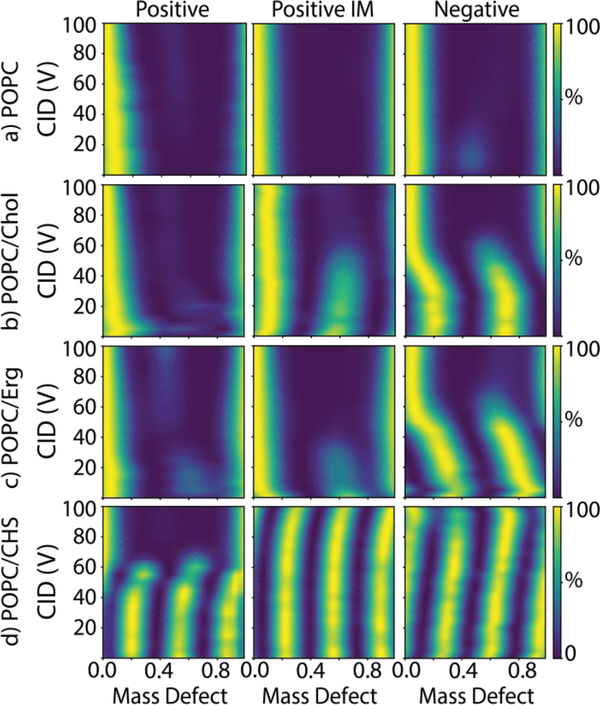
Figure 2:
The mass defect summed from all measured masses as a function of collision voltage for nanodiscs with a) POPC only, b) POPC/cholesterol, c) POPC/ergosterol, and d) POPC/CHS for spectra collected in positive mode with no additive (left), positive mode with added imidazole (center), and negative mode with no additive (right).
Similar to GM1, we expect cholesterol to be less likely than POPC to become positively charged and eject from the nanodisc by Coulombic repulsion. Instead, we believe that cholesterol is lost as a neutral due to weak intermolecular interactions with the bilayer. We were unable to observe any cholesterol ions ejected from the nanodisc during CID and only found signal for ejected POPC ions. It may be that gas-phase distortions in the bilayer—such as the collapse of nanodiscs at elevated collision voltage previously seen by ion mobility spectrometry[30]—cause cholesterol to be released. By stabilizing the nanodisc complex as a whole, we believe that charge reduction likely better preserves the structure of the bilayer and the weak intermolecular interactions that hold cholesterol in the nanodisc.
We also analyzed the same nanodiscs with negative ionization mode, which stabilizes POPC nanodiscs [17, 18]. Here, we found that the odd and the even series are the same height, indicating that more cholesterol is retained (Figures 2b and 3). Further evidence for cholesterol stability is observed in the shift in mass defect upon controlled ejection of cholesterol by CID. As the collision voltage increases, the even series shifts from a mass defect of 0.19 to 0.03, a shift of 0.16 (Figure S6). Because the mass defect of cholesterol is 0.5087 (386.66 Da / 760.08 Da), this indicates that an average of roughly 18 cholesterol molecules (18 × 0.5087 = 0.16) were incorporated into the nanodiscs with a total number of around 150 lipids, which is only slightly higher than the expected 90/10 molar ratio. These data confirm that cholesterol was successfully incorporated in the nanodisc but easily dissociates in the gas phase.
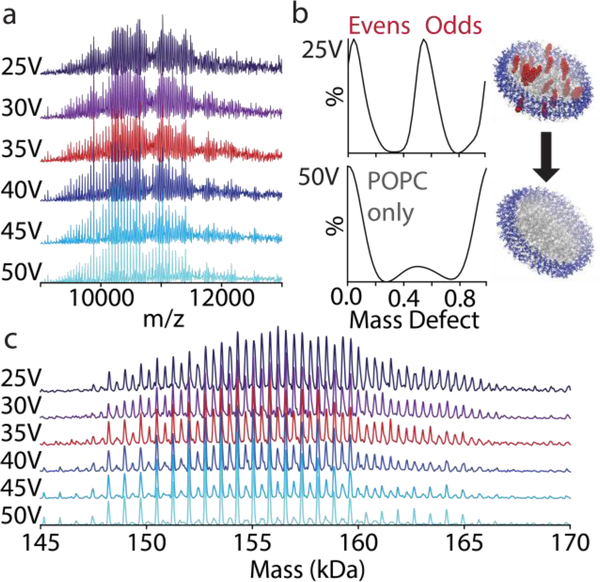
Figure 3:
Native mass spectra (a) in negative mode of MSP1D1(−) nanodiscs with 90/10 POPC/cholesterol from 25 to 50 V CID. The mass defect values summed over the whole spectrum (b) are extracted from the deconvolved zero-charge mass spectra (c). Under increasing CID, the mass defects shift from two series corresponding to nanodiscs with either even or odd numbers of cholesterol/nanodisc to nanodiscs with only background POPC.
Similar stability profiles were observed with ergosterol, the primary membrane sterol in fungi (Figure 2c). Ergosterol has a higher mass of 396.66 Da (Table S1), so the mass defect shift for 10% ergosterol nanodiscs is greater. Like cholesterol, a small shift was observed with imidazole addition, suggesting partial stabilization. Greater stabilization was observed with negative ionization mode.
Finally, we created nanodiscs using CHS, a derivative of cholesterol that adds an anionic succinate to the hydroxyl group of cholesterol. We predicted that the negative charge of the succinate would interact with the positive quaternary amine on POPC to form stronger intermolecular interactions that hinder dissociation. CHS has a neutral mass of 486.73 Da, so the mass defect of CHS is 0.64 (Table S1). Thus, three CHS molecules are similar in mass to two POPC molecules. Native MS of 90/10 POPC/CHS nanodiscs in positive mode with no additives showed three peak series (Figure 2d). Analogous to the even and odd series with cholesterol, each of these series corresponds to nanodiscs with numbers of CHS molecules that are congruent modulo 3. For example, one series has 0, 3, 6, etc. CHS molecules per nanodisc. Another series has 1, 4, 7, etc. The final series has 2, 5, 8, etc.
Because the mass defect of CHS is not perfectly 2/3 (Figure S3), each set of 3 CHS molecules will shift the series slightly by −0.08 (Table S2). This complicates assignment of the series. For example, the mass defect of one series begins at 0.58. This could be due to a mixture of 15 and 18 CHS molecules (mass defects of 0.61 and 0.53 respectively), but it could also be 4 CHS molecules (0.56). Thus, we cannot assign these peaks definitively but only say that CHS was incorporated and that there is not a significant population of empty nanodiscs that would favor one peak series over the other two (see Figure S3). As shown in Figure 2d, the CHS in positive mode undergoes a distinct dissociation to lose all CHS at around 60 V. For negative mode and imidazole experiments, CHS dissociates from the nanodisc at collision voltages above 100 V (Figures 2d and 4). Thus, like cholesterol and ergosterol, imidazole or negative mode stabilize CHS nanodiscs, but challenges in peak assignment prevent precise quantitation.
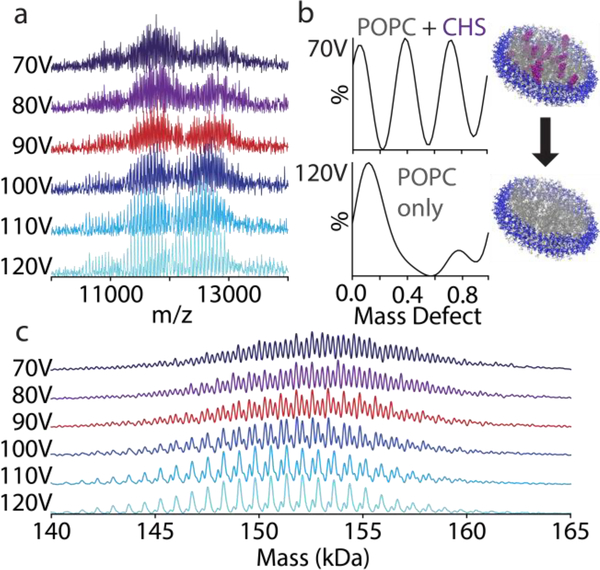
Figure 4:
Native mass spectra (a) in negative mode of MSP1D1(−) nanodiscs with 90/10 POPC/CHS from 70 to 120 V CID. The mass defect values summed over the whole spectrum (b) are extracted from the deconvolved zero-charge mass spectra (c). Under increasing CID, the mass defects shift from three series corresponding to nanodiscs with different numbers of CHS/nanodisc to a nanodisc with only background POPC.
Cardiolipin Nanodiscs
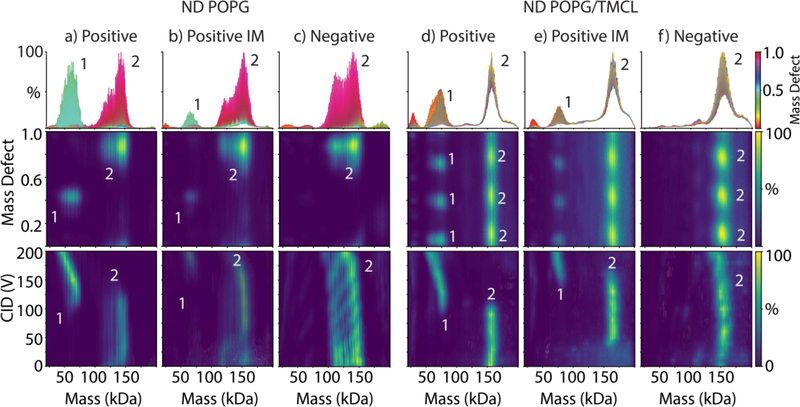
Building on the success in resolving more complex resonance patterns with POPC/CHS nanodiscs, we explored nanodiscs with mixtures of POPG and TMCL. Cardiolipin is an important lipid in bacterial and mitochondrial membranes, and prior native MS studies have found important interactions between membrane proteins and cardiolipin [36–41]. The neutral mass of TMCL is 1241.6 Da, and the mass of POPG is 749.00 Da (Table S1). Thus, the mass defect of TMCL with a reference lipid of POPG is 0.66 (1241.6 Da / 749.00 Da). Because the mass defect was similar to POPC/CHS, the spectra of POPG/TMCL showed a similar triple peak series (Figure 5d–f). Mass and mass defect analysis reveal approximately 9 TMCL and 135 POPG per nanodisc, which is only slightly higher than the expected ratio of 95/5.
Figure 5:
The zero-charge mass distribution summed over 0–200 V CID and colored by mass defect values (top), the mass defect values as a function of mass (middle), and the zero-charge mass distribution as a function of collision voltage (bottom) for nanodiscs with POPG alone (a–c) or POPG/TMCL (d–f) measured under positive mode with no additives (a, d), positive mode with added imidazole (b, e), and negative mode with no additives (c, f). The numbers of MSP per complex are annotated for key features. It is not clear what causes the bimodal mass distribution in POPG nanodiscs, but similar distributions have been previously observed [18].
Unlike POPC nanodiscs, which dissociate lipids continuously with elevated CID, POPG nanodiscs in positive mode tend to remain stable until they reach a critical collision voltage and break in half, going from two MSP belts per complex to only one with roughly half the mass (Figure 5a). Similarly, POPG/TMCL nanodiscs split in half with increasing CID in positive mode (Figure 5d). Addition of imidazole stabilizes both POPG and POPG/TMCL nanodiscs against splitting during CID (Figure 5e). Negative mode stabilizes both nanodiscs against splitting, but progressive loss of lipids does occur (Figure 5f).
Saposin A Lipid Nanoparticles
Finally, we explored the resonance approach with Saposin A lipid nanoparticles. MSP encircles nanodiscs with two belts, one for each leaflet of the bilayer. Saposin A, in contrast, covers both leaflets with a single monomer, but it does not encircle the lipid bilayer (Figure S7) [19]. Thus, it is more modular in size and can incorporate different numbers of SapA per complex depending on the assembly [20]. SapNPs have recently been employed for high-resolution cryo-electron microscopy of membrane proteins [42, 43]. Klassen and coworkers have shown that SapNPs are useful for presenting glycolipids for native MS [22–25] and have characterized the stoichiometries of complexes formed by addition of SapA to liposomes under acidic conditions [24]. Here, we reconstituted SapNPs from detergent solutions [20], similar to self-assembly of nanodiscs [44].
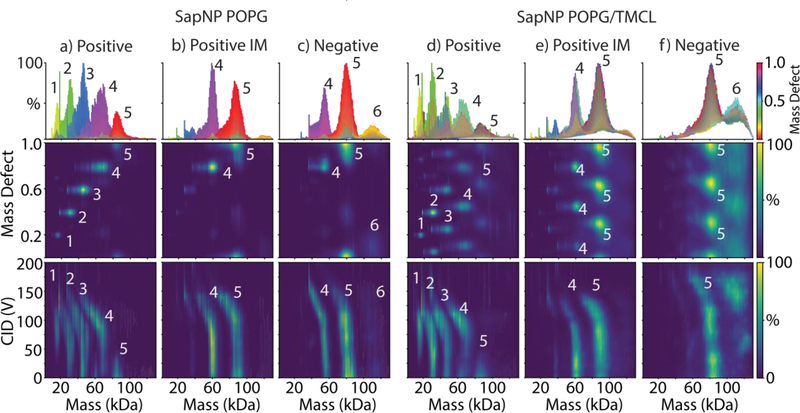
We determined stoichiometries of SapA with mass defect analysis. With POPG as the reference lipid, the mass defect of SapA is 0.19 (9131 Da / 749.00 Da). Thus, each added SapA shifts the mass defect by 0.19 (Tables S1 and S2). Positive ionization native MS of POPG SapNPs formed with a ratio of 12/1 lipid per SapA showed a mix of SapA stoichiometries beginning with 3, 4, or 5 SapA per complex and progressively dissociating to 2 or 1 SapA per complex with increasing collision voltage (Figure 6a). Similar to nanodiscs, adding imidazole (Figure 6b) or switching to negative ionization mode (Figure 6c) stabilized the SapNPs towards collisional dissociation. Stabilized SapNPs showed a mixture of 4 or 5 SapA per complex with a small population observed at 6 SapA per complex. Complexes with 5 SapA contained around 60 lipids per SapA, in agreement with the expected 12 to 1 ratio.
Figure 6:
The zero-charge mass distribution summed over 0–200 V CID and colored by mass defect values (top), the mass defect values as a function of mass (middle), and the zero-charge mass distribution as a function of collision voltage (bottom) for SapNPs with POPG alone (a–c) or POPG/TMCL (d–f) measured under positive mode with no additives (a, d), positive mode with added imidazole (b, e), and negative mode with no additives (c, f). The numbers of SapA per complex are annotated for key features.
SapNPs prepared with 95/5 POPG/TMCL showed similar stoichiometries and stabilities as POPG alone. The triplet pattern demonstrated the incorporation of cardiolipin, and the mass defect shift is consistent with a ratio of 95/5. Positive mode (Figure 6d) showed progressive dissociation of SapA from the complex and loss of cardiolipin. IM or negative mode stabilize the complexes towards dissociation of cardiolipin and SapA (Figure 6e and 6f). Thus, our approach of using resonant lipid masses and stabilization of lipoprotein complexes was successful for SapNPs as well as nanodiscs.
We also assembled SapNPs with POPC/cholesterol. However, we were unsuccessful in observing cholesterol incorporation at a 12/1 lipid/SapA ratio, even with stabilization by IM or negative mode. It is not clear whether cholesterol was not incorporated during self-assembly of the SapNPs or whether it was ejected during native MS. Further work will be necessary to unravel the structural or mechanistic differences.
Conclusions
We have demonstrated that combinations of lipids with resonant masses enable native MS analysis of intact nanodiscs and Saposin A lipoprotein nanoparticles with a wider range of different types of lipids, including glycolipids, sterols, and cardiolipin. Lipids that were close to 2, 1/2, 2/3, and 5/3 times the mass of the reference lipid were successfully resolved, and we expect that other lipid combinations where components are near multiples of 1, 2, or 3 of each other will be amenable to native MS. Furthermore, we have shown that negative ionization mode and addition of charge-reducing reagents such as imidazole stabilize lipoprotein complexes for native MS, particularly for labile lipids like cholesterol and ergosterol. Together, these approaches provide a framework for creating and analyzing lipoprotein complexes that more closely model native lipid combinations, which could prove useful in delivery of membrane proteins and peptides.
Resources
The compiled program and source code for the UniDec executable, Python API, and GUI are available online: https://github.com/michaelmarty/UniDec/Releases. Python scripts used to model nanodisc spectra are available for download with the source code.
Supplementary Material
Acknowledgements
The authors thank Maria Reinhardt-Szyba, Kyle Fort, and Alexander Makarov at Thermo Fisher Scientific for support on the UHMR Q-Exactive HF instrument. The pMSP1D1 plasmid was a gift from Stephen Sligar (Addgene plasmid #20061). The authors thank Christian Loew and Joanna Pieprzyk for providing the SapA plasmid and Elaine Marzluff for helpful discussions. This work was funded by the Bisgrove Scholar Award from Science Foundation Arizona, the American Society for Mass Spectrometry Research Award, and National Institute of General Medical Sciences and National Institutes of Health (Award Number R35 GM128624) to M.T.M. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1. Allison TM, Landreh M: Ion Mobility in Structural Biology. Advances in Ion MobilityMass Spectrometry: Fundamentals, Instrumentation and Applications, p. 161–195. Elsevier, (2019) [Google Scholar]
- 2. Calabrese AN, Radford SE: Mass spectrometry-enabled structural biology of membrane proteins. Methods. 147, 187–205 (2018) [DOI] [PubMed] [Google Scholar]
- 3. Kaur U, Johnson DT, Chea EE, Deredge D, Espino JA, Jones LM: Evolution of Structural Biology Through the Lens of Mass Spectrometry. Anal. Chem, DOI: 10.1021/acs.analchem.1028b05014 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marty MT, Hoi KK, Robinson CV: Interfacing Membrane Mimetics with Mass Spectrometry. Acc. Chem. Res 49, 2459–2467 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Calabrese AN, Watkinson TG, Henderson PJF, Radford SE, Ashcroft AE: Amphipols Outperform Dodecylmaltoside Micelles in Stabilizing Membrane Protein Structure in the Gas Phase. Anal. Chem 87, 1118–1126 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Watkinson TG, Calabrese AN, Giusti F, Zoonens M, Radford SE, Ashcroft AE: Systematic analysis of the use of amphipathic polymers for studies of outer membrane proteins using mass spectrometry. Int. J. Mass Spectrom 391, 54–61 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leney AC, McMorran LM, Radford SE, Ashcroft AE: Amphipathic Polymers Enable the Study of Functional Membrane Proteins in the Gas Phase. Anal. Chem 84, 9841–9847 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hopper JT, Yu YT, Li D, Raymond A, Bostock M, Liko I, Mikhailov V, Laganowsky A, Benesch JL, Caffrey M, Nietlispach D, Robinson CV: Detergent-free mass spectrometry of membrane protein complexes. Nat. Methods 10, 1206–1208 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hellwig N, Peetz O, Ahdash Z, Tascon I, Booth PJ, Mikusevic V, Diskowski M, Politis A, Hellmich Y, Hanelt I, Reading E, Morgner N: Native mass spectrometry goes more native: investigation of membrane protein complexes directly from SMALPs. Chem. Commun 54, 13702–13705 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chorev DS, Baker LA, Wu D, Beilsten-Edmands V, Rouse SL, Zeev-Ben-Mordehai T, Jiko C, Samsudin F, Gerle C, Khalid S, Stewart AG, Matthews SJ, Grünewald K, Robinson CV: Protein assemblies ejected directly from native membranes yield complexes for mass spectrometry. Science. 362, 829–834 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Dyck JF, Konijnenberg A, Sobott F: Native Mass Spectrometry for the Characterization of Structure and Interactions of Membrane Proteins In: Lacapere J-J (ed.) Membrane Protein Structure and Function Characterization: Methods and Protocols, p. 205–232. Springer New York, New York, NY, (2017) [DOI] [PubMed] [Google Scholar]
- 12. Reid DJ, Keener JE, Wheeler AP, Zambrano DE, Diesing JM, Reinhardt-Szyba M, Makarov A, Marty MT: Engineering Nanodisc Scaffold Proteins for Native Mass Spectrometry. Anal. Chem 89, 11189–11192 (2017) [DOI] [PubMed] [Google Scholar]
- 13. Zhang Y, Liu L, Daneshfar R, Kitova EN, Li C, Jia F, Cairo CW, Klassen JS: Protein-glycosphingolipid interactions revealed using catch-and-release mass spectrometry. Anal. Chem 84, 7618–7621 (2012) [DOI] [PubMed] [Google Scholar]
- 14. Han L, Morales LC, Richards MR, Kitova EN, Sipione S, Klassen JS: Investigating the Influence of Membrane Composition on Protein-Glycolipid Binding Using Nanodiscs and Proxy Ligand Electrospray Ionization Mass Spectrometry. Anal. Chem 89, 9330–9338 (2017) [DOI] [PubMed] [Google Scholar]
- 15. Li J, Richards MR, Kitova EN, Klassen JS: Delivering Transmembrane Peptide Complexes to the Gas Phase Using Nanodiscs and Electrospray Ionization. J. Am. Soc. Mass. Spectrom 28, 2054–2065 (2017) [DOI] [PubMed] [Google Scholar]
- 16. Marty MT, Hoi KK, Gault J, Robinson CV: Probing the Lipid Annular Belt by Gas-Phase Dissociation of Membrane Proteins in Nanodiscs. Angew. Chem. Int. Ed. Engl 55, 550–554 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Keener JE, Zambrano DE, Zhang G, Zak CK, Reid DJ, Deodhar BS, Pemberton JE, Prell JS, Marty MT: Chemical additives enable native mass spectrometry measurement of membrane protein oligomeric state within intact nanodiscs. J. Am. Chem. Soc 141, 1054–1061 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoi KK, Robinson CV, Marty MT: Unraveling the Composition and Behavior of Heterogeneous Lipid Nanodiscs by Mass Spectrometry. Anal. Chem 88, 6199–6204 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Popovic K, Holyoake J, Pomès R, Privé GG: Structure of saposin A lipoprotein discs. Proc. Natl. Acad. Sci 109, 2908–2912 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Flayhan A, Mertens HDT, Ural-Blimke Y, Martinez Molledo M, Svergun DI, Löw C: Saposin Lipid Nanoparticles: A Highly Versatile and Modular Tool for Membrane Protein Research. Structure. 26, 345–355.e345 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Frauenfeld J, Löving R, Armache J-P, Sonnen A, Guettou F, Moberg P, Zhu L, Jegerschöld C, Flayhan A, Briggs JAG, Garoff H, Löw C, Cheng Y, Nordlund P: A novel lipoprotein nanoparticle system for membrane proteins. Nat. Methods 13, 345–351 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leney AC, Rezaei Darestani R, Li J, Nikjah S, Kitova EN, Zou C, Cairo CW, Xiong ZJ, Prive GG, Klassen JS: Picodiscs for Facile Protein-Glycolipid Interaction Analysis. Anal. Chem 87, 4402–4408 (2015) [DOI] [PubMed] [Google Scholar]
- 23. Li J, Fan X, Kitova EN, Zou C, Cairo CW, Eugenio L, Ng KKS, Xiong ZJ, Prive GG, Klassen JS: Screening Glycolipids Against Proteins in Vitro Using Picodiscs and Catch-and-Release Electrospray Ionization-Mass Spectrometry. Anal. Chem 88, 4742–4750 (2016) [DOI] [PubMed] [Google Scholar]
- 24. Li J, Richards MR, Bagal D, Campuzano IDG, Kitova EN, Xiong ZJ, Prive GG, Klassen JS: Characterizing the Size and Composition of Saposin A Lipoprotein Picodiscs. Anal. Chem 88, 9524–9531 (2016) [DOI] [PubMed] [Google Scholar]
- 25. Li J, Han L, Li J, Kitova EN, Xiong ZJ, Prive GG, Klassen JS: Detecting Protein-Glycolipid Interactions Using CaR-ESI-MS and Model Membranes: Comparison of Pre-loaded and Passively Loaded Picodiscs. J. Am. Soc. Mass Spectrom 29, 1493–1504 (2018) [DOI] [PubMed] [Google Scholar]
- 26. Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG: Reconstitution of Membrane Proteins in Phospholipid Bilayer Nanodiscs In: Nejat D (ed.) Methods Enzymol, p. 211–231. Academic Press, San Diego, CA, (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lyons JA, Bøggild A, Nissen P, Frauenfeld J: Saposin-Lipoprotein Scaffolds for Structure Determination of Membrane Transporters In: Ziegler C (ed.) Methods Enzymol, p. 85–99. Academic Press, (2017) [DOI] [PubMed] [Google Scholar]
- 28. Reid DJ, Diesing JM, Miller MA, Perry SM, Wales JA, Montfort WR, Marty MT: MetaUniDec: High-Throughput Deconvolution of Native Mass Spectra. J. Am. Soc. Mass Spectrom 30, 118–127 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van de Waterbeemd M, Fort KL, Boll D, Reinhardt-Szyba M, Routh A, Makarov A, Heck AJ: High-fidelity mass analysis unveils heterogeneity in intact ribosomal particles. Nat. Methods 14, 283–286 (2017) [DOI] [PubMed] [Google Scholar]
- 30. Marty MT, Baldwin AJ, Marklund EG, Hochberg GK, Benesch JL, Robinson CV: Bayesian deconvolution of mass and ion mobility spectra: from binary interactions to polydisperse ensembles. Anal. Chem 87, 4370–4376 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Characterization of Compositional Heterogeneity in Intact Nanodisc Ions Containing Two Different Types of Lipids Using Fourier-Transformed Mass Spectra, Prell, J.S., Cleary, S.P.; ID 293709, June 4, Proceedings of the 66th ASMS Conference on Mass Spectrometry and Allied Topics, San Diego, Ca, June 3-June 7, 2018 [Google Scholar]
- 32. Yuan C, Furlong J, Burgos P, Johnston LJ: The size of lipid rafts: an atomic force microscopy study of ganglioside GM1 domains in sphingomyelin/DOPC/cholesterol membranes. Biophys. J 82, 2526–2535 (2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Simons K, Sampaio JL: Membrane organization and lipid rafts. Cold Sptring Harb. Perspect. Biol 3, a004697 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chini B, Parenti M: G-protein-coupled receptors, cholesterol and palmitoylation: facts about fats. J Mol Endocrinol. 42, 371–379 (2009) [DOI] [PubMed] [Google Scholar]
- 35. Goddard AD, Watts A: Regulation of G protein-coupled receptors by palmitoylation and cholesterol. BMC Biol. 10, 27–30 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marcoux J, Wang SC, Politis A, Reading E, Ma J, Biggin PC, Zhou M, Tao H, Zhang Q, Chang G, Morgner N, Robinson CV: Mass spectrometry reveals synergistic effects of nucleotides, lipids, and drugs binding to a multidrug resistance efflux pump. Proc. Natl. Acad. Sci 110, 9704–9709 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Patrick JW, Boone CD, Liu W, Conover GM, Liu Y, Cong X, Laganowsky A: Allostery revealed within lipid binding events to membrane proteins. Proc. Natl. Acad. Sci 115, 2976–2981 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Corey RA, Pyle E, Allen WJ, Watkins DW, Casiraghi M, Miroux B, Arechaga I, Politis A, Collinson I: Specific cardiolipin–SecY interactions are required for proton-motive force stimulation of protein secretion. Proc. Natl. Acad. Sci 115, 7967–7972 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schmidt V, Sidore M, Bechara C, Duneau JP, Sturgis JN: The lipid environment of Escherichia coli Aquaporin Z. Biochim Biophys Acta Biomembr. 1861, 431–440 (2019) [DOI] [PubMed] [Google Scholar]
- 40. Gupta K, Donlan JAC, Hopper JTS, Uzdavinys P, Landreh M, Struwe WB, Drew D, Baldwin AJ, Stansfeld PJ, Robinson CV: The role of interfacial lipids in stabilizing membrane protein oligomers. Nature. 541, 421–424 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martens C, Stein RA, Masureel M, Roth A, Mishra S, Dawaliby R, Konijnenberg A, Sobott F, Govaerts C, McHaourab HS: Lipids modulate the conformational dynamics of a secondary multidrug transporter. Nat. Struct. Mol. Biol 23, 744–751 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nguyen NX, Armache J-P, Lee C, Yang Y, Zeng W, Mootha VK, Cheng Y, Bai X.-c Jiang Y: Cryo-EM structure of a fungal mitochondrial calcium uniporter. Nature. 559, 570–574 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kintzer AF, Green EM, Dominik PK, Bridges M, Armache JP, Deneka D, Kim SS, Hubbell W, Kossiakoff AA, Cheng Y, Stroud RM: Structural basis for activation of voltage sensor domains in an ion channel TPC1. Proc. Natl. Acad. Sci 115, E9095–E9104 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bayburt TH, Grinkova YV, Sligar SG: Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2, 853–856 (2002) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.