Abstract
Objective:
The authors previously reported that the copy number of sequences encoding an Olduvai protein domain subtype (CON1) show a linear association with the severity of social deficits and communication impairment in individuals with autism. Here the authors use an improved measurement method and replicate this association in an independent population.
Method:
The authors obtained whole genome sequence data (WGS) and phenotype data on 215 individuals from the Autism Speaks MSSNG project. They derived copy number from WGS data using a modified sequence read depth technique. A linear mixed effects model was used to test association between Olduvai CON1 copy number and symptom severity measured by the Autism Diagnostic Interview – Revised. The authors then combined data from previous studies (N = 524) for final analyses.
Results:
The authors show a linear association between CON1 copy number and Social Diagnostic Score (SDS) (b = 0.24, p = 0.041) and Communication Diagnostic Score (CDS) (b = 0.23, p = 0.007). Using the combined data, the authors present strong associations of CON1 dosage with SDS (b = 0.18, p = 0.0008) and CDS (b = 0.13, p = 0.0009). The authors also implicate Olduvai subtypes found in two genes, NBPF1 and NBPF14 (R2 = 6.2%, p = 0.002). Associations were preferentially found in multiplex vs. simplex families.
Conclusions:
The finding of a third dose-dependent association between Olduvai sequences and autism severity, preferentially in multiplex families, provides strong evidence that this highly duplicated and under-examined protein domain family is playing an important role in inherited autism.
INTRODUCTION
Autism is a lifelong condition that is characterized by greatly impaired social and communicative abilities as well as increased repetitive behaviors and stereotyped interests. It is a common heritable condition that affects as many as 1 in 42 males and 1 in 189 females (1). Autism is an exceptionally heterogenous behaviorally defined condition that may be parsed into severity-based subtypes (2), but also may be best described as a continuum of symptom severity that progresses from minimal impact to a substantial one (3). For example, as many as 31% of individuals with autism have an intellectual disability, but also as many as 44% of children are identified with average and above average IQ (1). Similarly, traditional autism-like characteristics such as social reciprocity tend to follow a continuous distribution in individuals with autism (4).
Autism can also be divided genetically into multiplex and simplex forms. The former requires that families have at least two affected siblings while the latter requires only one affected child and one unaffected child. Increasing data is suggesting that the two forms may represent distinct conditions and involve different genetic etiologies (5,6). Even though it has been suggested that simplex autism is more common (7), the largest studies of autism reported so far do not appear to corroborate this view, and point to autism as primarily an inherited condition (8,9).
Important genetic components of autism are strongly suggested by heritability estimates which range from 50% to 90% (8,10). However, despite the high heritability, few genetic markers of substantial effects have been identified that can explain a large portion of the condition. Genome wide association studies (GWAS) have identified numerous single nucleotide polymorphisms (SNPs) of very small effect (11,12), and whole genome wide copy number variation (CNV) studies have identified de novo regions of very large effect in simplex children, yet these events appear to be rare (13,14).
Despite extensive efforts to identify the genetic factors underlying autism, a substantial portion of the expected genetic contribution to the disorder has not yet been identified. This missing heritability and the heterogeneity of the condition have led to hypotheses of polygenetic effects involving hundreds-to-thousands of small effect loci, and speculation that autism could be caused by numerous different alleles acting in summation or in a complex and unidentified pattern (15). However, evidence suggests this contribution may not substantive (16). Further, few genetic studies have accounted for the extensive heterogeneity of the condition, and most label all affected as the same in classic case control designs.
It has been proposed that the limited success of the efforts to identify the genetic factors behind autism may be because the critical genomic contributors are being missed by currently available genomic technologies (17). In this regard, highly duplicated, copy number polymorphic sequences are excellent candidates to fill such a role. Indeed, such sequences are not only missed by conventional genetic approaches, including exome sequencing, but because their copy number may vary continuously over a wide range, they have the potential to produce a broad spectrum of phenotypic variation.
One such candidate of high and variable copy number that has been absent from traditional genetic studies of autism is the Olduvai (formerly DUF1220) protein domain family (18,19) primarily encoded by NBPF genes (20). Olduvai domains are approximately 65 amino acids in the length and show the greatest human lineage-specific increase in copy number of any protein coding region in the genome (~300 copies in human with 165 being human-specific) (21,22). Olduvai copy number also shows a striking correlation with brain size and neuron number among anthropoid primates, and with brain size and cognitive aptitude in the human population (23–25). Interestingly, most human Olduvai copies map to 1q21, a genomic region in which duplications have been linked with autism (and macrocephaly) and reciprocal deletions to schizophrenia (and microcephaly) (26,27).
Olduvai copies have been parsed into six primary evolutionary subtypes based on sequence similarity: conserved (CON1–3) and human lineage-specific (HLS1–3) (21). The primary means of adding human-specific copies has been via amplification of the Olduvai triplet: a three-domain unit composed of an HLS1, HLS2 and HLS3 subtype (21). Olduvai copy number is also highly variable in the human population (ranging from 250–350 copies) but, because of the repetitive nature of the family, has not been directly studied in conventional genetic studies of autism. As such, Olduvai sequences represent a rich source of unexamined functional allelic variation.
Using an earlier method to follow Olduvai copy number (e.g. ddPCR), we previously demonstrated that, in multiplex families, the copy number of Olduvai subtype CON1 showed a linear association with the severity of the primary symptoms of autism (28), a finding that was subsequently replicated in a second population (29). However, CON1 subtypes, which range from approximately 60–85 diploid copies in humans, are found at >20 dispersed loci on chromosome 1, and ddPCR only measures the total number of copies in the genome. As a result, these previous studies were unable to provide information on which copies may be critical to the severity associations.
To address this limitation, we combined two recently developed resources: 1) availability of whole genome sequences for large numbers of individuals with autism (30,31) and 2) a higher resolution method for measuring Olduvai copy number (32). Using these tools, we here report a third linear association between CON1 dosage and the severity of the core symptoms of autism in an independent population. We also implicate gene-specific Olduvai subtypes as potential drivers of this association. Finally, we show that the Olduvai associations are preferentially found in multiplex vs. simplex families, and thus may be important to inherited forms of autism.
METHODS:
This study first sought to replicate previous work in an independent population using different assay methods, and once replication was identified data from all three studies were combined for an overall evaluation. Thirdly, a latent genotype was constructed from specific NBPF genes and then compared to a latent phenotype to identify important NBPF genes.
Replication Population
Individuals with phenotype data from the Autism Genetic Resource Exchange (33) and whole genome sequence data from the Autism Speaks MSSNG consortium (30,31) were used for the replication analysis. Individuals met autism criteria through the Autism Diagnostic Interview - Revised (ADI - R) and the Autism Diagnostic Observation Schedule (ADOS). While a large number of samples have been collected in the Autism Speaks MSSNG project, we were able to assemble 215 multiplex samples that were not included in our previous investigations and that had appropriate phenotype measures for replication. Non-white race ethnicity was included and identified by analyses of WGS using a consensus of google developed machine learning algorithms described further here (https://cloud.google.com/blog/products/gcp/genomic-ancestry-inference-with-deep-learning) and ADMIXTURE software on a subset of common SNPs (34). Given the small numbers of specific minorities non-white vs white ethnicity were collapsed into two categories. Population stratification was addressed through the addition of this race/ethnicity variable in multivariate regression analyses discussed further below.
Copy Number Measurement
Copy number of Olduvai domains in the replication population was obtained through a WGS read depth method (32). Briefly, the read depth of regions containing Olduvai domains was compared to the range of location of the regions based on hg38. The depth of reads compared to the breadth of the region of interest covered, corrected by GC content and copy number of ultra-conserved sequences, yielded a total copy number measurement of candidate Olduvai sequences per individual. This technique was validated by digital droplet PCR (ddPCR) and is explained further in a different report (32). Details on all aspects of whole genome sequencing and mapping for all data are described as part of the Autism Speaks MSSNG Project (31). To insure that cross-platform variation did not influence results, data from only one sequencing platform (illumina HiSeq X) was included. Briefly, sequences were generated using paired-end reads of 150-bp length on an Illumina HiSeq X platform following Illumina’s recommended protocol. Given high correlation among HLS1, HLS2 and HLS3 domains we utilize HLS1 measurements as representative of HLS2 and HLS3. The original reports that identified severity associations relied on ddPCR-derived copy number that determined total genomic copy number, e.g. for the CON1 subtypes. Read depth analysis of WGS data, on the other hand, allows specific subtype copy number to be identified within specific NBPF genes (32). In order to replicate previous work, Olduvai CON1 copy number derived from WGS data was summed across all NBPF genes and used as the primary explanatory variable in multivariate regression.
Replication Analysis
Linear multivariate regression with a random intercept for sibling family was used to test association between total Olduvai CON1 and autism symptom severity. The mixed model was fit with reduced maximum likelihood and tests of significance were two tailed t-tests. Severity was determined as previously by Social Diagnostic Score (SDS) and Communicative Diagnostic Score (CDS) from the ADI-R (28). Covariates included other Olduvai domain totals (CON2, CON3, and HLS1) as well as sex, age, Raven Matrices IQ, and race/ethnicity. Reduced models were developed though a backward selection procedure keeping only significant covariates (p < 0.05) and the suggestive interaction term sex by CON1 (p = 0.10 (SDS), and p = 0.02 (CDS)). Akaike Information Criterion (AIC) supported the use of a random effect with random intercept for sibling family. An interaction of CON1 by sex was explored given the biological plausibility. Residual diagnostics did not indicate departures from linear model assumptions.
Combined Analyses
Data from the three projects were combined for final evaluation. Associations of CON1 and HLS1 copy number with symptoms were tested with multivariate linear regression as previously described. Random effects, family ID and study, were explored but were not supported by AIC criteria and thus not included. Backward selection with appropriate, previously mentioned covariates was utilized and results from parsimonious models are presented. Interactions of copy number by sex and copy number by inheritance were examined and interaction p-values are presented where simplex effects were explored.
Latent Variable Analysis
We used Partial Least Squares Path Modeling (PLS-PM) to explore contributions of specific NBPF genes to a latent genotype and social severity phenotype. Bootstrap mean estimates and 95% confidence intervals were generated with 5000 iterations to validate PLS-PM associations. Gene and phenotype loading p - vales using z-statistics derived from the bootstrapped error were generated as recommended in other work (35), and are presented to aid in the evaluation of multiple comparisons during the latent variable construction. A conservative critical alpha in this stage of analyses is Bonferroni corrected p = 0.002 (0.05/30). This corresponded to 11 of the larger and variable NBPF genes (NBPF1, 4, 6, 8, 9, 10, 11, 12, 14, 19, and 20), by 2 clades (CON1 and HLS1) in addition to 8 phenotype measures described further below.
To avoid inflation of the association between latent variables PLS-PM model reduction was based on high levels of multicollinearity among Olduvai copy number and 95% bootstrap intervals of loadings that did not include 0. All analyses were conducted with R (https://cran.r-project.org/) the nlme package (36) and the plspm package (37).
Phenotype Information
This project utilized the ADI-R to provide the SDS and the CDS to replicate previous findings. The diagnostic scores contain numerous sub questions that are each organized into four subdomains that measure relevant social and communicative behaviors. These subdomains are then summed through an algorithm to create the diagnostic scores (e.g. SDS and CDS) and give clinical guidance in diagnosis. The four subdomains of the SDS include; SOC1, SOC2, SOC3, and SOC4, with questions probing items such as responding to other children, emotional sharing, seeking comfort and social smiling. The four subdomains of the verbal CDS included: COM1, COM2, COM3VT, and COM4 and explore language items such as stereotyped utterances, pronoun reversal and social use of language.
RESULTS:
Replication Population
Two hundred fifteen multiplex individuals were available for the replication analyses. See Table 1 for a phenotypic description of the replication population, which was predominantly male (77%) and of western European descent (77%). Sixty-eight sibling groups were used in these analyses and group size varied from 2 affected siblings to 4. The summed CON1 copy number ranged from 60.7 to 84.3 copies with a mean of 70.7 copies and followed a Gaussian distribution. Interestingly, CON1 copy number of NBPF1 had substantial variation and ranged from 8.5 to 28.5 copies. HLS1 copy number of NBPF14 was also variable and ranged from 22.9 to 46.2 copies (Figure 1).
Table1.
includes summary statistics of the phenotypes and characteristics of the population understudy.
Replication Population Characteristics
| Q1 | Mean | Q3 | |
|---|---|---|---|
| ADIR Social Score | 17 | 20.9 | 25 |
| ADIR Communication Score | 14 | 17.0 | 20 |
| ADIR Repetitive Behaviors | 5 | 6.6 | 8 |
| Raven Matrices IQ* | 95.0 | 107.0 | 115.5 |
| Age | 6.9 | 10.0 | 12.1 |
| Total CON1 copies | 68.1 | 70.7 | 73.0 |
| Total HLS1 | 188.5 | 200.2 | 212.1 |
| N (%) | |||
| Sex | male 165 (76.7%) | ||
| Predicted Ethnicity | AFR 3 (1.4%) | ||
| AMR 6 (2.8%) | |||
| EAS 4 (1.9%) | |||
| EUR 165 (76.7%) | |||
| OTH 37 (17.2%) | |||
| AFR 3 (1.4%) | |||
| OTH 37 (17.2%) | |||
22 missing
Figure 1. Olduvai copy number distribution in NBPF1 and NBPF14.
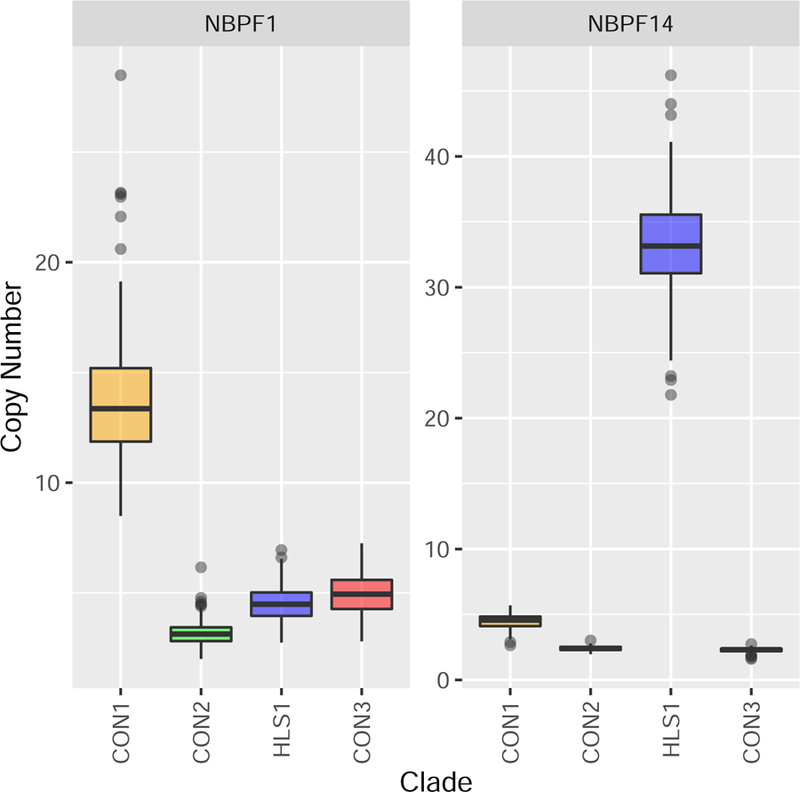
Figure 1 shows the distribution of copy number of clades within NBPF1 and NBPF14.
In the replication population incrementally increasing copy number of total CON1 was associated with incrementally increasing SDS in males (b = 0.25, 95%CI 0.02 – 0.49, p = 0.044) (interaction p = 0.10) (Table 2). A similar association was detected in males where incrementally increasing copies of total CON1 were associated with incrementally increasing severity of verbal communication diagnostic score (CDS) (b = 0.23, 95%CI 0.21 – 0.25, p = 0.008) (interaction p = 0.02) (Table 2). As presented in Table 2, these results are highly similar to the previous two reports.
Table 2.
Association Between CON1 copy number and symptoms in multiplex individuals
displays associations from all studies (this report, the findings from Davis et al 2014, and Davis et al 2015) between total CON1 copy number and social and communicative symptoms measured from the ADI-R in Multiplex children. The lack of association in Simplex individuals is also displayed.
| Symptom | B | Se | p-value |
|---|---|---|---|
| ADIR Social this report | 0.25 | 0.12 | 0.044 |
| ADIR Social 2015 | 0.24 | 0.11 | 0.036 |
| ADIR Social 2014 | 0.25 | 0.11 | 0.021 |
| ADIR Comm this report | 0.23 | 0.08 | 0.008 |
| ADIR Comm 2015 | 0.16 | 0.09 | 0.072 |
| ADIR Comm 2014 | 0.18 | 0.08 | 0.047 |
| Combined Population | |||
| ADIR Social | 0.18 | 0.05 | 0.0008 |
| ADIR Comm | 0.13 | 0.04 | 0.0009 |
| ADIR Social Simplex Strata (n=64) | −0.05 | 0.15 | 0.72 |
| ADIR Comm Simplex Strata (n=64) | −0.01 | 0.10 | 0.87 |
Combined Analyses
The combined population analysis included data for 524 individuals with 64 simplex and the remainder multiplex. In multiplex individuals, for each additional copy of CON1, SDS increased (i.e. severity increased) 0.18 points (95%CI 0.08 – 0.28, p = 0.0008), and CDS increased 0.13 points (95%CI 0.05 – 0.21, p = 0.0009) (Table 2) (SDS overall F-test = 5.16 on 4 and 517 df p = 0.0004, and CDS overall F-test = 6.11 on 2 and 515 df p = 0.002). Age was a significant predictor (bage = 0.18, 95%CI 0.06 – 0.30, p = 0.002) in the SDS analysis. Increasing copy number of HLS1 was protective in the model of CDS (bHLS = −0.02, 95%CI −0.04 – 0.0, p = 0.046). The importance of multiplex over simplex was reaffirmed with SDS and CDS, where no associations were identified in simplex (SDS bsimplex = −0.05, 95%CI −0.33 – 0.21, p = 0.72, interaction p = 0.11) and (CDS bsimplex = −0.02, 95%CI −0.23 – 0.19, p = 0.87, interaction p = 0.16).
Latent Genotype and Phenotype Analyses
PLS-PM analysis identified an important latent genotype and latent severity phenotype that were associated with each other (b = 0.24, 95%CI 0.14 – 0.34, p = 0.002), where the variation of the genetic latent variable explained a bootstrapped 6.2% (95% CI 0.02, 0.12) of the variation of the latent social severity phenotype (Table 3). The communication score domains and social domains had substantial positive loadings on the social latent variable where SOC1, SOC2, SOC3, SOC4 and COM2VT and COM4 were important (all p <0.0001) (Table 3). NBPF1 CON1 (positive loading = 0.75, 95% CI 0.19 – 1.00, p = 0.0004) and NBPF14 HLS1 (positive loading = 0.72, 95% CI 0.17 – 1.00, p = 0.0007) were important contributors to the latent genotype (Table 3).
Table 3a.
Association between latent genotype and latent phenotype.
displays the linear association between the latent genotype and the latent phenotype as well as the R-squared value between the two.
| B | Se | p-value | Bootstrap R- squared | ||
|---|---|---|---|---|---|
| Latent Genotype | 0.21 | 0.07 | 0.002 | 0.062 (95% 0.022 0.120) | |
| Table 3b. Bootstrap mean loadings and 95 % Confidence Interval | |||||
| displays loadings, bootstrapped confidence intervals, and p-values calculated from the confidence interval of important contributors to the latent genotype and phenotype. | |||||
| Loadings | Mean Bootstrap | Bootstrap | 95% CI | p-value* | |
| Latent Genotype | |||||
| NBPF1 CON1 | 0.75 | 0.19 | 1.00 | 0.0004 | |
| NBPF14 HLS1 | 0.72 | 0.21 | 1.00 | 0.0007 | |
| Latent Phenotype | |||||
| SOC1 | 0.73 | 0.50 | 0.84 | <0.0001 | |
| SOC2 | 0.66 | 0.44 | 0.79 | <0.0001 | |
| SOC3 | 0.74 | 0.53 | 0.84 | <0.0001 | |
| SOC4 | 0.68 | 0.42 | 0.82 | <0.0001 | |
| COMM2 | 0.61 | 0.40 | 0.80 | <0.0001 | |
| COMM4 | 0.74 | 0.60 | 0.85 | <0.0001 | |
p-value is derived from the bootstrapped 95% CI
DISCUSSION:
Third linear association between Olduvai copy number and autism severity
Here, using an independent population and a higher resolution measurement method, we present a third linear association between increasing Olduvai CON1 copy number and worsening of autism social and communicative symptoms. The patterns of associations found here are highly similar to those previously reported (28,29), with only minor differences: the associations here are found predominantly in males and, compared to previous studies, show an increased effect size in communication score. However, there are several reasons why such study-to-study variation can be expected: 1) autism is a complex condition with a wide range of phenotypic characteristics, 2) assessments of phenotypes can be expected to be imprecise, as they are primarily based only on caregiver reports (using algorithm scores from the ADI-R) rather than physiological measurements, and 3) highly accurate copy number measurement of highly duplicated sequences, such as those encoding Olduvai domains, is difficult and copy number determinations are sometimes inexact.
We also show that, when the findings of this study are combined with those from our two previous studies, the linear association between CON1 and autism severity is unusually strong: (n = 524; b = 0.18; se = 0.05; p = 0.0008 for ADIR Social and b = 0.13; se = 0.04; p = 0.0009 for ADIR Comm). Given the substantial challenges, mentioned above, related to accurate autism phenotyping and Olduvai genotyping, we believe it is remarkable that these studies have now identified similar associations in three independent groups. It is also noteworthy that, unlike other recent genetic studies of autism, the associations reported here have been detectable, and replicable, using only modest sample sizes. Taken together, such findings strongly support a significant role for Olduvai CON1 in the severity of social and communication impairment in autism.
Associations are found preferentially with Multiplex and not Simplex families
It is noteworthy that the significant linear associations with Olduvai copy number we report have appeared predominantly with multiplex families compared to simplex families. This trend fits with other accumulating evidence that points to the possibility that multiplex and simplex may be genetically and mechanistically distinct disorders (5). First, multiplex autism, which requires that at least two children have autism, is thought to better represent the inherited form of autism. In contrast, simplex is often due to de novo events (31,38), but not exclusively so (39). Second, the phenotypes of multiplex and simplex autism are typically distinct, with simplex phenotypes tending to be more severe than multiplex (6,40). Third, recent large-scale genome sequencing studies of simplex autism have not found genes that are involved in the typical core phenotypes of autism such as social deficits and communication impairment. Rather, such simplex studies are preferentially finding genes important to motor functions (41). In contrast, our studies, now confirmed in three independent populations, suggest an opposite trend: we preferentially find associations involving the core autism phenotypes (social and communication impairment) in multiplex and not simplex families. In this regard, it is worth pointing out that early gene finding studies of inherited autism met with minimal success, a fact that has been mentioned as a reason for initiating genomic studies of autism using simplex families (7). The findings we report here raise the possibility that the previous lack of success in identifying genetic contributors behind inherited autism may be because key sequences that are contributing to multiplex autism involve difficult to measure, highly duplicated, dynamic and copy number polymorphic sequences, such as those encoding the Olduvai family, and, as a result, have never been directly measured in other studies of autism (17).
Candidate Olduvai subtypes that may be driving the associations with autism severity
In our previous studies, we reported linear associations between autism and the copy number of the CON1 subtype of Olduvai. However, the measurement methods we used could not determine which of the many copies of CON1 (approximately 60–85 diploid copies) were underlying the association. To address this limitation, we employed a high-resolution sequence read depth method that, when applied to WGS datasets, allowed many of the different NBPF genes and Olduvai subtypes to be discriminated from one another.
To identify specific NBPF genes and Olduvai subtypes important to a social severity phenotype we examined our WGS read depth data utilizing a latent variable analytic method, partial least squares path modeling (PLS-PM). Numerous two by two comparisons are commonly conducted in genetic association studies (or potentially in this instance, numerous 1 continuous gene by 1 continuous phenotype comparisons are possible). These comparisons assume independence, and moreover, that the genotype has a unique effect on a specific measured phenotype. However, in autism many characteristics are highly correlated making traditional 2×2 statistical approaches inefficient and redundant. At the same time, simply summing characteristics or clinically grouping other combinations of symptoms will reduce the number of comparisons but may not best describe the phenotype as it relates to an important genotype. PLS-PM addresses these challenges through the construction of the latent genotype and phenotype variables, and then makes one statistical test between the two. In this case, of eight social metrics, two communication sub-scores were excluded and, of 22 NBPF genes and subtypes, two were retained, NBPF14-HLS1 and NBPF1-CON1. In this study the latent genetic variable explained 6.2% of the latent social phenotype which, in the context of genetic association studies of complex heterogenous conditions, is considerable. This unusually large effect size speaks to both the importance of Olduvai in autism and the importance of refined phenotypes in complex conditions.
Potential Importance of NBPF14-HLS1 and NBPF1- CON 1
NBPF14- HLS 1
The NBPF14 gene is located in the 1q21 region of chromosome 1 and is part of a genomic sequence that has been repeatedly associated with autism(26,27). NBPF14 is one of the four human NBPF genes that together encode the great majority of human-specific Olduvai copies. These additional human-specific copies have been primarily generated by tandem intragenic expansion of the Olduvai triplet composed of an HLS1, HLS2 and HLS3 subtype (21,22), These subtypes show high sequence similarity and, because additions/deletions of entire triplets is the predominant form of copy number variation in the expanded NBPF genes, the HLS1 subtype of NBPF14 can be viewed as a proxy for a full Olduvai triplet. These three-domain blocks are also highly copy number polymorphic in the human population, among both typically developing individuals and individuals with autism (28,32). Thus, their high level of variation in humans, the inability of conventional genomic methods to directly examine these coding sequences (17,42), the multiple studies implicating this genomic region in autism (43,44), and our studies that link Olduvai copy number variation to the severity of the core symptoms of multiplex autism (28,29), make sequences encoding NBPF14-related Olduvai triplets prime candidates to be involved in inherited forms of autism.
NBPF1 CON1:
The copy number of the NBPF1 gene among humans appears to be highly variable and the gene is thought to encode several CON1 domains (21,32). These characteristics fit with the idea that variation of CON1 in NBPF1 has not been monitored in most genomic studies of autism, and that NBPF1-related sequences (e.g. NBPF1L) may not be correctly mapped in the most recent human genome assembly (hg38). Given these observations, the possibility that NBPF1 CON1 may be involved in influencing autism severity should not be considered surprising.
Genomic trade-offs and links with neurogenesis
The Olduvai protein domain family has been linked with both evolutionary benefit (copy increase is associated with brain expansion and cognitive aptitude (23–25) as well as with disease (increases have been linked to autism and macrocephaly and decreases to schizophrenia and microcephaly (23,45). Such duality of effect has been incorporated into a cognitive genomic trade-off model that proposes that 1) Olduvai’s effect on brain size and neuron number may be dosage-dependent, e.g. more copies produce more neurons (45), and 2) variation in Olduvai copy number can occur in different ways, and it is which, where, how and when copies change that determines whether the consequences will be beneficial or harmful (17).
Evidence suggests the Olduvai trade-off may be related to variation in neurogenesis. For example, Olduvai has been shown to promote neural stem cell proliferation and exhibits a dosage-dependent association with gray matter volume and IQ in healthy individuals, both of which are related to neuron number (23–25). Olduvai copy number also shows a strong linear association with primate neuron number and brain size (but not body size) (23,24). Finally, human-specific Olduvai sequences have recently been linked with NOTCH-related pathways known to promote neurogenesis (unpublished data).
As previously suggested (45), the involvement of Olduvai in influencing neuron number, possibly via centrosomal-mediated effects, may also be relevant to its role in autism and schizophrenia. It is well established that how and when neurons are produced, and connections established involve processes that, if altered, can produce atypical neuronal density and growth. In this regard, excesses in neuron number along with accelerated brain growth have been associated with autism (46). In contrast, neuronal deficits may be important in schizophrenia where smaller gray matter regions (47) and reduced Olduvai copy number (48) have been reported. In short, as Olduvai increases autism severity and brain size increase while the opposite trend is true for schizophrenia. These observations add further support to the proposal that autism and schizophrenia may be opposites of a cognitive disease continuum (43) involving opposites in Olduvai dosage (17). Such findings raise the testable hypothesis that the associations of Olduvai dosage with the severity of autism and schizophrenia are due to Olduvai-driven abnormalities in neurogenesis.
Finally, the data reported here, linking Olduvai sequences with autism severity, using a different means of assessing copy number and different population, provides additional support for such a tradeoff model. Because of remaining technological limitations involving measurement of highly duplicated sequences, a more detailed description of the nature of Olduvai variation in humans, in either disease or non-disease populations, has not yet been reported. For similar reasons, it has been difficult to examine the potential involvement of Olduvai variation in autism risk. Given that much of the genetic contributors to inherited autism remain missing (17), and given the possibility that multiplex autism may be due to a single dominant transmitted trait (16), it is intriguing that variation in the ~300-member human Olduvai family has yet to be fully examined in inherited autism. Continued improvements in genome sequencing technology, that allow such sequences to be studied with greater precision, should permit these questions to be tested more rigorously in the future.
Figure 2. Olduvai latent genotype and latent social phenotype path model with loadings.
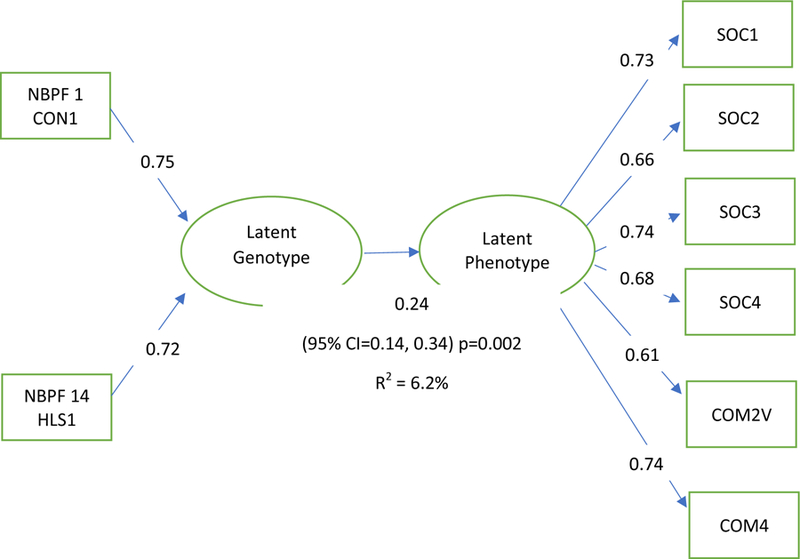
Figure 2 diagrams the formative path model of the latent genotype and latent phenotype relationship.
Acknowledgments
This work was supported by NIH grant R01 MH108684 (JMS) and Pilot Award 309230 from the Simons Foundation Autism Research Initiative (JMS).
We gratefully acknowledge the resources provide by the Autism Speaks MSSNG project and the Autism Genetic Resource Exchange (AGRE) Consortium as well as the participating families.
Footnotes
Previous presentation. None
Disclosures. All authors report no competing interests.
References
- 1.Baio J, Wiggins L, Christensen DL, Maenner MJ, Daniels J, Warren Z, et al. Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years — Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2014. MMWR Surveill Summ 2018. April 27;67(6):1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grzadzinski R, Huerta M, Lord C. DSM-5 and autism spectrum disorders (ASDs): an opportunity for identifying ASD subtypes. Mol Autism 2013. May 15;4(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spiker D, Lotspeich LJ, Dimiceli S, Myers RM, Risch N. Behavioral phenotypic variation in autism multiplex families: Evidence for a continuous severity gradient. Am J Med Genet 2002. March 8;114(2):129–36. [DOI] [PubMed] [Google Scholar]
- 4.Constantino JN, Todd RD. Autistic Traits in the General Population. Arch Gen Psychiatry 2003. May 1;60(5):524. [DOI] [PubMed] [Google Scholar]
- 5.Zhao X, Leotta A, Kustanovich V, Lajonchere C, Geschwind DH, Law K, et al. A unified genetic theory for sporadic and inherited autism. Proc Natl Acad Sci 2007. July 31;104(31):12831–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Virkud YV, Todd RD, Abbacchi AM, Zhang Y, Constantino JN. Familial aggregation of quantitative autistic traits in multiplex versus simplex autism. Am J Med Genet Part B Neuropsychiatr Genet 2009. April 5;150B(3):328–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Autism Risk Genes: Success with the Simplex Approach | Simons Foundation [Internet] [cited 2018 Jul 30]. Available from: https://www.simonsfoundation.org/2015/07/27/autism-risk-genessuccess-with-the-simplex-approach/
- 8.Sandin S, Lichtenstein P, Kuja-Halkola R, Larsson H, Hultman C, Reichenberg A. THe familial risk of autism. JAMA 2014;311(17):1770–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandin S, Lichtenstein P, Kuja-Halkola R, Hultman C, Larsson H, Reichenberg A. The Heritability of Autism Spectrum Disorder. JAMA 2017. September 26;318(12):1182–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tick B, Bolton P, Happé F, Rutter M, Rijsdijk F. Heritability of autism spectrum disorders: a metaanalysis of twin studies. J Child Psychol Psychiatry 2016. May;57(5):585–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB, et al. Most genetic risk for autism resides with common variation. Nat Genet 2014. August 20;46(8):881–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niemi MEK, Martin HC, Rice DL, Gallone G, Gordon S, Kelemen M, et al. Common genetic variants contribute to risk of rare severe neurodevelopmental disorders. Nature 2018. October 26;562(7726):268–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. Strong Association of De Novo Copy Number Mutations with Autism. Science (80- ) [Internet] 2007. April 20 ;316(5823):445–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy D, Ronemus M, Yamrom B, Lee Y ha, Leotta A, Kendall J, et al. Rare De Novo and Transmitted Copy-Number Variation in Autistic Spectrum Disorders. Neuron 2011;70:886–97. [DOI] [PubMed] [Google Scholar]
- 15.Klei L, Sanders SJ, Murtha MT, Hus V, Lowe JK, Willsey A, et al. Common genetic variants, acting additively, are a major source of risk for autism. Mol Autism 2012. October 15;3(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ronemus M, Iossifov I, Levy D, Wigler M. The role of de novo mutations in the genetics of autism spectrum disorders. Nat Rev Genet 2014. February 16;15(2):133–41. [DOI] [PubMed] [Google Scholar]
- 17.Sikela JM, Searles Quick VB. Genomic trade-offs: are autism and schizophrenia the steep price of the human brain? Hum Genet 2018. January 15;137(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Popesco MC, Maclaren EJ, Hopkins J, Dumas L, Cox M, Meltesen L, et al. Supporting Online Material for Human Lineage–Specific Hyperamplification, Selection, and Neuronal Expression of DUF1220 Domains. Publ 1 Sci 2006; 313. [DOI] [PubMed] [Google Scholar]
- 19.Sikela JM, van Roy F. Changing the name of the NBPF/DUF1220 domain to the Olduvai domain. F1000Research 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vandepoele K, Van Roy N, Staes K, Speleman F, Van Roy F. A novel gene family NBPF: Intricate structure generated by gene duplications during primate evolution. Mol Biol Evol 2005;22:2265–74. [DOI] [PubMed] [Google Scholar]
- 21.O’Bleness MS, Dickens CM, Dumas LJ, Kehrer-Sawatzki H, Wyckoff GJ, Sikela JM. Evolutionary history and genome organization of DUF1220 protein domains. G3 (Bethesda) 2012. September;2(9):977–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Bleness M, Searles VB, Dickens CM, Astling D, Albracht D, Mak ACY, et al. Finished sequence and assembly of the DUF1220-rich 1q21 region using a haploid human genome. BMC Genomics 2014;15:387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dumas LJ, O’Bleness MS, Davis JM, Dickens CM, Anderson N, Keeney JG, et al. DUF1220-domain copy number implicated in human brain-size pathology and evolution. Am J Hum Genet 2012. September 7;91(3):444–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keeney JG, Davis JM, Siegenthaler J, Post MD, Nielsen BS, Hopkins WD, et al. DUF1220 protein domains drive proliferation in human neural stem cells and are associated with increased cortical volume in anthropoid primates. Brain Struct Funct 2015. September;220(5):3053–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis JM, Searles VB, Anderson N, Keeney J, Raznahan A, Horwood LJ, et al. DUF1220 copy number is linearly associated with increased cognitive function as measured by total IQ and mathematical aptitude scores. Hum Genet 2014;134(1):67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino C a, Sahoo T, et al. Recurrent reciprocal 1 q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet 2008. December;40(12):1466–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mefford H, Sharp A, Baker C, Itsara A, Jiang Z, Buysse K, et al. Recurrent Rearrangements of Chromosome 1q21.1 and Variable Pediatric Phenotypes 2008;16(16). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis JM, Searles VB, Anderson N, Keeney J, Dumas L, Sikela JM. DUF1220 dosage is linearly associated with increasing severity of the three primary symptoms of autism. PLoS Genet 2014. March;10(3):e1004241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis JM, Searles Quick VB, Sikela JM. Replicated linear association between DUF1220 copy number and severity of social impairment in autism. Hum Genet 2015. June;134(6):569–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuen RKC, Merico D, Bookman M, L Howe J, Thiruvahindrapuram B, Patel RV, et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci 2017. April;20(4):602–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuen RKC, Thiruvahindrapuram B, Merico D, Walker S, Tammimies K, Hoang N, et al. Wholegenome sequencing of quartet families with autism spectrum disorder. Nat Med 2015. February;21(2):185–91. [DOI] [PubMed] [Google Scholar]
- 32.Astling DP, Heft IE, Jones KL, Sikela JM. High resolution measurement of DUF1220 domain copy number from whole genome sequence data. BMC Genomics 2017;18(1):614–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geschwind DH, Sowinski J, Lord C, Iversen P, Shestack J, Jones P, et al. The Autism Genetic Resource Exchange: A Resource for the Study of Autism and Related Neuropsychiatric Conditions. Am J Hum Genet 2001. August;69(2):463–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res 2009. September;19(9):1655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altman DG, Bland JM. How to obtain the P value from a confidence interval. BMJ 2011. August 8;343:d2304. [DOI] [PubMed] [Google Scholar]
- 36.Pinheiro Bates, DebRoy Sakar, Team RC. nlme: Linear and Nonlinear Mixed Effects Models [ Internet] 2018. Available from: https://cran.r-project.org/package=nlme.
- 37.Sanchez, Trinchera, Russolillo. plspm: Tools for Partial Least Squares Path Modeling (PLS-PM) [ Internet] 2017. Available from: https://github.com/gastonstat/plspm
- 38.Iossifov I, O/’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014. November 13;515(7526):216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K, et al. Excess of rare, inherited truncating mutations in autism. Nat Genet 2015. June 11;47(6):582–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Constantino JN, Zhang Y, Frazier T, Abbacchi AM, Law P. Sibling Recurrence and the Genetic Epidemiology of Autism. Am J Psychiatry 2010. November;167(11):1349–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buja A, Volfovsky N, Krieger AM, Lord C, Lash AE, Wigler M, et al. Damaging de novo mutations diminish motor skills in children on the autism spectrum. Proc Natl Acad Sci 2018. February 6;201715427. [DOI] [PMC free article] [PubMed]
- 42.Brahmachary M, Guilmatre A, Quilez J, Hasson D, Borel C, Warburton P, et al. Digital Genotyping of Macrosatellites and Multicopy Genes Reveals Novel Biological Functions Associated with Copy Number Variation of Large Tandem Repeats. Orr HT, editor. PLoS Genet 2014. June 19;10(6):e1004418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crespi BJ, Crofts HJ. Association testing of copy number variants in schizophrenia and autism spectrum disorders. J Neurodev Disord 2012. January;4(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Girirajan S, Dennis MY, Baker C, Malig M, Coe BP, Campbell CD, et al. Refinement and discovery of new hotspots of copy-number variation associated with autism spectrum disorder. Am J Hum Genet 2013;92:221–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dumas L, Sikela JM. DUF1220 domains, cognitive disease, and human brain evolution. Cold Spring Harb Symp Quant Biol 2009. January;74:375–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Courchesne E, Mouton PR, Calhoun ME, Semendeferi K, Ahrens-Barbeau C, Hallet MJ, et al. Neuron number and size in prefrontal cortex of children with autism. JAMA 2011. November 9;306(18):2001–10. [DOI] [PubMed] [Google Scholar]
- 47.Haijma SV, Van Haren N, Cahn W, Koolschijn PCMP, Hulshoff Pol HE, Kahn RS. Brain volumes in schizophrenia: A meta-analysis in over 18 000 subjects. Schizophr Bull 2013;39:1129–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Searles Quick V, Davis JM, Olincy A, Sikela J. DUF1220 copy number is associated with schizophrenia risk and severity: Implications for understanding autism and schizophrenia as related diseases. Transl Psychiatry 2015;5(12):e697. [DOI] [PMC free article] [PubMed] [Google Scholar]


