Middle East respiratory syndrome coronavirus (MERS-CoV) initially emerged in 2012 and has since been responsible for over 2,300 infections, with a case fatality ratio of approximately 35%. We have used the highly characterized model system of Saccharomyces cerevisiae to investigate novel functional interactions between viral proteins and eukaryotic cells that may provide new avenues for antiviral intervention. We identify a functional link between the MERS-CoV ORF4a proteins and the YDL042C/SIR2 yeast gene. The mammalian homologue of SIR2 is SIRT1, an NAD-dependent histone deacetylase. We demonstrate for the first time that SIRT1 is a proviral factor for MERS-CoV replication and that ORF4a has a role in modulating its activity in mammalian cells.
KEYWORDS: host-virus interaction, MERS-CoV, ORF4a, SIRT1, suppressor screen, virus-host interaction, yeast
ABSTRACT
Viral proteins must intimately interact with the host cell machinery during virus replication. Here, we used the yeast Saccharomyces cerevisiae as a system to identify novel functional interactions between viral proteins and eukaryotic cells. Our work demonstrates that when the Middle East respiratory syndrome coronavirus (MERS-CoV) ORF4a accessory gene is expressed in yeast it causes a slow-growth phenotype. ORF4a has been characterized as an interferon antagonist in mammalian cells, and yet yeast lack an interferon system, suggesting further interactions between ORF4a and eukaryotic cells. Using the slow-growth phenotype as a reporter of ORF4a function, we utilized the yeast knockout library collection to perform a suppressor screen where we identified the YDL042C/SIR2 yeast gene as a suppressor of ORF4a function. The mammalian homologue of SIR2 is SIRT1, an NAD-dependent histone deacetylase. We found that when SIRT1 was inhibited by either chemical or genetic manipulation, there was reduced MERS-CoV replication, suggesting that SIRT1 is a proviral factor for MERS-CoV. Moreover, ORF4a inhibited SIRT1-mediated modulation of NF-κB signaling, demonstrating a functional link between ORF4a and SIRT1 in mammalian cells. Overall, the data presented here demonstrate the utility of yeast studies for identifying genetic interactions between viral proteins and eukaryotic cells. We also demonstrate for the first time that SIRT1 is a proviral factor for MERS-CoV replication and that ORF4a has a role in modulating its activity in cells.
IMPORTANCE Middle East respiratory syndrome coronavirus (MERS-CoV) initially emerged in 2012 and has since been responsible for over 2,300 infections, with a case fatality ratio of approximately 35%. We have used the highly characterized model system of Saccharomyces cerevisiae to investigate novel functional interactions between viral proteins and eukaryotic cells that may provide new avenues for antiviral intervention. We identify a functional link between the MERS-CoV ORF4a proteins and the YDL042C/SIR2 yeast gene. The mammalian homologue of SIR2 is SIRT1, an NAD-dependent histone deacetylase. We demonstrate for the first time that SIRT1 is a proviral factor for MERS-CoV replication and that ORF4a has a role in modulating its activity in mammalian cells.
INTRODUCTION
Middle East respiratory syndrome coronavirus (MERS-CoV) first emerged in the Kingdom of Saudi Arabia in 2012 (1). To date, over 2,300 people have been infected by the virus, causing greater than 820 deaths and placing the case fatality ratio at approximately 35% (http://www.who.int/). MERS-CoV emerged from a zoonotic origin, with the original host thought to be bats and the current host reservoir being camels (2–5). The virus has yet to show sustained transmission between humans, but, with no currently approved antiviral therapeutics or vaccines, MERS-CoV is considered a “threat to global health” by the WHO. Identifying novel virus-host interactions is essential to guide future intervention strategies.
Coronaviruses have large (∼30-kb) positive-sense, single-stranded RNA genomes. Approximately two-thirds of this RNA is dedicated to the production of 16 nonstructural proteins which regulate replication of the viral genome. The remaining third encodes the structural and accessory proteins (6).The accessory proteins largely function to promote viral replication through modulating cellular responses to infection. One such protein, produced by MERS-CoV, is ORF4a. This accessory protein has been shown in vitro to bind double-stranded RNA and to disrupt the innate immune sensing pathways of RIG-I and MDA5 to suppress the interferon response (7–9). Additionally, ORF4a has been found to inhibit the PKR and stress granule response in cells (10–12), further interfering with cellular function to promote viral infection.
The yeast Saccharomyces cerevisiae have been used extensively throughout the history of eukaryotic cell biology, including its being the first eukaryotic genome to be fully sequenced (13). The genome is now highly annotated, due in part to experiments performed with the yeast knockout library collection (14, 15). The yeast knockout collection systematically knocked out each gene in S. cerevisiae with a known DNA cassette containing a unique “barcode” region. Therefore, by taking any yeast cell from within this library and PCR amplifying and sequencing the barcode region, it is possible to determine which gene has been deleted from that cell. The project managed to knock out ∼4,600 of the ∼6,000 yeast genes; deletion of the others resulted in an inability to produce viable yeast, and they were deemed essential for growth in glucose (Glu) media. Therefore, the yeast knockout collection represents a widely available, semi-genome-wide knockout library of yeast.
While extensively used to study eukaryotic cell biology, yeast species have also been used to look for factors involved in replication of RNA viruses (16–22). We and others have previously demonstrated that expression of certain proteins from viruses and bacteria in the yeast S. cerevisiae can inhibit growth (23–30). This growth phenotype can be leveraged to identify compounds with antiviral properties or functionally important residues of the viral proteins. In this study, we utilized the slow-growth phenotype to investigate novel genetic interactions between the viral protein and eukaryotic cells.
Through screening the MERS-CoV proteome, we identified various proteins that, when expressed in yeast, caused a slow-growth phenotype, including the protein encoded by the ORF4a accessory gene. Yeast lack an interferon response, and since ORF4a has largely been studied in experiments designed to disrupt these pathways, we hypothesized that ORF4a must have further functions in a eukaryotic cell that have yet to be analyzed. To investigate the cellular pathways that ORF4a interacts with in yeast, we took advantage of the yeast knockout library. We hypothesized that removal of genes involved in the ORF4a-mediated slow growth would cause a reversion of the slow-growth phenotype, allowing the identification of genetic suppressors. This screening approach identified the YDL042C/SIR2 yeast gene as a suppressor of ORF4a-mediated slow growth, and that gene became the focus of our study.
YDL042C/SIR2 is a sirtuin protein, first identified through its role in silencing the cryptic mating-type loci HML and HMR (31). Since that initial identification, sirtuins have been defined as NAD-dependent histone deacetylases (32) involved in regulating a multitude of cellular functions, including metabolism, apoptosis, stress responses, DNA repair, and gene expression (33–36). The mammalian homologue of SIR2, SIRT1, has been shown to have both proviral and antiviral roles, depending on the virus (37–43), making this an attractive candidate hit from our yeast-based screening for study in mammalian cells. Moreover, it has previously been suggested that resveratrol, a compound that activates sirtuins, inhibits MERS-CoV replication, and yet no mechanism was investigated in that work (44). Given the combination of SIRT1 being shown to have roles in replication of mammalian viruses and the fact that SIR2 was the most commonly identified gene in our suppressor screen, this sirtuin became the focus of the study. Other genes identified in yeast are yet to be examined in the mammalian context.
We found that chemical inhibition of SIRT1 or knockdown with either small interfering RNA (siRNA) or clustered regularly interspaced short palindromic repeat interference (CRISPRi) inhibited replication of MERS-CoV, demonstrating that SIRT1 is a proviral factor for viral replication. The sirtuin activator resveratrol was found to inhibit MERS-CoV replication, but this effect was not dependent on the presence of SIRT1, suggesting that other sirtuin proteins may have antiviral roles, while SIRT1 appears to have a proviral role. The yeast-based screening system demonstrated a functional link between ORF4a and SIR2. We did not observe a direct interaction between ORF4a and SIRT1 in mammalian cells. However, we do show that ORF4a can modulate SIRT1-mediated enhancement of NF-κB signaling, suggesting a functional link between the two proteins in mammalian cells. Our work demonstrates the utility of yeast for identifying novel interactions between viral proteins and eukaryotic cells and defines SIRT1 as a proviral factor for MERS-CoV replication.
RESULTS
MERS-CoV ORF4a causes a slow-growth phenotype in yeast, and YDL042C/SIR2 is a genetic suppressor.
Work performed in our laboratory and others previously found that certain proteins from various pathogens can cause a slow-growth phenotype in S. cerevisiae (23–30). This phenotype can be used to investigate functional aspects of these proteins in eukaryotic cells by the use of suppressor screens. In these screens, yeast genes are knocked out (as in this study) or overexpressed to identify genes that suppress the growth phenotype caused by the expression of the pathogen protein. We took advantage of the yeast model system to investigate cellular interactions of proteins from MERS-CoV.
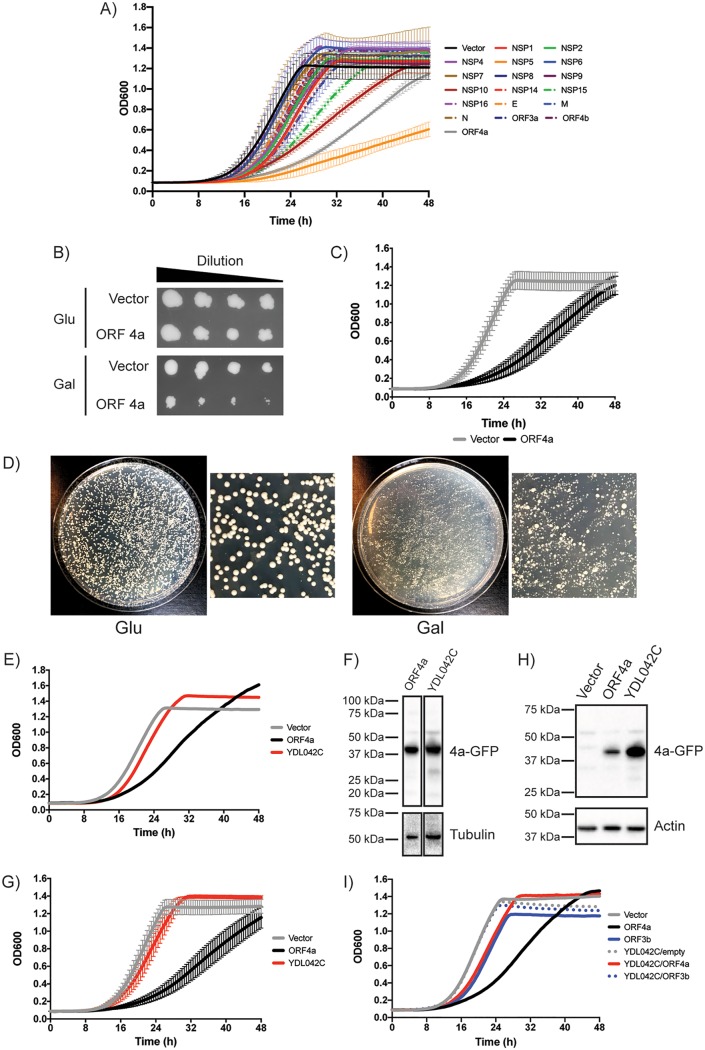
Genes from the MERS-CoV genome were cloned into a galactose (Gal)-inducible (GAL1) expression vector, based on pBR416 (45), with a C-terminal green fluorescent protein (GFP) tag, and were transformed into yeast. Grown in the presence of 2% glucose (Glu), the expression of viral genes was inhibited and the yeast strain containing this plasmid was able to grow similarly to yeast transformed with a vector control (Fig. 1B and data not shown). However, grown in the presence of 2% galactose (Gal), viral genes were expressed. We analyzed whether any of the MERS-CoV-encoded proteins could inhibit yeast growth by performing 48 h growth curves, measuring the optical density at 600 nm (OD600) on an automated plate reader as the readout for growth (Fig. 1A). We found that the following four MERS-CoV proteins inhibited growth of yeast: NSP5 (green dotted/dashed line in Fig. 1A), NSP10 (maroon line), NSP15 (orange line), and ORF4a (gray line). The other MERS-CoV genes examined had only a modest impact or no impact on growth, suggesting that expression of GFP-tagged proteins alone was not responsible for the slow growth observed. We decided to focus our further study on ORF4a as this accessory protein has been best described for roles in inhibiting the mammalian interferon (IFN) response, which is not conserved in yeast, therefore suggesting the possibility of hitherto-undescribed functions of ORF4a in eukaryotic cells. The growth inhibition caused by ORF4a was further assessed on solid media, where the levels of growth on Glu plates (no protein expression) were indistinguishable between vector control and ORF4a-expressing yeast whereas growth on Gal plates displayed growth inhibition of yeast expressing ORF4a (Fig. 1B). The results from an initial liquid-culture screening were also further confirmed (Fig. 1C).
FIG 1.
MERS-CoV ORF4a causes a slow-growth phenotype in yeast and a suppressor screen pulls out YDL042C/SIR2. (A) Yeast cells transformed with a galactose (Gal)-inducible plasmids to express MERS-CoV proteins were cultured for 2 days in media containing 2% raffinose to reach saturation. Yeast cells were diluted and cultured for 48 h in 2% Gal media at 30°C on a plate reader. Every 30 min, the plate was shaken for 30 s and the OD600 was read. The growth curve has OD600 plotted against time. Data are from one representative experiment performed using two separate colonies of yeast. Error bars represent deviations between results from the two colonies. (B) Yeast cells transformed with a Gal-inducible plasmid to express MERS-CoV ORF4a or a vector control were cultured for 2 days in media containing 2% raffinose to reach saturation. The OD600 was measured, yeast cells were diluted to an OD600 of 0.1 in 2% raffinose media, and then a 1:5 dilution series was made. This dilution series of the yeast was then replica plated on agar plates containing 2% glucose (Glu) or 2% Gal. (C) As described for panels A and B, vector control or ORF4a plasmid yeast cells were grown for 2 days in 2% raffinose to reach saturation. Yeast cells were diluted and cultured for 48 h in 2% Gal media at 30°C on a plate reader. Every 30 min, the plate was shaken for 30 s and the OD600 was read. The growth curve has OD600 plotted against time. Data are from three independent experiments, with error bars showing standard deviations. (D) The Gal-inducible ORF4a plasmid was transformed into a pool of the yeast knockout (YKO) library collection and plated onto agar plates containing Glu or Gal. A suppressor screen was performed by picking large colonies from the ORF4a YKO library on Gal plates and analyzing growth rates for colonies that no longer had the slow-growth phenotype caused by expression of ORF4a. The most commonly detected gene deletion was YDL042C/SIR2. (E) A representative graph for one of the large colonies picked from a Gal plate that was later determined to be ΔYDL042C/SIR2. (F) Western blot to demonstrate that the suppressor phenotype observed in ΔYDL042C/SIR2 colonies was not a result of a lack of ORF4a expression. (G) A known ΔYDL042C/SIR2 strain was picked from an arrayed collection of the YKO library and transformed with Gal-inducible ORF4a plasmid. Data show growth curves of these transformed yeast cells from 3 independent experiments (error bars represent standard deviations). (H) Western blot to confirm that ORF4a was still expressed in the transformed ΔYDL042C/SIR2 cells. (I) Control experiments testing whether ΔYDL042C/SIR2 yeast cells do or do not grow faster than wild-type yeast. SARS-CoV–ORF3b and an empty vector were transformed into yeast that were subsequently grown for 48 h in 2% raffinose media prior to growth to produce growth curves as described above.
We reasoned that the slow-growth phenotype induced by ORF4a was a result of the viral protein disrupting normal cellular function, allowing us to perform a suppressor screen. We hypothesized that knockout of yeast genes involved in the ORF4a-mediated slow growth might reduce the level of inhibition and therefore increase the growth rate. To test this hypothesis, the inducible ORF4a plasmid was transformed into a pooled collection of the yeast knockout library. This library consists of ∼4,600 nonessential gene knockouts. When this transformed library was plated onto Glu plates, the yeast colonies that formed were of similar sizes (Fig. 1D, Glu). However, plated to Gal plates to induce ORF4a expression, a broad range of colony sizes were observed (Fig. 1D, Gal). Of most interest were the large colonies, as these were thought to have been formed by yeast cells that contain deletions of genes that confer a suppressor phenotype on ORF4a-mediated slow growth. Large colonies were picked from Gal plates and were tested for growth in liquid culture and for continued expression of ORF4a (data not show). Suppressors were determined as knockout yeast that expressed ORF4a and showed enhanced growth compared to ORF4a-expressing wild-type (WT) yeast. The identity of the knockout was determined by PCR amplification and sequencing of the unique barcode regions that are used in the yeast knockout library.
For the screening process, ∼150 colonies were assessed for growth phenotypes and 56 were found to have suppressor phenotypes to various degrees (data not shown). These 56 colonies determined as hits were sequenced and found to represent 42 different genes (Table 1). The most commonly detected gene knockout was YDL042C/SIR2 (no other sirtuin genes were detected). Tested in liquid culture, ΔYDL042C/SIR2 colonies picked from Gal plates showed a suppressor phenotype. Panel E of Fig. 1 shows an example of a single colony that was picked from a galactose screening plate that had a suppressor phenotype. When the growth rate of the initial fast-growing colony was assessed, the identity of the gene knockout was unknown. Subsequent sequencing determined it to be ΔYDL042C. This colony represented one of the five ΔYDL042C colonies that were identified as suppressors in the screen. The colony displayed in Fig. 1E still maintained expression of ORF4a, such that the enhanced growth was not a consequence of a lack of expression of the viral protein (Fig. 1F). To validate the hypothesis that YDL042C/SIR2 is indeed a suppressor of ORF4a, a confirmed YDL042C/SIR2 deletion strain was transformed with the ORF4a expression plasmid. When the known transformed deletion strain was tested for growth (Fig. 1G) and protein expression (Fig. 1H), the suppressor phenotype was maintained. To further validate identification of YDL042C as a specific suppressor of ORF4a, ΔYDL042C yeast cells were also transformed with an empty vector or a vector expressing SARS-CoV ORF3b with a GFP tag (which does not cause a slow-growth phenotype). Empty vector and ORF3b-GFP ΔYDL042C yeast grew at rates similar to those seen with the vector control yeast, suggesting that the ΔYDL042C phenotype did not stimulate enhanced growth and therefore that the suppressor phenotype seen when ORF4a was present was specific (Fig. 1I). Combined, these data argue that YDL042C/SIR2 is a bona fide genetic suppressor of ORF4a-mediated slow growth.
TABLE 1.
Table of the yeast genes that were found in suppressor screening of the ORF4a YKO librarya
| Yeast gene name | Yeast protein name | No. of hits |
|---|---|---|
| YDL042C | SIR2 | 5 |
| YHR075C | PPE1 | 4 |
| YOL020W | TAT2 | 3 |
| YIL020C | HIS6 | 2 |
| YJR069C | HAM1 | 2 |
| YER174C | GRX4 | 2 |
| YNL300W | TOS6 | 2 |
| YMR305C | SCW10 | 2 |
| YGR181W | TIM13 | 1 |
| YOR223W | DSC3 | 1 |
| YDL051W | LHP1 | 1 |
| YLL001W | DNM1 | 1 |
| YNR060W | FRE4 | 1 |
| YAL014C | SYN8 | 1 |
| YCR010C | ADY2 | 1 |
| YOR248W | Dubious ORF | 1 |
| YHR073W | OSH3 | 1 |
| YMR210W | MGL2 | 1 |
| YPL003W | ULA1 | 1 |
| YLR020C | YEH2 | 1 |
| YLR426W | TDA5 | 1 |
| YLR151C | PCD1 | 1 |
| YGR142W | BTN2 | 1 |
| YHR018C | ARG4 | 1 |
| YJR094C | IME1 | 1 |
| YLR149C | YLR149C | 1 |
| YJR056C | YJR056C | 1 |
| YHR204W | MNL1 | 1 |
| YKL217W | JEN1 | 1 |
| YPL107W | YPL107W | 1 |
| YAL027W | SAW1 | 1 |
| YOR076C | SKI7 | 1 |
| YMR153C-A | Dubious ORF | 1 |
| YLR227C | YSH1 | 1 |
| YJR092W | BUD4 | 1 |
| YMR057C | Dubious ORF | 1 |
| YMR025W | CSI1 | 1 |
| YCL001W | RER1 | 1 |
| YKL076C | PSY1 | 1 |
| YHR046C | INM1 | 1 |
| YNR014W | YNR014W | 1 |
| YOR239W | ABP140 | 1 |
The data in the column labeled “No. of hits” indicate numbers of individual colonies that were found to have the same gene deletion. YKO, yeast knockout; ORF, open reading frame.
ORF4a and SIRT1 do not directly interact in mammalian cells.
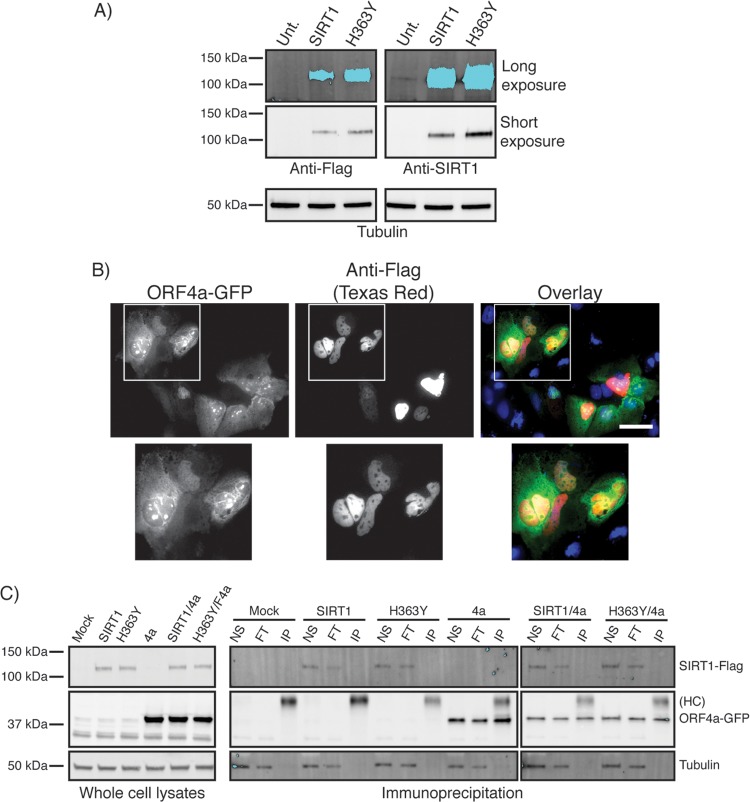
The mammalian homologue of yeast YDL042C/SIR2 is the histone deacetylase sirtuin protein SIRT1 (67% similarity at the protein level). We therefore hypothesized that ORF4a may interact with SIRT1 in mammalian cells to regulate MERS-CoV replication. Endogenous SIRT1 is localized to the nucleus in Huh7 cells, with areas that appear to have lower-intensity labeling (Fig. 2A). Transfected into Huh7 cells, a plasmid encoding ORF4a-GFP showed GFP signal in the cytosol, with additional puncta in the nucleus (Fig. 2B), in agreement with previously published data (7). The colabeling of these transfected cells with antibodies against SIRT1 showed both proteins to be in the nucleus. However, the nuclear puncta of ORF4a-GFP appeared to be in regions with lower levels of SIRT1 labeling (Fig. 2B). In cells infected with MERS-CoV, labeled with antisera to ORF4a, and colabeled with anti-SIRT1 antibody, ORF4a was founded in the nucleus and in cytosolic puncta, while SIRT1 was still localized to the nucleus (Fig. 2C). The more punctate appearance of cytosolic ORF4a in infected cells was possibly a result of lower expression levels compared to the overexpression construct. Overall, it appears that endogenous SIRT1 was localized to the nucleus, while GFP-tagged ORF4a and ORF4a expressed during infection were both localized in the nucleus and cytosol.
FIG 2.
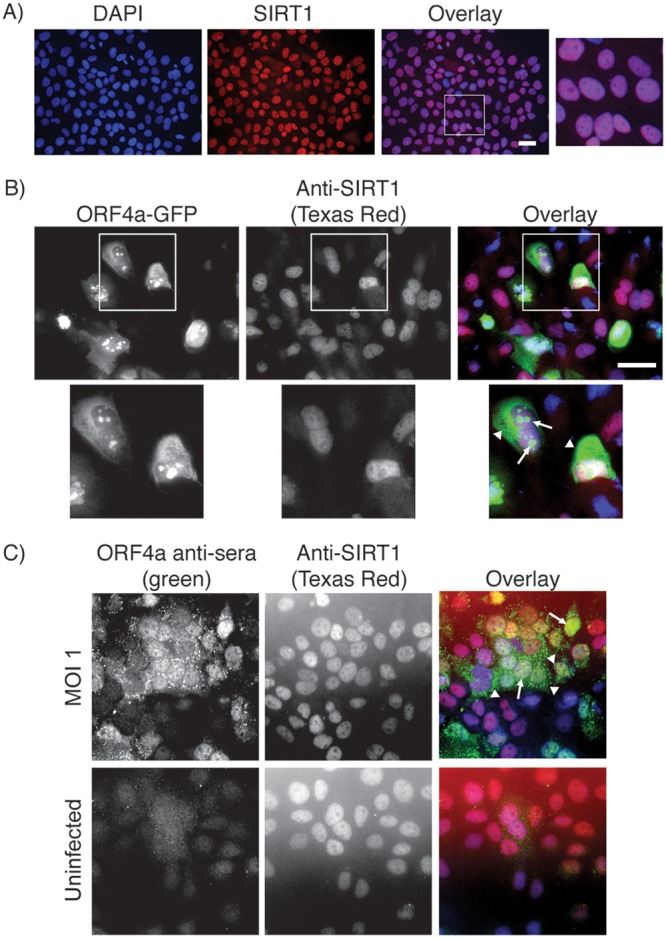
Localization of ORF4a and SIRT1 in Huh7 cells. (A) Huh7 cells grown on coverslips were subjected to immunofluorescence staining with anti-SIRT1 antibody (visualized with anti-mouse Texas Red). (B) Huh7 cells grown on coverslips were transfected with a plasmid expressing ORF4a-GFP and were subsequently subjected to immunofluorescence labeling for SIRT1 (visualized with anti-mouse Texas Red). Arrows point toward nuclear and arrowheads point toward cytosolic ORF4a localizations. (C) Huh7 cells were plated to coverslips 1 day prior to infection with MERS-CoV at an MOI of 1 (uninfected cells were grown as a control). Infection was allowed to proceed for 18 h prior to fixation and immunofluorescence labeling for SIRT1 (visualized with anti-mouse Texas Red) and ORF4a (visualized with anti-rabbit FITC). Arrows point toward nuclear and arrowheads point toward cytosolic ORF4a localizations. In all experiments, nuclei were labeled with DAPI. Scale bars = 50 μm.
To thoroughly test whether ORF4a and SIRT1 could interact in Huh7 cells, we acquired both WT and catalytically dead (H363Y) Flag-tagged expression plasmids of SIRT1 (Fig. 3A), reasoning that a higher level of expression of SIRT1 would allow us to detect even low levels of interaction. The overexpression constructs appeared to show a localization pattern similar to that seen with endogenous SIRT1 when expressed individually in cells (data not shown) and when cotransfected with ORF4a-GFP (Fig. 3B), suggesting that these are useful for investigating interaction of ORF4a and SIRT1 proteins. Once again, both SIRT1-Flag and ORF4a-GFP were seen in the nucleus, suggesting that they are in the same compartment of cells and therefore could potentially interact. To investigate this more directly, we used coimmunoprecipitation (co-IP) experiments. Cells were transfected with ORF4a-GFP alone or in combination with WT or H363Y SIRT1-Flag-expressing plasmids. Anti-GFP antibody was used to immunoprecipitate ORF4a-GFP, which could be detected in the IP fraction in blotting with anti-GFP antibody (Fig. 3C). In cells cotransfected with ORF4a-GFP and either of the SIRT1 constructs, while ORF4a-GFP could be precipitated, SIRT1 was detected only in the nonspecific and flowthrough fractions and never in the IP fraction (Fig. 3C). These data suggest that ORF4a and SIRT1 do not directly interact in Huh7 cells.
FIG 3.
ORF4a and SIRT1 do not directly interact in mammalian cells. (A) Whole-cell lysates from untransfected (Unt.) Huh7 cells or cells transfected with wild-type SIRT1 or H363Y mutant SIRT1-Flag-tagged plasmids were subjected to Western blotting with anti-SIRT1 or anti-Flag antibodies. Short and long exposures were performed to allow visualization of endogenous SIRT1 as indicated. (B) Huh7 cells grown on coverslips were cotransfected with ORF4a-GFP and SIRT1-Flag plasmid prior to being subjected to immunofluorescence labeling with anti-Flag antibody (visualized with anti-mouse Texas Red). Nuclei were labeled with DAPI. Scale bar = 50 μm. (C) Huh7 cells transfected with ORF4a-GFP or with wild-type or H363Y SIRT1-Flag or subjected to cotransfection were lysed and subjected to immunoprecipitation (IP) using anti-GFP antibody. Whole-cell lysate and nonspecific (NS), flowthrough (FT), and IP fractions were all analyzed (see Materials and Methods). Membranes were subjected to Western blotting with anti-GFP antibody and anti-SIRT1 antibody. “HC” denotes the IgG heavy chain.
SIRT1 is a proviral factor for MERS-CoV replication.
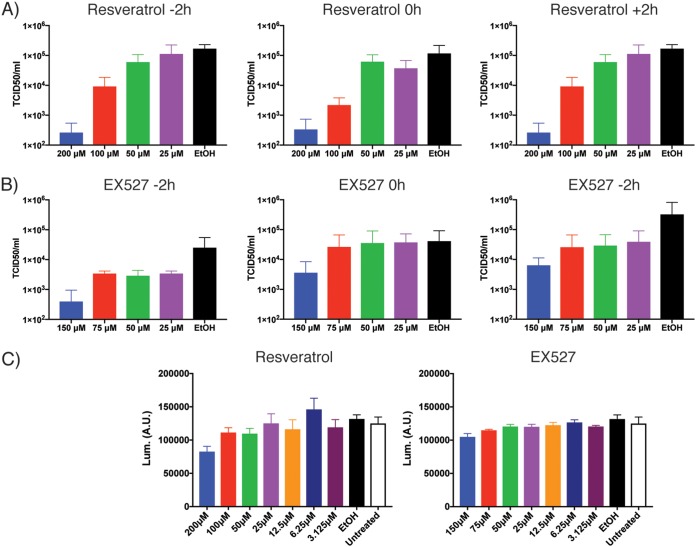
Yeast SIR2 was detected as a genetic suppressor of ORF4a-mediated slow growth, and yet ORF4a and SIRT1 were found to not directly interact in mammalian cells. The genetic suppression observed in yeast does not necessitate a direct interaction between the two proteins. We therefore hypothesized that ORF4a may modulate SIRT1 activity indirectly to regulate MERS-CoV replication. If this were the case, modulation of SIRT1 activity should alter the replication of MERS-CoV. It has previously been suggested that resveratrol, a chemical compound that broadly activates sirtuin proteins, can inhibit MERS-CoV replication (44). We looked to reproduce these results and indeed found that at higher concentrations, resveratrol was capable of inhibiting MERS-CoV replication as judged by assessing viral titer released into the supernatant (Fig. 4A). This inhibition was seen when the drug was added 2 h before, at the same time as, or 2 h after virus was added to the cells, suggesting that resveratrol was not inhibiting entry stages of infection. We additionally tested the impact of EX527, a chemical that inhibits SIRT1. These data showed that SIRT1 inhibition also resulted in inhibition of MERS-CoV replication (Fig. 4B). This result was again seen regardless of time of addition. Neither of these results appeared to be fully explainable by cell death, as only small differences were observed at the higher concentrations of drug in assessments of cell viability by CellTitre-Glo assay (Fig. 4C).
FIG 4.
Sirtuin-modulating drugs impact MERS-CoV infection. (A) Huh7 cells were treated with the indicated concentrations of resveratrol at −2, 0, or +2 h relative to infection with MERS-CoV at an MOI of 0.1. Cells were incubated for 24 h and supernatant collected. Virus TCID50 per milliliter in the supernatant was determined on Vero cells. Ethanol (EtOH) was the vehicle control for drug treatment. (B) As described for panel A, but cells were treated with EX527. For both panel A and panel B, data are from three independent experiments, with error bars representing standard deviations. (C) Cell viability was determined by CellTiter-Glo assay. Huh7 cells were treated with the indicated concentration of drug for 24 h prior to performance of the CellTiter-Glo assay to determine viability over a time period equivalent to that used in the MERS-CoV infection experiments. Error bars represent standard deviations of results of comparisons between 6 treatment wells from an experiment representing 3.
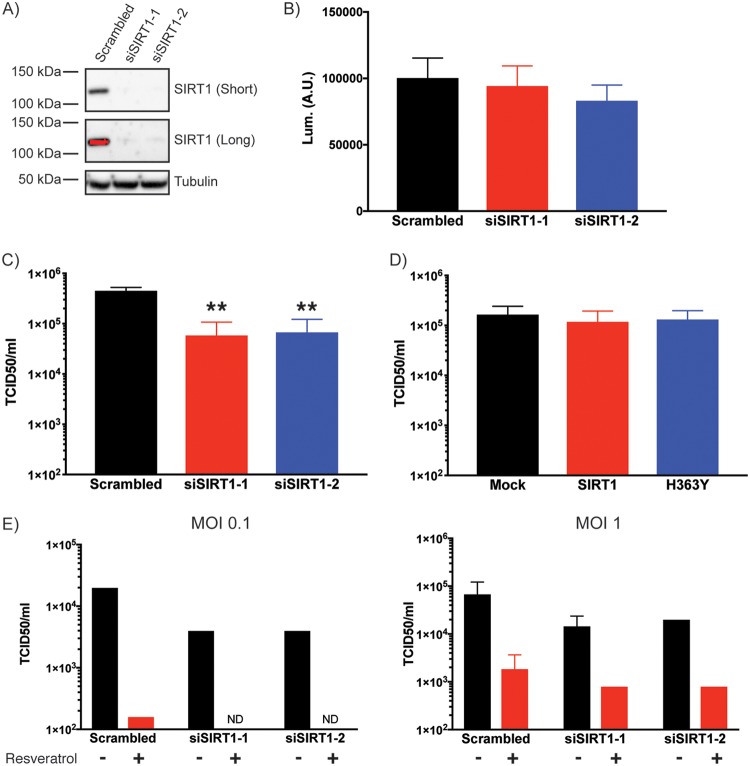
In order to investigate the role of SIRT1 more directly, siRNA was used to knock down expression of the protein. We used two siRNA sequences targeting SIRT1 (42). Both siRNA sequences were capable of knocking down SIRT1 expression (Fig. 5A), and both resulted in minor drops in cell viability as assessed by CellTitre-Glo assay (Fig. 5B). When these samples were infected with MERS-CoV, significant inhibition of infection was detected (Fig. 5C). While knockdown of SIRT1 was clearly capable of inhibiting MERS-CoV replication, overexpression of WT or H363Y SIRT1-Flag did not have any impact on virus production (Fig. 5D), suggesting that the endogenous levels of SIRT1 may be sufficient for replication.
FIG 5.
SIRT1 knockdown inhibits MERS-CoV replication. (A) Huh7 cells were transfected with one of two SIRT1-targeting siRNA sequences (siSIRT1-1 or siSIRT1-2) or scrambled siRNA as control. Cells were lysed and subjected to Western blotting for SIRT1. Results of long and short exposures of the blot are shown. (B) Cell viability of siRNA-transfected Huh7 cells was determined by CellTiter-Glo assay. Cells were plated 24 h prior to the assay to assess viability at a time that matched the time at which infections were performed. Lum., luminescence; A.U., arbitrary units. (C) siRNA-transfected cells were infected 24 h after plating with MERS-CoV was performed at an MOI of 0.1. Cells were incubated for 24 h prior to supernatant collection. Virus titer in the supernatant was determined by TCID50 assay on Vero cells. Data are from 3 independent experiments, with error bars representing standard deviations. **, P < 0.01 (by Student’s t test). (D) Huh7 cells were transfected with wild-type or H363Y mutant SIRT1 plasmids and infected with MERS-CoV at an MOI of 0.1. Virus titer in supernatant was determined as described for panel C. (E) siRNA-transfected cells were infected with MERS-CoV at an MOI of 0.1 or 1 and additionally treated with 200 μM resveratrol or ethanol (EtOH) vehicle control (added at the same time as the virus). Virus titer in the supernatant was determined as described for panels C and D. ND = not detected. MOI 1 was additionally used to allow detection of virus in the double treated cells.
These data suggest that SIRT1 is a proviral factor for MERS-CoV infection. However, this result is at odds with the data showing that resveratrol, a sirutin activator, inhibited infection. Resveratrol is a broad activator of sirtuin proteins, of which there are 7 in the human genome. We hypothesized that different sirtuin proteins may have different impacts on MERS-CoV infection. This was tested by transfecting cells with SIRT1 siRNA, treating with resveratrol, and infecting with MERS-CoV. Resveratrol was capable of inhibiting MERS-CoV infection even in the absence of SIRT1 (Fig. 5E). These data suggest that resveratrol inhibits MERS-CoV infection independently of SIRT1.
CRISPRi further demonstrates SIRT1 is a proviral factor for MERS-CoV replication.
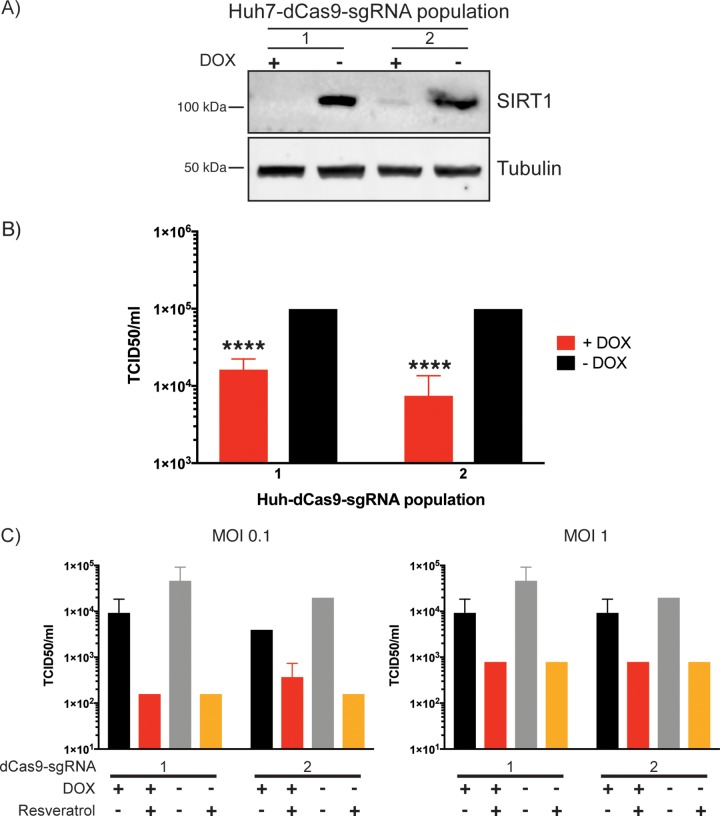
Our siRNA-mediated knockdown of SIRT1 in Fig. 5 suggests that SIRT1 is a proviral factor for MERS-CoV replication. In order to further assess the effect of SIRT1 on MERS-CoV replication, a CRISPRi approach was used (46). For this, Huh7 cells that stably express a catalytically dead version of Cas9 (dCas9) were produced through lentiviral transduction. The dCas9 can be inducibly expressed and binds to DNA as directed by the guide RNA but does not cleave, instead acting as a steric block to transcription. Moreover, a KRAB element on the dCas9 further enhances transcriptional repression. These stable Huh7-dCas9 cells were further transfected to stably express a short guide RNA (sgRNA) sequence to target the SIRT1 gene. Two lentiviral preparations were used to independently transfect two separate populations of Huh7 cells stably expressing the dCas9 construct (Huh7-dCas9 cells). The sgRNA is constitutively expressed, while the dCas9 is doxycycline (DOX) inducible; therefore, the stable cell populations can be divided into DOX-treated and untreated populations for internal controls of target gene knockdown. Both Huh7-dCas9-sgRNA cell populations demonstrated knockdown of SIRT1 under conditions of treatment with DOX for 7 days (Fig. 6A). When these doxycycline-treated cells were infected with MERS-CoV, there was a significant inhibition of replication in both populations (Fig. 6B). Knockdown of SIRT1 by both siRNA and CRISPRi inhibited MERS-CoV replication, showing this protein to be a proviral factor for replication.
FIG 6.
CRISPRi-mediated knockdown of SIRT1 inhibits MERS-CoV replication. (A) Two populations of Huh7 cells stably transfected with doxycycline (DOX)-inducible dCas9 were further transfected with two different lentiviral preparations containing a sgRNA sequence targeting SIRT1 to produce two independent populations of stable Huh7-dCas9-sgRNA cells. Each of the two populations was divided into two groups and either treated with DOX or grown in normal culture media for 7 days. Cells were lysed and samples subjected to Western blotting for SIRT1. (B) The stable Huh7-dCas9-sgRNA cells were grown for 7 days with or without DOX treatment and infected with MERS-CoV at an MOI of 0.1. For all cell groups, media were changed to normal growth media at the time of infection and samples were collected 24 h later to determine titer by TCID50 assay on Vero cells. Data are from three independent infections, with error bars representing standard deviations. ****, P < 0.001 (by Student’s t test comparing DOX-treated and untreated samples). (C) The two populations of stable dCas9-Huh7-sgRNA cells were cultured as described for panel B and infected with MERS-CoV at an MOI of 0.1 or 1 and simultaneously treated with 200 μM resveratrol or ethanol (EtOH) vehicle control. Virus titer in supernatant was determined as described for panel B. Data are from a single infection; error bars represent standard deviations in calculations of TCID50 per milliliter from three samples.
Using these CRISPRi cells, we also confirmed that resveratrol inhibited MERS-CoV replication even in the absence of SIRT1 (Fig. 6C), validating the siRNA results (Fig. 5E). The level of inhibition of MERS-CoV replication in control cells appears to have been slightly lower in this experiment; however, under conditions of testing over multiple infections (Fig. 6B), the inhibition results were statistically significant. Overall, it appears that even though the sirtuin activator resveratrol has antiviral activity with respect to MERS-CoV replication, SIRT1 is dispensable for this effect.
ORF4a modulates SIRT1 activity as assessed by an NF-κB pathway readout.
SIRT1 appears to function as a proviral factor for MERS-CoV replication (Fig. 5; see also Fig. 6). Our suppressor screen in yeast detected a genetic link between MERS-CoV ORF4a and SIR2 (Fig. 1), but we were unable to detect a direct interaction between ORF4a and mammalian SIRT1 (Fig. 3). We therefore speculated that ORF4a may have an indirect influence on SIRT1 activity in promoting MERS-CoV replication. SIRT1 has previously been shown to modulate NF-κB signaling through deacetylation of RelA/p65 (47). We therefore decided to use NF-κB signaling as a readout of SIRT1 activity to determine whether ORF4a alters this.
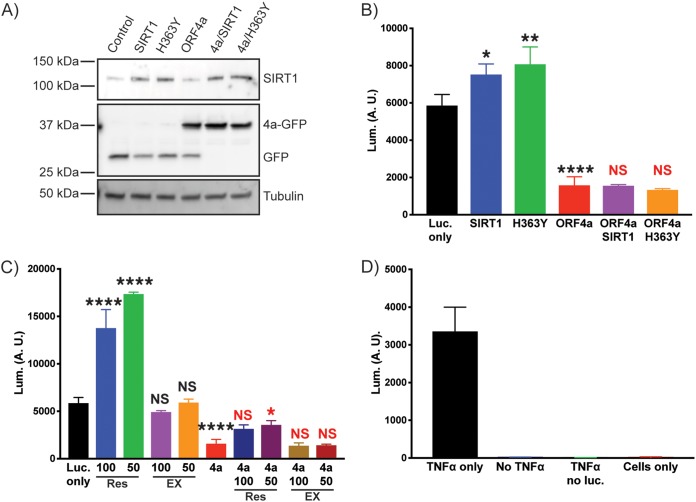
HEK293T cells were transfected with combinations of ORF4a, SIRT1 (WT or H363Y mutant), and a κB-luciferase plasmid (Fig. 7A). The following day, cells were treated with tumor necrosis factor alpha (TNF-α) for 6 h and lysed, and the amount of luciferase produced was determined. (Results of controls performed for analysis of background luminescence can be seen in Fig. 7D.) At odds with previously published data, we found that transient transfection of SIRT1 in our system significantly enhanced luciferase production following TNF-α treatment compared to the results seen with control transfected cells (Fig. 6; see also Fig. 7B). Cells transfected with H363Y SIRT1 also showed an enhanced level of luciferase production (Fig. 7B). When MERS-CoV ORF4a was transfected into cells, inhibition of luciferase production was observed (Fig. 7B). Importantly, when ORF4a was transfected into cells alongside SIRT1 (WT or H363Y), the luciferase levels were similar to those seen with ORF4a alone. These results suggest that SIRT1 no longer enhanced luciferase production in response to TNF-α when ORF4a was present, suggesting that ORF4a was altering this readout of SIRT1 activity (Fig. 7B).
FIG 7.
MERS-CoV ORF4a alters SIRT1 function in mammalian cells. (A) HEK293T cells were transfected with a κB-luciferase reporter plasmid and combinations of ORF4a-GFP, wild-type (WT) or H363Y mutant SIRT1 as labeled, and a GFP expressing plasmid to balance DNA levels (see Materials and Methods). Cells were lysed, and whole-cell lysates were Western blotted for SIRT1, GFP, and tubulin as a loading control. (B) As described for panel A, cells were transfected with various combinations of plasmids and then treated with TNF-α for 6 h prior to performing a firefly luciferase assay to assess production of luciferase from the κB promoter. Wild-type SIRT1 transfection significantly (P = 0.0132) enhanced luciferase production, as did H363Y SIRT1 (P = 0.0015), compared to cells with only the κB-luciferase plasmid (Luc. only). ORF4a significantly inhibited luciferase production (P = <0.0001) compared to the Luc.-only control. There was no significant difference when ORF4a and SIRT1 WT or H363Y were coexpressed in cells. Data are from analyses of three independent wells, with significance determined using one-way analysis of variance (ANOVA) with Bonferroni’s multiple-comparison test in Prism software. (C) Huh7 cells were transfected as described for panel A. Data from the Luc. only control and ORF4a data are the same as described for panel A; data are separated into two graphs for clarity. Cells were treated with resveratrol (Res) or EX527 (EX) at the indicated concentrations (in micromoles) for 2 h prior to addition of TNF-α and further incubation for 6 h. Luciferase production was determined as described for panel A. Drug treatment results are compared to those determined for the Luc. only cells (black annotation), and the results determined for ORF4a and drug are compared to those determined for ORF4a (red annotations) for significance testing (as described for panel B). Data are from three independent wells. (D) Control samples to assess background luminescence. Data are from experiments performed with the κB-luciferase reporter plasmid transfected along with a GFP expression plasmid or represent GFP expression only (no luc.) or no transfection. All samples were subjected to TNF-α treatment, with the exception of the no-TNF-α samples.
Treatment of cells with resveratrol also gave an enhanced level of NF-κB signaling in response to TNF-α, further suggesting a role for sirtuins in regulating this pathway in HEK293T cells (Fig. 7C). However, EX527 had no effect in our tests (Fig. 7C). A small but significant increase in NF-κB signaling was seen when ORF4a-transfected cells were treated with resveratrol, but luciferase production was largely blocked compared to the levels seen with cells without transfected ORF4a (Fig. 7B). These data further suggest that ORF4a is capable of modulating SIRT1-mediated enhancement of NF-κB signaling stimulated by TNF-α treatment. Overall, the results from the SIRT1 transfection and the resveratrol treatment suggest that ORF4a alters a SIRT1-mediated response in cells, suggesting a functional link between the two.
DISCUSSION
We have demonstrated that SIRT1 is a proviral cellular protein for MERS-CoV replication and that yeast cells represent a powerful tool to identify previously unknown virus-host interactions. Our initial work started with a suppressor screen in the yeast S. cerevisiae. We found that expression of MERS-CoV ORF4a in yeast caused slow growth. ORF4a functions as a double-stranded RNA binding protein that disrupts RIG-I, MDA5, PKR, and stress granule responses in infected cells to promote viral replication (7–12). ORF4a is well characterized as an inhibitor of the interferon response. Yeast species do not have an interferon response and yet show growth inhibition by ORF4a expression, suggesting further functional cellular interactions for the protein. To investigate the ways in which ORF4a might inhibit yeast growth, a suppressor screen was utilized. By screening the yeast knockout library for suppressors of ORF4a-induced growth attenuation, the YDL042C yeast gene was identified. The YDL042C gene encodes the SIR2 protein, and our work validated this as a bona fide suppressor of the the ORF4a-mediated slow-growth phenotype. The suppressor screen results suggest that there is a functional link between the two proteins but that the link does not rely on a direct physical interaction.
While yeast is a powerful starting point for investigating eukaryotic cell biology, in the context of MERS-CoV infection, it is important to establish relevance in a mammalian system. We did not find a direct interaction between ORF4a and SIRT1 in Huh7 cells by coimmunoprecipitation. However, when SIRT1 was inhibited either by EX527 treatment or by knockdown by siRNA or CRISPRi, MERS-CoV replication was inhibited. This establishes SIRT1 is a proviral factor for MERS-CoV replication.
Interestingly, our data also suggest that other sirtuin proteins may have antiviral roles in the context of MERS-CoV replication. Resveratrol, a compound derived from various plants, has been shown to activate sirtuin proteins (48) and was previously shown to inhibit MERS-CoV replication (44), a result that we replicated here. However, we demonstrated that knockdown of SIRT1 by siRNA or CRIPSRi had no impact on the resveratrol-mediated inhibition of MERS-CoV replication, suggesting that other members of the sirtuin family that are activated by the compound may have antiviral properties. Alternatively, resveratrol may have other effects on cell biology that inhibit MERS-CoV replication. Since SIRT1 was shown to be a proviral factor by our data and since resveratrol can activate SIRT1 function, it appears that the antiviral properties of resveratrol may outweigh the potential proviral function of SIRT1 or that the proviral role of SIRT1 is not based on its catalytic activity. Indeed, our experiments looking at the NF-κB pathway (discussed further below) suggest that SIRT1 may have activity in a catalytically dead form, arguing for a role in MERS-CoV replication that does not require enzymatic function. The role of other sirtuins in MERS-CoV replication and in how MERS-CoV modulates these to promote infection is the focus of future studies.
The yeast-based screening system identified a genetic interaction between ORF4a and SIR2, which led us to hypothesize that ORF4a may have a functional link with mammalian SIRT1 to regulate its function and promote MERS-CoV replication. SIRT1 has previously been shown to modulate NF-κB signaling in response to TNF-α (47). This work demonstrated a direct interaction between SIRT1 and p65/RelA and that SIRT1 inhibited the pathway by deacetylation of this NF-κB protein. We therefore reasoned that this modulation of the NF-κB pathway could be used as a readout to determine whether ORF4a expression alters SIRT1 function. Interestingly, we found that SIRT1 overexpression in HEK293T cells actually resulted in an enhanced NF-κB response following TNF-α treatment and that this enhancement occurred regardless of the level of catalytic activity of SIRT1. This discrepancy with previously published work could represent a result of the use of different cell lines. Our data suggest that SIRT1 promotes NF-κB signaling in HEK293T cells, perhaps by directly binding NF-κB and acting as a scaffold protein or by altering steps downstream in the signaling pathway, but that it does so independently of catalytic activity.
While we found the effects of SIRT1 on NF-κB to be different from those reported from previous studies in different cell lines, SIRT1 still altered the NF-κB response, allowing us to investigate the role of ORF4a in this pathway. Our experiments demonstrated that ORF4a was a potent suppressor of NF-κB signaling in HEK293T cells. When ORF4a and SIRT1 were coexpressed in cells, there was no longer an enhancement in luciferase production from the κB promoter. We cannot rule out the possibility that ORF4a may influence NF-κB signaling independently of SIRT1, but SIRT1 overexpression was not able to overcome ORF4a-mediated inhibition. These data, along with the genetic link observed in yeast, suggest that ORF4a and SIRT1 may have a functional connection without direct binding and that there is an additional node of regulation between the two proteins, which is under investigation. This work adds SIRT1 to the list of host factors that mediate MERS-CoV replication such as stress granule formation and host innate immune machinery (9–12). The magnitude of inhibition when those factors are affected is ∼1 log, demonstrating that, as seen in SIRT1, host machinery interacts with various virus replication steps to potentially hinder replication. All of these factors demonstrate novel therapeutic targets for future development.
Overall, we have demonstrated the utility of yeast for investigating the function of viral proteins in a highly tractable heterologous expression platform. We found that studies in yeast are relevant in a mammalian system, as the identification of yeast SIR2 as a suppressor was used to determine that the mammalian deacetylase, SIRT1, is a proviral factor for MERS-CoV replication. We currently do not understand how SIRT1 enhances MERS-CoV replication or the role that ORF4a plays in this. We hypothesize that acetylation of additional cellular or viral proteins during MERS-CoV replication is required for efficient replication and that ORF4a has evolved to modulate this activity. The multifunctional property of ORF4a has revealed novel SIRT1 functions and pathways that MERS-CoV and potentially other viruses utilize to impact their replication.
MATERIALS AND METHODS
Plasmids and compounds.
MERS-CoV ORF4a was cloned into a modified pRS413 plasmid containing a GAL1 promoter. The multicloning site of pRS413/GAL1 was replaced with a cassette containing Kpn1/EcoRI/Xma1/Sac1 (GGTACCGGGCCCCCCCTCGAGGTCGACGGTATCGATAAGCTTGATATCGAATTCCTGCAGCCCGGGGGATCCACTAGTTCTAGAGCGGCCGCCACCGCGGTGGAGCTC) to create pGAL1/URA3/MCS. MERS-CoV ORF4a was PCR amplified from MERS-CoV (EMC/2012) using a forward primer (GAATTCACCATGGATTACGTGTCTCT) and a reverse primer (CCCGGGGTTGGAGAATGGCTCCTCTT) and cloned into pCAGGS/GFP by the use of EcoRI/Xma1 to create pCAGGS/ORF4a/GFP. From this plasmid, the ORF4a-GFP insertion was cloned into pGAL1/URA3/MCS via EcoRI/Xma1 digestion. All other viral protein yeast plasmids were produced through the same approach. The primer sequences used can be provided upon request. The κB-luciferase plasmids were previously described (49). Wild-type and H363Y mutant Flag-SIRT1 plasmids were a gift from Michael Greenberg (Addgene plasmid no. 1791 and no. 1792) (50). The siRNAs targeting SIRT1 were purchased from Sigma using their Rosetta prediction for predesigned sequences. The SIRT1-siRNA-1 sense sequence used was CUGUGAAGCUGUACGAGGA(dT)(dT), and the SIRT1-siRNA-2 sense sequence used was GAAGUACAAACUUCUAGGA(dT)(dT) (these sequences were previously used in the study described in reference 42). The scrambled control siRNA used was Mission siRNA universal negative-control no. 1 (Sigma). pHAGE TRE dCas9-KRAB was a gift from Rene Maehr and Scot Wolfe (Addgene plasmid no. 50917) (51). pLenti SpBsmBI sgRNA puromycin vector was a kind gift from Rene Maehr (Addgene plasmid no. 62207) (52). Two rounds of site-directed mutagenesis (QuikChange site-directed mutagenesis kit; Agilent) were performed on pLenti SpBsmBI sgRNA Puro vector to incorporate extended hairpin and A-U flip, previously demonstrated to increase levels of Cas9 targeting (53). CRISPRi SIRT1 sgRNA (forward, ACACCGGCGCGTCGAGCGGGAGCAGG; reverse, TAAACCTGCTCCCGCTCGACGCGCCG) was cloned into pTCL36 using BsmB1. All constructs were verified by DNA sequencing. Resveratrol and EX527 were purchased from Sigma, and stocks made in 100% ethanol.
Yeast strains and growth.
The plasmid containing MERS-CoV ORF4a was transformed into the PDR1/PDR3 knockout strain derived from BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) as previously described (23, 24). For expression of MERS-CoV ORF4a or the vector control, yeast cells were transformed (per a protocol reported previously [54]) with the plasmid and maintained on Casamino Acids (CAA) media lacking uracil. For growth experiments, single colonies of yeast cells were picked from plates and grown for 2 days at 30°C to reach stationary-phase growth in CAA media containing 2% raffinose. Cultures were subsequently diluted in CAA media containing 2% galactose to induce gene expression. The optical density (OD600) of the cultures was analyzed using a Synergy HTX multimode plate reader. Plates were incubated at 30°C for 48 h. Every 30 min, plates were shaken for 30 s and the OD600 was measured. For dilution drop culture experiments, yeast cells were grown overnight in CAA media containing 2% glucose. The OD600 of these cultures was measured (using the Synergy HTX reader), samples were diluted in 2% glucose media to achieve an OD600 of 0.1, and a 5-fold dilution series was made. These diluted samples were subsequently plated to glucose-containing or galactose-containing agar plates using a multiplate replicator (Boekel Scientific) and incubated at 30°C for two (glucose) or three (galactose) days.
Yeast Western blotting sample preparation.
A sodium hydroxide extraction approach was used for Western blotting of proteins from yeast. For this, transformed yeast cells were grown for 2 days at 30°C in CAA media containing 2% galactose to reach stationary-phase growth. Samples of these cultures were then incubated for 10 min in 0.1 M NaOH. Cells were pelleted and resuspended in 1× Laemmli sample buffer (LSB) with β-mercaptoethanol. Samples were heated at 95°C for 5 min, and equal sample volumes were separated by SDS-PAGE and then subjected to Western blotting (see below for further details).
Mammalian cell culture.
Huh7 and HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Corning) supplemented with 10% (vol/vol) fetal calf serum (FCS; Sigma) and 1% (vol/vol) penicillin/streptomycin (pen/strep; Gemini Bioscience) (10,000 U/ml [10 mg/ml]). Vero cells were cultured in minimal essential medium (MEM; Corning) supplemented with 10% (vol/vol) FCS, 1% (vol/vol) pen/strep, and 1% (vol/vol) l-glutamine (Gibco) (2 mM final concentration). All cell lines were maintained at 37°C and 5% CO2.
Transient transfection.
For transient transfection, cells were plated in 6-well dishes 1 day prior to transfection to achieve approximately 70% to 90% confluence. Cells were transfected using Lipofectamine LTX and Plus regents (Thermo) per the manufacturer’s instructions (using 4 μg of DNA per well of transfection—for cotransfection experiments, this value was kept consistent such that 2 μg of each plasmid would be used). Cells were grown for 24 h after transfection prior to use.
Immunofluorescence microscopy.
For detection of SIRT1, cells were plated on glass coverslips 1 day prior to use. Cells were fixed by 15 min of incubation at room temperature (RT; the same temperature was used for all subsequent steps) with 4% (vol/vol) formaldehyde (Thermo) in phosphate-buffered saline (PBS). Cells were washed with PBS and excess formaldehyde quenched by 15 min of incubation with 50 mM NH4Cl–PBS. Cells were again washed and lysed in 0.1% (vol/vol) Triton X-100 (Sigma) and then incubated for 10 min. Samples were blocked for 10 min with 0.2% bovine serum albumin (BSA)–PBS. Primary antibodies were then incubated with the sample diluted in 0.2% BSA for 1 h. Samples were washed in 0.2% BSA three times for 5 min each time and were then incubated with secondary antibodies in 0.2% BSA for 45 min. Finally, coverslips were washed with PBS four times for 5 min each time and mounted to glass slides using Prolong Gold antifade reagent with DAPI (4′,6-diamidino-2-phenylindole; Invitrogen).
For the experiment looking at ORF4a localization with MERS-CoV infection, Huh7 cells were plated on coverslips 1 day prior to infection with MERS-CoV. Cells were infected for 18 h prior to being fixed with 4% PFA for 24 h per the standard operating practice (SOP) for the biosafety level 3 (BSL3) containment facility. Cells were then labeled for immunofluorescence microscopy as described above.
The antibodies used were as follows: mouse anti-SIRT1 (clone 1F3; Abcam) (1 mg/ml) and anti-mouse Texas Red and anti-rabbit fluorescein isothiocyanate (FITC; Vector Labs) (1.5 mg/ml). All antibodies were diluted 1:100 for use. Rabbit anti-ORF4a serum (a kind gift from Stanley Perlman; used as described previously [55]) was diluted 1:100 for use. Imaging was performed using an Echo Revolve microscope.
siRNA knockdown.
A double-siRNA-transfection approach was used to achieve knockdown. Cells were plated in a 6-well dish the morning of day 1 and allowed to settle on the plates for 3 to 4 h. Cells were then transfected with one of two SIRT1-targeting siRNAs (the sequences are described above in the “Plasmids and compounds” section and were the same as those reported in reference 42) or with scrambled siRNA (Mission siRNA universal negative-control no. 1 [Sigma]) using Oligofectamine reagent (Thermo) and Opti-MEM (Gibco). For a 6-well dish, 10 μl Oligofectamine and 20 μl Opti-MEM were mixed and the mixture was incubated for 5 min at RT and mixed with 160 μl Opti-MEM and 4 μl 50 μM siRNA. This transfection mixture was incubated for 20 min at RT. Media were removed from cells and replaced with 800 μl Opti-MEM, the transfection mixture was added, and the resulting mixture was incubated at 37°C/5% CO2 for 4 h. After this incubation period, a 1:1 volume of 20% FBS–DMEM was added to the cells. The following day, media were changed for 10% FBS–DMEM. Cells were collected on day 3 and replated for a second round of transfection using the same approach. Cells were then collected on day 5 and prepared for experimental use.
CRISPRi.
Huh7 cells were transduced with pHAGE TRE dCas9-KRAB lentiviruses, and the transduced cells were selected with Geneticin (Invitrogen) (0.5 mg/ml). Transduced cells were screened for inducible dCas9 expression to generate a doxycycline-inducible (1 μg/ml DOX; Sigma) gene repression cell line. This cell line was then further transduced with SIRT1 CRISPRi sgRNA lentiviruses (sequences for the sgRNA are provided above in the “Plasmids and compounds” section). SIRT1 CRISPRi sgRNAs were designed using the Broad Institute sgRNA design tool. Of three sgRNAs initially tested, only one resulted in clear and reproducible SIRT1 repression. In each of the tested sgRNA sequences, two independent lentivirus preparations (see below) were used for transduction on separate bulk populations of the Huh7-dCas9 inducible cell lines. Transduction of SIRT1 sgRNAs was selected for by the use of puromycin (Sigma) (1 μg/ml). The Huh-dCas9-sgRNA cells were continuously cultured in DMEM supplemented with 10% FBS, 1% pen/strep, 0.5 mg/ml neomycin, and 1 μg/ml puromycin. To achieve knockdown of SIRT1, these cells were cultured in growth medium containing 1 μg/ml DOX, with this medium being changed every 2 days over a 7-day period prior to use of the cells for experiments.
Lentivirus production and transduction.
All lentiviruses were produced in HEK293T cells. Lentivectors were cotransfected with vesicular stomatitis virus envelope glycoprotein (VSV-G)-expressing plasmid pMD2.G (a gift from Didier Trono; Addgene plasmid no. 12259) and packaging plasmid psPAX2 (a gift from Didier Trono; Addgene plasmid no. 12260) and were concentrated using PEG-it (System Biosciences). For transduction, cells were plated to achieve approximately 70% confluence in a 24-well dish. Lentivirus preparations were added to cells in media containing 4 μg/ml Polybrene (AmericanBio). Cells were then subjected to spinoculation at 1,000 rpm for 45 min and incubated overnight prior to addition of puromycin selection media.
Mammalian cell Western blot sample preparation.
Cells for Western blotting were plated 1 day prior to sample collection. Plates were placed on ice and washed twice with 4°C PBS. Cells were lysed on ice for 15 min by incubation with radioimmunoprecipitation assay (RIPA) buffer (Sigma) containing 1× complete protease inhibitor cocktail (cOmplete Mini; Roche). Lysate was collected and centrifuged at 21,000 × g for 10 min. Supernatant was collected and used for Western blotting or stored at −80°C prior to use. The protein content of the sample was quantified using a bicinchoninic acid (BCA) system (Thermo Scientific). A 40-μg volume of whole-cell lysate (WCL) was used in all Western blotting experiments.
Western blotting.
Samples were separated using 4% to 20% Mini-Protean TGX gels (Bio-Rad) and transferred to polyvinylidene difluoride (PVDF) membranes (Immoblion-FL; Millipore) using a Trans-Blot Turbo system (Bio-Rad) or a wet transfer approach (1 h of transfer at 350 mA). Membranes were blocked using AquaBlock (East Coast Bio) for 1 h at RT and were then incubated with primary antibody (diluted in AquaBlock) overnight at 4°C. The following day, membranes were washed with Tris-buffered saline with Tween 20 (TBST; 50 mM Tris, 150 mM NaCl, 0.05% Tween 20) and incubated with secondary antibodies diluted in AquaBlock for 1 h at RT. Membranes were further washed with TBST prior to imaging using a ChemiDoc imaging system (Bio-Rad). Blot visualization was achieved using either fluorescence or horseradish peroxidase (HRP)-conjugated secondary antibodies, detected by the use of Amersham ECL Prime reagent (GE Healthcare Life Sciences).
The primary antibodies used were as follows: mouse anti-SIRT1 (as described in the “Immunofluorescence microscopy” section), mouse anti-Flag (clone M2; Sigma) (1 mg/ml), mouse anti-tubulin (clone DMA1A; Sigma), rabbit anti-GFP (Sigma) (1 mg/ml), and mouse anti-actin (clone 1A4; Sigma). All primary antibodies were diluted 1:1,000 for Western blotting. The secondary antibodies used were as follows: goat anti-mouse HRP (Thermo Scientific) (0.8 mg/ml), goat anti-rabbit HRP (Thermo Scientific) (0.8 mg/ml), goat anti-mouse Alexa Fluor 546 (Life Technologies) (2 mg/ml), and goat anti-rabbit Alexa Fluor 633 (Life Technologies) (2 mg/ml). HRP-conjugated secondary antibodies were diluted 1:10,000, and fluorescent secondary antibodies were diluted 1:2,000.
Immunoprecipitation.
Cells were plated 1 day prior to use and lysed in RIPA buffer containing 1× complete protease inhibitor cocktail as described above (similarly to the co-IP protocols previously described in references 56 and 57, for example). Protein content in the whole-cell lysate was determined by BCA assay, following the instructions of the manufacturer (Thermo Scientific). WCL (450 μg) was used in the IP. Nonspecific binding to beads was cleared by incubating the 450 μg of WCL for 1 h at 4°C with protein A Magnetic Beads (CST). Beads were separated from lysate with a magnetic rack, and the supernatant was collected. The nonspecific fraction was prepared by adding 2× LSB with β-mercaptoethanol and heating at 95°C for 5 min. The collected supernatant was added to fresh magnetic beads and incubated with anti-GFP antibody (5 μg) overnight at 4°C. The following day, the beads were separated with a magnetic rack and the supernatant was collected as the flowthrough fraction. Beads were washed five times with RIPA buffer before addition of 2× LSB with β-mercaptoethanol and heating at 95°C for 5 min to prepare the immunoprecipitation fraction.
Viral infection.
MERS-CoV (Jordan strain—GenBank accession no. KC776174.1; MERS-CoV–Hu/Jordan-N3/2012) stocks were prepared by infection of Vero cells, and titers were determined by plaque assay using these cells (as described previously [58]). All experimental infections were performed with Huh7 cells at a multiplicity of infection (MOI) of 0.1 unless indicated otherwise. For quantification of virus production, medium samples were collected 24 h after infection, centrifuged in a table top centrifuge for 3 min at full speed, and stored at −80°C prior to use. TCID50 (50% tissue culture infective dose) assays of Vero cells were used to quantify the amounts of virus in the collected samples (58).
Luciferase assays.
For CellTiter-Glo (CTG) assays, cells were plated in opaque 96-well plates 1 day prior to use. For drug treatments, resveratrol or EX527 was added at the indicated concentrations and incubated for 24 h before CTG assays were performed. For siRNA samples, cells were plated and grown overnight and then used for CTG assays. A CellTiter-Glo luminescent cell viability assay (Promega) kit was used per the manufacturer’s instructions. Luminescence was read using a Synergy HTX multimode plate reader.
Reporter assays were used with κB luciferase plasmids as previously described (49). Briefly, HEK293T cells were plated 1 day prior to transient transfection as described above. These transfections were performed in 48-well plates using a total of 400 ng of DNA per well. At most, three plasmids were required for transfection (SIRT1/ORF4a/luciferase for example); in instances of fewer experimental proteins being needed, pReceive-GFP was used to balance the amounts of DNA. Cells were transfected for 18 h before further manipulation. In the cases of drug treatment, cells were pretreated with resveratrol or EX527 for 2 h and TNF-α stimulation was performed for 6 h using 10 ng per well (recombinant TNF-α was purchased from BioLegend). The luciferase assay was performed using a Pierce firefly luciferase glow assay (Thermo) kit, following the manufacturer’s instructions. Luminescence was read on the Synergy HTX reader as described above.
ACKNOWLEDGMENTS
We thank Kirsten Kulcsar and William Jackson for valuable comments and critical readings of the manuscript, Brendan Cormack for the yeast knockout library collection, and Stanley Perlman for the ORF4a antisera. We also thank Carolyn Frenkil for her generous donation that initiated this work.
REFERENCES
- 1.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus A, Fouchier R. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 2.Alagaili AN, Briese T, Mishra N, Kapoor V, Sameroff SC, de Wit E, Munster VJ, Hensley LE, Zalmout IS, Kapoor A, Epstein JH, Karesh WB, Daszak P, Mohammed OB, Lipkin WI. 2014. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. mBio 5:e00884-14. doi: 10.1128/mBio.00884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu DKW, Poon LLM, Gomaa MM, Shehata MM, Perera R, Abu Zeid D, El Rifay AS, Siu LY, Guan Y, Webby RJ, Ali MA, Peiris M, Kayali G. 2014. MERS coronaviruses in dromedary camels, Egypt. Emerg Infect Dis 20:1049–1053. doi: 10.3201/eid2006.140299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woo PC, Lau SK, Li KS, Tsang AK, Yuen K-Y. 2012. Genetic relatedness of the novel human group C betacoronavirus to Tylonycteris bat coronavirus HKU4 and Pipistrellus bat coronavirus HKU5. Emerg Microbes Infect 1:1. doi: 10.1038/emi.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boheemen SV, Graaf MD, Lauber C, Bestebroer TM, Raj VS, Zaki M, Zaki AM, Osterhaus A, Haagmans BL, Gorbalenya A, Snijder E, Fouchier R. 2012. Genomic characterization of newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio 3:e00473-12. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Wit E, van Doremalen N, Falzarano D, Munster VJ. 2016. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol 14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niemeyer D, Zillinger T, Muth D, Zielecki F, Horvath G, Suliman T, Barchet W, Weber F, Drosten C, Muller MA. 2013. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J Virol 87:12489–12495. doi: 10.1128/JVI.01845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y, Zhang L, Geng H, Deng Y, Huang B, Guo Y, Zhao Z, Tan W. 2013. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell 4:951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Batool M, Shah M, Patra MC, Yesudhas D, Choi S. 2017. Structural insights into the Middle East respiratory syndrome coronavirus 4a protein and its dsRNA binding mechanism. Sci Rep 7:11362. doi: 10.1038/s41598-017-11736-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siu K-L, Yeung ML, Kok K-H, Yuen K-S, Kew C, Lui P-Y, Chan C-P, Tse H, Woo PCY, Yuen K-Y, Jin D-Y. 2014. Middle East respiratory syndrome coronavirus 4a protein is a double-stranded RNA-binding protein that suppresses PACT-induced activation of RIG-I and MDA5 in the innate antiviral response. J Virol 88:4866–4876. doi: 10.1128/JVI.03649-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Rabouw HH, Langereis MA, Knaap RCM, Dalebout TJ, Canton J, Sola I, Enjuanes L, Bredenbeek PJ, Kikkert M, de Groot RJ, van Kuppeveld F. 2016. Middle East respiratory coronavirus accessory protein 4a inhibits PKR-mediated antiviral stress responses. PLoS Pathog 12:e1005982. doi: 10.1371/journal.ppat.1005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakagawa K, Narayanan K, Wada M, Makino S. 2018. Inhibition of stress granule formation by Middle East respiratory syndrome coronavirus 4a accessory protein facilitates viral translation, leading to efficient virus replication. J Virol 92:00902-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG. 1996. Life with 6000 genes. Science 274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 14.Giaever G, Nislow C. 2014. The yeast deletion collection: a decade of functional genomics. Genetics 197:451–465. doi: 10.1534/genetics.114.161620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, Arkin AP, Astromoff A, El Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Güldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kötter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 16.Kushner DB, Lindenbach BD, Grdzelishvili VZ, Noueiry AO, Paul SM, Ahlquist P. 2003. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc Natl Acad Sci U S A 100:15764–15769. doi: 10.1073/pnas.2536857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao L, Lindenbach B, Wang X, Dye B, Kushner D, He Q, Newton M, Ahlquist P. 2014. Genome-wide analysis of host factors in nodavirus RNA replication. PLoS One 9:e95799. doi: 10.1371/journal.pone.0095799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panavas T, Serviene E, Brasher J, Nagy PD. 2005. Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc Natl Acad Sci U S A 102:7326–7331. doi: 10.1073/pnas.0502604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naito T, Kiyasu Y, Sugiyama K, Kimura A, Nakano R, Matsukage A, Nagata K. 2007. An influenza virus replicon system in yeast identified Tat-SF1 as a stimulatory host factor for viral RNA synthesis. Proc Natl Acad Sci U S A 104:18235–18240. doi: 10.1073/pnas.0705856104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makarow M, Nevalainen LT, Kaariainen L. 1986. Expression of the RNA genome of an animal virus in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 83:8117–8121. doi: 10.1073/pnas.83.21.8117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serviene E, Shapka N, Cheng C-P, Panavas T, Phuangrat B, Baker J, Nagy PD. 2005. Genome-wide screen identifies host genes affecting viral RNA recombination. Proc Natl Acad Sci U S A 102:10545–10550. doi: 10.1073/pnas.0504844102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagy PD, Pogany J, Lin JY. 2014. How yeast can be used as a genetic platform to explore virus-host interactions: from “omics” to functional studies. Trends Microbiol 22:309–316. doi: 10.1016/j.tim.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Basu D, Walkiewicz MP, Frieman M, Baric RS, Auble DT, Engel DA. 2009. Novel influenza virus NS1 antagonists block replication and restore innate immune function. J Virol 83:1881–1891. doi: 10.1128/JVI.01805-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frieman M, Basu D, Matthews K, Taylor J, Jones G, Pickles R, Baric R, Engel DA. 2011. Yeast based small molecule screen for inhibitors of SARS-CoV. PLoS One 6:e28479. doi: 10.1371/journal.pone.0028479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oishi K, Yamayoshi S, Kawaoka Y. 2018. Identification of novel amino acid residues of influenza virus PA-X that are important for PA-X shutoff activity by using yeast. Virology 516:71–75. doi: 10.1016/j.virol.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barco A, Carrasco L. 1995. Poliovirus 2Apro expression inhibits growth of yeast cells. FEBS Lett 371:4–8. doi: 10.1016/0014-5793(95)00841-V. [DOI] [PubMed] [Google Scholar]
- 27.Oishi K, Yamayoshi S, Kozuka-Hata H, Oyama M, Kawaoka Y. 2018. N-terminal acetylation by NatB is required for the shutoff activity of influenza A virus PA-X. Cell Rep 24:851–860. doi: 10.1016/j.celrep.2018.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sisko JL, Spaeth K, Kumar Y, Valdivia RH. 2006. Multifunctional analysis of Chlamydia-specific genes in a yeast expression system. Mol Microbiol 60:51–66. doi: 10.1111/j.1365-2958.2006.05074.x. [DOI] [PubMed] [Google Scholar]
- 29.Slagowski NL, Kramer RW, Morrison MF, LaBaer J, Lesser CF. 2008. A functional genomic yeast screen to identify pathogenic bacterial proteins. PLoS Pathog 4:e9. doi: 10.1371/journal.ppat.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heidtman M, Chen EJ, Moy M-Y, Isberg RR. 2009. Large-scale identification of Legionella pneumophila Dot/Icm substrates that modulate host cell vesicle trafficking pathways. Cell Microbiol 11:230–248. doi: 10.1111/j.1462-5822.2008.01249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rine J, Herskowitz I. 1987. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics 116:9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imai S, Armstrong CM, Kaeberlein M, Guarente L. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 33.Michan S, Sinclair D. 2007. Sirtuins in mammals: insights into their biological function. Biochem J 404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi J-E, Mostoslavsky R. 2014. Sirtuins, metabolism, and DNA repair. Curr Opin Genet Dev 26:24–32. doi: 10.1016/j.gde.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haigis MC, Sinclair D. a. 2010. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol Mech Dis 5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bheda P, Jing H, Wolberger C, Lin H. 2016. The substrate specificity of sirtuins. Annu Rev Biochem 85:405–429. doi: 10.1146/annurev-biochem-060815-014537. [DOI] [PubMed] [Google Scholar]
- 37.Deng J-J, Kong K-Y, Gao W-W, Tang H-M, Chaudhary V, Cheng Y, Zhou J, Chan C-P, Wong D-H, Yuen M-F, Jin D-Y. 2017. Interplay between SIRT1 and hepatitis B virus X protein in the activation of viral transcription. Biochim Biophys Acta Gene Regul Mech 1860:491–501. doi: 10.1016/j.bbagrm.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 38.Zainal N, Chang C-P, Cheng Y-L, Wu Y-W, Anderson R, Wan S-W, Chen C-L, Ho T-S, AbuBakar S, Lin Y-S. 2017. Resveratrol treatment reveals a novel role for HMGB1 in regulation of the type 1 interferon response in dengue virus infection. Sci Rep 7:42998. doi: 10.1038/srep42998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han Y, Wang L, Cui J, Song Y, Luo Z, Chen J, Xiong Y, Zhang Q, Liu F, Ho W, Liu Y, Wu K, Wu J. 2016. SIRT1 inhibits EV71 genome replication and RNA translation by interfering with the viral polymerase and 5′UTR RNA. J Cell Sci 129:4534–4547. doi: 10.1242/jcs.193698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi Y, Li Y, Huang C, Ying L, Xue J, Wu H, Chen Z, Yang Z. 2016. Resveratrol enhances HBV replication through activating Sirt1-PGC-1α-PPARα pathway. Sci Rep 6:24744. doi: 10.1038/srep24744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W-Y, Ren J-H, Tao N-N, Ran L-K, Chen X, Zhou H-Z, Liu B, Li X-S, Huang A-L, Chen J. 2016. The SIRT1 inhibitor, nicotinamide, inhibits hepatitis B virus replication in vitro and in vivo. Arch Virol 161:621–630. doi: 10.1007/s00705-015-2712-8. [DOI] [PubMed] [Google Scholar]
- 42.Koyuncu E, Budayeva HG, Miteva YV, Ricci DP, Silhavy TJ, Shenk T, Cristea IM. 2014. Sirtuins are evolutionarily conserved viral restriction factors. mBio 5:e02249-14. doi: 10.1128/mBio.02249-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang H-M, Gao W-W, Chan C-P, Cheng Y, Deng J-J, Yuen K-S, Iha H, Jin D-Y. 2015. SIRT1 suppresses human T-cell leukemia virus type 1 transcription. J Virol 89:8623–8631. doi: 10.1128/JVI.01229-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin S-C, Ho C-T, Chuo W-H, Li S, Wang TT, Lin C-C. 2017. Effective inhibition of MERS-CoV infection by resveratrol. BMC Infect Dis 17:144. doi: 10.1186/s12879-017-2253-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mumberg D, Muller R, Funk M. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res 22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. 2013. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc 8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. 2004. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J 23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sinclair DA, Guarente L. 2014. Small-molecule allosteric activators of sirtuins. Annu Rev Pharmacol Toxicol 54:363–380. doi: 10.1146/annurev-pharmtox-010611-134657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matthews KL, Coleman CM, van der Meer Y, Snijder EJ, Frieman MB. 2014. The ORF4b-encoded accessory proteins of Middle East respiratory syndrome coronavirus and two related bat coronaviruses localize to the nucleus and inhibit innate immune signalling. J Gen Virol 95:874–882. doi: 10.1099/vir.0.062059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsy R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. 2004. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 51.Kearns NA, Genga RMJ, Enuameh MS, Garber M, Wolfe SA, Maehr R. 2014. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development 141:219–223. doi: 10.1242/dev.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pham H, Kearns NA, Maehr R. 2016. Transcriptional regulation with CRISPR/Cas9 effectors in mammalian cells. Methods Mol Biol 1358:43–57. doi: 10.1007/978-1-4939-3067-8_3. [DOI] [PubMed] [Google Scholar]
- 53.Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li G, Park J, Blackburn EH, Weissman JS, Qi LS, Huang B. 2013. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gietz RD, Woods RA. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350:87–96. doi: 10.1016/S0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 55.Comar CE, Goldstein SA, Li Y, Yount B, Baric RS, Weiss SR. 2019. Antagonism of dsRNA-induced innate immune pathways by NS4a and NS4b accessory proteins during MERS coronavirus infection. mBio 10:e00319-19. doi: 10.1128/mBio.00319-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nasrin N, Kaushik VK, Fortier E, Wall D, Pearson KJ, de Cabo R, Bordone L. 22 December 2009, posting date JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PLoS One doi: 10.1371/journal.pone.0008414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wong HH, Fung TS, Fang S, Huang M, Le MT, Liu DX. 2018. Accessory proteins 8b and 8ab of severe acute respiratory syndrome coronavirus suppress the interferon signaling pathway by mediating ubiquitin-dependent rapid degradation of interferon regulatory factor 3. Virology 515:165–175. doi: 10.1016/j.virol.2017.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Coleman CM, Frieman MB. 2015. Growth and quantification of MERS-CoV infection, p 15E.2.1–15E.2.9. In Current protocols in microbiology. John Wiley & Sons, Inc, Hoboken, NJ, USA. doi: 10.1002/9780471729259.mc15e02s37. [DOI] [PMC free article] [PubMed] [Google Scholar]