SEs play critical roles in cancer development. Since SEs assemble much bigger protein complexes on enhancers than typical enhancers (TEs), they are more sensitive than TEs to perturbations. Understanding the protein composition of SEs that are linked to key oncogenes may identify novel therapeutic targets. A genome-wide CRISPR screen specifically identified proteins essential for MYC ESE activity but not simian virus 40 (SV40) enhancer. These proteins not only were essential for the reporter activity but also were also important for MYC expression and LCL growth. Targeting these proteins may lead to new therapies for EBV-associated cancers.
KEYWORDS: CRISPR, EBV, MEF2C, MYC, super-enhancer, transcription factors
ABSTRACT
Super-enhancers (SEs) are clusters of enhancers marked by extraordinarily high and broad chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) signals for H3K27ac or other transcription factors (TFs). SEs play pivotal roles in development and oncogenesis. Epstein-Barr virus (EBV) super-enhancers (ESEs) are co-occupied by all essential EBV oncogenes and EBV-activated NF-κB subunits. Perturbation of ESEs stops lymphoblastoid cell line (LCL) growth. To further characterize ESEs and identify proteins critical for ESE function, MYC ESEs were cloned upstream of a green fluorescent protein (GFP) reporter. Reporters driven by MYC ESEs 525 kb and 428 kb upstream of MYC (525ESE and 428ESE) had very high activities in LCLs but not in EBV-negative BJAB cells. EBNA2 activated MYC ESE-driven luciferase reporters. CRISPRi targeting 525ESE significantly decreased MYC expression. Genome-wide CRISPR screens identified factors essential for ESE activity. TBP-associated factor (TAF) family proteins, including TAF8, TAF11, and TAF3, were essential for the activity of the integrated 525ESE-driven reporter in LCLs. TAF8 and TAF11 knockout significantly decreased 525ESE activity and MYC transcription. MEF2C was also identified to be essential for 525ESE activity. Depletion of MEF2C decreased 525ESE reporter activity, MYC expression, and LCL growth. MEF2C cDNA resistant to CRIPSR cutting rescued MEF2C knockout and restored 525ESE reporter activity and MYC expression. MEF2C depletion decreased IRF4, EBNA2, and SPI1 binding to 525ESE in LCLs. MEF2C depletion also affected the expression of other ESE target genes, including the ETS1 and BCL2 genes. These data indicated that in addition to EBNA2, TAF family members and MEF2C are essential for ESE activity, MYC expression, and LCL growth.
IMPORTANCE SEs play critical roles in cancer development. Since SEs assemble much bigger protein complexes on enhancers than typical enhancers (TEs), they are more sensitive than TEs to perturbations. Understanding the protein composition of SEs that are linked to key oncogenes may identify novel therapeutic targets. A genome-wide CRISPR screen specifically identified proteins essential for MYC ESE activity but not simian virus 40 (SV40) enhancer. These proteins not only were essential for the reporter activity but also were also important for MYC expression and LCL growth. Targeting these proteins may lead to new therapies for EBV-associated cancers.
INTRODUCTION
Epstein-Bar virus (EBV) is the first human DNA tumor virus identified from African Burkitt’s lymphoma over 50 years ago (1). EBV infection causes Burkitt’s lymphoma, Hodgkin’s lymphoma, HIV-related lymphomas, posttransplant lymphoproliferative diseases (PTLDs), nasopharyngeal carcinoma, and ∼10% of gastric cancers (2). During primary infection, EBV infects both oral epithelial cells and B cells. Immune surveillance efficiently eliminates EBV-infected cells and drives the EBV-infected cells into a latency state to establish lifelong infection (3).
In vitro, EBV efficiently transforms primary resting B lymphocytes (RBLs) to continuously proliferating lymphoblastoid cell lines (LCLs) (3). In LCLs, EBV expresses six EBV nuclear antigens (EBNAs), including EBNA1, EBNALP, EBNA2, EBNA3A, EBNA3B, and EBNA3C, three latent membrane proteins (LMPs), including LMP1, LMP2a, and LMP2b, noncoding RNAs, and microRNAs (miRNAs) (3). Genetic studies demonstrated that EBNALP, EBNA2, EBNA3A, EBNA3C, and LMP1 are essential for continuous LCL proliferation (4–11). EBNA2 and EBNALP are expressed immediately after EBV infection (12). EBNA2 activates both host and viral gene expression (13–15). EBNALP binds preferentially to promoters over enhancers and can coactivate with EBNA2 by removing transcription repressors (16–18) or by modulating the activity of the key transcription activator EP300 (19). EBNA3A and EBNA3C are each essential for LCL growth through repressing p16INK4A and p14ARF expression, thus preventing cell senescence (20–22). p16INK4A and p14ARF knockdown allows LCLs to grow in the absence of EBNA3A or EBNA3C (20). LMP1 mimics CD40 signaling to activate NF-κB (23). In vivo, LMP1 induces B-cell lymphoma (24, 25).
Overexpression of MYC in LCLs abrogated the requirement of EBNA2 and LMP1 for cell survival and proliferation (26). In LCLs and PTLDs, MYC expression is upregulated, in line with a high cell proliferation rate (27, 28). MYC small-molecule inhibitor 10058-F4 significantly suppresses LCL growth (29).
Super-enhancers (SEs) are clusters of enhancers bound by multiple transcription factors. SEs are associated with genes critical for cell growth and differentiation and are more sensitive to perturbation than typical enhancers (TEs) (30–32). Our previous work identified 187 EBV SEs (ESEs) that are bound by all the essential EBNAs and NF-κB subunits, with extraordinarily high and broad H3K27ac chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) signals (33). MYC is linked to two ESEs 428 kb and 525 kb upstream of the MYC transcription start site (TSS) (428ESE and 525ESE). Deletion of these two MYC ESEs by dual CRISPR-cas9 guide RNAs (gRNAs) decreases MYC expression and abrogates LCL growth (34). MYC ESE enhancer RNAs (eRNAs) are also important for MYC expression and LCL growth (35). However, little is known about full ESE proteomic composition.
To identify TFs essential for MYC ESE function, we first generated GM12878 LCLs stably expressing green fluorescent protein (GFP) driven by MYC ESE. Genome-wide CRISPR screen was then used to identify TFs that were essential for MYC ESE activity. TBP-associated factor (TAF) family proteins, including TAF3, TAF8, TAF11, and myocyte enhancer factor 2C (MEF2C), were significantly depleted in the screen. Further experiments confirmed that depletion of TAF8, TAF11, and MEF2C greatly decreased MYC ESE activity and downregulated MYC expression.
RESULTS
MYC 525ESE and 428ESE are active in LCLs.
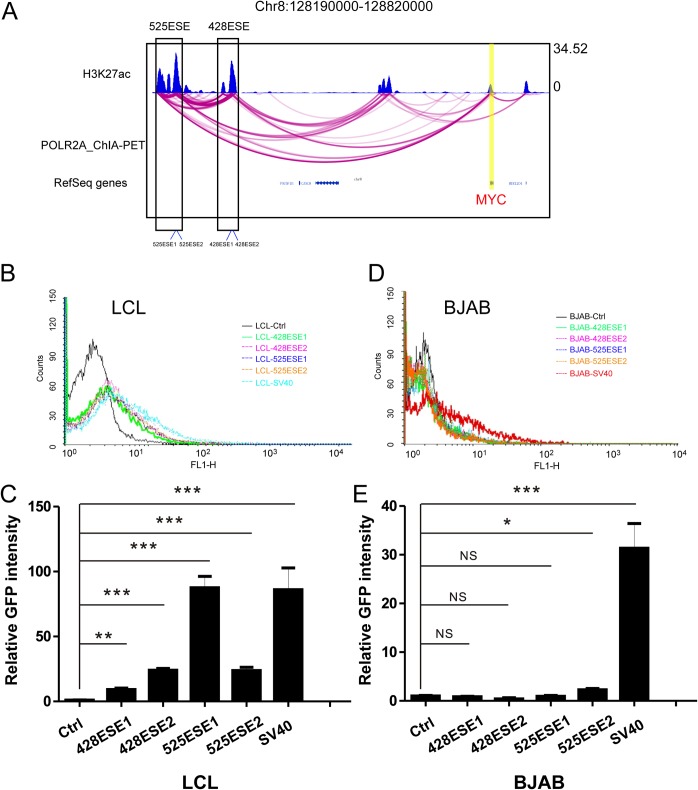
MYC 525ESE and 428ESE link to the MYC promoter by POLR2A chromatin interaction analysis followed by paired-end tag sequencing (ChIA-PET) (Fig. 1A). To compare the activities of these MYC ESEs in LCLs, we cloned different regions of 525ESE and 428SE upstream of a green fluorescent protein (GFP) reporter in pGreenFire-mCMV vector and packaged them into lentiviruses (Fig. 1A). Simian virus 40 (SV40) enhancer-driven GFP was used as a control. LCLs and EBV-negative BJAB cells were transduced with lentiviruses and selected for stable cell lines. Fluorescence-activated cell sorting (FACS) was used to determine the enhancer activity. ESEs activated the GFP reporter in LCLs (Fig. 1B). 428ESE1, 428ESE2, 525ESE1, and 525ESE2 activated the GFP reporter ∼9-, ∼24-, ∼87-, and ∼24-fold ((P < 0.01 and P < 0.001 [Fig. 1C]). ESEs did not activate the GFP reporter in BJAB cells, while SV40 enhancer greatly activated the GFP reporter in BJAB cells compared with empty vector control (P < 0.001 [Fig. 1D and E]). These data suggested that EBV proteins are required for MYC ESE activity.
FIG 1.
Identification of DNA elements critical MYC ESE activity. (A) MYC ESEs at the MYC locus. LCL H3K27ac ChIP-seq track is shown at the top. LCL POLR2A ChIA-PET track is in the middle. Each magenta line indicates a ChIA-PET interaction between MYC enhancer and the MYC TSS. ESEs 525 and 428 kb upstream of MYC TSS are indicated by black boxes. Vertical lines at the bottom of these boxes indicate the enhancer fragments cloned upstream of reporter vector. (B) GM12878 LCLs were transduced with lentiviruses expressing ESE-driven GFP/luciferase reporter. After selection, the GFP expression levels were determined by FACS. Ctrl, control. (C) Quantitation of GFP expression from LCLs transduced with indicated ESE-driven reporters where the control group was set to 1. ***, P < 0.001; **, P < 0.01. (D) EBV-negative BJAB cells were transduced with the same lentiviruses expressing ESE-driven GFP/luciferase reporters. GFP expression levels were determined by FACS. (E) Quantitation of GFP expression from BJAB cells transduced with indicated ESE-driven reporters where the control group was set to 1. ***, P < 0.001; *, P < 0.05; NS, not significant.
EBNA2 is essential for 525ESE1 activation.
525ESE1 activated the GFP reporter in LCLs the most. Therefore, we further investigated the unique features of this ESE.
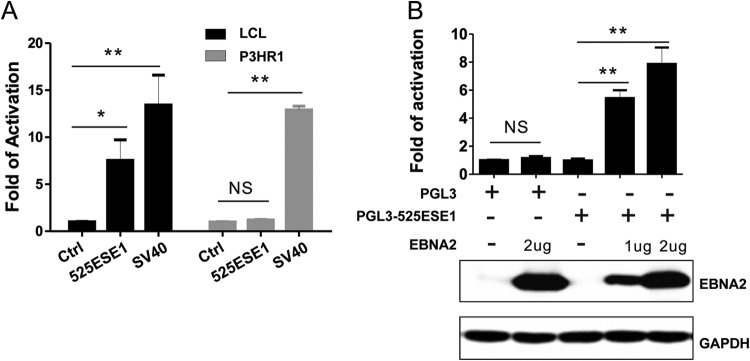
EBNA2 binds to 525ESE1 loci strongly (13). To test whether EBNA2 is an important transcriptional activator that upregulates 525ESE1 activity, pGreenFire-mCMV, pGreenFire-525ESE1, and pGreenFire-SV40 lentiviruses were used to infect P3HR1 cells. P3HR1 cells harbor a mutant EBV containing deletions for EBNA2 and the last two exons of EBNALP. Their GFP reporter activities were compared with that of LCLs. As expected, GFP was strongly activated by SV40 enhancer in both LCLs and P3HR1 cells (P < 0.01 [Fig. 2A]). GFP was activated by 525ESE1 only in LCLs (P < 0.05 [Fig. 2A]) and not in P3HR1 cells. To distinguish the effect of EBNA2 deletion from that of EBNALP deletion in P3HR1, 525ESE1 was cloned upstream of a minimum SV40 promoter in pGL3-promoter reporter plasmid. The reporter was transiently transfected into P3HR1 cells together with or without EBNA2 expression plasmid. Without EBNA2, the reporter had background level activity. Increasing amount of EBNA2 significantly increased the reporter activity (P < 0.01 [Fig. 2B]). These data indicated that EBNA2 is required but not sufficient for 525ESE1 activity.
FIG 2.
EBNA2 is important for MYC 525ESE1 activity. (A) Lentiviruses expressing 525ESE1-driven reporter were used to infect the GM12878 LCL and the P3HR1 Burkitt's lymphoma cells, which harbor mutant EBV with EBNA2 and the last two exons of EBNALP deleted. GFP expression levels were determined by FACS where the control was set to 1. NS, not significant. **, P < 0.01; *, P < 0.05; NS, not significant. (B) Luciferase reporter under the control of 525ESE1 was electroporated into BJAB cells in the presence or absence of EBNA2 expression plasmids. The empty expression plasmid control was set to 1. Renilla luciferase was used to normalize the transfection efficiency. EBNA2 expression levels were determined by Western blotting. **, P < 0.01; NS, not significant.
CRISPRi inhibition of 525ESE1 decreases MYC expression.
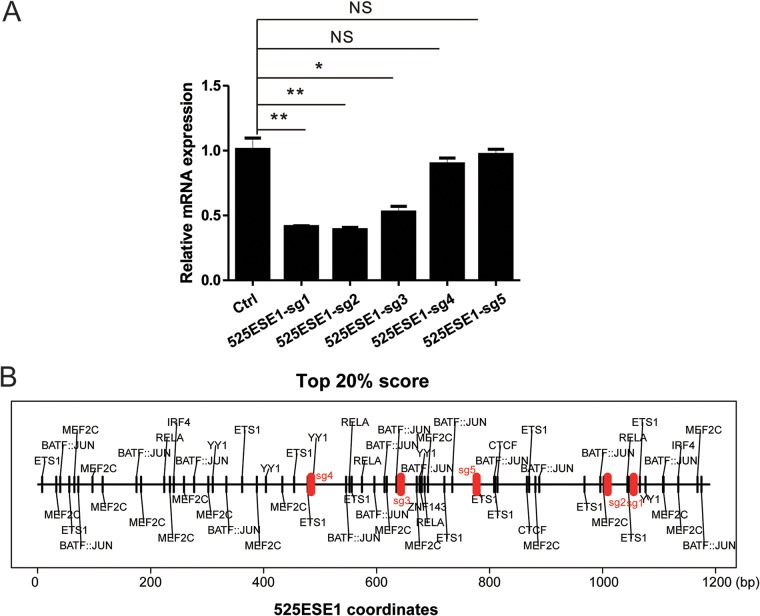
CRISPRi was used to characterize MYC ESEs in the context of an intact genome. Five single guide RNAs (sgRNAs) targeting different regions of 525ESE1 were designed and cloned into pLentiGuide-Puro vector. Lentiviruses were then packaged and used to infect LCLs stably expressing dCAS9-KRAB. MYC transcription levels were measured by quantitative reverse transcription-PCR (qRT-PCR). Among 5 sgRNAs tested, 3 significantly decreased MYC transcription (P < 0.05 [Fig. 3A]), supporting our previous finding where 525ESE was deleted (34). Motif analyses found numerous TF motifs enriched in 525ESE1 (Fig. 3B).
FIG 3.
525ESE1 CRISPRi represses MYC expression. (A) LCLs stably expressing a dCAS9-KRAB fusion proteins were transduced with lentiviruses expressing sgRNAs targeting different regions within 525ESE1. qRT-PCR was used to determine the MYC expression levels where control sgRNA was set to 1. **, P < 0.01; *, P < 0.05; NS, not significant. (B) Predicted TF binding sites within 525ESE1 are indicated by black vertical lines. sgRNA targets are indicated by red vertical lines.
CRISPR/Cas9 screens to identify host factors essential for 525ESE1 activity.
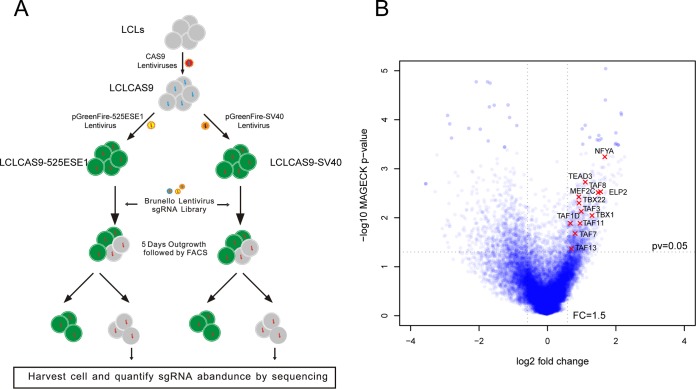
To identify additional host proteins essential for 525ESE1 activation, genome-wide CRISPR screens were used in LCLs expressing 525ESE1-driven GFP (Fig. 4A). As a genome-wide CRISPR screen will identify both genes essential for the reporter activity and LCL growth and survival, SV40 enhancer-driven GFP reporter was used as a control to exclude genes essential for LCL survival. LCLs stably expressing CAS9 and the GFP reporters were transduced with the Brunello library at a multiplicity of infection (MOI) of 0.3 (36). The Brunello library has 4 gRNAs per gene and a total of 76,441 gRNAs, including controls. Transduced cells were selected with puromycin for 3 days and grown for another 5 days. Cells that lost their GFP signals were collected by FACS. Genomic DNAs were prepared and sgRNAs were amplified by PCR. sgRNA abundance was quantitated by next-generation sequencing. MaGECK was used to identify depleted genes (37). By comparing the sgRNAs lost in LCLs with SV40-driven GFP and 525ESE1-driven GFP, we identified genes contributing to 525ESE1 activity (P < 0.05 [Fig. 4B and Data Set S1 in the supplemental material]).
FIG 4.
CRISPR screen for cell factors essential for ESE. (A) Schematic diagram of the genome wide CRISPR screen. (B) Differentially enriched genes essential for 525ESE1 function.
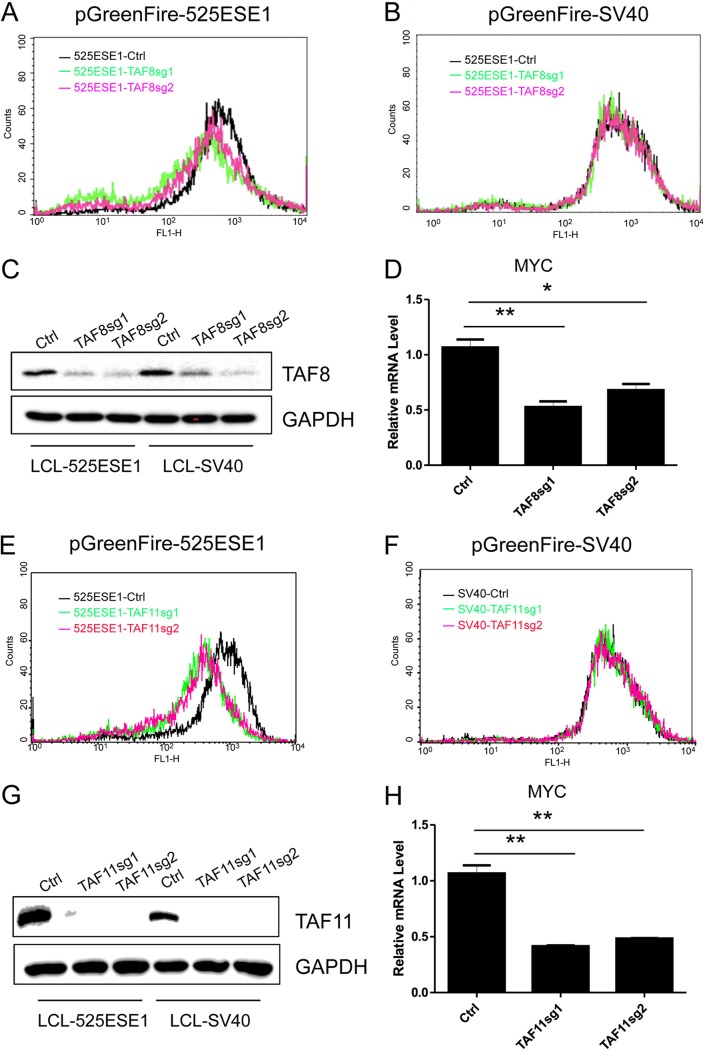
TAF family proteins are essential for 525ESE1 activity.
TBP-associated factors (TAFs) are components of the basal transcription machinery. TFIID serves as a scaffold for the assembly of preinitiation complex to initiate POLR2A-mediated transcription. A CRISPR screen found that TAF family proteins, including TAF3, TAF8, and TAF11, were selectively depleted in GFP-negative cells expressing pGreenFire-525ESE1 but not pGreenFire-SV40. These data indicated that TAF3, TAF8, and TAF11 were important for 525ESE1 activity, but not SV40 enhancer. To further confirm these findings, sgRNAs targeting TAF8 and TAF11 were cloned into plentiGuide-Puro vector. Packaged lentiviruses were used to infect LCLs stably expressing CAS9 and 525ESE1- or SV40 enhancer-driven reporters. TAF8 knockout greatly reduced GFP reporter activity in LCLs expressing 525ESE-driven GFP (Fig. 5A). TAF8 knockout did not affect SV40 driven reporter activity (Fig. 5B). Western blotting validated TAF8 depletion in both cell lines (Fig. 5C). MYC expression was evaluated by qRT-PCR. Both sgRNAs significant reduced MYC expression (P < 0.05 and P < 0.01 [Fig. 5D]). TAF11 knockout greatly reduced GFP reporter activity in LCLs expressing 525ESE-driven GFP (Fig. 5E). TAF11 knockout did not affect SV40-driven reporter (Fig. 5F). Western blotting validated TAF11 depletion in both cell lines (Fig. 5G). Both sgRNAs significantly reduced MYC expression as determined by qRT-PCR (P < 0.01 [Fig. 5H]).
FIG 5.
TAF8 and TAF11 are essential for 525ESE1 function. (A) LCLs stably expressing CAS9 and 525ESE1-driven GFP-luciferase reporter were transduced with lentiviruses expressing control sgRNA or sgRNA targeting TAF8. After puromycin selection, GFP levels were determined by FACS. (B) LCLs stably expressing CAS9 and SV40 enhancer-driven GFP-luciferase reporter were transduced with lentiviruses expressing control sgRNA or sgRNA targeting TAF8. GFP levels were determined by FACS. (C) TAF8 expression in LCLs expressing 525ESE1- or SV40 enhancer-driven reporters. (D) MYC expression levels determined by qRT-PCR following TAF8 knockout. The level of control sgRNA was set to 1. **, P < 0.01; *, P < 0.05. (E) LCLs expressing 525ESE1-driven reporter were transduced with lentiviruses expressing control sgRNA or sgRNA targeting TAF11. After puromycin selection, GFP levels were determined by FACS. (F) LCLs expressing SV40 enhancer-driven reporter were transduced with lentiviruses expressing control sgRNA or sgRNA targeting TAF11. GFP levels were determined by FACS. (G) TAF11 expression in LCLs expressing 525ESE1- or SV40 enhancer-driven reporters. (H) MYC expression levels were determined by qRT-PCR following TAF11 knockout. The level of control sgRNA was set to 1. **, P < 0.01.
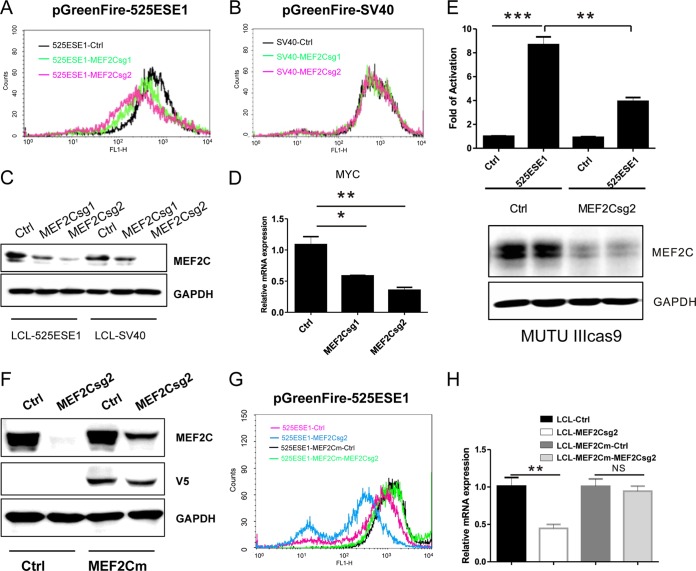
MEF2C is critical for 525ESE1 activity.
MEF2C is essential for LCL growth and survival (38). sgRNAs targeting MEF2C were depleted in the CRISPR screen in LCLs expressing 525ESE-driven GFP but not in LCLs expressing SV40 enhancer-driven GFP. To further validate this finding, two MEF2C sgRNAs were used to knock out MEF2C in LCLs expressing 525ESE1-driven GFP. MEF2C knockout greatly decreased 525ESE-driven reporter as determined by FACS (Fig. 6A). MEF2C knockout did not affect SV40 enhancer-driven reporter (Fig. 6B). Western blotting confirmed the reduction in MEF2C expression (Fig. 6C). Importantly, in line with decreased 525ESE1 reporter activity, MYC expression was also downregulated (P < 0.05 and P < 0.01 [Fig. 6D]). To evaluate if MEF2C is also important for 525ESE1 in other type III EBV latency cell lines that express all the EBNAs and LMPs, MEF2C sgRNA2 or control sgRNA was used to delete MEF2C in MUTU III cells stably expressing CAS9. Transient-transfection and reporter assays were used to evaluate if MEF2C was important in this cell line. Activities of 525ESE1-driven or control reporter were determined by firefly luciferase assay. Renilla luciferase was used as a control for transfection efficiency. 525ESE1-driven reporter activity was significantly higher than the control (P < 0.001 [Fig. 6E]). MEF2C knockout significantly reduced 525ESE1-driven reporter activity (P < 0.01 [Fig. 6E]). MEF2C depletion was validated by Western blotting (Fig. 6E). To exclude the possibility that the decreased 525ESE1 activity following MEF2C knockout was caused by CRISPR off-target effect, cDNA rescue was used. The PAM site for CRISPR CAS9 cutting in MEF2C cDNA was mutated without affecting the MEF2C amino acid sequence. CRISPR-resistant MEF2C cDNA was stably expressed in CAS9 LCLs harboring the reporters, as shown by Western blotting (Fig. 6F). In control LCLs, MEF2C sgRNA reduced 525ESE1-driven GFP expression. MEF2C CRISPR-resistant cDNA effectively restored GFP expression (Fig. 6G). Similarly, MEF2C sgRNA significantly reduced LCL endogenous MYC expression, as determined by qRT-PCR (P < 0.01). MEF2C CRISPR-resistant cDNA restored MYC expression (Fig. 6H). These data indicated that MEF2C is critically important for 525ESE1 activity and MYC expression.
FIG 6.
MEF2C is essential for 5252ESE1 function. (A) LCLs stably expressing CAS9 and 525ESE1-driven GFP-luciferase reporter were transduced with lentiviruses expressing control sgRNA or sgRNA targeting MEF2C. After puromycin selection, GFP levels were determined by FACS. (B) LCLs stably expressing CAS9 and SV40 enhancer-driven GFP-luciferase reporter were transduced with lentiviruses expressing control sgRNA or sgRNA targeting MEF2C. GFP levels were determined by FACS. (C) MEF2C expression in LCLs expressing 525ESE1- or SV40 enhancer-driven reporters. (D) MYC expression levels determined by qRT-PCR following MEF2C knockout. The level of control sgRNA was set to 1. **, P < 0.01; *, P < 0.05. (E) MEF2C expression was first knocked out by CRISPR in MUTU III cells. 525ESE1-driven reporter or control reporter was then electroporated into these cells. Luciferase activities were normalized by Renilla luciferase. Control reporter levels were set to 1. ***, P < 0.001; **, P < 0.01. (F) V5-tagged MEF2C cDNA with the 525ESE1 sgRNA2 PAM site mutated was expressed in LCLs. MEF2C sgRNA2 efficiently knocked out endogenous MEF2C but not MEF2C from rescue cDNA. (G) 525ESE1-driven GFP levels in cells with MEF2C knockout or cells expressing rescue cDNA. MEF2C knockout efficiently repressed MYC expression. (H) In cells expressing CRISPR-resistant MEF2C, knockout did not decrease MYC expression. MYC expression levels in control sgRNA treated cells were set to 1. **, P < 0.01; NS, not significant.
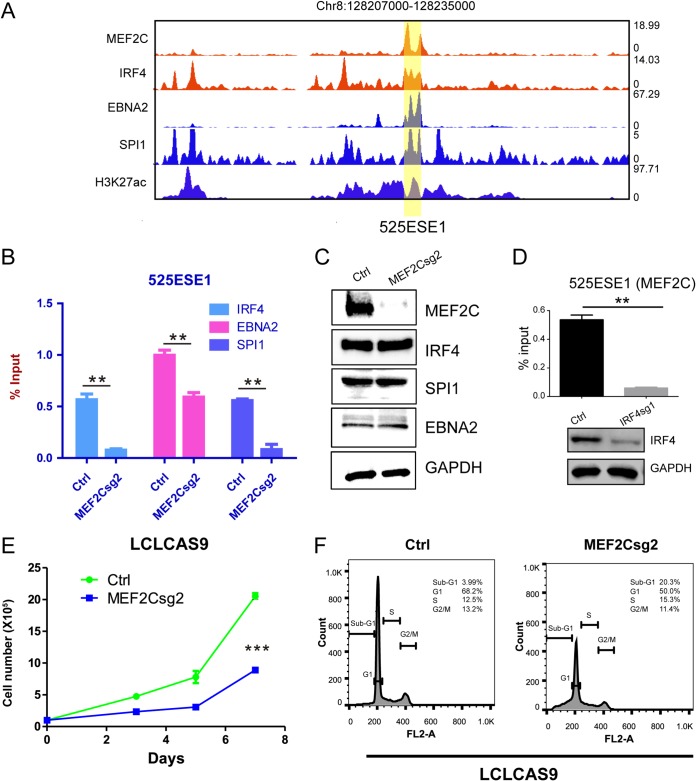
MEF2C knockout affects EBNA2, IRF4, and SPI1 binding to 525ESE1 and LCL cell growth and survival.
EBNA2 and multiple B-cell TFs, including IRF4 and SPI1, bind to 525ESE1 in LCLs (Fig. 7A). To determine if MEF2C can affect the DNA binding of other TFs, LCL MEF2C was knocked out. Antibodies against EBNA2, IRF4, and SPI1 were used to immunoprecipitate these TFs with 525ESE1. qPCR was used to quantitate precipitated DNA. MEF2C knockout significantly decreased EBNA2, IRF4, and SPI1 binding to 525ESE1 (P < 0.01 [Fig. 7B]). The protein levels of these TFs were unaffected by MEF2C knockout, as shown by Western blotting (Fig. 7C). These results suggested that in the absence of MEF2C, EBNA2, IRF4, and SPI1 bind to 525ESE1 at reduced levels. We also tested if IRF4 knockout affected MEF2C DNA binding by ChIP-qPCR. IRF4 knockout significantly reduced MEF2C DNA binding (P < 0.01 [Fig. 7D]). These data suggested that there may be synergistic effects for TFs to bind to MYC ESE. MEF2C knockout significantly reduced LCL growth and induced apoptosis (Fig. 7E and F).
FIG 7.
MEF2C knockout reduces IRF4, EBNA2, and SPI1 SE binding and LCL growth. (A) MEF2C, IRF4, EBNA2, SPI1, and H3K27ac ChIP-seq tracks at 525ESE1. (B) MEF2C CRISPR knockout was first done in LCLs. ChIP-qPCR was used to measure IRF4, EBNA2, and SPI1 binding to 525ESE1. **, P < 0.01. (C) Expression of MEF2C, IRF4, EBNA2, and SPI1 followed by MEF2C knockout. (D) IRF4 was knocked out in LCLs. MEF2C DNA binding was determined by ChIP-qPCR. **, P < 0.01. (E) LCL growth followed by MEF2C knockout. ***, P < 0.001. (F) LCL cell cycle distribution followed by MEF2C knockout.
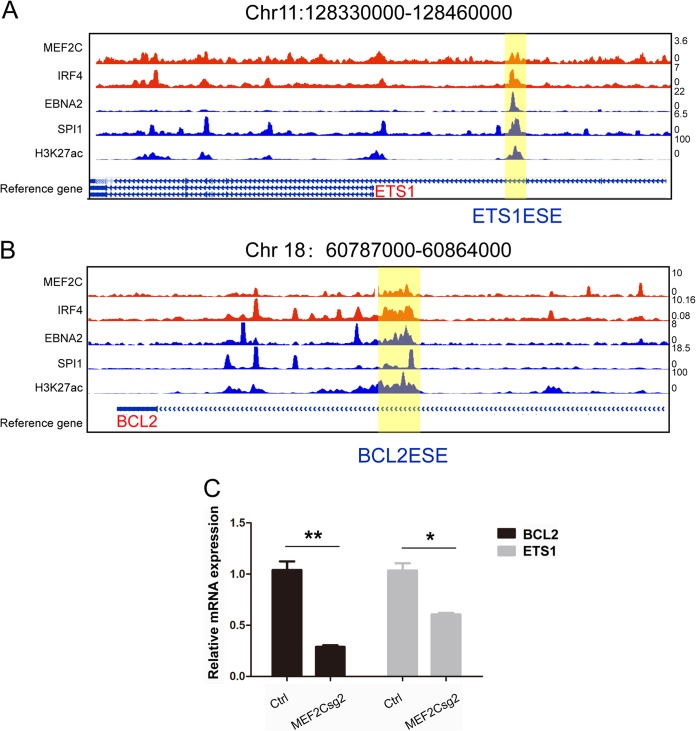
MEF2C knockout also affects ETS1 and BCL2 ESEs.
EBNA2, MEF2C, IRF4, and SPI1 also bind to ETS1 and BCL2 ESEs (Fig. 8A and B). qRT-PCR was used to determine if MEF2C knockout also affects the expression of these ESE-associated genes. MEF2C knockout significantly reduced the expression of ESE-associated ETS1 and BCL2 (P < 0.05 and P < 0.01 [Fig. 8C]).
FIG 8.
MEF2C knockout reduces ESE targets ETS1 and BCL2 expression. (A) MEF2C, IRF4, EBNA2, SPI1, and H3K27ac ChIP-seq tracks at ETS1 ESE. (B) MEF2C, IRF4, EBNA2, SPI1, and H3K27ac ChIP-seq tracks at BCL2 ESE. (C) Expression level of ETS1 and BCL2 followed by MEF2C knockout in LCLs by qRT-PCR. Levels of control sgRNA treated cells were set to 1. **, P < 0.01; *, P < 0.05.
DISCUSSION
SEs control the expression of key oncogenes, such as the MYC gene, in many different cancer types (30, 39). SEs can be acquired through multiple mechanisms. Cancer-specific mutations can acquire new TF binding sites, leading to SE assembly at these sites (40). Enhancer amplification can also generate SEs in cancer (41). DNA tumor viruses can assemble viral SEs (33). SE-activated genes are important in providing signals for cell cycle progression, survival, or metastasis (42). SEs are more sensitive to perturbations than TEs (30). BRD4 inhibitor JQ1 selectively targets SE functions, with much less effect on TEs. CDK7 inhibitor THZ1 can also efficiently block SE function (43). A more comprehensive understanding of the proteins specifically enriched in SEs will allow for identification of druggable targets.
MYC is one of the most important oncogenic drivers of cancer development. In LCLs, MYC is under the control of two ESEs hundreds of base pairs upstream of the MYC TSS. These ESEs loop to the MYC TSS (13, 33, 34). Deletions of these ESEs greatly decrease MYC expression and halt LCL growth. EBV TFs and multiple host TFs bind to these ESEs. However, little is known about the identities of proteins assembled at these ESE. Traditionally, affinity purification followed by mass spectrometry can be used to identify DNA binding proteins in vitro. However, it is difficult to determine if the proteins identified bind to the DNA specifically. Secondary assays are needed to determine if these bindings occurred in vivo. In this study, we developed a novel strategy to identify proteomic components of MYC ESEs using genome-wide CRISPR screen. We demonstrated that TAF family proteins, including TAF8, TAF11, TAF3, and MEF2C, were essential for MYC 525ESE activity and LCL MYC expression.
In our previous studies, CRISPR deletion of 525ESE and 428ESE greatly decreased MYC expression level and ceased LCL growth (34). To further validate the MYC ESE deletion effect and narrow down regions in 525ESE essential for its activity, CRISPRi was used to silence 525ESE1. Three CRISPRi gRNAs also effectively reduced MYC expression, indicating that the other two sgRNA target regions were not required for MYC activation. Results further support the notion that MYC ESE could indeed regulate MYC expression.
EBNA2 is a major EBV latency III transcription activator. Previous studies using a conditional EBNA2 system illustrated that in the presence of functional EBNA2, the interaction between MYC promoter and its upstream 525SE and 428SE were significantly higher than in LCLs with much reduced EBNA2 levels (13, 44). These data indicated that EBNA2 played a role in MYC ESEs looping to MYC promoter. In line with these studies, our results indicated that EBNA2 could directly affect 525ESE1 activity and MYC ESEs had background activities in cells that do not express EBNA2.
TAF family proteins are integral members of the general transcription factor complex TFIID. TFIID recognizes the core promoter of many genes and nucleates the assembly of a transcription preinitiation complex containing RNA polymerase II and other initiation factors (45). A CRISPR/Cas9 screen identified several TAF family proteins, including TAF8, TAF11, and TAF3, that were essential for MYC 525ESE1 activation. It is surprising that these general transcription factors were not required for SV40 enhancer activity. This finding further supports the notion that SEs are more sensitive to perturbation than TEs, as they might be composed of different TFs.
Myocyte enhancer factor 2 (MEF2) family TFs regulate the survival, proliferation, and differentiation of various cell types (46). Although MEF2 proteins were first identified as essential regulators in muscle cell differentiation during development, they are now implicated to be functional in many other cell types. For example, MEF2D has been shown to have proapoptotic function through regulating Nur77 expression during thymocyte development (47). MEF2C has been reported to regulate expression of CCND2 and the prosurvival factor BCL2L1. MEF2C is also required for B-cell proliferation and survival after antigen receptor stimulation (48, 49). Transitional B cells with low MEF2C expression failed to respond to BCR stimulation (50). Our CRISPR screens identified MEF2C as one of the TFs that regulate MYC ESE activity. It is still not known how MEF2C regulates ESE activity. We found that MEF2C depletion also impaired IRF4, SPI, and EBNA2 binding to the 525ESE1 site, suggesting a MEF2C cooperative role in ESE assembly.
Many B-cell transcription factors, including BATF, IRF4, and RBPJ, are essential for LCL growth (38). These genes might also be important for ESE function, as ChIP-seq data indicated that these genes also bind to MYC ESEs. Further study is needed to determine their roles in ESE functions.
MATERIALS AND METHODS
Plasmids and antibodies.
pGreenFire-mCMV plasmid was purchased from System Biosciences (catalog number TR010PA-N). pGreenFire-SV40, pGreenFire-525ESE1, pGreenFire-525ESE2, pGreenFire-428ESE1, and pGreenFire-428ESE2 were cloned by individually inserting SV40 enhancer and different fragments of MYC super-enhancers amplified by PCR into pGreenFire-mCMV vector. 525ESE1 was from chr8:128222111-128223116, 525ESE2 was from chr8:128222404-128223199, 428ESE1 was from chr8:128312976-128313979, and 428ESE2 was from chr8:128315031-128315410. pGL3-promoter vector was purchased from Promega (catalog number E1761), and pGL3-525ESE1 was obtained by infusing 525ESE1 into pGL3-promoter.
sgRNAs used to silence MYC ESEs were designed by using an online CRISPR guide tool from https://benchling.com/. sgRNAs targeting TAF8, TAF11, and MEF2C were selected from the Brunello CRISPR library pool based on sequencing results after the screen. sgRNAs were annealed and cloned into pLentiGuide-Puro according to the Zhang Lab protocol (http://www.genome-engineering.org/crispr/wp-content/uploads/2014/05/CRISPR-Reagent-Description-Rev20140509.pdf). All sgRNA sequences are listed in Table 1.
TABLE 1.
Primers used in this study
| Primer name | Sequence (5′–3′) |
|---|---|
| 525ESE1isg1 REV | aaacTTGTGCTTACGTTGCTCTGAC |
| 525ESE1isg1 FWD | CACCGTCAGAGCAACGTAAGCACAA |
| 525ESE1isg2 REV | aaacCAGGGGTGATGATCTGCCAC |
| 525ESE1isg2 FWD | CACCGTGGCAGATCATCACCCCTG |
| 525ESE1isg3 FWD | CACCGCTGATCAGATTTCAGCAGCG |
| 525ESE1isg3 REV | aaacCGCTGCTGAAATCTGATCAGC |
| 525ESE1isg4 FWD | CACCGGTAGGTACCATCTCACATG |
| 525ESE1isg4 REV | aaacCATGTGAGATGGTACCTACC |
| 525ESE1isg5 FWD | CACCGACCGGACAACTAAGGCCCAA |
| 525ESE1isg5 REV | aaacTTGGGCCTTAGTTGTCCGGTC |
| MEF2csg2m-F | GTATGGCAATCTACGAAACTCACCAGGTC |
| MEF2csg2m-R | CCGTTCCCTGCACTGGTG |
| TAF8sg1-F | caccgTGCATCCCAGAGGCGCGATG |
| TAF8sg1-R | aaacCATCGCGCCTCTGGGATGCAc |
| TAF8sg2-F | caccgGTGGTCACACTTGTTGAGAT |
| TAF8sg2-R | aaacATCTCAACAAGTGTGACCACc |
| MEF2Csg1-F | caccgACTCCTACTTTACCAGGACA |
| MEF2Csg1-R | aaacTGTCCTGGTAAAGTAGGAGTc |
| MEF2Csg2-F | caccgAGCAGACCTGGTGAGTTTCG |
| MEF2Csg2-R | aaacCGAAACTCACCAGGTCTGCTc |
| TAF11sg1-F | caccgCCGACAAAGGTGGAGAGACA |
| TAF11sg2-F | caccgCGACACCGATGGAATCCCAG |
| TAF11sg1-R | aaacTGTCTCTCCACCTTTGTCGGc |
| TAF11sg2-R | aaacCTGGGATTCCATCGGTGTCGc |
| 525ESE1-F | aaaattttatcgatgGAAAGGAATAACCTGCACATGAC |
| 525ESE1-R | actagttctagaattTGCTTACGTTGCTCTGAGATT |
| 428ESE1-R | actagttctagaattCAAGTATGTAGGTAGCACTGTGT |
| 428ESE2-F | aaaattttatcgatgCCTGCCTTGCTCTCTCAAT |
| 428ESE1-F | aaaattttatcgatgGAAAGAAGGCCTTTGTTGTGAG |
| 525ESE2-R | actagttctagaattAAATATCTGGCTGCAAACGAAA |
| 525ESE2-F | aaaattttatcgatgTTCCTTCCCACAGATATCAGATT |
| 428ESE2-R | actagttctagaattGCACTGCTCAGACAGGATAG |
| qRTCHIP525ESE-F | GAAAGGAATAACCTGCACATGAC |
| qRTCHIP525ESE-R | GGACGCCCACATATCTCTTC |
| qRTb-actin-F | AGAGCTACGAGCTGCCTGAC |
| qRTb-actin-R | AGCACTGTGTTGGCGTACAG |
| SV40enhancer-F | aaaattttatcgatgGGTGTGGAAAGTCCCC |
| SV40enhancer-R | actagttctagaattTAGCTCAGAGGCAGAGGC |
| PGL3-525ESE1-F | CTAGCCCGGGCTCGAGAAAGGAATAACCTGCACATGAC |
| PGL3-525ESE1-R | GATCGCAGATCTCGATGCTTACGTTGCTCTGAGATT |
| qRTBCL2-F | GGTGGGGTCATGTGTGTGG |
| qRTBCL2-R | CGGTTCAGGTACTCAGTCATCC |
| qRTETS1-F | GATAGTTGTGATCGCCTCACC |
| qRTETS1-R | GTCCTCTGAGTCGAAGCTGTC |
| qRTMYC-F | GGCTCCTGGCAAAAGGTCA |
| qRTMYC-R | CTGCGTAGTTGTGCTGATGT |
| IRF4sg1-F | CACCGGCAGGACTACAACCGCGAGG |
| IRF4sg1-R | aaacCCTCGCGGTTGTAGTCCTGCc |
Antibody against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was purchased from Abcam (catalog number ab8245), and MEF2C antibody was from Cell Signaling (catalog number 5030). EBNA2 antibody PE2 (51) and EBNALP antibody JF186 (52) were prepared by Bio X Cell. Anti-V5 antibody was purchased from Abcam (catalog number ab9116), and IRF4 (catalog number sc-48338) and SPI1 (catalog number sc-17824) antibodies were from Santa Cruz. TAF8 (catalog number PA5-69854) and TAF11 (catalog number PA5-40488) antibodies were purchased from Thermo Fisher.
Cell lines.
GM12878 is an EBV-transformed B-lymphoblastoid cell line (LCL); P3HR1 is a Burkitt’s lymphoma cell line. LCLs expressing dCAS9-KRAB fusion proteins were generated by transducing dCAS9-KRAB-expressing lentivirus into GM12878 LCLs followed by FACS sorting for mCherry. The Burkitt’s lymphoma cell line MUTU III with EBV type III latency was a gift from Jeff Sample. BJAB is an EBV-negative B-lymphoma cell line. All the B cells were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum (HyClone), 100 U/ml of streptomycin, and 100 mg/ml of penicillin (Gibco). HEK293T cells purchased from the ATCC were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum (HyClone), 100 U/ml of streptomycin, and 100 mg/ml of penicillin. All the cells were maintained at 37°C in a 5% CO2 humidified chamber. Cell selections were done with puromycin at 3 μg/ml, and hygromycin at 200 μg/ml. Cells were routinely confirmed to be negative for mycoplasma contamination by the MycoAlert mycoplasma detection kit (Lonza).
CRISPR library plasmid amplification and lentivirus generation.
The human CRISPR knockout pooled plasmid library (Brunello) used for the CRISPR screen was purchased from Addgene (catalog number 73178). The pooled plasmid library was amplified as follow: 50 ng of DNA was added to 25 μl of STBL4 electrocompetent cells. After mixing, cells were transferred to 0.1-cm cuvettes (Bio-Rad). Cells were electroporated 4 times at 1.2 kV and 25 μF on an ECM 630 (BTX). After electroporation, 1 ml of prewarmed super optimal broth with catabolite repression (SOC) was added to each cuvette and cells were transferred to 14-ml tubes with continuous shaking at 30°C for 1 h. After shaking, cells were spread into 20 prewarmed 10-cm round petri dishes. Cells were incubated for another 18 h at 30°C. Colonies were harvested by adding 2 ml of LB medium to each dish followed by scraping with a cell spreader. Maxiprep was done to extract plasmids from cells. To produce lentiviruses, four 15-cm petri dishes of HEK293T cells were seeded at ∼40% confluence the day before transfection. Transfection was done using TransIT-LT1 transfection reagent (catalog number MIR2306; Mirus Bio LLC). For each dish, 10 μg of lentiCRISPR plasmid library, 3 μg of pVSVg, and 8 μg of psPAX2 (Addgene) were first diluted in 600 μl of Opti-MEM (Life Technologies). A total of 80 μl of Mirus reagent was diluted in 320 μl of Opti-MEM, and after incubation at room temperature for 5 min, it was added to the DNA mixture and allow to incubate for another 20 min before being added to the cell culture. After 16 h, medium was replaced with 30 ml of RPMI 1640 medium supplemented with 30% fetal bovine serum (FBS) (Gibco). Lentiviruses were harvested twice, at 24 h and 48 h after medium change.
CRISPR/CAS9 loss-of-function screens.
Before genome-wide CRISPR screen, LCLs stably expressing CAS9 were transduced with lentiviruses expressing pGreenFire-SV40 or pGreenFire-525ESE1. Three days posttransduction, FACS was used to collect cells expressing GFP at high levels. Sorted cells were expanded in RPMI 1640 medium supplemented with 10% fetal bovine serum and 1 mg/ml of G418. A total of 150 million cells in each group were infected with CRISPR lentivirus library at a multiplicity of infection (MOI) of approximately 0.3. Infection was done in 12-well plates with spin infection at 300 × g for 2 h in the presence of 4 μg/ml of Polybrene. Plates were then returned to the incubator for another 6 h. After that, cells were harvested and transferred to seven T175 flasks at a concentration of 0.2 million/ml. Forty-eight hours later, cells were selected by puromycin (3 μg/ml) for another 3 days to eliminate uninfected cells. A total of 40 million cells were passaged every 72 h for a total of 5 days. Input was prepared 3 days after puromycin selection from 40 million cells. At day 5, 40 million cells were sorted by FACS for GFP. Three percent of cells losing GFP signal on the left side of FACS profiling were collected. Collected cells were used to extract genomic DNA using the blood and cell culture DNA maxikit (Qiagen) according to the manufacturer’s instructions. PCR was used to amplify sgRNAs for sequencing. Multiple 100-μl reaction volumes were used to amplify the library. Each reaction mixture consisted of 10 μg of genomic DNA, 10 μl of 10× reaction buffer, 8 μl of deoxynucleoside triphosphate (dNTP), 0.5 μl of 100 μM P5 primer mix, 1.5 μl of Ex Taq polymerase, 10 μl of 5 μM P7 primer, and distilled water (dH2O) to a total volume of 100 μl. PCR cycling conditions was set at an initial 1 min at 95°C followed by 30 s at 94°C, 30 s at 53°C, and 30 s at 72°C for 28 cycles and a final 10 min extension at 72°C. Amplified sgRNAs were purified with AMPure XP SPRI beads according to the manufacturer’s instructions (Beckman Coulter).
CRISPR/CAS9 screen data analysis.
After adapter trimming, multiple lanes of sequencing reads were pooled for each biological replicate of screen conditions. Trimmed sequencing reads were exactly mapped to the sgRNA sequence library (Brunello) to count sgRNA frequencies in each library. Significant genes exhibiting differential essentiality between 525 and SV40 conditions were tested using MAGeCK (37) software under default parameters with the sgRNA count table as input. MAGeCK tests positive and negative enriched genes separately within the same comparison. As a result, we merged separately tested P values into one volcano plot, shown in Fig. 4B.
Electroporation and reporter assays.
Electroporation was performed when cells were in log-phase growth. In brief, five million cells were electroporated with 2 μg of reporter plasmids, 0.2 μg of Renilla luciferase plasmid, and various expression plasmids using 4D-Nucleofector (Lonza) under program CM113. In all cases, the total amount of transfected DNA was held constant by adding empty vector. Cells were harvested 24 h after transfection, and luciferase activity was tested by using a dual-luciferase assay kit (Promega). Western blotting was performed to verify protein expression.
ChIP-qPCR.
Ten million cells were fixed with 1% formaldehyde. The cells were then lysed and lysates were sonicated with Bioruptor (Diagenode) with 30 s on, 30 s off for 45 cycles. Sonicated chromatin was diluted with ChIP dilution buffer and incubated with antibodies of interest or control antibodies. Protein-DNA complexes were precipitated with protein A beads. After precipitation, beads were washed extensively and eluted protein-DNA complexes were reverse cross-linked with NaCl. DNA was purified by using QIAquick spin columns (Qiagen). qPCR was used to quantify the DNA from the ChIP assay and normalize it to the percentage of input DNA. Primers used are listed in Table 1.
qRT-PCR.
Total mRNAs were extracted using a PureLink RNA minikit (Life Technologies). Two hundred nanograms of mRNA was used as the template for reverse transcription with iScript Reverse Transcription Supermix (Bio-Rad). cDNAs were then amplified on a CFX96 Touch real-time PCR detection system (Bio-Rad), and SYBR green (Thermo Fisher) was used to detect cDNA amplification. GAPDH was used to normalize gene expression. RNA relative expression was calculated using the threshold cycle (2−ΔΔCT) method. The value for the cells transduced with nontargeting sgRNA was set to 1.
Immunoblotting.
Cells were harvested and washed once with phosphate-buffered saline (PBS) before resuspension with PBS containing protease inhibitor (Sigma; catalog number 11836170001). SDS (3×) sample loading buffer was added to the cells and sonicated for 15 s. Lysates were boiled for 5 min. Whole-cell extracts were resolved by SDS-PAGE and proteins were transferred to nitrocellulose membranes. The membranes were blocked with 5% nonfat dried milk dissolved in Tris-buffered saline with Tween 20 (TBST) for 30 min. Primary antibodies targeting proteins of interest were incubated with membranes overnight at 4°C. Blots were then probed with horseradish peroxidase-coupled secondary antibodies (Cell Signaling Technology), visualized by enhanced chemiluminescence (Northern Lightning; Perkin Elmer), and imaged on a Carestream workstation.
cDNA rescue.
MEF2C cDNA in pDONR221 was purchased from DNASU (catalog number HsCD00079840). Site mutation was used to generate a silent point mutation in the protospacer-adjacent motif to abrogate CAS9 cutting. Mutated MEF2C was then cloned into PLX-TRC313 vector through Gateway cloning. CAS9-expressing LCLs containing constitutively active 525ESE1 or SV40 enhancer were used for MEF2C cDNA rescue. Cells were transduced with lentiviruses expressing MEF2C rescue cDNA and selected with hygromycin. cDNA expression was confirmed by immunoblotting. LCLs with stable control and rescue cDNA expression were then used in CRISPR experiments, as indicated. Primers used for MEF2C cDNA rescue are listed in Table 1.
ChIP-seq and ChIA-PET data sets.
The data sets used in the analyses are available through the following links (all for LCLs): MEF2C, https://de.cyverse.org/anon-files/iplant/home/mxteng/ENCFF845FPS.bigWig; IRF4, https://de.cyverse.org/anon-files/iplant/home/mxteng/ENCFF291ILI.bigWig; EBNA2, https://de.cyverse.org/anon-files/iplant/home/mxteng/1.bw; SPI1, https://de.cyverse.org/anon-files/iplant/home/mxteng/ENCFF793RKX.bigWig; H3K27ac, https://de.cyverse.org/anon-files/iplant/home/mxteng/ENCFF180LKW.bigWig; and RNAPII ChiA-PET, https://de.cyverse.org/anon-files/iplant/home/mxteng/GM12878_RNAPII_interaction.txt.gz.
Statistical analysis.
All experiments were done in biological triplicate. Paired Student’s t test was used to determine the statistical significance. In figures, P values are indicated as follows: *, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001.
Supplementary Material
ACKNOWLEDGMENTS
We thank Elliott Kieff for insightful discussions.
This work was funded by NIAID grant AI123420, NCI grant CA047006 (B.Z.), and NCI grant P30CA076292 (M.T.). B.E.G. is supported by an American Cancer Society Research Scholar grant, by a Burroughs Wellcome Career Award in Medical Sciences, and by NIAID grant AI137337.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00513-19.
REFERENCES
- 1.Epstein MA, Achong BG, Barr YM. 1964. Virus particles in cultured lymphoblasts from Burkitt’s lymphoma. Lancet i:702–703. doi: 10.1016/S0140-6736(64)91524-7. [DOI] [PubMed] [Google Scholar]
- 2.Cohen JI, Fauci AS, Varmus H, Nabel GJ. 2011. Epstein-Barr virus: an important vaccine target for cancer prevention. Sci Transl Med 3:107fs107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Longnecker RK, Kieff E, Cohen JI. 2013. Chapter 61, Epstein-Barr virus In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed (electronic), vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 4.Cohen JI, Picchio GR, Mosier DE. 1992. Epstein-Barr virus nuclear protein 2 is a critical determinant for tumor growth in SCID mice and for transformation in vitro. J Virol 66:7555–7559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammerschmidt W, Sugden B. 1989. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature 340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 6.Kaye KM, Izumi KM, Kieff E. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci U S A 90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mannick JB, Cohen JI, Birkenbach M, Marchini A, Kieff E. 1991. The Epstein-Barr virus nuclear protein encoded by the leader of the EBNA RNAs is important in B-lymphocyte transformation. J Virol 65:6826–6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomkinson B, Robertson E, Kieff E. 1993. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol 67:2014–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szymula A, Palermo RD, Bayoumy A, Groves IJ, Ba Abdullah M, Holder B, White RE. 2018. Epstein-Barr virus nuclear antigen EBNA-LP is essential for transforming naive B cells, and facilitates recruitment of transcription factors to the viral genome. PLoS Pathog 14:e1006890. doi: 10.1371/journal.ppat.1006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maruo S, Johannsen E, Illanes D, Cooper A, Kieff E. 2003. Epstein-Barr virus nuclear protein EBNA3A is critical for maintaining lymphoblastoid cell line growth. J Virol 77:10437–10447. doi: 10.1128/jvi.77.19.10437-10447.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maruo S, Wu Y, Ishikawa S, Kanda T, Iwakiri D, Takada K. 2006. Epstein-Barr virus nuclear protein EBNA3C is required for cell cycle progression and growth maintenance of lymphoblastoid cells. Proc Natl Acad Sci U S A 103:19500–19505. doi: 10.1073/pnas.0604919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alfieri C, Birkenbach M, Kieff E. 1991. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology 181:595–608. doi: 10.1016/0042-6822(91)90893-G. [DOI] [PubMed] [Google Scholar]
- 13.Zhao B, Zou J, Wang H, Johannsen E, Peng CW, Quackenbush J, Mar JC, Morton CC, Freedman ML, Blacklow SC, Aster JC, Bernstein BE, Kieff E. 2011. Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth. Proc Natl Acad Sci U S A 108:14902–14907. doi: 10.1073/pnas.1108892108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaiser C, Laux G, Eick D, Jochner N, Bornkamm GW, Kempkes B. 1999. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J Virol 73:4481–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grossman SR, Johannsen E, Tong X, Yalamanchili R, Kieff E. 1994. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc Natl Acad Sci U S A 91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harada S, Kieff E. 1997. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J Virol 71:6611–6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Portal D, Zhao B, Calderwood MA, Sommermann T, Johannsen E, Kieff E. 2011. EBV nuclear antigen EBNALP dismisses transcription repressors NCoR and RBPJ from enhancers and EBNA2 increases NCoR-deficient RBPJ DNA binding. Proc Natl Acad Sci U S A 108:7808–7813. doi: 10.1073/pnas.1104991108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Portal D, Zhou H, Zhao B, Kharchenko PV, Lowry E, Wong L, Quackenbush J, Holloway D, Jiang S, Lu Y, Kieff E. 2013. Epstein-Barr virus nuclear antigen leader protein localizes to promoters and enhancers with cell transcription factors and EBNA2. Proc Natl Acad Sci U S A 110:18537–18542. doi: 10.1073/pnas.1317608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang C, Zhou H, Xue Y, Liang J, Narita Y, Gerdt C, Zheng AY, Jiang R, Trudeau S, Peng CW, Gewurz BE, Zhao B. 2018. Epstein-Barr virus nuclear antigen leader protein coactivates EP300. J Virol 92:e02155-17. doi: 10.1128/JVI.02155-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maruo S, Zhao B, Johannsen E, Kieff E, Zou J, Takada K. 2011. Epstein-Barr virus nuclear antigens 3C and 3A maintain lymphoblastoid cell growth by repressing p16INK4A and p14ARF expression. Proc Natl Acad Sci U S A 108:1919–1924. doi: 10.1073/pnas.1019599108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skalska L, White RE, Franz M, Ruhmann M, Allday MJ. 2010. Epigenetic repression of p16(INK4A) by latent Epstein-Barr virus requires the interaction of EBNA3A and EBNA3C with CtBP. PLoS Pathog 6:e1000951. doi: 10.1371/journal.ppat.1000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skalska L, White RE, Parker GA, Turro E, Sinclair AJ, Paschos K, Allday MJ. 2013. Induction of p16(INK4a) is the major barrier to proliferation when Epstein-Barr virus (EBV) transforms primary B cells into lymphoblastoid cell lines. PLoS Pathog 9:e1003187. doi: 10.1371/journal.ppat.1003187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cahir-McFarland ED, Carter K, Rosenwald A, Giltnane JM, Henrickson SE, Staudt LM, Kieff E. 2004. Role of NF-kappa B in cell survival and transcription of latent membrane protein 1-expressing or Epstein-Barr virus latency III-infected cells. J Virol 78:4108–4119. doi: 10.1128/jvi.78.8.4108-4119.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang B, Kracker S, Yasuda T, Casola S, Vanneman M, Homig-Holzel C, Wang Z, Derudder E, Li S, Chakraborty T, Cotter SE, Koyama S, Currie T, Freeman GJ, Kutok JL, Rodig SJ, Dranoff G, Rajewsky K. 2012. Immune surveillance and therapy of lymphomas driven by Epstein-Barr virus protein LMP1 in a mouse model. Cell 148:739–751. doi: 10.1016/j.cell.2011.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minamitani T, Ma Y, Zhou H, Kida H, Tsai CY, Obana M, Okuzaki D, Fujio Y, Kumanogoh A, Zhao B, Kikutani H, Kieff E, Gewurz BE, Yasui T. 2017. Mouse model of Epstein-Barr virus LMP1- and LMP2A-driven germinal center B-cell lymphoproliferative disease. Proc Natl Acad Sci U S A 114:4751–4756. doi: 10.1073/pnas.1701836114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polack A, Hortnagel K, Pajic A, Christoph B, Baier B, Falk M, Mautner J, Geltinger C, Bornkamm GW, Kempkes B. 1996. c-myc activation renders proliferation of Epstein-Barr virus (EBV)-transformed cells independent of EBV nuclear antigen 2 and latent membrane protein 1. Proc Natl Acad Sci U S A 93:10411–10416. doi: 10.1073/pnas.93.19.10411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takano Y, Saegusa M, Ikenaga M, Okayasu I. 1997. Apoptosis and proliferative activity of non-Hodgkin’s lymphomas: comparison with expression of bcl-2, p53 and c-myc proteins. Pathol Int 47:90–94. doi: 10.1111/j.1440-1827.1997.tb03726.x. [DOI] [PubMed] [Google Scholar]
- 28.Korkolopoulou P, Patsouris E, Pangalis G, Tsenga A, Elemenoglou J, Thomas-Tsangli E, Spandidos D, Kittas C. 1993. A comparative assessment of proliferating cell nuclear antigen, c-myc p62, and nucleolar organizer region staining in non-Hodgkin’s lymphomas: a histochemical and immunohistochemical study of 200 cases. Hum Pathol 24:371–377. doi: 10.1016/0046-8177(93)90084-T. [DOI] [PubMed] [Google Scholar]
- 29.Gomez-Curet I, Perkins RS, Bennett R, Feidler KL, Dunn SP, Krueger LJ. 2006. c-Myc inhibition negatively impacts lymphoma growth. J Pediatr Surg 41:207–211. doi: 10.1016/j.jpedsurg.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 30.Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, Rahl PB, Sun HH, Yeda KT, Doench JG, Reichert E, Kung AL, Rodig SJ, Young RA, Shipp MA, Bradner JE. 2013. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell 24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou H, Schmidt SC, Jiang S, Willox B, Bernhardt K, Liang J, Johannsen EC, Kharchenko P, Gewurz BE, Kieff E, Zhao B. 2015. Epstein-Barr virus oncoprotein super-enhancers control B cell growth. Cell Host Microbe 17:205–216. doi: 10.1016/j.chom.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang S, Zhou H, Liang J, Gerdt C, Wang C, Ke L, Schmidt SCS, Narita Y, Ma Y, Wang S, Colson T, Gewurz B, Li G, Kieff E, Zhao B. 2017. The Epstein-Barr virus regulome in lymphoblastoid cells. Cell Host Microbe 22:561–573.e564. doi: 10.1016/j.chom.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang J, Zhou H, Gerdt C, Tan M, Colson T, Kaye KM, Kieff E, Zhao B. 2016. Epstein-Barr virus super-enhancer eRNAs are essential for MYC oncogene expression and lymphoblast proliferation. Proc Natl Acad Sci U S A 113:14121–14126. doi: 10.1073/pnas.1616697113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, Virgin HW, Listgarten J, Root DE. 2016. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, Liu XS. 2014. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma Y, Walsh MJ, Bernhardt K, Ashbaugh CW, Trudeau SJ, Ashbaugh IY, Jiang S, Jiang C, Zhao B, Root DE, Doench JG, Gewurz BE. 2017. CRISPR/Cas9 screens reveal Epstein-Barr virus-transformed B cell host dependency factors. Cell Host Microbe 21:580–591.e587. doi: 10.1016/j.chom.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ke L, Zhou H, Wang C, Xiong G, Xiang Y, Ling Y, Khabir A, Tsao GS, Zeng Y, Zeng M, Busson P, Kieff E, Guo X, Zhao B. 2017. Nasopharyngeal carcinoma super-enhancer-driven ETV6 correlates with prognosis. Proc Natl Acad Sci U S A 114:9683–9688. doi: 10.1073/pnas.1705236114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, Etchin J, Lawton L, Sallan SE, Silverman LB, Loh ML, Hunger SP, Sanda T, Young RA, Look AT. 2014. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 346:1373–1377. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Choi PS, Francis JM, Imielinski M, Watanabe H, Cherniack AD, Meyerson M. 2016. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genet 48:176–182. doi: 10.1038/ng.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hnisz D, Schuijers J, Lin CY, Weintraub AS, Abraham BJ, Lee TI, Bradner JE, Young RA. 2015. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol Cell 58:362–370. doi: 10.1016/j.molcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Zhang T, Kwiatkowski N, Abraham BJ, Lee TI, Xie S, Yuzugullu H, Von T, Li H, Lin Z, Stover DG, Lim E, Wang ZC, Iglehart JD, Young RA, Gray NS, Zhao JJ. 2015. CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell 163:174–186. doi: 10.1016/j.cell.2015.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wood CD, Veenstra H, Khasnis S, Gunnell A, Webb HM, Shannon-Lowe C, Andrews S, Osborne CS, West MJ. 2016. MYC activation and BCL2L11 silencing by a tumour virus through the large-scale reconfiguration of enhancer-promoter hubs. Elife 5:e18270. doi: 10.7554/eLife.18270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bieniossek C, Papai G, Schaffitzel C, Garzoni F, Chaillet M, Scheer E, Papadopoulos P, Tora L, Schultz P, Berger I. 2013. The architecture of human general transcription factor TFIID core complex. Nature 493:699–702. doi: 10.1038/nature11791. [DOI] [PubMed] [Google Scholar]
- 46.McKinsey TA, Zhang CL, Olson EN. 2002. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem Sci 27:40–47. doi: 10.1016/S0968-0004(01)02031-X. [DOI] [PubMed] [Google Scholar]
- 47.Youn HD, Liu JO. 2000. Cabin1 represses MEF2-dependent Nur77 expression and T cell apoptosis by controlling association of histone deacetylases and acetylases with MEF2. Immunity 13:85–94. doi: 10.1016/S1074-7613(00)00010-8. [DOI] [PubMed] [Google Scholar]
- 48.Khiem D, Cyster JG, Schwarz JJ, Black BL. 2008. A p38 MAPK-MEF2C pathway regulates B-cell proliferation. Proc Natl Acad Sci U S A 105:17067–17072. doi: 10.1073/pnas.0804868105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilker PR, Kohyama M, Sandau MM, Albring JC, Nakagawa O, Schwarz JJ, Murphy KM. 2008. Transcription factor Mef2c is required for B cell proliferation and survival after antigen receptor stimulation. Nat Immunol 9:603–612. doi: 10.1038/ni.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andrews SF, Dai X, Ryu BY, Gulick T, Ramachandran B, Rawlings DJ. 2012. Developmentally regulated expression of MEF2C limits the response to BCR engagement in transitional B cells. Eur J Immunol 42:1327–1336. doi: 10.1002/eji.201142226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young L, Alfieri C, Hennessy K, Evans H, O'Hara C, Anderson KC, Ritz J, Shapiro RS, Rickinson A, Kieff E, Cohen JI. 1989. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N Engl J Med 321:1080–1085. doi: 10.1056/NEJM198910193211604. [DOI] [PubMed] [Google Scholar]
- 52.Finke J, Rowe M, Kallin B, Ernberg I, Rosen A, Dillner J, Klein G. 1987. Monoclonal and polyclonal antibodies against Epstein-Barr virus nuclear antigen 5 (EBNA-5) detect multiple protein species in Burkitt's lymphoma and lymphoblastoid cell lines. J Virol 61:3870–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.