Abstract
The skyrocketing cost of health‐care demands that we question when to use multigene assay testing in the planning of treatment for breast cancer patients. A previously published algorithmic model gave recommendations for which cases to send out for Oncotype DX® (ODX) testing. This study is a multi‐institutional validation of that algorithmic model in 620 additional estrogen receptor positive breast cancer cases, with outcome data on 310 cases, named in this study as the Rochester Modified Magee algorithm (RoMMa). RoMMa correctly predicted 85% (140/164) and 100% (17/17) of cases to have a low‐ or high‐risk ODX recurrence score, respectively, consistent with the original publication. Applying our own risk stratification criteria, in patients who received appropriate hormonal therapy, only one of the 45 (2.0%) patients classified as low risk by our original algorithm have been associated with a breast cancer recurrence over 5‐10 years of follow‐up. Eight of 116 (7.0%) patients classified as low risk by ODX have been associated with a breast cancer recurrence with up to 11 years of follow‐up. In addition, 524 of 537 (98%) cases from our total population (n = 903) with an average modified Magee score ≤18 had an ODX recurrence score <26. Patients with an average modified Magee score ≤18 or >30 may not need to be sent out for ODX testing. By avoiding these cases sending out for ODX testing, the potential cost savings to the health‐care system in 2018 are estimated to have been over $100,000,000.
Keywords: algorithm, ER+ breast cancer, Oncotype DX®, recurrence, RoMMa
1. INTRODUCTION
The evolving era of precision medicine demands both cost‐efficient and cost‐effective strategies for diagnostic treatment algorithms. Challenges remain in accurately assessing which strategies are more cost‐effective for identifying hormone receptor‐positive breast cancer patients who will benefit from systemic chemotherapy.
Over the last decade, molecular approaches, including multigene assays for predicting prognosis and treatment response, have entered into the clinical arena of breast cancer care.1, 2, 3, 4, 5, 6, 7, 8 All of these multigene assays have been shown to have some prognostic and predictive value in certain subgroups of estrogen receptor (ER) positive breast cancer patients.3, 4, 9 Oncotype DX® (ODX) is the most widely used of these multigene assays. ODX is an expensive test, and although the test has been suggested to be cost‐effective,10, 11, 12, 13, 14, 15, 16 it may not be the most cost‐effective option in certain subsets of breast cancer patients.17, 18 A recent meta‐analyses by Wang et al18 suggests that a majority of published articles supporting the cost‐effectiveness of ODX were generally industry‐funded, and incorporated study designs that can increase the risk of bias. In a majority of these studies supporting the cost‐effectiveness of ODX, clinical characteristics commonly used to make chemotherapy decisions (ie, tumor size and grade) were not incorporated into simulation modeling. As such, these “supportive” studies might not reflect actual clinical practice.18
Several studies have suggested that standard clinical, histopathological, semi‐quantitative immunohistochemistry (IHC), and biomarker data can provide information similar to that provided by the ODX recurrence score (ODXRS).19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31 The IHC4 score23 uses semi‐quantitative information from the immunohistochemical assessment of ER, PR, HER‐2, and Ki‐67 (four of the genes measured in the ODX panel) to calculate a risk score using weighting factors and an algorithm. Recent studies have validated the use of the IHC4 score for identifying patients at low, moderate, or high risk of relapse following current endocrine therapy, and the IHC4+ C score (which includes clinical and additional pathologic variables) for identifying patients at low risk who potentially can avoid adjuvant radiotherapy.32, 33 The equations used for the IHC4 and IHC4+ C scores are available to the public free of charge.34
Klein and Dabbs et al19 published three linear equations (the new Magee equations) using different combinations of standard histopathological variables (Nottingham score [NS], ER, PR, HER‐2, Ki‐67, and tumor size). These new Magee equations are also available to the public free of charge (https://path.upmc.edu/onlineTools/mageeequations.html), and calculate a recurrence score, which was shown to correlate well with the ODXRS. We published an algorithm31 based on a modification of the new Magee equations, showing that this algorithm provides similar risk information to the ODX test.
The goal of this study was to further validate our original algorithm, using data from two separate institutions, and to examine outcome data from two separate institutions. We also present data which suggests further clinical utility of the average modified Magee equation, given the recent TAILORx findings that certain populations of patients with an ODXRS <26 may not benefit from additional systemic chemotherapy.35
2. MATERIALS AND METHODS
2.1. Patients and data retrieval
A total of 903 consecutive cases (889 patients) with ER+ invasive breast cancer were included in this study from the University of Rochester and the University of Louisville. Figures 1 and 2 highlight the cases that were used for the validation and outcome evaluations. All 903 cases were used for the evaluation of an ODXRS <26.
Figure 1.

Number of cases (patients) used for the validation and outcome evaluations
Figure 2.

Patients used for the outcome evaluations
Six hundred and twenty of 903 cases (606/889 patients) were included in the validation study (Figure 1). None of these cases were used in our original 2015 publication.31
Three hundred and ten of 903 cases (301/889 patients) were included in the outcome analysis (Figures 1 and 2). The outcome analysis included all patients who had at least 5 years of follow‐up data, and all patients who had a breast cancer recurrence.
Information on ER, PR, HER‐2, Ki‐67, and tumor size was extracted from the pathology report. The NS was calculated using the Nottingham modification of the Bloom‐Richardson system.36 Information on age, ethnicity, lymph node status, lymphovascular invasion, hormone therapy, chemotherapy, radiation therapy, recurrence status, and mortality were extracted from the medical record. Some type of hormonal therapy or therapies (Anastrozole, Exemestane, Tamoxifen, Lupron, and/or Letrozole) was known to have been received in 240 patients. Some type of systemic chemotherapy or therapies (Carboplatin, Cyclophosphamide, Docetaxel, Doxorubicin, 5‐flourouracil, Methotrexate, Paclitaxel, and/or Vinorelbine) was known to have been received in 59 patients. Some type of radiation therapy was known to have been received in 158 patients. All tumor H&E slides and IHC were reviewed by at least two board‐certified breast pathologists, with manual interpretation of ER, PR, HER‐2, and Ki‐67, using standard histological criteria for determining modified ER and PR H‐scores31 (FDA‐approved test kits [DAKO] ‐ ERα [clones ID5 and ER‐2‐123]; PR [clone PgR1294] pharmDxTM), and HER‐2 IHC scores (Rabbit anti‐human HER‐2 HercepTestTM). HER‐2 FISH was performed (FDA‐approved test kit [DAKO] ‐ HER‐2 IQFISH pharmDxTM) on all equivocal HER‐2 IHC results, and the FISH results were used in lieu of the IHC for these cases. Ki‐67 was evaluated by calculating the percentage of positive staining tumor cells on a single slide (Monoclonal mouse anti‐human Ki‐67 antigens [clone MIB‐1, code M7240]).
2.2. Study design
The algorithmic approach used has been previously described by Turner et al31 Briefly, an average modified Magee recurrence score was calculated, and all cases with an average modified Magee recurrence score of ≤12, or with a modified ER and PR H‐score ≥150 and a Ki‐67 <10%, were considered low risk. All cases with an average modified Magee recurrence score >30 were considered high risk. We compared our current results from the validation study with our results from the 2015 publication. We also examined different average modified Magee recurrence score groups (ie, groups with a score ≤9, groups with a score of 10, groups with a score of 11, etc) and their associated‐average ODXRS. Finally, we examined the association of breast cancer recurrence outcome data with the average modified Magee recurrence score, ODXRS, and clinicopathological data.
2.3. Statistical analysis
Available clinical and pathological data were summarized using percentages, descriptive statistics (mean, range, frequencies), and inferential statistics (chi‐square (χ2) test of independence, Pearson correlation, odds ratio, and t‐test). Each patients average modified Magee score was compared to their ODXRS using the ODXRS risk stratification categories [low (<18), intermediate (18‐30), or high (>30)]. All data analyses were performed using the statistical Analysis ToolPak (Microsoft Excel Office 2010 version 14.0.7015.100) except for the odds ratios and chi‐square (χ2) test of independence, which were performed with JavaStat 2‐way Contingency Table Analysis (revised version 7/23/2013 http://statpages.org/ctab2x2.html). For all results, a P‐value of <0.05 was considered significant. This study received IRB approval from the University of Rochester (IRB# RSRB00069270).
3. RESULTS
A summary of clinicopathological features in the patient population is detailed in Tables 1 and 2.
Table 1.
Demographic characteristics of validation casesa
| Mean age, years (range), n = 606 | 59.1 (28‐85) | |
|---|---|---|
| N (%) | ||
| Patientsb | Cases | |
| All | 606 (100)c | 620 (100)c |
| Ethnicity | ||
| White | 534 (88.1) | 547 (88.2) |
| African American | 50 (8.3) | 51 (8.2) |
| Hispanic | 8 (1.3) | 8 (1.3) |
| Asian | 8 (1.3) | 8 (1.3) |
| Unknown | 6 (1.0) | 6 (1.0) |
| Average modified Magee scored | ||
| Score ≤9 (low risk) | 12 (2.0) | 12 (1.9) |
| Score ≤12 (low risk) | 94 (15.5) | 97 (15.6) |
| Score >30 (high risk) | 17 (2.8) | 17 (2.7) |
| Nottingham score < 6; Modified ER/ PR H‐score ≥ 150;Ki‐67 < 10% (low risk)e | 64 (10.6) | 67 (10.8) |
| Oncotype DX<sup>®</sup> recurrence score | ||
| <11 (Oncotype DX® low risk) | 131 (21.6) | 134 (21.6) |
| 11‐17 (Oncotype DX® low risk) | 217 (35.8) | 218 (35.2) |
| 18‐30 (Oncotype DX® intermediate risk) | 209 (34.5) | 210 (33.9) |
| <26 (TAILORx lower risk group) | 478 (78.9) | 491 (79.2) |
| >30 (Oncotype DX® high risk) | 58 (9.6) | 58 (9.4) |
| Lymph Nodes | ||
| Negativef | 468 (77.2) | 477 (76.9) |
| Positive | 94 (15.5) | 95 (15.3) |
| Unknown | 46 (7.6) | 48 (7.8) |
| Available Nottingham score | 606 (100) | 620 (100) |
| Available ER‐H‐score | 606 (100) | 620 (100) |
| Available PR H‐score | 606 (100) | 620 (100) |
| Available Ki‐67 | 570 (94.1) | 584 (94.2) |
| Available tumor size | 604 (99.7) | 618 (99.7) |
| Available Her‐2 IHC | 606 (100) | 620 (100) |
| 0+ | 175 (28.9) | 180 (29.0) |
| 1+ | 268 (44.2) | 274 (44.2) |
| 2+ | 165 (27.2) | 165 (26.6) |
| 3+ | 1 (0.2) | 1 (0.2) |
| FISH | 171 (28.0) | 171 (27.6) |
Analyses of age, ethnicity, lymph node status, lymphovascular invasion, and years of follow‐up, include all patients. Analysis of average modified Magee score, ODXRS, NS, modified ER H‐score, modified PR H‐score, Ki‐67, tumor size, HER‐2 IHC, and HER‐2 fluorescence in situ hybridization (FISH), includes all case data.
May not equal 100%, as several patients have multiple tumors which fall into different risk stratification groups, and not all risk stratification groups are evaluated for the average modified Magee score.
469 cases from 455 different patients (393 invasive ductal carcinomas and 76 invasive lobular carcinomas) were identified from the pathology files at the University of Rochester Medical Center. Fourteen patients had two different tumors from two separate sites. 151 cases from 151 different patients (128 invasive ductal carcinomas and 23 invasive lobular carcinomas) were identified from the pathology files at the University of Louisville.
Average modified Magee scores >12 and ≤30 that do not meet low risk histologic criteria comprise the intermediate group (n = 478 [78.9%] patients, 487 [78.5%] cases). These cases would be recommended for Oncotype DX® testing.
Forty‐eight cases (47 patients) with an average modified Magee score ≤12 and meeting low risk histologic criteria; Nineteen cases (17 patients) with an average modified Magee score >12, but meeting low risk histologic criteria.
Includes patients with isolated tumor cells (n = 10).
Table 2.
Demographic data on outcome casesa
| Mean age, years (range), n = 301 | 57.2 (21‐84) | ||
|---|---|---|---|
| N (%) | Mean years of follow‐up (range) | ||
| Patientsb | Cases | ||
| 301 (100)c | 310 (100)c | 6.6 (2‐11) | |
| RoMMa risk stratificationd | |||
| Average modified Magee score ≤ 9 (low risk) | 13 (4.3) | 13 (4.2) | 7.4 (5‐10) |
| Average modified Magee score ≤ 12 (low risk) | 55 (18.3) | 57 (18.4) | 6.9 (5‐10) |
| Average modified Magee score > 30 (high risk) | 12 (4.0) | 12 (3.9) | 6.3 (3‐9) |
| Nottingham score < 6; Modified ER/ PR H‐score ≥ 150;Ki‐67 < 10% (low risk)e | 38 (12.6) | 39 (12.6) | 6.7 (5‐9) |
| Oncotype DX® recurrence score | |||
| <11 (Oncotype DX® low risk) | 50 (16.6) | 52 (16.8) | 6.8 (5‐11) |
| 11‐17 (Oncotype DX® low risk) | 119 (39.5) | 121 (39.0) | 6.7 (2‐11) |
| 18‐30 (Oncotype DX® intermediate risk) | 110 (36.5) | 111 (35.8) | 6.6 (2‐10) |
| <26 (TAILORx lower risk group) | 258 (85.7) | 266 (85.8) | 6.7 (2‐11) |
| >30 (Oncotype DX® high risk) | 24 (8.0) | 24 (7.7) | 6.0 (2‐10) |
| Lymph node status | |||
| Negativef | 236 (78.4) | 244 (78.7) | 6.7 (2‐11) |
| Positive | 52 (17.3) | 53 (17.1) | 6.0 (2‐10) |
| Unknown | 13 (4.3) | 13 (4.2) | 7.3 (5‐10) |
| Lymphovascular invasion statusg | |||
| Identified | 35 (11.9) | 35 (11.6) | 6.2 (2‐10) |
| Not identified | 258 (88.1) | 267 (88.4) | 6.7 (3‐11 ) |
Analyses of age, lymph node status, lymphovascular invasion, recurrence status, and years of follow‐up, include all patients. Analysis of average modified Magee score, ODXRS, NS, modified ER H‐score, modified PR H‐score, and Ki‐67 includes all case data.
May not equal 100%, as several patients have multiple tumors which fall into different risk stratification groups.
Nine patients had two different tumors from two separate sites.
Rochester Modified Magee Algorithm. Average modified Magee scores >12 and ≤30 that do not meet low risk histologic criteria comprise the intermediate group (n = 222 [73.8%] patients, 229 [73.9%] cases). These cases would be recommended for Oncotype DX® testing.
Twenty‐seven cases (26 patients) with an average modified Magee score ≤12 and meeting low risk histologic criteria; Twelve cases/patients with an average modified Magee score >12, but meeting low risk histologic criteria.
Includes isolated tumor cells (n = 7).
n = 293 patients, 302 cases; Not included ‐ four “suspicious” cases [mean years of follow up 5.5 (5‐6)], four unknown cases [mean years of follow up 6.5 (4‐10)].
3.1. Correlation and concordance between original publication and the validation population
Overall, the data in our validation population were remarkably consistent with the data in our original test population from the original publication31 (Figure 3A,B, Table 3, and Table S1). A Pearson correlation shows a significant correlation (P < 0.0001) between the original test population and the validation population when comparing the percentage of cases with each Magee score in the two groups (Figure S1 and Table S2). A Pearson correlation also shows a significant correlation (P < 0.0001) between the original test population and the validation population when comparing the percentage of cases in both a particular Magee score group and its correlating ODX risk group (Figure S2 Table 3 and Table S3).The chi‐squared (χ2) test of independence showed no significant difference between the observed frequencies in the original test population and the validation population (P = 0.351) when evaluating the number of cases with an average modified Magee score <18 or ≥18 (Table S4). The chi‐squared (χ 2) test of independence also showed no significant difference between the observed frequencies when evaluating cases with an average modified Magee score ≤18 in the original test population and the validation population (P = 0.559), that have an Oncotype DX score <26, or ≥26 (Table S5).
Figure 3.

A, Correlation of average modified Magee recurrence score and Oncotype DX® recurrence score from the original publication.31 B, Correlation of average modified Magee recurrence score and Oncotype DX® recurrence score in the validation population
Table 3.
Average modified Magee recurrence score groups and associated Oncotype DX® risk categories from the validation population (n = 620) and original test population31 (n = 283)
| Average modified Magee score (amMs)a |
Oncotype DX® recurrence score Validation population |
Oncotype DX® recurrence score Original population |
||||||
|---|---|---|---|---|---|---|---|---|
| High | Intermediate | Low | %b | High | Intermediate | Low | %b | |
| amMs <9 | 0 | 0 | 4 | 100.0% | 0 | 0 | 5 | 100.0% |
| amMs ≤10 | 0 | 2 | 22 | 91.7% | 0 | 2 | 21 | 91.3% |
| amMs ≤11 | 0 | 7 | 45 | 86.5% | 0 | 5 | 31 | 86.1% |
| amMs ≤12 | 0 | 13 | 84 | 86.6% | 0 | 9 | 43 | 82.7% |
| amMs ≤14 | 0 | 22 | 125 | 85.0% | 0 | 12 | 61 | 83.6% |
| amMs ≤15 | 1 | 36 | 206 | 84.8% | 0 | 26 | 96 | 78.7% |
| amMs <18 | 1 | 63 | 261 | 80.3% | 0 | 42 | 116 | 73.4% |
| NS <6 and ER/PR ≥150 AND Ki67 <10% | 0 | 11 | 56 | 83.6% | 0 | 4 | 34 | 89.5% |
| amMs >30 | 17 | 0 | 0 | 100.0% | 8 | 0 | 0 | 100.0% |
Cases with an available Ki‐67.
Percent Oncotype DX® low risk except for amMs >30 which would be percent Oncotype DX® high risk.
3.2. Cases with an average modified Magee recurrence score ≤18 and >18
Five hundred and twenty four of 537 (98%) cases with an average modified Magee score ≤18 had an ODXRS <26 (Table 4). One hundred and thirty three of 366 (36%) cases with an average modified Magee score >18 had an ODXRS ≥26 (Table 4). One hundred and thirty three of 146 (91%) cases with an ODXRS ≥26 had an average modified Magee score >18 (Table 4). An average modified Magee score is highly specific and predictive for an ODXRS <26 (Table 4).
Table 4.
Average modified Magee recurrence scores ≤18 and >18 and corresponding Oncotype DX® recurrence scores <26 and ≥26 (n = 903)
| Average modified Magee score |
Oncotype DX® recurrence score Total population (n = 903) |
|
|---|---|---|
| <26 | ≥26 | |
| ≤18 | 524 | 13 |
| >18 | 233 | 133 |
Sensitivity: 0.692.
Specificity: 0.911.
Positive predictive value: 0.976.
Negative predictive value: 0.363.
Odds ratio: 23.0 (P < 0.0001).
3.3. Outcome analysis
Eighteen of 301 (6%) patients in our outcome population have had a breast cancer recurrence (supplemental Table S6). Overall, two of 66 (3%) patients classified as low risk by our original algorithm recurred, and 10 of 156 (6%) patients classified as low risk by ODX recurred (Table 5). Two of the 18 patients who recurred did not receive hormonal therapy and these two patients were not included in our subsequent analysis of recurrence in “low risk” patients (Figure 2). Considering all the low risk patients who did recur except for the two that did not receive hormonal therapy (Table S6), and only the low risk patients who did not recur and who did not receive chemotherapy (Figure 2), none of the 28 (0%) patients recurred who were classified as low risk by our original algorithm who received radiation, five of the 65 (8%) patients recurred who were classified as low risk by ODX who received radiation, one of the 17 (6%) patients recurred who were classified as low risk by our original algorithm who did not receive radiation, and three of the 51 (6%) patients recurred who were classified as low risk by ODX who did not receive radiation. Seventeen of 18 (94%) patients who recurred had an average modified Magee score ≥13.5, and 13 of 18 (72%) patients who recurred had an average modified Magee score >18 (Table S6).
Table 5.
RoMMa risk stratification, Oncotype DX® risk stratification, and recurrence
| Recurrence | ||
|---|---|---|
| RoMMa risk stratification |
Yes n (%) |
No n (%) |
| Average Magee score ≤12 (low risk) | 1 (2) | 54 (98) |
| NS <6; Modified ER/ PR H‐score ≥150;Ki‐67 <10% (low risk)a | 1(3) | 37(97) |
| Oncotype DX® recurrence score | ||
| <11 (Oncotype DX® low risk) | 1 (2) | 48 (98) |
| 11‐17 (Oncotype DX® low risk) | 9 (8) | 108 (92) |
| 18‐30 (Oncotype DX® intermediate risk) | 5 (5) | 107 (95) |
| <26 (TAILORx lower risk group) | 12(5) | 246 (95) |
| >30 (Oncotype DX® high risk) | 3 (13) | 20(87) |
Twenty‐seven cases (26 patients) with modified Magee score ≤12 and meeting low risk RoMMa histologic criteria; Twelve cases/patients with modified Magee score >12, but meeting low risk RoMMa histologic criteria.
Patients who recurred had a significantly higher Ki‐67 (P < 0.0001) than patients who did not recur (Table S7). Overall, patients who recurred had a lower PR status (P = 0.12), statistically significant in patients with positive lymph nodes (P = 0.02) and in patients who did not receive chemotherapy (P = 0.02) (Table S7). Overall, neither NS (P = 0.45) nor ER status (P = 0.54) were significantly different between patients who did and did not recur (Table S7).
Patients who recurred had a significantly higher risk (OR = 6.2, P = 0.002) for having LVI (7/17[41%], supplemental Table S7) compared to patients who did not recur (28/276 [10%]). Patients who recurred also had a higher risk (OR = 2.0, P = 0.175) for LN involvement (5/18 [28%]) compared to patients who did not recur (47/270 [17%], Table S7), although this did not reach statistical significance.
All the patients with a known Ki‐67 who recurred (n = 13, Table S6) had some combination of a lowered modified PR H‐score (≤ 210), LN involvement, LVI, or a higher Ki‐67 (≥20). Twelve of the 18 patients who recurred in our population (67%) had an ODXRS of <26 (Table 5 and Table S6). Eleven of these 12 patients (92%) were 50 years or older at the time of diagnosis (Table S6).
3.4. Cost analysis
The list price reported in the Genomic Health 2017 Annual Report37 for the invasive breast carcinoma ODX test was $4,620. Using our previously published31 algorithmic approach (Figure 4) in our total population of cases between 2007 and 2018 (n = 903), 20.8% (n = 188) satisfied low risk algorithmic criteria, and 2.8% (n = 25) satisfied high risk algorithmic criteria. These 213 cases would potentially not have been sent out by our institutions for ODX testing, creating a combined potential cost savings of $984,060 for the University of Rochester and the University of Louisville. In that same 2017 annual report, “more than 126,740 ODX test reports were delivered for use for treatment planning.”37 The substantial majority (approximately 85%) of historical revenues from ODX testing comes from invasive breast carcinoma testing,31, 37, 38 suggesting that approximately 107,729 ER+ invasive breast cases were received for ODX testing in 2017. The ODX risk stratification of these 107,729 breast cancer cases was not available for this study; however, similar to our previous publication,31 we can envision a reasonable cost‐containment scenario. If we assume that the percentage of algorithmic low risk (20.8%) and high risk (2.5%) cases in our study is similar to the percentage of algorithmic low and high risk cases sent out for ODX testing, then approximately 25,424 breast cancer cases would potentially have been considered as cases not to send out for additional testing with ODX, resulting in an estimated cost savings to the health‐care system in 2018 of $117,459,083.
Figure 4.
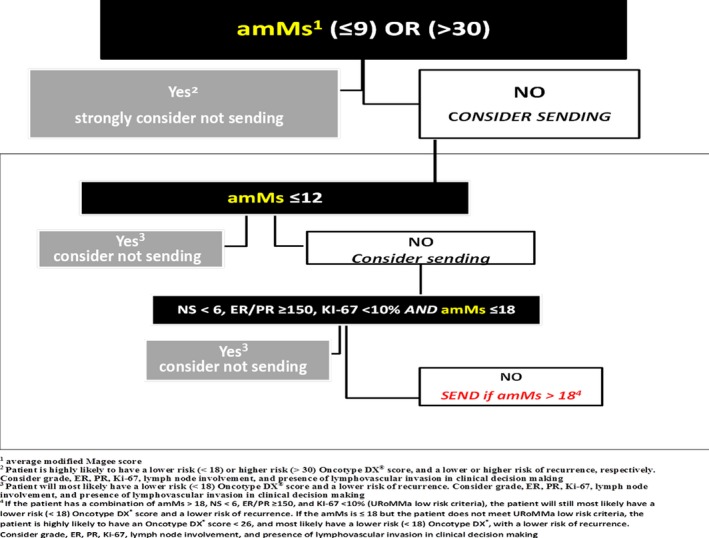
Rochester Modified Magee algorithm (RoMMa)
4. DISCUSSION
At the turn of the 21st century, Perou39 and Sorlie40 reported four significant groups of breast cancer subtypes using RNA expression profiling: luminal A (ER‐positive, HER‐2 negative), luminal B (ER‐positive, HER‐2 positive), HER2‐overexpressing (ER and PR negative, HER‐2 positive), and basal‐like. Prat et al41 later included a subtype of basal‐like breast cancer termed claudin‐low breast cancer. Basal‐like and claudin‐low breast cancers are predominantly ER PR and HER‐2 negative, or triple negative (TN). Compared to the luminal A subtype, women with luminal B tumors and HER‐2 overexpressing breast carcinoma have roughly a twofold increased adjusted risk of breast cancer mortality,42 and women with triple negative breast cancers have a poorer short‐term prognosis than all the other subtypes.43, 44 Hormonal therapy with systemic chemotherapy and targeted therapy with monoclonal antibodies (traztuzamab with or without pertuzamab) is the primary established treatment for patients with luminal B breast carcinoma; systemic chemotherapy and targeted therapy with monoclonal antibodies (traztuzamab with or without pertuzamab) is the primary established treatment for patients with HER‐2 overexpressing breast carcinoma; and, systemic chemotherapy is the primary established treatment for patients with TN breast carcinomas. Hormonal therapy is the primary established treatment for patients with luminal A breast carcinoma; however, a subset of luminal A breast carcinoma patients will also benefit from systemic chemotherapy. Challenges remain in accurately identifying which strategies are more cost‐effective and more cost‐efficient in identifying this unique subset of luminal A breast carcinoma patients who will also benefit from systemic chemotherapy.
While molecular approaches, including multigene assays, have been shown to have some prognostic and predictive value in certain subgroups of ER+ breast cancer patients,3, 4, 9 standard clinical practice has paid little attention to the suitability of breast tissue for molecular analysis, and current research suggests that alterations in the molecular integrity of breast tissue during the pre‐analytic stage may result in inaccurate results and potentially sub‐standard patient care.45 With so many new molecular assays available, on a case by case basis, there remains significant uncertainty on the part of many clinicians on how to best utilize this new molecular information, what incremental value these tests provide, or how best to integrate these assay results with the available clinicopathologic features of the patient's tumor.17, 18
ODX in particular is of considerable interest. Interestingly, four of the 16 cancer‐related genes measured by ODX (ER, PR, HER2, and Ki‐67) are routinely assessed by IHC as part of the diagnostic evaluation of breast cancer. Kim et al have recently proposed a model that accurately predicts ODX high‐ and low‐risk categories using Elston grade, ER, PR, HER‐2, and Ki‐67.46 Several studies have shown that linear regression equations which incorporate histopathological data, ER, PR, HER‐2, and Ki‐67, can also provide information that can be used to predict the ODXRS with a high degree of accuracy.19, 30, 31 We previously published data supporting the use of linear regression equations to risk stratify patients into low and high risks of recurrence,31, 47, 48 introducing an algorithmic approach using the modified Magee Equation 31 Hou et al have recently published data supporting our original study conclusions.49, 50 We now have additional data validating our original algorithm, which we are now calling the Rochester Modified Magee algorithm (RoMMa, Figure 4). Our validation study supports that our algorithmic approach is a cost‐effective, cost‐efficient alternative to ODX in risk stratifying certain breast cancer patients.
Consistent with our previously published data, in the current validation population, all the patients with an average modified Magee recurrence score >30 (n = 17), or an average modified Magee recurrence score <9 with an available Ki‐67 (n = 4), were correctly predicted to have a high or low ODXRS, respectively (Table 3). Our current results on a separate population of patients from two different institutions give further validation that our algorithm can be used by the clinician when considering which cases not to be sent out for ODX testing.
In our previous study31 there was no ‘two‐step’ discordance (a discordant high and low recurrence score using the ODX risk stratification criteria between a patients' average modified Magee recurrence score and ODXRS). There was a single case in our validation study with ‘two‐step’ discordance (Table S1). This case was high grade (NS = 8), with a Ki‐67 of 15%, and an average modified Magee score of 15.4. The ODXRS was 31. Using the RoMMa algorithmic risk stratification criteria (not the ODX risk stratification criteria), this case would have been RoMMa intermediate (and would have reflexed for ODX testing).
We examined the available outcome data of 301 patients from the original study (Table 2). In the original publication,31 11 patients with available follow‐up had an average modified Magee recurrence score ≤9 (with or without a Ki‐67). Nine of these 11 patients had a low ODX score. The other two patients had an intermediate ODX score of 19 and 20. None of these 11 patients have had a breast cancer recurrence. In the current validation population all the patients with an average modified Magee recurrence score ≤9 (n = 12) were correctly predicted to have a low ODXRS. As such, the original algorithm31 is slightly modified in that we consider patients who have an average modified Magee recurrence score ≤9 to be lowest risk (as opposed to just patients with an average modified Magee recurrence score < 9).
Bhargava et al found that 141 of 144 (98%) cases with new Magee equation scores < 18, or new Magee equation scores 18‐25 and mitosis score of 1, had an ODXRS < 26.51 Our data using the average modified Magee equation in larger population support these findings. In our population, 524 of 537 (98%) cases with an average modified Magee score ≤18 had an ODXRS <26 (Table 4). Based on this finding, we additionally modified the original algorithm to include consideration for an average modified Magee score ≤18 as “low risk” given the recent TAILORx findings.35
Sixty seven percent (12/18) of patients who recurred in our population had an ODXRS of <26 (Table 5 and Table S6). Eleven of these 12 patients (92%) were 50 years of age or older at the time of diagnosis. Three of these 11 patients did not receive any hormonal treatment, and only two of these 11 patients received any adjuvant systemic chemotherapy. The recent TAILORx results suggest that women with early breast cancer who are older than 50 years with an ODXRS <26 can be spared adjuvant chemotherapy.35 Our findings suggest that risk stratification with consideration for systemic chemotherapy is still important in these patients.
Our outcome data support that patients designated as low risk by our original algorithm (average modified Magee score of ≤12 or a combination of NS <6, ER/PR ≥150, and KI‐67 <10%) are unlikely to recur. Considering the population of low risk patients who recurred after receiving appropriate hormonal therapy and the population of low risk patients who did not recur and did not receive chemotherapy, one of the 45 (2%) patients classified as low risk by our original algorithm recurred, compared to eight of 116 (7%) patients classified as low risk by ODX who recurred (Figure 2).
Ninety four percent (17/18) of the patients who recurred had an average modified Magee score ≥13.5 (Table S6). Patients who recurred were more likely to have a lower PR, a higher Ki‐67, LN involvement, and LVI than were patients who did not recur (Table S7). Interestingly, neither the average NS nor ER status was significantly different in the recurrence and non‐recurrence populations (Table S7). Thirteen of the 18 patients who recurred had some combination of a lowered PR, a higher Ki‐67, LN involvement, or LVI (Table S6). In the five other patients who recurred (patient #1, #5, #8, #9, and #11), either the Ki‐67 and/or the LVI status was unknown. Our results support that the average modified Magee score, PR, Ki‐67, LN, and LVI status may be helpful in predicting patients with a higher risk of recurrence, and should be considered when risk stratifying breast cancer patients for systemic chemotherapy.
ODX testing was rapidly adopted into clinical care of breast cancer patients in 2004 without any randomized trials, based on small cohort studies which suggested that the ODXRS influenced patient preference and oncologist recommendations for chemotherapy.52 Although a number of studies have suggested that ODX is cost‐effective, with the cost of the test being offset primarily by reduction in the use of adjuvant chemotherapy,10, 11, 12, 13, 14, 15, 16, 52 these “supportive” studies have generally been industry funded, not based on real‐world population data, and may not reflect actual clinical practice.18, 52 A recent study based on real‐world population based data by Mittmann et al suggests that using the ODXRS to determine whether adjuvant chemotherapy should be added to endocrine therapy in ER+ lymph node‐negative breast cancer patients was approximately $3,000 more expensive per patient than not using the test.52 Our results suggest a potential estimated cost savings to the health‐care system in 2018 of over $100,000,000 if ODX testing was avoided in certain low‐ and high‐risk RoMMa patients. If all cases with an average Modified Magee score of ≤18 were also considered, the cost savings would undoubtedly be substantially higher.
The reproducibility and accuracy of histological grading and immunohistochemical reporting creates a concern for interpretation bias. The literature supports the reproducibility of ER, PR, HER‐2,53, 54, 55, 56, 57 and Ki‐67,58 if standardized criteria are used for their interpretation. We used standard criteria for the evaluation of ER, PR, HER‐2, and Ki‐67, and there was similar agreement in our study between the two institutions in the interpretation of the average modified Magee score relative to the ODX score (Table S1).
All of the patients without recurrence in our population had at least 5 years of follow‐up; however, only 16 of 301 (5%) of patients had 10 or more years of follow‐up. Although 5‐year follow‐up data are acceptable for evaluation of outcome in the published literature, 10 years or more of follow‐up is preferable. We continue to maintain our database of patients, with the goal of additional outcome evaluation that has 10 or more years of follow‐up. Access to larger database populations, such as ECOG, NSABP B‐14, NSABP B‐20, and SEER‐Medicare, would be helpful in providing additional longitudinal data with 10 or more years of follow‐up, which we believe will further validate our findings.
It is likely that variations in adherence to hormonal, systemic, and radiation therapies occurred within our population. A prospective study design or access to data obtained in a prospective fashion would help to eliminate this bias.
The 8th edition of the American Joint Commission on Cancer (AJCC) Cancer Staging manual59 determined it was appropriate to incorporate the ODXRS into staging for the subgroup of invasive breast carcinoma patients defined by Arm A of the TAILORx study,60 which includes ER+, HER‐2 negative, LN‐negative tumors that are 1.1‐5.0 cm in size (or 0.6‐1.0 cm with intermediate or high histologic or nuclear grade), and have an ODXRS <11. According to the AJCC recommendations, these patients should be placed into the same prognostic category as patients with pT1‐ N0 M0 (stage IA) breast cancers (AJCC Prognostic Stage Group I). Our results suggest that the likelihood of breast cancer recurrence with an average modified Magee score ≤12 is comparable with an ODXRS <11 (Table 5). One of the 49 (2%) patients with an ODXRS <11 recurred (Table 5 and Table S6). One of the 55 (2%) patients with an average modified Magee score ≤12 recurred, and this patient did not receive hormonal therapy (Table 5 and Table S6). However, neither RoMMa nor the average modified Magee score can reliably predict an ODXRS <11. It is worth noting, however, that ODX testing is not a requirement for staging, and Breaux et al61 have shown that an ODX score <11 changes the stage in only rare cases.
The large cost savings in our study is due to the assumption that none of the patients meeting low or high risk RoMMa criteria would receive ODX testing. It is likely that a number of these patients would still receive ODX testing based on many factors, not the least of these being patient and clinician concerns about not using a more accepted (and publicized) test; however, our study highlights the importance of considering other valid and less costly methods for assessing the risk of breast cancer recurrence. Additional studies testing the cost‐efficiency and cost‐effectiveness of integrating the RoMMa into clinical practice are necessary.
5. CONCLUSIONS
Our validation results continue to support that patients who satisfy our algorithmic low‐risk or high‐risk criteria are likely to have a low‐risk or high‐risk ODX, respectively.
Specifically, patients with an average modified Magee score of ≤9 are highly likely to be low risk by ODX criteria. Patients with an average modified Magee score ≤12 or, a combination of an average modified Magee score ≤18 with a NS < 6, ER/PR ≥150, and KI‐67 <10% (RoMMa low risk histologic criteria), are also very likely to be lower risk by ODX criteria. Patients with an average modified Magee score of >30 are highly likely to be high risk by ODX criteria. Consideration should be given to not sending out tissue for ODX testing in patients meeting low‐ or high‐risk RoMMa criteria, if treatment decisions will be made based on a low‐ or high‐risk ODXRS.
Patients with an average modified Magee score of ≤18 are highly likely to have an ODXRS <26. Given the recent TAILORx findings,35 we strongly recommend that patients with an average modified Magee score of ≤18 who meet low risk RoMMa histologic criteria not be sent out for ODX testing if the clinician is not considering chemotherapy for an ODXRS <26.
Our results also suggest that the likelihood of breast cancer recurrence in patients who satisfy RoMMA low‐risk criteria is comparable with outcomes in patients with an ODXRS <11, and better than an ODXRS <18.
We suggest a “stepwise” approach when risk stratifying breast cancer patients. One approach might be to use information from the RoMMa or similarly less expensive validated models to help identify cases with already available clinical and pathological metrics that will likely elicit information that is similar to ODX. In these cases ODX may not provide any additional significant clinical utility, and would likely not be cost‐effective or cost‐efficient. ODX testing could then be limited to cases where the assay results would potentially provide clinical utility beyond the available clinical and pathologic metrics. The potential cost savings to the health‐care system would be significant. Support for this “stepwise” approach will require additional validation of the RoMMa in multiple patient cohorts with outcome data to help insure that the information obtained is indeed generalizable to the broader breast cancer population.
CONFLICT OF INTERESTS
All authors have no conflict of interest.
AUTHOR CONTRIBUTIONS
Dr Turner: Conceptualization, project administration, data curation, methodology, formal analysis, original draft, writing, review, and editing. Dr Gimenez‐Sanders: Data curation, writing, review, and editing. Mr Soukiazian: Data curation, formal analysis, review. Dr Breaux: Data curation, review, and editing. Dr Skinner: Review and editing. Dr Shayne: Review and editing. Ms Soukaizian: Data curation. Dr Ling: Review. Dr Hicks: Conceptualization, methodology, review, and editing.
Supporting information
ACKNOWLEDGMENT
None.
Turner BM, Gimenez‐Sanders MA, Soukiazian A, et al. Risk stratification of ER‐positive breast cancer patients: A multi‐institutional validation and outcome study of the Rochester Modified Magee algorithm (RoMMa) and prediction of an Oncotype DX® recurrence score <26. Cancer Med. 2019;8:4176–4188. 10.1002/cam4.2323
REFERENCES
- 1. Allison KH. Molecular pathology of breast cancer: what a pathologist needs to know. Am J Clin Pathol. 2012;138:770‐780. [DOI] [PubMed] [Google Scholar]
- 2. Simpson PT, Reis‐Filho JS, Gale T, Lakhani SR. Molecular evolution of breast cancer. J Pathol. 2005;205:248‐254. [DOI] [PubMed] [Google Scholar]
- 3. Hicks DG, Turner B. Pathologic diagnosis, immunohistochemistry, multigene assays and breast cancer treatment: progress toward “precision” cancer therapy. Biotech Histochem. 2015;90(2):81‐92. [DOI] [PubMed] [Google Scholar]
- 4. Turner BM, Hicks DG. Pathologic diagnosis of breast cancer patients: evolution of the traditional clinical‐pathologic paradigm toward "precision" cancer therapy. Biotech Histochem. 2017;92(3):175‐200. [DOI] [PubMed] [Google Scholar]
- 5. van 't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530‐536. [DOI] [PubMed] [Google Scholar]
- 6. Sotiriou C, Wirapati P, Loi S, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262‐272. [DOI] [PubMed] [Google Scholar]
- 7. Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen‐treated, node‐negative breast cancer. New Engl J Med. 2004;351:2817‐2826. [DOI] [PubMed] [Google Scholar]
- 8. Paik S, Tang G, Shak S, et al. Gene expression and benefit of chemotherapy in women with node‐negative, estrogen receptor‐positive breast cancer. J Clin Onc. 2006;24:3726‐3734. [DOI] [PubMed] [Google Scholar]
- 9. Ishibe N, Schully S, Freedman A, Ramsey SD. Use of Oncotype DX® in Women with node‐positive breast cancer. PLoS Curr. 2011;3:RRN1249 10.1371/currents.RRN1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ward S, Scope A, Rafia R, et al. Gene expression profiling and expanded immunohistochemistry tests to guide the use of adjuvant chemotherapy in breast cancer management: a systematic review and cost‐effectiveness analysis. Health Technol Assess. 2013;17:4176‐302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rouzier R, Pronzato P, Chéreau E, Carlson J, Hunt B, Valentine WJ. Multigene assays and molecular markers in breast cancer: systematic review of health economic analyses. Breast Cancer Res Treat. 2013;139:621‐637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marrone M, Stewart A, Dotson WD. Clinical utility of gene‐expression profiling in women with early breast cancer: an overview of systematic reviews. Genet Med. 2015;17:519‐532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Klang SH, Hammerman A, Liebermann N, Efrat N, Doberne J, Hornberger J. Economic implications of 21‐gene breast cancer risk assay from the perspective of an Israeli‐managed health‐care organization. Value Health. 2010;13(4):381‐387. [DOI] [PubMed] [Google Scholar]
- 14. Hornberger J, Cosler LE, Lyman GH. Economic analysis of targeting chemotherapy using a 21‐gene RT‐PCR assay in lymph‐node‐negative, estrogen‐receptor‐positive, early‐stage breast cancer. Am J Manag Care. 2005;11(5):313‐324. [PubMed] [Google Scholar]
- 15. Lyman GH, Cosler LE, Kuderer NM, Hornberger J. Impact of a 21‐gene RT‐PCR assay on treatment decisions in early‐stage breast cancer: an economic analysis based on prognostic and predictive validation studies. Cancer. 2007;109(6):1011‐1018. [DOI] [PubMed] [Google Scholar]
- 16. Kondo M, Hoshi SL, Yamanaka T, Ishiguro H, Toi M. Economic evaluation of the 21‐gene signature (Oncotype DX) in lymph node‐negative/positive, hormone receptor‐positive early‐stage breast cancer based on Japanese validation study (JBCRG‐TR03). Breast Cancer Res Treat. 2011;127(3):739‐749. [DOI] [PubMed] [Google Scholar]
- 17. Hall PS, McCabe C, Stein RC, Cameron D. Economic evaluation of genomic test‐directed chemotherapy for early‐stage lymph node‐positive breast cancer. J Natl Cancer Inst. 2012;104(1):56‐66. [DOI] [PubMed] [Google Scholar]
- 18. Wang S‐Y, Dang W, Richman I, Mougalian SS, Evans SB, Gross CP. Cost‐effectiveness analyses of the 21‐gene assay in breast cancer: systematic review and critical appraisal. J Clin Oncol. 2018;36(16):1619‐1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Klein ME, Dabbs DJ, Shuai Y, et al. Prediction of the Oncotype DX® recurrence score: use of pathology‐generated equations derived by linear regression analysis. Mod Pathol. 2013;26:658‐664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Clark BZ, Dabbs DJ, Cooper KL, Bhargava R. Impact of progesterone receptor semiquantitative immunohistochemical result on Oncotype DX® recurrence score: a quality assurance study of 1074 cases. Appl Immunohistochem Mol Morphol. 2013;21:287‐291. [DOI] [PubMed] [Google Scholar]
- 21. Zbytek B, Cohen C, Wang J, et al. Nottingham‐defined mitotic score: comparison with visual and image cytometric phosphohistone H3 labeling indices and correlation with Oncotype DX® recurrence score. Appl Immunohistochem Mol Morphol. 2013;21:48‐53. [DOI] [PubMed] [Google Scholar]
- 22. Kraus JA, Dabbs DJ, Beriwal S, Bhargava R. Semi‐quantitative immunohistochemical assay versus Oncotype DX® qRT‐PCR assay for estrogen and progesterone receptors: an independent quality assurance study. Mod Pathol. 2012;25:869‐876. [DOI] [PubMed] [Google Scholar]
- 23. Cuzick J, Dowsett M, Pineda S, et al. Prognostic value of a combined estrogen receptor, progesterone receptor, Ki‐67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the Genomic Health recurrence score in early breast cancer. J Clin Oncol. 2011;29:4273‐4278. [DOI] [PubMed] [Google Scholar]
- 24. Biroschak JR, Schwartz GF, Palazzo JP, et al. Impact of Oncotype DX® on treatment decisions in ER‐positive, node‐negative breast cancer with histologic correlation. Breast J. 2013;19:269‐275. [DOI] [PubMed] [Google Scholar]
- 25. Lee JJ, Shen J. Is the Oncotype DX® assay necessary in strongly estrogen receptor‐positive breast cancers? Am Surg. 2011;77:1364‐1367. [PubMed] [Google Scholar]
- 26. Williams DJ, Cohen C, Darrow M, Page AJ, Chastain B, Adams AL. Proliferation (Ki‐67 and phosphohistone H3) and Oncotype DX® recurrence score in estrogen receptor‐positive breast cancer. Appl Immunohistochem Mol Morphol. 2011;19:431‐436. [DOI] [PubMed] [Google Scholar]
- 27. Sahebjam S, Aloyz R, Pilavdzic D, et al. Ki‐67 is a major, but not the sole determinant of Oncotype DX® recurrence score. Br J Cancer. 2011;105:1342‐1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Auerbach J, Kim M, Fineberg S. Can features evaluated in the routine pathologic assessment of lymph node‐negative estrogen receptor‐positive stage I or II invasive breast cancer be used to predict the Oncotype DX® recurrence score? Arch Pathol Lab Med. 2010;134:1697‐1701. [DOI] [PubMed] [Google Scholar]
- 29. Allison KH, Kandalaft PL, Sitlani CM, et al. Routine pathologic parameters can predict Oncotype DX® recurrence scores in subsets of ER positive patients: who does not always need testing? Breast Cancer Res Treat. 2012;131:413‐424. [DOI] [PubMed] [Google Scholar]
- 30. Tang P, Wang J, Hicks DG, et al. A lower Allred score for progesterone receptor is strongly associated with a higher recurrence score of 21‐gene assay in breast cancer. Cancer Invest. 2010;28:978‐982. [DOI] [PubMed] [Google Scholar]
- 31. Turner BM, Skinner KA, Tang P, et al. Use of modified Magee equations and histologic criteria to predict the Oncotype DX® recurrence score. Mod Pathol. 2015;28(7):921‐931. [DOI] [PubMed] [Google Scholar]
- 32. Bartlett JM, Christiansen J, Gustavson M, et al. Validation of the IHC4 breast cancer prognostic algorithm using multiple approaches on the multinational TEAM clinical trial. Arch Pathol Lab Med. 2016;140:66‐74. [DOI] [PubMed] [Google Scholar]
- 33. Lakhanpal R, Sestak I, Shadbolt B, et al. IHC4 score plus clinical treatment score predicts locoregional recurrence in early breast cancer. Breast. 2016;29:147‐152. [DOI] [PubMed] [Google Scholar]
- 34. Barton S, Zabaglo L, A'Hern R, et al. Assessment of the contribution of the IHC4+ C score to decision making in clinical practice in early breast cancer. Br J Cancer. 2012;106:1760‐1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sparano JA, Gray RJ, Makower DF, et al. Adjuvant chemotherapy guided by a 21‐gene expression assay in breast cancer. N Engl J Med. 2018;379(2):111‐112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I, the value of histological grade in breast cancer: experience from a large study with long‐term follow‐up. Histopathology. 1991;19:403‐410. [DOI] [PubMed] [Google Scholar]
- 37. Genomic Health Annual Report 2017. http://www.annualreports.com/Company/genomic-health-inc.
- 38. Genomic Health Annual Report 2012. http://www.annualreports.com/Company/genomic-health-inc.
- 39. Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747‐752. [DOI] [PubMed] [Google Scholar]
- 40. Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869‐10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Prat A, Parker JS, Karginova O, et al. Phenotypic and molecular characterization of the claudin‐low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Haque R, Ahmed SA, Inzhakova G, et al. Impact of breast cancer subtypes and treatment on survival: an analysis spanning two decades. Cancer Epidemiol Biomarkers Prev. 2012;21(10):1848‐1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple‐negative breast cancer: challenges and opportunities of a heterogeneous disease. Nature Rev Clin Oncol. 2016;13:674‐690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Malorni L, Shetty PB, De Angelis C, et al. Clinical and biologic features of triple‐negative breast cancers in a large cohort of patients with long‐term follow‐up. Breast Cancer Res Treat. 2012;136:795‐804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Turner BM, Moisini I, Hicks DG. Impact on clinical practice from a breast pathology perspective. Curr Pathobiol Rep. 2018;6:125 10.1007/s40139-018-0169-7. [DOI] [Google Scholar]
- 46. Kim H‐S, Umbricht CB, Illei PB, et al. Optimizing the use of gene expression profiling in early stage breast cancer. J Clin Oncol. 2016;34(36):4390‐4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Turner BM, Tang P, Hicks DG. Letter to the Editor. The value of algorithms predicting the Oncotype DX recurrence score should not be underestimated!.Breast Cancer Res Treat. 2017;164(1):249‐250. 10.1007/s10549-017-4236-2 [DOI] [PubMed] [Google Scholar]
- 48. Soukiazian A, Hicks DG, Turner BM. Reconsidering “low‐risk” criteria for breast cancer recurrence in hormone positive patients. Breast J. 2019;25(3):545–547. 10.1111/tbj.13275 [DOI] [PubMed] [Google Scholar]
- 49. Hou Y, Tozbikian G, Zynger DL, Li Z. Using the modified magee equation to identify patiens unlikely to benefit from the 21‐gene recurrence score assay (Oncotype DX assay). Am J Clin Pathol. 2017;147:541‐548. [DOI] [PubMed] [Google Scholar]
- 50. Hou Y, Zynger D, Li X, et al. Comparison of Oncotype DX with Modified Magee equation recurrence in low‐grade invasive carcinoma of breast. Am J Clin Pathol. 2017;148:5167‐5172. [DOI] [PubMed] [Google Scholar]
- 51. Bhargava R, Clark BZ, Dabbs D. Breast cancers with Magee equation score of less than 18, or 18–25 and mitosis score of 1, do not require Oncotype DX testing. Am J Clin Pathol. 2018;8:4176‐9. 10.1093/AJCP/AQY148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mittmann N, Earle CC, Cheng SY, et al. Study to determine the health system costs of using the 21‐gene assay. J Clin Oncol. 2018;36(3):238‐243. [DOI] [PubMed] [Google Scholar]
- 53. Harris LN, Ismaila N, McShane LM, et al; Society of Clinical Oncology . Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early‐stage invasive breast cancer: American Society of Clinical Oncology clinical practice guideline. J Clin. Oncol. 2016;34:1134‐1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hammond ME, Hayes DF, Dowsett M, et al; on behalf of the ASCO/CAP Hormone Receptor Testing in Breast Cancer Panel . American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen/progesterone receptors in breast cancer. J. Clin. Oncol. 2010;28:2784‐2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fitzgibbons PL, Hicks DG. Progress in implementing HER2 testing guidelines. Arch Pathol Lab Med. 2014;138:863‐864. [DOI] [PubMed] [Google Scholar]
- 56. Wolff AC, Hammond ME, Schwartz JN, et al; Society of Clinical Oncology; College of American Pathologists . American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118‐145. [DOI] [PubMed] [Google Scholar]
- 57. Wolff AC, Hammond ME, Hicks DG, et al; American Society of Clinical Oncology; College of American Pathologists . (2014) Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med. 2014;138:241‐256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Polley MY, Leung S, Gao D, et al; on behalf of the International Ki67 in Breast Cancer Working Group of the Breast International Group and North American Breast Cancer Group (BIG‐NABCG) . An international study to increase concordance in Ki67. Mod Pathol. 2015;28:778‐786. [DOI] [PubMed] [Google Scholar]
- 59. Hortobagyi GN, Connolly JL, Edge SB, et al. Breast In: Amin MB, Edge S, Greene F, eds. AJCC Cancer Staging Manual, 8th edn New York, NY: Springer International Publishing; 2016. [Google Scholar]
- 60. Sparano JA, Gray RJ, Makower DF, et al. Prospective validation of a 21‐gene expression assay in breast cancer. N Engl J Med. 2015;373(21):2005‐2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Breaux A, Turner B, Riley E, et al. Impact of the 21‐gene expression assay on staging estrogen receptor positive HER2 negative breast cancer. Clin Breast Cancer. 2019;19(1):E261‐E269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


