Abstract
Eukaryotic initiation factor 3 (EIF3) is one of the largest and most complex translation initiation factors, which consists of 13 subunits named eukaryotic translation initiation factor 3 subunit A (EIF3a) to EIF3m. EIF3a is the largest subunit of EIF3. Previous studies suggested that EIF3a is a housekeeping gene, recent results have found that EIF3a is closely related to the tumorigenesis and drug resistance. Circular RNAs (circRNAs) derived from biologically important gene can play an important role in gene regulation. However, the mechanism underlying circRNAs’ biological functions is not well understood yet. In this work, we screened 31 EIF3a‐derived circRNAs, in which two circEIF3as were identified to be correlated with cisplatin drug sensitivity in lung cancer. Two circEIF3as were found involved in RNA‐binding proteins‐mediated biological processes and may be related to translational regulation according to bioinformatics analyses. CircEIF3as, the transcriptional initiation factor EIF3a transcribed circRNAs, are associated with both drug sensitivity and translation regulation. These findings mean that they may have a functional synergy effect with EIF3a or be valuable therapeutic targets for treatment like EIF3a. This is the first study that exploits circRNAs screening from EIF3a in lung cancer, our findings provide a novel perspective on the function of EIF3a and circEIF3as in lung cancer.
Keywords: bioinformatics, circular RNA, EIF3a, lung cancer
1. INTRODUCTION
Translation is the second step in the process of protein biosynthesis, and translation initiation is the first step in the translation process. The eukaryotic initiation factor 3 (EIF3) family plays an important role in eukaryotic translation. Moreover, eukaryotic translation initiation factor 3 subunit A (EIF3a) is the largest subunit of EIF3 family, which is a key player in all steps of translation initiation.1
Circular RNAs (circRNAs) are a class of RNAs that have a closed loop structure and distinct from traditional linear RNAs. Up to date, investigating circRNAs functions has become a global hot spot. Previous studies showed that circRNA could function as miRNA sponges and transcriptional regulators. In addition, they can be translated into proteins or interact with RNA‐binding proteins (RBPs).2
There have been reports that some genes and the circRNAs transcribed from them have biological functions at the same time. It is reported that DLG associated protein 4 (DLGAP4) is directly linked with brain diseases,3 such as early‐onset cerebellar ataxia,4 and circDLGAP4 may serve as a novel clinical treatment target for acute ischemic stroke.5 Increasing evidence indicates that homeodomain‐interacting protein kinase 3 (HIPK3) is a kinase gene in huntingtin disease,6, 7 and a recent study has found that silencing circHIPK3 significantly inhibits human cell growth.8 In addition, circHIPK3 can mediate the retinal vascular dysfunction in diabetes mellitus.9 Studies show that the circRNA transcribed from eukaryotic translation initiation factor 3 subunit J (EIF3J) can affect the expression and function of EIF3J.10 Poly (A) binding protein interacting protein 2 (PAIP2) can limit productive cytomegalovirus replication,11 and its expression is influenced by the circRNA transcribed from PAIP2.10
Considering the regulatory effects of circRNAs on their parental genes with biologically important function, these circRNAs are believed to have some biological functions. Moreover, circRNAs have both functional similarities and differences compared with their parental genes, and can even affect the function of their parental genes. In this case, we speculate that the circRNAs transcripts derived from EIF3a also play an important biological role in humans.
2. MATERIALS AND METHODS
2.1. Cell culture
The A549 human lung cancer cell line was purchased from the Chinese Academy of Sciences (Shanghai, China) and the A549/DDP human lung cancer drug‐resistant cell line was obtained from the cell biology research laboratory and Modern Analysis Testing Center of Central South University (Changsha, China). All the cell lines were maintained as adherent cell cultures in RPMI 1640 medium (Gibco, Life Technologies, USA) supplemented with 10% fetal bovine serum (FBS, 10099‐141; Gibco, MA, USA). Besides, the A549/DDP cell line was cultured in medium with 2 mg/L cisplatin (Sigma, P4394) to maintain the drug‐resistant phenotype before experimentation. Cells were cultured at 37°C in a humidified atmosphere of 5% CO2.
2.2. Drug sensitivity assay
Cells were seeded in 96‐well plates (4,000 cells/well) and cultured with different concentrations of cisplatin for 48 hours at 37°C in a humidified atmosphere of 5% CO2, followed by incubation with CCK‐8 solution. Then the cells’ viability was detected by measuring the absorbance at 450 nm using an Eon plate reader. The half‐maximal inhibitory concentration (IC50) was calculated from the relative survival curves.
2.3. RNA extraction, RNA RNase R treatment, and real‐time quantitative RT‐PCR
Total RNA was extracted using TRIzol (Invitrogen, CA, USA) according to the instructions. Cytoplasmic and nuclear RNAs were extracted using Cytoplasmic and Nuclear RNA Purification Kit (Norgen, Canada) according to the manufacturer's instructions. RNA RNase R treatment was carried out for 10 minutes at 37°C using 1ul RNase R (20 U/UL). Primescript RT Reagent Kit with gDNA Eraser (Takara Bio Inc, Japan) was used for synthesizing cDNA according to the manufacturer's instructions. CircRNAs expression was assessed by real‐time quantitative RT‐PCR using SYBR Premix Dimer Eraser assay kits. Real‐time quantitative RT‐PCR was performed through the Roche LightCycler 480 PCR System.
2.4. RNA immunoprecipitation assay
RNA immunoprecipitation assay (RIP) was performed using EZMagna RIP Kit (Millipore, Billerica, MA) according to the manufacturer's instructions. A549 cell lysates were incubated with anti‐rabbit AGO2 antibodies (Abcam, Cambridge, MA) or anti‐rabbit IgG antibodies (Millipore, Billerica, MA).
2.5. Western blot analysis
After transfection for 48 hours, western blot assay was carried out using the standard method, and then transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Bedford, MA). After 2 hours of blocking, PVDF membranes were incubated with anti‐mouse β‐actin antibodies (Sigma‐Aldrich) and anti‐rabbit EIF3a antibodies (Cell Signaling Technology, USA) overnight at 4°C.
2.6. Differentially expressed gene analysis and survival analysis
We used THE HUMAN PROTEIN ATLAS (THPA) (https://www.proteinatlas.org/), THE CANCER GENOME ATLAS (TCGA) (https://cancergenome.nih.gov/) and ONCOMINE (https://www.oncomine.org) Database to identify the differential expression of EIF3a in various tissues and tumors. The prognostic value of EIF3a level in lung cancer and ovarian cancer are from KAPLAN‐MEIER PLOTTER (KMP) (http://kmplot.com/analysis/) Database.
2.7. Prediction for circEIF3as, circEIF3as‐RBPs axis and coding potential
CircEIF3as were predicted using CIRCBASE (http://circbase.org/) Database. The target RBPs of circEIF3as were predicted based on circRNA Interactome (https://circinteractome.nia.nih.gov/), RBPDB (http://rbpdb.ccbr.utoronto.ca/), catRAPID (http://service.tartaglialab.com/page/catrapid_group) and Cancer‐Specific CircRNA (http://gb.whu.edu.cn/CSCD/) Database. The graph of the circEIF3as‐RBPs axis was drawn with the Cytoscape 3_6_0. The Internal Ribosome Entry Site (IRES) and Open Reading Frame (ORF) of circEIF3as were predicted based on Circbank (http://www.circbank.cn/index.html) and ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/) Database.
2.8. Functional enrichment analysis
During functional enrichment analysis of circEIF3as, the online analysis tool‐Metascape (http://metascape.org/gp/index.html#/main/step1) was utilized to perform gene ontology (GO) enrichment analysis.
2.9. Statistical analysis
The SPSS 19.0 software was used for general statistical analysis. Survival rate was calculated using the Kaplan‐Meier method.
2.10. Data availability statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
3. RESULTS
3.1. The expression of EIF3a in tissues and tumors
In previous studies, EIF3a was thought to be associated with the occurrence, metastasis, prognosis and treatment of cancer. It has been reported that EIF3a has biological function in lung cancer,12 breast cancer,13 pancreas cancer,14 gastric cancer,15 oral cavity cancer,16 colon cancer,17 esophagus cancer,18 ovary cancer,19 urinary bladder cancer,20 cervix cancer.21 Besides these, EIF3a is also emerging as a new potential anticancer drug therapeutic target in clinic, which improves the cisplatin sensitivity in lung cancer 22, 23, 24 and ovarian cancer.19 We use THPA Database to compare the expression level of EIF3a in different tissues (Figure 1A), where EIF3a tissue data are reported as a mean transcript per million experiment samples. The color‐coding is based on tissue groups, which consists of tissues with common functional features. The expression level of EIF3a in 17 cancer types is generated by the TCGA Database, in which the cancer types are color‐coded according to the type of normal organ that the cancer originates from (Figure 1B). The differential expression of EIF3a in various types of tumors is illustrated in the data from ONCOMINE Database (Figure 1C), where the red/blue color indicates high/low expression, respectively, and the darker color means higher/lower level of expression.
Figure 1.
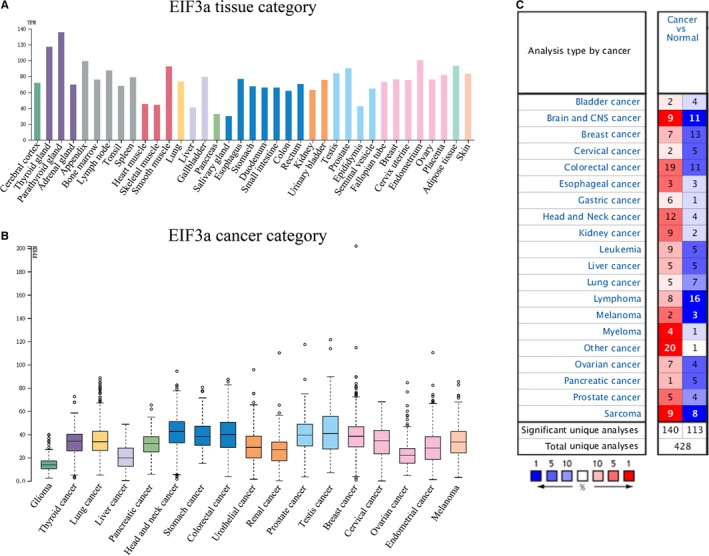
Panel A, EIF3a in different tissues, the data are obtained from THE HUMAN PROTEIN ATLAS (THPA) Database. Panel B, EIF3a in different cancers, the data are obtained from THE CANCER GENOME ATLAS (TCGA) Database. Panel C, The expression of EIF3a in various tumors, the data comes from ONCOMINE Database. EIF3a, eukaryotic translation initiation factor 3 subunit A
3.2. Association of prognostic value with EIF3a expression level
As shown in Figure 1, EIF3a is highly expressed in various tissues and cancers, and on the other hand, EIF3a could present as both biomarkers and potential anticancer drug target in lung cancer and ovarian cancer.19, 22, 23, 24 Therefore, lung cancer and ovarian cancer will be taken as an example to analyze the impact of EIF3a on the survival of patients who suffered from these cancers. The results suggest that EIF3a has a significant effect on the survival rate of patients with lung cancer, but has little effect on the prognosis of patients with ovarian cancer (Figure 2A,B). The results are collected from KMP Database.
Figure 2.
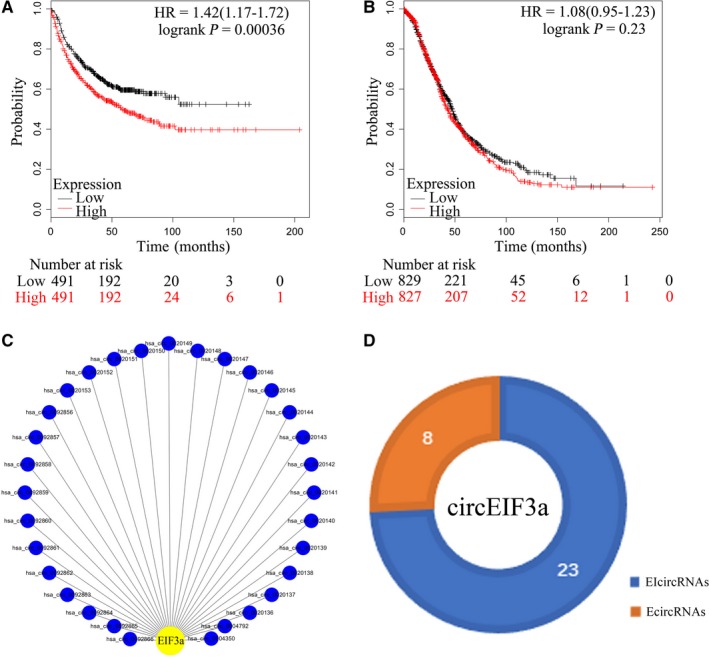
Panel A, The prognostic value of EIF3a level in lung cancer patients. Panel B, The prognostic value of EIF3a level in ovarian cancer patients. Panel C, Thirty‐one circRNAs derived from EIF3a. Panel D, The type and quantity of circEIF3as. EIF3a, eukaryotic translation initiation factor 3 subunit A. circRNA, circular RNA
3.3. The circRNAs screening from EIF3a
Eukaryotic translation initiation factor 3 subunit A is a highly conserved gene located at 10q26.11 with 22 exons, spanning a 46‐kb DNA region.25 Human EIF3a is a 170‐kDa protein consisting of 1382 amino acids. According to the record in the CIRCBASE Database, a total of 31 circRNAs are derived from EIF3a, including 8 exon‐originated circular RNAs and 23 exons‐introns‐originated circular RNAs (Figure 2C,D, Table 1).
Table 1.
Summary of circRNAs screening from EIF3a
| Circular RNA ID | Location | Genomic length | Spliced length | Type | |
|---|---|---|---|---|---|
| 1 | hsa_circ_0004350 | chr10:120832401‐120833449 | 1048 | 492 | EIcircRNA |
| 2 | hsa_circ_0004792 | chr10:120797749‐120797951 | 202 | 202 | EcircRNA |
| 3 | hsa_circ_0020136 | chr10:120794540‐120830597 | 36057 | 4618 | EIcircRNA |
| 4 | hsa_circ_0020137 | chr10:120796630‐120820356 | 23726 | 2692 | EIcircRNA |
| 5 | hsa_circ_0020138 | chr10:120797749‐120820356 | 22607 | 2501 | EIcircRNA |
| 6 | hsa_circ_0020139 | chr10:120809312‐120818909 | 9597 | 1215 | EIcircRNA |
| 7 | hsa_circ_0020140 | chr10:120810710‐120832565 | 21855 | 1942 | EIcircRNA |
| 8 | hsa_circ_0020141 | chr10:120810710‐120833449 | 22739 | 2270 | EIcircRNA |
| 9 | hsa_circ_0020142 | chr10:120818723‐120820840 | 2117 | 507 | EIcircRNA |
| 10 | hsa_circ_0020143 | chr10:120819113‐120819230 | 117 | 117 | EcircRNA |
| 11 | hsa_circ_0020144 | chr10:120819113‐120840334 | 21221 | 1589 | EIcircRNA |
| 12 | hsa_circ_0020145 | chr10:120820257‐120820356 | 99 | 99 | EcircRNA |
| 13 | hsa_circ_0020146 | chr10:120820257‐120820840 | 583 | 204 | EIcircRNA |
| 14 | hsa_circ_0020147 | chr10:120820257‐120833089 | 12832 | 1086 | EIcircRNA |
| 15 | hsa_circ_0020148 | chr10:120824910‐120825082 | 172 | 172 | EcircRNA |
| 16 | hsa_circ_0020149 | chr10:120824910‐120833089 | 8179 | 882 | EIcircRNA |
| 17 | hsa_circ_0020150 | chr10:120824910‐120833449 | 8539 | 1073 | EIcircRNA |
| 18 | hsa_circ_0020151 | chr10:120828957‐120830597 | 1640 | 409 | EIcircRNA |
| 19 | hsa_circ_0020152 | chr10:120832401‐120832565 | 164 | 164 | EcircRNA |
| 20 | hsa_circ_0020153 | chr10:120832401‐120833089 | 688 | 301 | EIcircRNA |
| 21 | hsa_circ_0092856 | chr10:120801769‐120802132 | 363 | 363 | EcircRNA |
| 22 | hsa_circ_0092857 | chr10:120809312‐120810833 | 1521 | 462 | EIcircRNA |
| 23 | hsa_circ_0092858 | chr10:120810710‐120816365 | 5655 | 237 | EIcircRNA |
| 24 | hsa_circ_0092859 | chr10:120810710‐120818909 | 8199 | 876 | EIcircRNA |
| 25 | hsa_circ_0092860 | chr10:120816251‐120816365 | 114 | 114 | EcircRNA |
| 26 | hsa_circ_0092861 | chr10:120816447‐120816552 | 105 | 105 | EcircRNA |
| 27 | hsa_circ_0092862 | chr10:120818723‐120830597 | 11874 | 1088 | EIcircRNA |
| 28 | hsa_circ_0092863 | chr10:120828957‐120833449 | 4492 | 901 | EIcircRNA |
| 29 | hsa_circ_0092864 | chr10:120830397‐120832565 | 2168 | 364 | EIcircRNA |
| 30 | hsa_circ_0092865 | chr10:120832542‐120833089 | 547 | 160 | EIcircRNA |
| 31 | hsa_circ_0092866 | chr10:120832952‐120833449 | 497 | 328 | EIcircRNA |
Abbreviation: EIF3a, eukaryotic translation initiation factor 3 subunit A.
3.4. Validation for the differential expression level and drug sensitivity of circEIF3as
The circEIF3as is verified by real‐time quantitative reverse transcription‐PCR (qRT‐PCR) in A549 and A549/DDP cell line. We designed two pairs of primers to each circEIF3a for qRT‐PCR (Table S1). The results show that only two circRNAs transcribed from EIF3a (hsa_circ_0004350, hsa_circ_0092857) have differential expression in the two cell lines, and the rest are without differential expression or even with no expression in the two cell lines (Figure 3A). The locus of hsa_circ_0004350 is on chromosome 10:120.832.401‐120.833.449 in humans, while hsa_circ_0092857 is located on chromosome 10:120.809.312‐120.810.833 in humans. Both two circRNAs contain three exons and two introns (Figure 3B,C). The data were obtained from UNIVERSITY OF CALIFORNIA SANTA CRUZ GENOME BROWSER Database. Next, we explored whether hsa_circ_0004350 and hsa_circ_0092857 could affect cisplatin resistance in lung cancer cells, we found that down‐regulating the expression of hsa_circ_0004350 and hsa_circ_0092857 could affect cisplatin resistance in lung cancer cells (Figure 3D).
Figure 3.
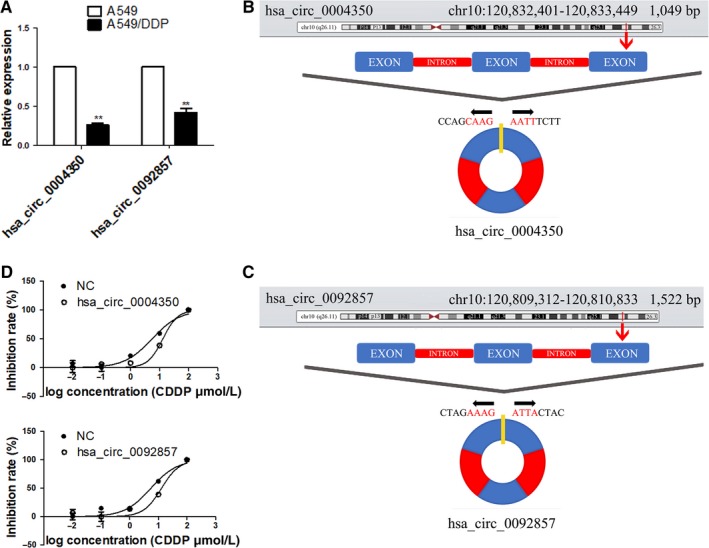
Panel A, The expression of hsa_circ_0004350 and hsa_circ_0092857 in A549 and A549/DDP cell line. (*P < 0.05; **P < 0.01). The diagram of position: hsa_circ_0004350 (panel B) and hsa_circ_0092857 (panel C). Panel D, The half‐maximal inhibitory concentration of hsa_circ_0004350 and hsa_circ_0092857
3.5. Possible biological functions of circEIF3as
Circular RNAs have different biological functions by acting as miRNA sponges or transcriptional regulators, and some of them are reported to have translation potential and can interact with RBPs. We conducted a series of experiments to determine the possible biological functions of circEIF3as. Compared with EIF3a mRNA, circEIF3as were resistant to RNase R digestion (Figure 4A). Subcellular fractionation assay indicated the cytoplasmic and nuclear enrichment and localization of circEIF3as (Figure 4B). RIP for AGO2 in A549 cells was performed, the results showed that circEIF3as were not accumulated in the AGO2 pellet (Figure 4C). In addition, we performed qRT‐PCR and western blot assays to investigate whether the circEIF3as can affect the expression of EIF3a (Figure 4D,E). In order to explore the translation potential of circEIF3as, we predicted the IRES and ORF via Circbank and ORF finder Database. Hsa_circ_0004350 has one IRES but no ORF, hsa_circ_0092857 has no IRES and ORF (Figure 4F). These data indicated that two potential circEIF3as were predominantly localized in the nucleus and were not accumulated in the AGO2 pellet. Furthermore, they had little effect on the expression of EIF3a and relatively low protein‐encoding potential. Taken together, we speculate that the two circEIF3as are involved in RBPs‐mediated biological processes.
Figure 4.
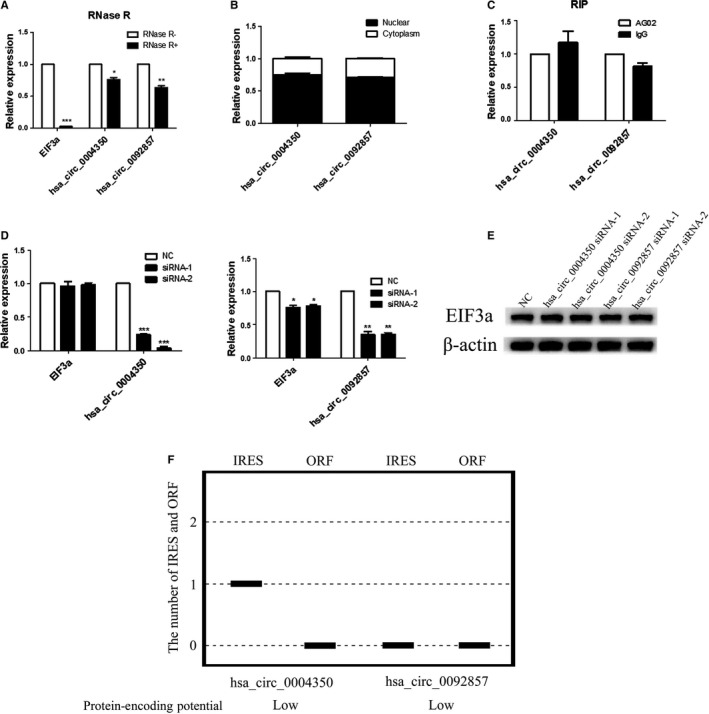
Panel A, After RNase R treatment, the expression levels of EIF3a and circEIF3as were determined by qRT‐PCR. Panel B, Subcellular fractionation assay was used to detect the localization of hsa_circ_0004350 and hsa_circ_0092857. Panel C, RIP assay indicating that hsa_circ_0004350 and hsa_circ_0092857 were not substantially accumulated in the AGO2 pellet. Hsa_circ_0004350 and hsa_circ_0092857 has little effect on the expression of EIF3a according to qRT‐PCR (panel D) and western blot (panel E) assays. Panel F, Protein‐encoding potential of circEIF3as. EIF3a, eukaryotic translation initiation factor 3 subunit A. EIF3a, eukaryotic translation initiation factor 3 subunit A; qRT‐PCR, quantitative reverse transcription‐PCR; RIP, RNA immunoprecipitation assay
3.6. Construction of the circEIF3as/RBPs axis regulation network
To explore the function of two potential circEIF3as, we predicted the circEIF3as/RBPs axis via circRNA Interactome, RBPDB, catRAPID, and Cancer‐Specific CircRNA Database. We used the four databases to determine the target RBPs for each circEIF3a and combined the RBPs of four databases as the target for each circEIF3a (Table S2). The network of circEIF3as/RBPs axis is illustrated using Cytoscape (Figure 5A).
Figure 5.
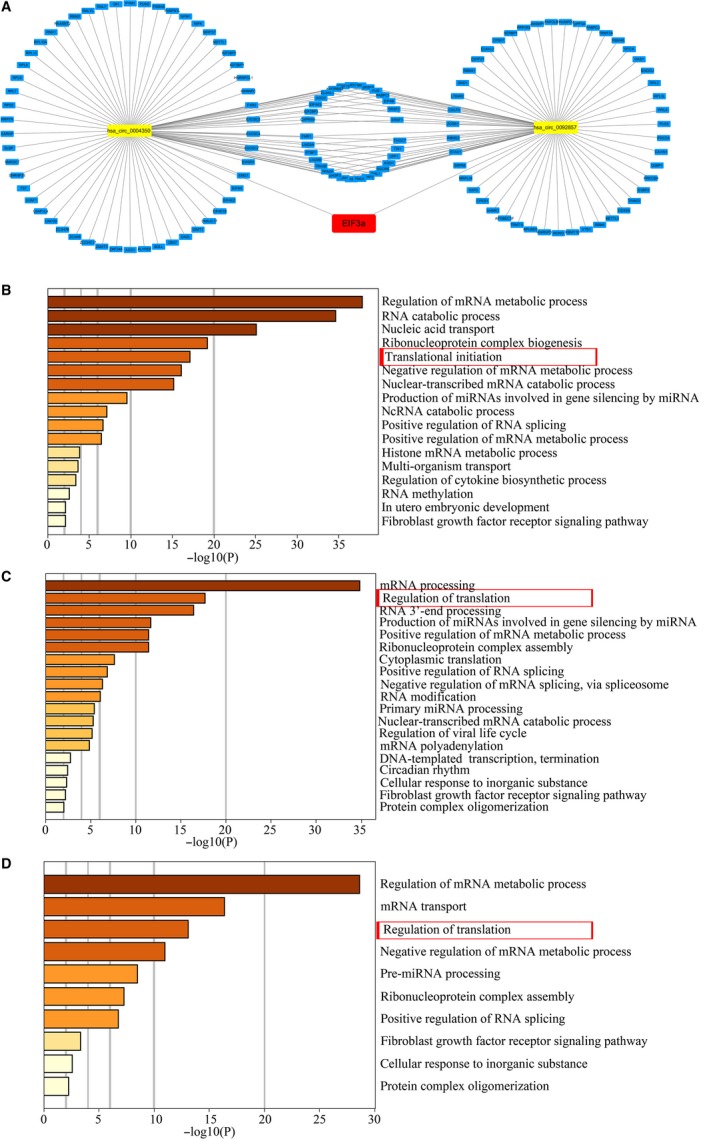
Panel A, The network of circEIF3as/RBPs axis. GO enrichment analysis of hsa_circ_0004350 (panel B) and hsa_circ_0092857 (panel C). Panel D, GO analysis on RBPs regulated by two circEIF3as. GO, gene ontology; RBPs, RNA‐binding proteins
3.7. Functional enrichment analysis of circEIF3as/RBPs
To identify the biological roles of hsa_circ_0004350 and hsa_circ_0092857, we annotated their functions by the GO enrichment analysis. We used Metascape to predict the biological functions of hsa_circ_0004350 (Figure 5B) and hsa_circ_0092857 (Figure 5C). It is interesting to note that hsa_circ_0004350 and hsa_circ_0092857 are significantly associated with translation regulation. Next, we performed GO analysis on the overlapping RBPs regulated by two circEIF3as, we found that these overlapping RBPs were also regulators of translation (Figure 5D). These results suggest that the two circEIF3as may have a function synergy effect with the parental EIF3a gene.
4. DISCUSSION
In the past few years, with the development of high‐throughput sequencing and bioinformatics analysis, circRNAs have gradually become a research topic, which were once considered as transcriptional noise. CircRNAs have also been recognized as transcriptional regulators and miRNAs sponges. In addition, they can interact with RBPs and some of them can even be translated into proteins.
As the largest subunit of EIF3, EIF3a plays a bridging role in the initiation of translation. Numbers of studies have shown that EIF3a is involved in the initiation of mRNA translation in yeast cells and mammalian cells,26 and also regulates the initiation of protein translation. In the meantime, EIF3a is involved in the regulation of cell cycle,27 and shows a significant correlation with cell growth 28 and differentiation.29 Recent studies have found that EIF3a is closely related to the occurrence, development, prognosis and chemotherapy efficacy of tumors.17 Moreover, EIF3a gene polymorphism is also associated with the susceptibility of malignant tumors.30
The above results indicate that EIF3a exerts a crucial influence on the physical fitness of human beings. EIF3a can be transcribed into both mRNA and circRNAs. Until now, there is no report about circRNAs transcribed from EIF3a. Therefore, we bravely assumed that circEIF3as has the biological function. We detected the expression of circEIF3as in the lung cancer cell line and lung cancer drug‐resistant cell line of human, the results showed that hsa_circ_0004350 and hsa_circ_0092857 had differential expression in two cell lines. Moreover, we found that down‐regulating the expression of hsa_circ_0004350 and hsa_circ_0092857 could affect cisplatin resistance in lung cancer cells. By using bioinformatical methods, we predicted the RBPs of these two circEIF3as in four databases. Next, we performed GO enrichment analysis of hsa_circ_0004350 and hsa_circ_0092857 to annotate the biological functions, we found that the two circEIF3as were related to translation regulation, showing that they may have a function synergy effect with EIF3a.
In conclusion, we systematically analyzed the expression of EIF3a in various tissues and tumors, and also discussed the prognostic value of EIF3a in lung cancer patients and ovarian cancer patients. Our study for the first time revealed the expression signatures of the circRNAs transcripts derived from EIF3a in lung cancer. Furthermore, we established the biological functions of circEIF3as with different bioinformatical analyses. The further studies on functions and mechanisms underlying hsa_circ_0004350 and hsa_circ_0092857 are being carried out in our laboratory. Our findings support that hsa_circ_0004350 and hsa_circ_0092857 are derived from the translation initiation factor‐EIF3a, moreover, they are involved in drug sensitivity and translational regulation. These results suggest that these two circEIF3as may have functional synergy with their parental gene‐EIF3a, and may also serve as potential targets for lung cancer treatment like EIF3a. These data provide the basis for further studies on the diagnosis, treatment and biological function of circEIF3as in lung cancer.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program of China (2016YFC1306900, 2016YFC0905002), National Natural Science Foundation of China (81573508), Open Foundation of Innovative Platform in Colleges and University of Hunan Province of China ([2015]54), Clinical Research Fund of Peking University Unamed‐Central South University Xiangya Hospital (xywm 2015I16), and The Strategy‐Oriented Special Project of Central South University in China (ZLXD2017003).
Huang M‐S, Yuan F‐Q, Gao Y, et al. Circular RNA screening from EIF3a in lung cancer. Cancer Med. 2019;8:4159–4168. 10.1002/cam4.2338
Funding information
This work was supported by the National Key Research and Development Program of China (2016YFC1306900, 2016YFC0905002), National Natural Science Foundation of China (81874327), Open Foundation of Innovative Platform in Colleges and University of Hunan Province of China ([2015]54), Clinical Research Fund of Peking University Unamed‐Central South University Xiangya Hospital (xywm 2015I16), and The Strategy‐Oriented Special Project of Central South University in China (ZLXD2017003).
REFERENCES
- 1. Yin J‐Y, Zhang J‐T, Zhang W, Zhou H‐H, Liu Z‐Q. eIF3a: a new anticancer drug target in the eIF family. Cancer Lett. 2018;412:81‐87. [DOI] [PubMed] [Google Scholar]
- 2. Huang M‐S, Zhu T, Li L, et al. LncRNAs and CircRNAs from the same gene: masterpieces of RNA splicing. Cancer Lett. 2017;415:49‐57. [DOI] [PubMed] [Google Scholar]
- 3. Rasmussen AH, Rasmussen HB, Silahtaroglu A. The DLGAP family: neuronal expression, function and role in brain disorders. Mol Brain. 2017;10(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Minocherhomji S, Hansen C, Kim H‐G, et al. Epigenetic remodelling and dysregulation of DLGAP4 is linked with early‐onset cerebellar ataxia. Hum Mol Genet. 2014;23(23):6163‐6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bai Y, Zhang Y, Han B, et al. Circular RNA DLGAP4 ameliorates ischemic stroke outcomes by targeting miR‐143 to regulate endothelial‐mesenchymal transition associated with blood‐brain barrier integrity. J Neurosci. 2017;38:32‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fu Y, Sun X, Lu B. HIPK3 modulates autophagy and HTT protein levels in neuronal and mouse models of Huntington disease. Autophagy. 2017;14:169‐170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yu M, Fu Y, Liang Y, et al. Suppression of MAPK11 or HIPK3 reduces mutant Huntingtin levels in Huntington's disease models. Cell Res. 2017;27(12):1441‐1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shan K, Liu C, Liu B‐H, et al. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. 2017;136(17):1629‐1642. [DOI] [PubMed] [Google Scholar]
- 10. Li Z, Huang C, Bao C, et al. Exon‐intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256‐264. [DOI] [PubMed] [Google Scholar]
- 11. McKinney C, Yu D, Mohr I. A new role for the cellular PABP repressor Paip2 as an innate restriction factor capable of limiting productive cytomegalovirus replication. Genes Dev. 2013;27(16):1809‐1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shen J, Yin J‐Y, Li X‐P, et al. The prognostic value of altered eIF3a and its association with p27 in non‐small cell lung cancers. PLoS ONE. 2014;9(4):e96008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bachmann F, Banziger R, Burger MM. Cloning of a novel protein overexpressed in human mammary carcinoma. Cancer Res. 1997;57(5):988‐994. [PubMed] [Google Scholar]
- 14. Wang S‐Q, Liu YU, Yao M‐Y, Jin J. Eukaryotic translation initiation factor 3a (eIF3a) promotes cell proliferation and motility in pancreatic cancer. J Korean Med Sci. 2016;31(10):1586‐1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen G, Burger MM. p150 overexpression in gastric carcinoma: the association with p53, apoptosis and cell proliferation. Int J Cancer. 2004;112(3):393‐398. [DOI] [PubMed] [Google Scholar]
- 16. Spilka R, Laimer K, Bachmann F, et al. Overexpression of eIF3a in squamous cell carcinoma of the oral cavity and its putative relation to chemotherapy response. J Oncol. 2012;2012:901956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haybaeck J, O'Connor T, Spilka R, et al. Overexpression of p150, a part of the large subunit of the eukaryotic translation initiation factor 3, in colon cancer. Anticancer Res. 2010;30(4):1047‐1055. [PubMed] [Google Scholar]
- 18. Chen G, Burger MM. p150 expression and its prognostic value in squamous‐cell carcinoma of the esophagus. Int J Cancer. 1999;84(2):95‐100. [DOI] [PubMed] [Google Scholar]
- 19. Zhang Y, Yu JJ, Tian Y, et al. eIF3a improve cisplatin sensitivity in ovarian cancer by regulating XPC and p27Kip1 translation. Oncotarget. 2015;6(28):25441‐25451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Spilka R, Ernst C, Bergler H, et al. eIF3a is over‐expressed in urinary bladder cancer and influences its phenotype independent of translation initiation. Cell Oncol (Dordr). 2014;37(4):253‐267. [DOI] [PubMed] [Google Scholar]
- 21. Dellas A, Torhorst J, Bachmann F, B�nziger R, Schultheiss E, Burger MM. Expression of p150 in cervical neoplasia and its potential value in predicting survival. Cancer. 1998;83(7):1376‐1383. [DOI] [PubMed] [Google Scholar]
- 22. Fang C, Chen YX, Wu NY, et al. MiR‐488 inhibits proliferation and cisplatin sensibility in non‐small‐cell lung cancer (NSCLC) cells by activating the eIF3a‐mediated NER signaling pathway. Sci Rep. 2017;7:40384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yin J‐Y, Meng X‐G, Qian C‐Y, et al. Association of positively selected eIF3a polymorphisms with toxicity of platinum‐based chemotherapy in NSCLC patients. Acta Pharmacol Sin. 2015;36(3):375‐384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu X, Han L, Yang H, et al. The A/G allele of eIF3a rs3740556 predicts platinum‐based chemotherapy resistance in lung cancer patients. Lung Cancer. 2013;79(1):65‐72. [DOI] [PubMed] [Google Scholar]
- 25. Ensinger C, Obrist P, Mikuz G, et al. Assignment1 of the p150 subunit of the eukaryotic initiation factor 3A gene (EIF3A) to human chromosome band 10q26 by in situ hybridisation. Cytogenet Cell Genet. 1998;83(1–2):74‐75. [DOI] [PubMed] [Google Scholar]
- 26. Saletta F, Suryo Rahmanto Y, Richardson DR. The translational regulator eIF3a: the tricky eIF3 subunit!. Biochim Biophys Acta. 2010;1806(2):275‐286. [DOI] [PubMed] [Google Scholar]
- 27. Dong Z, Liu Z, Cui P, et al. Role of eIF3a in regulating cell cycle progression. Exp Cell Res. 2009;315(11):1889‐1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dong Z, Zhang JT. EIF3 p170, a mediator of mimosine effect on protein synthesis and cell cycle progression. Mol Biol Cell. 2003;14(9):3942‐3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu Z, Dong Z, Yang Z, et al. Role of eIF3a (eIF3 p170) in intestinal cell differentiation and its association with early development. Differentiation. 2007;75(7):652‐661. [DOI] [PubMed] [Google Scholar]
- 30. Olson JE, Wang X, Goode EL, et al. Variation in genes required for normal mitosis and risk of breast cancer. Breast Cancer Res Treat. 2010;119(2):423‐430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).


