To the best of our knowledge, this is the first study that compares intra-individual evolution rates for FIV, SIV, and HIV following systematic review of the literature. Our findings have important implications for informing research strategies in the field of intra-individual virus dynamics for lentiviruses. We observed that FIV evolves more slowly than HIV and SIV at the intra-individual level and found that mutation rates may differ by gene sequence length but not by host, gene, strain, an experimental setting relative to a natural setting, or spillover host infection relative to primary host infection.
KEYWORDS: evolution rate, feline immunodeficiency virus, human immunodeficiency virus, lentivirus, simian immunodeficiency virus
ABSTRACT
Lentiviral replication mediated by reverse transcriptase is considered to be highly error prone, leading to a high intra-individual evolution rate that promotes evasion of neutralization and persistent infection. Understanding lentiviral intra-individual evolutionary dynamics on a comparative basis can therefore inform research strategies to aid in studies of pathogenesis, vaccine design, and therapeutic intervention. We conducted a systematic review of intra-individual evolution rates for three species groups of lentiviruses—feline immunodeficiency virus (FIV), simian immunodeficiency virus (SIV), and human immunodeficiency virus (HIV). Overall, intra-individual rate estimates differed by virus but not by host, gene, or viral strain. Lentiviral infections in spillover (nonadapted) hosts approximated infections in primary (adapted) hosts. Our review consistently documents that FIV evolution rates within individuals are significantly lower than the rates recorded for HIV and SIV. FIV intra-individual evolution rates were noted to be equivalent to FIV interindividual rates. These findings document inherent differences in the evolution of FIV relative to that of primate lentiviruses, which may signal intrinsic difference of reverse transcriptase between these viral species or different host-viral interactions. Analysis of lentiviral evolutionary selection pressures at the individual versus population level is valuable for understanding transmission dynamics and the emergence of virulent and avirulent strains and provides novel insight for approaches to interrupt lentiviral infections.
IMPORTANCE To the best of our knowledge, this is the first study that compares intra-individual evolution rates for FIV, SIV, and HIV following systematic review of the literature. Our findings have important implications for informing research strategies in the field of intra-individual virus dynamics for lentiviruses. We observed that FIV evolves more slowly than HIV and SIV at the intra-individual level and found that mutation rates may differ by gene sequence length but not by host, gene, strain, an experimental setting relative to a natural setting, or spillover host infection relative to primary host infection.
INTRODUCTION
A characteristic feature of lentiviruses is a high rate of molecular evolution relative to those of other viruses and parasites (1). Low fidelity of lentiviral reverse transcriptase (RT) during DNA synthesis leads to the accumulation of mutations, resulting in intra-host viral variants, often termed quasispecies (2–4). The accumulation of numerous mutations, especially in antigens targeted by neutralizing antibodies or therapeutic agents, is thought to provide a mechanism for immune escape and enhance the risk of disease progression (5). The generation and transmission of variant quasispecies further leads to high population-level (between-host) variation in viral sequences, which provides a challenge for vaccine design and the development of therapeutic agents (6).
A considerable body of research documents evolution rates of human immunodeficiency virus (HIV) and its analogues, simian immunodeficiency virus (SIV) and feline immunodeficiency virus (FIV). Shared similarities in lymphoid tropism, disease chronicity, and immunodeficiency outcomes support the use of SIV and FIV as evolutionary models for HIV/AIDS studies (7–10). It is accepted that HIV type 1 (HIV-1) arose in humans following contacts with nonhuman primates and epidemic HIV-1 resulted after human adaptation (11). HIV evolutionary analyses focus on naturally occurring infections, for obvious reasons, whereas SIV and FIV infections have been subject to naturally occurring (FIV) and experimental (SIV and FIV) evolution rate analyses. SIVs occur naturally in African primates but have been adapted for infection of Asian macaques both incidentally and purposefully (12). At least 27 of the 35 species of felids evaluated have been identified as seroreactive against FIV antibodies, and the FIVs studied characteristically demonstrate species specificity, though naturally occurring and experimental cross-species transmissions have been documented (12–16).
The majority of evolution rate analyses for lentiviruses focus on variation at the population level, rather than the individual level (6). In studies of population level evolution rates, one quasispecies sequence from an individual is identified for comparison to individual sequences from other individuals. In contrast, within single individuals, evolutionary estimates compare quasispecies sequences identified over serial collections. Studies of intra-individual evolution rates offer important insights into the viral and host-dependent selection pressures on replicating viral quasispecies (6, 17–19). Comparative analysis between host species can provide information on host restriction mechanisms that alter lentiviral genomics and pathogenicity during host switching and may reveal important biological differences between strains. Improved understanding of these key factors has the potential to inform strategies for estimating the risk of emerging lentiviral infections, as well as to aid in the assessment of viral pathogenesis and vaccine development (6).
The aim of this study was thus to describe patterns in intra-individual evolution rates of lentiviruses. We aimed to (i) systematically evaluate the intra-individual evolution rates among FIV, SIV, and HIV from the literature, (ii) compare evolution rates between strains, hosts, and viral genes within each lentiviral species, (iii) compare intra-individual evolution rates in experimental and natural settings, (iv) compare intra-individual evolution rates in spillover (nonadapted) versus primary (adapted) hosts, and (v) evaluate how gene sequence length influences intra-individual evolution rate estimates.
RESULTS
Intra-individual evolution rates are lower in FIV than in SIV and HIV.
We found significant differences among virus intra-individual evolution rates: the median values were 0.00129 substitutions/site/year for FIV, 0.0075 substitutions/site/year for HIV, and 0.0054 substitutions/site/year for SIV (F2, 382 = 21.04, P < 0.001) (Fig. 1). FIV has a significantly lower rate of evolution than HIV and SIV, but we did not detect a significant difference between the rates of evolution of HIV and SIV.
FIG 1.
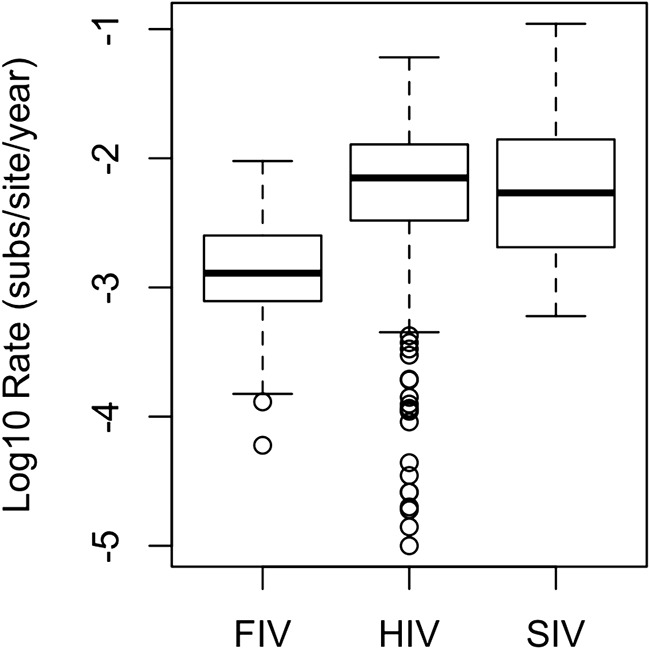
FIV has a significantly lower intra-individual evolution rate than primate lentiviruses. Log10 intra-individual evolution rates for FIV (n = 96), HIV (n = 218), and SIV (n = 71) are shown. Boxes represent the interquartile ranges of the data, and bars represent the median values. T bars represent the main bodies of data, and open circles represent outlying data points.
Intra-individual rates did not differ by gene, strain, or host within each of these retroviral species.
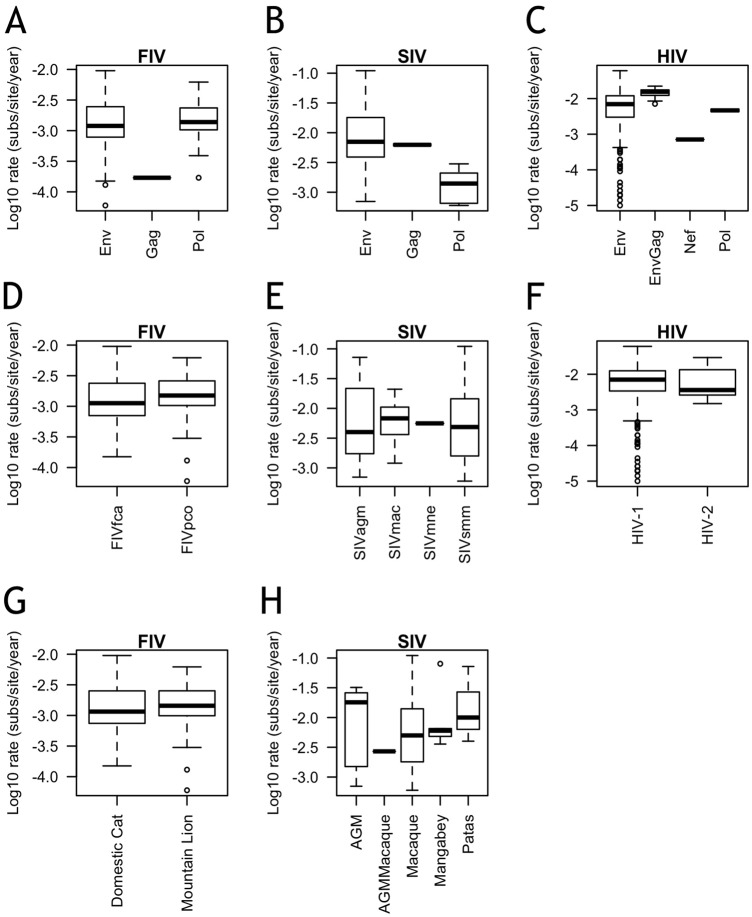
Evolution rates may vary between lentiviral genes (e.g., env and pol) (20–22), and intra-individual evolution rate estimates may also vary based on the length of gene sequenced (21). We found no difference in the evolution rates among FIV env (0.0012 substitutions/site/year), gag (0.00017 substitutions/site/year), or pol (0.0014 substitutions/site/year) (F3,92 = 0.709, P = 0.549) (Fig. 2A). In contrast, for SIV, there was a trend for env (0.0071 substitutions/site/year) and gag (0.0063 substitutions/site/year) to have higher intra-individual evolution rates than pol (0.0014 substitutions/site/year) (F2, 68 = 2.998, P = 0.057) (Fig. 2B). Similar to the findings for FIV, intra-individual evolution rates of HIV by viral gene did not differ between env (0.0070 substitutions/site/year), nef (0.0007 substitutions/site/year), pol (0.0047 substitutions/site/year), and, in one study that reported rates of env and gag together, envgag (0.0016 substitutions/site/year) (F2, 214 = 1.539, P = 0.205) (Fig. 2C).
FIG 2.
Few differences are noted in intra-individual evolution rates within a single host. Shown are log10 intra-individual evolution rates by gene (A, B, C), strain (D, E, F), and host species (G, H) for FIV (A, D, G), SIV (B, E, H), and HIV (C, F). Sample sizes for numbers of independent viral infections analyzed: for FIV, env, n = 72, gag, n = 1, and pol, n = 21; for SIV, env, n = 54, gag, n = 2, and pol, n = 15; and for HIV, env, n = 205, envgag, n = 10, nef, n = 1, and pol, n = 2. Sample sizes for individual infections with each strain: FIVfca, n = 54, FIVpco, n = 42, SIVagm, n = 11, SIVmac, n = 15, SIVmne, n = 1, SIVsmm, n = 44, HIV-1, n = 205, and HIV-2, n = 13. Sample sizes for host species: domestic cats, n = 56, mountain lions, n = 40, African green monkeys (AGM), n = 5, AGMMacaque, n = 1, macaques, n = 57, mangabeys, n = 5, and patas monkeys, n = 3. Boxes represent the interquartile ranges of the data, and bars represent the median values. T bars represent the main bodies of data, and open circles represent outlying data points.
There was no difference in intra-individual evolution rates between FIV strains FIVpco (0.0015 substitutions/site/year) and FIVfca (0.0011 substitutions/site/year) (F1, 94 = 0.065, P = 0.799) (Fig. 2D). There was also no difference among the intra-individual evolution rates of SIV strains SIVagm (0.0040 substitutions/site/year), SIVmac (0.0068 substitutions/site/year), SIVmne (0.0056 substitutions/site/year), and SIVsmm (0.0049 substitutions/site/year) (F3, 67 = 0.376, P = 0.771) (Fig. 2E). Furthermore, HIV-1 (0.0071 substitutions/site/year) and HIV-2 (0.0036 substitutions/site/year) did not differ from one another (F1, 216 = 0.077, P = 0.782) (Fig. 2F).
At the host species level, there was no difference between the intra-individual evolution rates of FIV in domestic cats (0.0012 substitutions/site/year) and in mountain lions (0.0014 substitutions/site/year) (F1, 94 = 0.001, P = 0.970) (Fig. 2G). Similarly, there was no difference among the evolution rates of SIV among African green monkeys (AGM; 0.0180 substitutions/site/year), macaques (0.0050 substitutions/site/year), sooty mangabeys (0.0060 substitutions/site/year), patas monkeys (0.0100 substitutions/site/year), and, in one study that reported averages of AGM and macaques, AGM/macaques (0.0027 substitutions/site/year), (F4, 66 = 1.347, P = 0.262) (Fig. 2H).
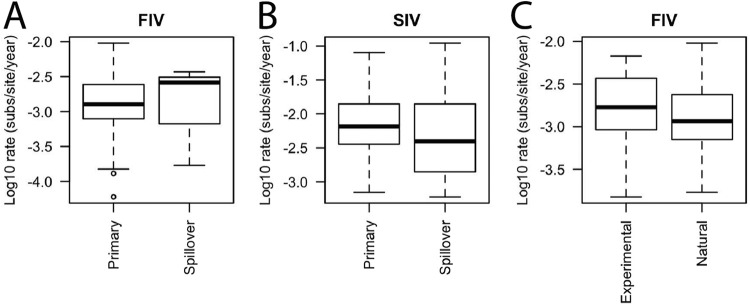
Intra-individual evolution rate does not differ in experimental versus natural infection or in primary or spillover hosts.
There were no significant differences noted in the intra-individual evolution rates of experimental FIV infections (0.0010 substitutions/site/year) compared to those of naturally acquired FIV infections (0.0013 substitutions/site/year) in domestic cats (F1, 54 = 0.683, P = 0.412) (Fig. 3C), nor were differences detected in the intra-individual evolution rates of FIV in primary hosts (0.0013 substitutions/site/year) and spillover host species (0.0026 substitutions/site/year) (F1, 94 = 0.056, P = 0.813; Fig. 3A). The SIV evolution rates in primary hosts (0.0065 substitutions/site/year) were also similar to those in spillover host species (0.0040 substitutions/site/year) (F1, 69 = 0.089, P = 0.766) (Fig. 3B).
FIG 3.
Evolution rates do not vary by primary or spillover host or in experimental versus natural FIV infection. Shown are log10 intra-individual evolution rates in primary hosts versus spillover hosts for SIV and FIV (A, B) and in experimental settings relative to natural settings for FIV in domestic cats (C). Sample sizes for primary hosts: FIV, n = 93, and SIV, n = 25. Numbers of spillover host infections: FIV, n = 3, and SIV, n = 46. Sample sizes for experimentally induced FIV infections versus naturally acquired FIV infections: n = 6 versus n = 90. Boxes represent the interquartile ranges of the data, and bars represent the median values. T bars represent the main bodies of data, and open circles represent outlying data points.
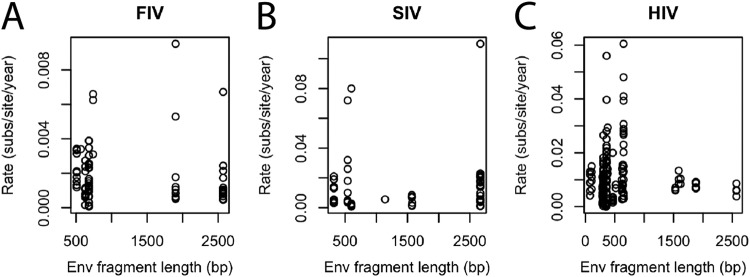
Gene sequence length influences intra-individual evolution rate estimates for HIV but not for FIV and SIV.
There was considerable variation among studies in the virus sequence lengths used to calculate intra-individual evolution rates (Fig. 4). For FIV env, the length of gene used to calculate intra-individual evolution rates did not predict the consistency of variance of rate estimates (F7, 64 = 0.541, P = 0.800) (Fig. 4A). The same was true for gene length and variance in evolution rate estimates for SIV env (F8, 45 = 0.476, P = 0.867) (Fig. 4B). However, for HIV, there was heterogeneity in the estimates of evolution rates for env (F18, 186 = 3.009, P < 0.001) (Fig. 4C), as intra-individual evolution rate estimates were less heterogeneous at short and long sequence lengths.
FIG 4.
Gene sequence length influences intra-individual evolution rate estimates of HIV but not of FIV or SIV. Relationships of gene lengths of env sequenced for FIV (A), SIV (B), and HIV (C) to intra-individual evolution rate estimates are shown.
DISCUSSION
Our systematic analysis of lentiviral intra-individual evolution documented several important findings. (i) FIV intra-individual evolution is approximately a log slower than the intra-individual evolution of SIV and HIV; (ii) within each lentiviral type, intra-individual evolution rates did not differ significantly by viral gene, host species, or viral strain; (iii) evolution rates were similar in experimental versus natural infections and in primary versus spillover host infections; and (iv) gene sequence length influenced heterogeneity in evolution rates in HIV but not in FIV and SIV. Evolution rates in this study include estimates using different phylogenetic models with various degrees of strictness with regard to the molecular clock parameters. Additionally, rates were either reported from specific individuals or averages of a cohort, depending on the study, and some sample size analyses were relatively small. All of these are limitations of this analysis. However, our findings representing a large synthesis of data collected over several decades by different laboratories ultimately resulted in remarkably consistent and unanticipated results.
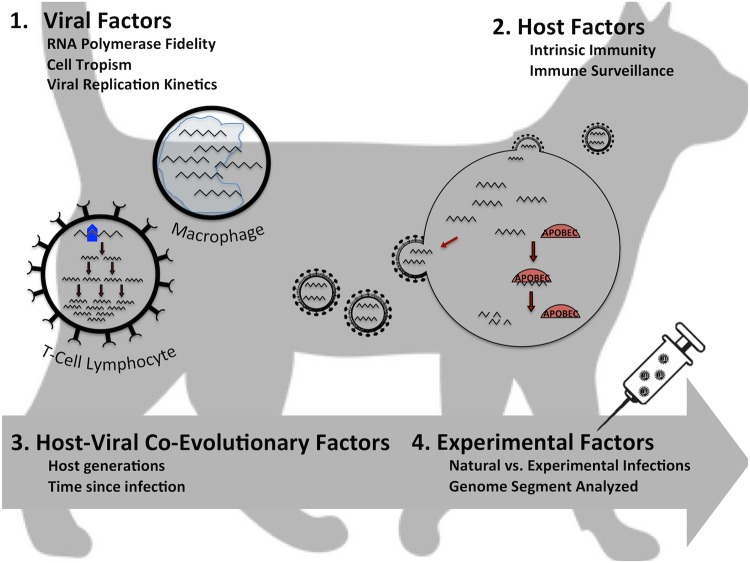
Differences in the ecology and immunology of host species (e.g., cat versus primate versus human) may be important drivers of intra-individual evolution rates (21, 23). Dual infection or coinfection status may accelerate mutation rates versus those in single infections by altering the immune landscape during infection (18). Viral replication kinetics and cell tropism also likely play a role in within-host evolution. Previous authors have thus attributed differences in intra-individual evolution rates to virus, host, or virus-host coevolution-dependent factors (Fig. 5). Our data indicate that viral factors may have the most important influence on intra-individual lentiviral evolution rates, since we noted the most remarkable difference in FIV compared to HIV and SIV. Had we found differences in evolution rates of SIV among primate species or FIV among felid species, we would have considered host effects to be contributory to these differences. If we had noted differences between primary and spillover hosts, we would have considered virus-host coevolution-dependent factors to be relevant to intra-individual evolution rates. Finally, differences between experimental and naturally acquired infections would have indicated a contribution of experimental differences, which could lead back to host or coevolutionary mechanisms.
FIG 5.
Putative host, virus, experimental, and coevolutionary factors that influence intra-individual evolution rates.
One potentially important factor contributing to lower evolution rates in FIV is the fidelity of its reverse transcriptase polymerase (24). The error rates of this enzyme have been shown to vary by retrovirus, with that of HIV being especially error prone (25). Others have previously noted that FIV reverse transcriptase may have higher fidelity than those of SIV and HIV, resulting in lower mutation rates (21, 26), which is supported by our analysis.
Viral kinetics and viral loads are similar between these FIV and primate lentiviral infections (12). There are slight differences in cell receptor use for viral entry, and viruses have similar cell tropisms but may infect different populations of immunocytes disproportionately (12); therefore, it is possible that the immunological microenvironment faced by FIV may be less hostile, requiring less replication for the infection to survive and transmit host-to-host.
Viruses with more recent origins may undergo higher evolution rates, as they are under novel selective pressures as they undergo an evolutionary arms race with the host (23, 27, 28). HIV and SIV are postulated to have more recent origins than FIV (23, 29), and it has been postulated that coadaptation of FIV with felids has resulted in FIV reaching an adaptive equilibrium (23, 30). More recently evolved HIV and SIV viruses may not yet have equilibrated to host immune system and intrinsic restriction factors, resulting in continually altering quasispecies compositions through natural selection (27). This hypothesis, however, is inconsistent with FIV mutation rates calculated during spillover infections, which would be hypothesized to accelerate following infection of a new host, suggesting other mechanisms are more likely to underlie our observations.
Mutation rates have also been found to vary over the much shorter time course of a single infection within an individual host (17). Analyses of viral quasispecies reveal that genetic diversity is reduced upon primary infection, with its generation of a viral bottleneck, and then increases as the infection proceeds and, finally, decreases late in the disease progression (17, 18). Some studies have indicated that evolution rates decrease with disease progression (17, 31), while others find higher evolution rates during accelerated disease (32). Time since infection was not a factor examined in this study, and it is possible that sampling time in relation to time of infection may contribute to our findings.
Host-dependent factors did not appear to affect intra-individual mutation rates. Even the differences between intra-host evolution rates of HIV-1 and HIV-2 were not significant. One mechanism leading to intra-host variation involves the cytosine deaminase APOBEC, a viral restriction factor that deaminates cytosine to uracil in mRNA, resulting in viral hypermutation and degradation of the virus particle (27). The host immune response includes the formation of neutralizing antibodies that bind to env proteins, preventing viral particle fusion and entry, resulting in adaptive mutation for viral variants that are not effectively neutralized (9). These antiviral responses would theoretically manifest as differences in evolution between hosts and across genes. Interestingly, we did not detect differences in intra-individual rates across gene segments, although there was a trend for SIV env and gag to have higher rates of evolution than SIV pol, corroborating previous findings (20–22). Because env is thought to be the primary target for host immune system evasion, we expected to observe higher rates of evolution than in other parts of the lentiviral genome, as has been reported from between-host evolutionary analyses (21). We did note that the length of the segment evaluated influences intra-individual rate estimates for HIV, but not for FIV and SIV. It is possible that gene sequence length may affect the precision of evolution rate estimates and that analysis of larger viral genome segments would result in more accurate estimates of substitution rates (21). We detected less scatter and, thus, a more precise rate estimate for longer gene sequence lengths for HIV (Fig. 4). Interestingly, shorter gene lengths also resulted in more precise rate estimates for HIV, with most individual heterogeneity noted at the intermediate lengths. One explanation is that many of the env estimates for HIV, which were intermediate in length, encompass the highly variable V3 region, whereas FIV and SIV estimates did not preferentially analyze this region (21).
The majority of analyses we reviewed were reported following analysis of proviral DNA rather than RNA sequences (Table 1). Since proviral DNA represents an integrated template following infection and may not be representative of replication-competent virus, it is possible that intra-individual variation dependent upon host-mediated immune pressures may not be accurately depicted in these analyses. Ideally, future analyses should compare the evolution of proviral DNA and circulating RNA virus to assess differences between these two compartments.
TABLE 1.
Use of proviral DNA versus viral RNA for evolution rate estimates
| Virus | No. of sequences of indicated type analyzeda
|
|||
|---|---|---|---|---|
| Proviral DNA | Viral RNA | Unknown | Total | |
| FIV | 93 | 3 | 0 | 96 |
| SIV | 71 | 0 | 0 | 71 |
| HIV | 130 | 84 | 4 | 218 |
| Total | 294 | 87 | 4 | 385 |
The majority of data were derived from proviral DNA rather than from viral RNA.
Because experimental infections are induced with an inoculum that typically has been serially passaged in vitro, we hypothesized that viral evolution rates would be higher following experimental inoculations, since tissue-propagated virus may accumulate mutations that render virus less infectious in the host environment (21). Our analyses, however, did not detect differences in experimental infections relative to naturally occurring infections.
The majority of manuscripts and data analyzed (70%) did not indicate that their authors had controlled for recombination events, and recombination was considered in fewer instances during FIV analysis than during HIV or SIV analysis (Table 2). While recombination effects could potentially skew intra-individual evolutionary estimates, depending upon the method of analysis used, several studies suggest that recombination may not have a large effect on evolution rate estimates (33, 34).
TABLE 2.
Controlling for recombination in evolution rate estimatesa
| Virus | No. of evolution rate estimates for which recombination was: |
Total | ||
|---|---|---|---|---|
| Excluded | Included | Not mentioned | ||
| FIV | 40 | 44 | 12 | 96 |
| SIV | 0 | 0 | 71 | 71 |
| HIV | 43 | 0 | 175 | 218 |
| Total | 83 | 44 | 258 | 385 |
Controlling for recombination was not noted in the majority of rate estimates.
Interestingly, it was observed for FIV that intra-individual rates were not dissimilar to evolution rates that have been reported for populations (i.e., interhost evolution) (20, 21, 35, 36). Follow-up studies comparing intra-individual and population rates could further our understanding of the interplay between intra-individual and interhost phylodynamics. Additionally, studies comparing synonymous and nonsynonymous evolution rates could aid in understanding how neutral or selective evolutionary processes relate to quasispecies diversity (37). Finally, data incorporated from the other well-studied lentiviruses, including caprine arthritis-encephalitis virus (CAEV), equine infectious anemia virus (EIAV), bovine immunodeficiency virus (BIV), and visna virus, could enable a more holistic synthesis of the lentiviruses (8).
To our knowledge, this is the first study that synthesizes intra-individual evolution rates for FIV, SIV, and HIV. We observed that FIV evolves more slowly than HIV and SIV at the intra-individual level. Overall, intra-individual rate estimates may differ by gene sequence length but not by host, gene, or strain, experimental relative to natural setting, or spillover relative to primary host infection. Future analyses of evolutionary selection pressures and individual versus population phylodynamics will be important follow-on studies to advance the knowledge of intra- and interspecific evolutionary events shaping lentiviral pathogenesis.
MATERIALS AND METHODS
In June 2018, we performed a systematic search of the literature documenting intra-individual evolution rates for FIV, SIV, and HIV. Papers were initially identified using the key words “intra-host,” “rate,” and “FIV,” “SIV,” or “HIV” in Web of Science. These manuscripts were reviewed to identify previously reported intra-individual evolution rates, as well as type of infection, viral strain and host species, genes and gene lengths analyzed, and other parameters noted in Tables 1 to 5. Citations in identified manuscripts that reported intra-individual evolution rates were additionally evaluated, permitting comparisons of additional published papers on intra-individual rates. Additional published works were identified using the Google Scholar search engine function “cited by.” This process was continued iteratively until no further new papers were recovered for FIV and SIV and after 218 HIV intra-host evolutionary estimates were identified (Table 3). We believe these data sets represent a comprehensive review of the FIV and SIV literature and a sufficiently comprehensive review of the HIV literature for a thorough systematic review and comparison across lentivirus families.
TABLE 3.
Distribution of papers and estimated evolution rates collected per virus for the studya
| Virus | No. of: |
|
|---|---|---|
| Papers | Evolution rates | |
| FIV | 9 | 96 |
| HIV | 18 | 218 |
| SIV | 8 | 71 |
| Total | 35 | 385 |
TABLE 4.
Evaluation of virus/host species combinations
| Virus, host species | No. of times evaluated | Classificationb |
|---|---|---|
| FIVpco, mountain lion | 20 | Primary |
| FIVfca, domestic cat | 74 | Primary |
| FIVpco, domestic cat | 2 | Spillover |
| SIVagm, AGMa | 5 | Primary |
| SIVagm, patas monkey | 3 | Spillover |
| SIVagm, AGM/macaque | 1 | Spillover |
| SIVagm, macaque | 2 | Spillover |
| SIVmac, macaque | 15 | Primary |
| SIVsmm, macaque | 39 | Spillover |
| SIVsmm, mangabey | 5 | Primary |
| SIVmne, macaque | 1 | Spillover |
| Total | 167 |
AGM, African green monkey.
Studies where host and viral species matched were classified as “primary,” and those that did not match were classified as “spillover.”
TABLE 5.
Spillover versus primary host pivot tablea
| Virus | Host classification |
|
|---|---|---|
| Spillover | Primary | |
| FIV | 2 | 94 |
| SIV | 46 | 25 |
FIV has a majority of primary host data, while SIV has a majority of spillover host data.
Intra-individual rates were identified through the use of serial samples collected from a single individual postinfection and descriptive terminology (e.g., individual or intra-host). Rates were reported as substitutions per site, or percent change, over a defined period of time. All rates were converted to substitutions per site per year. Metadata were collected on the virus strain, gene analyzed, number of nucleotides analyzed, and host species in which the rates were determined (Table 4). For SIV and FIV, additional metadata were collected on whether primary or spillover hosts were infected. “Primary” host was assigned to rates in which the strain of the virus matched the species. “Spillover” host was assigned to rates where the strain of the virus did not match the species in which it was evaluated (Table 5). Additionally, FIV data were analyzed to determine whether the infections resulted from natural or experimental infection with the virus. All SIV analyses were conducted following experimental infections, and obviously, all HIV analyses occurred from samples collected from HIV patients. Table 6 supplies a detailed summary of the previously described data collected from each paper for this study.
TABLE 6.
Summary of studies and data included for each study
| Reference | Virus | Evolution rate obtained (no. per study) | Gene(s) | Virus strain or type(s) | Host speciesa | Type ofb
: |
|
|---|---|---|---|---|---|---|---|
| Host | Infection | ||||||
| 21 | FIV | 40 | env and pol | FIVpco | ML | P | N |
| 39 | FIV | 24 | env | FIVfca | DC | P | N |
| 6 | FIV | 20 | env | FIVfca | DC | P | N |
| 23 | FIV | 3 | env | FIVfca | DC | P | N |
| 49 | FIV | 3 | env, pol, and gag | FIVfca | DC | P | N |
| 51 | FIV | 2 | env | FIVfca | DC | P | E |
| 50 | FIV | 2 | env | FIVfca | DC | P | E |
| 61 | FIV | 2 | Full genome | FIVpco | DC | S | E |
| 57 | SIV | 6 | env | SIVagm | AGM and patas monkey | P and S | NA |
| 47 | SIV | 5 | env | SIVagm | AGM and Mac | P and S | NA |
| 41 | SIV | 7 | env | SIVmac | Mac | P | NA |
| 22 | SIV | 39 | env and pol | SIVsmm | Mac | S | NA |
| 62 | SIV | 4 | env and gag | SIVsmm | Mang | P | NA |
| 58 | SIV | 1 | env | SIVmne | Mac | S | NA |
| 59 | SIV | 8 | env | SIVmac | Mac | P | NA |
| 43 | SIV | 1 | env | SIVsmm | Mang | P | NA |
| 54 | HIV | 6 | env | HIV-1 | Human | NA | NA |
| 55 | HIV | 12 | env | HIV-1 | Human | NA | NA |
| 65 | HIV | 12 | env | HIV-1 | Human | NA | NA |
| 66 | HIV | 7 | env | HIV-1 | Human | NA | NA |
| 44 | HIV | 2 | env | HIV-1 | Human | NA | NA |
| 53 | HIV | 4 | env | HIV-1 | Human | NA | NA |
| 48 | HIV | 12 | env | HIV-1 | Human | NA | NA |
| 46 | HIV | 38 | env and envgag | HIV-1 | Human | NA | NA |
| 52 | HIV | 2 | env | HIV-1 | Human | NA | NA |
| 64 | HIV | 4 | env and pol | HIV-1 | Human | NA | NA |
| 42 | HIV | 26 | env | HIV-1 | Human | NA | NA |
| 33 | HIV | 5 | env | HIV-1 | Human | NA | NA |
| 34 | HIV | 64 | env | HIV-1 | Human | NA | NA |
| 45 | HIV | 6 | env | HIV-1 | Human | NA | NA |
| 60 | HIV | 1 | nef | HIV-1 | Human | NA | NA |
| 63 | HIV | 8 | env | HIV-1 and -2 | Human | NA | NA |
| 56 | HIV | 8 | env | HIV-2 | Human | NA | NA |
| 40 | HIV | 1 | env | HIV-2 | Human | NA | NA |
ML, mountain lion; DC, domestic cat; AGM, African green monkey; Mac, macaque; Mang, mangabey.
P, primary host; S, spillover host; N, natural infection; E, experimental infection; NA, not applicable.
Intra-individual evolution rates were compared between viruses, host species, viral strains, viral genes, primary relative to spillover host species, and experimental relative to natural settings by analysis of variance (ANOVA) and Tukey’s post hoc test, with visual representation via box plots (38). Gene length was assessed using Levene’s test of homogeneity of variances with accompanying scatter plot (38). All analyses were undertaken using the R statistical software (38).
ACKNOWLEDGMENTS
Funding was provided by NSF-EID grant number 1413925. The funding agency had no role in study design, data collection and interpretation, or the decision to submit the work for publication. The authors declare that there is no conflict of interest.
Special thanks to Chris Kozakiewicz, Nick Fountain-Jones, Mary Nehring, Craig Miller, Elliott Chiu, and all members of the SVRG and Carver laboratories for their assistance in various parts of the project.
REFERENCES
- 1.Narayan O, Clements JE. 1989. Biology and pathogenesis of lentiviruses. J Gen Virol 70:1617–1639. doi: 10.1099/0022-1317-70-7-1617. [DOI] [PubMed] [Google Scholar]
- 2.Doolittle RF, Feng DF, Johnson MS, McClure MA. 1989. Origins and evolutionary relationships of retroviruses. Q Rev Biol 64:1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- 3.Gojobori T, Moriyama EN, Kimura M. 1990. Molecular clock of viral evolution, and the neutral theory. Proc Natl Acad Sci U S A 87:10015–10018. doi: 10.1073/pnas.87.24.10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown EW, Yuhki N, Packer C, O'Brien SJ. 1994. A lion lentivirus related to feline immunodeficiency virus: epidemiologic and phylogenetic aspects. J Virol 68:5953–5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phillips RE, Rowland-Jones S, Nixon DF, Gotch FM, Edwards JP, Ogunlesi AO, Elvin JG, Rothbard JA, Bangham CR, Rizza CR, McMichael AJ. 1991. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354:453–459. doi: 10.1038/354453a0. [DOI] [PubMed] [Google Scholar]
- 6.Bęczkowski PM, Hughes J, Biek R, Litster A, Willett BJ, Hosie MJ. 2015. Rapid evolution of the env gene leader sequence in cats naturally infected with feline immunodeficiency virus. J Gen Virol 96:893–903. doi: 10.1099/vir.0.000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bendinelli M, Pistello M, Lombardi S, Poli A, Garzelli C, Matteucci D, Ceccherini-Nelli L, Malvaldi G, Tozzini F. 1995. Feline immunodeficiency virus: an interesting model for AIDS studies and an important cat pathogen. Clin Microbiol Rev 8:87–112. doi: 10.1128/CMR.8.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller RJ, Cairns JS, Bridges S, Sarver N. 2000. Human immunodeficiency virus and AIDS: insights from animal lentiviruses. J Virol 74:7187–7195. doi: 10.1128/jvi.74.16.7187-7195.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller C, Abdo Z, Ericsson A, Elder J, VandeWoude S. 2018. Applications of the FIV model to study HIV pathogenesis. Viruses 10:E206. doi: 10.3390/v10040206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatziioannou T, Evans DT. 2012. Animal models for HIV/AIDS research. Nat Rev Microbiol 10:852–867. doi: 10.1038/nrmicro2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharp PM, Hahn BH. 2010. The evolution of HIV-1 and the origin of AIDS. Philos Trans R Soc Lond B Biol Sci 365:2487–2494. doi: 10.1098/rstb.2010.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.VandeWoude S, Apetrei C. 2006. Going wild: lessons from naturally occurring T-lymphotropic lentiviruses. Clin Microbiol Rev 19:728–762. doi: 10.1128/CMR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franklin SP, Troyer JL, Terwee JA, Lyren LM, Kays RW, Riley SP, Boyce WM, Crooks KR, Vandewoude S. 2007. Variability in assays used for detection of lentiviral infection in bobcats (Lynx rufus), pumas (Puma concolor), and ocelots (Leopardus pardalis). J Wildl Dis 43:700–710. doi: 10.7589/0090-3558-43.4.700. [DOI] [PubMed] [Google Scholar]
- 14.Lee J, Malmberg JL, Wood BA, Hladky S, Troyer R, Roelke M, Cunningham M, McBride R, Vickers W, Boyce W, Boydston E, Serieys L, Riley S, Crooks K, VandeWoude S. 2017. Feline immunodeficiency virus cross-species transmission: implications for emergence of new lentiviral infections. J Virol 91:e02134-16. doi: 10.1128/JVI.02134-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terwee JA, Yactor JK, Sondgeroth KS, Vandewoude S. 2005. Puma lentivirus is controlled in domestic cats after mucosal exposure in the absence of conventional indicators of immunity. J Virol 79:2797–2806. doi: 10.1128/JVI.79.5.2797-2806.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sprague WS, Troyer RM, Zheng X, Wood BA, Macmillian M, Carver S, VandeWoude S. 2018. Prior puma lentivirus infection modifies early immune responses and attenuates feline immunodeficiency virus infection in cats. Viruses 10:E210. doi: 10.3390/v10040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delwart EL, Pan H, Sheppard HW, Wolpert D, Neumann AU, Korber B, Mullins JI. 1997. Slower evolution of human immunodeficiency virus type 1 quasispecies during progression to AIDS. J Virol 71:7498–7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Padhi A, Ross H, Terwee J, Vandewoude S, Poss M. 2010. Profound differences in virus population genetics correspond to protection from CD4 decline resulting from feline lentivirus coinfection. Viruses 2:2663–2680. doi: 10.3390/v2122663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Diaz RS, Ho DD, Mosley JW, Busch MP, Mayer A. 1997. Host-specific driving force in human immunodeficiency virus type 1 evolution in vivo. J Virol 71:2555–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biek R, Drummond AJ, Poss M. 2006. A virus reveals population structure and recent demographic history of its carnivore host. Science 311:538–541. doi: 10.1126/science.1121360. [DOI] [PubMed] [Google Scholar]
- 21.Biek R, Rodrigo AG, Holley D, Drummond A, Anderson CR Jr, Ross HA, Poss M. 2003. Epidemiology, genetic diversity, and evolution of endemic feline immunodeficiency virus in a population of wild cougars. J Virol 77:9578–9589. doi: 10.1128/JVI.77.17.9578-9589.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson PR, Hamm TE, Goldstein S, Kitov S, Hirsch VM. 1991. The genetic fate of molecularly cloned simian immunodeficiency virus in experimentally infected macaques. Virology 185:217–228. doi: 10.1016/0042-6822(91)90769-8. [DOI] [PubMed] [Google Scholar]
- 23.Hayward JJ, Rodrigo AG. 2010. Molecular epidemiology of feline immunodeficiency virus in the domestic cat (Felis catus). Vet Immunol Immunopathol 134:68–74. doi: 10.1016/j.vetimm.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis DA, Bebenek K, Beard WA, Wilson SH, Kunkel TA. 1999. Uniquely altered DNA replication fidelity conferred by an amino acid change in the nucleotide binding pocket of human immunodeficiency virus type 1 reverse transcriptase. J Biol Chem 274:32924–32930. doi: 10.1074/jbc.274.46.32924. [DOI] [PubMed] [Google Scholar]
- 25.Preston BD, Dougherty JP. 1996. Mechanisms of retroviral mutation. Trends Microbiol 4:16–21. doi: 10.1016/0966-842X(96)81500-9. [DOI] [PubMed] [Google Scholar]
- 26.Menendez-Arias L. 2009. Mutation rates and intrinsic fidelity of retroviral reverse transcriptases. Viruses 1:1137–1165. doi: 10.3390/v1031137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daugherty MD, Malik HS. 2012. Rules of engagement: molecular insights from host-virus arms races. Annu Rev Genet 46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- 28.Lively CM. 1996. Host-parasite coevolution and sex: do interactions between biological enemies maintain genetic variation and cross-fertilization? Bioscience 46:107–114. doi: 10.2307/1312813. [DOI] [Google Scholar]
- 29.Wertheim JO, Worobey M. 2009. Dating the age of the SIV lineages that gave rise to HIV-1 and HIV-2. PLoS Comput Biol 5:e1000377. doi: 10.1371/journal.pcbi.1000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson MI, Holmes EC. 2007. The evolution of epidemic influenza. Nat Rev Genet 8:196–205. doi: 10.1038/nrg2053. [DOI] [PubMed] [Google Scholar]
- 31.Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, Learn GH, He X, Huang XL, Mullins JI. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol 73:10489–10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mikhail M, Wang B, Lemey P, Beckthold B, Vandamme AM, Gill MJ, Saksena NK. 2005. Role of viral evolutionary rate in HIV-1 disease progression in a linked cohort. Retrovirology 2:41. doi: 10.1186/1742-4690-2-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee HY, Perelson AS, Park SC, Leitner T. 2008. Dynamic correlation between intrahost HIV-1 quasispecies evolution and disease progression. PLoS Comput Biol 4:e1000240. doi: 10.1371/journal.pcbi.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemey P, Kosakovsky Pond SL, Drummond AJ, Pybus OG, Shapiro B, Barroso H, Taveira N, Rambaut A. 2007. Synonymous substitution rates predict HIV disease progression as a result of underlying replication dynamics. PLoS Comput Biol 3:e29. doi: 10.1371/journal.pcbi.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cano-Ortiz L, Junqueira DM, Comerlato J, Costa CS, Zani A, Duda NB, Tochetto C, Dos Santos RN, da Costa FVA, Roehe PM, Franco AC. 2017. Phylodynamics of the Brazilian feline immunodeficiency virus. Infect Genet Evol 55:166–171. doi: 10.1016/j.meegid.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 36.Fountain-Jones NM, Craft ME, Funk WC, Kozakiewicz C, Trumbo DR, Boydston EE, Lyren LM, Crooks K, Lee JS, VandeWoude S, Carver S. 2017. Urban landscapes can change virus gene flow and evolution in a fragmentation-sensitive carnivore. Mol Ecol 26:6487–6498. doi: 10.1111/mec.14375. [DOI] [PubMed] [Google Scholar]
- 37.Holmes EC. 2011. The evolution and emergence of RNA viruses. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 38.R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- 39.Bęczkowski P. 2012. Virus evolution in the progression of natural feline immunodeficiency virus infection. PhD thesis University of Glasgow, Glasgow, Scotland. [Google Scholar]
- 40.Borrego P, Marcelino JM, Rocha C, Doroana M, Antunes F, Maltez F, Gomes P, Novo C, Barroso H, Taveira N. 2008. The role of the humoral immune response in the molecular evolution of the envelope C2, V3 and C3 regions in chronically HIV-2 infected patients. Retrovirology 5:78. doi: 10.1186/1742-4690-5-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burns DP, Desrosiers RC. 1991. Selection of genetic variants of simian immunodeficiency virus in persistently infected rhesus monkeys. J Virol 65:1843–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carvajal-Rodriguez A, Posada D, Perez-Losada M, Keller E, Abrams EJ, Viscidi RP, Crandall KA. 2008. Disease progression and evolution of the HIV-1 env gene in 24 infected infants. Infect Genet Evol 8:110–120. doi: 10.1016/j.meegid.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Courgnaud V, Saurin W, Villinger F, Sonigo P. 1998. Different evolution of simian immunodeficiency virus in a natural host and a new host. Virology 247:41–50. doi: 10.1006/viro.1998.9217. [DOI] [PubMed] [Google Scholar]
- 44.Delwart EL, Sheppard HW, Walker BD, Goudsmit J, Mullins JI. 1994. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J Virol 68:6672–6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edo-Matas D, Lemey P, Tom JA, Serna-Bolea C, van den Blink AE, van 't Wout AB, Schuitemaker H, Suchard MA. 2011. Impact of CCR5delta32 host genetic background and disease progression on HIV-1 intrahost evolutionary processes: efficient hypothesis testing through hierarchical phylogenetic models. Mol Biol Evol 28:1605–1616. doi: 10.1093/molbev/msq326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edwards CT, Holmes EC, Wilson DJ, Viscidi RP, Abrams EJ, Phillips RE, Drummond AJ. 2006. Population genetic estimation of the loss of genetic diversity during horizontal transmission of HIV-1. BMC Evol Biol 6:28. doi: 10.1186/1471-2148-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fomsgaard A, Johnson PR, London WT, Hirsch VM. 1993. Genetic variation of the SIVagm transmembrane glycoprotein in naturally and experimentally infected primates. AIDS 7:1041–1047. doi: 10.1097/00002030-199308000-00003. [DOI] [PubMed] [Google Scholar]
- 48.Ganeshan S, Dickover RE, Korber BT, Bryson YJ, Wolinsky SM. 1997. Human immunodeficiency virus type 1 genetic evolution in children with different rates of development of disease. J Virol 71:663–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greene WK, Meers J, del Fierro G, Carnegie PR, Robinson WF. 1993. Extensive sequence variation of feline immunodeficiency virus env genes in isolates from naturally infected cats. Arch Virol 133:51–62. doi: 10.1007/BF01309743. [DOI] [PubMed] [Google Scholar]
- 50.Ikeda Y, Miyazawa T, Nishimura Y, Nakamura K, Tohya Y, Mikami T. 2004. High genetic stability of TM1 and TM2 strains of subtype B feline immunodeficiency virus in long-term infection. J Vet Med Sci 66:287–289. doi: 10.1292/jvms.66.287. [DOI] [PubMed] [Google Scholar]
- 51.Kraase M, Sloan R, Klein D, Logan N, McMonagle L, Biek R, Willett BJ, Hosie MJ. 2010. Feline immunodeficiency virus env gene evolution in experimentally infected cats. Vet Immunol Immunopathol 134:96–106. doi: 10.1016/j.vetimm.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 52.Lemey P, Rambaut A, Pybus OG. 2006. HIV evolutionary dynamics within and among hosts. AIDS Rev 8:125–140. [PubMed] [Google Scholar]
- 53.Li WH, Tanimura M, Sharp PM. 1988. Rates and dates of divergence between AIDS virus nucleotide sequences. Mol Biol Evol 5:313–330. doi: 10.1093/oxfordjournals.molbev.a040503. [DOI] [PubMed] [Google Scholar]
- 54.Liu SL, Schacker T, Musey L, Shriner D, McElrath MJ, Corey L, Mullins JI. 1997. Divergent patterns of progression to AIDS after infection from the same source: human immunodeficiency virus type 1 evolution and antiviral responses. J Virol 71:4284–4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lukashov VV, Kuiken CL, Goudsmit J. 1995. Intrahost human immunodeficiency virus type 1 evolution is related to length of the immunocompetent period. J Virol 69:6911–6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.MacNeil A, Sankale JL, Meloni ST, Sarr AD, Mboup S, Kanki P. 2007. Long-term intrapatient viral evolution during HIV-2 infection. J Infect Dis 195:726–733. doi: 10.1086/511308. [DOI] [PubMed] [Google Scholar]
- 57.Müller-Trutwin MC, Corbet S, Tavares MD, Hervé VM, Nerrienet E, Georges-Courbot MC, Saurin W, Sonigo P, Barré-Sinoussi F. 1996. The evolutionary rate of nonpathogenic simian immunodeficiency virus (SIVagm) is in agreement with a rapid and continuous replication in vivo. Virology 223:89–102. doi: 10.1006/viro.1996.0458. [DOI] [PubMed] [Google Scholar]
- 58.Overbaugh J, Rudensey LM, Papenhausen MD, Benveniste RE, Morton WR. 1991. Variation in simian immunodeficiency virus env is confined to V1 and V4 during progression to simian AIDS. J Virol 65:7025–7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pelletier E, Saurin W, Cheynier R, Letvin NL, Wain-Hobson S. 1995. The tempo and mode of SIV quasispecies development in vivo calls for massive viral replication and clearance. Virology 208:644–652. doi: 10.1006/viro.1995.1195. [DOI] [PubMed] [Google Scholar]
- 60.Plikat U, Nieselt-Struwe K, Meyerhans A. 1997. Genetic drift can dominate short-term human immunodeficiency virus type 1 nef quasispecies evolution in vivo. J Virol 71:4233–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poss M, Ross HA, Painter SL, Holley DC, Terwee JA, Vandewoude S, Rodrigo A. 2006. Feline lentivirus evolution in cross-species infection reveals extensive G-to-A mutation and selection on key residues in the viral polymerase. J Virol 80:2728–2737. doi: 10.1128/JVI.80.6.2728-2737.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rey-Cuille MA, Berthier JL, Bomsel-Demontoy MC, Chaduc Y, Montagnier L, Hovanessian AG, Chakrabarti LA. 1998. Simian immunodeficiency virus replicates to high levels in sooty mangabeys without inducing disease. J Virol 72:3872–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Skar H, Borrego P, Wallstrom TC, Mild M, Marcelino JM, Barroso H, Taveira N, Leitner T, Albert J. 2010. HIV-2 genetic evolution in patients with advanced disease is faster than that in matched HIV-1 patients. J Virol 84:7412–7415. doi: 10.1128/JVI.02548-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vrancken B, Rambaut A, Suchard MA, Drummond A, Baele G, Derdelinckx I, Van Wijngaerden E, Vandamme AM, Van Laethem K, Lemey P. 2014. The genealogical population dynamics of HIV-1 in a large transmission chain: bridging within and among host evolutionary rates. PLoS Comput Biol 10:e1003505. doi: 10.1371/journal.pcbi.1003505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wolfs TF, de Jong JJ, Van den Berg H, Tijnagel JM, Krone WJ, Goudsmit J. 1990. Evolution of sequences encoding the principal neutralization epitope of human immunodeficiency virus 1 is host dependent, rapid, and continuous. Proc Natl Acad Sci U S A 87:9938–9942. doi: 10.1073/pnas.87.24.9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wolinsky SM, Korber BT, Neumann AU, Daniels M, Kunstman KJ, Whetsell AJ, Furtado MR, Cao Y, Ho DD, Safrit JT. 1996. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science 272:537–542. [DOI] [PubMed] [Google Scholar]